Significance
The regulation of protein synthesis is a vital process in all cells. We have determined the structure of the BPI-inducible protein A (BipA), a GTPase involved in bacterial stress response, in various biologically relevant states, including bound to the ribosome in its active form. Our study provides the first structural basis to our knowedge for BipA working as a ribosome-dependent translational GTPase factor and serves as a crucial step for elucidating BipA functioning in vivo. Because no other stress response factor is known to interact with the A-site tRNA, our findings provide a novel mechanistic insight of translational regulation by BipA. Furthermore, the distinctive structural attributes of ribosome upon BipA binding can shed new light on the translational factor recruitment and GTPase activation mechanisms by the ribosome.
Keywords: BipA, ribosome, translational GTPase factors, X-ray crystallography, cryo-electron microscopy
Abstract
BPI-inducible protein A (BipA) is a member of the family of ribosome-dependent translational GTPase (trGTPase) factors along with elongation factors G and 4 (EF-G and EF4). Despite being highly conserved in bacteria and playing a critical role in coordinating cellular responses to environmental changes, its structures (isolated and ribosome bound) remain elusive. Here, we present the crystal structures of apo form and GTP analog, GDP, and guanosine-3′,5′-bisdiphosphate (ppGpp)-bound BipA. In addition to having a distinctive domain arrangement, the C-terminal domain of BipA has a unique fold. Furthermore, we report the cryo-electron microscopy structure of BipA bound to the ribosome in its active GTP form and elucidate the unique structural attributes of BipA interactions with the ribosome and A-site tRNA in the light of its possible function in regulating translation.
Bacterial protein synthesis involves four main translational GTPase (trGTPase) factors: initiation factor 2 (IF2), elongation factors Tu and G (EF-Tu and EF-G), and release factor 3 (RF3). These factors catalyze major steps in translation initiation, elongation (both decoding and mRNA–tRNA complex translocation), and termination, in a GTP-dependent manner. Several additional GTPase factors, including EF4 (formerly known as LepA), BipA, and RelA, have been revealed to be associated with ribosomes under stress conditions (1).
Both EF4 and BipA are paralogs of EF-G (1–5). Although EF4 is highly conserved in bacteria (4), deletion of ef4 gene causes no evident phenotype in Escherichia coli under optimal growth conditions (6). However, EF4 was shown to notably improve protein synthesis under stress conditions (7). Qin et al. (2) reported a unique function of EF4 promoting the back translocation of the elongation complex by one codon, hence presumably providing a second chance for EF-G to carry out a correct translocation.
BipA (BPI-inducible protein A) gene is highly conserved among bacterial and chloroplast genomes (4) and has been implicated in regulating a variety of cellular processes including bacterial virulence, symbiosis, various stress responses, resistance to host defenses, swarming motility, biofilm, and capsule formation (8–10). As is the case with EF4, BipA is not required under optimal growth conditions but becomes an essential factor for bacterial survival at low temperature, nutrient depletion, and various other stress conditions (1, 9). The diverse nature of these processes underscores the global regulatory properties of BipA. Similarity to classical trGTPases and EF4 led to the speculation that BipA affects translation through directly interacting with the ribosome. For example, wild-type (fully modified) ribosomes seem to depend on BipA for translation of specific mRNAs (11). Furthermore, as with EF4, overexpression of BipA inhibits transfer-messenger mRNA (tmRNA)-dependent peptide tagging activity of nonstop messages on ribosome (6). Thus, BipA likely functions as an elongation factor as well. Consistent with this notion, BipA is able to bind to 70S ribosome in a GTP-dependent manner and its GTPase activity is enhanced in the presence of ribosomes, a characteristic feature of classical trGTPase factors (5, 12). Salmonella enterica BipA has been shown to interact with either 70S ribosomes or 30S subunits depending on the relative abundance of GTP and of the stress alarmone guanosine-3′,5′-bisdiphosphate (ppGpp), respectively (12). In addition, a recent study links BipA to ribosome biogenesis because bipA gene deletion results in perturbed 50S subunit processing and assembly, particularly at low temperatures (13). Although the evidence for BipA involvement in ribosome biosynthesis and/or functioning in translation is mounting, its exact role remains elusive.
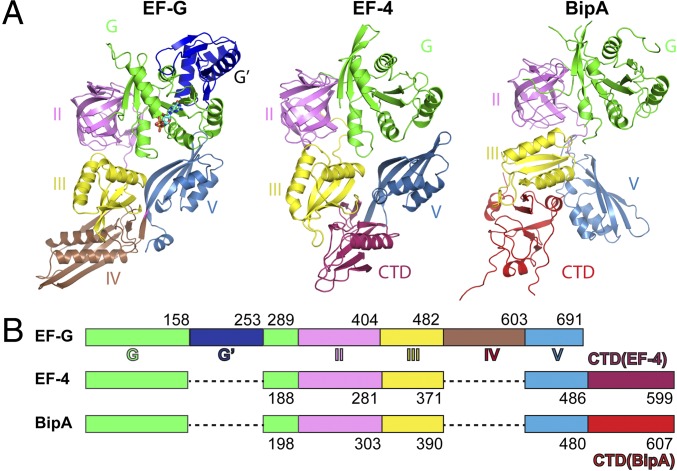
As a member of the ribosome-dependent trGTPase family, BipA is proposed to share structural similarity with EF4 and EF-G (4, 5). Indeed, all three consist of five domains, of which the N-terminal G domain (nucleotide-binding domain), the β-barrel domain (domain II), and the two α/β-domains (domains III and IV) are topologically equivalent (5) (Fig. 1). EF-G has G′ domain inserted into its G domain and a unique domain IV, whereas unique C-terminal domains (CTDs) are present in BipA and EF4 (Fig. 1). Despite the similarity, the three proteins have distinct functions probably attributed to their varied domain arrangements and ribosome-binding modes.
Fig. 1.
Comparison of domain arrangement and overall structure of EF-G, EF4, and BipA. (A) Structures of isolated EF-G and EF4 are obtained from Protein Data Bank (PDB ID codes: 2BM0 and 3CB4, respectively). Structure of BipA apo form is presented. Domain I (green), also known as the G domain, is the nucleotide-binding region. G′ domain insertion (dark blue) is a characteristic feature of the EF-G protein. Domain II (violet) contains the translation factor signature β-barrel motif. Domains III (yellow) and V (sky blue) contain α/β-motifs. EF-G has a unique domain IV (brown), whereas EF4 and BipA have unique C-terminal domains (warm pink and red, respectively). The same color scheme is used throughout this work. (B) Schematic diagram depicting the domain arrangement of EF-G, EF4, and BipA.
Extensive structural studies (14–22) of EF-G bound to ribosome have generated a wealth of atomic or near-atomic resolution information on how EF-G, in particular the positioning of its domain IV in ribosome decoding center, facilitates translocation. Mutagenesis study of EF-G revealed that the highly conserved loops I and II of domain IV disrupt the interactions between the decoding center and the codon–anticodon duplex that act as the barrier for mRNA–tRNA complex translocation (23). Structural studies have also shed light on the molecular basis of how EF4 reverses EF-G catalyzed translocation through its CTD reaching into the PTC and interacting with the acceptor stem of the peptidyl-tRNA in the P site (24, 25). In contrast, structures of neither the isolated BipA nor BipA bound to ribosome, which could illuminate the molecular basis of BipA functioning in protein translation, have been characterized yet. Hence, we aimed to structurally characterize the various biologically relevant states of BipA on and off the ribosome, toward a better understanding of the detailed function of BipA. Note that during the revision process, a paper was published reporting the structure of isolated BipA (26).
Results
Overall Structure of BipA.
We crystalized the E. coli full-length BipA protein and determined the structure of its nucleotide-free (apo) form (Table S1). The overall shape of the isolated BipA vaguely resembles the number “8” and is composed of five domains (Fig. 1). There is no structural counterpart to domain IV of EF-G; therefore, the fourth domain in BipA is renumbered as domain V because of its homology to EF-G domain V (Fig. 1), in a similar way as used for EF4 (3). Domain I (residues 1–198; see Fig. S1A for residue numbering and labeling of the secondary structure elements of BipA), also named G domain for nucleotide binding, is universally conserved among trGTPase proteins except for EF-G, which has an additional G′ domain insertion (Fig. 1). Domains II and III comprise residues 199–303 and 304–390, which form the typical β-barrel (all β-strands) and a α/β structural motif, respectively. Similar to domain III, domain V (residues 391–480) contains four-stranded β-sheets flanked by two α-helices on one side (Fig. 1). Domains III and V in BipA, EF4, and EF-G (including its unique domain IV) are rather similar in overall shape and α/β arrangement (3, 27). Following domain V, the C-terminal domain (CTD) is a structural feature observed only in two trGTPase families, BipA and EF4 (3). The CTD of BipA, consisting of residues 481–605, forms a unique motif with two crossed β-sheets (comprising of two and four β-strands, respectively) wrapped by three short α-helices forming a nearly equilateral triangle (Fig. 1). The loop (residues 547–553) protruding from the triangle was not modeled because of the poor density map. A DALI server (28) search revealed no other structures resembling the CTD of BipA, except for the structure of the C-terminal half of Vibrio parahaemolyticus BipA (PDB ID code 3E3X) with Z score of 17.8, implying a unique structural fold.
Table S1.
