Abstract
Breast cancer is a heterogeneous disease. Thanks to extensive efforts from research scientists and clinicians, treatment for breast cancer has advanced into the era of targeted medicine. With the use of several well-established biomarkers, such as hormone receptors (HRs) (i.e. estrogen receptor [ER] and progesterone receptor [PgR]) and Human Epidermal Growth Factor Receptor-2 (HER2), breast cancer patients can be categorized into multiple subgroups with specific targeted treatment strategies. Although therapeutic strategies for HR-positive (HR+) HER2-negative (HER2−) breast cancer and HR-negative (HR−) HER2-positive (HER2+) breast cancer are well-defined, HR+ HER2+ breast cancer is still an overlooked subgroup without tailored therapeutic options. In this review, we have summarized the molecular characteristics, etiology, preclinical tools and therapeutic options for HR+ HER2+ breast cancer. We hope to raise the attention of both the research and the medical community on HR+ HER2+ breast cancer, and to advance patient care for this subtype of disease.
Keywords: HR+ HER2+ breast cancer, ERα, HER2, the crosstalk between ER and HER2
1. Introduction
Since breast cancers are heterogeneous, targeted medicine improves the therapeutic effectiveness of breast cancer treatment care. Currently, the estrogen receptor (ER), progesterone receptor (PgR), Human Epidermal Growth Factor Receptor 2 (HER2) and Ki67 are used as routine biomarkers to diagnose and guide the use of adjuvant treatment options for breast cancer patients [1].
ER and PgR are grouped together into the class of hormone receptors (HRs). ER expression presents in approximately 75% of breast cancer patients [2]. ER-positive (ER+) breast cancers are susceptible to endocrine therapies (e.g., tamoxifen, aromatase inhibitors [AIs], luteinizing hormone releasing hormone analogues and bilateral oophorectomy), to interfere with ER signaling and/or block estrogen synthesis. A positive relationship between the ER Allred score and the response rate to endocrine therapies suggests that breast cancers patients with a higher ER level have better prognoses when they are subjected to endocrine therapies [3]. PgR expression is regulated through the transcriptional activity of ER. Therefore, PgR positivity is an indicator of ER signaling pathway integrity and good prognosis to endocrine therapy [4-6]. Ki67 is a nuclear protein that expresses throughout all phases of the cell cycle in the proliferating cells [7, 8]. High Ki67 expression is often associated with poor recurrence-free and disease-specific survival in HR-positive (HR+) breast cancers [9]. The Ki67 level can predict whether HR+ breast cancer patients would benefit from chemotherapeutic regimens: only high Ki67 tumors in ER+ patients receive clinical benefit from chemotherapy [10-12]. Also, the Ki67 level can be utilized as a pharmacodynamic endpoint to assess tumor response in a neoadjuvant setting.
Approximately 15-20% of breast carcinoma is HER2-positive (HER2+) [13]. Systemic treatment for HER2+ breast cancers involves a combination of chemotherapies and HER2-targeted blocking agents (e.g., trastuzumab, trastuzumab estansine [T-DM1], lapatinib and pertuzumab), regardless of HR status [14]. The Surveillance, Epidemiology and End Result Registry (SEER) conducted an assessment of incident rates of the major subtypes of breast cancer, based on 28% of the US population [15]. According to this study, 73% (36810) are HR+ HER2-negative (HER2−), 5% (2328) are HR-negative (HR−) HER2+, 12% (6912) are HR− HER2− and the remaining 10% (5240) are HR+ HER2+. Although it comprises one tenth of the total breast cancer population, the HR+ HER2+ subtype has drawn limited attention within the research community, compared to other subtypes.
Based on the transcriptional signature, breast carcinomas are classified as luminal A, luminal B, HER2-enriched or basal-like [16, 17]. HR+ carcinomas are classified as luminal type. This type is further subdivided into low-risk luminal A type, with high ER-regulated gene expression, and high-risk luminal B type, with low ER-regulated gene expression. In addition to ER activity, HER2 overexpression and high Ki67 proliferation index are also considered to be molecular characteristics of luminal B type [18]. The luminal A type is responsive to endocrine therapy, whereas the luminal B type is more aggressive and refractory to endocrine therapy. In luminal A breast cancers, the PgR positivity indicates that the ERα signaling pathway is active and predicts a good prognosis for endocrine therapy [4]. HER2+ and triple negative breast cancers (ER-negative [ER−] PgR-negative [PgR−] HER2− or HR− HER2−) are classified as HER2-enriched and basal-like, respectively. Gene expression profiling tools have become available commercially to provide predictive information to guide individualized treatment; these tools include the PAM50 (a 50-gene prediction analysis for microarrays), Oncotype DX (a 21-gene recurrence score) and MammaPrint (a 70-gene DNA microarray assay) [8, 19-22]. Despite the availability of comprehensive genetic profiling assays, the immunohistochemistry of ER, PgR, HER2 and Ki67 remains the gold standard to guide treatment plans.
HR+ HER2+ breast cancers are less characterized than other subtypes. About one eighth of the HR+ population are HER2+ and more than half of the HER2+ patients are HR+ [14, 15, 23]. Approximately 50% of luminal B type breast cancers are HR+ HER2+, which are more likely to be ER+ PgR− HER2+ [24, 25]. In clinical studies, the HR+ HER2+ subgroup displays a wider range of clinical endpoints (e.g., PFS and pCR) than other subtypes, implying the heterogeneity of this subtype. The five-year overall survival (OS) and disease free survival (DFS) of HR+ HER2+ patients are worse than that of HR+ HER2− but better than that of HR− HER2+ or HR− HER2− [19, 26, 27]. HR+ HER2+ breast cancers have higher chances of being diagnosed in younger populations and as higher-grade diseases, compared to other subtypes [15, 28]. These findings support the hypothesis that, not only the clinicopathological, but also biological characteristics of HR+ HER2+ tumors, are different from other subtypes of breast tumors. Therefore, HR+ HER2+ breast cancers should be recognized as a distinct subcategory of breast cancers. It is important to understand the molecular features of HR+ HER2+ breast cancers and tailor therapeutic strategies specific to this subtype.
In this review, we will summarize the molecular circuits and functional signaling units of the ER and HER2 signaling pathways. We will further illustrate how the coexistence of active ER and HER2 pathways affects the molecular characteristics of HR+ HER2+ breast cancers and will clarify the potential for drug resistance that occurs due to the signaling crosstalk between ER and HER2. In addition, we will describe preclinical studies that dissect the molecular features and etiology of this subtype of breast cancers and elaborate strategies for optimizing the treatment of HR+ HER2+ breast cancer patients.
2. Estrogen Receptor (ER)
2.1 ER signaling pathway
In ER+ breast cancer cells, estrogen stimulates cell proliferation and survival via the genomic and non-genomic action of ERs. These actions involve the two classical nuclear receptors, ERα and ERβ, as well as G-protein coupled receptor GPER, which is also as known as GPR-30 [29, 30]. ERα and ERβ are encoded by ESR1 and ESR2, respectively. Among the ERs, ERα is the major player contributing to breast malignancies, whereas ERβ plays a prominent role in prostate cancer [29, 31]. In breast tissue, ERβ tends to coexpress with ERα and suppresses its function in cell proliferation [30, 32]. GPER, located in the plasma membrane, binds to estrogen and involves in early estrogen signaling events [33, 34]. Several reports suggest GPER promotes cell migration and proliferation in breast carcinomas [35-38]. However, the function of GPR30 is still controversial and needs to be clarified. Herein, we will mainly focus on ERα.
ERα encodes a 66 kDa ligand-dependent nuclear receptor that contains one DNA-binding domain and two transcriptional activation function (AF) domains, AF1 and AF2 [39, 40]. AF2 contains the binding site for estrogen and is activated in response to ligand binding, whereas AF1 is estrogen ligand-independent and can be activated through phosphorylation by growth factors (e.g., epidermal growth factor [EGF] and insulin-like growth factor 1 [IGF-1]) [41, 42]. ERα contains both nuclear localization sequences and nuclear export sequences, suggesting that it functions both inside and outside of the nucleus [42]. In addition to the nucleus, the localization of ERα has also been documented in the plasma membrane, cytoplasm and mitochondria [43, 44].
