Significance
Human memory is a highly heritable and complex trait that is the outcome of several neurobiologically distinct mental processes, such as learning, memory maintenance, and emotional modulation of memory strength. Such processes, which are not always amenable to direct observation, can be dissociated using computational models. Here we provide converging evidence linking computational model parameters and related behavioral and neuroimaging phenotypes to sets of biologically related genes in several populations of healthy young and elderly adults. Our findings suggest that distinct genetic profiles underlie specific mental processes of human episodic memory. Understanding the genetic and neural bases of such processes is essential for the development of novel therapeutic approaches that could lead to a better treatment of neuropsychiatric disorders.
Keywords: learning and memory, computational model, gene set enrichment analysis, functional neuroimaging, frontal lobe
Abstract
Episodic memory performance is the result of distinct mental processes, such as learning, memory maintenance, and emotional modulation of memory strength. Such processes can be effectively dissociated using computational models. Here we performed gene set enrichment analyses of model parameters estimated from the episodic memory performance of 1,765 healthy young adults. We report robust and replicated associations of the amine compound SLC (solute-carrier) transporters gene set with the learning rate, of the collagen formation and transmembrane receptor protein tyrosine kinase activity gene sets with the modulation of memory strength by negative emotional arousal, and of the L1 cell adhesion molecule (L1CAM) interactions gene set with the repetition-based memory improvement. Furthermore, in a large functional MRI sample of 795 subjects we found that the association between L1CAM interactions and memory maintenance revealed large clusters of differences in brain activity in frontal cortical areas. Our findings provide converging evidence that distinct genetic profiles underlie specific mental processes of human episodic memory. They also provide empirical support to previous theoretical and neurobiological studies linking specific neuromodulators to the learning rate and linking neural cell adhesion molecules to memory maintenance. Furthermore, our study suggests additional memory-related genetic pathways, which may contribute to a better understanding of the neurobiology of human memory.
Human memory is a highly heritable and complex trait that is the outcome of numerous neurocognitive processes (1, 2). Although many genetic variations associated with human memory performance have been identified (3–9), most were based on general memory scores, such as the number of recalled items. Such scores may serve as good correlates of overall memory ability, but they offer little insight into the genetic underpinnings of specific memory-related mental processes, such as encoding, memory maintenance, modulation by emotional arousal, or guessing strategies. Importantly, neurobiological and molecular profiles of such processes are known to be partly distinct (10–12). Because some of the mental processes involved in memory are not readily amenable to direct observation, computational modeling can be used to make inferences about them (13, 14). Model-based analyses provided insights into neurocomputational mechanisms of reward-based learning and decision making (15–17), related model parameters such as the learning rate to genetic polymorphisms (18, 19), and provided a computational explanation for the inverted-U-shaped relation between stress intensity and behavioral performance (20). As a relatively recent development, the model-based analysis approach has largely been missing from studies of human episodic memory and genome-wide association studies (GWAS).
GWAS already had identified a number of associations between single genetic markers and cognitive traits, such as episodic memory (7–9) and modulation of memory strength by negative emotional arousal (21). However, because many such traits are complex and multigenic, it is unlikely that a single genetic variant will explain a sufficient amount of behavioral variability. As a statistically more powerful and biologically meaningful alternative, gene set-based methods of linking sets of biologically related genes with traits of interest have been developed (22, 23). They test whether a group of genes (gene set) is more enriched in associations with a studied phenotype than would be expected by chance. Such studies identified gene sets related to neuropsychiatric phenotypes (24, 25) and to cognition in healthy individuals, including working memory (26) and emotionally aversive memory (27).
In the present study, we performed a gene set enrichment analyses (GSEA) of specific parameters of episodic memory performance estimated from a computational model of verbal memory in young adults (21). We found four distinct gene sets associated with different computational parameters in independent samples, three of which also were linked to related measures of picture memory in young adults and verbal memory in elderly individuals. The L1 cell adhesion molecule (L1CAM) interactions gene set also was associated with differences in picture recognition-related brain activity in a number of frontal cortical areas.
Results
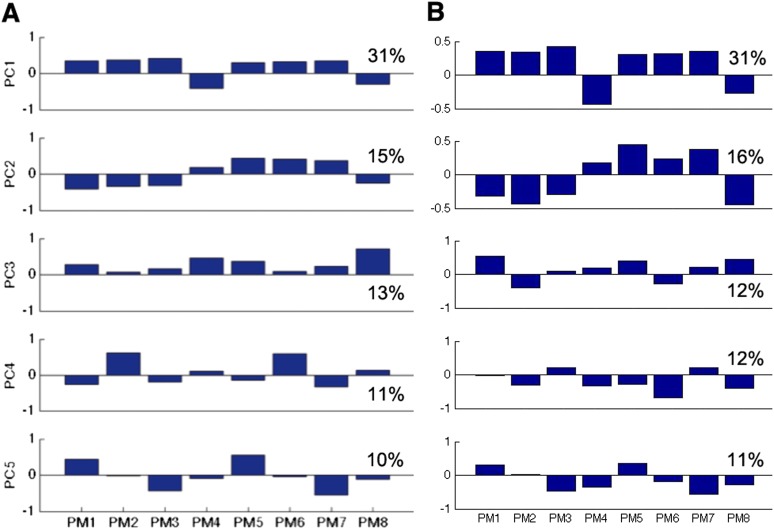
The verbal memory task contained neutral, positive, and negative words, which the participants had to recall freely first after their presentation and later after an ∼5 min delay. Participants in the discovery sample (n = 1,239; Table 1) and the replication sample (n = 526) performed the identical verbal memory task. Eight performance measures (PM1–8; Table 2) indicated the number of correctly recalled words in each valence category (i.e., neutral, positive, and negative) and confabulative errors (words not on the learning list) in the immediate and the delayed recall. Because performance measures were partly correlated, the effective dimensionality of data was smaller: principal component analysis (PCA) revealed five major components (Fig. S1).
Table 1.
Samples and core phenotypes used
| Sample name (location) | Phenotypes of interest | Sample size |
| Discovery (Zurich and Basel) | Computational model parameters | 1,239 |
| Replication (Basel) | Computational model parameters | 526 |
| Words/pictures* (Basel) | Free recall of pictures | 493 |
| Pictures/fMRI (Basel) | Free recall of pictures | 835 |
| Picture recognition | 822 | |
| Working memory | 825 | |
| fMRI data on picture recognition | 795 | |
| fMRI data on working memory | 797 | |
| AgeCoDe elderly (Bonn) | Free recall of words | 743 |
This is a subsample of the Replication sample. All other samples are independent.
Table 2.
Description of performance measures (PM1–8) in the verbal memory task and their population statistics
| Performance measure | Discovery sample, mean ± SEM | Replication sample, mean ± SEM |
| PM1: Positive words correctly recalled immediately | 7.666 ± 0.042 | 7.645 ± 0.058 |
| PM2: Negative words correctly recalled immediately | 8.318 ± 0.036 | 8.364 ± 0.050 |
| PM3: Neutral words correctly recalled immediately | 7.392 ± 0.047 | 7.345 ± 0.065 |
| PM4: Mistakes made immediately after encoding | 1.302 ± 0.038 | 1.164 ± 0.050 |
| PM5: Positive words correctly recalled after 5 min | 2.948 ± 0.043 | 3.055 ± 0.061 |
| PM6: Negative words correctly recalled after 5 min | 2.645 ± 0.041 | 2.882 ± 0.059 |
| PM7: Neutral words correctly recalled after 5 min | 2.448 ± 0.041 | 2.618 ± 0.056 |
| PM8: Mistakes made 5 min after encoding | 1.681 ± 0.048 | 1.727 ± 0.071 |
Fig. S1.
PCA of eight performance measures (PM1–PM8, described in Table 2) in the discovery sample (A) (modified from ref. 21) and the replication sample (B). Percentages denote variance explained by each component. The first five principal components (PC1–5) explain more than 80% of the variance in the data; their loadings suggest that the first component (PC1) can be approximately related to general learning ability and the second one (PC2) to delayed memory recall (compared with immediate recall). The latter three components could be interpreted only in the discovery sample: PC3 related approximately to errors, PC4 to the recall of negative words, and PC5 to the recall of positive minus neutral words.
Computational Modeling.
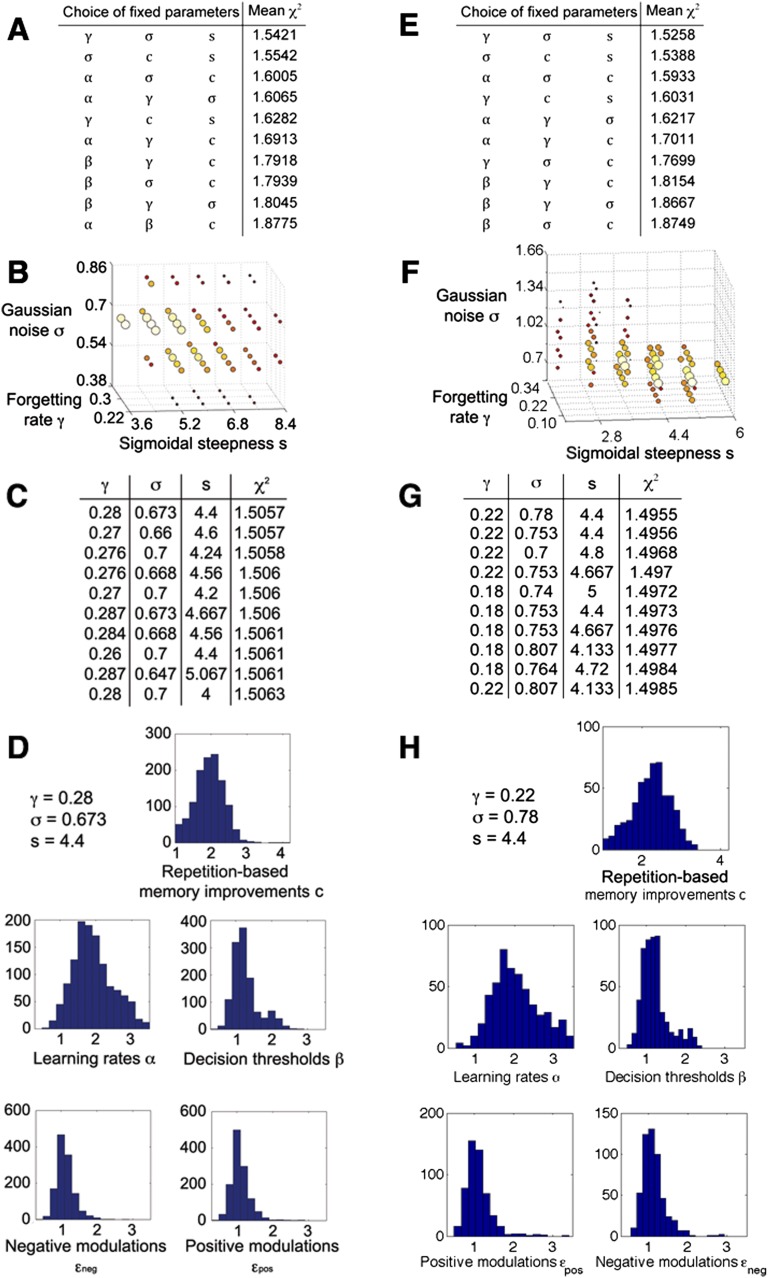
To study specific cognitive processes relevant for memory performance, we used a simple computational model of learning in the verbal task (21). Our model had the following parameters, which we aimed to infer from the behavioral data: the learning rate α and the Gaussian noise σ, the repetition-based memory improvement c (related to memory maintenance) and the forgetting rate γ, the modulation of memory strength by positive or negative emotional arousal εpos and εneg, the decision threshold β, and the sigmoidal steepness s (for a detailed description see Materials and Methods). Because the effective dimensionality of the behavioral data was lower than the number of model parameters, we empirically selected five of these parameters (α, β, c, εpos, εneg) to be estimated individually, keeping the three remaining ones fixed. After the estimation (Fig. S2), more than 99% of the resulting individual parameter sets passed the χ2 test of goodness-of-fit [mean χ2(discovery) = 1.5057 and χ2(replication) = 1.4955], indicating that the model was sufficiently flexible. High correlations [mean ρSpearman(discovery) = 0.981 and ρSpearman(replication) = 0.986] among the 10 best parameter sets (hill climbing end points in stage 2 of the estimation) suggested high estimation reliability, and the presence of 99.8% of individual parameter sets within the middle 90% of the value ranges (except the bound of c = 1, because repetition should not weaken memories) indicated that parameter estimation bounds did not constrain the results.
Fig. S2.
Parameter estimation results for the discovery sample (A–D) (modified from ref. 21) and the replication sample (E–H). (A and E) Stage 1: Based on a moderately accurate estimation of model parameters, two selections of fixed parameters (γ, σ, s and σ, c, s) are chosen for hill climbing. The best 10 of all 56 possible three-of-eight combinations are shown. (B and F) Stage 2: The hill-climbing results of estimating three fixed parameters for the best model (with Gaussian noise σ, sigmoidal steepness s, and forgetting rate γ fixed) are shown, with bigger circles and lighter colors indicating better goodness-of-fit; the 10 best hill-climbing points (biggest light yellow circles) were selected for evaluating averages of all their possible combinations. (C and G) Stage 3: The 10 combinations with the best goodness-of-fit (χ2) are displayed. Overall, 267 of 1,023 combinations had better χ2 than the best hill-climbing point (χ2 = 1.522) for the discovery sample, as did 77 of 1,023 combinations for the replication sample (χ2 = 1.507), suggesting that averaging parameters helps overcome gaps in step size and leads to refined parameter values. (D and H) Histograms of the best-fitting individual parameters show distributions with the following means: c = 1.95, α = 1.93, β = 1.27, εneg = 1.12, εpos = 1.09 for the discovery sample and c = 2.23, α = 2.04, β = 1.22, εneg = 1.13, εpos = 1.11 for the replication sample.
