Abstract
More than 375 genes have been identified that are involved in regulating skin pigmentation, and those act during development, survival, differentiation and/or responses of melanocytes to the environment. Many of those genes have been cloned and disruptions of their functions are associated with various pigmentary diseases, however many remain to be identified. We have performed a series of microarray analyses of hyperpigmented compared to less pigmented skin to identify genes responsible for those differences. The rationale and goal for this study was to perform a meta-analysis on those microarray databases to identify genes that may be significantly involved in regulating skin phenotype either directly or indirectly that might not have been identified due to subtle differences by any of those individual studies alone. The meta-analysis demonstrates that 1,271 probes representing 921 genes are differentially expressed at significant levels in the 5 microarray datasets compared, which provides new insights into the variety of genes involved in determining skin phenotype. Immunohistochemistry was used to validate 2 of those markers at the protein level (TRIM63 and QPCT) and we discuss the possible functions of those genes in regulating skin physiology.
INTRODUCTION
The regulation of pigmentation in human skin has many important implications, including its role in photoprotection from UV damage, its cosmetic and social roles and its roles in various pigmentary diseases. A large number of genes are involved in regulating mammalian pigmentation, and those act during development, survival, differentiation and/or responses of melanocytes to the environment. Historically, pigment genes were initially identified from spontaneous mutations that resulted in visible phenotypic changes, usually in mice, but also in many other species including humans. Before the era of gene cloning, about 65 pigment genes had been identified (Silvers, 1979), but since that time there has been a rapid increase in the number of known pigment genes, exceeding 100 by the year 2000 (Bennett and Lamoreux, 2003) and at this time, >375 pigment genes are known, of which ~170 have been cloned [curated database at: http://www.espcr.org/micemut/]. Many of those genes and the functions of their encoded proteins have been characterized, and in many cases mutations in those genes have been associated with human pigmentary diseases and/or variations in normal pigmentation. Gene expression profiling has become increasingly common and useful to identify genes involved in regulating normal skin and hair physiology as well as those involved in skin diseases such as psoriasis, keloids and age spots by various types of cells in the skin (Smith et al., 2008; Calles et al., 2010; Choi et al., 2010; Mitsui et al., 2012; Peters et al., 2013; Pollock et al., 2014; Inkeles et al., 2014).
Several specific genes (e.g. TYR, OCA2, MC1R and SLC24A5) have been reported to be the major genes involved in the regulation of human skin, hair and eye color (Nordlund et al., 2006; Hearing and Merlino, 2009; Yamaguchi and Hearing, 2009; Sturm and Duffy, 2012; Baxter and Pavan, 2013) but there is no doubt that many other genes are involved, and that many genes involved in regulating the phenotype of human skin remain to be discovered. In general, mutations in many of the known pigment genes cause the loss of color in the affected tissues, i.e. they are associated with hypopigmentation, but relatively little is known about genes associated with increased pigmentation. Genes involved with hypopigmentary diseases, such as albinism, white spotting and Hermansky-Pudlak syndrome, have been identified, and the characterization of the functions of their encoded products and why mutations in those genes cause the loss of pigmentation, has provided important information about molecular mechanisms involved in the regulation of melanocyte function. Interestingly, there are a number of hyperpigmentary conditions of the skin, such as UV-induced melanosis, post-inflammatory hyperpigmentation and senile lentigines (age spots), but factors and genes involved in those are relatively poorly understood. To address that, we have begun a series of microarray analyses of normally pigmented skin compared with hyperpigmented lesions on the same individuals to identify genes that are responsible for those disruptions (Choi et al., 2010; Coelho et al., 2015a; Coelho et al., 2015b). In a related study, we have also used microarray analysis to compare lightly pigmented (Caucasian) to moderately pigmented (Asian) and darkly pigmented (African) skin (Yin et al., 2014), which identified a number of genes involved in regulating constitutive skin color.
Those analyses have identified several key genes that are correlated with hyperpigmented skin but due to the relatively small sample sizes of those clinical studies, many other genes seem to be subtly regulated but not consistently enough to reach statistical significance. The rationale and goal for this study was to perform a meta-analysis on all those microarray databases to increase the statistical power and to identify important genes that are significantly involved in regulating skin pigmentation and phenotype that might not have been identified by any of those individual studies alone.
We now report that 1,271 probes representing 921 genes were differentially expressed in the 5 microarray datasets from 5 different models of skin hyperpigmentation compared by meta-analysis. Among the 1,271 differential probes, 118 never showed a significant p value in any of the individual studies. Further, as proof of principle, we used immunohistochemistry to validate 2 of those markers at the protein level (TRIM63 and QPCT). Although this study focused on the relevance to skin pigmentation, since the biopsies included all epidermal cell populations, these data also provide insights into other aspects of skin physiology.
