Significance
As one of the most important growth-promoting hormones, auxin regulates many aspects of plant growth and development. Understanding auxin action has long been a challenging task because of the complexity of the hormone transport involved in auxin response. Despite tremendous progress made in Arabidopsis, auxin response and transport are poorly understood in crop plants, which impedes the application of hormone knowledge in agricultural improvement. This study not only identifies a novel positive regulator of plant growth in rice and demonstrates its significant role in improving seed size and grain yield, it also illustrates the specific involvement of the plasma membrane-associated protein in regulating auxin response and transport, thus illuminating a new strategy for enhancing crop productivity.
Keywords: grain size, Big Grain1, auxin, biomass, grain yield
Abstract
Grain size is one of the key factors determining grain yield. However, it remains largely unknown how grain size is regulated by developmental signals. Here, we report the identification and characterization of a dominant mutant big grain1 (Bg1-D) that shows an extra-large grain phenotype from our rice T-DNA insertion population. Overexpression of BG1 leads to significantly increased grain size, and the severe lines exhibit obviously perturbed gravitropism. In addition, the mutant has increased sensitivities to both auxin and N-1-naphthylphthalamic acid, an auxin transport inhibitor, whereas knockdown of BG1 results in decreased sensitivities and smaller grains. Moreover, BG1 is specifically induced by auxin treatment, preferentially expresses in the vascular tissue of culms and young panicles, and encodes a novel membrane-localized protein, strongly suggesting its role in regulating auxin transport. Consistent with this finding, the mutant has increased auxin basipetal transport and altered auxin distribution, whereas the knockdown plants have decreased auxin transport. Manipulation of BG1 in both rice and Arabidopsis can enhance plant biomass, seed weight, and yield. Taking these data together, we identify a novel positive regulator of auxin response and transport in a crop plant and demonstrate its role in regulating grain size, thus illuminating a new strategy to improve plant productivity.
Because it is one of the most important staple food crops cultivated worldwide, improvement of grain yield is a major focus of rice-breeding programs (1). Grain size is one of the determining factors of grain yield (2, 3). A number of quantitative trait loci (QTLs) controlling rice grain size have been identified in recent years (4–11). However, functional mechanisms of these genes remain largely unknown. Because QTLs usually have important functions in determining grain size, many of them have been widely selected in breeding processes or existed in modern elite varieties, and a certain QTL could be only applicable in certain varieties (12). Thus, exploration of new grain size-associated genes and elucidation of their functional mechanisms have great significance for further improvement of rice yield (12).
Seed size, as well as other organ size, is controlled by various plant hormones, such as auxin, brassinosteroid, and cytokinin (10, 13, 14). A number of studies have demonstrated that auxin plays a vital role in organ size determination by affecting cell division, cell expansion, and differentiation (15–17). Auxin exists predominantly as indole-3-acetic acid (IAA) in plants, and genetic studies of its biosynthetic genes in Arabidopsis have demonstrated that IAA regulates many aspects of plant growth and development, including stem elongation, lateral branching, vascular development, and tropic growth responses (18, 19). Combined with biochemical studies, the tryptophan (Trp)-dependent IAA biosynthesis pathway has been clearly established involving the YUCCA family flavin monooxgenases (20). Importantly, the two-step pathway is highly conserved throughout the plant kingdom (21). Until very recently, the Trp-independent auxin biosynthetic pathway was elucidated as contributing to early embryogenesis in Arabidopsis (22). Primary auxin signaling is a rapid process initiated from the hormone perception by receptor TIR1, an F-box protein, followed by degradation of the negative regulator AUX/IAA proteins, and further release the downstream auxin response factors (ARFs) (23–26). However, how the ARFs work in plants remains elusive. Auxin transport, generally referring to the cell-to-cell transportation of the hormone directed basipetally from shoots to roots in vascular tissues, plays a critical role in auxin response (18). The transport involves a number of membrane-associated proteins, such as PINs (protein inhibitor of nNOS), AUX1 (AUXIN TRANSPORTER PROTEIN 1), and ABCBs (ATP-BINDING CASSETTE, SUB-FAMILY B PROTEINS) as efflux or influx carriers (27–30). Disruption of auxin transport induced by either gene mutations or chemical inhibitor treatment will lead to diverse development defects, such as decreased lateral organ initiation and defective tropic growth responses (27, 31–34).
In this study, we identify a rice mutant, named big grain1-D (Bg1-D) because it is a dominant mutant having extralarge grain size. BG1 encodes a novel plasma membrane-associated protein, and is specifically induced by auxin treatment. We show that BG1 is a new positive regulator of auxin response involved in auxin transport, and demonstrate that manipulation of BG1 expression can greatly improve grain size and plant productivity.
Results
Bg1-D Shows Increased Grain Size Phenotype.
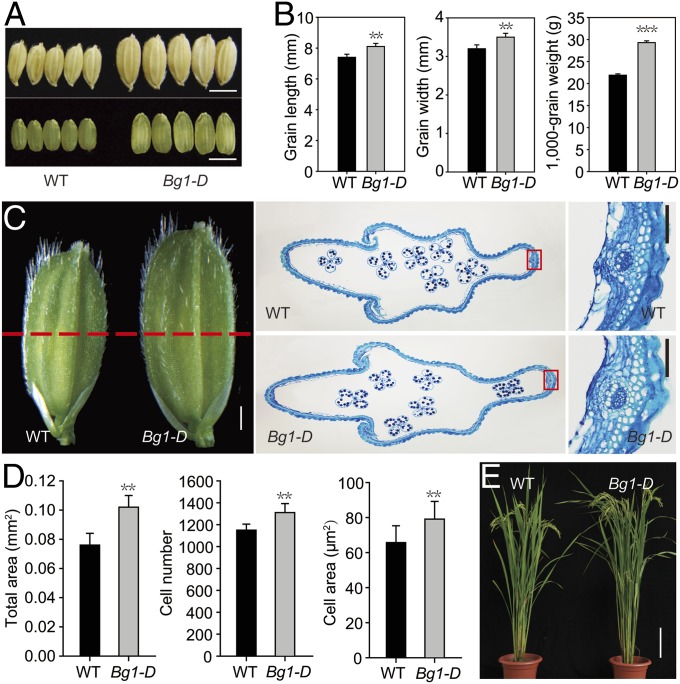
We have identified a number of rice mutants with altered grain size from our T-DNA insertion population. Among them, Bg1-D has the most obviously increased grain size (Fig. 1A). Compared with WT (Nipponbare, Oryza sativa L. ssp. japonica), the 1,000-grain weight of Bg1-D has increased about 33.8%, with the grain length and grain width increased about 15.2% and 17.0%, respectively (Fig. 1B). Observation of spikelet hull by scanning electron microscopy showed that the epidermis cells of both palea and lemma in Bg1-D are much longer than that of the WT (Fig. S1). Careful examination of the hull cross-section before flowering revealed significant increases of both number and area of the parenchyma cells in the Bg1-D mutant (Fig. 1 C and D). Consistent with this, a number of genes associated with cell cycle and cell expansion were up-regulated in the panicles of the Bg1-D mutant (Fig. S2).
