Abstract
The lagging annotation of bacterial genomes and the inherent genetic complexity of many phenotypes is hindering the discovery of new drug targets and the development of new antimicrobials and vaccines. Here we present the method Tn-seq, with which it has become possible to quantitatively determine fitness for most genes in a microorganism and to screen for quantitative genetic interactions on a genome-wide scale and in a high-throughput fashion. Tn-seq can thus direct studies in the annotation of genes and untangle complex phenotypes. The method is based on the construction of a saturated transposon insertion library. After library selection, changes in frequency of each insertion mutant are determined by sequencing of the flanking regions en masse. These changes are used to calculate each mutant's fitness. The method was originally developed for the Gram-positive bacterium Streptococcus pneumoniae, a causative agent of pneumonia and meningitis, but has now been applied to several different microbial species.
Keywords: transposon sequencing, Tn-seq, Streptococcus pneumoniae, Vibrio cholerae, genome-wide fitness, genetic interactions, transposon mutagenesis, massively parallel sequencing
Introduction
Gram-positive bacterium Streptococcus pneumoniae is a common occupant of the nasopharynx and the causative agent of a variety of diseases ranging from otitis media to pneumonia, bacteremia, and meningitis. Each year, 1.5 million people succumb to infection with S. pneumoniae (Tuomanen et al., 2004). Due to increasing antibiotic resistance across many bacterial species and because the drug pipeline is running on empty, we are rapidly closing in on a pre-antibiotics era in terms of treatment options (Taubes, 2008). Vaccines are a promising alternative for the future to protect against bacterial infections. Against S. pneumoniae, there are currently two different polysaccharide-based vaccines available that protect ∼60% of vaccinees. However, they are expensive, and the polysaccharide approach is limiting, as S. pneumoniae serotypes against which the vaccines are directed can be replaced by other serotypes that are not covered by the vaccine.
For S. pneumoniae, as well as for many other pathogens, it is key to gain a better understanding of the link between genotype and phenotype not only in order to be able to understand processes such as colonization, persistence, and virulence, but also to be able to identify new drug or vaccine antigen targets. One of the problems, however, is the lagging annotation of bacterial genomes. For instance, more than 30% of the genes for S. pneumoniae are of unknown function, and it is reasonable to assume that most genes to which functions have been ascribed are only partially understood. In addition, microorganisms are complex systems, and a phenotype does not simply arise as the product of the individual genetic components; rather, a phenotype is a complex trait that results from these components interacting.
To aid in this quest, we have developed a widely applicable high-throughput tool for gene disruption, called Tn-seq, which has great potential for elucidating gene function, linking genotypes to phenotypes, and resolving complex pathways in microorganisms. Tn-seq is a robust and sensitive method that enables the determination of each gene's contribution to fitness in a specific environment, through massive parallel sequencing. The approach is based on the assembly of a saturated transposon insertion library. Upon growth of the library under a test condition of interest (in vitro or in vivo), insertion mutants with a lower fitness decrease in frequency in the population, while other mutants, depending on the effect of the transposon insertion, remain the same or increase in frequency. Changes in frequency are determined by sequencing the transposon flanking regions en masse on an Illumina HiSeq. Subsequently, fitness of every individual gene knockout in a bacterial genome can be calculated. Due to the quantitative nature of massive parallel sequencing, fitness of a gene knockout can be quantitatively determined and is a direct measure of the growth rate of the bacterial clone containing it. For instance, a mutant with a fitness of 0.5 translates to a two-fold higher doubling time than the wild-type (van Opijnen et al., 2009; van Opijnen and Camilli 2013).
By applying Tn-seq to the Gram-positive bacterium S. pneumoniae, a causative agent of pneumonia and meningitis, we have shown that besides determining a gene's contribution to fitness, it is also possible to identify genes essential for growth, conditionally important genes, regultoray relationships through genetic interactions (van Opijnen et al., 2009; van Opijnen and Camilli 2012), non coding RNAs (Mann et al., 2012) and genes important for in vivo colonization of the nasopharynx and for survival in the lung and the blood in mouse models of infection (van Opijnen and Camilli 2012; Mann et al., 2012). Importantly, Tn-seq is not limited to S. pneumoniae or Gram-positive bacteria, but can be applied to any microorganism for which a suitable means of generating a library of marked (e.g. transposon or suicide plasmid integration) mutations is available (van Opijnen and Camilli 2013). Here we also describe use of Tn-seq in the Gram-negative bacterium Vibrio cholerae, the causative agent of cholera. In addition, we describe a more general method of preparing transposon junctions for massively parallel sequencing that can be used to perform Tn-seq with any insertional element.
Basic Protocol 1 describes the purification of Mariner transposase. Basic Protocol 2 describes the construction of a mini-Mariner transposon library in S. pneumoniae. Basic Protocol 3 provides methods for making S. pneumoniae starter cultures, and then creating competent cells and transforming them with linear fragments of genomic DNA from the transposon library. Basic Protocol 4 describes creation of the S. pneumoniae transposon library starter cultures and their use in selection to determine each gene's fitness in a specific environment. It also addresses sample preparation for Illumina sequencing that is specific to the transposon used, which is used in the determination of fitness. Basic Protocol 5 describes the construction of a mTn5 transposon library in Vibrio cholerae. Basic Protocol 6 describes a more general method of Tn-seq sample preparation for Illumina sequencing that can be applied to any transposon or other insertional element.
Caution
S. pneumoniae and V. cholerae are Biosafety Level 2 (BSL-2) pathogens. Follow all appropriate guidelines and regulations for the use and handling of pathogenic microorganisms. See unit 1a.1 and other pertinent resources (APPENDIX 1B) for more information. S. pneumoniae is a human pathogen, capable of causing a variety of diseases ranging from otitis media to pneumonia, bacteremia, and meningitis, whereas V. cholerae is a human pathogen capable of causing severe secretory diarrhea following oral ingestion.
Note
S. pneumoniae is grown on blood agar plates or statically in rich liquid medium such as Todd Hewitt Broth supplemented with 0.5% yeast extract (THY) or Brain Heart Infusion (BHI). Liquid cultures and agar plates are incubated overnight in an incubator at 37°C with a 5% CO2 atmosphere or in a candle jar. V. cholerae is grown on Luria-Bertani (LB) agar plates or in LB broth with aeration, both at 37°C.
Basic Protocol 1
Purification of Mariner Transposase
The protocol describes the purification of a maltose binding protein (MBP)-Himar1 Mariner MarC9 transposase fusion protein essentially as described (Lampe et al., 1996). MarC9 is a hyperactive derivative of the transposase (Lampe et al, 1999). The fusion protein is over-expressed using the E. coli (pMalC9) strain. The fusion protein is purified by affinity chromatography using an amylose resin column. The purified transposase is then stored as single use aliquots in the freezer.
Materials
Complete, Mini, EDTA-free protease inhibitor cocktail (Roche, cat. no.1 836 170)
Column Buffer (CB): 1× Complete EDTA-free protease inhibitor cocktail, 20 mM Tris-HCl (pH 7.4), 200 mM NaCl, 1 mM EDTA.
Transposase Wash Buffer (TWB): 1× Complete EDTA-free protease inhibitor cocktail, 20 mM Tris-HCl (pH 7.4), 200 mM NaCl, 1 mM EDTA, 2 mM Dithiothreitol (DTT), 10% glycerol
Transposase Elution Buffer (TEB): 1× Complete EDTA-free protease inhibitor cocktail, 20 mM Tris-HCl (pH 7.4), 200 mM NaCl, 1 mM EDTA, 2 mM DTT, 10% Glycerol, 10 mM maltose
E. coli (pMalC9) (Lampe et al., 1999) (available from the authors on request; andrew.camilli@tufts.edu) isopropyl-β-D-thiogalactopyranoside (IPTG)
amylose resin (New England Biolabs)
Purification
Grow a 5 ml overnight culture of E. coli (pMalC9) in LB broth supplemented with 100 μg/ml ampicillin at 37°C with aeration.
Inoculate 80 ml of LB broth supplemented with 100 μg/ml ampicillin with 0.8 ml of the overnight culture. Grow with aeration at 37°C until the OD AT 600 NM of the cell culture is ca. 0.5.
Induce protein production by the addition of IPTG to a final concentration of 0.3 mM. Continue incubation at 37°C with aeration for 2 hr.
Harvest the cells by centrifugation at 8000 × g for 10 min at 4°C. Discard the supernatant and resuspend the cell pellet in 10 ml of cold CB. If necessary the cell suspension can be stored at -20°C at this point.
Break open the cells by any reliable method. For example, bead-beating, or a French Press. Failure to recover protein or poor yields often result from a failure to disrupt the cells. Disruption of cells can be assessed by examining the cells before and after breakage by phase-contrast microscopy.
Clarify the lysate by pelleting insoluble material at 15,000 - 20,000 × g for 10 minutes 4 °C.
Discard the pellets and transfer the supernatant to a 15 ml centrifuge tube on ice.
Thoroughly resuspend the amylose resin and aliqot 500 μl into a 1.5 ml tube. Wash the resin three times with cold TWB by pelleting the resin at 10,000 × g in a microfuge for 30 seconds, discarding the supernatant, and resuspending the pellet in 1 ml of cold TWB. Finally, resuspend the resin in 1 ml of cold TWB.
Transfer the resuspended amylose resin to the clarified cell lysate in the 15 ml tube on ice. Incubate the resin and lysate together at 4°C on a rocking platform for one hr to bind the MBP-transposase protein to the resin.
Pellet the resin in the 15 ml tube at 10,000 × g for 5 minutes at 4°C. Carefully remove the supernatant and discard it. Resuspend the pellet in 2 ml of cold TWB and transfer the resuspended resin into two 1.5 ml microcentrifuge tubes.
Wash the resin a total of four times as in step 8, using cold TWB. Remove the final supernatant from each tube.
Elute the bound protein by resuspending each resin pellet in 200 μl of cold TEB. Incubate on ice for 5 minutes with mixing of the tube by gentle vortexing every 1-2 minutes.
