Abstract
Background
Depressive symptoms are common in older adults and associated with poor outcomes. While circadian genes have been implicated in depression, the relationship between circadian genes and depressive symptoms in older adults is unclear.
Methods
A cross-sectional genetic association study of 529 single nucleotide polymorphisms (SNPs) representing 30 candidate circadian genes was performed in two population-based cohorts: Osteoporotic Fractures in Men Study (MrOS, n=1270, age 76.58±5.61 years) and the Study of Osteoporotic Fractures (SOF) in women (n=1740, 84.05±3.53 years) and a meta-analysis was performed. Depressive symptoms were assessed with the Geriatric Depression Scale categorizing participants as having “none-few symptoms” (0-2), “some depressive symptoms” (>2<6), or “many depressive symptoms” (≥6).
Results
We found associations meeting multiple testing criteria for significance between the PER3 intronic SNP rs12137927 and decreased odds of reporting “some depressive symptoms” in the SOF sample (OR 0.61, CI 0.48-0.78, df=1, Wald chi-square −4.04, p=0.000054) and the meta-analysis (OR 0.61, CI 0.48-0.78, z= −4.04, p=0.000054) and between the PER3 intronic SNPs rs228644 (OR 0.74, CI 0.63-0.86, z= 3.82, p-value=0.00013) and rs228682 (OR 0.74, CI 0.86 0.63, z= 3.81, p-value=0.00014) and decreased odds of reporting “some depressive symptoms” in the meta-analysis compared to endorsing none-few depressive symptoms. The RORA intronic SNP rs11632098 was associated with greater odds of reporting “many depressive symptoms” (OR 2.16, CI 1.45-3.23, df=1, Wald chi-square 3.76, p=0.000168) in the men. In the meta analysis the association was attenuated and nominally significant (OR 1.63, CI 1.24-2.16, z=3.45, p=0.00056).
Conclusions
PER3 and RORA may play important roles in the development of depressive symptoms in older adults.
Keywords: Depression, older adults, PER3, RORA, circadian, gene
Background
Depressive syndromes are common in older adults and represent a major public health concern. Major depressive disorder (MDD) is affects 3-10% of older adults in primary care settings (1-4) and “subthreshold” depressive syndromes (SubD, e.g. dysthymia, minor depression and subsyndromal depression) are even more common (9-24%) (3, 5-8). Both MDD and SubD are more prevalent in long-term care settings (9). Depression is a critical issue in this age group because of its association with adverse outcomes including functional impairment (10), medical illnesses (11), hospitalizations (12), and disability (13). In older adults first-line pharmacotherpies targeting monoamines often fail to adequately treat symptoms (14). A better understanding of the biological pathways contributing depression in older adults could lay the groundwork for the development of more effective treatment strategies such as novel interventions targeting other aspects of the underlying pathophysiology of depression. A better understanding of the role of contributing genes in the pathophysiology of depression could also help to identify subgroups of older adults at higher risk for depression who may be candidates for indicated prevention strategies.
Clinical and molecular studies support a role for chronobiological disturbances in the pathophysiology of depression (15-17). Circadian rhythm disturbances (e.g. disruptions in the daily cycles of body temperature, and rest and activity) occur in mood disorders (18, 19). Depression is associated with changes in protein expression in the superchiasmatic nucleus (SCN; the master circadian pacemaker) (20) and enzymes controlling catecholamine metabolism are regulated by circadian genes (21, 22). Associations between polymorphisms in the human circadian genes, PER3 (23), PER2 (24), AANAT (25), CRY1 (26), CRY2 (24), NPAS2 (26), ASMT (27), TIMELESS (28), and RORA (29) and depression have been reported.
Aging is associated with changes in circadian rhythms (30-32). Few studies have investigated the relationship between chronobiological disturbances and depressive symptoms in older adults. We previously reported a cross-sectional graded association between desynchronization of circadian activity rhythms and greater levels of depressive symptoms in a large cohort of community-dwelling older women, the Study of Osteoporotic Fractures (SOF)(33). This association was significant even for women endorsing sub-threshold levels of depressive symptoms. In this study, we examine the relationship between circadian gene polymorphisms and levels of depressive symptoms in the same cohort and a cohort of community-dwelling older men. We hypothesized that there would be associations between circadian gene polymorphisms and depressive symptom level, including subthreshold levels of depressive symptoms.
Methods
Participants
Data were collected from participants of two large, multi-center, cohort studies, SOF and the Osteoporotic Fractures in Men (MrOS) Study. The studies included participants who were ≥65 years old and excluded participants who required assistance from another person for ambulation or had undergone bilateral hip replacement. Details have been published(34). Data were collected with written informed consent and approval by review boards of participating institutions.
In SOF, the 9,704 older, primarily Caucasian women making up the original cohort were recruited from four locations (Baltimore, MD, the Monongahela Valley near Pittsburgh, PA; Minneapolis, MN; Portland, OR) between 1986 and 1988. Follow-up visits were conducted every two years. The current cross-sectional analyses focused on self-identified white women participating in SOF Visit 8 (2002-2004) which included assessment of circadian gene polymorphisms. There were 4,727 participants at Visit 8 (84% of active survivors). Of these women, 1731 self-identified white women who had DNA extracted, SNPs genotyped, and completed a Geriatric Depression Scale (GDS) were included in these analyses.
In MrOS, 5994 older men were recruited from six locations (Birmingham, AL; the Monongahela Valley near Pittsburgh, PA; Minneapolis, MN; Palo Alto, CA; San Diego, CA; and Portland, OR) between 2000 and 2002 (35, 36). The MrOS Sleep Study (2003-2005), recruited 3,135 MrOS participants at the time of their first two-year follow up (68%) and included assessment of circadian gene polymorphisms. Of the 2,859 men who did not participate, 344 had died, 36 had already stopped participating in MrOs, 332 were not invited because recruitment goals were met, 150 were ineligible and 1,997 refused. Among participants of the MrOS Sleep Study, 2,480 self-identified white men who had DNA extracted, SNPs genotyped, and completed a GDS were included in this cross-sectional analyses.
Directly genotyped circadian gene polymorphisms
A custom Illumina Golden Gate assay (Illumina, San Diego, CA, USA) designed to genotype polymorphisms in circadian genes was developed in a collaborative effort by investigators at the California Pacific Medical Center and the University of California San Diego as previously described(37). Candidate genes were selected based on a review of published human association studies and studies done in common model organisms (37).
TagSNPs were selected using Tagger (38) (r2≥0.8, minor allele frequency (MAF)≥0.01) with HapMap CEU Phase II (release 22) genotype data in the candidate gene regions including 10 kb upstream and downstream of transcript boundaries. Using these Tagger settings, 314 tagSNPs would have been selected from the 798 SNPs found within the 761 kb of genomic DNA that includes the RORA gene and 20 kb of flanking DNA. To reduce our RORA genotyping burden, the linkage disequilibrium (LD) threshold in Tagger was lowered to 0.6 and a maximum of 100 tagSNPs were selected for genotyping, resulting in 81% of the 798 RORA SNPs being captured at r2≥0.6. In total, 658 SNPs within the candidate gene regions were selected for genotyping. Genotypes were called using Beadstudio software. Genotype concordance rate was >0.99 (8% of MrOS samples and 3 SOF samples were plated in duplicate). Samples with <90% SNP call rate were excluded. SNPs with missing frequency >0.05 and MAF<0.01 were excluded. Autosomal SNPs with an HWE exact P-value <8×105 (Bonferroni-corrected P-value for 658 SNPs) were excluded (39). Among the all-male MrOS samples, X-linked SNPs with heterozygous genotypes were excluded. Among the 658 genotyped SNPs, 529 in MrOS and 508 in SOF passed QC filters as previously described (37). To correct for residual population stratification in these self-identified European American cohorts, 195 independent, autosomal SNPs were used in multidimensional scaling analyses (MDS) as implemented in PLINK (40). The resulting first two MDS components were included as covariates in regression models.
