Proteins catalyze crucial reactions via unstable, high-energy chemical intermediates. In the absence of physiological substrates, activated redox cofactors become ticking time bombs, capable of producing oxidative damage to the protein. In PNAS, Gray and Winkler (1) propose that chains of tryptophan (Trp) and tyrosine (Tyr) residues may serve as escape routes for potentially damaging, highly oxidizing electron holes (oxidizing equivalents) that are generated in enzymatic reactions. The aromatic residues are suggested to provide charge-shuttling pathways that lead out to the protein surface from buried active sites. Once outside the protein, the holes can be safely scavenged by cellular reductants.
Since at least the 1960s, aromatic amino acids were postulated to play a special role in biological redox chemistry as electron hole carriers (2). Circadian rhythm photochemistry, photosynthetic water splitting, nucleic acid biosynthesis, and cell signaling are now well understood to engage redox-active Trp and Tyr residues as essential parts of their function (3). Could Tyr and Trp also play a protective role during enzymatic reactions (such as the oxidation of aliphatic C–H bonds) that require high-energy intermediates (4, 5)? The answer will be dictated by the set of time scales at play: Hole hopping from the activated redox cofactor to the first Trp/Tyr must be slower than catalysis (to avoid disabling the enzyme’s valuable catalytic function) but faster than oxidative protein damage. Gray and Winkler’s (1) query of the Protein Data Bank (e.g., they searched for Trp/Tyr pairs within a 5-Å edge-to-edge cutoff distance) provides a critical starting point for more detailed investigations of these hole-transfer chains.
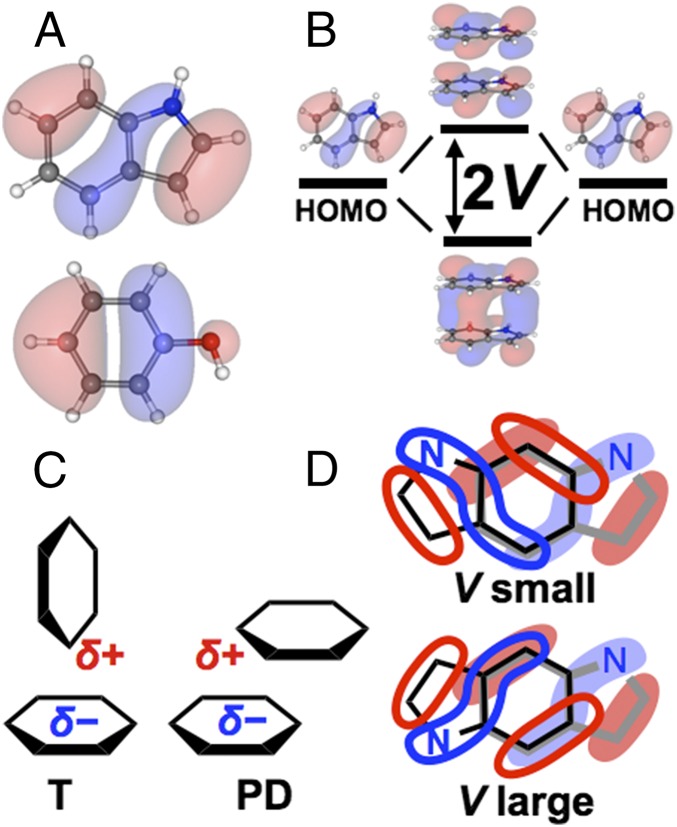
Electron-transfer (ET) rates in biochemical reactions are usually proportional to the square of the electronic coupling interaction energy (V, Fig. 1B) between the electron donor and acceptor, facilitated by the protein medium, or by direct van der Waals (vdW) contact between redox species. V decays rapidly with distance, and the steepness of this decay depends on the structure of the intervening medium (9). ET pathways and relative donor/acceptor orientations influence wave function overlap, and thus determine V and the ET rate. The relative Trp and Tyr orientations fine-tune the hole-hopping rates. Two orientations are common among protein aromatics (Fig. 1C), the so-called parallel-displaced (PD) and the T-shape (T) orientations, where ring interactions capture quadrupolar electrostatic stabilization (10). PD geometries may use strong stacking interactions (akin to sigma bonding between p-orbitals) to maximize V; however, the strength of such interactions depends on orbital symmetry, as shown in Fig. 1D for indole pairs. Indeed, the greater than 10-fold stronger V for Tyr90-Trp231 in Fig. 2 arises from its PD geometry, compared with the T geometry of the other pairs.
