Significance
The bacteriophage T4 tail is a complex nanomachine that undergoes a succession of structural changes as it infects a bacterium. We analyzed cryo-electron microscopic images of T4 at different stages of infection. Three-dimensional visualization of key intermediates revealed unprecedented structural details, allowing a better understanding of this fundamental and highly efficient process. Contrary to common descriptions, most long tail fibers are folded back against the virion before infection, and not all interact with the cell before the short tail fibers irreversibly bind. Sheath contraction drives the tail tube only into the periplasm where, unexpectedly, the cytoplasmic membrane bulges outwards to fuse with the tail tube. Fusion does not require the proton motive force, which only becomes necessary for genome translocation.
Keywords: phage T4, cryo-ET, structure, infected cell, membrane curvature
Abstract
The first stages of productive bacteriophage infections of bacterial host cells require efficient adsorption to the cell surface followed by ejection of phage DNA into the host cytoplasm. To achieve this goal, a phage virion must undergo significant structural remodeling. For phage T4, the most obvious change is the contraction of its tail. Here, we use skinny E. coli minicells as a host, along with cryo-electron tomography and mutant phage virions, to visualize key structural intermediates during initiation of T4 infection. We show for the first time that most long tail fibers are folded back against the tail sheath until irreversible adsorption, a feature compatible with the virion randomly walking across the cell surface to find an optimal site for infection. Our data confirm that tail contraction is triggered by structural changes in the baseplate, as intermediates were found with remodeled baseplates and extended tails. After contraction, the tail tube penetrates the host cell periplasm, pausing while it degrades the peptidoglycan layer. Penetration into the host cytoplasm is accompanied by a dramatic local outward curvature of the cytoplasmic membrane as it fuses with the phage tail tip. The baseplate hub protein gp27 and/or the ejected tape measure protein gp29 likely form the transmembrane channel for viral DNA passage into the cell cytoplasm. Building on the wealth of prior biochemical and structural information, this work provides new molecular insights into the mechanistic pathway of T4 phage infection.
The Hershey and Chase experiment (1), which showed that most DNA of bacteriophage T2 entered an infected cell whereas most virion proteins remained outside, provided the final evidence necessary for acceptance of DNA being the genetic material. This experiment also set the stage for understanding the mechanism of phage infection. However, the apparent complexity of the infection process, the realization that some virion proteins do enter the infected cell (2), and a lack of suitable experimental approaches have heretofore prevented a detailed description.
The T4 virion comprises a capsid containing a 170-kb dsDNA genome, a collar region that displays short whiskers called whisker antigen control (Wac) (also known as fibritin), a contractile tail with a complex baseplate that harbors short tail fibers (STFs), and a set of side or long tail fibers (LTFs) (Fig. 1D) (3). After recognition of a host, the tail transmits a signal to the head for genome ejection and provides the channel through which the DNA moves.
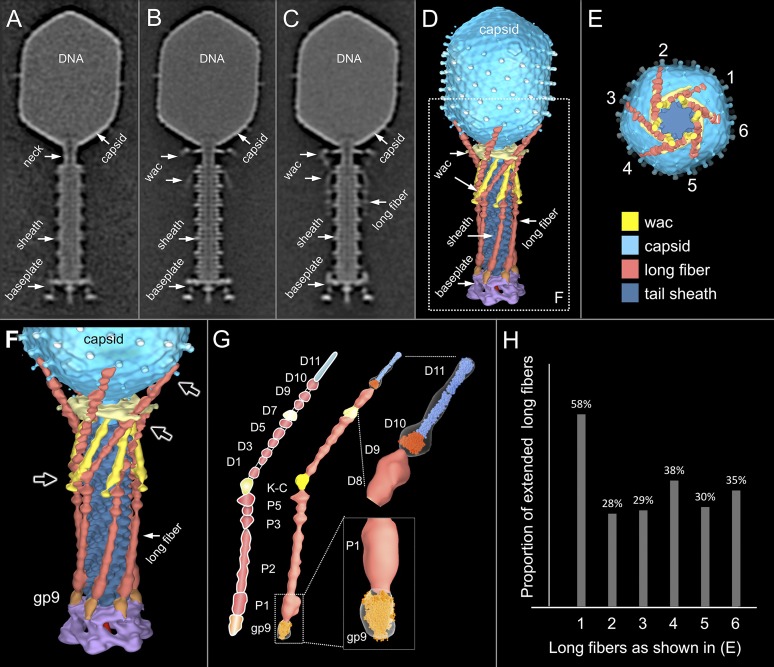
Fig. 1.
Comparative structural analysis of infective T4 virions. Asymmetric 3D reconstructions are shown as central slices of Wac-minus (A), fiberless (X4E mutant) (B), and WT (C) virions. In the absence of Wac, LTFs are seen only as short stubs emanating from the baseplate due to their flexibility when extended. A 3D reconstruction of WT T4 (D), enlarged in F, highlighting the six (orange) LTFs attached (open arrows) to the whiskers and collar of the sixfold symmetrical Wac (yellow) and the fivefold symmetrical capsid vertex. (G) Domain structure of an LTF (4) aligned with our cryo-ET reconstruction. For clarity, not all subdomains are named on the figure. Subdomains P1–P5 correspond to a trimer of gp34, K-C to gp35, and part of gp36, and subdomains D1–D11 in the distal half-fiber are comprised of trimeric gp36 and gp37. Crystal structures of gp9 (PDB ID code 1S2E) (18) and the gp37 fiber tip (PDB ID code 2XGF) (57) are fitted into the density map. Classification of each LTF reveals the symmetry mismatch viewed along the tail sheath axis, where fiber 1 is set to interact with a capsid edge (E). (H) Classification was also used to evaluate the density of each LTF and thus to estimate the distribution of extended fibers on a WT virion. Also, see Figs. S1–S3.
T4 LTF can be divided into proximal and distal half-fibers, angled about 20° (4, 5). The LTFs of mature virions are displayed in two conformations: “up” or retracted and “down” or extended states (6). Although it was long thought that extended fibers were required for rapid adsorption and that retracted fibers were found only under adverse conditions (e.g., low pH, low ionic strength, and low temperature) for infection, it was eventually concluded that individual fibers were in a dynamic equilibrium between retracted and extended states (7).
Twelve copies of Wac form the collar and whiskers, which are assembled just below the head–tail junction (8–13). wac is a nonessential gene; wac mutants form small plaques at normal efficiencies, but the burst is decreased because attachment of the LTF to the baseplate is inefficient (14). Wac binds to the “kneecap” (K-C) or hinge region joining the proximal and distal half-fibers and promotes proximal half-fiber attachment to the baseplate protein gp9 (3, 15). Consequently, the LTFs of wac mutant particles are extended under conditions where those of WT are retracted (6).
Six gp9 trimers bind to the upper edge of the baseplate; the trimeric C-terminal domain acts as a rigid body that swivels around the axis of its N-terminal domain. This arrangement allows the LTFs, which are coaxially attached to the C-terminal domain of gp9, to change their orientation relative to the remainder of the virion (16–19). Recent cryo-electron microscopic (cryo-EM) single particle reconstructions of extended tail and urea-treated, contracted tail T4 virions, coupled with X-ray crystallography of individual proteins, have provided a wealth of structural details (3).
The six kinked LTFs, extending away from the tail baseplate, contact Escherichia coli B lipopolysaccharide (LPS) (20) or K-12 OmpC (21) on the cell surface. Binding triggers a hexagon→star conformational transition of the baseplate, whose diameter increases from 40 to 60 nm (22, 23). This conformational change unpins the STFs, which rotate downward and bind tightly to the lipid A-KDO region of LPS (3, 24, 25). The change in baseplate conformation also triggers contraction of the tail sheath, which, in mature virions, is in a metastable conformation (26, 27). Contraction can be initiated in vitro by 2–3 M urea although the contracted sheath is resistant to 7M (28). It is commonly assumed that the tail tube directly penetrates the cytoplasm via a “syringe-like” mechanism (1, 29) although electron microscopy (EM) revealed that it reaches only the periplasmic face of the cytoplasmic membrane (22, 23). However, this pioneering study necessarily relied on the use of fixatives, thin sectioning, and stains, which can induce artifacts that complicate data interpretation.
Cell wall degradation is an important step during the initiation of T4 infection, in particular at low temperature (30). During virion assembly, the distal end of the tail tube interacts with the baseplate central hub complex, a membrane-piercing device consisting of trimers of gp27 and gp5, terminated by a monomeric gp5.4 needle (31). Gp5 is cleaved during phage assembly to yield the N-terminal gp5* (32), which contains lysozyme activity, and the triple-stranded β-helix gp5C (33). The overall complex is thus (gp27-gp5*-gp5C)3-gp5.4. It is thought that gp5C (and presumably gp5.4) dissociates as the complex penetrates the outer membrane (34), exposing the full lysozyme activity of gp5*. Gp5* also likely dissociates and diffuses away as the tail tube enters the periplasm because high multiplicities of infection lead to lysis-from-without, a phenomenon where excessive lysozyme activity lyses the cell before any phage development (35). It is not known whether gp27 dissociates from the tail tube together with gp5*.
An energized membrane is required for phage DNA ejection into the cell cytoplasm (36–38). It is not clear, however, what step requires energy. The membrane potential (∆pH was shown to be unimportant) (39) is required for the transient efflux of K+ ions from the cytoplasm (40) immediately after infection, suggesting that formation of a transmembrane channel requires energy. Efflux could also be associated with a fusion of the inner and outer membranes (41, 42), which would then create a channel across the entire cell envelope. Alternatively, an energized membrane could simply allow it to come closer to the tip of the T4 tail tube (43).
Cryo-electron tomography (cryo-ET) provides 3D structures of cells and subcellular complexes in a near-native and frozen-hydrated state at near nanometer resolution (44–46). Cryo-ET is being increasingly used to study phage–host interactions (47–52). We recently developed skinny E. coli minicell as a host to study phage infection in situ (53, 54). Despite their lack of a chromosome, minicells are metabolically active and can support phage infection (54–56). This study examines T4 at the initiation of infection, using high-throughput cryo-ET and subtomogram averaging. Together with mutant phage virions, we determine a series of high-resolution reconstructions of infecting T4 virions at successive stages of adsorption and genome penetration.
Results
Most Long Tail Fibers of Infective T4 Virions Are Folded Against the Tail Sheath.
