Abstract
About one third of cancers harbor activating mutations in rat sarcoma viral oncogene homolog (RAS) oncogenes. In melanoma, aberrant neuroblastoma-RAS (NRAS) signaling fuels tumor progression in about 20% of patients. Current therapeutics for NRAS driven malignancies barely impact overall survival. To date, pathway interference downstream of mutant NRAS seems to be the most promising approach. In this study, data revealed that mutant NRAS induced Plk1 expression, and pharmacologic inhibition of Plk1 stabilized the size of NRAS mutant melanoma xenografts. The combination of MEK and Plk1 inhibitors resulted in a significant growth reduction of NRAS mutant melanoma cells in vitro, and regression of xenografted NRAS mutant melanoma in vivo. Independent cell cycle arrest and increased induction of apoptosis underlies the synergistic effect of this combination. Data further suggest that the p53 signaling pathway is of key importance to the observed therapeutic efficacy. This study provides in vitro, in vivo and first mechanistic data, that a MEK/Plk1 inhibitor combination might be a promising treatment approach for patients with NRAS driven melanoma. Since mutant NRAS signaling is similar across different malignancies, this inhibitor combination could also offer a previously unreported treatment modality for NRAS mutant tumors of other cell origins.
Introduction
Mutations in the Neuroblastoma Rat Sarcoma viral oncogene homolog (NRAS) gene account for up to 20% of driving oncogenes in melanoma, making NRAS an enticing target for treatment (Jakob et al. 2012; Fedorenko et al. 2013). Although small molecule inhibitors directed against the constitutively active protein would be ideal, selectively targeting mutant RAS in vivo has thus far proven to be impossible (Eskandarpour et al. 2005; Jaiswal et al. 2009; Kelleher and McArthur 2012). Current therapeutics barely impact overall survival, emphasizing the need for improved treatment modalities.
Recent advances in the treatment of NRAS mutant melanoma arise from interfering with key downstream signaling cascades of RAS, such as the mitogen activated protein kinase (MAPK), PI3K and Ral pathways as well as cell cycle regulator proteins. The MAPK pathway is critical for anchorage independent growth and survival of melanoma cells (Mishra et al. 2010; Atefi et al. 2011; Greger et al. 2012; Posch et al. 2013; Rebecca et al. 2014). Still, single inhibitor treatment, targeting this pathway only marginally improved overall survival (Ascierto et al. 2013). MAPK reactivation and increased signaling through other pro-survival cascades such as the PI3K/mammalian target of rapamycin (mTOR) and/or cell cycle pathways cause resistance to treatment after only months of therapy (Catalanotti et al. 2013; Long et al. 2014). Accordingly, current research focuses on the development of effective inhibitor combinations (Kwong et al. 2012; Posch et al. 2013).
In this study, we show that the expression of the mitotic regulator, Polo-like kinase 1 (Plk1) is increased in a large panel of NRAS mutant melanoma cells. It has been established previously, that Plk1 directly contributes to malignant transformation and is over expressed in various cancers, including melanoma (Wolf et al. 1997; Knecht et al. 1999; Gray et al. 2004; Jalili et al. 2011). Still, Plk1 inhibition alone did not meet preclinical expectations in recent clinical trials (Lin et al. 2014; Stadler et al. 2014).
The induction of Plk1 by mutant NRAS and the importance of the MAPK pathway for tumor cell homeostasis, provided the rationale to investigate the combination of a MEK and a Plk1 inhibitor for the treatment of NRAS mutant melanoma. This study provides first evidence that combined MEK and Plk1 inhibitor treatment induces apoptosis and synergistically inhibits NRAS mutant melanoma in vitro and in vivo.
Results
Plk1 is over-expressed in melanoma cells bearing NRAS(Q61) mutations
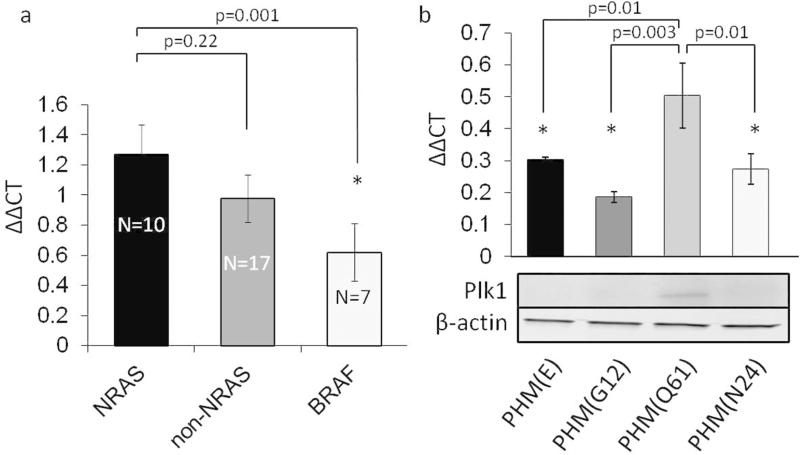
To study Plk1 mRNA expression in melanoma, we performed real time qPCR in 23 human melanoma cell lines bearing the most common melanoma driving mutations. Cell lines are known to harbor mutations in v-Raf murine sarcoma viral oncogene homolog B (BRAF), NRAS, C-KIT and guanine nucleotide-binding protein G(q) subunit alpha/11 (GNAQ/GNA11). One cell line was wild type for the above mentioned mutations. We found Plk1 expression in all 23 cell lines. On average, NRAS mutant cell lines expressed high levels of Plk1 mRNA, particularly compared to BRAF mutant melanoma cells (Fig. 1a). C-KIT, GNAQ and GNA11 mutant melanoma cell lines also displayed increased Plk1 levels compared to BRAF lines tested (table S1). To investigate a potential correlation between NRAS mutations and Plk1 expression levels, we stably transduced primary human melanocytes (PMH) with different NRAS vectors. PHM bearing a NRAS(Q61) mutation showed significantly increased Plk1 mRNA and protein levels compared to PHM transduced with NRAS(G12), non-mutagenic NRAS(N24), or empty vector controls (E) (Fig. 1b).
Figure 1. NRAS mutant cell lines express high levels of Plk1 mRNA.