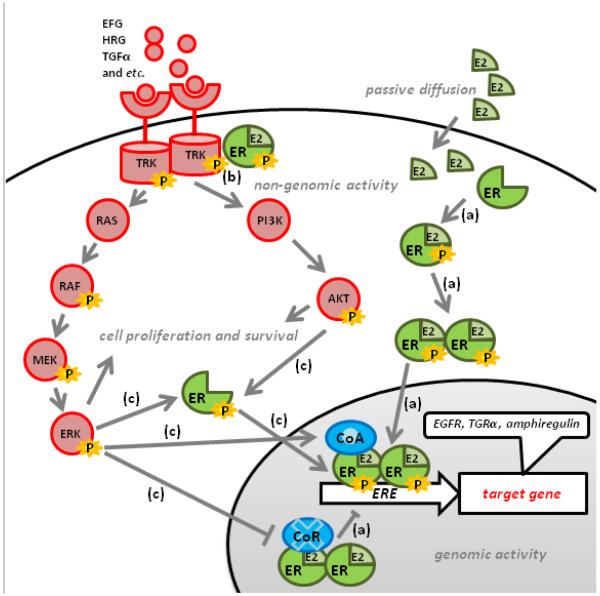
ERα function has been classified into two categories: genomic and non-genomic action. For its genomic action, ERα binds to estrogen and gets activated in the cytoplasm. After activation, ERα dimerizes and is translocated into the nucleus, where it binds to the estrogen response element (ERE) on its target genes, and activates and/or inhibits estrogen response genes (Fig. 1a). Ligand binding induces conformational changes in ERα that favor the recruitment of coregulators, coactivators and corepressors. ERα target genes include the following: growth factors, growth factor receptors, transcription factors (e.g., c-Myc, c-Fos and c-Jun) and cell cycle components (e.g., cyclin D1 and p21). These target genes are involved in promoting cell proliferation and survival [40, 42]. Also, ERα can be activated in an estrogen-independent manner, via its AF1 domain. For example, ERα can be phosphorylated and activated by mitogen-activated protein kinase (MAPK) through the induction of the growth factors EGF and IGF-1 (Fig. 1c) [45]. When ERα is activated in an estrogen-independent manner, it interacts with coactivators as well, and can subsequently activate target gene transcription (Fig. 1c). The transcriptional activity of ERα is modulated by its interaction with coactivators and corepressors and influenced by both the abundance of and the ratio between coactivators and corepressors (Fig. 1) [46-49]. In addition, ERα can also bind to activator protein-1 (AP-1) and specificity protein-1 (SP-1) to regulate genes that do not contain EREs, such as IGF1 [42, 50]. The non-genomic actions of ERα are typically more transient than the genomic actions [33]. ERα interacts with signaling components and complexes, including HER2, outside of the nucleus to regulate cell proliferation and survival (Fig. 1b) [51]. For example, in the presence of estrogen, the ERα interacts with the IGF1 receptor (IGF-1R), leading to the activation of IGF-1R and its downstream signaling via MAPK [52]. The interaction between ERα and other signaling molecules, as well as the activity of these signaling pathways in the cytoplasm, modulates ERα localization and activity [49, 51]. Post-translational modifications (e.g., phosphorylation, methylation, ubiquitination) to ERα and its signaling partners can also alter the dynamics of these signaling crosstalks.
Fig. 1. The signaling crosstalk between ERα and HER2 in HR+ HER2+ breast cancer.
(a) ERα binds to estrogen and gets activated in the cytoplasm. After activation, ERαs dimerize and are translocated into the nucleus, where they bind to the estrogen response element (ERE) on ERα target genes. The recruitment of coactivators (CoA) and corepressors (CoR) to ERα facilitates the transcriptional activity of ERα. The activated ERα induces the expression of growth factor receptor ligands, such as TGFα and amphiregulin, and membrane tyrosine receptor kinases (e.g., HER1 and HER2) to augment the activity of the HER2 signaling pathway. (b) The non-genomic action of ERα occurs transiently outside the nucleus. The estrogen-activated ERα in the plasma membrane and/or cytosol can interact with and activate HER2, and subsequently activate a downstream signaling cascade. In addition, the overexpression of HER2 sequesters ERα in the cytosol, which augments the non-genomic activity of ERα. (c)The downstream signaling output of the HER pathway diverges into two major signaling axes: PI3K/AKT (AKT signaling module) and RAS/RAF/MEK/ERK (MAPK signaling module). These two signaling pathways are involved in cell proliferation and survival. The activation of the AKT and MAPK signaling modules phosphorylates and activates ERα. Furthermore, the activation of ERK can enhance the function of CoAs (e.g., AIB1) but diminish the function of CoRs (e.g., SMRT).
In addition to the 66kDa ERα (ERα-66), ERα encodes another variant that is 36 kDa (ERα-36), which is expressed through alternative splicing and different promoter use. ERα-36 retains the DNA-binding domain and ligand-binding domain but lacks both of the transactivation domains [53]. It is predominantly localized in plasma membrane and cytosol, and functions at non-genomic level [53]. It antagonizes the function of ERα-66, resulting in resistance to endocrine therapies [54].
Genetic lesions (e.g., point mutations) also occur to ERα at low frequency during breast cancer malignancies. The ligand-binding domain AF2 is a mutation hotspot in ERα [55]. ERαK303R increases the binding affinity of ERα to its coactivators, conferring cells with hypersensitivity to estrogen and a proliferative advantage [56]. ERαY537N and ERαY537S exhibit constitutive transactivation activity in an estrogen-independent manner, resulting in endocrine resistance [57-59].
2.2 ER-targeted therapies
Estrogen is produced by aromatase, a cytochrome P450 superfamily member that catalyzes the conversion of testosterone to estradiol. Prior to menopause, the major internal source of estrogen is the ovary. After menopause, estrogen is mainly produced in nonglandular tissues, such as subcutaneous fat and plasma, and the overall estrogen level is reduced [60].
The first-line treatment for HR+ patients is endocrine therapy in both the adjuvant and metastatic situation. Endocrine therapies either block the ligand-binding action of ERα by acting as estrogen antagonists or deprive cells of estrogen input. Studies implicate that mutations and/or loss of ERα is one of the molecular causes of endocrine resistance [49].
In premenopausal women, the selective estrogen receptor modulator (SERM) tamoxifen (Nolvadex/Istubal/Valodex, AstraZeneca), acting as an estrogen antagonist, is widely used to interfere with ER function via disrupting the ligand-receptor interaction. However, tamoxifen also possesses estrogen agonist activity [61]. The interaction between tamoxifen and ERα leads to ERα dimerization and DNA binding, resulting in some ERα signaling activity. Furthermore, the presence of ERα coactivators and corepressors modulates the agonistic versus antagonistic activities of SERMs [46, 49].
In post-menopausal women, AIs are prescribed as a first-line treatment to inhibit the activity of aromatase and block estrogen production, thereby depriving cells of ERα signaling input. AIs are classified as steroidal inhibitors, such as exemestane (Aromasin, Pfizer), and non-steroidal inhibitors, such as letrozole (Femara, Novartis) and anastrozole (Arimidex, AstraZeneca) [62]. Typically, AIs have more influence on the genomic rather than the nongenomic activity of ERα.
Fulvestrant (ICI 182,780/Faslodex, AstraZeneca) is another approved estrogen antagonist and a so-called ‘pure’ anti-estrogen. It is currently utilized in advanced metastatic breast cancer patients who are not responsive to other endocrine therapies, such as AIs and tamoxifen [63]. The interaction of fulvestrant and ERα sequesters the ERα in the cytosol and triggers its premature degradation, which removes both the genomic and non-genomic activity of ERα rather than blocking estrogen signaling input.
3. Human Epidermal Growth Factor Receptor 2 (HER2)
3.1 HER1-4 family of receptor tyrosine kinase (RTK)
Overexpression and/or amplification of HER2, also known as HER2/neu or epidermal growth factor receptor ErbB-2, results in aggressive tumor growth and poor clinical outcomes [64]. HER2 encodes a transmembrane RTK that perceives extracellular signals (e.g., growth factors) to initiate a signaling cascade that mediates cell proliferation and survival.s RTK family members are generally composed of an extracellular ligand binding domain, a transmembrane domain and a functional intracellular tyrosine kinase domain. Together with other HER family members HER1 (EGF receptor, EGFR), HER3 and HER4, HER2 stimulates cell proliferation, inhibits apoptosis, enhances cellular adhesion and migration, and promotes angiogenesis [65]. Hetero- and/or homo-dimerization among these family members allows the binding of various peptide ligands, such as the HER ligand heregulin (HRG), EGF, Amphiregulin (AR) and transforming growth factor α (TGFα), leading to tyrosine phosphorylation and activation of the downstream signaling cascades (Fig. 1) [27, 66, 67]. Among these four RTK HERs, HER2 lacks the ligand binding domain, relying on its dimerization partners to perceive the signal, and HER3 lacks the intracellular kinase domain, relying on its dimerization partners for activation [8, 27].
The HER2-HER3 heterodimer plays a dominant role in cell proliferation and angiogenesis in HER2+ breast cancer [68]. The overexpression of HER2 and HER3 are highly correlated in breast cancer patient samples [66]. The function of HER1 is mainly documented in lung cancer rather than breast cancer [69]. In HER2+ breast cancer patients, HER1-positive (HER1+) patients have worse progression-free survival (PFS) and OS than HER1-negative (HER1−) patients, indicating the adverse effect of HER1 in HER2+ breast cancers [70]. The coexpression of HER1 and HER2 correlates with high-grade tumors [27]. In contrast to HER1, HER2 and HER3, which are all associated with poor prognosis, HER4 is associated with better prognosis in breast cancers [71].
3.2 HER2 signaling pathway
The downstream signaling output of the HER pathway diverges into two major signaling axes: 1) the phosphatidylinositol 3-kinase (PI3K)/AKT/mammalian target of rapamycin (mTOR) and 2) RAS/RAF/mitogen-activated and extracellular signal-regulated kinase kinase (MEK)/extracellular signal-regulated kinase (ERK) pathways (Fig. 1).