Fig. 1.
Phenotype of Bg1-D mutant. (A) Grain morphology of WT and Bg1-D. (Scale bar, 0.5 cm.) (B) Statistical data of the grain length, grain width, and 1,000-grain weight in WT and Bg1-D. (C) Cross-sections of the spikelet hulls of WT and Bg1-D. Dashed line indicates the position of the cross-section. (Scale bar, 1 mm.) Magnified views in the boxes are shown. (Scale bar, 0.1 mm.) (D) Statistical data of the total area, cell number and cell area in the outer parenchyma layer of the spikelet hulls of WT and Bg1-D. (E) Gross morphology of 4-mo-old plants of WT and Bg1-D grown in paddy field. (Scale bar, 20 cm.) Means ± SD are given in B (n = 20) and D (n = 15). **P < 0.01. ***P < 0.001 (t test).
Despite the greatly enlarged grain size, Bg1-D also exhibits enhanced growth of other tissues at both the vegetative and reproductive stages (Fig. 1E and Fig. S3). One-week-old seedlings of the Bg1-D mutant have obviously increased length of leaf and root compared with WT (Fig. S3 A–D). When plants enter reproductive stage, the leaves of Bg1-D are longer and wider than those of the WT plants, and the Bg1-D also has increased plant height (Fig. 1E, Fig. S3E, and Table S1). In addition, the Bg1-D panicles are much larger than those of the WT (Fig. S3 F and G).
We also compared the fresh and dry weights of seeds between the Bg1-D and WT, and found that the weight of Bg1-D endosperms was markedly increased compared with that of WT at 15 days after fertilization, and the difference reached a maximum at 25 days after fertilization (Fig. S4). Thus, the significant overall improvement of the grain weight in the Bg1-D mutant is attributed to the increases in both hull size and endosperm volume.
Molecular Cloning of BG1.
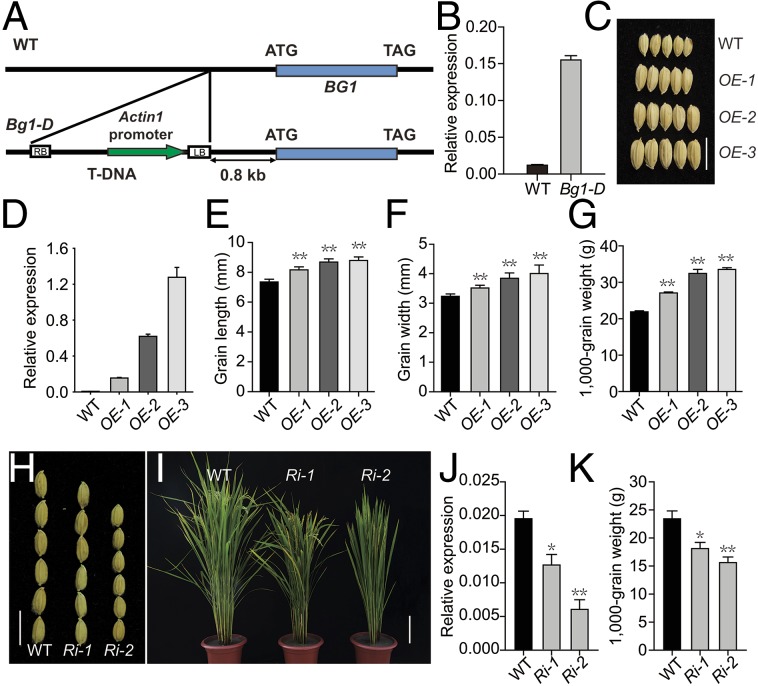
Genetic analyses showed that F1 progenies of the reciprocal crosses between Bg1-D and WT have big grain phenotype (Fig. S5A). In addition, the phenotype of Bg1-D cosegregates with the T-DNA insertion in the F2 population, and the segregation ratio is ∼3:1 (Fig. S5B), suggesting that the mutant phenotype is caused by a T-DNA insertion, which activates the expression of BG1. We cloned the T-DNA 3′-flanking sequence by the site-finding PCR method based on thermal asymmetric interlaced PCR (35, 36). Sequence analysis revealed that a T-DNA was inserted at 0.8 kb upstream of the gene Os03g0175800 (Fig. 2A). Additional gene-expression analysis showed that, whereas other adjacent genes of the insertion site have comparable transcript levels in the mutant and WT, this gene has increased expression ∼10-fold in the Bg1-D mutant (Fig. 2B). Thus, Os03g0175800 is potentially the gene responsible for the Bg1-D mutant phenotypes and was designated as BG1.
Fig. 2.
Cloning of BG1 gene and verification of BG1 function. (A) Schematic map of the T-DNA flanking region in Bg1-D mutant. The T-DNA contains rice ACTIN1 promoter near the left border (LB), which is 0.8 kb upstream of BG1 (Os03g0175800). (B) The expression level of BG1 in WT and Bg1-D. (C) Grain morphology of WT and BG1-overexpressing plants (OEs). (Scale bar, 1 cm.) (D) Expression level of BG1 in WT and OEs. (E–G) Statistical data of the grain length (E), grain width (F), and 1,000-grain weight (G) in WT and OEs. (H) Grain morphology. (Scale bar, 1 cm.) (I) Gross plant morphology. (Scale bar, 20 cm.) (J) BG1 expression level. (K) 1,000-grain weight in WT and BG1-knockdown plants (Ris). Means ± SD are given in E and F (n = 20) and G and K (n = 5) *P < 0.05, **P < 0.01 (t test).
BG1 Is a Novel Positive Regulator That Controls Grain Size.
