Mutations in the genes encoding four of the phagocyte NADPH oxidase components, p22-phox, p47-phox, p67-phox and 40-phox, cause the autosomal recessive forms of chronic granulomatous disease (CGD). These four forms of the disease collectively account for approximately one-third of all CGD cases. Many new mutations have been identified in these four genes since publication of the first updated version of the tables with these mutations (1). The remaining two-thirds of cases are caused by mutations in the X-linked gene for gp91-phox, CYBB; these mutations have been tabulated previously in this journal (2). The incidence of CGD as a whole is between 1 in 200,000 and 1 in 250,000 individuals.
The protein p22-phox is one of two membrane-bound subunits of cytochrome b558 (the other is gp91-phox), and mutations in the p22-phox gene (CYBA, located at 16q24, OMIM *608508) account for about 6% of CGD (Table 1). Also about 6% of CGD cases are caused by mutations in the gene for p67-phox (NCF2, 1q25, OMIM *608515), a cytosolic component of the superoxide-generating NADPH oxidase system (Table 2). The most common form of autosomal recessive CGD (about 20% of all cases) is caused by mutations in the gene for p47-phox (NCF1, 7q11.23, OMIM *608512), a second cytosolic component of the enzyme (Table 3). Only one patient has been described with mutations in NCF4 (22q13.1, OMIM *601488), the gene encoding p40-phox, the third cytosolic NADPH oxidase component (Table 4). The type, position and number of the mutations in these four genes is depicted in figure 1. Tables 5-8 list apparently benign polymorphisms that have been identified in the CYBA, NCF2, NCF1 and NCF4 genes, respectively. It is important to realize that SNPs and other sequence variants available on the internet are not necessarily functionally neutral.
Table 1.
Mutations in the p22-phox gene CYBA.
| Nucleotide change | Mutation | Amino acid or mRNA change | CGD type | Reference | Families (alleles)a | Accession number(s)g | |
|---|---|---|---|---|---|---|---|
| g. del>10 kb | Deletion | NAb | A22° | [28] | 1(2) | ||
| c.2T>A | Missense | p.Met1Lys | A22? | unpubl. d | 1(2) c | * | |
| c.7C>T | Nonsense | p.Gln3X | A22° | [29] unpubl. | 5(10) | H0023 H0026 |
|
| c.26G>A | Nonsense | p.Trp9X | A22° | [30] | 1(1) | H0017 | |
| c.27G>A | Nonsense | p.Trp9X | A22° | [31] | 1(2) | H0027 | |
| c.58+4_+7delAGTG e | Splice site | ins. 79 bp in intron 1 p.Ile20SerfsX77 |
A22° | [31,32] unpubl. | 5(9) | H0028 H0039 |
|
| g.exon2_3del | Deletion | p.Ile20ArgfsX6 | A22° | [30] | 1(2) | ||
| g.exon2_5del | Deletion | p.Ile20SerfsX68 | A22° | unpubl. | 1(2) c | * | |
| c.70G>A | Missense | p.Gly24Arg | A22° | [16,29–31,33] unpubl. | 9(14) | H0021 H0024 H0025 H0028 H0030 H0034 |
|
| c.71G>A | Missense | p.Gly24Glu | A22° | [16] | 1(2) | * | |
| c.74G>T | Missense | p.Gly25Val | A22° | [30] | 1(1) | H0017 | |
| c.77delT | Deletion | p.Ile26ThrfsX48 | A22° | unpubl. | 1(1) | * | |
| c.107G>A | Nonsense | p.Trp36X | A22° | [30] | 1(1) | H0021 | |
| g.exon3_5del | Deletion | p.Ile43MetfsX68 | A22° | [34] | 1(2) | * | |
| g.exon3_6del | Deletion | NA | A22° | [33] | 1(2) | * | |
| c.152T>A | Missense | p.Leu51Gln | A22° | unpubl. | 1(1) | * | |
| c.155T>C | Missense | p.Leu52Pro | A22° | [30] | 1(2) | H0014 | |
| c.158A>T | Missense | p.Glu53Val | A22° | [35] | 1(1) | H0005 | |
| c.164C>G | Missense | p.Pro55Arg | A22° | [16] | 1(2) | * | |
| c.166dupC | Insertion | p.Arg56ProfsX157 | A22° | [30,33] unpubl. | 5(8) | H0019 H0020 |
|
| c.171delG | Deletion | p.Lys58ArgfsX16 | A22° | unpubl. | 1(2) | * | |
| c.171dupG | Insertion | p.Lys58GlufsX155 | A22° | [16,35] unpubl. | 5(9) | H0005 H0032 H0036 H0037 |
|
| c.173delA # | Deletion | p.Lys58ArgfsX16 | A22° | [34] | 1(2) | H0041 | * |
| c.203+1G>T e | Splice site | del. exon 3 p.Ile43_Trp68delinsMet |
A22° | [30] | 1(2) | H0016 | |
| c.204–2A>G e | Splice site | del. exon 4? p.Gly69_Leu96del? |
A22° | [36] | 1(2) | * | |
| c.223delG # | Deletion | p.Ala75ProfsX3 | A22° | [34] | 1(2) | H0042 | * |
| c.246delC | Deletion | p.Phe83LeufsX108 | A22° | [28,30] | 2(3) | H0001 H0015 |
|
| c.261C>G | Nonsense | p.Tyr87X | A22° | unpubl. | 1(2) | * | |
| c.261C>A | Nonsense | p.Tyr87X | A22° | unpubl. | 1(2) | * | |
| c.268C>T | Missense | p.Arg90Trp | A22° | [30] unpubl. | 8(14) | H0018 H0019 H0020 |
|
| c.268C>G | Missense | p.Arg90Gly | A22° | unpubl. | 1(2) | * | |
| c.269G>A | Missense | p.Arg90Gln | A22° | [28,37] | 2(3) | H0001 H0006 H0007 H0008 |
|
| c.269G>C | Missense | p.Arg90Pro | A22° | unpubl. | 1(2) | * | |
| c.281A>G | Missense | p.His94Arg | A22° | [37] | 1(2) | H0012 | |
| c.287+1G>A e | Splice site | del. exon 4 p.Gly69_Leu96del | A22° | [37] unpubl. | 2(4) | H0009 H0035 |
|
| c.287+1G>T e | Splice site | del. exon 4 p.Gly69_Leu96del | A22° | unpubl. | 1(2) | * | |
| c.287+2T>C e | Splice site | del. exon 4? p.Gly69_Leu96del? |
A22° | unpubl. | 1(1) f | * | |
| c.288–6_296del16 e | Splice site | del. exon5 p.Leu97ArgfsX68 |
A22° | unpubl. | 1(1) | * | |
| c.288–15_308del36 e | Splice site | intron4ins179/exon5del21 | A22° | [38] | 1(2) | H0038 | * |
| c.288G>T | Splice site | del. exon 5 p.Leu97ArgfsX68 |
A22° | unpubl. | 1(2) | * | |
| c.295_301delGTGCCCG | Deletion | p.Val99ProfsX90 | A22° | [13,17,19] unpubl. | 5(10) c | H0040 | * |
| c.339C>A | Nonsense | p.Cys113X | A22° | [33] | 1(2) | * | |
| c.354C>A | Missense | p.Ser118Arg | A22° | [28,30] unpubl. | 4(8) | H0003 H0010 H0022 |
|
| c.369+1G>C e | Splice site | del. exon 5 p.Leu97ArgfsX68 |
A22° | [6,39] | 1(2) | H0002 | |