Imputed circadian gene polymorphisms
Analysis of imputed circadian SNPs was used to provide additional information to fine map association signals detected in the analyses of directly genotyped SNPs. Imputation was performed using MACH v. 1.0.16 (http://www.sph.umich.edu/csg/abecasis/MaCH/). Phased haplotypes of 60 unrelated HapMap II CEU founders were used as the reference data (for autosomes: CEU_r22_ nr.b36_fwd.phased; http://hapmap.ncbi.nlm.nih.gov/downloads/phasing/2007-08_rel22/phased/ , for X chromosome and the pseudo-autosomal regions PAR1/PAR2: CEU_r21_nr_fwd_phased; http://hapmap.ncbi.nlm.nih.gov/downloads/phasing/2006-07_phaseII/phased/). Imputations were based on 537,371 genotyped SNPs in common with the reference data (G-C and A-T SNPs were excluded). To allow imputation with MACH for the haploid non-par regions of the male X chromosome, the phased haplotypes were duplicated. A two-step imputation approach was used: step 1 estimated per SNP error and per interval crossover rates at 50 iterations from a random sample of 200 genotyped European-American individuals, step 2 used these model parameters to estimate allele dosages and genotypes based on a maximum likelihood approach. A total of 488,335 SNPs were imputed, with 4446 high-confidence SNPs remaining after removal of SNPs in low LD (r2 < 0.3) with genotyped markers. Allelic imputation accuracy rates were estimated by masking 20% of the genotyped data and showed a <5% error rate. Adding SNPs not used for imputation purpose resulted in a total of 5076 genotyped and imputed SNPs available for association analyses. Of these, 93 imputed SNPs within the PER3 gene region and 898 imputed SNPs within the RORA gene region were utilized in this study to fine map significant associations observed for directly genotyped SNPs in these genes.
Depressive Symptoms
Depressive symptoms were assessed using the Geriatric Depression Scale (GDS), a 15-item validated self-report questionnaire commonly used for assessment of depressive symptoms in older adults (41). Participants were categorized into three groups [0-2 (no/few depressive symptoms), >2 to<6 (some depressive symptoms), ≥6 (many depressive symptoms)] according to depressive symptoms reported at follow-up. To remain consistent with prior publications using this strategy (42), the GDS ≥6 group was referred to as “depressed.” A standard cut-off of ≥6 on the GDS has been shown to have a sensitivity of 91% and specificity of 65% for diagnosis of a major depressive episode compared with the Diagnostic and Statistical Manual of Mental Disorders, Fourth Edition (43).
Other Sample Characteristics
Demographic information was recorded at the baseline SOF or MrOS assessment. At SOF and MrOS follow-up visits including SOF Visit 8 and MrOS sleep study (MrOS Visit 2), participants completed questionnaires regarding health status, alcohol consumption, exercise, and medical history. Reported medical conditions included stroke, diabetes, Parkinson’s disease, chronic obstructive pulmonary disease, congestive heart failure, myocardial infarction, thyroid disease, hypertension, and cancer. During a clinic interview participants were asked whether they had problems with 6 instrumental activities of daily living (IADLs). Height and weight were measured and BMI was calculated as weight in kilograms divided by height in meters squared. Medications taken daily or almost daily during the prior 30 days were recorded and categorized according to a computerized coding dictionary (44). Cognition was assessed by administration of the Modified Mini-Mental State Exam (3MS)(45) in the MrOs cohort or the Mini-Mental State Exam (MMSE) (46) in the SOF cohort. This information is presented in Table 1.
Table 1. Characteristics of the population.
Key characteristics of the SOF and MrOS samples are shown. SOF: Study of Osteoporotic Fractures; MrOS Osteoporotic Fractures in Men Study.
| Characteristic | MrOS | SOF | p-value (df) |
|---|---|---|---|
| Participants included in analysis | 2480 | 1731 | |
| ^ Geriatric Depression Scale total score, mean(±SD) | 1.76 +/− 2.14 | 2.74 +/− 2.86 | <0.0001 (2) |
| No or few depressive symptoms (0-2), n(%) | 1875 (75.6) | 1040 (60.08) | <0.0001 |
| Some depressive symptoms (>2 - <6), n(%) | 448 (18.06) | 439 (25.36) | <0.0001 |
| Many depressive symptoms (≥6), n(%) | 157 (6.33) | 252 (14.56) | <0.0001 |
| ‡ Age (years), mean(±SD) | 76.58 (±5.61) | 84.04 (±3.51) | <0.0001 (4209) |
| ‡ Body mass index (kg/m2) | 27.25 (±3.79) | 26.72 (±4.81) | 0.0003 (4209) |
| ^ Self-reported health status excellent or good, n(%) | 2166 (87.34) | 1268 (73.25) | <0.0001 (1) |
| ^ Greater than 12 years education, n(%) | 1390 (56.05) | 649 (37.49) | <0.0001 (1) |
| ^ Current smoker, n(%) | 46 (1.85) | 46 (2.64) | 0.0796 (1) |
|
†
Average number alcoholic beverages per week,
mean(±SD) |
1.93 (±1.72) | 0.92 (±2.66) | <0.0001 |
| ^ Current antidepressant use, n(%) | 146 (5.89) | 163 (10.53) | <0.0001 (1) |
| † Instrumental activities of daily living, mean(±SD) | 0.36 (±0.83) | 3.09 (±4.04) | <0.0001 |
| † Number medical conditions, mean (±SD) | 1.07 (±0.99) | 1.5 (±1.17) | <0.0001 |
Compared using Chi-squared tests
Compared using t-tests
Compared using non parametric Mann-Whitney tests
Statistical Analyses
Associations between directly genotyped circadian SNPs and depressive symptom levels were evaluated using multinomial logistic regression models to estimate odds ratios (OR) and 95% confidence intervals (CI) for falling into the “some depressive symptoms” or the “depressed” groups compared with the group with no or few depressive symptoms. Multinomial regression analysis was performed in each cohort separately, for all 520 circadian SNPs to identify gene regions of interest with significant SNP associations. To correct for multiple hypothesis testing in the presence of LD, the effective number of independent SNPs using our QC-filtered genotype data was estimated and a multiple-testing significance threshold of 1.7×10−4 was adopted.(47) Subsequently associations between imputed SNPs in the gene regions of interest and depressive symptom levels were evaluated using logistic regression models as described above. Results for all three modes of inheritance (additive, dominant, and recessive) are reported here for SNPs with significant (p-value <0.00017) or nominally significant (p-value<0.05) associations under the dominant inheritance mode. For the meta-analysis, results from the two cohorts were combined by fixed-effect meta-analysis using inverse variance weighting of effect estimates. P-values from the 2DF test in each cohort were combined by a Z-statistic based approach weighted by the square root of the sample size. Heterogeneity between studies was assessed using the I2 statistic and the P-value from the Q-test. All SNPs presented had a p-value > 0.05 for heterogeneity. Association analysis was performed using PLINK (http://pngu.mgh.harvard.edu/~purcell/plink/), meta-analysis was performed using METAL,(48) and power calculations were performed using QUANTO (49). Two gene regions of interest were identified; PER3 and RORA and regional association plots including imputed SNPs were generated using LocusZoom (http://csg.sph.umich.edu/locuszoom). Plots shown are for best-fit models (dominant mode of inheritance) for outcomes where significant associations were found (“some depressive symptoms” category for PER3 region and “depressed” category for RORA region).
Results
Characteristics of the population
The MrOS sample was about 7.5 years younger, had a higher average BMI, and were more likely to report good or excellent health, less iADL impairment, fewer medical problems, and greater alcohol consumption (Table 1). Average scores on cognitive screening assessments (MrOS: mean (+/−standard deviation) 3MS score 93.14 (+/−5.54); SOF: mean MMSE score 27.97(+/−2.04) were in the normal range.
The average GDS score was in the “no or few depressive symptoms” range (1.76+/−2.14) in the MrOs group versus the “some depressive symptoms” range (2.74+/−2.86) in the SOF group. A greater proportion of the SOF sample were categorized as having “some depressive symptoms” (25.36% vs. 18.06%) or “depressed” (14.56% vs. 6.33%). The percentage of patients endorsing use of antidepressant medications was higher in the SOF sample (10.52% vs. 5.89%).
PER3 SNP Associations with Depressive Symptom Level
There were significant associations between several directly genotyped SNPs at the PER3 locus and decreased odds of reporting “some depressive symptoms” in models testing a dominant mode of inheritance (Figure 1, Table 2). There was a significant association between the PER3 intronic SNP rs12137927 and decreased odds of reporting “some depressive symptoms” in the SOF sample (ORDOM 0.61, CI 0.48-0.78, df=1, Wald chi-square −4.04, p=0.000054) but not the MrOS sample. This association was also significant in the meta-analysis (ORDOM 0.61, CI 0.48-0.78, z=4.04, p=0.000054). The PER3 intronic SNPs rs228644 (ORDOM 0.74, CI 0.63-0.86, z=3.82, p-value=0.00013) and rs228682 (ORDOM 0.74, CI 0.86-0.63, z=3.81 p-value=0.00014) were also both associated with decreased odds of endorsing “some depressive symptoms” in the meta-analysis. There were no significant associations between PER3 SNPs and the “depressed” category for directly genotyped PER3 SNPs.
