Abstract
Bartonella bacilliformis is the etiologic agent of Carrión's disease or Oroya fever. B. bacilliformis infection represents an interesting model of human host specificity. The notable differences in clinical presentations of Carrión's disease suggest complex adaptations by the bacterium to the human host, with the overall objectives of persistence, maintenance of a reservoir state for vectorial transmission, and immune evasion. These events include a multitude of biochemical and genetic mechanisms involving both bacterial and host proteins. This review focuses on proteins involved in interactions between B. bacilliformis and the human host. Some of them (e.g., flagellin, Brps, IalB, FtsZ, Hbp/Pap31, and other outer membrane proteins) are potential protein antigen candidates for a synthetic vaccine.
1. Introduction
Bartonella bacilliformis is a member of the alpha-2 subgroup of Proteobacteria and is the etiologic agent of Carrión's disease or Oroya fever in humans. Bartonellosis has been historically described in Peru, Ecuador, and Colombia [1, 2], with Peru considered the most important endemic area in the world. This disease often, but not always, presents in a biphasic manner. The primary, life-threatening acute phase is characterized by a septic state worsened by a severe hemolytic anemia (bacteria invade the erythrocytes which are destroyed by the spleen) [3]. This phase, known as Oroya fever, has a high fatality rate (up to 88%) if not treated and is more common in children [4]. Fever, pallor, general malaise, myalgia, headache, jaundice, and hepato/splenomegaly are the main symptoms of Oroya fever [1]. Some patients develop, as a consequence of the septic state, a transient immunosuppression that results in the onset of opportunistic infections such as toxoplasmosis, tuberculosis, salmonellosis, shigellosis, histoplasmosis, malaria, and pneumocystosis [1, 5, 6]. In the secondary chronic phase, known as verruga peruana (Peruvian wart), the bacteria invade the endothelial cells producing wart-like hemangiomatous lesions of the skin and mucous membranes 4 to 8 weeks after the onset of Oroya fever. The duration of the eruptive phase is 3 to 6 months. Lesions are classified as miliary (small reddish papules < 3 mm in diameter), mular (nodular tumors > 5 mm), and diffuse subdermal nodules [7]. Inhabitants, especially schoolchildren, in endemic regions often develop the eruptive phase as the sole manifestation of the disease [8]. Humans are the only known natural reservoir for B. bacilliformis [7, 9]. B. bacilliformis is believed to be transmitted to humans by bites of phlebotomine sand flies living in the high mountain valleys of the Andes [10].
Since B. bacilliformis was first described in 1905 by Alberto Barton; numerous research studies have been reported, including clinical and experimental studies. In the last 25 years, arguably the most important studies have been conducted to explain the physiopathology of the disease.
The notable differences in clinical presentations during a B. bacilliformis infection suggest a complex adaptation to the human host with the objectives of persistence, maintenance of a reservoir state for vector-based transmission, and immune evasion. The bacterium-host relationship evokes a multitude of biochemical and genetic mechanisms involving both bacterial and host proteins.
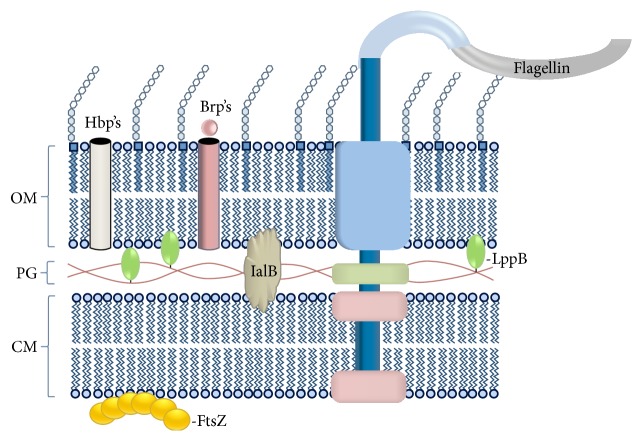
This review focuses on proteins involved in B. bacilliformis' interactions with the human host. We hypothesize that certain proteins would be ideal protein antigen candidates to be included in a chemically synthesized vaccine to block B. bacilliformis interactions with host cells (see Figure 1).
Figure 1.