RESULTS
Selection of subsets of microarray datasets based on pigment gene expression features
Details of these microarray studies and a summary of the 5 different models of skin hyperpigmentation (UV, LLP, PIH, AS and ES) are provided in SI Methods. The 5 datasets included in the meta-analysis are summarized in Table S1. Table S2 shows information about each raw data file, and in total, 90 chips were included in the meta-analysis. Since several of these 5 studies examined gene expression at multiple time points, the rationale for choosing a specific time point and control in each of those studies is detailed in.SI Methods
Meta-analysis results
Identification of meta-genes
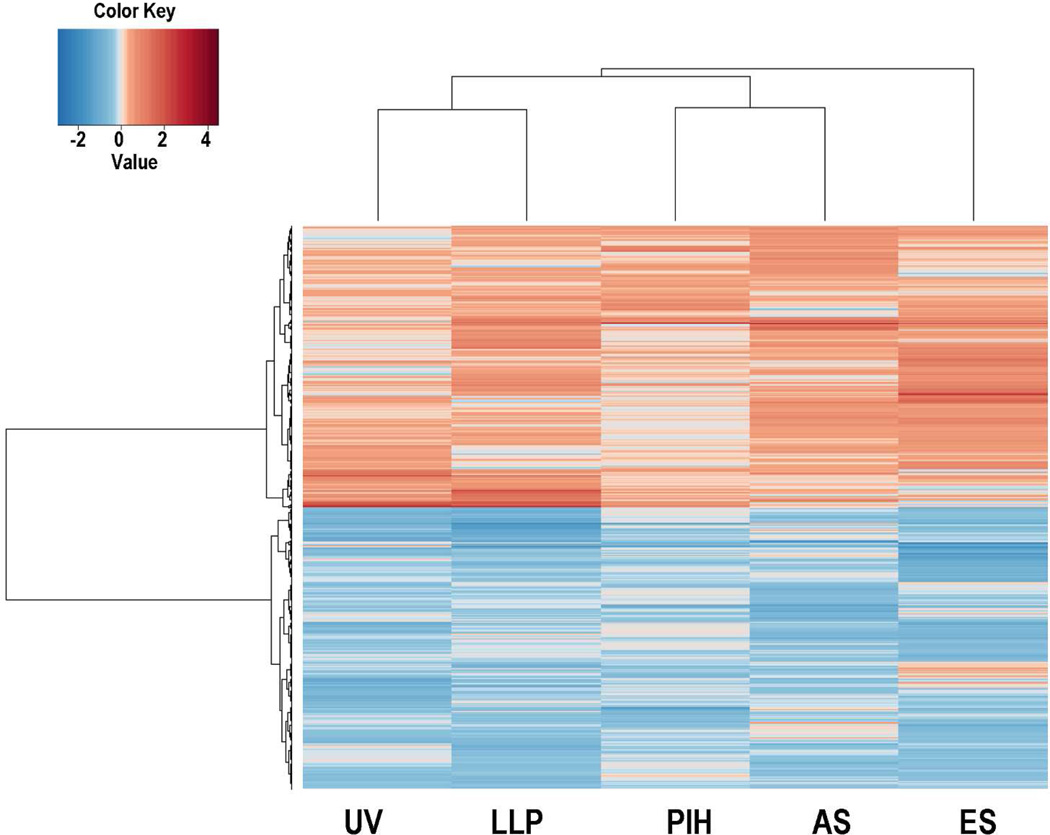
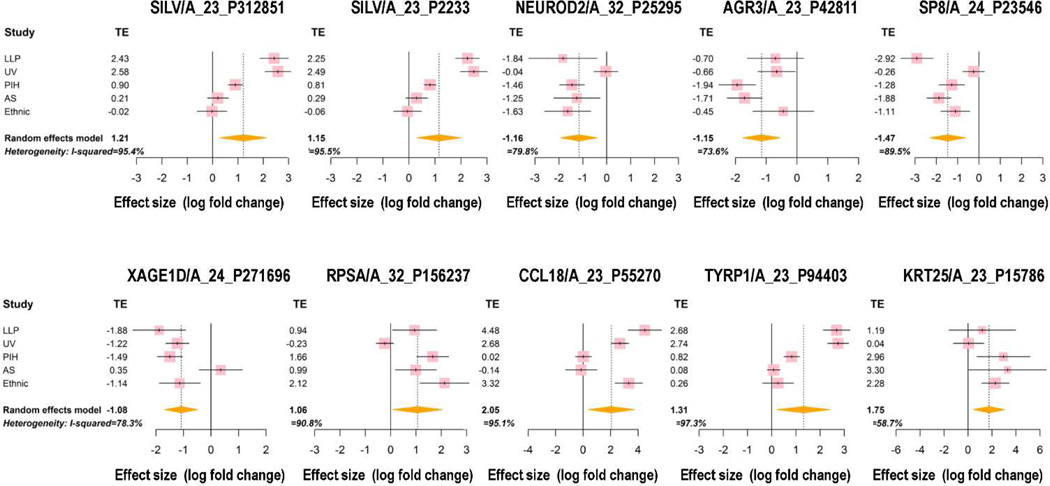
In order to reduce the false discovery rate of microarray data analysis, a gene filtering step was implemented before the differential analysis. Within the 41,000 probes on the Agilent-014850 Whole Human Genome Microarray, 27,761 probes were selected based on variation and intensity. Meta-analysis was applied on those 27,761 probes and the statistics of I2 were calculated to measure heterogeneity for each gene (Figure S3). Statistical analysis revealed that 1,271 of the 27,761 probes fulfilled the criteria for meta-gene selection (Table S3). They represent 921 genes and were distributed relatively equally as up- or down-regulated genes; a heatmap of those gene probes is shown in Figure 1. Those genes were ranked by their absolute value of fold change, and the top 10 meta-genes are shown in Figure 2. Two SILV gene probes and one TYRP1 gene probe were found in the top 10 list, both of which are important pigment-specific genes, which suggests that these other genes might also play important roles in regulating skin pigmentation. The top 25 up-regulated gene probes and the top 25 down-regulated gene probes are listed in Table 1.
Figure 1. Heatmap of differentially expressed meta-genes showing and the equal distribution of down-regulated and up-regulated genes.
UV represents the UV data set which compared UV-exposed skin with unexposed skin at two weeks. LLP represents the LLP data set which compared UV-exposed skins with unexposed skin at day 25. PIH represents the PIH data set which compared Post-Inflammatory Hyperpigmented skin with non-treated skins at 6 weeks. AS represents the age spot data set which compared age spots with perilesional control areas. ES represents the Ethnic Skin data set which compared African skin with Caucasian skin.
Figure 2. Forest plots of the Top 10 meta-genes.
The confidence interval (CI) for each study is represented by a horizontal line and the treatment effect (TE) is represented by a square. The size of the square corresponds to the weight of the study in the meta-analysis. The confidence interval for summary effect is represented by a diamond.