The full-length cDNA of BG1 was amplified by 5′ RACE, which is 1,190 bp in length and contains an ORF of 933 bp encoding 330 amino acids. To verify that activation of BG1 is responsible for the mutant phenotype, we overexpressed BG1 cDNA under the control of the rice ACTIN1 promoter in WT and found that the BG1-overexpressing plants (BG1-OE) showed obviously increased grain size and other phenotypes, such as enhanced plant height, longer leaves, and larger panicles, which resemble the Bg1-D mutant phenotypes (Fig. 2C and Fig. S6 A and B). Importantly, severity of the phenotypes was apparently correlated with the BG1 expression level; that is, plants with higher BG1 expression have bigger grain size as well as increased 1,000-grain weight (Fig. 2 D–G), demonstrating that activation of BG1 is the cause for the Bg1-D mutant phenotypes.
Suppression of BG1 by RNA interference (BG1-Ri) led to obviously opposite phenotypes, including reduced plant height and decreased panicle length and grain size, as well as reduced 1,000-grain weight (Fig. 2 H–K and Fig. S6 C and D). The phenotype severity is also consistent with the BG1 expression level, suggesting that the native BG1 indeed controls grain size and plant growth in rice.
Molecular Characterization of BG1.
BLAST search of the protein databases discovered a number of orthologous proteins of BG1 in various higher plants. Phylogenetic analysis showed that the proteins can be classified into two subgroups: dicot- and monocot-specific groups (Fig. S7). However, none of them has been functionally characterized, including four Arabidopsis BG1-like proteins. Moreover, no known functional domain in these proteins could be found, suggesting that BG1 is a novel plant-specific regulator controlling organ size.
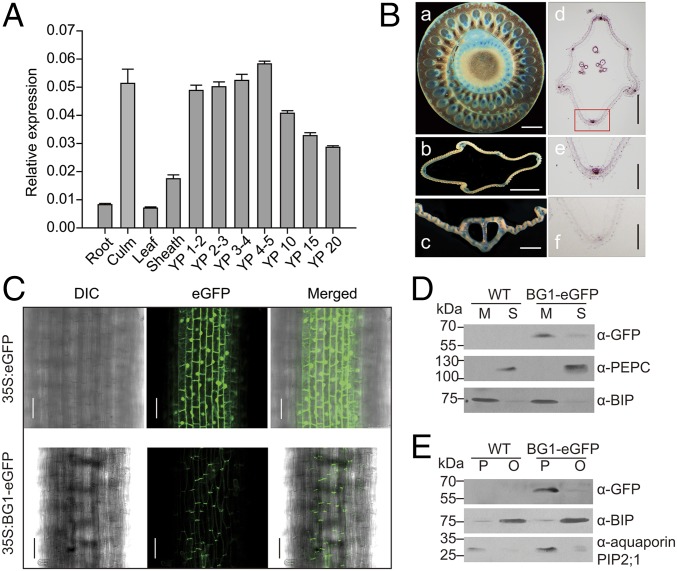
Quantitative RT-PCR analysis showed that BG1 transcripts can be detected in various tissues, but the expression is relatively stronger in culms and panicles, and the level is gradually decreased as the panicle matures (Fig. 3A). We also analyzed the BG1 promoter activity in various tissues of PROBG1:GUS transgenic plants. GUS staining revealed that the BG1 promoter is specifically active in the vascular tissues of leaves, culms, and hulls (Fig. 3B). Furthermore, RNA in situ hybridization of developing hulls demonstrated the preferential expression of BG1 in hull vascular tissues with a gradual decrease as the development progressed (Fig. 3B and Fig. S8E).
Fig. 3.
BG1 expression pattern and protein subcellular localization. (A) Expression of BG1 in various organs analyzed by quantitative analysis. Root, culm, leaf, and leaf sheath were harvested from 2-mo-old WT plants. Young panicles (YP) in different lengths (indicated as numbers, cm) were included for the analysis. (B) GUS staining of various tissues of PROBG1:GUS transgenic plants and RNA in situ hybridization. (a) Cross-section of young culm. (Scale bar, 1 mm.) (b) Cross-section of hull. (Scale bar, 500 μm.) (c) Cross-section of leaf. (Scale bar, 1 mm.) (d) In situ localization of BG1 mRNA in the hull from 5-cm length young panicle. (Scale bar, 500 μm.) (e) Magnified views of the boxed area in d. (Scale bar, 200 μm.) (f) Negative control. (Scale bar, 200 μm.) (C) Confocal observation of root of the transgenic plants with 35S:eGFP (Upper) and 35S:BG1-eGFP (Lower). (Scale bar, 50 μm.) DIC indicates the differential interference contrast phase. (D) Microsomes separation and detection of BG1 by anti-GFP antibody. M, microsomes; S, soluble fraction. Blotting signals with anti-PEPC and anti-BIP were used as controls. (E) Further plasma membrane isolation and detection of BG1 by anti-GFP antibody. P, plasma membrane fraction; O, other fractions. Blotting signals with anti-BIP and antiaquaporin PIP2;1 were used as controls.
BG1 Is Localized to the Plasma Membrane.
Because BG1 encodes a novel protein without known sequence characteristics, we first investigated the protein subcellular localization. A 35S:BG1-eGFP vector containing eGFP-tagged BG1 was introduced into WT rice, and transgenic plants displayed obvious phenotypes similar to the Bg1-D mutant (Fig. S8 A–C), indicating that the fused BG1-eGFP is functional. GFP fluorescence observation using roots as material showed that the protein is predominantly localized to the plasma membrane (Fig. 3C). Consistent with this finding, we also obtained a similar result in rice protoplasts transformed with the same vector (Fig. S8D). To confirm these results, we separated the microsomes of the transgenic plants and then detected the proteins by anti-GFP antibodies. Immunoblotting results showed that BG1-eGFP can be obviously detected in the microsomal pellets, but only a very faint signal was observed in the soluble fraction (Fig. 3D). An additional two-phase partitioning assay demonstrated that the BG1-eGFP protein was highly enriched in the plasma membrane fraction, but not in the other membrane’s fractions (Fig. 3E).
BG1 Is a Primary Auxin Response Gene.