Crystallographic data and refinement statistics
| apo | GDPCP | GDP | ppGpp | |
| Data collection | ||||
| Cell parameters, Å | a = 242, | a = 237.5, | a = 242.0, | a = 244.3, |
| b = 242, | b = 237.5, | b = 242.0, | b = 244.3, | |
| c = 242, | c = 237.5 | c = 242.0 | c = 244.3 | |
| Space group | I41 | I4132 | I4132 | I4132 |
| Resolution range,* Å | 50.00–3.30 (3.51–3.30) | 48.49–3.66 (3.86–3.66) | 47.45–3.80 (4.00–3.80) | 47.91–4.00 (4.24–4.00) |
| Redundancy | 17.3 (15.1) | 18.4 (17.3) | 17.8 (17.8) | 18.2 (18.0) |
| Observed reflections >1σ | 601,153 | 236,914 | 189,332 | 196,017 |
| Unique reflections | 86,230 (5,081) | 12,870 (1,802) | 10,596 (1,654) | 10,792 (1,675) |
| Completeness, % | 96.1 (90.1) | 99.5 (97.2) | 99.6 (98.7) | 99.6 (99.2) |
| Overall (I/ σ (I)) | 17.1 (1.1) | 14.3 (2.1) | 14.96 (1.31) | 14 (0.82) |
| CC(1/2) | 99.9 (87.2) | 99.9 (79.2) | 99.9 (46.2) | 99.9 (60.2) |
| Rsym,† % | 18.9 (301.9) | 14.1 (145) | 17.2 (228) | 16.3 (386) |
| Refinement‡ | ||||
| Resolution range, Å | 49.4–3.3 | 48.49–3.7 | 47.45–3.80 | 47.91–4.00 |
| Rwork§ (no. of reflections) | 0.26 (85198) | 0.24 (11,598) | 0.25 (10,584) | 0.25 (9,974) |
| Rfree¶ (no. of reflections) | 0.28 (9,472) | 0.30 (940) | 0.31 (1,176) | 0.32 (809) |
| rmsd bond lengths, Å | 0.013 | 0.010 | 0.010 | 0.010 |
| rmsd bond angles, o | 1.34 | 1.70 | 1.52 | 1.44 |
| Average B factors, Å2 | ||||
| Main chain | 131.10 | 150.60 | 238.8 | 290 |
| Side chain | 140.00 | 157.80 | 246.4 | 297 |
The high-resolution bin details are in the parentheses.
Rsym = ∑|Ii – <I>|/|Ii| where Ii is the intensity of the ith measurement, and <I> is the mean intensity for that reflection.
Reflections with I > σ was used in the refinement.
Rwork = |Fobs – Fcalc|/|Fobs|, where Fcalc and Fobs are the calculated and observed structure factor amplitudes, respectively.
Rfree = as for Rwork, but for 5–10% of the total reflections chosen at random and omitted from refinement.
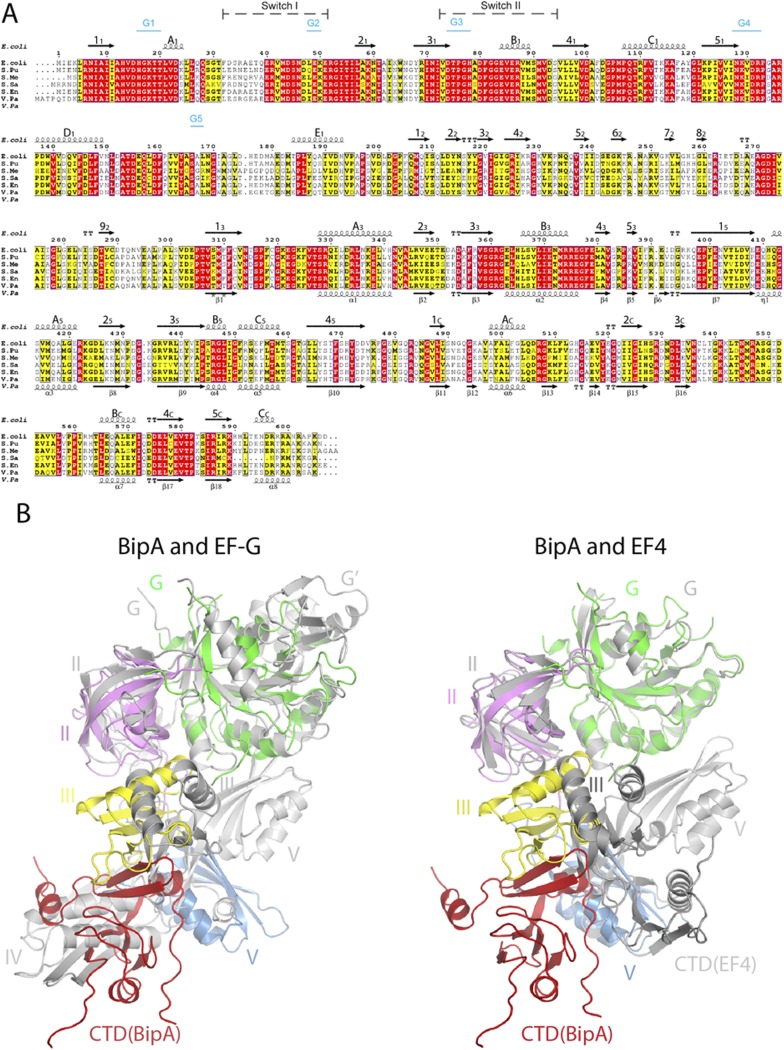
Fig. S1.
BipA sequence alignment and structure comparison with EF-G and EF4. (A) Sequence alignment of BipA from various bacteria (E. coli, Escherichia coli; S. Pu, Pseudomonas putida; S. Me, Sinorhizobium meliloti; S. Sa, Suaeda salsa; S. En, Salmonella enterica subspecies Enterica typhimunium; V. Pa, Vibrio parahaemolyticus) was performed by using ClustalW. Conserved and semiconserved residues are highlighted in red and yellow, respectively. Secondary structure elements of full-length E. coli (shown above the sequence alignment) and C-terminal half of Vibrio parahaemolyticus (V. Pa) BipA proteins (shown below the sequence) are derived from the apo form BipA structure presented in this work, and the structure deposited in PDB (3E3X). α-Helices (springs) and β-sheets (arrows) are labeled. Switch regions and G-motifs are indicated as well. (B) Comparison of the overall structure of BipA with EF-G and EF4. BipA structure in apo form is superimposed (by aligning the G domains) with EF-G (PDB ID code: 2BMO) and EF4 (PDB ID code: 3CB4). BipA domains are colored as in Fig. 1. EF-G and EF4 are shown in gray cartoon.
Excluding the unique domains, the structures of the individual domains of BipA, EF-G, and EF4 are similar (Fig. 1), as expected from their sequence similarity. However, structural studies indicate that although their overall shape is roughly similar, the spatial arrangement of the domains within EF-G, EF4, and BipA proteins are different (Fig. 1). The orientation of domains III and V in EF4 with respect to its G domain differs from the one seen in EF-G, with domain III rotated by ∼10° and domain V rotated by ∼10° and shifted by 15 Å (3). Compared (by aligning the G domains) with domain III of EF-G and EF4, the domain III of BipA makes a clockwise rotation by ∼82° and ∼70°, respectively (Fig. S1B). Whereas the β-sheet in domain III of EF-G and EF4 has a similar positioning pointing toward domain IV (EF-G) or CTD (EF4), the rotation of domain III in BipA results in ∼13 Å movement of the β-sheet toward domain II (Fig. S1B). The most striking difference is observed in domain V, which directly contacts the G domain in EF-G and EF4, but rotates almost by 90° in BipA positioning it more than 20 Å away from the G domain. Unexpectedly, the CTD of BipA, which has a equivalent domain in EF4 but is absent in EF-G, occupies a similar position as domain IV of EF-G, but has little spatial overlap with the CTD of EF4, except for its BC helix (residues 565–571) overlapping with a loop in EF4, demonstrating a distinctive domain arrangement of BipA. Hence, the global conformations of BipA, EF4, and EF-G are rather different because of their distinct domain arrangement and possibly underlying their diverse function in protein synthesis.
The Structure of BipA in Complex with GDPCP, GDP, and ppGpp.
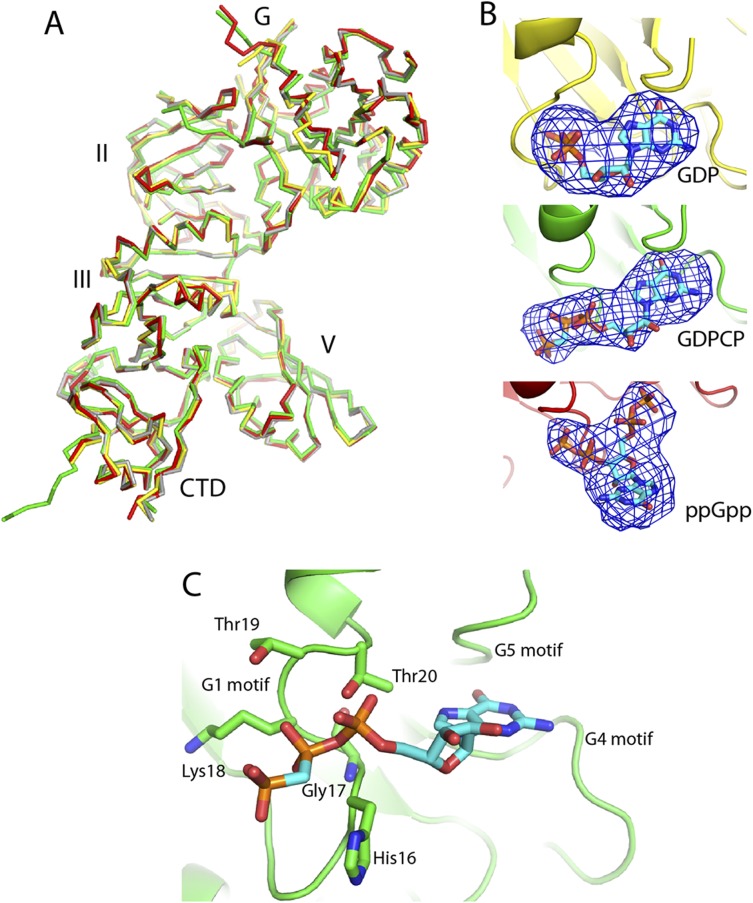
GTPases are called molecular switches for their ability to interconvert between guanosine 5′-diphosphate (GDP)- and guanosine 5′-triphosphate (GTP)-bound states. Cycling between these states facilitates periodic interactions with their cognate binding partners. In case of trGTPases, this partner is the ribosome. An intriguing feature of BipA is that it has been reported to exhibit different modes of ribosome binding when in complex with ppGpp or GTP, namely binding to 30S subunits and 70S ribosomes, respectively (5, 12). Therefore, we obtained the structures of BipA in complex with GDPCP (a nonhydrolysable GTP analog), GDP, and ppGpp by using the cocrystallization method (Table S1). Despite the unbiased difference Fourier electron density map unambiguously demonstrating the presence of the various ligands (Fig. S2B), comparison of these structures revealed no notable difference in the overall conformation of BipA (Fig. S2A). This finding is not entirely surprising given that the structure of EF-G in nucleotide-free form (PDB ID code 1ELO) is almost identical to that of GTP- (2BV3) and GDP- (2BM0) bound forms (27, 29, 30). Nevertheless, the structure of isolated BipA bound to GDPCP (a GTP analog) allowed us to delineate the nucleotide-binding site (SI Results).
