Significance
Phytochemical diversity is a key component of functional diversity. Challenges in quantifying phytochemical diversity have limited our understanding of the causes and consequences of variation in phytochemical diversity across plant species and families. Here we show that phytochemical diversity across dozens of plant species predicts herbivore diversity, herbivore specialization, phototoxicity, herbivory, and attack by natural enemies. Our approach and findings provide a framework for future investigations focused on uncovering chemical underpinnings of trophic interactions at realistic ecological, geographic, and taxonomic scales, and have implications for the conservation of functional and taxonomic diversity.
Keywords: diversity, plant defense, diet breadth, tritrophic, herbivore
Abstract
What are the ecological causes and consequences of variation in phytochemical diversity within and between plant taxa? Despite decades of natural products discovery by organic chemists and research by chemical ecologists, our understanding of phytochemically mediated ecological processes in natural communities has been restricted to studies of either broad classes of compounds or a small number of well-characterized molecules. Until now, no studies have assessed the ecological causes or consequences of rigorously quantified phytochemical diversity across taxa in natural systems. Consequently, hypotheses that attempt to explain variation in phytochemical diversity among plants remain largely untested. We use spectral data from crude plant extracts to characterize phytochemical diversity in a suite of co-occurring plants in the tropical genus Piper (Piperaceae). In combination with 20 years of data focused on Piper-associated insects, we find that phytochemical diversity has a direct and positive effect on the diversity of herbivores but also reduces overall herbivore damage. Elevated chemical diversity is associated with more specialized assemblages of herbivores, and the cascading positive effect of phytochemistry on herbivore enemies is stronger as herbivore diet breadth narrows. These results are consistent with traditional hypotheses that predict positive associations between plant chemical diversity, insect herbivore diversity, and trophic specialization. It is clear from these results that high phytochemical diversity not only enhances the diversity of plant-associated insects but also contributes to the ecological predominance of specialized insect herbivores.
The Anthropocene has been characterized by huge losses of biodiversity caused by rapid global change, including habitat loss, fragmentation, invasive species, and climate change. Ecologists struggle to understand not only the consequences of diversity loss but also how to quantify ecologically relevant dimensions of diversity, including genetic, taxonomic, and functional diversity. Although it has been difficult to measure, phytochemical diversity (i.e., richness and abundance of plant compounds) is a key axis of functional diversity (1) that affects associated trophic levels and is likely driving other aspects of biodiversity (2–4). Variation in phytochemical or metabolic diversity in plants, which is further downstream than genomic, transcriptomic, or proteomic diversity (5, 6), potentially reflects variation in response to a diversity of natural enemies, including specialist and generalist insect herbivores (7, 8). Furthermore, phytochemistry is one of the most relevant traits to measure when determining functional roles of plants in natural and managed communities (9).
Considering the importance of phytochemical diversity for numerous natural processes, it is not surprising that a broad range of ecological and evolutionary hypotheses has been proposed to explain their role in interactions between plants and herbivores. From a coevolutionary perspective, the concept of an arms race between plants and herbivores, yielding increasing diversity of plant secondary compounds (3), has long been an appealing theoretical framework for evolutionary biologists, and is still a theoretical cornerstone of chemical ecology (Table 1). Additionally, the screening hypothesis, which has received less attention, suggests that phytochemical diversity is maintained because it increases a plant’s likelihood of containing a potent compound or a precursor to a potent compound that is effective against a particular type of natural enemy, cumulatively creating a selective advantage against a diverse assemblage of natural enemies (2). The screening hypothesis also posits that phytochemical diversity provides effective combinations of compounds that work synergistically against a particular type of natural enemy (10, 11). The coevolutionary and screening hypotheses are not mutually exclusive, and both rely on assumptions that there are strong correlations between phytochemical diversity, plant diversity, and consumer diversity (Table 1). These hypotheses on the causes and consequences of phytochemical diversity also predict that mixtures of secondary metabolites within a plant species may differentially impact herbivores with different diet breadths (12, 13) and thus structure plant-specific herbivore assemblages. Specialist herbivores that share a coevolutionary history with plants are often adapted to specific compounds, and thus phytochemically diverse plants may have higher richness and abundance of specialists. Generalist herbivores that lack adaptations to particular plant taxa may be less prevalent on chemically diverse and well-defended plants; on the other hand, extremely generalized herbivores might be well-adapted to a broad array of defenses and be able to persist on plants with high phytochemical diversity.
Table 1.
Predicted consequences of increased phytochemical diversity based on long-standing general hypotheses or models
| Predictions from hypotheses linking phytochemical diversity, plants, herbivores, and enemies | Relevant path coefficients | What support is provided by results from the Piper system? |
| Divergent phytochemistry hypothesis: Higher diversity of chemical defense within a plant community is associated with increased herbivore diversity and increased specialization in diet breadth (7, 8, 28, 29). | All path coefficients are relevant. | The overall results provide strong support for this general hypothesis, along with clear mechanisms for increasing herbivore diversity via increased specialization. |
| Screening hypothesis: Phytochemical diversity is maintained by high taxonomic and guild diversity of associated herbivores; increases in phytochemical diversity cause reduced total herbivory (2). | IA, IB | Results from this study are consistent with the screening hypothesis, with a strong effect of phytochemical diversity reducing herbivory and increasing toxicity. |
| Bottom-up cascade: Increases in plant diversity (including functional or phytochemical diversity) cause greater diversity at upper trophic levels through direct and indirect mechanisms (45). | IA, IB, IVA, IVB | Results provide good support for the bottom-up hypothesis and that phytochemical diversity (rather than toxicity) increases rates of parasitoid success. |
| Hypothesized mode of action: Phytochemical diversity of phototoxic compounds (associated with downfield NMR spectra) reduces the diversity of associated herbivores as well as overall herbivory (25, 26, 46, 47). | IC, ID, IE | Although further support for this hypothesis will require experimental work, the indirect evidence for this hypothesis is clear. |
| Extreme diet breadth hypothesis: Greater diversity of chemical defense is associated with more restricted diets for specialist herbivores and broader diets for generalist herbivores (19, 20). | IIA, IIB, IIC, IID | Results provide moderate support for this hypothesis. Path coefficients were not large for these specific associations, but local diet breadth is a complex variable affected by a mix of local biotic interactions and evolutionary history. |
| Coevolution is dead hypothesis: Specialization in herbivorous insects does not evolve in response to phytochemistry (48). | IA, IB, IIB, IIC | Results do not support the prediction of no relationship between phytochemistry, diet breadth, and herbivory (49). |
| Null hypothesis: Phytochemical diversity does not affect herbivory, herbivore diversity, phototoxicity, mean consumer diet breadth, or densities of upper trophic levels. | Null path model | This model is not supported by the Piper data. |
The predictions are listed in order of decreasing support from the empirical data, and the key references for the major hypotheses (indicated in bold) that have generated these predictions are noted. The specific hypotheses tested were assessed with a priori path models (I, II, III, IV), and the support for these hypotheses is denoted by corresponding path coefficients (A–E).
These hypotheses that address the origin and maintenance of phytochemical diversity (2, 14–16) have remained largely untested simply because researchers have been unable to rigorously quantify phytochemical diversity in natural systems (17). Indeed, our understanding of phytochemically mediated ecological processes in natural communities has been restricted to either broad classes of compounds or a small number of well-characterized molecules (18, 19).
