Significance
The vertebral column provides essential structural and protective functions. The total number of vertebral elements and their specific morphologies are remarkably reproducible within a given species, yet can be tailored to the requirements of separate vertebrate species. Major genetic determinants driving formation of the vertebral column are known, but how they are regulated to achieve a highly reproducible structure remains to be fully elucidated. In this report, we show that the miR-196 family of microRNAs are essential in defining correct vertebral number and vertebral identity in mouse. We reveal the molecular landscape controlled, either directly or indirectly, by miR-196 activity, to demonstrate that miR-196 impacts many key developmental signalling pathways and reinforces a timely trunk-to-tail Hox code transition.
Keywords: Hox genes, microRNA, miR-196, axial patterning, vertebral specification
Abstract
The Hox genes play a central role in patterning the embryonic anterior-to-posterior axis. An important function of Hox activity in vertebrates is the specification of different vertebral morphologies, with an additional role in axis elongation emerging. The miR-196 family of microRNAs (miRNAs) are predicted to extensively target Hox 3′ UTRs, although the full extent to which miR-196 regulates Hox expression dynamics and influences mammalian development remains to be elucidated. Here we used an extensive allelic series of mouse knockouts to show that the miR-196 family of miRNAs is essential both for properly patterning vertebral identity at different axial levels and for modulating the total number of vertebrae. All three miR-196 paralogs, 196a1, 196a2, and 196b, act redundantly to pattern the midthoracic region, whereas 196a2 and 196b have an additive role in controlling the number of rib-bearing vertebra and positioning of the sacrum. Independent of this, 196a1, 196a2, and 196b act redundantly to constrain total vertebral number. Loss of miR-196 leads to a collective up-regulation of numerous trunk Hox target genes with a concomitant delay in activation of caudal Hox genes, which are proposed to signal the end of axis extension. Additionally, we identified altered molecular signatures associated with the Wnt, Fgf, and Notch/segmentation pathways and demonstrate that miR-196 has the potential to regulate Wnt activity by multiple mechanisms. By feeding into, and thereby integrating, multiple genetic networks controlling vertebral number and identity, miR-196 is a critical player defining axial formulae.
A defining feature of the vertebrate animals is the presence of a segmented vertebral column. Species are uniquely characterized by the total number of vertebrae that form and by the regionalization of these vertebra along the anterior-to-posterior axis into groups with distinct morphologies (e.g., cervical, thoracic, lumbar, and sacral). The genetic determinants of vertebral number and vertebral identity have largely been considered as separate; thus how, or even whether, these processes are molecularly integrated remains to be clearly elucidated.
Vertebral precursors, known as somites, arise by continued expansion and segmentation of a region of the caudal embryo, the presomitic mesoderm (PSM) (1). Expansion of the PSM requires a self-renewing axial progenitor population that initially resides in the node-streak border of the epiblast and subsequently repositions to the tailbud (2–6). These progenitors provide a source of cells that, following ingression through the primitive streak, populate the PSM and other derivatives to drive posterior elongation. Key players in this process include genes involved in Wnt and Fgf signaling, in addition to the Cdx transcription factors, as evidenced by severe axis truncations when each are mutated (7). Balancing the expansion of this cell population, cells of the anterior PSM bud off to form somites with a rhythmic periodicity inherent to each species. The eventual exhaustion of progenitor self-renewal capacity is thought to halt axis elongation, the timing of which is a critical factor in establishing species-specific vertebral number (8).
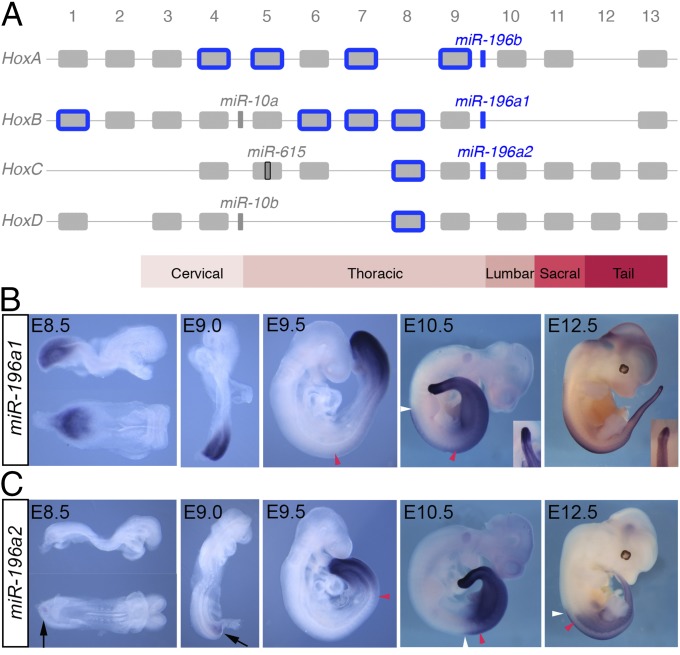
Within vertebral precursors, specific combinations of Hox transcription factors impart positional information that governs vertebral identity (9). In mammals, the 39 Hox genes are clustered at four separate genomic loci (HoxA, HoxB, HoxC, and HoxD), with each gene classified into 1 of 13 paralogous groups dependent on sequence similarities and relative positions within the respective clusters (Fig. 1A). These genes are expressed in partially overlapping domains during embryonic development, with a spatiotemporal collinearity that reflects genomic ordering (10, 11). Exhaustive analysis of Hox mouse mutants over more than 20 years has revealed individual and cumulative Hox function in conferring specific positional identities to the forming vertebral column (9). For instance, the central/trunk Hox proteins (paralogs 5–8) primarily pattern thoracic vertebra, whereas Hox 11 paralogs pattern sacral and caudal vertebra (12) and position the sacrum (13, 14).
Fig. 1.
Unique and overlapping expression patterns of miR-196 paralogs in mouse. (A) Mouse Hox clusters, with the position of Hox-embedded microRNAs depicted. Predicted Hox targets of the miR-196 family are indicated in blue. (B and C) Detection of eGFP transcripts in miR-196a1GFP/+ (B) and miR-196a2GFP/+ (C) embryos demonstrates spatiotemporal expression differences for these identical miRNAs. Embryonic age indicated: red and white arrowheads indicate the anterior boundary of somitic and neural expression, respectively. A discrete band of reduced eGFP signal in the anterior PSM of later stage 196a1GFP/+ embryos is shown in Insets. Weak ventral eGFP signal in miR-196a2GFP/+ embryos at E8.5 and E9.0 is indicated with black arrows.
In addition to transcripts encoding for the Hox proteins, transcription within the genomic Hox clusters produce noncoding regulatory RNAs, including several microRNAs (miRNAs) (Fig. 1A) (15). In mice, these include the miR-10 family, which is found throughout most bilaterian animals; miR-615, which his found in eutherian mammals; and the miR-196 family, which is found in vertebrates and tunicates. Three murine miR-196 paralogs exist (referred to as 196a1, 196a2, and 196b), each with essentially identical targeting potential (16, 17). The three miR-196 paralogs exhibit deep conservation across all vertebrate lineages analyzed to date, both in terms of their genomic positioning upstream of Hox9 paralogs and in their extensive predicted targeting of Hox 3′ UTRs primarily of the trunk region (Fig. 1A) (17–19). In an early developmental context, in vivo validation of these interactions has focused primarily on a single Hox target, Hoxb8 (17, 20–24), with no evidence for additional Hox target regulation observed in miR-196 knockdown studies in zebrafish (24). Thus, the extent to which collective Hox output is regulated by miR-196, either in terms of the number of genes affected or the relative levels of regulation, is unknown.
The extent to which the developmental modules that define total vertebral number are integrated with those that impart positional information has not been well established, although these processes can be uncoupled (25–27). A function for Hox proteins in establishing total vertebral number has been largely dismissed because, with the exception of Hoxb13−/− (28), Hox knockouts (KOs) do not phenotypically support such a role (9). However, ectopic trunk Hox activity can, under certain conditions, drive axis elongation (29). Conversely, posterior Hox activity slows axis elongation and terminates the main body axis (29, 30), suggesting an alternative view of Hox activity in this context. In this light, phenotypic observations following reduced activity of miR-196, a repressor of Hox activity, are quite remarkable. Knockdown studies in chick and zebrafish support a role for miR-196 in regulating vertebral identity (22, 24). Additionally, miR-196 morphant zebrafish exhibit an extended vertebral column, with what appears to be an “insertion” of a rib-bearing precaudal element (24). How this latter phenotype arises developmentally is not known and is difficult to reconcile with derepression of trunk Hox target genes alone (31, 32). These knockdown approaches could not shed light on individual paralog contributions for this highly related miRNA family, and importantly, the molecular networks downstream of miR-196, which have the potential to drive phenotypic alterations, remain uncharacterized.
Here, we generated individual KO alleles for each of the three miR-196 family members in mice. These tools have allowed us to build an entire allelic deletion series to reveal the individual and additive roles of miR-196 paralogs in patterning vertebral identity at many axial levels and in controlling the total number of vertebrae. We characterized the detailed molecular landscape controlled by miR-196 activity in the early embryo to show that miR-196 regulates, and therefore has the ability to integrate, multiple key signaling pathways to drive developmental processes.
Results
Differential Transcription of miR-196a1 and miR-196a2 in the Developing Embryo.