Table 1.
| a. Top 25 gene probes in the up-regulated meta-gene list | ||||||
|---|---|---|---|---|---|---|
| ProbeID | Fold Change |
P value* | Primary Accession |
UniGeneID | GeneSymbo | GeneName |
| A_23_P55270 | 4.14 | 1.77E-02 | NM_002988 | Hs.143961 | CCL18 | chemokine (C-C motif) ligand 18 (pulmonary and activation-regulated) |
| A_23_P15786 | 3.36 | 8.77E-03 | NM_181534 | Hs.55412 | KRT25 | keratin 25 |
| A_23_P94403 | 2.48 | 1.93E-02 | NM_000550 | Hs.270279 | TYRP1 | tyrosinase-related protein 1 |
| A_23_P312851 | 2.32 | 1.06E-02 | NM_006928 | Hs.95972 | SILV | silver homolog (mouse) |
| A_23_P2233 | 2.23 | 8.20E-03 | NM_006928 | Hs.95972 | SILV | silver homolog (mouse) |
| A_32_P156237 | 2.09 | 3.41E-02 | BC010054 | RPSA | ribosomal protein SA | |
| A_23_P302595 | 1.94 | 3.27E-03 | NM_152738 | Hs.173337 | C6orf218 | chromosome 6 open reading frame 218 |
| A_24_P151005 | 1.93 | 3.53E-02 | NM_030967 | Hs.247934 | KRTAP1-1 | keratin associated protein 1-1 |
| A_23_P45560 | 1.90 | 2.55E-03 | NM_000273 | Hs.74124 | GPR143 | G protein-coupled receptor 143 |
| A_32_P31144 | 1.90 | 8.63E-03 | THC2711870 | |||
| A_23_P79108 | 1.89 | 3.09E-02 | NM_138813 | Hs.306212 | ATP8B3 | ATPase, class I, type 8B, member 3 |
| A_32_P165047 | 1.89 | 2.00E-02 | THC2675163 | |||
| A_32_P194062 | 1.87 | 4.28E-02 | NM_000372 | Hs.503555 | TYR | tyrosinase (oculocutaneous albinism IA) |
| A_24_P367247 | 1.85 | 5.66E-04 | NM_181607 | Hs.61552 | KRTAP19-1 | keratin associated protein 19-1 |
| A_23_P2831 | 1.85 | 2.20E-02 | NM_003991 | Hs.82002 | EDNRB | endothelin receptor type B |
| A_32_P191441 | 1.85 | 2.02E-02 | NM_001004298 | Hs.587663 | C10orf90 | chromosome 10 open reading frame 90 |
| A_23_P129225 | 1.83 | 3.63E-02 | NM_002420 | Hs.155942 | TRPM1 | transient receptor potential cation channel, subfamily M, member 1 |
| A_24_P196878 | 1.82 | 4.08E-04 | AF289567 | Hs.684029 | ||
| A_23_P93641 | 1.79 | 1.47E-06 | NM_020299 | Hs.116724 | AKR1B10 | aldo-keto reductase family 1, member B10 (aldose reductase) |
| A_23_P209735 | 1.78 | 4.18E-03 | NM_025139 | Hs.471610 | ARMC9 | armadillo repeat containing 9 |
| A_23_P159406 | 1.77 | 4.10E-02 | NM_003125 | Hs.1076 | SPRR1B | small proline-rich protein 1B (cornifin) |
| A_23_P347610 | 1.77 | 9.98E-03 | NM_012206 | Hs.129711 | HAVCR1 | hepatitis A virus cellular receptor 1 |
| A_24_P129341 | 1.76 | 7.41E-06 | NM_020299 | Hs.116724 | AKR1B10 | aldo-keto reductase family 1, member B10 (aldose reductase) |
| A_23_P114983 | 1.74 | 1.62E-02 | NM_032588 | Hs.279709 | TRIM63 | tripartite motif-containing 63 |
| A_24_P277657 | 1.74 | 3.49E-02 | NM_006877 | Hs.484741 | GMPR | guanosine monophosphate reductase |
| b. Top 25 gene probes in the down-regulated meta-gene list | ||||||
|---|---|---|---|---|---|---|
| ProbeID | Fold Change |
P value* | Primary Accession |
Uni GeneID |
Gene Symbol |
GeneName |
| A_24_P23546 | −2.77 | 6.46E-04 | NM_198956 | Hs.195922 | SP8 | Sp8 transcription factor |
| A_32_P25295 | −2.24 | 3.29E-03 | NM_006160 | Hs.322431 | NEUROD2 | neurogenic differentiation 2 |
| A_23_P42811 | −2.22 | 2.26E-04 | NM_176813 | Hs.100686 | AGR3 | anterior gradient homolog 3 (Xenopus laevis) |
| A_24_P271696 | −2.12 | 3.99E-04 | NM_133431 | Hs.112208 | XAGE1D | X antigen family, member 1D |
| A_24_P184803 | −2.02 | 7.73E-03 | NM_004086 | Hs.21016 | COCH | coagulation factor C homolog, cochlin (Limulus polyphemus) |
| A_32_P183765 | −1.97 | 6.41E-10 | NM_005235 | Hs.390729 | ERBB4 | v-erb-a erythroblastic leukemia viral oncogene homolog 4 (avian) |
| A_23_P22398 | −1.90 | 2.78E-03 | NM_080676 | Hs.661576 | MACROD2 | MACRO domain containing 2 |
| A_24_P142305 | −1.89 | 8.63E-05 | NM_000517 | Hs.654744 | HBA2 | hemoglobin, alpha 2 |
| A_23_P26457 | −1.87 | 2.87E-04 | NM_000517 | Hs.654744 | HBA2 | hemoglobin, alpha 2 |
| A_24_P184799 | −1.80 | 2.82E-02 | NM_004086 | Hs.21016 | COCH | coagulation factor C homolog, cochlin (Limulus polyphemus) |
| A_32_P10936 | −1.75 | 1.76E-02 | NM_004061 | Hs.113684 | CDH12 | cadherin 12, type 2 (N-cadherin 2) |
| A_24_P363711 | −1.73 | 3.23E-02 | NM_001926 | Hs.711 | DEFA6 | defensin, alpha 6, Paneth cell-specific |
| A_32_P61298 | −1.72 | 5.86E-04 | AK054921 | Hs.571748 | CDR1 | cerebellar degeneration-related protein 1, 34kDa |
| A_23_P84063 | −1.71 | 3.00E-02 | NM_016522 | Hs.504352 | HNT | neurotrimin |
| A_24_P251950 | −1.70 | 3.84E-04 | NM_174914 | Hs.348941 | UGT3A2 | UDP glycosyltransferase 3 family, polypeptide A2 |
