Abstract
Obesity and type 2 diabetes (T2D) are common and complex metabolic diseases, which are caused by an interchange between environmental and genetic factors. Recently, a number of large-scale genome-wide association studies (GWAS) have improved our knowledge of the genetic architecture and biological mechanisms of these diseases. Currently, more than ~250 genetic loci have been found for monogenic, syndromic, or common forms of T2D and/or obesity-related traits. In this review, we discuss the implications of these GWAS for obesity and T2D, and investigate the overlap of loci for obesity-related traits and T2D, highlighting potential mechanisms that affect T2D susceptibility.
Keywords: Obesity, Body mass index, Fat distribution, Waist-to-hip ratio, Adiponectin, Leptin, Non-alcoholic fatty liver disease, Pericardial fat, Subcutaneous fat, Visceral fat, Fat percent, Type 2 diabetes, Genome-wide association study, Insulin resistance, Beta-cell function, Sexual dimorphism
Introduction
Type 2 diabetes (T2D) is a common metabolic disease of increased plasma glucose levels to which individuals are predisposed to by a combination of genes and environmental factors. The hyperglycemia typically results from decreased insulin sensitivity (insulin resistance) in insulin-dependent tissues (such as skeletal muscle, liver and adipose tissues), which leads to hyperinsulinemia. Subsequently, when the pancreatic beta cells are not capable of producing the amount of insulin required to maintain normal glycemic status, which may be caused by beta-cell dysfunction and/or reduced beta-cell mass, chronic hyperglycemia and T2D occur (reviewed in [1]).
Overall obesity is defined when a person’s body mass index (BMI, weight (in kilograms) divided by height (in meters) squared) is ≥30 kg/m2 [2]. Directly measured fat percent (fat%, measured by bioimpedance (BI), dual-energy X-ray absorptiometry (DXA), computerized tomography (CT), or magnetic resonance imaging (MRI)) is a more accurate indicator of adiposity, which also takes the amount of lean and fat mass into account [3]. Other specific measures of individual fat depots and fat distribution include waist circumference (WC), hip circumference (HC), waist-to-hip ratio (WHR), and subcutaneous and visceral adipose tissue (SAT and VAT) [4, 5]. Levels of adiponectin secreted from adipose tissue [6–13], ectopic fat depots such as pericardial fat [14], and non-alcoholic fatty liver disease (NAFLD) [15] are also obesity-related traits.
The prevalence of obesity and T2D is currently escalating worldwide as a consequence of a sedentary lifestyle and increased consumption of high-energy content food [2]. Between 1980 and 2014, the worldwide prevalence of obesity more than doubled. In 2014, 11 % of men (>205 million) and 15 % of women (>297 million) in the world were obese, compared with 5 % for men and 8 % for women in 1980 [16]. The overall prevalence of obesity is at least four times higher in high-income countries compared to that in low-income countries. A similar accompanying increase in the prevalence of T2D is seen as obesity is a risk factor for T2D [17, 18]. In 2014, it was estimated that there are 387 million people living with diabetes (ages 20–79) with a worldwide prevalence of 8.3 %, and ~90 % of these are individuals with T2D. By 2035, this number is expected to increase by 205 million. It is estimated that 77 % of people with diabetes live in low- and middle-income countries [19].
Genetic, but also environmental, factors interact to cause both obesity and T2D as shown by familial aggregation [20–23], family and twin studies on obesity (heritability (h2)~40–70 %) [22, 24, 25] and T2D (h2~26–69 %) [21, 26]. Beyond a sedentary lifestyle, socioeconomic status, poor nutrition, infections and differences in the gut flora have also been added to the list of potential environmental triggers of obesity and T2D [27]. Genetic and environmental evidence is also provided by numerous animal studies. Rodent models for T2D, such as the Lepob and Zucker mice strains rely on the mutations in genes encoding leptin or its receptor to develop T2D via obesity (reviewed in [28]). Evidence of both environmental and genetic effects in an animal model has been shown to exist in the Agouti Avy mouse, where the obesity phenotype is inherited through an epigenetic effect that is dependent on the maternal diet [29].
Early evidence for the genetic effect in obesity and diabetes was found through linkage studies of monogenic forms of these diseases segregating as Mendelian disorders, in which mutations occurring in a gene lead to extreme and early-onset forms of these conditions. For obesity, these include genes functioning in the leptin-melanocortin pathway, such as the leptin (LEP) and melanocortin 4 receptor (MC4R) genes (reviewed in [30, 31]). Similarly, monogenic forms of diabetes are caused by mutations in genes such as GCK, HNF4A and HNF1A with allelic series causing maturity onset of the young (MODY) (reviewed in [32]). Linkage studies have been subsequently accompanied by larger and statistically more powerful genome-wide association studies (GWAS) that are designed to dissect the genetic architecture of common complex traits in a hypothesis-free way [33]. GWAS are useful for identifying common genetic variants (i.e. single-nucleotide polymorphisms (SNPs) with a minor allele frequency (MAF) >5 %) that affect a trait outcome or increase the risk of a disease of interest by comparing frequencies of alleles in thousands of individuals, or between cases and healthy controls, respectively. Many variants associated with complex traits and diseases have been discovered so far through the GWAS approach. SNPs that reach genome-wide significance (p < 5 × 10−8 after correction for multiple-testing, 0.05/1,000,000 independent tests among common variants in the human genome) are specifically targeted for replication and further functional experiments [33]. These associations are important for unraveling biological mechanisms and pathways that might lend themselves to informing about new therapeutic targets.
In this article, we review the current GWAS of obesity-related traits and consider the overlap with T2D-associated loci in order to gain insights into the genetic susceptibility and potential mechanisms that lead to increased risk of T2D.
Overview of Genetics of T2D and Obesity-Related Traits in the GWAS Era
To date, T2D GWAS efforts including samples of European [34, 35, 36•, 37, 38], East Asian [39–45], South Asian [46, 47], Mexican/Mexican American [48] and African American [49] descent have delivered 76 robust susceptibility loci [50••]. The majority of these T2D associations appear to act through beta-cell function-related pathways. In contrast, a handful of T2D-associated loci seem to primarily operate through insulin resistance (reviewed in [51]). These associated loci generally have small effect sizes and only explain ~6 and ~10–20 % of the variance in disease susceptibility and the heritability, respectively [36•]. Thus, much of the genetic contribution to the disease remains to be discovered.
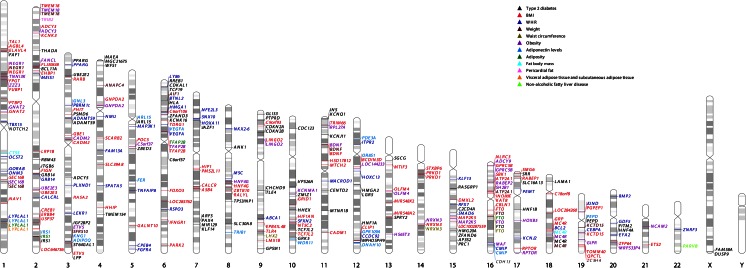
Overall ~185 loci associated with obesity traits have been identified in large-scale GWAS efforts; analyses in Europeans found associations between 77 loci and BMI [52–57, 58••], 48 loci and WHR (adjusted for BMI, WHRadjBMI) [59–61, 62••], and three loci and body fat% [3]. Furthermore, 13 loci were associated with extreme and/or early-onset obesity [63–68], one locus with VAT in women and one locus with VAT/SAT ratio [4], five loci with NAFLD [15], one locus with pericardial fat [14], and seven loci with clinical classes of obesity [69] (Fig. 1). The most recent meta-analysis by the Genetic Investigation of Anthropometric Traits (GIANT) consortium involved 125 studies of European (up to 322,154 individuals) and non-European (up to 17,072 individuals) ancestry [58••, 62••]. Inclusion of non-European ethnicities in these analyses revealed additional genetic associations with 10 BMI loci and a WHRadjBMI locus [58••, 62••]. Ten more associations with BMI were discovered in the secondary analyses of this recent GIANT study [58••]. In other non-European GWAS of BMI and WHRadjBMI, eight additional loci were identified [70–72].
Fig. 1.
Genome-wide hits for T2D and obesity-related traits loci. GWAS data were obtained from GWAS catalog (http://www.ebi.ac.uk/gwas/, accessed 26 June 2015). Search terms used included obesity, type 2 diabetes, adiposity, waist, fat, body mass index, non-alcoholic liver, adiponectin, weight and adipose. P value threshold for association was p < 5 × 10−8. Associations are labeled with corresponding trait colors
Since the heritability of BMI is 7 % higher at younger ages and increases with the mean age in childhood studies (+1.2 % per year) [73], GWAS of children, adolescents and young adults have been carried out identifying three loci associated with childhood BMI [66, 74–76]. To date, ~2.7 % of the phenotypic variation in BMI was explained by the 97 associations in populations of European and non-European ancestry. Furthermore, common genetic variation (MAF > 5 %) accounted for ~21 % of BMI variation.