(a) Relative expression of Plk1 in 23 human melanoma cell lines. NRAS mutant melanoma cell lines show higher Plk1 expression compared to non-NRAS and, in particular, to BRAF(V600) mutant cell lines. (b) Plk1 expression in stably transduced primary human melanocytes (PHM) expressing the indicated NRAS mutations or an empty vector control (E). NRAS(Q61) mutant cells show a significant increase of Plk1 expression on mRNA and protein level compared to empty vector controls, non-oncogenic NRAS(N24) mutants and NRAS(G12) mutant cells. Bars represent the mean relative expression of Plk1. β-actin served as an endogenous control. (N=3; * indicates p<0.05; mean±SEM)
Small molecule inhibitors of Plk1 and MEK reduce growth of NRAS mutant melanoma cell lines in vitro
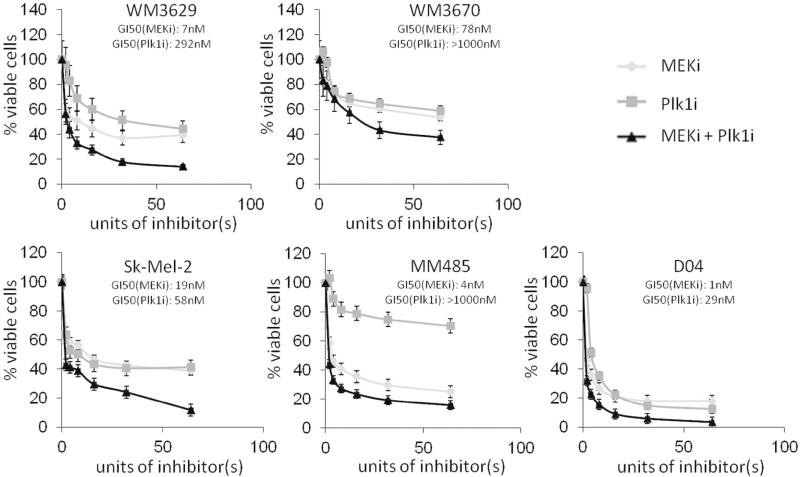
In this study we used JTP-74057 (trametinib) for selective MEK inhibition and BI6727 (volasertib) for selective Plk1 inhibition. BI6727 has previously been shown to be a highly potent and selective inhibitor of Plk1 (IC50=0.87nM), while failing to show any inhibitory activity in a panel of >50 kinases up to a drug concentration of 10NM (Rudolph et al, 2009). Both inhibitors have previously been demonstrated to be more potent than several other small molecule compounds currently available for selective inhibition of the desired targets (Schmit et al. 2009; Posch et al. 2013). Inhibitor concentrations to reach 50% growth inhibition (GI50) varied across cell lines (Fig. 2, S1), with several cells displaying GI50 in the low nanomolar range. All NRAS mutant cells, except WM1366, required higher concentrations of the Plk1 inhibitor compared to the MEK inhibitor for equipotent growth inhibition (Fig. 2, S1, table S2). WM3629 cells had a mutation in the Plk1 gene at 633(C>T) (table S1); However, WM3629 cells showed a decrease in cell viability after Plk1 inhibition comparable to Plk1 wild type cells.
Figure 2. Dose dependent reduction of cell viability with inhibitors of MEK and Plk1.
Dose response curves of 5 NRAS mutant melanoma cell lines (WM3629, WM3670, Sk-Mel-2, MM485 and D04) using inhibitors of MEK, Plk1 or their combination. All cell lines were sensitive to MEK and Plk1 inhibition as well as the combination of both inhibitors. (GI50 = concentration at 50% growth inhibition; N=3; mean±SD; one unit of inhibitor represents 1nM of the MEK inhibitor JTP/74057 and 8nM for the Plk1 inhibitor BI6727)
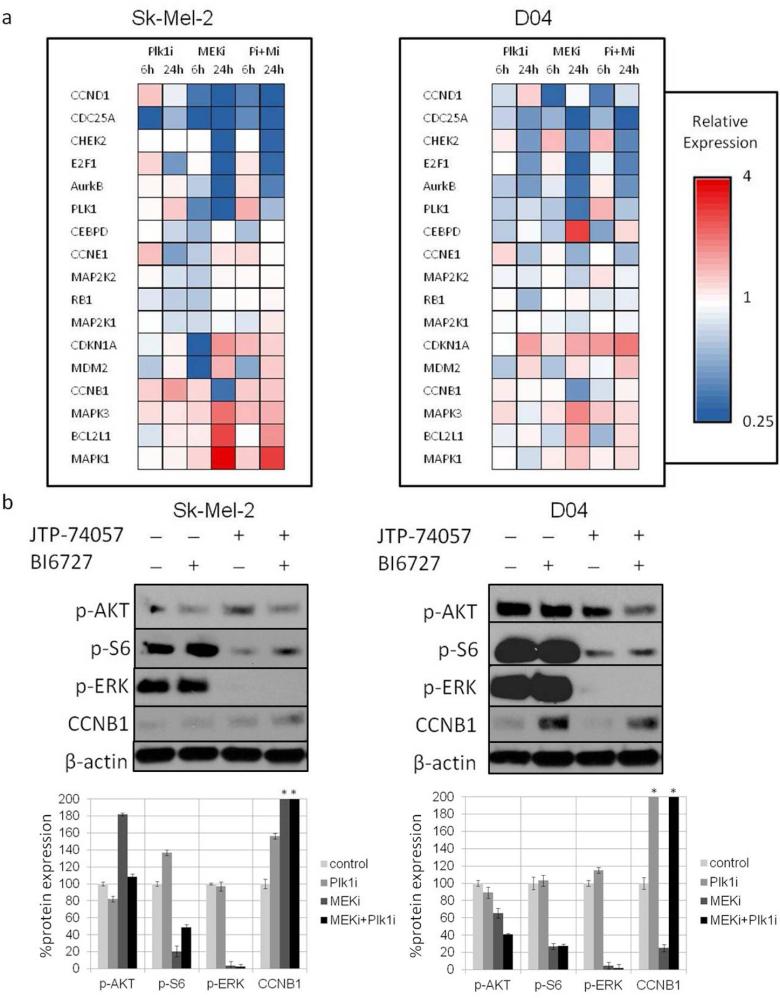
Signaling changes following inhibitor treatment were examined by a custom mRNA array and immunoblotting (Fig. 3). Plk1 inhibition affected several genes involved in the cell cycle regulation, with a reduction of E2F1, CDC25A, CCNE1 and RB1 expression observed after 24h of incubation. The MEK inhibitor (MEKi) reduced several cell cycle related genes and induced mRNA levels of CEBPD, which has been reported to be involved in cell cycle regulation and programmed cell death (Balamurugan and Sterneck 2013). We observed an increase of MAPK1 and MAPK3 mRNA levels by the MEKi (Fig. 3a) and elevated mRNA levels of BCL2L1, an inhibitor of cell death. Immunoblot analyses showed comparable regulation in select targets at the protein level (Fig. S2). Additionally, we found reduced p-S6 and extinguished p-ERK protein levels after MEKi treatment. p-AKT was induced in Sk-Mel-2 cells after MEKi treatment, as previously described (Posch et al. 2013). Plk1 inhibition resulted in a modest to pronounced increase of CCNB1 protein levels in Sk-Mel-2 and D04 cells, respectively (Fig. 3a and 3b).
