To the Editor:
Cutaneous squamous cell carcinoma (cSCC) is the second most common form of cancer with approximately 700,000 cSCCs diagnosed in the USA annually1. Despite a generally good prognosis, 2-6% of cSCCs metastasize leading to approximately 3900-8800 deaths annually2,3,4. There are no targeted therapies or biomarkers for metastatic cSCC. The goal of this study was to determine whether there were differentially expressed microRNAs (miRNAs) in metastatic cSCCs relative to non-metastatic primary cSCCs in order to identify candidates for therapeutic intervention.
All human studies were approved by the OSU Institutional Review Board. We collected formalin-fixed paraffin-embedded tissue samples from individuals with metastatic cSCC or nonmetastatic cSCC for whom at least 5 years of follow-up data was available. Inclusion criteria included being immunocompetent, having sufficient tumor tissue and histologically normal skin available for RNA extraction, and having a pathologist confirmed primary cutaneous SCC of non-lip origin. Individuals with metastases were excluded if it was not clear which primary tumor gave rise to the metastasis or if the primary tumor was not available. Both regional and distant metastases were included.
Tissue samples were reviewed by a pathologist and areas of normal skin, primary tumor and metastatic tumor were marked for coring. Tissue cores for tumors were taken from regions with greater than 70% tumor cells and non-necrotic regions. RNA was isolated from tissue cores using an Ambion RecoverAll Kit, and concentration was measured by Nanodrop. After selecting the highest quality RNA samples, we performed miRNA expression analysis in 48 RNA samples using the NanoString nCounter miRNA panel of approximately 800 miRNAs. We profiled 30 matched samples from individuals with metastatic cSCC including ten normal skin (NM), ten primary cSCCs/one recurrent cSCC (TM) and nine metastatic cSCC RNA samples (MM). For comparison we profiled RNA from nine pairs (18 samples) of matched normal skin (NN) and non-metastatic primary cSCCs (TN). MicroRNA data are available at the Gene Expression Omnibus data repository (http://www.ncbi.nlm.nih.gov/geo/), accession number GSE55768.
Expression data for each sample was normalized to the entire miRNA dataset using the global sum of six positive controls and then quantile normalization method was performed on all miRNAs5. Linear models and paired T-tests compared non-metastatic primary tumors to primary tumors or metastases. Approximately 225 miRNAs were expressed in the skin. Differential expression significance was determined by controlling the expected false positive numbers across the 225 expressed miRNAs6. We used a nominal p-value of 0.01 to allow the expected false positive number of 2.25 for each comparison, and we considered 1.5-fold as biological significance. A heat map of unsupervised clustering of the seven most differentially expressed miRNAs between MM and TM/TN was generated using TIGR Multiexperimental Viewer (Figure 1).
Figure 1. Heat map of differential expressed miRNAs.
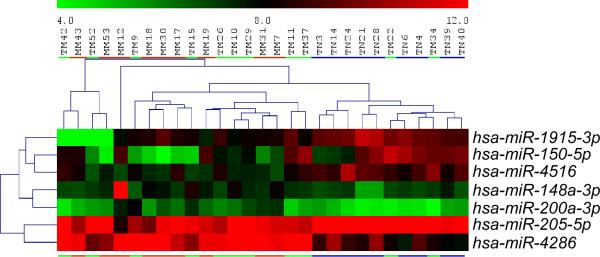
A heat map of unsupervised clustering of the seven most differentially expressed miRNAs between MM and primary tumors TM/TN was generated using TIGR Multiexperimental Viewer. miRNA names are indicated on the right side. MM samples (towards the left) are indicated by an orange line. TM are shown in green and TN (towards the right) are indicated by a blue line.
Expression of multiple miRNAs showed significant differences between MM and TM/TN tumors. These included up-regulation of miR-4286, miR-200a-3p and miR-148-3p and down-regulation of miR-1915-3p, miR-205-5p, miR-4516 and miR-150-5p (Table 1). Statistically significant differences were not found between paired MM and TM samples. However, there were 14 miRNAs showing significant differences between TM and TN including miR-4286, miR-421, miR-4516, and miR-574-5p. miRNAs, miR-135b, miR-21, miR-145, miR-100, and miR-214, which were previously shown to exhibit aberrant expression in cSCCs relative to normal skin were observed in a comparison of TM/TN and NM/NN 7,8. Additionally, multiple miRNAs previously associated with metastasis in other tumor types showed differential expression between the MM versus NM and NN; these include miR-4286, miR-135b, miR-21-5p, and miR-203 9,10.
