Summary
The third variable (V3) loop and the CD4 binding site (CD4bs) of the HIV-1 envelope are frequently targeted by neutralizing antibodies (nAbs) in infected individuals. In chronic infection, HIV-1 escape mutants repopulate the plasma, and V3 and CD4bs nAbs emerge that can neutralize heterologous tier 1 easy-to-neutralize, but not tier 2 difficult-to-neutralize HIV-1 isolates. However, neutralization sensitivity of autologous plasma viruses to this type of nAb response has not been studied. We describe the development and evolution in vivo of antibodies distinguished by their target specificity for V3and CD4bs epitopes on autologous tier 2 viruses but not on heterologous tier 2 viruses. A surprisingly high fraction of autologous circulating viruses was sensitive to these antibodies. These findings demonstrate a role for V3 and CD4bs antibodies in constraining the native envelope trimer in vivo to a neutralization-resistant phenotype, explaining why HIV-1 transmission generally occurs by tier 2 neutralization-resistant viruses.
Introduction
Inducing antibodies with neutralization breadth is a primary goal of HIV-1 vaccine development (Mascola and Haynes, 2013). Broadly neutralizing antibodies (bnAbs) can block infection in macaques; however, current HIV-1 envelope (Env) immunogens induce only nAbs that inhibit easy-to-neutralize (tier 1) HIV-1 strains. In contrast, bnAbs that potently neutralize difficult-to-neutralize (tier 2) HIV-1 strains associated with HIV-1 transmission are not induced by current vaccines (Mascola and Haynes, 2013).
Initial autologous nAbs in HIV-1-infected subjects are restricted to neutralizing the infecting transmitted/founder (T/F) virus (Derdeyn et al., 2014). Epitopes frequently targeted by these nAbs are the third constant region-variable loop 4 (C3-V4) domain, the base of the third variable (V3) loop, the first and second variable loop (V1V2) regions, and the CD4 binding site (CD4bs). In chronic HIV-1 infection, virus escape mutants repopulate the plasma virus pool, and neutralization breadth accrues to varying degrees in different persons (Hraber et al., 2014). V3 and CD4bs nAbs arise that can neutralize heterologous tier 1 but not tier 2 HIV-1 isolates (Montefiori et al., 2012; Moore et al., 1994), although heterologous tier 2 neutralization is seen with some CD4bs (Scheid et al., 2011) and V3 (Gorny et al., 2009; Hioe et al., 2010) antibodies. However, neutralization sensitivity of autologous plasma viruses to this type of V3 and CD4bs nAb response has not been studied. Here, we isolated from two chronically HIV-1-infected persons V3 and CD4bs neutralizing monoclonal antibodies (mAbs) with breadth for tier 1 but not tier 2 heterologous viruses and tested their ability to neutralize a large panel of autologous viruses. We also isolated CD4bs bnAbs with tier 2 breadth and tested their ability to neutralize the same panel of autologous viruses that includes escape mutants. Surprisingly, we found a group of V3 and CD4bs tier 1 heterologous virus-nAbs that neutralized a proportion of autologous tier 2 viruses. These nAbs differ from more common tier 1 virus-reactive antibodies that neutralize autologous and heterologous tier 1 viruses exclusively. We suggest that the autologous tier 2-reactive V3 and CD4bs nAbs described here play a previously underappreciated role in neutralizing autologous tier 1 and tier 2 primary viruses, thus continuously selecting autologous viruses for neutralization-resistance. This additional level of immune surveillance against HIV-1 Env trimers with relaxed or “open” conformations likely contributes to the finding that T/F viruses exhibit tier 2 (or greater) neutralization resistance, a finding relevant to HIV-1 vaccine research.
Results
Restricted or broad nAbs from chronically infected persons
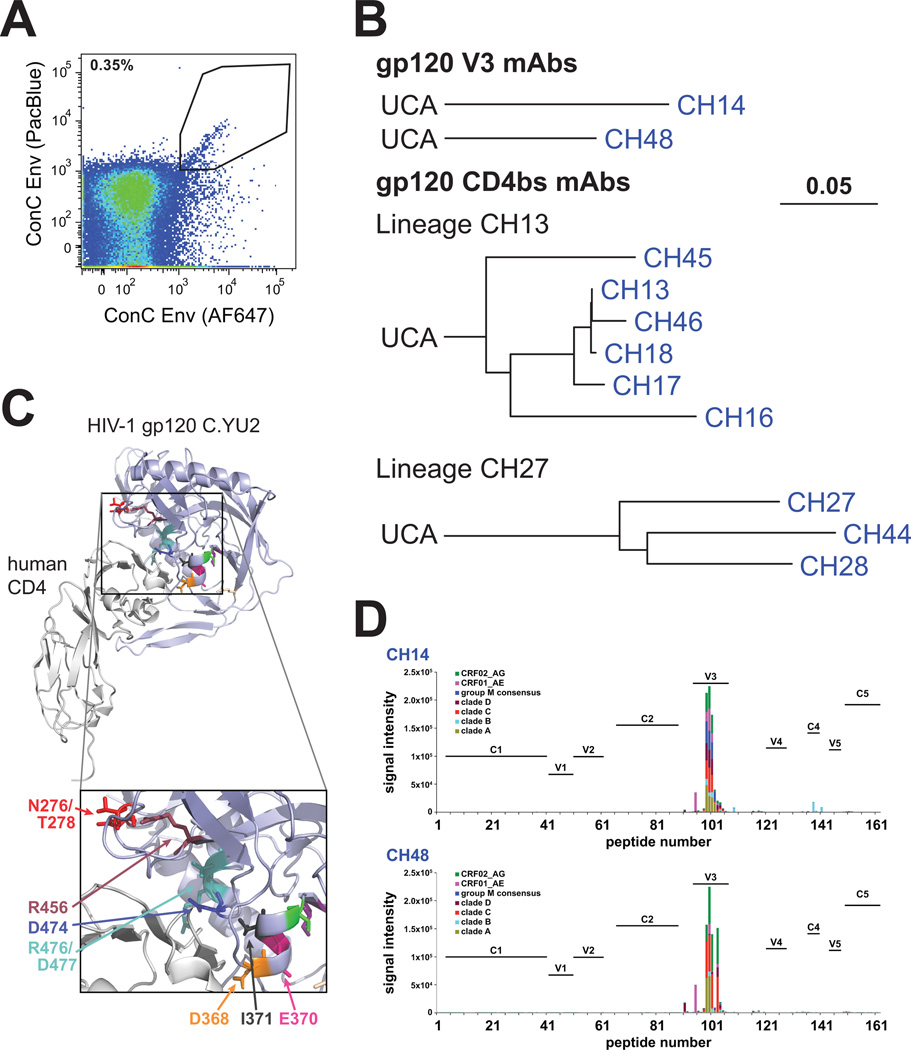
We isolated two B cell clonal lineages (CH13, CH27) and single mAbs from chronically clade C HIV-1 infected African person CH0457 known to have plasma broad neutralizing activity (Tomaras et al., 2011), using antigen-specific memory B cell sorting of peripheral blood mononuclear cells (PBMC) (Fig. 1AB; Table S1). Epitope mapping with virus mutants showed CH13 lineage mAbs bound to the CD4bs (Fig. 1C; Tables S2–S3); lineage members neutralized 8/8 tier 1,but 0/40 tier 2 heterologous HIV-1 Env pseudoviruses (Fig. 2). Two other mAbs, CH14 and CH48, were not clonally related and both mapped to the HIV-1 Env V3 loop (Fig. 1D; Table S4). Like CD4bs clonal lineage CH13, V3 mAbs CH14 and CH48 neutralized tier 1 but not tier 2 heterologous HIV-1 strains (Fig. 2).