Figure 3. Regulation of cell cycle genes, and p53/p21 signaling members with inhibitors of MEK and Plk1.
(a) Hierarchical clustering of mRNA levels in 2 NRAS mutant lines in response to the respective inhibitors after 6h and 24h of incubation. The expression patterns were similar in both cell lines. Several cell cycle related genes were affected by the single inhibitors and the MEK/Plk1 combination. (b) Immunoblot analyses of the human melanoma lines Sk-Mel-2 and D04 and corresponding bar graphs for relative protein expression. Reduced phospho-protein levels of ERK and S6 in MEK inhibitor treated conditions. Induction of p-AKT in Sk-Mel-2 cells after MEK inhibition. Reduction of p-AKT, p-ERKT and p-S6 as well as induction of CCNB1 by the combination treatment. (*greater than 200%; mean±SD; N=3)
Pharmacologic targeting of MEK and Plk1 synergistically inhibits NRAS mutant melanoma cells
Whereas the effects of single inhibitor treatment on cell viability varied across cell lines, the combination of a MEK and Plk1 inhibitor (Plk1i) potently reduced growth in all 10 human NRAS melanoma cells tested (Fig. 2 and S1). Several different ratios of the two components were assayed for synergism. Calculating the combination index (CI), which indicates whether a certain combination is synergistic, additive, or antagonistic (table S3), revealed synergistic growth inhibition with several different ratios in select cell lines; however, all NRAS mutant lines were synergistically inhibited with a ratio of MEK:Plk1=1:8 (Fig. S3). Non-transformed melanocytes as well as a BRAF mutant cell line showed no further decrease in cell viability by the addition of the Plk1i to the MEKi (Fig. S4). Table S2 summarizes GI50, CI values and the dose reduction index (DRI), which indicates how much the dose of each drug in a synergistic combination may be reduced at a given effect level, compared with the doses of each drug alone (Chou and Talalay 1984).
Signaling changes due to combination inhibitor treatment were assessed by a custom mRNA array. Cell cycle related genes CCND1, CDC25A, CHEK2, E2F1, AurkB, and Plk1 were potently reduced after 24h of incubation with the MEK/Plk1 combination. Following 24h incubation with the MEK/Plk1 combination, induction of MAPK3 was less pronounced compared to MEKi treatment alone. Interestingly, when comparing the two time points (6h, 24h), we observed induction of MDM2 and CDKN1A at 24h in both the MEK and Plk1 treatment groups, which persisted in the combinational group, suggesting regulation of p53/p21 signaling. Additionally, Plk1 inhibition attenuated mRNA levels of the pro-survival gene BCL2L1 in the combinational therapy when compared to the observed induction of BCL2L1 after single MEKi treatment. Furthermore, we observed an earlier and stronger induction of pro-apoptotic signals, such as caspase3/7 with the MEK/Plk1 combination (Fig. 3a, 3b, 4a, 4b, S5).
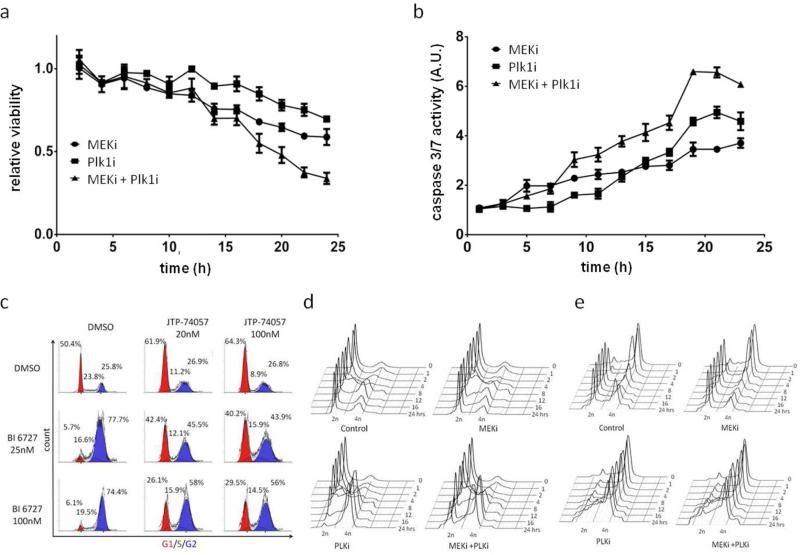
Figure 4. Induction of apoptosis and cell cycle arrest with the MEK/Plk1 combination.
(a) Earlier and pronounced decrease of cell viability over time and (b) earlier and stronger induction of caspase 3/7 activity with the MEK/Plk1 combination. (c) G0/G1 arrest in MEKi treated groups and G2/M arrest in BI6727 treated cells. The combination of both inhibitors caused a dose dependent dual G1 (red) and G2/M (blue) arrest. (d) Double thymidine block and time course analyses of cell cycle progression showed a G1 arrest with the MEKi. Progressing cells in the combinational treatment group succumb to G2/M arrest induced by the Plk1i. (e) The thymidine-nocodazole block revealed continuous G2/M arrest after Plk1i treatment. (N=3; mean±SD; MEKi(JTP/74017)=20nM; Plk1i(BI6727)=100nM)
Inhibition of MEK and Plk1 results in independent, dual cell cycle arrests in a phase dependent manner
Since several cell cycle related genes were affected by the MEK/Plk1 combination, we investigated cell cycle profiles by flow cytometry. Sk-Mel-2 cells were incubated with varying doses of a MEKi, a Plk1i, or DMSO control in both mono and combination therapies (Fig. 4c). In support of our results showing decreased transcription of cyclin D, we observed a cell cycle arrest at G0/G1 in both MEKi treated groups. In contrast, the Plk1i treated groups showed a G2/M arrest. Interestingly, when cells are treated with the MEK/Plk1 combination, they arrest in both G1 and G2/M phases in a dose dependent manner (Fig. 4c). Protein levels of cyclins D, E, and B in Sk-Mel-2 cells were similar to the transcript data, showing reduced, slightly decreased and increased expression, respectively (Fig. S2). Taken together, these data suggest that the drugs are capable of inducing cell cycle arrest at distinct phases, with a potential overlap during DNA replication.