GSEA.
We first performed a GWAS with the estimated model parameters (α, β, c, εpos, εneg) with 1,239 individuals in the discovery sample (for quality control and statistics, see Materials and Methods). Based on the resulting P values of association with these parameters, we performed a GSEA using MAGENTA (23). Of the 1,411 gene sets used for input, we found false-discovery rate (FDR)-corrected enrichment for seven gene sets (Table 3). A second GSEA was performed for these gene sets in an independent sample of 526 individuals (Table 1), again using GWAS-derived P values of association with the estimated model parameters. Of the seven gene sets significantly enriched in the discovery sample, four largely nonoverlapping gene sets were also enriched in the replication sample (Table 3): amine compound solute-carrier (SLC) transporters associated with the learning rate α, L1CAM interactions associated with the repetition-based memory improvement c, and collagen formation and transmembrane receptor protein tyrosine kinase activity associated with εneg. Although behavioral measures of verbal memory were correlated with model parameters (Table S1), none of the gene sets reported above would have been detected if these behavioral measures had served as starting points for GSEA (using the same multiple testing correction threshold as for the model parameters); however, five of the seven gene sets identified in the discovery sample were nominally associated with the relevant behavioral measures.
Table 3.
Associations between computational model parameters for the verbal task and different gene sets
| Model parameter | Database | Gene set (number of genes in gene set) | Discovery sample (n = 1,239) | Replication sample (n = 526) | |
| Pnominal | PFDR | Pnominal | |||
| Learning rate α | Reactome | Amine compound SLC transporters (27) | 2.0 × 10−4 | 0.0405 | 0.0031 |
| Decision threshold β | KEGG | Axon guidance (129) | 4.4 × 10−5 | 0.0067 | 0.405 |
| Repetition-based memory improvement c | Reactome | L1CAM interactions (86) | 3.0 × 10−6 | 0.0011 | 0.0231 |
| Interaction between L1 and ankyrins (23) | 1.0 × 10−3 | 0.0365 | 0.176 | ||
| Modulation by negative emotional arousal εneg | Reactome | Collagen formation (58) | 1.0 × 10−4 | 0.0129 | 0.0025 |
| Gene Ontology | Transmembrane receptor protein tyrosine kinase activity (43) | 4.1 × 10−5 | 0.0124 | 0.0212 | |
| Transmembrane receptor protein kinase activity (51) | 1.0 × 10−4 | 0.0398 | 0.059 | ||
Only the gene sets surviving MAGENTA FDR correction in the discovery sample are shown. The replicated gene sets are shown in bold. Interaction between L1 and ankyrins is a subset of L1CAM interactions, and transmembrane receptor protein tyrosine kinase activity is a subset of transmembrane receptor protein kinase activity. L1CAM interactions contains four genes [EGFR, FGFR1, and neuropilin 1 and 2 (NRP1, and NRP2)] shared with transmembrane receptor protein tyrosine kinase activity.
Table S1.
Correlations of estimated model parameters with related behavioral measures of verbal task performance in the discovery sample and picture task performance in the replication (words/pictures) sample
| Model parameter | Relevant phenotype in the verbal task | Correlation, Spearman’s ρ | Relevant free recall phenotype in the picture task | Correlation, Spearman’s ρ |
| Learning rate α | Percentage of correctly remembered words in the immediate recall | 0.722 | Percentage of correctly remembered pictures | 0.152 |
| Decision threshold β | Number of mistakes in the delayed recall | −0.825 | Number of mistakes in the picture recall | −0.204 |
| Repetition-based memory improvement c | Percentage of correctly remembered words in the delayed recall | 0.607 | Percentage of correctly remembered pictures | 0.098 |
| Modulation by positive emotional arousal εpos | Percentage of correctly remembered positive minus neutral words in the delayed recall | 0.737 | Percentage of correctly remembered positive minus neutral pictures | 0.133 |
| Modulation by negative emotional arousal εneg | Percentage of correctly remembered negative minus neutral words in the delayed recall | 0.708 | Percentage of correctly remembered negative minus neutral pictures | 0.074 |
Correlations with verbal task performance are quite high because computational modeling was performed for the verbal task. Correlations with picture task performance suggest that the percentage of correctly remembered pictures in the free recall is related more to the learning rate α than to the repetition-based memory improvement c and hence is used for the replication of gene sets associated with α. All correlations are statistically significant.
We next examined if the four replicated gene sets showed enrichment in associations with related, albeit less specific, behavioral measures in different tasks and in additional populations (Table 1). Computational modeling was not practical for these tasks because of the low dimensionality of the behavioral data. In the sample of nondemented elderly individuals from the German Study on Aging, Cognition and Dementia in Primary Care Patients (AgeCoDe) (n = 743), participants performed a word-list verbal recall task from the Consortium to Establish a Registry for Alzheimer’s Disease (CERAD) neuropsychological test battery (28). In a picture task, performed by two populations of healthy young adults, participants were presented neutral, positive, and negative pictures, which they had to recall freely 10 min after presentation. After a longer delay, participants of the Basel pictures/functional MRI (fMRI) sample also were asked to perform picture recognition: provided with both previously shown and new pictures, they had to answer whether they had seen each picture. For all of these tasks, we selected behavioral measures most closely related to each computational parameter. For the learning rate α, immediate recall is more appropriate than delayed recall, because the latter also accounts for forgetting. However, because immediate recall was not available in the picture task, we used free delayed recall (see also Table S1). For the repetition-based memory improvement c, only delayed recall was appropriate, because immediate recall did not account for memory maintenance. In the picture task both free recall and recognition measured memory over delays; however, because we also aimed to study the neural correlates of memory maintenance using functional neuroimaging, which were available only for picture recognition data, we chose the latter phenotype. For negative emotional modulation factor εneg, only picture data were available; therefore we chose the difference between freely recalled negative and neutral pictures as a relevant phenotype. Based on the GWAS results of these phenotypes (population statistics are shown in Table 4), we used MAGENTA for testing the enrichment of the four gene sets associated with different computational parameters of the (original) verbal task. We found that amine compound SLC transporters were associated with immediate recall of words in the elderly sample and with free recall of pictures in both young samples; L1CAM interactions were associated with delayed recall of words in the elderly sample and with picture recognition in the pictures/fMRI sample; collagen formation was associated with free recall of negative minus neutral pictures in the words/pictures sample; and there were no significant associations with transmembrane receptor protein tyrosine kinase activity (Table 5). Because the arousal of negative pictures [2.36 (mean) ± 0.06 (SEM)] was higher than that of positive pictures (1.94 ± 0.04; Student t test; P = 0.000002), the association of collagen formation with picture recall may be mediated by arousal rather than by negative valence. We also checked if sex was correlated with any of the significant SNPs (one per gene) in any of the significantly enriched gene sets (Tables 3 and 5). No significant correlations, corrected for the number of SNPs per gene set, were found (Ps > 0.05).
Table 4.
Population statistics of relevant phenotypes for the picture task and for the word task used in the German elderly sample
| Phenotype | Data sample | Mean ± SEM |
| Percentage of correctly remembered pictures in the free recall | Words/pictures (n = 493) | 39.58 ± 0.49 |
| Percentage of correctly remembered negative pictures minus neutral pictures in the free recall | Words/pictures (n = 493) | 17.17 ± 0.59 |
| Percentage of correctly remembered pictures in the free recall | Pictures/fMRI (n = 835) | 42.57 ± 0.39 |
| Percentage of correctly remembered negative pictures minus neutral pictures in the free recall | Pictures/fMRI (n = 835) | 17.67 ± 0.45 |
| Picture recognition | Pictures/fMRI (n = 822) | |
| Percentage of correctly recollected pictures* | 80.74 ± 0.55 | |
| Percentage of previously seen pictures rated as familiar | 15.68 ± 0.49 | |
| Percentage of previously seen pictures rated as new | 3.57 ± 0.15 | |
| Percentage of new pictures rated as known | 1.02 ± 0.07 | |
| Percentage of new pictures rated as familiar | 9.41 ± 0.28 | |
| Percentage of new pictures rated as new | 89.57 ± 0.31 | |
| Number of correctly remembered words in the immediate recall | AgeCoDe elderly (n = 743) | 19.55 ± 0.13 |
| Number of correctly remembered words in the delayed recall | AgeCoDe elderly (n = 743) | 5.88 ± 0.07 |
Previously seen pictures rated as known.
Table 5.
Association between the four replicated gene sets and other related phenotypes
| Database | Gene set (number of genes in gene set, associated model parameter) | Relevant phenotype | Data sample | Association P value |
| Reactome | Amine compound SLC transporters (27, α) | Percentage of correctly remembered pictures in the free recall | Words/pictures | 0.0302 |
| Pictures/fMRI | 0.0001 | |||
| Number of correctly remembered words in the immediate recall | AgeCoDe elderly | 0.032 | ||
| L1CAM interactions (86, c) | Percentage of correctly recollected pictures in picture recognition | Pictures/fMRI | 0.0419 | |
| Number of correctly remembered words in the delayed recall | AgeCoDe elderly | 0.0068 | ||
| Collagen formation (58, εneg) | Percentage of correctly remembered negative pictures minus neutral pictures in the free recall | Words/pictures | 0.0206 | |
| Pictures/fMRI | 0.145 | |||
| Gene Ontology | Transmembrane receptor protein tyrosine kinase activity (43, εneg) | Percentage of correctly remembered negative pictures minus neutral pictures in the free recall | Words/pictures | 0.147 |
| Pictures/fMRI | 0.153 |
Neural Correlates of the Discovered Gene Sets.
Because the amine compound SLC transporters and L1CAM interactions gene sets were associated with free recall and recognition of pictures, respectively, in the pictures/fMRI sample, we examined neural correlates of these associations using functional neuroimaging. Participants in this sample performed the encoding and recognition parts of the picture task in the fMRI scanner; free recall was performed outside the scanner. We studied association with amine compound SLC transporters (related to the learning rate α) by using two encoding contrasts: all pictures vs. scrambled controls and the difference due to memory (Dm, i.e., subsequently remembered vs. not remembered pictures; 29). Because neural correlates of memory maintenance may not be apparent during encoding, we examined the association with L1CAM interactions (related to the repetition-based memory improvement c) using the recognition contrast of previously seen vs. new pictures, a task that engages the brain areas involved in memory maintenance and retrieval, such as prefrontal cortex and medial temporal lobe (30, 31). To capture multiallelic effects of each gene set on the differences in brain activity, we generated individual multilocus genetic scores (Materials and Methods). For L1CAM interactions, the score was built using 28 significant SNPs from the same number of genes (one most significant SNP per gene; Table S2); the genetic score for amine compound SLC transporters comprised 14 SNPs.
Table S2.