| A_23_P37856 | −1.70 | 5.74E-03 | NM_000558 | Hs.449630 | HBA1 | hemoglobin, alpha 1 |
| A_23_P51767 | −1.66 | 7.68E-09 | NM_001765 | Hs.132448 | CD1C | CD1c molecule |
| A_24_P307964 | −1.66 | 5.76E-05 | NM_001012415 | Hs.120464 | SOHLH1 | spermatogenesis and oogenesis specific basic helix-loop-helix 1 |
| A_23_P52499 | −1.66 | 5.17E-03 | NM_003054 | Hs.654476 | SLC18A2 | solute carrier family 18 (vesicular monoamine), member 2 |
| A_23_P36531 | −1.65 | 2.00E-04 | NM_004616 | Hs.170563 | TSPAN8 | tetraspanin 8 |
| A_23_P64161 | −1.64 | 2.80E-02 | NM_178127 | Hs.318370 | ANGPTL5 | angiopoietin-like 5 |
| A_23_P203558 | −1.62 | 1.42E-02 | NM_000518 | Hs.523443 | HBB | hemoglobin, beta |
| A_23_P134347 | −1.62 | 3.88E-03 | NM_019029 | Hs.233389 | CPVL | carboxypeptidase, vitellogenic-like |
| A_23_P114883 | −1.62 | 8.43E-04 | NM_002023 | Hs.519168 | FMOD | fibromodulin |
| A_23_P258769 | −1.61 | 2.55E-02 | NM_002121 | Hs.485130 | HLA-DPB1 | major histocompatibility complex, class II, DP beta 1 |
P value is from meta analysis
Figure S4 shows forest plots of the SOX2 and MYB genes, where none of the 5 individual datasets showed a significant change in the expression of those two genes while meta-analysis assigned a significant p value to them. SOX2 has been shown to be an upstream regulator of the pigmentation master gene, the transcription factor MITF (Cimadamore et al., 2012). It has also been shown that a MYB-like transcription factor regulates petal pigmentation in a flowering peach ‘Genpei’ bearing variegated and fully pigmented flowers (Uematsu et al., 2014). Among the 4,076 significant probes identified by raw p value in the meta-analysis, 959 never had a p<0.05 in any one of the 5 datasets. Among the 1,271 meta-gene probes, 118 never had a significant p value in an individual dataset but were significant by meta-analysis (Table S4). These results provide strong evidence that meta-analysis has a greater power to detect differentially expressed genes (DEGs).
Comparison of meta-genes with known color genes
The curated website of color genes (http://www.espcr.org/micemut/) provides information about known genes that regulate pigmentation in mice and in their human and zebrafish homologues. We compared our meta-gene list with the 171 known color genes that have been cloned at this time; 160 of those genes are represented on the microarray chips used. Twenty-two common gene names were found and are shown in Table S5.
Proteins encoded by the meta-genes exist in human melanosomes
Melanosomes exist in 4 distinct stages as they become increasingly mature and are filled with melanin pigment before they are transferred to neighboring keratinocytes through melanocyte dendrites. A proteomics study (Chi et al., 2006) has identified the components of the different stages of melanosome maturation, and that information is listed in the Protein Information Resource (PIR), an integrated public bioinformatics resource (http://pir.georgetown.edu/cgi-bin/textsearch_iprox.pl?data=mnt). One of the major goals of this study was to identify previously unknown melanosomal proteins that might be involved in melanogenesis. We compared our meta-gene list with known melanosomal proteins at various stages of maturation and identified 23 common genes, which suggests that 23 of the meta-genes identified encode melanosomal proteins (Table S6). We confirmed the specific expression of QPCT in melanocytes as detailed below.
Bioinformatic analysis
GO term enrichment analysis
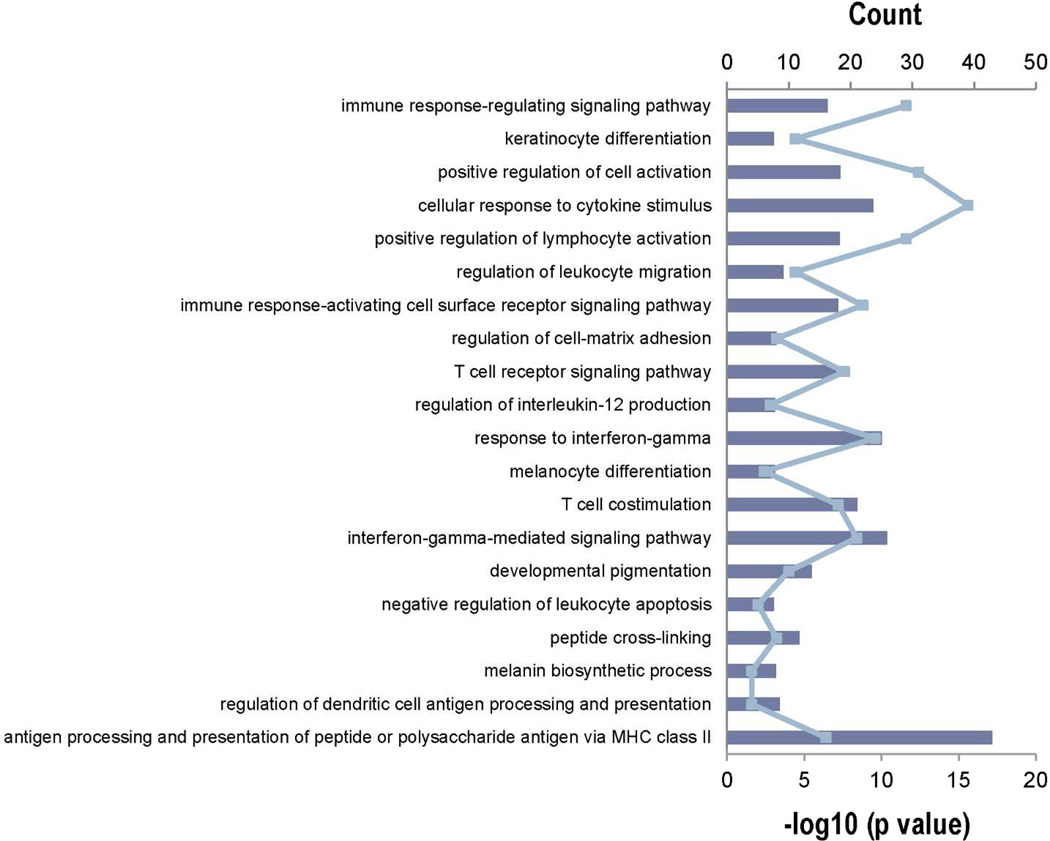
To identify functional relationships in DEGs, the R package GOstats based on the Gene Ontology database was used. Hypergeometric testing was used to identify significantly enriched GO terms. The 1,271 meta-gene probes were analyzed for enrichment of functional GO terms in comparison to the reference gene set (also called the "gene universe"), which is derived from all gene probes on the Agilent whole human genome chip. A p≤0.001 was used as cut off for the final list. Tables S7a, S7b and S7c list the Enriched Biological Processes, Cellular Components and Molecular Functions, respectively, which were ranked by their Odds Ratio. The Top 20 biological processes are shown in Figure 3, which includes GO:0042438 (Melanin Biosynthetic Process), GO:0048066 (Developmental Pigmentation) and GO:0030318 (Melanocyte Differentiation). The fact that those 3 pigment-related categories were scored so highly is indicative of the relevance of these genes to mammalian pigmentation.
