Significance
The human indigenous microbial communities (microbiota) play critical roles in health and may be especially important for mother and fetus during pregnancy. Using a case-control cohort of 40 women, we characterized weekly variation in the vaginal, gut, and oral microbiota during and after pregnancy. Microbiota membership remained relatively stable at each body site during pregnancy. An altered vaginal microbial community was associated with preterm birth; this finding was corroborated by an analysis of samples from an additional cohort of nine women. We also discovered an abrupt change in the vaginal microbiota at delivery that persisted in some cases for at least 1 y. Our findings suggest that pregnancy outcomes might be predicted by features of the microbiota early in gestation.
Keywords: 16S rRNA gene, pregnancy, preterm birth, microbiome, premature labor
Abstract
Despite the critical role of the human microbiota in health, our understanding of microbiota compositional dynamics during and after pregnancy is incomplete. We conducted a case-control study of 49 pregnant women, 15 of whom delivered preterm. From 40 of these women, we analyzed bacterial taxonomic composition of 3,767 specimens collected prospectively and weekly during gestation and monthly after delivery from the vagina, distal gut, saliva, and tooth/gum. Linear mixed-effects modeling, medoid-based clustering, and Markov chain modeling were used to analyze community temporal trends, community structure, and vaginal community state transitions. Microbiota community taxonomic composition and diversity remained remarkably stable at all four body sites during pregnancy (P > 0.05 for trends over time). Prevalence of a Lactobacillus-poor vaginal community state type (CST 4) was inversely correlated with gestational age at delivery (P = 0.0039). Risk for preterm birth was more pronounced for subjects with CST 4 accompanied by elevated Gardnerella or Ureaplasma abundances. This finding was validated with a set of 246 vaginal specimens from nine women (four of whom delivered preterm). Most women experienced a postdelivery disturbance in the vaginal community characterized by a decrease in Lactobacillus species and an increase in diverse anaerobes such as Peptoniphilus, Prevotella, and Anaerococcus species. This disturbance was unrelated to gestational age at delivery and persisted for up to 1 y. These findings have important implications for predicting premature labor, a major global health problem, and for understanding the potential impact of a persistent, altered postpartum microbiota on maternal health, including outcomes of pregnancies following short interpregnancy intervals.
The human body harbors diverse, complex, and abundant microbiota whose composition is determined largely by body site but also by host genetics, environmental exposures, and time (1, 2). The microbiota plays critical roles in health and in disease, including nutrient acquisition, immune programming, and protection from pathogens (3). Normal pregnancy represents a unique, transient, and dynamic state of altered anatomy, physiology, and immune function. Preterm birth, i.e., before 37 wk of gestation, occurs in 11% of pregnancies and is the leading cause of neonatal death (4). In both term and preterm pregnancies, the interplay between the microbiota and the host remains poorly understood.
Approximately 25% of preterm births are associated with occult microbial invasion of the amniotic cavity (5). Evidence suggests that the most common source of invading microbes is the host microbiota. In studies of amniotic fluid from women with preterm labor and either intact or ruptured membranes, 16S ribosomal RNA (rRNA) sequences of known vaginal, gut, and oral indigenous bacterial species have been recovered in 15–50% of cases, and their relative abundances have correlated directly with markers of inflammation and inversely with time to delivery (6–9). Preterm birth also is associated with bacterial vaginosis, a community-wide alteration of the vaginal microbiota (10, 11) that increases the risk of preterm birth approximately twofold (12, 13).
Several studies have examined the vaginal microbiota during pregnancy using cultivation-independent techniques (14–19). Collectively, these studies found the vaginal communities of pregnant women to be dominated by Lactobacillus species and characterized by lower richness and diversity than in nonpregnant women but with higher stability. Of the two studies that evaluated pregnancy outcomes, one found preterm birth to be linked with higher intracommunity (alpha) diversity in the vagina (16), but the other found no significant association between preterm birth and any specific community type or microbial taxon (17).
Other (nonvaginal) body sites have been even less well studied in the setting of pregnancy. The subgingival crevice has been investigated only with cultivation (20, 21) or with taxon-specific molecular approaches (22). Two studies of the fecal microbiota reported differences in bacterial community structure between the first and third trimesters (23, 24); in each study, however, samples were collected at only two time points. These limited findings support the need for longitudinal investigations of the microbiota at multiple body sites during pregnancy.
As part of a larger ongoing study, we examined a total of 49 women who were divided into two groups, each of which included controls (term deliveries) and cases (preterm deliveries). We characterized the temporal dynamics of microbiota composition based on prospective weekly sampling during pregnancy from four body sites: vagina, distal gut (stool), saliva, and tooth/gum, as well as after delivery. Our data reveal microbiota compositional stability during pregnancy at all body sites, a diverse vaginal community state early during pregnancy in women who subsequently delivered prematurely, and a dramatic shift in vaginal microbiota composition at the time of delivery that in some cases persisted for the maximum duration of postpartum sampling (1 y).
Results
We studied 49 women, 40 of whom contributed samples for a discovery dataset (11 of these 40 women delivered preterm) and nine of whom contributed samples for a validation dataset (four of these nine women delivered preterm). Demographic and baseline clinical characteristics of the study population appear in SI Appendix, Table S1. The first group of 40 subjects provided 3,767 clinical specimens (SI Appendix, Fig. S1) that were analyzed at a mean depth of 5,125 filtered, high-quality pyrosequencing reads per sample (19,306,851 total reads). This discovery dataset was used to characterize temporal dynamics of the microbiota at four body sites (vaginal, stool, saliva, tooth/gum) and to identify stereotypic community features associated with preterm birth. The second group of nine subjects provided 246 vaginal specimens that were analyzed at a mean depth of 203,391 filtered, high-quality Illumina reads per sample (50,034,186 total reads); this validation dataset was used to test features of the vaginal community identified in the discovery dataset as being associated with preterm birth. SI Appendix, Table S2 presents additional details of the 15 women in total who delivered preterm (before gestational week 37).