Composition of the L1CAM interactions genetic score: SNP IDs, tagged genes, statistics of association with picture recognition in the pictures/fMRI sample, reference allele with direction of association with picture recognition memory, major allele
| SNP rsID | Gene symbol | Association P value | Reference allele | Direction of association | Major allele |
| rs9459771 | RPS6KA2 | 0.0011 | T | + | C |
| rs11715984 | DLG1 | 0.0017 | G | + | A |
| rs13258436 | KCNQ3 | 0.0020 | T | + | C |
| rs197721 | ITGA9 | 0.0049 | T | + | G |
| rs7153298 | RPS6KA5 | 0.0071 | T | − | T |
| rs17210194 | CNTN6 | 0.0110 | G | − | G |
| rs3506 | AP2B1 | 0.0113 | T | + | T |
| rs17547685 | SH3GL2 | 0.0130 | T | − | T |
| rs17333297 | NRCAM | 0.0149 | G | − | A |
| rs6983315 | FGFR1 | 0.0185 | G | + | G |
| rs11009157 | ITGB1 | 0.0195 | G | − | G |
| rs4948381 | ANK3 | 0.0235 | G | + | A |
| rs442103 | DPYSL2 | 0.0247 | G | − | G |
| rs10504047 | ANK1 | 0.0254 | G | − | C |
| rs1478438 | ITGA1 | 0.0267 | T | − | T |
| rs7781264 | EGFR | 0.0281 | T | + | C |
| rs645078 | RPS6KA4 | 0.0311 | C | − | A |
| rs11647503 | CSNK2A2 | 0.0318 | T | + | C |
| rs949215 | LAMA1 | 0.0318 | C | − | C |
| rs6580842 | SCN8A | 0.0335 | G | + | A |
| rs883501 | NRP2 | 0.0335 | T | + | A |
| rs1421933 | ITGA2 | 0.0352 | G | − | G |
| rs17115275 | NCAM1 | 0.0352 | G | − | A |
| rs7643664 | ALCAM | 0.0360 | C | + | C |
| rs16943748 | CLTC | 0.0369 | G | + | G |
| rs6793943 | SCN5A | 0.0407 | T | + | C |
| rs11240351 | CNTN2 | 0.0407 | T | + | T |
| rs1331325 | NRP1 | 0.0407 | T | − | C |
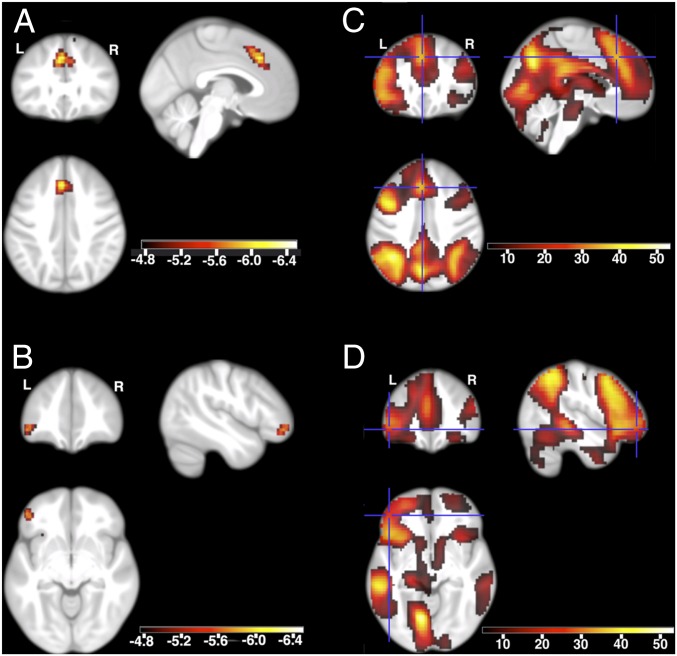
Genetic score-dependent analysis revealed no whole-brain familywise error (FWE)-corrected correlations with differences in brain activity for amine compound SLC transporters. However, the L1CAM interactions genetic score was negatively correlated at the whole-brain FWE-corrected level with differences in the activity of numerous frontal regions: large clusters in the left superior frontal cortex and the left inferior frontal gyrus (Fig. 1, A and B and Table 6) and small clusters in the bilateral orbitofrontal cortices and insulae (Table 6). The results were very similar if the two most significant SNPs per gene were used to build a genetic score (Table S3) or if only correctly rated pictures were included in the contrast (recollected previously seen pictures vs. new pictures rated as new). We also performed a genetic score-independent analysis of brain activity, which revealed substantial differences in activity at all cluster peaks dependent on the L1CAM interactions genetic score (Fig. 1, C and D), suggesting that higher values of the genetic score reflect smaller increases in frontal activity during the recognition of previously seen pictures as compared with the viewing of new pictures. To ensure that the observed effect specifically reflected differences in the episodic memory of previously seen pictures, we also used the contrast between recollected previously seen pictures vs. previously seen pictures rated as familiar or new (n = 786). Of the 238 voxels significant in the contrast between previously seen vs. new pictures, the great majority also passed small-volume correction (SVC) for the latter contrast [162 voxels in superior frontal cortex, peak coordinates (0, 17, 48), T = −4.93; four voxels in left lateral orbitofrontal cortex, peak coordinates (−30, 22, −8), T = −5.07], with the peak voxels also surviving whole-brain correction.
Fig. 1.
Functional neuroimaging of picture recognition (the contrast between previously seen vs. new pictures). (A and B) Differences in brain activity dependent on the L1CAM interactions genetic score. (C and D) Genotype-independent differences in brain activity. Color-coded t values are shown. The maps are centered at (−3, 28, 40) in the left superior frontal cortex (A and C) and at (−47, 44, −8) in the left inferior frontal gyrus (B and D). Only voxels surviving whole-brain FWE correction are shown (P whole-brain FWE-corrected < 0.05; |T| > 4.77; n = 795). Activations are overlaid on sections of the study-specific group template.
Table 6.
Association between the L1CAM interactions genetic score and brain activity differences during picture recognition
| Regional correspondence of the maximum | Left/right | No. of voxels | Peak MNI coordinates | Peak statistics | Empirical P value | |||
| X | Y | Z | T | Pnominal | ||||
| Superior frontal cortex (79%), caudal anterior cingulate cortex (2%) | Left | 187 | −3 | 28 | 40 | −6.19 | 9.4 × 10−10 | 0.0001 |
| Pars orbitalis cortex (34%), pars triangularis cortex (28%), rostral middle frontal cortex (16%) | Left | 39 | −47 | 44 | −8 | −5.70 | 1.6 × 10−8 | 0.0021 |
| Lateral orbitofrontal cortex (20%), insula (23%) | Right | 6 | 30 | 22 | 0 | −5.16 | 3.2 × 10−7 | 0.018 |
| Lateral orbitofrontal cortex (73%), insula (8%) | Left | 4 | −28 | 22 | −4 | −4.87 | 1.4 × 10−6 | 0.053 |
Previously seen vs. new pictures fMRI contrast was used. Only clusters with at least three voxels surviving whole-brain FWE correction are shown (P whole-brain FWE-corrected < 0.05; |T| > 4.77; n = 795). Region names are in accordance with the FreeSurfer nomenclature; probabilities are in accordance with the in-house atlas. For each cluster we also provide empirical P values of the peak voxel based on 10,000 randomly generated genetic scores (null distribution).
Table S3.
Association between differences in brain activity during picture recognition and the L1CAM interactions genetic score that uses the two most significant SNPs per gene
| Regional correspondence of the maximum | Left/right | No. of voxels | Peak MNI coordinates | Peak statistics | |||
| X | Y | Z | T | Pnominal | |||
| Superior frontal cortex (80%) | Left | 76 | −3 | 30 | 40 | −5.77 | 1.1 × 10−8 |
| Pars orbitalis cortex (56%), pars triangularis cortex (22%), rostral middle frontal cortex (4%) | Left | 23 | −47 | 41 | −12 | −5.42 | 8.0 × 10−8 |
| Pars orbitalis cortex (35%), rostral middle frontal cortex (1%) | Right | 23 | 41 | 44 | −12 | −5.37 | 1.0 × 10−7 |
| Lateral orbitofrontal cortex (20%), insula (23%) | Right | 21 | 30 | 22 | 0 | −5.32 | 1.3 × 10−7 |
| Superior frontal cortex (84%) | Right | 3 | 11 | 28 | 60 | −5.02 | 6.2 × 10−7 |
The fMRI contrast between previously seen vs. new pictures was used. Only clusters with at least three voxels surviving whole-brain FWE correction are shown (P whole-brain FWE-corrected < 0.05; |T| > 4.77; n = 795). Region names are in accordance with the FreeSurfer nomenclature (71); probabilities are in accordance with the in-house atlas.
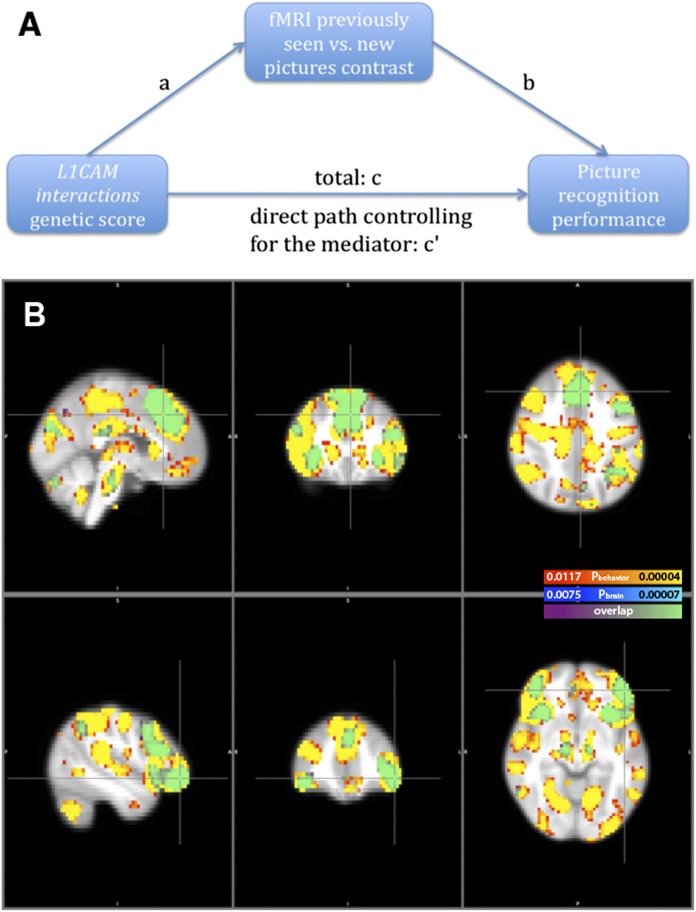
Because the differences in brain activity at all cluster peaks dependent on the genetic score of L1CAM interactions also were negatively associated with recognition performance (Table S4, n = 795), we performed a mediator analysis to exclude the possibility that the association between the genetic score and the differences in brain activity could be explained fully by the correlated behavioral phenotype. We examined whether the differences in activity in these brain regions were mediating the correlation between the genetic score and picture recognition performance. We found that all four clusters acted as significant mediators of the genetic score–behavior relationship (for more details about mediation, see Materials and Methods and Fig. S3).
Table S4.
Association between differences in brain activity during picture recognition (fMRI contrast of previously seen vs. new pictures) and the percentage of correctly recollected pictures
| Regional correspondence of the maximum | Left/right | MNI coordinates at the maximum | T value | ||
| X | Y | Z | |||
| Superior frontal cortex (79%), caudal anterior cingulate cortex (2%) | Left | −3 | 28 | 40 | −12.13 |
| Pars orbitalis cortex (34%), pars triangularis cortex (28%), rostral middle frontal cortex (16%) | Left | −47 | 44 | −8 | −9.11 |
| Lateral orbitofrontal cortex (20%), insula (23%) | Right | 30 | 22 | 0 | −9.08 |
| Lateral orbitofrontal cortex (73%), insula (8%) | Left | −28 | 22 | −4 | −9.65 |
Test statistics are shown at the maxima of clusters associated with the L1CAM interactions genetic score (Table 6). They indicate that differences in brain activity at these clusters are also associated with picture recognition performance in the same direction as the L1CAM interactions genetic score. Region names are in accordance with the FreeSurfer nomenclature (71); probabilities are in accordance with the in-house atlas.
Fig. S3.
Scheme and results of the mediation analysis. (A) The overall predictor–outcome relationship is denoted as “c,” and the direct effect controlling for the mediator is denoted as “c′.” The a × b effect tests the significance of mediation (c–c′). For more details see Materials and Methods. (B) Mediation results at (−3, 28, 40) and (−47, 44, −8), depicting voxels for which the contrast in brain activation related to the previously seen vs. new pictures was considered a significant mediator of the relationship between the genetic score and recognition performance (blue scale), at a significance threshold of P(FDR) = 0.0075. The value of this threshold for whole-brain correction might be unusually high because of correlations between the genetic score and the behavioral phenotype. As a control, random shuffling of fMRI images between subjects was performed 10 times, always leading to no voxels surviving the FDR significance threshold. We also found that behavioral phenotype was a significant mediator of the relationship between the genetic score and differences in brain activity. Results are shown based on the red-to-yellow scale, superimposed on the original ones, at a significance threshold of P(FDR) = 0.0117. Effect sizes with neuroimaging and behavioral phenotypes as mediators were comparable in the four clusters associated with the genetic score. For the coronal and horizontal sections left in the picture refer to the right side of the brain and vice versa.
As two additional controls, we first extracted average beta values for the two largest clusters, whose activity was associated with the picture recognition phenotype at the whole-brain FWE-corrected level [cluster 1: peak coordinates (0, 28, 40), T = −12.36, number of voxels = 2,917; cluster 2: peak coordinates (−47, 41, 16), T = −10.57, number of voxels = 1,541] and subjected them to GSEA using MAGENTA. The L1CAM interactions gene set was significantly enriched in both cases, with nominal P = 0.013 for each cluster. Second, we generated 10,000 genetic scores based on randomly selected SNPs (28 SNPs per genetic score, as for L1CAM interactions) and the picture recognition phenotype, using the same procedure as for the real genetic scores. None of the random scores showed statistically significant, whole-brain-corrected correlation with the neuroimaging phenotype (the contrast between previously seen vs. new pictures) after correction for the number of genetic scores (threshold Pnominal = 2.2 × 10−10). Based on the null distribution computed with the 10,000 random scores, we also provide empirical P values for the four clusters, which describe the probabilities of their peak voxel significance being reached for the most significant voxel across the whole brain using the random scores (Table 6).