| c.369+1G>A e | Splice site | del exon 5 p.Leu97ArgfsX68 |
A22° | [34] | 1(2) | H0045 H0046 |
* |
| c.370G>T | Missense | p.Ala124Ser | A22° | [17] | 1(2) | * | |
| c.371C>T | Missense | p.Ala124Val | A22° | [31] | 1(1) | H0030 | |
| c.373G>A | Missense | p.Ala125Thr | A22° | [34] | 1(2) | H0047 | * |
| c.385G>A | Missense | p.Glu129Lys | A22? | unpubl. | 1(2) | * | |
| c.385_388delGAGC | Deletion | p.Glu129SerfsX61 | A22° | [34] | 2(4) | H0043 H0044 |
* |
| c.399delC | Deletion | p.Ile134SerfsX57 | A22? | unpubl. | 1(2) c | * | |
| c.408delC | Deletion | p.Lys137SerfsX54 # | A22° | [33] | 1(2) | * | |
| c.467C>A | Missense | p.Pro156Gln | A22+ | [40] | 1(2) | H0011 | |
| c.472_484del | Deletion | p.Pro160AlafsX27 | A22? | [1] | 1(2) c | H0031 | |
| c.571_604del | Deletion | p.Thr191ProfsX | A22° | [31] | 1(2) | ||
| Mutations in CYBA | |||||||
|
| |||||||
| Number of different alleles | Total number of alleles | ||||||
|
| |||||||
| Deletions | 16 alleles | (29.1%) | 42 alleles | (24.3%) | |||
| Nonsense mutations | 7 alleles | (12.7%) | 20 alleles | (11.6%) | |||
| Splice site mutations | 11 alleles | (20.0%) | 29 alleles | (16.8%) | |||
| Missense mutations | 19 alleles | (34.6%) | 65 alleles | (37.5%) | |||
| Insertions | 2 alleles | (3.6%) | 17 alleles | (9.8%) | |||
| Total 55 different allelic mutations | Total 87 families with 173 identified alleles in the 96 patients | ||||||
Number of different families with patients with this mutation (number of alleles carrying this mutation).
Not applicable.
One patient presumed homozygous for this mutation.
Unpublished data from the authors' laboratories.
Position of introns in CYBA: intron 1 c.58_59; intron 2 c.128_129; intron 3 c.203_204; intron 4 c.287_288; intron 5 c.369_370.
Patient is heterozygous for this mutation and for an unidentified mutation in the other allele.
Accession number in database at http://www.uta.fi/imt/bioinfo/CYBAbase/.
New mutations since ref. [1].
Corrected after consultation of the authors.
Table 2.
Mutations in the p67-phox gene NCF-2.
| Nucleotide change | Mutation | Amino acid or mRNA change | CGD type | Families (alleles) | Reference | Accession number h | |||
|---|---|---|---|---|---|---|---|---|---|
| c.–547–?_174+?del g | Deletion | p.Met1_Lys58del | A67° | 1(2) | Unpubl. | * | |||
| c–274–?_174 + ?del∼400 g | Deletion | p.Met1_Lys58del | A67° | 1(2) | Unpubl. | * | |||
| c–274–?_174 + ?del∼400 g | Deletion | p.Met1_Lys58del | A67° | 1(2) | Unpubl. | * | |||
| c.1A>G | Missense | p.Met1Val | A67° | 1(1) | Unpubl. | * | |||
| c.29G>A | Nonsense | p.Trp10X | A67° | 1(2) | Unpubl. | * | |||
| c.55_63delAAGAAGGAC a | Deletion | p.Lys19_Asp21del | A67° | 4(4) | [41–43] unpubl. | M0004 M0015 |
|||
| c.125A>G | Missense | p.Asn42Ser | A67° | 1(2) | Unpubl. | * | |||
| c.130G>C | Missense | p.Gly44Arg | A67? | 2(4) b | [1,43] | ||||
| c.130G>T | Missense | p.Gly44Cys | A67° | 1(2) | Unpubl. | * | |||
| c.172_174delAAG | Deletion | p.Lys58del | A67+ | 2(3) c | [44,45] unpubl. | M0009 | |||
| c.175–1G>A g | Splice site | del. exon 3? p.Ala59IlefsX2? |
A67° | 1(2) | Unpubl. | * | |||
| c.196C>T | Nonsense | p.Arg66X | A67° | 3(5) | [42] | ||||
| c.229C>T | Nonsense | p.Arg77X | A67° | 4(7) | [46], unpubl. | * | |||
| c.230G>A | Missense | p.Arg77Gln | A67° | 3(3) | [42], unpubl. | M0016 | |||
| c.233G>A | Missense | p.Gly78Glu | A67° | 1(2) | [47] | M0002 | |||
| c.257+2T>C g | Splice site | del. exon3 p.Ala59IlefsX2 |
A67° | 2(4) | [13,48] | M0010 M0020 |
|||
| c.258–?_366 + ?del∼1100 g | Deletion | p.Tyr87CysfsX22 | A67? | 1(2) | Unpubl. | * | |||
| c.279C>G d | Missense | p.Asp93Glu | A67° | 4(8) | [46], unpubl. | * | |||
| c.287_289delAAG | Deletion | p.Glu96del | A67− | 1(2) | Unpubl. | * | |||
| c.298C>T | Nonsense | p.Gln100X | A67° | 5(5) | [42], unpubl. | M0016 | |||
| c.304C>T | Nonsense | p.Arg102X | A67° | 6(11) b | [16,41,46,48] unpubl. | M0001 | |||
| c.305G>C | Missense | p.Arg102Pro | A67+? | 1(1) | Unpubl. | * | |||
| c.323A>T | Missense | p.Asp108Val | A67−? | 1(2) | [43] | * | |||
| c.364_366+2delGAGGT g | Splice site | del. exon 4? p.Tyr87CysfsX2? |
A67° | 1(2) | [17] | * | |||
| c.366+1G>A g | Splice site | del. exon 3_4 p.Ala59_Glu122del |
A67° | 8(12) | [17,41–43] unpubl. | M0011 M0018 |
|||
| c.366+1G>C e, g | Splice site | del. exon 4? p.Tyr87CysfsX22? |
A67° | 4(8) | Unpubl. | * | |||
| c.366+2401_502–527 del1380 g | Deletion | del. exon 5 p.Val123_Trp167del |
A67° | 4(8) | [50] | * | |||
| c.383C>T | Missense | p.Ala128Val | A67° | 1(2) | [42] | M0013 | |||
| c.398_399dupAG | Insertion | p.Lys134ArgfsX12 | A67° | 1(2) | [51] | M0006 | |||
| c.409T>A | Missense | p.Trp137Arg | A67− | 1(2) | Unpubl. | * | |||
| c.419C>G | Missense | p.Ala140Asp | A67° | 1(1) | Unpubl. | * | |||
| c.[479A>T; 481A>G] | Dbl. missense | p.AspLys160_161 ValGlu |
A67° | 1(1) f | [52] | M0012 | |||
| c.482delA | Deletion | p.Lys161ArgfsX16 | A67? | 1(2) b | Unpubl. | * | |||
| c.488_501delTGGAGTGTGTCTGG g | Deletion/splice site | del.exon 5 p.Val123_Trp167del |
A67° | 1(1) | Unpubl. | * | |||
| c.502–1G>T g | Splice site | del exon 6? p.Lys168_Thr203del? |
A67° | 1(2) | Unpubl. | * | |||
| c.505C>G | Missense | p.Gln169Glu | A67? | 1(2) | Unpubl. | * | |||
| c.550C>T | Nonsense | p.Arg184X | A67? | 1(2) | Unpubl. | * | |||
| c.551G>C | Missense | p.Arg184Pro | A67° | 1(2) | Unpubl. | * | |||
| c.576C>T | Nonsense | p.Gln192X | A67? | 1(2) b | [53] | * | |||
| c.586_588delAAG | Deletion | p.Lys196del | A67+ | 1(2) | Unpubl. | * | |||
| c.605C>T | Missense | p.Ala202Val | A67− | 2(4) | [46] unpubl. | * | |||