In the present study, we quantified phytochemical diversity using 1H-NMR spectroscopy and metabolic profiling for 22 species in the chemically diverse tropical plant genus Piper (Fig. 1), and quantified the effects of chemical diversity on herbivore diversity, herbivory, biological function (phototoxicity), herbivore specialization, and attack by parasitoids (flies and wasps, major sources of mortality for caterpillars). In accordance with ecological diversity metrics, we define phytochemical diversity as the richness, relative abundance, and molecular complexity of secondary metabolites within a plant taxon or community. We tested the following specific ecological hypotheses using path models (Table 1): (i) Local increases in phytochemical diversity cause increases in associated herbivore diversity (1); (ii) effects of phytochemical diversity have divergent effects on the diet breadth of relatively specialized and generalized herbivore assemblages (20); (iii) increases in overall toxicity are a consequence of greater phytochemical diversity (17) and decreases damage by herbivores; (iv) increases in phytochemical diversity increase rates of attack by parasitoids (10, 11, 21); and (v) changes in phytochemical diversity and parasitism are a consequence of plant density (Table 1).
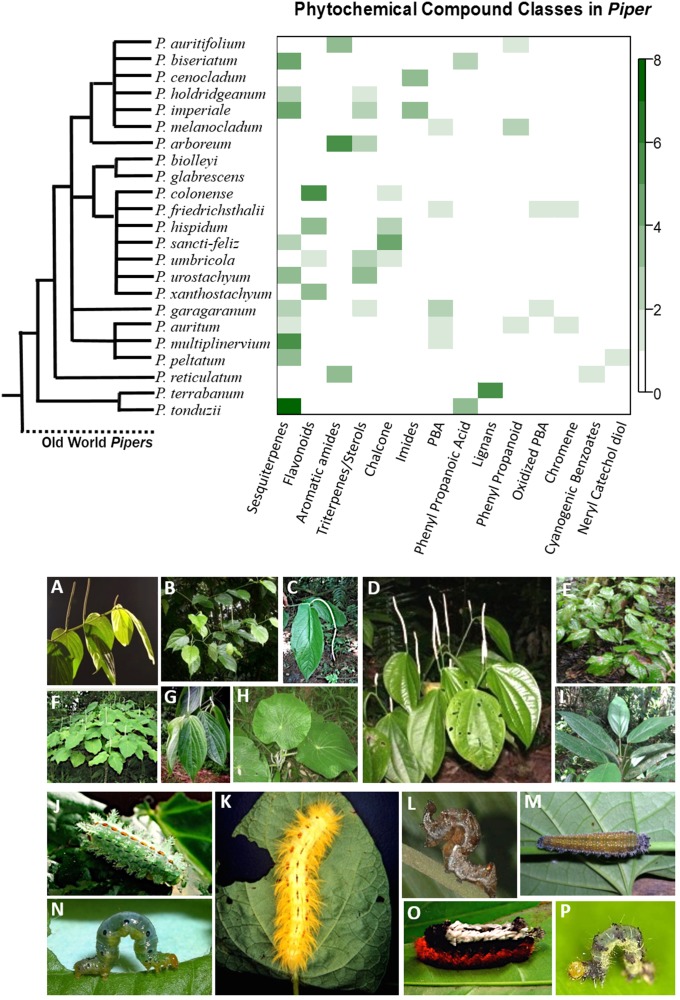
Fig. 1.
Photographs include examples of Piper species and herbivores included in this study. (Top) The heat map summarizes the number of compounds in each class that have been discovered in the corresponding species; absent classes are shown in white, and more abundant compounds are depicted by darker colors. (A) Piper santi-felicis. (B) Piper multiplinervium. (C) Piper cenocladum. (D) Piper reticulatum. (E) Piper holdrigeanum. (F) Piper auritum. (G) Piper xanthostachym. (H) Piper peltatum. (I) Piper melanocladum. (J) Euclea plumgma (Limacodidae, host: 1 Piper sp. and 6 other species from different families). (K) Apatelodes erotina (Apatelodidae, hosts: 5 Piper spp. and 18 other species from different families). (L) Gonodonta latimacula (Erebidae, hosts: 3 Piper spp.). (M) Consul parariste jansoni (Nymphalidae, hosts: 3 Piper spp.). (N) Eois picalis (Geometridae, host: 1 Piper sp.). (O) Tarchon felderi (Apatelodidae, hosts: 8 Piper spp. and 40 other species from different families). (P) Eois nympha (Geometridae, hosts: 6 Piper spp.).
Determining an appropriate measure of phytochemical diversity has been a particular challenge when comparing plant taxa that produce many different compound classes and do not necessarily share identical metabolites (22). The structural resolution provided by 1H-NMR spectral data is optimal for comparing phytochemical diversity between plant taxa and evaluating its relationship to other ecological or functional variables (23, 24). For this study, we developed an index from analyses of downfield regions (5.0–14.0 ppm) of crude 1H-NMR spectra, providing a measure of phytochemical diversity that is reflective of both inter- and intramolecular complexity of metabolite mixtures (including both complex mixtures and complex molecular structures; SI Materials and Methods). Using this approach to assess phytochemical diversity, we made species-level comparisons of the role of phytochemical diversity in ecological interactions. From a combination of long-term datasets (Materials and Methods), we calculated the herbivore community diversity associated with each Piper species (1 − D) as well as the average standing leaf area removed by herbivores for each species. One common mechanism of Piper antiherbivore defense is phototoxicity, which is defined as enhanced toxicity at higher light exposure (25). To further examine mechanisms by which phytochemical diversity affects insects, we used published data (26) to calculate an index of phototoxicity for each Piper species in our dataset. We combined these data in a series of a priori specified path models, which included a null model (SI Materials and Methods) and hypothesized relationships between phytochemical diversity, herbivore diversity, herbivory, herbivore diet breadth, parasitism rates, and phototoxicity (Table 1).
SI Materials and Methods
1H-NMR and LC-MS Comparisons.
Many techniques have been applied to measure metabolomic characteristics of various plant and animal tissues; however, until now, the application of these techniques to measuring chemical diversity has not been developed, nor compared. To evaluate our approach to measuring phytochemical diversity, we compared the goodness of fit between models using different analytical data recorded from identical methanolic plant extracts, including full 1H-NMR spectra (0–14.0 ppm), the downfield region of the 1H-NMR spectra (5–14 ppm), and LC-MS traces (Fig. S4). For LC-MS data, we analyzed the crude extracts on a micrOTOF-Q II (Bruker) coupled to a Shimadzu fast HPLC. The LC system consisted of two analytical pumps (model LC-20AD), SIL-20AHT autoinjector, SPD-20A UV/vis detector, CTO-20A column oven, and CBM-20A controller. For the mobile phase, we used MeOH:H2O (0.2% formic acid) with a gradient of 0–2 min 20% MeOH, from 2 to 30 min 20–100% MeOH, and finally 100% MeOH in 35 min, using a flux of 200 µL/min. We used a Phenomenex Luna 5-μm (PFP2) column (100 Å, 150 × 2 mm), and kept the oven at 40 °C and the UV detector at 254 and 280 nm. The mass spectra were run in both electrospray positive and negative mode, with a nebulization at 4 bar, drying gas at 8 L/min, capillary voltage at 4,500 and 3,500 V, respectively, for positive and negative mode, and drying temperature at 200 °C. Collision cell and quadrupole energy was set to 10 and 5 eV, respectively. All crude data were converted to network common data form (NetCDF) format to be analyzed. Peaks were picked using the first derivative peak picker and integrated, and the maximum abundance ion was extracted for each peak using OpenChrom software (https://www.openchrom.net/).
