Significance
The photoreceptor phytochrome-B (phyB) cycles between its active Pfr [far red light (FRL)-absorbing state λmax, 730 nm] and inactive Pr [red light (RL)-absorbing state λmax, 660 nm] forms and regulates as red/far red light-activated/inactivated molecular switch plant growth and development. Here we show that conjugation of small ubiquitin-like modifier to the photoreceptor inhibits interaction of phyB Pfr with its immediate signaling partner PHYTOCHROME INTERACTING FACTOR 5 (PIF5). The impaired interaction of these proteins negatively affects photomorphogenic responses; thus, SUMOylation similar to phosphorylation plays a role in desensitizing phyB-mediated signaling. OVERLY TOLERANT TO SALT 1 and 2 (OTS1 and OTS2) are involved in regulating phyB action as these SUMO proteases mediate deconjugation of SUMO from phyB.
Keywords: photoreceptor, phytochrome, sumoylation, signaling, photomorphogenesis
Abstract
The red/far red light absorbing photoreceptor phytochrome-B (phyB) cycles between the biologically inactive (Pr, λmax, 660 nm) and active (Pfr; λmax, 730 nm) forms and functions as a light quality and quantity controlled switch to regulate photomorphogenesis in Arabidopsis. At the molecular level, phyB interacts in a conformation-dependent fashion with a battery of downstream regulatory proteins, including PHYTOCHROME INTERACTING FACTOR transcription factors, and by modulating their activity/abundance, it alters expression patterns of genes underlying photomorphogenesis. Here we report that the small ubiquitin-like modifier (SUMO) is conjugated (SUMOylation) to the C terminus of phyB; the accumulation of SUMOylated phyB is enhanced by red light and displays a diurnal pattern in plants grown under light/dark cycles. Our data demonstrate that (i) transgenic plants expressing the mutant phyBLys996Arg-YFP photoreceptor are hypersensitive to red light, (ii) light-induced SUMOylation of the mutant phyB is drastically decreased compared with phyB-YFP, and (iii) SUMOylation of phyB inhibits binding of PHYTOCHROME INTERACTING FACTOR 5 to phyB Pfr. In addition, we show that OVERLY TOLERANT TO SALT 1 (OTS1) de-SUMOylates phyB in vitro, it interacts with phyB in vivo, and the ots1/ots2 mutant is hyposensitive to red light. Taken together, we conclude that SUMOylation of phyB negatively regulates light signaling and it is mediated, at least partly, by the action of OTS SUMO proteases.
Plants are sessile organisms; thus, they have to adapt to the ever-changing environment by modifying their growth and developmental programs. To respond to changes in ambient light conditions, plants evolved a battery of photoreceptors including the blue/UVA absorbing cryptochromes and phototropins (1, 2), the red/far red absorbing phytochromes (phys) (3), and the UVB-specific photoreceptor UVR8 (4). The red light/far red light (RL/FRL) absorbing phys exist as dimers, and each monomer has a covalently linked open tetrapyrrole chain as chromophore. They are synthesized in their biologically inactive Pr form (RL-absorbing state; λmax, 660 nm) and converted into the biologically active Pfr (FRL-absorbing state; λmax, 730 nm) by RL. Subsequent FRL treatment converts the Pfr form back into Pr. The Pr and Pfr conformers have partially overlapping absorption spectra; thus, phys cycle between their Pfr/Pr forms, and the ratio of Pr/Pfr forms is determined by the RL/FRL content of the incipient sunlight (5). In Arabidopsis five phys have been identified (phyA, phyB, phyC, phyD, and phyE), and among these, phyB has been shown to be especially important after seedling establishment (6).
The overwhelming majority of molecular events underlying phyB-controlled photomorphogenesis take place in the nucleus. Light in a quality- and quantity-dependent fashion induces translocation of phyB Pfr in the nuclei (7, 8), where it interacts with downstream acting regulatory proteins including PHYTOCHROME-INTERACTING-FACTORS (PIFs) (9). The very early steps of phyB signaling include (i) inactivation or alteration of the substrate specificity of CONSTITUTIVE PHOTOMORPHOGENIC 1 (COP1) that targets proteins to degradation (10), (ii) degradation and/or modulation of the transcriptional activity of negative regulatory PIF TFs (11), and (iii) induction of transcriptional cascades that modulate the expression of 2,500–3,000 genes of the Arabidopsis genome (12).
The number of phyB Pfr molecules quantitatively determines physiological responses. Thus, it is evident that any molecular process that modifies (i) the abundance of phyB Pfr such as ubiquination of the photoreceptor followed by its subsequent degradation (13, 14), (ii) the accessibility (8), and (iii) the capacity of phyB Pfr to interact with its signaling partners alters the flux of RL-induced signaling. Very recently it was shown that the Ser86 (15) and/or Tyr104 amino acid residues (16) of phyB are phosphorylated in planta. These reports established that phosphorylation of S86 accelerates thermal relaxation (light-independent reversion of Pfr into Pr, also called dark reversion) of phyB, whereas light-induced phosphorylation of Tyr104 inhibits binding of PIF3 to phyB Pfr. Taken together, these results clearly demonstrated that multiple phosphorylation of phyB negatively regulates the action of this photoreceptor.
Reversible conjugation of small ubiquitin-like modifier (SUMO) to the target proteins represents an emerging posttranslational modification (17–20). Work performed in mammalian and yeast cells showed that (i) reversible SUMOylation of proteins regulates a broad spectrum of cellular processes and (ii) addition of SUMO either modifies the activity of the target protein and/or promotes interaction with specific proteins harboring the SUMO-interacting motif (21, 22). It has also been established that in plants, like in mammalian and yeast cells, a SUMOylation pathway exists for SUMOylation and de-SUMOylation of proteins (23, 24). More specifically, it was shown that in Arabidopsis thaliana (i) expression of isoforms of SUMO1, -2, -3, and -5 can be detected (25); (ii) conjugation of SUMO to target proteins occurs via the same sequential biochemical steps as in yeast (26, 27); (iii) these reactions are catalyzed by the E1 heterodimer (SAE1a/b and SAE2), a single E2 (SCE1), and two E3 (SIZ1 and MMS2/HPY2) enzymes as well as E4-type SUMO ligases (28); whereas (iv) de-conjugation of SUMO substrates is performed by ubiquitin-like SUMO-specific proteases including ESD4 and OTS1/2 (29–34). It appears that in plants reversible conjugation of SUMO to target proteins is particularly important in regulating responses to abiotic environmental stresses including heat shock (35), phosphate starvation (36), drought (37), and salt stress (31), but SUMOylation of specific proteins also modifies responses to ABA signaling (38) and phytopathogen infection (39) and modulates flowering time (33).
Light is arguably the most important environmental factor regulating plant growth and development, yet until now, in vitro and in planta studies have not indicated SUMOylation of any component of photoreceptor-controlled light signaling cascades. Here we report that (i) the photoreceptor phyB is reversibly SUMOylated in planta, (ii) SUMOylated phyB preferentially accumulates in light, (iii) binding of PIF5 to the SUMOylated phyB Pfr is significantly reduced, and (iv) transgenic plants expressing the mutant phyBLys996Arg photoreceptor are hypersensitive to RL. Furthermore, we show that the ots1/ots2 mutant displays hyposensitivity to RL and enhanced phyB SUMOylation, whereas OTS1 de-SUMOylates phyB in vitro and binds to phyB in planta. Taken together, these data suggest that OTS1/OTS2 is involved in mediating the reversible SUMOylation of phyB and that SUMOylation of phyB desensitizes RL-induced signaling, similarly to phosphorylation of the photoreceptor.
Results
PhyB Is SUMOylated in Plants Grown in Light/Dark Cycles.