Figure 3. Top 20 enriched biological process Gene Ontology (GO) terms identified by GOstats R package.
Enriched GO terms rank by odds ration after screened by cut off p value of 0.001. The blue line shows the number of genes that is common between the tested biological process and the uploaded gene set. The blue bars are the –log10 of the p value as determined by GOstats.
Upstream regulators
The overall goal of this study was to identify genes that play important roles in hyperpigmentation of the skin that might not have been identified in the individual studies. One of the drawbacks of microarray technology is that the sensitivity and precision of this technology might not provide accurate results regarding transcription factors with low RNA levels. Therefore, we used another approach to explore upstream regulators of pigmentation. The 1,271 meta-gene probes were loaded into IPA (Ingenuity Pathway Analysis), which can identify cascades of upstream transcriptional regulators and enzymes, and thereby explain the observed gene expression changes in our meta dataset. For upstream regulators, we focused only on transcription factors and enzymes, not on microRNAs or drugs. For each potential transcriptional regulator, 2 statistical measures, an overlap p value and an activation Z score, were computed. If a large number of target genes of a specific transcription factor was found in our DEG list, it is reasonable to hypothesize that transcription factor might play an important role in hyperpigmentation. Table S8a shows the top 10 activated upstream regulators and Table S8b shows the top 10 inhibited upstream regulators. Note that MITF is the most activated factor in the list while TNF is the most inhibited; both of those are key regulators of mammalian pigmentation. TRIM63 was another transcription factor identified in this study that might play an important role in regulating skin pigmentation as discussed further below.
Clustering
Biological insights can be captured by examining clusters of DEGs, that is, groups of genes which are up- or down- regulated simultaneously. The Mfuzz package in R was employed to explore common gene expression patterns in the 1,271 meta-gene probes. Figure S5a shows the profiles of the 40 clusters obtained from the Mfuzz package when m=1.25. Note that many of the most important known pigment genes (ARMC9, EDNRB, GPR143, MC1R, PAX3, PMEL, SOX5, TYR and TYRP1) are found in cluster 26 (Table S9). The genes in cluster 26 were up-regulated dramatically in the UV and LLP datasets (Figure S5b), were up-regulated moderately in the PIH dataset, but had only minor alterations in the Age Spot (AS) and Ethnic skin (ES) datasets. In Figure S5b (left), the vertical axis represents the standardized expression change of Cluster 26. In Figure S5b (right), the fold change of each gene in each dataset is shown on the y axis. Cluster analysis identifies groups of genes that perform similar or complementary functions to known genes, i.e. genes in a cluster respond similarly to a given experimental condition. Cluster 26 is of particular interest since a number of known key regulators of pigmentation that function at various cellular levels to regulate melanin production are in that group, suggesting that other members of that group might also be involved in regulating skin pigmentation.
Using the top 50 meta-genes to classify skin samples
If the meta-genes identified are feature genes that represent the different phenotypes of pigmentation, then they should be useful to classify hyperpigmented samples from normal samples or to classify hypopigmented samples from normal samples. Specimens from pigmentation disorders, such as from vitiligo skin, would be ideal to test the classification ability of the meta-genes.
The GEO microarray database was searched using the keywords "Homo sapiens skin vitiligo". Ten records were retrieved with one dataset describing gene expression profiles in vitiligo lesional skin (Rashighi et al., 2014). In that study, total RNA was isolated from 10 human samples from formalin-fixed, paraffin-embedded skin specimens, 5 from vitiligo patients and 5 from controls. Control skins were age- and site-matched excisions without pathology.
All meta-genes were ranked by absolute values of fold change. The Top 50 genes were selected for this classification test. Sample distances were calculated by the Euclidean method and sample clustering was performed by the Ward.D method; the results are shown in Figure S6. All 5 vitiligo samples clustered together and showed a similar heatmap pattern. We also used a PCA plot (Figure S6) to display the data features of these 10 human skin samples. All vitiligo samples were located at the top left of the 2d surface, which is distinct from the normal skin area (lower part of the 2d surface).