| c.714-? _924+?dup∼1100 g | Insertion | p.Glu309GlyfsX15 | A67° | 2(3) f | [54] unpubl. | * | |||
| c.714–1G>T g | Splice site | del. exon 9? p.Ala239ArgfsX59? |
A67° | 1(1) | Unpubl. | * | |||
| c.728delA | Deletion | p.Glu243GlyfsX28 | A67° | 1(1) | [41] | * | |||
| c.767_768dupAA | Insertion | p.Glu257LysfsX15 | A67° | 1(2) | Unpubl. | * | |||
| c.799_800delGT | Deletion | pVal267LeufsX8 | A67° | 1(2) | [17] | * | |||
| c.835_836delAC | Deletion | p.Thr279GlyfsX16 | A67° | 2(4) b | [42] unpubl. | M0017 | |||
| c.855+1G>A g | Splice site | del. exon 8_9 p.Ala224_Gln285del |
A67° | 1(2) | [55] | M0007 | |||
| c.1026G>A g | Splice site | del. exon 12 p.Gln335SerfsX38 |
A67° | 2(2) | [49] | * | |||
| c.1034dupA | Insertion | p.Leu346AlafsX36 | A67? | 1(1) | Unpubl. | * | |||
| c.1099C>T | Nonsense | p.Gln367X | A67? | 1(2) b | Unpubl. | * | |||
| c.1171_1175delAAGCT | Deletion | p.Lys391GlufsX9 | A67° | 6(12) | [16,19,41] | M0005 | |||
| c.1179–2A>T g | Splice site | del. exon 14? p.Ser393ArgfsX54? |
A67° | 1(2) | Unpubl. | * | |||
| c.1256A>T | Missense | p.Asn419Ile | A67° | 1(2) | [13] | M0019 | * | ||
| Mutations in NCF2 | |||||||||
|
| |||||||||
| Number of different alleles | Total number of alleles | ||||||||
|
| |||||||||
| Deletions | 14 alleles | (25.9%) | 48 alleles | (28.1%) | |||||
| Nonsense mutations | 8 alleles | (14.8%) | 36 alleles | (21.0%) | |||||
| Splice site mutations | 11 alleles | (20.4%) | 38 alleles | (22.2%) | |||||
| Missense mutations | 17 alleles | (31.5%) | 41 alleles | (24.0%) | |||||
| Insertions | 4 alleles | (7.4%) | 8 alleles | (4.7%) | |||||
| Total 54 different allelic mutations | Total 83 families with 171 identified alleles in the 95 patients | ||||||||
Always in combination with c.1183C>T (polymorphism, rs13306575) on the same allele.
One patient presumed homozygous for this mutation.
One patient is heterozygous for this mutation and for an undefined deletion of 11-13 kb in the other allele [44,45].
Always in combination with c.366+1G>C on the same allele.
Always in combination with c.279C>G on the same allele.
One patient is heterozygous for this mutation and for an unidentified mutation in the other allele [54].
Positions of introns in NCF2: intron 1 c.–510_–274 in 5′ UTR; intron 2 c.174_175; intron 3 c.257_258; intron 4 c.366_367; intron 5 c.501_502; intron 6 c.609_610; intron 7 c.669_670; intron 8 c.713_714; intron 9 c.855_856; intron 10 c.924_925; intron 11 c.1000_1001; intron 12 c.1026_1027; intron 13 c.1178_1179; intron 14 c.1290_1291; intron 15 c.1468_1469.
Accession number in database at http://www.uta.fi/imt/bioinfo/NCF2base/.
Table 3.
Mutations in the p47-phox gene NCF1.
| Nucleotide change | Mutation | Amino acid or mRNA change | CGD type | Families (alleles) | Reference | Accession number(s) a | |
|---|---|---|---|---|---|---|---|
| c.72+1G>A e | Splice site | del. exon 1? p.Met1_Tyr24del? |
A47° | 1(1) | [8] | N0031 | |
| c.72+3G>T e | Splice site | del. exon 1? p.Met1_Tyr24del? |
A47° | 1(1)] | [8 | N0032 | |
| c.75_76delGT | Deletion | p.Tyr26HisfsX26 | A47° | >300 homozygous, 20 heterozygous b | [3–19] unpubl. | N0004–7,9–23, 27–29, 31–35, 40–63, 69,70 | |
| c.125G>A | Missense | p.Arg42Gln | A47° | 3(3) | [8] unpubl. | N0034N0035 | |
| c.153+1G>A e | Splice site | c.153+1_+73insc p.Lys52MetX24 |
A47° | 1(1) | [14] | N0068 | * |
| c.154–283_451+821 del2858 e | Deletion | del. exon 3_5 p.Lys52ThrfsX82 |
A47° | 1(1) | [14] | N0061 | * |
| c.271C>T | Nonsense | p.Gln91X | A47° | 1(1) | [14] | N0027 | * |
| c.333T>A | Nonsense | p.Cys111X | A47° | 1(1) | [9,14] | N0028 N0056 N0057 N0058 |
|
| c.353_354delCC insAA | Deletion/insertion | p.Phe118X | A47° | 1(1) | [10,14] | N0059 | * |
| c.417_451+650 del685 e | Deletion | del. exon 5 p.Thr133HisfsX66 |
A47° | 1(1) | unpubl. | * | |
| c.502delG | Deletion | p.Glu168ArgfsX19 | A47° | 1(1) | [5] | N0002 | |
| c.574G>A e | Splice site | del. exon 6+7 d p.Asp151_Ala227del |
A47° | 4(7) unpubl. | [8, 14] | N0036 N0064 N0065 N0066 N0067 |
|
| c.579G>A | Nonsense | p.Trp193X | A47° | 17(31) | [14,16,56] unpubl. | N0024 N0025 N0026 N0037 N0038 N0039 N0060 N0068 |
* |
| c.604C>T | Nonsense | p.Arg202X | A47° | 1(1) | [17] | * | |
| c.612G>A | Nonsense | p.Trp204X | A47? | 1(2) f | [57] | * | |
| c.678T≫G | Nonsense | p.Tyr226X | A47° | 1(2) | [17] | * | |
| c.682+1G>C e | Splice site | del. exon 7 p.Trp193_Gly228del |
A47° | 1(1) | [14] | N0062 N0063 |
* |
| c.730G>A | Missense | p.Glu244Lys | A47° | 1(1) | unpubl. | * | |
| c.734_748del15 | Deletion | p.Val245_Glu249del | A47° | 1(2) | unpubl. | * | |
| c.784G>A | Missense | p.Gly262Ser | A47° | 1(1) | [8] | N0033 | |
| c.789G>C | Missense | p.Trp263Cys | A47° | 1(1) | unpubl. | * | |
| c.811delG | Deletion | p.Val271SerfsX105 | A47° | 1(1) | [8] | ||
| c.838delC | Deletion | p.Leu280CysfsX96 | A47° | 1(1) | [18] | * | |
| Mutations in NCF1 | |||||||
|
| |||||||
| Number of different alleles | Total number of alleles | ||||||
|
| |||||||
| Deletions | 7 alleles | (30.4%) | 7 alleles | (11.1%) | |||
| Nonsense mutations | 6 alleles | (26.1%) | 38 alleles | (60.3%) | |||
| Splice site mutations | 5 alleles | (21.7%) | 11 alleles | (17.5%) | |||
| Missense mutations | 4 alleles | (17.4%) | 6 alleles | (9.5%) | |||
| Deletion/insertions | 1 allele | (4.4%) | 1 allele | (1.6%) | |||
| Total 23 different allelic mutations (including delta-GT) | Total 42 families with 6 (other than delta-GT) in 3 identified alleles the 53 patients | ||||||
Accession number in database at http://www.uta.fi/imt/bioinfo/NCF1base/.