Fig. S2.
Comparison of BipA apo, GDP-, GDPCP-, and ppGpp-bound forms and the structure of isolated BipA nucleotide binding site. (A) GDP- (yellow), GDPCP- (green), and ppGpp- (red) bound BipA are aligned with apo form (gray) and shown in ribbon representation. (B) The unbiased difference Fourier electron density map is shown for ligands GDP, GDPCP, and ppGpp. Carbon, oxygen, nitrogen, and phosphate atoms are colored cyan, red, blue, and orange, respectively. (C) Structure of the nucleotide-binding site of the GDPCP–BipA complex. GDPCP interactions with BipA G1 motif residues are shown. G4 and G5 motifs are indicated as well. G2 motif region is disordered in current structure.
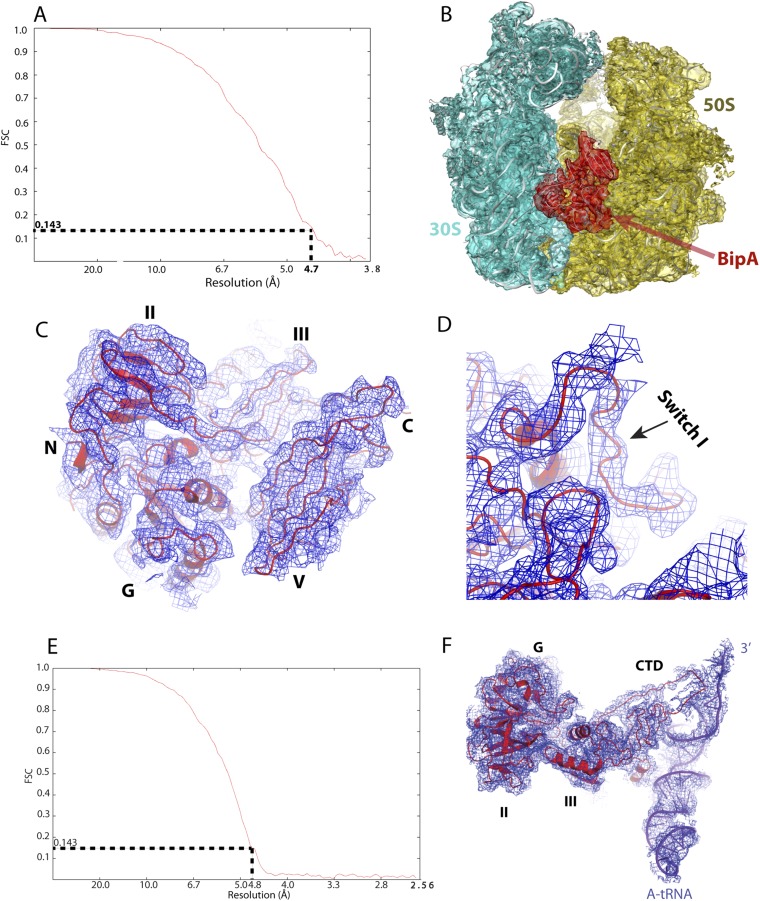
Overall Structure of BipA Bound to the Ratcheted Ribosome.
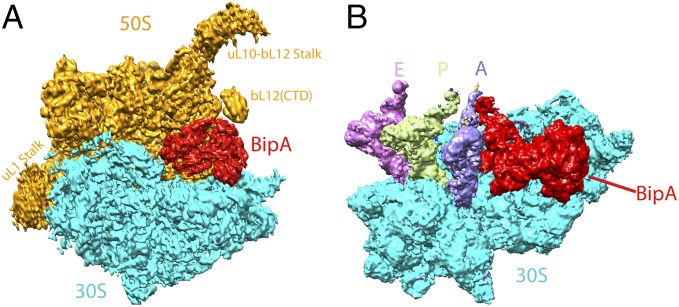
The cryo-EM single-particle reconstitution clearly shows extra density present in the ribosome factor binding site that can be used for modeling of the ribosome bound BipA structure at 4.7 Å resolution (Fig. 2A and Fig. S3). Compared with the structure of the EF-G–bound ribosome (canonical POST state) (14), the present structure shows that the 30S body is rotated counterclockwise by ∼6° with respect to the 50S subunit, and the 30S head is swiveled toward the L1 stalk by ∼5.5°, demonstrating an intermediate ratcheted state of ribosome. The overall structure of BipA–ribosome complex resembles that of EF-G in its GTP state (PRE state) bound to ribosome (18), albeit a slight change in the degree of ribosome rotation. However, our structure differs from the EF4-ribosome structure (25), where a clockwise rotation of the 30S subunit by ∼5° was observed.
Fig. 2.
Overall structure of BipA bound to ribosome. (A) Overall view of the GTP form BipA–ribosome complex. BipA protein, 50S, and 30S subunits are shown as cryo-EM density in red, orange, and cyan, respectively. Structural landmarks of 50S are labeled for clarity. (B) Structure of BipA with ribosome containing tRNAs. BipA (red), 30S subunit (cyan), A- (purple blue), P- (limon), and E- (violet) site tRNAs are shown as cryo-EM density. For clarity, the 50S subunit is not shown.
Fig. S3.
Quality of single-particle reconstitution of BipA–ribosome complex cryo-EM data. (A and E) Estimation of cryo-EM map resolution by Fourier shell correlation. Fourier shell correlation (FSC) curve for 3D reconstruction of the BipA–ribosome complex (A) and BipA-tRNA–ribosome complex (E). (B) Ribosome-bound BipA (red mesh) with corresponding cryo-EM reconstitution density for the 30S (cyan mesh) and 50S (dark orange) subunits. (C) Cryo-EM map (blue mesh) for BipA, in which helices and beta-strands are clearly visible. The red cartoon is modeled BipA. (D) Close-up view of cryo-EM map (blue mesh) for the switch I region. The switch I region of BipA is ordered upon binding to ribosome. (F) Cryo-EM map (blue mesh) for the BipA and A-tRNA showing the interactions. CTD of BipA becomes structured upon interaction with the A-tRNA.
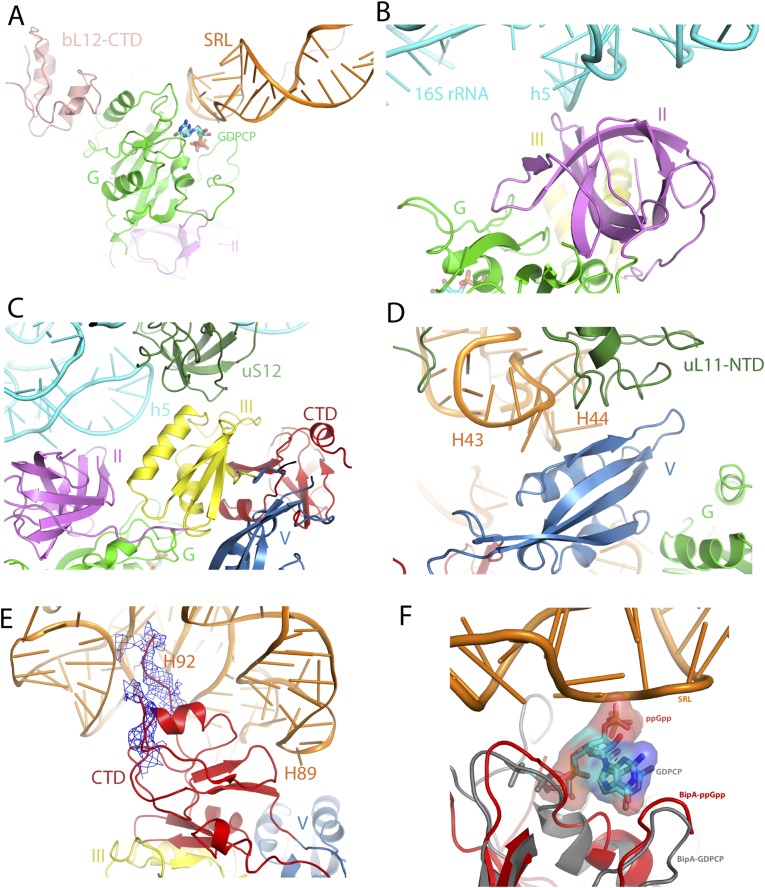
By contacting both the 50S and 30S subunits (Fig. 2A and Fig. S4A), BipA is held in a similar pocket as previously observed for EF-G bound to the ribosome (14). In addition to interacting with the universally conserved sarcin-ricin loop (SRL) in a similar way as observed for EF-Tu and EF-G (14, 31), the G domain of BipA directly contacts ribosomal protein bL12 (naming of ribosomal proteins is as proposed in ref. 32) of the uL10-bL12 stalk (Fig. S4A). The interaction with BipA stabilizes one copy of the otherwise flexible CTD of bL12 proteins in a fashion reminiscent of the one seen in the most complete 70S ribosome (uL10-bL12 stalk) model (14). G domain interactions with the ribosome will be covered below in more detail. Domain II, comprising a β-barrel fold typical for RNA binding, interacts with 16S rRNA helices 5 and 15 (h5 and h15; 30S subunit 16S and 50S subunit 23S rRNA helices are labeled throughout the text with h and H, respectively) in the shoulder of the 30S subunit (Fig. S4B). Interestingly, domain III contacts directly all of the other domains of BipA that surround it. The ribosomal protein uS12 in the shoulder of the 30S subunit also interacts with domain III, thereby closing the gap around it (Fig. S4C). In domain V, helix A5 (residues 411–421), β-strand 25 (residues 427–434), and the connecting loop establish contacts with the uL11 region (with both the uL11 protein and the stem loops of H43 and H44) of the 50S subunit (Fig. S4D). Finally, the CTD occupies the A site of the 50S subunit, and its β-turn (residues 527–532) interacts with H89 (Fig. S4E), which is an important element of ribosome because it connects the PTC and the elongation factor binding site (33). Furthermore, the distal loop residues (552–555 and beyond 555) appear to touch the hairpin loop of H92 at the PTC. Note that the hairpin loop of H92, so-called “A-loop,” binds the 3′-CCA end of A-site aminoacyl-tRNA (aa-tRNA) and helps to position the aminoacyl group in the PTC during the protein synthesis (34). Taken together, these results provide a structural explanation for the biochemical data (5) that these regions (more specifically, Lys427, Lys434, and Arg436 in domain V and His527 and Arg529 in CTD) in BipA make a significant contribution to its association with the ribosome.