Figure 1. Clonal lineages derived from CH0457.
A: The kite-shaped gate shows a diagonal of gp120ConC core+/+ memory B cells that were sorted as single cells. Antigen-specific cell frequency was similar in both samples; wk 8 shown.
B: IgG1 gp120 V3 mAbs (CH14, CH48) were not related to other isolated mAbs. Lineage CH13 had 6 IgG1 mAbs (VH1~69/JH3; VK1~39/JK4); mean heavy chain (HC) mutation frequency = 9.8%. Lineage CH27 had 3 mAbs—2 IgA2 (CH27, CH28) and a IgG1 (CH44) (VH3~66/JH2; VK3~20/JK1); mean HC mutation frequency = 15.7%. Trees plotted on the same scale.
C: Residues critical for lineage CH13 and lineage CH27 mAb binding (Tables S2–3) highlighted in the structure of gp120C.YU2 (CD4 light gray, gp120 light blue, mAb 17b not shown). Mapped residues are largely located within the CD4-gp120 contact surface. V1/V2 not shown because it was absent in the crystal structure.
D. CH14 and CH48 binding to peptides reflective of multiple HIV-1 clades. Both bound V3 loop peptides (residues 301–325) across multiple clades; no binding found for other epitopes.
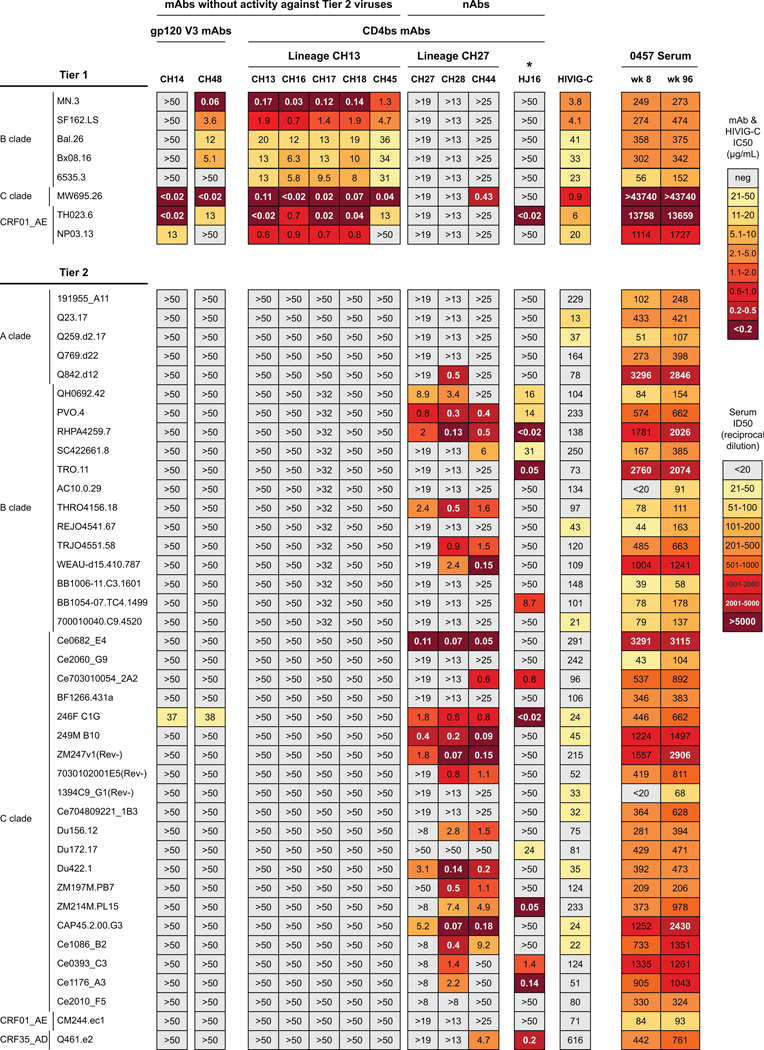
Figure 2. Heterologous neutralization by mAbs from CH0457.
Neutralization of mAbs against tier 1 and tier 2 viruses from diverse clades shown in colored boxes (EC50). Control (HIVIG-C) and CH0457 serum from wks 8 and 96 shown; serum shown as reciprocal ED50. Lineage CH13 mAbs and non-lineage mAbs (CH14, CH48) potently neutralized tier 1 viruses but only neutralized one tier 2 virus (C.246F_C1G). Lineage CH27 neutralized one tier 1 virus but 23/40 (58%) tier 2 viruses. CH0457 serum from wks 8 and 96 neutralized all tier 1 viruses at >1:20, and 37/40 (93%) and 31/40 (78%) of tier 2 viruses, respectively.