One patient is a compound heterozygote for this mutation and for an undefined chromosomal microdeletion on the other allele [58].
Activation of cryptic donor splice site in intron 2, leading to incorporation of 73 nucleotides from the 5′ side of intron 2 into mRNA, including the mutated G>A at position +1 of intron 2. At the protein level, this mutation predicts incorporation of 24 aberrant amino acids after His51, followed by a stop codon at position 76 [14].
In addition, these patients show evidence of mRNA for p47phox from which the last 22 bp at the 3′ region of exon 6 has been skipped (r.552_574del), as well as mRNA in which intron 6 has been included and in which the mutated exon 6 is expressed (r.[intron6+1_exon6–1ins; 574 g>a]) [14].
Positions of introns in NCF1: intron 1 c.72_73; intron 2 c.153_154; intron 3 c. 229_230; intron 4 c.395_396; intron 5 c.451_452; intron 6 c.574_575; intron 7 c.682_683; intron 8 c.801_802; intron 9 c.905_906; intron 10 c.1051_1052.
Patient presumed to be homozygous for the mutation.
Table 4.
Mutations in the p40-phox gene NCF4.
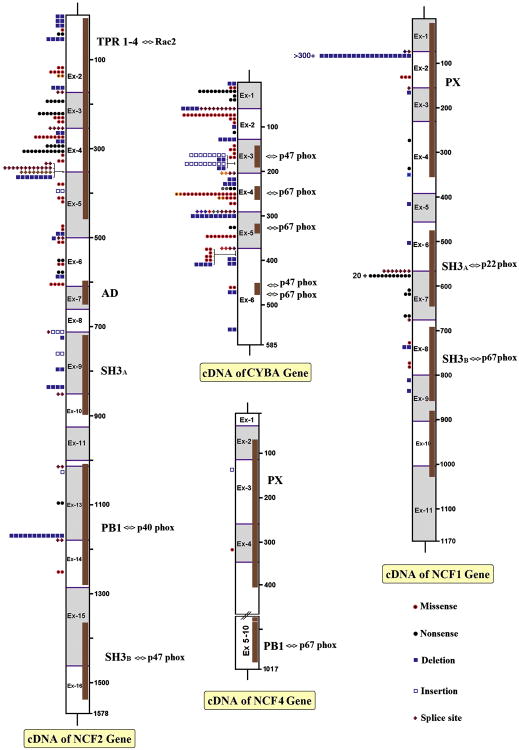
Figure 1. Schematic overview of mutations in NCF2, CYBA, NCF4 and NCF1.
For each cDNA, the exon positions and the corresponding protein domains have been depicted. For some of the protein domains, their interaction with other proteins has been indicated. The PX domains interact with phosphatidyl-inositol-phosphates. The type of mutations (explained in the right hand corner), their position and number of mutated alleles are indicated. Splice site mutations are given at the exon borders.
Table 5.
Polymorphisms in the p22-phox gene CYBA.
| Polymorphic nucleotide | Amino acid change | Reference |
|---|---|---|
| c.59–37A/G | NA | [30] |
| c.36A/G | p.Glu12Glu | [60] |
| c.179A/C | p.Lys60Thr | [30] |
| c.214C/T | p.His72Tyr | [28,60] |
| c.288–138ins50 | NA | [13] |
| c.381T/C | p.Arg127Arg | [60] |
| c.403G/A | p.Glu135Lys | [30] |
| c.480G/A | p.Pro160Pro | [30,37,60] |
| c.512A/G | p.Glu171Gly | [60] |
| c.521C/T | p.Ala174Val | [28,30,31,60] |
| c.579G/T | p.Glu193Asp | [60] |
| c.612A/G (+24 of 3′ UT region) | NA | [37,60] |
Table 8.
Polymorphisms in the p40-phox gene NCF4.
| Polymorphic nucleotidea | Amino acid change | Reference |
|---|---|---|
| c.32+1258G/T | N.A. | [61] |
| c.33–1101T/C | N.A. | [61] |
| c.33–728T/C | N.A. | [61] |
| c.118–360G/A | N.A. | [61] |
| c.342+202G/C | N.A. | [61] |
| c.342+342G/T | N.A. | [61] |
| c.342+1326G/A | N.A. | [61] |
| c.343–1378A/G | N.A. | [61] |
| c.343–339A/G | N.A. | [61] |
| c.528+16G/A | N.A. | [61] |
| c.627+711G/A | N.A. | [61] |
| c.627+1040G/T | N.A. | [61] |
| c.628–1193G/A | N.A. | [61] |
| c.758+57A/T | N.A. | [61] |
Positions of introns in NCF4: intron 1 c.32_33; intron 2 c.117_118; intron 3 c.271_272; intron 4 c.342_343; intron 5 c.470_471; intron 6 c.528_529; intron 7 c.627_628; intron 8 c.758_759; intron 9 c.824_825.
Unlike the other autosomal recessive and X-linked forms of the disease, in which there is a large heterogeneity among mutations, a single defect accounts for the vast majority of cases of p47-phox-deficiency. Of ∼250 patients investigated worldwide at the DNA level, all but 53 patients in 42 families appear to be homozygous for a dinucleotide (GT) deletion (ΔGT) at the start of exon 2 (3-19). Of the 42 families with exceptions, 20 had patients who were compound heterozygotes for the GT deletion and one additional mutation, and the others had patients with mutations other than ΔGT on both alleles of NCF1 (20 homozygous, 2 compound heterozygous). The ΔGT-bearing allele of NCF1 is therefore the most common CGD-causing allele in the population, carried by approximately 1 in 250 individuals. The reason for this predominance is that most normal individuals have two p47-phox pseudogenes, each of which co-localizes with the functional gene to 7q11.23 and carries ΔGT. Recombination events between NCF1 and these highly homologous pseudogenes lead to the incorporation of ΔGT into NCF1 (7, 20).