Diversity and Composition of Bacterial Communities of the Vagina, Distal Gut, Saliva, and Tooth/Gum Are Relatively Stable During Pregnancy.
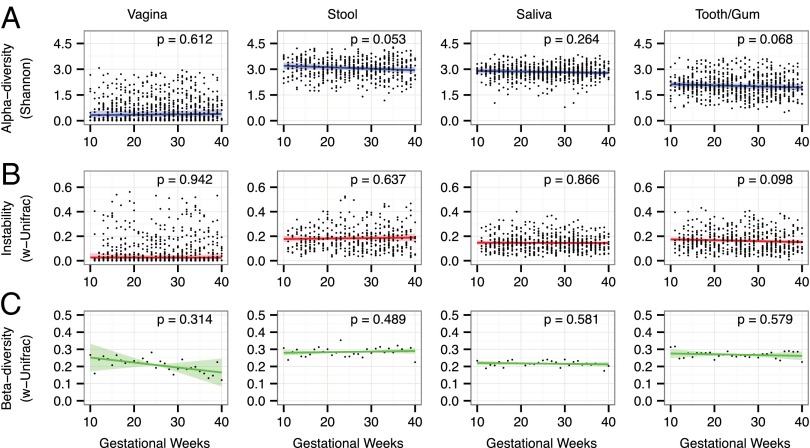
We evaluated spatial and temporal trends in the structure of the bacterial communities of each body site during pregnancy in a group of 40 women by using a linear mixed-effects (LME) model to regress alpha diversity measures against gestational time while accounting for the subject-structure of these longitudinal data by treating the subject as a random effect. No significant trend in the Shannon diversity index was found for any of the four body sites during the course of gestation (P > 0.05, t test) (Fig. 1A). This finding was robust to other alpha diversity measures (SI Appendix, Fig. S2). Similarly, no significant trends over gestational time were observed in the weighted UniFrac distance between same-subject communities sampled in consecutive weeks (i.e., weekly instability) for any of the four body sites (P > 0.05, t test) (Fig. 1B). This finding also was robust to the choice of distance measure (SI Appendix, Fig. S3). There was no particular week (or series of weeks) of pregnancy in which large, coordinated shifts occurred.
Fig. 1.
Human-associated bacterial communities are stable during pregnancy. Based on the data from the first group of 40 women, the estimated trends of alpha diversity, weekly instability (week-to-week variation within subjects), and beta diversity with gestational time are insignificant (P > 0.05) at all body sites. (A) Shannon diversity is plotted against gestational time for specimens taken from the vagina, stool, saliva, and tooth/gum. Blue lines indicate the linear mixed-effects regression of diversity on time with grouping by subject. Shading indicates the 95% confidence interval (CI). Because vaginal diversity and stability data were highly skewed, they were log-transformed before fitting to improve normality. (B) Weighted-UniFrac distance between same-subject samples taken 1 wk apart is plotted against gestational time. Red lines indicate the lme regression, and the shaded area indicates the 95% CI. (C) Average weighted-UniFrac distance between different-subject samples taken within the same gestational week is plotted against gestational time. The green lines indicate the linear fit, and the shading indicates the 95% CI as estimated by a permutation bootstrap (SI Appendix, SI Methods).
The average pairwise-weighted UniFrac distance between communities in different subjects was used as an estimate of beta diversity. We evaluated the significance of beta diversity trends with a permutation test in which the null-distribution of our test statistic was estimated from the ensemble of randomly time-reversed subjects (SI Appendix, SI Methods). We found no significant trend with time at any of the four body sites (P > 0.05, permutation test) (Fig. 1C). Similarly, this finding was robust to the choice of distance measure for quantifying beta diversity (SI Appendix, Fig. S4).
If consecutive weekly instability were gradual and consistent, then microbiota composition near the end of pregnancy should be distinct from that near the start, although other types of dynamics (e.g., oscillations) might not give rise to this distinction. However, as did the statistical summaries of these communities, the average taxonomic composition remained constant over gestational time. This compositional stability was illustrated by nonmetric multidimensional scaling ordination of Bray–Curtis distances based on relative operational taxonomic unit (OTU) abundances, which revealed significant overlap between communities sampled early in pregnancy and those sampled late in pregnancy at all body sites (SI Appendix, Fig. S5). Furthermore, almost none of the most prevalent OTUs (those present in >25% of samples) exhibited significant shifts in relative abundance between early and late in pregnancy (SI Appendix, Table S3).
Taken together, these results suggest that the progression of pregnancy is not associated with a dramatic remodeling of the diversity and composition of a woman’s indigenous microbiota.
Vaginal Community State Types of Pregnancy.