To evaluate the specificity of our imaging genetics finding, we examined if the correlation between the L1CAM interactions genetic score and differences in frontal activity also could be observed during encoding (the subsequent memory contrast, Dm). No such correlation was found at either the whole-brain FWE-corrected level or when applying SVC for 238 voxels significant in the recognition contrast, suggesting either that the relation between L1CAM interactions and differences in frontal activity was not evident during encoding and developed later or that it was specific to the cognitive demands of the recognition task. Because the participants in the pictures/fMRI sample also performed a working memory task (N-back), we examined if the reported association with L1CAM interactions was related to working memory. The MAGENTA analysis performed using P values of GWAS of N-back performance revealed significant enrichment for L1CAM interactions (P = 0.023, n = 825). The multilocus genetic score based on N-back performance was negatively correlated with the one based on picture recognition performance (ρSpearman = −0.140, P = 5.9 × 10−5, n = 815), suggesting that, with regard to this gene set, better working memory was related to worse picture recognition. Because common frontal mechanisms may mediate both effects, we examined the neural correlates of working memory using fMRI (2-back vs. 0-back contrast, n = 797) and found no correlation between differences in brain activity and the working memory-based L1CAM interactions genetic score at either the whole-brain or SVC levels of significance (number of voxels for the latter, 238). We also examined if the multilocus genetic score based on N-back performance was related to differences in brain activity in the recognition of previously seen vs. new pictures. Again no significant voxels were found at either whole-brain FWE-corrected or SVC thresholds, suggesting that there was no neural evidence implicating L1CAM interactions in working memory or common mechanisms with picture recognition.
Discussion
In this study we show robust associations between multiple gene sets and different computational parameters of verbal memory as well as other relevant neurobehavioral traits in four samples of young and elderly individuals (Table 1). Specifically, amine compound SLC transporters were associated with the learning rate α in two samples of young individuals performing the verbal task, with free recall of pictures in two samples of young individuals, and with immediate recall of words in an elderly sample. L1CAM interactions were linked to the repetition-based memory improvement c (reflecting memory maintenance) in two young samples, to picture recognition and large clusters of differences in recognition-related frontal activity in a third young sample, and to delayed recall of words in the elderly sample. Finally, collagen formation and transmembrane receptor protein tyrosine kinase activity were associated with modulation of verbal memory strength by negative emotional arousal (εneg) in two young samples, with the former gene set also linked to free recall of negative minus neutral pictures in young individuals.
At this point, it is important to specify the term replication in the context of gene set-based association studies. In contrast to SNP-based studies, in which the object of replication is the significant SNP (or variants in high linkage disequilibrium), the object of replication in gene set-based studies is the significant gene set. Across different samples, replicated gene sets typically are composed of different tagging SNPs (32, 33), a phenomenon related to the allelic and locus heterogeneity of complex traits (22, 34). In our study we did not observe substantial tagging SNP overlap between replicated gene sets across samples (Tables S5 and S6).
Table S5.
Tagging SNPs of the amine compound SLC transporters gene set
| Gene symbol | Tagging SNP in the discovery sample | Tagging SNP in the replication sample and its LD with the discovery SNP | Tagging SNP in the pictures/fMRI sample and its LD with the discovery SNP | Tagging SNP in the words/pictures sample and its LD with the discovery SNP | Tagging SNP in the AgeCoDe sample and its LD with the discovery SNP | ||||
| SLC6A11 | rs6442209 | rs746278 | 0.0054 | rs9868537 | 0.1251 | rs1601370 | 0.0192 | rs966030 | 0.0055 |
| SLC14A2 | rs7232775 | rs7226686 | 0.0068 | rs7232775 | 1 | rs17670646 | 0.0845 | rs4890292 | 0.0256 |
| SLC6A20 | rs17279437 | rs7653640 | 0.0077 | rs2251109 | 0.097 | rs17213127 | 0.0058 | rs11718841 | 0.017 |
| SLC6A2 | rs734980 | rs1861646 | 0.004 | rs36016 | 0.0106 | rs879522 | 0.0118 | rs10521329 | 0.0006 |
| SLC44A3 | rs7550014 | rs11165265 | 0.0191 | rs3849308 | 0.0297 | rs859064 | 0.3058 | rs7550014 | 1 |
| SLC6A6 | rs2164576 | rs17308574 | 0.016 | rs3773167 | 0.0162 | rs4685160 | 0.1064 | rs6774615 | 0.0685 |
| SLC6A7 | rs3776087 | rs9885318 | 0.2705 | rs3776087 | 1 | rs4601032 | 0.0252 | rs3776087 | 1 |
| SLC6A18 | rs13361701 | rs6554678 | 0.0822 | rs13361701 | 1 | rs7704728 | 0.0489 | rs6554677 | 0.1335 |
| SLC18A2 | rs363334 | rs363224 | 0.0007 | rs363223 | 0.0006 | rs363278 | 0.0023 | rs363271 | 0 |
For each gene listed in Table 7, the corresponding tagging (i.e., the most significant) SNP in each sample is shown together with its linkage disequilibrium (LD) r2 value with the tagging SNP from the discovery sample. Gene descriptions, chromosome numbers, and association P values are provided in Table 7.
Table S6.
Tagging SNPs of the L1CAM interactions gene set
| Gene symbol | Tagging SNP in the discovery sample | Tagging SNP in the replication sample and its LD with the discovery SNP | Tagging SNP in the pictures/fMRI sample and its LD with the discovery SNP | Tagging SNP in the AgeCoDe sample and its LD with the discovery SNP | |||
| RPS6KA2 | rs2281143 | rs1894660 | 0.0009 | rs9459771 | 0.0005 | rs9459777 | 0.0008 |
| KCNQ3 | rs2597348 | rs2721905 | 0.1884 | rs13258436 | 0.0091 | rs1457784 | 0.0006 |
| DPYSL2 | rs7829347 | rs17322275 | 0.0244 | rs442103 | 0.0083 | rs9314325 | 0.0935 |
| ITGA9 | rs399703 | rs197763 | 0.8689 | rs197721 | 0.057 | rs6782856 | 0 |
| ANK3 | rs11593565 | rs4948254 | 0.0053 | rs4948381 | 0.001 | rs12355908 | 0.0107 |
| CNTN6 | rs1353825 | rs9880762 | 0.0007 | rs17210194 | 0.0102 | rs3772295 | 0.1717 |
| SH3GL2 | rs10491540 | rs1998247 | 0.0414 | rs17547685 | 0.0092 | rs3808776 | 0.0101 |
| SCN2A | rs353128 | rs13025009 | 0 | rs13023748 | 0.1728 | rs10497258 | 0.0004 |
| FGFR1 | rs11777067 | rs2288696 | 0.0958 | rs6983315 | 0.3326 | rs2978073 | 0.003 |
| NRCAM | rs6466228 | rs425013 | 0.0114 | rs17333297 | 0.0019 | rs404287 | 0.0313 |
| ITGA1 | rs870992 | rs7718758 | 0.0224 | rs1478438 | 0.0518 | rs2938793 | 0 |
| LAMA1 | rs11661841 | rs10163962 | 0.1539 | rs949215 | 0.0009 | rs8084092 | 0.0085 |
| EGFR | rs6593210 | rs11238349 | 0.0017 | rs7781264 | 0.0123 | rs4947986 | 0.0606 |
| NCAM1 | rs17115275 | rs884110 | 0.0109 | rs17115275 | 1 | rs1245135 | 0.0038 |
| SPTBN1 | rs6545424 | rs10166797 | 0.0988 | rs6715538 | 0.0069 | rs17343939 | 0.1586 |
| CSNK2A2 | rs1393203 | rs37358 | 0.0706 | rs11647503 | 0.0063 | rs2550362 | 0.0203 |
| ANK1 | rs7823955 | rs11784321 | 0.0266 | rs10504047 | 0.5597 | rs2304872 | 0.0052 |
| ITGA2 | rs153141 | rs1421940 | 0.6648 | rs1421933 | 0.0069 | rs1421933 | 0.0282 |
| CNTN1 | rs1119060 | rs10784951 | 0.0219 | rs12829787 | 0.0237 | rs7970805 | 0.0116 |
| NRP2 | rs849581 | rs849570 | 0.7338 | rs883501 | 0.0027 | rs3771000 | 0.0004 |
| NRP1 | rs2506150 | rs1331312 | 0.0144 | rs1331325 | 0.005 | rs2243668 | 0.0008 |
The correspondence of significant genes between different samples and phenotypes shows that for amine compound SLC transporters genes encoding GABA, norepinephrine, choline, and vesicular amine transporters are significant in multiple samples, including the discovery sample (Table 7 and Table S5). These genes have been implicated in epilepsy, substance abuse, attention deficit hyperactivity disorder, schizophrenia, and neurodegenerative disorders (35–37). On a neurocomputational level, acetylcholine plays an important role in the encoding of new memories, may act as a switch between encoding and retrieval (38), and has been suggested as a neural correlate of the learning rate (39). Variations in several dopaminergic genes have been linked to the learning rate in the reinforcement learning framework (18, 19) and in economic games (40). Therefore, our results support the notion that the relations between certain neuromodulators and the learning rate generalize for various types of learning models.
Table 7.
Correspondence of significant amine compound SLC transporters genes among samples
| Gene name | Chromosome | Description | Discovery sample | Replication sample | Pictures/fMRI sample | Words/pictures sample | AgeCoDe sample |
| SLC6A11 | 3 | GABA transporter 3 | 0.0184 | 0.0167 | 0.0084 | 0.0004 | 0.0707 |
| SLC14A2 | 18 | Urea transporter 2 | 0.0179 | 0.0159 | 0.0141 | 0.0049 | 0.0619 |
| SLC6A20 | 3 | X transporter protein 3 | 0.0059 | 0.1808 | 0.0092 | 0.2848 | 0.0024 |
| SLC6A2 | 16 | Norepinephrine transporter | 0.0491 | 0.1976 | 0.0141 | 0.0097 | 0.0162 |
| SLC44A3 | 1 | Choline transporter-like protein 3 | 0.0184 | 0.0529 | 0.0235 | 0.0047 | 0.2009 |
| SLC6A6 | 3 | Taurine transporter | 0.0287 | 0.0371 | 0.0437 | 0.0932 | 0.0235 |
| SLC6A7 | 5 | Brain-specific l-proline transporter | 0.0184 | 0.0275 | 0.0295 | 0.1263 | 0.1680 |
| SLC6A18 | 5 | Sodium channel-like protein | 0.0377 | 0.0482 | 0.0417 | 0.0234 | 0.1872 |
| SLC18A2 | 10 | Vesicular amine transporter 2 | 0.0491 | 0.1651 | 0.0388 | 0.0205 | 0.1940 |
Only genes with significant SNPs in the discovery sample and at least two other samples are shown (the SNP rsIDs and linkage disequilibrium between them are provided in Table S5). The P values of association with relevant phenotypes (Table 5) are provided. Genes are sorted by the product of these values.
For L1CAM interactions, genes encoding fibroblast growth factor receptor (FGFR), ankyrin at node of Ranvier (ANK3), contactins, integrins, and important neural cell adhesion molecules (NRCAM and NCAM1) were significant in multiple samples including the discovery sample (Table 8 and Table S6). These molecules have been implicated in neuronal development and signaling and are essential for synaptic plasticity and its stability in many different brain regions, spatial learning, and memory consolidation (41–44), supporting our discovered association with a memory maintenance-related computational parameter. Some of them, e.g., ANK3 and NCAM1, also have been related to neurodevelopmental and psychiatric disorders (45) and to chronic stress-related cognitive disturbances (41).
Table 8.
Correspondence of significant L1CAM interactions genes between samples
| Gene name | Chromosome | Description | Discovery sample | Replication sample | Pictures/fMRI sample | AgeCoDe sample |
| RPS6KA2 | 6 | Ribosomal S6 kinase 3 | 0.0005 | 0.0177 | 0.0011 | 0.0020 |
| KCNQ3 | 8 | Potassium channel subunit Kv7.3 | 0.0017 | 0.0044 | 0.0020 | 0.0048 |
| DPYSL2 | 8 | Collapsin response mediator protein 2 | 0.0059 | 0.0062 | 0.0247 | 0.0009 |
| ITGA9 | 3 | Integrin alpha-9 | 0.0027 | 0.0008 | 0.0049 | 0.0858 |
| ANK3 | 10 | Ankyrin 3, node of Ranvier | 0.0023 | 0.0197 | 0.0235 | 0.0013 |
| CNTN6 | 3 | Contactin 6 | 0.0009 | 0.0131 | 0.0110 | 0.0235 |
| SH3GL2 | 9 | Endophilin A1 BAR domain | 0.0032 | 0.0031 | 0.0130 | 0.0578 |
| SCN2A | 2 | Sodium channel, type II, α SUBUNIT | 0.0024 | 0.0072 | 0.0676 | 0.0087 |
| FGFR1 | 8 | Fibroblast growth factor receptor 1 | 0.0125 | 0.0018 | 0.0185 | 0.0260 |
| NRCAM | 7 | Neuronal cell adhesion molecule | 0.0004 | 0.0409 | 0.0149 | 0.0578 |
| ITGA1 | 5 | Integrin, alpha 1 | 0.0079 | 0.0345 | 0.0267 | 0.0024 |
| LAMA1 | 18 | Laminin, alpha 1 | 0.0310 | 0.0879 | 0.0318 | 0.0003 |
| EGFR | 7 | Epidermal growth factor receptor | 0.0011 | 0.0328 | 0.0281 | 0.0459 |
| NCAM1 | 11 | Neural cell adhesion molecule 1 | 0.0144 | 0.0037 | 0.0352 | 0.0840 |
| SPTBN1 | 2 | Beta-II spectrin | 0.0273 | 0.0045 | 0.1314 | 0.0162 |
| CSNK2A2 | 16 | Casein kinase II subunit alpha | 0.0059 | 0.0807 | 0.0318 | 0.0190 |
| ANK1 | 8 | Ankyrin 1, erythrocytic | 0.0152 | 0.0114 | 0.0254 | 0.0788 |
| ITGA2 | 5 | Integrin, alpha 2 | 0.0161 | 0.0312 | 0.0352 | 0.0247 |
| CNTN1 | 12 | Contactin 1/F3 | 0.0140 | 0.0290 | 0.0632 | 0.0235 |
| NRP2 | 2 | Neuropilin 2 | 0.0266 | 0.0181 | 0.0335 | 0.0504 |
| NRP1 | 10 | Neuropilin 1 | 0.0240 | 0.0275 | 0.0407 | 0.0823 |
Only genes with significant SNPs in the discovery sample and at least two other samples are shown (the SNP rsIDs and linkage disequilibrium between them are provided in Table S6). P values of association with relevant phenotypes (Table 5) are provided. Genes are sorted by the product of these values.