PI3K/AKT/mTOR signaling plays critical roles in protein synthesis, cell proliferation, cell survival and cellular metabolism [8]. It is also involved in endocrine resistance in breast cancer. The activation signals from RTK HERs are perceived by the central hub PI3K, which is composed of a regulatory subunit (p85) and a catalytic subunit (p110) (Fig.1) [72, 73]. The catalytic subunit has three isoforms p110α, p110β and p110δ, which are encoded by PIK3CA, PIK3CB and PIK3CD, respectively. PIK3CA mutations are common aberrations in breast carcinomas [8]. PI3K, in turn, phosphorylates the serine/threonine kinase AKT, which can be repressed by Phosphatase and TENsin homolog deleted on chromosome ten (PTEN), a tumor suppressor gene (Fig. 1) [8, 72, 74]. The loss of PTEN is another common event in breast cancer malignancies [75, 76]. Subsequently, the serine/threonine protein kinase mTOR serves as a downstream target of PI3K and AKT [77]. Functioning as the catalytic kinase subunit in two regulatory complexes, mTOR plays pivotal roles in cell metabolism, growth, proliferation and survival [78, 79]. mTORC1 contains mTOR, proline-rich AKT substrate 40 (PRAS40), Raptor, mLST8, and DEPTOR. AKT phosphorylates PRAS40 and also activates mTORC1 via tuberous sclerosis 1/2 (TSC1/2). mTORC1 mediates cell metabolism, cell size and protein translation and synthesis via its two major downstream substrates, 40S ribosomal protein S6 kinase 1 (S6K1) and eukaryotic inhibition factor 4E binding protein (4EBP1) [80]. mTORC2 contains mTORC, mLST8, Rictor and DEPTOR. Acting upstream, mTORC2 activates AKT [81]. Although the function of mTORC2 is not as well-studied as mTORC1, it has been shown the inhibition of mTORC2 affects cell metabolism and cancer cell growth [82].
Similarly, the activation signals from RTK HERs also emanate via the RAS/RAF/MEK/ERK signaling axis into the nucleus (Fig. 1) [83]. The oncoprotein RAS, a small GTPase, is one of the major players in growth factor-regulated cell survival in breast cancer and can link HER activation to cancer growth [84]. In response to activation, RAS facilitates the heterodimerization and activation of the intracellular kinase RAF to initiate the signaling cascade (Fig. 1). The phosphorylation of RAF transmits the signal to MEK1 and MEK2, which further activates ERK (Fig.1). ERK1 and ERK2, also known as MAPK3 and MAPK1, are translocated into the nucleus to initiate the transcriptional events of dysregulated cell cycle progression, proliferation, survival and invasion.
3.3 HER2-targeted therapies
Approximately 15-20% of breast cancer patients contain HER2 overexpression/amplification, which is associated with poor prognosis and a high recurrence rate and mortality [85]. The availability of HER2-targeted therapies has improved their clinical outcome. Currently, the combination of trastuzumab, pertuzumab, and a chemotherapy agent taxane is prescribed as the first-line treatment in advanced HER2+ breast cancer. T-DM1 is recommended as the second-line treatment. Lapatinib is used in the trastuzumab-exposed recurrent or metastatic breast cancer patients.
The humanized monoclonal antibody trastuzumab (Herceptin, Genentech) binds to the extracellular domain of HER2, which prevents dimerization, blocks the activation of its intracellular tyrosine kinase domain, reduces the cleavage of the extracellular domain of HER2, recruits immune effector cells and induces endocytosis for HER2 destruction [86]. Trastuzumab disrupts the ligand-independent HER2-HER3 complex and inhibits the activation of AKT, resulting in the inhibition of cell proliferation [87]. Trastuzumab-dependent inhibition on HER2-overexpression cell proliferation correlates with the deactivation of AKT and its downstream target PRAS40 [87]. The overexpression of HER1 is associated with poor overall survival in HER2+ metastatic breast cancer patients who received trastuzumab [88]. The loss of PTEN and presence of the PI3KCA activating mutations (E542K, E545K and H1047R) are tumorigenic and render cells trastuzumab resistant [89-91].
T-DM1 (Kadcyla, Genentech) is an antibody-drug conjugate (ADC), composed of trastuzumab, a stable thioether linker, and a cytotoxic drug DM-1 (a derivative of maytansine) [92]. It is also known as ado-trastuzumab emtansine. Trastuzumab allows T-DM1 to be delivered specifically to HER2 overexpression cells, while DM-1 enters and destroys the cells by targeting tubulin. Preclinical studies indicate actions from both trastuzumab and DM1 provide antitumor activity in trastuzumab- and lapatinib-insensitive research models. The specific cytotoxicity to HER2 overexpressing cells limits damages to normal cells, resulting in a lower toxicity profile and a broader therapeutic index for T-DM1 [92, 93].
Pertuzumab (Perjeta, Genentech), another FDA-approved humanized monoclonal antibody, binds to the extracellular domain of HER2 and inhibits ligand-dependent HER2-HER3 dimerization [64, 87, 94].
Lapatinib (Tykerb, GlaxoSmithKline) is a selective and reversible dual tyrosine kinase inhibitor against both HER1 and HER2, suppressing PI3K/AKT/mTOR and RAS/RAF/MEK/ERK signaling transmission [64]. Lapatinib treatment inhibits the phosphorylation of HER1, HER2 and HER3 as well as the activation of AKT and ERK [95]. It prevents the ubiquitination of HER2, accumulates HER2 on the cell surface, stabilizes its dimerization and inhibits its activation. The accumulation of inactive HER1 dimers by lapatinib could sequester ligands without signaling activation [96].
In preclinical studies, the sensitivity of trastuzumab-resistant cells to lapatinib suggests that lapatinib behaves more efficiently than trastuzumab to suppress the activation of HER2 signaling pathways [97, 98]. However, trastuzumab is the current major HER2 blocking agent in the clinic. The use of individual HER inhibitors often results in incomplete blockade of downstream signaling and drug resistance because of their selective actions on specific HER dimmers [99]. Therefore, a combination of inhibitors targeted to multi-HER inhibition and/or downstream signaling hubs would produce better clinical outcomes. The combination of lapatinib and trastuzumab synergistically inhibits tumor growth in vitro, implying a complementary mechanism of action between trastuzumab and lapatinib [96]. One explanation is that the accumulation of HER2 by lapatinib enhances trastuzumab-dependent cellular cytotoxicity [96]. Clinical studies, such as the NeoALTTO trial, suggest that the dual HER2-targeted therapy, the combination of trastuzumab and lapatinib, provides better clinical outcomes [100].
4. The signaling cross-talk between ERα and HER2 in HR+ HER2+ breast cancers
ERα action can regulate the activity of the HER2 signaling pathway at multiple levels: 1) At the genomic level, activated ERα induces the expression of growth factor receptor ligands, such as TGFα and AR, and RTK HERs (e.g., HER1 and HER2) to augment the activity of the HER2 signaling pathway (Fig. 1a) [42, 101, 102]. 2) At the non-genomic level, the estrogen-activated membrane ERα can interact with and activate HER2 to initiate the downstream signaling cascade (Fig. 1b) [51, 103, 104]. At the same time, HER2 signaling pathways regulate ERα via several manners: 1) EGF reduces ERα expression via HER1 [105]. 2) Through growth factor signaling pathways, ERα can also be activated in an estrogen-independent manner: the activation of AKT and MAPK signaling modules can phosphorylate and activate ERα (Fig. 1c). 3) Furthermore, the activation of MAPK can activate the ERα coactivator AIB1. On the other hand, the activation of MAPK via HER1 also reduces the binding affinity of the ERα corepressor SMRT to its nuclear receptor ERα and sequesters SMRT in the cytoplasm (Fig. 1c) [106]. These post-translational modifications enhance the genomic activity of ERα but reduce its dependency on estrogen [107-109]. 4) In addition, the overexpression of HER2 sequesters ERα in the cytosol, which augments the non-genomic activity of ERα (Fig. 1) [51, 110]. Therefore, the crosstalk between ERα and HER2 signaling forms a positive-feedback molecular circuit to synergize the HR+ HER2+ tumor growth [40].
Overexpression of HER2 promotes tamoxifen resistance and the addition of trastuzumab can resensitize the tamoxifen response [111, 112]. It has been shown that there are concurrent high levels of HER2 and AIB1, ERα coactivator, in the tamoxifen-resistant tumors [113]. Also, the long-term acquired resistance to fulvestrant, an ERα antagonist, results in elevated HER1 signaling and MAPK activation [114]. Furthermore, the activation of RTK HER signaling pathways releases cell growth from the dependence on estrogen by reducing the expression of ER and/or activating ER without estrogen, leading to endocrine resistance [49]. The activation of p70S6K, the downstream target of PI3K/AKT/mTOR, and RAS/MAPK, has also been suggested to correlate with tamoxifen resistance [115].
On the other hand, alteration of the ERα signaling pathway also plays a role in de novo and acquired resistance to HER2-targeted regimens [98]. Several groups have demonstrated that the therapeutic usage of HER2 blocking agents, such as lapatinib, upregulates ERα, the ERα regulators FOXO3a and caveolin1, and ER-regulated genes, such as PgR and Bcl-2, in HR+ HER2+ cell lines [98, 116]. Once lapatinib resistance is established, lapatinib can still exert inhibitory effects on the activation of RTK HERs but fails to decrease the phosphorylation of AKT and ERK [95]. In a lapatinib-resistant cell line harboring functional HRs, the expression of ER and PgR are elevated, compared to the parental cell lines. Resistance to lapatinib is associated with the upregulation of FOXO3a and the ERα coactivator AIB1. The acquired lapatinib resistance resensitizes the cells to fulvestrant. Meanwhile, estrogen deprivation and fulvestrant resensitize the cells to lapatinib. It suggests that cells can rely on ERs to escape the inhibitory effect of HER2 blocking agents when the HER2 signaling pathway is handicapped [98]. Wang et al. report that the ER signaling pathway is initially utilized as an escape pathway for cell proliferation in response to the establishment of lapatinib resistance. However, the HER2 pathway can be reactivated in later stages of lapatinib resistance and dominate the growth of resistant cells [98].