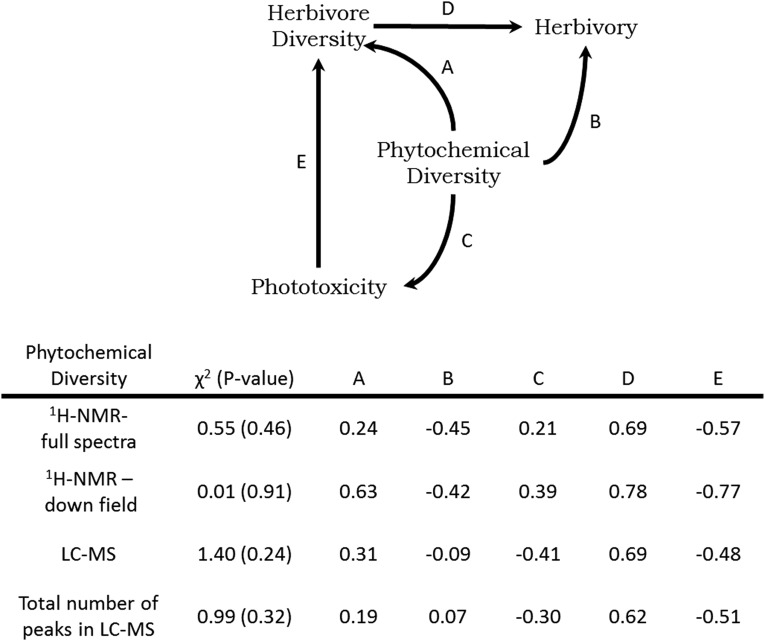
Fig. S4.
Summary of path models using different measures of phytochemical diversity: diversity quantified from 1H-NMR using the full spectra (0.5–14.0 ppm), diversity quantified from the downfield region (5.0–14.0 ppm), LC-MS spectra, and a sum of the total number of peaks from LC-MS. The table indicates model fit (χ2) and the standardized path coefficients for associated causal relationships.
Fig. S1.
Null model with no causal assumptions regarding the effects of phytochemical diversity on ecological interactions associated with 14 Piper species. All pairwise relationships were unresolved (double-headed arrows; indicating a correlation rather than a causal relationship) except path D, which was necessary to provide 1 degree of freedom. Model fit: χ2 = 1.640, df = 1, P = 0.20.
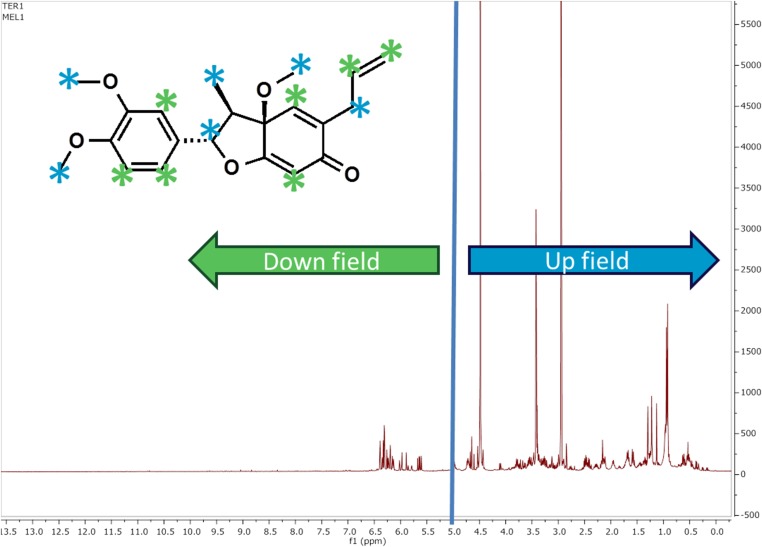
Fig. S2.
Example of a 1H-NMR spectrum with downfield and upfield regions marked. Protons associated with the downfield and upfield regions are indicated in the structure by the color-coordinated asterisks.
Fig. S3.
Path model based on predicted relationships between phytochemical diversity (including both upfield diversity and downfield diversity) and associated arthropod parameters for Piper species (model fit: χ2 = 2.415, df = 3, P = 0.49). Relationship strengths are indicated by the standardized path coefficient; blue lines indicate positive effects, and red lines indicate negative effects.
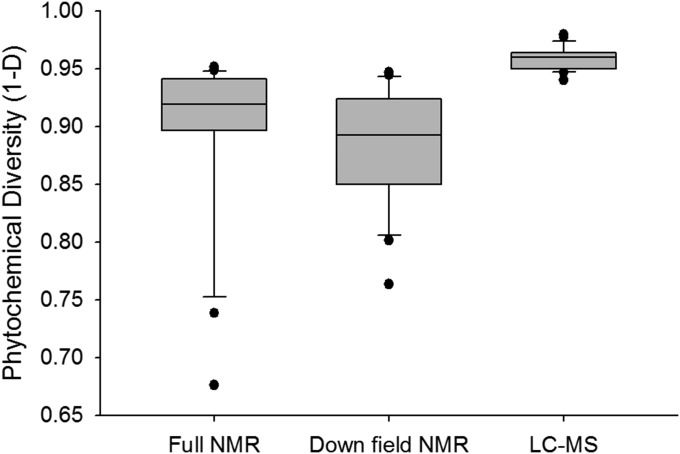
From the LC-MS data, we calculated diversity as 1 − D [where D = Σ (n/N)2, n is the area of a peak at a specific retention time, and N is the total usable peak area]. In addition, we compared the model using the total number of peaks as an indicator of the number of compounds in the phytochemical mixtures (Fig. S4). From these four measures, the model using the downfield region fit the data best, whereas the models that used LC-MS or peak counting yielded the poorest fit to the data. The sign and magnitude of paths A–C changed depending on the method used to measure phytochemical diversity. The 1H-NMR–based approaches were consistent with each other and provided the best-fitting models, suggesting that intermolecular and intramolecular complexity are both important components of chemical diversity and have an important effect on biological function. Although LC-MS–based approaches provide rapid measures of the intermolecular complexity of a chemical mixture, redundancy of retention times and molecular formula can limit both its application to the measurement and comparison of chemical diversity. The comparison of Piper phytochemical diversity variance calculated from 1H-NMR and LC-MS revealed lower variance for LC-MS–based chemical diversity measurements than measurements using 1H-NMR (Fig. S5). Across 22 species of Piper the mean number of peaks in the LC-MS data per species was 29.09, with relatively low variation between species (±1.76 SE); thus, Piper species tend to have a similar number of peaks with similar relative concentrations. Based on our experience with structural analysis from the crude 1H-NMR spectra and natural products isolation of these 22 species, we found compounds from over 9 compound classes including prenylated benzoic acids, chromenes, chalcones, amides, sesquiterpenes, flavonoids, phytols, phenylpropanoids, and catechols. The chemical diversity measurements made using LC-MS and full 1H-NMR analysis did not accurately reflect the chemical variation that is observed across the species. Redundancies in the aliphatic 1H-NMR spectral region present in the full 1H-NMR spectra and retention times and m/z in our LC-MS evaluation greatly reduce the measurement of chemical variation and significantly reduce the resolution that can be achieved through these analyses. Overall, this analysis suggests that chemical diversity measured using the downfield region of the crude 1H-NMR spectrum provides the optimal resolution for this type of analysis.