Overlap Between GWAS of Obesity-Related Traits and T2D
Body Mass Index
The first overall obesity GWAS [54] identified a robust association between BMI and SNPs in the first intron of the fat mass and obesity-associated (FTO) gene that has been widely replicated since [70, 72, 77–80]. FTO variants had previously been associated with T2D (p < 5 × 10−8), but this association disappeared after adjusting for BMI, which showed that FTO association with T2D is largely due its effect on BMI [54]. In line with this, the effect of FTO variants on 23 cardiometabolic traits, in addition to T2D, is mainly mediated through BMI [81]. The FTO locus is not only strongly associated with T2D risk [50••] and higher BMI [52] but also increased fasting insulin and homeostatic model estimated insulin resistance (HOMA-IR; p = 9.5 × 10−5), which is in agreement with insulin resistance playing a part in the FTO association with T2D via increased BMI [52]. The FTO protein has been characterized as a 2-oxoglutarate- and Fe(II)-dependent demethylase, possibly involved in mRNA modification, and it is highly expressed in the brain [82, 83]. However, a recent study suggested that the obesity-associated FTO variants affect expression of IRX3, but not FTO, in the human brain, which may mean that FTO is not the causal gene in this region. Functional experiments also supported this finding; body weight of Irx3-deficient mice was reduced by 25–30 %, and hypothalamic expression of a dominant-negative form of Irx3 resulted in the same metabolic phenotype as the Irx3-deficient mice [84]. The precise biological role of the BMI-associated variants at the FTO locus is still unclear and remains to be disentangled.
Following the identification of obesity variants in FTO, a robust and replicated association between BMI and variants ~188 kb upstream of MC4R was reported [55, 58••, 85–87]. Previous studies have shown that mutations causing MC4R inactivation lead to severe and monogenic forms of obesity [31]. Low frequency variants in MC4R were identified in morbid obese individuals (BMI > 40 kg/m2) and were associated with obesity [88, 89]. MC4R is a neural G-protein-coupled melanocortin receptor that is highly expressed in the brain [90]. It plays an important role in the regulation of energy balance, specifically in the regulation of energy intake via the control of satiety and energy expenditure (reviewed in [91]).
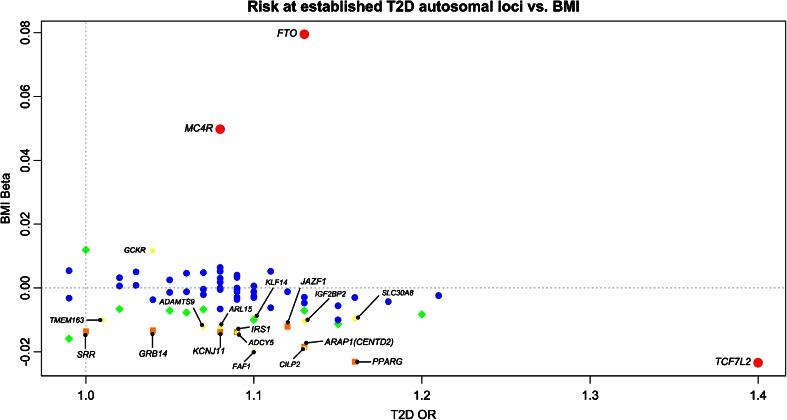
The MC4R locus was associated with both T2D (rs12970134, odds ratio (OR) = 1.08, 95 % confidence interval (CI) = 1.03–1.12, European p = 0.0002, trans-ethnic p = 2.6 × 10−8) [50••] and BMI (beta = 0.05, 95 % CI = 0.043–0.057, p = 4.7 × 10−47) [58••] (Fig. 2). In addition, another SNP (rs571312) at the same locus, in strong linkage disequilibrium with rs12970134 (r2 = 0.87, D′ = 0.96, HapMap2, Utah Residents with European ancestry population (CEU)), was associated with increased fasting insulin (p = 5.2 × 10−5), HOMA-IR (p = 7.6 × 10−5) and T2D (p = 0.0004), which is in agreement with insulin resistance playing a part in the MC4R association with T2D through BMI [52]. However, in an exome sequencing study of 6760 Pima Indians, mutations decreasing MC4R activity were detected and these individuals with MC4R defects had increased T2D risk, partially independent of BMI in childhood (BMI-adjusted hazard rate ratio = 3.3, 95 % CI = 1.2–9.2, p = 0.03). This effect might be due to an increased rate of weight gain compared to adulthood and MC4R affecting downstream insulin signaling [92, 93]. Nevertheless, the effect of MC4R variants on T2D risk was completely attributable to BMI in adulthood [94]. Thus, taking into account the changing physiology and hormonal levels during different stages of life would be valuable when considering the biology behind traits and diseases such as BMI and T2D.
Fig. 2.
Risk at T2D autosomal loci [50••] vs. BMI [58••]. P value thresholds for association with BMI (y-axis) are p < 5 × 10−8 (red), 5 × 10−8 ≤ p < 10−4 (orange), 10−4 ≤ p < 0.01 (yellow), 0.01 ≤ p < 0.05 (green) and p ≥ 0.05 (blue). Red, orange and yellow associations are labeled with corresponding gene names
The genetic association between T2D and variants in transcription factor 7-like 2 (TCF7L2) was first discovered in a candidate gene study [95]. This association was later detected in GWAS, and it is the strongest known association with the disease to date among common variants (rs7903146, OR = 1.4, p = 1.9 × 10−59) [34, 35, 36•, 37, 38, 50••]. The same lead SNP was also identified in the recent GIANT meta-analysis of BMI (beta = −0.023, p = 1.1 × 10−11) [58••] (Fig. 2). It is interesting that the T allele of rs7903146 increases T2D risk while decreasing BMI, opposing the idea that increased BMI leads to insulin resistance and T2D. In comparison to FTO and MC4R variants, TCF7L2 variants have a much larger effect on T2D risk and a smaller effect on BMI, which might indicate that the TCF7L2 variants act via T2D to affect BMI (Fig. 2). TCF7L2 is a transcription factor functioning in WNT signaling, which is crucial for cell proliferation, motility, normal embryogenesis, and regulation of myogenesis and adipogenesis (reviewed in [96]). Although the causal variant is still unclear, the T2D risk allele appears to act via lowering the levels of insulin secretion and influencing beta-cell function (reviewed in [51, 96, 97]).
In Fig. 2, 17 genes associated with both T2D [50••] and BMI (orange: p < 5 × 10−8 ≤ p < 10−4 for BMI; yellow: 10-4 ≤ p < 0.01 for BMI) [58••] are shown. These associations provide insights into the genetic overlap of T2D and BMI. For instance, the ARL15 (rs702634) T2D risk allele was associated with increased fasting insulin (BMI-adjusted, p = 5 × 10–12), HOMA-IR (p = 0.02) and triglyceride levels (p = 0.01) as well as decreased high-density lipoprotein (HDL) levels and BMI (p = 5.6 × 10−5) [50••]. These associations implicate that ARL15 variants may play a role in insulin resistance leading to T2D susceptibility independently of BMI.
GWAS of Fat Percent
In a meta-analysis of 15 GWAS with 36,626 individuals of European and Indian Asian descent informative for fat% (as measured by BI and/or DXA), three loci near FTO, SPRY2 and IRS1 were identified [3]. All of these loci were previously associated with T2D [42, 98]. The fat%-decreasing allele of rs2943650 near IRS1 was associated with increased risk of T2D as that allele decreased subcutaneous fat but not visceral fat, which is more health damaging (reviewed in [99]). The T2D risk allele of another IRS1 variant, rs2943640 (r2 = 0.97, D′ = 1.00, HapMap2, CEU), was also nominally associated with decreased BMI (beta = −0.014, p = 1.1 × 10−5; Fig. 2) [58••]. Furthermore, another variant (rs2943634), strongly correlated with the T2D and fat%-associated rs2943650 (r2 = 0.80, D′ = 0.96, HapMap2, CEU), was associated with fasting insulin levels (beta = 0.025, p = 2.5 × 10−14) [34, 100, 101]. Insulin receptor substrate 1 encoded by IRS1 is an important member of the insulin signaling cascade functioning as a docking protein and activating downstream signaling when phosphorylated by the insulin receptor [102]. Given the essential function of IRS1 in insulin signaling and the association of IRS1 variants with T2D as well as fat% and BMI, this gene is likely to be involved in fat distribution, adipocyte biology and/or insulin resistance [98].
GWAS of Extreme/Early-Onset Obesity
In the polygenic form of extreme/early-onset obesity, mutations in more than one gene may play a role in susceptibility. Individuals with extreme/early-onset obesity are likely to be enriched for genetic variants predisposing the general population to obesity. Out of five GWAS, only three studies identified novel loci that were not discovered by the previous GWAS of BMI [64, 65, 68]. Except for FTO and MC4R variants, which affect T2D susceptibility through their effect on BMI, none of these loci overlap with the known T2D loci (Fig. 1) [50••].