The second mAb clonal lineage, termed CH27 (Fig. 1B), had 1 IgG1 (CH44) and 2 IgA2 (CH27, CH28) members (Table S1). All lineage members (CH27, CH28, CH44) neutralized 40% (range 25–48%) of 40 tier 2 heterologous HIV-1 strains (Fig. 2) and preferentially neutralized tier 2 but not tier 1 heterologous viruses. HJ16 is a CD4bs bnAb isolated from another infected person and like the CH27 lineage mAbs, HJ16 neutralizes multiple tier 2 but not tier 1 viruses. Mutation of Env at N276 conferred resistance to HJ16 (Balla-Jhagjhoorsingh et al., 2013) and CH27 lineage mAbs were sensitive to mutations at N276 and T278 (Table S3). CH27, CH44 and HJ16 cross-blocked each other for Env binding (Fig. S1), showing that CH27 lineage bnAbs were similar to HJ16 (Fig. 2). Serum from chronically-infected person CH0457 taken at weeks (wks) 8 and 96 of study were tested against the same panel of heterologous viruses (Fig. 2). Neutralization titers and breadth were similar at the two chronic infection time points (R2=0.95, Pearson’s correlation p<2.2×10−16). The neutralization pattern of CH27 lineage bnAbs accounted for part of serum breadth, but some viruses in the panel were neutralized by serum and not the isolated mAbs (eg, C.Ce2010_F5 in Fig. 2) indicating that the isolated mAbs only partially recapitulated serum neutralization and that other bnAbs might have been present in this person.
From a second person, CH505, previously described to have a CD4bs bnAb lineage (represented by CH103 in Fig. 3) (Liao et al., 2013), we isolated by limiting dilution memory B cell culture two V3 nAbs (DH151, DH228; Table S4) from 176 and 41 wks after transmission, respectively (Fig. 3). DH151 and DH228 neutralization was similarly restricted to a subset of tier 1 heterologous viruses and they neutralized 0/16 tier 2 heterologous viruses (Fig. 3).
Figure 3. Heterologous neutralization by mAbs from CH505.
V3 mAbs DH151 and DH228 from CH505 tested against heterologous HIV isolates. Tier 1 isolates (2/4) neutralized; 0/16 tier 2 isolates neutralized.
Virus evolution in chronically infected person CH0457
We amplified 209 CH0457 env gene sequences by single genome amplification (SGA) from 10 time points over two years during chronic infection (wks of chronic infection study 0, 2, 4, 8, 12, 16, 24, 48, 72, 96 post-enrollment). A mean of 21 (range 12–35) SGA env sequences were analyzed for each time point. Phylogenetic analysis showed that Env sequences continuously evolved over time; Envs from wks 48, 72 and 96 were more divergent versus earlier Envs (wk 0–16) (Fig. S2). Furthermore, within-subject phylogeny maintained a persistent minority clade that represented a small fraction (mean 14%) of Envs sampled at any given time point (Fig, 4; Fig. S2) throughout the study. Consensus of this clade differed at 85/888 (9.6%) aligned Env amino acid positions from the main clade consensus. Analysis of sequences from CH0457 relative to the database indicated that despite the genetic distance, sequences from this minor persistant clade were more closely related to other sequences from CH0457 than to heterologous strains (not shown); we further validated the relationship of the two clades by identifying transition states between the two clades in archival DNA proviral sequences (Fig. S2). The minor clade contained members from many sampling time points across two years of infection; a Slatkin-Madison test strongly supported compartmentalization (p=0.0001). Interestingly, these two divergent virus populations persisted during the 96 wks of sampling without apparent recombination between the two clades, although some recombination within the lineages was evident.
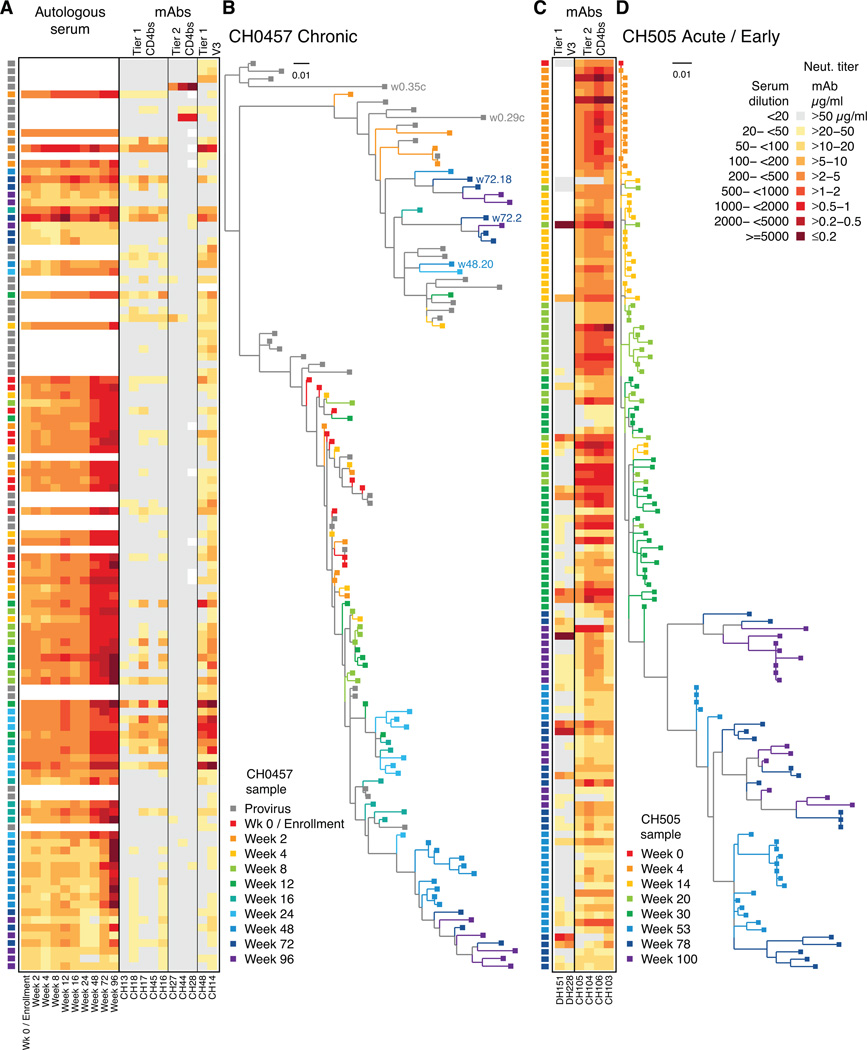
Figure 4. Autologous neutralization and Env sequence phylogenies.