To reveal the individual expression patterns, and therefore potential for functional redundancy, of identical miRNAs 196a1 and 196a2, we generated eGFP knock-in alleles termed 196a1GFP and 196a2GFP (Fig. S1). Expression of reporter mRNA reflects sites of active transcription, although it does not reveal additional posttranscriptional regulation that endogenous miRNAs may undergo. Whole mount in situ hybridization analysis of reporter mRNA indicated that both miRNAs were expressed specifically in the posterior embryonic derivatives of all three germ layers and revealed striking differences in their spatiotemporal kinetics that have not previously been delineated (Fig. 1 B and C). miR-196a1 is expressed throughout the posterior growth zone at embryonic day (E)8.5 (Fig. 1B). Strong expression is maintained in the PSM until the end of axis elongation, with a discrete band of low expression in the anterior PSM from E10.5 (Fig. 1B, Inset). The anterior boundary of somitic and neural expression extends to approximately somite 13/14 [prevertebra (pv) 9, thoracic (T) 2] at E9.5 with a caudal shift in somitic tissue and a rostral shift in neural tissue as development proceeds. This expression profile indicates that 196a1 exhibits a classic collinear profile relative to the adjacent Hox gene, Hoxb9 (anterior limit at E9.5, pv3) (33). miR-196a2 expression is temporally delayed relative to 196a1 (Fig. 1C), with faint expression ventral to the PSM at E8.5–9.0. Strong expression is then observed throughout the PSM and neural plate at E9.5. A stable anterior somitic limit at approximately somite 21/22 (pv17, T10) and neural limit 2 somites rostral to this is established soon after, consistent with its positioning between Hoxc9 and Hoxc10 (34). This analysis revealed both unique and overlapping expression patterns of 196a1 and 196a2, suggesting these identical miRNAs might have both unique functions where individually expressed and either redundant or additive functions at sites of coexpression.
Fig. S1.
Generation of miR-196a1GFP and miR-196a2GFP knock-in mouse lines. (A and B) Knock-in targeting strategy for miR-196a1GFP (A) and miR-196a2GFP (B). (C and D) Correct targeting was confirmed by Southern blot analysis of the miR-196a1 (C) and miR-196a2 (D) loci. The Southern blot strategy for each knock-in allele is indicated (A and B), with the position of each Southern probe indicated with a blue box.
Genetic Deletion of miR-196 Leads to Altered Vertebral Identity.
The collective function of miR-196 family members has yet to be assessed in mammals. Moreover, the dissection of paralog contributions to overall miR-196 activity has not been achieved in any system. To address this, we generated straight KO alleles at each of the three murine miR-196 loci (Fig. S2), allowing us to create the complete allelic series of single, double, and triple miR-196 KO embryos. Skeletal analysis across this allelic series allowed us to demonstrate an essential requirement for miR-196 activity in patterning the midthoracic, the thoraco-lumbar transition, and lumbo-sacral regions, with both paralog-specific and additive effects revealed.
Fig. S2.
Generation of miR-196a1−/−, miR-196a2−/−, and miR-196b−/− KO mouse lines. (A) Generalized targeting strategy used by the Wellcome Trust Sanger Institute to create miRNA KO ES cells (74). Before ES cell injection, correct targeting was confirmed in house by Southern blot analysis of the miR-196a1−/− (B), miR-196a2−/− (C), and miR-196b−/− (D) loci. The general Southern blot strategy is indicated in blue in A.
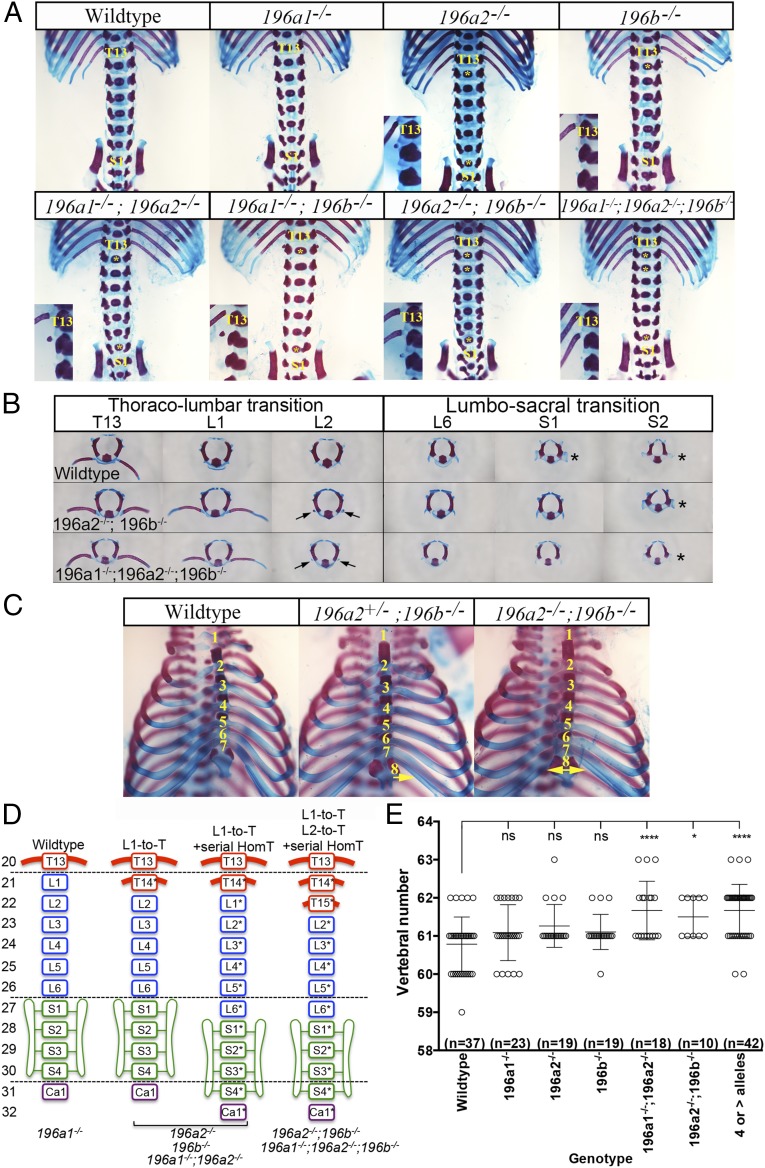
Removal of individual miR-196 paralogs alone revealed partially penetrant homeotic patterning defects (Fig. 2A and Table S1). In 196a2 or 196b single-mutant embryos, the presence of an ectopic rudimentary rib nubbin on the first lumbar vertebra indicated an anterior homeotic transformation of this element (Fig. 2A). Additionally, in approximately one quarter of cases, we observed anterior homeotic transformations encompassing all subsequent lumbar and sacral elements, resulting in a posterior displacement of the sacrum (schematized in Fig. 2D). Although this latter phenotype could be interpreted as an insertion of a thoracic element, the repositioned last lumbar vertebrae (L6* in Fig. 2D) was often asymmetric, with both lumbar and sacral characteristics (Table S1), which supports the interpretation of serial identity changes, beginning at L1 and encompassing all subsequent elements. We did not observe a similar L1-to-T anterior homeotic transformation in 196a1 single-mutant embryos, which for the most part exhibited no overt vertebral alterations (Fig. 2A). However, at very low penetrance (Table S1), 196a1 single-mutant embryos displayed an anterior displacement of the sacrum, with or without a reduction in rib length of the last thoracic element (T13), suggesting these paralogs may have an opposing role at this axial level.
Fig. 2.
miR-196 paralogs function in establishing vertebral identity and number in mouse. (A) Identification of vertebral patterning defects in individual and compound miR-196 loss-of-function E18.5 embryos. Genotypes indicated. The positions of the 13th thoracic element (T13) and first sacral element (S1) are labeled. (Inset) Thoraco-lumbar junction. (B) Individual vertebra analysis to demonstrate identity alterations at the thoraco-lumbar and lumbo-sacral junctions. Genotypes indicated. The position of a rib-like nubbin on lumbar elements is marked with arrow. The position of sacral process is marked with an asterisk. (C) Rib fusion defects observed following loss of miR-196 alleles, genotypes indicated. Fusion of the eighth rib to the sternum was unilateral or bilateral as indicated with arrows. (D) Summary of patterning defects identified across the miR-196 allelic series. An asterisk indicates a homeotic transformation of that vertebral element. (E) Quantification of vertebral number in single and compound mir-196 loss-of-function E18.5 embryos identifies a role for miR-196 in controlling axis length in mouse. Statistical comparison of vertebral number relative to WT was performed using a permutation test, with P values corrected for multiple hypothesis testing using the Bonferroni method: *P < 0.05, ***P < 0.001, and ****P < 0.0001.
Table S1.