Fig. S4.
BipA interactions with the ribosome. (A) G-domain interaction with ribosome 50S subunit 23S rRNA sarcin-ricin loop (SRL) and the CTD of r-protein bL12. (B) Domain II interaction with 16S rRNA h5. (C) Domain III interaction with 30S protein uS12. (D) Domain V interaction with 50S subunit H43, H44, and r-protein uL11. (E) CTD interaction with 50S H89 and H92. The cryo-EM reconstitution density map in close vicinity to the nonmodeled residues is shown in blue mesh. (F) Structural comparison of the ppGpp and GTP forms of BipA. Isolated BipA-ppGpp (red) and ribosome-bound BipA-GDPCP (gray) structures were superposed based on G domain. The 23S rRNA SRL is shown in orange.
GTPase Activation Site of Ribosome-Bound BipA.
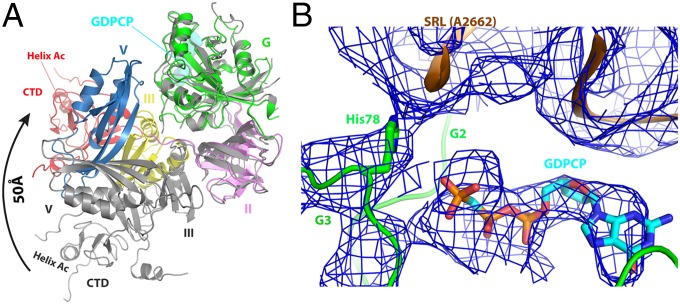
The overall conformation of ribosome-bound EF4 is similar to that of the nucleotide-free (apo) isolated EF4 (3, 25). In contrast, large conformational changes take place in EF-G upon binding to ribosome (14). In case of BipA, the overall conformation of its ribosome-bound form is remarkably different from that of isolated BipA with or without GDPCP, GDP, or ppGpp (Fig. 3A). Superposing of ribosome-bound and isolated BipA based on G domain, which is highly conserved and comprises the nucleotide-binding site, shows a large conformational change for domains III, V, and CTD, whereas little change occurs for domain II (Fig. 3A), except for the shift of the tip of β-sheets 12 and 22 (residues 206–215), and β-strand 42 (residues 224–228) by ∼5.5 Å. Taking the C terminus of helix A3 (residues 328–341) as the pivot, the entire domain III makes a counterclockwise reorientation by more than 30° relative to domain G, upon ribosome binding. As for domain V, it rotates by almost 90° toward the G domain, thereby establishing direct contacts between them (Fig. 3A). Finally, the most striking conformational change is the rearrangement of the CTD with the distal helix AC (residues 498–504) moving ∼50 Å (Fig. 3A). Similar to the two conformations (elongated and compact) recently revealed for EF-G (20), the structure of isolated BipA (nucleotide-bound or free) exhibits an elongated conformation, and the ribosome-bound BipA a compact one.
Fig. 3.
Structure of ribosome-bound BipA in GTP form. (A) Conformational changes and domain rearrangements in BipA upon ribosome binding. Ribosome-bound BipA (colored as in Fig. 1) and isolated BipA (gray) are aligned based on G domain. (B) BipA G domain switch I (G2 motif) and switch II (G3 motif) region interactions with GDPCP and 50S subunit SRL. The A2662 residue of SRL and the proposed catalytic His78 residue of BipA G3 motif are highlighted. The corresponding cryo-EM reconstitution density map is shown in blue mesh.
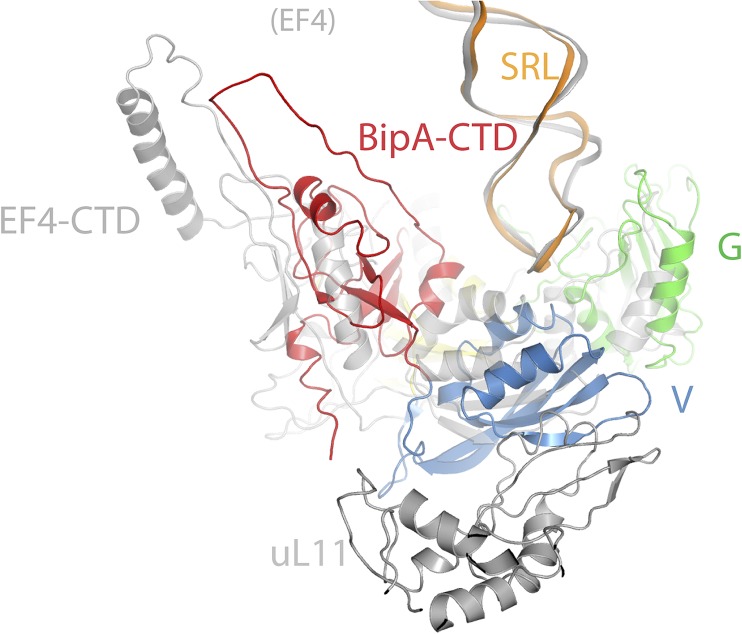
The switch I region of BipA, which was disordered in all of the isolated structures as mentioned above, can be visualized in the presence of the ribosome (Fig. S3D), allowing us to explore the structural basis of GTPase activation by BipA. In addition to interacting with the GTP analog (GDPCP), the switch I region (Phe32 and Asp33) is stabilized by bilateral contacts with the SRL of the 50S subunit and h8 of the 30S subunit (Fig. 3B), resulting in an ordered structure. GDPCP binds to the same pocket in domain G as in the aforementioned isolated BipA complexes with nucleotides (Fig. S2C). The involvement of two additional BipA motifs, namely G2 and G3, in nucleotide binding is newly established in the presence of ribosome (Fig. 3B). Notably, switch I region comprises the G2 motif, and the G3 motif is part of the switch II region, where the proposed catalytic His78 residue is located (Fig. S1A).
The SRL of 23S rRNA projects into the cleft surrounded by domains G, III, V, and CTD of BipA. Similar to that observed in structure of EF-G bound to the ribosome (18), the SRL is involved in the formation of the nucleotide-binding pocket by partially occluding the entrance. The universally conserved SRL has been demonstrated to play a central role in trGTPase activation through both biochemical and structural studies (15, 18, 35, 36). In particular, it was proposed that nucleotide A2662 in the SRL is crucial for placing the catalytic His84 residue in EF-Tu and His87 in EF-G into their activated positions (15, 18, 36). Our cryo-EM structure clearly shows that the SRL (most likely its A2662 residue) directly contacts the proposed catalytic His78 residue in BipA, thereby placing it within interacting distance to the GDPCP nucleotide, demonstrating an activated conformation (Fig. 3B).
Interaction with the uL10-b12 Stalk.
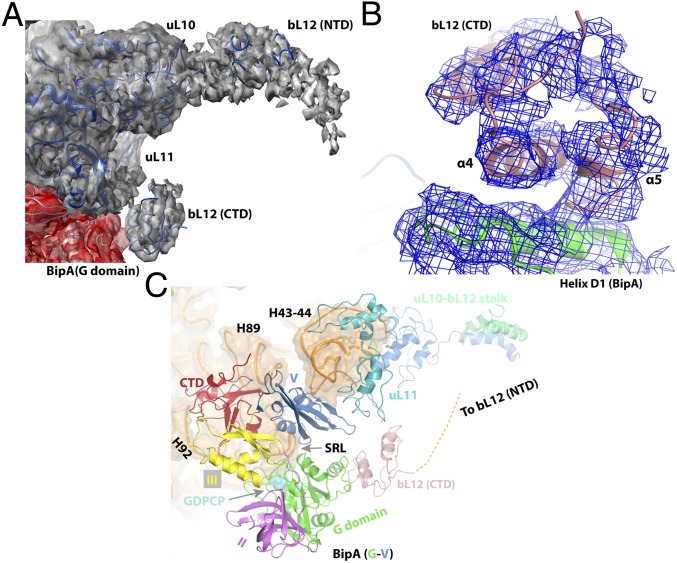
The “GTPase-associated center” (GAC) is responsible for recruiting translational factors and stimulating their GTPase activity. In addition to the SRL, a key component of the GAC, the other two crucial regions are the ribosomal uL10-bL12 stalk (comprised of uL10 protein and 4–6 copies of bL12) and its base comprising the uL11 region (uL11 protein and 23S rRNA helices H43 and H44) (37). Both the uL10-bL12 stalk and the uL11 region are extremely mobile elements of the ribosome. This dynamic feature is essential for ribosome functioning and, consequently, any mutation within ribosome or in an exogenous compound binding to the ribosome and impairing the flexibility of this region, would be deleterious to cell. The thiopeptide antibiotics (thiostrepton, nosiheptide, and micrococcin) cause ribosome dysfunction by binding to the uL11 region, blocking its flexibility, and changing its oscillation behavior (37). However, the remarkable flexibility is a major obstacle for elucidating their structure. Structural information is sparse for the uL10-bL12 stalk in particular, because the only insight comes from the structure of EF-G bound to the 70S ribosome, where one C-terminal domain (CTD) of bL12 interacting with EF-G domain G′ was observed (14, 18, 38). Given that the G′ domain is unique to EF-G, it is of significant interest to find out whether and how the CTD of bL12 interacts with the universally conserved G domain of other trGTPases.
Unexpectedly, our structure of BipA in complex with ribosome clearly shows the entire uL11 region and one copy of CTD of bL12 (Fig. 4A). Furthermore, a large density that corresponds to the N-terminal domain (NTD) of bL12 associated with the long helix in uL10 (Fig. 4A) is also visible. The CTD of bL12 comes into contact with the G domain of BipA, unlike the interaction mode with the G′ domain of EF-G (14, 18). This newly observed interaction interface involves two helices, α4 and α5, in bL12 and helix D1 in BipA (Fig. 4B). In addition, the CTD of bL12 is within interacting distance to uL11 protein that, in turn, contacts BipA domain V (Fig. 4C). Given that the proteins bL12 and uL10 interact with each other and with the G domain of BipA, which, in turn, interacts with the SRL, the BipA protein links together all three components of the GAC (Fig. 4C). Considering that the G domain is highly conserved, this newly observed interaction between the CTD of bL12 and the G domain could be universal to all trGTPase proteins that lack the G′ domain, such as IF2, EF-Tu, RF3, and EF4. Such a network of contacts could rationalize the biochemical data on EF-Tu, namely that the CTD of bL12 has an important role in translational factor binding and GTPase activity, yet none of the conserved residues in CTD by themselves are critical for GTPase activation (39).