An asymmetric reconstruction of a WT T4 virion was generated after cryo-ET and subtomogram averaging (Fig. 1 C and D). Considering the known symmetry mismatch between the capsid and the tail sheath, we did not apply any rotational symmetry in structure determination. Overall, our structure displays the same features as reported by single particle cryo-EM (16). However, it clearly shows that the LTF interacts extensively with the virion (Fig. 1 C and D). To better understand the LTF structure and its interactions with Wac and the baseplate, we also made asymmetric reconstructions of Wac (Fig. 1A) and fiberless (Fig. 1B) mutant virions by cryo-ET and subtomogram averaging. Pair-wise difference maps allowed density in the WT structure to be specifically assigned to Wac and the LTF and also identified where the LTF attaches to gp9 on the baseplate (Fig. 1 F and G and Fig. S1 D and E).
Fig. S1.
Long tail fibers are essential for tail contraction. Slices of tomograms of fiberless X4E virions in the presence of cells are shown in A and B. Contracted tails are not seen. The average map is shown in a central slice (C) and a surface rendering (D). A close-up view shows the hexagonal baseplate and gp9 (E).
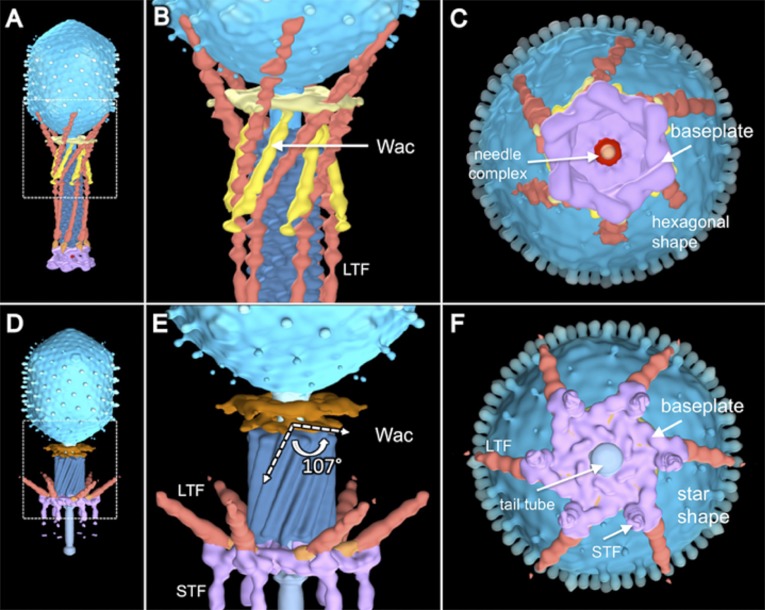
Each LTF consists of a 70-nm-long proximal trimeric gp34 half-fiber (Fig. 1G, P1–P5) attached to the baseplate gp9, a kneecap (K-C), comprised of gp35 and part of the trimeric gp36, and a 75-nm-long distal trimeric gp37 half-fiber (D1–D11) (4, 5, 57). Our asymmetric reconstruction of WT T4 shows that the proximal half-fibers are folded back and bent around the sheath, forming about one-quarter of a right-handed helix (Fig. 1 D and F). The K-C region interacts with the tip of the Wac whisker domain, the Wac collar with the D7 domain of each distal half-fiber, and the D10–D11 subdomains with the capsid. Binding of the LTF to the sheath should provide some stability against premature contraction. In the absence of Wac, the little defined fiber density in the averaged structure suggests that extended LTFs are mobile (Fig. 1A). The Wac collar and whiskers of fiberless virions have similar conformations as WT (Fig. 1 B and C and Fig. S1).
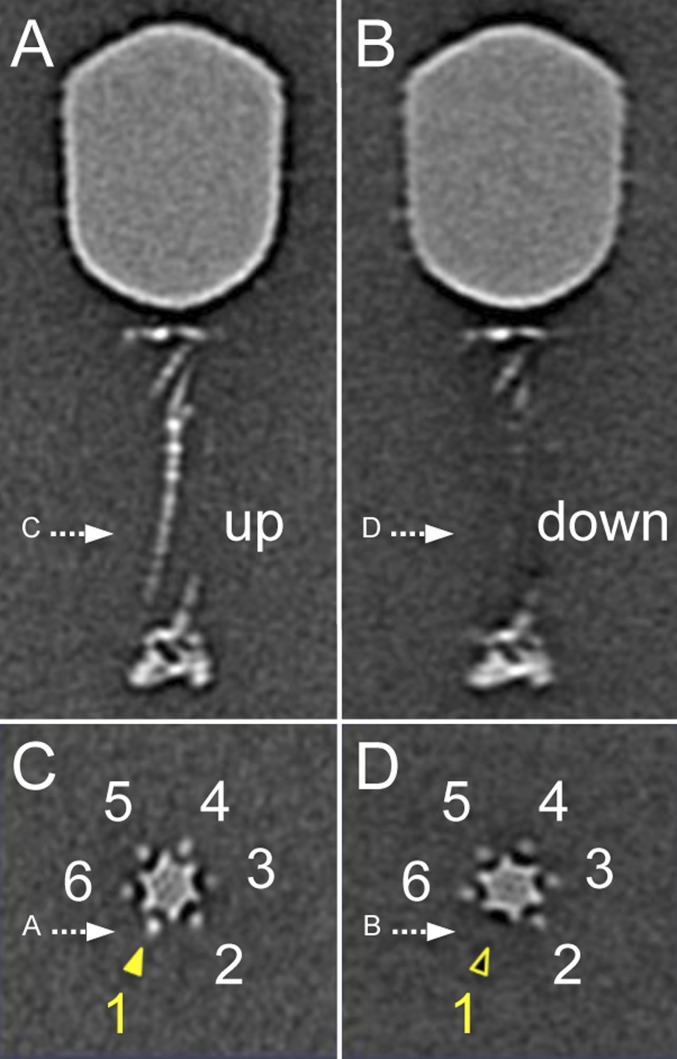
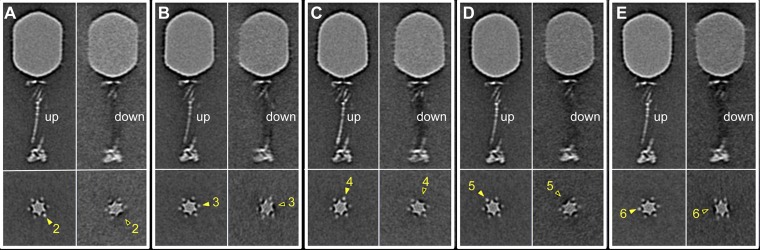
The six LTFs are symmetrically anchored around the baseplate whereas their tips bind the capsid. Consequently, a symmetry mismatch is unavoidable, and any interaction between the fiber tips and the capsid decoration proteins Hoc and Soc cannot apply to all fibers. The symmetry mismatch can be viewed along the tail sheath axis, where fiber 1 is set to interact with a capsid edge (Fig. 1E). Each capsid-bound LTF has a slightly different conformation than adjacent LTFs. Additional specific local classification for each LTF reveals two different conformations: retracted (“up”) and capsid-bound, and extended (“down”) (Figs. S2 and S3). Surprisingly, not all fibers are bound to the capsid at any given time. Most capsids had three or four LTFs bound, with the fiber bound to a capsid edge twice as likely to be extended as one bound to a capsomer face. Very few virions had all six LTFs either extended or bound to the capsid. The distribution of fibers on a mature WT virion is shown in Fig. 1H.
Fig. S2.
Six LTFs, numbered from 1 to 6, as shown in C. Multivariate statistical analysis and classification based on the density of LTF 1 show two distinctive classes. LTF 1 is in the “up” (retracted) position in class 1 (A and C) whereas it is in the “down” (extended) position and not visible in class 2 (B and D).
Fig. S3.
Characterization of LTFs 2–6 (A–E) also show two distinctive classes.
Initial Stages of T4 Infection Visualized in Situ by Cryo-ET.
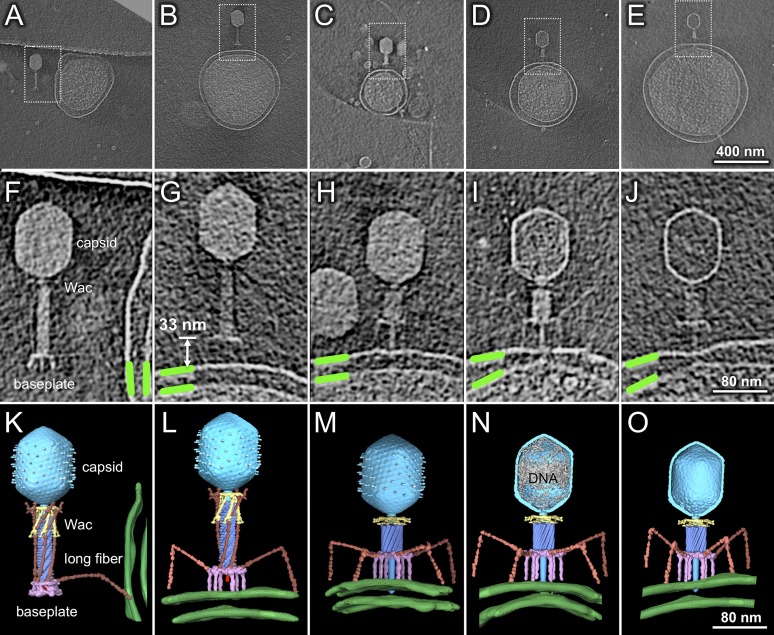
To capture representative phage structures during infection initiation, we prepared frozen-hydrated specimens at various times after infection (Table S1). Cryo-ET reveals five distinct conformations of the virions (Fig. 2), with their frequencies roughly correlating to infection time, strongly suggesting that they are normal intermediates. Time 0 is when phage was added to minicells. Within 30 s, some virions have contacted the outer membrane, often through only one LTF (Fig. 2 A, F, and K and Movie S1). The distance between the baseplate and cell surface is highly variable. After 1 min, although many virions had adsorbed and undergone sheath contraction, a few particles were captured at earlier stages. Some particles had moved closer to the cell with a hexagonal baseplate ∼10–20 nm above the outer membrane; these particles showed no STFs and most LTFs were still retracted, bound to the sheath, Wac, and capsid (Fig. S4 A–C). We also captured a few particles where the baseplate had undergone the hexagon→star transition (Fig. S4 E–G and compare Movie S2 with Movie S1) but where tail sheath contraction had not yet occurred. Notably, these particles had less than a full complement of both STFs and LTFs bound to the cell, the plane of the baseplate was not always parallel to that of the outer membrane, and the distance between them was variable. A rare structure shows the baseplate fixed ∼33 nm above and parallel to the membrane; all STFs, but not all LTFs, are bound to the cell, and the sheath is still extended (Fig. 2 B, G, and L). Together, these data provide direct visual confirmation of the generally accepted pathway (3, 27), that changes in the baseplate trigger STF release and that both occur before tail sheath contraction. However, it is significant that these same tomograms show one or two LTFs still bound to the tail sheath (Fig. 2 B, G, and L, Fig. S4 E–G, and Movie S2).