Various protein antigens of B. bacilliformis and their locations in the cell. Brp's: Bartonella repeat proteins, FtsZ: cell division protein, Hbp's: hemin-binding proteins, IalB: invasion-associated locus B protein, LppB: lipoprotein B, OM: outer membrane, PG: peptidoglycan, and CM: cytosolic membrane.
Flagellin. B. bacilliformis cells are highly motile by lophotrichous flagella, appendages consisting of ~42 kDa flagellin protein subunits that are resistant to protease or trypsin treatment [11]. B. bacilliformis was reported to possess 1 to 10 polar flagella 3 to 10 μm in length [11]. In addition to blood flow within the circulatory system, flagella provide the bacterium with a high degree of mobility and may serve to propel the pathogen during its search for erythrocytes [12].
Bacterial motility seems to be required for internalization, since entry of nonmotile B. bacilliformis was not observed in the experiments of Mernaugh and Ihler [13].
Antiserum to flagella significantly reduced B. bacilliformis association with red cells as compared to controls [11], and there was poor adherence of nonmotile variants and flagellin-minus (bald) mutants [14, 15]. These data suggest that flagella may possess adhesive qualities and/or they increase bacteria-host cell interactions [11]. In view of the significant decrease in erythrocyte infection due to antiflagellin antibodies, this protein is a promising antigen for use in a potential subunit vaccine.
Brps. Bartonella species invade a variety of eukaryotic cells by employing trimeric autotransporter adhesion proteins (TAAs) [16]. The TAAs all use a type V secretion pathway and consist of a passenger domain and a beta domain used to deliver the passenger component out of the cell via the outer membrane. B. bacilliformis possesses three brp genes encoding potential Bartonella repeat proteins (Brps) [17]. B. bacilliformis Brp proteins share common domains and structural features with the TAAs. While the biological role of B. bacilliformis Brp proteins has not yet been investigated, they may be involved in similar biological processes as the TAA proteins of B. henselae and B. quintana, including autoagglutination, adhesion to host cells and extracellular matrix proteins (fibronectin and collagens), inhibition of phagocytosis, and induction of a proangiogenic response in host cells [10]. Immune-mediated inhibition of these hypothetical functions could possibly confer protection against the pathogen.
IalB. The invasion-associated locus B (IalB) protein (see Figure 2) was originally discovered by screening a B. bacilliformis genomic library for erythrocyte-invasive strains of host E. coli [18]. IalB has a significant effect on enhancing B. bacilliformis' erythrocyte invasion. It is possible that environmental changes during passage of the bacterium from sand fly to humans (e.g., changes in temperature, iron availability, and pH) have an effect on ialB gene expression [19]. In other research, IalB protein levels were correlated with changes in ialB mRNA and changes of temperature/pH of the growth media. The greatest amount of ialB was present in B. bacilliformis that had been acid-shocked (pH 5), grown at 20°C, or temperature downshifted to 20°C from 30°C. As with ialB mRNA levels, IalB protein amounts in B. bacilliformis decreased in response to basic pH (pH 8.0) or a temperature upshift to 37°C [20].
Figure 2.
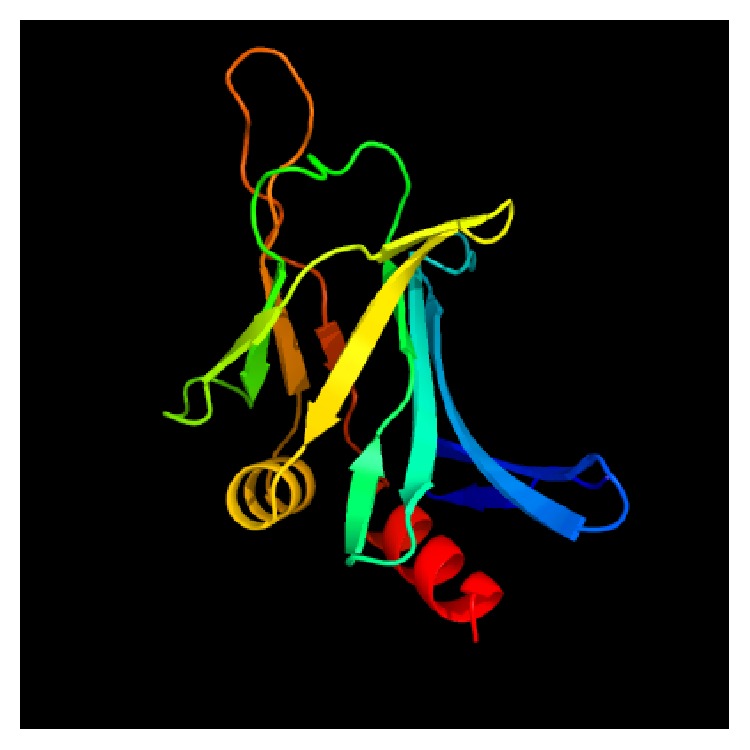
Crystal structure of the invasion-associated locus B protein; 144 residues (77% of Bartonella bacilliformis IalB sequence) modeled with 100.0% confidence by the single highest scoring template. Program Phyre2 at http://www.sbg.bio.ic.ac.uk/phyre2/html/page.cgi?id=index. Model based on B. henselae IalB crystal template c3dtdi_.