Additional information about the tabulated mutations and about CGD in general can be found in recent reviews (21-25) and in the cited literature. In the following tables we have used the standard notation for differentiating the various phenotypes of CGD (e.g., A22°, A22+, A67°, A67+, A67 −, A47°, A40° and A40+). In this nomenclature the first letter refers to the mode of inheritance (autosomal recessive), the numeral indicates the phox component affected, and the superscript symbol indicates whether the protein is absent (°), diminished (−) or normal (+), based on immunoblot analysis. When this information is unavailable, that has been indicated as (?). The respective proteins can be non-functional, exert residual activity, or in case of (−) be fully functional. Online Mendelian Inheritance in Man (OMIM) numbers for A22, A67, A47 CGD are #233690, #233710, and #233700, respectively. Mutations added since the last updated versions of Tables 1-3 were published (1) are marked with an asterisk in the right hand column. The nucleotide numbering system we have used is based on the cDNA sequence and follows the convention that +1 is the A of the ATG initiator codon. This differs from the numbering of the GenBank sequences; for p22-phox (GenBank accession nos. M21186 and J03774) subtract 28 from the GenBank sequence number to make the initiator A +1; for p67-phox (accession no. M32011) subtract 67 from the GenBank numbering; for p47-phox (GenBank accession nos. M25665 and M26193) subtract 12 from the GenBank numbering, and for p40-phox (accession no. NM_000631) subtract 184 from the GenBank numbering. The notation of the mutations and polymorphisms follows the recommendations of the Human Genome Variation Society (26) (see also www.hgvs.org/mutnomen). Where possible we have cross-referenced the mutations listed here with those in three CGD databases that list CGD patients by accession number. These databases contain additional biochemical, genetic and clinical information and are available at http://www.uta.fi/imt/bioinfo/CYBAbase/ (or NCF1base/, or NCF2base/). In addition, information can also be found in the HGMD database at http://www.hgdm.cf.ac.uk/ac.search.php. The consequences of the mutations for protein composition have been checked with the Mutalyzer program (www.lovd.nl/mutalyzer) (27).
Table 6.
Polymorphisms in the p67-phox gene NCF2.
| Polymorphic nucleotide | Amino acid change | Reference |
|---|---|---|
| c.–185G/A | NA | [41,42] |
| c.–181G/A | NA | [41,42] |
| c.–24C/T | NA | [41,42] |
| c.235A/G | p.Met79Val | [41] |
| c.542A/G | p.Lys181Arg | [41,42,51] |
| c.606G/A | p.Ala202Ala | [42] |
| c.895C/T | p.Leu299Leu | [41,47,51] |
| c.925–21G/A | NA | [41] |
| c.983G/A | p.Arg328Lys | [41,47,51] |
| c.1105G/A | p.Gly369Arg | [42] |
| c.1167C/A | p.His389Gln | [41,42] |
| c.1183C/T | p.Arg395Trp | [41–43] |
Table 7.
Polymorphisms in the p47-phox gene NCF1.
| Polymorphic nucleotidea | Amino acid change | Reference |
|---|---|---|
| c.66G/C | p.Glu22His | Unpubl. |
| c.73G/A | p.Val25Met | Unpubl. |
| c.345C/T | p.Leu115Leu | [8] |
| c.468C/T | p.Ile156Ile | Unpubl. |
| c.558A/G | p.Val186Val | Unpubl. |
| c.621G/A | p.Ala206Ala | Unpubl. |
| c.825C/T | p.Phe275Phe | Unpubl. |
| c.849A/G | p.Ser283Ser | Unpubl. |
| c.936C/T | p.His312His | Unpubl. |
Identification of polymorphic sites in NCF1 is complicated by the p47-phox pseudogenes, which contain several differences from the functional gene; the referenced polymorphism was identified after amplification of NCF1 with primers that do not bind to the pseudogenes [8]. More synonymous polymorphisms can be expected to be introduced into NCF1 by recombination with the pseudogenes [20].
Acknowledgments
We thank the CGD Research Trust, London, UK, for financial support. We thank Katrin Höhne (Univ. Children's Hospital, Dresden, Germany), for excellent assistance. We are grateful to Cécile Martel, Michelle Mollin and Sylvain Beaumel for their constant and excellent technical assistance and we sincerely thank all the physicians collaborating with and trusting in the CGD diagnosis and research Center in Grenoble, France.
This project has been funded in part with federal funds from the National Cancer Institute, National Institutes of Health, under Contract No. HHSN261200800001E. The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the U.S. Government.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Cross AR, Noack D, Rae J, Curnutte JT, Heyworth PG. Hematologically important mutations: The autosomal recessive forms of chronic granulomatous disease (first update) Blood Cells Mol Dis. 2000;22:268–270. doi: 10.1006/bcmd.2000.0333. [DOI] [PubMed] [Google Scholar]
- 2.Heyworth PG, Curnutte JT, Rae J, Noack D, Roos D, van Koppen E, Cross AR. Hematologically important mutations: X-linked chronic granulomatous disease (second update) Blood Cells Mol Dis. 2001;23:443–450. doi: 10.1006/bcmd.2000.0347. [DOI] [PubMed] [Google Scholar]
- 3.Casimir CM, Bu-Ghanim HN, Rodaway ARF, Bentley DL, Rowe P, Segal AW. Autosomal recessive chronic granulomatous disease caused by deletion at a dinucleotide repeat. Proc Natl Acad Sci USA. 1991;88:2753–2757. doi: 10.1073/pnas.88.7.2753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Iwata M, Nunoi H, Yamazaki H, Nakano T, Niwa H, Tsuruta S, Ohga S, Ohmi S, Kanegasaki S, Matsuda I. Homologous dinucleotide (GT or TG) deletion in Japanese patients with chronic granulomatous disease with p47-phox deficiency. Biochem Biophys Res Commun. 1994;199:1372–1377. doi: 10.1006/bbrc.1994.1382. [DOI] [PubMed] [Google Scholar]