Fig. 1.
The electronic coupling interaction energy (V) between indole and phenol pairs depends on orbital overlap. (A) Highest occupied molecular orbital (HOMO) of indole and phenol. (B) V between degenerate orbitals (here, indole HOMOs) is one-half the energy splitting between bonding and antibonding HOMO combinations. (C) Prevalent T and PD geometries between aromatics. (D) Two PD indole pairs (black and gray, within each pair), aligned by six-membered benzyl rings, with overlaid HOMOs. Orbital lobes that overlap an equal amount of positive (red) and negative (blue) amplitude have negligible contribution to V. Destructive interference between orbital lobes of T-shaped orientations are responsible for the weak V values (1–4 meV) shown in Fig. 2.
Fig. 2.
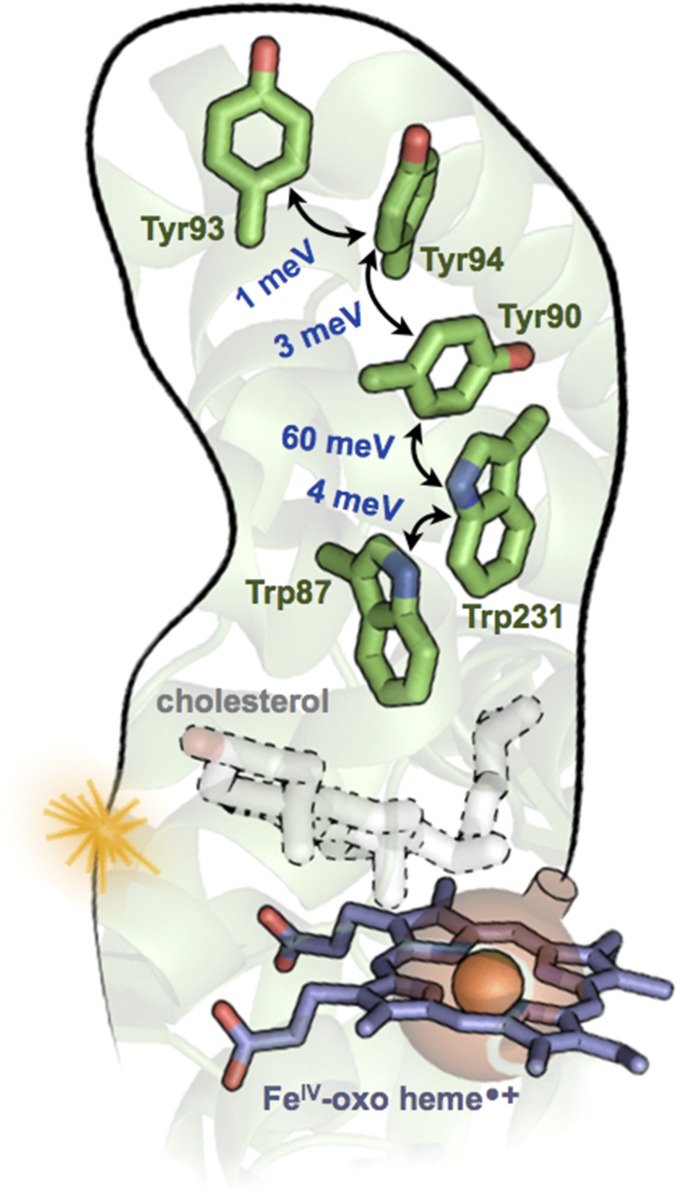
A chain of Trp and Tyr residues in the cytochrome P450 protein CYP11A1 (PDB ID code 3N9Y) discussed by Gray and Winkler (1). DFT-derived V values are shown in blue [the method in refs. 6 and 7 was applied using NWChem (8), with the M06-HF functional, the D3 dispersion correction, the cc-pVTZ basis set for C and H atoms, and aug-cc-pVTZ for N and O]. Each hopping rate is proportional to V2. The greater than 10-fold stronger V between Tyr90–Trp231 arises from its PD geometry, compared with the T geometry of the other pairs. Because each pair of residues is within vdW contact, no protein medium is included in the computations of V. The Fe(IV)-oxo heme radical cation (CpdI) of P450, shown as a bomb ready to explode, converts cholesterol to pregnenolone. If unable to react with cholesterol, CpdI might be defused by the nearby Trp87 (followed by hole hopping to the protein surface at Tyr93) before damaging the protein.