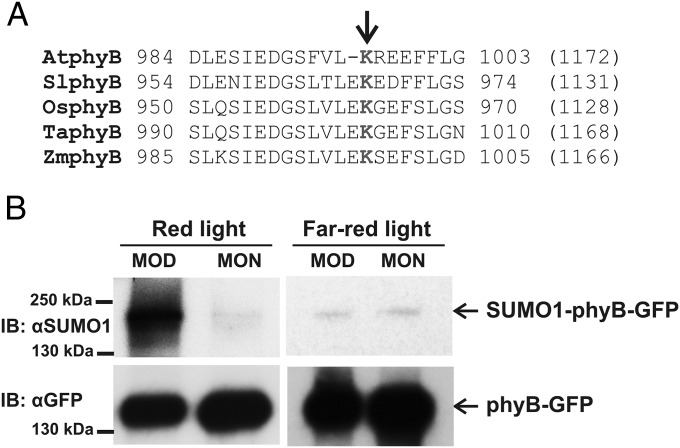
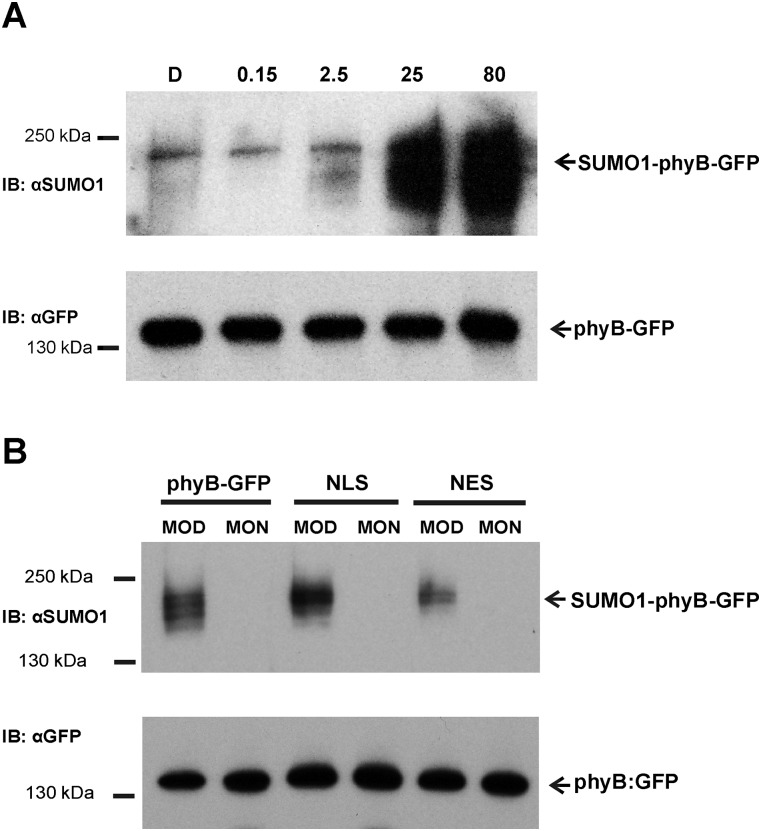
Experimentally determined SUMO attachment sites contain the ΨKXE sequence, where Ψ (Psi) is a large hydrophobic amino acid and K represents the lysine that binds SUMO (40). The ever-increasing number of identified SUMOylation sites in eukaryotic cells made it possible to construct reliable sequence analysis programs to identify, in silico, sites that are potential targets for SUMOylation in vivo (41). We used the SUMOsp 2.0 software to search for potential SUMOylation site(s) in phyB of A. thaliana (42). This search predicted the presence of two potential SUMOylation sites, K996 (high probability) in the C terminus and Lys475 (low probability) in the N terminus of the photoreceptor in AtphyB (Table S1). Fig. 1A shows the sequence context for the Lys996 in Arabidopsis as well as in phyB sequences of other plants species. To test if phyB is indeed SUMOylated in planta, first we analyzed SUMOylation of AtphyB-YFP in Nicotiana benthamiana leaves transiently coexpressing HA-SUMO and phyB-YFP or the phyBLys475Arg or phyBLys996Arg mutants. Our data demonstrate that AtphyB is SUMOylated and that mutation of Lys996 but not that of Lys475 inhibits SUMOylation of the photoreceptor in tobacco leaves (Fig. S1). To confirm that phyB is indeed subject of SUMOylation, we grew transgenic phyB-9 Arabidopsis plants expressing the 35S:PHYB-YFP transgene in light/dark (L/D) cycles, isolated the phyB-YFP fusion protein by affinity purification, and determined its SUMOylation status by using an antibody specific to AtSUMO-1. Accumulation of SUMOylated phyB-YFP reached high levels in 12 h RL/12 h D (Fig. 1B), and it was around the limit of detection in 12 h FRL/12 h D cycles (Fig. 1C). This figure also demonstrates that accumulation of the SUMOylated phyB in RL/D cycles is light regulated, as it accumulates to higher levels at the middle of day (MOD) and decreases to lower levels by the middle of the night (MON). The abundance of phyB does not significantly change in the 35S:PHYB-YFP plants grown under these conditions (Fig. S2A); thus, we conclude that phyB is SUMOylated in a reversible fashion in RL/D cycles. We also monitored SUMOylation of fusion proteins containing truncated forms of phyB including phyB(1–651)-YFP (43) and phyB(1–991)-YFP. The abundance of the truncated fusion proteins is similar to that of phyB-GFP, yet SUMOylation of these fusion proteins was below detection level in plants grown in RL/D cycles (Fig. S2B). Taken together, these data show that phyB is SUMOylated in vivo, the SUMOylated form of phyB accumulates to high levels in RL, and in harmony with data obtained by the transient SUMOylation assays, the site of reversible SUMOylation of phyB is located at the C-terminal domain of the photoreceptor.
Table S1.
Predicted SUMOylation sites of PHYB proteins of different species
| PhyB | Position | Type | Score | Cutoff |
| AtphyB | K996 | I | 0.991 | 0.1 |
| SlphyB | K967 | II | 2.7 | 2.26 |
| OsphyB | K1006 | II | 2.57 | 2.26 |
| TaphyB | K1001 | II | 2.57 | 2.26 |
| ZmphyB | K998 | II | 3.13 | 2.26 |
All data are predicted by the SUMOsp 2.0 software. For full species names and GenBank accession numbers, see Fig. 1A.
Fig. 1.
The photoreceptor phyB is SUMOylated in seedlings grown in RL/D cycles. (A) Analysis of A. thaliana (At, NP_179469), Solanum lycopersicum (Sl, XP_010318997), Oryza sativa (Os, EEE58928), Triticum aestivum (Ta, BAP91202), and Zea mays (Zm, NP_001168077) phyB protein sequences by SUMOsp 2.0 software identified sequence motifs as potential targets for SUMOylation. The Lys996 amino acid of the conserved SUMOylation site for AtphyB is indicated by an arrow. Numbers at the beginning and at the end of the presented sequences indicate the corresponding amino acid positions, and numbers in brackets show the length of PHYB proteins from different species. (B) phyB-9 seedlings expressing the 35S:PHYB-GFP transgene were grown on MS medium in 12 h RL (25 µmol m−2·s−1)/12 h D cycles and 12 h FRL (10 µmol m−2·s−1)/12 h D cycles for 5 d. Samples were harvested on day 6, at MOD and MON. phyB-GFP was immunoprecipitated by using GFP-Trap agarose beads (IP:αGFP), and samples containing identical amounts of the fusion protein were analyzed. phyB-GFP was detected by anti-GFP (IB:αGFP), whereas AtSUMO1-conjugated phyB-GFP was visualized using anti-SUMO1 antibody (IB:αSUMO1).
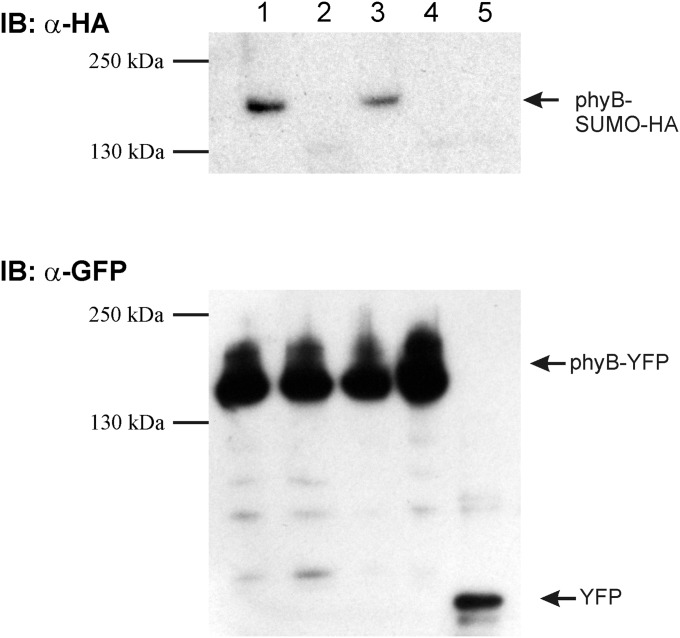
Fig. S1.
The phyB photoreceptor is SUMOylated at residue K996. phyB-YFP (lane 1), phyBLys996Arg-YFP (lane 2), phyBLys475Arg-YFP (lane 3), phyBLys996Arg/Lys475Arg-YFP (lane 4), and YFP (lane 5) were coexpressed with HA-SUMO1 in N. benthamiana tissues. Next phyB fusion proteins were immunoprecipitated using anti-GFP beads, and identical amounts of immunoprecipitated proteins were resolved on two separate SDS/PAGE gels and blotted onto PVDF membranes. Western blotting with anti-GFP and anti-HA antibodies showed the presence of YFP and HA-SUMO1.
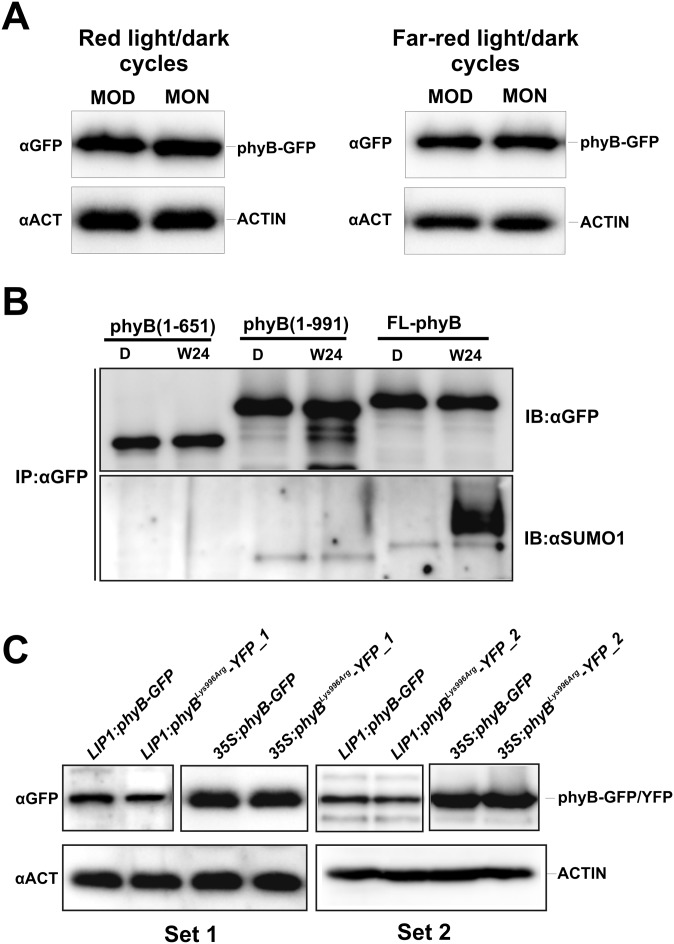
Fig. S2.
(A) The abundance of phyB-GFP does not change significantly in seedlings grown under 12h L/12h D cycles. Transgenic phyB-9 seedlings expressing the 35S:PHYB-GFP transgene were grown in 12 h WL or RL/12 h D cycles for 5 d. Samples were harvested at MOD and MON of day 6. Total protein extracts were separated by SDS/PAGE. phyB-GFP and ACTIN proteins were detected using anti-GFP and anti-ACTIN antibodies, respectively. (B) Truncated phyB-YFP fusion proteins lacking Lys996 are not SUMOylated. Seedlings expressing full-length phyB (FL-phyB) and truncated phyB(1–651) or phyB(1–991) proteins fused to GFP or YFP were grown in darkness for 4 d (D) and transferred to WL for 24 h (W24). Fusion proteins were immunoprecipitated using GFP-Trap agarose beads (IP:αGFP). Samples containing comparable amounts of the different fusion proteins were separated by SDS/PAGE. Replicate Western blots were probed with anti-GFP (IB:αGFP) or anti-SUMO1 (IB:αSUMO1) antibodies. (C) Abundance of phyB-GFP and phyBLys996Arg-YFP fusion proteins expressed under the control of LIP1 and 35S promoters is identical pair-wise. Transgenic phyB-9 seedlings expressing the phyB-GFP fusion proteins were grown in darkness for 5 d. Total protein extracts were separated by SDS/PAGE. phyB-GFP and ACTIN proteins were detected using anti-GFP and anti-ACTIN antibodies, respectively. Results were obtained from independent phyBLys996Arg-YFP–expressing transgenic lines, 1 and 2 (Set 1 and Set 2). Separately framed images of the upper panels were produced from different blots.
