To The Editor
The shape of the mitochondrial network results from the cumulative activity of two opposing processes: fusion and fission (Mishra and Chan, 2014). These processes collaborate to ensure homeostatic maintenance of mitochondrial function, cellular bioenergetics, and commitment to mitosis (Nasrallah and Horvath, 2014). While the contributions of aberrant mitochondrial dynamics in neurodegenerative and cardiometabolic diseases are established, little is known about the contribution of mitochondrial dynamics in cancer development, prognosis, or treatment.
Recently, a role for dynamin related protein 1 (DRP1) was revealed in oncogenic RAS-induced cellular transformation, and in cellular responses to oncogenic MAPK inhibition (e.g., BRAFV600E inhibition with PLX-4032)(Bollag et al., 2010; Serasinghe et al., 2015). DRP1 is a large cytosolic GTPase that induces fission of the mitochondrial network (Yoon et al., 2001; Smirnova et al., 2001). For example, when DRP1 is phosphorylated at serine 616 (DRP1S616℗), DRP1 localizes to mitochondria, undergoes oligomerization, and initiates membrane scission (Mishra and Chan, 2014). DRP1S616℗ is directly induced by ERK1/2 within the BRAFV600E pathway leading to chronic mitochondrial fission, cancer-associated mitochondrial dysfunction, and resistance to targeted therapies (Serasinghe et al., 2015). In melanoma, DRP1S616℗ status dichotomized wild type BRAF (BRAFWt) from BRAFV600E disease, suggesting a mechanistic contribution of DRP1S616℗ in BRAFV600E melanoma (Serasinghe et al., 2015). Based on these observations, we were interested in determining if DRP1S616℗ was prevalent in all BRAFV600E skin lesions (e.g., nevi), or if DRP1S616℗ was indicative of BRAFV600E melanoma.
To investigate this question, we performed IHC for the BRAFV600E and DRP1S616℗ status on a cohort of tissues. Benign nevi (68 samples; Figures 1a & S1a), dysplastic nevi (40 samples; Figures 1b & S1b), primary melanomas (187 samples; Figures 1c & S1c), and nevi derived from patients eventually diagnosed with melanoma (46 sets; Figure 1d) were stained. BRAFV600E and DRP1S616℗ scoring methods were developed (0, 1+ = negative; 2+, 3+ = positive) based on standard histopathological analyses within the Mount Sinai Medical Center and relevant literature (Figures S1a-c) (Pearlstein et al., 2014; Serasinghe et al., 2015). As control, we examined tissues stained with no primary antibody and rabbit IgG to ensure specificity (Figures S2a-d & S3a-b); and we also examined total DRP1, which was minimally expressed in normal skin, benign nevi, BRAFWt melanoma, and BRAFV600E melanoma (Figures S2a-d & S3a-b).
Figure 1. Increased DRP1S616℗ is associated with the incidence of BRAFV600E melanoma.
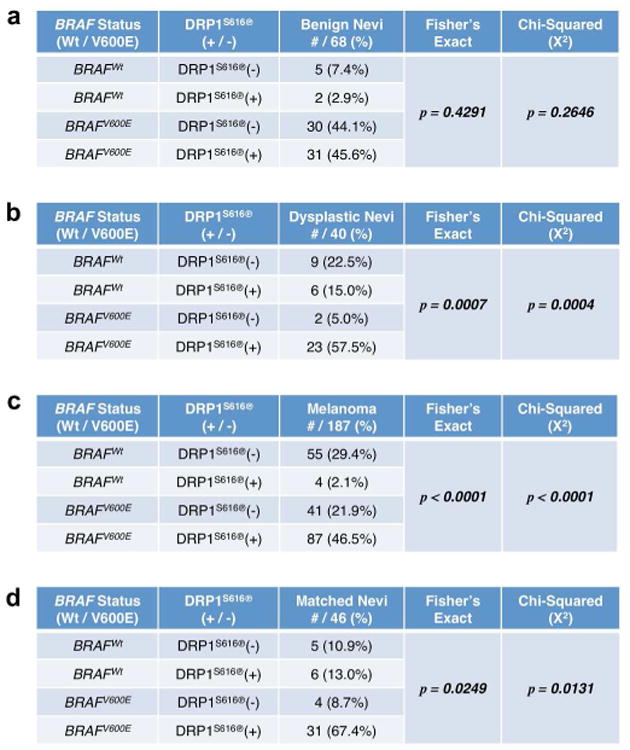
(ad) IHC was performed to detect the status of BRAFV600E and DRP1S616℗ in benign nevi (68 samples, a), dysplastic nevi (40 samples, b), primary melanoma (187 samples, c), and nevi from patients that developed melanoma (46 samples, d). Fisher's Exact and Chi-Squared Tests determined statistical significance.