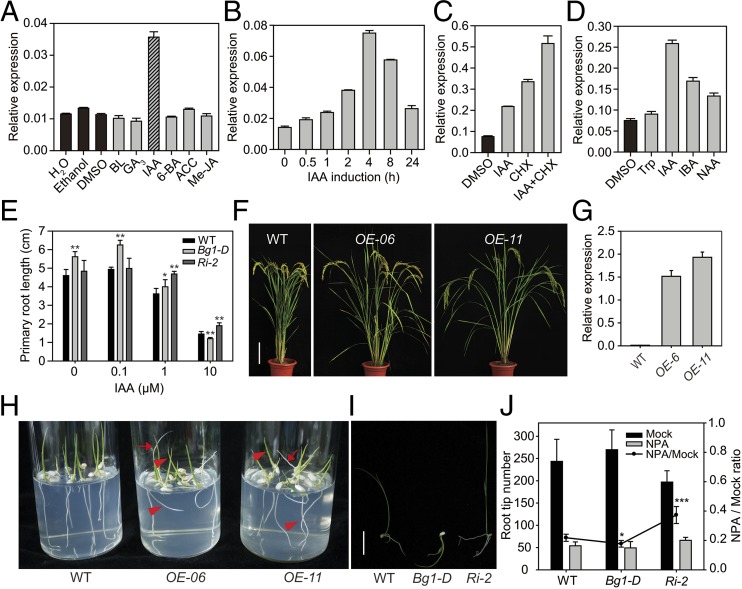
To investigate whether phytohormones affect BG1 transcription, we analyzed BG1 expression in WT seedlings under the treatment of various hormones. After 2-h application of hormones, we found that BG1 is specifically induced by IAA, but not by others (Fig. 4A). An additional time-course analysis revealed that the BG1 transcript level was rapidly induced within 30 min of IAA application and the expression reached the maximum with ∼eightfold of that without hormone treatment after 4-h treatment (Fig. 4B). The IAA-induced accumulation still occurred in the presence of cycloheximide (CHX), a protein synthesis inhibitor, suggesting that the induction does not rely on protein synthesis (Fig. 4C). Moreover, IBA (indole-3-butyric acid) and NAA (naphthylacetic acid) (two other auxin compounds) can also induce the expression of BG1, but to a less extent, whereas tryptophan (a structurally similar molecule to IAA) failed to induce BG1 accumulation (Fig. 4D), showing no hormonal activity. These results strongly suggest that BG1 is a novel specific and primary auxin response gene. Consistent with this, we found that Bg1-D had increased sensitivity to IAA in term of root elongation/inhibition and, in contrast, BG1-Ri plants had greatly decreased sensitivity to IAA (Fig. 4E and Fig. S9A), demonstrating that BG1 is involved in auxin response.
Fig. 4.
BG1 is a specific auxin response gene. (A) Relative expression level of BG1 under different phytohormone treatments. BL, brassinolide; GA3, gibberellic acid; IAA, indole-3-acetic acid; 6-BA, 6-benzylaminopurine; ACC, 1-aminocyclo-propane-1-carboxylic acid; JA, jasmonic acid. (B) Time-course response of BG1 to 10 μM IAA treatment. (C) Effect of CHX on BG1 expression. (D) BG1 response to different auxin compounds. (E) Effects of different IAA concentrations on primary root length in WT, Bg1-D, and Ri-2. (F) Gross morphology of WT and two independent BG1-overexpression lines (OEs) at the reproductive stage. (Scale bar, 20 cm.) (G) BG1 expression levels in the plants shown in F. (H) Growth of the plants on agar medium. Arrows indicate the upward growth of roots. Arrowheads indicate slanting growth of shoots and roots. (I) Growth of WT, Bg1-D, and Ri-2 under 0.1 μM NPA treatment. (Scale bar, 1 cm.) (J) Statistical data of root tip number of WT, Bg1-D, and Ri-2 under 0.1 μM NPA treatment. Means ± SD are given in E (n = 20) and J (n = 10). *P < 0.05, **P < 0.01, ***P < 0.001 (t test). Zhonghua 11 was used as WT in F–H.
BG1 Is Involved in Auxin Transport.
In BG1-overexpression plants, a number of lines with high expression levels of BG1 showed obviously enlarged tiller angles (Fig. 4 F and G), a phenotype associated with perturbed auxin transport (33). Combined with the membrane localization of the BG1 protein and the induction of BG1 by auxin treatment, these results strongly imply that BG1 is involved in auxin transport. Strikingly, we further observed that these BG1-overexpressing lines have obvious defects in tropic growth at the young seedling stage. When their seeds were sowed on agar culture media, with a high frequency, both the roots and the shoots grew in a slanting direction, and in some cases the roots even grew upward, resembling the NPA (N-1-naphthylphthalamic acid, an auxin transport inhibitor)-treated phenotype (Fig. 4H and Fig. S9B). To test whether the Bg1-D mutant also has altered gravitropism response, we treated the Bg1-D mutant, WT, and BG1-Ri plants with 0.1 μM NPA and then compared their phenotypes (Fig. 4I and Fig. S9C). In plant shoots, whereas a few of the WT plants had slightly oblique growth under low concentration of NPA, BG1-Ri plants had normal growth. In contrast, ∼35% of Bg1-D mutant shoots grew downward into the culture medium. In roots, NPA strongly inhibited the lateral root initiation in all of the three plants. However, compared with those without treatment, the root tip numbers of WT, Bg1-D, and BG1-Ri plants were repressed to 21.8%, 17.7%, and 37.3%, respectively (Fig. 4J), indicating that the NPA effect is more remarkable on Bg1-D but less on BG1-Ri than on WT. In conclusion, Bg1-D has increased sensitivity to NPA, whereas BG1-Ri has decreased sensitivity. Taken together, these results strongly suggest that BG1 is involved in auxin transport.