Table 1.
MicroRNAs showing differential expression between metastatic tumors (MM) and all primary cSCCs (TM and TN)
| miRNA | Fold MM/TM and TN | p-value |
|---|---|---|
| hsa-miR-4286 | 3.2 | 0.002 |
| hsa-miR-200a-3p | 2.2 | 0.0005 |
| hsa-miR-148a-3p | 2.1 | 0.004 |
| hsa-miR-1915-3p | 0.53 | 0.001 |
| hsa-miR-205-5p | 0.48 | 0.002 |
| hsa-miR-4516 | 0.45 | 0.0003 |
| hsa-miR-150-5p | 0.23 | 0.001 |
In summary, several miRNAs show differential expression between MM and TM/TN; these may be useful as biomarkers to predict metastasis or as potential therapeutic targets. As the sample size was small, additional studies are warranted to confirm these findings.
Acknowledgments
Funding: This work was supported in part by a Pelotonia Idea Grant, the National Institutes of Health (R03 CA173788), and the American Cancer Society (RSG-07-083-01-MGO).
Footnotes
Conflict of Interest: The authors have no conflicts to disclose
References
- 1.Rogers HW, Weinstock MA, Harris AR, et al. Incidence estimate of nonmelanoma skin cancer in the United States, 2006. Arch Dermatol. 2010;146:283–287. doi: 10.1001/archdermatol.2010.19. [DOI] [PubMed] [Google Scholar]
- 2.Schmults CD, Karia PS, Carter JB, Han J, Qureshi AA. Factors predictive of recurrence and death from cutaneous squamous cell carcinoma. A 10-year, single-institution cohort study. JAMA Dermatol. 2013;149:541–547. doi: 10.1001/jamadermatol.2013.2139. [DOI] [PubMed] [Google Scholar]
- 3.Brantsch KD, Meisner C, Schönfisch B, et al. Analysis of risk factors determining prognosis of cutaneous squamous cell carcinoma: a prospective study. Lancet Oncol. 2008;9:713–720. doi: 10.1016/S1470-2045(08)70178-5. [DOI] [PubMed] [Google Scholar]
- 4.Karia PS, Han J, Schmults CD. Cutaneous squamous cell carcinoma: estimated incidence of disease, nodal metastasis, and deaths from disease in the United States, 2012. J Am Acad Dermatol. 2013;68:957–966. doi: 10.1016/j.jaad.2012.11.037. [DOI] [PubMed] [Google Scholar]
- 5.Rao Y, Lee Y, Jarjoura D, et al. A comparison of normalization techniques for microRNA microarray data. Stat Appl Genet Mol Biol. 2008;7 doi: 10.2202/1544-6115.1287. Article22. [DOI] [PubMed] [Google Scholar]
- 6.Gordon A, Glazko G, Qiu X, Yakovlev A. Control of the mean number of false discoveries, Bonferroni and stability of multiple testing. Annals of applied statistics. 2007;1:179–190. [Google Scholar]
- 7.Sand M, Skrygan M, Georgas D, et al. Microarray analysis of microRNA expression in cutaneous squamous cell carcinoma. J Dermatol Sci. 2012;68:119–126. doi: 10.1016/j.jdermsci.2012.09.004. [DOI] [PubMed] [Google Scholar]
- 8.Bruegger C, Kempf W, Spoerri I, et al. MicroRNA expression differs in cutaneous squamous cell carcinomas and healthy skin of immunocompetent individuals. Exp Dermatol. 2013;22:426–428. doi: 10.1111/exd.12153. [DOI] [PubMed] [Google Scholar]
- 9.Sand M, Skrygan M, Sand D, et al. Comparative microarray analysis of microRNA expression profiles in primary cutaneous malignant melanoma, cutaneous malignant melanoma metastases, and benign melanocytic nevi. Cell Tissue Res. 2013;351:85–98. doi: 10.1007/s00441-012-1514-5. [DOI] [PubMed] [Google Scholar]
- 10.Aigner A. MicroRNAs (miRNAs) in cancer invasion and metastasis: therapeutic approaches based on metastasis-related miRNAs. J Mol Med. 2011;89:445–457. doi: 10.1007/s00109-010-0716-0. [DOI] [PubMed] [Google Scholar]