Removal of miR-196 family members causes vertebral defects
| Genotype | Number analyzed | C1 or C2 malformation | C7 to T Transformation | Number of sternal ribs | Transitional vertebra | Reduced T13 rib(s) | L1 (WT) | L1 to T | L1+L2 to T | Vertebral position of first sacral element | |||||||
| 7 (WT) | 7/8 | 8 | T10 (WT) | T11 | 26 | 26/27 | 27 (WT) | 27/28 | 28 | ||||||||
| WT (C57Bl6 J) | 18 | 2 | 18 | 18 | 18 | 1 | 17 | ||||||||||
| WT (C57Bl6 J/N) | 30 | 30 | 30 | 30 | 30 | ||||||||||||
| 196a1 GFP/+ | 20 | 1 | 20 | 20 | 1 | 19 | 2 | 3 | 18 | ||||||||
| 196a1 GFP/GFP | 12 | 12 | 12 | 1 | 11 | 3 | 1 | 8 | |||||||||
| 196a1+/− | 24 | 1 | 24 | 24 | 24 | 24 | |||||||||||
| 196a1−/− | 16 | 16 | 16 | 1 | 15 | 2 | 1 | 12 | |||||||||
| 196a2 GFP/+ | 8 | 1 | 7 | 1 | 7 | 1 | 7 | 1 | 7 | 1 | |||||||
| 196a2 GFP/GFP | 14 | 14 | 3 | 11 | 1 | 13 | 10 | 2 | 2 | ||||||||
| 196a2+/− | 15 | 15 | 14 | 1 | 15 | 15 | |||||||||||
| 196a2−/− | 7 | 7 | 7 | 3 | 4 | 6 | 1 | ||||||||||
| 196b+/− | 38 | 3 | 38 | 38 | 37 | 1 | 38 | ||||||||||
| 196b−/− | 26 | 5 | 26 | 26 | 13 | 13 | 22 | 2 | 2 | ||||||||
| a1-GFP;a2-GFP | |||||||||||||||||
| Het; Het | 13 | 1 | 12 | 1 | 12 | 1 | 11 | 2 | 13 | ||||||||
| Het;Mut | 3 | 2 | 1 | 1 | 2 | 0 | 3 | 0 | 3 | ||||||||
| Mut; Het | 8 | 7 | 1 | 5 | 3 | 7 | 1 | 8 | |||||||||
| Mut;Mut | 18 | 15 | 3 | 0 | 18 | 0 | 18 | 13 | 5 | ||||||||
| a1-GFP;b−/− | |||||||||||||||||
| Het; Het | 15 | 15 | 15 | 15 | 15 | ||||||||||||
| Het;Mut | 13 | 12 | 1 | 13 | 4 | 9 | 9 | 2 | 2 | ||||||||
| Mut;Het | 10 | 10 | 10 | 10 | 9 | 1 | |||||||||||
| Mut;Mut | 5 | 4 | 1 | 5 | 1 | 4 | 3 | 2 | |||||||||
| a2-GFP;b−/− | |||||||||||||||||
| Het;Het | 3 | 3 | 1 | 2 | 1 | 1 | 1 | ||||||||||
| Het;Mut | 19 | 10 | 16 | 2 | 1 | 17 | 2 | 0 | 19 | 0 | 1 | 18 | |||||
| Mut;Het | 2 | 1 | 1 | 0 | 2 | 0 | 1 | 1 | 0 | 2 | |||||||
| Mut;Mut | 13 | 6 | 6 | 7 | 4 | 9 | 0 | 13 | 0 | 13 | |||||||
| a1-GFP;a2-GFP;b−/− | |||||||||||||||||
| Het;Het;Het | 11 | 2 | 10 | 1 | 11 | 4 | 7 | 5 | 2 | 4 | |||||||
| Het;Het;Mut | 12 | 4 | 5 | 3 | 6 | 6 | 0 | 12 | 0 | 12 | |||||||
| Het;Mut;Het | 7 | 6 | 1 | 1 | 6 | 0 | 7 | 0 | 7 | ||||||||
| Het;Mut;Mut | 8 | 2 | 4 | 2 | 2 | 6 | 0 | 8 | 0 | 8 | |||||||
| Mut;Het;Het | 7 | 1 | 1 | 7 | 5 | 2 | 2 | 5 | 3 | 4 | |||||||
| Mut;Het;Mut | 6 | 1 | 4 | 2 | 3 | 3 | 1 | 4 | 1 | 1 | 1 | 4 | |||||
| Mut;Mut;Het | 3 | 3 | 1 | 2 | 0 | 3 | 0 | 3 | |||||||||
| Mut;Mut;Mut | 3 | 1 | 3 | 0 | 3 | 0 | 3 | 0 | 3 | ||||||||
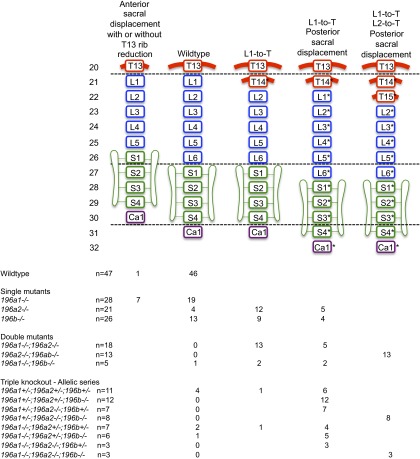
Summary of vertebral malformations and vertebral transformations identified in single and compound miR-196 KO mice. See SI Discussion for further details.
We hypothesized that the penetrance and severity of the phenotypes observed after mutating single miR-196 paralogs could be enhanced by combining these mutations. Indeed, 196a2−/−;196b−/− double-mutant skeletons exhibited a fully penetrant phenotype, with two pairs of supernumerary ribs and anterior homeotic transformation of all subsequent elements (Fig. 2 A, B, and D). Relative to this double mutant phenotype, triple KO embryos, 196a1−/−;196a2−/−;196b−/−, displayed no additional patterning defects (Fig. 2 A and D, complete dataset summarized in Fig. S3).
Fig. S3.
Summary of vertebral patterning alterations observed in miR-196 single and compound mutant mice. A summary of the main patterning defects observed in miR-196 mutant mice, homeotic transformation (HomT) of the WT axial formulae, is marked in with an asterisk. The numbers of skeletons analyzed for each genotype and their phenotypic spectrum are indicated. See SI Discussion for further details.
We also hypothesized that combining these mutations might reveal additional defects not observed in single mutants. Indeed, all double-mutant skeletons, or skeletons with a triple KO combination of four or more alleles removed, exhibited a partially penetrant increase in the number of ribs attached to the sternum (Table S1 and Fig. 2C), indicating a transformation of the eighth thoracic element to a more anterior identity. Together, our analysis showed that (i) 196a2 and 196b have single and additive effects in patterning the thoraco-lumbar transition and in positioning the sacrum, with a possible opposing role or 196a1 at this axial level, and (ii) 196a1, 196a2, and 196b act redundantly to pattern the midthoracic region, with phenotypic alterations observed only when two or more paralogs are removed. As such, our work has provided the first genetic proof, to our knowledge, for miR-196 as a homeotic family of genes and revealed identity changes at multiple axial levels.
Genetic Deletion of miR-196 Leads to an Increase in Vertebral Number.
Homeotic transformations do not alter the number of vertebrae, simply their identity. It was therefore surprising that, in zebrafish, miR-196 has been shown to constrain total vertebral number (24). We assessed whether this was an evolutionarily conserved function of miR-196 and found that the three murine miR-196 paralogs constrain total vertebral number in a redundant fashion. Wild-type (WT) C57BL6J/N mice exhibit small variations in the total number of vertebrae (Fig. 2E). Compared with the WT mean, we observed a statistically significant increase of approximately one vertebral element in various allelic combinations, including 196a1−/−;196a2−/−, 196a2−/−;196b−/−, and triple KO combinations with four or more alleles deleted (Fig. 2E). Depending on the exact allelic combination, this additional element was patterned as a thoracic (e.g., in 196a2−/−;196b−/− mice) or a postsacral (e.g., in 196a1−/−;196a2−/− mice) element. Together, these results indicate that miR-196–mediated control of vertebral number and patterning of segment identity are separable processes. All three miR-196 paralogs contribute additively to establishing vertebral number within mouse. Layered on top of this, individual miR-196 paralogs have a differential impact on positional identity and ultimate axial formulae, likely as a result of their differential spatiotemporal kinetics (Fig. 1 B–K) relative to target mRNAs.
Transcriptome Alterations Are Detected Following Allelic Removal of miR-196 Activity.