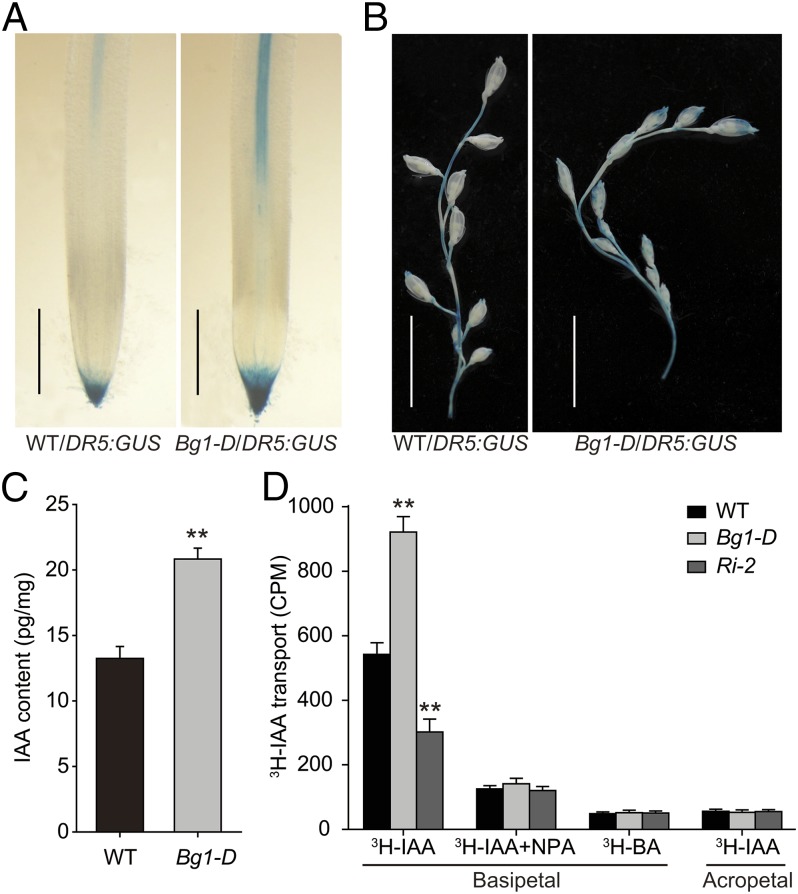
Bg1-D Exhibits Altered IAA Distribution and Transport.
DR5:GUS has been widely used as a reporter to study endogenous auxin distribution (37). To study whether activation of BG1 caused altered auxin distribution, we crossed the DR5:GUS plants to Bg1-D, and then analyzed the DR5 promoter activity in the double mutant by GUS staining. Compared with the DR5:GUS plants where the GUS signal was mainly detected at the apex area in both roots and spikelets, the GUS expression in the Bg1-D mutant was more intense and covered a much broader area, indicating that the Bg1-D mutant had enhanced auxin distribution in these tissues (Fig. 5 A and B). Consistent with this finding, the IAA level was markedly increased in the panicles of the Bg1-D mutant compared with WT (Fig. 5C). In addition, we directly measured the basipetal IAA transport in the coleoptiles of dark-grown seedlings of WT, Bg1-D, and BG1-Ri plants. Compared with the WT, the Bg1-D mutant had an ∼50% increase in the basipetal IAA transport, whereas BG1-Ri had decreased transport (Fig. 5D), further confirming that BG1 is involved in auxin transport.
Fig. 5.
Altered auxin distribution and transport in Bg1-D mutant. (A and B) Comparison of DR5 promoter activity in root (A) and spikelet (B) of WT and Bg1-D by GUS staining. (Scale bar, 1 mm in A and 1 cm in B). (C) Quantification of IAA content in panicles of WT and Bg1-D. (D) Measurement of auxin transport ability in WT, Bg1-D, and Ri-2 using coleoptile as materials. Means ± SD are given in C (n = 3) and D (n = 15). **P < 0.01 (t test).
Manipulation of BG1 Increases Plant Biomass and Grain Yield.
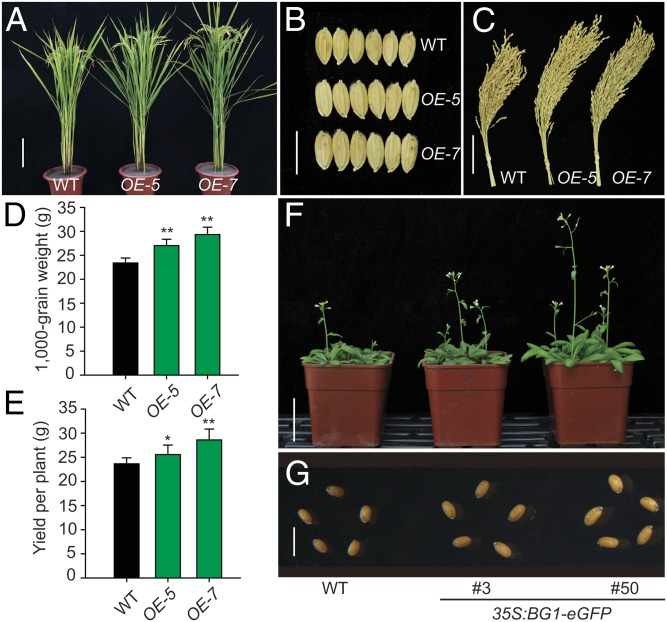
Because activation of BG1 leads to a marked increase in grain size and grain weight, we tested whether BG1 can be applied to improve grain yield. We found that most of the BG1-overexpressing plants, including the Bg1-D mutant, have insignificant change of grain yield per plant because of a lower percentage of full matured grains compared with the WT plants, which may be caused by ectopic expression of BG1 under a strong constitutive promoter. To deal with this problem, we introduced the BG1 gene into WT plants using a 3.6-kb genomic fragment containing the 2-kb native promoter and the entire coding region of BG1, as well as a 667 bp downstream sequence. Consistent with the predominant role of BG1 in panicle development, the transgenic plants showed obviously increased grain size and larger panicle without other obviously negative effects (Fig. 6 A–C). Statistical analysis showed that the 1,000-grain weight of two lines, OE-5 and OE-7, increased 15.48% and 25.38%, respectively, and the grain yield per plant increased 6.69% and 16.65%, respectively, compared with WT (Fig. 6 D and E). An additional field trial test showed that the actual yield per plot (15 m2) of both transgenic lines had increased about 15.1% and 20.8%, respectively (Table S2).
Fig. 6.
Manipulation of BG1 improves plant yield. (A) Gross morphology of WT and two independent genomic BG1 transgenic plants, OE-5 and OE-7. (Scale bar, 20 cm.) (B) Grain morphology of WT and the transgenic plants. (Scale bar, 1 cm.) (C). Comparison of the harvested panicles of one plant individual in WT and the transgenic plants. (Scale bar, 5 cm.) (D and E) 1,000-grain weight D (n = 10) and yield per plant E (n = 25) of WT and the transgenic plants. Means ± SD were given, *P < 0.05, **P < 0.01 (t test). (F and G) Phenotypes of BG1-overexpressing plants in Arabidopsis. (Scale bar, 5 cm in F and 1 mm in G).
Despite the increased grain weight, overexpression of BG1 also significantly enhanced plant biomass, with the highest about 18% increase of dry weight in three analyzed lines (Fig. S10A). In contrast, the BG1-Ri lines showed greatly decreased biomass. In addition, we also generated a 35S:BG1 vector and introduced it into Arabidopsis (Columbia, Col-0), and found that overexpression of BG1 in Arabidopsis also resulted in greatly increased seed size and enhanced growth at both vegetative (Fig. S10 B and C) and reproductive (Fig. 6 F and G) stages, suggesting that BG1 is also functional in dicots. Considering that orthologs of BG1 exist in many other plant species, including sorghum, Medicago, maize, and soybean (Fig. S7), BG1 may have extensively practical roles in improving plant biomass and grain productivity in various species.