Considering the key roles of ER-mediated and HER2-mediated pathways in breast cancers, the bi-directional signaling crosstalk between these two pathways has been suggested to provide escape mechanisms for either endocrine therapies or HER2-targeted therapies in HR+ HER2+ breast cancers, resulting in both de novo and acquired resistance to these treatments [49, 95, 98, 107, 116]. The combination of different regimens targeting different signaling pathways prolongs the development of resistance and inhibits cancer recurrence. Therefore, it is critical to block this signaling crosstalk by targeting the ER and HER2 pathway simultaneously in order to reach the optimal therapeutic efficacy for HR+ HER2+ breast cancers.
5. Preclinical tools for HR+ HER2+ breast cancers
The low success rate for translating research hypotheses into clinical success has been a problematic issue. Oncology drug discovery has one of the highest failure rates on clinical trials among all the therapeutic areas [117]. So far, approximately only one-third of the drugs tested in preclinical models show activity in phase II clinical trials [118]. In order to deliver accurate preclinical data to support late stage clinical trials, early findings should be reproduced in multiple research models. To tailor therapeutic treatment strategies to HR+ HER2+ breast cancer patients, adequate preclinical cell lines and suitable animal models would help advance our understanding of the biology of HR+ HER2+ breast tumors and test the therapeutic efficacy of treatment regimen candidates.
The utilization of in vitro cell lines can facilitate molecular dissection to identify druggable targets via various molecular, genetic and bioinformatic techniques. The common cell lines utilized in the context of HR+ HER2+ breast cancer preclinical study include MDA-MB-361, MCF7 HER2 and BT474. Unsurprisingly, each cell line responds to therapeutic regimens in unique ways, due to the differences in their composition of molecular signaling pathways [87, 98, 111, 119-121]. The identification of additional suitable in vitro cell lines would provide a better preclinical research system that represents HR+ HER2+ breast cancers in the clinic. Although all three cell lines mentioned above can form xenografts in immunodeficient mice, tumor heterogeneity cannot be recapitulated with these tools [98, 122].
Recently, the application of patient-derived xenografts (PDXs) in preclinical studies has begun to open the door to mimicking human disease on the bench. Breast cancer PDXs are generated by engrafting patient surgical resections and/or biopsies of breast tumor tissue into immunodeficient mice. PDX models not only recapitulate the tumor heterogeneity but also reflect the genetic, molecular and histological characteristics of the original tumors from patients [57, 123, 124]. Therefore, the PDX model is a powerful tool to test drug efficacy in the preclinical setting before human clinical trials. However, breast cancer PDXs with ER expression are difficult to establish [125]. So far, few groups have successfully used it as a tool to test anticancer agents in HR+ HER2+ breast cancers.
6. The clinical implications of HR+ HER2+ breast cancers
Regardless of HR status, patients with HER2 overexpression/amplification are subjected to a combination of HER2-targeted therapies and chemotherapy as the standard first-line therapy [14, 126]. Clinical studies of NeoALTTO, TBCRC and NeoSphere have reported that HR+ HER2+ patients have a lower pathological complete response (pCR) rate to neoadjuvant anti-HER2 agents plus chemotherapy, compared to HR− HER2+ patients [100, 127, 128]. These results reinforce the need for improvements to the current standard treatment strategy for HR+ HER2+ breast cancers. In agreement with preclinical data showing that HER2 overexpression induces endocrine resistance in ER+ cells, HER2 expression has adverse effects on ER+ breast cancer patients treated with endocrine therapies alone, such as tamoxifen and letrozole [129-131]. The rationale for applying endocrine therapies plus HER2 blocking agents in HR+ HER2+ breast cancers is supported by preclinical observations that either the ERα or HER2 signaling pathway can initially drive cell proliferation in HR+ HER2+ breast cancers [98].
In the phase II clinical trial TBCRC006, some of the ER+ HER2+ patients derived clinical benefit from neoadjuvant treatment of using a combination of lapatinib, trastuzumab and letrozole, suggesting that the simultaneous eradication of the HER2 and ERα pathways inhibits ER+ HER2+ tumor growth [132]. The phase III trial TAnDEM has been conducted to evaluate the therapeutic efficacy of a combination of hormonal agents and trastuzumab without chemotherapy in HR+ HER2+ metastatic breast cancers. The result shows postmenopausal women receiving a combination of trastuzumab and anastrozole have better PFS than groups that received anastrozole alone [133]. Another phase III trial also indicates that the addition of lapatinib to letrozole leads to better PFS in HR+ HER2+ breast cancers in a metastatic setting [134]. However, the OS was not significantly improved by the combination of hormonal agents and HER2-blocking agents in these studies. Based on these results, Giordano et al. suggested that a combination of endocrine therapies and HER2 blocking agents may be an option for HR+ HER2+ patients [14]. Although the combination of endocrine therapies and HER2+ targeting agents has an improved toxicity profile, the use of HER2 blocking agents plus chemotherapy is still preferred for HR+ HER2+ patients, due to its OS benefit. Until now, there has been no available clinical trial addressing whether the replacement of chemotherapy with endocrine therapies in the standard treatment provides a favorable clinical benefit to HR+ HER2+ breast cancer patients.
In luminal B type breast cancers, the survival benefits, such as breast cancer-related survival (BCS) and OS, for ER+ PgR− HER2+ cancers are lower, compared to ER+ PgR positive (PgR+) HER2+ cancers, suggesting the prognostic value of PgR in luminal B type [28]. Studies suggest that the hyperactivation of growth factor signaling pathways, through PI3K/AKT/mTOR, deactivates PgR expression [135]. This speculation is consistent with the finding by Arpino et al. showing that HR+ HER2+ breast cancers have higher chances of being PgR− [25]. In HR+ HER2+ patients treated with tamoxifen, poor clinical outcome correlates with a high expression level of HER2 and/or HER1 in the ER+/PR− subgroup but not the ER+/PR+ group, suggesting the PgR negativity is an indicator of the dominant role of HER1/HER2 pathway [25, 39]. On the other hand, Vici et al. propose that ER+ PgR+ HER2+ tumors with high expression of HRs behave similarly to the HR+ HER2− subtype in the clinic, suggesting that certain HR+ HER2+ patients may be over-treated by the standard treatment [136]. Therefore, the expression levels of ER and/or PgR may serve as predictive biomarkers to guide the choice of treatment regimens and predict prognosis in HR+ HER2+ breast cancers.
7. Perspectives on HR+ HER2+ breast cancers
Although the combination of endocrine therapies and HER2 blocking agents is recommended for the HR+ HER2+ subtype, the standard treatment regimen that uses HER2 blocking agents plus chemotherapy is preferred in the current clinical setting [14]. In HR+ HER2+ breast cancer, the crosstalk between the ER and HER2 signaling pathway results in new regulatory mechanisms that require targeted treatment. Major concerns have been raised regarding the biology and treatment of HR+ HER2+ breast cancers:
a) Does HR+ HER2+ breast cancer behave uniquely in the clinic compared to other well-defined subtypes? According to epidemiology studies and treatment prognosis reports, HR+ HER2+ breast cancers are distinguishable from other subtypes. The biology of this subtype is not simply the combined phenotypes of HR+ HER2− and HR−HER2+ cancers. The signaling crosstalk between ER and HER2 grants a positive-feedback molecular circuit to synergize the HR+ HER2+ tumor growth.
b) Should the HR+ HER2+ subtype be treated differently from HR− HER2+ patients, considering the active ER signaling pathway? Does the paradigm of chemotherapy plus HER2-targeted regimens provide the best clinical outcomes for all HR+ HER2+ patients? Using the standard treatment, HR+ HER2+ patients end up with worse prognoses, as compared to their HR− HER2+ counterparts. The crosstalk between ER and HER2 signaling desensitizes either endocrine therapies or HER2-targeted therapies alone in HR+ HER2+ breast cancers, suggesting that interventions targeting both pathways simultaneously could improve clinical outcome for HR+ HER2+ patients.
c) Is there a subgroup of HR+ HER2+ patients who are over-treated using the standard paradigm? Can the expression levels of ER and/or PgR serve as predictive indicators to guide the choice of treatment regimens and predict prognosis? HR+ HER2+ patients with higher ER expression are less sensitive to chemotherapy or HER2 blocking agents [137]. Vici et al. suggests that HR+ HER2+ with high ERα expression may be treatable with endocrine therapy alone. The addition of HER2 targeted agents may not provide additional clinical benefit. On the other hand, the PgR positivity suggests the less dominant role of HER2 signaling pathway in HR+ HER2+ breast cancers, which suggests the addition of endocrine therapies rather than chemotherapies to HER2 blocking agents can be beneficial for ER+ PgR+ HER2+ breast cancer patients. Therefore, the expression level of PgR can be considered as a benchmark of whether HR+ HER2+ patients should be subjected to the combination of endocrine therapies and HER2 blocking agents or the combination of chemotherapies and HER2 blocking agents. Evaluation of both the ER and the PgR level may avoid overtreatment of patients. Therefore, there is an urgent need to define suitable criteria, based on the expression levels of ER and PgR. This will allow clinicians to identify the subset of HR+ HER2+ patients that should receive a combination of endocrine therapies and HER2-targeted therapies or even endocrine therapy alone, instead of the standard treatment. In addition, due to the plasticity and heterogeneity of breast cancer, the alteration of the status of ER, PgR and HER2 may occur, as tumors progress [1]. These changes should also be taken into consideration for fine-tuning therapeutic strategies in HR+ HER2+ breast cancers.
d) Are there any additional potential druggable targets for HR+ HER2+ breast cancers? The activation of PI3K/AKT/mTOR has a prominent role in resistance to both endocrine therapies and HER2 blocking agents. Preclinical studies have shown PI3K/AKT inhibitors have inhibitory effects on cells with acquired resistance to either endocrine therapies or HER2-targeted therapies, suggesting that activation of PI3K/AKT/mTOR may serve as a common escape mechanism for endocrine therapies and HER2 blocking agents. Also, the PI3K/AKT/mTOR pathway may play a role as the downstream signal hub in the ER and HER2 signaling crosstalk. Therefore, in addition to the combination of endocrine therapy and HER2-targeted therapy, drugs interfering with the pathways downstream of this cross-talk may also benefit HR+ HER2+ patients.