Transgenic phyB-9 plants expressing the mutant phyBLys996Arg-YFP are hypersensitive to RL.
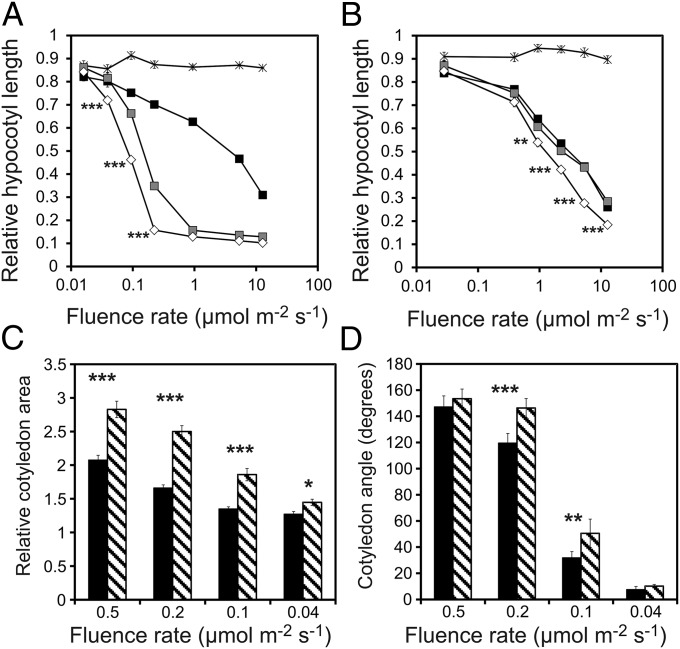
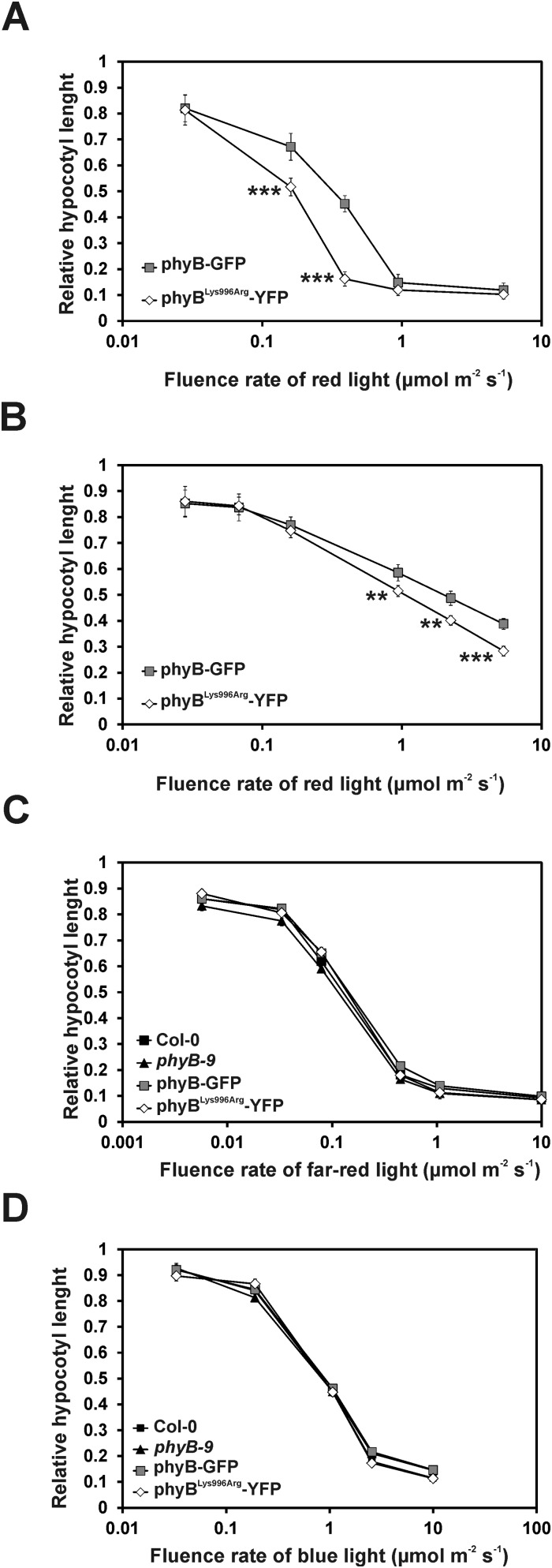
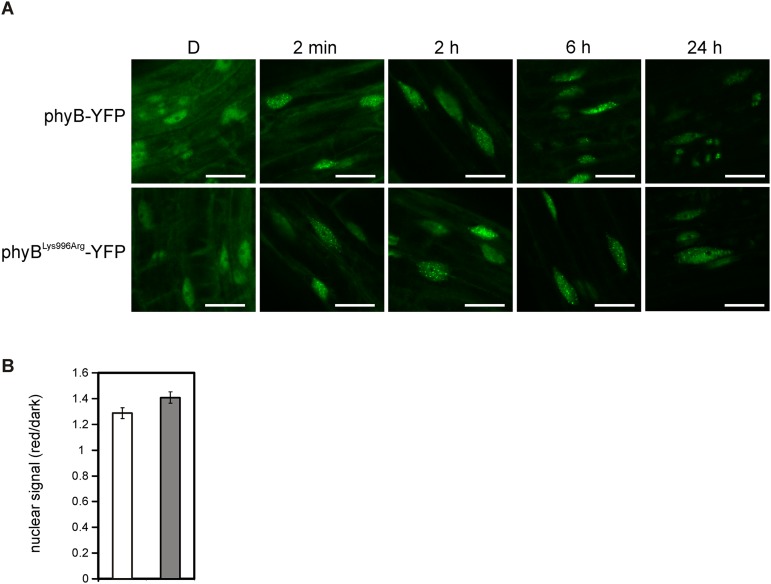
To determine the physiological importance of the predicted SUMOylation site in phyB, we generated transgenic lines expressing, under the control of the viral 35S promoter, the phyB-GFP and phyBLys996Arg-YFP fusion proteins at comparable levels (Fig. S2C, Set 1). Analysis of photomorphogenic responses in RL demonstrated that the transgenic 35S:PHYB-GFP lines displayed characteristic overexpression phenotypes compared with wild-type (Col-0) seedlings including fluence rate-dependent inhibition of hypocotyl elongation (Fig. 2A and Fig. S3A). Importantly, we also show that the 35S:PHYBLys996Arg-YFP lines displayed greater hypersensitivity to continuous RL not only for hypocotyl elongation (Fig. 2A) but cotyledon expansion (Fig. 2C) and cotyledon opening (Fig. 2D), indicating enhanced signaling by the mutant photoreceptor. In contrast, these lines did not display exaggerated responsiveness to FRL or blue light (BL) (Fig. S3 C and D). To recapitulate these data in plants containing phyB at physiological levels, we generated transgenic phyB-9 plants that expressed phyB-YFP or phyBLys996Arg-YFP fusion proteins under the control of the moderately active LIP1 (LIGHT INSENSITIVE PERIOD 1) promoter (44). Expression of the LIP1:PHYB-YFP transgene did not result in an overexpression phenotype but restored RL fluence rate-dependent inhibition of hypocotyl elongation similar to the wild-type (Col-0) level (Fig. 2B). Furthermore, we found that LIP1:PHYBLys996Arg-YFP seedlings are also hypersensitive to RL (Fig. 2B and Fig. S3B) compared with LIP1-PHYB-YFP, although the abundance of the fusion proteins is identical (Fig. S2C). Thus, we conclude that the mutant phyBLys996Arg-YFP, overexpressed or expressed at the physiological level, invariably displays enhanced RL-induced signaling. To test if enhanced signaling by the mutant phyBLys996Arg-YFP was brought about by increased/accelerated translocation of the mutant photoreceptor into the nucleus, we determined RL-dependent nuclear accumulation of the various phyB fusion proteins in planta. The data obtained indicate that the velocity of import into the nuclei and the formation as well as the size of phyB-YFP and phyBLys996Arg-YFP–associated photobodies do not differ in planta (Fig. S4).
Fig. 2.
Seedlings expressing the mutant phyBLys996Arg-YFP are hypersensitive to RL. Relative fluence rate-dependent inhibition of hypocotyl elongation of wild-type Col-0 (black square), phyB-9 (star symbol), and transgenic phyB-9 plants expressing phyB-GFP (gray square) or phyBLys996Arg-YFP (white diamond) under the control of the constitutively active (A) viral 35S promoter or (B) the Arabidopsis LIP1 promoter is shown. Seedlings were grown in darkness or at the indicated fluence rates of RL for 4 d. Hypocotyl lengths of irradiated seedlings were measured, and the average value of 25–30 seedlings was calculated and normalized to the hypocotyl length of D-grown plants. (C) Transgenic phyB-9 seedlings expressing 35S:PHYB-GFP (black columns) or 35S:PHYBLys996Arg-YFP (striped columns) were grown at the indicated fluence rates of RL for 4 d. Cotyledon areas of irradiated seedlings were measured, and the average value was normalized to the cotyledon area of D-grown plants. (D) Cotyledon angle of D- or RL-grown seedlings analyzed in Fig. 3C was measured. All experiments shown in Fig. 2 were repeated three times. Error bars indicate SE. Asterisks show statistically significant differences between phyB-GFP or phyBLys996Arg-YFP expressor plants as determined by Student’s t test (*P < 0.05; **P < 0.01; ***P < 0.001).