Fig. S5.
Box plots of phytochemical diversity calculated from full 1H-NMR spectra (0–14.0 ppm), the downfield region of the 1H-NMR spectrum (5–14 ppm), and LC-MS traces. Medians are represented by lines within the box plots, the boxes represent upper (75th) and lower (25th) quartiles, and the whiskers show the minimum and maximum observations of the data.
Structural Equation Models for Phytochemical Diversity Mediating Ecological Interactions.
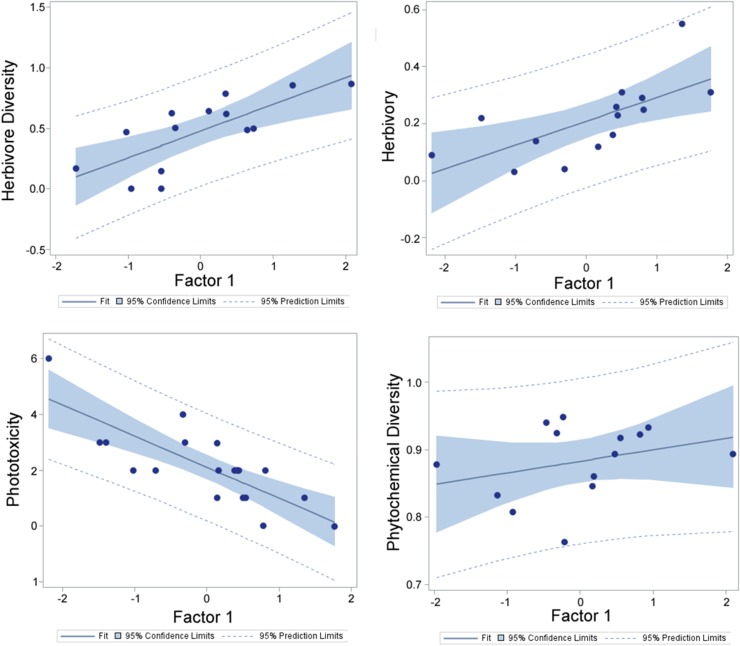
To validate the results we found in model I, we ran four factor analyses with varimax rotations (PROC FACTOR; SAS Institute) for the variables in model I, each time omitting one variable. Factor 1 from each of these analyses was treated as a latent variable and was included, along with the one omitted variable, in a linear regression. For example, in the factor analysis of phytochemical diversity, herbivory, and phototoxicity, factor 1, which explains 57.62% of the variance, was analyzed in a linear regression with herbivore diversity. This tests the hypothesis that a latent variable associated with phytochemical diversity, herbivory, and phototoxicity is responsible for changes in herbivore diversity. Using this approach, factor 1 was a significant predictor of herbivory, herbivore diversity, and phototoxicity (Fig. S6; F1,12 = 10.28, P = 0.0075; F1,12 = 15.41, P = 0.002; F1,17 = 28.26, P < 0.0001, respectively). However, factor 1 was not a significant predictor of phytochemical diversity (F1,12 = 1.31, P = 0.28). This approach corroborates the causal results from model I, in which phytochemical diversity predicts ecological interactions but not vice versa; thus, our data suggest that for the Piper system, variation in phytochemical diversity plays an important role in determining herbivory, herbivore diversity, and phototoxicity but that these variables do not affect chemical diversity. Additionally, these data provide support for the screening hypothesis, in that phytochemical diversity is maintained independent of ecological interactions.
Fig. S6.
Linear regressions of herbivore diversity, herbivory, phototoxicity, and phytochemical diversity and their respective factor from a factor analysis.
Results
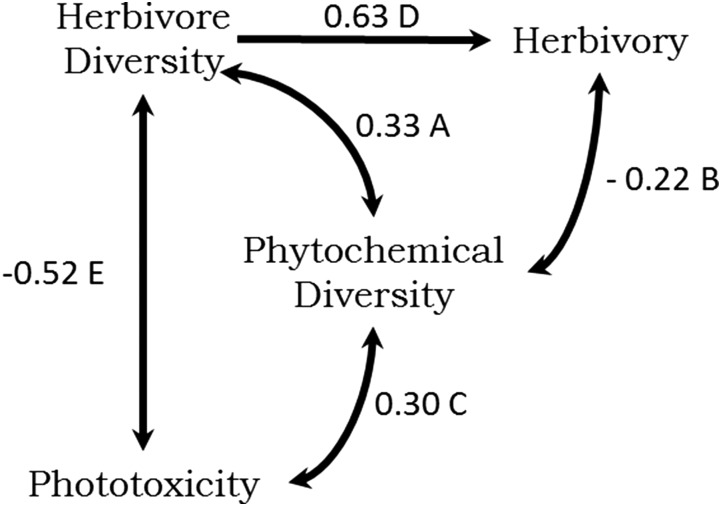
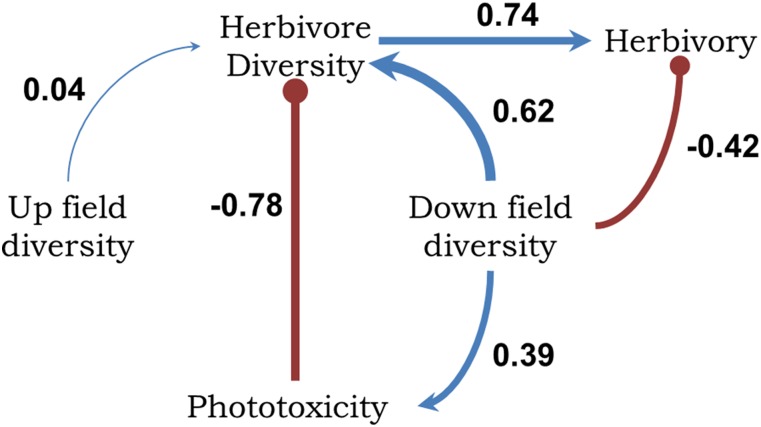
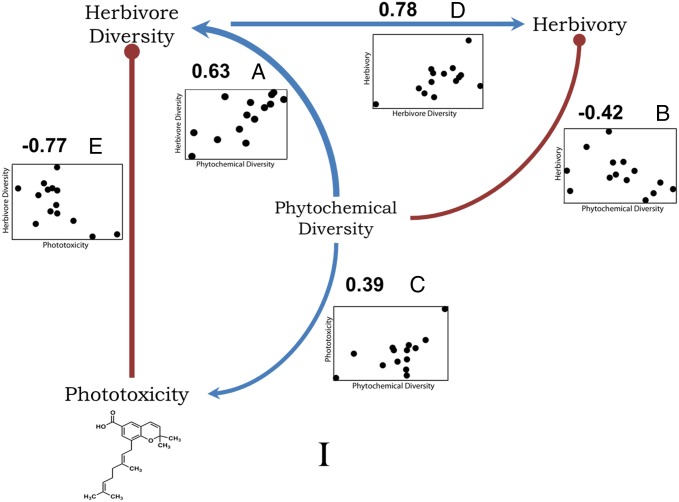
Path model I incorporates hypothesized causal relationships between phytochemical diversity, herbivore diversity, herbivory, and phototoxicity (Fig. 2) and was a significant fit to the data; this model included the 14 Piper species for which all relevant variables were available (model fit: χ2 = 0.012, df = 1, P = 0.914; P values closer to 1 indicate a good fit). Phytochemical diversity was positively associated with phototoxicity and had a strong positive effect on herbivore diversity and a negative relationship with herbivory, which suggests that our measure of chemical diversity is an effective predictor of ecological interactions. Interestingly, when the direct effects of phototoxicity were examined (factoring out the association between phytochemical and herbivore diversity), phototoxicity negatively affected herbivore diversity, suggesting a potential mode of defense against a diverse suite of herbivores. The positive chemistry–herbivore association (path A, Fig. 2) is consistent with general coevolutionary hypotheses that plants with high phytochemical diversity will have a high diversity of herbivores (27). Path model I (Fig. 2) fits the data substantially better than a null model in which there were no assumptions of directional, causal relationships (i.e., unresolved causal structure; model fit: χ2 = 1.640, df = 1, P = 0.20). Additional analyses in SI Materials and Methods further support the relationships tested by model I.