- 5.Volpp BD, Lin Y. In vitro molecular reconstitution of the respiratory burst in B lymphoblasts from p47-phox-deficient chronic granulomatous disease. J Clin Invest. 1993;91:201–207. doi: 10.1172/JCI116171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Roos D, De Boer M, Kuribayashi F, Meischl C, Weening RS, Segal AW, Åhlin A, Nemet K, Hossle JP, Bernatowska-Matuszkiewicz E, Middleton-Price H. Mutations in the X-linked and autosomal recessive forms of chronic granulomatous disease. Blood. 1996;87:1663–1681. [PubMed] [Google Scholar]
- 7.Roesler J, Curnutte JT, Rae J, Barrett D, Patiño P, Chanock SJ, Görlach A. Recombination events between the p47-phox gene and its highly homologous pseudogenes are the main cause of autosomal recessive chronic granulomatous disease. Blood. 2000;95:2150–2156. [PubMed] [Google Scholar]
- 8.Noack D, Rae J, Cross AR, Ellis BA, Newburger PE, Curnutte JT, Heyworth PG. Autosomal recessive chronic granulomatous disease caused by defects in NCF1, the gene encoding the phagocyte p47-phox: mutations not arising in the NCF1 pseudogenes. Blood. 2001;97:305–311. doi: 10.1182/blood.v97.1.305. [DOI] [PubMed] [Google Scholar]
- 9.Vihinen M, Arredondo-Vega FX, Casanova JL, Etzioni A, Giliani S, Hammarström L, Hershfield MS, Heyworth PG, Hsu AP, Lähdesmäki A, Lappalainen I, Notarangelo LD, Puck JM, Reith W, Roos D, Schumacher RF, Schwarz K, Vezzoni P, Villa A, Väliaho J, Smith CIE. Primary immunodeficiency mutation databases. In: Hall JC, Dunlap JC, Friedmann T, Giannelli F, editors. Advances in Genetics. Vol. 43. 2000. in press. [DOI] [PubMed] [Google Scholar]
- 10.Jurkowska M, Kurenko-Deptuch M, Bal J, Roos D. The search for a genetic defect in Polish patients with chronic granulomatous disease. Arch Immunol Ther Exp. 2004;52:441–446. [PubMed] [Google Scholar]
- 11.Prando-Andrade C, Agudelo-Florez P, Lopez JA, Aparecida de Souza Paiva M, Costa-Carvalho BT, Condino-Neto A. Autosomal chronic granulomatous disease: case report and mutation analysis of two Brazilian siblings. J Pediatr (Rio J) 2004;80:425–428. [PubMed] [Google Scholar]
- 12.Agudelo-Flórez P, Cardoso Prando-Andrade C, López JA, Costa-Carvalho BT, Quezada A, Espinosa FJ, Aparacida de Souza Paiva M, Roxo P, Grumach A, Abe Jacob C, Salles Carneiro-Sampaio MM, Newburger PE, Condino-Neto A. Chronic granulomatous Disease in Latin American patients: clinical spectrum and molecular genetics. Pediatr Blood Cancer. 2006;46:243–252. doi: 10.1002/pbc.20455. [DOI] [PubMed] [Google Scholar]
- 13.El Kares R, Barbouche MR, Elloumi-Zghal H, Bejaoui M, Chemli J, Mellouli F, Tebib N, Abdelmoula MS, Boukthir S, Fitouri Z, M'Rad S, Bouslama K, Touiri H, Abdelhak S, Dellagi MK. Genetic and mutational heterogeneity of autosomal recessive chronic granulomatous disease in Tunesia. J Hum Genet. 2006;51:887–895. doi: 10.1007/s10038-006-0039-8. [DOI] [PubMed] [Google Scholar]
- 14.Roos D, de Boer M, Köker MY, Dekker J, Singh-Gupta V, Åhlin A, Palmblad J, Sanal Ö, Kurenko-Deptuch M, Jolles S, Wolach B. Chronic granulomatous disease caused by mutations other than the common GT deletion in NCF1, the gene encoding the p47phox component of the phagocyte NADPH oxidase. Hum Mutat. 2006;27:1218–1229. doi: 10.1002/humu.20413. [DOI] [PubMed] [Google Scholar]
- 15.Köker MY, Sanal Ö, de Boer M, Teczan İ, Metin A, Ersoy F, Roos D. Mutations of chronic granulomatous disease in Turkish families. Eur J Clin Invest. 2007;37:589–595. doi: 10.1111/j.1365-2362.2007.01828.x. [DOI] [PubMed] [Google Scholar]
- 16.Wolach B, Gavrieli R, de Boer M, Gottesman G, Ben-Ari J, Rottem M, Schlesinger Y, Etzioni A, Roos D. Chronic granulomatous disease in Israel: functional and molecular studies of 38 patients. Clin Immunol. 2008;129:103–114. doi: 10.1016/j.clim.2008.06.012. [DOI] [PubMed] [Google Scholar]
- 17.Kannengiesser C, Gérard B, El Benna J, Henri D, Kroviarski Y, Chollet-Martin S, Gougerot-Pocidalo MA, Elbim C, Grandchamp B. Molecular epidemiology of chronic granulomatous disease in a series of 80 kindreds: identification of 31 novel mutations. Hum Mutat. 2008;29:E132–E149. doi: 10.1002/humu.20820. [DOI] [PubMed] [Google Scholar]
- 18.Van de Vosse E, van Wengen A, van Geelen JA, de Boer M, Roos D, van Dissel JT. A novel mutation in NCF1 in an adult CGD patient with a liver abscess as First presentation. J Hum Genet. 2009;54:313–316. doi: 10.1038/jhg.2009.24. [DOI] [PubMed] [Google Scholar]
- 19.Bakri FG, Martel C, Khuri-Bulos N, Mahafza A, El-Khateeb MS, Al-Wahadneh AM, Hayajneh WA, Hamamy HA, Maquet E, Molin M, Stasia MJ. First report of clinical, functional, and molecular investigation of chronic granulomatous disease in nine Jordanian families. J Clin Immunol. 2009;29:215–230. doi: 10.1007/s10875-008-9243-y. [DOI] [PubMed] [Google Scholar]
- 20.Görlach A, Lee PL, Roesler J, Hopkins PJ, Christensen B, Green ED, Chanock SJ, Curnutte JT. A p47-phox pseudogene carries the most common mutation causing p47-phox-deficient chronic granulomatous disease. J Clin Invest. 1997;100:1907–1918. doi: 10.1172/JCI119721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Roos D, Kuijpers TW, Curnutte JT. Chronic granulomatous disease. In: Ochs HD, Smith CIE, Puck JM, editors. Primary immunodeficiency diseases, a molecular and genetic approach. 2nd. Oxford University Press; New York: 2007. pp. 525–549. [Google Scholar]
- 22.Stasia MJ, Li XJ. Genetics and immunopathology of chronic granulomatous disease. Semin Immunopathol. 2008;30:209–235. doi: 10.1007/s00281-008-0121-8. [DOI] [PubMed] [Google Scholar]
- 23.Martire B, Rondelli R, Soresina A, Pignata C, Broccoletti T, Finocchi A, Rossi P, Gattorno M, Rabusin M, Azzari C, Dellepiane RM, Pietrogrande MC, Trizzino A, DiBartolomeo P, Martino S, Carpino L, Cossu F, Locatelli F, Maccario R, Pierani P, Putti MC, Stabile A, Notarangelo LD, Ugazio AG, Plebani A, De Mattia D. Clinical features, long-term follow-up and outcome of a large cohort of patients with chronic granulomatous disease: an Italian multicenter study. Clin Immunol. 2008;126:155–164. doi: 10.1016/j.clim.2007.09.008. [DOI] [PubMed] [Google Scholar]