Neutralization heat map of autologous serum and mAbs shown (AC). Each row in the neutralization panel (A) of 84 CH0457 pseudoviruses and phylogeny tree (B) is a distinct Env isolate spanning wk 0 (enrollment; red) to wk 96 after enrollment (purple). PBMC provirus sequences are grey. Autologous serum neutralization (reciprocal dilution) and isolated mAbs (µg/mL) shown for (A) lineage CH13 mAbs (tier 1 CD4bs), lineage CH27 mAbs (tier 2 CD4bs), and CH14 and CH48 (tier 1 V3); and (C) CH505 mAbs DH151 and DH228 (tier 1 V3) and lineage CH103 (tier 2 CD4bs). (D) CH505 Env phylogeny spans transmission (wk 0, red) through wk 100 (purple). A high fraction of circulating viruses were sensitive to autologous mAbs.
Neutralization of autologous viruses by bnAbs and tier 1 virus-nAbs
We made 84 pseudoviruses from these env sequences (Fig. 4B; mean 8 per time point; range 7–11) to assay against CH0457 serum (Fig. 4A). Later serum time points (wk 72–96) potently neutralized early viruses (wk 0–48) but not later viruses, indicating that autologous nAbs were continuously elicited during chronic infection in CH0457 (Fig. 4AB).
Assay of CH27 CD4bs lineage bnAbs against these 84 autologous pseudoviruses showed that 5 were weakly neutralized by 1/3 lineage CH27 bnAbs (range 32–50 µg/mL), while 79 (94%) resisted CH27 lineage bnAbs (Fig. 4A; Fig. S3A). This suggested that the autologous virus pool in CH0457 had already escaped from pressure exerted by the CH27 lineage bnAbs, with viral escape occurring during chronic infection prior to study enrollment.
To seek definitive evidence of selection by CH27 lineage bnAbs, we amplified proviral env genes archived in PBMC from the earliest time point (wk 0) in this study. Like plasma-derived Env pseudoviruses, most PBMC-derived Env pseudoviruses resisted lineage CH27 bnAbs (Fig. 4AB). However, two cell-derived Env pseudoviruses (w0.35c, w0.29c) were highly sensitive to CH27 lineage bnAbs, thus documenting CH27 bnAb lineage-mediated escape (Fig. 4A; Fig. S3B). Remarkably, both sensitive viruses were members of the persistent minority clade (Fig. 4B; Fig. S2).
We tested CH0457 tier 1 virus-nAbs (5 CH13 lineage CD4bs mAbs, 2 V3 mAbs CH14, CH48) against autologous HIV-1 pseudoviruses and found that the V3 and CD4bs mAbs neutralized autologous viruses throughout the 2-year study period, including PBMC-archived viruses (Fig. 4AB; Fig. S3AB). Remarkably, the CD4bs lineage CH13 tier 1-nAbs neutralized 52/84 (62%) and 11/34 (32%) of autologous plasma and PBMC viruses, respectively, while V3 tier 1 virus-nAbs (CH14, CH48) neutralized 67/84 (80%) plasma and 28/34 (82%) PBMC viruses. Tier 1 virus-nAb potency ranged from 0.06–50 µg/mL, with potency of ≤2 µg/mL for 21/257 (8%) autologous virus neutralization assays. Sensitivity to CH13 lineage and V3 mAbs peaked at wk 24 after enrollment; by wk 48 most viruses resisted the V3 mAbs (Fig. 4A; Fig. S3A) suggesting escape from these nAbs. Across all time points, 32/84 (38%) autologous pseudoviruses resisted the CD4bs nAbs while 17/84 (20%) resisted the V3 mAbs. CH0457 Env sequences did not show accumulation of Env mutations at nAb contact sites suggested by epitope mapping (Tables S2–S4). Other studies have shown that V3 in chronic infection either did not change (Nyambi et al., 2008) or accumulated sensitivity mutations over time (Haldar et al., 2011), and V3 epitopes otherwise sensitive to mAbs can be occluded by other regions of Env (Upadhyay et al., 2014). Thus, both common CD4bs and V3 tier 1 virus-nAbs can select virus escape mutants that shape the autologous virus reservoir towards neutralization resistance.
Among viruses sampled between wks 48–96, only 3 were moderately sensitive to the tier 1-virus nAb CD4bs CH13 lineage (w48.20, w72.2, w72.18), with the rest weakly sensitive or completely resistant. These CH0457 viruses were all within the persistent minor clade (3/10 in minor clade vs. 0/17 in dominant clade; Fisher’s exact test p=0.04). That for both CD4bs lineages the sensitive viruses persisted longest in the minor but not the dominant virus clade raises the possibility that viruses in the minor clade may be emerging from an immunologically protected site (eg, brain or CD4 T cell latent pool) where antibody pressure would be limited.
To determine if autologous virus neutralization by autologous tier-1 virus nAbs was unique to CH0457, we studied two V3 tier 1 virus-nAbs (DH151, DH228; Table S4) isolated from a second HIV-1-infected African person, CH505, 176 and 41 wks after transmission, respectively; CH505 also developed a CD4bs clonal lineage (termed CH103) at 136 wks after transmission (Liao et al., 2013). CH505 was studied earlier during infection versus CH0457, thus Env selection by bnAbs was still ongoing and many autologous Env pseudoviruses were only partially resistant to the CH103 bnAb lineage (Fig. S3C) (Gao et al., 2014). Both CH505 V3 mAbs (CH151, CH228) neutralized some heterologous tier 1 viruses but 0/16 tier 2 viruses (Fig. 3). However, V3 mAbs DH151 and DH228 neutralized 45/96 (47%, IC50 range 0.03–50 µg/mL) autologous CH505 viruses (Fig. 4C; Fig. S3C) and potently neutralized 7/96 (7.3 %) viruses at ≤2 µg/mL. Interestingly, the T/F virus from CH505 resisted both V3 nAbs but became sensitive by wk 14 (Fig. 4CD; Fig. S3C) showing some CH505 autologous viruses had Env V3 epitope exposure by wk 14 after infection. As in CH0457, we did not find Env mutations accumulating at binding sites of these mAbs but we did find autologous CH505 viruses sensitive to V3 mAbs at all time points studied. Thus, CH505 V3 mAbs DH151 and DH228 had no neutralizing activity against heterologous tier 2 viruses but neutralized autologous CH505 viruses, indicating that this phenomenon was not limited to the chronically HIV-1-infected person CH0457.