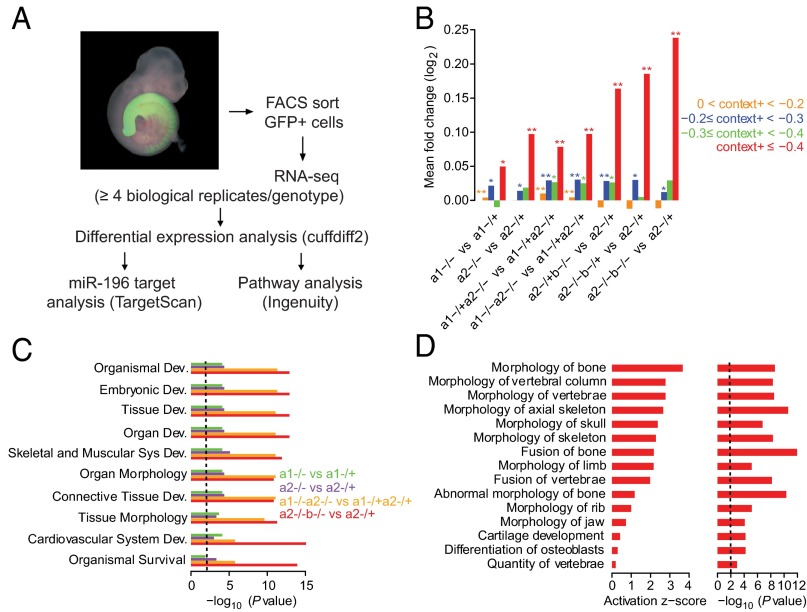
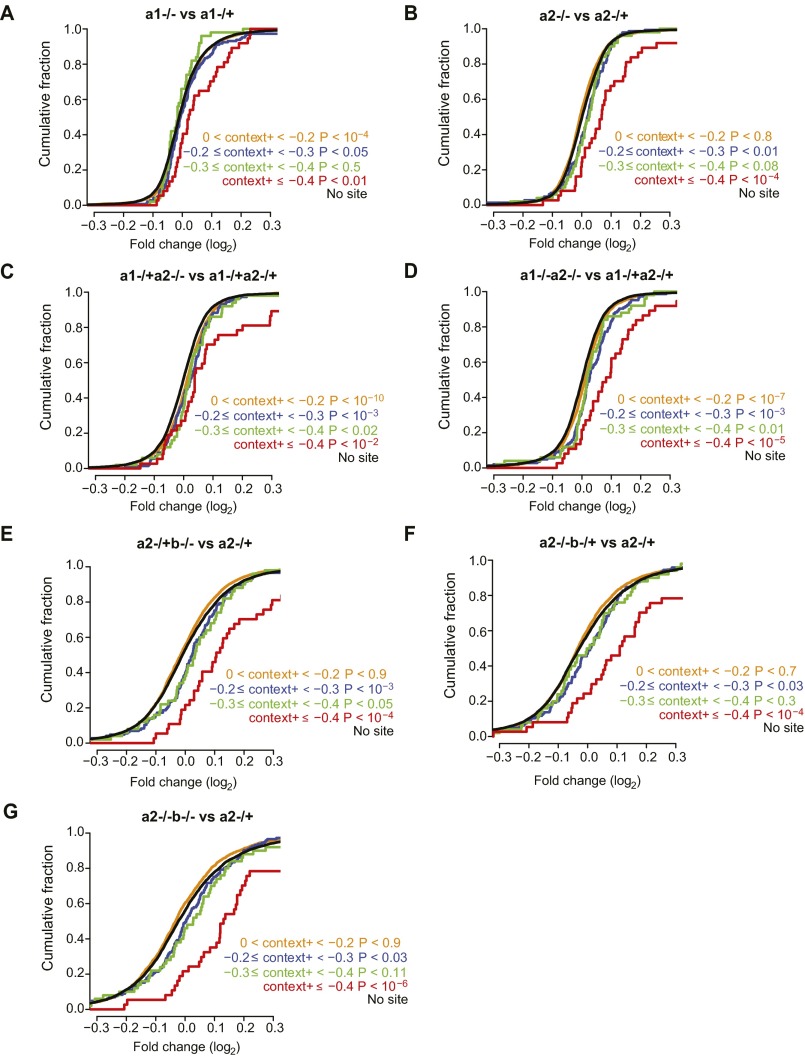
To elucidate the molecular mechanism and targets downstream of miR-196, we examined the response of mRNAs to the loss of mir-196 alleles in E9.5 embryos. To focus these molecular analyses on the relevant cells, i.e., those cells that normally express miR-196, we used only embryos with at least one eGFP knock-in allele and performed RNA-seq on RNA isolated from cells that were GFP positive (Fig. 3A). With mRNA profiled across 10 genotypes (Table S2), we then compared mRNA changes as increasing numbers and differing combinations of alleles were deleted (Dataset S1). We first examined the effect of allelic miR-196 deletion on predicted miR-196 target genes. Using the total context+ score from TargetScan 6.2, which considers the number and type of miRNA binding sites, as well as additional features to predict the genes most effectively targeted by each miRNA (35), we observed that the top predicted targets of miR-196 exhibited significant derepression on the loss of additional miR-196 alleles (Fig. 3B and Fig. S4). The derepression of these predicted targets increased with the number of additional alleles deleted (Fig. 3B), revealing miR-196 dosage sensitivity. The direct interaction between miR-196 and its target transcripts could occur in any of the three germ layer derivatives in which miR-196 was expressed, and indeed, an unbiased analysis of all differentially expressed genes revealed statistically altered molecular signatures reflecting this (Fig. S5). Of particular interest, we observe statistical enrichment in genes controlling skeletal morphology (Fig. 3 C and D and Fig. S5), indicating the presence of a molecular signature consistent with the vertebral abnormalities observed at the phenotypic level.
Fig. 3.
Whole transcriptome analysis of miR-196 mutant cells reveals a dysregulation of miRNA targets and skeletal genes. (A) Overview of the experimental and computational strategy used to identify global transcriptome alterations following loss of miR-196 function. (B) Mean fold changes of genes associated with predicted targets of miR-196, partitioned into four context+ intervals according to predicted miRNA targeting efficacy (0 < context+ < −0.2, n = 2,112; −0.2 ≤ context+ < −0.3, n = 145; −0.3 ≤ context+ < −0.4, n = 50; context+ ≤ −0.4, n = 37), across seven genotype comparisons. Statistical comparison of observed up-regulation of genes relative to genes with no miRNA target site, as evaluated by a one-sided Kolmogorov–Smirnov (K-S) test; *P < 0.05, **P < 0.001. (C) Top 10 significant categories related to gene development and function associated with differentially expressed genes. (D) Top 15 categories related to skeletal and muscular development activated in the 196a2−/−;196b−/− vs. 196a2−/+ comparison, with corresponding activation z-scores and P values. An activation z-score is a measurement of the consistency between the observed pattern of up- and down-regulation of genes in a category and the predicted activation or inhibition pattern in networks stored in the Ingenuity Knowledgebase relative to a random pattern (83). P values in C and D are Benjamini–Hochberg corrected P values, with dashed black lines indicating a significance threshold of 0.01.
Table S2.
RNA-seq library statistics
| Biological replicate no. | Sample | Sample code | Percent mapping | Total reads | Uniquely mapping in genome | Multiply mapping in genome | Total mapping | Total unique reads |
| 1 | a1+/− | 196a1_C13_Emb1_BC1RD4ACXX_ATCACG | 97.71% | 34,302,537 | 28,566,828 | 4,950,941 | 33,517,769 | 15,834,590 |
| 2 | a1+/− | 196a1_C13_Emb2_BC1RD4ACXX_CGATGT | 97.57% | 36,493,576 | 30,250,080 | 5,357,562 | 35,607,642 | 16,306,036 |
| 3 | a1+/− | 196a1_C13_Emb5_BC1RD4ACXX_TGACCA | 97.75% | 36,635,013 | 30,478,680 | 5,330,328 | 35,809,008 | 16,745,799 |
| 4 | a1+/− | 196a1_C48_Emb4_BC1RD4ACXX_CAGATC | 97.75% | 35,209,752 | 29,640,772 | 4,775,981 | 34,416,753 | 16,768,115 |
| 5 | a1+/− | 196a1_C48_Emb8_BC1RD4ACXX_TAGCTT | 97.69% | 36,435,824 | 30,610,577 | 4,984,636 | 35,595,213 | 16,688,316 |
| 1 | a1−/− | 196a1_C13_Emb4_BC1RD4ACXX_TTAGGC | 97.68% | 36,001,696 | 29,923,398 | 5,244,430 | 35,167,828 | 16,653,752 |
| 2 | a1−/− | 196a1_C48_Emb2_BC1RD4ACXX_ACAGTG | 97.70% | 42,097,844 | 35,276,234 | 5,854,056 | 41,130,290 | 18,871,066 |
| 3 | a1−/− | 196a1_C48_Emb3_BC1RD4ACXX_GCCAAT | 97.70% | 37,268,415 | 31,395,191 | 5,015,979 | 36,411,170 | 17,424,219 |
| 4 | a1−/− | 196a1_C48_Emb5_BC1RD4ACXX_ACTTGA | 97.77% | 37,050,074 | 31,201,935 | 5,020,156 | 36,222,091 | 17,868,027 |
| 5 | a1−/− | 196a1_C48_Emb7_BC1RD4ACXX_GATCAG | 97.68% | 36,052,009 | 30,258,424 | 4,956,997 | 35,215,421 | 16,500,172 |
| 1 | a2+/− | 196a2_C11_Emb5_BC1RD4ACXX_AGTCAA | 97.83% | 36,930,544 | 31,149,691 | 4,979,609 | 36,129,300 | 16,968,075 |
| 2 | a2+/− | 196a2_C21_Emb1_BC1RD4ACXX_AGTTCC | 97.82% | 39,407,655 | 33,253,041 | 5,297,002 | 38,550,043 | 17,863,925 |
| 3 | a2+/− | 196a2_C21_Emb2_BC1RD4ACXX_ATGTCA | 97.90% | 39,450,701 | 33,305,209 | 5,318,122 | 38,623,331 | 17,718,849 |
| 4 | a2+/− | 196a2_C21_Emb3_BC1RD4ACXX_CCGTCC | 97.77% | 41,768,476 | 35,190,696 | 5,645,960 | 40,836,656 | 18,489,309 |
| 5 | a2+/− | 196a2_C21_Emb8_BC1RD4ACXX_GTTTCG | 97.86% | 38,940,440 | 33,162,700 | 4,946,120 | 38,108,820 | 18,744,345 |
| 1 | a2−/− | 196a2_C11_Emb2_BC1RD4ACXX_GGCTAC | 97.83% | 37,181,569 | 31,143,092 | 5,229,789 | 36,372,881 | 16,802,724 |
| 2 | a2−/− | 196a2_C11_Emb3_BC1RD4ACXX_CTTGTA | 97.70% | 37,111,314 | 31,132,310 | 5,127,021 | 36,259,331 | 16,864,379 |
| 3 | a2−/− | 196a2_C21_Emb4_BC1RD4ACXX_GTCCGC | 97.79% | 37,134,604 | 31,332,133 | 4,982,472 | 36,314,605 | 17,290,622 |
| 4 | a2−/− | 196a2_C21_Emb5_BC1RD4ACXX_GTGAAA | 97.84% | 37,816,495 | 32,062,267 | 4,937,324 | 36,999,591 | 18,291,836 |
| 5 | a2−/− | 196a2_C21_Emb6_BC1RD4ACXX_GTGGCC | 97.81% | 37,591,637 | 31,674,555 | 5,093,143 | 36,767,698 | 17,367,253 |
| 1 | a1+/−a2+/− | 196a1a2_B83_Emb1_BC1RD4ACXX_CGTACG | 97.72% | 39,919,329 | 33,339,075 | 5,668,668 | 39,007,743 | 18,240,196 |
| 2 | a1+/−a2+/− | 196a1a2_B83_Emb2_BC1RD4ACXX_GAGTGG | 97.80% | 36,338,514 | 30,588,692 | 4,948,951 | 35,537,643 | 17,293,410 |
| 3 | a1+/−a2+/− | 196a1a2_B83_Emb3_BC1RD4ACXX_ACTGAT | 97.77% | 40,965,107 | 34,438,035 | 5,611,690 | 40,049,725 | 18,569,309 |