Presumably, B. bacilliformis undergoes a temperature shift from 37°C to 20°C when it is ingested by a sand fly, as the blood meal in the insect cools to ambient temperature. Concurrent with this temperature downshift, bacterial ialB expression would presumably be upregulated for erythrocyte invasion following transfer to another human host when the insect subsequently feeds. A change of the pH also occurs in the sand fly midgut (pH increases slightly to pH 7.4), where ialB expression would presumably be upregulated. When bacteria pass from the sand fly vector to a human host, the event would be signalled by a rapid upshift in temperature to 37°C and an increase in pH to 7.4. The ialB gene expression would be downregulated but not abrogated under these conditions. Undoubtedly, other bacterial factors are involved in erythrocyte adherence and invasion. IalB was localized to the inner membrane of B. bacilliformis, where it is unlikely to directly interact with erythrocytes. In contrast, the IalB homolog of B. henselae was localized to both inner and outer membrane fractions of the bacterium [21]. It is unknown if this protein is a transporter or signal transducer involved in human erythrocyte invasion, and its mechanism of action remains elusive [20].
FtsZ. The FtsZ protein of B. bacilliformis (FtsZBb) is a ~75 kDa protein and a structural homolog of B. henselae's cell division protein, FtsZ [20]. FtsZBb is strongly immunogenic in humans and was one of two major antigens (i.e., 75 kDa and 65 kDa proteins) identified in early work [9, 22]. In addition, the C terminus of FtsZBb contains a 256-amino acid region that is unusual and found in other related bacteria such as Rhizobium meliloti. As a result, the FtsZBb is almost twice as large as the majority of the FtsZ homologs, perhaps contributing to its antigenicity [17]. Interestingly, the ftsZ gene has been included in MLST schemes to study clonal and evolutionary relatedness of B. bacilliformis strains [23].
2. Miscellaneous Outer Membrane Proteins
Several B. bacilliformis outer membrane proteins have been identified by a variety of means. Minnick identified 14 proteins ranging from 11.2 to 75.3 kDa. Three of these proteins (31.5, 42, and 45 kDa) were prominent immunoprecipitants when they were exposed to rabbit hyperimmune serum [24]. Iwaki-Egawa and Ihler identified six B. bacilliformis proteins that mediate bacterial interactions with erythrocytes (100, 92, 84, 46, 37, and 12 kDa) [25].
A 43 kDa immunogenic lipoprotein of B. bacilliformis was identified through screening of a genomic DNA lambda library. The protein is a homolog to the LppB proteins of Haemophilus spp. and the NlpD protein of E. coli and was thus designated LppB [24]. The LppB lipoprotein is likely to use the same biosynthetic pathway as other bacterial lipoproteins. The protein's prominent immunogenicity was demonstrated using five convalescent sera from patients with Oroya fever or verruga peruana [26].
Buckles and McGinnis Hill demonstrated that B. bacilliformis was able to bind to several human erythrocyte proteins, including α and β subunits of spectrin, band 3 protein, glycophorin A, and glycophorin B [27]. Band 3 is a major transmembrane glycoprotein of the erythrocyte membrane, and it may be one of the possible erythrocyte receptors of B. bacilliformis [27]. In addition, band 3 has been suggested to be involved Plasmodium invasion of erythrocytes [28, 29].