- 24.Jones LB, McGrogan P, Flood TJ, Gennery AR, Morton L, Thrasher A, Goldblatt D, Parker L, Cant AJ. Chronic granulomatous disease in the United Kingdom and Ireland: a comprehensive national patient-based registry. Clin Exp Immunol. 2008;152:211–218. doi: 10.1111/j.1365-2249.2008.03644.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Van den Berg JM, van Koppen E, Åhlin A, Belohradsky BH, Bernatowska E, Corbeel L, Espanõl T, Fischer A, Kurenko-Deptuch M, Mouy R, Petropoulou T, Roesler J, Seger R, Stasia MJ, Valerius NH, Weening RS, Wolach B, Roos D, Kuijpers TW. Chronic granulomatous disease: the European experience. PLoS ONE. 2009;4:e5234. doi: 10.1371/journal.pone.0005234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Den Dunnen JT, Antonarakis SE. Mutation nomenclature extensions and suggestions to describe complex mutations: a discussion. Hum Mutat. 2000;15:7–12. doi: 10.1002/(SICI)1098-1004(200001)15:1<7::AID-HUMU4>3.0.CO;2-N. [DOI] [PubMed] [Google Scholar]
- 27.Wildeman M, van Ophuizen E, den Dunnen JT, Taschner PE. Improving sequence variant descriptions in mutation databases and literature using the Mutalyzer sequence variation nomenclature checker. Hum Mutat. 2008;29:6–13. doi: 10.1002/humu.20654. [DOI] [PubMed] [Google Scholar]
- 28.Dinauer MC, Pierce EA, Bruns GAP, Curnutte JT, Orkin SH. Human neutrophil cytochrome b light chain (p22-phox). Gene structure, chromosomal location, and mutations in cytochrome-negative autosomal recessive chronic granulomatous disease. J Clin Invest. 1990;86:1729–1737. doi: 10.1172/JCI114898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yamada M, Ariga T, Kawamura N, Ohtsu M, Imajoh-Ohmi S, Ohshika E, Tatsuzawa O, Kobayashi K, Sakiyama Y. Genetic studies of three Japanese patients with p22-phox-deficient chronic granulomatous disease: detection of a possible common mutant CYBA allele in Japan and a genotype-phenotype correlation in these patients. Br J Haematol. 2000;108:511–517. doi: 10.1046/j.1365-2141.2000.01857.x. [DOI] [PubMed] [Google Scholar]
- 30.Rae J, Noack D, Heyworth PG, Ellis BA, Curnutte JT, Cross AR. Molecular analysis of nine new families with chronic granulomatous disease caused by mutations in CYBA, the gene encoding p22-phox. Blood. 2000;96:1106–1112. [PubMed] [Google Scholar]
- 31.Ishibashi F, Nunoi H, Endo F, Matsuda I, Kanegasaki S. Statistical and mutational analysis of chronic granulomatous disease in Japan with special reference to gp91-phox and p22-phox deficiency. Hum Genet. 2000;106:473–481. doi: 10.1007/s004390000288. [DOI] [PubMed] [Google Scholar]
- 32.De Boer M, Hartl D, Wintergerst U, Belohradsky BH, Roos D. A donor splice site mutation in intron 1 of CYBA, leading to chronic granulomatous disease. Blood Cells Mol Dis. 2005;35:365–369. doi: 10.1016/j.bcmd.2005.08.002. [DOI] [PubMed] [Google Scholar]
- 33.Köker MY, van Leeuwen K, de Boer M, Çelmeli F, Metin A, Özgür TT, Sanal Ö, Roos D. Six different CYBA mutations including three novel mutations in ten families from Turkey, resulting in autosomal recessive chronic granulomatous disease. Eur J Clin Invest. 2009;39:311–319. doi: 10.1111/j.1365-2362.2009.02093.x. [DOI] [PubMed] [Google Scholar]
- 34.Teimourian S, Zomorodian E, Badalzadeh M, Pouya A, Kannengiesser C, Mansouri D, Cheraghi T, Parvaneh N. Characterization of six novel mutations in CYBA: the gene causing autosomal recessive chronic granulomatous disease. Br J Haematol. 2008;141:848–851. doi: 10.1111/j.1365-2141.2008.07148.x. [DOI] [PubMed] [Google Scholar]
- 35.Hossle JP, De Boer M, Seger RA, Roos D. Identification of allele-specific p22-phox mutations in a compound heterozygous patient with chronic granulomatous disease by mismatch PCR and restriction enzyme analysis. Hum Genet. 1994;93:437–442. doi: 10.1007/BF00201671. [DOI] [PubMed] [Google Scholar]
- 36.Bagg A, Gonzales-Peralta R, Petrovic A, Sleasman JW. Novel CYBA gene mutation in a patient with chronic granulomatous disease associated with autoimmune hepatitis. J Allergy Clin Immunol. 2007;119(Suppl. 1):S16. [Google Scholar]
- 37.De Boer M, De Klein A, Hossle JP, Seger R, Corbeel L, Weening RS, Roos D. Cytochrome b558-negative, autosomal recessive chronic granulomatous disease: two new mutations in the cytochrome b558 light chain of the NADPH oxidase (p22-phox) Am J Hum Genet. 1992;51:1127–1135. [PMC free article] [PubMed] [Google Scholar]
- 38.Stasia MJ, Bordigoni P, Martel C, Morel F. A novel and unusual case of chronic granulomatous disease in a child with homozygous 36-bp deletion in the CYBA gene (A22°) leading to the activation of a cryptic splice site in intron 4. Hum Genet. 2002;110:444–450. doi: 10.1007/s00439-002-0720-8. [DOI] [PubMed] [Google Scholar]
- 39.Porter CD, Parkar MH, Kinnon C. Identification of a donor splice site mutation leading to loss of p22-phox exon 5 in autosomal chronic granulomatous disease. Hum Mutat. 1996;7:374. doi: 10.1002/humu.1380070402. [DOI] [PubMed] [Google Scholar]
- 40.Dinauer MC, Pierce EA, Erickson RW, Muhlebach TJ, Messner H, Orkin SH, Seger RA, Curnutte JT. Point mutation in the cytoplasmic domain of the neutrophil p22-phox cytochrome b subunit is associated with a nonfunctional NADPH oxidase and chronic granulomatous disease. Proc Natl Acad Sci USA. 1991;88:11231–11235. doi: 10.1073/pnas.88.24.11231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Patiño PJ, Rae J, Noack D, Erickson RW, Ding J, Garcia de Olarte D, Curnutte JT. Molecular characterization of autosomal recessive chronic granulomatous disease caused by a defect of the NADPH oxidase component p67-phox. Blood. 1999;94:2505–2514. [PubMed] [Google Scholar]
- 42.Noack D, Rae J, Cross AR, Munoz J, Salmen S, Mendoza JA, Rossi N, Curnutte JT, Heyworth PG. Autosomal recessive chronic granulomatous disease caused by novel mutations in NCF-2, the gene encoding the p67-phox component of phagocyte oxidase. Hum Genet. 1999;105:460–467. doi: 10.1007/s004399900152. [DOI] [PubMed] [Google Scholar]