Among collagen formation genes significant in both discovery and replication samples, TLL2 was linked to mouse avoidance behavior and human bipolar disorder (46), and ADAMTS14 was linked to multiple sclerosis (47). Collagens are essential for connective tissue and mobility, but they also have been implicated in anxiety- and fear-related behaviors in humans and mice (48–50). Interestingly, we found that the collagen formation gene set was significantly enriched (P = 0.005) in the AgeCoDe elderly sample with Geriatric Depression Scale score (51) as phenotype. Finally, for the transmembrane receptor protein tyrosine kinase activity genes significant in both discovery and replication samples, we did not find references implicating them in emotional memory, anxiety, or affective disorders.
Despite the convergence of our results, particularly for amine compound SLC transporters and L1CAM interactions, the exact processes and gene systems influencing the learning rate and memory maintenance cannot be inferred directly, because GSEA are fundamentally based on gene sets defined in different databases [Reactome, Kyoto Encyclopedia of Genes and Genomes (KEGG), Gene Ontology, BioCarta] that are in the process of being refined and extended. Our use of related behavioral phenotypes rather than computational model parameters in the picture task and the AgeCoDe elderly sample also is a notable limitation. Because a variety of relevant behavioral measures are necessary for fitting nontrivial computational models, it is essential for modelers and experimentalists to collaborate during the process of task design. Furthermore, we note that none of our previously reported GWAS SNPs associated with memory performance, such as those of KIBRA (7) and CTNNBL1 (9), was significantly associated with the model parameters. Memory recall performance and model parameters are not identical phenotypes; thus SNPs significantly associated with a model parameter can be missed when a behavioral measure serves as a starting phenotype, and vice versa.
Functional neuroimaging results revealed a robust negative correlation between the L1CAM interactions genetic score and the differences in activity in left superior and inferior frontal gyri, which have been implicated in the recognition of words and pictures and in working memory in healthy individuals (31, 52–54) and Alzheimer’s patients (55, 56). The superior frontal gyrus has been implicated in cognitive control (57), and the inferior frontal gyrus has been implicated in retrieval attempt and effort (58, 59) and depth of memory (60). Reverse inference analysis using Neurosynth (a database of fMRI coordinates and associated keywords automatically extracted from the literature; 61) showed that peak coordinates of the largest cluster dependent on the L1CAM interactions genetic score (−3, 28, 40) were related mostly to the term “response conflict” (Z-score = 5.03, posterior probability = 0.82). Thus, one explanation for the negative correlation between the differences in the activity in these regions and the genetic score is that these frontal activity patterns reflect increased effort or conflict between different options in the case of poorer recognition performance and less effort/conflict in the case of good memory. The negative correlation between the L1CAM interactions genetic score and the differences in activity in the left superior and inferior frontal gyri also may indicate that the reported regions code specifically for familiarity rather than recollection of pictures, as suggested by dual-process models (62, 63). However, because of the ceiling effect in our picture recognition data (with less than 4% of previously seen pictures rated as new and more than 80% as recollected), the percentages of correctly recollected pictures were strongly inversely correlated with percentages of previously seen pictures rated as familiar (ρSpearman = −0.96). Therefore, based on our data, it was impossible to dissociate recollection and familiarity effectively at either the behavioral or neural level (where contrasts based on familiar pictures were significantly underpowered), which remains an important question for future studies.
In conclusion, by computationally dissecting episodic memory performance into specific components, we were able to identify distinct genetic profiles that underlie specific mental processes of human episodic memory.
Materials and Methods
Participants, Data Preprocessing, and GWAS Quality Control.
We had three samples of healthy, young Swiss subjects: the discovery sample (used for associations with model parameters in the verbal task) consisted of the Zurich subsample (192 males, 514 females, age mean ± SD = 21.92 ± 2.95 y), and the Basel subsample (261 males, 504 females, age 22.47 ± 3.62 y). The Basel words/pictures sample (200 males, 410 females, age 22.48 ± 3.51 y) was used for the direct replication of associations with model parameters in the verbal task and for testing associations with performance in the picture task; the Basel pictures/fMRI sample (404 males, 588 females, age 22.47 ± 3.35 y) was used for testing associations with performance in the picture task and studying their neural correlates with neuroimaging. Participants’ physical and mental health was assessed by standard questionnaires and telephone interviews. Indications for exclusion were the recent use of medications influencing the central nervous system, pregnancy, visual system disorders, hormonal irregularities, a body mass index outside the 18.0–35.0 range (20.0–35.0 for men), recent or regular use of cannabis, excessive use of alcohol, use of other psychoactive drugs, history of mental illness, cancer, brain surgery, and for the fMRI sample, the presence of metal implants and claustrophobia. Written informed consent was obtained from the subjects after a complete description of the study. The ethics committees of the Cantons of Zurich and Basel, Switzerland approved the study protocols.
A total of 930,856 SNPs were genotyped. For association testing, markers with call rate less than 0.95, with minor allele frequency less than 0.05, and with Hardy–Weinberg equilibrium P < 0.05 were excluded. Population stratification was assessed by analyzing all genomewide, array-based autosomal SNPs passing the quality-control (QC) criteria with EIGENSTRAT (64). PCA was applied to each population (Zurich words subsample, Basel words subsample, Basel words/pictures sample, and Basel pictures/fMRI sample) to reduce genetic variation to a few dimensions. For PCA, default parameters were used (i.e., definition of 10 principal components in five iterations; the outlier criterion was 6 SDs). EIGENSTRAT identified a number of individuals deviating from a large, genetically homogenous population cluster: 112 in the Zurich words subsample, 68 in the Basel words subsample, 55 in the Basel words/pictures sample, and 112 in the Basel pictures/fMRI sample. These outliers were removed from further analyses. Because learning and memory abilities deteriorate with age (65), we also excluded the outliers (individuals with age more than 2 SDs from the mean in each population) from further analyses: 18 in the Zurich words subsample, 44 in the Basel words subsample, 29 in the Basel words/pictures sample, and 47 in the Basel pictures/fMRI sample. After outliers based on population stratification and age were excluded, the following numbers of participants remained for the final analysis: 1,239 in the discovery sample (582 in the Zurich words subsample, 657 in the Basel words subsample), 526 in the Basel words/pictures sample (picture data available for 493 subjects), and 835 in the Basel pictures/fMRI sample [picture recognition data available for 822 subjects and fMRI data for 795 subjects (for the remaining subjects the fMRI data were corrupted); working memory data were available for 825 subjects, and fMRI data were available for 797 subjects; picture recognition and working memory data were available for 815 subjects].
GWAS Statistics and GSEA.
All GWAS were run under the assumption of an additive model and using the Spearman rank correlation test. Sex chromosomes were excluded from the analysis. The resulting P values of autosomal SNPs in the GWAS (one list per sample and phenotype) served as input for the GSEA, which was performed using MAGENTA (23). Briefly, the method first maps SNPs to genes (each SNP can be counted toward only one gene) and then assigns each gene an SNP association score (i.e., the maximum SNP P value within ± 0 kb of the annotated gene; 26). By applying a stepwise multiple linear regression, the analysis is corrected for the following confounders: gene size, number of SNPs, number of independent SNPs, number of recombination hotspots, linkage disequilibrium, and genetic distance (measured in centimorgans). Last, a gene set enrichment-like statistical test is applied to determine whether a gene set is enriched for highly ranked P values compared with a gene set of identical size randomly drawn from the genome. Based on recommendations in the literature (23, 26), we used a 75th percentile cutoff to determine the significance of a gene, because that cutoff has the optimal power for weak genetic effects that are expected for complex, polygenic traits. The gene sets used were extracted and curated from the MSigDB v3.1 database (www.broadinstitute.org/gsea/msigdb), including gene sets from different online databases (KEGG, Gene Ontology, BioCarta, and Reactome). We used a gene set size of 20 to 200 genes to avoid both overly narrow and overly broad functional gene set categories (26), resulting in 1,411 gene sets to be analyzed. Furthermore, to reduce bias, genes mapping in the high linkage disequilibrium (LD) extended major histocompatibility complex were excluded from the respective gene sets (66). FDR multiple testing correction was used to correct for the number of gene sets.
Memory Testing—the Verbal Task.
Subjects viewed six series of five semantically unrelated nouns presented at a rate of one word/s with the instruction to learn the words for immediate free recall after each series. The words were taken from the collections of Hager and Hasselhorn (67) and consisted of 10 neutral words such as “angle,” 10 positive words such as “happiness,” and 10 negative words such as “poverty.” The order of words was pseudorandom, with each group of five words containing no more than three words per valence category. In addition, subjects underwent an unexpected delayed free-recall test of the learned words after ∼5 min. The free recall of a word was considered successful only if it was spelled correctly or a with single-letter typo that did not make it become a different valid word. The relevant performance measures (PMs) are described in Table 2. In the Basel words/pictures sample, the picture task was performed more than 3 h after the end of the verbal task. Word arousal ratings were available in the Zurich sample (on a scale of 1–5), and the mean arousal ratings did not differ significantly between positive and negative words: arousal(positive) = 3.41 (mean) ± 0.44 (SEM); arousal(negative) = 4.10 ± 0.16; Student t test; P = 0.16.
Computational Model for the Verbal Task.
To dissociate specific mental processes involved in learning and memory, we used a computational model to describe individual performance in the verbal memory task (21). Depending on how well individuals remember a word, they may or may not try to write it down in the free recall, and if they try, their recall may or may not be correct. The probability that the attempted recall is correct depends on memory strength m of each word as follows: , with sigmoidal steepness s and center of the sigmoid m50% = 1. The decision to attempt the recall of weak memories depends on the participant’s willingness to risk making errors. We modeled this decision-making aspect using the decision threshold β, where recall was attempted for words with memory strength m > β but not for those with m < β. During the encoding, initial memory strength for each word was assigned as m = α × ε + N(0, σ), with α being the learning rate, ε the emotional modulation of memory strength (ε = εneg for negative words, ε = εpos for positive words, ε = 1 for neutral words), and N(0, σ) the Gaussian noise with mean 0 and SD σ, reflecting randomness in learning different words. Because the memory strength of words that have been recalled correctly in the immediate recall is likely to increase through repetition (i.e., the participants writing them down), we multiplied the memory strength m of immediately recalled words by the repetition-based memory improvement c (c≥1). To model forgetting during the 5-min delay, we multiplied all memory strengths by the forgetting rate γ (γ<1). It is important to note that, because no external measure could distinguish the repetition-based memory improvement from the forgetting rate in our experiments, these two parameters were closely related and together reflected memory maintenance.
The described model had eight parameters: α, β, γ, εpos, εneg, s, c, and σ. Because our behavioral phenotype consisted of only eight partly correlated measures per individual, it was impossible to estimate all eight parameters reliably. Because the PCA indicated five substantial components (Fig. S1), we chose to keep five of the eight parameters in the model flexible (different between individuals) and the three remaining ones fixed (same for all individuals). The selection of the most appropriate model (i.e., which parameters would be flexible and which fixed) was performed empirically, based on the corresponding mean goodness-of-fit values. Such selection ensured that closely related parameters, such as c and γ, would not both be flexible, whereas relatively independent parameters, which can account for more variance, would.
Estimation and Evaluation of Best-Fitting Model Parameters.
To estimate the best-fitting model parameters, we computed the expected values of all performance measures (PM1–8; Table 2) as a function of eight model parameters (α, β, γ, εpos, εneg, s, c, σ). We numerically computed integrals over probability distributions of memory strength m, because this method was more efficient and robust than simulating the model with random numbers and computing averages over multiple simulation runs. As a control, we also simulated the model stochastically: The averages of PMs over 100,000 simulations were almost exactly the same as those using the expected value-based method.
To evaluate how well the model with a particular set of parameters fits individual behavioral performance, we used the following goodness-of-fit function (20, 21, 68):
where PMiexp and PMimod are experimental and modeled performance measures of that individual, respectively, and (σiexp)2 is the variance in the experimental data of PMi.
With χ2 as the objective function to minimize, we performed the estimation of best-fitting parameters in several stages (21):
-
i)
Model selection: To determine which five parameters should be flexible, we evaluated all 56 possible five-of-eight combinations. Because of the high computational cost of running 56 full estimation procedures, at this stage we performed only a moderately accurate estimation of the three fixed parameters.
-
ii)
Using the two best models, we performed a more refined estimation of fixed parameters, thereby improving the χ2 values. We note that although the improvements in χ2 values were substantial, they were small compared with the differences between the initial χ2 values of the two best models and those of the other, worse models; therefore, it is very unlikely that any of those other models would become comparatively better with refinement.