Fig. 2.
Path model I based on predicted relationships between phytochemical diversity and associated arthropod parameters for Piper species. Standardized path coefficients are noted. Positive relationships are shown in blue with arrowheads indicating causality; negative relationships are indicated in red with bullet heads. Plots of the partial correlations for each path are shown with the dependent variable on the x axis and the response variable on the y axis. The data support hypothesized causal relationships between insect herbivore species diversity, phytochemical diversity, leaf area lost to herbivores, and phototoxicity in 14 Piper species (model fit: χ2 = 0.012, df = 1, P = 0.914).
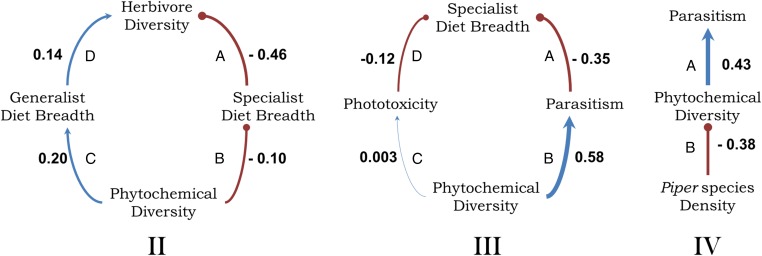
To address predicted relationships between phytochemical diversity, diet breadth, and parasitism rates, we separated the herbivore community into “specialists” (those that feed exclusively on Piper) and “generalists” (feeding on Piper plus host plants in other genera or families). In a model that included 22 species of Piper, we found that communities of Piper specialists were more diverse on the more phytochemically diverse hosts (Fig. 3; model fit: χ2 = 0.1047, df = 2, P = 0.95). However, Piper phytochemical diversity also had a positive association with the diet breadth of generalists, such that the most chemically diverse hosts were attacked by the most extremely generalized herbivores, feeding in some cases on up to 30 plant species. This suggests that among generalists, only those that have adapted to feed on a very broad variety of plant species can feed on Piper species with high chemical diversity. Thus, both highly generalized and highly specialized herbivores contribute to the overall increase in herbivore diversity with increasing phytochemical diversity. These results are consistent with the recent suggestion that disruptive selection on diet breadth could be responsible for the predominance of specialized herbivores in communities that also include a predictable component of more generalized species (20).
Fig. 3.
Summary of the best-fitting path models based on predicted relationships between phytochemical diversity and herbivore diet breadth (model II) and parasitism rates (models III and IV) for Piper species. Each model used subsets of Piper host species for which all relevant data were available. Standardized path coefficients are noted. Positive relationships are indicated in blue with arrowheads indicating causality; negative relationships are indicated in red with bullet heads. Model II includes different diet breadths and quantifies associations between herbivore diversity, phytochemical diversity, and diet breadth of the herbivore community for 22 Piper species (model fit: χ2 = 0.1047, df = 2, P = 0.95). Greater phytochemical diversity drives greater levels of specialization and generalization, both of which contribute to higher herbivore diversity. For the subset of data that includes only Piper specialist caterpillars, model III quantifies associations between phytochemical diversity, phototoxicity, parasitism rates, and the number of Piper species that are hosts to the herbivore community for 13 Piper species (model fit: χ2 = 0.1676, df = 2, P = 0.92). In model IV, local Piper species density is included in a model of phytochemical diversity and specialist parasitism rates (20 species; model fit: χ2 = 0.040, df = 1, P = 0.84).
Because the effects of plant defense potentially cascade up to higher trophic levels, we calculated parasitism rates by wasps and flies from the long-term rearing dataset for specialist caterpillar species. A direct effect of phototoxicity on herbivores (25) and an indirect effect on herbivores mediated through susceptibility to parasitism (10, 11) represent mechanisms by which phytochemical diversity might affect ecological interactions, and thus both were included in our model. We found that herbivores feeding on phytochemically diverse species had higher parasitism rates and were more specialized (Fig. 3; model fit: χ2 = 0.1676, df = 2, P = 0.92). Our results corroborate the established strong positive effect of phytochemical diversity on parasitism and diet breadth (21). Furthermore, the changes in herbivore and parasitoid communities observed in our study are not simply a consequence of changes in plant density (e.g., higher herbivore and parasite loads associated with greater plant abundance). In fact, the strongest effect of increased plant density was a direct negative effect on phytochemical diversity and an indirect negative effect on parasitism of herbivores (Fig. 3, model IV). The least abundant plant species had higher phytochemical diversity and higher specialist parasitism rates.
Discussion
The best-resolved systems studying relationships between phytochemistry and herbivores are the tropical plant genera Bursera (8, 28, 29) and Inga (7) and their associated herbivore communities. In both of these systems, the diversity of measured defensive compounds has been examined across different ecological and evolutionary scales to understand the relationship between herbivore diversity and specialization. There appears to be an escalation of defense over time (8, 28, 29) as well as increased beta diversity of plant defenses within communities characterized by higher herbivore diversity and specialization (7, 8, 28, 29). Similarly, for Piper species in Costa Rica, phytochemical diversity had strong, measurable relationships with plant and arthropod communities. Numerous hypotheses have been developed to explain the origins and ecological consequences of chemical diversity in natural systems, and generally rely on the assumption that plants maintain diverse mixtures of metabolites to defend against a variety of enemies through action on different physiological or biochemical targets. Our results demonstrate that chemically diverse plants benefit due to reduced herbivore damage, potentially through the exploitation of unique modes of toxicity, such as phototoxicity, or through the synergistic action of phytochemical mixtures.
Other studies aimed at investigating the role of phytochemical diversity in plant–herbivore interactions across taxa have focused on volatiles, such as terpenoids (8, 27), or compounds that are unique to a specific clade, such as the cardenolides in Asclepias species (30) and salicylates in Salix species (31). However, most plants produce defensive compounds from multiple metabolic pathways that are considered to be different “classes” of defense. The Piper phytochemical diversity reported here includes both intra- and intermolecular phytochemical diversity across taxa, and these compounds are produced by a mix of biosynthetic pathways. Our results also support the preliminary evidence of a link between phytochemical diversity and insect species richness found by Jones and Lawton (32), which was based on limited phytochemical data; such limitations have been alleviated by extensive advances in spectroscopy and increased interdisciplinary collaborations (33).