Pellet the resin at 10,000 × g for 2 minutes at 4°C. Remove the supernatants and transfer to a single clean 1.5 ml tube. Repeat the centrifugation to pellet any residual resin. Transfer the supernatant to a clean tube on ice. This is the purified transposase. Aliqout this solution into ca. 10 μl volumes in microfuge tubes or smaller PCR tubes and freeze at -70°C.
The typical concentration of MBP-transposase after using the procedure outlined above is approximately 100 μg/ml. To assess the purity of the final product, we recommend analysis on an 8% SDS-PAGE gel. If low yields are obtained, failure to lyse the cells is typically at fault. The molecular weight of the MBP-transposase fusion protein is 83 kDa. The purified transposase stored at -70°C or colder retains activity for at least one year.
Basic Protocol 2
Library Construction Via in Vitro Transposition
The protocol describes the construction of a transposon library in S. pneumoniae. Through in vitro transposition mediated by the transposase MarC9, the mini-transposon magellan6 (which is a derivative of the Himar1 Mariner transposon), encoding spectinomycin resistance, is introduced into bacterial genomic DNA linear fragments. Magellan6 is provided on a plasmid, pMagellan6, in the reaction, and transposes by a “cut-and-paste” mechanism into the genomic DNA. The transposition event leaves single-strand gaps at each transposon-chromosome junction, which are repaired with T4 DNA polymerase and E. coli DNA ligase.
Materials
S. pneumoniae genomic DNA (obtained via Qiagen Blood & Tissue Kit)
pMagellan6 DNA (available from the authors on request; andrew.camilli@tufts.edu)
2× Buffer A (see recipe)
3 M sodium acetate, pH 5.2 (APPENDIX 2A)
70% and 100% ethanol
MarC9: mariner transposase from (Basic Protocol 1) (add 0.5 μl volume of appropriate concentration; appropriate concentration should be experimentally assessed)
10× Buffer B (see recipe)
10× (10 mg/ml) bovine serum albumin (BSA; New England Biolabs)
2 mM dNTPs (APPENDIX 2A)
3 U/μl T4 DNA polymerase (New England Biolabs)
2.6 mM nicotine adenine dinucleotide (NAD)
10 U/μl E. coli DNA ligase (New England Biolabs)
30° and 75°C heating blocks
12°C and 16°C cooling blocks
Additional reagents and equipment for measuring DNA concentration (Gallagher and Desjardins, 2006)
Carry out transposition
Combine 1 μg S. pneumoniae genomic DNA, 1 μg pMagellan6 (plasmid), 10 μl 2× Buffer A, and sufficient distilled water for an end volume of 19.5 μl.
-
Add 0.5 μl of MarC9 at the appropriate concentration (empirically determined).
Make sure MarC9 is stored at −70°C or colder and that the transposase is divided into small aliquots, as thawing more than once lowers transposition efficiency.
Incubate at 30°C for 1 hr in a heating block.
Heat inactivate at 75°C for 10 min and transfer to ice for 5 min in a heating block.
Precipitate DNA by adding 1/10 volume of 3 M sodium acetate, pH 5.2, and 2 volumes of 100% ethanol at 4°C, incubating 20 min at 4°C, microcentrifuging at maximum speed, 4°C, to form a pellet, and washing the pellet with room temperature 70% ethanol. Air dry pellet and dissolve in 12.5 μl water.
-
Measure DNA concentration (Gallagher and Desjardins, 2006) using 1.5 μl of the DNA solution.
A yield in the range of 50 to 150 ng/μl should be obtained.
Repair transposon junctions
-
7. Mix the following:
2 μl 10× Buffer B
2 μl 10× BSA
1 μl 2 mM dNTPs
3 μl H2O
1 μl 3 U/μl T4 DNA polymerase
11 μl DNA from step 5.
8. Incubate at 12°C for 20 min, then inactivate the reaction at 75°C for 10 min.
-
9. Add 0.2 μl 2.6 mM NAD and 1 μl 10 U/μl E. coli DNA ligase and incubate at 16°C overnight.
If a very small-volume pipettor such as a Rainin P2 is not available, 1 μl of 0.52 mM NAD can be used; however, the final volume of the ligation reaction will be slightly altered.
Basic Protocol 3
Starter Cultures and Transformation of S. pneumoniae
This protocol describes how to make starter cultures of S. pneumoniae and how these are used to make competent cells. Competent cells are transformed with linear fragments of genomic DNA containing the magellan6 insertions to yield a transposon insertion library. Upon transformation, the magellan6 insertions are integrated into the genome by double cross-over homologous recombination. Transformants are selected on medium containing antibiotic to which the transposon encodes resistance. Because in vitro transposition and subsequent transformation of S. pneumoniae are both low-frequency events, the overwhelming majority of transformants harbor single transposon insertions. The resulting library can be used in any environment of interest (in vivo or in vitro) to gain insight into each gene's contribution to fitness (survival and/or growth) in that specific environment.
Though this protocol takes advantage of the natural competence property of S. pneumoniae, non-competent microbial species for which in vivo transposition methods are available may also be subjected to Tn-seq, as in the example of V. cholerae below. In addition, although other transposons (e.g., Tn5) or insertion elements (e.g., suicide vector) can be used with Tn-seq, the protocol described below is specific to magellan6. A more general method for purifying the transposon-chromosome junctions for sequencing, which can work with any transposon or insertional element, is provided in Basic Protocol 6, below.
Of significant importance in the protocol described here is the elimination of the step of plating transformed bacterial cells in soft agar layers, which has been the standard method for selecting S. pneumoniae transformants. Instead, transformed cells are outgrown for a short period of time to allow expression of the transposon drug marker, and then spread on the surface of antibiotic-containing blood agar plates. This change in procedure results in a dramatic increase in the dynamic range of the experiments, i.e., bacteria containing transposon insertions that have a fitness ≥0.05 are recovered (van Opijnen and Camilli, 2012). This is in stark contrast to the use of the original soft agar “layering” method where insertion mutants are recovered that have a fitness ≥0.5 (van Opijnen et al., 2009).
Materials
S. pneumoniae strain of choice (e.g., D39 or TIGR4)
Sheep's blood agar plates (with and without 200 μg/ml spectinomycin; see recipe)
Todd Hewitt yeast broth (THY; pH 7.3): 30 g TH base (Becton Dickinson, cat. no. 249240) per liter
Oxyrase (Oxyrase, cat. no. OB-0010)
THY (see above) containing 12% (v/v) glycerol
1 N HCl
20% (w/v) glycine (in distilled H2O)
1 N NaOH
8% (w/v) bovine serum albumin (BSA)
1 M CaCl2
350 ng/μl competence stimulating peptide (CSP) appropriate to the S. pneumoniae strain used (e.g., CSP1 for D39, CSP2 for TIGR4) (AnaSpec)
Transposon library DNA (Basic Protocol 1)
37°C, 5% CO2 incubator
Spectrophotometer
Large (16-mm diameter) and small (13-mm diameter) glass culture tubes
Centrifuge
Heat block at 37°C
Create starter cultures for transformation
Streak an S. pneumoniae strain on a sheep's blood agar plate (without spectinomycin) and incubate overnight at 37°C with 5% CO2.
Pick a single colony and grow in 250 μl THY and 5 μl Oxyrase in a 1.5 ml microcentrifuge tube for 3 hr.
Plate 100 μl on a blood agar plate (without spectinomycin) and incubate overnight at 37°C with 5% CO2.
On the next day, resuspend all colonies from plate's surface in 3 ml THY, transfer to a large culture tube, and make volume up to 10 ml THY containing 5 μl/ml Oxyrase. Incubate for 2 to 3 hr at 37°C with 5% CO2 until the OD at 600 nm is ∼0.3.
Centrifuge 8 min at 3000 × g, room temperature.
Discard the supernatant and resuspend the cell pellet in 5 ml THY containing 12% glycerol.
Aliquot into twenty 1.5-ml microcentrifuge tubes and store the starter cultures up to 9 months at −70°C or colder.
Carry out transformation
it is very important to have fast-growing starter cultures; the faster a starter culture grows, the higher the transformation efficiency becomes. For best results, a starter culture should reach the appropriate OD within 2 hr of seeding the growth medium.
8. Thaw a starter culture and transfer to 7.5 ml THY in a large glass culture tube (do not add antibiotics or Oxyrase).
9. Add 97.5 μl 1 N HCl and 18.8 μl 20% glycine. Allow to grow for 1.5 to 2 hr and measure the OD at 600 nm (should be between 0.03 and 0.1). Note that all OD measurements below are also at 600 nm.
10. Transfer a volume of the culture into a small culture tube and add prewarmed THY to final volume of 1 ml so that the OD is adjusted to ∼0.03 (e.g., if OD of the original culture is 0.06 then add 500 μl THY to 500 μl culture).
11. Place culture tube in heating block at 37°C.
-
12. Add the following in order, briefly and gently vortexing after each addition:
10 μl 1 N NaOH (the volume depends on the culture volume used at step 10, e.g. if 500 μl culture is used, 10 μl NaOH is added, the amount in μl to use is thus ‘culture volume’/50).
25 μl 8% BSA
1 μl 1 M CaCl.
Note: Make sure to add these components separately. They will precipitate out if mixed beforehand.
13. Add 1.5 μl CSP, immediately start the timer, and place the culture tube back into 37°C incubator with 5% CO2.
-
14. Precisely 14 min after adding CSP2 or 15 min after adding CSP1, add 22 μl DNA from Basic Protocol 2, step 9.
The type of CSP and incubation time depends on the S. pneumoniae strain used. For instance use CSP1 and 15 min of incubation for strain D39, and use CSP2 and 14 min of incubation for TIGR4 (Pozzi et al., 1996; Hsieh et al., 2006).