-
iii)
For the final refinement, we evaluated the averages of all 210 − 1 = 1,023 combinations of the 10 best parameter sets for each model, thereby further improving the χ2 values. Finally, parameter sets from the model with the best goodness-of-fit were used for GWAS.
In all parameter estimation steps the search was performed in the following ranges: (α, β, εpos, εneg, σ) ∈ (0.3, 3.5), c ∈ (1, 4.2), γ ∈ (0, 0.8), and s ∈ (0, 16). In choosing the ranges we balanced two partially opposing aims: to keep them as similar as possible to avoid bias to the estimation results and to keep them as close as possible to the likely distribution of each parameter to maximize estimation accuracy. The default range of 0.3–3.5 ensured that fewer than 1% of estimated parameter values were near the boundaries. Other ranges were used either because of fundamental constraints (c ≥1 and γ<1) or because the likely spread of parameter values was very different from the default. To evaluate how well the model fits individual data, we used the χ2 test with ν = 8 − 5 = three degrees of freedom (five flexible parameters and eight PMs). Values of P > 0.05 indicate no statistical difference between modeled and observed PMs, meaning that the model fits the data well.
The Picture Task and Controls.
After receiving general information about the study and giving their informed consent, participants were instructed and trained on the picture task as well as the N-back working memory task. They performed the encoding of pictures for 20 min and then performed the N-back task for 10 min.
In the picture task, stimuli consisted of 72 pictures that were selected from the International Affective Picture System (IAPS; 69) and from in-house standardized picture sets that allowed us to equate the pictures for visual complexity and content (e.g., human presence). Participants were instructed to view the pictures passively and subsequently rate them according to emotional valence/arousal. Pictures received from IAPS were classified according to the IAPS valence rating; the remaining eight neutral pictures were rated based on an in-house valence rating. Positive stimuli were selected initially to match the arousal ratings of negative stimuli based on data from a pilot study in 20 subjects. Examples of pictures are as follows: erotica, sports, and appealing animals for the positive valence; bodily injury, snake, attack scenes for the negative valence; and neutral faces, household objects, and buildings for the neutral condition. On the basis of normative valence scores (from 1 to 9), pictures were assigned to the emotionally negative (2.3 ± 0.6), emotionally neutral (5.0 ± 0.3), and emotionally positive (7.6 ± 0.4) conditions, resulting in 24 pictures for each emotional valence. Four additional pictures showing neutral objects were used to control for primacy and recency effects in memory. Two of these pictures were presented in the beginning and two at the end of the picture task. They were not included in the analysis. In addition, 24 scrambled pictures were used. The background of the scrambled pictures contained the color information of all pictures used in the experiment (except primacy and recency pictures), overlaid with a crystal and distortion filter (Adobe Photoshop CS3). In the foreground, a mostly transparent geometrical object (rectangle or ellipse of different sizes and orientations) was shown. The pictures were presented for 2.5 s in a quasi-randomized order so that no more than four pictures in the same category occurred consecutively. A fixation-cross appeared on the screen for 500 ms before each picture presentation. The stimulus onset time was jittered within 3 s [1 repetition time (TR)] per valence category with regard to the scan onset. After a picture was presented, participants subjectively rated the picture according to valence (negative, neutral, positive) and arousal (large, medium, small) on a three-point scale (self assessment manikin) by pressing a button with a finger of their dominant hand. For scrambled pictures, participants rated the form (vertical, symmetric or horizontal) and size (large, medium, small) of the geometrical object in the foreground. A maximum 6 s was allowed for rating each picture. Participants were not told that they should remember the pictures for later recall.
The participants also performed the 0- and 2-back versions of the N-back task (70). The task consisted of 12 blocks (six 0-back, six 2-back) in which 14 test stimuli (letters) were presented. The 0-back condition required participants to respond to the occurrence of the letter “x” in a sequence of letters (e.g., N-l-X-g ...), and the 2-back condition required subjects to compare the currently presented letter with the penultimate letter to decide whether they are identical or not (e.g., S-f-s-g ...) Performance was recorded as the number of correct responses, and the difference between the 2-back and the 0-back conditions served as a control phenotype of working memory.
After finishing the tasks, participants performed free recall of the pictures, which required them to write down a short description (a few words) of the previously seen pictures. Remembered primacy and recency pictures and training pictures were excluded from the analysis. No time limit was set for this task. Two trained investigators independently rated the descriptions for recall success (interrater reliability >99%). Depending on the computational model parameter with which each gene set was associated, different behavioral phenotypes were used. For the learning rate α, the relevant phenotype was the percentage of pictures remembered in the free recall (Table S1). For the negative emotional modulation parameter εneg, the relevant phenotype was the percentage of negative pictures minus the percentage of neutral pictures remembered in the free recall.
Multilocus Genetic Scores.
To capture the multiallelic effect of gene sets associated with different model parameters, we generated individual multilocus genetic scores for each of these gene sets. The score comprised SNPs from an equal number of genes (i.e., one most significant SNP per gene) that were associated with the relevant phenotype of the picture task (or the control N-back task) in the pictures/fMRI sample. The score was computed by summing up the individual number of reference alleles over all of these SNPs, weighted by the direction of effect on the phenotype with “1” (the reference allele positively correlated with the phenotype) or “−1” (the reference allele negatively correlated with the phenotype). Each missing SNP was assigned the average value of that SNP over all participants of the sample for whom it was not missing. There were 0.41% of SNPs missing for L1CAM interactions, and 0.69% were missing for amine compound SLC transporters. To test the robustness of the fMRI results, we also built a genetic score using two most significant SNPs per gene.
Construction of a Population-Average Anatomical Probabilistic Atlas.
Automatic segmentation of 1,000 subjects’ T1-weighted images, including the participants from the current study, was used to build a population-average probabilistic anatomical atlas. More precisely, each participant's T1-weighted image first was automatically segmented into cortical and subcortical structures using FreeSurfer (version 4.5, surfer.nmr.mgh.harvard.edu; 71). Labeling of the cortical gyri was based on the Desikan–Killiany Atlas (72), yielding 35 regions per hemisphere.
The segmented T1 image then was normalized to the study-specific anatomical template space using the subject’s previously computed warp field and was affine-registered to the MNI space. Nearest-neighbor interpolation was applied to preserve labeling of the different structures. The normalized segmentations finally were averaged across subjects to create a population-average probabilistic atlas. Each voxel of the template consequently could be assigned a probability of belonging to a given anatomical structure, based on the individual information from 1,000 subjects.
Functional Neuroimaging and Picture Recognition.
Participants of the Basel pictures/fMRI sample performed the picture and N-back tasks in the scanner. They were positioned in the scanner after training. The participants received earplugs and headphones to reduce scanner noise. Their heads were fixated in the coil using small cushions, and they were told not to move their heads. Functional MRIs were acquired during the performance of the picture task for ∼20 min, followed by the 10-min N-back task. After that task, participants were taken from the scanner to a different room for free recall of the presented pictures. Pictures were presented in the scanner using MR-compatible LCD goggles (Visuastim XGA; Resonance Technology). Eye correction was used when necessary.
Approximately 80 min after the presentation of the last picture during encoding, participants were repositioned in the scanner and performed a recognition task for 20 min. The recognition task consisted of two sets of stimuli that were either new (i.e., not presented before) or old (i.e., presented during the picture encoding task). Each of the two sets contained 72 pictures (24 pictures of each valence). During recognition, the pictures were presented for 1 s in a quasi-randomized order so that no more than four pictures of the same category appeared consecutively. A fixation cross appeared on the screen for 500 ms before each picture presentation. The stimulus onset time was jittered within 3 s (1 TR) per valence and previously seen/new category with regard to the scan onset. After picture presentation, participants subjectively rated the picture as recollected, familiar, or new by pressing a button with a finger of their dominant hand. Picture rating was possible in a time window of maximum 3 s.
Depending on the computational model parameter with which each gene set was found to be associated, different fMRI contrasts were used. For the learning rate α, we used two encoding contrasts: pictures of all valence categories vs. scrambled controls and subsequently remembered pictures (of all valence categories) vs. subsequently not-remembered pictures; the latter contrast is known as difference due to memory (Dm; 29). For the repetition-based memory improvement c, which reflects memory maintenance, the relevant behavioral phenotype was the percentage of recollected pictures that had been seen previously, and the appropriate fMRI contrast was previously seen pictures vs. new pictures during recognition. Recognition data were used for this parameter because free recall was not performed in the scanner, and memory maintenance processes may not be reflected during the encoding. To ensure that the effects observed using contrast between the previously seen vs. new pictures reflected the episodic memory component of previously seen, not new, pictures, we also used the contrast of previously seen pictures that were recollected vs. those rated as familiar or new. To investigate neural correlates of working memory, the 2-back vs. 0-back contrast was used.
The contrasts described above were calculated individually using a fixed-effects model (first-level analysis). Because we were using such contrasts, we controlled for all factors that were constant between the two conditions, whether of neuronal or nonneuronal origin. The resulting contrast parameters then were used for genotype-dependent analyses in a random effects model (second-level analysis). Specifically, we used a regression model to analyze gene-dose–dependent differences in brain activity, with the multilocus genetic score, based on the related behavioral phenotype, as covariate. We controlled for the effect of sex by including it as a covariate. The significance threshold for score-dependent analysis was set at P < 0.05, FWE-corrected for multiple comparisons in the whole brain. We also performed second-level analysis with the related behavioral phenotype itself as a covariate.
Mediation Analysis.
To test whether the observed effect of the L1CAM interactions gene set on behavioral performance was mediated by the activation of specific brain regions, we ran a whole-brain mediation analysis using the Multilevel Mediation and Moderation toolbox (wagerlab.colorado.edu/wiki/doku.php/help/mediation/m3_mediation_fmri_toolbox; 73). We looked for brain regions whose activation during recognition (the contrast between previously seen vs. new pictures) mediated the association between the multilocus genetic score (called the “predictor”) and recognition performance. To be considered as mediator, a voxel had to satisfy the following criteria: the predictor variable must be related to the mediator (path a); the mediator must be directly related to behavior, controlling for the predictor (path b); the mediation effect must be significant (effect a*b), meaning that the predictor–behavior relationship is reduced significantly by including the mediator in the model. Bootstrapping was used to obtain corresponding P values, and an FDR threshold of q < 0.05 across all effects of interest (a, b, and a*b coefficients) was applied to correct for multiple comparisons. Furthermore, sex was included as covariate.
Because causality between correlated brain activity and behavior cannot be inferred, we also tested using the same procedure to determine whether the association between the multilocus genetic score and differences in brain activity during recognition (the contrast between previously seen vs. new pictures) was mediated by picture recognition.
Sample from the German AgeCoDe Study.
The sample consisted of elderly participants in the German AgeCoDe study, an ongoing primary care-based prospective longitudinal study on the early detection of mild cognitive impairment and dementia established by the German Competence Network Dementia. The sampling frame and sample selection process of the AgeCoDe study have been described in detail previously (74). After the application of the selection criteria (SI Materials and Methods), a total of 1,244 subjects remained in the sample. Sufficient DNA samples for genomewide genotyping were available for 782 subjects. Additionally, 39 subjects were excluded because of sex-check inconsistencies, low call rate, or extreme values of heterozygosity rate, which may indicate a genotyping bias. The final sample comprised 743 subjects (241 males and 502 females; mean age: 79.53 ± 3.21 y). As part of the structured clinical interviews, subjects were presented a list of 10 words from the German version of the CERAD Word List Learning Task (28) three times (presentation per word: 2 s); each trial was presented in a different order. Participants were asked to read each word aloud as it was presented. After each trial, subjects were asked to recall freely as many words as possible. The number of correctly remembered items over the three trials (immediate recall) served as a relevant phenotype for the gene set associated with the learning rate α, whereas the number of correctly recalled items after a 10-min delay (filled with other questions of the structured interview) was relevant for the gene set associated with the repetition-based memory improvement c.
SI Materials and Methods
Array-Based SNP Genotyping.