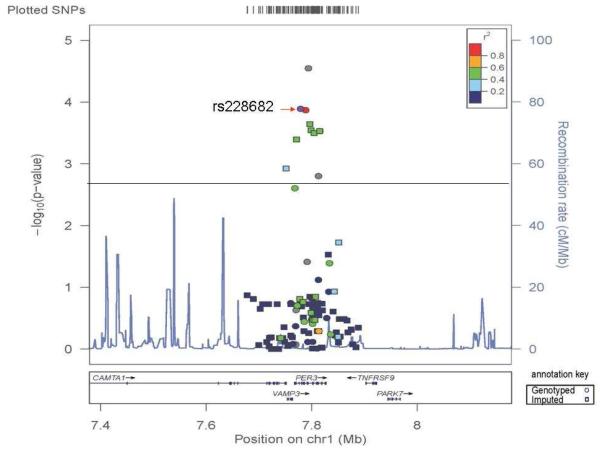
FIGURE 1. A regional association plot is shown for directly genotyped and imputed PER3 SNPs and the “some depressive symptoms” category.
Data shown is for models testing the dominant mode of inheritance (i.e. the best-fit model) in the meta-analysis. Circles represent directly genotyped SNPs and squares represent imputed SNPs. SNP: single nucleotide polymorphism, Chr: chromosome.
Table 2. Genetic Association of Circadian SNPs and Depressive Symptom Level.
Associations between directly genotyped circadian SNPs and different levels of depressive symptoms are shown. Odds ratios (OR) and 95% confidence intervals (CI) are given for each depressive symptom level category compared with the GDS 0-2 group. MAF is the weighted average between MAFs for the SOF and MrOS cohorts. The p-values for associations passing multiple testing criteria for significance (p-value <0.00017) are bolded and underlined. Nominally significant p-values (≥0.00017 but ≤0.05) are underlined. Non-significant p-values > 0.05 are in normal font; MAF: Minor allele frequency; SOF: Study of Osteoporotic Fractures; MrOS Osteoporotic Fractures in Men Study.
| Directly Genotyped SNPs |
Some Depressive Symptoms (2>GDS<6) | Depressed (GDS≥6) | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
| ||||||||||||
| SNP | Gene | Location | Alleles | MAF | Mode | MrOS (n=448) | SOF (n=439) | *Meta (n=887) | MrOS (n=157) | SOF (n=252) | Meta (n=409) | |
| rs12137927 | PER3 | Intron | C/T | 0.219 | Add | OD (95%CI) | 0.86 (0.72-1.03) | 0.69 (0.57-0.85) | 0.69 (0.56-0.84) | 0.98 (0.74-1.29) | 0.85 (0.67-1.09) | 1.1 (0.92-1.33) |
| Test statistic (df) | −1.59( 1df ) | −3.5( 1df ) | 1.43 | −0.15( 1df ) | −1.29( 1df ) | 1.06 | ||||||
| P-value | 0.111 | 0.0004624 | 0.1541 | 0.8787 | 0.1984 | 0.2882 | ||||||
|
|
||||||||||||
| Dom | OD (95%CI) | 0.80 (0.65-1) | 0.61 (0.48-0.78) | 0.61 (0.48-0.78) | 0.98 (0.7-1.36) | 0.78 (0.58-1.04) | 0.86 (0.69-1.06) | |||||
| Test statistic (df) | −1.99( 1df ) | −4.04( 1df ) | 4.04 | −0.14( 1df ) | −1.69( 1df ) | 1.36 | ||||||
| P-value | 0.04619 | 0.00005423 | 0.00005405 | 0.8921 | 0.09096 | 0.1726 | ||||||
|
|
||||||||||||
| Rec | OD (95%CI) | 1.03 (0.65-1.62) | 0.91 (0.53-1.58) | 1.1 (0.63-1.89) | 0.96 (0.46-2.02) | 1.13 (0.6-2.14) | 0.95 (0.59-1.54) | |||||
| Test statistic (df) | 0.11( 1df ) | −0.33( 1df ) | 0.12 | −0.11( 1df ) | 0.37( 1df ) | 0.21 | ||||||
| P-value | 0.9122 | 0.7439 | 0.9012 | 0.9143 | 0.7112 | 0.8334 | ||||||
|
|
||||||||||||
| 2DF | Test statistic (df) | −1.43( 2df ) | −2.21( 2df ) | 3.68 | 0.02( 2df ) | −1.42( 2df ) | 0.07 | |||||
| P-value | 0.7945 | 0.3394 | 0.00023 | 0.9004 | 0.9676 | 0.9422 | ||||||
|
| ||||||||||||
| rs228644 | PER3 | Intron | G/A | 0.406 | Add | OD (95%CI) | 0.81 (0.7-0.94) | 0.9 (0.76-1.06) | 0.85 (0.76-0.95) | 0.95 (0.75-1.2) | 1.18 (0.96-1.45) | 1.07 (0.92-1.25) |
| Test statistic (df) | −2.7( 1df ) | −1.24( 1df ) | 2.85 | −0.44( 1df ) | 1.55( 1df ) | 0.88 | ||||||
| P-value | 0.006372 | 0.214 | 0.004369 | 0.6575 | 0.1217 | 0.3813 | ||||||
|
|
||||||||||||
| Dom | OD (95%CI) | 0.71 (0.57-0.88) | 0.77 (0.61-0.97) | 0.74 (0.63-0.86) | 0.98 (0.69-1.39) | 1.22 (0.9-1.66) | 1.11 (0.88-1.39) | |||||
| Test statistic (df) | −3.16( 1df ) | −2.2( 1df ) | 3.82 | −0.11( 1df ) | 1.26( 1df ) | 0.87 | ||||||
| P-value | 0.001593 | 0.02753 | 0.0001346 | 0.9102 | 0.207 | 0.3842 | ||||||
|
|
||||||||||||
| Rec | OD (95%CI) | 0.85 (0.65-1.13) | 1.08 (0.8-1.47) | 0.95 (0.78-1.17) | 0.85 (0.55-1.34) | 1.26 (0.88-1.81) | 1.08 (0.81-1.43) | |||||
| Test statistic (df) | −1.04( 1df ) | 0.52( 1df ) | 0.41 | −0.67( 1df ) | 1.25( 1df ) | 0.55 | ||||||
| P-value | 0.2982 | 0.6021 | 0.6837 | 0.5052 | 0.2117 | 0.584 | ||||||
|
|
||||||||||||
| 2DF | Test statistic (df) | −1.57( 2df ) | −2.24( 2df ) | 2.97 | 0.52( 2df ) | −0.07( 2df ) | 0.53 | |||||
| P-value | 0.02885 | 0.5515 | 0.003 | 0.5835 | 0.1241 | 0.5967 | ||||||
|
| ||||||||||||
| rs228682 | PER3 | Intron | T/C | 0.407 | Add | OD (95%CI) | 0.81 (0.7-0.94) | 0.9 (0.76-1.06) | 0.85 (0.76-0.95) | 0.97 (0.77-1.23) | 1.18 (0.96-1.46) | 0.92 (0.79-1.08) |
| Test statistic (df) | −2.66( 1df ) | −1.24( 1df ) | 2.84 | −0.34( 1df ) | 1.61( 1df ) | 0.98 | ||||||
| P-value | 0.006536 | 0.2138 | 0.004443 | 0.7306 | 0.1082 | 0.3253 | ||||||
|
|
||||||||||||
| Dom | OD (95%CI) | 0.71 (0.57-0.88) | 0.77 (0.61-0.97) | 0.74 (0.86-0.63) | 1.01 (0.71-1.43) | 1.23 (0.9-1.67) | 0.89 (0.7-1.12) | |||||
| Test statistic (df) | −3.16( 1df ) | −2.22( 1df ) | 3.81 | −0.11( 1df ) | 1.31( 1df ) | 0.91 | ||||||
| P-value | 0.001779 | 0.02631 | 0.0001414 | 0.9102 | 0.1895 | 0.3636 | ||||||
|
|
||||||||||||