Waist Circumference and Waist-to-Hip Ratio
WHRadjBMI is a measure of fat distribution that indicates the amount of metabolically adverse visceral fat [61], while taking into account the metabolically protective role of gluteal fat [103, 104]. Both WC and WHR are associated with T2D risk independently of BMI [17, 18], and are correlated with the gold standard MRI measurement of central adiposity (i.e. visceral fat, r2 = 0.6 and r2 = 0.5, respectively). However, when targeting genetic associations independent of BMI, WHRadjBMI is a better measure of central fat distribution given the strong correlation between WC and BMI (WC-BMI r2 = 0.9, WHR-BMI r2 = 0.6) [59].
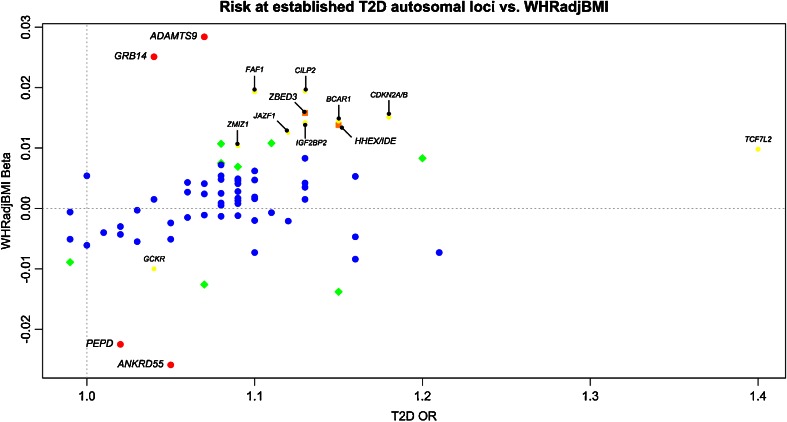
A number of variants strongly associated (p < 5 × 10−8) with T2D risk exhibit opposite directions of effect on BMI and WHRadjBMI (Figs. 2 and 3). For instance, while the T2D risk allele in GCKR is associated with increased BMI (rs780094, C allele, beta = 0.01, p = 0.0002) [58••] (Fig. 2), the same variant has an opposite effect on WHRadjBMI (rs780094, beta = −0.01, p = 0.004) [62••] (Fig. 3). Interestingly, sexual dimorphism was also observed in WHRadjBMI, with a statistically significant (p < 0.05) GCKR association only in women (rs780094, beta = −0.015, p = 0.001). Glucokinase regulatory protein (GCKR) regulates glucokinase (GCK), which is a crucial enzyme for glucose metabolism in the liver and glucose-stimulated insulin secretion from pancreatic beta cells. It was previously observed that over-expression of GCKR in the liver significantly improved insulin sensitivity and glucose tolerance in mice resulting in decreased leptin and increased triglyceride levels [105]. This finding may provide a possible explanation for the observed genetic association; the effect of GCKR variants may act through leptin to increase BMI, while independently affecting central fat distribution.
Fig. 3.
Risk at T2D autosomal loci [50••] vs. WHRadjBMI [62••]. P value thresholds for association with WHRadjBMI (y-axis) are p < 5 × 10−8 (red), 5 × 10−8 ≤ p < 10−4 (orange), 10−4 ≤ p < 0.01 (yellow), 0.01 ≤ p < 0.05 (green) and p ≥ 0.05 (blue). Red, orange and yellow associations are labeled with corresponding gene names
Similarly, a pattern of sexual dimorphism was detected for >~40 % (20/49) of the WHRadjBMI lead SNPs, while no pronounced gender difference was found in the BMI and T2D studies [50••, 58••, 62••]. The stronger associations with WHRadjBMI in women were identified in genes that are known to be involved in insulin resistance and/or lipid traits [61, 62••]. One of these genes is GRB14 (growth factor receptor-bound protein 14; rs3923113, beta = 0.025, p = 1.0 × 10−12) [62••], which encodes a protein functioning in the regulation of insulin signaling. It binds to insulin receptors, leading to inhibition of their catalytic activity [106]. Female-specific effects of GRB14 variants were also detected in previous studies of WHRadjBMI and blood lipids [61, 107, 108]. In addition, a gender-specific association with T2D risk was observed with a stronger association in women (rs3923113; ORmen = 1.05, p = 0.005; ORwomen = 1.11, p = 1.8 × 10−9) [36•]. Furthermore, another WHRadjBMI-associated GRB14 variant (rs10195252, r2 = 0.79, D′ = 1.00, HapMap2, CEU) also exhibited association with decreased BMI (beta = −0.010, p = 0.002), HC (beta = −0.021, p = 3 × 10−9), HDL (Z-score = −2.6, p = 0.008), increased low-density lipoprotein (LDL) (Z-score = 3.5, p = 4.5 × 10−4), triglycerides (Z-score = 5.8, p = 7.4 × 10−9), fasting insulin (Z-score = 4.6, p = 5 × 10−6) and HOMA-IR (Z-score = 4.8, p = 1.9 × 10−6). SNP rs10195252 was associated with expression of GRB14 in SAT as well, indicating that GRB14 could indeed be the effector transcript in this locus [61].
GRB14 is an interesting example of a gene with T2D risk alleles causing increased WHRadjBMI and decreased BMI (Figs. 2 and 3). Associations of T2D risk alleles with increased fasting insulin and HOMA-IR implicate GRB14 variants playing a role in insulin resistance [109]. In rodents and humans, expression of GRB14 in adipose tissue was negatively correlated with insulin sensitivity. In addition, prolonged fasting and metformin treatment in mice significantly decreased Grb14 expression in peri-epididymal adipose tissue [110]. Furthermore, improved glucose homeostasis and enhanced insulin signaling were observed in Grb14-deficient mice [111]. These findings provide evidence for the importance of GRB14 regulation in insulin resistance and show that complete understanding of its regulation is essential for identification of new therapeutic pathways in obesity and T2D [112].
ADAMTS9 is a member of the ADAMTS (a disintegrin and metalloproteinase with thrombospondin motifs) family of proteins involved in cleaving proteoglycans, controlling organ maturation and development as well as inhibiting angiogenesis. Its expression is high in all fetal tissues, adult heart and skeletal muscle [113]. Similar to GRB14, ADAMTS9 is also implicated in insulin sensing [61], and variants within this gene show a stronger WHRadjBMI association in women (rs6795735, beta = 0.025, p = 9.8 × 10−14 [61]; rs2371767, pmen = 0.008, pwomen = 7.1 × 10−23 [108]; rs6795735-rs2371767 r2 = 0.31, D′ = 1.00, HapMap2, CEU). Furthermore, the WHRadjBMI-associated variant (rs6795735) was also nominally associated with decreased HDL (rs6795735, Z-score = −2.5, p = 0.01) and T2D risk (OR = 1.12, p = 0.002), but not with BMI (Fig. 3) [50••, 61]. ADAMTS9 seems to play a role in insulin resistance in peripheral tissues [114]. Although a possible association of T2D risk allele with beta-cell function has also been reported [115], it was not detected in larger GWAS [36•].
ANKRD55-MAP3K1 is another T2D susceptibility locus (rs459193, OR = 1.05, p = 0.03 ) [50••] that was also associated with WHRadjBMI (beta = −0.026, p = 1.6 × 10−11) [62••] but not overall obesity (Fig. 3). Due to lack of association of the lead SNP (rs459193) with ANKRD55 expression, ANKRD55 may not be the functional gene in this region. The neighboring gene MAP3K1, with known functions in insulin signaling, has been suggested as a mediator of the biological effect [36•, 101], but that remains to be validated.
PEPD encodes peptidase D, which is an enzyme functioning in the recycling of proline and potentially in collagen production. Variants near PEPD were associated with fasting insulin (rs731839, BMI-adjusted beta = 0.015, p = 5.1 × 10−12) [100, 101] and adiponectin levels (rs731839, beta = −0.03, p = 8 × 10−12) [7], which is directionally consistent with the function of adiponectin in regulating insulin sensitivity. Furthermore, the intronic variant rs3786897 (rs3786897-rs731839 r2 = 0.34, D′ = 1.00, HapMap2, CEU) was nominally associated with T2D susceptibility (European OR = 1.02, p = 0.3, 12,171 cases and 56,862 controls; East Asian OR = 1.17, p = 3.5 × 10−7, 6952 cases and 11,865 controls; trans-ethnic p = 3.3 × 10−4 ) [50••] and WHRadjBMI (beta = −0.022, p = 9.7 × 10−11; Fig. 3) [62••]. Interestingly, SNP rs8182584 is strongly correlated with the fasting insulin and adiponectin-associated variant at this locus (rs731839-rs8182584 r2 = 0.82, D′ = 0.92, HapMap2, CEU) and was also associated with reduced expression levels of PEPD in adipose tissue (beta = −0.13, p = 9.96 × 10−10) [7]. The effect of PEPD variants may be mediated through insulin and/or adiponectin pathways. Even though PEPD associations with T2D are ethnically heterogeneous, identification of a possible role of PEPD in susceptibility to T2D and obesity may provide crucial insights into biological mechanisms of these conditions.