Fig. 4.
Structure of the uL10-bL12 stalk region of the BipA–ribosome complex. (A) Cryo-EM density (gray) corresponding to the uL10 and uL11 proteins, as well as the NTD and CTD of bL12 protein. (B) bL12 CTD interaction interface (salmon) with BipA G domain (green). The corresponding cryo-EM reconstitution density map is shown in blue mesh. (C) Ribosomal components of the GTPase-associated center and BipA. Binding of BipA stabilizes the CTD of bL12, part of the dynamic uL10-bL12 stalk, thereby bridging together the GAC components. BipA is colored as in Fig. 1.
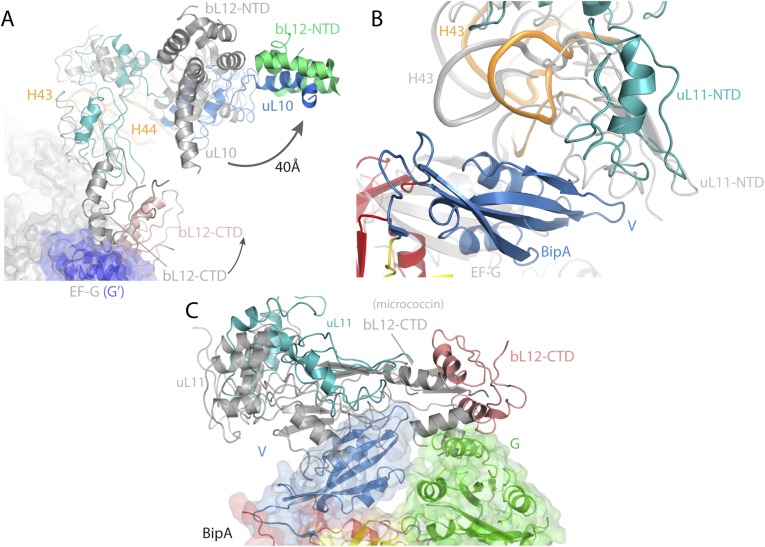
Comparison of the POST complex (14) with the present complex (by aligning the 23S rRNAs) reveals the different location of the CTD of bL12 protein (Fig. S5A). Namely, the bL12 CTD of BipA–ribosome complex would clash with the G′ domain of EF-G. In line with the direction of the CTD movement, the stalk protein uL10 and its binding partner, the NTD of bL12, are displaced by ∼40 Å away from the uL11 region (Fig. S5A). Compared with domain V of EF-G, domain V of BipA is located closer to the stalk base, which results in a large conformational change of the uL11 protein and 23S rRNA helices H43 and H44 to avoid a structural clash (Fig. S5B). Both micrococcin and thiostrepton bound to the uL11 region of ribosome induce a conformational change in the NTD of uL11 protein close to domain V of BipA that would cause a steric clash (37) (Fig. S5C). Interestingly, the location of the CTD of bL12 in the micrococcin structure is different from its location in the present structure (Fig. S5C) and in that with EF-G (14). Indeed, the CTD of bL12 is remarkably mobile and the transient nature of its positioning with respect to trGTPase protein greatly affect its activation. Thus, as proposed (37), micrococcin probably promotes a stable interaction between the CTD of bL12 and the NTD of uL11, such that the CTD becomes optimally positioned to contact the G′ subdomain of EF-G leading to GTPase activity stimulation.
Fig. S5.
Conformational changes of the uL10-bL12 stalk region depending on the presence of different binding partners and ligands. (A) Comparison of the orientation of bL12 protein CTD and NTD, as well as the uL10 protein, and 23S rRNA helices H43 and H44 in BipA–ribosome in the absence of tRNA (this work) and EF-G–posttranslocational ribosome (14) complexes. (B) Comparison of the orientation of domain V, NTD of uL11, and 23S rRNA H43 in the same complexes as in A. (C) Comparison of the orientation of the uL11 and the CTD of bL12 protein in BipA–ribosome complex (without tRNA) with the one observed in ribosome in complex with micrococcin (37).
Positioning of the CTD of BipA in Ribosomal A Site.
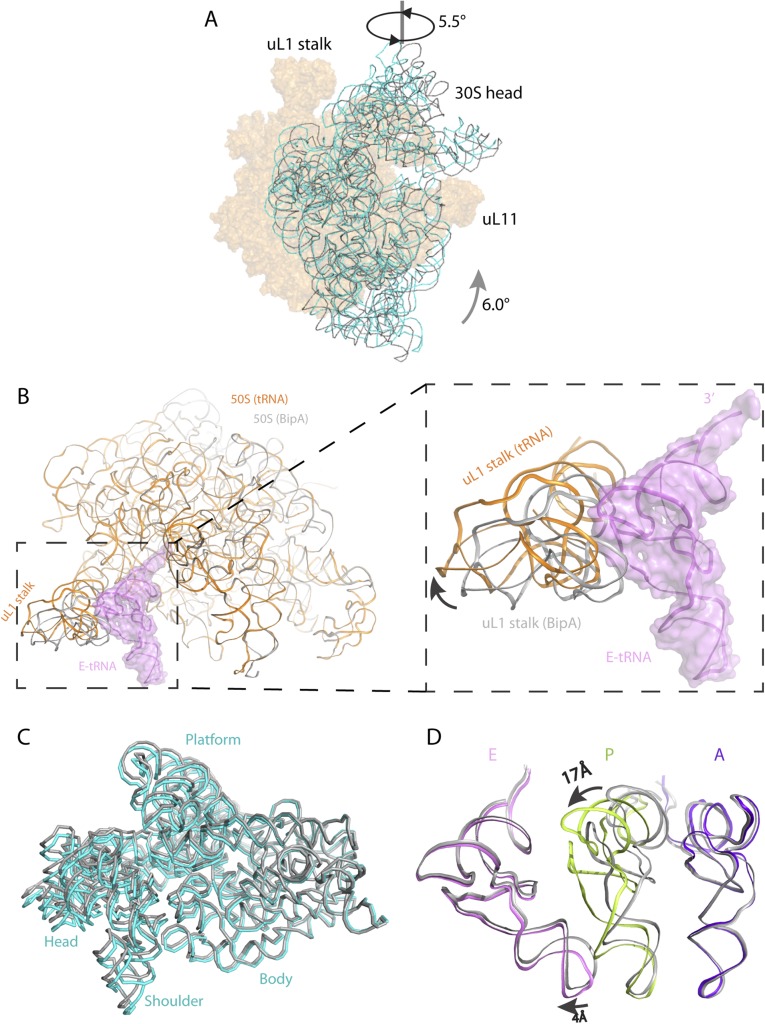
Furthermore, we determined the cryo-EM structure of BipA bound to the ribosome containing tRNAs and mRNA, in which the well-ordered A-, P-, and E-site tRNAs were visualized (Fig. 2B and Fig. S3F). The overall structure of BipA bound to ribosome with tRNAs/mRNA is similar to the aforementioned complex except for: (i) lesser 30S head swiveling; and (ii) shifting of the uL1 stalk in 50S away from the E-site tRNA resulting in an open form (Fig. S6 B and C). In particular, the overall conformation and the positioning within ribosome of BipA in both structures are almost identical. While the uL10-bL12 stalk is more defined in the absence of tRNAs/mRNA, the BipA–ribosome–tRNAs/mRNA complex can provide a valuable insight into BipA interaction with tRNA. Superposing the present structure with that of ribosome containing A-, P- and E-site tRNA (34) by aligning the 23S rRNAs shows that the A- and E-site tRNAs take a similar positioning, except for a ∼4-Å shift of the anticodon loop of E-site tRNA following the 30S head swiveling in the present complex (Fig. S6D). In contrast, a large conformational change in the P-site tRNA was observed, with a ∼17-Å shift of the TΨC loop compared with its classical position in ribosome (Fig. S6D).
Fig. S6.
Comparison of the BipA–ribosome complex with the POST complex (EF-G bound) and with the BipA–tRNA–ribosome complex. (A) Head swiveling and body rotation of the 30S subunit relative to the 50S subunit in the BipA–ribosome complex. The 16S rRNA (cyan) shows a counterclockwise rotation by 6° of the 30S body and an orthogonal swiveling by 5.5° of the 30S head toward the uL1 stalk (viewed from the solvent side of the 30S subunit), compared with 16S rRNA (gray) in the POST complex (EF-G bound to the posttranslocational ribosome in canonical state trapped by fusidic acid) (14). The two structures are aligned on 23S rRNA. (B) The uL1 stalk in 50S of tRNA/mRNA ribosome complex (orange) moves away from the position of E-site tRNA (colored violet with surface), resulting in an open form. The 50S and uL1 stalk in the BipA–ribosome complex is colored gray. (C) Comparison of 30S subunits. The body of 16S rRNA of the BipA–tRNA–ribosome complex is almost identical to that without tRNA, whereas the head swivels less, with a ∼2° of 30S head swiveling in the former complex and a ∼5.5° in the latter, respectively. The 16S rRNA in the BipA–tRNA–ribosome complex and the BipA–ribosome complex are colored cyan and gray, respectively. (D) Comparison of A-, P-, and E-site tRNAs in our structure with that in classical state (34). The A- and E- site tRNAs are in a similar positioning in ribosome, despite that a 4Å shift of anticodon loop was observed in E-site tRNA following 30S head swiveling. A large shift (∼17 Å) was observed in the TΨC loop compared our structure with that in the classical P-site tRNA. The A-, P-, and E-site tRNAs in current structure are colored purple blue, limon, and violet, respectively, whereas those of the classical state are colored gray.