| Rec | OD (95%CI) | 0.85 (0.64-1.13) | 1.09 (0.8-1.47) | 1.05 (0.85-1.29) | 0.89 (0.57-1.38) | 1.27 (0.88-1.83) | 0.91 (0.69-1.2) | |||||
| Test statistic (df) | −0.97( 1df ) | 0.54( 1df ) | 0.34 | −0.49( 1df ) | 1.29( 1df ) | 0.69 | ||||||
| P-value | 0.3336 | 0.5858 | 0.7347 | 0.6265 | 0.1963 | 0.493 | ||||||
|
|
||||||||||||
| 2DF | Test statistic (df) | −1.64( 2df ) | −2.28( 2df ) | 2.93 | 0.35( 2df ) | −0.06( 2df ) | 0.67 | |||||
| P-value | 0.03285 | 0.5587 | 0.0034 | 0.6797 | 0.1107 | 0.502 | ||||||
|
| ||||||||||||
| rs10519084 | RORA | Intron | A/G | 0.2832 | Add | OD (95%CI) | 1.06 (0.90-1.25) | 1.03 (0.86-1.22) | 1.04 (0.93-1.78) | 0.73 (0.55-0.96) | 0.73 (0.58-0.93) | 0.73 (0.61-0.87) |
| Test statistic (df) | 0.71( 1df ) | 0.28( 1df ) | 0.70 | −2.26( 1df ) | −2.61( 1df ) | 3.46 | ||||||
| P-value | 0.48 | 0.7797 | 0.4826 | 0.024 | 0.0091 | 0.00055 | ||||||
|
|
||||||||||||
| Dom | OD (95%CI) | 1.10 (0.89-1.35) | 1.00 (0.80-1.25) | 1.05 (0.90-1.22) | 0.68 (0.51-0.90) | 0.66 (0.47-0.93) | 0.67 (0.54-0.84) | |||||
| Test statistic (df) | 0.86( 1df ) | 0( 1df ) | 0.62 | −2.39( 1df ) | −2.66( 1df ) | 3.58 | ||||||
| P-value | 0.3923 | 0.9974 | 0.5333 | 0.01687 | 0.017 | 0.00035 | ||||||
|
|
||||||||||||
| Rec | OD (95%CI) | 1.02 (0.69-1.49) | 1.14 (0.77-1.68) | 1.07 (0.82-1.41) | 0.73 (0.37-1.42) | 0.71 (0.40-1.24) | 0.72 (0.46-1.10) | |||||
| Test statistic (df) | 0.08( 1df ) | 0.64( 1df ) | 0.51 | −0.93( 1df ) | −1.21( 1df ) | 1.53 | ||||||
| P-value | 0.9342 | 0.5213 | 0.6124 | 0.3528 | 0.2255 | 0.1272 | ||||||
|
|
||||||||||||
| 2DF | Test statistic (df) | 0.52( 2df ) | −0.61( 2df ) | 0.12 (1df) | −0.71( 2df ) | −0.58( 2df ) | 2.14 | |||||
| P-value | 0.7655 | 0.5752 | 0.9079 | 0.1728 | 0.08452 | 0.0323 | ||||||
|
| ||||||||||||
| rs11632098 | RORA | Intron | T/C | 0.0708 | Add | OD (95%CI) | 1.05 (0.77-1.42) | 1.07 (0.80-1.43) | 1.06 (0.86-1.31) | 1.93 (1.34-2.80) | 1.27 (0.89-1.80) | 1.55 (1.20-2.00) |
| Test statistic (df) | 0.47( 1df ) | 0.32( 1df ) | 0.56 | 3.49( 1df ) | 1.31( 1df ) | 3.35 | ||||||
| P-value | 0.6368 | 0.7518 | 0.5744 | 0.00049 | 0.19 | 0.00081 | ||||||
|
|
||||||||||||
| Dom | OD (95%CI) | 1.06 (0.77-1.45) | 1.10 (0.80-1.52) | 1.08 (0.86-1.35) | 2.16 (1.45-3.23) | 1.26 (0.86-1.85) | 1.63 (1.24-2.16) | |||||
| Test statistic (df) | 0.36( 1df ) | 0.59( 1df ) | 0.67 | 3.76( 1df ) | 1.17( 1df ) | 3.45 | ||||||
| P-value | 0.7223 | 0.5539 | 0.5058 | 0.000168 | 0.2426 | 0.00056 | ||||||
|
|
||||||||||||
| Rec | OD (95%CI) | 0.32 (0.04-2.58) | 1.46 (0.45-4.75) | 1.01 (0.36-2.82) | 0.94 (0.12-7.63 ) | 1.96 (0.52-7.39 ) | 1.59 (0.52-4.87 ) | |||||
| Test statistic (df) | 0.63( 1df ) | −1.07( 1df ) | 0.02 | −0.05( 1df ) | 0.99( 1df ) | 0.81 | ||||||
| P-value | 0.5308 | 0.287 | 0.9848 | 0.957 | 0.3228 | 0.4208 | ||||||
|
|
||||||||||||
| 2DF | Test statistic (df) | −0.46( 2df ) | 1.26( 2df ) | 0.16 | 1.34( 2df ) | −0.39( 2df ) | 0.58 | |||||
| P-value | 0.5257 | 0.2946 | 0.8747 | 0.9399 | 0.3049 | 0.5614 | ||||||
Test statistic: Wald test for SOF and MrOs analyses; Z-statistic based approach weighted by the square root of the sample size for meta-analysis
A secondary analysis was performed examining associations between directly genotyped PER3 SNPs and reporting “some or many” depressive symptoms (GDS>2) compared with “none or few” (GDS 0-2) depressive symptoms (Table 3). There was a significant association between rs13137927 and “some or many” depressive symptoms in the SOF sample (ORDOM 0.67, CI 0.55-0.82, df=1, Wald chi-square −3.88, p-value=0.000104) but not in the MrOS sample. This association was also significant in the meta-analysis (ORDOM 0.76, CI 0.66 0.87, z=3.95, p-value=0.000078). There were nominal associations between rs228644 (ORDOM 0.77, CI 0.63-0.93, df=1, Wald chi-square −2.71, p-value=0.0068) and rs228682 (ORDOM 0.77, CI 0.64-0.94, df=1, Wald chi-square −2.71, p-value=0.009) and the “some or many” depressive symptoms category in the MrOS sample but not in the SOF sample. These associations were nominally significant in the meta-analysis (Table 3).
Table 3. Genetic Association of PER3 SNPs and Depressive Symptoms GDS ≥2.
Associations between directly genotyped Per3 SNPs and geriatric depression scale (GDS) score ≥2 are shown. Odds ratios (OR) and 95% confidence intervals (CI) are given for having depressive symptoms compared with the GDS 0-2 group. MAF is the weighted average between MAFs for the SOF and MrOS cohorts. The p-values for associations passing multiple testing criteria for significance (p-value <0.00017) are bolded and underlined. Nominally significant p-values (≥0.00017 but ≤0.05) are underlined. Non-significant p-values > 0.05 are in normal font; MAF: Minor allele frequency; SOF: Study of Osteoporotic Fractures; MrOS Osteoporotic Fractures in Men Study.