At any given tropical forest site, there are typically a few plant genera that contribute disproportionately to species richness; Piper is an example of such a “species swarm” (34, 35), making it a good system for understanding the fascinating relationships between diversity of phytochemicals, plants, and insects. Species swarms are potential components of what Becerra (29) hypothesizes is a positive feedback loop between high phytochemical diversity within a community and insect herbivore specialization that ultimately yields high plant and insect diversity. Empirical data related to Becerra’s hypothesis are difficult to generate, but our study is consistent with the predictions that specialist herbivores are favored in communities with high levels of phytochemical diversity and that community overdispersion of plant chemical defense is a product of pressure from specialist herbivores (7, 28). However, such a feedback loop, if it exists, does not operate in a trophic vacuum; natural enemies also affect herbivore specialization because predators avoid chemically defended specialist herbivores (36) and parasitoids prefer to attack specialists, which provide enemy-free space (37). Thus, it is likely that the impacts of plant chemical diversity on herbivore specialization reported here also affect communities of natural enemies by increasing the diet breadth of extreme generalists and decreasing the diet breadth of specialists. The positive association between Piper phytochemical diversity and levels of parasitism is consistent with the safe haven (38) hypothesis, which predicts that plant chemical defense will indirectly increase parasitism rates by providing enemy-free space to parasitoids in the form of well-defended specialist hosts that have sequestered plant defensive compounds. Understanding the effects of a higher diversity of sequestered compounds on parasitoids and predators or the direct effects of phytochemical diversity on natural enemies is an important area of future research in the ecology and evolution of tritrophic interactions.
Our results examining diversity associated with Piper provide mechanistic insight into previous studies (7, 8, 27–29, 38) documenting positive relationships between plant and arthropod diversity and also constitute methodologically rigorous support for the screening hypothesis. Our approach and findings provide a framework for investigations focused on the chemical underpinnings of trophic interactions at realistic ecological, geographic, and taxonomic scales. As global change continues to drive rapid loss of the unique natural products that comprise phytochemical diversity, particularly in the tropics, understanding the role of this component of biodiversity should continue to be a priority for the sciences.
Materials and Methods
Study System.
Piper species are characterized by a high diversity of plant secondary metabolites from over 15 classes of compounds. The 112 Piper species worldwide that have been investigated for phytochemicals yielded 667 different compounds distributed as follows: 190 alkaloids/amides, 49 lignans, 70 neolignans, 97 terpenes, 39 propenylphenols, 15 steroids, 18 kavapyrones, 17 chalcones/dihydrochalcones, 16 flavones, 6 flavanones, 4 piperolides (cinnamylidone butenolides), and 146 compounds that do not fit into the major categories of secondary metabolites (39–41). Constitutive secondary metabolites have been found in all parts of the plant, and there are countless demonstrations of strong biological activity, including synergistic effects (on herbivores and pathogens) for all classes of Piper compounds, especially the amides (10, 39).
Study Site.
La Selva Biological Station, Heredia Province, Costa Rica (10° 26′ N 83° 59′ W) consists of just over 1,600 ha of forest and clearings located at the confluence of the Sarapiquí and Puerto Viejo rivers on the eastern Caribbean slope in Costa Rica. The elevation varies between 35 and 140 m, with mean annual precipitation of ∼4,200 mm with a mild dry season that lasts only 2 mo. From this site, we have 20 y of data on caterpillars and parasitoids associated with over 400 host plant species from the station and surrounding areas, including nearby forest patches in Braulio Carrillo National Park, Tirimbina Reserve, and Bijagual Reserve.
Ecological Data.
Over the last 20 y, our team has collected externally feeding or shelter-building immature Lepidoptera from host plants throughout La Selva and nearby forest fragments (42, 43). This team included G. L. Gentry, H. Garcia Lopez, G. Vega Chavarria, L.A.D., A.M.S., and dozens of graduate and undergraduate students. Data collection was augmented by Earthwatch volunteers who worked on this project in Costa Rica every July and December. Larvae were collected using a plot method and general collecting. Ten-meter-diameter, circular plots were chosen using the following criteria: Each plot must be at least 10 m from a trail or road, and must contain at least 1 of the 40 focal genera used in our project (44) that are common in the study area and for which we have the most data. The plots were divided into four equal wedges, and one person spent 30 min looking for caterpillars on all of the plants within each wedge using beat sheets or visual searches. Once the circle had been searched, leaf abundance of any of the 30 focal genera present in the plot was estimated. General collecting involved all externally feeding or leaf-rolling caterpillar species encountered in forest patches throughout La Selva and surrounding reserves. Every collection event (the act of finding a caterpillar) received a unique voucher code that links the specimen to the host species in the database. From these plot data, we calculated the average plant density of each Piper species from 151 plots haphazardly distributed across the forest site.
All collected caterpillars were reared individually in clear plastic bags or glass jars in our rearing barn at ambient temperature and humidity. Every 2 d, we replaced the foliage in each container, and added vermiculite, soil, or rotten wood to the containers for caterpillar species that require a substrate for pupation. All pupae were checked daily to collect any adult Lepidoptera or parasitoids that have emerged from the pupae. They were allowed to fully harden before being stored in a freezer or in 75% aqueous ethanol before pinning and identification. All species of caterpillars were photographed with a Canon EOS D30 digital camera (with macro lens) and described in a standardized manner. Adult lepidopteran specimens were pinned and curated using standard techniques (44). Voucher specimens were examined by taxonomic authorities and are housed in collections of the taxonomists’ preference to facilitate further description and systematic studies.
From this database, 4,447 individual caterpillars were included from 45 species of Piper. Unidentified individuals and singletons were removed for the analysis. Herbivore diversity was calculated for each Piper species as a Simpson’s index of diversity [1 − D; where D = Σ (n/N)2, n is the total number of caterpillars reared of one species, and N is the total number of caterpillars reared on the host species]. Herbivore diet breadth was determined by calculating the number of host plant species each herbivore species was reared from. We split these into two groups: Piper specialists, which feed exclusively on Piper species, and generalists, which feed on Piper and other species from other families. Piper specialists include lepidopteran larvae in the genus Eois (Larentiinae, Geometridae), which are extreme Piper specialists: Each species feeds on one or two Piper host species. We calculated a mean diet breadth for the specialist and generalist herbivore communities of each Piper species.
Herbivory.
We measured herbivory on all species of Piper encountered at La Selva and surrounding forest fragments in a variety of studies, including pilot studies, long-term experiments, and observational studies; the methods are reviewed in refs. 40, 42, and 43. The method used was identical for all studies and is briefly outlined here. Piper individuals were located in primary forest patches. Because many Piper species reproduce vegetatively through fragmentation (40), individuals for which herbivory was recorded were separated by at least 5 m. On each plant, we examined recently expanded mature leaves and used a translucent grid to measure leaf area and the percent area removed from each leaf by herbivores. In addition, damage was quantified for known Piper herbivores (which allowed for determination of herbivore diet breadth), including Atta cephalotes (Hymenoptera: Formicidae: Myrmicinae), tettigoniids (Orthoptera), chrysomelids and curculionids (Coleoptera), Quadrus cerealis caterpillars (Lepidoptera: Hesperiidae), and Eois (Lepidoptera: Geometridae). Each type of herbivore damage is easily identified based on distinctive patterns of damage (42).
Chemical Data.