Therefore, the identification of additional diagnostic and prognostic biomarkers as well as potential druggable targets could further facilitate the formulation of personalized treatments for this HR+ HER2+ group, which is 10% of the total breast cancer patient population.
Acknowledgements
We thank Dr. Nancy Linford providing us suggestions on manuscript writing and proofreading. The research was supported by the City of Hope Excellence Award and the National Cancer Institute (P30 CA033572).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Reference
- 1.Karlsson E, et al. Breast cancer during follow-up and progression - A population based cohort on new cancers and changed biology. Eur J Cancer. 2014 doi: 10.1016/j.ejca.2014.08.014. [DOI] [PubMed] [Google Scholar]
- 2.Anderson WF, et al. Estrogen receptor breast cancer phenotypes in the Surveillance, Epidemiology, and End Results database. Breast Cancer Res Treat. 2002;76(1):27–36. doi: 10.1023/a:1020299707510. [DOI] [PubMed] [Google Scholar]
- 3.Ellis MJ, et al. Letrozole is more effective neoadjuvant endocrine therapy than tamoxifen for ErbB-1− and/or ErbB-2-positive, estrogen receptor-positive primary breast cancer: evidence from a phase III randomized trial. J Clin Oncol. 2001;19(18):3808–16. doi: 10.1200/JCO.2001.19.18.3808. [DOI] [PubMed] [Google Scholar]
- 4.Bardou VJ, et al. Progesterone receptor status significantly improves outcome prediction over estrogen receptor status alone for adjuvant endocrine therapy in two large breast cancer databases. J Clin Oncol. 2003;21(10):1973–9. doi: 10.1200/JCO.2003.09.099. [DOI] [PubMed] [Google Scholar]
- 5.Clark GM, et al. Progesterone receptors as a prognostic factor in Stage II breast cancer. N Engl J Med. 1983;309(22):1343–7. doi: 10.1056/nejm198312013092240. [DOI] [PubMed] [Google Scholar]
- 6.Cui X, et al. Biology of progesterone receptor loss in breast cancer and its implications for endocrine therapy. J Clin Oncol. 2005;23(30):7721–35. doi: 10.1200/JCO.2005.09.004. [DOI] [PubMed] [Google Scholar]
- 7.Gerdes J, et al. Production of a mouse monoclonal antibody reactive with a human nuclear antigen associated with cell proliferation. Int J Cancer. 1983;31(1):13–20. doi: 10.1002/ijc.2910310104. [DOI] [PubMed] [Google Scholar]
- 8.Kourea HP, Zolota V, Scopa CD. Targeted pathways in breast cancer: molecular and protein markers guiding therapeutic decisions. Curr Mol Pharmacol. 2015;7(1):4–21. doi: 10.2174/187446720701150105170830. [DOI] [PubMed] [Google Scholar]
- 9.Cheang MC, et al. Ki67 index, HER2 status, and prognosis of patients with luminal B breast cancer. J Natl Cancer Inst. 2009;101(10):736–50. doi: 10.1093/jnci/djp082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nitz U, et al. Final analysis of the prospective WSG-AGO EC-Doc versus FEC phase III trial in intermediate-risk (pN1) early breast cancer: efficacy and predictive value of Ki67 expression. Ann Oncol. 2014;25(8):1551–7. doi: 10.1093/annonc/mdu186. [DOI] [PubMed] [Google Scholar]
- 11.Ellis MJ, et al. Outcome prediction for estrogen receptor-positive breast cancer based on postneoadjuvant endocrine therapy tumor characteristics. J Natl Cancer Inst. 2008;100(19):1380–8. doi: 10.1093/jnci/djn309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dowsett M, et al. Assessment of Ki67 in breast cancer: recommendations from the International Ki67 in Breast Cancer working group. J Natl Cancer Inst. 2011;103(22):1656–64. doi: 10.1093/jnci/djr393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wolff AC, et al. Recommendations for human epidermal growth factor receptor 2 testing in breast cancer: American Society of Clinical Oncology/College of American Pathologists clinical practice guideline update. J Clin Oncol. 2013;31(31):3997–4013. doi: 10.1200/JCO.2013.50.9984. [DOI] [PubMed] [Google Scholar]
- 14.Giordano SH, et al. Systemic therapy for patients with advanced human epidermal growth factor receptor 2-positive breast cancer: American Society of Clinical Oncology clinical practice guideline. J Clin Oncol. 2014;32(19):2078–99. doi: 10.1200/JCO.2013.54.0948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Howlader N, et al. US Incidence of Breast Cancer Subtypes Defined by Joint Hormone Receptor and HER2 Status. J Natl Cancer Inst. 2014;106(5) doi: 10.1093/jnci/dju055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Perou CM, et al. Molecular portraits of human breast tumours. Nature. 2000;406(6797):747–52. doi: 10.1038/35021093. [DOI] [PubMed] [Google Scholar]
- 17.Sorlie T, et al. Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc Natl Acad Sci U S A. 2001;98(19):10869–74. doi: 10.1073/pnas.191367098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang MH, et al. Estrogen receptor-positive breast cancer molecular signatures and therapeutic potentials (Review) Biomed Rep. 2014;2(1):41–52. doi: 10.3892/br.2013.187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Parker JS, et al. Supervised risk predictor of breast cancer based on intrinsic subtypes. J Clin Oncol. 2009;27(8):1160–7. doi: 10.1200/JCO.2008.18.1370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Paik S, et al. A multigene assay to predict recurrence of tamoxifen-treated, node-negative breast cancer. N Engl J Med. 2004;351(27):2817–26. doi: 10.1056/NEJMoa041588. [DOI] [PubMed] [Google Scholar]
- 21.Nielsen TO, et al. A comparison of PAM50 intrinsic subtyping with immunohistochemistry and clinical prognostic factors in tamoxifen-treated estrogen receptor-positive breast cancer. Clin Cancer Res. 2010;16(21):5222–32. doi: 10.1158/1078-0432.CCR-10-1282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.van de Vijver MJ, et al. A gene-expression signature as a predictor of survival in breast cancer. N Engl J Med. 2002;347(25):1999–2009. doi: 10.1056/NEJMoa021967. [DOI] [PubMed] [Google Scholar]
- 23.Konecny G, et al. Quantitative association between HER-2/neu and steroid hormone receptors in hormone receptor-positive primary breast cancer. J Natl Cancer Inst. 2003;95(2):142–53. doi: 10.1093/jnci/95.2.142. [DOI] [PubMed] [Google Scholar]
- 24.Parise CA, Caggiano V. Breast Cancer Survival Defined by the ER/PR/HER2 Subtypes and a Surrogate Classification according to Tumor Grade and Immunohistochemical Biomarkers. J Cancer Epidemiol. 2014;2014:469251. doi: 10.1155/2014/469251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Arpino G, et al. Estrogen receptor-positive, progesterone receptor-negative breast cancer: association with growth factor receptor expression and tamoxifen resistance. J Natl Cancer Inst. 2005;97(17):1254–61. doi: 10.1093/jnci/dji249. [DOI] [PubMed] [Google Scholar]
- 26.Onitilo AA, et al. Breast cancer subtypes based on ER/PR and Her2 expression: comparison of clinicopathologic features and survival. Clin Med Res. 2009;7(1-2):4–13. doi: 10.3121/cmr.2009.825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Witton CJ, et al. Expression of the HER1-4 family of receptor tyrosine kinases in breast cancer. J Pathol. 2003;200(3):290–7. doi: 10.1002/path.1370. [DOI] [PubMed] [Google Scholar]
- 28.Cancello G, et al. Progesterone receptor loss identifies Luminal B breast cancer subgroups at higher risk of relapse. Ann Oncol. 2013;24(3):661–8. doi: 10.1093/annonc/mds430. [DOI] [PubMed] [Google Scholar]
- 29.Nass N, Kalinski T. Tamoxifen resistance: From cell culture experiments towards novel biomarkers. Pathol Res Pract. 2015 doi: 10.1016/j.prp.2015.01.004. [DOI] [PubMed] [Google Scholar]
- 30.Musgrove EA, Sutherland RL. Biological determinants of endocrine resistance in breast cancer. Nat Rev Cancer. 2009;9(9):631–43. doi: 10.1038/nrc2713. [DOI] [PubMed] [Google Scholar]
- 31.Christoforou P, Christopoulos PF, Koutsilieris M. The role of estrogen receptor beta in prostate cancer. Mol Med. 2014;20:427–34. doi: 10.2119/molmed.2014.00105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hayashi SI, et al. The expression and function of estrogen receptor alpha and beta in human breast cancer and its clinical application. Endocrine-Related Cancer. 2003;10(2):193–202. doi: 10.1677/erc.0.0100193. [DOI] [PubMed] [Google Scholar]