| Directly Genotyped SNPs | Combined Subthreshold/Depressed (GDS≥2) | |||||||
|---|---|---|---|---|---|---|---|---|
|
| ||||||||
| SNP | Location | Alleles | MAF | Mode | MrOS (n=605) | SOF (n=691) | Meta (n=1296) | |
| rs12137927 | Intron | C/T | 0.224 | Add | OD (95%CI) | 0.89 (0.76-1.04) | 0.75 (0.63-0.89) | 0.83 (0.73-0.93) |
| Test statistic (df) | −1.45( 1df ) | −3.26( 1df ) | 3.27 | |||||
| P-value | 0.1478 | 0.001119 | 0.001075 | |||||
|
|
||||||||
| Dom | OD (95%CI) | 0.84 (0.70-1.018) | 0.67 (0.54-0.82) | 0.75 (0.66-0.87) | ||||
| Test statistic (df) | −1.78( 1df ) | −3.88( 1df ) | 3.95 | |||||
| P-value | 0.07564 | 0.000104 | 0.00007769 | |||||
|
|
||||||||
| Rec | OD (95%CI) | 1.01 (0.67-1.51) | 0.99 (0.62-1.57) | 1.001 (0.73-1.36) | ||||
| Test statistic (df) | 0.03( 1df ) | −0.05( 1df ) | 0.01 | |||||
| P-value | 0.9747 | 0.962 | 0.9949 | |||||
|
|
||||||||
| 2DF | Test statistic (df) | −1.19(2df ) | −2.39( 1df ) | 0.67 | ||||
| P-value | 0.769 | 0.4903 | 0.5043 | |||||
|
| ||||||||
| rs228644 | Intron | G/A | 0.412 | Add | OD (95%CI) | 0.84 (0.74-0.96) | 0.99 (0.86-1.143) | 0.91 (0.83-1.00) |
| Test statistic (df) | −2.49( 1df ) | −0.11( 1df ) | 1.91 | |||||
| P-value | 0.01266 | 0.909 | 0.056 | |||||
|
|
||||||||
| Dom | OD (95%CI) | 0.76 (0.63-0.93) | 0.90 (0.73-1.10) | 0.82(0.72-0.95) | ||||
| Test statistic (df) | −2.71( 1df ) | −1.04( 1df ) | 2.69 | |||||
| P-value | 0.00683 | 0.2998 | 0.007235 | |||||
|
|
||||||||
| Rec | OD (95%CI) | 0.86(0.67-1.10) | 1.16(0.89-1.50) | 0.99(0.83-1.18) | ||||
| Test statistic (df) | −1.22( 1df ) | 1.1( 1df ) | 0.15 | |||||
| P-value | 0.2229 | 0.2694 | 0.8782 | |||||
|
|
||||||||
| 2DF | Test statistic (df) | −1.05(2df ) | −1.84(2df ) | 1.86 | ||||
| P-value | 0.03261 | 0.7345 | 0.0632 | |||||
|
| ||||||||
| rs228682 | Intron | T/C | 0.412 | Add | OD (95%CI) | 0.85(0.74-0.97) | 0.99(0.86-1.15) | 0.91(0.83-1.007) |
| Test statistic (df) | −2.41( 1df ) | −0.09( 1df ) | 1.83 | |||||
| P-value | 0.01583 | 0.9323 | 0.068 | |||||
|
|
||||||||
| Dom | OD (95%CI) | 0.77 (0.63-0.93) | 0.90(0.73-1.10) | 0.83(0.72-0.95) | ||||
| Test statistic (df) | −2.71( 1df ) | −1.02( 1df ) | 2.68 | |||||
| P-value | 0.00683 | 0.3059 | 0.007425 | |||||
|
|
||||||||
| Rec | OD (95%CI) | 0.87(0.68-1.12) | 1.164(0.90-1.51) | 0.99(0.83-1.20) | ||||
| Test statistic (df) | −1.08( 1df ) | 1.14( 1df ) | 0.09 | |||||
| P-value | 0.2821 | 0.2532 | 0.93 | |||||
|
|
||||||||
| 2DF | Test statistic (df) | −1.19(2df ) | −1.86(2df ) | 1.80 | ||||
| P-value | 0.04204 | 0.7087 | 0.07187 | |||||
Test statistic: Wald test for SOF and MrOs analyses; Z-statistic based approach weighted by the square root of the sample size for meta-analysis
There were six imputed PER3 SNPs that were associated with decreased odds (All ORDOM 0.63, CI 0.50-0.80, df=1, Wald chi-square 3.78-3.83, p=0.00015) of “some depressive symptoms” in the SOF sample but not in the MrOS sample (Figure 1, Supplementary Table). In the meta-analysis these associations were attenuated and did not reach multiple testing criteria for significance. One imputed PER3 SNP (rs11581279) was significantly associated with increased odds of falling into the “depressed” category under the additive mode of inheritance (ORADD 1.81, CI1.33-2.45, df=1, Wald chi-square 3.80, p-value=0.00014) while the association was nominally significant under the dominant mode of inheritance (ORDOM 1.92, CI 1.33-2.77, df=1, Wald chi-square 3.49, p-value=0.00049). This association was attenuated and did not reach multiple testing criteria for significance in the meta-analysis. There were no significant associations for either directly genotyped or imputed PER3 SNPs in models testing the recessive mode of inheritance.
RORA SNP Associations with Depressive Symptom Level
In the MrOS sample, the directly genotyped RORA intronic SNP rs11632098 was associated with greater odds of falling into the “depressed” category (Table 2; ORDOM 2.16, CI 1.45-3.23,df=1, Wald chi-square 3.76, p=0.000168). This association was not significant in the SOF cohort (Supplemental Table) and was attenuated and nominally significant (ORDOM 1.63, CI 1.24-2.16, z=3.45, p=0.00056) in the meta-analysis. There were also nominally significant associations between the intronic SNP rs10519084 and the “depressed” category (odds ratios 0.66-0.73, df=1, Wald chi-square −2.66 to −2.39, p-values 0.0035 0.024) in the SOF and MrOS samples and in the meta-analysis in models testing the additive and dominant mode of inheritance.
Sixteen imputed RORA SNPs were significantly associated with the “depressed” category and 2 were nominally associated with the “depressed” category in the MrOS cohort only (Supplementary Table). There were no significant associations between imputed RORA SNPs and depressive symptom level in the SOF cohort.
Discussion
These analyses identified associations between SNPs in two circadian genes, PER3 and RORA, and levels depressive symptoms in two large cohorts of older adults. These data add to a growing body of evidence implicating chronobiological pathways e in the pathophysiology of depression (50, 51). Because older adults are particularly vulnerable to depression risk factors (e.g. medical comorbidities, cerebrovascular changes, dementia, and decreased socialization), the etiology of depression in late-life is often multi-factorial. None-the-less, our data suggests that circadian genes continue to exert an effect on mood in late-life.
We identified a significant association between one directly genotyped PER3 SNP and 0.61-fold decreased odds of reporting “some depressive symptoms” in the SOF group and nominally significant associations between two additional directly genotyped SNPs and 0.71-0.77 fold decreased odds of endorsing “some depressive symptoms” in both SOF and MrOS samples. These findings are supported by the meta-analysis in which the associations between all three SNPs and the “some depressive symptoms” category met multiple testing criteria for significance and the additional significant associations identified in the imputated PER3 SNP set. No significant association was found between these PER3 SNPs and the “depressed” category. One possible explanation for this is that these particular PER3 gene polymorphisms influence milder forms of depression. However, it is notable that the “depressed” category included fewer women. It is therefore possible that the power to detect such associations in models testing associations between SNPs and the “depressed” outcome” was insufficient. In a secondary analysis, similar associations were found between the directly genotyped PER3 SNPs and “some to many” depressive symptoms (GDS>2). Thus, PER3 variants may be associated with decreased risk for depressive symptoms, including subthreshold levels of depressive symptoms, in older adults.
The period family is a core component of the molecular biological clock machinery and PER3 plays a role in sleep-wake cycle and circadian phenotypes (52) (53-55). The literature regarding PER3 gene variants and mood disorders is mixed. Studies have reported significant associations between PER3 variants and mood disorder characteristics such as age of onset, response to pharmacotherapy, and circadian mood oscillations (23, 56, 57). Other studies found only suggestive associations (58, 59), or no significant associations (26). Associations between circadian gene polymorphisms and subthreshold levels of depressive symptoms have not previously been reported in older adults. In this age group SubD is twice as common as MDD (3, 5-7) and associated with adverse outcomes including functional impairment (60), disability (61), physical decline (62), and progression to MDD (5). Attention is warranted to increasing knowledge about SubD and to developing treatment interventions.
We identified one directly genotyped RORA SNP and 16 imputed RORA SNPS that were significantly associated with 2.16-2.34 fold increased odds being categorized “depressed” in the men but not in the women. There was also an association for one directly genotyped RORA SNP and 0.66-0.68 fold decreased odds of falling into the “depressed” category in both cohorts and in the meta-analysis. However the later association did not meet multiple testing criteria for significance.
RORA belongs to the NR1 subfamily of nuclear hormone receptors. RORA is involved in regulation of circadian rhythms, neurodevelopment, neuroprotection, and regulation of steroid hormones. Disruption of any of these processes could potentially contribute to the pathophysiology of depression. These results are consistent with other studies linking RORA with depression-related variables including diagnoses (24), depression-associated personality traits (29), and response to citalopram (63). In contrast, a recent GWAS analysis found no associations reaching significance at the genome-wide multiple testing level between RORA SNPs and MDD or dysthymia (64). Other studies found associations between RORA variants and bipolar disorder (65), post-traumatic stress disorder (66), attention deficit hyperactivity disorder (67), and autism (68). Accordingly, it been suggested that polymorphisms in RORA may increase non-specific vulnerability to mental illness. We found associations between RORA polymorphisms and depressive symptoms in the older male, but not the older female, cohort. The data raise the possibility that RORA variants could contribute to the development of depressive symptoms particularly in older men. However, the analyses presented here were not designed to examine gender differences and gender is confounded by study (i.e. SOF vs. MrOS). Another candidate gene study in which an association between depression with early morning wakening and an interaction between SNPs in TIMELESS and RORA was observed in males but not females (28). These observations could be explained, in part, by the fact that RORA is a hormone-dependent transcription factor that is differentially regulated by male and female sex hormones (69). Future studies could be designed specifically to determine whether associations can be replicated in other cohorts and whether RORA’s role is gender specific.