Discussion
Several lines of evidence support the involvement of BG1 in auxin transport. First, BG1 is localized to plasma membrane, expresses in vascular tissues, and is induced by auxin treatment. These characteristics are reminiscent of typical auxin transport genes, such as PINs (28, 38). Second, Bg1-D has increased sensitivities to both auxin and NPA, whereas BG1-Ri has decreased sensitivities. Severe BG1-overexpressing lines have obviously defective gravitropism growth. Third, Bg1-D has obviously enhanced basipetal auxin transport, whereas BG1-Ri has decreased auxin transport. Therefore, it is very likely that BG1 plays an important role in controlling plant growth and development through regulating auxin transport. In Arabidopsis, auxin transport regulates a number of processes associated with cell expansion or division, including hypocotyl elongation, shade avoidance, and stamen filament elongation (39–42). In rice, overexpression of a small auxin-up RNA (SAUR) gene SAUR39 reduces auxin transport, resulting in abnormal shoot and root growth and altered shoot morphology compared with WT plants (43). In addition, mutation of the NARROW LEAF1 (NAL1) gene reduces polar auxin transport activity, leading to reduction in leaf size and stem length (44). Recently, NAL1 has been identified as an important QTL regulating grain yield by three independent studies (45–47). Near-isogenic lines carrying a gain-of-function allele of NAL1, as well as the gene overexpressors, have increased leaf length, panicle size, and grain productivity. Importantly, this allele has been selected in high-yield rice-breeding process. Although the detailed mechanism of NAL1 function remains unclear, these results suggest a potential of modulating auxin transport in improving grain yield. Our study provides direct evidence that strengthened auxin transport caused by activation of BG1 leads to enhanced growth, whereas suppression of BG1 weakens auxin transport, resulting in reduced growth. Interestingly, although Bg1-D exhibits an overall enhanced growth in different tissues, the most significant change is the increased grain size and grain weight. Because grain weight depends to a large extent on the hull development and endosperm maturation, the role of auxin transport in coordination of this process needs to be further investigated.
So far, the detailed relationship between auxin transport and auxin signaling remains unclear. It appears that auxin transport determines its distribution, where auxin exerts its function through auxin signaling. However, abundant studies have demonstrated that a regional gradient of hormone concentrations, resulted by polar auxin transport and local auxin biosynthesis, is important for auxin-mediated developmental processes (48, 49). Consistent with this idea, BG1-Ri plants have obviously decreased IAA sensitivity. In addition, transcripts of both BG1 and PINs are greatly induced by auxin treatment, and it has been shown that PINs up-regulation is dependent on AUX/IAA-mediated signaling pathway (38). We also found there are two auxin response elements in the BG1 promoter, implying that BG1 might be regulated by ARF. Thus, the auxin transport and auxin signaling may form a regulatory loop to stimulate each other, leading to a rapid and efficient auxin response (50, 51).
Because BG1 has no obvious membrane localization peptide sequence and hydrophobic regions, it seems unlikely that BG1 directly transports auxin. BG1 may interact with other membrane-localized auxin transporters, such as members of PIN, AUX1/LAX, or ABCB families (28, 29, 49), to modulate their activities or to facilitate the auxin transport. Several factors involved in vesicular trafficking or phosphorylation modification were reported to regulate PIN function (52, 53). A BIG protein has also been found to modulate PIN localization (54). Interestingly, whereas both BG1 and PIN appear to be involved in auxin transport, it is unexpected that some BG1-overexpressing plants have a similar phenotype as OsPIN1-knockdown plants, which also have enlarged tiller angles as well as enhanced NPA sensitivity (55). One possibility is that BG1 functions in a way different from PINs to regulate auxin transport. Mutation of LAZY1 also leads to enlarged tiller angles with enhanced basipetal auxin transport (33). Further investigation on the relationship between BG1 and these transport-associated components is essential to elucidate the BG1 functional mechanism.
Although auxin was discovered more than 80 y ago and has been demonstrated to play important roles in many aspects of plant growth and development, in rice both auxin signaling and transport are poorly understood. Thus, identification of a novel auxin transport regulator in rice has great significance for understanding of auxin biology in monocotyledon. Although the mechanism for BG1 regulation of auxin transport remains elusive, this study establishes a connection between auxin transport and grain size, and demonstrates the feasibility of modulating auxin transport to enhance grain productivity, thus illuminating a new strategy to improve plant yield.
Materials and Methods
The Bg1-D mutant was identified from our T-DNA insertion population with japonica Nipponbare background (56). Details of experimental procedures, including plant cultivation, gene-expression analysis, histological analysis, RNA in situ hybridization, subcellular localization, IAA measurement, and auxin transport assay, are described in SI Materials and Methods. Primers used in this study are listed in Tables S3–S7.
Supplementary Material
Acknowledgments
This work was supported by grants from National Natural Science Foundation of China (91335203), the Ministry of Agriculture of China (2014ZX08009-003-003, 2014ZX08001-004), and the Ministry of Science and Technology of China (2012CB944800).
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1512748112/-/DCSupplemental.