Within a subset of orientations (e.g., T or PD), additional geometric effects between aromatic pairs may be used to control V. For example, by placing the nodal planes that pass through the indole six-member ring (Fig. 1A) at 90° to each other, the indole–indole coupling can be reduced from ∼165 meV (Fig. 1D, Bottom) to <1 meV (Fig. 1D, Top), substantially modulating the hole escape kinetics. Although aromatic residues play a known role in structural stability and substrate binding, might the modulation of V be a key factor in the evolution of hole-hopping chains?
Cytochrome P450
Gray and Winkler (1) identified a five-residue Trp/Tyr chain in the mitochondrial cholesterol metabolizing cytochrome P450, CYP11A1 (Fig. 2). Each Trp or Tyr is in vdW contact with its neighbor. During the conversion of cholesterol to pregnenolone, the heme of CYP11A1 likely cycles through a highly oxidizing intermediate known as compound I (CpdI). What are the consequences of CpdI formation in the absence of cholesterol? Could the Trp/Tyr chain defuse the CpdI bomb before it damages the protein? Fig. 2 stresses an important functional point: The deactivation of CpdI by Trp/Tyr chains should not outcompete catalysis when substrate is bound. (Indeed, cholesterol is closer to the heme iron than is Trp87: 4.3 vs. 9.6 Å.)
The Trp and Tyr orientations in CYP11A1 sample characteristic T and PD geometries (Fig. 1C). Fig. 2 shows that the magnitude of V is sensitive to the orientation: V ranges from 1 meV (T) to 60 meV (PD). Such differences in V would manifest as 103- to 104-fold differences in charge transfer rates between aromatic pairs. Still, protein fluctuations are expected to influence the mean squared values of V and, consequently, the transport rates.
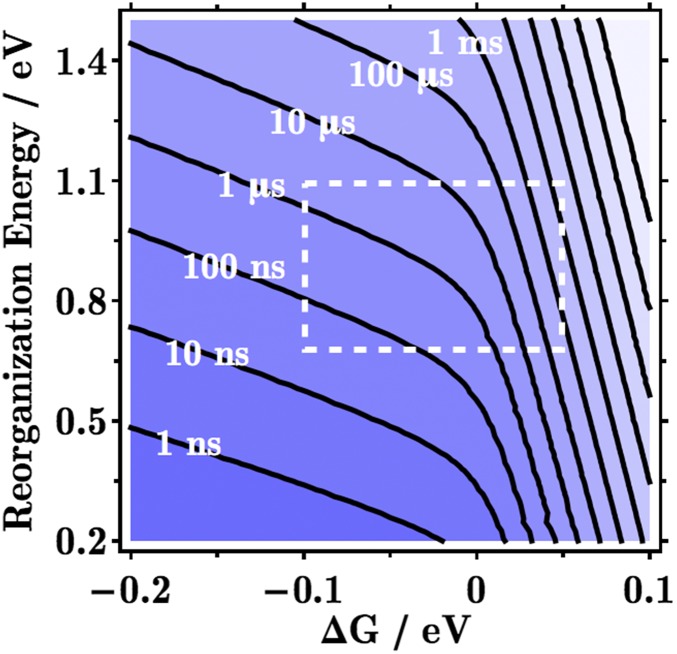
Using the values of V from Fig. 2 in a simple kinetic mechanism ) with rates governed by the nonadiabatic ET theory (11), the mean first passage time (MFPT) (12) from CpdI to Tyr93 is given in Fig. 3. For a particular free energy difference (ΔG) and reorganization energy (λ) of each step, the MFPT is the characteristic time scale for hole migration from CpdI to the protein surface. For example, when ΔG = 0 and λ = 0.8 eV, the MFPT is ∼1 μs [very similar to the 3-μs value reported by Gray and Winkler (1)]. For these parameters, turnover of CpdI during catalysis must proceed in the submicrosecond range, in order for the enzyme to function. In the absence of substrate, might Trp87 maneuver to within vdW contact of CpdI, thereby facilitating rapid hole transfer to the protein surface?