| 4 | a1+/−a2+/− | 196a1a2_B83_Emb4_BC1RD4ACXX_ATTCCT | 97.71% | 39,335,305 | 33,057,525 | 5,378,346 | 38,435,871 | 18,245,843 |
| 1 | a1+/−a2−/− | 196a1a2_B83_Emb6_BC1RD4ACXX_CGATGT | 97.74% | 33,629,245 | 28,203,748 | 4,664,743 | 32,868,491 | 16,076,041 |
| 2 | a1+/−a2−/− | 196a1a2_B86_Emb1_BC1RD4ACXX_TGACCA | 97.76% | 37,910,867 | 31,669,765 | 5,393,711 | 37,063,476 | 17,679,960 |
| 3 | a1+/−a2−/− | 196a1a2_B86_Emb3_BC1RD4ACXX_ACAGTG | 97.83% | 36,297,105 | 30,633,363 | 4,875,078 | 35,508,441 | 17,010,432 |
| 4 | a1+/−a2−/− | 196a1a2_B86_Emb6_BC1RD4ACXX_ACTTGA | 97.94% | 41,689,263 | 35,353,365 | 5,477,345 | 40,830,710 | 18,748,568 |
| 1 | a1−/−a2−/− | 196a1a2_B83_Emb5_BC1RD4ACXX_ATCACG | 97.75% | 32,683,698 | 27,451,914 | 4,495,476 | 31,947,390 | 15,602,679 |
| 2 | a1−/−a2−/− | 196a1a2_B83_Emb8_BC1RD4ACXX_TTAGGC | 97.80% | 36,892,210 | 31,024,386 | 5,055,624 | 36,080,010 | 17,593,692 |
| 3 | a1−/−a2−/− | 196a1a2_B86_Emb4_BC1RD4ACXX_GCCAAT | 97.80% | 40,072,358 | 33,580,863 | 5,609,205 | 39,190,068 | 18,461,128 |
| 4 | a1−/−a2−/− | 196a1a2_B86_Emb5_BC1RD4ACXX_CAGATC | 97.78% | 37,070,960 | 31,287,599 | 4,960,190 | 36,247,789 | 17,706,694 |
| 1 | a2+/−b−/− | a2_b-C32_Emb2_AC2G5EACXX_CGTACG | 97.76% | 27,174,181 | 22,718,480 | 3,846,079 | 26,564,559 | 13,569,105 |
| 2 | a2+/−b−/− | a2_b-C32_Emb4_AC2G5EACXX_GAGTGG | 97.68% | 25,749,887 | 21,595,570 | 3,557,403 | 25,152,973 | 13,189,719 |
| 3 | a2+/−b−/− | a2_b-C34_Emb5_AC2G5EACXX_ATTCCT | 97.72% | 25,510,534 | 21,342,113 | 3,586,357 | 24,928,470 | 13,176,571 |
| 4 | a2+/−b−/− | a2_b-C42_Emb7_AC2G5EACXX_ACTGAT | 97.70% | 26,635,248 | 22,247,374 | 3,775,533 | 26,022,907 | 13,628,103 |
| 1 | a2−/−b+/− | a2_b-C22_Emb3_AC2G5EACXX_GTCCGC | 97.65% | 27,601,840 | 23,057,570 | 3,894,948 | 26,952,518 | 14,082,089 |
| 2 | a2−/−b+/− | a2_b-C22_Emb5_AC2G5EACXX_GTGAAA | 96.91% | 25,966,607 | 20,989,945 | 4,173,614 | 25,163,559 | 12,739,183 |
| 3 | a2−/−b+/− | a2_b-C22_Emb8_AC2G5EACXX_ATGTCA | 97.76% | 26,612,265 | 22,267,198 | 3,750,241 | 26,017,439 | 13,589,972 |
| 4 | a2−/−b+/− | a2_b-C22_Emb9_AC2G5EACXX_GTTTCG | 97.82% | 28,241,022 | 23,815,069 | 3,811,572 | 27,626,641 | 14,220,558 |
| 1 | a2−/−b−/− | a2_b-C22_Emb10_AC2G5EACXX_CCGTCC | 97.71% | 27,530,655 | 23,198,701 | 3,701,516 | 26,900,217 | 14,130,807 |
| 2 | a2−/−b−/− | a2_b-C22_Emb2_AC2G5EACXX_AGTCAA | 97.69% | 25,080,001 | 21,014,009 | 3,486,723 | 24,500,732 | 13,153,346 |
| 3 | a2−/−b−/− | a2_b-C22_Emb4_AC2G5EACXX_AGTTCC | 97.66% | 26,682,021 | 22,202,689 | 3,855,872 | 26,058,561 | 13,674,466 |
| 4 | a2−/−b−/− | a2_b-C22_Emb7_AC2G5EACXX_GTGGCC | 97.70% | 26,344,878 | 21,945,717 | 3,793,538 | 25,739,255 | 13,405,178 |
Summary of read mapping statistics associated with the 44 RNA-seq samples generated in this study, including total number of reads sequenced per sample and total mapped to the mouse genome (mm10).
Fig. S4.
Predicted miRNA target genes are up-regulated on the loss of miR-196. (A–G) Cumulative density plots of the fold changes of genes predicted as targets of miR-196, partitioned into four context+ intervals according to increasing predicted miRNA targeting efficacy (0 < context+ < −0.2, n = 2,112; −0.2 ≤ context+ < −0.3, n = 145; −0.3 ≤ context+ < −0.4, n = 50; context+ ≤ −0.4, n = 37), and genes with no predicted target site (n = 6,924), across seven genotype comparisons. The P values indicate a statistical comparison of the observed derepression of genes relative to genes with no miRNA target site, as evaluated by a one-sided K-S test.
Fig. S5.
Significant functional categories associated with differentially expressed genes. (A–G) All significant categories related to gene development and function associated with differentially expressed genes, across seven genotype comparisons. All P values are Benjamini–Hochberg corrected, with dashed black lines indicating a significance threshold of 0.01.
Hox Cluster Expression Dynamics Are Altered in miR-196 Mutant Embryos.
It was not known exactly how many of the 10 predicted murine miR-196 Hox target genes are in fact bona fide targets in an in vivo developmental context, nor was it known the relative level of regulation that these predicted targets undergo. When specifically interrogating our transcriptome datasets to assess effects on Hox gene expression, a significant and dose-dependent up-regulation of predicted miR-196 Hox targets was observed (Fig. 4A), which paralleled the dose-dependent patterning defects (Fig. 2A). Comparison of 196a2−/−;196b−/− vs. 196a2+/−profiles identified 7 of 10 predicted miR-196 Hox targets as significantly derepressed in double-mutant cells at this developmental stage. Those predicted Hox targets exhibiting no significant derepression in our analysis included Hoxb1, Hoxa4, and Hoxa5. The most highly derepressed Hox targets were Hoxc8 and Hoxa7, both of which harbor multiple predicted miR-196 binding sites in their 3′ UTRs, and Hoxb8, which exhibits unusually extensive complementarity to miR-196 (17, 23). Further, the measurement of differential expression (Fig. 4A) was almost certain to be an underestimate, because our strategy used eGFP-positive control samples in which at least one miR-196 allele had been removed. Whole mount in situ hybridization (WISH) further revealed that the derepression of Hoxb8 and Hoxc8 target transcripts in 196a2−/−;196b−/− E9.5 embryos relative to WT manifested as a posterior expansion of endogenous expression domains in both the PSM and neural tube (Fig. 4 B and C; n = 3/3 per genotype, respectively). In light of previous reports (31, 32), this failure in timely clearance of the trunk Hox program from more posterior locations is likely to drive supernumerary rib formation observed in miR-196 mutant embryos.
Fig. 4.
Loss of miR-196 function alters global Hox signatures. (A) Extensive Hox gene dysregulation is identified following loss of miR-196. Quantitative expression analysis of all 39 Hox genes in cells isolated from E9.5 mutant embryos; genotype comparisons are color coded. Hox genes with one or more predicted miR-196 target binding sites are indicated in red. Filled circles at the tips of fold changes represent a statistically significant change at q < 0.05. (B and C) WISH analysis of miR-196a2GFP/GFP;miR-196b−/− E9.5 embryos relative to WT identifies a caudal expansion of Hoxb8 (B; n = 3/3) and Hoxc8 (C; n = 3/3). The PSM is indicated with a red line/arrowhead and neural tube with a white arrowhead.