Fig. S3.
phyBLys996Arg-YFP–expressing seedlings are hypersensitive to RL. Transgenic lines, independent of those presented in Fig. 2, express (A) the viral 35S promoter-driven phyBLys996Arg-YFP or phyB-GFP or (B) these chimera proteins under the control of the Arabidopsis LIP1 promoter in phyB-9 background (see set 2 in Fig. S1C). For experimental setup and a detailed explanation, see the Fig. 2 legend. phyBLys996Arg-YFP–expressing seedlings are not hypersensitive to BL or FRL. Wild-type Col-0, phyB-9, and transgenic phyB-9 seedlings expressing the 35S:PHYB-GFP or 35S:PHYBLys996Arg-YFP transgenes were grown in darkness or at the indicated fluence rates of (C) FRL or (D) BL for 4 d. Fluence rate-dependent relative inhibition of hypocotyl lengths to the corresponding D controls is shown. The average of 25–30 seedlings is plotted for each data point; error bars indicate SE.
Fig. S4.
R-induced translocation, accumulation, and complex formation of phyB-YFP and phyBLys996Arg-YFP in the nucleus are identical. (A) Four-day-old seedlings expressing the LIP1:PHYB-YFP (upper row) or the LIP1:PHYBLys996Arg-YFP (lower row) transgene were grown in D and subsequently were irradiated by 1 µmol m−2·s−1 RL for 2 min, 2 h, 6 h, or 24 h. Images using confocal laser scanning microscope were taken on the hypocotyl cells located at the upper part of the hypocotyls. White bars represent 25 µm. (B) Nuclear fluorescence of transgenic seedlings expressing LIP1:PHYB-YFP (white bar) or the LIP1:PHYBLys996Arg-YFP (gray bar) was calculated from confocal laser scanning microscopy images taken of 4-d-old etiolated seedlings treated or not treated with 1 µmol m−2·s−1 RL for 6 h. The ratio of mean gray values derived from nonirradiated/irradiated seedlings from three independent experiments are plotted; error bars indicate the SE.
The Lys996Arg mutation reduces accumulation of SUMOylated phyB-YFP.
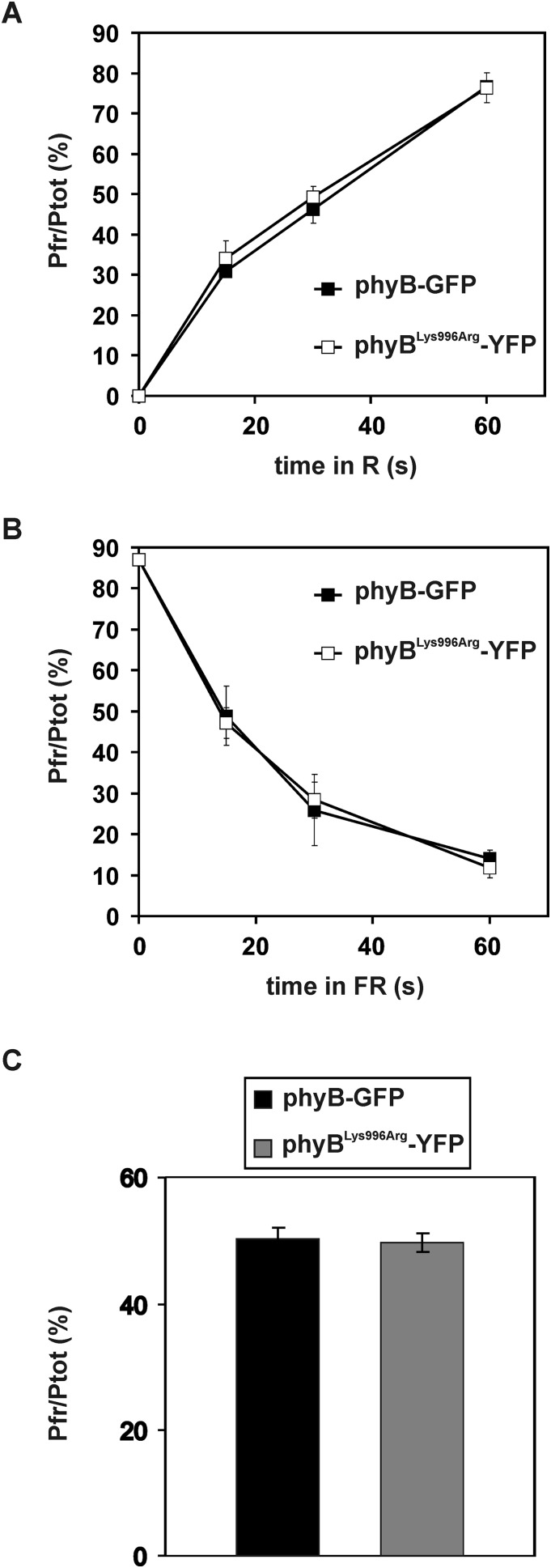
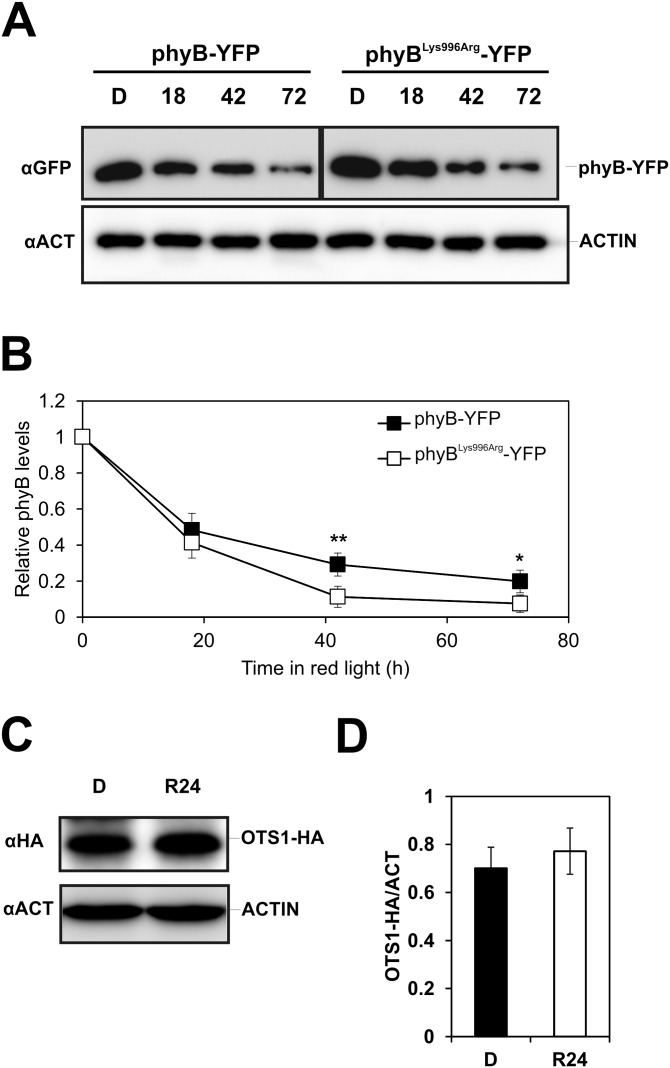
To test if enhanced RL-induced signaling of the mutant phyBLys996Arg-YFP photoreceptor was indeed brought about by its altered SUMOylation, we performed two lines of experiments. First we determined whether the Lys/Arg substitution affected rates of photoconversion and/or dark reversion of phyB. Our data demonstrate that the basic photochemical properties of the mutant phyB do not significantly differ from those of phyB (Fig. S5). Next we compared the accumulation of SUMOylated phyB-GFP and phyBLys996Arg-YFP in planta. We found that amounts of detectable SUMOylated phyB-GFP and phyBLys996Arg-YFP are low in etiolated seedlings, whereas RL treatment elevates the level of SUMOylated phyB-GFP but not that of phyBLys996Arg-YFP (Fig. 3). Accumulation of SUMOylated phyB-GFP increases slowly during the first 3–6 h of irradiation and reaches a substantially higher level after 24 h of treatment with RL (Fig. 3), and elevated intensities of RL are more effective to induce accumulation of SUMOylated phyB-GFP (Fig. S6A). Exposure of etiolated seedlings to white light (WL) also differentially modulates accumulation of SUMOylated phyB-GFP and phyBLys996Arg-YFP (Fig. 4). Importantly, this figure also illustrates that the constitutively active phyBTyr276His-YFP photoreceptor is hyper-SUMOylated in a light-independent fashion in etiolated seedlings. In addition, we found that nuclear pool of phyB is more prone to SUMOylation by comparing the level of SUMOylation of phyB in phyB-GFP–, phyB-GFP-nuclear localization signal (NLS)–, and phyB-GFP-nuclear export signal (NES)–expressing transgenic lines (Fig. S6B). Taken together, these data suggest that SUMOylated phyB preferably accumulates in its Pfr form in the nucleus, its accumulation is RL fluence rate-dependent, and perturbation of the putative SUMOylation site significantly reduces the detectable amount of SUMOylated phyB-GFP Pfr leading to enhanced signaling.
Fig. S5.
Photoconversion and D reversion of phyB are not altered by the Lys996Arg mutation. Arabidopsis seedlings expressing phyB-GFP or phyBLys996Arg-YFP mutant in phyB-9 mutant background were grown for 4 d in darkness and preirradiated with RL for 3 h to deplete endogenous phyA before photoconversion measurement. (A) Pr to Pfr photoconversion was measured after irradiation with RL (5 µmol m−2·s−1) for 15, 30, or 60 s. The amount of Pfr is shown relative to the total phy amount (Ptot). Error bars indicate the SE of at least three independent experiments. (B) Pfr to Pr photoconversion was measured after irradiation with FRL (20 µmol m−2·s−1) for 15, 30, or 60 s. The amount of Pfr is shown relative to the total phy amount (Ptot). Error bars indicate the SE of at least three independent experiments. (C) After the 3 h RL treatment, seedlings were transferred to darkness for 1 h. The amount of Pfr is shown relative to the total phy amount (Ptot). Error bars indicate the SE of at least four independent experiments.
Fig. 3.
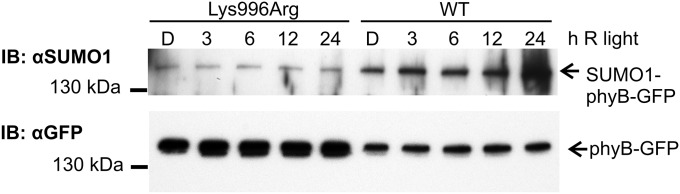
RL-induced accumulation of the SUMOylated phyBLys996Arg mutant photoreceptor is reduced. phyB-9 seedlings expressing phyB-GFP (WT) or phyBLys996Arg-YFP (Lys996Arg) fusion proteins were grown in darkness for 4 d (D) and subsequently transferred to RL (12.5 µmol m−2·s−1) for the indicated time. Sample preparation and detection of SUMOylation were performed as described in Fig. 1.