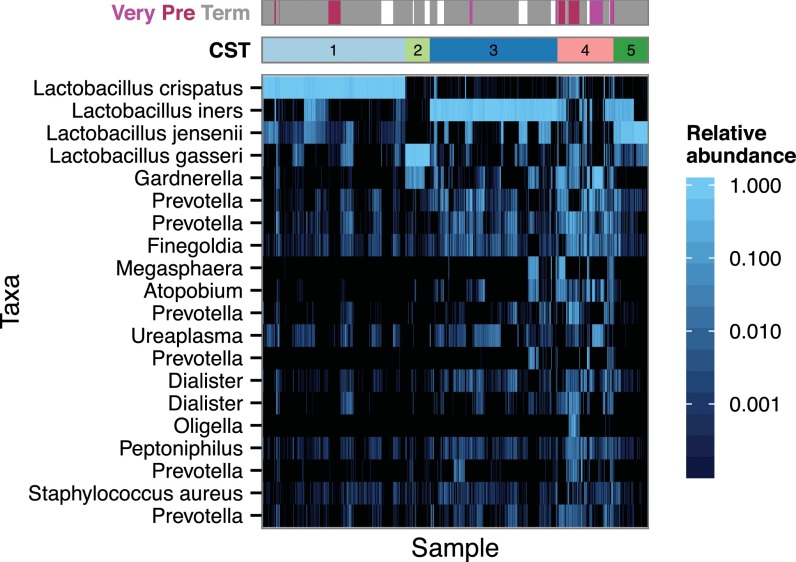
With the data from the 40 women in the first subject group, we applied de novo clustering, based on relative OTU abundances, to the samples from each body site to explore community structure and to reduce dimensionality. For the vaginal communities, this analysis yielded a set of four community state types (CSTs) comprising highly uneven communities dominated by different species of Lactobacillus and a fifth CST that was characterized by much greater evenness and taxonomic diversity (Fig. 2). These CSTs corresponded well to those described by Ravel et al. (11, 25) and thus were numbered concordantly: CST 1, Lactobacillus crispatus-dominant; CST 2, Lactobacillus gasseri-dominant; CST 3, Lactobacillus iners-dominant; CST 4, diverse community; and CST 5, Lactobacillus jensenii-dominant. The pregnancy-associated communities at the other body sites (stool, saliva, and tooth/gum) could not be represented by a small number of discrete CSTs.
Fig. 2.
Heat map of the fractional abundance of the 20 most abundant OTUs in the vaginal communities of 40 women sampled longitudinally during pregnancy. Clustering on the abundance profiles of individual samples (n = 761) using the partitioning around medoids algorithm identified six CSTs. CSTs 1, 2, 3, and 5 were characterized by dominant Lactobacillus species that typically account for >90% of the community: L. crispatus, L. jensenii, L. iners, and L. gasseri, respectively. CST 4 was significantly more diverse. Pregnancy outcomes are indicated by the bar at the top: term delivery (gray), >37 gestational weeks; preterm (maroon), <36 wk; very preterm (pink), <32 wk; marginal delivery during the 37th gestational week (white).
The Dynamic Network of the Vaginal Communities During Pregnancy Reveals Strong Variation in CST Stability and Interconnectedness.
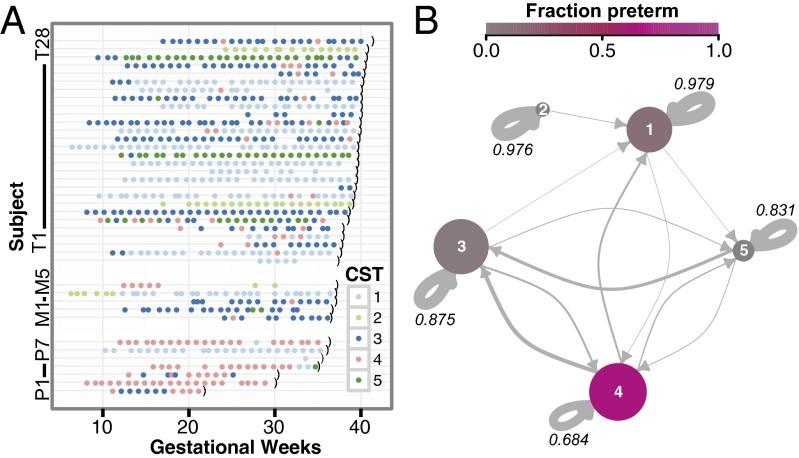
Intraindividual vaginal community states were generally stable on the time scale of weeks (Fig. 3A). However, substantial interindividual variability was observed with respect to both the most prevalent CST and the frequency of inter-CST transitions. Some subjects (e.g., T11) stably maintained a single CST throughout gestation, whereas other subjects (e.g., T6) exhibited relatively frequent transitions between CSTs. Notably, the frequency of interstate transitions did not appear to be associated with either healthy term delivery or preterm delivery.
Fig. 3.
Dynamics of the vaginal community during pregnancy. (A) Vaginal CST time course of the 40 subjects from the first subject group. Color indicates CST as shown in the key; the black parenthesis indicates delivery. Subjects P1–P7 delivered preterm (before gestational week 37); subjects M1–M5 were considered marginal (gestational week 37); subjects T1–T28 delivered at term (>37 gestational weeks). Marginal subjects were excluded when calculating associations with preterm birth. (B) Dynamics of the vaginal communities were approximated as a Markov chain with subject-independent transition probabilities between CSTs. Arrow weights are proportional to the maximum-likelihood-estimate of the week-to-week transition probabilities between states. Node sizes scale with the number of subjects in which the CST was seen. Color indicates the strength of the association with preterm birth (i.e., the proportion of the specimens from the CST that came from subjects who delivered preterm). The self-transition probabilities are shown numerically.
Because vaginal communities exhibited interstate transitions, we represented vaginal CST dynamics as a Markov chain. Fig. 3B presents a Markov chain generated by inferring inter-CST transition probabilities from our data (SI Appendix, Table S4). Our model indicated that the four Lactobacillus-dominated CSTs (CSTs 1, 2, 3, and 5) were more stable (had higher self-transition probabilities) than the diverse CST (4). This finding is qualitatively similar to the observations of Gajer et al. of CSTs in nonpregnant women (25); however, the Lactobacillus-dominated CSTs were more stable in our cohort (SI Appendix, SI Discussion).
The structure of the observed inter-CST transition patterns also is of interest. CST 2 (L. gasseri-dominated) had the fewest connections. Indeed, in our cohort, CST 2 was not observed to be reachable from any other CST, and when CST 2 transitioned to another state, it transitioned only to CST 1 (L. crispatus-dominated). In contrast, CST 4 was the most interconnected and was the only state exhibiting bidirectional transitions with three other CSTs (all except CST 2).
The High-Diversity Vaginal CST Was Associated with Preterm Birth.