15. Place back in 5% CO2 incubator at 37°C for 45 min.
Prepare transposon library starter cultures
16. Transfer 1 ml of transformed culture into a microcentrifuge tube and centrifuge 8 min at 3000 × g, room temperature.
-
17. Discard 900 μl of supernatant, resuspend the bacterial cell pellet in the leftover 100 μl of THY, and plate on a blood agar plate containing 200 μg/ml spectinomycin.
A cell pellet is often not visible.
18. Incubate overnight at 37°C with 5% CO2.
19. Resuspend colonies in 3 ml THY, transfer to a culture tube, and bring to 10 ml with THY containing 200 μg/ml spectinomycin and 5 μl/ml Oxyrase.
20. Incubate for 30 min to 1 hr to an OD of 0.3.
21. Spin down cells by centrifuging 8 min at 3000 × g, room temperature, discard supernatant, and resuspend pellet in 5 ml THY containing 12% glycerol.
22. Aliquot into 20 microcentrifuge tubes and store the transposon library starter cultures at −80°C.
Basic Protocol 4
Magellan6 Transposon Library Selection and Illumina Sequencing Sample Preparation
This protocol describes how a transposon library starter culture can be used to determine each gene's fitness in a specific environment. In short, a starter culture is thawed and briefly cultured (30 min), a sample is taken for DNA isolation (t1), and a part of the culture is used to seed an environment of interest. This latter culture is incubated for several hr during which the library of strains is selected upon (e.g., for growth in a liquid medium), then another sample is taken for DNA isolation (t2). Next, the DNA from both time points is prepared for Illumina sequencing. Here we use THY as an example of a selective environment. In this example, we are thus determining which genes are important and which are dispensable for growth in THY. However, this environment can be changed to any environment of interest including infection in a mouse model.
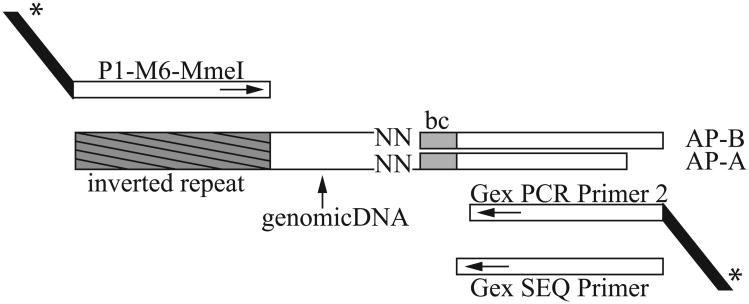
A key step in this protocol is digestion of the DNA with the type IIS restriction enzyme MmeI. (FIGURE 1) The inverted repeat sequences present at the left and right ends of magellan6 were engineered to contain an MmeI recognition sequence. MmeI cuts 20 bp downstream of its recognition site, leaving a random two base 3′ overhang. MmeI thus generates DNA fragments, of which a subset contain the magellan6 inverted repeat and ∼20 bp of flanking DNA. By subsequently ligating on an adapter with a random two base 5′ overhang, DNA fragments with the same spacing between adapter and magellan6 inverted repeat are created. These fragments can be amplified in a PCR reaction in an unbiased manner and then sequenced en masse.
Figure 1.
A detailed schema of how the magellan6-specific primers and adapter combine to result in a 120-bp DNA fragment that can be sequenced on an Illumina HiSeq. AP-B (AP-B_bc-ACAC) and AP-A (AP-A_bc-ACAC) are two oligos that make up the adapter. AP-A has a random two base overhang (NN), which is employed to ligate the adapter onto the genomic DNA (MmeI leaves a random two-base overhang). Note that to prevent self-ligation, AP-B is slightly longer than AP-A. In addition, the adapter contains a 4-base barcode (bc) which enables mixing of different samples in a single flow cell lane. With primers P1-M6-MmeI and Gex PCR Primer 2 a 120-bp fragment is generated that contains Illumina specific sequences (the black tails with asterisk) that are necessary for sequencing. The Gex Sequencing primer is used for sequencing once samples are loaded on the flow cell (also see Tables 1 and 2 for oligo details).
Materials
Transposon library starter culture (Basic Protocol 3)
Todd Hewitt yeast broth (THY; pH 7.3): 30 g TH base (Becton Dickinson, cat. no. 249240) per liter
Spectinomycin (Sigma-Aldrich)
Oxyrase (Oxyrase, cat. no. OB-0010)
listed later
Adapter oligonucleotides (Table 1)
Table 1. Adapter oligonucleotide sequences for processing magellan6 insertion libraries.
| ADAPTER | DNA SEQUENCE 5′ TO 3′ | PURIFICATION | PLATFORMa |
|---|---|---|---|
| AP-A_bc-ACACb | GTT CAG AGT TCT ACA GTC CGA CGA TCA CAC NN | PAGE | GAII |
| AP-B_bc-ACACc | 5Phos/GTG T GA TCG TCG GAC TGT AGA ACT CTG AAC CTG TC/3Phos | PAGE | GAII |
| ADBC-F-INDX1ad | ACA CGA CGC TCT TCC GAT CTG GAA CTC CT A CTG AC NN | PAGE | Hiseq |
| ADBC-F-INDX1b | ACA CGA CGC TCT TCC GAT CTT AGC ACA TG C TGA CT NN | PAGE | Hiseq |
| ADBC-F-INDX1c | ACA CGA CGC TCT TCC GAT CTC CTT GAG GA T GAC TG NN | PAGE | Hiseq |
| ADBC-F-INDX1d | ACA CGA CGC TCT TCC GAT CTA TCG TGT AC G ACT GA NN | PAGE | Hiseq |
| ADBC-R-INDX1a | 5PHOS/GTC AGT AGG AGT TCC AGA TCG GAA GAG CGT CGT GTA GGG A/3PHOS | PAGE | Hiseq |
| ADBC-R-INDX1b | 5PHOS/AGT CAG CAT GTG CTA AGA TCG GAA GAG CGT CGT GTA GGG A/3PHOS | PAGE | Hiseq |
| ADBC-R-INDX1c | 5PHOS/CAG TCA TCC TCA AGG AGA TCG GAA GAG CGT CGT GTA GGG A/3PHOS | PAGE | Hiseq |
| ADBC-R-INDX1d | 5PHOS/TCA GTC GTA CAC GAT AGA TCG GAA GAG CGT CGT GTA GGG A/3PHOS | PAGE | Hiseq |
The Illumina sequencing platform the adapters are optimized for.
The underlined sequence represents the barcode sequence (ACAC). The “NN” two-base random nucleotide overhang allows for ligation to the ends of MmeI-digested DNA fragments.
The underlined sequence is the barcode sequence (GTGT; the complement of ACAC). The phosphorylated 5′-end allows for ligation to the dephosphorylated genomic DNA fragments. Note that AP-B_bc-ACAC is five nucleotides longer on the 3′ end than AP-A_bc-ACAC, which prevents adapter-adapter ligation. In addition, the 3′ phosphorylation of AP-B_bc-ACAC prevents the formation of adapter-adapter products.
Due to several technological differences between the Illumina Genome Analyzer II and Hiseq platforms, we redesigned and optimized adapters for the Hiseq platform. The bold sequence is something we refer to as the ‘initiation’ sequence. Since the Hiseq platform is less accurate in the first cycles (it reaches its highest accuracy after the first 7-9 cycles) 9 nucleotides were incorporated in the adapter that are not used in the analysis. Additionally, the Hiseq is very sensitive to repetitive sequences in the first sequenced bases. Therefore these adapters are most optimal when used in ‘blocks of 4’, for instance adapters 1a, 1b, 1c and 1d, together have a perfect 1:1:1:1 distribution of each nucleotide in the first 9 nucleotides, which greatly improves cluster separation by the Hiseq platform. Directly behind this initiation sequence, a 6-nucleotide barcode is incorporated (underlined), which is used for multiplexing. The longer barcode is necessary for the HiSeq platform because of the decreased accuracy of sequencing relative to the Genome Analyzer II.
1 mM Tris Cl, pH 8.3 (APPENDIX 2A)
MmeI restriction enzyme (New England Biolabs, cat. no. R0637S)
32 mM S-adenosyl methionine (SAM; New England Biolabs)
CutSmart buffer (New England Biolabs)
Calf intestinal phosphatase (CIP; New England Biolabs, cat. no. M0290S)
24:23:1 (v/v/v) phenol:chloroform:isoamyl alcohol
100% and 70% ethanol
400 U/μl T4 DNA ligase (New England Biolabs) and T4 DNA ligase buffer
25 mM dNTP mix
PFU Ultra polymerase and PFU Ultra buffer (Stratagene)
10 μM PCR primers (Table 2)
Table 2. PCR and Illumina sequencing primers for processing magellan6 insertion libraries.
| Primers | Primer sequence 5′ to 3′ | Purification | Platform |
|---|---|---|---|
| P1_M6_MmeIa, b | CAA GCA GAA GAC GGC ATA CGA AGA CCG GGG ACT TAT CAT CCA ACC TGT | Standard desalting | GAII, Hiseq |
| Gex PCR Primer 2b | AAT GAT ACG GCG ACC ACC GAC AGG TTC AGA GTT CTA CAG TCC GA | Standard desalting | GAII |
| Gex Sequencing Primerc | CGA CAG GTT CAG AGT TCT ACA GTC CGA CGA TC | Standard desalting | GAII |
| ADPT-Tnseq-PCRPrimer d | AAT GAT ACG GCG ACC ACC GAG ATC TAC ACT CTT TCC CTA CAC GAC GCT CTT CCG ATCT | Standard desalting | Hiseq |
| Genomic DNA sequencing primer e | ACA CTC TTT CCC TAC ACG ACG CTC TTC CGA TCT | Standard desalting | Hiseq |
The underlined sequence of primer P1-M6-MmeI is the complement of the Magellan inverted repeat.
The italicized regions of primers P1-M6-MmeI and Gex PCR Primer 2 are Illumina specific sequences necessary for annealing to the oligos present in the flow cell.
This primer is identical to the Illumina small RNA primer.