Fig. 3.
Contour lines of the mean first passage time for a hole to escape the heme to the protein surface via the Trp/Tyr chain of CYP11A1 in Fig. 2, as a function of the free energy difference (ΔG) and the reorganization energy (λ) of each hopping step (using the Marcus nuclear factor). For given ΔG and λ values, catalysis must be faster than the time scale shown here, and oxidative protein damage must be slower, in order for the Trp/Tyr chain to be competent to defuse the redox bomb (CpdI). Biological values of ΔG and λ typically fall within the dashed box. We used the square barrier tunneling model with parameters chosen by Gray and Winkler (1) for the CpdI to Trp87 step: kET = 1013 exp[− β (R-R0) ] exp [− (ΔG + λ)2/(4 λ kBT) ], with R = 9.6 Å, R0 = 3 Å, β = 1.1 Å-1, and kBT = 1/40 eV.
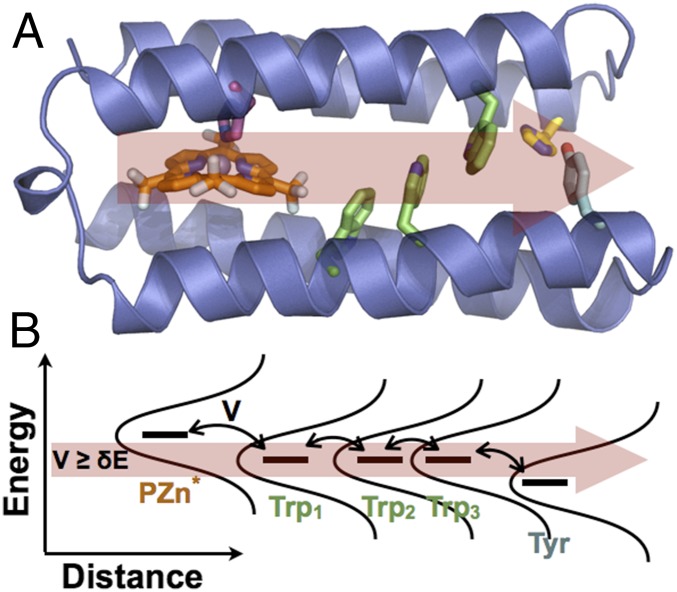
Emerging flickering resonance models for charge movement through redox chains in vdW contact may be applicable to Trp and Tyr hole-transfer chains as well (9, 13). The energy levels of Trp/Tyr stacks will fluctuate in and out of energetic degeneracy (Fig. 4B). These flickering resonances can enable ballistic charge flow through such chains (13). Could the proposed chemistry of these vdW contact aromatic chains enable resonant charge transport away from the fragile active sites to the protein surface?
Fig. 4.
(A) De novo protein design for vectorial hole transfer (red arrow) via Trp (green) and Tyr (blue) residues. Electronic excitation of a photooxidant (orange) triggers hole hopping. (B) Flickering resonance model for charge transport through the protein in A. The red arrow demarks the energy width (δE) that is comparable to the size of the coupling interactions (V) between hole-transfer partners that is required for a functional flickering resonance pathway.