Table S1.
Data summary
| T4 virions | Condition | No. of tomograms used for 3D reconstructions | No. of subtomograms (contracted tail) | No. of subtomograms (extended tail) | Total no. of subtomograms |
| WT | Less than 1-min infection | 23 | 24 | — | — |
| 12* | None | — | — | ||
| 3-min infection | 76 | 246 | — | — | |
| 10-min infection | 99 | 419 | 1,958 | 5,366 | |
| 60-min infection | 146 | 945 | — | — | |
| 10-min infection after pretreatment with FCCP† | 96 | 1,774 | — | — | |
| Wac minus | Free virions | 22 | 367 | 267 | 634 |
| Fiberless (X4E)‡ | 3-min infection | 94 | None | 2,749 | 2,749 |
| gp9 minus‡ | Free virions | 104 | 1,998 | None | 1,998 |
| Total no. | N/A | 660 | 5,773 | 4,974 | 10,747 |
E. coli infected with T4 for different times were analyzed. Each tomogram, which typically contained one or more adsorbed T4 virions, was subdivided into subtomograms containing a single virion. Subtomograms were 3D-averaged to improve the resolution and to reduce the signal-to-noise ratio. —, not applicable.
K2 direct electron detector data.
Cells were treated with 30 μg/mL p-trifluoromethoxycarbonylcyanide phenylhydrazone (FCCP) for 3 min before infection at 37 °C.
X4E and the gene 9 mutant virions were frozen directly.
Fig. 2.
Distinct virion conformations during infection initiation. Pictured are 3D tomograms, shown as central slices, of individual virions after 30 s (A and F), 1 min (B and G), 3 min (C and H), 5 min (D and I), and 10 min (E and J) of infection. Boxed areas in A–E are enlarged in panels F–J and also rendered in 3D in K–O; the outer and inner membranes (green) were segmented manually. The baseplate (purple) changes conformation from hexagonal (A, F, and K) to star (B–E, G–J, and L–O), releasing the STF. The capsid is in cyan, tail sheath in blue, Wac in yellow, and LTF in orange. DNA remaining in the capsid in N is in gray.
Fig. S4.
Transient intermediates showing a star-shaped baseplate with extended tail. (A–C) Three virions close to the cell surface with a hexagonal baseplate 10–17 nm from the outer membrane. (E–G) Virions with the baseplate in a star configuration, an extended tail sheath, and released short tail fibers. The outer membrane (OM) is highlighted by the green line. Yellow and red arrows, respectively, indicate some STFs and LTFs. The schematic models highlight the structural changes in the baseplate (D and H). (Scale bar: 30 nm.) (F) One slice of the 3D tomogram shown in Movie S2.
The hexagon→star conformational change of the baseplate during infection was first described in detail in a seminal study (22, 23). Fig. S5C shows an end-on view of our asymmetric reconstruction of a mature WT T4 virion. The baseplate is in its metastable hexagonal configuration; the tip of the tail needle complex is clearly evident in its center. The diameter of the Wac collar is almost the same as the baseplate and is barely visible, but the distal tips of the LTFs, which are bound to the capsid, are resolved. In Fig. S5F, the baseplate has expanded into the star configuration, with an enlarged central hole. The six STFs are now visible, but the tip of the tail needle complex has disappeared and is replaced by gp27, which caps the end of the tail tube. The proximal half-fibers of the LTFs are still visible but are no longer bound to the capsid because of sheath contraction and the conformational change in Wac. Our observations are fully consistent with earlier, traditional EM data (22, 23).
Fig. S5.
Conformational change of Wac after tail sheath contraction. The 3D structure of a WT virion with an extended tail (A); a portion is enlarged in B. The hexagonal baseplate is shown in a bottom view (C). After sheath contraction, Wac undergoes a large conformational change: side view (D, enlarged in E). Wac whiskers have bent upwards about 107°. Immediately before sheath contraction, the baseplate undergoes a large conformational change from the hexagon (C) to the star configuration (F). See Initial Stages of T4 Infection Visualized in Situ by Cryo-ET for details.
At least one LTF must interact with the cell surface to promote baseplate reorganization and tail contraction because, in their absence, no contracted tails are seen (Fig. S1). Although less than a complete complement of cell-bound LTFs seems sufficient to trigger conformational changes in the baseplate, in our study, such structures could also be a consequence of infecting skinny minicells, where some LTFs must rotate significantly more than usual to contact the highly curved outer membrane of our skinny minicells. However, T4 has been shown to exhibit a definite preference for adsorbing to cell poles (58), where the membrane is also highly curved.
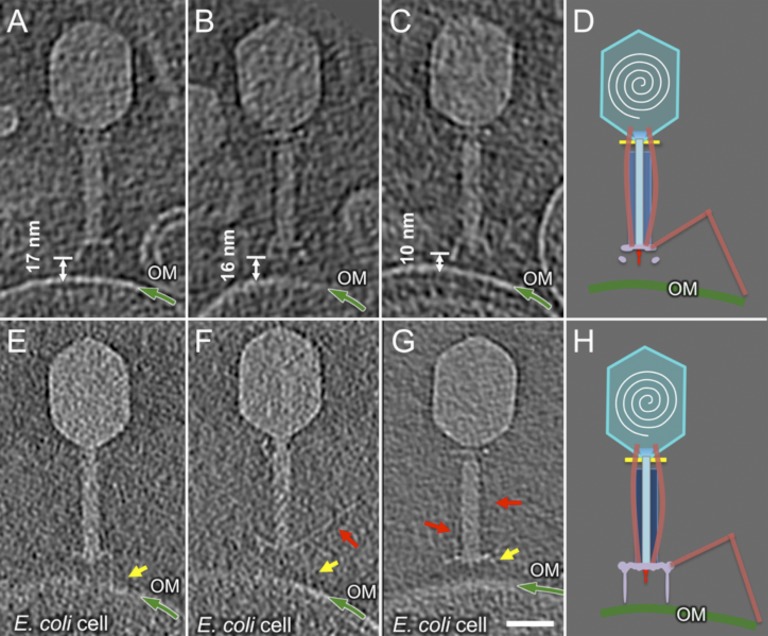
After 3 min of infection, sheath contraction has caused the tail tube to penetrate the outer membrane. A few tubes were captured before they completely crossed the periplasm; the DNA of these particles is still in the head (Fig. 2 C, H, and M) (described in The Cytoplasmic Membrane Bulges Outwards in Fusing with the Phage Tail Tip). However, by 5 min of infection, the majority of tail tubes seem fused to the cytoplasmic membrane, and some virions have at least partially ejected their DNA (Fig. 2 D, I, and N). Density in these tomograms is contiguous between the tail tube and the cytoplasm, and DNA is presumably passing through a transmembrane channel. There is a surprising and significant reorganization of the cytoplasmic membrane after tail tube insertion. The membrane bulges outwards, locally reducing the width of the periplasm, allowing the tail tube to fuse with the membrane and to make a channel for DNA ejection. After 10 min of infection, most infecting virions have completed DNA ejection (Fig. 2 E, J, and O), but the cytoplasmic membrane has not returned to its original conformation. The ejection machinery of the infecting T4 particle apparently does not spontaneously disassemble after the genome has entered the cell cytoplasm.
Conformational Changes in Wac After Sheath Contraction.
There are 12 copies of Wac per virion (11); 6 form the collar and 6 the whiskers, but all 12 interact with the LTF when the tail sheath is extended. Comparison of the structures shown in Fig. 1, Fig. 3, and Fig. S5 clearly reveals that, although the Wac collar does not change significantly in structure after tail contraction, the whiskers bend by ∼107°. These observations are consistent with an earlier study on Wac, where a steric clash between the whiskers domain and the contracted sheath was also revealed (11). The whiskers bend upwards, either directly pushed by the top of the contracting sheath or by the upward movement of the K-C region of those LTFs that are still retracted and are thus still bound to both the whiskers and the contracting sheath (Fig. S5). Either mechanism ultimately abrogates binding of the K-C region to the Wac whiskers, of the distal half-fiber D7 domain to the Wac collar, and of the tip domain with the capsid. Sheath contraction therefore ensures that all LTFs become extended, where they are free to contact the cell surface.
Fig. 3.
T4 structures after sheath contraction show different amounts of DNA remaining in the capsid and reorganization of the cytoplasmic membrane during T4 infection. The 3D structures are shown as central slices of T4 phage with full DNA (A), partial DNA (B), and no DNA inside the head (C). Their 3D maps are shown in G, H, and I, respectively. A portal-like structure (white arrow) is clearly visible in the DNA free head (C and I). The 3D classification reveals three distinct tail-membrane conformations. In the first class, the inner and outer membranes are still ∼30 nm apart, and the tail tube abuts the cell wall (D and J). In the second class, the inner membrane has moved closer to the tail tube (E and K). In the third class, the tail tube seems to be fused with the inner membrane (F and L). The outer (OM) and inner (IM) membranes are in green. The tip of the ejection machinery in D–F is highlighted by yellow arrows.
The Cytoplasmic Membrane Bulges Outwards in Fusing with the Phage Tail Tip.
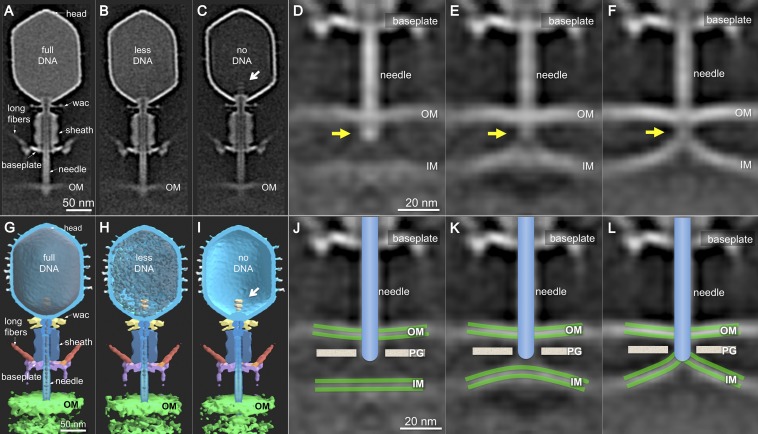
Almost all virions with contracted tails are oriented perpendicular to the outer membrane because of the tight, irreversible binding of the six STFs to the cell surface. Three distinct capsid structures are easily observed: full of DNA (Fig. 3 A and G), partially full (Fig. 3 B and H), and empty (Fig. 3 C and I), all key intermediates of infection. Partially filled capsids are commonly captured by cryo-ET, strongly suggesting that T4 genome translocation into the cell is slower than the estimated 10 kb/s (59, 60).