References
- 1.Sakamoto T, Matsuoka M. Identifying and exploiting grain yield genes in rice. Curr Opin Plant Biol. 2008;11(2):209–214. doi: 10.1016/j.pbi.2008.01.009. [DOI] [PubMed] [Google Scholar]
- 2.Ikeda M, Miura K, Aya K, Kitano H, Matsuoka M. Genes offering the potential for designing yield-related traits in rice. Curr Opin Plant Biol. 2013;16(2):213–220. doi: 10.1016/j.pbi.2013.02.002. [DOI] [PubMed] [Google Scholar]
- 3.Xing Y, Zhang Q. Genetic and molecular bases of rice yield. Annu Rev Plant Biol. 2010;61:421–442. doi: 10.1146/annurev-arplant-042809-112209. [DOI] [PubMed] [Google Scholar]
- 4.Song XJ, Huang W, Shi M, Zhu MZ, Lin HX. A QTL for rice grain width and weight encodes a previously unknown RING-type E3 ubiquitin ligase. Nat Genet. 2007;39(5):623–630. doi: 10.1038/ng2014. [DOI] [PubMed] [Google Scholar]
- 5.Weng J, et al. Isolation and initial characterization of GW5, a major QTL associated with rice grain width and weight. Cell Res. 2008;18(12):1199–1209. doi: 10.1038/cr.2008.307. [DOI] [PubMed] [Google Scholar]
- 6.Shomura A, et al. Deletion in a gene associated with grain size increased yields during rice domestication. Nat Genet. 2008;40(8):1023–1028. doi: 10.1038/ng.169. [DOI] [PubMed] [Google Scholar]
- 7.Ishimaru K, et al. Loss of function of the IAA-glucose hydrolase gene TGW6 enhances rice grain weight and increases yield. Nat Genet. 2013;45(6):707–711. doi: 10.1038/ng.2612. [DOI] [PubMed] [Google Scholar]
- 8.Zhang X, et al. Rare allele of OsPPKL1 associated with grain length causes extra-large grain and a significant yield increase in rice. Proc Natl Acad Sci USA. 2012;109(52):21534–21539. doi: 10.1073/pnas.1219776110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Qi P, et al. The novel quantitative trait locus GL3.1 controls rice grain size and yield by regulating Cyclin-T1;3. Cell Res. 2012;22(12):1666–1680. doi: 10.1038/cr.2012.151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zuo J, Li J. Molecular genetic dissection of quantitative trait loci regulating rice grain size. Annu Rev Genet. 2014;48(1):99–118. doi: 10.1146/annurev-genet-120213-092138. [DOI] [PubMed] [Google Scholar]
- 11.Wang S, et al. Control of grain size, shape and quality by OsSPL16 in rice. Nat Genet. 2012;44(8):950–954. doi: 10.1038/ng.2327. [DOI] [PubMed] [Google Scholar]
- 12.Miura K, Ashikari M, Matsuoka M. The role of QTLs in the breeding of high-yielding rice. Trends Plant Sci. 2011;16(6):319–326. doi: 10.1016/j.tplants.2011.02.009. [DOI] [PubMed] [Google Scholar]
- 13.Gonzalez N, Beemster GT, Inzé D. David and Goliath: What can the tiny weed Arabidopsis teach us to improve biomass production in crops? Curr Opin Plant Biol. 2009;12(2):157–164. doi: 10.1016/j.pbi.2008.11.003. [DOI] [PubMed] [Google Scholar]
- 14.Li J, Nie X, Tan JL, Berger F. Integration of epigenetic and genetic controls of seed size by cytokinin in Arabidopsis. Proc Natl Acad Sci USA. 2013;110(38):15479–15484. doi: 10.1073/pnas.1305175110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schruff MC, et al. The AUXIN RESPONSE FACTOR 2 gene of Arabidopsis links auxin signalling, cell division, and the size of seeds and other organs. Development. 2006;133(2):251–261. doi: 10.1242/dev.02194. [DOI] [PubMed] [Google Scholar]
- 16.Hu Y, Xie Q, Chua NH. The Arabidopsis auxin-inducible gene ARGOS controls lateral organ size. Plant Cell. 2003;15(9):1951–1961. doi: 10.1105/tpc.013557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mizukami Y, Fischer RL. Plant organ size control: AINTEGUMENTA regulates growth and cell numbers during organogenesis. Proc Natl Acad Sci USA. 2000;97(2):942–947. doi: 10.1073/pnas.97.2.942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Teale WD, Paponov IA, Palme K. Auxin in action: Signalling, transport and the control of plant growth and development. Nat Rev Mol Cell Biol. 2006;7(11):847–859. doi: 10.1038/nrm2020. [DOI] [PubMed] [Google Scholar]
- 19.Zhao Y. Auxin biosynthesis and its role in plant development. Annu Rev Plant Biol. 2010;61:49–64. doi: 10.1146/annurev-arplant-042809-112308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dai X, et al. The biochemical mechanism of auxin biosynthesis by an Arabidopsis YUCCA flavin-containing monooxygenase. J Biol Chem. 2013;288(3):1448–1457. doi: 10.1074/jbc.M112.424077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhao Y. Auxin biosynthesis: A simple two-step pathway converts tryptophan to indole-3-acetic acid in plants. Mol Plant. 2012;5(2):334–338. doi: 10.1093/mp/ssr104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang B, et al. Tryptophan-independent auxin biosynthesis contributes to early embryogenesis in Arabidopsis. Proc Natl Acad Sci USA. 2015;112(15):4821–4826. doi: 10.1073/pnas.1503998112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chapman EJ, Estelle M. Mechanism of auxin-regulated gene expression in plants. Annu Rev Genet. 2009;43:265–285. doi: 10.1146/annurev-genet-102108-134148. [DOI] [PubMed] [Google Scholar]
- 24.Kepinski S, Leyser O. The Arabidopsis F-box protein TIR1 is an auxin receptor. Nature. 2005;435(7041):446–451. doi: 10.1038/nature03542. [DOI] [PubMed] [Google Scholar]
- 25.Dharmasiri N, Dharmasiri S, Estelle M. The F-box protein TIR1 is an auxin receptor. Nature. 2005;435(7041):441–445. doi: 10.1038/nature03543. [DOI] [PubMed] [Google Scholar]
- 26.Kim J, Harter K, Theologis A. Protein-protein interactions among the Aux/IAA proteins. Proc Natl Acad Sci USA. 1997;94(22):11786–11791. doi: 10.1073/pnas.94.22.11786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bennett MJ, et al. Arabidopsis AUX1 gene: A permease-like regulator of root gravitropism. Science. 1996;273(5277):948–950. doi: 10.1126/science.273.5277.948. [DOI] [PubMed] [Google Scholar]
- 28.Gälweiler L, et al. Regulation of polar auxin transport by AtPIN1 in Arabidopsis vascular tissue. Science. 1998;282(5397):2226–2230. doi: 10.1126/science.282.5397.2226. [DOI] [PubMed] [Google Scholar]