GWAS of Other Obesity-Related Traits: Abdominal Subcutaneous and Visceral Adipose Tissue, Non-alcoholic Fatty Liver Disease and Pericardial Fat
Similar to WHRadjBMI associations, sexual dimorphism was observed in genetic associations with SAT and VAT, highlighting the importance of physiological and hormonal difference in susceptibility to obesity and T2D in men and women [4]. In a GWAS of NAFLD, associations with five loci were identified [15]. One of these loci, the previously reported WHRadjBMI locus at LYPLAL1, was also associated with VAT/SAT ratio in women (rs4846567, p = 0.0004) [4] and T2D (rs2820446, p = 2.3 × 10−6; rs4846567-rs2820446 r2 = 1.00, D′ = 1.00, HapMap2, CEU) [50••]. Women are known to have more subcutaneous fat but less visceral fat compared to men [5]. Given the protective role of subcutaneous fat in T2D susceptibility, it is plausible to observe more men with T2D. Globally, the prevalence of T2D is higher in men, but the reasons for this observation may not be limited to the amount of subcutaneous fat in men [116]. In a GWAS of pericardial fat, only one locus (TRIB2) reached genome-wide significance, but this locus is also devoid from associations with T2D and other obesity traits (Fig. 1). This lack of overlap between loci associated with T2D, the more commonly used obesity trait measures (BMI, WHRadjBMI, etc.) and the other obesity-related traits such as pericardial fat may indicate that there is a different genetic architecture for pericardial fat and potentially for other ectopic fat depots. Anthropometric and more specifically measured traits might be more distinct than the close relationships between these phenotypes indicate, or these observations most likely reflect that there is a power difference in detection of loci between these studies [14].
GWAS of Adiponectin Levels
Adiponectin is an adipokine secreted by adipocytes increasing insulin sensitivity [117–119]. Adiponectin levels are positively correlated with HC and inversely correlated with WC, WHR, body fat%, BMI, T2D and coronary heart disease [120–124]. Levels of adiponectin are highly heritable (30–70 %) [125–127], and a number of GWAS have been performed to identify genes affecting adiponectin levels [6–13]. The previously discussed T2D/BMI-associated ARL15 locus also showed an independent association with adiponectin levels (rs702634-rs4311394 r2 = 0.09, D′ = 0.90, HapMap2, CEU). The lead SNP at the ARL15 locus (rs4311394) was associated with lower adiponectin levels (p = 2.9 × 10−8), increased T2D risk (OR = 1.11, p = 3.2 × 10−3) and coronary heart disease (OR = 1.12, p = 8.5 × 10−6) [13]. The function of ARL15 is not known, but ARL15 expression is more pronounced in skeletal muscle [13]. Interestingly, glucose is disposed in skeletal muscle in an insulin-dependent manner, and adiponectin trafficking is essential for insulin sensitivity and glucose transport in muscle. ARL15 is structurally similar to proteins functioning in intracellular vesicle trafficking, and it was suggested that it might play a role in insulin signaling and glucose transport [128, 129]. Therefore, effects of ARL15 variants may be mediated via insulin resistance and/or adiponectin trafficking [13].
Conclusions
T2D loci appear to affect susceptibility to T2D via two main mechanisms: (1) through insulin resistance, i.e. insulin sensitivity (measured by fasting insulin and HOMA-IR) and/or (2) through a beta-cell dysfunction (measured by fasting glucose and homeostatic model estimated beta-cell function). In addition, these loci, in general, also exhibit overlapping associations with obesity-related traits and blood lipid levels (HDL, LDL, triglycerides), which might explain the phenotypic overlap with obesity and cardiovascular diseases. However, these associations are often heterogeneous and variants may have opposite directions of effect for different obesity-related traits, reflecting the intricate biology behind them. For instance, most T2D risk alleles seem to be associated with a decrease in BMI, except for the variants in FTO, MC4R and GCKR, two of which are known to affect T2D susceptibility through BMI [50••]. In contrast, most T2D risk variants are associated with increased WHRadjBMI (Figs. 2 and 3). This observation might indicate distinct mechanisms by which (1) WHRadjBMI- and BMI-increasing alleles act on T2D risk, and (2) T2D risk alleles act on BMI. Increased BMI and central adiposity (defined by increased WHRadjBMI) are known to be health damaging and raising T2D risk via insulin resistance. However, there seems to be a second mechanism where risk alleles (e.g. TCF7L2 variants) predominantly act via T2D and decrease BMI, not vice versa. More targeted genetic and functional studies are necessary to explore these mechanisms and biological pathways implicated (reviewed in [130, 131]).
The heritability of obesity and T2D is not entirely explained by all the loci discovered so far [36•, 50••, 52, 58••, 61, 62••]. More studies with larger sample sizes, in different ethnicities, employing various approaches such as rare variant analysis, exome sequencing, studies of epigenetics and gene-environment interactions are necessary to help explain the missing heritability. Identifying actual functional variants may also increase the phenotypic variance explained by the known loci. Identification of novel loci and functional variants is also required to gain a better understanding of the genetic architecture of body shape, fat depots and T2D. Discovery of additional overlapping genetic associations could provide important insights into the role played by obesity in susceptibility to T2D.
Beyond filling out the gaps in the heritability estimates, deciphering biological mechanisms and pathways that mediate effects leading to susceptibility to obesity and T2D is essential for development of new therapeutic strategies, including lifestyle changes. It is noticeable that genes within loci that are BMI- and WHRadjBMI-associated display different gene expression patterns; they have higher expression levels in the hypothalamus and adipose/peripheral tissues, respectively [52, 61]. These initial observations were further supported by the evidence from Data-driven Expression Prioritized Integration for Complex Traits (DEPICT) analyses in the recent GIANT BMI and WHRadjBMI meta-analyses [58••, 62••]. For WHRadjBMI, significant pathways and gene sets included adiponectin signaling, insulin sensitivity and regulation of glucose levels, skeletal growth, transcriptional regulation and those functioning in the development of metabolically active tissues such as adipose, liver and muscle [62••]. In contrast, highlighted pathways and gene sets for BMI included those functioning in the central nervous system involved in synaptic function, long-term potentiation and neurotransmitter signaling [58••].
Monogenic obesity genes in the leptin-melanocortin pathway provide the link between adipose tissue and the hypothalamus, which are critical sites for balancing the energy need of the body. Genes functioning in the leptin-melanocortin pathway such as those encoding leptin (LEP), leptin receptor (LEPR), melanocortin 4 receptor (MC4R), pro-opiomelanocortin (POMC) and brain-derived neurotrophic factor (BDNF) have been implicated in the monogenic form of obesity (reviewed in [30]). Leptin is a hormone produced by adipocytes that play an important role in food intake and weight regulation. Increased leptin signaling in the hypothalamus leads to decreased food intake via MC4R and POMC-derived peptide alpha-melanocyte stimulating hormone (alpha-MSH). Many of the monogenic obesity genes lie within loci that are also associated with T2D [50••]. The overlap between monogenic obesity genes and obesity genes identified via GWAS (e.g. MC4R and BDNF) might imply a role of hypothalamic dysfunction affecting the regulation of energy balance in polygenic obesity, which can drive T2D.
Gender-specific effects are observed for anthropometric traits, particularly for waist-related phenotypes, and understanding their biological influences is crucial [61, 108]. For instance, variants in and around PPARG have been associated with T2D, monogenic obesity and WHRadjBMI. Of these, the WHRadjBMI association exhibited sexual dimorphism with a significantly stronger effect in women (betawomen = 0.035, betamen = 0.005) [62••]. In parallel with that, gender differences were detected in response to PPARG-agonist therapy in patients with T2D which might indicate different mechanisms for insulin resistance in men and women [132]. Even though biological functions of associated loci are not clear for many genes, gender-specific effects are detected during/after puberty and are potentially attributable to sex hormones [133]. In addition, distribution of body fat also affects metabolic pathways, and body fat has an endocrinological role producing hormones such as estrogen, progesterone, leptin and adiponectin, which affect the regulation of energy balance in the hypothalamus and insulin sensitivity [134]. The understanding of sexual dimorphisms is likely to improve exploration of metabolic disease processes and design of better therapeutic approaches.
In summary, the recent GWAS of obesity-related traits and T2D show considerable overlap in associated loci. These identified associations point to potential mechanisms through which obesity traits affect T2D susceptibility.
Acknowledgments
CML is funded by Wellcome Trust (086596/Z/08/Z and 086596/Z/08/A) and the Li Ka Shing Foundation.