The mechanism by which alterations in the PER3 and RORA genes could impact depressive symptoms remains unclear. The “phase-shift hypothesis” posits that depression is caused by a misalignment of circadian phase compared to the sleep-wake cycle (70). We previously reported a cross sectional association between less robust circadian rest-activity rhythms but no association between circadian rest-activity rhythm timing and depressive symptoms in SOF (33). Further, no significant associations were found between the PER3 and RORA SNPs reported here and circadian rest-activity rhythm characteristics in the SOF or MrOS cohorts (37). Therefore the identified gene polymorphisms may impact depression through alternate mechanisms. For example, the “neuroinflammatory hypothesis” of depression argues that inflammatory cytokines, elevated in the setting of stress or illness can contribute to depression through a combination of several mechanisms: 1) influencing regulation of neurotransmitter (especially monoamine) metabolism in the brain, 2) promoting deleterious effects on cells in the brain including decreased neurotrophic support, decreased neurogenesis, and induction of apoptosis in microglia, and dysregulation of glial/neuronal interactions with chronic inflammation (71). Both RORA and Per3 may play roles in down regulation of pro-inflammatory cytokines such as IL-6. More investigation is required to determine how these genes could mediate their effects on mood.
Two strengths of this study are the inclusion of models testing for associations with the “some depressive symptoms” category which is likely to represent subthreshold depression and the use of two large cohorts of participants who weren’t selected on the basis of mood symptoms or circadian characteristics. Our study avoids many pitfalls commonly associated with earlier candidate gene studies (72). The Clock gene pathway, from which the candidate genes were selected, is well established and we systematically surveyed common genetic variation in our set of candidate genes rather than choosing individual variants to test within genes. Multiple test correction was applied using modern techniques that take LD into account. Population stratification was accounted for by restricting the analyses to self-identified Caucasian participants and adjusting for genetic ancestry using components from multidimensional scaling analyses. One possible disadvantage of the candidate gene approach is that genetic associations not represented in our candidate gene pool would not be detected.
A limitation of the study is that the sample size for the “depressed” category was smaller reducing the power to detect significant associations in models including that category as an outcome. Therefore the lack of identified associations between PER3 SNPs and the “depressed” category could represent a false-negative result. Furthermore, the sample size was not adequate to determine whether circadian SNPs were associated with increased or decreased odds of falling into the “depressed” versus the “some depressive symptoms” category. Because each study enrolled only women or men, it was not possible to evaluate gender differences in associations between SNPs and depressive symptoms. Another limitation of the study is the use of a questionnaire rather than a diagnostic interview to assess depressive symptoms. The cut-off of GDS total score ≥6 has been validated in comparison to a diagnosis of MDD according DSM-IV criteria but the score range used for the “some depressive symptom” category has not been validated compared with DSM diagnoses (e.g. minor depression). The analysis was restricted to older Caucasian participants and results may not be generalizable to other populations.
In summary PER3 and RORA may play important roles in the pathophysiology of depression in older adults.
Supplementary Material
FIGURE 2. A regional association plot is shown for directly genotyped and imputed RORA SNPs and the “depressed” category.
Data shown is for models testing the dominant mode of inheritance (i.e. the best-fit model) in the meta-analysis. Circles represent directly genotyped SNPs and squares represent imputed SNPs. SNP: single nucleotide polymorphism, Chr: chromosome.
ACKNOWLEDGEMENTS
The authors would like to thank Daniel Kripke MD for thoughtful discussion related to these analyses.
Funding: Dr. Maglione was supported by T32MH019934-18, and Dr. Nievergelt by R01 MH093500 and R01 AG030474-01A2. The Osteoporotic Fractures in Men (MrOS) Study is supported by: U01 AG027810, U01 AG042124, U01 AG042139, U01 AG042140, U01 AG042143, U01 AG042145, U01 AG042168, U01 AR066160, and UL1 TR000128. The National Heart, Lung, and Blood Institute (NHLBI) provides funding for the MrOS Sleep ancillary study “Outcomes of Sleep Disorders in Older Men” under the following grant numbers: R01 HL071194, R01 HL070848, R01 HL070847, R01 HL070842, R01 HL070841, R01 HL070837, R01 HL070838, and R01 HL070839. The Study of Osteoporotic Fractures (SOF) is supported by: R01 AG005407, R01 AR35582, R01 AR35583, R01 AR35584, R01 AG005394, R01 AG027574, R01 AG027576, and R01 AG026720.
Footnotes
FINANCIAL DISCLOSURES
Disclosures: This was not an industry supported study. Dr. Ancoli-Israel has been a consultant or on the advisory board of Astra Zeneca, Aptalis Pharma, Arena, Ferring Pharmaceuticals Inc., Merck, NeuroVigil, Inc., Orphagen Pharmaceuticals, and Purdue Pharma LP. Drs. Maglione, Nievergelt, Evans, Stone, Yaffe, Redline, Tranah and Neeta Parimi all indicated no financial conflicts of interest.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Borson S, Barnes RA, Kukull WA, Okimoto JT, Veith RC, Inui TS, et al. Symptomatic depression in elderly medical outpatients. I. Prevalence, demography, and health service utilization. J Am Geriatr Soc. 1986;34(5):341–7. doi: 10.1111/j.1532-5415.1986.tb04316.x. [DOI] [PubMed] [Google Scholar]
- 2.Schulberg HC, Mulsant B, Schulz R, Rollman BL, Houck PR, Reynolds CF., 3rd Characteristics and course of major depression in older primary care patients. Int J Psychiatry Med. 1998;28(4):421–36. doi: 10.2190/G23R-NGGN-K1P1-MQ8N. [DOI] [PubMed] [Google Scholar]
- 3.Lyness JM, King DA, Cox C, Yoediono Z, Caine ED. The importance of subsyndromal depression in older primary care patients: prevalence and associated functional disability. J Am Geriatr Soc. 1999;47(6):647–52. doi: 10.1111/j.1532-5415.1999.tb01584.x. [DOI] [PubMed] [Google Scholar]
- 4.Steffens DC, Skoog I, Norton MC, Hart AD, Tschanz JT, Plassman BL, et al. Prevalence of depression and its treatment in an elderly population: the Cache County study. Arch Gen Psychiatry. 2000;57(6):601–7. doi: 10.1001/archpsyc.57.6.601. [DOI] [PubMed] [Google Scholar]
- 5.Judd LL, Akiskal HS. The clinical and public health relevance of current research on subthreshold depressive symptoms to elderly patients. Am J Geriatr Psychiatry. 2002;10(3):233–8. [PubMed] [Google Scholar]
- 6.Judd LL, Akiskal HS, Maser JD, Zeller PJ, Endicott J, Coryell W, et al. A prospective 12-year study of subsyndromal and syndromal depressive symptoms in unipolar major depressive disorders. Arch Gen Psychiatry. 1998;55(8):694–700. doi: 10.1001/archpsyc.55.8.694. [DOI] [PubMed] [Google Scholar]
- 7.Judd LL, Akiskal HS, Maser JD, Zeller PJ, Endicott J, Coryell W, et al. Major depressive disorder: a prospective study of residual subthreshold depressive symptoms as predictor of rapid relapse. J Affect Disord. 1998;50(2-3):97–108. doi: 10.1016/s0165-0327(98)00138-4. [DOI] [PubMed] [Google Scholar]
- 8.Olfson M, Broadhead WE, Weissman MM, Leon AC, Farber L, Hoven C, et al. Subthreshold psychiatric symptoms in a primary care group practice. Arch Gen Psychiatr. 1996;53(10):880–6. doi: 10.1001/archpsyc.1996.01830100026004. [DOI] [PubMed] [Google Scholar]
- 9.Adams KB, Moon H. Subthreshold depression: characteristics and risk factors among vulnerable elders. Aging Ment Health. 2009 Sep;13(5):682–92. doi: 10.1080/13607860902774501. [DOI] [PubMed] [Google Scholar]
- 10.Bruce ML. Depression and disability in late life: directions for future research. Am J Geriatr Psychiatry. 2001;9(2):102–12. [PubMed] [Google Scholar]
- 11.Frasure-Smith N, Lesperance F. Depression--a cardiac risk factor in search of a treatment. JAMA. 2003;289(23):3171–3. doi: 10.1001/jama.289.23.3171. 18. [DOI] [PubMed] [Google Scholar]
- 12.Prina AM, Deeg D, Brayne C, Beekman A, Huisman M. The Association between depressive symptoms and non-psychiatric hospitalisation in older adults. PLoS One. 2012;7(4):e34821. doi: 10.1371/journal.pone.0034821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Alexopoulos GS, Vrontou C, Kakuma T, Meyers BS, Young RC, Klausner E, et al. Disability in geriatric depression. Am J Psychiatry. 1996 Jul;153(7):877–85. doi: 10.1176/ajp.153.7.877. [DOI] [PubMed] [Google Scholar]