To determine if the induced cell cycle arrest is dependent on the current phase progression of the cell, we synchronized Sk-Mel-2 cells in late G1 using a thymidine block. Analyses revealed that cells treated with MEKi had an observable release from G1 at eight hours, but to a lesser extent than that of the control condition (Fig. 4d). Additionally, MEKi treated cells maintained a larger G1 population compared to the other treatment groups throughout the duration of the experiment, suggesting MEK inhibition is capable of inducing an arrest beyond the restriction point (Fig. S6). Further evidence of G1 arrest was seen by comparing the combination with the Plk1i treatment groups in the 18-24 hour ranges. Importantly, the progressing population in the combinational treatment group succumbs to G2/M arrest induced by the Plk1i.
A second synchronization at G2/M was achieved by using a thymidine-nocodazole block (Fig. 4e). Following release, the control cells progressed through mitosis and returned to a G0/G1 state at two to four hours post synchronization, with a second cell division cycle occurring in the 18-24 hour range. However, the Plk1i treated cells failed to release from mitosis and maintained a G2/M arrest. Comparing the combinational group to Plk1i alone at 24 hours after release from G2/M synchronization, there was a measurable, but not significant increase in the G1 population in MEK/Plk1 treated cells (Fig. S6). Altogether, these findings point into the direction that cells escaping Plk1i mediated arrest maintain sensitivity to MEK inhibition.
Reduction of cell viability by the MEK/Plk1 combination is most effective in cells expressing p53
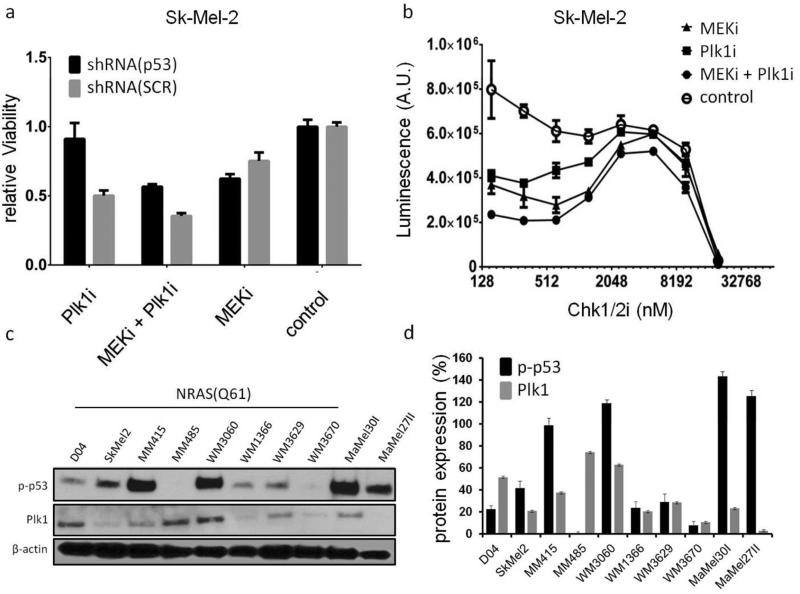
In our study, results of the custom mRNA array revealed regulation of genes in the p21/p53 pathway when cells were treated with the MEK/Plk1 combination. After 24 hours, mRNA levels of the p53 transcriptional targets MDM2 and CDKN1A were increased in both Sk-Mel-2 and D04 lines (Fig. 3a). Conversely, MM485 cells with no detectable levels of phospho-p53 had decreased levels of both transcripts (Fig. S7). Thus, we studied the role of the p53 pathway by interfering with p53 itself and via inhibition of Chk proteins (Fig. 5a and 5b). shRNA mediated knockdown of p53 in Sk-Mel-2 cells (Sk-Mel-2shRNA(p53)) reduced p53 protein levels (Fig. S8) and impaired Plk1i mediated reduction of cell viability (Fig. 5a).
Figure 5. p-53 signaling affects Plk1 and MEK/Plk1 inhibitor efficacy.
a. Stable knockdown of p53 reduced the activity of Plk1 and MEK/Plk1 inhibition. p53 interference had no effect on viability in cells treated with the MEKi only, or on vehicle treated cells. (b) Additional Inhibition of Chk1/2 markedly reduced viability in MEK/Plk1 treated compared to single inhibitor or vehicle treated controls in a biphasic manner. (c) Immunoblot analyses of p-p53 and Plk1 in all 10 NRAS mutant cell lines used in this study. All cells, except MM485, express p-p53. Plk1 protein expression was pronounced in NRAS(Q61) mutant cells. (d) Relative expression of the respective proteins compared to β/actin. (N=3; mean±SD; SCR=scramble control shRNA; MEKi(JTP-74057)=20nM; Plk1i(BI6727)=100nM; Chk1/2i=PF-0477736)
As displayed in panels 5c and 5d, MM485 cells did not express detectable levels of p-p53 protein. In line with the important role of p53 in Sk-Mel-2 cells, MM485 cells also showed low sensitivity to single Plk1i treatment (Fig. 2). p53 interference did not affect MEKi mediated reduction of cell viability. The efficacy of the MEK/Plk1 combination was also impaired by shRNA mediated p53-knockdown (Fig. 5a). This effect is, at least partly, a result of reduced Plk1i-mediated inhibition of cell viability.
Due to the critical role of p53 in the DNA damage response and its known activation by the checkpoint proteins Chk1 and Chk2, we investigated the effect of MEKi and Plk1i on cell viability in the presence of the Chk1/2 inhibitor PF-0477736 (Chk1/2i). Sk-Mel-2 cells treated with MEKi and Plk1i individually or in combination demonstrated a biphasic response to increasing Chk1/2i concentrations (Fig. 5b). This pattern could be the result of Chki being sufficient to mitigate checkpoint activation induced by Plk1i and, to a lesser extent, by MEKi in the low micromolar ranges, while higher concentrations result in increased genotoxicity and the complete abrogation of the DNA damage checkpoints. This hypothesis is supported by reports showing that Plk1 is involved in checkpoint adaptation with DNA damage checkpoint inhibition resulting in the sensitization of cancer cells to DNA damaging agents (Yoo et al. 2004; Jiang et al. 2009; Anderson et al. 2011). In support of a Chki specific effect, we were unable to recapitulate these results using other kinase inhibitors, such as CDK4i or receptor tyrosine kinase inhibitors added to the MEK/Plk1 combination (data not shown).