The observed CSTs exhibited substantially different strengths of association with preterm birth (Fig. 3B and SI Appendix, Table S5). In particular, the Lactobacillus-poor CST 4 community exhibited a much stronger association with preterm delivery than did any of the Lactobacillus-dominated CSTs (1–3, 5).
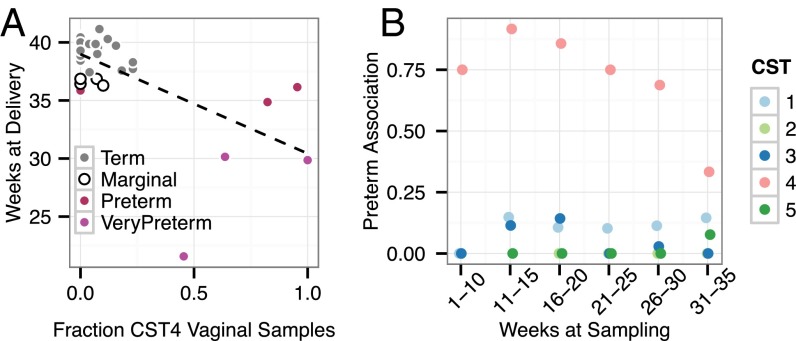
We next explored the relationship of preterm birth with temporal features of the CST 4 community. We found the duration and proportion of time during which a woman’s vaginal community remained in CST 4 to be associated with preterm birth. Fig. 4A demonstrates that CST 4 prevalence is correlated with an earlier gestational age at delivery (P = 1.1 × 10−4, Pearson; P = 0.015, Spearman). This correlation remained significant after correcting for the effect of white or nonwhite race (P = 2.5 × 10−4, Pearson; P = 0.046, Spearman). This association between CST 4 and preterm birth was present at every time window during gestation, suggesting that the value of CST 4 in predicting preterm birth begins early in pregnancy (Fig. 4B).
Fig. 4.
The high-diversity vaginal CST 4 was associated with earlier deliveries and a higher likelihood of preterm birth in the first group of 40 women. (A) Gestational age at delivery is plotted against the fraction of vaginal specimens assigned to the high-diversity CST 4 for the 33 subjects for whom at least 10 vaginal specimens were collected. The dashed line indicates the linear fit. Increased prevalence of the diverse vaginal CST was significantly correlated with earlier delivery (P = 1.1 × 10−4, Pearson; P = 0.0147, Spearman). (B) The fraction of specimens collected from subjects who delivered preterm is shown by specimen CST and the gestational period during which the specimens were collected. CST 4 specimens collected at any time during pregnancy were associated with a higher proportion of preterm birth.
Gardnerella and Ureaplasma Abundances Stratify Preterm Risk for Women with the High-Diversity Vaginal CST.
We tested CST 4 samples from the first group of subjects (n = 40) for associations between the relative abundances of individual taxa and preterm delivery. The nonindependence of samples from the same subject, combined with heterogeneity in the number and timing of samples from different subjects in our study, complicated this comparison. Therefore, we tested under the two extreme but contrary models of sample dependence—complete sample independence and complete dependence of samples within subjects—with the recognition that these two models bound the actual solution.
When CST 4 samples were treated independently, both Ureaplasma (Padj = 5 × 10−34, Benjamini–Hochberg–corrected Wald test) and Gardnerella (Padj = 1.5 × 10−13) had strong positive associations with preterm birth. When CST 4 samples within subjects were treated completely nonindependently (by merging the CST 4 samples from each subject before testing) only the association with Gardnerella remained significant (Padj = 0.054). Although caution is warranted because of the small number of subjects, these findings suggest that in the setting of low Lactobacillus abundance, a high abundance of Gardnerella in particular may increase the risk of preterm birth. In addition, Ureaplasma deserves further investigation as a risk factor (SI Appendix, SI Discussion).
Evaluation of a Second Group of Nine Women for Putative Preterm Risk Factors Identified in Vaginal Communities from the First Group of 40 Women.
We used the data from an additional group of nine women as a validation set for three preterm risk factors identified in our discovery dataset: (i) CST 4 (for which <50% Lactobacillus was used as a proxy); (ii) CST 4 with abundant Gardnerella; and (iii) CST 4 with abundant Ureaplasma. In this group of subjects, four women had multiple CST 4 samples collected during pregnancy; two of these women delivered preterm. Given that four of nine women delivered preterm, this is a neutral result for CST 4 alone when used as a simple binary trait for classification. However, when women with CST 4 vaginal communities were evaluated for high abundance of either Gardnerella or Ureaplasma, both community profiles were associated with preterm birth. Two women with multiple CST 4 samples had abundant Ureaplasma, and both delivered preterm. One of these two women also had abundant Gardnerella (SI Appendix, Fig. S6). Although these numbers are very small, these findings corroborate the microbial community features identified in our discovery dataset as exhibiting the strongest associations with preterm birth.
Effect of Sampling Frequency on Study Findings.
We assessed whether sampling less frequently than weekly would have affected overall study findings. We found that most vaginal CSTs in the discovery dataset had persistence times of many weeks: CST 1, 48.1 wk; CST 2, 40.5 wk; CST 3, 7.5 wk; CST 5, 5.4 wk; however, CST 4 had a much shorter persistence time, 2.6 wk. Therefore, a longer sampling interval would have disproportionately impacted the ability to detect excursions into CST 4 and to estimate the proportion of time that a woman spent in CST 4, the state most strongly associated with preterm birth (Figs. 3 and 4A and SI Appendix, Table S5).
The communities at other body sites did not cluster into CSTs, so persistence times could not be extracted in the same manner. However, the average distance between communities was assessed as a function of their separation in time, or lag (SI Appendix, Fig. S7). In the oral communities the increased similarity between samples collected closely in time decayed on a roughly monthly time scale. The results for the stool community were less clear.
Delivery Was Accompanied by a Strong Disruption of Vaginal Community Structure in Most Women.