- 43.Yu G, Hong DK, Dionis KY, Rae J, Heyworth PG, Curnutte JT, Lewis DB. The continuing diagnostic challenge of autosomal recessive chronic granulomatous disease. Clin Immunol. 2008;128:117–126. doi: 10.1016/j.clim.2008.05.008. [DOI] [PubMed] [Google Scholar]
- 44.Åhlin A, De Boer M, Roos D, Leusen J, Smith CIE, Sundin U, Rabbani H, Palmblad J, Elinder G. Prevalence, genetics and clinical presentation of chronic granulomatous disease in Sweden. Acta Paediatr. 1995;84:1386–1394. doi: 10.1111/j.1651-2227.1995.tb13575.x. [DOI] [PubMed] [Google Scholar]
- 45.Leusen JHW, De Klein A, Hilarius PM, Åhlin A, Palmblad J, Smith CIE, Diekmann D, Hall A, Verhoeven AJ, Roos D. Disturbed interaction of p21-rac with mutated p67-phox causes chronic granulomatous disease. J Exp Med. 1996;184:1243–1249. doi: 10.1084/jem.184.4.1243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Köker MY, Sanal Ö, van Leeuwen K, de Boer M, Metin A, Patıroğlu T, Özgür TT, Tezcan I, Roos D. Four different NCF2 mutations in six families from Turkey and an overview of NCF2 gene mutations. Eur J Clin Invest. 2009;39:942–951. doi: 10.1111/j.1365-2362.2009.02195.x. [DOI] [PubMed] [Google Scholar]
- 47.De Boer M, Hilarius-Stokman PM, Hossle JP, Verhoeven AJ, Graf N, Kenney RT, Seger R, Roos D. Autosomal recessive chronic granulomatous disease with absence of the 67-kD cytosolic NADPH oxidase component: identification of mutation and detection of carriers. Blood. 1994;83:531–536. [PubMed] [Google Scholar]
- 48.Tanugi-Cholley LC, Issartel JP, Lunardi J, Freycon F, Morel F, Vignais PV. A mutation located at the 5′ splice junction sequence of intron 3 in the p67-phox gene causes the lack of p67-phox mRNA in a patient with chronic granulomatous disease. Blood. 1995;85:242–249. [PubMed] [Google Scholar]
- 49.Khan HA, Good RA, Tangsinmankong N, Rae J, Noack D, Heyworth P, Day NK, Bahna S. P67-phox deficient chronic granulomatous disease due to heterozygous defects in exons 4 and 12 of the NCF2 gene. J Allergy Clin Immunol. 2002;109(Suppl. 1):S278. [Google Scholar]
- 50.Gentsch M, Kaczmarczyk A, van Leeuwen K, de Boer M, Kaus-Drobek M, Dagher MC, Kaiser P, Arkwright P, Gahr M, Rösen-Wolff A, Bochtler M, Secord E, Saifi M, Maddalena A, Dbaibo G, Bustamante J, Casanova JL, Roos D, Roesler J. Alu-repeat-induced deletions within the NCF2 gene cause p67-phox-deficient chronic granulomatous disease (CGD) Hum Mutat. 2009 Dec 1; doi: 10.1002/humu.21156. Epub ahead of print. [DOI] [PubMed] [Google Scholar]
- 51.Nunoi H, Iwata M, Tatsuzawa S, Onoe Y, Shimizu S, Kanegasaki S, Matsuda I. AG dinucleotide insertion in a patient with chronic granulomatous disease lacking cytosolic 67-kD protein. Blood. 1995;86:329–333. [PubMed] [Google Scholar]
- 52.Bonizzato A, Russo MP, Donini M, Dusi S. Identification of a double mutation (D160V-K161E) in the p67-phox gene of a chronic granulomatous disease patient. Biochem Biophys Res Commun. 1997;231:861–863. doi: 10.1006/bbrc.1997.6204. [DOI] [PubMed] [Google Scholar]
- 53.Al-Muhsen S, Al-Hemidan A, Al-Sheri A, Al-Harbi A, Al-Ghonaium A, Al-Saud B, Al-Mousa H, Al-Dhekri H, Arnaout R, Al-Mohsen I, Alsmadi O. Ocular manifestations in chronic granulomatous disease in Saoudi Arabia. J Am Ass Pediatr Ophtalmol Strabismus. 2009;13:396–399. doi: 10.1016/j.jaapos.2009.05.011. [DOI] [PubMed] [Google Scholar]
- 54.Borgato L, Bonizzato A, Lunardi C, Dusi S, Andrioli G, Scarperi A, Corrocher R. A 1.1-kb duplication in the p67-phox gene causes chronic granulomatous disease. Hum Genet. 2001;108:504–510. doi: 10.1007/s004390100526. [DOI] [PubMed] [Google Scholar]
- 55.Aoshima M, Nunoi H, Shimazu M, Shimizu S, Tatsuzawa O, Kenney RT, Kanegasaki S. Two-exon skipping due to a point mutation in p67-phox-deficient chronic granulomatous disease. Blood. 1996;88:1841–1845. [PubMed] [Google Scholar]
- 56.De Boer M, Singh V, Dekker J, Di Rocco M, Goldblatt D, Roos D. Prenatal diagnosis in two families with autosomal, p47phox-deficient chronic granulomatous disease due to a novel point mutation in NCF1. Prenatal Diagn. 2002;22:235–240. doi: 10.1002/pd.296. [DOI] [PubMed] [Google Scholar]
- 57.Jakobsen MA, Pedersen SS, Barington T. Detection of non-ΔGT NCF1 mutations in chronic granulomatous disease. Genet Test Mol Biomarkers. 2009;13:505–510. doi: 10.1089/gtmb.2009.0016. [DOI] [PubMed] [Google Scholar]
- 58.Kabuki T, Kawai T, Kin Y, Joh K, Ohashi H, Kosho T, Yachie A, Kanegane H, Miyawaki T, Oh-ishi T. A case of Williams syndrome with p47-phox-deficient chronic granulomatous disease. Nihon Rinsho Meneki Gakkai Kaishi. 2003;26:299–303. doi: 10.2177/jsci.26.299. [DOI] [PubMed] [Google Scholar]
- 59.Matute JD, Arias AA, Wright NAM, Wrobel I, Waterhouse CCM, Li XJ, Marchal CC, Stull ND, Lewis DB, Steele M, Kellner JD, Yu W, Meroueh SO, Nauseef WM, Dinauer MC. A new genetic subgroup of chronic granulomatous disease with autosomal recessive mutations in p40phox and selective defects in neutrophil NADPH oxidase activity. Blood. 2009 doi: 10.1182/blood-2009-07-231498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bedard K, Attar H, Bonnefont J, Jaquet V, Borel C, Plastre O, Stasia MJ, Antonarakis SE, Krause KH. Three common polymorphisms in the CYBA gene form a haplotype associated with decreased ROS formation. Hum Mutat. 2009;30:1123–1133. doi: 10.1002/humu.21029. [DOI] [PubMed] [Google Scholar]
- 61.Olsson LM, Lindqvist AK, Källberg H, Padyukov L, Burkhardt H, Alfredsson L, Klareskog L, Holmdahl R. A case-control study of rheumatoid arthritis identifies an associated single nucleotide polymorphism in the NCF4 gene, supporting a role for the NADPH-oxidase complex in autoimmunity. Arthritis Res Ther. 2007;9:R98. doi: 10.1186/ar2299. [DOI] [PMC free article] [PubMed] [Google Scholar]