Interestingly, in some tomographic reconstructions, the cytoplasmic membrane bulges out of its plane. Multivariate statistical analysis revealed three distinct classes of tail–membrane conformations (Fig. 3 D–F). In the first, which corresponds to only ∼2% of phages with contracted tails, the inner and outer membranes are still the same distance found in uninfected cells (Fig. 3 D and J). The tail tube complex is abutting the cell wall, which is likely being degraded by gp5*. The second shows ∼24 nm of the tail tube inside the periplasm, not quite reaching the cytoplasmic membrane (Fig. 3 E and K). Thus, the tail tube does not directly penetrate the membrane, an observation first made using stained thin sections of T4-infected cells (22). The third shows the tail tube fused with a significantly remodeled inner membrane (Fig. 3 F and L), which now displays a large convex curvature (∼16 nm).
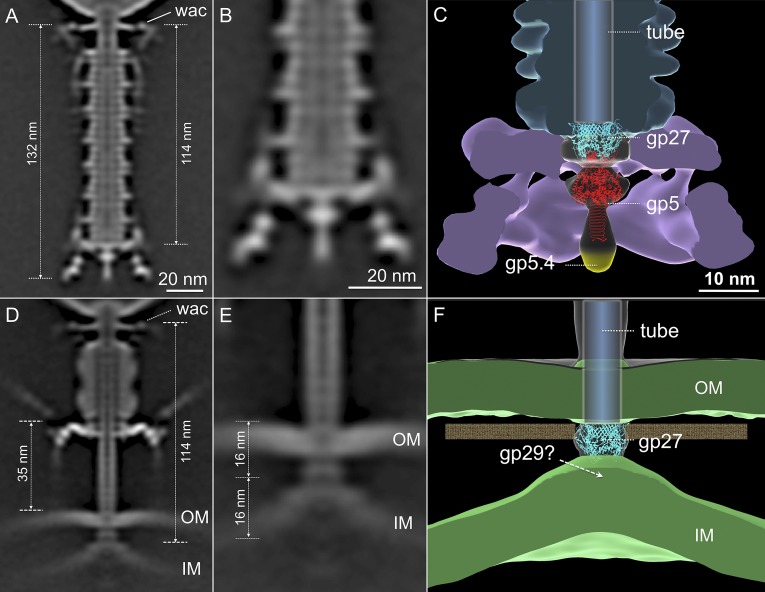
Because the virion is anchored to the cell surface through the STF and baseplate, sheath contraction pushes the tail tube and needle complex through the outer membrane. Measuring the distance between the Wac collar and the tip of the T4 ejection machinery in uncontracted and contracted states reveals that the latter have lost a tip structure of about 16 nm (Fig. 4). The length of the (gp27–gp5*–gp5C)3 complex is 19 nm, with the gp5C β–helix comprising the distal 11 nm (61). Adsorbed phages with contracted tails have therefore lost gp5C and gp5.4, and probably also gp5*, the dissociation of which would then allow free digestion of the peptidoglycan cell wall.
Fig. 4.
A transmembrane channel model in T4. The hub needle complex structure (PDB ID code 1K28) is placed into the map (C) that is derived from free T4 virions (A; enlarged in B). gp27, gp5, and the needle tip gp5.4 are colored cyan, red, and yellow, respectively. Before contraction (A), the tail is 132 nm in length. After contraction (D), the tail tube complex is about 114 nm, with the distal 16 nm inside the cell. The tail tube reaches the outer surface of the inner membrane (IM), which has undergone a striking conformational change, moving outwards about 16 nm (E). Accounting for the dissociation of gp5* and gp5C-gp5.4, we propose that gp27 abuts the bulging membrane (F), perhaps being held there through its interaction with the ejected tape measure protein gp29, which may be contributing to the electron density of the membrane.
Fitting the crystal structure of the gp5–gp27 complex (PDB ID code 1K28) into our cryo-ET maps (Fig. 4) is consistent with the dissociation of gp5* from the ejection machinery. However, it is less clear which T4 protein fuses with the cytoplasmic membrane to form the DNA translocation channel. The atomic structure of the trimeric gp27 fits well into the density observed in our cryo-ET reconstructions, and the trimer forms a hollow cylinder through which the genome could be ejected (61). However, gp29, the tape measure that determines tail length (62), must also be ejected from the tail tube in order for the T4 genome to enter the cell. Gp29 may simply be released into the periplasm before membrane fusion or, if gp27 forms a complete transmembrane channel, into the cytoplasm. Alternatively, gp29 may be contributing to the cryo-ET density of the bulged cytoplasmic membrane. Elaboration of this idea is presented in Discussion.
Deenergizing the Cytoplasmic Membrane Inhibits DNA Injection but Has No Impact on Membrane Reorganization.
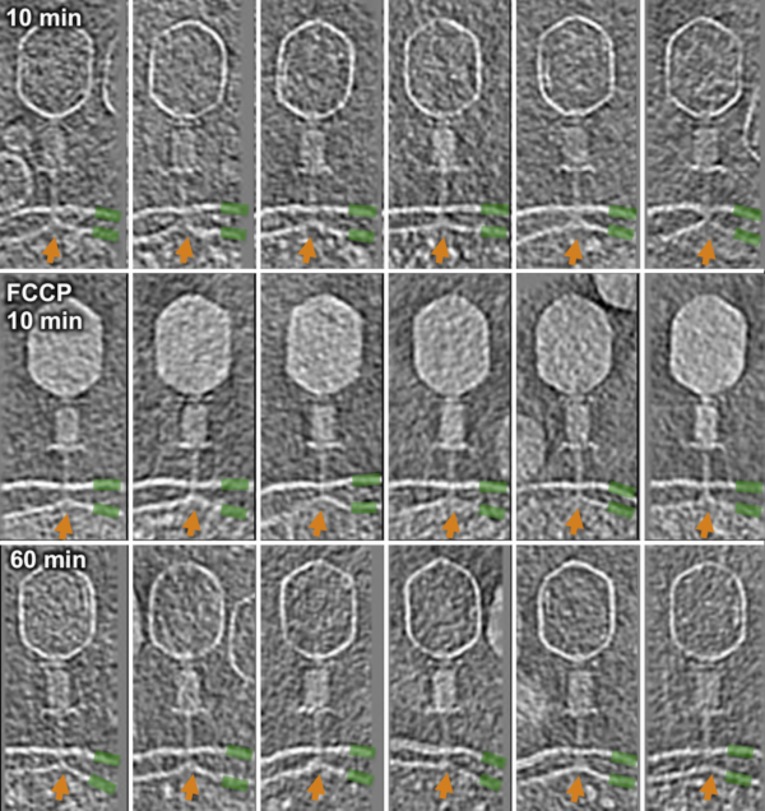
A membrane potential is necessary for translocation of DNA from the phage head into the cell cytoplasm (36–39). However, it is not known how inhibition is manifest and whether it has any impact on membrane reorganization. We therefore treated E. coli minicells at 37 °C with the ionophore p-trifluoromethoxycarbonylcyanide phenylhydrazone (FCCP) before infection. After 10 min, most adsorbed virions still contained all their DNA (Fig. S6). In contrast, in untreated cells, the majority of virions were empty. Importantly, even in the presence of FCCP, the cytoplasmic membrane still blebs out from its normal plane to fuse with the T4 tail tube. The membrane potential thus has no impact on tail sheath contraction or on reorganization of the inner membrane. Although the deenergized membrane clearly fuses with the T4 ejection nanomachine, because capsids remain full of DNA, no transmembrane channel can have been created. These observations are consistent with others’ data, made using different approaches, showing that the membrane potential is required to make an ion-permeable channel across the membrane (40), and thus it is also necessary for DNA translocation into the cytoplasm (36–39).
Fig. S6.
The proton motive force is required for genome ejection. After 10 min of a normal infection, DNA is ejected in the majority of T4 virions (Top). After 10-min infection in the presence of FCCP, DNA remains in the capsid (Middle). After 60 min of infection (no FCCP, but in the presence of rifampicin to inhibit phage gene expression), membrane curvature, highlighted by orange arrows, remains visible (Bottom). The outer and inner membranes are highlighted with green bars.
Discussion
The contractile tail of T4 is an extraordinary nanomachine that attaches to the host bacterium and undergoes major conformational changes in its baseplate. These changes allow the release of the STF, which bind irreversibly to the cell. The baseplate also transmits a signal to the head to initiate genome ejection. Recent cryo-EM and X-ray crystallography studies on T4 have provided a wealth of structural details of the capsid and its double-stranded DNA genome (for reviews, see refs. 3, 19, and 63). What is less well understood, and is the focus of this study, is the mechanism of T4 adsorption and penetration of the E. coli cell envelope. It is both remarkable and gratifying that the model of T4 infection that emerged from decades of research is so similar to what we now, for the first time, to our knowledge, observe directly in 3D. In particular, the structure of T4 particles contracted by urea seems identical to T4 irreversibly bound to a cell. It is similarly noteworthy that the classic EM study of T4 infection (22, 23), where stains, fixatives, and thin sectioning were necessary tools, also provided accurate 2D snapshots of the infection process. Nevertheless, high-throughput cryo-ET of skinny E. coli minicells, together with subtomogram averaging and correspondence analysis that provide high-resolution reconstructions of cell–virus complexes, reveals new paradigms of the initiation of myophage infection.
Long Tail Fibers of Rapidly Adsorbing Phage Are in Two Distinct Conformations.
Most schematics show T4 landing on the outer membrane of E. coli, much like a lunar module: All LTFs are extended, and the baseplate descends only after all LTFs have bound their receptor. However, our cryo-ET data of rapidly adsorbing WT T4 virions show that the six LTFs are in different orientations. In the “up” or retracted position, the proximal half-fiber is wrapped around and bound to the tail sheath, also interacting with the Wac fibritin protein and the capsid. On average, two or three fibers are in the extended (“down”) position in freshly purified phages. No chemical energy is expended in maintaining a dynamic equilibrium between extended and retracted conformations, and, unless external factors alter the equilibrium state, mature phage particles remain stable.