Leaf samples from each species were collected from La Selva Biological Research Station, Costa Rica. The samples were dried with silica and transported to the University of Nevada, Reno. Leaf samples were ground to a fine powder, and 2 g was transferred to a screwcap test tube and combined with 10 mL methanol. The samples were sonicated for 10 min and filtered to separate the leaf material from the supernatant. This step was repeated a second time, and the supernatants were combined and transferred to a preweighed 20-mL scintillation vial. The solvent was removed under reduced pressure at 30 °C and prepared for NMR analysis. The 1H-NMR spectrum of a crude extract captures the molecular complexity of a broad range of compound classes with a range of molecular weights and polarity. The advantages of NMR in these types of comparisons include quantitative results, the ability to observe a wide range of compound classes in a single analysis, nondestructive analysis, reproducibility, and superior structural resolution.
Data Acquisition and Analysis.
The NMR spectral data were processed using MestReNova software (Mestrelab Research). Spectra from the crude extracts were aligned using the solvent peak, baseline-corrected, phase-corrected, normalized to the total area of 100, and binned every 0.04 ppm from 0.5 to 14 ppm. The data from each species were exported and combined into one dataset for analysis. We calculated the diversity indices [1 − D; where D = Σ (n/N)2, n is the integral at a specific binned frequency range, and N is the total number of binned frequency ranges] and tested a priori path models (PROC CALIS) using SAS software (SAS Institute).
For 1H-NMR spectra, upfield resonances are commonly associated with aliphatic protons in a molecule, whereas downfield resonances primarily represent protons that are connected to deshielded carbon atoms (those influenced by an electronegative atom, such as a heteroatom or halogen) and protons associated with unsaturated functional groups, including aromatic and alkenyl chromophores, which could be associated with any phototoxicity. The overlap and ubiquity of peaks in the aliphatic region are less diagnostic for measures of chemical diversity. In comparison, the downfield region of the 1H-NMR spectrum is indicative of intramolecular and intermolecular complexity and is the most important area for any structural determination process using 1H-NMR spectroscopy. Thus, to estimate phytochemical diversity, we calculated the diversity of the upfield (0.5–4.9 ppm) and downfield (5.0–14.0 ppm) regions of the 1H-NMR spectra separately (SI Materials and Methods). Before testing the model in Fig. 1, we tested a similar model that included both upfield and downfield diversity (SI Materials and Methods) and found that upfield diversity was a very weak indicator of herbivore diversity and confirmed our expectation that downfield diversity is a better predictor of ecological and biological interactions. Often downfield is considered to be 3 ppm and above, and when we split upfield and downfield at 3 ppm, the data were a good fit to the model (χ2 =1.6329, df = 3, P = 0.65); however, chemistry had a weaker effect in the model. The path coefficients for upfield and downfield diversity to herbivore diversity were 0.21 and 0.18, respectively.
Acknowledgments
Thanks to M. Kato and L. Yamaguchi for contributions, edits, and helpful comments on the manuscript and assistance with LC-MS. A special thanks to Humberto Garcia for his dedication and hard work in the field, and the La Selva Biological Station, Organization of Tropical Studies. The authors were supported by grants from the National Science Foundation (DEB 1442103, DEB 1145609, DEB 1020509), Earthwatch Institute, and Strategic Environmental Research and Development Program (12-CR-11330136-084).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The data reported in this paper have been deposited in caterpillars.unr.edu/lsacatold/Families.htm.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1504977112/-/DCSupplemental.
References
- 1.Dyer LA, Parchman TL, Jeffrey CS, Richards LA. New dimensions of tropical diversity: An inordinate fondness for insect molecules, taxa, and trophic interactions. Curr Opin Insect Sci. 2014;2:14–19. doi: 10.1016/j.cois.2014.06.001. [DOI] [PubMed] [Google Scholar]
- 2.Firn RD, Jones CG. Natural products—A simple model to explain chemical diversity. Nat Prod Rep. 2003;20(4):382–391. doi: 10.1039/b208815k. [DOI] [PubMed] [Google Scholar]
- 3.Ehrlich PR, Raven PH. Butterflies and plants: A study in coevolution. Evolution. 1964;18:586–608. [Google Scholar]
- 4.Wilson JS, et al. Host conservatism, host shifts and diversification across three trophic levels in two Neotropical forests. J Evol Biol. 2012;25(3):532–546. doi: 10.1111/j.1420-9101.2011.02446.x. [DOI] [PubMed] [Google Scholar]
- 5.Crutsinger GM, et al. Plant genotypic diversity predicts community structure and governs an ecosystem process. Science. 2006;313(5789):966–968. doi: 10.1126/science.1128326. [DOI] [PubMed] [Google Scholar]
- 6.Crawford KM, Rudgers JA. Genetic diversity within a dominant plant outweighs plant species diversity in structuring an arthropod community. Ecology. 2013;94(5):1025–1035. doi: 10.1890/12-1468.1. [DOI] [PubMed] [Google Scholar]
- 7.Kursar TA, et al. The evolution of antiherbivore defenses and their contribution to species coexistence in the tropical tree genus Inga. Proc Natl Acad Sci USA. 2009;106(43):18073–18078. doi: 10.1073/pnas.0904786106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Becerra JX. Insects on plants: Macroevolutionary chemical trends in host use. Science. 1997;276(5310):253–256. doi: 10.1126/science.276.5310.253. [DOI] [PubMed] [Google Scholar]
- 9.Loranger J, et al. Predicting invertebrate herbivory from plant traits: Polycultures show strong nonadditive effects. Ecology. 2013;94(7):1499–1509. doi: 10.1890/12-2063.1. [DOI] [PubMed] [Google Scholar]
- 10.Richards LA, Dyer LA, Smilanich AM, Dodson CD. Synergistic effects of amides from two Piper species on generalist and specialist herbivores. J Chem Ecol. 2010;36(10):1105–1113. doi: 10.1007/s10886-010-9852-9. [DOI] [PubMed] [Google Scholar]
- 11.Richards LA, et al. Synergistic effects of iridoid glycosides on the survival, development and immune response of a specialist caterpillar, Junonia coenia (Nymphalidae) J Chem Ecol. 2012;38(10):1276–1284. doi: 10.1007/s10886-012-0190-y. [DOI] [PubMed] [Google Scholar]
- 12.Dyer LA, et al. Ecological causes and consequences of variation in defensive chemistry of a neotropical shrub. Ecology. 2004;85(10):2795–2803. [Google Scholar]
- 13.Lankau RA. Specialist and generalist herbivores exert opposing selection on a chemical defense. New Phytol. 2007;175(1):176–184. doi: 10.1111/j.1469-8137.2007.02090.x. [DOI] [PubMed] [Google Scholar]