3. Hemin-Binding Proteins (Hbp's)
The B. bacilliformis hemin-binding proteins (Hbp's) are homologs of the bacteriophage-associated, Pap31, protein of Bartonella henselae [30] and the five hemin-binding proteins of B. quintana [31, 32]. B. bacilliformis possesses three, tandem hbp-encoding genes in its chromosome. In addition to serving as a receptor for hemin and associating with the phage coat, data from work with B. henselae suggest that these proteins may also serve as adhesins for fibronectin, heparin, and host cells such as endothelial cells [33]. The Hbp (Pap31) proteins are highly expressed antigens in growing cultures of B. bacilliformis and are immunologically dominant, making them ideal antigens for use in enzyme-linked immunosorbent assays (ELISAs) and in immunoblots. In fact, use of this single protein antigen permits the use of ELISA, Western blot methods, or rapid lateral flow assay for the diagnosis of a B. bacilliformis infection [34]. To what extent B. bacilliformis Hbp's are immunoreactive in humans is unknown. However, the HbpE homolog of B. quintana is a commonly recognized antigen in sera from trench fever patients [35]. This observation suggests that one or more B. bacilliformis Hbp's may also be seroreactive in humans and thus useful subunit vaccine candidates.
4. Conclusions
B. bacilliformis infection represents an interesting model of human host specificity by a bacterium. While we now know more about the biology of the bacterium and the pathogenesis of Carrión's disease, we still lack adequate knowledge to select specific protein candidates to generate a subunit vaccine formulation. Studies regarding comparative genomics and proteomics are sorely lacking, especially considering the significant impact that bartonelosis outbreaks represent for the affected populations in South America. Carrión's disease is an emerging infectious disease that is not amenable to eradication with antibiotics or pesticides. Thus, the most effective intervention strategy to improve public health will be the development of a vaccine. Advances in B. bacilliformis genomics and proteomics hold the promise a rational vaccine design in the near future [36].
Conflict of Interests
The authors declare that there is no conflict of interests regarding the publication of this paper.
References
- 1.Maguiña C., Gotuzzo E. BARTONELLOSIS: new and old. Infectious Disease Clinics of North America. 2000;14(1):1–22. doi: 10.1016/s0891-5520(05)70215-4. [DOI] [PubMed] [Google Scholar]
- 2.Alexander B. A review of bartonellosis in Ecuador and Colombia. American Journal of Tropical Medicine and Hygiene. 1995;52(4):354–359. doi: 10.4269/ajtmh.1995.52.354. [DOI] [PubMed] [Google Scholar]
- 3.Henríquez C., Hinojosa J. C., Ventosilla P., et al. Report of an unusual case of persistent bacteremia by Bartonella bacilliformis in a splenectomized patient. American Journal of Tropical Medicine and Hygiene. 2004;71(1):53–55. [PubMed] [Google Scholar]
- 4.Huarcaya E., Maguiña C., Torres R., Rupay J., Fuentes L. Bartonelosis (Carrion's Disease) in the pediatric population of Peru: an overview and update. Brazilian Journal of Infectious Diseases. 2004;8(5):331–339. doi: 10.1590/s1413-86702004000500001. [DOI] [PubMed] [Google Scholar]
- 5.Gray G. C., Johnson A. A., Thornton S. A., et al. An epidemic of Oroya fever in the Peruvian Andes. American Journal of Tropical Medicine and Hygiene. 1990;42(3):215–221. doi: 10.4269/ajtmh.1990.42.215. [DOI] [PubMed] [Google Scholar]
- 6.Maguiña Vargas C., Ugarte-Gil C., Breña Chávez P., et al. Actualización de la enfermedad de Carrión. Revista Medica Herediana. 2008;19(1):36–41. [Google Scholar]
- 7.Maguiña C., Guerra H., Ventosilla P. Bartonellosis. Clinics in Dermatology. 2009;27(3):271–280. doi: 10.1016/j.clindermatol.2008.10.006. [DOI] [PubMed] [Google Scholar]