Reversible Adsorption.
The variable conformation of the T4 LTF is comparable with that observed for phage T7 (53), which was suggested to randomly “walk” over the cell surface to find an optimal site for irreversible adsorption and infection. A similar process may occur with T4; each cell contains about 105 copies of OmpC (64), but T4 exhibits a distinct preference for infection at cell poles (58), suggesting that it moves over the cell surface from its initial adsorption site before committing to infection. Maintaining most fibers in their retracted conformation allows faster particle diffusion in 3D space, and reversible adsorption and “walking” on a surface provides a reduction of dimensionality (65) into a 2D search for a preferred cellular receptor. This receptor may be at a pole or at an invagination of a future division site (58). Phage λ has also been shown to move over the cell surface to a cell pole (66). Sf6, which is endowed with tailspikes rather than LTFs, does not prefer to infect poles of WT cells; however, on cells lacking the outer membrane proteins OmpA and OmpC, to which the phage adsorbs more slowly, Sf6 has been shown to move over the cell surface (67). The tight binding of Sf6 to WT cells must prevent any subsequent movement. Movement over the cell surface process actually requires weak binding, a condition that is supported by in vitro measurements of T4 LTF and isolated E. coli B LPS (68). We are unaware of similar studies using the E. coli K-12 receptor OmpC.
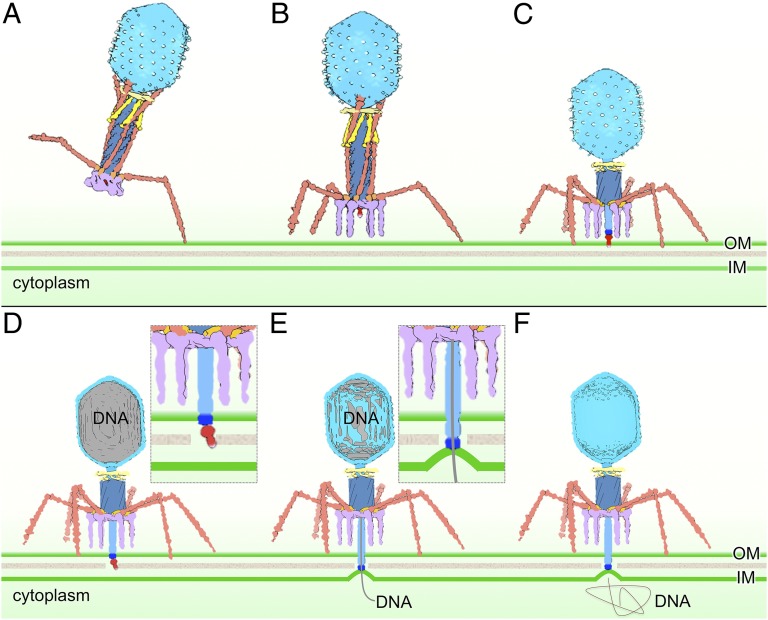
The first specific phage–cell complex during infection involves a single phage LTF bound to its receptor (Fig. 5A and Movie S1). Binding temporarily prevents both diffusion of the phage and the return of that fiber to the retracted state. A second LTF may then extend and contact the cell before the first has dissociated. Repetition of this process using different, transiently extended LTF can then allow the phage to walk randomly over the cell surface without committing to infection. Virions with two adsorbed LTFs are shown in Movies S3 and S4. During this stage, the baseplate should remain in its hexagonal configuration. Recognition of a preferred adsorption site disfavors further movement, and any differential movement of the phage particle relative to the bacterium—by convection, Brownian motion, or even cell motility—can strain the connection between the proximal half-fiber gp34 and gp9 in the baseplate beyond what occurs in free particles. The more fibers that are bound, the greater the strain that can be imposed on the baseplate, potentially destabilizing its hexagonal conformation.
Fig. 5.
A schematic model of T4 infection initiation. When the phage particle is free in solution, an extended LTF (brown) does not have a fixed orientation. A subset, probably one to three, of the LTFs binds to host receptors (A). Strain in LTF–gp9 baseplate junctions can trigger conformational changes in the baseplate (purple) that release a subset of STF (B). The baseplate of this transient intermediate rapidly transitions into its star configuration, triggering contraction of the tail and a conformational change in the Wac (yellow) collar, which releases the remaining LTF (C). Tail contraction pushes the needle through the outer membrane (OM), after which gp5* and gp5C3-gp5.4 (red) dissociate (D). Gp27 (dark blue), now the distal end of the ejection machinery, contacts the inner membrane (IM), which is bulged out from its normal plane. Bulging may be caused by the tape measure protein gp29 that must exit the tail tube before DNA can leave the capsid (E). Phage DNA is fully released into the cytoplasm (F). See Discussion for a more detailed description.
Irreversible Adsorption.
If all six LTFs are bound, the phage baseplate will naturally be oriented parallel to the cell surface, and, after baseplate expansion, all six STFs will rotate down from the baseplate plane and bind irreversibly to their receptor. However, such a coordinated process cannot be obligatory. Our cryo-ET data reveal infecting particles with only a subset of both LTFs and STFs bound to a cell and with the baseplate not yet parallel to the cell surface. Only a few particles have been captured at this stage, which thus presumably reflects a transient intermediate. A subset of weakly bound LTFs will not necessarily orient the baseplate parallel to the cell, and we suggest that the orientation is fixed by the irreversible binding of the STFs. When two or three adjacent LTFs bind to a cell, they may locally destabilize the baseplate enough to unpin a subset of the STFs (Fig. 5B). Tight binding by only a single STF to its receptor should prohibit any further movement of the entire particle. Unpinning of an STF will further destabilize the baseplate; it cannot return to a hexagonal conformation and will rapidly complete its transition to star, releasing additional STFs that, when they bind a receptor, will then orient the phage particle ready for DNA ejection.
Baseplate Expansion and Tail Sheath Contraction.
Our cryo-ET data show that all six STFs bind to the cell whereas some LTFs remain retracted against the sheath, with their tips still on the capsid, and that the baseplate transitions from hexagon→star before sheath contraction. Baseplate expansion creates a central hole through which the hub needle complex and tail tube can pass. The few virions we observed where all STFs have bound but the sheath is still extended suggest that movement of the baseplate hub complex is rapidly followed by sheath contraction. Contraction is likely delayed because those LTFs remaining retracted are bound through their proximal fiber domains to the sheath, which should stabilize the extended conformation. Interactions of the K-C and D7 domains of the distal half-fiber with Wac should provide additional stability, and binding of the fiber tips to the capsid should also resist the rotation associated with a contracting sheath (3, 28).
Sheath contraction causes the Wac whiskers to change conformation (Fig. 5C) and releases the remaining LTF. Because the phage is firmly anchored to the cell, sheath contraction also pushes the tail tube complex through the outer membrane, where gp(5*-5C)3-gp5.4 dissociate, likely as free gp5* and gp5C3-gp5.4 (Fig. 5D). Gp5*-catalyzed degradation of the cell wall facilitates penetration by the tail tube, which is now terminated by the gp27 trimer.
The major difference between this scheme and those previously described in more detail (3, 19, 63) is that, rather than six receptor-bound LTFs triggering a single coordinated event that includes baseplate expansion, release of all STFs, and their binding to the cell, our data (Fig. 2 B, G, and L, Fig. S4 E and F, and Movie S2) are more consistent with a series of reactions that are initiated after only a subset of the LTFs has bound at a preferred site of adsorption. Binding locally destabilizes the baseplate and allows release of only a subset of the STFs, which bind irreversibly to the cell. The baseplate then continues its transition to the star configuration, allowing the tail tube to begin penetration of the outer membrane, and finally allowing sheath contraction and release of the remaining LTFs.
This scheme is consistent with observations that only three LTFs are necessary for infection (69, 70) although binding of three adjacent LTFs is more likely to locally destabilize the baseplate and release a subset of the STFs than three symmetrically oriented LTFs. The reduced emphasis on the role of receptor-bound LTFs in triggering irreversible adsorption and baseplate expansion not only is directly suggested by our cryo-ET data but also better explains the observation that mutant particles lacking the peripheral baseplate protein gp9 are infective, even on E. coli B/4, a strain normally resistant to infection by T4 (71). Virions that lack gp9 are obligatorily fiberless because gp9 connects the LTF to the baseplate. Although the lack of LTF normally prevents adsorption, the absence of gp9 destabilizes the baseplate; less of a trigger is then needed to initiate the hexagon to star transition and to allow STFs to be released for irreversible adsorption. A comparable destabilization can explain expanded host range mutants of T4 that have mutations affecting various baseplate components (60). In contrast, although mutant particles lacking STF still adsorb to cells through their LTFs, after baseplate expansion and sheath contraction, they are released from the cell as DNA-filled noninfective particles (72).
Membrane Reorganization, Membrane Potential, and Genome Ejection.
The classic model for phage T4 DNA ejection is via a syringe-like mechanism, in which the phage genome is directly injected into the host cytoplasm after contraction of the phage tail and an “uncorking” process, being driven by the release of pressure within the phage head (1, 29). However, internal pressures cannot transfer a complete phage genome into the cell, and even the importance of such pressures during infection is in question (73, 74). Furthermore, despite contraction of the tail sheath, the tail tube does not directly penetrate the cytoplasmic membrane. Ejection into the periplasm is highly improbable in primary T4 infections because the DNA would then be susceptible to the same degradation that is suffered by superinfecting phage genomes.
Our cryo-ET data clearly show that the cytoplasmic membrane bulges from its normal plane by ∼16 nm to fuse with the ejection nanomachine (Fig. 5E) and that bulging does not require an energized membrane. In contrast, opening a channel across the membrane (Fig. 5 E and F) to allow DNA transport does require a membrane potential (40).
Our asymmetric reconstructions of the fusion complex are consistent with the intact gp27 trimer being sandwiched between the tail tube and the cytoplasmic membrane, but structural predictions do not reveal any propensity of gp27 to bind the likely inner membrane receptor, phosphatidylglycerol (75, 76). However, the gp29 tail tape measure protein has a strongly predicted transmembrane segment near the middle of its amino acid sequence. Gp29 has an essential role in assembling the central hub of the baseplate (77), and thus it interacts with gp27; gp29 also anchors the sheath to the baseplate (62). Gp29 extends the length of the tail tube, possibly interacting with gp3, the tail tube terminator (78), and it is therefore in position to transmit a signal, emanating from conformational changes in the baseplate, into the head that initiates genome release. Gp29 must leave the tail tube to allow DNA ejection, and the conformational changes associated with baseplate expansion likely also result in the ejection of gp29. We suggest that at least part of the electron density associated with the bulged membrane during genome ejection into the cytoplasm is due to gp29 and that this protein forms the transmembrane channel.