- 33.Cato AC, Nestl A, Mink S. Rapid actions of steroid receptors in cellular signaling pathways. Sci STKE. 2002;2002(138):re9. doi: 10.1126/stke.2002.138.re9. [DOI] [PubMed] [Google Scholar]
- 34.Gao F, et al. GPR30 activation opposes estrogen-dependent uterine growth via inhibition of stromal ERK1/2 and estrogen receptor alpha (ERalpha) phosphorylation signals. Endocrinology. 2011;152(4):1434–47. doi: 10.1210/en.2010-1368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lucki NC, Sewer MB. Genistein stimulates MCF-7 breast cancer cell growth by inducing acid ceramidase (ASAH1) gene expression. J Biol Chem. 2011;286(22):19399–409. doi: 10.1074/jbc.M110.195826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kang K, et al. Tectoridin, a poor ligand of estrogen receptor alpha, exerts its estrogenic effects via an ERK-dependent pathway. Mol Cells. 2009;27(3):351–7. doi: 10.1007/s10059-009-0045-8. [DOI] [PubMed] [Google Scholar]
- 37.Pupo M, et al. Bisphenol A induces gene expression changes and proliferative effects through GPER in breast cancer cells and cancer-associated fibroblasts. Environ Health Perspect. 2012;120(8):1177–82. doi: 10.1289/ehp.1104526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Scaling AL, Prossnitz ER, Hathaway HJ. GPER mediates estrogen-induced signaling and proliferation in human breast epithelial cells and normal and malignant breast. Horm Cancer. 2014;5(3):146–60. doi: 10.1007/s12672-014-0174-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Arpino G, et al. Crosstalk between the estrogen receptor and the HER tyrosine kinase receptor family: molecular mechanism and clinical implications for endocrine therapy resistance. Endocr Rev. 2008;29(2):217–33. doi: 10.1210/er.2006-0045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schiff R, et al. Cross-talk between estrogen receptor and growth factor pathways as a molecular target for overcoming endocrine resistance. Clin Cancer Res. 2004;10:331S–6S. doi: 10.1158/1078-0432.ccr-031212. 1 Pt 2. [DOI] [PubMed] [Google Scholar]
- 41.Ali S, et al. Modulation of transcriptional activation by ligand-dependent phosphorylation of the human oestrogen receptor A/B region. EMBO J. 1993;12(3):1153–60. doi: 10.1002/j.1460-2075.1993.tb05756.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lee AV, Cui X, Oesterreich S. Cross-talk among estrogen receptor, epidermal growth factor, and insulin-like growth factor signaling in breast cancer. Clin Cancer Res. 2001;7(12 Suppl):4429s–4435s. discussion 4411s-4412s. [PubMed] [Google Scholar]
- 43.Levin ER, Pietras RJ. Estrogen receptors outside the nucleus in breast cancer. Breast Cancer Res Treat. 2008;108(3):351–61. doi: 10.1007/s10549-007-9618-4. [DOI] [PubMed] [Google Scholar]
- 44.Razandi M, et al. Proximal events in signaling by plasma membrane estrogen receptors. J Biol Chem. 2003;278(4):2701–12. doi: 10.1074/jbc.M205692200. [DOI] [PubMed] [Google Scholar]
- 45.Kato S. Estrogen receptor-mediated cross-talk with growth factor signaling pathways. Breast Cancer. 2001;8(1):3–9. doi: 10.1007/BF02967472. [DOI] [PubMed] [Google Scholar]
- 46.Schiff R, et al. Breast cancer endocrine resistance: how growth factor signaling and estrogen receptor coregulators modulate response. Clin Cancer Res. 2003;9:447S–54S. 1 Pt 2. [PubMed] [Google Scholar]
- 47.Chen JD, Evans RM. A transcriptional co-repressor that interacts with nuclear hormone receptors. Nature. 1995;377(6548):454–7. doi: 10.1038/377454a0. [DOI] [PubMed] [Google Scholar]
- 48.McKenna NJ, Lanz RB, O'Malley BW. Nuclear receptor coregulators: cellular and molecular biology. Endocr Rev. 1999;20(3):321–44. doi: 10.1210/edrv.20.3.0366. [DOI] [PubMed] [Google Scholar]
- 49.Osborne CK, Schiff R. Mechanisms of endocrine resistance in breast cancer. Annu Rev Med. 2011;62:233–47. doi: 10.1146/annurev-med-070909-182917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kushner PJ, et al. Estrogen receptor pathways to AP-1. J Steroid Biochem Mol Biol. 2000;74(5):311–7. doi: 10.1016/s0960-0760(00)00108-4. [DOI] [PubMed] [Google Scholar]
- 51.Yang Z, Barnes CJ, Kumar R. Human epidermal growth factor receptor 2 status modulates subcellular localization of and interaction with estrogen receptor alpha in breast cancer cells. Clin Cancer Res. 2004;10(11):3621–8. doi: 10.1158/1078-0432.CCR-0740-3. [DOI] [PubMed] [Google Scholar]
- 52.Kahlert S, et al. Estrogen receptor alpha rapidly activates the IGF-1 receptor pathway. J Biol Chem. 2000;275(24):18447–53. doi: 10.1074/jbc.M910345199. [DOI] [PubMed] [Google Scholar]
- 53.Su X, et al. ER-alpha36: a novel biomarker and potential therapeutic target in breast cancer. Onco Targets Ther. 2014;7:1525–33. doi: 10.2147/OTT.S65345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhang X, Wang ZY. Estrogen receptor-alpha variant, ER-alpha36, is involved in tamoxifen resistance and estrogen hypersensitivity. Endocrinology. 2013;154(6):1990–8. doi: 10.1210/en.2013-1116. [DOI] [PubMed] [Google Scholar]
- 55.Baselga MP, Rugo Hope, Chen David, Burris Howard A., Campone Mario, Noguchi Shinzaburo, Perez Alejandra, Deleu Inas, Shtivelband Mikhail, Provencher Louise, Derti Adnan, Huang Alan, McDonald Rob, Kalfoglou Creton, Robinson Douglas, Taran Tetiana, Sahmoud Tarek, Lebwohl David, Hortobagyi Gabriel N. Assessment of genetic alterations using next-generation sequencing in postmenopausal women with hormone receptor-positive, HER2-negative advanced breast cancer: results from the BOLERO-2 phase III trial. Ann. Oncol. 2013;24:iii25–iii26. [Google Scholar]
- 56.Fuqua SA, et al. A hypersensitive estrogen receptor-alpha mutation in premalignant breast lesions. Cancer Res. 2000;60(15):4026–9. [PubMed] [Google Scholar]
- 57.Li S, et al. Endocrine-therapy-resistant ESR1 variants revealed by genomic characterization of breast-cancer-derived xenografts. Cell Rep. 2013;4(6):1116–30. doi: 10.1016/j.celrep.2013.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tremblay GB, et al. Ligand-independent activation of the estrogen receptors alpha and beta by mutations of a conserved tyrosine can be abolished by antiestrogens. Cancer Res. 1998;58(5):877–81. [PubMed] [Google Scholar]
- 59.Zhang QX, et al. An estrogen receptor mutant with strong hormone-independent activity from a metastatic breast cancer. Cancer Res. 1997;57(7):1244–9. [PubMed] [Google Scholar]
- 60.Mauriac L, Smith I. Aromatase inhibitors in early breast cancer treatment. Semin Oncol. 2003;30(4 Suppl 14):46–57. doi: 10.1016/s0093-7754(03)00304-x. [DOI] [PubMed] [Google Scholar]
- 61.Frasor J, et al. Selective estrogen receptor modulators: discrimination of agonistic versus antagonistic activities by gene expression profiling in breast cancer cells. Cancer Res. 2004;64(4):1522–33. doi: 10.1158/0008-5472.can-03-3326. [DOI] [PubMed] [Google Scholar]
- 62.Johnston SR, Dowsett M. Aromatase inhibitors for breast cancer: lessons from the laboratory. Nat Rev Cancer. 2003;3(11):821–31. doi: 10.1038/nrc1211. [DOI] [PubMed] [Google Scholar]
- 63.Woode DR, et al. Effect of Berry Extracts and Bioactive Compounds on Fulvestrant (ICI 182,780) Sensitive and Resistant Cell Lines. Int J Breast Cancer. 2012;2012:147828. doi: 10.1155/2012/147828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ahmed S, Sami A, Xiang J. HER2-directed therapy: current treatment options for HER2-positive breast cancer. Breast Cancer. 2015 doi: 10.1007/s12282-015-0587-x. [DOI] [PubMed] [Google Scholar]
- 65.Yarden Y, Sliwkowski MX. Untangling the ErbB signalling network. Nat Rev Mol Cell Biol. 2001;2(2):127–37. doi: 10.1038/35052073. [DOI] [PubMed] [Google Scholar]
- 66.Hynes NE, Lane HA. ERBB receptors and cancer: the complexity of targeted inhibitors. Nat Rev Cancer. 2005;5(5):341–54. doi: 10.1038/nrc1609. [DOI] [PubMed] [Google Scholar]
- 67.Derynck R. Transforming growth factor alpha. Cell. 1988;54(5):593–5. doi: 10.1016/s0092-8674(88)80001-1. [DOI] [PubMed] [Google Scholar]
- 68.Marmor MD, Yarden Y. Role of protein ubiquitylation in regulating endocytosis of receptor tyrosine kinases. Oncogene. 2004;23(11):2057–70. doi: 10.1038/sj.onc.1207390. [DOI] [PubMed] [Google Scholar]