Validation of meta-genes using immunohistochemistry
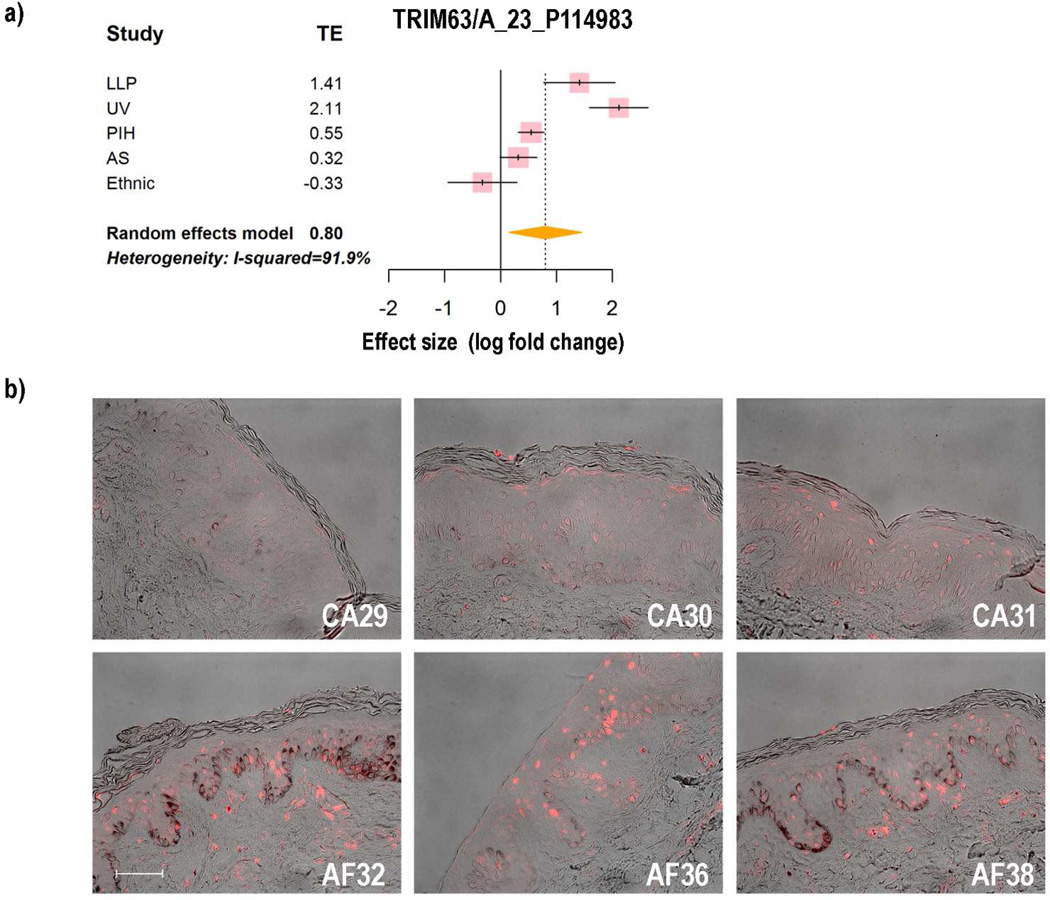
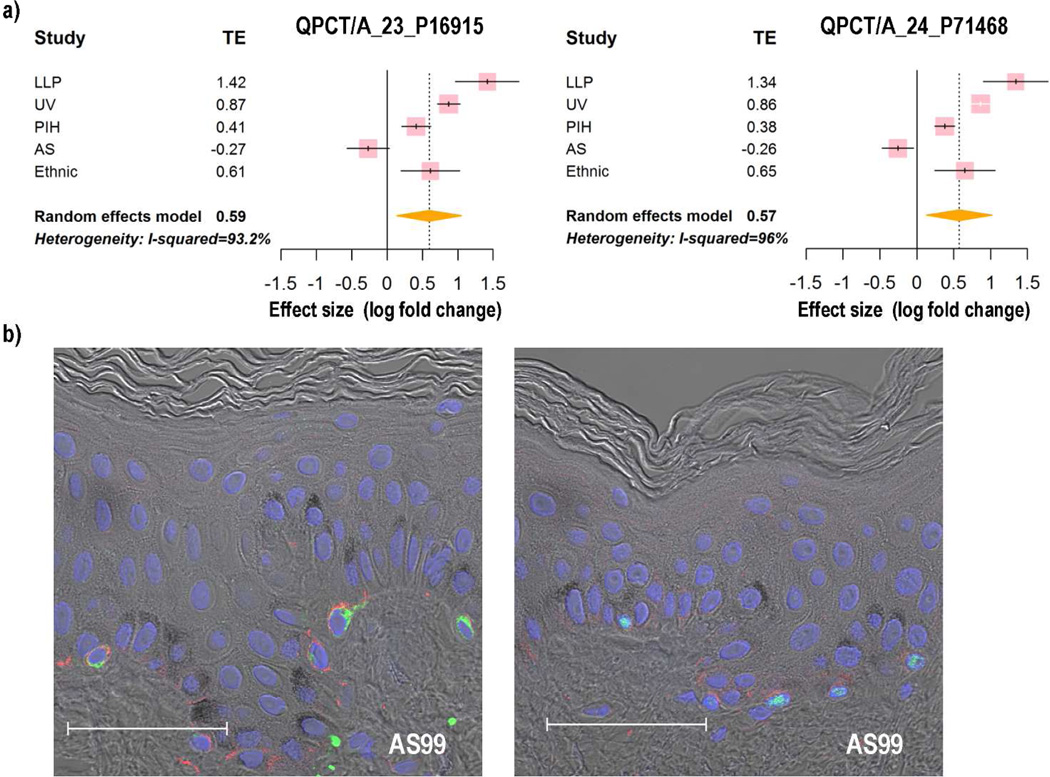
To confirm the expression patterns of some of the DEGs identified, we used immunohistochemistry to characterize the localization of their encoded proteins in African American, Asian and Caucasian skin. We tested one of the genes (TRIM63) encoding a transcription factor that fell in cluster 26, where the pigment genes were enriched. TRIM63 was up-regulated in the LLP, UV and PIH microarray datasets with fold changes of 2.66, 4.33 and 1.46, respectively, while it was not changed significantly in ES and AS datasets. Meta-analysis revealed that the summarized effect of TRIM63 is 1.74 (Figure 4a), which means that compared to normal samples, the gene expression level of TRIM63 in hyperpigmented samples is 1.74 times higher. Immunohistochemical staining (Figure 4b) showed a stronger signal for TRIM63 in African American skin than in Caucasian skin, indicating that more TRIM63 protein is expressed in more pigmented skin than in lighter skin, which is consistent with the meta-analysis. Also, we tested another meta-gene, named QPCT, which had been identified in the melanosome proteomics database. Meta-analysis showed that the gene expression level of QPCT in hyperpigmented samples is 1.51 times higher than in the less pigmented samples (Figure 5a). Immunohistochemical staining showed that QPCT and the melanocyte-specific marker MART1 co-localize in the same cells (Figure 5b, left), which suggests that QPCT is a melanocyte-specific gene that might play an important role in pigmentation. This cellular localization was confirmed using another melanocyte-specific marker, MITF (Figure 5b, right).
Figure 4. Expression patterns of TRIM63.
a) Forest plot of TRIM63; b) Immunohistochemical staining of TRIM63 in Caucasian skin specimens (top row) and in African American skin specimens (bottom row). Bar = 50 µm.