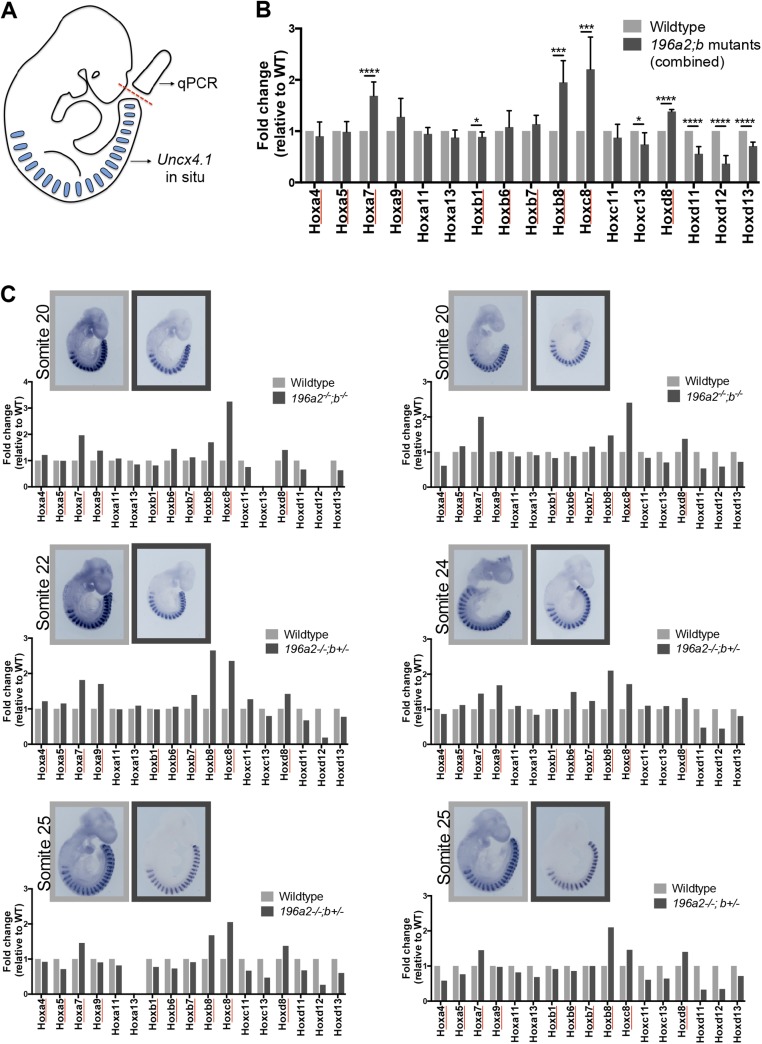
Importantly, we also identified a dose-dependent down-regulation of posterior Hox genes following progressive removal of miR-196 alleles (Fig. 4A). This down-regulation was particularly evident for Hoxd10-d13 genes and was also significant for posterior genes of the HoxA and HoxC clusters. To confirm that these molecular alterations indeed represented a delay in the activation of a posterior Hox program, rather than an overall developmental delay in mutant embryos, we performed quantitative PCR analysis of 17 Hox genes in somite-matched WT and 196a2;196b mutant embryos (somite range, 20–25). This analysis revealed an altered Hox signature in mutant embryos (Fig. S6) that was remarkably consistent with that identified by RNA-seq (Fig. 4A). Although the absence of predicted miR-196 sites within these posterior Hox mRNAs, together with the direction of the regulation (down instead of up with diminished miRNA), indicated that this regulation was indirect, it was nonetheless notable for three reasons. First, given the potential for phenotypic dominance of posterior over anterior Hox protein output (e.g., rib suppression role of Hox10 paralogs (12, 36), a timely activation of a posterior developmental program in miR-196 mutants would be expected to suppress supernumerary rib formation. Second, these posterior Hox proteins, particularly Hoxd11 and Hoxa11, are known to position the lumbo-sacral junction (13, 37, 38), providing a molecular explanation for how the sacrum was repositioned in miR-196 mutants. Finally, in addition to understanding vertebral identity defects, these molecular alterations may provide important experimental support for a proposed model whereby maintenance of tailbud cell divisions, and therefore total vertebral number, is promoted by trunk Hox proteins and antagonized by caudal Hox proteins (28–30). Our results place mir-196 activity at this critical junction, coordinating a reproducible trunk-to-tail Hox code transition. We suggest that a delay in Hox code transition could contribute to the formation of an additional vertebral element observed following genetic removal of miR-196 activity in mice. Such a role for miR-196 is likely to be broadly conserved across vertebrate species, as supported by regionalized vertebral expansion observed in miR-196 morphant zebrafish (24).
Fig. S6.
miR-196 activity regulates the trunk-to-tail Hox code transition. (A) Schematic of experimental procedure. (B) Combined analysis of Hox gene expression across all biological replicates. Hox gene expression values within WT samples were set to 1. Predicted direct Hox targets of miR-196 are underlined with red. Error bars represent SEM. Reported P values are from the Student t test: *P < 0.05, **P < 0.005, ***P < 0.0005, and ****P < 0.0001. (C) Hox gene expression analysis for each individual biological replicate; somite staging of embryos was performed based on Uncx4.1 expression as shown. Hox expression values within a WT sample were set to 1. See SI Materials and Methods for experimental details.
Identification of Additional Direct Targets of miR-196.
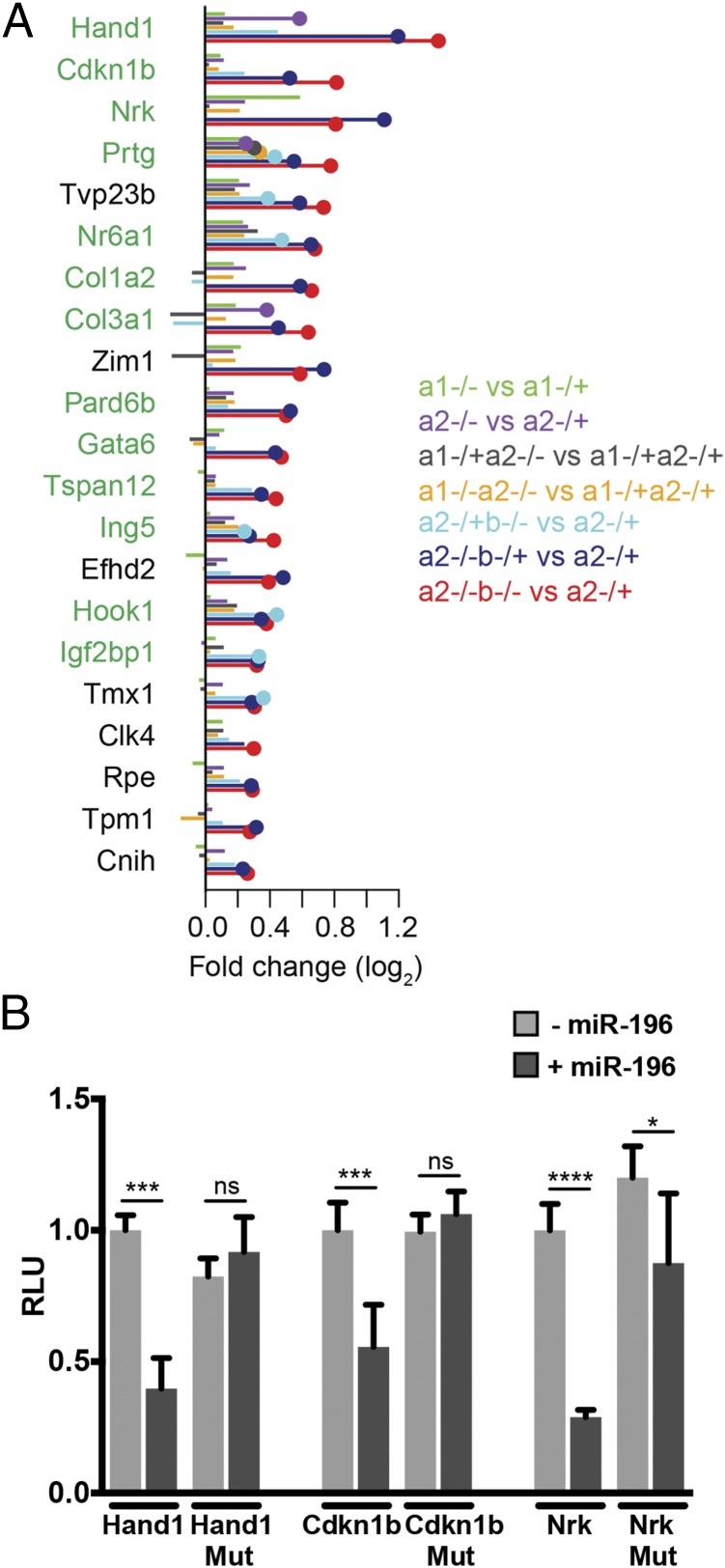
The statistical enrichment of Hox genes among all miR-196 predicted targets (18) prioritized these mRNAs for immediate analysis. However, microRNAs can simultaneously repress extensive suites of target genes (16). To provide experimental support for additional direct targets of miR-196 that have the potential to function in this developmental context, we identified the most highly up-regulated genes in our RNA-seq dataset that either contained a conserved binding site or were predicted to respond strongly to the miRNA (i.e., context+ score ≤ –0.2; Fig. 5A). For the top three evolutionarily conserved miR-196 target genes identified, we assessed whether regulation of their expression by miR-196 required direct binding to sites within their 3′ UTR. Using a luciferase-based reporter assay system in cell culture, miR-196 was shown to repress each of the target genes in a sequence-specific manner (Fig. 5B). Of particular interest within this set was the cell adhesion molecule (Prtg) involved in the ingression of PSM progenitors (39) and an orphan nuclear receptor (Nr6a1) essential for somitogenesis in mice (40) and one of the very few genes that has be associated with variation of vertebral number (41). These experimentally supported miR-196 targets highlight important avenues for future investigation, not only with respect to axial patterning and elongation but also the many other developmental processes (20, 21, 24) and pathological conditions (42, 43) involving miR-196.
Fig. 5.