Our benign nevi collection demonstrated no dysplasia at the time of diagnosis, had no known relationship to melanoma, and DRP1S616℗ was not significantly related to BRAF status (Figures 1a). While melanoma progression is not absolutely understood, dysplastic nevi are often considered precursors to disease and increase the risk of developing melanoma (Goldstein and Tucker, 2013). Indeed, analysis of a dysplastic nevi collection from the Mount Sinai Dermatopathology Division which contained a subset of tissues derived from patients eventually diagnosed with melanoma revealed that approximately 79.3% (23/29 cases) of tissues that are DRP1S616℗ positive are also BRAFV600E, and 92% (23/25 cases) of BRAFV600E dysplastic nevi display DRP1S616℗ (Figure 1b). Within the melanoma panel (59 BRAFWt, 128 BRAFV600E), we observed DRP1S616℗ in 91 samples; the vast majority (87/91 cases, 95.6%) was in BRAFV600E tumors (Figure 1c). In contrast, only 4 out of 59 BRAFWt tumors were positive for DRP1S616℗. Fisher's Exact (p<0.0001) and Chi-Squared (p<0.0001) analyses revealed that these relationships are highly significant. We also analyzed an additional larger cohort of nevi (containing benign and dysplastic) that are all matched to patients with melanoma, and similar relationships were obtained (Figure 1d). Together, these data suggest that DRP1S616℗ is significantly related to BRAFV600E status in dysplastic nevi and human melanoma, with correlations most striking in BRAFV600E melanoma.
Literature and the above data suggest that DRP1S616℗ may contribute to the survival of BRAFV600E disease (Serasinghe et al., 2015). Indeed, silencing oncogenic MAPK signaling via the pharmacological inhibition of BRAFV600E or MEK (with PLX-4032 or GSK-1120212, respectively) decreased DRP1S616℗ by western blot and immunofluorescence, but not DRP1Total, in BRAFV600E melanoma cells (Figures 2a & S4a). The oncogenic MAPK pathway often reactivates following the inhibition of BRAFV600E or MEK, and this confounds interpreting a direct pro-survival role for DRP1S616℗ (Holderfield et al., 2014). Therefore, we examined DRP1S616℗ contributions in the proliferation and survival of BRAFV600E melanoma cell lines by DRP1 loss of function experiments using RNAi and a small molecule (mDIVI-1) that durably inhibits mitochondrial fission by blocking the DRP1 GTPase (Cassidy-Stone et al., 2008). A375 cells were infected with RNAi lentivirus against Drp1, and monitored for proliferation. Loss of Drp1 expression correlated with decreased proliferation and clonogenic survival (Figures 2b-e). Next, A375 cells were treated with mDIVI-1, evaluated by fluorescent microscopy for expected changes to mitochondrial shape (i.e., mitochondrial fusion = DRP1 inhibition), and then scored for apoptotic responses. Indeed, the inhibition of DRP1 function by mDIVI-1 led to a marked decrease in DRP1-dependent mitochondrial fission (Figure 2f) and dose-dependent apoptosis (Figure 2g). In contrast, the BRAFWt melanoma line MeWo displayed minimal DRP1S616℗ and blunted pro-apoptotic responses to mDIVI-1 treatment (Figures S5a-b). We also treated these cells with staurosporine to ensure they had intact pro-apoptotic signaling (Figure S5c).
Figure 2. Inhibition of DRP1 suppresses BRAFV600E melanoma cell growth and survival.