The frequency of Trp/Tyr chains in proteins noted by Gray and Winkler (1), as well as the established role played by these amino acids in biocatalysis, warrant the mechanistic study of how biological hole transport pathways function in detail. In this regard, de novo protein design (Fig. 4) allows a systematic exploration of how the protein environment may tune hole propagation through designed hole transfer chains. Practically, hole-transfer chains may be valuable design elements to ensure the stability and function of critical enzymes in metabolically engineered organisms that overproduce oxidized metabolites, for example, in the generation of biofuels.
Redox cofactors are essential components in life-sustaining enzymatic reactions, but the high oxidation states accessed by these cofactors may damage and ultimately disable the protein machinery (1). The role proposed for Trp/Tyr chains in minimizing the occurrence of oxidative damage suggests that evolution may have found a compromise between catalytic function and protection: If the time scale for oxidative damage defines the fuse length of the redox bomb, Trp/Tyr chains may let the fuse burn short without explosive consequences.
Acknowledgments
This work was supported by National Institutes of Health Grants GM-48043 and GM-71628.
Footnotes
The authors declare no conflict of interest.
See companion article on page 10920.
References
- 1.Gray HB, Winkler JR. Hole hopping through tyrosine/tryptophan chains protects proteins from oxidative damage. Proc Natl Acad Sci USA. 2015;112:10920–10925. doi: 10.1073/pnas.1512704112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Winfield ME. Electron transfer within and between haemoprotein molecules. J Mol Biol. 1965;12(3):600–611. doi: 10.1016/s0022-2836(65)80314-x. [DOI] [PubMed] [Google Scholar]
- 3.Migliore A, Polizzi NF, Therien MJ, Beratan DN. Biochemistry and theory of proton-coupled electron transfer. Chem Rev. 2014;114(7):3381–3465. doi: 10.1021/cr4006654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Winkler JR, Gray HB. Long-range electron tunneling. J Am Chem Soc. 2014;136(8):2930–2939. doi: 10.1021/ja500215j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Winkler JR, Gray HB. Could tyrosine and tryptophan serve multiple roles in biological redox processes? Phil Trans R Soc A. 2015;373(2037):20140178. doi: 10.1098/rsta.2014.0178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Migliore A. Full-electron calculation of effective electronic couplings and excitation energies of charge transfer states: Application to hole transfer in DNA π-stacks. J Chem Phys. 2009;131(11):114113. doi: 10.1063/1.3232007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Migliore A. Nonorthogonality problem and effective electronic coupling calculation: Application to charge transfer in π-stacks relevant to biochemistry and molecular electronics. J Chem Theory Comput. 2011;7(6):1712–1725. doi: 10.1021/ct200192d. [DOI] [PubMed] [Google Scholar]
- 8.Valiev M, et al. NWChem: A comprehensive and scalable open-source solution for large scale molecular simulations. Comput Phys Commun. 2010;181(9):1477–1489. [Google Scholar]
- 9.Beratan DN, et al. Charge transfer in dynamical biosystems, or the treachery of (static) images. Acc Chem Res. 2015;48(2):474–481. doi: 10.1021/ar500271d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.McGaughey GB, Gagné M, Rappé AK. π-Stacking interactions. Alive and well in proteins. J Biol Chem. 1998;273(25):15458–15463. doi: 10.1074/jbc.273.25.15458. [DOI] [PubMed] [Google Scholar]
- 11.Nitzan A. Chemical Dynamics in Condensed Phases: Relaxation, Transfer and Reactions in Condensed Molecular Systems. Oxford Univ Press; New York: 2006. [Google Scholar]
- 12.Polizzi NF, Skourtis SS, Beratan DN. Physical constraints on charge transport through bacterial nanowires. Faraday Discuss. 2012;155:43–62, discussion 103–114. doi: 10.1039/c1fd00098e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang Y, Liu C, Balaeff A, Skourtis SS, Beratan DN. Biological charge transfer via flickering resonance. Proc Natl Acad Sci USA. 2014;111(28):10049–10054. doi: 10.1073/pnas.1316519111. [DOI] [PMC free article] [PubMed] [Google Scholar]