Fig. S6.
Accumulation of SUMOylated phyB-GFP is greatly increased at higher fluence rates of RL and reaches higher levels in the nuclear phyB-GFP pool. (A) Transgenic phyB-9 seedlings expressing the phyB-GFP fusion protein under the control of the viral 35S promoter were germinated in and grown in darkness for 4 d. Seedlings were next irradiated for 24 h with the indicated fluence rates of RL (units in µmol m−2·s−1). After harvesting, phyB-GFP proteins were immunoprecipitated using anti–GFP-Trap agarose beads. Samples containing identical amounts of the different fusion proteins were separated by SDS/PAGE. Replicate Western blots were probed with anti-GFP (IB:αGFP) or anti-SUMO1 (IB:αSUMO1) antibodies. (B) Transgenic phyB-9 seedlings expressing the phyB-GFP fusion protein or phyB-GFP-NLS (NLS) or phyB-GFP-NES (NES) under the control of the 35S promoter were grown in 12 h WL/12 h D cycles for 7 d before harvesting. Samples were harvested on day 8, at MOD and MON. phyB-GFP was immunoprecipitated by using GFP-Trap agarose beads, and samples containing identical amounts of the fusion proteins were analyzed. phyB-GFP, phyB-GFP-NLS, and phyB-GFP-NES molecules were detected by anti-GFP (IB:αGFP), whereas their AtSUMO1 conjugated forms were visualized using anti-SUMO1 antibody (IB:αSUMO1).
Fig. 4.
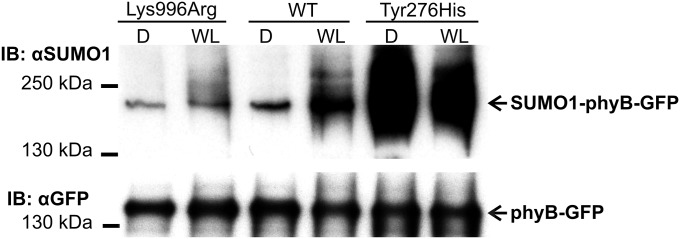
The SUMOylated form of the constitutively Pfr phyBTyr276His mutant accumulates to equally high levels in darkness and light. Transgenic phyB-9 seedlings expressing phyB-GFP (WT), phyBLys996Arg-YFP (Lys996Arg), or phyBTyr276His–YFP (Tyr276His) were grown in darkness for 4 d (D) and transferred to WL (50 µmol m−2·s−1) for 24 h. Sample preparation and detection of SUMOylation was performed as described in Fig. 1.
Binding of PIF5 to phyB Pfr is inhibited by SUMOylation of the photoreceptor.
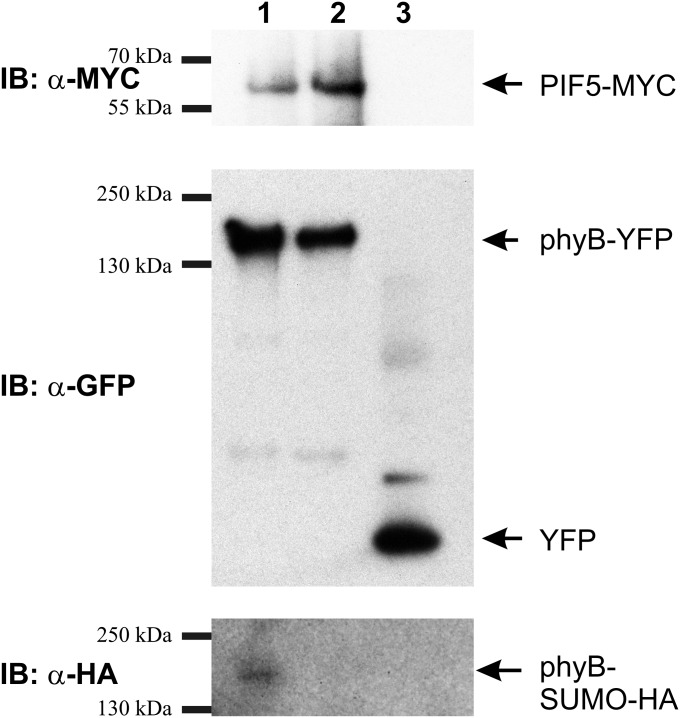
PIF1 and PIF3–PIF8 represent a subgroup of the bHLH (basic-helix–loop–helix) type transcription factor family that binds to phyB Pfr (45). PIFs negatively regulate signaling by phyB (46, 47) and function as a signaling hub to integrate light and hormone-induced signaling cascades (9, 48). Binding of PIFs to phyB initiates phosphorylation and subsequent degradation (49) or inactivation of these TFs (11, 50) and results in altered transcription of hundreds of genes in seedlings exposed to RL. It has been shown that compromised binding of PIFs to phyB Pfr results in reduced sensitivity to RL (49). To test whether SUMOylation of phyB interferes with PIF5 binding to phyB Pfr, we performed pull-down assays from infiltrated N. benthamiana leaves transiently coexpressing phyB-YFP or phyBLys996Arg-YFP with PIF5-MYC and HA-SUMO1 proteins. Our data demonstrate that binding of PIF5 to phyB-GFP (Fig. 5, lane 1) is reduced compared with phyBLys996Arg-YFP (Fig. 5, lane 2), although the amount of phyB-YFP and phyBLys996Arg-YFP is identical. This figure also demonstrates that Lys996Arg substitution inhibits SUMOylation of the receptor. We also note that in these assays the level of SUMOylated phyB-YFP is likely to be lowered by conjugation of N. benthamiana SUMO1. Taken together, these results suggest that binding of PIF5 to phyB Pfr is inhibited by the SUMOylation of the photoreceptor.
Fig. 5.
SUMOylation of phyB inhibits binding of the transcription factor PIF5. The figure shows results obtained by YFP pull-down of phyB-YFP, phyBLys996Arg-YFP, and YFP proteins from N. benthamiana tissue coinfiltrated with PIF5-MYC and HA-SUMO1. phyB fusion proteins were immunoprecipitated using anti-GFP beads. Next phyB-YFP (lane 1), phyBLys996Arg-YFP (lane 2), and YFP (lane 3) immunoprecipitated proteins were resolved on three separate SDS/PAGE gels and blotted onto PVDF membranes to detect the presence of phyB-YFP, phyBLys996Arg-YFP, YFP, PIF5-MYC, and HA-SUMO1.
Binding of PIFs to phyB Pfr has been known to induce degradation of not only the transcription factors but also phyB (13, 45), and a more recent report described the components and molecular mechanism that mediate mutual degradation of these proteins (14). SUMOylation of phyB appears to inhibit binding of PIF5 (Fig. 5); therefore, we determined how differential binding of PIFs to phyB-YFP and phyBLys996Arg-YFP affects RL-induced degradation of these fusion proteins. Fig. S7 shows that after prolonged treatment with RL the abundance of phyBLys996Arg-YFP is moderately but significantly lowered compared with phyB-YFP, indicating that SUMOylation of phyB inhibits RL-induced, PIF-mediated degradation of the photoreceptor. Taken together, our data demonstrate that SUMOylation of phyB negatively regulates RL-induced signaling, which can be explained, at least partly, by the compromised interaction of the SUMOylated phyB Pfr with PIF5 as well as with other PIFs.
Fig. S7.
Comparison of the stability of phyBLys996Arg-YFP and phyB-YFP in RL illuminated seedlings. (A) Transgenic phyB-9 seedlings expressing phyB-YFP or phyBLys996Arg-YFP fusion proteins under the control of the LIP1 promoter were grown in darkness for 4 d and transferred to RL for 18 h, 42 h, or 72 h before harvesting. Total protein extracts were separated by SDS/PAGE. phyB-YFP and ACTIN proteins were detected using anti-GFP and anti-ACTIN antibodies, respectively. (B) Quantification of chemiluminescent signals was done by the MetaMorph software. GFP-specific values were normalized to the corresponding ACTIN-specific signals. Values relative to the D levels are shown. Error bars represent SE values calculated from three independent datasets. An asterisk indicates statistically significant difference determined by Student’s t test (*P < 0.05; **P < 0.01). Stability/abundance of OTS1 is not regulated by light. (C) Transgenic Col-0 seedlings expressing the OTS1-HA fusion proteins under the control of the 35S promoter were grown in D and exposed to RL. Western blot hybridization was performed as described above except that OTS1 was detected using anti-HA antibodies. (D) Quantitative analysis was performed as described above.
The SUMO proteases OTS1/OTS2 are involved in de-SUMOylation of phyB.