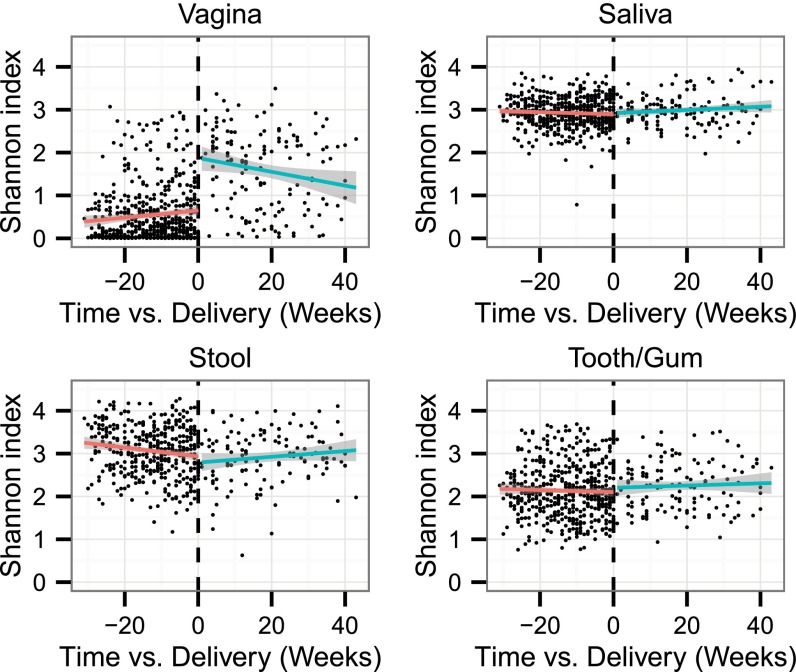
Twenty-five of the 40 women in the first subject group provided at least one postpartum sample, and several women provided samples for up to 1 y after delivery (SI Appendix, Fig. S1). We found that delivery typically was accompanied by a significant change in vaginal community structure, the clearest feature being a sharp increase in diversity after delivery (Fig. 5 and SI Appendix, Fig. S8). However, this increase was not universal, and some individual women exhibited a variable course or even an overall decrease in diversity (SI Appendix, Fig. S8). Delivery mode (i.e., vaginal versus cesarean section) did not affect this phenomenon (SI Appendix, Fig. S9). The role of other potential factors, such as active breast feeding and the use of contraceptives or antibiotics, could not be evaluated because those data were not collected after delivery. Stool and oral communities did not demonstrate similar degrees of change upon delivery.
Fig. 5.
Alpha diversity pre- and postdelivery at each body site. The Shannon index is plotted versus the time at which each specimen was collected relative to delivery for the subjects in the first group of 40 subjects who provided at least one postdelivery sample. Linear fits are shown for the predelivery (red) and postdelivery (green) specimens, considered separately. Significant shifts to higher diversity were observed postdelivery in vaginal communities (P = 5.2 × 10−6 paired Wilcoxon test) and, to a lesser extent, in the tooth/gum communities (P = 0.014) but not in saliva or stool.
The postdelivery shift in vaginal community diversity appeared to be particularly associated with significant declines in abundances of the most prevalent Lactobacillus species (SI Appendix, Fig. S10 and Table S6). Lactobacilli were replaced by a diverse mixture of bacteria, including Peptoniphilus, Prevotella, and Anaerococcus, which are typically anaerobic (SI Appendix, Fig. S11 and Table S6). The postdelivery changes in the vaginal communities caused them to become significantly more similar to stool communities (P < 10−3, paired Wilcoxon test; SI Appendix, Fig. S12). Analogous changes in similarity were not observed when comparing the vaginal communities with either of the oral communities. We found no evidence for systematic interactions between the communities at different body sites during pregnancy. The validation dataset from the second group of subjects corroborated the finding from our discovery dataset of a postdelivery shift in vaginal community diversity (SI Appendix, Fig. S13).
Discussion
Pregnancy is a time of dramatic host environmental remodeling as a mother accommodates and nourishes her developing fetus. Although it is believed that the indigenous microbiota plays a fundamental role in both shaping and responding to the host environment, there are relatively few data on human microbiota structure at multiple body sites with high temporal resolution during this critical time. The importance of the microbiota for maternal and fetal health is suggested by links between conditions of altered microbial community structure in the vagina (e.g., bacterial vaginosis) (12, 13) or in the oral cavity (e.g., periodontitis) (26) and preterm birth. In addition, studies of amniotic fluid from women with preterm labor have revealed bacterial species indigenous to the vagina, oral cavity, and gut (6–9).
Using prospectively collected samples, we analyzed the microbiota from a total of 49 women in two case-control analyses of nonoverlapping subject groups. First, we characterized the temporal dynamics of microbiota structure and composition during pregnancy in 40 women based on weekly sampling from four relevant body sites: vagina, distal gut (stool), saliva, and tooth/gum. Of note, in this discovery dataset we found that the structure and composition of the microbiota are relatively stable overall but that some site-specific communities can vary considerably over time (e.g., disparate interstate transition rates of vaginal CSTs). Despite the small number of women with premature deliveries in this study, we identified candidate vaginal community compositional profiles in early pregnancy that were significantly associated with an elevated rate of subsequent preterm birth. We subsequently analyzed vaginal samples from nine additional women and corroborated the association between the high-risk community profiles and preterm birth.
In nonpregnant women, longitudinal studies of the vaginal microbiota have demonstrated stability when viewed at a weekly timescale, punctuated by occasional daily fluctuations (25, 27–32). These studies also documented significant variability among individuals with respect to the typical vaginal CST and its level of stability. Certain types of communities, such as that dominated by L. crispatus, are more stable than others. Menses can disrupt vaginal community composition, albeit transiently (25).