The MEK/Plk1 inhibitor combination regresses human NRAS mutant melanoma xenografts
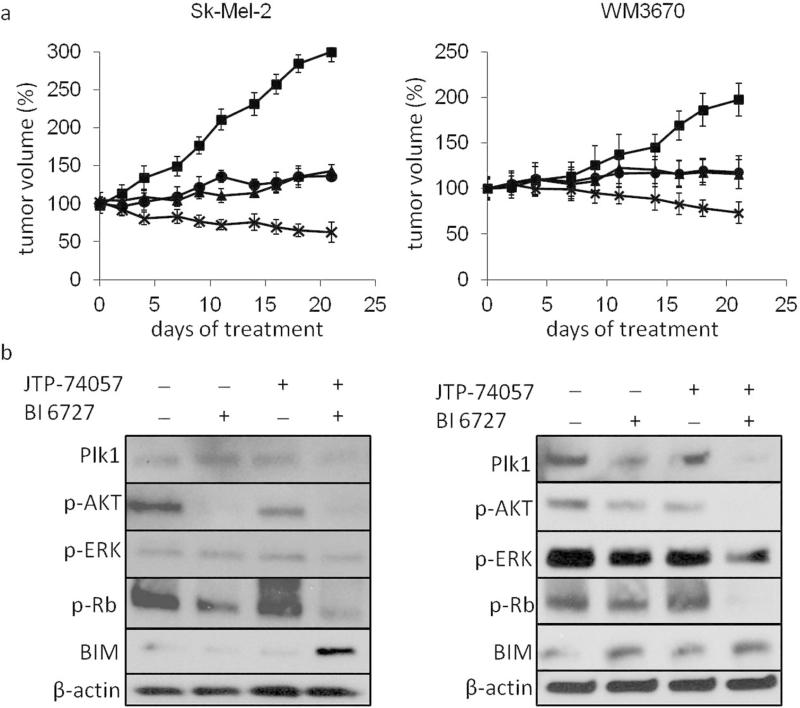
To test if the combination of a MEKi and a Plk1i is also effective in vivo, we established mouse melanoma xenografts using the human melanoma cell lines Sk-Mel-2 and WM3670. Single drugs or their combination were administered by oral gavage. The MEKi was given at a dose of 2mg/kg/day leading to tumor size stabilization as demonstrated previously (Posch et al. 2013). The Plk1i was given 3 times per week to reach a total amount of 50mg/kg/week and reduced tumor growth. Strikingly, combined MEK/Plk1 inhibition resulted in significant tumor shrinkage in all cell lines tested (Fig. 6a). No side effects of the combination therapy were observed. Immunoblot analyses of extracted tumor proteins revealed reduction of p/ERK, Plk1, p/AKT and p-RB. We further observed induction of the pro-apoptotic proteins BIM and BAX and a reduction of the anti-apoptotic protein Bcl-2 as well as increased caspase 3/7 activity in tumors treated with the combination of MEK/Plk1 (Fig. 6b, S9).
Figure 6. Tumor size reduction of NRAS mutant xenografts with the MEK/Plk1 combination.
(a) Treatment of established mouse xenografts showed tumor size reduction after initiation of treatment (day 0) with the combination of a MEKi (2 mg*kg−1*d−1) and Plk1i (50 mg*kg−1*week−1), but not with either MEKi or Plk1i alone. (cell lines: Sk-Mel-2, WM3670; MEKi: JTP-74057; Plk1i: BI6727; n=4) Results are displayed as the mean change in tumor volume at indicated time points ± SD. (b) Corresponding immunoblot analyses of mouse tumors after 3 weeks of treatment with the indicated conditions.
Discussion
It is well established that specific mutations and defined signaling aberrations in cancer contribute to differences in the biological behavior of malignancies. Mutant RAS genes are found in up to one third of human cancer (Bos 1989; Repasky et al. 2004; Schubbert et al. 2007) with NRAS mutations being the most prevalent in melanoma (Omerovic et al. 2007). Therapeutic modalities such as radiation therapy or chemotherapy for patients with NRAS mutant melanoma barely impact overall survival. Since direct inhibition of mutant NRAS is at present limited to preclinical models, pharmacologic interference with important RAS downstream signaling cascades seems to be the most promising treatment approach (Kwong et al. 2012; Posch et al. 2013; Vujic et al. 2014). One of the dominant signaling pathways in NRAS mutant melanoma is the MAPK pathway. Indeed, inhibition of the MAPK cascade with the MEK inhibitor MEK162 was tested in patients with NRAS mutant melanoma with encouraging results. However, responses were short and the development of resistance inevitable (Ascierto et al. 2013). This can, at least in part, be explained by the activation of other prosurvival pathways such as the PI3K/mTOR pathway after MEK inhibition (Fig. 3) (Posch et al. 2013). Thus, the search for effective co-extinction targets to MAPK inhibition is an ongoing and important area of research (Kwong et al. 2012; Ascierto et al. 2013; Posch et al. 2013).
Our data indicates that NRAS(Q61) mutations in PHM significantly increase Plk1 expression. This observation might be linked to higher oncogenicity of NRAS(Q61) compared to NRAS(G12) mutations in melanocytic cells (Burd et al. 2014). Comparing Plk1 levels of 6 NRAS(Q61) with 4 NRAS(G12) mutant melanoma cell lines also showed a trend towards higher Plk1 mRNA and protein expression in NRAS(Q61) cells; however, these analyses were not statistically significant (p=0.08; p=0.052; Fig. S10). Hence, we investigated if the a MEK/Plk1 inhibitor combination has antitumor activity in NRAS(G12) and NRAS(Q61) mutant melanoma cells.
Since the discovery of Plk proteins and proof of their biological importance in cancer maintenance, several compounds have been developed to target these molecules (Gumireddy et al. 2005; Lansing et al. 2007; Steegmaier et al. 2007; Beria et al. 2010). Among the 5 mammalian Plk family members, selective Plk1 depletion appears to be most desirable. Plk1 plays an important role throughout mitosis by influencing activation of CDK1/cyclin B, centrosome maturation, spindle formation, kinetochore assembly and regulation of microtubule nucleation as well as chromosome segregation and execution of cytokinesis (Lane and Nigg 1996; Seong et al. 2002; Sumara et al. 2004; van Vugt et al. 2004; Cholewa et al. 2014). Results presented in this study suggest that NRAS mutations are linked to high Plk1 expression. This is supported by findings in a model of transduced PHM as well as in a representative collection of melanoma lines. Additionally, when evaluating clinical data from The Cancer Genome Atlas (TCGA) of cutaneous melanoma, mutations in NRAS and increased expression of Plk1 co-occur (p=0.009) (Cerami et al. 2012; Gao et al. 2013). Even though further research is needed to address potential connections of RAS signaling and Plk1 expression, our results support current knowledge, highlighting the importance of Plk1 for viability of RAS activated malignancies (Luo et al. 2009; Wang et al. 2013; Yim and Erikson 2014). Still, cancer therapy with Plk1 inhibitors alone is ineffective clinically (Luo et al. 2009). It is believed that the somewhat low response rates might be explained by undesired co-targeting of Plk2 and Plk3, which attenuates the growth inhibitory effects of Plk1 abrogation (Strebhardt 2010).