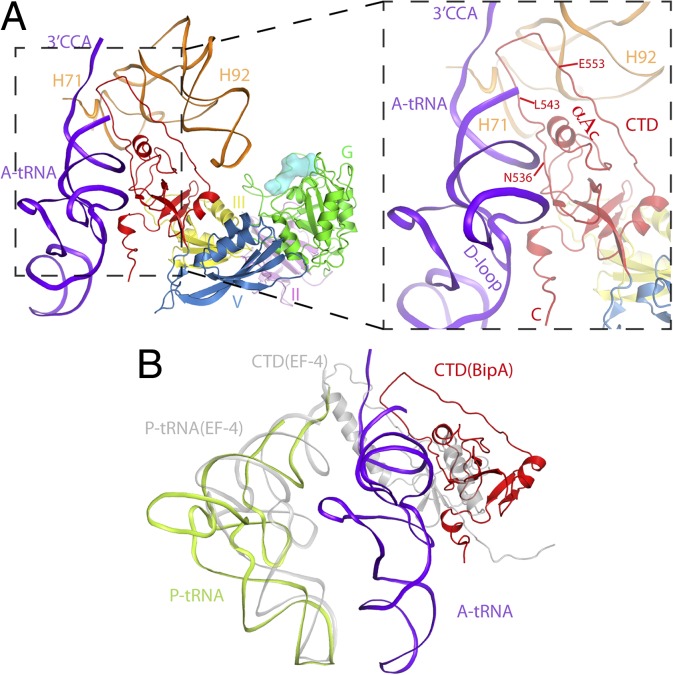
When bound to ribosome, the CTD of BipA positions close to the PTC (Fig. 5A) in the A site. As expected, binding of the A-site tRNA stabilizes its loop region (residues 543–553) (Figs. S3F and S4E). In addition to interacting with 23S rRNA as aforementioned (Fig. S4E), the CTD of BipA makes multiple contacts with the A-site tRNA (Fig. 5A). The helix AC and the loop region (residues 536–539) of BipA come into close vicinity and directly contact the acceptor stem of the tRNA. Moreover, the C-terminal region (residues 595–602) of BipA establishes a strong interaction interface with the D-loop region of the tRNA. This observation is consistent with the C-terminal sequence of BipA family members being rich in basic Arg/Lys residues (Fig. S1A) capable of preferentially binding nucleic acid, and with the biochemical data demonstrating a significant role of this C-terminal helix of BipA in ribosome binding (5). Finally, the newly modeled residues L543–E553 project deeply into the PTC region and are sandwiched by the 5′ and 3′ ends of tRNA and H71 and H92 of 23S rRNA (peptide transfer region) (Fig. 5A). In our structure, the loop 536–542 of BipA is within interacting distance to both the acceptor stem of tRNA and 23S rRNA. This finding is consistent with the previous report that the highly conserved Asn536, Lys541, and Lys542 residues in this region are important for ribosome binding (5).
Fig. 5.
Positioning of the CTD of BipA and its interactions with the A-site tRNA in the ribosome–BipA–tRNA complex. (A) BipA bound to tRNA ribosome complex reveals that the CTD of BipA interacts with the acceptor stem region of A-tRNA (shown in purple blue) and 23S rRNA (H71 and H92). The loop residues (L543–E553) deeply projects into peptidyl transfer region in the A-site, surrounded by both 3′- and 5′-ends of tRNA as well as H71 and H92 of 23S rRNA. (B) Comparison of BipA–tRNA–ribosome with A-site tRNA and EF4-ribosome with P-site tRNA complexes. CTD of BipA and EF4 (25) occupy A and P sites, respectively.
Although the EF4-ribosome structure shows that the CTD of EF4 also reaches into the PTC, it interacts with the acceptor stem of the P-site tRNA instead (25). Superposing this structure with ours by aligning the 23S rRNA reveals that the SRL and the G domain superimpose reasonably well, but the uL11 region and other domains of BipA/EF4 have different orientations, and that the uL11 protein of EF4–ribosome complex would clash with the domain V of BipA (Fig. S7). Interestingly, the tip of EF4 CTD comprising a helix-turn-helix (HTH) motif projects deeply into the PTC in the P site and is not compatible with A-site tRNA, whereas the tip of BipA CTD extends toward the PTC in the A site (Fig. 5B). The present structure demonstrates small counterclockwise rotation (particularly for 30S head swiveling) of the ribosome and both A- and P-site tRNA in a classical state, implying a new intermediate state. It appears that this state occurs in between peptidyl transfer and hybrid state formation for EF-G binding. In contrast, the binding of EF4 induces a clockwise-ratcheted ribosome presumably underlying its unique function in back translocation (2, 25). Nevertheless, both BipA and EF4 confer a growth advantage to bacteria under stress conditions, and appear to regulate translation of specific but distinct subsets of mRNAs (1). Perhaps the diverse stress–response functions of BipA and EF4 result from the varied location of their CTD in PTC (A site and P site, respectively), leading to interfering with different targets.
Fig. S7.
Comparison of BipA–tRNA–ribosome and EF4–ribosome (25) structures revealing a different orientation of domain V and CTD as well as r-protein uL11, whereas 23S rRNA including the SRL superposed reasonably well. Domain G, domain V, and CTD of BipA in the BipA–tRNA–ribosome complex is colored green, sky blue, and red, respectively. EF-4 (including CTD) and uL11 protein from the EF4–ribosome complex are colored gray for comparison. SRLs are shown as cartoon in orange and gray for the BipA–ribosome and EF-4–ribosome complexes, respectively.
Discussion
Both EF-Tu and EF-G bound to the ribosome in their activated states demonstrate a direct interaction between the SRL and the catalytic histidine positioned into the active site, a crucial part of GTPase activation process (15, 18, 36). Consistent with these observations, our structure of GDPCP-BipA bound to the ribosome reveals an activated state with catalytic histidine (His78) positioned within interaction distance to the SRL (Fig. 3B), further corroborating BipA as a classic trGTPase (5, 12). In addition to the SRL and the trGTPase factor, efficient activation of trGTPases also requires other GAC components. Indeed, a novel positioning of the CTD domain of bL12, interacting with domain G of BipA (Fig. 4B), is revealed. Such an interaction is proposed to be universal among ribosome-bound trGTPase lacking the G′ domain and to provide a structural basis for the role of the CTD domain of bL12 in trGTPase factor recruitment and GTPase activation (39). Moreover, we observed a reorientation of the uL11 region and the uL10-bL12 stalk to accommodate BipA (Fig. S5 A and B). Upon binding to the ribosome, a large conformational changes take place in BipA, resulting in a compact conformation (Fig. 3A) with domain III at the core contacting all of the other domains of BipA and the ribosome (Fig. S4C). The formation of such a strategic domain arrangement of BipA appears to be facilitated by the precise ratcheting of the ribosome, thereby achieving the active state with the positioning of SRL, the uL10-bL12 stalk, and the stalk base (Fig. 4C), reminiscent of the one proposed for EF-G (18). Taken together, our results provide structural insight into how BipA in its active-state bridges all GAC components together to an overall orientation optimal for the BipA-dependent GTPase activation in the ribosome.
Despite the mounting evidence for BipA classification as a ribosome-dependent trGTPase, its precise cellular function remains unclear. The two main views regarding its role are: (i) similar to classical trGTPases and EF4, BipA has a regulatory role in protein translation; and (ii) BipA is a ribosome assembly factor reminiscent of GTPases like Era, EngA, and CgtAE (11, 13). In support of the first view, BipA shares structural similarity with trGTPases EF-G and EF4 (Fig. 1). In addition, our cryo-EM reconstitution showed that BipA binding site on ribosome (Fig. 2) overlaps with that of EF-G, EF-Tu, and EF4. The fact that EF-G can displace BipA from the ribosome may indicate that BipA is present transiently and/or under specialized stress conditions such as low temperature (40). Furthermore, according to our model, BipA is in active form with its proposed catalytic residue and bound GTP analog positioned close to the SRL of 23S rRNA (Fig. 3B) in an intermediately ratcheted ribosome (Fig. S6A). This observation qualifies BipA to be a bona fide translational factor. In particular, the structure of BipA bound to ribosome with A- and P-site tRNAs reveals a new intermediate state, which perhaps implies a function for BipA in positioning of A-site tRNA or in preventing translocation by EF-G, in response to stress. However, when the effect of BipA deletion on ribosome biogenesis was studied, phenotypes often associated with defective ribosome assembly, such as altered subunit ratios and accumulation of 50S precursor particles with partially processed 23S rRNA, were observed (13). This finding suggests that BipA is involved in the production of 50S subunit. These two proposed functions of BipA may not be mutually exclusive. For example, BipA may be involved in the regulation of translation of specific mRNAs whose products act as assembly factors. Indeed, BipA has been reported to be involved in the expression of stress response protein (10, 13).
Because BipA ribosome-binding mode has been reported to differ depending on the cellular levels of GTP and ppGpp (12), we cocrystallized BipA with ppGpp as well. The electron density in the difference Fourier map clearly demonstrates that ppGpp is located in the nucleotide-binding pocket, as expected (Fig. S2B). Aligning the G domain of ppGpp-BipA structure with that of the ribosome-bound BipA, reveals that the protrusion of the additional diphosphate moiety at the 3′ hydroxyl of ppGpp results in a steric clash with the SRL of 50S (Fig. S4F). Alike, ppGpp binding to IF2 interfers with binding to its interaction partners (41). This finding could provide a structural insight for the observation that BipA associates with either the 70S ribosome or the 30S subunit depending on the relative intracellular abundance of GTP and ppGpp (12).
Methods
For cryo-EM grid preparation, the ribosomes were incubated at room temperature with BipA alone or with tRNAs/mRNA, before being used for cryo-EM by vitrification. Electron micrographs were collected automatically by using FEI Arctica microscope with a back-thinned FEI Falcon II direct electron detector. A total of 658 and 1,531 micrographs were collected for BipA-bound ribosome without and with tRNA/mRNA complex, yielding 127,048 and 158,784 particles, respectively. Two-dimensional classification was performed to discard bad particles, followed by 3D classification to guide sorting of ribosome particles containing BipA. The final map of BipA-bound ribosome, based on 61,165 particles, reached a resolution of 4.7 Å (Fig. S3A), whereas that for BipA–tRNA–ribosome complex, based on 77,127 particles, reached a resolution of 4.8 Å (Fig. S3E). Detailed methods for BipA and ribosome preparation, image processing, and model building, as well as BipA crystallization and structure determination, are provided in SI Methods. Data refinement statistics are included in Table S1.