The primer to use in combination with P1_M6_MmeI when using the Hiseq adapters, and thus it replaces the Gex PCR Primer 2, which is used in combination with the GAII adapters.
This primer is identical to the Illumina genomic DNA sequencing primer.
SeaKem LE agarose
50-bp DNA ladder
QIAquick Gel Extraction Kit (Qiagen cat. no. 28704)
Large (16 mm diameter) glass culture tubes
Spectrophotometer
37°C, 5% CO2 incubator
Heat block
Bioanalyser (Agilent)
16°C cooling block
PCR thermal cycler
Additional reagents and equipment for isolation of genomic DNA from bacteria (DNeasy Blood & Tissue Kit [Qiagen]; also see Wilson, 1997), phenol/chloroform/isoamyl alcohol precipitation of DNA (Moore and Dowhan, 2002), and agarose gel electrophoresis (Voytas, 2000)
Select library
Thaw a transposon library starter culture and transfer to 4 ml THY (add spectinomycin to 200 μg/ml, and 20 μl Oxyrase). Allow to grow for 30 to 60 min and measure OD, which should be ∼ 0.1.
Seed 10 ml THY at an OD of 0.005 and incubate at 37°C with 5% CO2 (the selection). In addition, take a small sample to enumerate colony-forming units (CFU) in the culture at t1.
-
Spin down the remaining culture that was used to seed the selective culture 8 min at 3000 × g room temperature, and discard supernatant. Isolate genomic DNA using the DNeasy Blood & Tissue Kit (also see Wilson, 1997) immediately, or store cell pellet at −20°C for later DNA isolation.
Make sure the genomic DNA is clean, which increases the efficiency of MmeI restriction digestion (step 8). In addition, it is essential to perform the phenol-chloroform step of Basic Protocol 4 (step 10); omitting this step will result in failure of the following steps.
This represents the t1 DNA.
-
When the selective culture reaches an OD of approximately 0.6, take a small sample to enumerate CFU, spin down the rest of the culture 8 min at 3000 × g, room temperature, discard supernatant, and isolate genomic DNA using the DNeasy Blood & Tissue Kit (also see Wilson, 1997).
Make sure the genomic DNA is clean, which increases the efficiency of MmeI restriction digestion (step 8). In addition, it is essential to perform the phenol-chloroform step of Basic Protocol 4 (step 10); omitting this step will result in failure of the following steps.
This represents the t2 DNA.
Prepare sample for sequencing
-
5. Prepare the adapter by bringing both oligonucleotides (Table 1) to 0.2 mM with 1 mM Tris·Cl, pH 8.3. Mix equal volumes of each in a microcentrifuge tube and place in a heat block at 96°C for 2 min.
See Figure 1 and Tables 1 and 2 for details on how the adapter and primers combine and result in DNA molecules that can be sequenced on an Illumina HiSeq. Note that the two oligonucleotides that form the adapter contain a four-nucleotide barcode (index) sequence (see Table 2). By varying this sequence, multiple samples can be mixed into a single Illumina flow cell lane, which can be separated after sequencing based on the barcode. For instance, by varying the barcode for t1 and t2, these two samples can be mixed and sequenced in the same flow cell lane.
6. To anneal the two oligonucleotides, place the part of the heat block that holds the microcentrifuge tubes on the bench and let it cool to room temperature (∼10 to 20 min).
7. Store the annealed ds adapter at −20°C until use.
-
8. Digest genomic DNA from t1 and t2 with the following mixture at 37°C for 2.5 hr:
3 μg S. pneumoniae DNA
3 μl MmeI (6 units)
0.44 μl 32 mM SAM
20 μl Buffer 4
H2O to 200 μl.
9. Add 2 μl CIP and incubate at 37°C for 1 hr.
-
10. Extract the DNA with phenol:chloroform:isoamyl alcohol, precipitate with 95% ethanol, wash pellet with 70% ethanol, air dry on the bench for 10 minutes and resuspend in 25 μl distilled water.
Extraction and precipitation of DNA are detailed in Moore and Dowhan (2002). Note that extraction and precipitation of the DNA greatly improves the efficiency of the next two steps.
-
11. Ligate adapter by mixing:
25 μl DNA from step 10
1 μl annealed adapter from step 7
1 μl 400 U/μl T4 DNA ligase
3 μl T4 DNA ligase buffer
Incubate at 16°C overnight.
-
12. Prepare the following PCR mix to amplify the 120-bp target:
5 μl PFU Ultra buffer
0.5 μl 25 mM dNTPs
1 μl 10 μM primer P1_M6_MmeI (Table 2)
1 μl 10 μM Gex PCR Primer 2 (Table 2)
0.5 μl PFU Ultra
40.5 μl H2O
1.5 μl ligation mix from step 11.
-
13. Carry out the following PCR program:
1 cycle: 30 sec 95°C (initial denaturation) 18-22 cycles: 10 sec 95°C (denaturation) 25 sec 55°C (annealing) 5 sec 72°C (extension) 1 cycle: 10 min 72°C (final extension). The goal is to perform as few cycles as possible in order to remain in the linear range of the PCR.
14. Load and run all of the PCR reaction products on a 1.8% agarose gel (Voytas, 2000) together with a 50-bp DNA ladder.
15. Excise the 120-bp band under low UV light, avoiding primer and primer-dimer bands.
16. Purify the DNA from the excised gel slice (e.g., using a QIAquick Gel Extraction Kit) and elute in 50 μl distilled water.
-
17. Determine yield and assess quality of the DNA on a Bioanalyzer (Agilent).
The yield should be ∼3 ng/μl and the Bioanalyzer should show a distinct peak in the 120-bp size range.
-
18. Sequence the t1 and t2 samples on an Illumina HiSeq according to the manufacturer's protocol.
After sequencing, reads are mapped to the genome and the change in the number of reads per insertion over time (the change from t1 to t2) can be used to accurately determine fitness for each gene. A detailed description of this method and data analysis can be found elsewhere (van Opijnen et al., 2009).
Basic Protocol 5
In Vivo Transposition Library Construction
The protocol describes the construction of a mini-Tn5 (mTn5) transposon library in V. cholerae using conjugation (via bacterial mating) essentially as previously described (Merrell et al, 2002). The mTn5, which encodes resistance to kanamycin, is delivered via conjugal transfer of the suicide plasmid pUTmTn5Km2 from an E. coli donor strain (Sm10λpir) into V. cholerae. After transfer, the plasmid is incapable of replicating in V. cholerae, however the Tn5 transposase encoded on the plasmid can mediate transposition of the mTn5 (also on the plasmid)into the V. cholerae genome.
Materials
V. cholerae recipient strain, resistant to Streptomycin
E. coli donor strain Sm10λpir (pUTmTn5Km2) (Merrell et al, 2002) (available from the authors on request; andrew.camilli@tufts.edu)
LB broth
LB agar plates with no antibiotics or supplemented with 100 μg/mL Streptomycin and 130 μg/mL Kanamycin
80% glycerol
Replica plating cylinder and velvet squares
Mating
Grow donor and recipient strains to late-exponential phase (OD at 600 nm = 0.5-1) at 37°C with aeration in LB broth supplemented with Kanamycin or Streptomycin, respectively.
Pellet 2 mL of each culture at 10,000 × g, discard supernatant and wash cells once with LB to remove any traces of antibiotics. Resuspend cells in 0.5 mL LB broth, and mix together at a 1:1 ratio based on OD at 600 nm.
Spread an appropriate amount of mixture, determined empirically to generate 500-1000 transposition mutant colonies per plate (determined in step 4 below), onto 40 LB agar lacking antibiotics. Incubate plates at 37°C for 1 hr to allow bacterial mating.
Selection and pooling of transposition mutants
-
4. Replica plate each mating plate onto LB agar supplemented with Streptomycin and Kanamycin to select for transposition mutants. Incubate plates 16 h at 37°C.
Incubating longer than 16 h or plating too much of the mating mixture will increase background on the selection plates. If background is problematic, then replica plate the selection plates (after 16 h of growth) to new antibiotic containing plates and incubate these at 16 h at 37°C.
5. Pool transposition mutants by flooding each plate with 2 mL of LB broth and resuspending colonies with a glass plating rod. Collect the fluid from each plate and add to a 50 mL conical tube. After adding the pooled colonies from each plate, the entire pooled library mixture should be vortexed to fully resuspend the cells. Add glycerol to final concentration of 15% (v/v) and vortex again to mix. Dispense many 1 mL aliquots of library into microfuge tubes or cryovials and freeze at -70°C or colder for long-term storage.
A yield in the range of 20,000 to 40,000 transposon insertion mutants should be obtained. Because each mutant arose from an independent mating event, each is considered an independent transposition event. Note that insertions into essential genes or essential intergenic sequences will be absent from the final library. In addition, insertions that slow the growth of V. cholerae in LB broth and/or on LB agar will be under-represented in the final library.
One disadvantage of this method of generating a transposon insertion library is that approximately 20% of the exconjugates will have the suicide plasmid randomly integrated into the recipient genome instead of a true transposon hop. These are identifiable because they contain the antibiotic resistance gene on the plasmid backbone (bla in this case) and thus are resistant to that antibiotic. All plasmid integration mutants will yield identical transposon-plasmid junctional sequence upon sequencing, and thus can be ignored since this sequence will not map to the bacterial genome and thus is filtered out bioinformatically.
Basic Protocol 6
General method for Tn-seq sample preparation for Illumina Sequencing
This protocol describes how the transposon junctions from a V. cholerae mTn5 library are prepared for massively parallel sequencing on the Illumina HiSeq. Selection on the library and determination of gene fitness using the Tn-seq data are done essentially as described above for the S. pneumoniae magellan6 transposon library.
While the protocol above is specific for magellan6, the method below, which is a variation of Homopolymer Tail Mediated Ligation-PCR (HTML-PCR) (Klein et al., 2012; Lazinski et al., 2013), can be used with essentially any transposon or insertional element. We describe use of this method for the V. cholerae mTn5 library described above.