Figure 5. Expression patterns of QPCT.
a) Forest plot of QPCT; b) Immunohistochemical staining of Asian skin showing the colocalization of QPCT (red) in melanocytes labeled with MART1 (left, green) and MITF (right, green). Bar = 50 µm.
DISCUSSION
Due to the small sample size for each of our 5 microarray studies, the power of each study alone may not be sufficient to detect significant differences in gene expression, and thereby the statistical conclusions may not be consistent throughout similar or relative biological studies. Meta-analysis provides an ideal opportunity to combine gene effect sizes in various studies. With the exponential growth of microarray data in public databases and the rapid development of computing ability, increasing numbers of microarray meta-analyses are being performed to seek mechanisms underlying biological processes. To better understand the gene expression patterns involved in skin hyperpigmentation, we applied meta-analysis on 5 distinct hyperpigmentation datasets that represent 5 different models for skin hyperpigmentation, each with a limited number of clinical specimens. Among the 4 most commonly used differential gene detection methods of meta-analysis (‘combine p values’, ‘combine effect sizes’, ‘combine ranks’ and ‘directly merge after normalization’), we decided to use ‘combine effect sizes’ (gene alteration: log fold change) since we were most interested in genes that were consistently up- or down-regulated in all hyperpigmented conditions. The methods of ‘combine p value’ and ‘combine ranks’ are not able to tell genes with discordance automatically. Further, we selected the random effect model to ‘combine effect sizes’ from various studies since the 5 datasets we used employed 5 different types of skin hyperpigmentation. There was heterogeneity in those studies and genes won’t share common effect sizes among those studies. Although the 5 datasets used were all from the Agilent whole human genome array platform, we did not use ‘directly merge after normalization’ because the 5 studies were carried out sequentially at different times. There are substantial batch effects among the studies, even within some individual studies, such as the PIH and LLP studies. The microarray chips were hybridized in different batches. Additionally, in the UV, LLP, PIH and AS datasets, the samples were paired, which means that the hyperpigmented samples and the corresponding control samples were taken from the same subjects, while in the ES dataset, the samples were not paired and were from unrelated African and Caucasian subjects. Therefore, we used unpaired t tests to compare the ES dataset, and paired t tests to compare the other datasets. The gene effect sizes in each study were calculated respectively based on the data features of each study, and then were summarized by the random effect model. The advantage of meta-analysis for the hyperpigmentation microarray data is evidenced by the list of meta-genes which contains a large number of known pigment genes such as TYR, TYRP1 and SILV. The gene alteration pattern determined by the meta-analysis is more reliable.
Some genes with significant differences in one study but with non-significant changes in another study were identified as DEGs by the meta-analysis. For instance, TRIM63 has been shown to be up-regulated after repetitive UV treatment by microarray analysis and by immunohistochemical staining (Choi et al., 2010). However, it was not significantly changed when African American skin was compared with Caucasian skin in the ES dataset. Through meta-analysis, we found that the summary effect of TRIM63 is statistically significant, and immunohistochemical staining confirmed those results on ethnic skin specimens. Therefore, in the case of inconsistent results obtained in different studies, meta-analysis provides an ideal opportunity to summarize information and obtain a better understanding of how genes work during similar biological conditions. It is also clear that levels of functional proteins do not always correlate with their mRNA levels. The involvement of other proteins in our TOP25 list has been documented in other studies from our group, e.g. SOX7 in the LLP study (Coelho et al., 2015b) and NEUROD2 in the PIH study (Ebsen et al., in preparation).
The dynamics of changes in skin pigmentation over time has become clear as a result of these different approaches to study the various hyperpigmented phenotypes of human skin. Dramatic changes in increased skin pigmentation can be elicited within days (by UV exposure), weeks (by PIH), months (by LLP), years (in Age Spots) and a lifetime (Ethnic Skin color). The gene expression changes reported in our various microarray studies reflect those dynamics, with very rapid changes in gene and protein expression levels that lead directly to melanin synthesis (e.g. TYR and other melanosomal constituents) within a few days following UV exposure, to changes in factors regulating melanosome distribution and keratinocyte-related factors during PIH, to long-term more subtle changes in melanocyte function and skin architecture that result in LLP. Even longer term changes result in the development of age spots that take years to develop and reflect changes in melanosome distribution in the lower epidermis and towards the dermis, and of course the constitutive differences in skin pigmentation seen in light- versus darkly-pigmented skin. It is clear that a wide variety of genes are involved in those changes of pigmentation at each level, and that those include functional changes in various types of cells in the skin that influence melanin production and distribution. Although this study focused on skin pigmentation, since the biopsies included all epidermal cell populations, these data also provide insights on other aspects of skin physiology.
The individual microarray studies identified a number of specific targets in each system analyzed, but the relatively small number of specimens in each clinical study limited the statistical analysis. The combined meta-analysis of all 5 studies effectively increases the sample size and allows a number of additional genes to reach statistically different expression levels that were not identified in any of the individual studies. The DEGs identified provide a wealth of information about processes involved in skin hyperpigmentation and will no doubt provide many useful novel targets for those conditions. It should be noted that even relatively small changes in melanin production and distribution can have dramatic effects on visible skin pigmentation so changes in gene expression that still fall below the level of significance used in our analyses might still prove important in the physiological regulation of skin pigmentation.
MATERIALS AND METHODS
Studies included in the meta-analysis
Our laboratories have established 5 models (UV, LLP, PIH, AS and ES) to investigate genome-wide gene expression profiles in hyperpigmented human skin samples using the Agilent-014850 Whole Human Genome Microarray 4x44K platform. In order to avoid large measurement variances from different hybridization platforms and from different species, only studies on human skin samples using Agilent microarrays were included in this meta-analysis. Using key words 'Homo sapiens[organism] AND skin AND (GPL6480 OR GPL4133)', we searched databases available in Gene Expression Omnibus (GEO) at the National Center for Biotechnology Information (NCBI) and Arrayexpress at the European Bioinformatics Institute (EBI). Only one study was identified as a genome-wide profiling of a human skin pigmentary disorder, i.e. a dataset (GSE21429) we submitted to GEO in Apr, 2010. Therefore, we decided to perform this meta-analysis based only on 5 microarray datasets from our own laboratories. A brief introduction to our 5 individual studies is shown in Suppl Info Methods and detailed microarray data file information is listed in Table S1.