Identification of additional putative direct (non-Hox) miR-196 targets. (A) List of the most highly up-regulated genes and their associated fold changes in seven genotype comparisons that either (i) contain a conserved miR-196 binding site or (ii) are predicted to respond strongly to the miRNA (i.e., have a context+ score ≤ −0.2). Genes with one or more conserved miR-196 target binding sites are indicated in green. (B) In vitro luciferase analysis confirms sequence-specific regulation of three experimentally supported target genes of miR-196. Renilla luciferase intensity values have been normalized to their respective Firefly values (RLU). Controls (WT 3′ UTR construct without miR-196b) were set to 1. MUT, mutated 3′ UTR construct destroying miR-196 binding site. Error bars represent SD. P values, Student t test: *P < 0.05, ***P < 0.0005, and ****P < 0.0001.
miR-196 Activity Is Required for Signaling Pathways Associated with Axis Elongation, Segmentation, and the Trunk-to-Tail Transition.
miR-196 activity has been shown to negatively regulate retinoic acid pathway activity in the context of pectoral fin formation (24), but regulation of additional developmental signaling pathways in the early embryo, either directly or indirectly, has not been systematically assessed. On further interrogation of our RNA-seq data, we found altered molecular signatures of both axis elongation and somite segmentation across many allelic comparisons (Fig. 6). We observed a clear up-regulation of the Wnt negative feedback inhibitor Dkk1 (44). In addition, the collective down-regulation of numerous direct and indirect downstream targets of Wnt signaling (45–49) (Fig. 6), and the prediction of diminished β-catenin/CTNNB1 activity following global pathway analysis (Fig. S7), indicated an overall reduction in Wnt activity in mutant embryos. Wnt and Fgf signaling positively reinforce one another in the mouse tailbud (50–52), and consistent with diminished Wnt activity in miR-196 mutants, we also observed a down-regulation of the Fgf8 ligand and numerous Fgf downstream effectors (Fig. 6). We observed a robust down-regulation of Notch signaling components and anterior PSM genes Mesp2, Epha4, and Ripply2, likely as a consequence of diminished Wnt activity acting via the Notch ligand Dll1 (52–54). Interestingly, these molecular alterations described for miR-196 mutant embryos resembled alterations observed following removal of all mature miRNAs in the mesoderm lineage (55), which in the latter case resulted in a caudal displacement of the hindlimb by three somites.
Fig. 6.
Loss of miR-196 function alters signaling pathways known to control segmentation and axis elongation. Quantitative expression analysis of pathways known to control segmentation and axial extension in cells isolated from E9.5 mutant embryos, genotype comparisons are color-coded. Filled circles at the tips of fold changes represent a statistically significant change at q < 0.05.
Fig. S7.
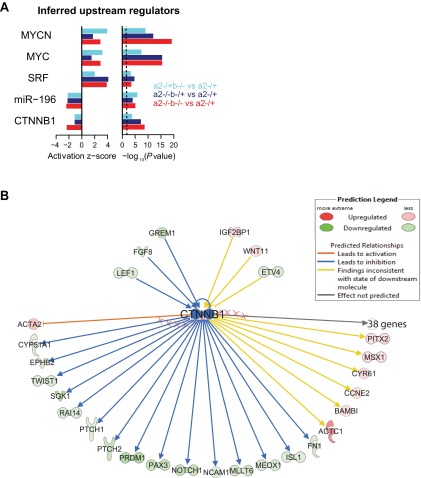
Inference of upstream regulators reveals a down-regulation of Wnt activity. (A) Upstream regulators inferred by Ingenuity Pathway Analysis as being dysregulated based on the behavior of differentially expressed genes in three genotype comparisons. Activation z-scores and P values are computed as described in Fig. 3D. As a positive control, miR-196 is correctly inferred as the most significant miRNA to have diminished activity. β-Catenin/CTNNB1 (Wnt) activity is predicted to also diminish with the loss of miR-196; in contrast, MYCN, MYC, and SRF activity is predicted to become activated. (B) Network of upstream and downstream interactions in the Ingenuity Knowledgebase that were used to infer decreased Wnt activity in the 196a2−/−;196b−/− vs. 196a2−/+ comparison. Genes are shaded according to their observed up- or down-regulation in this comparison.
Finally, a coordinated temporal delay in the trunk-to-tail Hox code transition has been observed in mice null for Gdf11 (56), which as heterozygotes, bear striking phenotypic resemblance to 196a2−/−;196b−/− or miR-196 triple KO mouse embryos. We therefore specifically interrogated our RNA-seq data to assess the levels of Gdf11 and its direct downstream effector Isl1 (57). In 196a2−/−;196b−/− embryos, which exhibit 100% penetrant L-to-T transformation and sacral displacement, we observed a statistically significant reduction in Gdf11 and Isl1 levels by 15% (Dataset S1). As mentioned, this is likely to be an underestimate of the level of regulation, given the experimental strategy used. The requirement for Gdf11 in defining presacral vertebral number is dose dependent (56). The exact threshold requirement for Gdf11 signaling is not known, and it remains to be determined whether subtle down-regulation of Gdf11 contributes to phenotypic alterations observed in miR-196 mutant mice. Together, our transcriptome analyses revealed multiple developmental networks that require miR-196 activity for appropriate control of gene expression and suggest intriguing avenues for future experimental exploration.
miR-196 Has the Potential to Modulate Wnt Signaling by Multiple Mechanisms.
Vertebral progenitors in the epiblast and tailbud are sensitive to the levels of Wnt signaling. Genetic removal of the Wnt3a ligand (58), or conversely, ectopic activation of Wnt3a in the epiblast (59), result in severe axis truncation posterior to the forelimb. Wnt3a expression has been shown to decrease as progenitor cells commit to a paraxial mesoderm fate (60, 61), and sustained Wnt activity disrupts somite formation (51) and somite polarity (59), dependent on timing and method of activation. These observations indicate that careful titration of Wnt levels is essential throughout the process of somite formation. Our data suggest that miR-196 activity is required in maintaining precise levels of Wnt activity (Fig. 6). Mechanistically, this could be achieved in at least two ways. First, miR-196 could directly target genes in the Wnt pathway. Specifically, the potent Wnt antagonist Dkk1 harbors a single predicted miR-196 site within its 3′ UTR, and Dkk1 expression was up-regulated following removal of miR-196 activity (Fig. 6). Using WISH, we confirmed increased expression of Dkk1 in 196a1−/−;196a2−/− embryos relative to 196a1+/−;196a2+/− (Fig. 7A; n = 2/2 per genotype). To test whether miR-196 can act directly to repress Dkk1, we used a luciferase-based reporter assay system in cell culture to show that, indeed, miR-196 negatively regulates the Dkk1 3′ UTR in a sequence-specific manner (Fig. 7B). However, the repression in the reporter assay was more modest than that observed in vivo using RNA-seq (Fig. 6), and Dkk1 is not a conserved target of miR-196, suggesting that indirect regulation by miR-196 also plays a role. Second, miR-196 control over Wnt activity might work in part via Hox intermediates, which have the potential to either activate or repress Wnt signaling (29, 30). We recently showed using chick in vivo electroporation and imaging that the collinear activation of a subset of Hox9-13 posterior Hox genes within paraxial mesoderm progenitors translates into a graded increase in Wnt repression and a slowing down of axis elongation (30). One Hox gene that was found to significantly repress Wnt activity using this in vivo luciferase-based Wnt reporter assay was the miR-196 target Hoxa9. We therefore went on to test whether additional miR-196 Hox targets have the ability to repress Wnt activity in this context. We coelectroporated a Wnt/β-catenin reporter (BATLuc) and a CMV-Renilla construct in paraxial mesoderm progenitors together with an expression vector containing either Venus or Hoxb1, Hoxa5, Hoxa7, Hoxb7, Hoxb8, and Hoxc8. Of these six Hox genes tested, four (Hoxa7, Hoxb7, Hoxb8, and Hoxc8) showed strong repression of luciferase activity, whereas two (Hoxb1 and Hoxa5) did not (Fig. 7C). Interestingly, the two Hox genes do not influence Wnt/β-catenin reporter activity in early chick paraxial mesoderm progenitors are the same Hox genes that show no indication of direct regulation by miR-196 in E9.5 mouse tissue (Fig. 4A). Together, these data demonstrate that miR-196 has the potential to directly and indirectly regulate the precise levels of Wnt activity in the developing embryo.
Fig. 7.
miR-196 has the potential to regulate Wnt signaling by both direct and indirect mechanisms. (A) WISH analysis confirms increased Dkk1 in 196a1−/− ;196a2−/− E9.5 embryos relative to 196a1−/+;196a2−/+ (n = 2/2 for each genotype). (B) In vitro luciferase assay confirms sequence-specific regulation of Dkk1 by miR-196 . Renilla luciferase intensity values have been normalized to their respective Firefly values (RLU). Controls (WT 3′ UTR construct without miR-196b) were set to 1. MUT, mutated 3′ UTR construct destroying miR-196 binding site. (C) Luciferase assay measuring Wnt/β-catenin activity after overexpression of BATLuc together with CMV-Renilla and either control, Hoxb1, Hoxa5, Hoxa7, Hoxb7, Hoxb8, or Hoxc8; n = 4–9 samples per gene assessed. Firefly luciferase intensity values have been normalized to their respective Renilla values (RLU). Control values were set to 1. In B and C, error bars represent SD. Reported P values are from the Student t test: *P 0.05, **P < 0.005, ***P < 0.0005, and ****P < 0.0001.