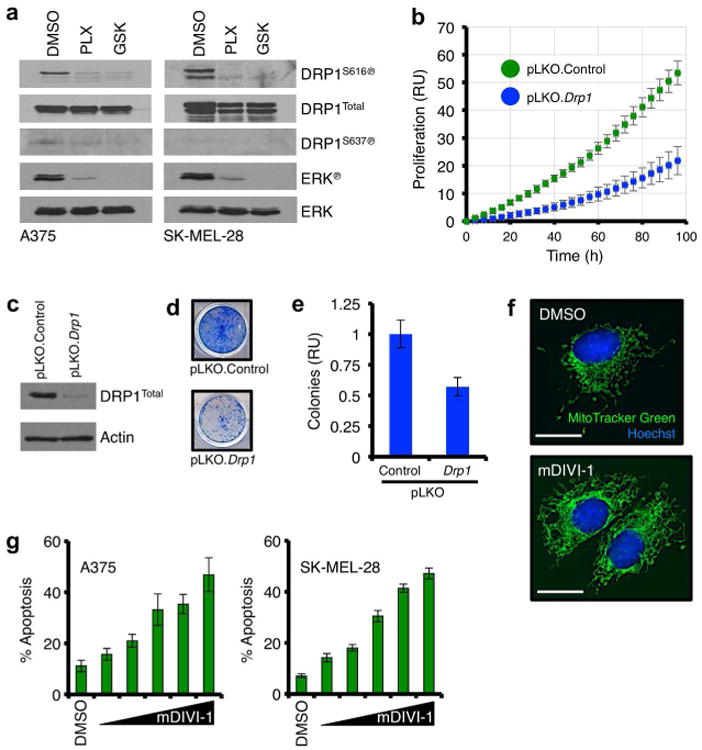
(a) A375 and SK-MEL-28 cells were treated with PLX-4032 (1 μM) or GSK-1120212 (10 nM) for 8 hours, and lysates were western blotted for indicated proteins. ERK℗ is shown as a positive control for drug sensitivity. Multiple DRP1 isoforms explain the presence of additional bands in the SK-MEL-28 DRP1 blots. (b) A375 cells were infected with control or Drp1 RNAi, and proliferation was quantified for 96 hours. (c) A375 cells were infected with control or Drp1 RNAi, and lysates were western blotted for indicated proteins. (d) A375 cells were infected with control or Drp1 RNAi, cultured for 12 days, and stained. (e) Colony formation in d was quantified. (f) A375 cells were treated with mDIVI-1 (10 μM) for 8 hours, and loaded with MitoTracker Green and Hoechst 33342 before live cell imaging. (g) A375 and SK-MEL-28 cells were treated with mDIVI-1 (0, 5, 10, 25, 50, 100 μM) for 48 hours before AnnexinV-FITC analysis. All data are representative of at least triplicate experiments, and reported as ± S.D., as required. Scale bars = 25 μm.
Altogether, these data suggest that the induction of DRP1S616℗ in dysplastic nevi (and nevi derived from patients eventually diagnosed with melanoma) and primary melanoma is a potential contributing factor to BRAFV600E disease; and examining DRP1S616℗ status may be a useful progression biomarker along with BRAFV600E to determine which lesions are most likely to develop into disease. DRP1S616℗ is undetectable in normal skin, and the frequency of genomic alterations to DRP1 is only ∼10% in cancer (Figures S2a-b & S6a-b) (Cerami et al., 2012; Gao et al., 2013, Serasinghe et al., 2015). However, the activation of DRP1 by oncogenic MAPK signaling that occurs during cellular transformation is regulated in the majority of samples (Serasinghe et al., 2015). In addition, DRP1S616℗ is markedly enhanced in BRAFV600E positive dysplastic nevi and melanomas. This activation correlates with the survival of BRAFV600E cancer cells following treatment with targeted therapies (Serasinghe et al., 2015). While there is also a subset of BRAFV600E positive lesions that are negative for DRP1S616℗ (Figure 1c), recent studies suggest there are alternative mechanisms to induce mitochondrial hyper-fragmentation and subsequent apoptotic resistance through the mitochondrial dynamics machinery (Renault et al., 2015). Collectively, these efforts suggest that studying DRP1, and potentially other proteins involved in orchestrating mitochondrial dynamics and function, may offer a unique perspective to better understand melanoma development, diagnosis, and treatment.
Supplementary Material
Supplemental Figure S1. BRAFV600E and DRP1S616℗ status scoring system in benign nevi, dysplastic nevi, and primary BRAFV600E melanoma.
(a - c) IHC was performed to detect the status of BRAFV600E and DRP1S616℗; and examples of scoring are shown for each tissue (benign nevi, a; dysplastic nevi, b; primary BRAFV600E melanoma, c). Scale bars = 200 μm.
Supplemental Figure S2. Control stainings in normal skin, benign nevi, and BRAFWt melanoma.
(a - d) IHC was performed with no primary antibody, rabbit IgG, total DRP1, and DRP1S616℗ in designated tissues (normal skin, a; benign nevi, b; BRAFWt melanoma, c – 20×, d – 40×). Scale bars = 200 μm.
Supplemental Figure S3. Control stainings in BRAFV600E melanoma.
(a - b) IHC was performed with no primary antibody, rabbit IgG, total DRP1, and DRP1S616℗ in BRAFV600E melanoma (a – 20×, b – 40×). Scale bars = 200 μm.
Supplemental Figure S4. Pharmacological inhibition of BRAFV600E or MEK reduces DRP1S616℗ but not DRP1Total.