Blood was drawn by using 2 × 9 mL EDTA tubes (Sarstedt). Saliva was collected with the Oragene DNA sample collection kit (DNA Genotek Inc.). DNA isolation was done according to standard protocols. Samples were processed as described in the Genome-Wide Human SNP Nsp/Sty 6.0 User Guide (Affymetrix). Briefly, the genomic DNA concentration was determined by using a Nano-Drop ND-1000 and was adjusted to 50 ng/µL in water. DNA (250 ng) was digested in parallel with 10 units of StyI and NspI restriction enzymes (New England Biolabs) for 2 h at 37 °C. Enzyme-specific adaptor oligonucleotides then were ligated onto the digested ends with T4 DNA Ligase for 3 h at 16 °C. After adjustment to 100 µL with water, 10 µL of the diluted ligation reactions were subjected to PCR. Three PCR reactions of 100 µL were performed for StyI-digested products, and four PCR reactions were performed for NspI-digested products. PCR was performed with Titanium Taq DNA Polymerase (Clontech) in the presence of 4.5 μM PCR primer 002 (Affymetrix), 350 μM each dNTP (Clontech), 1 M G-C Melt (Clontech), and 1× Titanium Taq PCR Buffer (Clontech). Cycling parameters were as follows: initial denaturation at 94 °C for 3 min, amplification at 94 °C for 30 s, 60 °C for 45 s, and extension at 68 °C for 15 s, repeated a total of 30 times, with a final extension at 68 °C for 7 min. Reactions then were verified to migrate at an average size of 200–1,100 bp using 2% TBE gel electrophoresis. PCR products were combined and purified with the Filter Bottom Plate (Seahorse Bioscience) using Agencourt Magnetic Beads (Beckman Coulter). Purified PCR products were quantified on a Zenyth 200rt microplate reader (Anthos Labtec). On average, 4–5 µg/µL were obtained for each sample. From this stage on, the SNP Nsp/Sty 5.0/6.0 Assay Kit (Affymetrix) was used. Around 250 µg of purified PCR products were fragmented using 0.5 units of DNaseI at 37 °C for 35 min. Fragmentation of the products to an average size of less than 180 bp was verified using 4% Tris-borate-EDTA gel electrophoresis. Following fragmentation, the DNA was end-labeled with 105 units of terminal deoxynucleotidyl transferase at 37 °C for 4 h. The labeled DNA then was hybridized onto a Genome-Wide Human SNP 6.0 Array at 50 °C for 18 h at 60 rpm (Affymetrix GeneChip Hybridization Oven 645, Rotisserie 00-0331). The hybridized array was washed, stained, and scanned according to the manufacturer’s (Affymetrix) instructions using Affymetrix GeneChip Command Console (AGCC version 3.0.1.1214). Generation of SNP calls and Array QC were performed using the command line programs of the Affymetrix Power Tools package (version: apt-1.14.2). According to the manufacturer’s recommendation, Contrast QC was chosen as the QC metric, using the default value of greater or equal than 0.4. The mean call rate for all samples averaged >98.5%. All samples passing QC criteria were subsequently genotyped using the Birdseed (v2) algorithm.
fMRI Data Acquisition and Preprocessing.
Measurements were performed on a Siemens MAGNETOM Verio 3-T whole-body MR unit equipped with a 12-channel head coil. Functional time series were acquired with a single-shot echo-planar sequence using parallel imaging (GRAPPA). We used the following acquisition parameters: echo time (TE) = 35 ms, field of view (FOV) = 22 cm; acquisition matrix = 80 × 80; interpolated to 128 × 128; voxel size: 2.75 × 2.75 × 4 mm3; GRAPPA acceleration factor R = 2.0. Using a midsagittal scout image, 32 contiguous axial slices placed along the anterior–posterior commissure (AC–PC) plane covering the entire brain with a TR = 3,000 ms (α = 82°) were acquired using an ascending interleaved sequence. A high-resolution T1-weighted anatomical image was acquired using a magnetization prepared gradient echo sequence (MPRAGE, TR = 2,000 ms; TE = 3.37 ms; TI = 1,000 ms; flip angle = 8°; 176 slices; FOV= 256 mm).
Preprocessing and data analysis were performed using SPM8 (Statistical Parametric Mapping, Wellcome Trust Centre for Neuroimaging; www.fil.ion.ucl.ac.uk/spm/) implemented in MATLAB R2011b (MathWorks Inc.). Volumes were slice-time corrected to the first slice and realigned using the “register to mean” option. A mean image was generated from the realigned series and coregistered to the structural image, ensuring that functional and structural images were spatially aligned.
The functional images and the structural images were spatially normalized by applying DARTEL, which leads to an improved registration between subjects. Normalization incorporated the following steps: (i) Structural images of each subject were segmented using the “New Segment” procedure in SPM8. (ii). The resulting gray- and white-matter images were used to derive a study-specific group template. The template was computed from a large population of 1,000 subjects that included the 795 subjects from this study. (iii) An affine transformation was applied to map the group template to MNI space. (iv) Subject-to-template and template-to-MNI transformations were combined to map the functional images to MNI space. The functional images were smoothed with an isotropic 8-mm FWHM Gaussian filter.
For each subject, analyses were conducted in the framework of the general linear model (GLM). Regressors modeling the onsets and duration of stimulus events were convolved with a canonical hemodynamic response function. More precisely, the model comprised regressors for button presses modeled as stick/delta functions, picture presentations modeled with an epoch/boxcar function (duration: encoding: 2.5 s; retrieval: 1 s), and rating scales modeled with an epoch/boxcar function of variable duration (depending on when the subsequent button press occurred). Serial correlations were removed using a first-order autoregressive model, and a high-pass filter (128 s) was applied to remove low-frequency noise. Six movement parameters were also entered as nuisance covariates. Pictures accounting for possible primacy and recency effects were excluded from statistical analysis.
fMRI Group Statistics.
EPI sequences suffer from signal loss in the presence of magnetic field inhomogeneities that can occur close to air–tissue boundaries. The normalization procedure applied in DARTEL accurately transforms both voxels with signal and voxels with signal loss to MNI space. In SPM8, signal loss at an MNI coordinate in a functional image of only one subject leads to the exclusion of the voxel at this coordinate from the group-level analysis. Therefore, the probability of a voxel being excluded increases with sample size. GLM Flex circumvents this problem by allowing a variable number of subjects at each voxel (Martinos Center, Massachusetts General Hospital; mrtools.mgh.harvard.edu). The minimum number of subjects per voxel was set to 400.
German AgeCoDe Sample: Selection of Participants.
Participants were recruited between January 2003 and November 2004 in six German study centers (Bonn, Duesseldorf, Hamburg, Leipzig, Mannheim, and Munich) via general practitioners (GPs) connected with the respective study sites. Inclusion criteria were age of 75 y or older, absence of dementia (according to the GP’s judgment), and at least one contact with the GP within the last 12 mo. Exclusion criteria were GP consultations by home visits only, residence in a nursing home, presence of a severe illness with an anticipated fatal outcome within 3 mo, insufficient German language abilities, deafness or blindness, lack of ability to provide an informed consent, and status as being only an occasional patient of the participating GP. A total of 3,327 subjects were contacted successfully and assessed with structured clinical interviews at their homes. A total of 110 individuals were excluded after the first interview because of the presence of dementia or an actual age below 75 y (falsely classified as age 75 y or older in the sample-selection process). For the present analyses, data from baseline and three follow-up measurements at 18-mo intervals were available. In a primary care-based sample of older individuals, conditions can be present that affect cognition and the reliability of neuropsychological tests. To generate a sample of healthy elderly individuals, we further used the following selection criteria at baseline: age between 75 and 90 y, German as native language, at least a school-leaving certificate, absence of severe hearing or vision impairments, absence of insufficient test motivation as judged by the interviewer, absence of disturbing factors during neuropsychological testing, and absence of all the following comorbid conditions: Parkinson’s disease, epilepsy, alcohol abuse, stroke, multiple sclerosis, evidence of depression (a score of 6 or higher on the Geriatric Depression Scale), traumatic brain injury with unconsciousness of more than 30 min, visible neurological malfunctions, and dementia according to Diagnostic and Statistical Manual of Mental Disorders-IV criteria. In addition, we excluded subjects who converted to dementia before the third follow-up (i.e., within 4.5 y after baseline) or for whom neuropsychological test data were not available at baseline and all follow-up visits.
Acknowledgments
We thank the AgeCoDe study group for their help in data collection: Heinz-Harald Abholz, Christian Brettschneider, Cadja Bachmann, Horst Bickel, Wolfgang Blank, Hendrik van den Bussche, Sandra Eifflaender-Gorfer, Marion Eisele, Annette Ernst, Angela Fuchs, Kathrin Heser, Hanna Kaduszkiewicz, Teresa Kaufeler, Mirjam Köhler, Hans-Helmut König, Alexander Koppara, Carolin Lange, Tobias Luck, Melanie Luppa, Manfred Mayer, Edelgard Mösch, Julia Olbrich, Michael Pentzek, Jana Prokein, Anna Schumacher, Janine Stein, Susanne Steinmann, Franziska Tebarth, Klaus Weckbecker, Dagmar Weeg, Jochen Werle, Siegfried Weyerer, Birgitt Wiese, and Thomas Zimmermann. This work was funded by Swiss National Science Foundation Sinergia Grants CRSI33_130080 (to D.J.-F.d.Q.) and CRSII1_136227 (to A.P.), by the European Community‘s Seventh Framework Programme (FP7/2007–2013) under Grant Agreement 602450 (IMAGEMEND), by the German Research Network on Dementia (German Federal Ministry of Education and Research Grants 01GI0102, 01GI0420, 01GI0422, 01GI0423, 01GI0429, 01GI0431, 01GI0433, and 01GI0434), and by the German Research Network on Degenerative Dementia (German Federal Ministry of Education and Research Grants 01GI0710, 01GI0711, 01GI0712, 01GI0713, 01GI0714, 01GI0715, and 01GI0716).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1500860112/-/DCSupplemental.
References
- 1.Green AE, et al. Using genetic data in cognitive neuroscience: From growing pains to genuine insights. Nat Rev Neurosci. 2008;9(9):710–720. doi: 10.1038/nrn2461. [DOI] [PubMed] [Google Scholar]
- 2.Papassotiropoulos A, de Quervain DJ. Genetics of human episodic memory: Dealing with complexity. Trends Cogn Sci. 2011;15(9):381–387. doi: 10.1016/j.tics.2011.07.005. [DOI] [PubMed] [Google Scholar]
- 3.Egan MF, et al. The BDNF val66met polymorphism affects activity-dependent secretion of BDNF and human memory and hippocampal function. Cell. 2003;112(2):257–269. doi: 10.1016/s0092-8674(03)00035-7. [DOI] [PubMed] [Google Scholar]
- 4.Kölsch H, et al. Gene polymorphisms in prodynorphin (PDYN) are associated with episodic memory in the elderly. J Neural Transm. 2009;116(7):897–903. doi: 10.1007/s00702-009-0238-5. [DOI] [PubMed] [Google Scholar]
- 5.de Quervain DJ, et al. A deletion variant of the alpha2b-adrenoceptor is related to emotional memory in Europeans and Africans. Nat Neurosci. 2007;10(9):1137–1139. doi: 10.1038/nn1945. [DOI] [PubMed] [Google Scholar]
- 6.de Quervain DJ, et al. A functional genetic variation of the 5-HT2a receptor affects human memory. Nat Neurosci. 2003;6(11):1141–1142. doi: 10.1038/nn1146. [DOI] [PubMed] [Google Scholar]
- 7.Papassotiropoulos A, et al. Common Kibra alleles are associated with human memory performance. Science. 2006;314(5798):475–478. doi: 10.1126/science.1129837. [DOI] [PubMed] [Google Scholar]
- 8.Milnik A, et al. Association of KIBRA with episodic and working memory: A meta-analysis. Am J Med Genet B Neuropsychiatr Genet. 2012;159B(8):958–969. doi: 10.1002/ajmg.b.32101. [DOI] [PubMed] [Google Scholar]
- 9.Papassotiropoulos A, et al. A genome-wide survey and functional brain imaging study identify CTNNBL1 as a memory-related gene. Mol Psychiatry. 2013;18(2):255–263. doi: 10.1038/mp.2011.148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kandel ER. The molecular biology of memory storage: A dialogue between genes and synapses. Science. 2001;294(5544):1030–1038. doi: 10.1126/science.1067020. [DOI] [PubMed] [Google Scholar]
- 11.McGaugh JL. Memory--a century of consolidation. Science. 2000;287(5451):248–251. doi: 10.1126/science.287.5451.248. [DOI] [PubMed] [Google Scholar]
- 12.Panizzon MS, et al. Genetic architecture of learning and delayed recall: A twin study of episodic memory. Neuropsychology. 2011;25(4):488–498. doi: 10.1037/a0022569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Luksys G, Sandi C. Neural mechanisms and computations underlying stress effects on learning and memory. Curr Opin Neurobiol. 2011;21(3):502–508. doi: 10.1016/j.conb.2011.03.003. [DOI] [PubMed] [Google Scholar]