- 8.Kosek M., Lavarello R., Gilman R. H., et al. Natural history of infection with Bartonella bacilliformis in a nonendemic population. Journal of Infectious Diseases. 2000;182(3):865–872. doi: 10.1086/315797. [DOI] [PubMed] [Google Scholar]
- 9.Knobloch J. Analysis and preparation of Bartonella bacilliformis antigens. American Journal of Tropical Medicine and Hygiene. 1988;39(2):173–178. doi: 10.4269/ajtmh.1988.39.173. [DOI] [PubMed] [Google Scholar]
- 10.Minnick M. F., Anderson B. E., Lima A., Battisti J. M., Lawyer P. G., Birtles R. J. Oroya fever and verruga peruana: bartonelloses unique to South America. PLoS Neglected Tropical Diseases. 2014;8(7) doi: 10.1371/journal.pntd.0002919.e2919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Scherer D. C., Deburon-Connors I., Minnick M. F. Characterization of Bartonella bacilliformis flagella and effect of antiflagellin antibodies on invasion of human erythrocytes. Infection and Immunity. 1993;61(12):4962–4971. doi: 10.1128/iai.61.12.4962-4971.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Minnick M. F., Mitchell S. J., McAllister S. J. Cell entry and the pathogenesis of Bartonella infections. Trends in Microbiology. 1996;4(9):343–347. doi: 10.1016/0966-842x(96)10055-x. [DOI] [PubMed] [Google Scholar]
- 13.Mernaugh G., Ihler G. M. Deformation factor: an extracellular protein synthesized by the Bartonella bacilliformis that deforms erythrocyte membranes. Infection and Immunity. 1992;60(3):937–943. doi: 10.1128/iai.60.3.937-943.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Benson L. A., Kar S., McLaughlin G., Ihler G. M. Entry of Bartonella bacilliformis into erythrocytes. Infection and Immunity. 1986;54(2):347–353. doi: 10.1128/iai.54.2.347-353.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Battisti J. M., Minnick M. F. Development of a system for genetic manipulation of Bartonella bacilliformis . Applied and Environmental Microbiology. 1999;65(8):3441–3448. doi: 10.1128/aem.65.8.3441-3448.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Riess T., Andersson S. G. E., Lupas A., et al. Bartonella adhesin a mediates a proangiogenic host cell response. The Journal of Experimental Medicine. 2004;200(10):1267–1278. doi: 10.1084/jem.20040500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kaiser P. O., Riess T., O'Rourke F., Linke D., Kempf V. A. J. Bartonella spp.: throwing light on uncommon human infections. International Journal of Medical Microbiology. 2011;301(1):7–15. doi: 10.1016/j.ijmm.2010.06.004. [DOI] [PubMed] [Google Scholar]
- 18.Mitchell S. J., Minnick M. F. Characterization of a two-gene locus from Bartonella bacilliformis associated with the ability to invade human erythrocytes. Infection and Immunity. 1995;63(4):1552–1562. doi: 10.1128/iai.63.4.1552-1562.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Coleman S. A., Minnick M. F. Establishing a direct role for the Bartonella bacilliformis invasion-associated locus B (IalB) protein in human erythrocyte parasitism. Infection and Immunity. 2001;69(7):4373–4381. doi: 10.1128/iai.69.7.4373-4381.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Coleman S. A., Minnick M. F. Differential expression of the invasion-associated locus B (ialB) gene of Bartonella bacilliformis in response to environmental cues. Microbial Pathogenesis. 2003;34(4):179–186. doi: 10.1016/s0882-4010(03)00005-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chenoweth M. R., Greene C. E., Krause D. C., Gherardini F. C. Predominant outer membrane antigens of Balgonella henselae . Infection and Immunity. 2004;72(6):3097–3105. doi: 10.1128/iai.72.6.3097-3105.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Padmalayam I., Anderson B., Kron M., Kelly T., Baumstark B. The 75-kilodalton antigen of Bartonella bacilliformis is a structural homolog of the cell division protein FtsZ. Journal of Bacteriology. 1997;179(14):4545–4552. doi: 10.1128/jb.179.14.4545-4552.