Compliance with Ethics Guidelines
ᅟ
Conflict of Interest
Tugce Karaderi, Alexander W. Drong, and Cecilia M. Lindgren declare that they have no conflict of interest.
Human and Animal Rights and Informed Consent
This article does not contain any studies with human or animal subjects performed by any of the authors.
Footnotes
This article is part of the Topical Collection on Genetics
Contributor Information
Tugce Karaderi, Email: tugce.karaderi@well.ox.ac.uk.
Alexander W. Drong, Email: alexd@well.ox.ac.uk
Cecilia M. Lindgren, Email: celi@broadinstitute.org
References
Papers of particular interest, published recently, have been highlighted as: • Of importance •• Of major importance
- 1.Kahn SE, Hull RL, Utzschneider KM. Mechanisms linking obesity to insulin resistance and type 2 diabetes. Nature. 2006;444(7121):840–6. doi: 10.1038/nature05482. [DOI] [PubMed] [Google Scholar]
- 2.Tsigos C, Hainer V, Basdevant A, et al. Management of obesity in adults: European clinical practice guidelines. Obes Facts. 2008;1(2):106–16. doi: 10.1159/000126822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kilpelainen TO, Zillikens MC, Stancakova A, et al. Genetic variation near IRS1 associates with reduced adiposity and an impaired metabolic profile. Nat Genet. 2011;43(8):753–60. doi: 10.1038/ng.866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fox CS, Liu Y, White CC, et al. Genome-wide association for abdominal subcutaneous and visceral adipose reveals a novel locus for visceral fat in women. PLoS Genet. 2012;8(5):e1002695. doi: 10.1371/journal.pgen.1002695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fox CS, Massaro JM, Hoffmann U, et al. Abdominal visceral and subcutaneous adipose tissue compartments: association with metabolic risk factors in the Framingham Heart Study. Circulation. 2007;116(1):39–48. doi: 10.1161/CIRCULATIONAHA.106.675355. [DOI] [PubMed] [Google Scholar]
- 6.Chung CM, Lin TH, Chen JW, et al. A genome-wide association study reveals a quantitative trait locus of adiponectin on CDH13 that predicts cardiometabolic outcomes. Diabetes. 2011;60(9):2417–23. doi: 10.2337/db10-1321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dastani Z, Hivert MF, Timpson N, et al. Novel loci for adiponectin levels and their influence on type 2 diabetes and metabolic traits: a multi-ethnic meta-analysis of 45,891 individuals. PLoS Genet. 2012;8(3):e1002607. doi: 10.1371/journal.pgen.1002607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Heid IM, Henneman P, Hicks A, et al. Clear detection of ADIPOQ locus as the major gene for plasma adiponectin: results of genome-wide association analyses including 4659 European individuals. Atherosclerosis. 2010;208(2):412–20. doi: 10.1016/j.atherosclerosis.2009.11.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jee SH, Sull JW, Lee JE, et al. Adiponectin concentrations: a genome-wide association study. Am J Hum Genet. 2010;87(4):545–52. doi: 10.1016/j.ajhg.2010.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Morisaki H, Yamanaka I, Iwai N, et al. CDH13 gene coding T-cadherin influences variations in plasma adiponectin levels in the Japanese population. Hum Mutat. 2012;33(2):402–10. doi: 10.1002/humu.21652. [DOI] [PubMed] [Google Scholar]
- 11.Wu Y, Gao H, Li H, et al. A meta-analysis of genome-wide association studies for adiponectin levels in East Asians identifies a novel locus near WDR11-FGFR2. Hum Mol Genet. 2014;23(4):1108–19. doi: 10.1093/hmg/ddt488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wu Y, Li Y, Lange EM, et al. Genome-wide association study for adiponectin levels in Filipino women identifies CDH13 and a novel uncommon haplotype at KNG1-ADIPOQ. Hum Mol Genet. 2010;19(24):4955–64. doi: 10.1093/hmg/ddq423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Richards JB, Waterworth D, O'Rahilly S, et al. A genome-wide association study reveals variants in ARL15 that influence adiponectin levels. PLoS Genet. 2009;5(12):e1000768. doi: 10.1371/journal.pgen.1000768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fox CS, White CC, Lohman K, et al. Genome-wide association of pericardial fat identifies a unique locus for ectopic fat. PLoS Genet. 2012;8(5):e1002705. doi: 10.1371/journal.pgen.1002705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Speliotes EK, Yerges-Armstrong LM, Wu J, et al. Genome-wide association analysis identifies variants associated with nonalcoholic fatty liver disease that have distinct effects on metabolic traits. PLoS Genet. 2011;7(3):e1001324. doi: 10.1371/journal.pgen.1001324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.World Health Organization FSN-Oao, 2015. http://www.who.int/mediacentre/factsheets/fs311/en/.
- 17.Carey VJ, Walters EE, Colditz GA, et al. Body fat distribution and risk of non-insulin-dependent diabetes mellitus in women. The Nurses’ Health Study. Am J Epidemiol. 1997;145(7):614–9. doi: 10.1093/oxfordjournals.aje.a009158. [DOI] [PubMed] [Google Scholar]
- 18.Wang Y, Rimm EB, Stampfer MJ, et al. Comparison of abdominal adiposity and overall obesity in predicting risk of type 2 diabetes among men. Am J Clin Nutr. 2005;81(3):555–63. doi: 10.1093/ajcn/81.3.555. [DOI] [PubMed] [Google Scholar]
- 19.International Diabetes Federation. IDF Diabetes Atlas update poster teB, Belgium: International Diabetes Federation, 2014. http://www.idf.org/diabetesatlas.
- 20.Meigs JB, Cupples LA, Wilson PW. Parental transmission of type 2 diabetes: the Framingham Offspring Study. Diabetes. 2000;49(12):2201–7. doi: 10.2337/diabetes.49.12.2201. [DOI] [PubMed] [Google Scholar]
- 21.Poulsen P, Kyvik KO, Vaag A, et al. Heritability of type II (non-insulin-dependent) diabetes mellitus and abnormal glucose tolerance—a population-based twin study. Diabetologia. 1999;42(2):139–45. doi: 10.1007/s001250051131. [DOI] [PubMed] [Google Scholar]
- 22.Stunkard AJ, Foch TT, Hrubec Z. A twin study of human obesity. JAMA. 1986;256(1):51–4. doi: 10.1001/jama.1986.03380010055024. [DOI] [PubMed] [Google Scholar]
- 23.Glans F, Elgzyri T, Shaat N, et al. Immigrants from the Middle-East have a different form of type 2 diabetes compared with Swedish patients. Diabet Med. 2008;25(3):303–7. doi: 10.1111/j.1464-5491.2007.02366.x. [DOI] [PubMed] [Google Scholar]
- 24.Maes HH, Neale MC, Eaves LJ. Genetic and environmental factors in relative body weight and human adiposity. Behav Genet. 1997;27(4):325–51. doi: 10.1023/A:1025635913927. [DOI] [PubMed] [Google Scholar]
- 25.Zillikens MC, Yazdanpanah M, Pardo LM, et al. Sex-specific genetic effects influence variation in body composition. Diabetologia. 2008;51(12):2233–41. doi: 10.1007/s00125-008-1163-0. [DOI] [PubMed] [Google Scholar]
- 26.Almgren P, Lehtovirta M, Isomaa B, et al. Heritability and familiality of type 2 diabetes and related quantitative traits in the Botnia Study. Diabetologia. 2011;54(11):2811–9. doi: 10.1007/s00125-011-2267-5. [DOI] [PubMed] [Google Scholar]
- 27.Moreno-Indias I, Cardona F, Tinahones FJ, et al. Impact of the gut microbiota on the development of obesity and type 2 diabetes mellitus. Front Microbiol. 2014;5:190. doi: 10.3389/fmicb.2014.00190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chatzigeorgiou A, Halapas A, Kalafatakis K, et al. The use of animal models in the study of diabetes mellitus. In Vivo. 2009;23(2):245–58. [PubMed] [Google Scholar]
- 29.Morgan HD, Sutherland HG, Martin DI, et al. Epigenetic inheritance at the agouti locus in the mouse. Nat Genet. 1999;23(3):314–8. doi: 10.1038/15490. [DOI] [PubMed] [Google Scholar]
- 30.Farooqi IS, O'Rahilly S. Mutations in ligands and receptors of the leptin-melanocortin pathway that lead to obesity. Nat Clin Pract Endocrinol Metab. 2008;4(10):569–77. doi: 10.1038/ncpendmet0966. [DOI] [PubMed] [Google Scholar]
- 31.Farooqi IS, Yeo GS, Keogh JM, et al. Dominant and recessive inheritance of morbid obesity associated with melanocortin 4 receptor deficiency. J Clin Invest. 2000;106(2):271–9. doi: 10.1172/JCI9397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schwitzgebel VM. Many faces of monogenic diabetes. J Diabetes Investig. 2014;5(2):121–33. doi: 10.1111/jdi.12197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Manolio TA. Genomewide association studies and assessment of the risk of disease. N Engl J Med. 2010;363(2):166–76. doi: 10.1056/NEJMra0905980. [DOI] [PubMed] [Google Scholar]