In combination with MEK inhibition, however, we observe significant cell death in vitro and tumor shrinkage as well as induction of apoptosis in vivo (Fig. 6). The importance of cell cycle regulation in NRAS mutant melanoma has previously been shown. Recent findings using MEK/CDK4,6 inhibitor combinations support this notion, with promising (pre)clinical results (Kwong et al. 2012). However, several NRAS mutant cells and clinical tumors do not respond to treatment with MEK/CDK4,6 inhibitors. This might be explained by recent findings suggesting that NRAS mutation status may only determine response to this combination, when evaluated in tandem with aberrations in CDKN2A (Dong 2013). Data presented in the present study reveal, however, that the MEK/Plk1 inhibitor combination reduces cell growth independent of CDKN2A and Plk1 mutations (Fig. 2, S1, table S1).
Mounting evidence suggests that Plk1 affects p53 via direct binding and subsequent inhibition of its pro-apoptotic function (Ando et al. 2004). Accordingly, our findings show that the efficacy of Plk1 inhibition is related to p53 expression, because i) functional shRNA mediated knockdown of p53 in Sk-Mel-2 cells reduced the inhibitory effects of Plk1 and MEK/Plk1 treatment, and ii) cells with high p-p53(Ser15) protein expression showed a trend towards requiring less Plk1 inhibitor to reach GI50 (Fig. S11) compared to cells with low or non-detectable p-p53(Ser15) protein expression (Tsvetkov et al. 2003; Ando et al. 2004; Strebhardt 2010).
In addition to RNA silencing of p53, pharmacologic Chk1/2 inhibition also influenced the efficacy of combined MEK/Plk1 inhibition. However, it should be mentioned that even in cells with low p53 protein expression, MEK/Plk1 treatment effectively reduced cell growth, suggestive of other factors contributing to the inhibitory activity of this combination. While studies investigating the role of Plk1 in p53 signaling and its function outside of mitosis are scarce (Cholewa et al. 2013), possible redundancy with p53 family members, p63 and p73, need to be considered. In HeLa cells p73 is capable of inducing p21 expression in p53-inactivated cells following Plk1 inhibition, which results in greater sensitivity to additional therapeutics (Kreis et al. 2009). Furthermore, it has been reported that Plk1 phosphorylates and thus suppresses p63 mediated cell death in liver tumor cells (Komatsu et al. 2009). Taken together, our data offers important insights into the Plk1-p53 axis, which warrants further investigation.
In addition to the induction of pro-apoptotic signals in vitro and in vivo, MEK/Plk1 also caused dual cell cycle arrest. The MEKi-induced G1 arrest can potentially be attributed to the reduction in cyclin D expression. Cyclin D is required for cell cycle reentry and has previously been shown to be abrogated in response to MEK inhibition (Modi et al. 2012; Atefi et al. 2015). Plk1i induces a G2/M arrest through aberrant spindle pole formation, which ultimately results in mitotic catastrophe and subsequent apoptosis (Schmit et al. 2009). Results of this study suggest that cells escaping G1 arrest induced by MEK inhibition, or cells escaping G2/M arrest resulting from Plk1 inhibition, maintain their susceptibility to the other drug in combination.
In this study, we provide first in vitro and in vivo results, that a MEK/Plk1 combination might be a promising approach for the treatment of NRAS driven melanoma. Of note, and in support of signaling similarities across several NRAS mutant malignancies (Vujic et al. 2014), this inhibitor combination also reduced viability in one NRAS mutant neuroblastoma and one NRAS mutant lung cancer cell line in vitro (Fig. S12). This is particularly important, as effective therapeutic modalities for patients with NRAS mutant tumors of various etiologies are limited.
Material and Methods
Cell culturing, growth inhibition experiments and immunoblotting were preformed as previously described (Posch et al. 2013). Detailed material and methods as well as information on lentiviral production, transduction, cell cycle analyses, the custom mRNA array and qPCR including primer sequences (table S4) can be found in the electronic supporting information. Delta CT and deltadelta CT values of the custom mRNA array are provided in table S5. Xenograft studies were performed in CrTac:NCr-Foxn1nu mice. Inhibitor treatment was administered by oral gavage when tumors reached a volume of 80/100mm^3. All animal studies were approved by IACUC/LARC of the University of California San Francisco (AN086990).
Supplementary Material
Acknowledgements
The authors thank Boris Bastian for providing the cell lines used in this study. The Genome Analysis Core Facility, UCSF is acknowledged for their support in gene expression analysis. This study was supported by National Cancer Institute of the National Institutes of Health under award number K08CA155035 and the Melanoma Research Alliance Young Investigator Award. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. This work was also partially supported by funding from the National Institute of Health (T32 ES007015-35 to BDC; R01AR059130, R01CA176748 to NA) and the Department of Veterans Affairs (VA Merit Review Award 1I01BX001008 to NA). The authors are also grateful to Timothy Dattels, the Max Kade Foundation and the René Touraine Foundation for their generous support.
Footnotes
Conflict of interest: The authors have no conflict of interest.