SI Results
Following the section The Structure of BipA in Complex with GDPCP, GDP, and ppGpp in the main text, we take the nucleotide-binding site of BipA–GDPCP complex as an example. Here, we describe the nucleotide-binding site based on the structure of isolated BipA bound to GDPCP (a GTP analog). The GTP-binding pocket typically involves five G motifs (G1–G5) (18). G1 (residues 16–20 in BipA; Fig. S1A) mainly contributes to the interaction with alpha-beta phosphate part of GDPCP, in particular the side-chain hydroxyl group of Thr20 and the main-chain N atom of Gly17 (Fig. S2C). It appears that G1 interaction with GDPCP in BipA is weaker than the one with GDP or with a GTP analog in EF-G, where G1 has more interface with nucleotide (29, 42). In addition, BipA G4 and G5 motifs are involved in the recognition of the guanine nucleobase. Despite the unbiased difference map clearly showing the GDPCP nucleotide, the G2 motif of BipA switch I region is disordered in the present structure, as was the case with the switch I of isolated EF-G in complex with a GTP analog (29). Additionally, G3 motif, located in switch II and containing the proposed catalytic His78 residue, is too far to directly interact with the nucleotide, suggesting that the observed nucleotide-binding site represents the post-GTP hydrolysis state of BipA. Indeed, the entire switch I and switch II regions become ordered upon ribosome binding of BipA trapped in GTP state and are interacting with the nucleotide as described later.
SI Methods
Preparation of Ribosomes and BipA.
T. thermophilus ribosome with a C-terminal truncation of the L9 protein was purified as described (14, 43). E. coli BipA gene was cloned and transformed into E. coli BL21 (DE3) Rosetta T1R cell line, BipA protein was overexpressed with an N-terminal His6-tag. Protein was purified by using three chromatographic steps. Briefly, BipA was first purified by affinity chromatography using Ni-NTA column. His tag was then removed by TEV protease cleavage, after which the untagged protein was collected and further purified by gel filtration and ion exchange chromatography. The protein was finally concentrated to 142 μM and stored in buffer (25 mM Tris⋅HCl pH 8.0, 50 mM NaCl, 10 mM MgCl2, and 5 mM 2-mercaptoethanol) at −80 °C.
BipA Crystallization and Structure Determination.
Crystallization trials of apo BipA, BipA-GDPCP, BipA-GDP, and BipA-ppGpp (with a molar ration of protein to ligand 1:5) were performed by using commercially available screens. The crystallization experiments were conducted by using 96-well format on a Phoenix robot (Art Robbins Instruments) at 20 °C. The initial crystal hits were obtained at 20% (wt/vol) PEG 3350, 0.2 M potassium sodium tartrate tetrahydrate (apo BipA) and at 20% (wt/vol) PEG 3350, 0.2 M sodium fluoride (BipA-GDPCP, BipA-GDP, and BipA-ppGpp). Further optimization of crystallization conditions resulted in diffraction quality crystals. Crystals were directly picked from drop and flash-frozen in liquid N2. Diffraction data were collected at BL13B1 (National Synchrotron Radiation Research Centre, Taiwan) and PX I (Swiss Light Source, Switzerland) beamlines. Apo BipA structure was solved by molecular replacement using the program Phaser (44). First, the C-terminal domain was solved by using the structure of the C-terminal domain of BipA (PDB ID code: 3E3X) as model. Then the program Molrep-auto MR was used to search the N-terminal domain by using EF4 structure (PDB ID code: 3CB4) as model. Ligand-bound structures were solved by using the program Molrep-auto MR in CCP4 suite and apo BipA as model.
When necessary, the model was manually built in COOT (45) and refinement was performed with Refmac (46). At the final stage of refinement, well-ordered water molecules were included. All structure-related figures reported in this manuscript were prepared by using PyMol (The PyMOL Molecular Graphics System).
BipA–Ribosome Complex Formation.
The 70S ribosome and GDPCP state BipA complex was prepared in buffer G (5 mM Hepes pH 7.5, 10 mM MgAc, 50 mM KCl, 10 mM NH4Cl, and 6 mM 2-mercaptoethanol). Briefly, 20 μM BipA was mixed with 100 μM GDPCP and incubated at room temperature for 30 min. Subsequently, BipA-GDPCP and ribosome (4 μM) with 200 μM neomycin samples were mixed together and incubated for 30 min at 37 °C. Reactions mixture was layered onto 200 µL of sucrose (1.1 M) cushion in buffer G. Ribosomes were pelleted in a Beckmann MLA 130 rotor at 600,000 × g for 3 h (4 °C). Pelleted ribosomes were analyzed by using precast SDS/PAGE (Bio-Rad) and Coomassie staining.
Cryo-EM Grid Preparation and Data Collection.
The BipA–ribosome complex was reconstituted in the following order. Ribosomes (170 nM) were incubated with 200 μM neomycin at room temperature. In parallel, 6.8 µM BipA was incubated with 100 µM GDPCP at room temperature. The two reactions were mixed together and allowed to form a complex at 37 °C for 30 min. As for BipA-bound ribosome with tRNAs/mRNA complex, its preparation, including the mRNA Z4C, tRNAPhe (A-site) and tRNAfMet (P-site), is similar as described (47), except for different molar concentration: 150 nM ribosome with 200 μM neomycin, 300 nM mRNA, 300 nM of the two tRNAs, 7.5 µM BipA, and 100 µM GDPCP.
Four-microliter aliquots of the reconstituted BipA–ribosome complex were incubated for 30 s on a glow-discharged 2-nm carbon-coated holey grids (Quantifoil R2/2). Grids were blotted for 3 s in 100% humidity at 4 °C and flash frozen in liquid N2 cooled liquid ethane by using an FEI Vitrobot. Grids were loaded to an FEI Arctica electron microscope operated at 200 kV. Data were acquired by using FEI’s automated data acquisition software (EPU). In total, 658 micrographs for BipA-ribosome and 1,531 micrographs for BipA–ribosome with tRNA/mRNA complex were recorded in a movie mode as a set of seven frames (total dose of 20 electrons) on a back-thinned FEI Falcon II direct electron detector. BipA–ribosome data were recorded at a calibrated magnification of 73,684 resulting in a pixel size of 1.9 Å, whereas that of BipA-ribosome with tRNA/mRNA complex was recorded at a calibrated magnification of 109,375 resulting in a pixel size of 1.28 Å. Both the data were recorded at defocus values from 1.5 to 4.0 μM. CTF parameters were estimated by using CTFFIND3 (48). Particles were picked by using EMAN2 e2boxer.py (49) and processed with Relion (50).
Cryo-EM Image Processing and Modeling.
For BipA–ribosome complex (without tRNA), a total of 127,048 particles were used for reference-free 2D classification to discard the bad particles. After 2D classifications, 107,341 particles selected were used for further 3D classification by using a 60-Å filtered empty 70S ribosome (free of tRNAs and factors) as a reference to sort out the heterogeneity in dataset. Three-dimensional classification resulted in sorting three classes of particles namely as the 50S subunit, poorly aligned 70S with weak density of BipA and 70S with strong density of BipA (61,165). For the BipA–ribosome with tRNA/mRNA complex, a total of 158,734 particles were subjected to reference-free 2D classification, resulting in 147,221 good particles. For tRNAs complex, the 3D classification resulted in sorting three classes of particles namely 50S; empty 70S and BipA; and the 70S, BipA, and 3 tRNAs complex (77,127 particles).
Particles (61,165) corresponding to the strong BipA density in 70S and 77,127 particles corresponding to the 70S, BipA, and 3 tRNAs complex were used for their respective final reconstruction with statistical movie processing to correct beam-induced particle movements in Relion. Final reconstructions for BipA–ribosome and BipA–ribosome with tRNA/mRNA complex yielded maps of 4.7 Å and 4.8 Å resolution, respectively, as determined with gold standard FCS criteria in Relion (Fig. S3 A and E). Initial docking of 30S, 50S subunits (PDB ID code 4V4Y) and BipA X-ray structures into cryo-EM maps was performed in Chimera (51).
Acknowledgments
We thank Jean Yuch-Cheng (National Synchrotron Radiation Research Centre) and T. Tomizaki and M. Wang (Swiss Light Source) for their help with X-ray data collection. This work was supported by the Singapore National Research Foundation Grant NRF-RF2009-RF001-267 (to Y.-G.G.), AcRF Tier II Grants 2014-T2-1-083 (to Y.-G.G.) and Tier I 2014-T1-001-019 (to S.B.) from the Ministry of Education of Singapore.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The atomic coordinates have been deposited in the Protein Data Bank, www.pdb.org [PDB ID codes 5A9V (apo BipA), 5A9W (BipA-GDPCP), 5A9X (BipA-GDP), 5A9Y (BipA-ppGpp), 5A9Z (BipA-70S), and 5AA0 (BipA-70S-tRNA)]. The cryo-EM density maps have been deposited in the Electron Microscopy Data Bank (EMD) with accession nos. 6396 (BipA-70S) and 6397 (BipA-70S-mRNA-tRNAs).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1513216112/-/DCSupplemental.