- 14.Corrado G, Doya K. Understanding neural coding through the model-based analysis of decision making. J Neurosci. 2007;27(31):8178–8180. doi: 10.1523/JNEUROSCI.1590-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schweighofer N, et al. Low-serotonin levels increase delayed reward discounting in humans. J Neurosci. 2008;28(17):4528–4532. doi: 10.1523/JNEUROSCI.4982-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Daw ND, O’Doherty JP, Dayan P, Seymour B, Dolan RJ. Cortical substrates for exploratory decisions in humans. Nature. 2006;441(7095):876–879. doi: 10.1038/nature04766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Behrens TE, Woolrich MW, Walton ME, Rushworth MF. Learning the value of information in an uncertain world. Nat Neurosci. 2007;10(9):1214–1221. doi: 10.1038/nn1954. [DOI] [PubMed] [Google Scholar]
- 18.Frank MJ, Doll BB, Oas-Terpstra J, Moreno F. Prefrontal and striatal dopaminergic genes predict individual differences in exploration and exploitation. Nat Neurosci. 2009;12(8):1062–1068. doi: 10.1038/nn.2342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Frank MJ, Moustafa AA, Haughey HM, Curran T, Hutchison KE. Genetic triple dissociation reveals multiple roles for dopamine in reinforcement learning. Proc Natl Acad Sci USA. 2007;104(41):16311–16316. doi: 10.1073/pnas.0706111104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Luksys G, Gerstner W, Sandi C. Stress, genotype and norepinephrine in the prediction of mouse behavior using reinforcement learning. Nat Neurosci. 2009;12(9):1180–1186. doi: 10.1038/nn.2374. [DOI] [PubMed] [Google Scholar]
- 21.Luksys G, et al. BAIAP2 is related to emotional modulation of human memory strength. PLoS One. 2014;9(1):e83707. doi: 10.1371/journal.pone.0083707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang K, Li M, Hakonarson H. Analysing biological pathways in genome-wide association studies. Nat Rev Genet. 2010;11(12):843–854. doi: 10.1038/nrg2884. [DOI] [PubMed] [Google Scholar]
- 23.Segrè AV, Groop L, Mootha VK, Daly MJ, Altshuler D. DIAGRAM Consortium MAGIC investigators Common inherited variation in mitochondrial genes is not enriched for associations with type 2 diabetes or related glycemic traits. PLoS Genet. 2010;6(8):e1001058. doi: 10.1371/journal.pgen.1001058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Holmans P, et al. Wellcome Trust Case-Control Consortium Gene ontology analysis of GWA study data sets provides insights into the biology of bipolar disorder. Am J Hum Genet. 2009;85(1):13–24. doi: 10.1016/j.ajhg.2009.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.O’Dushlaine C, et al. International Schizophrenia Consortium Molecular pathways involved in neuronal cell adhesion and membrane scaffolding contribute to schizophrenia and bipolar disorder susceptibility. Mol Psychiatry. 2011;16(3):286–292. doi: 10.1038/mp.2010.7. [DOI] [PubMed] [Google Scholar]
- 26.Heck A, et al. Converging genetic and functional brain imaging evidence links neuronal excitability to working memory, psychiatric disease, and brain activity. Neuron. 2014;81(5):1203–1213. doi: 10.1016/j.neuron.2014.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Papassotiropoulos A, et al. Human genome-guided identification of memory-modulating drugs. Proc Natl Acad Sci USA. 2013;110(46):E4369–E4374. doi: 10.1073/pnas.1314478110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Welsh KA, et al. The consortium to establish a registry for Alzheimer’s disease (CERAD). Part V. A normative study of the neuropsychological battery. Neurology. 1994;44(4):609–614. doi: 10.1212/wnl.44.4.609. [DOI] [PubMed] [Google Scholar]
- 29.Paller KA, Wagner AD. Observing the transformation of experience into memory. Trends Cogn Sci. 2002;6(2):93–102. doi: 10.1016/s1364-6613(00)01845-3. [DOI] [PubMed] [Google Scholar]
- 30.Frankland PW, Bontempi B. The organization of recent and remote memories. Nat Rev Neurosci. 2005;6(2):119–130. doi: 10.1038/nrn1607. [DOI] [PubMed] [Google Scholar]
- 31.Leveroni CL, et al. Neural systems underlying the recognition of familiar and newly learned faces. J Neurosci. 2000;20(2):878–886. doi: 10.1523/JNEUROSCI.20-02-00878.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang K, et al. Diverse genome-wide association studies associate the IL12/IL23 pathway with Crohn Disease. Am J Hum Genet. 2009;84(3):399–405. doi: 10.1016/j.ajhg.2009.01.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Karlsson EK, et al. Natural selection in a bangladeshi population from the cholera-endemic ganges river delta. Sci Transl Med. 2013;5(192):192ra86. doi: 10.1126/scitranslmed.3006338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mooney MA, Nigg JT, McWeeney SK, Wilmot B. Functional and genomic context in pathway analysis of GWAS data. Trends Genet. 2014;30(9):390–400. doi: 10.1016/j.tig.2014.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lee TS, et al. GAT1 and GAT3 expression are differently localized in the human epileptogenic hippocampus. Acta Neuropathol. 2006;111(4):351–363. doi: 10.1007/s00401-005-0017-9. [DOI] [PubMed] [Google Scholar]
- 36.Biederman J, et al. Sexually dimorphic effects of four genes (COMT, SLC6A2, MAOA, SLC6A4) in genetic associations of ADHD: A preliminary study. Am J Med Genet B Neuropsychiatr Genet. 2008;147B(8):1511–1518. doi: 10.1002/ajmg.b.30874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lin Z, et al. High regulatability favors genetic selection in SLC18A2, a vesicular monoamine transporter essential for life. FASEB J. 2010;24(7):2191–2200. doi: 10.1096/fj.09-140368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hasselmo ME. The role of acetylcholine in learning and memory. Curr Opin Neurobiol. 2006;16(6):710–715. doi: 10.1016/j.conb.2006.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Doya K. Metalearning and neuromodulation. Neural Netw. 2002;15(4-6):495–506. doi: 10.1016/s0893-6080(02)00044-8. [DOI] [PubMed] [Google Scholar]
- 40.Set E, et al. Dissociable contribution of prefrontal and striatal dopaminergic genes to learning in economic games. Proc Natl Acad Sci USA. 2014;111(26):9615–9620. doi: 10.1073/pnas.1316259111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sandi C. Stress, cognitive impairment and cell adhesion molecules. Nat Rev Neurosci. 2004;5(12):917–930. doi: 10.1038/nrn1555. [DOI] [PubMed] [Google Scholar]
- 42.Maness PF, Schachner M. Neural recognition molecules of the immunoglobulin superfamily: Signaling transducers of axon guidance and neuronal migration. Nat Neurosci. 2007;10(1):19–26. doi: 10.1038/nn1827. [DOI] [PubMed] [Google Scholar]
- 43.Lüthi A, Laurent JP, Figurov A, Muller D, Schachner M. Hippocampal long-term potentiation and neural cell adhesion molecules L1 and NCAM. Nature. 1994;372(6508):777–779. doi: 10.1038/372777a0. [DOI] [PubMed] [Google Scholar]
- 44.Puzzo D, et al. F3/Contactin promotes hippocampal neurogenesis, synaptic plasticity, and memory in adult mice. Hippocampus. 2013;23(12):1367–1382. doi: 10.1002/hipo.22186. [DOI] [PubMed] [Google Scholar]
- 45.Iqbal Z, et al. Homozygous and heterozygous disruptions of ANK3: At the crossroads of neurodevelopmental and psychiatric disorders. Hum Mol Genet. 2013;22(10):1960–1970. doi: 10.1093/hmg/ddt043. [DOI] [PubMed] [Google Scholar]
- 46.de Mooij-van Malsen JG, et al. Cross-species genetics converge to TLL2 for mouse avoidance behavior and human bipolar disorder. Genes Brain Behav. 2013;12(6):653–657. doi: 10.1111/gbb.12055. [DOI] [PubMed] [Google Scholar]
- 47.Goertsches R, Comabella M, Navarro A, Perkal H, Montalban X. Genetic association between polymorphisms in the ADAMTS14 gene and multiple sclerosis. J Neuroimmunol. 2005;164(1-2):140–147. doi: 10.1016/j.jneuroim.2005.04.005. [DOI] [PubMed] [Google Scholar]
- 48.Bulbena A, Gago J, Sperry L, Bergé D. The relationship between frequency and intensity of fears and a collagen condition. Depress Anxiety. 2006;23(7):412–417. doi: 10.1002/da.20160. [DOI] [PubMed] [Google Scholar]
- 49.Kakoi C, Udo H, Matsukawa T, Ohnuki K. Collagen peptides enhance hippocampal neurogenesis and reduce anxiety related behavior in mice. Biomed Res. 2012;33(5):273–279. doi: 10.2220/biomedres.33.273. [DOI] [PubMed] [Google Scholar]
- 50.Mallorquí-Bagué N, et al. Neuroimaging and psychophysiological investigation of the link between anxiety, enhanced affective reactivity and interoception in people with joint hypermobility. Front Psychol. 2014;5:1162. doi: 10.3389/fpsyg.2014.01162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mcnally RJ. Memory and anxiety disorders. Philos Trans R Soc Lond B Biol Sci. 1997;352(1362):1755–1759. doi: 10.1098/rstb.1997.0158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rugg MD, Fletcher PC, Chua PM, Dolan RJ. The role of the prefrontal cortex in recognition memory and memory for source: An fMRI study. Neuroimage. 1999;10(5):520–529. doi: 10.1006/nimg.1999.0488. [DOI] [PubMed] [Google Scholar]
- 53.Kuchinke L, Fritzemeier S, Hofmann MJ, Jacobs AM. Neural correlates of episodic memory: Associative memory and confidence drive hippocampus activations. Behav Brain Res. 2013;254:92–101. doi: 10.1016/j.bbr.2013.04.035. [DOI] [PubMed] [Google Scholar]
- 54.Feredoes E, Postle BR. Prefrontal control of familiarity and recollection in working memory. J Cogn Neurosci. 2010;22(2):323–330. doi: 10.1162/jocn.2009.21252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Xu G, et al. The influence of parental history of Alzheimer’s disease and apolipoprotein E epsilon4 on the BOLD signal during recognition memory. Brain. 2009;132(Pt 2):383–391. doi: 10.1093/brain/awn254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yetkin FZ, Rosenberg RN, Weiner MF, Purdy PD, Cullum CM. FMRI of working memory in patients with mild cognitive impairment and probable Alzheimer’s disease. Eur Radiol. 2006;16(1):193–206. doi: 10.1007/s00330-005-2794-x. [DOI] [PubMed] [Google Scholar]
- 57.Egner T, Hirsch J. The neural correlates and functional integration of cognitive control in a Stroop task. Neuroimage. 2005;24(2):539–547. doi: 10.1016/j.neuroimage.2004.09.007. [DOI] [PubMed] [Google Scholar]
- 58.Badre D, Wagner AD. Left ventrolateral prefrontal cortex and the cognitive control of memory. Neuropsychologia. 2007;45(13):2883–2901. doi: 10.1016/j.neuropsychologia.2007.06.015. [DOI] [PubMed] [Google Scholar]
- 59.Taylor SF, et al. A functional neuroimaging study of motivation and executive function. Neuroimage. 2004;21(3):1045–1054. doi: 10.1016/j.neuroimage.2003.10.032. [DOI] [PubMed] [Google Scholar]
- 60.Iidaka T, Matsumoto A, Nogawa J, Yamamoto Y, Sadato N. Frontoparietal network involved in successful retrieval from episodic memory. Spatial and temporal analyses using fMRI and ERP. Cereb Cortex. 2006;16(9):1349–1360. doi: 10.1093/cercor/bhl040. [DOI] [PubMed] [Google Scholar]
- 61.Yarkoni T, Poldrack RA, Nichols TE, Van Essen DC, Wager TD. Large-scale automated synthesis of human functional neuroimaging data. Nat Methods. 2011;8(8):665–670. doi: 10.1038/nmeth.1635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Yonelinas AP. Components of episodic memory: The contribution of recollection and familiarity. Philos Trans R Soc Lond B Biol Sci. 2001;356(1413):1363–1374. doi: 10.1098/rstb.2001.0939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wixted JT, Mickes L. A continuous dual-process model of remember/know judgments. Psychol Rev. 2010;117(4):1025–1054. doi: 10.1037/a0020874. [DOI] [PubMed] [Google Scholar]
- 64.Price AL, et al. Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet. 2006;38(8):904–909. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- 65.Nilsson LG. Memory function in normal aging. Acta Neurol Scand Suppl. 2003;179:7–13. doi: 10.1034/j.1600-0404.107.s179.5.x. [DOI] [PubMed] [Google Scholar]
- 66.Horton R, et al. Gene map of the extended human MHC. Nat Rev Genet. 2004;5(12):889–899. doi: 10.1038/nrg1489. [DOI] [PubMed] [Google Scholar]
- 67.Hager W, Hasselhorn M. Handbuch Deutschsprachiger Wortnormen. Hogrefe; Göttingen, Germany: 1994. p. 441. [Google Scholar]
- 68.Press WH, Flannery BP, Teukolsky SA, Vetterling WT. Numerical Recipes in C: The Art of Scientific Computing. Cambridge Univ Press; Cambridge, UK: 1992. p. 994. [Google Scholar]
- 69.Lang PJ, Bradley MM, Cuthbert BN. International Affective Picture System (IAPS): Affective Ratings of Pictures and Instruction Manual. University of Florida; Gainesville, FL: 2008. [Google Scholar]
- 70.Gevins A, Cutillo B. Spatiotemporal dynamics of component processes in human working memory. Electroencephalogr Clin Neurophysiol. 1993;87(3):128–143. doi: 10.1016/0013-4694(93)90119-g. [DOI] [PubMed] [Google Scholar]
- 71.Fischl B, et al. Whole brain segmentation: Automated labeling of neuroanatomical structures in the human brain. Neuron. 2002;33(3):341–355. doi: 10.1016/s0896-6273(02)00569-x. [DOI] [PubMed] [Google Scholar]
- 72.Desikan RS, et al. An automated labeling system for subdividing the human cerebral cortex on MRI scans into gyral based regions of interest. Neuroimage. 2006;31(3):968–980. doi: 10.1016/j.neuroimage.2006.01.021. [DOI] [PubMed] [Google Scholar]
- 73.Wager TD, Davidson ML, Hughes BL, Lindquist MA, Ochsner KN. Prefrontal-subcortical pathways mediating successful emotion regulation. Neuron. 2008;59(6):1037–1050. doi: 10.1016/j.neuron.2008.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Luck T, et al. AgeCoDe group Mild cognitive impairment in general practice: Age-specific prevalence and correlate results from the German study on ageing, cognition and dementia in primary care patients (AgeCoDe) Dement Geriatr Cogn Disord. 2007;24(4):307–316. doi: 10.1159/000108099. [DOI] [PubMed] [Google Scholar]