Materials
Transposon library (Basic Protocol 5)
Terminal deoxnucleotidyl Transferase (Promega)
Terminal deoxnucleotidyl Transferase buffer (Promega)
9.5 mM dCTP
0.5 mM ddCTP
25 mM dNTP mix
Easy A Cloning Enzyme /buffer (Agilent)
30 μM PCR primers (Table 3)
Table 3. PCR and Illumina sequencing primers for processing mTn5 insertion libraries.
| Primers | Primer sequence 5′ to 3′ | Purification |
|---|---|---|
| olj491a | ACC TGC AGG CAT GCA AGC TTC GGC C | Standard desalting |
|
| ||
| olj376b,c | GTG ACT GGA GTT CAG ACG TGT GCT CTT CCG ATC TGG GGG GGG GGG GGG GG | Standard desalting |
| olj492a,c | AAT GAT ACG GCG ACC ACC GAG ATC TAC ACT CTT TCG CGG CCG CAC TTG TGT ATA AGA GT | HPLC |
| Barcode Primerc,d | CAA GCA GAA GAC GGC ATA CGA GAT NNN NNN GTG ACT GGA GTT CAG ACG TGT GCT CTT CCG ATC T | HPLC |
|
| ||
| Custom Sequencing Primer HK89e | ACA CTC TTT CGC GGC CGC ACT TGT GTA TAA GAG TCA G | Standard desalting |
Primers olj491 and olj492 are nested primers specific to one end of mTn5; the underlined sequence is complementary to mTn5.
The underlined sequence of primer olj376 is complementary to the poly-dC tail.
The italicized regions of primer olj492 and the barcode primer are Illumina specific sequences necessary for annealing to the oligos present in the flow cell.
The NNNNNN represents the reverse complement of the barcode and is varied with each sample.
Specific to the mTn5 inverted repeat end sequence.
GelGreen DNA stain (Biotium)
Branson high intensity cuphorn sonifier (Branson)
Performa DTR Gel Filtration Cartridge (Performa)
PCR thermal cycler
Additional reagents and equipment for isolation of genomic DNA from bacteria (DNeasy Blood & Tissue Kit [Qiagen]; also see Wilson, 1997), DNA purification (QIAquick PCR Purification kit [Qiagen] and QIAquick PCR Purification kit [Qiagen]), and agarose gel electrophoresis (Voytas, 2000)
Prepare sample for sequencing
1. Dissolve all oligonucleotides (Table 3) to 30 μM with pure water or 1 mM Tris·Cl, pH 8.3. Store the oligonucleotides at −20°C until use.
2. Purify genomic DNA from transposon libraries using DNeasy Blood & Tissue kit.
3. Partially digest genomic DNA enzymatically with DNAse I (New England Biolabs)or Fragmentase (New England Biolabs), or mechanically shear via sonication or nebulization. We use sonication as follows: Sonicate between 1 and 10 μg of genomic DNA in a 50 μl volume in 1.5 ml microcentrifuge tubes in a pre-chilled Branson high intensity cuphorn sonifier for 2 minutes at 50% intensity with a 10 seconds ON/5 seconds OFF duty cycle. Pellet contents. Repeat sonication for 1 minute. Run 5 μl on a 2% agarose gel with GelGreen stain to confirm that the DNA has been sheared to between 100 and 600 bp. If fragments are still too large, then do more sonication.
3. Remove short DNA fragments below 100 bp by purifying the sheared DNA using the Qiagen QIAquick PCR Purification kit.
- 4. Prepare the following mix in order to add poly-dC tails to the 3′ ends:
- Make a small volume (e.g. 20 μl) of 9.5 mM dCTP, 0.5 mM ddCTP.
- Add 1 ul of the dCTP/ddCTP mix to 4 μl 5× TdT Promega reaction buffer
- Add 0.5 μl of TdT enzyme (Promega) and mix the reaction by pipeting three times, and pellet contents by a brief centrifugation
- Let reaction proceed at 37°C for 1 hr, then heat inactivate at 75°C for 20 minutes.
-
5. Remove small molecules (eg. buffer components) by passing the reaction mix through a Performa DTR Gel Filtration Cartridge as follows.
Pre-spin column for 3 min at 850 RCF in table top microfuge to remove column water
move column to new collection tube
pipet reaction mix directly onto middle of top of column
centrifuge for 3 min at 850 RCF
Dispose of column and save flow-through
-
6. Prepare the following mix to do the first of a nested PCR to amplify the transposon junctions:
34.2 μl pure water
5 μl DNA from step 5 above
3 μl30 μM primer olj376 (Table 3)
1 μl 30 μM primer olj491 (Table 3)
0.8 μl 25 mM dNTPs
5 μl 10× Easy A buffer
1 μl Easy A Cloning Enzyme
- 7. Carry out the following PCR program:
1 cycle: 1 min 95°C (initial denaturation) 15 cycles: 30 sec 95°C (denaturation) 30 sec 52°C (annealing) 2 min 72°C (extension) 1 cycle: 2 min 72°C (final extension) -
6. Prepare the following mix to do the second of the nested PCR in order to further amplify the transposon junctions and also add a barcode sequence:
40.7 μl pure water
0.5 μl of first PCR reaction from step above
1.0 μl 30 μM primer olj492 (Table 3)
1.0 μl 30 μM barcode primer (Table 3)
0.8 μl 25 mM dNTPs
5.0 μl 10× Easy A buffer
1.0 μl Easy A Cloning Enzyme
- 7. Carry out the following PCR program:
1 cycle: 1 min 95°C (initial denaturation) 25 cycles: 30 sec 95°C (denaturation) 30 sec 58°C (annealing) 2 min 72°C (extension) 1 cycle: 2 min 72°C (final extension) -
8. Purify the PCR products using the QIAquick PCR Purification kit and elute with 50 μl. Run 5 μl of each purified sample on a 2% agarose gel with GelGreen stain.
The second PCR products should run as a smear of DNA fragments ranging in size from 140 bp to greater than 700 bp with the peak of intensity centered between 250 bp and 400 bp.
-
9. Determine yield and assess quality of the DNA on a Bioanalyzer (Agilent).
The sample concentration should be ca. 50 to 250 ng/μl
-
10. Multiplex equivalent amounts and sequence on the Illumina HiSeq using the custom sequencing primer HK89. The standard Illumina Multiplexing Index Sequencing Primer should be used to read the barcode.
After sequencing, reads are demultiplexed and then mapped to the genome and gene fitness is determined as described above.
Reagents and Solutions
Use deionized, distilled water in all recipes and protocol steps. For common stock solutions, see APPENDIX 2A; for suppliers, see SUPPLIERS APPENDIX.
Buffer A, 2×
5 μl 80% (v/v) glycerol
4 μl 20 mM DTT
4 μl 10 mg/ml BSA (New England Biolabs)
8 μl 5× salt (see recipe)
Prepare fresh just before use
Buffer B, 10×
500 mMTris·Cl, pH 7.8 (APPENDIX 2A)
100 mM MgCl2
10 mM DTT
Store up to 1 year at −20°C
Salt, 5×
100 μl 125 mM HEPES, pH 7.9
11.5 μl 5 M NaCl
5.75 μl 1 M MgCl2
Prepare fresh just before use
Sheep's blood agar plates
42.5 g blood agar base number 2 (Sigma, cat. no. B1676; prepare according to manufacturer's instructions)
1 liter H2O
Autoclave with submerged magnetic stir bar
Cool to ∼50°C
With stirring on magnetic hot-plate stirrer, add 50 ml pre-warmed (37°C) defibrinated sheep's blood (Northeast Laboratory Services, http://www.nelabservices.com/)
Add 200 μg/ml spectinomycin (Sigma-Aldrich) if required in protocol step
Pour into sterile 100 × 15–mm plates
Store up to 2 months at 4°C in the dark
Commentary
Background Information
Understanding how a genotype leads to a specific phenotype is key to understanding how an organism functions and survives. For some time now, it has been clear that the link between genotype and phenotype is not a linear one—rather, interactions among and between genetic components and the environment create a complex pathway leading to a specific phenotype. The result is intriguing in that through these interactions a robust organism emerges that is able to withstand many kinds of genetic and environmental disturbances. Such interactions are described in terms of epistasis, which is a phenomenon whereby the effects of a given gene on a biological trait are masked or enhanced by one or more other genes (Kitano, 2004; Moore, 2005). In the last decade, such dependencies between genes have been extensively explored in Saccharomyces cerevisiae by identifying synthetic genetic interactions through systematically knocking out all possible double combinations of genes. This method has identified specific networks of genes responsible for important pathways such as DNA integrity (Ooi et al., 2003; Pan et al., 2006), mRNA export (Hieronymus et al., 2004), drug resistance (Begley et al., 2002; Giaever et al., 2004; Dudley et al., 2005; Parsons et al., 2006), and phosphorylation (Fiedler et al., 2009).
Three approaches have been developed to identify genetic interactions in S. cerevisiae: synthetic genetic array (SGA) technology (Tong et al., 2001), diploid-based synthetic lethality analysis on microarrays (dSLAM; Pan et al., 2004), and epistatic miniarray profile (E-MAP). Where SGA and dSLAM are able to map synthetic sick/lethal (SSL) relationships, E-MAP is more sensitive and able to measure a larger range of epistatic interactions, including positive interactions (Schuldiner et al., 2005; Collins et al., 2006, 2007; Roguev et al. 2008; Wilmes et al., 2008). These methods all make use of robotic automation to construct double mutants by mating. A query strain, with a drug-resistance marker replacing a gene of interest, is crossed to an arrayed collection of single-gene deletion strains of opposite mating type marked with a different selectable marker. After meiosis, sporulation, and selection for double mutants, genetic interactions can be identified by measuring colony size. Meiotic assortment is not applicable to bacteria. However, two analogous methods have been developed for E. coli: GIANT coli and eSGA, both of which make use of conjugation to drive genetic exchange of a marked query gene mutation from an E. coli Hfr donor strain into a genome-wide, high-density-arrayed collection of E. coli F- recipient single-gene mutants. Robotic pinning, dual-marker selection, and colony imaging are used to identify colony growth defects in the resulting double mutants. However, the applicability of these methods to a wide range of microorganisms is greatly limited, primarily because there are arrayed collections of single-gene knockouts for only a handful of species. A more generally applicable method to identify genetic interactions in bacteria is TraSH (Joshi et al., 2006), as well as related genomic footprinting methods (Girgis et al., 2007). TraSH is based on the construction of a transposon library, the generation of probes from outward reading T7 promoters located at each end of the transposon, and subsequent detection by microarray. However, the accuracy of this and related methods is somewhat limited, and the use of a microarray restricts its use to the strain for which the microarray is developed.