This idea lends itself to a speculative explanation for the membrane curvature. Gp29 interacts with the hub protein gp27 during initiation of baseplate assembly, and gp27 lies at the tip of the tail tube after it penetrates the periplasm. Assuming the interaction is maintained, gp29 would remain anchored by gp27 as it exits the tail tube in the extended conformation of a tape measure. Insertion of the transmembrane segment of gp29 into the cytoplasmic membrane, perhaps followed by refolding of the remainder of the protein, could then pull the membrane out of its normal plane toward the tip of the tail tube.
What remains completely unresolved from this work is how the channel across the cytoplasmic membrane is sealed after genome translocation, a process that must occur to prevent complete dissipation of the proton motive force. Even long after infection, the cytoplasmic membrane remains bulged out, fused with the T4 ejection apparatus. T4 ghosts, particles with ruptured heads that have lost their genome and all internal proteins (79), kill cells because they cannot seal the transmembrane channel. This result suggests that a T4 protein normally makes the seal. The early imm gene product, the immunity protein, has been proposed to bind to sites of DNA translocation on the cytoplasmic membrane (80) and theoretically could provide one mechanism. But imm is nonessential, necessitating that an alternative process be available. Furthermore, T4 protein synthesis is not even required for sealing (81). One possibility is the gene 2 protein, which is bound to the genome ends (and thus not present in ghosts), and which prevents degradation by the RecBCD nuclease after DNA ejection (82). Perhaps the trailing copy of gp2 can seal the channel as the last of the genome enters the cell cytoplasm.
T4 Infection Initiation and Other Contractile Systems.
The combination of high-throughput cryo-ET, 3D subtomogram analysis, and our use of skinny minicells has allowed visualization of intermediate structures at high resolution during phage T4 infection. T4 is a member of the Myoviridae, and many features of the T4 infection process are likely to pertain to other contractile-tailed phages. However, other phages may exhibit a simpler infection scheme. Only the T-even family are known to possess a complex baseplate that harbors a second set of fibers (STFs) used for adsorption (3, 19, 63).
Interestingly, T4 shares many core structural components with many bacterial contractile systems, including Pseudomonas aeruginosa type VI secretion (T6SS) and R-type pyocins (3, 19, 63, 83). Whereas contraction of the T4 sheath pushes the tail tube complex through the outer membrane, and into the inner membrane to create a channel for both DNA and proteins, sheath contraction of the T6SS launches the tube out of the host and then through the plasma membrane of a target eukaryotic cell or the entire cell envelope of a target bacterium to deliver virulence factor proteins or toxins. Cell killing by R-type pyocins is more directly comparable with phage infection, except that their channel into the cell cytoplasm allows transport only of protons or other cations. Nevertheless, our cryo-ET study of T4 infection initiation likely has broad implications in understanding the mechanisms of bacterial contractile systems.
Materials and Methods
Phage and Bacteria.
Phage T4 and the amber mutants 9amE17, 12amN69, X4E (34amB25, 34amA455, 35amB252, 37amN52, 37amB280, 38amB262), and wac-amE727J were propagated on E. coli B40 argI40 supD at 37 °C. Defective virions were prepared by infecting E. coli B at a multiplicity of infection of 4–5; after 9 min, cells were superinfected at the same multiplicity of infection. Lysates were clarified by low-speed centrifugation, and phages were then concentrated by centrifugation at 50,000 × g for 30 min in a Beckman SW28 rotor. The pellet was resuspended in 10 mM Tris, pH 7.6, 10 mM MgCl2, 0.1 M NaCl, and 0.02% gelatin and clarified again by brief centrifugation, and the phages were stored at 4 °C before use.
E. coli K12 WM4011 [BW25113 ∆yhdE::cat mreB-A125V Δmin(CDE)::kan fadR13::Tn10 (pBAD30-flhDC)] has previously been described (53). It produces motile skinny (∼0.3-μm diameter) minicells at high frequency. Bacteria were grown at 37 °C in tryptone broth with 0.2% l-arabinose to late log phase and centrifuged at 10,000 × g for 5 min to remove most large cells, and the supernatant was centrifuged at 41,000 × g for 15 min to harvest minicells. Minicells were resuspended at ∼108/mL in tryptone broth; as judged by light microscopy, most were motile.
Infection, Cryo-ET Data Collection, and 3D Reconstructions.
Minicells were infected at 37 °C at a multiplicity of 5–10. Infections longer than 10 min were initiated after treating cells with rifampicin (200 µg/mL) to prevent phage gene expression. At various times, samples were taken to prepare frozen-hydrated EM specimens on holey carbon grids. To collapse the membrane potential, cells were treated with 30 μg/mL p-trifluoromethoxycarbonylcyanide phenylhydrazone (FCCP; Sigma) for 3 min before infection and then infected with phage for 10 min at 37 °C.
Infected cell cultures were mixed with 15-nm colloidal gold (as fiducial markers) and then deposited onto freshly glow-discharged holey carbon grids. Grids were blotted briefly with filter paper and then rapidly frozen in liquid ethane. Frozen-hydrated specimens were imaged at −170 °C using a Polara G2 electron microscope (FEI Company) equipped with a field emission gun and a 16-megapixel CCD camera (TVIPS; GMBH) or a K2 direct electron detector (Gatan). The microscope was operated at 300 kV with a magnification of 23,000×, resulting in an effective pixel size of 7.8 Å after 2 × 2 binning. Using the FEI “batch tomography” program, low-dose, single-axis tilt series were collected from each minicell at −4 to −6 µm defocus with a cumulative dose of ∼100 e/Å2 distributed over 87 images and covering an angular range of −64° to +64°, using increments of 1.5°. To better visualize early infection intermediates (less than 1 min), we also used SerialEM (84) and dose fractionation mode to collect 12 tilt series on a direct electron detector (Gatan). Tilted images were automatically aligned and reconstructed using a combination of the IMOD (85) and RAPTOR (86) packages. In total, 672 reconstructions were generated and used for further processing.
Subtomogram Averaging and Correspondence Analysis.
Conventional imaging analysis, including 4 × 4 × 4 binning, contrast inversion, and low-pass filtering enhanced the contrast of binned tomograms (87). Phage particles (10,748) were manually selected from 660 reconstructions and were separated into two groups: phages with extended tails and phages with contracted tails. The orientation of each particle was initially estimated from the head and tail coordinates, thereby providing two of the three Euler angles. To accelerate image analysis, 4 × 4 × 4 binned subtomograms (100 × 100 × 100 voxels) and 2 × 2 × 2 binned subtomograms (200 × 200 × 200 voxels) were generated. A global average of all of the extracted 4 × 4 × 4 binned subtomograms was performed after application of the two Euler angles previously determined. After a translational alignment based on the global average, multivariate statistical analysis and hierarchical ascendant classification were used to analyze the structures of the tail tube and fibers as described (53). Specifically, we used a local mask around each retracted LTF for multivariate statistical analysis. For each LTF, two distinct conformations (retracted and extended) were found (Figs. S2 and S3). Based on this information, we calculated the distribution of an extended conformation for each LTF. Class averages were computed by averaging Fourier coefficients so missing regions were taken into account explicitly (88, 89). Fourier shell correlation between the two independent reconstructions was used to estimate the resolution of the averaged structures (Table S2). EM maps have been deposited in the EM Data Bank with accession numbers EMD 2774 and 6078–6083.
Table S2.
Resolution, as estimated by Fourier shell correlation
| WT | Wac-minus virions | Fiberless (X4E) virions | Gp12-minus (contracted tail) virions | gp9-minus virions | WT after DNA ejection | WT after sheath contraction, before DNA ejection | |
| EMDB code | 2774 | 6078 | 6079 | 6080 | 6081 | 6083 | 6082 |
| Resolution, Å | 36 | 38 | 31 | 40 | 31 | 33 | 31 |
| Subtomograms | 1,958 | 267 | 2,749 | 1,985 | 1,998 | 1,275 | 1,904 |
Three-Dimensional Visualization.
We used IMOD (85) to take snapshots of 2D slices from 3D tomograms. We used UCSF Chimera (90) for the surface rendering of 3D averaged structures. We used Amira (Visage Imaging) for surface rendering of 3D reconstructions of E. coli minicells infected by phages.
Supplementary Material
Acknowledgments
We thank Shuji Kanamaru and Fumio Arisaka, who kindly provided T4 mutants. We thank Petr Leiman for stimulating discussions and the anonymous reviewers for their constructive comments and suggestions. This work was supported by National Institute of General Medical Sciences (NIGMS) Grant GM110243 (to J.L. and I.J.M.) and Welch Foundation Grant AU-1714 (to J.L.). W.M. was supported by NIGMS Grant GM61074. The direct electron detector was funded by NIH Award S10OD016279.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: EM maps have been deposited in the EM Data Bank (accession nos. 2774 and 6078–6083).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1501064112/-/DCSupplemental.