- 14.Grotewold E. Plant metabolic diversity: A regulatory perspective. Trends Plant Sci. 2005;10(2):57–62. doi: 10.1016/j.tplants.2004.12.009. [DOI] [PubMed] [Google Scholar]
- 15.Lewinsohn E, Gijzen M. Phytochemical diversity: The sounds of silent metabolism. Plant Sci. 2009;176(2):161–169. [Google Scholar]
- 16.Romeo JT, Saunders JA, Barbosa P. Phytochemical Diversity and Redundancy in Ecological Interactions. Plenum; New York: 1996. [Google Scholar]
- 17.Prince EK, Pohnert G. Searching for signals in the noise: Metabolomics in chemical ecology. Anal Bioanal Chem. 2010;396(1):193–197. doi: 10.1007/s00216-009-3162-5. [DOI] [PubMed] [Google Scholar]
- 18.Gershenzon J, Fontana A, Burrow M, Wittstock U, Degenhardt J. In: The Ecology of Plant Secondary Metabolites: From Genes to Global Processes. Iason GR, Dicke M, Hartley SE, editors. Cambridge Univ Press; Cambridge, UK: 2012. pp. 56–77. [Google Scholar]
- 19.Ayres MP, Clausen TP, MacLean SF, Jr, Redman AM, Reichardt PB. Diversity of structure and antiherbivore activity in condensed tannins. Ecology. 1997;78(6):1696–1712. [Google Scholar]
- 20.Forister ML, et al. The global distribution of diet breadth in insect herbivores. Proc Natl Acad Sci USA. 2015;112(2):442–447. doi: 10.1073/pnas.1423042112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Smilanich AM, Dyer LA, Chambers JQ, Bowers MD. Immunological cost of chemical defence and the evolution of herbivore diet breadth. Ecol Lett. 2009;12(7):612–621. doi: 10.1111/j.1461-0248.2009.01309.x. [DOI] [PubMed] [Google Scholar]
- 22.Hilker M. New synthesis: Parallels between biodiversity and chemodiversity. J Chem Ecol. 2014;40(3):225–226. doi: 10.1007/s10886-014-0402-8. [DOI] [PubMed] [Google Scholar]
- 23.Krishnan P, Kruger NJ, Ratcliffe RG. Metabolite fingerprinting and profiling in plants using NMR. J Exp Bot. 2005;56(410):255–265. doi: 10.1093/jxb/eri010. [DOI] [PubMed] [Google Scholar]
- 24.Macel M, Van Dam NM, Keurentjes JJB. Metabolomics: The chemistry between ecology and genetics. Mol Ecol Resour. 2010;10(4):583–593. doi: 10.1111/j.1755-0998.2010.02854.x. [DOI] [PubMed] [Google Scholar]
- 25.Berenbaum M. Phototoxicity of plant secondary metabolites: Insect and mammalian perspectives. Arch Insect Biochem Physiol. 1995;29(2):119–134. doi: 10.1002/arch.940290204. [DOI] [PubMed] [Google Scholar]
- 26.Downum KR, Swain LA, Faleiro LJ. Influence of light on plant allelochemicals: A synergistic defense in higher plants. Arch Insect Biochem Physiol. 1991;17(4):201–211. [Google Scholar]
- 27.Iason GR, O’Reilly-Wapstra JM, Brewer MJ, Summers RW, Moore BD. Do multiple herbivores maintain chemical diversity of Scots pine monoterpenes? Philos Trans R Soc Lond B Biol Sci. 2011;366(1569):1337–1345. doi: 10.1098/rstb.2010.0236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Becerra JX. The impact of herbivore-plant coevolution on plant community structure. Proc Natl Acad Sci USA. 2007;104(18):7483–7488. doi: 10.1073/pnas.0608253104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Becerra JX. On the factors that promote the diversity of herbivorous insects and plants in tropical forests. Proc Natl Acad Sci USA. 2015;112(19):6098–6103. doi: 10.1073/pnas.1418643112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Agrawal AA, Salminen JP, Fishbein M. Phylogenetic trends in phenolic metabolism of milkweeds (Asclepias): Evidence for escalation. Evolution. 2009;63(3):663–673. doi: 10.1111/j.1558-5646.2008.00573.x. [DOI] [PubMed] [Google Scholar]
- 31.Volf M, Hrcek J, Julkunen-Tiitto R, Novotny V. To each its own: Differential response of specialist and generalist herbivores to plant defence in willows. J Anim Ecol. 2015;84(4):1123–1132. doi: 10.1111/1365-2656.12349. [DOI] [PubMed] [Google Scholar]
- 32.Jones CG, Lawton JH. Plant chemistry and insect species richness of British umbellifers. J Anim Ecol. 1991;60:767–777. [Google Scholar]
- 33.Dyer LA. New synthesis—Back to the future: New approaches and directions in chemical studies of coevolution. J Chem Ecol. 2011;37(7):669. doi: 10.1007/s10886-011-9979-3. [DOI] [PubMed] [Google Scholar]
- 34.Gentry AH. Neotropical floristic diversity: Phytogeographical connections between Central and South America, Pleistocene climatic fluctuations, or an accident of the Andean orogeny? Ann Mo Bot Gard. 1982;69(3):557–593. [Google Scholar]
- 35.Gentry AH. 1989. Speciation in tropical forests. Tropical Forests: Botanical Dynamics, Speciation and Diversity, eds Holm-Nielsen LB, Nielsen IC, Balslev H (Academic, San Diego), pp 113–134.
- 36.Dyer LA. Effectiveness of caterpillar defenses against three species of invertebrate predators. J Res Lepid. 1997;34:48–68. [Google Scholar]
- 37.Gentry GL, Dyer LA. On the conditional nature of neotropical caterpillar defenses against their natural enemies. Ecology. 2002;83(11):3108–3119. [Google Scholar]
- 38.Lampert EC, Dyer LA, Bowers MD. Caterpillar chemical defense and parasitoid success: Cotesia congregata parasitism of Ceratomia catalpae. J Chem Ecol. 2010;36(9):992–998. doi: 10.1007/s10886-010-9840-0. [DOI] [PubMed] [Google Scholar]
- 39.Dyer LA, Dodson CD, Richards J. In: Piper. A Model Genus for Studies of Evolution, Chemical Ecology, and Trophic Interactions. Dyer LA, Palmer AN, editors. Kluwer Academic; Boston: 2004. pp. 117–139. [Google Scholar]
- 40.Parmar VS, et al. Phytochemistry of the genus Piper. Phytochemistry. 1997;46(4):597–673. [Google Scholar]
- 41.Kato MJ, Furlan M. Chemistry and evolution of the Piperaceae. Pure Appl Chem. 2007;79(4):529–538. [Google Scholar]
- 42.Dyer LA, Letourneau DK, Chavarria GV, Amoretti DS. Herbivores on a dominant understory shrub increase local plant diversity in rain forest communities. Ecology. 2010;91(12):3707–3718. doi: 10.1890/08-1634.1. [DOI] [PubMed] [Google Scholar]
- 43.Fincher RM, et al. Inter- and intraspecific comparisons of antiherbivore defenses in three species of rainforest understory shrubs. J Chem Ecol. 2008;34(4):558–574. doi: 10.1007/s10886-008-9432-4. [DOI] [PubMed] [Google Scholar]
- 44.Dyer LA, et al. Host specificity of Lepidoptera in tropical and temperate forests. Nature. 2007;448(7154):696–699. doi: 10.1038/nature05884. [DOI] [PubMed] [Google Scholar]
- 45.Dyer LA, Letourneau DK. Can climate change trigger massive diversity cascades in terrestrial ecosystems? Diversity. 2013;5(3):479–504. [Google Scholar]
- 46.Philogène BJR, et al. Berberine: A naturally occurring phototoxic alkaloid. J Chem Ecol. 1984;10(1):115–123. doi: 10.1007/BF00987648. [DOI] [PubMed] [Google Scholar]
- 47.Downum KR. Light-activated plant defence. New Phytol. 1992;112(3):401–420. doi: 10.1111/j.1469-8137.1992.tb00068.x. [DOI] [PubMed] [Google Scholar]
- 48.Bernays E, Graham M. On the evolution of host specificity in phytophagous arthropods. Ecology. 1988;69(4):866–892. [Google Scholar]
- 49.Rausher MD. Is coevolution dead? Ecology. 1988;69(4):898–901. [Google Scholar]