An overview of Tn-seq
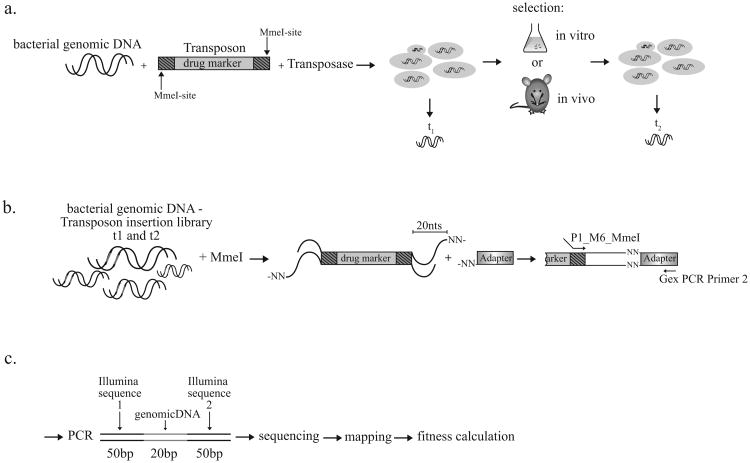
With the development of Tn-seq it has become possible to quantitatively determine fitness for most genes in a microorganism and to screen for quantitative genetic interactions on a genome-wide scale and in a high-throughput fashion (van Opijnen and Camilli 2013). In S. pneumoniae, for which the method was originally developed, Tn-seq works as follows (Fig. 2A-C). A gene disruption library is constructed by first transposing the mini-transposon magellan6 into bacterial genomic DNA in vitro using the transposase MarC9 and subsequently transforming bacteria with the transposed DNA. The result is a pool of strains where each bacterium contains a single transposon insertion in its genome. DNA is isolated from a portion of the bacterial pool (t1); another portion is used to seed a culture on which selection is performed (in vitro or in vivo); after selection, bacteria are recovered and DNA is isolated again (t2). To accomplish sequencing of only the regions flanking the transposon, a single nucleotide in the transposon's inverted repeats was mutated, thereby introducing an MmeI restriction site. DNA from both time points is digested with MmeI, which cuts 20 bp downstream of its recognition site leaving a two-base overhang to which an adapter is ligated. A PCR is performed with one primer having complementarity to the adapter and the other complementary to the magellan6 inverted repeat sequence. The resulting PCR product is 120 bp long, with 16 bp of bacterial specific DNA flanked by Illumina specific sequences, which enable sequencing. After sequencing, different samples are split based on barcode sequence; reads are mapped to the genome and counted for each insertion, thus allowing fitness to be calculated (van Opijnen and Camilli 2013).
Figure 2.
Flowchart depicting Tn-seq using the mini-transposon magellan6, from library construction to massively parallel sequencing of transposon-chromosome junctions. (A) A gene-disruption library is constructed by transposing magellan6 into bacterial genomic DNA in vitro and subsequently transforming a bacterial population with the transposed DNA. The result is a pool of strains where each bacterium contains a single transposon randomly inserted in its genome. DNA is isolated from a portion of the bacterial pool (t1); another portion is used to seed a culture on which selection is performed (in vitro or in vivo); after selection, bacteria are recovered and DNA is isolated again (t2). (B) To accomplish sequencing of only the regions flanking magellan6 insertions, DNA from both time points is digested with MmeI, which binds a sequence in the magellan6 terminal inverted repeats but cuts 20 bp downstream, leaving a two-base overhang to which an adapter is ligated. A PCR is performed with one primer having complementarity to the adapter and the other complementary to the inverted repeat sequence. (C) The resulting PCR product is 120 bp long, with approximately 20 bp of bacterial specific DNA flanked by Illumina specific sequences needed for sequencing. After sequencing, different samples are split based on barcode sequence, and the bacteria-specific reads are mapped to the genome and counted for each insertion, thus allowing fitness to be calculated.
Because Mariner transposons work in many organisms including bacteria, the magellan6-based method of Tn-seq has applicability to a variety of bacterial species. For bacteria that are naturally competent for transformation like S. pneumoniae, the in vitro transposition method illustrated here can be used. However, many bacteria are not naturally competent, and instead require a method of in vivo transposition such as by conjugal transfer of a transposon delivery vector, like the mTn5 method illustrated here. V. cholerae is naturally competent, so we had a choice of using either in vitro or in vivo transposition.
One disadvantage of Mariner transposons is that they have insertion site preference for TA dinucleotides. This prevents the ability to use Mariner transposons for the interrogation of certain regions of the genome such as small genes or promoters, which might lack TA dinucleotides. However, a number of other transposons can be used in bacteria that do not have such insertion site specificity, including the Tn5 derivative illustrated here.
Obtaining a robust fitness value and sequencing multiple samples in a single flow cell lane
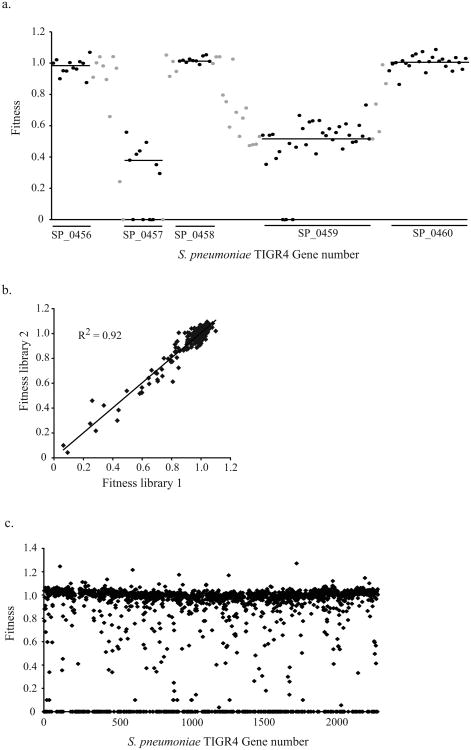
The application of massively parallel sequencing in Tn-seq makes it possible to determine the exact location of each transposon insertion in a complex transposon insertion strain library and simultaneously follow the quantitative change during selection of each of these insertion strains in the population over time. The quantitative change is then used to determine the fitness effect of each transposon insertion. However, what makes Tn-seq so robust is that a gene's fitness is based on the average effect of multiple independent transposon insertions into a gene. In Figure 3A, fitness effects of multiple independent transposon insertions from four independent libraries and their overall means are plotted for the region covering genes SP0456 to SP0460, illustrating how robust fitness values are obtained for individual genes.
Figure 3.
Tn-seq fitness details. (A) A gene's fitness is based on the average effect of multiple independent transposon insertions into a gene. On the x axis, the genomic region for S. pneumoniae TIGR4 is plotted covering genes SP0456 to SP0460. Black data points depict the fitness of a single transposon insertion within the coding region of a gene. A black line depicts the average fitness of a specific gene calculated from the individual insertions into that gene. For instance, the average fitness for genes SP0456, SP0458, and SP0460 is around 1 (same as wild type), while insertions into genes SP0457 and SP0459 are clearly disadvantageous and give average fitness values well below 1. The gray data points depict fitness from insertions into intergenic regions between genes. (B) Reproducibility between independently generated libraries, consisting of 10,000 transposon insertion each is high (R2 = 0.92). (C) Fitness for all genes across the S. pneumoniae genome.
In this light, it is important to consider the number of transposon insertions and samples that are sequenced in a single flow cell lane. When we first started to perform Tn-seq, we generally obtained 6 × 106 reads per flow cell lane and combined three libraries, containing 10,000 insertion strains each, in a single lane. This means that, on average, we obtained 200 reads per insertion, which gives good resolution to determine small changes in frequency of an insertion mutant. Now, however, due partly to experience and largely to improvements in the Illumina sequencing platform, we generally obtain 1.6 × 108 reads, which enables us to combine up to 24 independent libraries each containing 10,000 mutants. In other experiments, libraries of much high complexity, on the order of 100,000 mutants, can be utilized, which results in much higher insertion density and thus more detailed interrogation of the genome. Thus, the cost effectiveness of Tn-seq has increased by more than 10-fold in just a few years.
Library construction improvement
Since the original publication of Tn-seq (van Opijnen et al., 2009), we have made a significant improvement to the transformation protocol during construction of the transposon insertion library in S. pneumoniae. Originally, the transformation protocol for S. pneumoniae called for plating the transformants in a soft agar overlay on top of a blood agar plate, and then overlaying that with a layer containing antibiotic to select for transformants. Embedded colonies appear after overnight growth, each of which contains a transposon insertion. Transformants then have to be separated from the soft agar, which is accomplished by chopping up the agar, diluting it in THY, and then incubating for several hr. We hypothesized that due to out-competition during these early steps of library construction, mutants with fitness values less than 0.5 were not identified, and changes to the protocol that reduce the number of generations prior to selection at t1 would increase the dynamic range of the method. The hypothesis turned out to be correct and the solution extremely simple. We found that, after a short outgrowth period in liquid medium, the transformed cells can be directly spread onto a blood agar plate containing the selective antibiotic without loss of transformation efficiency. The next morning, antibiotic-resistant transformants can simply be washed off of the surface of these plates (see steps 16 to 19 in Basic Protocol 3).