References
- 1.Starosta AL, Lassak J, Jung K, Wilson DN. The bacterial translation stress response. FEMS Microbiol Rev. 2014;38(6):1172–1201. doi: 10.1111/1574-6976.12083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Qin Y, et al. The highly conserved LepA is a ribosomal elongation factor that back-translocates the ribosome. Cell. 2006;127(4):721–733. doi: 10.1016/j.cell.2006.09.037. [DOI] [PubMed] [Google Scholar]
- 3.Evans RN, Blaha G, Bailey S, Steitz TA. The structure of LepA, the ribosomal back translocase. Proc Natl Acad Sci USA. 2008;105(12):4673–4678. doi: 10.1073/pnas.0801308105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Margus T, Remm M, Tenson T. Phylogenetic distribution of translational GTPases in bacteria. BMC Genomics. 2007;8:15. doi: 10.1186/1471-2164-8-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.deLivron MA, Makanji HS, Lane MC, Robinson VL. A novel domain in translational GTPase BipA mediates interaction with the 70S ribosome and influences GTP hydrolysis. Biochemistry. 2009;48(44):10533–10541. doi: 10.1021/bi901026z. [DOI] [PubMed] [Google Scholar]
- 6.Shoji S, Janssen BD, Hayes CS, Fredrick K. Translation factor LepA contributes to tellurite resistance in Escherichia coli but plays no apparent role in the fidelity of protein synthesis. Biochimie. 2010;92(2):157–163. doi: 10.1016/j.biochi.2009.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pech M, et al. Elongation factor 4 (EF4/LepA) accelerates protein synthesis at increased Mg2+ concentrations. Proc Natl Acad Sci USA. 2011;108(8):3199–3203. doi: 10.1073/pnas.1012994108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Neidig A, et al. TypA is involved in virulence, antimicrobial resistance and biofilm formation in Pseudomonas aeruginosa. BMC Microbiol. 2013;13:77. doi: 10.1186/1471-2180-13-77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pfennig PL, Flower AM. BipA is required for growth of Escherichia coi K12 at low temperature. Mol Genet Genomics. 2001;266(2):313–317. doi: 10.1007/s004380100559. [DOI] [PubMed] [Google Scholar]
- 10.Kiss E, Huguet T, Poinsot V, Batut J. The typA gene is required for stress adaptation as well as for symbiosis of Sinorhizobium meliloti 1021 with certain Medicago truncatula lines. Mol Plant Microbe Interact. 2004;17(3):235–244. doi: 10.1094/MPMI.2004.17.3.235. [DOI] [PubMed] [Google Scholar]
- 11.Krishnan K, Flower AM. Suppression of DeltabipA phenotypes in Escherichia coli by abolishment of pseudouridylation at specific sites on the 23S rRNA. J Bacteriol. 2008;190(23):7675–7683. doi: 10.1128/JB.00835-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.deLivron MA, Robinson VL. Salmonella enterica serovar Typhimurium BipA exhibits two distinct ribosome binding modes. J Bacteriol. 2008;190(17):5944–5952. doi: 10.1128/JB.00763-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Choudhury P, Flower AM. Efficient assembly of ribosomes is inhibited by deletion of bipA in Escherichia coli. J Bacteriol. 2015;197(10):1819–1827. doi: 10.1128/JB.00023-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gao YG, et al. The structure of the ribosome with elongation factor G trapped in the posttranslocational state. Science. 2009;326(5953):694–699. doi: 10.1126/science.1179709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tourigny DS, Fernández IS, Kelley AC, Ramakrishnan V. Elongation factor G bound to the ribosome in an intermediate state of translocation. Science. 2013;340(6140):1235490. doi: 10.1126/science.1235490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pulk A, Cate JH. Control of ribosomal subunit rotation by elongation factor G. Science. 2013;340(6140):1235970. doi: 10.1126/science.1235970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhou J, Lancaster L, Donohue JP, Noller HF. Crystal structures of EF-G-ribosome complexes trapped in intermediate states of translocation. Science. 2013;340(6140):1236086. doi: 10.1126/science.1236086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen Y, Feng S, Kumar V, Ero R, Gao YG. Structure of EF-G-ribosome complex in a pretranslocation state. Nat Struct Mol Biol. 2013;20(9):1077–1084. doi: 10.1038/nsmb.2645. [DOI] [PubMed] [Google Scholar]
- 19.Zhou J, Lancaster L, Donohue JP, Noller HF. How the ribosome hands the A-site tRNA to the P site during EF-G-catalyzed translocation. Science. 2014;345(6201):1188–1191. doi: 10.1126/science.1255030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lin J, Gagnon MG, Bulkley D, Steitz TA. Conformational changes of elongation factor G on the ribosome during tRNA translocation. Cell. 2015;160(1-2):219–227. doi: 10.1016/j.cell.2014.11.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ramrath DJ, et al. Visualization of two transfer RNAs trapped in transit during elongation factor G-mediated translocation. Proc Natl Acad Sci USA. 2013;110(52):20964–20969. doi: 10.1073/pnas.1320387110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Brilot AF, Korostelev AA, Ermolenko DN, Grigorieff N. Structure of the ribosome with elongation factor G trapped in the pretranslocation state. Proc Natl Acad Sci USA. 2013;110(52):20994–20999. doi: 10.1073/pnas.1311423110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu G, et al. EF-G catalyzes tRNA translocation by disrupting interactions between decoding center and codon-anticodon duplex. Nat Struct Mol Biol. 2014;21(9):817–824. doi: 10.1038/nsmb.2869. [DOI] [PubMed] [Google Scholar]
- 24.Connell SR, et al. A new tRNA intermediate revealed on the ribosome during EF4-mediated back-translocation. Nat Struct Mol Biol. 2008;15(9):910–915. doi: 10.1038/nsmb.1469. [DOI] [PubMed] [Google Scholar]
- 25.Gagnon MG, Lin J, Bulkley D, Steitz TA. Crystal structure of elongation factor 4 bound to a clockwise ratcheted ribosome. Science. 2014;345(6197):684–687. doi: 10.1126/science.1253525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fan H, Hahm J, Diggs S, Perry JJ, Blaha G. 2015. Structural and functional analysis of BipA, a regulator of virulence in enteropathogenic Escherichia coli. J Biol Chemjbc.M115.659136. [DOI] [PMC free article] [PubMed]
- 27.AEvarsson A, et al. Three-dimensional structure of the ribosomal translocase: Elongation factor G from Thermus thermophilus. EMBO J. 1994;13(16):3669–3677. doi: 10.1002/j.1460-2075.1994.tb06676.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Holm L, Rosenström P. Dali server: Conservation mapping in 3D. Nucleic Acids Res. 2010;38(Web Server issue):W545–W549. doi: 10.1093/nar/gkq366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hansson S, Singh R, Gudkov AT, Liljas A, Logan DT. Crystal structure of a mutant elongation factor G trapped with a GTP analogue. FEBS Lett. 2005;579(20):4492–4497. doi: 10.1016/j.febslet.2005.07.016. [DOI] [PubMed] [Google Scholar]
- 30.Czworkowski J, Wang J, Steitz TA, Moore PB. The crystal structure of elongation factor G complexed with GDP, at 2.7 A resolution. EMBO J. 1994;13(16):3661–3668. doi: 10.1002/j.1460-2075.1994.tb06675.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schmeing TM, et al. The crystal structure of the ribosome bound to EF-Tu and aminoacyl-tRNA. Science. 2009;326(5953):688–694. doi: 10.1126/science.1179700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ban N, et al. A new system for naming ribosomal proteins. Curr Opin Struct Biol. 2014;24:165–169. doi: 10.1016/j.sbi.2014.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Burakovsky DE, et al. The structure of helix 89 of 23S rRNA is important for peptidyl transferase function of Escherichia coli ribosome. FEBS Lett. 2011;585(19):3073–3078. doi: 10.1016/j.febslet.2011.08.030. [DOI] [PubMed] [Google Scholar]
- 34.Voorhees RM, Weixlbaumer A, Loakes D, Kelley AC, Ramakrishnan V. Insights into substrate stabilization from snapshots of the peptidyl transferase center of the intact 70S ribosome. Nat Struct Mol Biol. 2009;16(5):528–533. doi: 10.1038/nsmb.1577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hausner TP, Atmadja J, Nierhaus KH. Evidence that the G2661 region of 23S rRNA is located at the ribosomal binding sites of both elongation factors. Biochimie. 1987;69(9):911–923. doi: 10.1016/0300-9084(87)90225-2. [DOI] [PubMed] [Google Scholar]
- 36.Voorhees RM, Schmeing TM, Kelley AC, Ramakrishnan V. The mechanism for activation of GTP hydrolysis on the ribosome. Science. 2010;330(6005):835–838. doi: 10.1126/science.1194460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Harms JM, et al. Translational regulation via L11: Molecular switches on the ribosome turned on and off by thiostrepton and micrococcin. Mol Cell. 2008;30(1):26–38. doi: 10.1016/j.molcel.2008.01.009. [DOI] [PubMed] [Google Scholar]
- 38.Datta PP, Sharma MR, Qi L, Frank J, Agrawal RK. Interaction of the G′ domain of elongation factor G and the C-terminal domain of ribosomal protein L7/L12 during translocation as revealed by cryo-EM. Mol Cell. 2005;20(5):723–731. doi: 10.1016/j.molcel.2005.10.028. [DOI] [PubMed] [Google Scholar]
- 39.Diaconu M, et al. Structural basis for the function of the ribosomal L7/12 stalk in factor binding and GTPase activation. Cell. 2005;121(7):991–1004. doi: 10.1016/j.cell.2005.04.015. [DOI] [PubMed] [Google Scholar]
- 40.Owens RM, et al. 2004. A dedicated translation factor controls the synthesis of the global regulator Fis. EMBO J 23(16):3375–3385, and retraction (2007) 26(21):4607.
- 41.Milon P, et al. The nucleotide-binding site of bacterial translation initiation factor 2 (IF2) as a metabolic sensor. Proc Natl Acad Sci USA. 2006;103(38):13962–13967. doi: 10.1073/pnas.0606384103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hansson S, Singh R, Gudkov AT, Liljas A, Logan DT. Structural insights into fusidic acid resistance and sensitivity in EF-G. J Mol Biol. 2005;348(4):939–949. doi: 10.1016/j.jmb.2005.02.066. [DOI] [PubMed] [Google Scholar]
- 43.Selmer M, Gao YG, Weixlbaumer A, Ramakrishnan V. Ribosome engineering to promote new crystal forms. Acta Crystallogr D Biol Crystallogr. 2012;68(Pt 5):578–583. doi: 10.1107/S0907444912006348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.McCoy AJ, et al. Phaser crystallographic software. J Appl Cryst. 2007;40(Pt 4):658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Emsley P, Cowtan K. Coot: Model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60(Pt 12 Pt 1):2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 46.Winn MD, Murshudov GN, Papiz MZ. Macromolecular TLS refinement in REFMAC at moderate resolutions. Methods Enzymol. 2003;374:300–321. doi: 10.1016/S0076-6879(03)74014-2. [DOI] [PubMed] [Google Scholar]
- 47.Feng S, Chen Y, Gao YG. Crystal structure of 70S ribosome with both cognate tRNAs in the E and P sites representing an authentic elongation complex. PLoS One. 2013;8(3):e58829. doi: 10.1371/journal.pone.0058829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mindell JA, Grigorieff N. Accurate determination of local defocus and specimen tilt in electron microscopy. J Struct Biol. 2003;142(3):334–347. doi: 10.1016/s1047-8477(03)00069-8. [DOI] [PubMed] [Google Scholar]
- 49.Tang G, et al. EMAN2: An extensible image processing suite for electron microscopy. J Struct Biol. 2007;157(1):38–46. doi: 10.1016/j.jsb.2006.05.009. [DOI] [PubMed] [Google Scholar]
- 50.Scheres SH. RELION: Implementation of a Bayesian approach to cryo-EM structure determination. J Struct Biol. 2012;180(3):519–530. doi: 10.1016/j.jsb.2012.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pettersen EF, et al. UCSF Chimera–a visualization system for exploratory research and analysis. J Comput Chem. 2004;25(13):1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]