- 14.Lenze EJ, Sheffrin M, Driscoll HC, Mulsant BH, Pollock BG, Dew MA, et al. Incomplete response in late-life depression: getting to remission. Dialogues Clin Neurosci. 2008;10(4):419–30. doi: 10.31887/DCNS.2008.10.4/jlenze. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Germain A, Kupfer DJ. Circadian rhythm disturbances in depression. Hum Psychopharmacol. 2008;23(7):571–85. doi: 10.1002/hup.964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.McClung CA. Circadian rhythms and mood regulation: insights from pre-clinical models. Eur Neuropsychopharmacol. 2011;21(Suppl 4):S683–93. doi: 10.1016/j.euroneuro.2011.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Monteleone P, Martiadis V, Maj M. Circadian rhythms and treatment implications in depression. Prog Neuropsychopharmacol Biol Psychiatry. 2011;35(7):1569–74. doi: 10.1016/j.pnpbp.2010.07.028. [DOI] [PubMed] [Google Scholar]
- 18.Lamont EW, Legault Coutu D, Cermakian N, Boivin DB. The role of circadian clock genes in mental disorders. Dialogues Clin Neurosci. 2007;9(3):333–42. doi: 10.31887/DCNS.2007.9.3/elamont. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Linkowski P, Mendlewicz J, Leclercq R, Brasseur M, Hubain P, Golstein J, et al. The 24-hour profile of adrenocorticotropin and cortisol in major depressive illness. J Clin Endocrinol Metab. 1985;61(3):429–38. doi: 10.1210/jcem-61-3-429. [DOI] [PubMed] [Google Scholar]
- 20.Hofman MA, Swaab DF. Alterations in circadian rhythmicity of the vasopressin-producing neurons of the human suprachiasmatic nucleus (SCN) with aging. Brain Res. 1994;651(1-2):134–42. doi: 10.1016/0006-8993(94)90689-0. [DOI] [PubMed] [Google Scholar]
- 21.Hampp G, Albrecht U. The circadian clock and mood-related behavior. Commun Integr Biol. 2008;1(1):1–3. doi: 10.4161/cib.1.1.6286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hampp G, Ripperger JA, Houben T, Schmutz I, Blex C, Perreau-Lenz S, et al. Regulation of monoamine oxidase A by circadian-clock components implies clock influence on mood. Curr Biol. 2008;18(9):678–83. doi: 10.1016/j.cub.2008.04.012. [DOI] [PubMed] [Google Scholar]
- 23.Artioli P, Lorenzi C, Pirovano A, Serretti A, Benedetti F, Catalano M, et al. How do genes exert their role? Period 3 gene variants and possible influences on mood disorder phenotypes. Eur Neuropsychopharmacol. 2007;17(9):587–94. doi: 10.1016/j.euroneuro.2007.03.004. [DOI] [PubMed] [Google Scholar]
- 24.Lavebratt C, Sjoholm LK, Partonen T, Schalling M, Forsell Y. PER2 variantion is associated with depression vulnerability. Am J Med Genet B Neuropsychiatr Genet. 2010;153B(2):570–81. doi: 10.1002/ajmg.b.31021. [DOI] [PubMed] [Google Scholar]
- 25.Soria V, Martinez-Amoros E, Escaramis G, Valero J, Crespo JM, Gutierrez-Zotes A, et al. Resequencing and association analysis of arylalkylamine N-acetyltransferase (AANAT) gene and its contribution to major depression susceptibility. J Pineal Res. 2010;49(1):35–44. doi: 10.1111/j.1600-079X.2010.00763.x. [DOI] [PubMed] [Google Scholar]
- 26.Soria V, Martinez-Amoros E, Escaramis G, Valero J, Perez-Egea R, Garcia C, et al. Differential association of circadian genes with mood disorders: CRY1 and NPAS2 are associated with unipolar major depression and CLOCK and VIP with bipolar disorder. Neuropsychopharmacology. 2010;35(6):1279–89. doi: 10.1038/npp.2009.230. [Research Support, Non-U.S. Gov’t]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Galecki P, Szemraj J, Bartosz G, Bienkiewicz M, Galecka E, Florkowski A, et al. Single-nucleotide polymorphisms and mRNA expression for melatonin synthesis rate-limiting enzyme in recurrent depressive disorder. J Pineal Res. 2010;48(4):311–7. doi: 10.1111/j.1600-079X.2010.00754.x. [DOI] [PubMed] [Google Scholar]
- 28.Utge SJ, Soronen P, Loukola A, Kronholm E, Ollila HM, Pirkola S, et al. Systematic analysis of circadian genes in a population-based sample reveals association of TIMELESS with depression and sleep disturbance. PLoS One. 2010;5(2):e9259. doi: 10.1371/journal.pone.0009259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Terracciano A, Tanaka T, Sutin AR, Sanna S, Deiana B, Lai S, et al. Genome-wide association scan of trait depression. Biol Psychiatry. 2010;68(9):811–7. doi: 10.1016/j.biopsych.2010.06.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Carrier J, Monk TH, Buysse DJ, Kupfer DJ. Amplitude reduction of the circadian temperature and sleep rhythms in the elderly. Chronobiol Int. 1996 Nov;13(5):373–86. doi: 10.3109/07420529609012661. [DOI] [PubMed] [Google Scholar]
- 31.Hofman MA, Swaab DF. Living by the clock: the circadian pacemaker in older people. Ageing Res Rev. 2006;5(1):33–51. doi: 10.1016/j.arr.2005.07.001. [DOI] [PubMed] [Google Scholar]
- 32.Munch M, Cajochen C, Wirz-Justice A. Sleep and circadian rhythms in ageing. Z Gerontol Geriatr. 2005;38(Suppl 1):I21–3. doi: 10.1007/s00391-005-1106-z. [DOI] [PubMed] [Google Scholar]
- 33.Maglione JE, Ancoli-Israel S, Peters KW, Paudel ML, Yaffe K, Ensrud KE, et al. Depressive Symptoms and Circadian Activity Rhythm Disturbances in Community-Dwelling Older Women. Am J Geriatr Psychiatry. 2013;22(4):349–61. doi: 10.1016/j.jagp.2012.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cummings SR, Black DM, Nevitt MC, Browner W, Cauley J, Ensrud K, et al. Bone density at various sites for prediction of hip fractures. The Study of Osteoporotic Fractures Research Group. Lancet. 1993;341(8837):72–5. doi: 10.1016/0140-6736(93)92555-8. 9. [DOI] [PubMed] [Google Scholar]
- 35.Blank JB, Cawthon PM, Carrion-Petersen ML, Harper L, Johnson JP, Mitson E, et al. Overview of recruitment for the osteoporotic fractures in men study (MrOS) Contemp Clin Trials. 2005;26(5):557–68. doi: 10.1016/j.cct.2005.05.005. [DOI] [PubMed] [Google Scholar]
- 36.Orwoll E, Blank JB, Barrett-Connor E, Cauley J, Cummings S, Ensrud K, et al. Design and baseline characteristics of the osteoporotic fractures in men (MrOS) study--a large observational study of the determinants of fracture in older men. Contemp Clin Trials. 2005;26(5):569–85. doi: 10.1016/j.cct.2005.05.006. [DOI] [PubMed] [Google Scholar]
- 37.Evans DS, Parimi N, Nievergelt CM, Blackwell T, Redline S, Ancoli-Israel S, et al. Common genetic variants in ARNTL and NPAS2 and at chromosome 12p13 are associated with objectively measured sleep traits in the elderly. Sleep. 2013;36(3):431–46. doi: 10.5665/sleep.2466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.de Bakker PI, Yelensky R, Pe’er I, Gabriel SB, Daly MJ, Altshuler D. Efficiency and power in genetic association studies. Nat Genet. 2005 Nov;37(11):1217–23. doi: 10.1038/ng1669. [DOI] [PubMed] [Google Scholar]
- 39.Wigginton JE, Cutler DJ, Abecasis GR. A note on exact tests of Hardy-Weinberg equilibrium. Am J Hum Genet. 2005;76(5):887–93. doi: 10.1086/429864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, et al. PLINK: a tool set for whole-genome association and population based linkage analyses. Am J Hum Genet. 2007;81(3):559–75. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Brink TL. Clinical Gerontology: A Guide to Assessment and Intervention. Howarth Press; New York: 1986. [Google Scholar]
- 42.Paudel ML, Taylor BC, Diem SJ, Stone KL, Ancoli-Israel S, Redline S, et al. Association between depressive symptoms and sleep disturbances in community-dwelling older men. J Am Geriatr Soc. 2008;56(7):1228–35. doi: 10.1111/j.1532-5415.2008.01753.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Almeida OP, Almeida SA. Short versions of the geriatric depression scale: a study of their validity for the diagnosis of a major depressive episode according to ICD-10 and DSM-IV. Int J Geriatr Psychiatry. 1999;14(10):858–65. doi: 10.1002/(sici)1099-1166(199910)14:10<858::aid-gps35>3.0.co;2-8. [DOI] [PubMed] [Google Scholar]
- 44.Pahor M, Chrischilles EA, Guralnik JM, Brown SL, Wallace RB, Carbonin P. Drug data coding and analysis in epidemiologic studies. Eur J Epidemiol. 1994;10(4):405–11. doi: 10.1007/BF01719664. [DOI] [PubMed] [Google Scholar]
- 45.Teng EL, Chui HC. The Modified Mini-Mental State (3MS) examination. J Clin Psychiatry. 1987;48(8):314–8. [PubMed] [Google Scholar]
- 46.Folstein MF, Folstein SE, McHugh PR. “Mini-mental state”. A practical method for grading the cognitive state of patients for the clinician. J Psychiatr Res. 1975;12(3):189–98. doi: 10.1016/0022-3956(75)90026-6. [DOI] [PubMed] [Google Scholar]
- 47.Nyholt DR. A simple correction for multiple testing for single-nucleotide polymorphisms in linkage disequilibrium with each other. Am J Hum Genet. 2004;74(4):765–9. doi: 10.1086/383251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Willer CJ, Li Y, Abecasis GR. METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics. 2010;26(17):2190–1. doi: 10.1093/bioinformatics/btq340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gauderman W, Morrison J. QUANTO 1.1: A computer program for power and sample size calculations for genetic-epidemiology studies. 2006 http://hydra.usc.edu/gxe.