References
- Anderson VE, Walton MI, Eve PD, et al. CCT241533 is a potent and selective inhibitor of CHK2 that potentiates the cytotoxicity of PARP inhibitors. Cancer Res. 2011;71:463–72. doi: 10.1158/0008-5472.CAN-10-1252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ando K, Ozaki T, Yamamoto H, et al. Polo-like kinase 1 (Plk1) inhibits p53 function by physical interaction and phosphorylation. J Biol Chem. 2004;279:25549–61. doi: 10.1074/jbc.M314182200. [DOI] [PubMed] [Google Scholar]
- Ascierto PA, Schadendorf D, Berking C, et al. MEK162 for patients with advanced melanoma harbouring NRAS or Val600 BRAF mutations: a non-randomised, open-label phase 2 study. Lancet Oncol. 2013;14:249–56. doi: 10.1016/S1470-2045(13)70024-X. [DOI] [PubMed] [Google Scholar]
- Atefi M, von Euw E, Attar N, et al. Reversing melanoma cross-resistance to BRAF and MEK inhibitors by co-targeting the AKT/mTOR pathway. PloS One. 2011;6:e28973. doi: 10.1371/journal.pone.0028973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atefi M, Titz B, Avramis E, et al. Combination of Pan-RAF and MEK inhibitors in NRAS mutant melanoma. Mol Cancer. 2015;14:27. doi: 10.1186/s12943-015-0293-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balamurugan K, Sterneck E. The Many Faces of C/EBP? and their Relevance for Inflammation and Cancer. Int J Biol Sci. 2013;9:917–33. doi: 10.7150/ijbs.7224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beria I, Ballinari D, Bertrand JA, et al. Identification of 4,5-dihydro-1H-pyrazolo[4,3-h]quinazoline derivatives as a new class of orally and selective Polo-like kinase 1 inhibitors. J Med Chem. 2010;53:3532–51. doi: 10.1021/jm901713n. [DOI] [PubMed] [Google Scholar]
- Bos JL. ras oncogenes in human cancer: a review. Cancer Res. 1989;49:4682–9. [PubMed] [Google Scholar]
- Burd CE, Liu W, Huynh MV, et al. Mutation-Specific RAS Oncogenicity Explains NRAS Codon 61 Selection in Melanoma. Cancer Discov. 2014;4:1418–29. doi: 10.1158/2159-8290.CD-14-0729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catalanotti F, Solit DB, Pulitzer MP, et al. Phase II trial of MEK inhibitor selumetinib (AZD6244, ARRY-142886) in patients with BRAFV600E/K-mutated melanoma. Clin Cancer Res Off J Am Assoc Cancer Res. 2013;19:2257–64. doi: 10.1158/1078-0432.CCR-12-3476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerami E, Gao J, Dogrusoz U, et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2:401–4. doi: 10.1158/2159-8290.CD-12-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cholewa BD, Liu X, Ahmad N. The role of polo-like kinase 1 in carcinogenesis: cause or consequence? Cancer Res. 2013;73:6848–55. doi: 10.1158/0008-5472.CAN-13-2197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cholewa BD, Pellitteri-Hahn MC, Scarlett CO, et al. Large-Scale Label-Free Comparative Proteomics Analysis of Polo-Like Kinase 1 Inhibition via the Small-Molecule Inhibitor BI 6727 (Volasertib) in BRAF(V600E) Mutant Melanoma Cells. J Proteome Res. 2014 doi: 10.1021/pr5002516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou TC, Talalay P. Quantitative analysis of dose-effect relationships: the combined effects of multiple drugs or enzyme inhibitors. Adv Enzyme Regul. 1984;22:27–55. doi: 10.1016/0065-2571(84)90007-4. [DOI] [PubMed] [Google Scholar]
- Dong J. Overcoming Resistance to BRAF and MEK Inhibitors by Simultaneous Suppression of CDK4. In: Duc H, editor. Melanoma - Early Detect. Treat. InTech.; 2013. [Google Scholar]
- Eskandarpour M, Kiaii S, Zhu C, et al. Suppression of oncogenic NRAS by RNA interference induces apoptosis of human melanoma cells. Int J Cancer J Int Cancer. 2005;115:65–73. doi: 10.1002/ijc.20873. [DOI] [PubMed] [Google Scholar]
- Fedorenko IV, Gibney GT, Smalley KSM. NRAS mutant melanoma: biological behavior and future strategies for therapeutic management. Oncogene. 2013;32:3009–18. doi: 10.1038/onc.2012.453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao J, Aksoy BA, Dogrusoz U, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal. 2013;6:pl1. doi: 10.1126/scisignal.2004088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray PJ, Bearss DJ, Han H, et al. Identification of human polo-like kinase 1 as a potential therapeutic target in pancreatic cancer. Mol Cancer Ther. 2004;3:641–6. [PubMed] [Google Scholar]
- Greger JG, Eastman SD, Zhang V, et al. Combinations of BRAF, MEK, and PI3K/mTOR inhibitors overcome acquired resistance to the BRAF inhibitor GSK2118436 dabrafenib, mediated by NRAS or MEK mutations. Mol Cancer Ther. 2012;11:909–20. doi: 10.1158/1535-7163.MCT-11-0989. [DOI] [PubMed] [Google Scholar]
- Gumireddy K, Reddy MVR, Cosenza SC, et al. ON01910, a non-ATP-competitive small molecule inhibitor of Plk1, is a potent anticancer agent. Cancer Cell. 2005;7:275–86. doi: 10.1016/j.ccr.2005.02.009. [DOI] [PubMed] [Google Scholar]
- Jaiswal BS, Janakiraman V, Kljavin NM, et al. Combined targeting of BRAF and CRAF or BRAF and PI3K effector pathways is required for efficacy in NRAS mutant tumors. PloS One. 2009;4:e5717. doi: 10.1371/journal.pone.0005717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakob JA, Bassett RL, Jr, Ng CS, et al. NRAS mutation status is an independent prognostic factor in metastatic melanoma. Cancer. 2012;118:4014–23. doi: 10.1002/cncr.26724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jalili A, Moser A, Pashenkov M, et al. Polo-like kinase 1 is a potential therapeutic target in human melanoma. J Invest Dermatol. 2011;131:1886–95. doi: 10.1038/jid.2011.136. [DOI] [PubMed] [Google Scholar]
- Jiang H, Reinhardt HC, Bartkova J, et al. The combined status of ATM and p53 link tumor development with therapeutic response. Genes Dev. 2009;23:1895–909. doi: 10.1101/gad.1815309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelleher FC, McArthur GA. Targeting NRAS in melanoma. Cancer J Sudbury Mass. 2012;18:132–6. doi: 10.1097/PPO.0b013e31824ba4df. [DOI] [PubMed] [Google Scholar]
- Knecht R, Elez R, Oechler M, et al. Prognostic significance of polo-like kinase (PLK) expression in squamous cell carcinomas of the head and neck. Cancer Res. 1999;59:2794–7. [PubMed] [Google Scholar]
- Komatsu S, Takenobu H, Ozaki T, et al. Plk1 regulates liver tumor cell death by phosphorylation of TAp63. Oncogene. 2009;28:3631–41. doi: 10.1038/onc.2009.216. [DOI] [PubMed] [Google Scholar]
- Kreis N-N, Sommer K, Sanhaji M, et al. Long-term downregulation of Polo-like kinase 1 increases the cyclin-dependent kinase inhibitor p21(WAF1/CIP1). Cell Cycle Georget Tex. 2009;8:460–72. doi: 10.4161/cc.8.3.7651. [DOI] [PubMed] [Google Scholar]