Genetic interactions, different environments, strains and species
In addition to its utility in determining the fitness contribution of genes, Tn-seq is well suited for subsequently indentifying genome-wide genetic interactions. To identify interactions between a gene of interest (query gene) and the rest of the genome, the query gene is deleted, and subsequently a saturated transposon library is introduced in this query gene deletion background. The fitness of each double mutant is determined by Tn-seq, and compared to the expected fitness value, which is the product of the individual fitness values previously determined using Tn-seq in the wild-type background. A double mutant fitness value that deviates from the expected value reveals a genetic interaction (Avery and Wasserman, 1992; Drees et al., 2005; Collins et al., 2007; Fiedler et al., 2009; van Opijnen et al., 2009).
When performed in a systematic manner, it is possible to untangle complex phenotypes, regulatory relationships, and assign genes of unknown function to molecular pathways through interaction-profile association with known genes (Tong et al., 2001; Tong, 2004; St. Onge et al., 2007; Carter et al., 2009; van Opijnen et al., 2009; van Opijnen and Camilli 2012). Moreover, Tn-seq can be used to assign a role to intergenic regions (van Opijnen and Camilli 2013), to discover noncoding RNAs (Mann et al., 2012), and it can be easily applied to different environments (e.g., different growth media, chemical or physical stresses, or infection models), as well as to different strains of a particular species. The Himar1 Mariner transposon, illustrated here, can be used in many other microorganisms including E. coli (Lampe et al., 1999), Staphylococcus aureus (Bae et al., 2008), Haemophilus influenzae (Akerley et al., 1998), and Mycobacterium tuberculosis (Sassetti et al., 2003), Bacteroidetes thetaiotaomicron (Goodman et al 2009). However, other transposons, such as the mTn5 illustrated here, or insertional elements such as suicide plasmids, can be used in a wide variety of microorganisms. We illustrated two methods of preparing the Tn-chromosomal junctions for sequencing, one specific for the MmeI mutant form of the Mariner transposon, magellan6, and the more general HTML-PCR based method that can be applied to any transposon or insertional element. We also like to point the reader to three other similar methods that were developed simultaneously with Tn-seq: INseq for Bacteroidetes thetaiotaomicron (Goodman et al., 2009), HITS for Haemophilus influenza (Gawronksi et al., 2009) and TraDISH for Salmonella enterica serovar Typhi (Langridge et al., 2009).
Critical Parameters and Troubleshooting
The most important point to consider when performing Tn-seq in any microorganism is whether or not the selection condition will impose a ‘bottleneck’ that will result in the stochastic loss of insertion strains regardless of their fitness. If the bottleneck is severe, in our experience, greater than a 15% loss of library complexity, then there will be commensurate noise in the data that will greatly limit the ability to identify short genes, which will have few insertions to begin with, or genes having subtle or intermediate fitness alterations: Instead, only long genes or genes with very severe fitness alterations will be identified. If a severe bottleneck is present, then steps to eliminate or reduce the bottleneck should be taken. Alternatively, libraries of lower complexity may be used, although each library will sample only a subset of the genome. If a bottleneck cannot be avoided, we present two methods of data analysis. In the first, the size of the bottleneck is estimated by determining the proportion of insertion mutants that disappeared from a set of neutral insertions (a transposon into a neutral site, for example, in degenerate genes, should not have an impact on fitness). Subsequently, to correct for the bottleneck, for each gene the same proportion of insertions is removed from the set of insertions that were lost. In the second, all insertions with a gene are computationally pooled, and a simple ratio of output reads divided by input reads is used to calculate fitness for the gene.
An important point to consider when performing Tn-seq in S. pneumoniae is whether the presence of a capsule is necessary for the experiments to be successful. If the capsule is of no interest, an acapsular strain is preferred, for the simple reason that transformation efficiency is at least an order of magnitude more efficient, and therefore large transposon libraries are more easily obtained. However, by scaling up the number of transformations, we have also been successful in making large transposon libraries in capsular strains.
In Basic Protocol 3, step 8, it is very important to have fast-growing starter cultures; the faster a starter culture grows, the higher the transformation efficiency becomes. For best results, a starter culture should reach the appropriate OD within 2 hr of seeding the growth medium.
In Basic Protocol 4, make sure the genomic DNA is clean, which increases the efficiency of MmeI restriction digestion (step 8). In addition, it is essential to perform the phenol-chloroform step of Basic Protocol 4 (step 10); omitting this step will result in failure of the following steps.
In Basic Protocol 5, we illustrate an in vivo transposition method based on conjugation. However, another commonly used method of in vivo transposition not illustrated here is to induce transposition from a replicating plasmid within the organism of study.
In Basic Protocol 6, the two nested PCR primers olj491 and olj492, and custom sequencing primer HK89, are specific to the mTn5 transposon used. Therefore, when using this method on libraries generated with other insertional elements, these primers must be redesigned accordingly.
Anticipated Results
Robust fitness effects for individual genes are obtained because they are measured by averaging across multiple independent transposon insertions in each gene (Fig. 3A). Due to improvements made to the transformation protocol during construction of the transposon insertion library provided in this unit, the dynamic range of Tn-seq in S. pneumoniae is significantly increased. Figure 3B shows a Pearson correlation of two independent libraries (10,000 insertion strains each) that were constructed with the improved transformation protocol, which illustrates that reproducibility is very high (R2 = 0.92) and mutants with fitness <0.5 are observed. In addition, Figure 3C shows the distribution of fitness for all genes in the S. pneumoniae TIGR4 genome based on four independent libraries (containing 10,000 insertion mutants each), which also confirms that with the improved transformation protocol mutants with fitness <0.5 are identified.
Time Considerations
The complete procedure for performing Tn-seq in S. pneumoniae, excluding purifying the Mariner transposase and massively parallel sequencing, can be performed in 8 days: Basic Protocol 2 (transposon library construction) takes 1 day. Basic Protocol 3 takes 4 days total, including incubation times: making and freezing the starter cultures for the transformation, (2 days), carrying out the transformation (1 day), and preparing and freezing the transposon library starter cultures (1 day). Basic Protocol 4 takes 3 days: selecting the library (1 day) and preparing the library for sequencing (2 days). The complete procedure for performing Tn-seq in V. cholerae, excluding sequencing, can be performed in 5 days: Basic Protocol 5 (transposon library construction and selection) takes 3 days. Basic Protocol 6 (preparing the library for sequencing) takes 1-2 days. The sequencing can take anywhere between 2 to 4 days, depending on the Illumina massively parallel sequencing platform used.
Supplementary Material
Acknowledgments
This work was supported by U.S. National Institutes of Health grant AI055058 and the Howard Hughes Medical Institute. T.v.O. was supported by a postdoctoral fellowship from the Netherlands Organization for Scientific Research (Rubicon-NWO) and the Charles H. Hood Foundation. A.C. is an investigator of the Howard Hughes Medical Institute.
Literature Cited
- Akerley BJ, Rubin EJ, Camilli A, Lampe DJ, Robertson HM, Mekalanos JJ. Systematic identification of essential genes by in vitro Mariner mutagenesis. Proc Natl Acad Sci USA. 1998;95:8927–8932. doi: 10.1073/pnas.95.15.8927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avery L, Wasserman S. Ordering gene function: The interpretation of epistasis in regulatory hierarchies. Trends Genet. 1992;8:312–316. doi: 10.1016/0168-9525(92)90263-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bae T, Glass EM, Schneewind O, Missiakas D. Generating a collection of insertion mutations in the Staphylococcus aureus genome using bursa aurealis. Methods Molec Biol. 2008;416:103–116. doi: 10.1007/978-1-59745-321-9_7. [DOI] [PubMed] [Google Scholar]
- Begley TJ, Rosenbach AS, Ideker T, Samson LD. Damage recovery pathways in Saccharomyces cerevisiae revealed by genomic phenotyping and interactome mapping. Molec Cancer Res. 2002;1:103–112. [PubMed] [Google Scholar]
- Carter GW, Galas DJ, Galitski T. Maximal extraction of biological information from genetic interaction data. PLoS Comput Biol. 2009;5:e100347. doi: 10.1371/journal.pcbi.1000347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins S, Schuldiner M, Krogan N, Weissman J. A strategy for extracting and analyzing large-scale quantitative epistatic interaction data. Genome Biol. 2006;7:R63. doi: 10.1186/gb-2006-7-7-r63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins S, Miller K, Maas N, Roguev A, Fillingham J, Chu C, Schuldiner M, Gebbia M, Recht J, Shales M, Ding H, Xu H, Han J, Ingvarsdottir K, Cheng B, Andrews B, Boone C, Berger S, Hieter P, Zhang Z, Brown G, Ingles C, Emili A, Allis C, Toczyski D, Weissman J, Greenblatt J, Krogan N. Functional dissection of protein complexes involved in yeast chromosome biology using a genetic interaction map. Nature. 2007;446:806–810. doi: 10.1038/nature05649. [DOI] [PubMed] [Google Scholar]