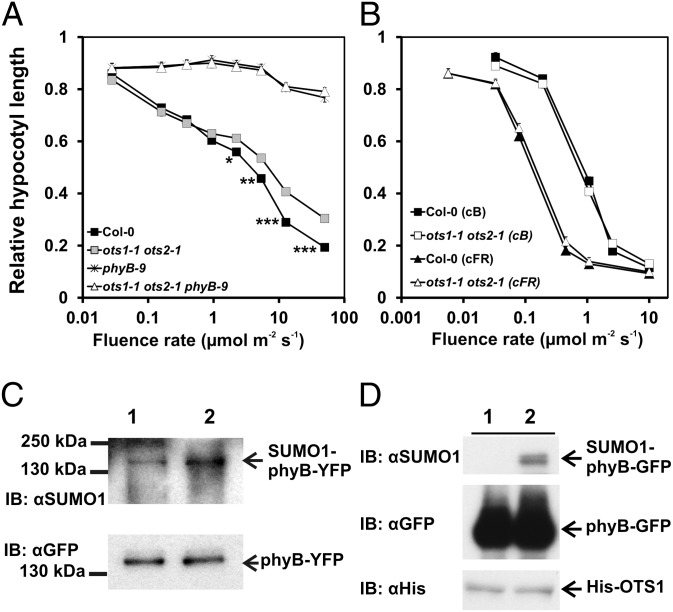
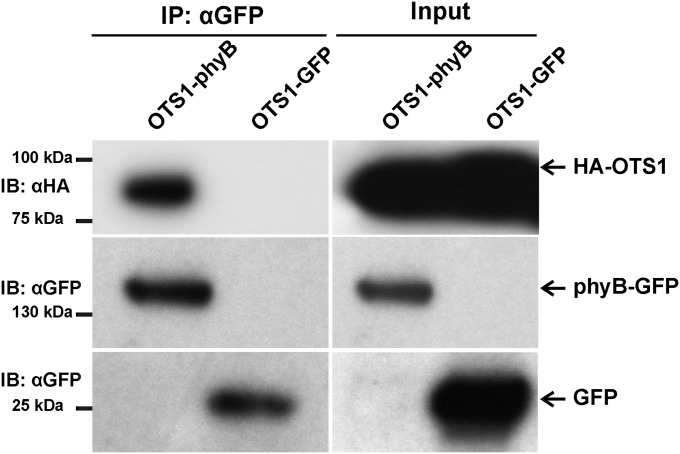
SUMOylation of proteins is a reversible process mediated by enzymes that catalyze conjugation/deconjugation of SUMO to/from the target proteins. Recent research identified a number of genes encoding SUMO proteases in Arabidopsis (for a review, see ref. 27). We have launched a program to systematically test if any of these proteases is required for de-SUMOylation of physs, including phyB. So far we have identified two SUMO proteases, OTS1 and OTS2, that appear to play an active role in de-SUMOylation of phyB. We found that the ots1/ots2 double mutant is hyposensitive to RL (Fig. 6A). Hyposensitivity of the ots1/ots2 mutant is specific to RL, as the double mutant displayed normal responsiveness to BL and FRL (Fig. 6B), whereas the triple ots1/ots2/phyB-9 mutant was completely insensitive to RL, similarly to phyB-9 (Fig. 6A). To validate the involvement of OTS1/OTS2 in regulating RL-induced signaling, we determined the accumulation of SUMOylated phyB-YFP in transgenic phyB-9 and ots1/ots2/phyB-9 plants expressing phyB-YFP. To ensure that phyB-YFP is expressed at identical levels, we introgressed the LIP1:PHYB-YFP transgene from phyB-9 (Fig. 2B) into an ots1/ots2/phyB-9 background. The abundance level of OTS1/OTS2 is not regulated by light (Fig. S7 C and D), and Fig. 6C demonstrates that the lack of active OTS1/OTS2 proteases increased the accumulation of SUMOylated phyB-YFP. Next we performed two lines of experiments to demonstrate that OTS1 is directly involved in de-SUMOylating phyB. First, we tested if recombinant OTS1 can deconjugate SUMO from phyB in vitro, and next we investigated if phyB interacts with OTS1 in planta. Our data clearly demonstrate that OTS1 cleaves off SUMO from plant-derived SUMOylated phyB (Fig. 6D) and that OTS1 binds to phyB when transiently coexpressed in N. benthamiana leaves (Fig. S8). Taken together, we conclude that OTS1/OTS2 act redundantly to mediate, at least partly, reversible SUMOylation of phyB.
Fig. 6.
The SUMO proteases OTS1 and OTS2 mediate de-SUMOylation of phyB. (A) The ots1/ots2 mutant is hyposensitive to RL. Fluence rate-dependent relative inhibition of hypocotyl elongation of Col-0 (black square), phyB-9 (star symbol), ots1/ots2 (gray square), and ots1/ots2/phyB-9 (white triangle) seedlings grown in RL is shown. For additional details, see the Fig. 2 legend. (B) The ots1/ots2 mutant exhibits normal photomophogenesis in FRL or BL. Hypocotyl elongation inhibition in FRL-grown (Col-0, black triangle; ots1/ots2, white triangle) and BL-grown (Col-0, black square; ots1/ots2, white square) seedlings is shown. For additional details, see the Fig. 2 legend. (C) Accumulation of the SUMOylated form of phyB is enhanced in transgenic seedlings lacking functional OTS1/OTS2 SUMO proteases. Transgenic phyB-9 (lane 1) and ots1/ots2/phyB-9 (lane 2) seedlings expressing LIP1:PHYB-YFP were grown under 12 h L/12 h D cycles for 5 d, and samples were harvested on the sixth day at MOD. Accumulation of the SUMOylated phyB-YFP was detected as described in Fig. 1. (D) Recombinant OTS1 deSUMOylates phyB-GFP in vitro. SUMOylated phyB-GFP protein was affinity purified from total protein extracts with anti-GFP antibody and incubated with an identical amount of recombinant OTS1 (His:OTS1) (lane 1) or catalytically inactive OTS1 (His:OTS1CS) (lane 2) proteins. After incubation (typically 4 h at room temperature), the beads were reapplied to the column [the flow-through was used to determine 6xHis-OTS1 levels with anti-Histidine tag antibodies (IB:αHis)] and washed, and bound proteins were eluted with SDS/PAGE loading buffer and separated by electrophoresis, blotted, and probed with anti-GFP (IB:αGFP) and anti-SUMO1 (IB:αSUMO1) antibodies.
Fig. S8.
OTS1 binds to phyB in planta. The Agrobacterium cultures containing the combination of constructs (phyB-GFP/HA-OTS1 or GFP/HA-OTS1) were infiltrated into N. benthamiana leaves. Three days after the Agrobacterium infiltration, the expression of the fusion proteins was confirmed by immunoblotting with anti-HA and anti-GFP antibodies (input panels). Coimmunoprecipitation assays performed with anti-GFP antibodies showed that HA-OTS1 immunoprecipitated with phyB-GFP but not with GFP alone, indicating that OTS1 interacts with phyB.
Discussion
PhyB regulates growth and development of plants throughout their entire life cycle. PhyB exerts it regulatory function in a RL/FRL reversible fashion via a complex signaling network consisting of an ever-growing number of components of which numerous proteins, including PIFs, directly interact with the biologically active Pfr form of the photoreceptor. Binding of the negative regulatory PIF to phyB Pfr initiates either (i) phosphorylation of these TFs (49) followed by mutual degradation of these proteins (14) or (ii) inactivation of PIFs via a not fully understood mechanism (11). These events then culminate in altered transcription of about 15% of the genome during the initial phase of light to dark transition in Arabidopsis (12).
Here we report that phyB is SUMOylated at its C terminus, and Lys996 is critical for the efficient conjugation of SUMO to phyB both in transient assays (Fig. S1) and in stably transformed transgenic lines. Accumulation of SUMOylated phyB-YFP follows a diurnal pattern in seedlings grown under L/D cycles (Fig. 1) and it is increased by R or WL in a fluence rate-dependent fashion (Figs. 3 and 4). In contrast, SUMOylation of phyBLys996Arg-YFP remains invariably low under all conditions tested, whereas SUMOylation of truncated phyB-YFP fusion proteins [phyB(1–651)-YFP-NLS, phyB(1–990)-GFP] lacking Lysine996 is below detection level in planta (Fig. S1). Transgenic phyBLys996Arg-YFP lines similar to phyB(1–651)-GUS-GFP-NLS (51) or phyB(1–991)-GFP (52) display hypersensitivity to RL compared with phyB-YFP. Taken together, these data indicate that reduced SUMOylation of phyB enhances RL-induced signaling; thus, SUMOylation inhibits phyB action. We note that despite its low abundance we can readily detect SUMOylated phyBLys996Arg-YFP in etiolated seedlings (Figs. 3 and 4). At present we do not know if this is caused by conjugation of SUMO onto phyB at a second site or whether this is functionally relevant.
SUMOylation of phyB interferes with the conformation-specific interaction of phyB and the negative regulatory factor PIF5 (Fig. 5). We postulate that SUMOylation of phyB at its C-terminal domain directly inhibits interaction of these proteins. This model is supported by recent findings demonstrating that (i) mutations in the C-terminal domain of phyB inhibit binding of PIF3 (53) and that (ii) mutations that fully inactivate signaling by the N-terminal domain of phyB and binding of PIF3 in vitro only partially impair the activity of the full-length photoreceptor (54). Thus, it is plausible to assume that the interaction of PIFs with phyB Pfr is mediated by multiple binding sites and conjugation of the bulky SUMO to the C-terminal domain of phyB substantially weakens its interaction with PIFs. However, it has been recently shown that phosphorylation of phyBTyr104 also inhibits binding of PIF3 to phyB Pfr (16), whereas phosphorylation of phyBSer86 accelerates thermal reversion (also called D reversion) of phyB Pfr into Pr and thereby prevents interaction of these proteins (15). Thus, it is also possible that SUMOylation interferes with binding of kinases/phosphatases to phyB and thus indirectly via modifying phosphorylation status of phyB alters the interaction of phyB Pfr with PIFs. Binding of PIFs to phyB Pfr has been shown to be essential for mutual degradation of these proteins (14). Our data illustrate that extended RL treatment moderately decreases the abundance of phyBLys996Arg-YFP compared with phyB-YFP. This observation can be readily explained by compromised interaction of phyB-YFP and PIFs, which is brought about by increased SUMOylation of phyB-YFP compared with the phyBLys996Arg-YFP mutant.
We show that SUMOylation of phyB is a reversible process and propose that the OTS1 and OTS2 SUMO proteases play a significant role in deconjugating SUMO from phyB. This conclusion is supported by data demonstrating that phyB-YFP is hyper-SUMOylated in ots1/ots2/phyB-9 compared with phyB-9 (Fig. 6C), OTS1 de-SUMOylates phyB in vitro (Fig. 6D), OTS1 binds to phyB in planta (Fig. S8), and the ots1/ots2 double mutant is hyposensitive to RL (Fig. 6A). However, this mutant exhibits normal BL and FRL-induced physiological responses, indicating that other photoreceptors are not targets of SUMOyltion or not deSUMOylated by OTS1/OTS2. SUMOylated phyB is hardly detectable in etiolated or FRL irradiated seedlings, whereas it accumulates to substantially higher levels in RL and WL. These data and the fact that the constitutively active phyBTyr276His mutant is highly SUMOylated both in WL and darkness indicate that SUMOylated phyB preferentially accumulates in its Pfr form. This can be explained either by conformation-specific SUMOylation and/or by de-SUMOylation of phyB. OTS1 binds to phyB Pfr in vivo (Fig. S8), removes SUMO from phyB Pfr in vitro (Fig. 6D), and its abundance is not regulated by light (Fig. S7 C and D). Thus, we conclude that OTS1 is able to de-SUMOylate phyB Pfr as well as phyB Pr, as the abundance of SUMOylated phyB rapidly declines during the D phase in seedlings grown under L/D cycles (Fig. 1B). These data suggest that light-regulated expression and/or modification of yet unknown SUMO proteases or conjugating enzymes or their conformation-specific interaction with phyB Pfr are responsible for the higher level accumulation of SUMOylated phyB Pfr compared with that of phyB Pr.