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chaloner G. L., Ventosilla P., Birtles R. J., Picardeau M. Multi-locus sequence analysis reveals profound genetic diversity among isolates of the human pathogen Bartonella bacilliformi . PLoS Neglected Tropical Diseases. 2011;5(7):p. e1248. doi: 10.1371/journal.pntd.0001248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Minnick M. F. Identification of outer membrane proteins of Bartonella bacilliformis . Infection and Immunity. 1994;62(6):2644–2648. doi: 10.1128/iai.62.6.2644-2648.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Iwaki-Egawa S., Ihler G. M. Comparison of the abilities of proteins from Bartonella bacilliformis and Bartonella henselae to deform red cell membranes and to bind to red cell ghost proteins. FEMS Microbiology Letters. 1997;157(1):207–217. doi: 10.1016/s0378-1097(97)00478-3. [DOI] [PubMed] [Google Scholar]
- 26.Padmalayam I., Kelly T., Baumstark B., Massung R. Molecular cloning, sequencing, expression, and characterization of an immunogenic 43-kilodalton lipoprotein of Bartonella bacilliformis that has homology to NlpD/LppB. Infection and Immunity. 2000;68(9):4972–4979. doi: 10.1128/iai.68.9.4972-4979.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Buckles E. L., McGinnis Hill E. Interaction of Bartonella bacilliformis with human erythrocyte membrane proteins. Microbial Pathogenesis. 2000;29(3):165–174. doi: 10.1006/mpat.2000.0381. [DOI] [PubMed] [Google Scholar]
- 28.Roggwiller E., Bétoulle M. E. M., Blisnick T., Breton C. B. A role for erythrocyte band 3 degradation by the parasite gp76 serine protease in the formation of the parasitophorous vacuole during invasion of erythrocytes by Plasmodium falciparum . Molecular and Biochemical Parasitology. 1996;82(1):13–24. doi: 10.1016/0166-6851(96)02714-4. [DOI] [PubMed] [Google Scholar]
- 29.Deng H. K., Le Rhun D., Le Naour E., Bonnet S., Vayssier-Taussat M. Identification of Bartonella Trw host-specific receptor on erythrocytes. PLoS ONE. 2012;7(7) doi: 10.1371/journal.pone.0041447.e41447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bowers T. J., Sweger D., Jue D., Anderson B. Isolation, sequencing and expression of the gene encoding a major protein from the backteriophage associated with Bartonella henselae . Gene. 1998;206(1):49–52. doi: 10.1016/s0378-1119(97)00580-5. [DOI] [PubMed] [Google Scholar]
- 31.Minnick M. F., Sappington K. N., Smitherman L. S., Andersson S. G. E., Karlberg O., Carroll J. A. Five-member gene family of Bartonella quintana . Infection and Immunity. 2003;71(2):814–821. doi: 10.1128/iai.71.2.814-821.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Carroll J. A., Coleman S. A., Smitherman L. S., Minnick M. F. Hemin-binding surface protein from Bartonella quintana . Infection and Immunity. 2000;68(12):6750–6757. doi: 10.1128/iai.68.12.6750-6757.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dabo S. M., Confer A. W., Anderson B. E., Gupta S. Bartonella henselae Pap31, an extracellular matrix adhesin, binds the fibronectin repeat III13 module. Infection and Immunity. 2006;74(5):2513–2521. doi: 10.1128/IAI.74.5.2513-2521.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Taye A., Chen H., Duncan K., et al. Production of recombinant protein Pap31 and its application for the diagnosis of Bartonella bacilliformis infection. Annals of the New York Academy of Sciences. 2005;1063:280–285. doi: 10.1196/annals.1355.045. [DOI] [PubMed] [Google Scholar]
- 35.Boonjakuakul J. K., Gerns H. L., Chen Y.-T., et al. Proteomic and immunoblot analyses of Bartonella quintana total membrane proteins identify antigens recognized by sera from infected patients. Infection and Immunity. 2007;75(5):2548–2561. doi: 10.1128/iai.01974-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Doolan D. L., Apte S. H., Proietti C. Genome-based vaccine design: the promise for malaria and other infectious diseases. International Journal for Parasitology. 2014;44(12):901–913. doi: 10.1016/j.ijpara.2014.07.010. [DOI] [PubMed] [Google Scholar]