Autologous virus neutralization sensitivity
To assess autologous virus susceptibility to heterologous nAbs, we assayed CH0457 pseudoviruses and found 73/84 (87%) sensitive to heterologous VRC01-like CD4bs bnAb CH31 (Wu et al., 2011) and 55/62 (89%) sensitive to loop binding CD4bs bnAb CH106 (Liao et al., 2013) (Fig. S3A). Glycan-dependent bnAb HJ16 neutralized only 5/72 (7%) viruses, consistent with escape from the HJ16-like lineage CH27 nAbs (Fig. S1, Table S3).
To assess overall neutralization sensitivity, we tested 84 CH0457 Env pseudoviruses against pooled serum (HIVIG-C) and a subset against well-characterized HIV-1 patient sera (Seaman et al., 2010) (Fig. S3D). The data suggested that CH0457 viruses sensitive to autologous tier 1 virus-nAbs (CH13 lineage, CH14, CH48) had exposed V3 and CD4bs epitopes. We analyzed a subset of Env pseudoviruses (10 sensitive / 10 resistant to autologous V3 and CD4bs tier 1 virusnAbs) against heterologous V3 and CD4bs mAbs known to not neutralize tier 2 viruses (Gorny et al., 2009; Montefiori et al., 2012) (Fig. S4A). Viruses sensitive to autologous tier 1 virus-nAbs were neutralized by heterologous V3 and CD4bs nAbs, suggesting that V3 loop and CD4bs epitopes were trimer-surface exposed. Viruses resistant to autologous nAbs also resisted heterologous nAbs (Fig. S4A). Sensitive viruses also had intermediate sensitivity to a panel of neutralization typing sera (Fig. S4A) consistent with an intermediate (tier 1B) (Seaman et al., 2010) phenotype (Fig. S4B). Autologous viruses from CH505 tested with a similar panel also showed predominant tier 1B neutralization sensitivity (Fig. S4C). These data showed that viruses from chronic infection in African persons CH0457 and CH505 could be neutralized by common V3 and CD4bs tier 1 virus-nAbs that lacked tier 2 heterologous virus-neutralizing activity.
Discussion
This study shows that a subset of common CD4bs and V3 antibodies that neutralize tier 1 but not tier 2 heterologous viruses have the capacity to neutralize autologous tier 2 viruses and select for virus escape mutants. This observation is critical to understanding how neutralization-resistant T/F viruses are selected and how antibodies with tier 1 virus neutralization are induced.
The initial autologous nAb response in acute HIV-1 infection is specific for autologous virus with little or no tier 1 heterologous breadth (Richman et al., 2003; Wei et al., 2003). In chronic infection breadth develops for heterologous tier 1 viruses and viruses can evolve in response to those nAbs (Haldar et al., 2011; Kelly et al., 2005; Nyambi et al., 2008). However, autologous virus-neutralizing mAbs isolated from HIV-1-infected people have been strictly strain-specific with no detectable heterologous tier 1 or tier 2 HIV-1 neutralization (Derdeyn et al., 2014) (DC Montefiori, personal communication). This study to examines the dynamics of this evolution by combining virus SGA with mAb isolation to deconvolute the polyclonal antibody response in chronically infected persons and demonstrate autologous nAbs with both heterologous tier 1 and autologous tier 2 neutralizing activity. We did not isolate all bnAbs or tier 1 nAbs from these persons. However, we have shown ongoing and repeated emergence of viruses sensitive to common CD4bs and V3 nAbs. In particular, this class of nAbs does not appear to exert immune pressure to the same degree as bnAbs where we found evidence of complete viral escape. It is possible that when autologous nAbs begin to show heterologous tier 1 breath they may be developing into bnAbs as occurred in the CH103 CD4bs lineage (Liao et al., 2013), but at this time it is not known which antibody lineages will mature to breadth.
We found autologous virus isolates were variably neutralized by mAbs against the CD4bs and V3 loop (Fig. S4AC). Recent work has shown that sensitive Envs spend more time in conformationally open states that permit binding of antibodies without neutralization breadth (Munro et al., 2014). Our data show that these sensitive viruses arise during chronic HIV-1 infection and are likely the antigenic stimulus inducing common antibodies that neutralize tier 1 heterologous viruses. When we tested common CD4bs and V3 nAbs isolated from the same subjects against their autologous viruses, the mAbs neutralized both the sensitive viruses and a subset of autologous isolates resistant to heterologous tier 1 virus-nAbs. We speculate that neutralization of resistant viruses was possible because the mAbs and viruses co-evolved in the same HIV-1-infected persons and the antibodies gained the ability to bind autologous viruses. During HIV-1 infection, virus quasispecies evolve that have different degrees of Env reactivity, including viruses with high intrinsic reactivity (tier 1A) (Seaman et al., 2010) that are more reactive with soluble CD4 and nAbs (Haim et al., 2011). Our study indicates that autologous viruses with low Env reactivity (tier 1B or tier 2) (Haim et al., 2011; 2013) can act as templates for antibody evolution, promoting development of nAbs that bind and neutralize autologous virus Envs with low reactivity. Such nAbs could broadly react with high reactivity heterologous tier 1A Envs but would be expected to bind poorly to low reactivity heterologous tier 2 Envs.
Autologous nAbs that arise in acute HIV-1 infection can exert immune pressure and influence T/F virus evolution; the initial autologous-specific nAb response appears within the first year of infection and is associated with development of resistant viruses in virtually all infected people (Richman et al., 2003; Wei et al., 2003). In addition, tier-1 virus nAbs of the type studied here were shown to exert immune pressure in a humanized mouse HIV-1 infection model (Klein et al., 2014). Our study is unique in showing autologous tier 1 virus-nAb evolution over time and the ability of those nAbs to combine with tier 2-virus-nAbs to shape the neutralization sensistivity of the autologous virus pool.