- 69.Milanezi F, Carvalho S, Schmitt FC. EGFR/HER2 in breast cancer: a biological approach for molecular diagnosis and therapy. Expert Rev Mol Diagn. 2008;8(4):417–34. doi: 10.1586/14737159.8.4.417. [DOI] [PubMed] [Google Scholar]
- 70.Nieto Y, et al. Prognostic significance of overexpression and phosphorylation of epidermal growth factor receptor (EGFR) and the presence of truncated EGFRvIII in locoregionally advanced breast cancer. J Clin Oncol. 2007;25(28):4405–13. doi: 10.1200/JCO.2006.09.8822. [DOI] [PubMed] [Google Scholar]
- 71.Tovey SM, et al. Outcome and human epidermal growth factor receptor (HER) 1-4 status in invasive breast carcinomas with proliferation indices evaluated by bromodeoxyuridine labelling. Breast Cancer Res. 2004;6(3):R246–51. doi: 10.1186/bcr783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Paplomata E, O'Regan R. The PI3K/AKT/mTOR pathway in breast cancer: targets, trials and biomarkers. Ther Adv Med Oncol. 2014;6(4):154–66. doi: 10.1177/1758834014530023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Cantley LC. The phosphoinositide 3-kinase pathway. Science. 2002;296(5573):1655–7. doi: 10.1126/science.296.5573.1655. [DOI] [PubMed] [Google Scholar]
- 74.Maehama T, Dixon JE. The tumor suppressor, PTEN/MMAC1, dephosphorylates the lipid second messenger, phosphatidylinositol 3,4,5-trisphosphate. J Biol Chem. 1998;273(22):13375–8. doi: 10.1074/jbc.273.22.13375. [DOI] [PubMed] [Google Scholar]
- 75.Cancer Genome Atlas N. Comprehensive molecular portraits of human breast tumours. Nature. 2012;490(7418):61–70. doi: 10.1038/nature11412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Samuels Y, Velculescu VE. Oncogenic mutations of PIK3CA in human cancers. Cell Cycle. 2004;3(10):1221–4. doi: 10.4161/cc.3.10.1164. [DOI] [PubMed] [Google Scholar]
- 77.Wander SA, Hennessy BT, Slingerland JM. Next-generation mTOR inhibitors in clinical oncology: how pathway complexity informs therapeutic strategy. J Clin Invest. 2011;121(4):1231–41. doi: 10.1172/JCI44145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Lawlor MA, Alessi DR. PKB/Akt: a key mediator of cell proliferation, survival and insulin responses? J Cell Sci. 2001;114:2903–10. doi: 10.1242/jcs.114.16.2903. Pt 16. [DOI] [PubMed] [Google Scholar]
- 79.Brunet A, et al. Akt promotes cell survival by phosphorylating and inhibiting a Forkhead transcription factor. Cell. 1999;96(6):857–68. doi: 10.1016/s0092-8674(00)80595-4. [DOI] [PubMed] [Google Scholar]
- 80.Holz MK. The role of S6K1 in ER-positive breast cancer. Cell Cycle. 2012;11(17):3159–65. doi: 10.4161/cc.21194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Roskoski R., Jr. The ErbB/HER family of protein-tyrosine kinases and cancer. Pharmacol Res. 2014;79:34–74. doi: 10.1016/j.phrs.2013.11.002. [DOI] [PubMed] [Google Scholar]
- 82.Sarbassov DD, et al. Prolonged rapamycin treatment inhibits mTORC2 assembly and Akt/PKB. Mol Cell. 2006;22(2):159–68. doi: 10.1016/j.molcel.2006.03.029. [DOI] [PubMed] [Google Scholar]
- 83.Giltnane JM, Balko JM. Rationale for targeting the Ras/MAPK pathway in triple-negative breast cancer. Discov Med. 2014;17(95):275–83. [PubMed] [Google Scholar]
- 84.Saini KS, et al. Targeting the PI3K/AKT/mTOR and Raf/MEK/ERK pathways in the treatment of breast cancer. Cancer Treat Rev. 2013;39(8):935–46. doi: 10.1016/j.ctrv.2013.03.009. [DOI] [PubMed] [Google Scholar]
- 85.Harris L, et al. American Society of Clinical Oncology 2007 update of recommendations for the use of tumor markers in breast cancer. J Clin Oncol. 2007;25(33):5287–312. doi: 10.1200/JCO.2007.14.2364. [DOI] [PubMed] [Google Scholar]
- 86.Hudis CA. Trastuzumab--mechanism of action and use in clinical practice. N Engl J Med. 2007;357(1):39–51. doi: 10.1056/NEJMra043186. [DOI] [PubMed] [Google Scholar]
- 87.Junttila TT, et al. Ligand-independent HER2/HER3/PI3K complex is disrupted by trastuzumab and is effectively inhibited by the PI3K inhibitor GDC-0941. Cancer Cell. 2009;15(5):429–40. doi: 10.1016/j.ccr.2009.03.020. [DOI] [PubMed] [Google Scholar]
- 88.Gallardo A, et al. Increased signalling of EGFR and IGF1R, and deregulation of PTEN/PI3K/Akt pathway are related with trastuzumab resistance in HER2 breast carcinomas. Br J Cancer. 2012;106(8):1367–73. doi: 10.1038/bjc.2012.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Vogt PK, et al. Cancer-specific mutations in phosphatidylinositol 3-kinase. Trends Biochem Sci. 2007;32(7):342–9. doi: 10.1016/j.tibs.2007.05.005. [DOI] [PubMed] [Google Scholar]
- 90.Fujita T, et al. PTEN activity could be a predictive marker of trastuzumab efficacy in the treatment of ErbB2-overexpressing breast cancer. Br J Cancer. 2006;94(2):247–52. doi: 10.1038/sj.bjc.6602926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Nagata Y, et al. PTEN activation contributes to tumor inhibition by trastuzumab, and loss of PTEN predicts trastuzumab resistance in patients. Cancer Cell. 2004;6(2):117–27. doi: 10.1016/j.ccr.2004.06.022. [DOI] [PubMed] [Google Scholar]
- 92.LoRusso PM, et al. Trastuzumab emtansine: a unique antibody-drug conjugate in development for human epidermal growth factor receptor 2-positive cancer. Clin Cancer Res. 2011;17(20):6437–47. doi: 10.1158/1078-0432.CCR-11-0762. [DOI] [PubMed] [Google Scholar]
- 93.Teicher BA, Doroshow JH. The promise of antibody-drug conjugates. N Engl J Med. 2012;367(19):1847–8. doi: 10.1056/NEJMe1211736. [DOI] [PubMed] [Google Scholar]
- 94.Agus DB, et al. Targeting ligand-activated ErbB2 signaling inhibits breast and prostate tumor growth. Cancer Cell. 2002;2(2):127–37. doi: 10.1016/s1535-6108(02)00097-1. [DOI] [PubMed] [Google Scholar]
- 95.Liu L, et al. Novel mechanism of lapatinib resistance in HER2-positive breast tumor cells: activation of AXL. Cancer Res. 2009;69(17):6871–8. doi: 10.1158/0008-5472.CAN-08-4490. [DOI] [PubMed] [Google Scholar]
- 96.Scaltriti M, et al. Lapatinib, a HER2 tyrosine kinase inhibitor, induces stabilization and accumulation of HER2 and potentiates trastuzumab-dependent cell cytotoxicity. Oncogene. 2009;28(6):803–14. doi: 10.1038/onc.2008.432. [DOI] [PubMed] [Google Scholar]
- 97.Nahta R, et al. Lapatinib induces apoptosis in trastuzumab-resistant breast cancer cells: effects on insulin-like growth factor I signaling. Mol Cancer Ther. 2007;6(2):667–74. doi: 10.1158/1535-7163.MCT-06-0423. [DOI] [PubMed] [Google Scholar]
- 98.Wang YC, et al. Different mechanisms for resistance to trastuzumab versus lapatinib in HER2-positive breast cancers--role of estrogen receptor and HER2 reactivation. Breast Cancer Res. 2011;13(6):R121. doi: 10.1186/bcr3067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Arpino G, et al. Treatment of human epidermal growth factor receptor 2-overexpressing breast cancer xenografts with multiagent HER-targeted therapy. J Natl Cancer Inst. 2007;99(9):694–705. doi: 10.1093/jnci/djk151. [DOI] [PubMed] [Google Scholar]
- 100.Baselga J, et al. Lapatinib with trastuzumab for HER2-positive early breast cancer (NeoALTTO): a randomised, open-label, multicentre, phase 3 trial. Lancet. 2012;379(9816):633–40. doi: 10.1016/S0140-6736(11)61847-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Bates SE, et al. Expression of transforming growth factor alpha and its messenger ribonucleic acid in human breast cancer: its regulation by estrogen and its possible functional significance. Mol Endocrinol. 1988;2(6):543–55. doi: 10.1210/mend-2-6-543. [DOI] [PubMed] [Google Scholar]
- 102.Saeki T, et al. Regulation by estrogen through the 5'-flanking region of the transforming growth factor alpha gene. Mol Endocrinol. 1991;5(12):1955–63. doi: 10.1210/mend-5-12-1955. [DOI] [PubMed] [Google Scholar]
- 103.Pedram A, Razandi M, Levin ER. Nature of functional estrogen receptors at the plasma membrane. Mol Endocrinol. 2006;20(9):1996–2009. doi: 10.1210/me.2005-0525. [DOI] [PubMed] [Google Scholar]
- 104.Shou J, et al. Mechanisms of tamoxifen resistance: increased estrogen receptor-HER2/neu cross-talk in ER/HER2-positive breast cancer. J Natl Cancer Inst. 2004;96(12):926–35. doi: 10.1093/jnci/djh166. [DOI] [PubMed] [Google Scholar]
- 105.Stoica A, et al. Regulation of estrogen receptor-alpha gene expression by epidermal growth factor. J Endocrinol. 2000;165(2):371–8. doi: 10.1677/joe.0.1650371. [DOI] [PubMed] [Google Scholar]