- 34.Dupuis J, Langenberg C, Prokopenko I, et al. New genetic loci implicated in fasting glucose homeostasis and their impact on type 2 diabetes risk. Nat Genet. 2010;42(2):105–16. doi: 10.1038/ng.520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kong A, Steinthorsdottir V, Masson G, et al. Parental origin of sequence variants associated with complex diseases. Nature. 2009;462(7275):868–74. doi: 10.1038/nature08625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.•.Morris AP, Voight BF, Teslovich TM, et al. Large-scale association analysis provides insights into the genetic architecture and pathophysiology of type 2 diabetes. Nat Genet. 2012;44(9):981–90. doi: 10.1038/ng.2383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Voight BF, Scott LJ, Steinthorsdottir V, et al. Twelve type 2 diabetes susceptibility loci identified through large-scale association analysis. Nat Genet. 2010;42(7):579–89. doi: 10.1038/ng.609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zeggini E, Scott LJ, Saxena R, et al. Meta-analysis of genome-wide association data and large-scale replication identifies additional susceptibility loci for type 2 diabetes. Nat Genet. 2008;40(5):638–45. doi: 10.1038/ng.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cho YS, Chen CH, Hu C, et al. Meta-analysis of genome-wide association studies identifies eight new loci for type 2 diabetes in East Asians. Nat Genet. 2012;44(1):67–72. doi: 10.1038/ng.1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Li H, Gan W, Lu L, et al. A genome-wide association study identifies GRK5 and RASGRP1 as type 2 diabetes loci in Chinese Hans. Diabetes. 2013;62(1):291–8. doi: 10.2337/db12-0454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Qi L, Cornelis MC, Kraft P, et al. Genetic variants at 2q24 are associated with susceptibility to type 2 diabetes. Hum Mol Genet. 2010;19(13):2706–15. doi: 10.1093/hmg/ddq156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shu XO, Long J, Cai Q, et al. Identification of new genetic risk variants for type 2 diabetes. PLoS Genet. 2010;6(9):e1001127. doi: 10.1371/journal.pgen.1001127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tsai FJ, Yang CF, Chen CC, et al. A genome-wide association study identifies susceptibility variants for type 2 diabetes in Han Chinese. PLoS Genet. 2010;6(2):e1000847. doi: 10.1371/journal.pgen.1000847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yamauchi T, Hara K, Maeda S, et al. A genome-wide association study in the Japanese population identifies susceptibility loci for type 2 diabetes at UBE2E2 and C2CD4A-C2CD4B. Nat Genet. 2010;42(10):864–8. doi: 10.1038/ng.660. [DOI] [PubMed] [Google Scholar]
- 45.Takeuchi F, Serizawa M, Yamamoto K, et al. Confirmation of multiple risk loci and genetic impacts by a genome-wide association study of type 2 diabetes in the Japanese population. Diabetes. 2009;58(7):1690–9. doi: 10.2337/db08-1494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kooner JS, Saleheen D, Sim X, et al. Genome-wide association study in individuals of South Asian ancestry identifies six new type 2 diabetes susceptibility loci. Nat Genet. 2011;43(10):984–9. doi: 10.1038/ng.921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tabassum R, Chauhan G, Dwivedi OP, et al. Genome-wide association study for type 2 diabetes in Indians identifies a new susceptibility locus at 2q21. Diabetes. 2013;62(3):977–86. doi: 10.2337/db12-0406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Parra EJ, Below JE, Krithika S, et al. Genome-wide association study of type 2 diabetes in a sample from Mexico City and a meta-analysis of a Mexican-American sample from Starr County Texas. Diabetol. 2011;54(8):2038–46. doi: 10.1007/s00125-011-2172-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Palmer ND, McDonough CW, Hicks PJ, et al. A genome-wide association search for type 2 diabetes genes in African Americans. PLoS One. 2012;7(1):e29202. doi: 10.1371/journal.pone.0029202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.••.Replication DIG, Meta-analysis C. Genome-wide trans-ancestry meta-analysis provides insight into the genetic architecture of type 2 diabetes susceptibility. Nat Genet. 2014;46(3):234–44. doi: 10.1038/ng.2897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Florez JC. Newly identified loci highlight beta cell dysfunction as a key cause of type 2 diabetes: where are the insulin resistance genes? Diabetologia. 2008;51(7):1100–10. doi: 10.1007/s00125-008-1025-9. [DOI] [PubMed] [Google Scholar]
- 52.Speliotes EK, Willer CJ, Berndt SI, et al. Association analyses of 249,796 individuals reveal 18 new loci associated with body mass index. Nat Genet. 2010;42(11):937–48. doi: 10.1038/ng.686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Scuteri A, Sanna S, Chen WM, et al. Genome-wide association scan shows genetic variants in the FTO gene are associated with obesity-related traits. PLoS Genet. 2007;3(7):e115. doi: 10.1371/journal.pgen.0030115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Frayling TM, Timpson NJ, Weedon MN, et al. A common variant in the FTO gene is associated with body mass index and predisposes to childhood and adult obesity. Science. 2007;316(5826):889–94. doi: 10.1126/science.1141634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Loos RJ, Lindgren CM, Li S, et al. Common variants near MC4R are associated with fat mass, weight and risk of obesity. Nat Genet. 2008;40(6):768–75. doi: 10.1038/ng.140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Willer CJ, Speliotes EK, Loos RJ, et al. Six new loci associated with body mass index highlight a neuronal influence on body weight regulation. Nat Genet. 2009;41(1):25–34. doi: 10.1038/ng.287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Thorleifsson G, Walters GB, Gudbjartsson DF, et al. Genome-wide association yields new sequence variants at seven loci that associate with measures of obesity. Nat Genet. 2009;41(1):18–24. doi: 10.1038/ng.274. [DOI] [PubMed] [Google Scholar]
- 58.••.Locke AE, Kahali B, Berndt SI. Genetic studies of body mass index yield new insights for obesity biology. Nature. 2015;518(7538):197–206. doi: 10.1038/nature14177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lindgren CM, Heid IM, Randall JC, et al. Genome-wide association scan meta-analysis identifies three loci influencing adiposity and fat distribution. PLoS Genet. 2009;5(6):e1000508. doi: 10.1371/journal.pgen.1000508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Heard-Costa NL, Zillikens MC, Monda KL, et al. NRXN3 is a novel locus for waist circumference: a genome-wide association study from the CHARGE Consortium. PLoS Genet. 2009;5(6):e1000539. doi: 10.1371/journal.pgen.1000539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Heid IM, Jackson AU, Randall JC, et al. Meta-analysis identifies 13 new loci associated with waist-hip ratio and reveals sexual dimorphism in the genetic basis of fat distribution. Nat Genet. 2010;42(11):949–60. doi: 10.1038/ng.685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.••.Shungin D, Winkler TW, Croteau-Chonka DC. New genetic loci link adipose and insulin biology to body fat distribution. Nature. 2015;518(7538):187–96. doi: 10.1038/nature14132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hinney A, Nguyen TT, Scherag A, et al. Genome wide association (GWA) study for early onset extreme obesity supports the role of fat mass and obesity associated gene (FTO) variants. PLoS One. 2007;2(12):e1361. doi: 10.1371/journal.pone.0001361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Meyre D, Delplanque J, Chevre JC, et al. Genome-wide association study for early-onset and morbid adult obesity identifies three new risk loci in European populations. Nat Genet. 2009;41(2):157–9. doi: 10.1038/ng.301. [DOI] [PubMed] [Google Scholar]