HIV-1 vaccine efficacy trials have not convincingly shown a protective effect of vaccine-elicited tier 1 virus-nAbs, although in secondary analyses, tier 1 nAbs correlated with decreased risk of infection in VAX004 (Gilbert et al., 2005) and RV144 (Haynes et al., 2012). The only vaccine study to date that demonstrated any protection (RV144) did not elicit tier 2 nAbs (Montefiori et al., 2012). Secondary analyses of RV144 found that V2-specific antibodies correlated with decreased transmission risk (Gottardo et al., 2013; Zolla-Pazner et al., 2014); as did antibody dependent cellular cytotoxicity-mediating antibodies (Haynes et al., 2012) and V3 antibodies (Gottardo et al., 2013) in the context of other responses such as low Env-IgA (Haynes et al., 2012). Recent studies of HIV-1 Env evolution over the pandemic indicate that T/F viruses are evolving to be more resistant to bnAbs (Bouvin-Pley et al., 2014); thus a successful vaccine may have to contend with an ever increasing degree of resistance among HIV-1 strains. However, we found that some tier 1 virus-nAbs can neutralize autologous tier 1B and tier 2 HIV-1 Envs with which they co-evolved (Fig. 4; Fig. S3AC). If a vaccine elicited a sufficiently large and diverse pool of tier 1 virus-nAbs, some degree of protection might be possible.
Antibodies and autologous viruses are passed together in bodily fluids associated with HIV-1 transmission. The present study suggests that viruses with a more sensitive phenotype would be passed along with nAbs against those viruses, while resistant viruses would be passed without nAbs against them. This situation could cause a selection bias to favor infection by viruses resistant to the autologous nAb response. Thus, the observation that T/F viruses isolated from recently-infected people are of a more resistant phenotype could be partly explained by the restricted tier 1 autologous virus-nAbs described here.
In addition, there is a setting where these kinds of nAbs could be transferred to a susceptible host prior to virus exposure—mother-to-child transmission (MTCT). Maternal IgG is actively transferred to the fetus (Malek et al., 1996) and maternally-derived nAbs could plausibly prevent newborn infection. Thus, V3- or CD4bs-directed nAbs of the type described here could correlate with decreased transmission risk for MTCT. A recent analysis has shown that a correlate of MTCT transmission risk is low plasma tier 1 virus-nAbs (Permar et al., 2015). Boosting to achieve high levels of V3 and CD4bs autologous nAbs in pregnant women might be a way to reduce intrapartum and peripartum HIV-1 transmission to infants that occurs despite peripartum treatment with anti-retroviral drugs. Thus, tier1 virus-nAbs can exert selection pressure on autologous virus variants and determine intrinsic Env reactivity and sensitivity to neutralization. Whether these common antibodies can be harnessed for protection in the setting of vaccination for MTCT remains to be determined.
Experimental Procedures
Flow cytometry, memory B cell cultures and mAb isolation
The gp120ConC core protein was produced and used in flow cytometry on PBMC from CH0457 using a two-color technique as described (Gray et al., 2011). IgG+ memory cells from CH505 isolated from PBMC were cultured as described (Gao et al., 2014). In both cases, Ig genes were amplified from RNA from isolated cells, expression cassettes made and mAbs expressed as described (Gao et al., 2014).
Antibody reactivity by binding antibody multiplex assay (BAMA), enzyme-linked immunosorbent assay (ELISA) and peptide microarray
Expressed mAbs were assayed for reactivity to HIV-1 antigens using a standardized custom BAMA using Luminex (Tomaras et al., 2011). BAMA was performed with a panel of HIV-1 antigens (gp140ConC, gp120ConC full length, gp140ConB, gp140ConG, gp140JR.FL); mAbs with a blank-bead-subtracted value >2000 units and >1000× mAb IgG in µg/mL were evaluated further. Binding was confirmed on purified mAbs.
ELISA was performed as described (Tomaras et al., 2011); testing was considered positive if the optical density at 405 nm was >0.3 units and >4-fold over background. Epitope mapping by cross-clade peptide microarray was performed as described (Tomaras et al., 2011).
Amplification of full-length env genes and generation of pseudoviruses
After recovery of viral RNA or proviral DNA, 3′ half genomes from plasma or PBMC were amplified by single genome amplication (SGA) as described (Salazar-Gonzalez et al., 2009). Sequence contigs from each env SGA were assembled and edited using Sequencher 4.7 (Gene Codes; Ann Arbor, MI). Pseudoviruses were made by adding the CMV promoter to the 5′ end of each SGA-amplified env gene then the pPCR product was cotransfected with the env-deficient HIV-1 backbone pSG3Δenv into 293T cells.
Neutralization assay in TZM-bl cells
Neutralizing antibody assays in TZM-bl cells were performed as described (Montefiori et al., 2012). For some isolates, amino acid substitutions were introduced by oligonucleotide-directed PCR mutagenesis. Neutralization of pseudoviruses containing Env mutations was compared to wild-type pseudovirus. A ≥15-fold increase in IC50 from wild-type to mutant was considered positive.
Statistical analysis
Statistical tests were performed in SAS, version 9.2 (SAS Institute, Cary, NC) or in R, version 2.15.2 (R Foundation for Statistical Computing, Vienna, Austria). The statistical test is noted when p values are presented.
Supplementary Material
Acknowledgements
We regret that due to space limitations we were unable to cite many excellent papers of our colleagues.
HJ16 was generously donated by Davide Corti (Institute for Research in Biomedicine, Bellinzona, Switzerland).
Support provided by a Collaboration for AIDS Vaccine Discovery grant to BFH from the Bill and Melinda Gates Foundation, the Center For HIV/AIDS Vaccine Immunology (CHAVI; grant U19 AI067854) and the Center For HIV/AIDS Vaccine Immunology-Immunogen Discovery grant (CHAVI-ID; grant UM1 AI100645). Further support provided by CAVD funding for the CTVIMC, grant number OPP1032325; the Duke Center for AIDS Research Flow Cytometry and Virology cores (CFAR; P30-AI-64518); NIH grant P01 AI 100151 (SZP) and funds from the Department of Veterans Affairs.