In pregnant women, vaginal microbial community dynamics have been less well studied, and dynamics at nonvaginal body sites have hardly been studied at all. Two recent studies by Romero and colleagues, with a sampling frequency of every 4 wk until week 24 of gestation and then every 2 wk until the last prenatal visit, provide the most meaningful comparisons with our findings (17, 18). One study of 22 women with healthy pregnancies reported the vaginal microbiota to be more stable during pregnancy than in the nonpregnant state (18). The other study, involving 72 women with healthy term pregnancies, described changes in the composition of the vaginal microbiota (e.g., an increase in the relative abundance of Lactobacillus species and a decrease in anaerobic species) as a function of gestational age (17). In contrast to this latter study, we found no significant change in the taxonomic structure of the vaginal microbial communities as pregnancy progressed. Importantly, because we obtained few samples early in the first trimester, we cannot preclude the possibility of significant changes in the vaginal microbial community at the beginning of pregnancy.
Two longitudinal studies (16, 19) and two cross-sectional studies (14, 15) also used culture-independent methods to characterize vaginal communities sampled from one or more trimesters. In aggregate their findings suggested that the vaginal microbiota during healthy pregnancy mostly resembles the nonpregnant microbiota, but with an even higher prevalence of Lactobacillus species.
The results of our analysis of the distal gut microbiota differ from those from Koren et al. (24) who reported a “remodeling of the gut microbiome” based on analysis of a single time point from each of the first and third trimesters in 91 pregnant women. Although Koren et al. reported that 16 of the women in their study took probiotic supplements during pregnancy, and that seven used antibiotics, the authors could not relate changes in beta diversity in individual women to their use of either supplements or antibiotics (24). The differences in study findings may reflect the fact that many of their subjects received a dietary intervention between those two time points.
Vaginal Community Structure.
We found, as have others (11, 17, 18), that vaginal communities exhibited distinct patterns of assembly distinguished by the dominance of a particular Lactobacillus species (i.e., CST 1, 2, 3, and 5) or by a non-Lactobacillus–dominated diverse community (CST 4). The robustness of CST assignment across different studies using various methodologies supports CSTs as valid descriptors of vaginal microbial communities that can facilitate comparisons across pregnancy status and studies. The relative stability of each Lactobacillus-dominated CST in our study was similar to that reported in nonpregnant women (25). We also found L. crispatus to be associated with increased stability of the vaginal community.
Vaginal Community Signatures of Preterm Birth.
We found the high-diversity vaginal CST 4 to be associated with an increased incidence of preterm birth. Indeed, CST 4 exhibited both dose–response (Fig. 4A) and temporal (Fig. 4B) relationships with preterm birth. This finding is consistent with previous associations between either high-diversity vaginal communities (16) or bacterial vaginosis (12, 13) and increased risk for preterm birth. Of note, if we had collected specimens less frequently than weekly, we would have missed a number of excursions to CST 4 and hence would have been hindered in associating this state with preterm birth.
Further analysis of taxa abundances within CST4 suggested that high abundance of Gardnerella combined with low abundance of Lactobacillus might be particularly predictive of preterm birth. Although this finding was based on a relatively small number of subjects, it remained significant even when the most conservative approach was taken to test the independence of samples from the same subject. The finding was confirmed with a validation dataset from nine subjects in which the one woman who had high Gardnerella in conjunction with low Lactobacillus delivered preterm. Our analysis also suggested that a high abundance of Ureaplasma combined with a low abundance of Lactobacillus is associated with preterm birth. Both Gardnerella and Ureaplasma have been implicated by a large body of literature as having potential roles in the pathogenesis of preterm delivery (SI Appendix, SI Discussion).
In their recent longitudinal study, Romero and colleagues (17) found no significant difference in taxon abundances or diversity of vaginal communities between term and preterm pregnancies. One possible reason for the contradictory results is the difference in the racial composition of our study cohorts. Previous work has demonstrated significant differences between vaginal microbial communities associated with race (11). In addition, the study by Romero et al. included only births before gestational week 34. Our cohort was clinically more heterogeneous and included some women whose labor onset was nonspontaneous (e.g., caused by preterm premature rupture of membranes) (SI Appendix, Table S2). Another potentially important difference between our study and previous work, especially when considering patterns related to Gardnerella abundance, is the use of different PCR primers (18, 25).
Vaginal Community Alteration After Delivery.
We found that most women experienced a significant, abrupt, and durable alteration in the vaginal community beginning with the first sample collected postdelivery. Compared with samples from other body sites in the postpartum period, the vaginal communities became more similar to the gut communities. The mechanisms underlying this potential convergence are unknown. The persistence of the disturbance, as well as its presence after both vaginal and caesarean deliveries, argues against simple translocation of stool communities to the vagina as the primary mechanism.
The potential impact on maternal health of the postdelivery shift of the vaginal community also is unknown but includes the possibility that a persistently altered vaginal community might affect the outcome of a subsequent pregnancy if conception occurs too soon after delivery. A short interpregnancy interval (e.g., <12 mo) is associated with an increased risk of preterm birth. Whether an altered postdelivery vaginal community plays a contributing role warrants further study.
Materials and Methods
To enhance the reproducibility of our results, the reader will find all the code necessary to reproduce the analyses and figures presented in this article available at statweb.stanford.edu/∼susan/papers/PNASRR.html. This website includes pointers to the R markdown files, the output in html, and to the data in the form of OTU count tables. OTU tables and associated data are provided in Datasets S1 and S2.
Study Population and Sampling Procedures.
Pregnant women age 18 y or older presenting to the obstetrical clinics of the Lucille Packard Children’s Hospital at Stanford University for prenatal care were enrolled, and two case-control groups were selected. The first consisted of 40 women, 11 of whom delivered preterm; the second consisted of nine women, four of whom delivered preterm. Specimens from the vagina, stool, saliva, and tooth/gum were self-collected by participants weekly from the time of study enrollment until delivery and monthly from the time of delivery for up to 12 mo. For further information, see SI Appendix, SI Methods and Tables S1 and S2.