(a) A375 cells were treated with PLX-4032 (1 μM) or GSK-1120212 (10 nM) for 8 hours. Cells were fixed and stained for DRP1S616℗ (FITC) and nuclei (DAPI; inset). Scale bars = 25 μm.
Supplemental Figure S5. The BRAFWt melanoma cell line MeWo does not engage apoptosis upon mDIVI-1 treatment, and fails to reduce DRP1S616℗ upon PLX-4032 or GSK-1120212 treatment despite intact pro-apoptotic signaling.
(a) MeWo cells were treated with PLX-4032 (1 μM), GSK-1120212 (25 nM), or mDIVI-1 (50 μM) for 24 hours before AnnexinV-FITC analysis. All data are representative of at least triplicate experiments, and reported as ± S.D., as required. (b) MeWo cells were treated with PLX-4032 (1 μM) or GSK-1120212 (10 nM) for 8 hours, and lysates were western blotted for indicated proteins. ERK℗ is shown as a positive control for drug sensitivity. PLX-4032 activates BRAFWt, leading to increased ERK℗. Multiple DRP1 isoforms explain the presence of additional bands in the DRP1Total blots. (c) A375, SK-MEL-28, and MeWo cells were treated with 1 μM staurosporine (STS) for 24 hours before AnnexinV-FITC analysis. All data are representative of at least triplicate experiments, and reported as ± S.D., as required.
Supplemental Figure S6. DRP1 alterations in BRAFWt and BRAFV600E tumors.
(a) Graphical representation of DRP1 alterations in BRAFWt and BRAFV600E tumors. The cBioPortal (www.cbioporal.org; TCGA Skin Cutanenous Melanoma subset - 374 samples - selected samples are shown) results shown here are in whole or part based upon data generated by the TCGA Research Network (www.cancergenome.nih.gov) (Cerami et al., 2012; Gao et al., 2013). (b) Data from A presented according to BRAF status.
Acknowledgments
This work was supported by: NIH grant CA157740 (to J.E.C.), the JJR Foundation (to J.E.C.), the William A. Spivak Fund (to J.E.C.), the Fridolin Charitable Trust (to J.E.C.), an American Cancer Society Research Scholar Award (to J.E.C.), and an Irma T. Hirschl/Monique Weill-Caulier Trust Research Award (to J.E.C.). This work was also supported in part by two research grants (5-FY11-74 and 1-FY13-416) from the March of Dimes Foundation (to J.E.C.), an Einstein Research Fellowship (to S.Y.W.), an American Skin Association Medical Students Grant (to S.Y.W.), an American Federation for Aging Research MSTAR Grant (to J.C.S.), and the Developmental Research Pilot Project Program within the Department of Oncological Sciences at Mount Sinai (to E.B., J.T.C., and J.E.C.). We would also like to thank Joanna Dong for assistance.
Abbreviations
- BRAF
v-Raf murine sarcoma viral oncogene homolog B
- DRP1
dynamin related protein 1
- ERK
extracellular signal-regulated kinase
- FFPE
formalin-fixed paraffin embedded
- FITC
fluorescein isothiocyanate
- GTP
guanosine-5′-triphosphosphate
- IHC
immunohistochemistry
- MEK
mitogen-activated protein kinase/ERK kinase
- RAS
rat sarcoma
Footnotes
Conflict of Interest: The authors state no conflict of interest.