Independent of the precise mechanism mediating reversible SUMOylation of phyB, we note that SUMOylation has been shown to play a pervasive role in the regulation of stress responses including heat (35), salt (31), and ABA signaling (38). Interestingly, it was shown that OTS1/OTS2 is destabilized by salt stress, and overexpression of OTS1/OTS2 significantly alters salt tolerance of the transgenic plants (31). Because OTS1/OTS2 is involved in mediating reversible SUMOylation of the photoreceptor, we speculate that SUMOylation of phyB might play an important role in the integration of light, salt, and other stress-induced responses.
Materials and Methods
Plant Material and Growth Conditions.
Col-0 and phyB-9 (55) mutant plants were used in this work. Plants expressing PHYB-YFP or N651-GFP in phyB-9 background have been described (7, 15, 43). The cDNA fragment corresponding to amino acid residues 1–991 of the PHYB protein was inserted as an XbaI-SmaI fragment between the 35S promoter and the YFP-NOS terminator of pPCVB (47). The Lys996Arg and Tyr276His mutations were created by a site-directed mutagenesis kit (Stratagene). Plants were transformed by the Agrobacterium-mediated floral dip method (56). Basta-resistant plants were selected and grown to maturation in the greenhouse. Homozygous T3 plants grown at 22 °C were used in all assays. WL was provided by cool-white fluorescent tubes at a 25 µmol m−2·s−1 fluence rate. RL (λmax, 667 nm) or FRL (λmax, 730 nm) was provided by LED light sources (Quantum Devices Inc.) at 25 or 10 µmol m−2·s−1 fluence rates, respectively. For diurnal conditions, plants were grown in 12 h WL (25 μmol m−2·s−1)/12 h D or 12 h RL (25 μmol m−2·s−1)/12 h D cycles for 7 d before harvesting.
Transient Assays in N. benthamiana.
PIF5 cDNA was cloned in pGWB17 so that expressed proteins had a C-terminal 4xMYC tag, and SUMO1 was cloned in pEG201 so that expressed proteins had a N-terminal HA tag. Gene constructs were transiently expressed in N. benthamiana plants using Agrobacterium-mediated transformation (57). The protein samples extracted (57) were mixed with 50 µL anti-GFP (Chromotek anti-GFP beads) and incubated on ice for 15 min. The beads were centrifuged down at 10,000 × g for 1 h and washed three times with 1 mL of cold IP buffer. After the last wash, 50 µL of preheated (95 °C) 1× SDS loading buffer was used to elute the immuno-complex and analyzed on 8–10% SDS/PAGE using immunoblotting methods with Clontech (clone, JL-8) anti-GFP (Abcam) anti-Myc, and anti-HA antibodies.
Western Blot Analysis, Hypocotyl Length Measurement.
For details, see SI Materials and Methods.
SI Materials and Methods
Western Blot Hybridization.
For the immunoblot analysis, 4-d-old D-grown seedlings were exposed to various treatments and harvested at different time points. After protein extraction (31), the samples were separated by SDS/PAGE and blotted onto PVDF membranes. After blocking with 5% (wt/vol) milk in PBST, the upper part of the membrane was probed with anti-GFP monoclonal antibody (Clontech, Living colors, clone JL-8) and the lower part with monoclonal anti-actin IgG (Sigma-Aldrich, clone 10-B3) for 1.5 h, anti-SUMO1 antibody (31) for 3 h, anti-His antibody (Millipore, 05–949) for 2 h, and anti-HA antibody (Abcam, ab9110) for 2 h, followed by anti-mouse/anti-rabbit IgG-HRP conjugate (Thermoscientific) for 1.5 h. The membranes were treated with Immobilon Western Chemiluminescent HRP substrate (Millipore), and luminescent signals were detected using a liquid nitrogen-cooled charge-coupled device camera (Micromax; Roper Scientific). Digital images were analyzed and signals were quantified using the Metamorph software package (Universal Imaging).
Hypocotyl Length, Cotyledon Area, and Angle Measurements.
Seedlings were grown on wet filter paper for 4 d under the indicated irradiation circumstances and subsequently were positioned on agar plates. Scanned seedling images were analyzed using MetaMorph Software.
Confocal microscopy.
Confocal laser scanning microscopy was performed on 4-d-old seedlings using Leica SP5 AOBS confocal laser scanning microscope (Leica) on a DMI6000 microscope base. Microscope configuration was as follows: objective lens, HC PL APO 20× (N.A. 0.7); sampling speed, 100 Hz; line averaging, 3×; pinhole, 200 µm; excitation, 514 nm laser; and spectral emission detector for YFP, 534–589 nm. Brightness and contrast settings were uniformly done on the corresponding image pairs. YFP images were pseudocolored green.
Photoconversion kinetics of phyB wild-type and mutant versions in A. thaliana.
Four-day-old etiolated A. thaliana seedlings expressing phyB-GFP or mutant versions of phyB-YFP in phyB-9 mutant background were pretreated with RL (LED; λmax, 660 nm; 20 µmol m−2·s−1) for 3 h to deplete phyA. Then filter papers with seedlings were placed on an ice/water mix. For Pr to Pfr photoconversion kinetics, seedlings were irradiated with FRL (LED; λmax, 740 nm; 100 µmol m−2 s−1) for 90 s to revert all Pfr to Pr. Subsequently seedlings were irradiated with RL (LED; λmax, 660 nm; 5 µmol m−2·s−1) for 15, 30, or 60 s. For Pfr to Pr photoconversion kinetics, seedlings were irradiated with RL (LED; λmax, 660 nm; 20 µmol m−2·s−1) for 90 s to establish the maximal Pfr/Ptot ratio (87%). Subsequently seedlings were irradiated with FRL (LED; λmax, 740 nm; 20 µmol m−2·s−1) for 15, 30, or 60 s. Seedlings were harvested and placed into cuvettes, and Pfr/Ptot ratios were measured immediately after the light treatment in a dual wavelength spectrophotometer.
Acknowledgments
Work performed in Edinburgh and Szeged was supported by Biotechnology and Biological Sciences Research Council Grant BB/K006975/1 and Hungarian Scientific Research Fund Grants K-108559 and NN-110636 (to F.N.). A.V. was supported by a Bolyai János Scholarship of the Hungarian Academy of Sciences. Work in the A.S. laboratory in Durham was supported by the Biotechnology and Biological Sciences Research Council and the European Research Council consolidator grant scheme.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1415260112/-/DCSupplemental.
References
- 1.Liu H, Liu B, Zhao C, Pepper M, Lin C. The action mechanisms of plant cryptochromes. Trends Plant Sci. 2011;16(12):684–691. doi: 10.1016/j.tplants.2011.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Goyal A, Szarzynska B, Fankhauser C. Phototropism: At the crossroads of light-signaling pathways. Trends Plant Sci. 2013;18(7):393–401. doi: 10.1016/j.tplants.2013.03.002. [DOI] [PubMed] [Google Scholar]
- 3.Quail PH. Phytochromes. Curr Biol. 2010;20(12):R504–R507. doi: 10.1016/j.cub.2010.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rizzini L, et al. Perception of UV-B by the Arabidopsis UVR8 protein. Science. 2011;332(6025):103–106. doi: 10.1126/science.1200660. [DOI] [PubMed] [Google Scholar]
- 5.Bae G, Choi G. Decoding of light signals by plant phytochromes and their interacting proteins. Annu Rev Plant Biol. 2008;59:281–311. doi: 10.1146/annurev.arplant.59.032607.092859. [DOI] [PubMed] [Google Scholar]
- 6.Sharrock RA, Clack T. Patterns of expression and normalized levels of the five Arabidopsis phytochromes. Plant Physiol. 2002;130(1):442–456. doi: 10.1104/pp.005389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kircher S, et al. Nucleocytoplasmic partitioning of the plant photoreceptors phytochrome A, B, C, D, and E is regulated differentially by light and exhibits a diurnal rhythm. Plant Cell. 2002;14(7):1541–1555. doi: 10.1105/tpc.001156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen M, Schwab R, Chory J. Characterization of the requirements for localization of phytochrome B to nuclear bodies. Proc Natl Acad Sci USA. 2003;100(24):14493–14498. doi: 10.1073/pnas.1935989100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Leivar P, Quail PH. PIFs: Pivotal components in a cellular signaling hub. Trends Plant Sci. 2011;16(1):19–28. doi: 10.1016/j.tplants.2010.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Huang X, Ouyang X, Deng XW. Beyond repression of photomorphogenesis: Role switching of COP/DET/FUS in light signaling. Curr Opin Plant Biol. 2014;21:96–103. doi: 10.1016/j.pbi.2014.07.003. [DOI] [PubMed] [Google Scholar]
- 11.Park E, et al. Phytochrome B inhibits binding of phytochrome-interacting factors to their target promoters. Plant J. 2012;72(4):537–546. doi: 10.1111/j.1365-313X.2012.05114.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ma L, et al. Light control of Arabidopsis development entails coordinated regulation of genome expression and cellular pathways. Plant Cell. 2001;13(12):2589–2607. doi: 10.1105/tpc.010229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Leivar P, et al. The Arabidopsis phytochrome-interacting factor PIF7, together with PIF3 and PIF4, regulates responses to prolonged red light by modulating phyB levels. Plant Cell. 2008;20(2):337–352. doi: 10.1105/tpc.107.052142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ni W, et al. A mutually assured destruction mechanism attenuates light signaling in Arabidopsis. Science. 2014;344(6188):1160–1164. doi: 10.1126/science.1250778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Medzihradszky M, et al. Phosphorylation of phytochrome B inhibits light-induced signaling via accelerated dark reversion in Arabidopsis. Plant Cell. 2013;25(2):535–544. doi: 10.1105/tpc.112.106898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nito K, Wong CC, Yates JR, 3rd, Chory J. Tyrosine phosphorylation regulates the activity of phytochrome photoreceptors. Cell Reports. 2013;3(6):1970–1979. doi: 10.1016/j.celrep.2013.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Johnson ES. Protein modification by SUMO. Annu Rev Biochem. 2004;73:355–382. doi: 10.1146/annurev.biochem.73.011303.074118. [DOI] [PubMed] [Google Scholar]