DNA Extraction, 16S rDNA Amplification, and Amplicon Sequencing.
After extraction of genomic DNA, the V3–V5 region of the 16S rRNA gene was PCR-amplified from 3,767 vaginal, stool, saliva, and tooth/gum specimens from the 40 women in the first group and analyzed with pyrosequencing as a discovery dataset. The V4 region was amplified from 246 vaginal specimens from the nine women in the second group and analyzed with Illumina-based sequencing as a validation dataset.
Sequence Filtering, OTU Clustering, and Chimera Removal.
The two datasets were analyzed similarly but separately. Initial quality processing was performed using QIIME version 1.7 (qiime.org), followed by global trimming to 350 bases and filtering. Raw Illumina read-pairs were quality filtered, merged, and de-multiplexed. OTU clustering at a 97% sequence identity threshold was performed using the UPARSE algorithm. After OTU clustering, removal of chimeric sequences was performed in a stringent two-step process. See SI Appendix, SI Methods for details.
Bioinformatics Approach and Statistical Analysis.
Statistical analyses were performed using ‘R’ language and environment [R 2014, https://www.r-project.org version 3.1.1, including phyloseq (33)] and other packages. See SI Appendix, SI Methods for details.
Evaluating Trends with Gestational Time.
Changes in stability and diversity over the course of pregnancy were evaluated by LME modeling using the nlme::lme function in R. The subject was included as a random effect for both the intercept and the slope of the estimated fit. In all cases, the analysis of trends with gestational time was repeated with multiple measures of stability and diversity.
Evaluating Differential Abundance.
DESeq2 (34) was used to perform paired (by subject) two-class testing for differential relative abundance and to perform unpaired testing for associations between differential abundance within CST 4 specimens and preterm delivery. OTUs were considered significantly differentially abundant between classes if their adjusted P value was <0.1 and if the estimated fold change was >1.5 or <1/1.5.
Clustering into CSTs.
First, the Bray–Curtis distance between all samples was calculated. This distance matrix was denoised by extracting the most significant Principal Coordinates Analysis (PCoA) eigenvectors. The partitioning around medoids algorithm (pam in R) was applied to these PCoA distances. The number of clusters (k = 5) was determined from the gap statistic (SI Appendix, Fig. S14) (35).
Estimating Vaginal CST Transition Rates.
Before vaginal CST transition rates were estimated, the dataset was restricted to pairs of consecutive samples collected 4–10 d apart. This set of 652 paired samples had time-separations of 4–10 d (mean 6.96 d) and a first-quartile, third-quartile, and median of 7 d. The 1-wk transition rate was quantified as the maximum-likelihood estimate from this set of paired samples.
Supplementary Material
Acknowledgments
We thank the study participants and Cele Quaintance, Nick Scalfone, Chris Paiji, Kat Sanders, Katie Cumnock, Ana Laborde, March of Dimes Prematurity Research Center study coordinators, the nursing staff in the obstetrical clinics and the labor and delivery unit of Lucille Packard Children’s Hospital, and Stephen Cornell and participants of the Newton Institute Workshop on Metagenomics. This research was supported by the March of Dimes Prematurity Research Center at Stanford University, NIH National Center for Advancing Translational Science Clinical and Translational Science Award UL1 TR001085, the Stanford Child Health Research Institute, NIH Grant R01 GM086884 (to S.P.H. and B.J.C.), National Science Foundation Division of Mathematical Sciences Grant 1162538 (to S.P.H. and B.J.C.), and the Thomas C. and Joan M. Merigan Endowment at Stanford University (D.A.R.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: Raw sequence data have been deposited at the Sequence Read Archive (SRP no. 288562).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1502875112/-/DCSupplemental.
References
- 1.Goodrich JK, et al. Human genetics shape the gut microbiome. Cell. 2014;159(4):789–799. doi: 10.1016/j.cell.2014.09.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ursell LK, et al. The interpersonal and intrapersonal diversity of human-associated microbiota in key body sites. J Allergy Clin Immunol. 2012;129(5):1204–1208. doi: 10.1016/j.jaci.2012.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dethlefsen L, McFall-Ngai M, Relman DA. An ecological and evolutionary perspective on human-microbe mutualism and disease. Nature. 2007;449(7164):811–818. doi: 10.1038/nature06245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Blencowe H, et al. Born too soon: The global epidemiology of 15 million preterm births. Reprod Health. 2013;10(Suppl 1):S2. doi: 10.1186/1742-4755-10-S1-S2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Romero R, Dey SK, Fisher SJ. Preterm labor: One syndrome, many causes. Science. 2014;345(6198):760–765. doi: 10.1126/science.1251816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.DiGiulio DB, et al. Microbial prevalence, diversity and abundance in amniotic fluid during preterm labor: A molecular and culture-based investigation. PLoS One. 2008;3(8):e3056. doi: 10.1371/journal.pone.0003056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.DiGiulio DB, et al. 2010. Prevalence and diversity of microbes in the amniotic fluid, the fetal inflammatory response, and pregnancy outcome in women with preterm pre-labor rupture of membranes. Am J Reprod Immunol 64(1):38–57.