- Drees BL, Thorsson V, Carter GW, Rives AW, Raymond MZ, Avila-Campillo I, Shannon P, Galitski T. Derivation of genetic interaction networks from quantitative phenotype data. Genome Biol. 2005;6:R38. doi: 10.1186/gb-2005-6-4-r38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dudley AM, Janse DM, Tanay A, Shamir R, Church GM. A global view of pleiotropy and phenotypically derived gene function in yeast. Mol Syst Biol. 2005;1:1–11. doi: 10.1038/msb4100004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiedler D, Braberg H, Mehta M, Chechik G, Cagney G, Mukherjee P, Silva AC, Shales M, Collins SR, von Wageningen S, Kemmeren P, Holstege FC, Weissman JS, Keogh M, Koller D, Shokat KM, Krogan NJ. Functional organization of the S. cerevisiae phosphorylation network. Cell. 2009;136:952–963. doi: 10.1016/j.cell.2008.12.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallagher SR, Desjardins PR. Quantitation of DNA and RNA by absorption and fluorescence spectroscopy. Curr Protoc Mol Biol. 2006;76:A.3D.1–A.3D.21. doi: 10.1002/0471142727.mba03ds76. [DOI] [PubMed] [Google Scholar]
- Gawronski JD, Wong SMS, Giannoukos G, Ward DV, Akerley BJ. Tracking insertion mutants within libraries by deep sequencing and a genome-wide screen for Haemophilus genes required in the lung. Proc Natl Acad Sci USA. 2009;106:16422–7. doi: 10.1073/pnas.0906627106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giaever G, Flaherty P, Kumm J, Proctor M, Nislow C, Jaramillo DF, Chu AM, Jordan MI, Arkin AP, Davis RW. Chemogenomic profiling: Identifying the functional interactions of small molecules in yeast. Proc Natl Acad Sci USA. 2004;101:793–798. doi: 10.1073/pnas.0307490100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girgis H, Liu Y, Ryu W, Tavazoie S. A comprehensive genetic characterization of bacterial motility. PLoS Genet. 2007;3 doi: 10.1371/journal.pgen.0030154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodman AL, et al. Identifying genetic determinants needed to establish a human gut symbiont in its habitat. Cell Host Microbe. 2009;6:279–89. doi: 10.1016/j.chom.2009.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hieronymus H, Yu M, Silver PS. Genome-wide mRNA surveillance is coupled to mRNA export. Genes Dev. 2004;18:2652–2662. doi: 10.1101/gad.1241204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsieh YC, Wang JT, Lee WS, Hsueh PR, Shao PL, Chang LY, Lu CY, Lee CY, Huang FY, Huang LM. Serotype competence and penicillin resistance in Streptococcus pneumoniaex. Emerging Infect Dis. 2006;12:1709–1714. doi: 10.3201/eid1211.060414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joshi SM, Pandey AK, Capite N, Fortune SM, Rubin EJ, Sassetti CM. Characterization of mycobacterial virulence genes through genetic interaction mapping. Proc Natl Acad Sci USA. 2006;103:11760–11765. doi: 10.1073/pnas.0603179103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamp HD, Patimalla-Dipali B, Lazinski DW, Wallace-Gadsden F, Camilli A. Gene fitness landscapes of Vibrio cholerae at important stages of its life cycle. PLoS Pathogens. 2013 doi: 10.1371/journal.ppat.1003800. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitano H. Biological robustness. Nat Rev Genet. 2004;5:826–837. doi: 10.1038/nrg1471. [DOI] [PubMed] [Google Scholar]
- Klein BA, Tenorio EL, Lazinski DW, Camilli A, Duncan MJ, Hu LT. Identification of essential genes of the periodontal pathogen Porphyromonas gingivalis. BMC Genomics. 2012;13:578. doi: 10.1186/1471-2164-13-578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langridge GC, et al. Simultaneous assay of every Salmonella Typhi gene using one million transposon mutants. Genome Research. 2009;19:2308–2316. doi: 10.1101/gr.097097.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lampe D, Churchill M, Robertson H. A purified Mariner transposase is sufficient to mediate transposition in vitro. EMBO J. 1996;15:5470–5479. [PMC free article] [PubMed] [Google Scholar]
- Lampe DJ, Akerley BJ, Rubin EJ, Mekalanos JJ, Robertson HM. Hyperactive transposase mutants of the Himar1 Mariner transposon. Proc Natl Acad Sci USA. 1999;96:11428–11433. doi: 10.1073/pnas.96.20.11428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazinski DW, Camilli A. Homopolymer tail-mediated ligation PCR: a streamlined and highly efficient method for DNA cloning and library construction. Biotechniques. 2013;54:25–34. doi: 10.2144/000113981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merrell DS, Hava DL, Camilli A. Identification of novel factors involved in colonization and acid tolerance of Vibrio cholerae. Mol Microbiol. 2002;43:1471–1491. doi: 10.1046/j.1365-2958.2002.02857.x. [DOI] [PubMed] [Google Scholar]
- Mann B, van Opijnen T, Wang J, Obert C, Wang YD, Carter R, McGoldrick DJ, Ridout G, Camilli A, Tuomanen EI, Rosch JW. Control of virulence by small RNAs in Streptococcus pneumoniae. Plos Pathogens. 2012 Jul;8(7):e1002788. doi: 10.1371/journal.ppat.1002788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore JP. A global view of epistasis. Nat Genet. 2005;37:13–14. doi: 10.1038/ng0105-13. [DOI] [PubMed] [Google Scholar]
- Moore D, Dowhan D. Purification and concentration of DNA from aqueous solutions. Curr Protoc Mol Biol. 2002;59:2.1.1–2.1.10. doi: 10.1002/0471142727.mb0201as59. [DOI] [PubMed] [Google Scholar]
- Ooi S, Shoemaker D, Boeke J. DNA helicase gene interaction network defined using synthetic lethality analyzed by microarray. Nat Genet. 2003;35:277–286. doi: 10.1038/ng1258. [DOI] [PubMed] [Google Scholar]
- Pan X, Yuan DS, Xiang D, Wang X, Sookhai-Mahadeo S, Bader JS, Hieter P, Spencer F, Boeke JD. A robust toolkit for functional profiling of the yeast genome. Mol Cell. 2004;16:487–496. doi: 10.1016/j.molcel.2004.09.035. [DOI] [PubMed] [Google Scholar]
- Pan X, Ye P, Yuan DS, Wang X, Bader JS, Boeke JD. A DNA integrity network in the yeast Saccharomyces cerevisiae. Cell. 2006;124:1069–1081. doi: 10.1016/j.cell.2005.12.036. [DOI] [PubMed] [Google Scholar]
- Parsons A, Lopez A, Givoni I, Williams D, Gray C, Porter J, Chua G, Sopko R, Brost R, Ho C. Exploring the mode-of-action of bioactive compounds by chemical-genetic profiling in yeast. Cell. 2006;126:611–625. doi: 10.1016/j.cell.2006.06.040. [DOI] [PubMed] [Google Scholar]
- Pozzi G, Masala L, Iannelli F, Manganelli R, Havarstein LS, Piccoli L, Simon D, Morrison DA. Competence for genetic transformation in encapsulated strains of Streptococcus pneumoniae: Two allelic variants of the peptide pheromone. J Bacteriol. 1996;178:6087–6090. doi: 10.1128/jb.178.20.6087-6090.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roguev A, Bandyopadhyay S, Zofall M, Zhang K, Fischer T, Collins S, Qu H, Shales M, Park HO, Hayles J, Hoe KL, Kim DU, Ideker T, Grewal SI, Weissman J, Krogan N. Conservation and rewiring of functional modules revealed by an epistasis map in fission yeast. Science. 2008;322:405–410. doi: 10.1126/science.1162609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sassetti C, Boyd DH, Rubin EJ. Genes required for mycobacterial growth defined by high density mutagenesis. Mol Microbiol. 2003;48:77–84. doi: 10.1046/j.1365-2958.2003.03425.x. [DOI] [PubMed] [Google Scholar]
- Schuldiner M, Colllins S, Thompson N, Denic V, Bhamidpati A, Punna T, Ihmels J, Andrews B, Boone C, Greenblatt J. Exploration of the function and organization of the yeast early secretory pathway through an epistatic miniarray profile. Cell. 2005;123:507–519. doi: 10.1016/j.cell.2005.08.031. [DOI] [PubMed] [Google Scholar]
- St Onge RP, Mani R, Oh J, Proctor M, Fung E, Davis R, Nislow C, Roth F, Giaever G. Systematic pathway analysis using high-resolution fitness profiling of combinatorial gene deletions. Nat Genet. 2007;39:199–206. doi: 10.1038/ng1948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taubes G. The bacteria fight back. Science. 2008;321:356–361. doi: 10.1126/science.321.5887.356. [DOI] [PubMed] [Google Scholar]
- Tong A. Global mapping of the yeast genetic interaction network. Science. 2004;303:808–813. doi: 10.1126/science.1091317. [DOI] [PubMed] [Google Scholar]
- Tong AH, Evangelista M, Parsons A, Xu H, Bader GD, Page N, Robinson M, Raghibizadeh S, Hogue CW, Bussey H, Andrews B, Tyers M, Boone C. Systematic genetic analysis with ordered arrays of yeast deletion mutants. Science. 2001;294:2364–2368. doi: 10.1126/science.1065810. [DOI] [PubMed] [Google Scholar]
- Tuomanen L, Mitchel TJ, Morrison DA, Spratt BG. The Pneumococcus. ASM Press; Washington, D.C: 2004. [Google Scholar]
- Van Opijnen T, Bodi KL, Camilli A. Tn-seq: high-throughput parallel sequencing for fitness and genetic interaction studies in microorganisms. Nat Methods. 2009;6:767–772. doi: 10.1038/nmeth.1377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Opijnen T, Camilli A. A fine scale phenotype-genotype virulence map of a bacterial pathogen. Genome Research. 2012;22:2541–2551. doi: 10.1101/gr.137430.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Opijnen T, Camilli A. Transposon insertion sequencing: a new tool for systems-level analysis of microorganisms. Nature Reviews Microbiology. 2013 Jul 11;(7):435–442. doi: 10.1038/nrmicro3033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voytas D. Agarose gel electrophoresis. Curr Protoc Mol Biol. 2000;51:2.5A.1–2.5A.9. doi: 10.1002/0471142727.mb0205as51. [DOI] [PubMed] [Google Scholar]
- Wilmes GM, Bergkessel M, Bandyopadhyay S, Shales M, Braberg H, Cagney G, Collins S, Whitworth GB, Kress TL, Weissman J, Ideker T, Guthrie C, Krogan N. A genetic interaction map of RNA-processing factors reveals links between Sem1/DssI-containing complexes and mRNA export and splicing. Mol Cell. 2008;32:735–746. doi: 10.1016/j.molcel.2008.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson K. Preparation of genomic DNA from bacteria. Curr Protoc Mol Biol. 1997;27:2.4.1–2.4.5. doi: 10.1002/0471142727.mb0204s56. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.