References
- 1.Hershey AD, Chase M. Independent functions of viral protein and nucleic acid in growth of bacteriophage. J Gen Physiol. 1952;36(1):39–56. doi: 10.1085/jgp.36.1.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hershey AD. An upper limit to the protein content of the germinal substance of bacteriophage T2. Virology. 1955;1(1):108–127. doi: 10.1016/0042-6822(55)90009-x. [DOI] [PubMed] [Google Scholar]
- 3.Leiman PG, et al. Morphogenesis of the T4 tail and tail fibers. Virol J. 2010;7:355. doi: 10.1186/1743-422X-7-355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cerritelli ME, Wall JS, Simon MN, Conway JF, Steven AC. Stoichiometry and domainal organization of the long tail-fiber of bacteriophage T4: A hinged viral adhesin. J Mol Biol. 1996;260(5):767–780. doi: 10.1006/jmbi.1996.0436. [DOI] [PubMed] [Google Scholar]
- 5.Ward S, et al. Assembly of bacteriophage T4 tail fibers. II. Isolation and characterization of tail fiber precursors. J Mol Biol. 1970;54(1):15–31. doi: 10.1016/0022-2836(70)90443-2. [DOI] [PubMed] [Google Scholar]
- 6.Conley MP, Wood WB. Bacteriophage T4 whiskers: A rudimentary environment-sensing device. Proc Natl Acad Sci USA. 1975;72(9):3701–3705. doi: 10.1073/pnas.72.9.3701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kellenberger E, Stauffer E, Häner M, Lustig A, Karamata D. Mechanism of the long tail-fiber deployment of bacteriophages T-even and its role in adsorption, infection and sedimentation. Biophys Chem. 1996;59(1-2):41–59. doi: 10.1016/0301-4622(95)00117-4. [DOI] [PubMed] [Google Scholar]
- 8.Coombs DH, Eiserling FA. Studies on the structure, protein composition and aseembly of the neck of bacteriophage T4. J Mol Biol. 1977;116(3):375–405. doi: 10.1016/0022-2836(77)90076-6. [DOI] [PubMed] [Google Scholar]
- 9.Letarov A, Manival X, Desplats C, Krisch HM. gpwac of the T4-type bacteriophages: Structure, function, and evolution of a segmented coiled-coil protein that controls viral infectivity. J Bacteriol. 2005;187(3):1055–1066. doi: 10.1128/JB.187.3.1055-1066.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Efimov VP, et al. Fibritin encoded by bacteriophage T4 gene wac has a parallel triple-stranded alpha-helical coiled-coil structure. J Mol Biol. 1994;242(4):470–486. doi: 10.1006/jmbi.1994.1595. [DOI] [PubMed] [Google Scholar]
- 11.Fokine A, et al. The molecular architecture of the bacteriophage T4 neck. J Mol Biol. 2013;425(10):1731–1744. doi: 10.1016/j.jmb.2013.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yanagida M, Ahmad-Zadeh C. Determination of gene product positions in bacteriophage T4 by specific antibody association. J Mol Biol. 1970;51(2):411–421. doi: 10.1016/0022-2836(70)90151-8. [DOI] [PubMed] [Google Scholar]
- 13.Dickson RC, Barnes SL, Eiserling FA. Structural proteins of bacteriophage T4. J Mol Biol. 1970;53(3):461–474. doi: 10.1016/0022-2836(70)90077-x. [DOI] [PubMed] [Google Scholar]
- 14.Wood WB, Conley MP. Attachment of tail fibers in bacteriophage T4 assembly: Role of the phage whiskers. J Mol Biol. 1979;127(1):15–29. doi: 10.1016/0022-2836(79)90455-8. [DOI] [PubMed] [Google Scholar]
- 15.Terzaghi BE, Terzaghi E, Coombs D. The role of the collar/whisker complex in bacteriophage T4D tail fiber attachment. J Mol Biol. 1979;127(1):1–14. doi: 10.1016/0022-2836(79)90454-6. [DOI] [PubMed] [Google Scholar]
- 16.Kostyuchenko VA, et al. The tail structure of bacteriophage T4 and its mechanism of contraction. Nat Struct Mol Biol. 2005;12(9):810–813. doi: 10.1038/nsmb975. [DOI] [PubMed] [Google Scholar]
- 17.Kostyuchenko VA, et al. Three-dimensional structure of bacteriophage T4 baseplate. Nat Struct Biol. 2003;10(9):688–693. doi: 10.1038/nsb970. [DOI] [PubMed] [Google Scholar]
- 18.Kostyuchenko VA, et al. The structure of bacteriophage T4 gene product 9: The trigger for tail contraction. Structure. 1999;7(10):1213–1222. doi: 10.1016/s0969-2126(00)80055-6. [DOI] [PubMed] [Google Scholar]
- 19.Leiman PG, Chipman PR, Kostyuchenko VA, Mesyanzhinov VV, Rossmann MG. Three-dimensional rearrangement of proteins in the tail of bacteriophage T4 on infection of its host. Cell. 2004;118(4):419–429. doi: 10.1016/j.cell.2004.07.022. [DOI] [PubMed] [Google Scholar]
- 20.Prehm P, Jann B, Jann K, Schmidt G, Stirm S. On a bacteriophage T3 and T4 receptor region within the cell wall lipopolysaccharide of Escherichia coli B. J Mol Biol. 1976;101(2):277–281. doi: 10.1016/0022-2836(76)90377-6. [DOI] [PubMed] [Google Scholar]
- 21.Montag D, Hashemolhosseini S, Henning U. Receptor-recognizing proteins of T-even type bacteriophages: The receptor-recognizing area of proteins 37 of phages T4 TuIa and TuIb. J Mol Biol. 1990;216(2):327–334. doi: 10.1016/S0022-2836(05)80324-9. [DOI] [PubMed] [Google Scholar]
- 22.Simon LD, Anderson TF. The infection of Escherichia coli by T2 and T4 bacteriophages as seen in the electron microscope. I. Attachment and penetration. Virology. 1967;32(2):279–297. doi: 10.1016/0042-6822(67)90277-2. [DOI] [PubMed] [Google Scholar]
- 23.Simon LD, Anderson TF. The infection of Escherichia coli by T2 and T4 bacteriophages as seen in the electron microscope. II. Structure and function of the baseplate. Virology. 1967;32(2):298–305. doi: 10.1016/0042-6822(67)90278-4. [DOI] [PubMed] [Google Scholar]
- 24.Riede I. Receptor specificity of the short tail fibres (gp12) of T-even type Escherichia coli phages. Mol Gen Genet. 1987;206(1):110–115. doi: 10.1007/BF00326544. [DOI] [PubMed] [Google Scholar]
- 25.Heller K, Ölschläger T, Schwartz H. Infection of LPS mutants of Escherichia coli B by phage T6. FEMS Microbiol Lett. 1983;17(1-3):1–6. [Google Scholar]
- 26.Amos LA, Klug A. Three-dimensional image reconstructions of the contractile tail of T4 bacteriophage. J Mol Biol. 1975;99(1):51–64. doi: 10.1016/s0022-2836(75)80158-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Coombs DH, Arisaka F. T4 tail structure and function. In: Karam JD, editor. Molecular Biology of Bacteriophage T4. American Society for Microbiology; Washington, DC: 1994. pp. 259–281. [Google Scholar]
- 28.Aksyuk AA, et al. The tail sheath structure of bacteriophage T4: A molecular machine for infecting bacteria. EMBO J. 2009;28(7):821–829. doi: 10.1038/emboj.2009.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Stent GS. Molecular Biology of Bacterial Viruses. Freeman; San Francisco: 1963. [Google Scholar]
- 30.Kanamaru S, Ishiwata Y, Suzuki T, Rossmann MG, Arisaka F. Control of bacteriophage T4 tail lysozyme activity during the infection process. J Mol Biol. 2005;346(4):1013–1020. doi: 10.1016/j.jmb.2004.12.042. [DOI] [PubMed] [Google Scholar]
- 31.Shneider MM, et al. PAAR-repeat proteins sharpen and diversify the type VI secretion system spike. Nature. 2013;500(7462):350–353. doi: 10.1038/nature12453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mosig G, Lin GW, Franklin J, Fan WH. Functional relationships and structural determinants of two bacteriophage T4 lysozymes: A soluble (gene e) and a baseplate-associated (gene 5) protein. New Biol. 1989;1(2):171–179. [PubMed] [Google Scholar]
- 33.Kanamaru S, Gassner NC, Ye N, Takeda S, Arisaka F. The C-terminal fragment of the precursor tail lysozyme of bacteriophage T4 stays as a structural component of the baseplate after cleavage. J Bacteriol. 1999;181(9):2739–2744. doi: 10.1128/jb.181.9.2739-2744.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nishima W, Kanamaru S, Arisaka F, Kitao A. Screw motion regulates multiple functions of T4 phage protein gene product 5 during cell puncturing. J Am Chem Soc. 2011;133(34):13571–13576. doi: 10.1021/ja204451g. [DOI] [PubMed] [Google Scholar]
- 35.Kao SH, McClain WH. Roles of bacteriophage T4 gene 5 and gene s products in cell lysis. J Virol. 1980;34(1):104–107. doi: 10.1128/jvi.34.1.104-107.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Labedan B, Goldberg EB. Requirement for membrane potential in injection of phage T4 DNA. Proc Natl Acad Sci USA. 1979;76(9):4669–4673. doi: 10.1073/pnas.76.9.4669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Furukawa H, Yamada H, Mizushima S. Interaction of bacteriophage T4 with reconstituted cell envelopes of Escherichia coli K-12. J Bacteriol. 1979;140(3):1071–1080. doi: 10.1128/jb.140.3.1071-1080.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kalasauskaite EV, Kadisaite DL, Daugelavicius RJ, Grinius LL, Jasaitis AA. Studies on energy supply for genetic processes: Requirement for membrane potential in Escherichia coli infection by phage T4. Eur J Biochem. 1983;130(1):123–130. [PubMed] [Google Scholar]
- 39.Labedan B, Heller KB, Jasaitis AA, Wilson TH, Goldberg EB. A membrane potential threshold for phage T4 DNA injection. Biochem Biophys Res Commun. 1980;93(2):625–630. doi: 10.1016/0006-291x(80)91124-9. [DOI] [PubMed] [Google Scholar]
- 40.Boulanger P, Letellier L. Characterization of ion channels involved in the penetration of phage T4 DNA into Escherichia coli cells. J Biol Chem. 1988;263(20):9767–9775. [PubMed] [Google Scholar]
- 41.Tarahovsky YS, Khusainov AA, Daugelavichus R, Bakene E. Structural changes in Escherichia coli membranes induced by bacteriophage T4 at different temperatures. Biophys J. 1995;68(1):157–163. doi: 10.1016/S0006-3495(95)80170-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tarahovsky YS, Khusainov AA, Deev AA, Kim YV. Membrane fusion during infection of Escherichia coli cells by phage T4. FEBS Lett. 1991;289(1):18–22. doi: 10.1016/0014-5793(91)80899-e. [DOI] [PubMed] [Google Scholar]
- 43.Furukawa H, Kuroiwa T, Mizushima S. DNA injection during bacteriophage T4 infection of Escherichia coli. J Bacteriol. 1983;154(2):938–945. doi: 10.1128/jb.154.2.938-945.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lucić V, Förster F, Baumeister W. Structural studies by electron tomography: From cells to molecules. Annu Rev Biochem. 2005;74:833–865. doi: 10.1146/annurev.biochem.73.011303.074112. [DOI] [PubMed] [Google Scholar]