- 106.Hong SH, Privalsky ML. The SMRT corepressor is regulated by a MEK-1 kinase pathway: inhibition of corepressor function is associated with SMRT phosphorylation and nuclear export. Mol Cell Biol. 2000;20(17):6612–25. doi: 10.1128/mcb.20.17.6612-6625.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Montemurro F, Di Cosimo S, Arpino G. Human epidermal growth factor receptor 2 (HER2)-positive and hormone receptor-positive breast cancer: new insights into molecular interactions and clinical implications. Ann Oncol. 2013;24(11):2715–24. doi: 10.1093/annonc/mdt287. [DOI] [PubMed] [Google Scholar]
- 108.Kato S, et al. Activation of the estrogen receptor through phosphorylation by mitogen-activated protein kinase. Science. 1995;270(5241):1491–4. doi: 10.1126/science.270.5241.1491. [DOI] [PubMed] [Google Scholar]
- 109.Font de Mora J, Brown M. AIB1 is a conduit for kinase-mediated growth factor signaling to the estrogen receptor. Mol Cell Biol. 2000;20(14):5041–7. doi: 10.1128/mcb.20.14.5041-5047.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Kumar R, et al. A naturally occurring MTA1 variant sequesters oestrogen receptor-alpha in the cytoplasm. Nature. 2002;418(6898):654–7. doi: 10.1038/nature00889. [DOI] [PubMed] [Google Scholar]
- 111.Benz CC, et al. Estrogen-dependent, tamoxifen-resistant tumorigenic growth of MCF-7 cells transfected with HER2/neu. Breast Cancer Res Treat. 1992;24(2):85–95. doi: 10.1007/BF01961241. [DOI] [PubMed] [Google Scholar]
- 112.Witters LM, et al. Enhanced anti-proliferative activity of the combination of tamoxifen plus HER-2-neu antibody. Breast Cancer Res Treat. 1997;42(1):1–5. doi: 10.1023/a:1005798224288. [DOI] [PubMed] [Google Scholar]
- 113.Osborne CK, et al. Role of the estrogen receptor coactivator AIB1 (SRC-3) and HER-2/neu in tamoxifen resistance in breast cancer. J Natl Cancer Inst. 2003;95(5):353–61. doi: 10.1093/jnci/95.5.353. [DOI] [PubMed] [Google Scholar]
- 114.McClelland RA, et al. Enhanced epidermal growth factor receptor signaling in MCF7 breast cancer cells after long-term culture in the presence of the pure antiestrogen ICI 182,780 (Faslodex) Endocrinology. 2001;142(7):2776–88. doi: 10.1210/endo.142.7.8259. [DOI] [PubMed] [Google Scholar]
- 115.Beelen K, et al. Phosphorylated p-70S6K predicts tamoxifen resistance in postmenopausal breast cancer patients randomized between adjuvant tamoxifen versus no systemic treatment. Breast Cancer Res. 2014;16(1):R6. doi: 10.1186/bcr3598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Xia W, et al. A model of acquired autoresistance to a potent ErbB2 tyrosine kinase inhibitor and a therapeutic strategy to prevent its onset in breast cancer. Proc Natl Acad Sci U S A. 2006;103(20):7795–800. doi: 10.1073/pnas.0602468103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Begley CG, Ellis LM. Drug development: Raise standards for preclinical cancer research. Nature. 2012;483(7391):531–3. doi: 10.1038/483531a. [DOI] [PubMed] [Google Scholar]
- 118.Johnson JI, et al. Relationships between drug activity in NCI preclinical in vitro and in vivo models and early clinical trials. Br J Cancer. 2001;84(10):1424–31. doi: 10.1054/bjoc.2001.1796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Lasfargues EY, Coutinho WG, Redfield ES. Isolation of two human tumor epithelial cell lines from solid breast carcinomas. J Natl Cancer Inst. 1978;61(4):967–78. [PubMed] [Google Scholar]
- 120.Reddel RR, et al. Differential sensitivity of human breast cancer cell lines to the growth-inhibitory effects of tamoxifen. Cancer Res. 1985;45(4):1525–31. [PubMed] [Google Scholar]
- 121.Subik K, et al. The Expression Patterns of ER, PR, HER2, CK5/6, EGFR, Ki-67 and AR by Immunohistochemical Analysis in Breast Cancer Cell Lines. Breast Cancer (Auckl) 2010;4:35–41. [PMC free article] [PubMed] [Google Scholar]
- 122.Garcia-Garcia C, et al. Dual mTORC1/2 and HER2 blockade results in antitumor activity in preclinical models of breast cancer resistant to anti-HER2 therapy. Clin Cancer Res. 2012;18(9):2603–12. doi: 10.1158/1078-0432.CCR-11-2750. [DOI] [PubMed] [Google Scholar]
- 123.DeRose YS, et al. Tumor grafts derived from women with breast cancer authentically reflect tumor pathology, growth, metastasis and disease outcomes. Nat Med. 2011;17(11):1514–20. doi: 10.1038/nm.2454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Kabos P, et al. Patient-derived luminal breast cancer xenografts retain hormone receptor heterogeneity and help define unique estrogen-dependent gene signatures. Breast Cancer Res Treat. 2012;135(2):415–32. doi: 10.1007/s10549-012-2164-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Marangoni E, Poupon MF. Patient-derived tumour xenografts as models for breast cancer drug development. Curr Opin Oncol. 2014;26(6):556–61. doi: 10.1097/CCO.0000000000000133. [DOI] [PubMed] [Google Scholar]
- 126.Slamon DJ, et al. Use of chemotherapy plus a monoclonal antibody against HER2 for metastatic breast cancer that overexpresses HER2. N Engl J Med. 2001;344(11):783–92. doi: 10.1056/NEJM200103153441101. [DOI] [PubMed] [Google Scholar]
- 127.Robidoux A, et al. Lapatinib as a component of neoadjuvant therapy for HER2-positive operable breast cancer (NSABP protocol B-41): an open-label, randomised phase 3 trial. Lancet Oncol. 2013;14(12):1183–92. doi: 10.1016/S1470-2045(13)70411-X. [DOI] [PubMed] [Google Scholar]
- 128.Gianni L, et al. Efficacy and safety of neoadjuvant pertuzumab and trastuzumab in women with locally advanced, inflammatory, or early HER2-positive breast cancer (NeoSphere): a randomised multicentre, open-label, phase 2 trial. Lancet Oncol. 2012;13(1):25–32. doi: 10.1016/S1470-2045(11)70336-9. [DOI] [PubMed] [Google Scholar]
- 129.Prat A, Baselga J. The role of hormonal therapy in the management of hormonal-receptor-positive breast cancer with co-expression of HER2. Nat Clin Pract Oncol. 2008;5(9):531–42. doi: 10.1038/ncponc1179. [DOI] [PubMed] [Google Scholar]
- 130.Houston SJ, et al. Overexpression of c-erbB2 is an independent marker of resistance to endocrine therapy in advanced breast cancer. Br J Cancer. 1999;79(7-8):1220–6. doi: 10.1038/sj.bjc.6690196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Lipton A, et al. Serum HER-2/neu and response to the aromatase inhibitor letrozole versus tamoxifen. J Clin Oncol. 2003;21(10):1967–72. doi: 10.1200/JCO.2003.09.098. [DOI] [PubMed] [Google Scholar]
- 132.Rimawi MF, et al. Multicenter phase II study of neoadjuvant lapatinib and trastuzumab with hormonal therapy and without chemotherapy in patients with human epidermal growth factor receptor 2-overexpressing breast cancer: TBCRC 006. J Clin Oncol. 2013;31(14):1726–31. doi: 10.1200/JCO.2012.44.8027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Kaufman B, et al. Trastuzumab plus anastrozole versus anastrozole alone for the treatment of postmenopausal women with human epidermal growth factor receptor 2-positive, hormone receptor-positive metastatic breast cancer: results from the randomized phase III TAnDEM study. J Clin Oncol. 2009;27(33):5529–37. doi: 10.1200/JCO.2008.20.6847. [DOI] [PubMed] [Google Scholar]
- 134.Schwartzberg LS, et al. Lapatinib plus letrozole as first-line therapy for HER-2+ hormone receptor-positive metastatic breast cancer. Oncologist. 2010;15(2):122–9. doi: 10.1634/theoncologist.2009-0240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Cui X, et al. Progesterone crosstalks with insulin-like growth factor signaling in breast cancer cells via induction of insulin receptor substrate-2. Oncogene. 2003;22(44):6937–41. doi: 10.1038/sj.onc.1206803. [DOI] [PubMed] [Google Scholar]
- 136.Vici P, et al. Triple positive breast cancer: A distinct subtype? Cancer Treat Rev. 2015;41(2):69–76. doi: 10.1016/j.ctrv.2014.12.005. [DOI] [PubMed] [Google Scholar]
- 137.Vici P, et al. Outcomes of HER2-positive early breast cancer patients in the pre-trastuzumab and trastuzumab eras: a real-world multicenter observational analysis. The RETROHER study. Breast Cancer Res Treat. 2014;147(3):599–607. doi: 10.1007/s10549-014-3133-1. [DOI] [PMC free article] [PubMed] [Google Scholar]