We thank Daniel M. Kozink, Florence Perrin and Abby J. Cooper for expert technical support.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Supplemental information includes additional Experimental Procedures, four tables and four figures.
Author Contributions
MAM, FG, MB, GDT, SZP, JRM, BHa, GMS, JGS, HXL, DCM, BTK and BFH designed the studies and wrote/edited the paper. MAM, TCG, JDA, DJM and JFW isolated mAbs by sorting. FG, AK and BHo isolated env genes by SGA and made viruses. SMX, RP, KEL, AF, SMA, GF, XS, GDT, ESG, NLT, LM, HXL, DCM and BFH assayed viruses and/or mAbs. KKH, XL and MB isolated mAbs by culture. MAM, FG, AK, BHo, MB, AF, NAV, SMA, GF, XS, GDT, ESG, NLT, LM, SZP, MKG, JGS, HXL, DCM, PTH, BTK and BFH analyzed data. GK, MSC, NES and SK recruited the subjects.
References
- Balla-Jhagjhoorsingh SS, Corti D, Heyndrickx L, Willems E, Vereecken K, Davis D, Vanham G. The N276 glycosylation site is required for HIV-1 neutralization by the CD4 binding site specific HJ16 monoclonal antibody. PLoS ONE. 2013;8:e68863. doi: 10.1371/journal.pone.0068863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouvin-Pley M, Morgand M, Meyer L, Goujard C, Moreau A, Mouquet H, Nussenzweig M, Pace C, Ho D, Bjorkman PJ, et al. Drift of the HIV-1 envelope glycoprotein gp120 toward increased neutralization resistance over the course of the epidemic: a comprehensive study using the most potent and broadly neutralizing monoclonal antibodies. J Virol. 2014;88:13910–13917. doi: 10.1128/JVI.02083-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derdeyn CA, Moore PL, Morris L. Development of broadly neutralizing antibodies from autologous neutralizing antibody responses in HIV infection. Curr Opin HIV AIDS. 2014;9:210–216. doi: 10.1097/COH.0000000000000057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao F, Bonsignori M, Kumar A, Xia S-M, Lu X, Cai F, Hwang K-K, Song H, Zhou T, Lynch RM, et al. Cooperation of B Cell Lineages in Induction of HIV-1-Broadly Neutralizing Antibodies. Cell. 2014;158:481–491. doi: 10.1016/j.cell.2014.06.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert PB, Peterson ML, Follmann D, Hudgens MG, Francis DP, Gurwith M, Heyward WL, Jobes DV, Popovic V, Self SG, et al. Correlation between immunologic responses to a recombinant glycoprotein 120 vaccine and incidence of HIV-1 infection in a phase 3 HIV-1 preventive vaccine trial. J Infect Dis. 2005;191:666–677. doi: 10.1086/428405. [DOI] [PubMed] [Google Scholar]
- Gorny MK, Wang X-H, Williams C, Volsky B, Revesz K, Witover B, Burda S, Urbanski M, Nyambi P, Krachmarov C, et al. Preferential use of the VH5-51 gene segment by the human immune response to code for antibodies against the V3 domain of HIV-1. Mol Immunol. 2009;46:917–926. doi: 10.1016/j.molimm.2008.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottardo R, Bailer RT, Korber BT, Gnanakaran S, Phillips J, Shen X, Tomaras GD, Turk E, Imholte G, Eckler L, et al. Plasma IgG to linear epitopes in the V2 and V3 regions of HIV-1 gp120 correlate with a reduced risk of infection in the RV144 vaccine efficacy trial. PLoS ONE. 2013;8:e75665. doi: 10.1371/journal.pone.0075665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray ES, Madiga MC, Hermanus T, Moore PL, Wibmer CK, Tumba NL, Werner L, Mlisana K, Sibeko S, Williamson C, et al. The Neutralization Breadth of HIV-1 Develops Incrementally over Four Years and Is Associated with CD4+ T Cell Decline and High Viral Load during Acute Infection. J Virol. 2011;85:4828–4840. doi: 10.1128/JVI.00198-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haim H, Salas I, McGee K, Eichelberger N, Winter E, Pacheco B, Sodroski J. Modeling virus- and antibody-specific factors to predict human immunodeficiency virus neutralization efficiency. Cell Host Microbe. 2013;14:547–558. doi: 10.1016/j.chom.2013.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haim H, Strack B, Kassa A, Madani N, Wang L, Courter JR, Princiotto A, McGee K, Pacheco B, Seaman MS, et al. Contribution of intrinsic reactivity p 20 of 22 of the HIV-1 envelope glycoproteins to CD4-independent infection and global inhibitor sensitivity. PLoS Pathog. 2011;7:e1002101. doi: 10.1371/journal.ppat.1002101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haldar B, Burda S, Williams C, Heyndrickx L, Vanham G, Gorny MK, Nyambi P. Longitudinal study of primary HIV-1 isolates in drug-naïve individuals reveals the emergence of variants sensitive to anti-HIV-1 monoclonal antibodies. PLoS ONE. 2011;6:e17253. doi: 10.1371/journal.pone.0017253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haynes BF, Gilbert PB, McElrath MJ, Zolla-Pazner S, Tomaras GD, Alam SM, Evans DT, Montefiori DC, Karnasuta C, Sutthent R, et al. Immune-correlates analysis of an HIV-1 vaccine efficacy trial. N Engl J Med. 2012;366:1275–1286. doi: 10.1056/NEJMoa1113425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hioe CE, Wrin T, Seaman MS, Yu X, Wood B, Self S, Williams C, Gorny MK, Zolla-Pazner S. Anti-V3 monoclonal antibodies display broad neutralizing activities against multiple HIV-1 subtypes. PLoS ONE. 2010;5:e10254. doi: 10.1371/journal.pone.0010254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hraber P, Seaman MS, Bailer RT, Mascola JR, Montefiori DC, Korber BT. Prevalence of broadly neutralizing antibody responses during chronic HIV-1 infection. Aids. 2014;28:163–169. doi: 10.1097/QAD.0000000000000106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly HR, Urbanski M, Burda S, Zhong P, Konings F, Nanfack J, Tongo M, Kinge T, Achkar J, Nyambi P. Neutralizing antibody patterns and viral escape in HIV-1 non-B subtype chronically infected treatment-naive individuals. Hum Antibodies. 2005;14:89–99. [PubMed] [Google Scholar]