- 45.Milne JLS, Subramaniam S. Cryo-electron tomography of bacteria: Progress, challenges and future prospects. Nat Rev Microbiol. 2009;7(9):666–675. doi: 10.1038/nrmicro2183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tocheva EI, Li Z, Jensen GJ. Electron cryotomography. Cold Spring Harb Perspect Biol. 2010;2(6):a003442. doi: 10.1101/cshperspect.a003442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Böhm J, et al. FhuA-mediated phage genome transfer into liposomes: A cryo-electron tomography study. Curr Biol. 2001;11(15):1168–1175. doi: 10.1016/s0960-9822(01)00349-9. [DOI] [PubMed] [Google Scholar]
- 48.Chang JT, et al. Visualizing the structural changes of bacteriophage Epsilon15 and its Salmonella host during infection. J Mol Biol. 2010;402(4):731–740. doi: 10.1016/j.jmb.2010.07.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Guerrero-Ferreira RC, et al. Alternative mechanism for bacteriophage adsorption to the motile bacterium Caulobacter crescentus. Proc Natl Acad Sci USA. 2011;108(24):9963–9968. doi: 10.1073/pnas.1012388108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Liu X, et al. Structural changes in a marine podovirus associated with release of its genome into Prochlorococcus. Nat Struct Mol Biol. 2010;17(7):830–836. doi: 10.1038/nsmb.1823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Dai W, et al. Visualizing virus assembly intermediates inside marine cyanobacteria. Nature. 2013;502(7473):707–710. doi: 10.1038/nature12604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sun L, et al. Icosahedral bacteriophage ΦX174 forms a tail for DNA transport during infection. Nature. 2014;505(7483):432–435. doi: 10.1038/nature12816. [DOI] [PubMed] [Google Scholar]
- 53.Hu B, Margolin W, Molineux IJ, Liu J. The bacteriophage t7 virion undergoes extensive structural remodeling during infection. Science. 2013;339(6119):576–579. doi: 10.1126/science.1231887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Liu J, Chen CY, Shiomi D, Niki H, Margolin W. Visualization of bacteriophage P1 infection by cryo-electron tomography of tiny Escherichia coli. Virology. 2011;417(2):304–311. doi: 10.1016/j.virol.2011.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Reeve JN. Bacteriophage infection of minicells: A general method for identification of “in vivo” bacteriophage directed polypeptide biosynthesis. Mol Gen Genet. 1977;158(1):73–79. doi: 10.1007/BF00455121. [DOI] [PubMed] [Google Scholar]
- 56.Roozen KJ, Fenwick RG, Jr, Curtiss R., 3rd Synthesis of ribonucleic acid and protein in plasmid-containing minicells of Escherichia coli K-12. J Bacteriol. 1971;107(1):21–33. doi: 10.1128/jb.107.1.21-33.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bartual SG, et al. Structure of the bacteriophage T4 long tail fiber receptor-binding tip. Proc Natl Acad Sci USA. 2010;107(47):20287–20292. doi: 10.1073/pnas.1011218107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Edgar R, et al. Bacteriophage infection is targeted to cellular poles. Mol Microbiol. 2008;68(5):1107–1116. doi: 10.1111/j.1365-2958.2008.06205.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Grinius L. Energy Transduction and Gene Transfer in Chemotrophic Bacteria: Macromolecules on the Move. Harwood Academic; Zurich: 1987. [Google Scholar]
- 60.Goldberg E, Grinius L, Letellier L. Recognition, attachment, and injection. In: Karam JD, editor. Molecular Biology of Bacteriophage T4. American Society for Microbiology; Washington, DC: 1994. pp. 347–356. [Google Scholar]
- 61.Kanamaru S, et al. Structure of the cell-puncturing device of bacteriophage T4. Nature. 2002;415(6871):553–557. doi: 10.1038/415553a. [DOI] [PubMed] [Google Scholar]
- 62.Abuladze NK, Gingery M, Tsai J, Eiserling FA. Tail length determination in bacteriophage T4. Virology. 1994;199(2):301–310. doi: 10.1006/viro.1994.1128. [DOI] [PubMed] [Google Scholar]
- 63.Leiman PG, Shneider MM. Contractile tail machines of bacteriophages. Adv Exp Med Biol. 2012;726:93–114. doi: 10.1007/978-1-4614-0980-9_5. [DOI] [PubMed] [Google Scholar]
- 64.Henning U, Höhn B, Sonntag I. Cell envelope and shape of Escherichia coli K12: The ghost membrane. Eur J Biochem. 1973;39(1):27–36. doi: 10.1111/j.1432-1033.1973.tb03099.x. [DOI] [PubMed] [Google Scholar]
- 65.Adam G, Delbrück M. Reduction of dimensionality in biological diffusion processes. In: Rich A, Davidson N, editors. Structural Chemistry and Molecular Biology. Freeman; San Francisco: 1968. pp. 198–215. [Google Scholar]
- 66.Rothenberg E, et al. Single-virus tracking reveals a spatial receptor-dependent search mechanism. Biophys J. 2011;100(12):2875–2882. doi: 10.1016/j.bpj.2011.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Parent KN, et al. OmpA and OmpC are critical host factors for bacteriophage Sf6 entry in Shigella. Mol Microbiol. 2014;92(1):47–60. doi: 10.1111/mmi.12536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wilson JH, Luftig RB, Wood WB. Interaction of bacteriophage T4 tail fiber components with a lipopolysaccharide fraction from Escherichia coli. J Mol Biol. 1970;51(2):423–434. doi: 10.1016/0022-2836(70)90152-x. [DOI] [PubMed] [Google Scholar]
- 69.Wood WB, Henninger M. Attachment of tail fibers in bacteriophage T4 assembly: Some properties of the reaction in vitro and its genetic control. J Mol Biol. 1969;39(3):603–618. doi: 10.1016/0022-2836(69)90148-x. [DOI] [PubMed] [Google Scholar]
- 70.Crawford JT, Goldberg EB. The function of tail fibers in triggering baseplate expansion of bacteriophage T4. J Mol Biol. 1980;139(4):679–690. doi: 10.1016/0022-2836(80)90054-6. [DOI] [PubMed] [Google Scholar]
- 71.Crowther RA. Mutants of bacteriophage T4 that produce infective fibreless particles. J Mol Biol. 1980;137(2):159–174. doi: 10.1016/0022-2836(80)90323-x. [DOI] [PubMed] [Google Scholar]
- 72.King J. Assembly of the tail of bacteriophage T4. J Mol Biol. 1968;32(2):231–262. doi: 10.1016/0022-2836(68)90007-7. [DOI] [PubMed] [Google Scholar]
- 73.Molineux IJ, Panja D. Popping the cork: Mechanisms of phage genome ejection. Nat Rev Microbiol. 2013;11(3):194–204. doi: 10.1038/nrmicro2988. [DOI] [PubMed] [Google Scholar]
- 74.Panja D, Molineux IJ. Dynamics of bacteriophage genome ejection in vitro and in vivo. Phys Biol. 2010;7(4):045006. doi: 10.1088/1478-3975/7/4/045006. [DOI] [PubMed] [Google Scholar]
- 75.Baumann L, Benz WC, Wright A, Goldberg EB. Inactivation of urea-treated phage T4 by phosphatidylglycerol. Virology. 1970;41(2):356–364. doi: 10.1016/0042-6822(70)90088-7. [DOI] [PubMed] [Google Scholar]
- 76.Benz WC, Goldberg EB. Interactions between modified phage T4 particles and spheroplasts. Virology. 1973;53(1):225–235. doi: 10.1016/0042-6822(73)90481-9. [DOI] [PubMed] [Google Scholar]
- 77.Kikuchi Y, King J. Genetic control of bacteriophage T4 baseplate morphogenesis. III. Formation of the central plug and overall assembly pathway. J Mol Biol. 1975;99(4):695–716. doi: 10.1016/s0022-2836(75)80180-x. [DOI] [PubMed] [Google Scholar]
- 78.Vianelli A, et al. Bacteriophage T4 self-assembly: Localization of gp3 and its role in determining tail length. J Bacteriol. 2000;182(3):680–688. doi: 10.1128/jb.182.3.680-688.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Duckworth DH. Biological activity of bacteriophage ghosts and “take-over” of host functions by bacteriophage. Bacteriol Rev. 1970;34(3):344–363. doi: 10.1128/br.34.3.344-363.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Lu MJ, Stierhof YD, Henning U. Location and unusual membrane topology of the immunity protein of the Escherichia coli phage T4. J Virol. 1993;67(8):4905–4913. doi: 10.1128/jvi.67.8.4905-4913.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Silver S, Levine E, Spielman PM. Cation fluxes and permeability changes accompanying bacteriophage infection of Escherichia coli. J Virol. 1968;2(8):763–771. doi: 10.1128/jvi.2.8.763-771.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Lipinska B, Rao AS, Bolten BM, Balakrishnan R, Goldberg EB. Cloning and identification of bacteriophage T4 gene 2 product gp2 and action of gp2 on infecting DNA in vivo. J Bacteriol. 1989;171(1):488–497. doi: 10.1128/jb.171.1.488-497.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Ge P, et al. Atomic structures of a bactericidal contractile nanotube in its pre- and postcontraction states. Nat Struct Mol Biol. 2015;22(5):377–382. doi: 10.1038/nsmb.2995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Mastronarde DN. Automated electron microscope tomography using robust prediction of specimen movements. J Struct Biol. 2005;152(1):36–51. doi: 10.1016/j.jsb.2005.07.007. [DOI] [PubMed] [Google Scholar]
- 85.Kremer JR, Mastronarde DN, McIntosh JR. Computer visualization of three-dimensional image data using IMOD. J Struct Biol. 1996;116(1):71–76. doi: 10.1006/jsbi.1996.0013. [DOI] [PubMed] [Google Scholar]
- 86.Amat F, et al. Markov random field based automatic image alignment for electron tomography. J Struct Biol. 2008;161(3):260–275. doi: 10.1016/j.jsb.2007.07.007. [DOI] [PubMed] [Google Scholar]
- 87.Liu J, Wright ER, Winkler H. 3D visualization of HIV virions by cryoelectron tomography. Methods Enzymol. 2010;483:267–290. doi: 10.1016/S0076-6879(10)83014-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Winkler H, et al. Tomographic subvolume alignment and subvolume classification applied to myosin V and SIV envelope spikes. J Struct Biol. 2009;165(2):64–77. doi: 10.1016/j.jsb.2008.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Winkler H. 3D reconstruction and processing of volumetric data in cryo-electron tomography. J Struct Biol. 2007;157(1):126–137. doi: 10.1016/j.jsb.2006.07.014. [DOI] [PubMed] [Google Scholar]
- 90.Pettersen EF, et al. UCSF Chimera: A visualization system for exploratory research and analysis. J Comput Chem. 2004;25(13):1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.