- 65.Scherag A, Dina C, Hinney A, et al. Two new Loci for body-weight regulation identified in a joint analysis of genome-wide association studies for early-onset extreme obesity in French and German study groups. PLoS Genet. 2010;6(4):e1000916. doi: 10.1371/journal.pgen.1000916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Paternoster L, Evans DM, Nohr EA, et al. Genome-wide population-based association study of extremely overweight young adults—the GOYA study. PLoS One. 2011;6(9):e24303. doi: 10.1371/journal.pone.0024303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wang K, Li WD, Zhang CK, et al. A genome-wide association study on obesity and obesity-related traits. PLoS One. 2011;6(4):e18939. doi: 10.1371/journal.pone.0018939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wheeler E, Huang N, Bochukova EG, et al. Genome-wide SNP and CNV analysis identifies common and low-frequency variants associated with severe early-onset obesity. Nat Genet. 2013;45(5):513–7. doi: 10.1038/ng.2607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Berndt SI, Gustafsson S, Magi R, et al. Genome-wide meta-analysis identifies 11 new loci for anthropometric traits and provides insights into genetic architecture. Nat Genet. 2013;45(5):501–12. doi: 10.1038/ng.2606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wen W, Cho YS, Zheng W, et al. Meta-analysis identifies common variants associated with body mass index in East Asians. Nat Genet. 2012;44(3):307–11. doi: 10.1038/ng.1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Liu CT, Monda KL, Taylor KC, et al. Genome-wide association of body fat distribution in African ancestry populations suggests new loci. PLoS Genet. 2013;9(8):e1003681. doi: 10.1371/journal.pgen.1003681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Monda KL, Chen GK, Taylor KC, et al. A meta-analysis identifies new loci associated with body mass index in individuals of African ancestry. Nat Genet. 2013;45(6):690–6. doi: 10.1038/ng.2608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Elks CE, den Hoed M, Zhao JH, et al. Variability in the heritability of body mass index: a systematic review and meta-regression. Front Endocrinol (Lausanne) 2012;3:29. doi: 10.3389/fendo.2012.00029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Bradfield JP, Taal HR, Timpson NJ, et al. A genome-wide association meta-analysis identifies new childhood obesity loci. Nat Genet. 2012;44(5):526–31. doi: 10.1038/ng.2247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Graff M, Ngwa JS, Workalemahu T, et al. Genome-wide analysis of BMI in adolescents and young adults reveals additional insight into the effects of genetic loci over the life course. Hum Mol Genet. 2013;22(17):3597–607. doi: 10.1093/hmg/ddt205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Namjou B, Keddache M, Marsolo K, et al. EMR-linked GWAS study: investigation of variation landscape of loci for body mass index in children. Front Genet. 2013;4:268. doi: 10.3389/fgene.2013.00268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Cho YS, Go MJ, Kim YJ, et al. A large-scale genome-wide association study of Asian populations uncovers genetic factors influencing eight quantitative traits. Nat Genet. 2009;41(5):527–34. doi: 10.1038/ng.357. [DOI] [PubMed] [Google Scholar]
- 78.Dina C, Meyre D, Gallina S, et al. Variation in FTO contributes to childhood obesity and severe adult obesity. Nat Genet. 2007;39(6):724–6. doi: 10.1038/ng2048. [DOI] [PubMed] [Google Scholar]
- 79.Li H, Kilpelainen TO, Liu C, et al. Association of genetic variation in FTO with risk of obesity and type 2 diabetes with data from 96,551 East and South Asians. Diabetologia. 2012;55(4):981–95. doi: 10.1007/s00125-011-2370-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Okada Y, Kubo M, Ohmiya H, et al. Common variants at CDKAL1 and KLF9 are associated with body mass index in East Asian populations. Nat Genet. 2012;44(3):302–6. doi: 10.1038/ng.1086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Fall T, Hagg S, Magi R, et al. The role of adiposity in cardiometabolic traits: a Mendelian randomization analysis. PLoS Med. 2013;10(6):e1001474. doi: 10.1371/journal.pmed.1001474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Gerken T, Girard CA, Tung YC, et al. The obesity-associated FTO gene encodes a 2-oxoglutarate-dependent nucleic acid demethylase. Science. 2007;318(5855):1469–72. doi: 10.1126/science.1151710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Tung YC, Ayuso E, Shan X, et al. Hypothalamic-specific manipulation of Fto, the ortholog of the human obesity gene FTO, affects food intake in rats. PLoS One. 2010;5(1):e8771. doi: 10.1371/journal.pone.0008771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Smemo S, Tena JJ, Kim KH, et al. Obesity-associated variants within FTO form long-range functional connections with IRX3. Nature. 2014;507(7492):371–5. doi: 10.1038/nature13138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Been LF, Nath SK, Ralhan SK, et al. Replication of association between a common variant near melanocortin-4 receptor gene and obesity-related traits in Asian Sikhs. Obesity (Silver Spring) 2010;18(2):425–9. doi: 10.1038/oby.2009.254. [DOI] [PubMed] [Google Scholar]
- 86.Cheung CY, Tso AW, Cheung BM, et al. Obesity susceptibility genetic variants identified from recent genome-wide association studies: implications in a Chinese population. J Clin Endocrinol Metab. 2010;95(3):1395–403. doi: 10.1210/jc.2009-1465. [DOI] [PubMed] [Google Scholar]
- 87.Hester JM, Wing MR, Li J, et al. Implication of European-derived adiposity loci in African Americans. Int J Obes (Lond) 2012;36(3):465–73. doi: 10.1038/ijo.2011.131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Vaisse C, Clement K, Guy-Grand B, et al. A frameshift mutation in human MC4R is associated with a dominant form of obesity. Nat Genet. 1998;20(2):113–4. doi: 10.1038/2407. [DOI] [PubMed] [Google Scholar]
- 89.Young EH, Wareham NJ, Farooqi S, et al. The V103I polymorphism of the MC4R gene and obesity: population based studies and meta-analysis of 29 563 individuals. Int J Obes (Lond) 2007;31(9):1437–41. doi: 10.1038/sj.ijo.0803609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Liu H, Kishi T, Roseberry AG, et al. Transgenic mice expressing green fluorescent protein under the control of the melanocortin-4 receptor promoter. J Neurosci. 2003;23(18):7143–54. doi: 10.1523/JNEUROSCI.23-18-07143.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Girardet C, Butler AA. Neural melanocortin receptors in obesity and related metabolic disorders. Biochim Biophys Acta. 2014;1842(3):482–94. doi: 10.1016/j.bbadis.2013.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Chai B, Li JY, Zhang W, et al. Melanocortin-4 receptor activation inhibits c-Jun N-terminal kinase activity and promotes insulin signaling. Peptides. 2009;30(6):1098–104. doi: 10.1016/j.peptides.2009.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Chai B, Li JY, Zhang W, et al. Melanocortin-4 receptor activation promotes insulin-stimulated mTOR signaling. Peptides. 2010;31(10):1888–93. doi: 10.1016/j.peptides.2010.06.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Thearle MS, Muller YL, Hanson RL, et al. Greater impact of melanocortin-4 receptor deficiency on rates of growth and risk of type 2 diabetes during childhood compared with adulthood in Pima Indians. Diabetes. 2012;61(1):250–7. doi: 10.2337/db11-0708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Grant SF, Thorleifsson G, Reynisdottir I, et al. Variant of transcription factor 7-like 2 (TCF7L2) gene confers risk of type 2 diabetes. Nat Genet. 2006;38(3):320–3. doi: 10.1038/ng1732. [DOI] [PubMed] [Google Scholar]
- 96.Weedon MN. The importance of TCF7L2. Diabet Med. 2007;24(10):1062–6. doi: 10.1111/j.1464-5491.2007.02258.x. [DOI] [PubMed] [Google Scholar]
- 97.Gloyn AL, Braun M, Rorsman P. Type 2 diabetes susceptibility gene TCF7L2 and its role in beta-cell function. Diabetes. 2009;58(4):800–2. doi: 10.2337/db09-0099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Rung J, Cauchi S, Albrechtsen A, et al. Genetic variant near IRS1 is associated with type 2 diabetes, insulin resistance and hyperinsulinemia. Nat Genet. 2009;41(10):1110–5. doi: 10.1038/ng.443. [DOI] [PubMed] [Google Scholar]
- 99.Loos RJ. Genetic determinants of common obesity and their value in prediction. Best Pract Res Clin Endocrinol Metab. 2012;26(2):211–26. doi: 10.1016/j.beem.2011.11.003. [DOI] [PubMed] [Google Scholar]
- 100.Manning AK, Hivert MF, Scott RA, et al. A genome-wide approach accounting for body mass index identifies genetic variants influencing fasting glycemic traits and insulin resistance. Nat Genet. 2012;44(6):659–69. doi: 10.1038/ng.2274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Scott RA, Lagou V, Welch RP, et al. Large-scale association analyses identify new loci influencing glycemic traits and provide insight into the underlying biological pathways. Nat Genet. 2012;44(9):991–1005. doi: 10.1038/ng.2385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.White MF. The IRS-signalling system: a network of docking proteins that mediate insulin action. Mol Cell Biochem. 1998;182(1-2):3–11. doi: 10.1023/A:1006806722619. [DOI] [PubMed] [Google Scholar]