- 50.McClung CA. Circadian rhythms and mood regulation: insights from pre-clinical models. Eur Neuropsychopharmacol. 2012;21(Suppl 4):S683–93. doi: 10.1016/j.euroneuro.2011.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.McClung CA. Circadian genes, rhythms and the biology of mood disorders. Pharmacol Ther. 2007;114(2):222–32. doi: 10.1016/j.pharmthera.2007.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Dijk DJ, Archer SN. PERIOD3, circadian phenotypes, and sleep homeostasis. Sleep Med Rev. 2010;14(3):151–60. doi: 10.1016/j.smrv.2009.07.002. [DOI] [PubMed] [Google Scholar]
- 53.Archer SN, Robilliard DL, Skene DJ, Smits M, Williams A, Arendt J, et al. A length polymorphism in the circadian clock gene Per3 is linked to delayed sleep phase syndrome and extreme diurnal preference. Sleep. 2003;26(4):413–5. doi: 10.1093/sleep/26.4.413. [DOI] [PubMed] [Google Scholar]
- 54.Lazar AS, Slak A, Lo JC, Santhi N, von Schantz M, Archer SN, et al. Sleep, diurnal preference, health, and psychological well-being: a prospective single allelic variation study. Chronobiol Int. 2012;29(2):131–46. doi: 10.3109/07420528.2011.641193. [DOI] [PubMed] [Google Scholar]
- 55.Viola AU, Chellappa SL, Archer SN, Pugin F, Gotz T, Dijk DJ, et al. Interindividual differences in circadian rhythmicity and sleep homeostasis in older people: effect of a PER3 polymorphism. Neurobiol Aging. 2012;33(5):1010, e17–27. doi: 10.1016/j.neurobiolaging.2011.10.024. [DOI] [PubMed] [Google Scholar]
- 56.Benedetti F, Dallaspezia S, Colombo C, Pirovano A, Marino E, Smeraldi E. A length polymorphism in the circadian clock gene Per3 influences age at onset of bipolar disorder. Neurosci Lett. 2008;445(2):184–7. doi: 10.1016/j.neulet.2008.09.002. [DOI] [PubMed] [Google Scholar]
- 57.Dallaspezia S, Lorenzi C, Pirovano A, Colombo C, Smeraldi E, Benedetti F. Circadian clock gene Per3 variants influence the postpartum onset of bipolar disorder. Eur Psychiatry. 2011;26(3):138–40. doi: 10.1016/j.eurpsy.2010.11.009. [DOI] [PubMed] [Google Scholar]
- 58.Kripke DF, Nievergelt CM, Tranah GJ, Murray SS, Rex KM, Grizas AP, et al. FMR1, circadian genes and depression: suggestive associations or false discovery? J Circadian Rhythms. 2013;11(1):3. doi: 10.1186/1740-3391-11-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Nievergelt CM, Kripke DF, Barrett TB, Burg E, Remick RA, Sadovnick AD, et al. Suggestive evidence for association of the circadian genes PERIOD3 and ARNTL with bipolar disorder. Am J Med Genet B Neuropsychiatr Genet. 2006;141B(3):234–41. doi: 10.1002/ajmg.b.30252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Cooklin AR, Giallo R, Rose N. Parental fatigue and parenting practices during early childhood: an Australian community survey. Child: care, health and development. 2011;38(5):654–64. doi: 10.1111/j.1365-2214.2011.01333.x. [DOI] [PubMed] [Google Scholar]
- 61.Bruguerolle B, Simon N. Biologic rhythms and Parkinson’s disease: a chronopharmacologic approach to considering fluctuations in function. Clin Neuropharmacol. 2002;25(4):194–201. doi: 10.1097/00002826-200207000-00002. [DOI] [PubMed] [Google Scholar]
- 62.Ros L, Latorre JM, Aguilar MJ, Serrano JP, Navarro B, Ricarte JJ. Factor structure and psychometric properties of the center for epidemiologic studies depression scale (CES-D) in older populations with and without cognitive impairment. Int J Aging Hum Dev. 72(2):83–110. doi: 10.2190/AG.72.2.a. [DOI] [PubMed] [Google Scholar]
- 63.Garriock HA, Kraft JB, Shyn SI, Peters EJ, Yokoyama JS, Jenkins GD, et al. A genomewide association study of citalopram response in major depressive disorder. Biol Psychiatry. 2010;67(2):133–8. doi: 10.1016/j.biopsych.2009.08.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Miller MW, Wolf EJ, Logue MW, Baldwin CT. The retinoid-related orphan receptor alpha (RORA) gene and fear-related psychopathology. J Affect Disord. 2013;151(2):702–8. doi: 10.1016/j.jad.2013.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Le-Niculescu H, Patel SD, Bhat M, Kuczenski R, Faraone SV, Tsuang MT, et al. Convergent functional genomics of genome-wide association data for bipolar disorder: comprehensive identification of candidate genes, pathways and mechanisms. Am J Med Genet B Neuropsychiatr Genet. 2009;150B(2):155–81. doi: 10.1002/ajmg.b.30887. [DOI] [PubMed] [Google Scholar]
- 66.Logue MW, Baldwin C, Guffanti G, Melista E, Wolf EJ, Reardon AF, et al. A genome-wide association study of post-traumatic stress disorder identifies the retinoid-related orphan receptor alpha (RORA) gene as a significant risk locus. Mol Psychiatry. 2012;18(8):937–42. doi: 10.1038/mp.2012.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Neale BM, Lasky-Su J, Anney R, Franke B, Zhou K, Maller JB, et al. Genome-wide association scan of attention deficit hyperactivity disorder. Am J Med Genet B Neuropsychiatr Genet. 2008;147B(8):1337–44. doi: 10.1002/ajmg.b.30866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Nguyen A, Rauch TA, Pfeifer GP, Hu VW. Global methylation profiling of lymphoblastoid cell lines reveals epigenetic contributions to autism spectrum disorders and a novel autism candidate gene, RORA, whose protein product is reduced in autistic brain. FASEB J. 2010;24(8):3036–51. doi: 10.1096/fj.10-154484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Sarachana T, Xu M, Wu RC, Hu VW. Sex hormones in autism: androgens and estrogens differentially and reciprocally regulate RORA, a novel candidate gene for autism. PLoS One. 2011;6(2):e17116. doi: 10.1371/journal.pone.0017116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Stephenson KM, Schroder CM, Bertschy G, Bourgin P. Complex interaction of circadian and non-circadian effects of light on mood: shedding new light on an old story. Sleep Med Rev. 2012;16(5):445–54. doi: 10.1016/j.smrv.2011.09.002. [DOI] [PubMed] [Google Scholar]
- 71.Miller AH, Maletic V, Raison CL. Inflammation and its discontents: the role of cytokines in the pathophysiology of major depression. Biol Psychiatry. 2009;65(9):732–41. doi: 10.1016/j.biopsych.2008.11.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ioannidis JP, Tarone R, McLaughlin JK. The false-positive to false-negative ratio in epidemiologic studies. Epidemiology. 2011;22(4):450–6. doi: 10.1097/EDE.0b013e31821b506e. [DOI] [PubMed] [Google Scholar]
- 73.Ioannidis JP. Why most discovered true associations are inflated. Epidemiology. 2008;19(5):640–8. doi: 10.1097/EDE.0b013e31818131e7. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.