References
- Bollag G, Hirth P, Tsai J, et al. Clinical efficacy of a RAF inhibitor needs broad target blockade in BRAF-mutant melanoma. Nature. 2010;467:596–599. doi: 10.1038/nature09454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cassidy-Stone A, Chipuk JE, Ingerman E, et al. Chemical inhibition of the mitochondrial division dynamin reveals its role in Bax/Bak-dependent mitochondrial outer membrane permeabilization. Dev Cell. 2008;14:193–204. doi: 10.1016/j.devcel.2007.11.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerami E, Gao J, Dogrusoz U, et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discovery. 2012;2:401–4. doi: 10.1158/2159-8290.CD-12-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao J, Aksoy BA, Dogrusoz U, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Science Signaling. 2013;6:pl1. doi: 10.1126/scisignal.2004088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldstein AM, Tucker MA. Dysplastic nevi and melanoma. Cancer Epidemiol Biomarkers Prev. 2013;22:528–532. doi: 10.1158/1055-9965.EPI-12-1346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holderfield M, Deuker MM, McCormick F, et al. Targeting RAK kinases for cancer therapy: BRAF-mutated melanoma and beyond. Nat Rev Cancer. 2014;14:455–467. doi: 10.1038/nrc3760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishra P, Chan DC. Mitochondrial dynamics and inheritance during cell division, development and disease. Nat Rev Mol Cell Biol. 2014;15:634–46. doi: 10.1038/nrm3877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nasrallah CM, Horvath TL. Mitochondrial dynamics in the central regulation of metabolism. Nat Rev Endocrinol. 2014;10:650–658. doi: 10.1038/nrendo.2014.160. [DOI] [PubMed] [Google Scholar]
- Pearlstein MV, Zedek DC, Ollila DW, et al. Validation of the VE1 immunostain for the BRAF V600E mutation in melanoma. J Cutan Pathol. 2014;41:723–732. doi: 10.1111/cup.12364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renault TT, Floros KV, Elkholi R, et al. Mitochondrial shape governs BAX-induced membrane permeabilization and apoptosis. Mol Cell. 2015;57(1):69–82. doi: 10.1016/j.molcel.2014.10.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serasinghe MN, Wieder SY, Renault TT, et al. Mitochondrial division is requisite to RAS-induced transformation and is targeted by oncogenic MAPK pathway inhibitors. Mol Cell. 2015;57(3):521–36. doi: 10.1016/j.molcel.2015.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smirnova E, Griparic L, Shurland DL, et al. Dynamin-related protein Drp1 is required for mitochondrial division in mammalian cells. Mol Biol Cell. 2001;12:2245–2256. doi: 10.1091/mbc.12.8.2245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon Y, Pitts KR, McNiven MA. Mammalian dynamin-like protein DLP1 tubulates membranes. Mol Biol Cell. 2001;12:2894–2905. doi: 10.1091/mbc.12.9.2894. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Figure S1. BRAFV600E and DRP1S616℗ status scoring system in benign nevi, dysplastic nevi, and primary BRAFV600E melanoma.
(a - c) IHC was performed to detect the status of BRAFV600E and DRP1S616℗; and examples of scoring are shown for each tissue (benign nevi, a; dysplastic nevi, b; primary BRAFV600E melanoma, c). Scale bars = 200 μm.
Supplemental Figure S2. Control stainings in normal skin, benign nevi, and BRAFWt melanoma.
(a - d) IHC was performed with no primary antibody, rabbit IgG, total DRP1, and DRP1S616℗ in designated tissues (normal skin, a; benign nevi, b; BRAFWt melanoma, c – 20×, d – 40×). Scale bars = 200 μm.
Supplemental Figure S3. Control stainings in BRAFV600E melanoma.
(a - b) IHC was performed with no primary antibody, rabbit IgG, total DRP1, and DRP1S616℗ in BRAFV600E melanoma (a – 20×, b – 40×). Scale bars = 200 μm.
Supplemental Figure S4. Pharmacological inhibition of BRAFV600E or MEK reduces DRP1S616℗ but not DRP1Total.
(a) A375 cells were treated with PLX-4032 (1 μM) or GSK-1120212 (10 nM) for 8 hours. Cells were fixed and stained for DRP1S616℗ (FITC) and nuclei (DAPI; inset). Scale bars = 25 μm.
Supplemental Figure S5. The BRAFWt melanoma cell line MeWo does not engage apoptosis upon mDIVI-1 treatment, and fails to reduce DRP1S616℗ upon PLX-4032 or GSK-1120212 treatment despite intact pro-apoptotic signaling.
(a) MeWo cells were treated with PLX-4032 (1 μM), GSK-1120212 (25 nM), or mDIVI-1 (50 μM) for 24 hours before AnnexinV-FITC analysis. All data are representative of at least triplicate experiments, and reported as ± S.D., as required. (b) MeWo cells were treated with PLX-4032 (1 μM) or GSK-1120212 (10 nM) for 8 hours, and lysates were western blotted for indicated proteins. ERK℗ is shown as a positive control for drug sensitivity. PLX-4032 activates BRAFWt, leading to increased ERK℗. Multiple DRP1 isoforms explain the presence of additional bands in the DRP1Total blots. (c) A375, SK-MEL-28, and MeWo cells were treated with 1 μM staurosporine (STS) for 24 hours before AnnexinV-FITC analysis. All data are representative of at least triplicate experiments, and reported as ± S.D., as required.
Supplemental Figure S6. DRP1 alterations in BRAFWt and BRAFV600E tumors.
(a) Graphical representation of DRP1 alterations in BRAFWt and BRAFV600E tumors. The cBioPortal (www.cbioporal.org; TCGA Skin Cutanenous Melanoma subset - 374 samples - selected samples are shown) results shown here are in whole or part based upon data generated by the TCGA Research Network (www.cancergenome.nih.gov) (Cerami et al., 2012; Gao et al., 2013). (b) Data from A presented according to BRAF status.