- 18.Saracco SA, Miller MJ, Kurepa J, Vierstra RD. Genetic analysis of SUMOylation in Arabidopsis: Conjugation of SUMO1 and SUMO2 to nuclear proteins is essential. Plant Physiol. 2007;145(1):119–134. doi: 10.1104/pp.107.102285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lois LM. Diversity of the SUMOylation machinery in plants. Biochem Soc Trans. 2010;38(Pt 1):60–64. doi: 10.1042/BST0380060. [DOI] [PubMed] [Google Scholar]
- 20.Elrouby N, Coupland G. Proteome-wide screens for small ubiquitin-like modifier (SUMO) substrates identify Arabidopsis proteins implicated in diverse biological processes. Proc Natl Acad Sci USA. 2010;107(40):17415–17420. doi: 10.1073/pnas.1005452107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Song J, Durrin LK, Wilkinson TA, Krontiris TG, Chen Y. Identification of a SUMO-binding motif that recognizes SUMO-modified proteins. Proc Natl Acad Sci USA. 2004;101(40):14373–14378. doi: 10.1073/pnas.0403498101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kerscher O. SUMO junction-what’s your function? New insights through SUMO-interacting motifs. EMBO Rep. 2007;8(6):550–555. doi: 10.1038/sj.embor.7400980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hannich JT, et al. Defining the SUMO-modified proteome by multiple approaches in Saccharomyces cerevisiae. J Biol Chem. 2005;280(6):4102–4110. doi: 10.1074/jbc.M413209200. [DOI] [PubMed] [Google Scholar]
- 24.Park HC, et al. Identification and molecular properties of SUMO-binding proteins in Arabidopsis. Mol Cells. 2011;32(2):143–151. doi: 10.1007/s10059-011-2297-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.van den Burg HA, Kini RK, Schuurink RC, Takken FL. Arabidopsis small ubiquitin-like modifier paralogs have distinct functions in development and defense. Plant Cell. 2010;22(6):1998–2016. doi: 10.1105/tpc.109.070961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kurepa J, et al. The small ubiquitin-like modifier (SUMO) protein modification system in Arabidopsis. Accumulation of SUMO1 and -2 conjugates is increased by stress. J Biol Chem. 2003;278(9):6862–6872. doi: 10.1074/jbc.M209694200. [DOI] [PubMed] [Google Scholar]
- 27.Castro PH, Tavares RM, Bejarano ER, Azevedo H. SUMO, a heavyweight player in plant abiotic stress responses. Cell Mol Life Sci. 2012;69(19):3269–3283. doi: 10.1007/s00018-012-1094-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tomanov K, et al. Arabidopsis PIAL1 and 2 promote SUMO chain formation as E4-type SUMO ligases and are involved in stress responses and sulfur metabolism. Plant Cell. 2014;26(11):4547–4560. doi: 10.1105/tpc.114.131300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chosed R, Mukherjee S, Lois LM, Orth K. Evolution of a signalling system that incorporates both redundancy and diversity: Arabidopsis SUMOylation. Biochem J. 2006;398(3):521–529. doi: 10.1042/BJ20060426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Colby T, Matthäi A, Boeckelmann A, Stuible HP. SUMO-conjugating and SUMO-deconjugating enzymes from Arabidopsis. Plant Physiol. 2006;142(1):318–332. doi: 10.1104/pp.106.085415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Conti L, et al. Small ubiquitin-like modifier proteases OVERLY TOLERANT TO SALT1 and -2 regulate salt stress responses in Arabidopsis. Plant Cell. 2008;20(10):2894–2908. doi: 10.1105/tpc.108.058669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ishida T, Yoshimura M, Miura K, Sugimoto K. MMS21/HPY2 and SIZ1, two Arabidopsis SUMO E3 ligases, have distinct functions in development. PLoS One. 2012;7(10):e46897. doi: 10.1371/journal.pone.0046897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Murtas G, et al. A nuclear protease required for flowering-time regulation in Arabidopsis reduces the abundance of SMALL UBIQUITIN-RELATED MODIFIER conjugates. Plant Cell. 2003;15(10):2308–2319. doi: 10.1105/tpc.015487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Castaño-Miquel L, et al. Diversification of SUMO-activating enzyme in Arabidopsis: Implications in SUMO conjugation. Mol Plant. 2013;6(5):1646–1660. doi: 10.1093/mp/sst049. [DOI] [PubMed] [Google Scholar]
- 35.Miller MJ, Barrett-Wilt GA, Hua Z, Vierstra RD. Proteomic analyses identify a diverse array of nuclear processes affected by small ubiquitin-like modifier conjugation in Arabidopsis. Proc Natl Acad Sci USA. 2010;107(38):16512–16517. doi: 10.1073/pnas.1004181107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Miura K, et al. The Arabidopsis SUMO E3 ligase SIZ1 controls phosphate deficiency responses. Proc Natl Acad Sci USA. 2005;102(21):7760–7765. doi: 10.1073/pnas.0500778102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Catala R, et al. The Arabidopsis E3 SUMO ligase SIZ1 regulates plant growth and drought responses. Plant Cell. 2007;19(9):2952–2966. doi: 10.1105/tpc.106.049981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Miura K, et al. Sumoylation of ABI5 by the Arabidopsis SUMO E3 ligase SIZ1 negatively regulates abscisic acid signaling. Proc Natl Acad Sci USA. 2009;106(13):5418–5423. doi: 10.1073/pnas.0811088106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lee J, et al. Salicylic acid-mediated innate immunity in Arabidopsis is regulated by SIZ1 SUMO E3 ligase. Plant J. 2007;49(1):79–90. doi: 10.1111/j.1365-313X.2006.02947.x. [DOI] [PubMed] [Google Scholar]
- 40.Rodriguez MS, Dargemont C, Hay RT. SUMO-1 conjugation in vivo requires both a consensus modification motif and nuclear targeting. J Biol Chem. 2001;276(16):12654–12659. doi: 10.1074/jbc.M009476200. [DOI] [PubMed] [Google Scholar]
- 41.Lamoliatte F, et al. Large-scale analysis of lysine SUMOylation by SUMO remnant immunoaffinity profiling. Nat Commun. 2014;5:5409. doi: 10.1038/ncomms6409. [DOI] [PubMed] [Google Scholar]
- 42.Ren J, et al. Systematic study of protein sumoylation: Development of a site-specific predictor of SUMOsp 2.0. Proteomics. 2009;9(12):3409–3412. doi: 10.1002/pmic.200800646. [DOI] [PubMed] [Google Scholar]
- 43.Palágyi A, et al. Functional analysis of amino-terminal domains of the photoreceptor phytochrome B. Plant Physiol. 2010;153(4):1834–1845. doi: 10.1104/pp.110.153031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kevei E, et al. Arabidopsis thaliana circadian clock is regulated by the small GTPase LIP1. Curr Biol. 2007;17(17):1456–1464. doi: 10.1016/j.cub.2007.07.018. [DOI] [PubMed] [Google Scholar]
- 45.Khanna R, et al. A novel molecular recognition motif necessary for targeting photoactivated phytochrome signaling to specific basic helix-loop-helix transcription factors. Plant Cell. 2004;16(11):3033–3044. doi: 10.1105/tpc.104.025643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kim J, et al. Functional characterization of phytochrome interacting factor 3 in phytochrome-mediated light signal transduction. Plant Cell. 2003;15(10):2399–2407. doi: 10.1105/tpc.014498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bauer D, et al. Constitutive photomorphogenesis 1 and multiple photoreceptors control degradation of phytochrome interacting factor 3, a transcription factor required for light signaling in Arabidopsis. Plant Cell. 2004;16(6):1433–1445. doi: 10.1105/tpc.021568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Leivar P, Monte E. PIFs: Systems integrators in plant development. Plant Cell. 2014;26(1):56–78. doi: 10.1105/tpc.113.120857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Al-Sady B, Ni W, Kircher S, Schäfer E, Quail PH. Photoactivated phytochrome induces rapid PIF3 phosphorylation prior to proteasome-mediated degradation. Mol Cell. 2006;23(3):439–446. doi: 10.1016/j.molcel.2006.06.011. [DOI] [PubMed] [Google Scholar]
- 50.Li L, et al. Linking photoreceptor excitation to changes in plant architecture. Genes Dev. 2012;26(8):785–790. doi: 10.1101/gad.187849.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Matsushita T, Mochizuki N, Nagatani A. Dimers of the N-terminal domain of phytochrome B are functional in the nucleus. Nature. 2003;424(6948):571–574. doi: 10.1038/nature01837. [DOI] [PubMed] [Google Scholar]
- 52.Krall L, Reed JW. The histidine kinase-related domain participates in phytochrome B function but is dispensable. Proc Natl Acad Sci USA. 2000;97(14):8169–8174. doi: 10.1073/pnas.140520097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kircher S, et al. Light quality-dependent nuclear import of the plant photoreceptors phytochrome A and B. Plant Cell. 1999;11(8):1445–1456. doi: 10.1105/tpc.11.8.1445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Oka Y, Matsushita T, Mochizuki N, Quail PH, Nagatani A. Mutant screen distinguishes between residues necessary for light-signal perception and signal transfer by phytochrome B. PLoS Genet. 2008;4(8):e1000158. doi: 10.1371/journal.pgen.1000158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Reed JW, Nagpal P, Poole DS, Furuya M, Chory J. Mutations in the gene for the red/far-red light receptor phytochrome B alter cell elongation and physiological responses throughout Arabidopsis development. Plant Cell. 1993;5(2):147–157. doi: 10.1105/tpc.5.2.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Clough SJ, Bent AF. Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998;16(6):735–743. doi: 10.1046/j.1365-313x.1998.00343.x. [DOI] [PubMed] [Google Scholar]
- 57.Ewan R, et al. Deubiquitinating enzymes AtUBP12 and AtUBP13 and their tobacco homologue NtUBP12 are negative regulators of plant immunity. New Phytol. 2011;191(1):92–106. doi: 10.1111/j.1469-8137.2011.03672.x. [DOI] [PubMed] [Google Scholar]