- Kwong LN, Costello JC, Liu H, et al. Oncogenic NRAS signaling differentially regulates survival and proliferation in melanoma. Nat Med. 2012;18:1503–10. doi: 10.1038/nm.2941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lane HA, Nigg EA. Antibody microinjection reveals an essential role for human polo-like kinase 1 (Plk1) in the functional maturation of mitotic centrosomes. J Cell Biol. 1996;135:1701–13. doi: 10.1083/jcb.135.6.1701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lansing TJ, McConnell RT, Duckett DR, et al. In vitro biological activity of a novel small-molecule inhibitor of polo-like kinase 1. Mol Cancer Ther. 2007;6:450–9. doi: 10.1158/1535-7163.MCT-06-0543. [DOI] [PubMed] [Google Scholar]
- Lin C-C, Su W-C, Yen C-J, et al. A phase I study of two dosing schedules of volasertib (BI 6727), an intravenous polo-like kinase inhibitor, in patients with advanced solid malignancies. Br J Cancer. 2014;110:2434–40. doi: 10.1038/bjc.2014.195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Lei M, Erikson RL. Normal cells, but not cancer cells, survive severe Plk1 depletion. Mol Cell Biol. 2006;26:2093–108. doi: 10.1128/MCB.26.6.2093-2108.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long GV, Fung C, Menzies AM, et al. Increased MAPK reactivation in early resistance to dabrafenib/trametinib combination therapy of BRAF-mutant metastatic melanoma. Nat Commun. 2014;5:5694. doi: 10.1038/ncomms6694. [DOI] [PubMed] [Google Scholar]
- Luo J, Emanuele MJ, Li D, et al. A genome-wide RNAi screen identifies multiple synthetic lethal interactions with the Ras oncogene. Cell. 2009;137:835–48. doi: 10.1016/j.cell.2009.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishra PJ, Ha L, Rieker J, et al. Dissection of RAS downstream pathways in melanomagenesis: a role for Ral in transformation. Oncogene. 2010;29:2449–56. doi: 10.1038/onc.2009.521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Modi PK, Komaravelli N, Singh N, et al. Interplay between MEK-ERK signaling, cyclin D1, and cyclin-dependent kinase 5 regulates cell cycle reentry and apoptosis of neurons. Mol Biol Cell. 2012;23:3722–30. doi: 10.1091/mbc.E12-02-0125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Omerovic J, Laude AJ, Prior IA. Ras proteins: paradigms for compartmentalised and isoform-specific signalling. Cell Mol Life Sci CMLS. 2007;64:2575–89. doi: 10.1007/s00018-007-7133-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Posch C, Moslehi H, Feeney L, et al. Combined targeting of MEK and PI3K/mTOR effector pathways is necessary to effectively inhibit NRAS mutant melanoma in vitro and in vivo. Proc Natl Acad Sci U S A. 2013 doi: 10.1073/pnas.1216013110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rebecca VW, Alicea GM, Paraiso KHT, et al. Vertical inhibition of the MAPK pathway enhances therapeutic responses in NRAS-mutant melanoma. Pigment Cell Melanoma Res. 2014 doi: 10.1111/pcmr.12303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Repasky GA, Chenette EJ, Der CJ. Renewing the conspiracy theory debate: does Raf function alone to mediate Ras oncogenesis? Trends Cell Biol. 2004;14:639–47. doi: 10.1016/j.tcb.2004.09.014. [DOI] [PubMed] [Google Scholar]
- Schmit TL, Zhong W, Setaluri V, et al. Targeted depletion of Polo-like kinase (Plk) 1 through lentiviral shRNA or a small-molecule inhibitor causes mitotic catastrophe and induction of apoptosis in human melanoma cells. J Invest Dermatol. 2009;129:2843–53. doi: 10.1038/jid.2009.172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schubbert S, Shannon K, Bollag G. Hyperactive Ras in developmental disorders and cancer. Nat Rev Cancer. 2007;7:295–308. doi: 10.1038/nrc2109. [DOI] [PubMed] [Google Scholar]
- Seong Y-S, Kamijo K, Lee J-S, et al. A spindle checkpoint arrest and a cytokinesis failure by the dominant-negative polo-box domain of Plk1 in U-2 OS cells. J Biol Chem. 2002;277:32282–93. doi: 10.1074/jbc.M202602200. [DOI] [PubMed] [Google Scholar]
- Stadler WM, Vaughn DJ, Sonpavde G, et al. An open-label, single-arm, phase 2 trial of the Polo-like kinase inhibitor volasertib (BI 6727) in patients with locally advanced or metastatic urothelial cancer. Cancer. 2014;120:976–82. doi: 10.1002/cncr.28519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steegmaier M, Hoffmann M, Baum A, et al. BI 2536, a potent and selective inhibitor of polo-like kinase 1, inhibits tumor growth in vivo. Curr Biol CB. 2007;17:316–22. doi: 10.1016/j.cub.2006.12.037. [DOI] [PubMed] [Google Scholar]
- Strebhardt K. Multifaceted polo-like kinases: drug targets and antitargets for cancer therapy. Nat Rev Drug Discov. 2010;9:643–60. doi: 10.1038/nrd3184. [DOI] [PubMed] [Google Scholar]
- Sumara I, Giménez-Abián JF, Gerlich D, et al. Roles of polo-like kinase 1 in the assembly of functional mitotic spindles. Curr Biol CB. 2004;14:1712–22. doi: 10.1016/j.cub.2004.09.049. [DOI] [PubMed] [Google Scholar]
- Tsvetkov L, Xu X, Li J, et al. Polo-like kinase 1 and Chk2 interact and co-localize to centrosomes and the midbody. J Biol Chem. 2003;278:8468–75. doi: 10.1074/jbc.M211202200. [DOI] [PubMed] [Google Scholar]
- Van Vugt MATM, van de Weerdt BCM, Vader G, et al. Polo-like kinase-1 is required for bipolar spindle formation but is dispensable for anaphase promoting complex/Cdc20 activation and initiation of cytokinesis. J Biol Chem. 2004;279:36841–54. doi: 10.1074/jbc.M313681200. [DOI] [PubMed] [Google Scholar]
- Vujic I, Posch C, Sanlorenzo M, et al. Mutant NRAS Q61 shares signaling similarities across various cancer types – potential implications for future therapies. Oncotarget. 2014;5 doi: 10.18632/oncotarget.2326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Kaiser CE, Frett B, et al. Targeting mutant KRAS for anticancer therapeutics: a review of novel small molecule modulators. J Med Chem. 2013;56:5219–30. doi: 10.1021/jm3017706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf G, Elez R, Doermer A, et al. Prognostic significance of polo-like kinase (PLK) expression in non-small cell lung cancer. Oncogene. 1997;14:543–9. doi: 10.1038/sj.onc.1200862. [DOI] [PubMed] [Google Scholar]
- Yim H, Erikson RL. Plk1-targeted therapies in TP53- or RAS-mutated cancer. Mutat Res Rev Mutat Res. 2014 doi: 10.1016/j.mrrev.2014.02.005. [DOI] [PubMed] [Google Scholar]
- Yoo HY, Kumagai A, Shevchenko A, et al. Adaptation of a DNA replication checkpoint response depends upon inactivation of Claspin by the Polo-like kinase. Cell. 2004;117:575–88. doi: 10.1016/s0092-8674(04)00417-9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.