- 103.Snijder MB, Dekker JM, Visser M, et al. Associations of hip and thigh circumferences independent of waist circumference with the incidence of type 2 diabetes: the Hoorn Study. Am J Clin Nutr. 2003;77(5):1192–7. doi: 10.1093/ajcn/77.5.1192. [DOI] [PubMed] [Google Scholar]
- 104.Snijder MB, Dekker JM, Visser M, et al. Trunk fat and leg fat have independent and opposite associations with fasting and postload glucose levels: the Hoorn study. Diabetes Care. 2004;27(2):372–7. doi: 10.2337/diacare.27.2.372. [DOI] [PubMed] [Google Scholar]
- 105.Slosberg ED, Desai UJ, Fanelli B, et al. Treatment of type 2 diabetes by adenoviral-mediated overexpression of the glucokinase regulatory protein. Diabetes. 2001;50(8):1813–20. doi: 10.2337/diabetes.50.8.1813. [DOI] [PubMed] [Google Scholar]
- 106.Desbuquois B, Bereziat V, Authier F, et al. Compartmentalization and in vivo insulin-induced translocation of the insulin-signaling inhibitor Grb14 in rat liver. FEBS J. 2008;275(17):4363–77. doi: 10.1111/j.1742-4658.2008.06583.x. [DOI] [PubMed] [Google Scholar]
- 107.Teslovich TM, Musunuru K, Smith AV, et al. Biological, clinical and population relevance of 95 loci for blood lipids. Nature. 2010;466(7307):707–13. doi: 10.1038/nature09270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Randall JC, Winkler TW, Kutalik Z, et al. Sex-stratified genome-wide association studies including 270,000 individuals show sexual dimorphism in genetic loci for anthropometric traits. PLoS Genet. 2013;9(6):e1003500. doi: 10.1371/journal.pgen.1003500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Harder MN, Ribel-Madsen R, Justesen JM, et al. Type 2 diabetes risk alleles near BCAR1 and in ANK1 associate with decreased beta-cell function whereas risk alleles near ANKRD55 and GRB14 associate with decreased insulin sensitivity in the Danish Inter99 cohort. J Clin Endocrinol Metab. 2013;98(4):E801–6. doi: 10.1210/jc.2012-4169. [DOI] [PubMed] [Google Scholar]
- 110.Cariou B, Capitaine N, Le Marcis V, et al. Increased adipose tissue expression of Grb14 in several models of insulin resistance. FASEB J. 2004;18(9):965–7. doi: 10.1096/fj.03-0824fje. [DOI] [PubMed] [Google Scholar]
- 111.Cooney GJ, Lyons RJ, Crew AJ, et al. Improved glucose homeostasis and enhanced insulin signalling in Grb14-deficient mice. EMBO J. 2004;23(3):582–93. doi: 10.1038/sj.emboj.7600082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Goenaga D, Hampe C, Carre N, et al. Molecular determinants of Grb14-mediated inhibition of insulin signaling. Mol Endocrinol. 2009;23(7):1043–51. doi: 10.1210/me.2008-0360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Clark ME, Kelner GS, Turbeville LA, et al. ADAMTS9, a novel member of the ADAM-TS/ metallospondin gene family. Genomics. 2000;67(3):343–50. doi: 10.1006/geno.2000.6246. [DOI] [PubMed] [Google Scholar]
- 114.Boesgaard TW, Gjesing AP, Grarup N, et al. Variant near ADAMTS9 known to associate with type 2 diabetes is related to insulin resistance in offspring of type 2 diabetes patients—EUGENE2 study. PLoS One. 2009;4(9):e7236. doi: 10.1371/journal.pone.0007236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Trombetta M, Bonetti S, Boselli ML, et al. PPARG2 Pro12Ala and ADAMTS9 rs4607103 as “insulin resistance loci” and “insulin secretion loci” in Italian individuals. The GENFIEV study and the Verona Newly Diagnosed Type 2 Diabetes Study (VNDS) 4. Acta Diabetol. 2013;50(3):401–8. doi: 10.1007/s00592-012-0443-9. [DOI] [PubMed] [Google Scholar]
- 116.Wild S, Roglic G, Green A, et al. Global prevalence of diabetes: estimates for the year 2000 and projections for 2030. Diabetes Care. 2004;27(5):1047–53. doi: 10.2337/diacare.27.5.1047. [DOI] [PubMed] [Google Scholar]
- 117.Hivert MF, Sullivan LM, Fox CS, et al. Associations of adiponectin, resistin, and tumor necrosis factor-alpha with insulin resistance. J Clin Endocrinol Metab. 2008;93(8):3165–72. doi: 10.1210/jc.2008-0425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Hung J, McQuillan BM, Thompson PL, et al. Circulating adiponectin levels associate with inflammatory markers, insulin resistance and metabolic syndrome independent of obesity. Int J Obes (Lond) 2008;32(5):772–9. doi: 10.1038/sj.ijo.0803793. [DOI] [PubMed] [Google Scholar]
- 119.Wannamethee SG, Tchernova J, Whincup P, et al. Associations of adiponectin with metabolic and vascular risk parameters in the British Regional Heart Study reveal stronger links to insulin resistance-related than to coronory heart disease risk-related parameters. Int J Obes (Lond) 2007;31(7):1089–98. doi: 10.1038/sj.ijo.0803544. [DOI] [PubMed] [Google Scholar]
- 120.Lindsay RS, Funahashi T, Hanson RL, et al. Adiponectin and development of type 2 diabetes in the Pima Indian population. Lancet. 2002;360(9326):57–8. doi: 10.1016/S0140-6736(02)09335-2. [DOI] [PubMed] [Google Scholar]
- 121.Pischon T, Girman CJ, Hotamisligil GS, et al. Plasma adiponectin levels and risk of myocardial infarction in men. JAMA. 2004;291(14):1730–7. doi: 10.1001/jama.291.14.1730. [DOI] [PubMed] [Google Scholar]
- 122.Spranger J, Kroke A, Mohlig M, et al. Adiponectin and protection against type 2 diabetes mellitus. Lancet. 2003;361(9353):226–8. doi: 10.1016/S0140-6736(03)12255-6. [DOI] [PubMed] [Google Scholar]
- 123.Weyer C, Funahashi T, Tanaka S, et al. Hypoadiponectinemia in obesity and type 2 diabetes: close association with insulin resistance and hyperinsulinemia. J Clin Endocrinol Metab. 2001;86(5):1930–5. doi: 10.1210/jcem.86.5.7463. [DOI] [PubMed] [Google Scholar]
- 124.Smith J, Al-Amri M, Sniderman A, et al. Leptin and adiponectin in relation to body fat percentage, waist to hip ratio and the apoB/apoA1 ratio in Asian Indian and Caucasian men and women. Nutr Metab (Lond) 2006;3:18. doi: 10.1186/1743-7075-3-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Comuzzie AG, Funahashi T, Sonnenberg G, et al. The genetic basis of plasma variation in adiponectin, a global endophenotype for obesity and the metabolic syndrome. J Clin Endocrinol Metab. 2001;86(9):4321–5. doi: 10.1210/jcem.86.9.7878. [DOI] [PubMed] [Google Scholar]
- 126.Menzaghi C, Trischitta V, Doria A. Genetic influences of adiponectin on insulin resistance, type 2 diabetes, and cardiovascular disease. Diabetes. 2007;56(5):1198–209. doi: 10.2337/db06-0506. [DOI] [PubMed] [Google Scholar]
- 127.Vasseur F, Helbecque N, Dina C, et al. Single-nucleotide polymorphism haplotypes in the both proximal promoter and exon 3 of the APM1 gene modulate adipocyte-secreted adiponectin hormone levels and contribute to the genetic risk for type 2 diabetes in French Caucasians. Hum Mol Genet. 2002;11(21):2607–14. doi: 10.1093/hmg/11.21.2607. [DOI] [PubMed] [Google Scholar]
- 128.Gillingham AK, Munro S. The small G proteins of the Arf family and their regulators. Annu Rev Cell Dev Biol. 2007;23:579–611. doi: 10.1146/annurev.cellbio.23.090506.123209. [DOI] [PubMed] [Google Scholar]
- 129.Hofmann I, Thompson A, Sanderson CM, et al. The Arl4 family of small G proteins can recruit the cytohesin Arf6 exchange factors to the plasma membrane. Curr Biol. 2007;17(8):711–6. doi: 10.1016/j.cub.2007.03.007. [DOI] [PubMed] [Google Scholar]
- 130.Billings LK, Florez JC. The genetics of type 2 diabetes: what have we learned from GWAS? Ann N Y Acad Sci. 2010;1212:59–77. doi: 10.1111/j.1749-6632.2010.05838.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.McCarthy MI. Genomics, type 2 diabetes, and obesity. N Engl J Med. 2010;363(24):2339–50. doi: 10.1056/NEJMra0906948. [DOI] [PubMed] [Google Scholar]
- 132.Arnetz L, Dorkhan M, Alvarsson M, et al. Gender differences in non-glycemic responses to improved insulin sensitivity by pioglitazone treatment in patients with type 2 diabetes. Acta Diabetol. 2014;51(2):185–92. doi: 10.1007/s00592-013-0457-y. [DOI] [PubMed] [Google Scholar]
- 133.Wells JC. Sexual dimorphism of body composition. Best Pract Res Clin Endocrinol Metab. 2007;21(3):415–30. doi: 10.1016/j.beem.2007.04.007. [DOI] [PubMed] [Google Scholar]
- 134.Kershaw EE, Flier JS. Adipose tissue as an endocrine organ. J Clin Endocrinol Metab. 2004;89(6):2548–56. doi: 10.1210/jc.2004-0395. [DOI] [PubMed] [Google Scholar]