- 8.Gardella C, et al. Identification and sequencing of bacterial rDNAs in culture-negative amniotic fluid from women in premature labor. Am J Perinatol. 2004;21(6):319–323. doi: 10.1055/s-2004-831884. [DOI] [PubMed] [Google Scholar]
- 9.Han YW, Shen T, Chung P, Buhimschi IA, Buhimschi CS. Uncultivated bacteria as etiologic agents of intra-amniotic inflammation leading to preterm birth. J Clin Microbiol. 2009;47(1):38–47. doi: 10.1128/JCM.01206-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fredricks DN, Fiedler TL, Marrazzo JM. Molecular identification of bacteria associated with bacterial vaginosis. N Engl J Med. 2005;353(18):1899–1911. doi: 10.1056/NEJMoa043802. [DOI] [PubMed] [Google Scholar]
- 11.Ravel J, et al. Vaginal microbiome of reproductive-age women. Proc Natl Acad Sci USA. 2011;108(Suppl 1):4680–4687. doi: 10.1073/pnas.1002611107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hillier SL, et al. The Vaginal Infections and Prematurity Study Group Association between bacterial vaginosis and preterm delivery of a low-birth-weight infant. N Engl J Med. 1995;333(26):1737–1742. doi: 10.1056/NEJM199512283332604. [DOI] [PubMed] [Google Scholar]
- 13.Leitich H, et al. Bacterial vaginosis as a risk factor for preterm delivery: A meta-analysis. Am J Obstet Gynecol. 2003;189(1):139–147. doi: 10.1067/mob.2003.339. [DOI] [PubMed] [Google Scholar]
- 14.Aagaard K, et al. A metagenomic approach to characterization of the vaginal microbiome signature in pregnancy. PLoS One. 2012;7(6):e36466. doi: 10.1371/journal.pone.0036466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hernández-Rodríguez C, et al. Vaginal microbiota of healthy pregnant Mexican women is constituted by four Lactobacillus species and several vaginosis-associated bacteria. Infect Dis Obstet Gynecol. 2011;2011:851485. doi: 10.1155/2011/851485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hyman RW, et al. Diversity of the vaginal microbiome correlates with preterm birth. Reprod Sci. 2014;21(1):32–40. doi: 10.1177/1933719113488838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Romero R, et al. The vaginal microbiota of pregnant women who subsequently have spontaneous preterm labor and delivery and those with a normal delivery at term. Microbiome. 2014;2:18. doi: 10.1186/2049-2618-2-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Romero R, et al. The composition and stability of the vaginal microbiota of normal pregnant women is different from that of non-pregnant women. Microbiome. 2014;2(1):4. doi: 10.1186/2049-2618-2-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Verstraelen H, et al. Longitudinal analysis of the vaginal microflora in pregnancy suggests that L. crispatus promotes the stability of the normal vaginal microflora and that L. gasseri and/or L. iners are more conducive to the occurrence of abnormal vaginal microflora. BMC Microbiol. 2009;9:116. doi: 10.1186/1471-2180-9-116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jensen J, Liljemark W, Bloomquist C. The effect of female sex hormones on subgingival plaque. J Periodontol. 1981;52(10):599–602. doi: 10.1902/jop.1981.52.10.599. [DOI] [PubMed] [Google Scholar]
- 21.Kornman KS, Loesche WJ. The subgingival microbial flora during pregnancy. J Periodontal Res. 1980;15(2):111–122. doi: 10.1111/j.1600-0765.1980.tb00265.x. [DOI] [PubMed] [Google Scholar]
- 22.Adriaens LM, Alessandri R, Spörri S, Lang NP, Persson GR. Does pregnancy have an impact on the subgingival microbiota? J Periodontol. 2009;80(1):72–81. doi: 10.1902/jop.2009.080012. [DOI] [PubMed] [Google Scholar]
- 23.Collado MC, Isolauri E, Laitinen K, Salminen S. Distinct composition of gut microbiota during pregnancy in overweight and normal-weight women. Am J Clin Nutr. 2008;88(4):894–899. doi: 10.1093/ajcn/88.4.894. [DOI] [PubMed] [Google Scholar]
- 24.Koren O, et al. Host remodeling of the gut microbiome and metabolic changes during pregnancy. Cell. 2012;150(3):470–480. doi: 10.1016/j.cell.2012.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gajer P, et al. Temporal dynamics of the human vaginal microbiota. Sci Transl Med. 2012;4(132):132ra52. doi: 10.1126/scitranslmed.3003605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Siqueira FM, et al. Intrauterine growth restriction, low birth weight, and preterm birth: Adverse pregnancy outcomes and their association with maternal periodontitis. J Periodontol. 2007;78(12):2266–2276. doi: 10.1902/jop.2007.070196. [DOI] [PubMed] [Google Scholar]
- 27.Brotman RM, Ravel J, Cone RA, Zenilman JM. Rapid fluctuation of the vaginal microbiota measured by Gram stain analysis. Sex Transm Infect. 2010;86(4):297–302. doi: 10.1136/sti.2009.040592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chaban B, et al. Characterization of the vaginal microbiota of healthy Canadian women through the menstrual cycle. Microbiome. 2014;2:23. doi: 10.1186/2049-2618-2-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hickey RJ, et al. 2013. Effects of tampons and menses on the composition and diversity of vaginal microbial communities over time. BJOG 120(6):695–704; discussion 704–696.
- 30.Santiago GL, et al. Longitudinal study of the dynamics of vaginal microflora during two consecutive menstrual cycles. PLoS One. 2011;6(11):e28180. doi: 10.1371/journal.pone.0028180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Srinivasan S, et al. Temporal variability of human vaginal bacteria and relationship with bacterial vaginosis. PLoS One. 2010;5(4):e10197. doi: 10.1371/journal.pone.0010197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Thoma ME, et al. Longitudinal changes in vaginal microbiota composition assessed by gram stain among never sexually active pre- and postmenarcheal adolescents in Rakai, Uganda. J Pediatr Adolesc Gynecol. 2011;24(1):42–47. doi: 10.1016/j.jpag.2010.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.McMurdie PJ, Holmes S. phyloseq: An R package for reproducible interactive analysis and graphics of microbiome census data. PLoS One. 2013;8(4):e61217. doi: 10.1371/journal.pone.0061217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15(12):550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tibshirani R, Walter G, Hastie T. Estimating the number of clusters in a data set via the gap statistic. J R Stat Soc, B. 2001;63:411–423. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.