- 29.Péret B, et al. AUX/LAX genes encode a family of auxin influx transporters that perform distinct functions during Arabidopsis development. Plant Cell. 2012;24(7):2874–2885. doi: 10.1105/tpc.112.097766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Balzan S, Johal GS, Carraro N. The role of auxin transporters in monocots development. Front Plant Sci. 2014;5:393. doi: 10.3389/fpls.2014.00393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Reed RC, Brady SR, Muday GK. Inhibition of auxin movement from the shoot into the root inhibits lateral root development in Arabidopsis. Plant Physiol. 1998;118(4):1369–1378. doi: 10.1104/pp.118.4.1369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Noh B, Bandyopadhyay A, Peer WA, Spalding EP, Murphy AS. Enhanced gravi- and phototropism in plant mdr mutants mislocalizing the auxin efflux protein PIN1. Nature. 2003;423(6943):999–1002. doi: 10.1038/nature01716. [DOI] [PubMed] [Google Scholar]
- 33.Li P, et al. LAZY1 controls rice shoot gravitropism through regulating polar auxin transport. Cell Res. 2007;17(5):402–410. doi: 10.1038/cr.2007.38. [DOI] [PubMed] [Google Scholar]
- 34.Okada K, Ueda J, Komaki MK, Bell CJ, Shimura Y. Requirement of the auxin polar transport system in early stages of Arabidopsis floral bud formation. Plant Cell. 1991;3(7):677–684. doi: 10.1105/tpc.3.7.677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liu YG, Mitsukawa N, Oosumi T, Whittier RF. Efficient isolation and mapping of Arabidopsis thaliana T-DNA insert junctions by thermal asymmetric interlaced PCR. Plant J. 1995;8(3):457–463. doi: 10.1046/j.1365-313x.1995.08030457.x. [DOI] [PubMed] [Google Scholar]
- 36.Wang H, et al. Computation-assisted SiteFinding-PCR for isolating flanking sequence tags in rice. Biotechniques. 2011;51(6):421–423. doi: 10.2144/000113787. [DOI] [PubMed] [Google Scholar]
- 37.Ulmasov T, Murfett J, Hagen G, Guilfoyle TJ. Aux/IAA proteins repress expression of reporter genes containing natural and highly active synthetic auxin response elements. Plant Cell. 1997;9(11):1963–1971. doi: 10.1105/tpc.9.11.1963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Vieten A, et al. Functional redundancy of PIN proteins is accompanied by auxin-dependent cross-regulation of PIN expression. Development. 2005;132(20):4521–4531. doi: 10.1242/dev.02027. [DOI] [PubMed] [Google Scholar]
- 39.Jensen PJ, Hangarter RP, Estelle M. Auxin transport is required for hypocotyl elongation in light-grown but not dark-grown Arabidopsis. Plant Physiol. 1998;116(2):455–462. doi: 10.1104/pp.116.2.455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lin R, Wang H. Two homologous ATP-binding cassette transporter proteins, AtMDR1 and AtPGP1, regulate Arabidopsis photomorphogenesis and root development by mediating polar auxin transport. Plant Physiol. 2005;138(2):949–964. doi: 10.1104/pp.105.061572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chae K, et al. Arabidopsis SMALL AUXIN UP RNA63 promotes hypocotyl and stamen filament elongation. Plant J. 2012;71(4):684–697. doi: 10.1111/j.1365-313X.2012.05024.x. [DOI] [PubMed] [Google Scholar]
- 42.Keuskamp DH, et al. Blue-light-mediated shade avoidance requires combined auxin and brassinosteroid action in Arabidopsis seedlings. Plant J. 2011;67(2):208–217. doi: 10.1111/j.1365-313X.2011.04597.x. [DOI] [PubMed] [Google Scholar]
- 43.Kant S, Bi YM, Zhu T, Rothstein SJ. SAUR39, a small auxin-up RNA gene, acts as a negative regulator of auxin synthesis and transport in rice. Plant Physiol. 2009;151(2):691–701. doi: 10.1104/pp.109.143875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Qi J, et al. Mutation of the rice Narrow leaf1 gene, which encodes a novel protein, affects vein patterning and polar auxin transport. Plant Physiol. 2008;147(4):1947–1959. doi: 10.1104/pp.108.118778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Takai T, et al. A natural variant of NAL1, selected in high-yield rice breeding programs, pleiotropically increases photosynthesis rate. Sci Rep. 2013;3:2149. doi: 10.1038/srep02149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Fujita D, et al. NAL1 allele from a rice landrace greatly increases yield in modern indica cultivars. Proc Natl Acad Sci USA. 2013;110(51):20431–20436. doi: 10.1073/pnas.1310790110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhang GH, et al. LSCHL4 from Japonica Cultivar, which is allelic to NAL1, increases yield of indica super rice 93-11. Mol Plant. 2014;7(8):1350–1364. doi: 10.1093/mp/ssu055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Vanneste S, Friml J. Auxin: A trigger for change in plant development. Cell. 2009;136(6):1005–1016. doi: 10.1016/j.cell.2009.03.001. [DOI] [PubMed] [Google Scholar]
- 49.Petrásek J, Friml J. Auxin transport routes in plant development. Development. 2009;136(16):2675–2688. doi: 10.1242/dev.030353. [DOI] [PubMed] [Google Scholar]
- 50.Nick P, Han MJ, An G. Auxin stimulates its own transport by shaping actin filaments. Plant Physiol. 2009;151(1):155–167. doi: 10.1104/pp.109.140111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Benjamins R, Scheres B. Auxin: The looping star in plant development. Annu Rev Plant Biol. 2008;59:443–465. doi: 10.1146/annurev.arplant.58.032806.103805. [DOI] [PubMed] [Google Scholar]
- 52.Geldner N, et al. The Arabidopsis GNOM ARF-GEF mediates endosomal recycling, auxin transport, and auxin-dependent plant growth. Cell. 2003;112(2):219–230. doi: 10.1016/s0092-8674(03)00003-5. [DOI] [PubMed] [Google Scholar]
- 53.Friml J, et al. A PINOID-dependent binary switch in apical-basal PIN polar targeting directs auxin efflux. Science. 2004;306(5697):862–865. doi: 10.1126/science.1100618. [DOI] [PubMed] [Google Scholar]
- 54.Gil P, et al. BIG: A calossin-like protein required for polar auxin transport in Arabidopsis. Genes Dev. 2001;15(15):1985–1997. doi: 10.1101/gad.905201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Xu M, Zhu L, Shou H, Wu P. A PIN1 family gene, OsPIN1, involved in auxin-dependent adventitious root emergence and tillering in rice. Plant Cell Physiol. 2005;46(10):1674–1681. doi: 10.1093/pcp/pci183. [DOI] [PubMed] [Google Scholar]
- 56.Ma Y, et al. Molecular analysis of rice plants harboring a multi-functional T-DNA tagging system. J Genet Genomics. 2009;36(5):267–276. doi: 10.1016/S1673-8527(08)60114-9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.