Details of the Microarray studies included in the meta-analysis and selection of subsets used, Microarray Data Preprocessing, Statistical Analysis, Bioinformatics Analysis, Immunohistochemical Analysis and Statistical Language and Packages are provided in Supplemental Information Methods.
Supplementary Material
ACKNOWLEDGEMENTS
This work was supported in part by the Intramural Research Program of the National Cancer Institute and in part by the Office of Science and the Center for Devices and Radiological Health, Food and Drug Administration.
Abbreviations
- AS
age spot
- ES
ethnic skin
- FDR
false discovery rate
- LLP
long-lasting pigmentation
- PIH
post-inflammatory hyperpigmentation
- UV
ultraviolet
Footnotes
CONFLICT OF INTEREST
The authors declare no conflict of interest.
REFERENCES
- Baxter LL, Pavan WJ. The etiology and molecular genetics of human pigmentation disorders. Wiley Interdiscip Rev Dev Biol. 2013;2:379–392. doi: 10.1002/wdev.72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett DC, Lamoreux ML. The color loci of mice - a genetic century. Pigment Cell Res. 2003;16:333–344. doi: 10.1034/j.1600-0749.2003.00067.x. [DOI] [PubMed] [Google Scholar]
- Calles C, Schneider M, Macaluso F, et al. Infrared A radiation influences the skin fibroblast transcriptome: mechanisms and consequences. J Invest Dermatol. 2010;130:1524–1536. doi: 10.1038/jid.2010.9. [DOI] [PubMed] [Google Scholar]
- Chi A, Valencia JC, Hu ZZ, et al. Proteomic and bioinformatic characterization of the biogenesis and function of melanosomes. J Proteome Res. 2006;5:3135–3144. doi: 10.1021/pr060363j. [DOI] [PubMed] [Google Scholar]
- Choi W, Miyamura Y, Wolber R, et al. Regulation of human skin pigmentation in situ by repetitive UV exposure - Molecular characterization of responses to UVA and/or UVB. J Invest Dermatol. 2010;130:1685–1696. doi: 10.1038/jid.2010.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cimadamore F, Shah M, Amador-Arjona A, et al. SOX2 modulates levels of MITF in normal human melanocytes, and melanoma lines in vitro. Pigment Cell Melanoma Res. 2012;25:533–536. doi: 10.1111/j.1755-148X.2012.01012.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coelho SG, Yin L, Ebsen D, et al. Photobiological implications of melanin photoprotection after UVB-induced tanning of human skin but not UVA-induced tanning. Pigment Cell Melanoma Res. 2015a;28:210–216. doi: 10.1111/pcmr.12331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coelho SG, Yin L, Valencia JC, et al. UV exposure modulates hemidesmosome plasticity contributing to long-term pigmentation in human skin. J Pathol. 2015b;236:17–29. doi: 10.1002/path.4497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hearing VJ, Merlino G. Melanocyte biology and melanomagenesis. In: Balch CM, Houghton AN, Sober AJ, Soong S-J, Atkins MB, Thompson JF, editors. Cutaneous Melanoma. 5th ed. St Louis: Quality Medical Publishing; 2009. pp. 821–845. [Google Scholar]
- Inkeles MS, Scumpia PO, Swindell WR, et al. Comparison of molecular signatures from multiple skin diseases identifies mechanisms of immunopathogenesis. J Invest Dermatol. 2014;135:151–159. doi: 10.1038/jid.2014.352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitsui H, Suarez-Farinas M, Belkin DA, et al. Combined use of laser capture microdissection and cDNA microarray analysis identifies locally expressed disease-related genes in focal regions of psoriasis vulgaris skin lesions. J Invest Dermatol. 2012;132:1615–1626. doi: 10.1038/jid.2012.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nordlund JJ, Boissy RE, Hearing VJ, Oetting WS, King RA, Ortonne JP. The Pigmentary System: Physiology and Pathophysiology. 2 ed. Edinburgh: Blackwell Science; 2006. [Google Scholar]
- Peters EM, Liezmann C, Spatz K, et al. Profiling mRNA of the graying human hair follicle constitutes a promising state-of-the-art tool to assess its aging: an exemplary report. J Invest Dermatol. 2013;133:1150–1160. doi: 10.1038/jid.2012.462. [DOI] [PubMed] [Google Scholar]
- Pollock RA, Abji F, Liang K, et al. Gene expression differences between psoriasis patients with and without inflammatory arthritis. J Invest Dermatol. 2014;135:620–623. doi: 10.1038/jid.2014.414. [DOI] [PubMed] [Google Scholar]
- Rashighi M, Agarwal P, Richmond JM, et al. CXCL10 is critical for the progression and maintenance of depigmentation in a mouse model of vitiligo. Sci Transl Med. 2014;6:223ra23. doi: 10.1126/scitranslmed.3007811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silvers WK. The Coat Colors of Mice: A model for mammalian gene action and interaction. Basel: Springer-Verlag; 1979. [Google Scholar]
- Smith JC, Boone BE, Opalenik SR, et al. Gene profiling of keloid fibroblasts shows altered expression in multiple fibrosis-associated pathways. J Invest Dermatol. 2008;128:1298–1310. doi: 10.1038/sj.jid.5701149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sturm RA, Duffy DL. Human pigmentation genes under environmental selection. Genome Biol. 2012;13:248. doi: 10.1186/gb-2012-13-9-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uematsu C, Katayama H, Makino I, et al. Peace, a MYB-like transcription factor, regulates petal pigmentation in flowering peach 'Genpei' bearing variegated and fully pigmented flowers. J Exp Bot. 2014;65:1081–1094. doi: 10.1093/jxb/ert456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi Y, Hearing VJ. Physiological factors that regulate skin pigmentation. BioFactors. 2009;35:193–199. doi: 10.1002/biof.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin L, Coelho SG, Ebsen D, et al. Epidermal gene expression and ethnic pigmentation variations among individuals of Asian, European and African ancestry. Exp Dermatol. 2014;23:731–735. doi: 10.1111/exd.12518. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.