- Klein F, Nogueira L, Nishimura Y, Phad G, West AP, Halper-Stromberg A, Horwitz JA, Gazumyan A, Liu C, Eisenreich TR, et al. Enhanced HIV-1 immunotherapy by commonly arising antibodies that target virus escape variants. Journal of Experimental Medicine. 2014;211:2361–2372. doi: 10.1084/jem.20141050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao H-X, Lynch R, Zhou T, Gao F, Alam SM, Boyd SD, Fire AZ, Roskin KM, Schramm CA, Zhang Z, et al. Co-evolution of a broadly neutralizing HIV-1 antibody and founder virus. Nature. 2013;496:469–476. doi: 10.1038/nature12053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malek A, Sager R, Kuhn P, Nicolaides KH, Schneider H. Evolution of maternofetal transport of immunoglobulins during human pregnancy. Am. J. Reprod. Immunol. 1996;36:248–255. doi: 10.1111/j.1600-0897.1996.tb00172.x. [DOI] [PubMed] [Google Scholar]
- Mascola JR, Haynes BF. HIV-1 neutralizing antibodies: understanding nature's pathways. Immunol Rev. 2013;254:225–244. doi: 10.1111/imr.12075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montefiori DC, Karnasuta C, Huang Y, Ahmed H, Gilbert P, de Souza MS, mclinden R, Tovanabutra S, Laurence-Chenine A, Sanders-Buell E, et al. Magnitude and breadth of the neutralizing antibody response in the RV144 and Vax003 HIV-1 vaccine efficacy trials. Journal of Infectious Diseases. 2012;206:431–441. doi: 10.1093/infdis/jis367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore JP, Sattentau QJ, Wyatt R, Sodroski J. Probing the structure of the human immunodeficiency virus surface glycoprotein gp120 with a panel of p 21 of 22 monoclonal antibodies. J Virol. 1994;68:469–484. doi: 10.1128/jvi.68.1.469-484.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munro JB, Gorman J, Ma X, Zhou Z, Arthos J, Burton DR, Koff WC, Courter JR, Smith AB, Kwong PD, et al. Conformational dynamics of single HIV-1 envelope trimers on the surface of native virions. Science. 2014;346:759–763. doi: 10.1126/science.1254426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nyambi P, Burda S, Urbanski M, Heyndrickx L, Janssens W, Vanham G, Nadas A. Neutralization patterns and evolution of sequential HIV type 1 envelope sequences in HIV type 1 subtype B-infected drug-naive individuals. AIDS Res Hum Retroviruses. 2008;24:1507–1519. doi: 10.1089/aid.2008.0154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Permar SR, Fong Y, Vandergrift N, Fouda GG, Gilbert P, Parks R, Jaeger FH, Pollara J, Martelli A, Liebl BE, et al. Maternal HIV-1 envelope-specific antibody responses and reduced risk of perinatal transmission. J Clin Invest. 2015;125:2702–2706. doi: 10.1172/JCI81593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richman DD, Wrin T, Little SJ, Petropoulos CJ. Rapid evolution of the neutralizing antibody response to HIV type 1 infection. Proceedings of the National Academy of Sciences. 2003;100:4144–4149. doi: 10.1073/pnas.0630530100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salazar-Gonzalez JF, Salazar MG, Keele BF, Learn GH, Giorgi EE, Li H, Decker JM, Wang S, Baalwa J, Kraus MH, et al. Genetic identity, biological phenotype, and evolutionary pathways of transmitted/founder viruses in acute and early HIV-1 infection. J Exp Med. 2009;206:1273–1289. doi: 10.1084/jem.20090378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheid JF, Mouquet H, Ueberheide B, Diskin R, Klein F, Oliveira TYK, Pietzsch J, Fenyo D, Abadir A, Velinzon K, et al. Sequence and Structural Convergence of Broad and Potent HIV Antibodies That Mimic CD4 Binding. Science. 2011;333:1633–1637. doi: 10.1126/science.1207227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seaman MS, Janes H, Hawkins N, Grandpre LE, Devoy C, Giri A, Coffey RT, Harris L, Wood B, Daniels MG, et al. Tiered categorization of a diverse panel of HIV-1 Env pseudoviruses for assessment of neutralizing antibodies. J Virol. 2010;84:1439–1452. doi: 10.1128/JVI.02108-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomaras GD, Binley JM, Gray ES, Crooks ET, Osawa K, Moore PL, Tumba N, Tong T, Shen X, Yates NL, et al. Polyclonal B cell responses to conserved neutralization epitopes in a subset of HIV-1-infected individuals. J Virol. 2011;85:11502–11519. doi: 10.1128/JVI.05363-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Upadhyay C, Mayr LM, Zhang J, Kumar R, Gorny MK, Nadas A, Zolla-Pazner S, Hioe CE. Distinct mechanisms regulate exposure of neutralizing epitopes in the V2 and V3 loops of HIV-1 envelope. J Virol. 2014;88:12853–12865. doi: 10.1128/JVI.02125-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei X, Decker JM, Wang S, Hui H, Kappes JC, Wu X, Salazar-Gonzalez JF, Salazar MG, Kilby JM, Saag MS, et al. Antibody neutralization and escape by HIV-1. Nature. 2003;422:307–312. doi: 10.1038/nature01470. [DOI] [PubMed] [Google Scholar]
- Wu X, Zhou T, Zhu J, Zhang B, Georgiev I, Wang C, Chen X, Longo NS, Louder M, McKee K, et al. Focused evolution of HIV-1 neutralizing antibodies revealed by structures and deep sequencing. Science. 2011;333:1593–1602. doi: 10.1126/science.1207532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zolla-Pazner S, DeCamp A, Gilbert PB, Williams C, Yates NL, Williams WT, Howington R, Fong Y, Morris DE, Soderberg KA, et al. Vaccine-induced IgG antibodies to V1V2 regions of multiple HIV-1 subtypes correlate with decreased risk of HIV-1 infection. PLoS ONE. 2014;9:e87572. doi: 10.1371/journal.pone.0087572. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.