Discussion
Our work demonstrates the essential role for murine miR-196 in regulating vertebral identity across different levels of the body axis and reveals evolutionary conservation in the role of miR-196 in constraining total vertebral number. Importantly, our strategy has allowed us to comprehensively dissect paralog contribution to resultant phenotypes, allowing us to distinguish a patterning role for miR-196 from its role in modulating vertebral number. Moreover, we characterized the detailed molecular landscape controlled by miR-196 activity in the early embryo to show that miR-196 regulates, and therefore has the ability to integrate, multiple key signaling pathways to drive developmental processes.
miR-196 Activity Is Essential for Vertebral Identity.
Despite the clear potential for functional redundancy between miR-196 paralogs (17), homeotic transformation of vertebral elements could be observed at low penetrance following removal of an individual miR-196 paralog (e.g., 196a2−/− or 196b−/− single mutants). With increasing loss of miR-196 family members (e.g., 196a2−/−;196b−/− double mutants), fully penetrant vertebral phenotypes were observed that were equivalent in severity to many single and compound Hox mutants (14, 62). Vertebral identity changes were observed at sites where loss-of-function phenotypes have previously been described for many of the direct Hox target genes (62), reinforcing the view that miR-196 acts within endogenous Hox domains rather than simply as a fail-safe mechanism to clear an anterior developmental program at more posterior locations (22). Paradoxically, the 196a2−/−;196b−/− or triple KO phenotypes are remarkably similar to either Hoxc8−/− or Hoxc8−/−;Hoxd8−/− skeletons, with eight ribs attached to the sternum, L1-to-T transformation, and a posterior displacement of the sacrum (62). However, with respect to number of sternal rib attachments and L1-to-T transformation, Hoxc8 loss-of-function and gain-of-function mutant mice exhibit identical phenotypes (31, 62). These data indicate that exquisite regulation of a quantitative Hox code is essential in defining vertebral identity at this axial location. Interestingly, deletion of Hoxb8 rescues many defects observed in Hoxc8-null mice, highlighting that there are aspects of a qualitative Hox code that we are yet to understand. Nonetheless, similar to Hoxc8, ectopic Hoxb8 expression results in supernumerary rib formation throughout the lumbar region (32), supporting the view that a collective up-regulation of direct Hox target genes drives homeotic alterations of the midthoracic to upper lumbar region in miR-196 mutant mice.
A shift in the position of the sacrum observed in miR-196 mutant embryos was not easily reconcilable with the function of miR-196 in directly repressing trunk Hox target genes (31, 32). However, we show that in addition to direct Hox gene regulation, miR-196 indirectly regulates the expression levels or temporal activation of many caudal Hox genes, including those whose protein products are known to control positioning of the sacrum, such as Hoxa10, Hoxd10, and Hoxd11 (13, 14, 63, 64). The mechanisms leading to a delay in posterior Hox gene activation in miR-196 mutant mice are currently unknown. A similar coordinated temporal shift in the trunk-to-tail Hox code has been demonstrated in Gdf11−/− mice (56), which show conservation in the types of vertebral transformations we observe here in miR-196 mutant embryos. In this context, Gdf11 appears to work via retinoic acid signaling (57, 65), and whether altered Gdf11 and retinoic acid signaling contribute to miR-196 phenotypic alterations remains to be tested.
miR-196 Activity Constrains Total Vertebral Number.
Total vertebral number of a given species is highly reproducible, and mutations that extend the vertebral column of model organisms are very rare. Among vertebrate species, however, great diversity in vertebral number has arisen. Cross-species comparison (8) or direct genetic perturbation (25, 27) demonstrates that the periodicity of segmentation clock oscillation relative to the rate of PSM growth is the central parameter in defining vertebral number. It remains to be determined how an additional vertebral element seen here in miR-196 mutant mice, or in miR-196 morphant zebrafish (24), are generated at a cellular level (i.e., does the clock tick faster or does it tick at the same rate for longer). Our analysis does, however, reveal molecular alterations in miR-196 mutant embryos that have the potential to affect vertebral number.
First, altered expression of Notch, Wnt, and Fgf pathways could alter the periodicity of segment formation (1). However, diminished Wnt and Fgf would be predicted to increase somite size (27, 66–68), which if axis elongation was unaltered, would lead to a reduction in vertebral number. Further work is required to clarify any functional role for miR-196 in the molecular networks coordinating segmentation.
Second, we showed that miR-196 activity can modulate the expression levels of many Hox genes, either directly or indirectly. It is well documented that Hox genes control mesodermal ingression, thus regulating cell injection into the PSM (30, 69). The rate of PSM growth is not uniform along the anterior-posterior axis (8), with a switch to PSM shortening occurring at about the trunk-to-tail transition in most amniotes. This switch correlates with activation of a posterior Hox code (Hox9 onward), and a subset of posterior Hox genes slows axis elongation by controlling the ingression of PM progenitors via Wnt repression (30). We show here that the ability to repress Wnt signaling is not exclusive to posterior Hox genes but that Hox7/8 paralogs also down-regulate Wnt signaling in the chick epiblast. The repression of Wnt by posterior Hox genes as a means to slow down and terminate axis elongation (29, 30) is consistent with the known function of Wnt3a in driving axis elongation (58). The repression of Wnt by trunk Hox genes is less intuitive because, at least on a Cdx2/4 mutant background, ectopic Hoxb8 has been shown to maintain Wnt expression in the tailbud (29). However, the importance of precise Wnt levels in the early steps of axis formation, and of cellular context, are beginning to be appreciated (59, 70) and may underlie such discrepancies. Exactly how miR-196 activity integrates within the genetic networks controlling PM progenitor maintenance and differentiation remains to be fully elucidated. At E9.5, this progenitor population represents quite a minor fraction of miR-196–expressing cells. Therefore, the extent to which molecular alterations we identified in miR-196 mutants, including reduced Wnt/Fgf signaling, reflect an altered PM progenitor molecular status requires clarification at a cellular resolution.
Although a heterochronic shift in the trunk-to-tail Hox code transition could be predicted to vary vertebral number, morphological evidence for this has been scarce. Analysis of total vertebral number in Gdf11−/− mice, which exhibit a dramatic heterochronic shift in Hox code, is hampered by caudal truncation (56). Although ectopic trunk Hox expression (Hoxa5 and Hoxb8) has the ability to rescue axis truncation defects of a genetically engineered mutant (29), they do not appear to increase vertebral number on a WT background (29, 31). This lack of phenotype is possibly due to the fact that posterior prevalence still holds; caudal Hox genes and miR-196 would be expressed at the usual time and place to regulate and terminate axis elongation. In the case of miR-196 KOs, the cumulative effect on both trunk and caudal Hox gene expression could permit continued maintenance of progenitor divisions while delaying commencement of axis elongation slow down, resulting in increased vertebral number.
Together, our results highlight an essential requirement for miR-196 activity in reinforcing a timely trunk-to-tail Hox code transition and reproducibility of axial formulae. Given the ancestral role of Hox activity in species that use a posterior growth zone (71), and the recurrent acquisition of miRNAs within the Hox clusters across metazoan taxa (15, 17, 72, 73), variation in Hox-miRNA interactions may represent an important mechanism for the evolution of animal body plans.
Materials and Methods
miR-196a1GFP and miR-196a2GFP Knock-In Construction.
A 72- (miR-196a1) or 52-bp (miR-196a2) genomic fragment encompassing each mature miRNA sequence was replaced with a cassette containing eGFP fused to the rabbit β-globin 3′ UTR followed by flippase recognition target (FRT)-flanked PGKem7-Neomycin. A Kozak sequence was inserted upstream of the eGFP start codon. Targeting constructs were generated using a 129/Sv sequence and electroporated into J1 embryonic stem cells. Correctly targeted ES cells were identified and used to generate germ-line transmitting knock-in lines. Before analysis, the Neomycin selection cassette was removed by crossing to a ubiquitous FLPe-deleter mouse line. Resulting lines were bred onto a C57BL/6J background and confirmed as isogenic by SNP genotyping.
miR-196a1−/−, miR-196a2−/−, and miR-196b−/− Generation.
Previously targeted ES cells at each of the three miR-196 loci have been generated (74). Correctly targeted JM8A3 ES cells were reconfirmed by Southern blot and used to generate germ-line transmitting KO lines. Before analysis, the puDeltaTK selection cassette was removed by crossing to a ubiquitous Cre-deleter mouse. Resulting lines are on a mixed C57BL/6J and C57BL/6N background.
Mouse Skeletal Preparation and Analysis.
Skeletal preparation was performed on E18.5 embryos or p0 postnatal pups as previously described (75).
In Situ Hybridization.
Whole mount in situ hybridization was performed as previously described (76).
FACS Sorting and RNA-seq Sample Preparation.
Freshly dissected E9.5 embryos were dissociated in 0.25% trypsin/2% (vol/vol) chick serum, neutralized in DMEM + 10% (vol/vol) FBS, and washed into PBS + 2% (vol/vol) FBS for FACS sorting. GFP-positive cells were FACS sorted directly into RNeasy lysis buffer buffer (Qiagen) and RNA isolated using RNEasy with added on-column DNase treatment (Qiagen). RNA quality was assessed using a Bioanalyser, and 200 ng per individual embryo was used as input for RNA-seq library generation (unstranded Illumina TruSeq Kit). Libraries were multiplexed and sequenced using an Illumina HiSEq. 2000 instrument, generating 50-bp single end reads.
RNA-seq and Category Enrichment Analysis.
Quantification of the transcriptome using RNA-seq data was performed as previously described (77). Raw reads were aligned to the latest build of the mouse genome (mm10) using STAR v. 2.3.1n (options–outFilterType BySJout–outFilterMultimapScoreRange 0–readMatesLengthsIn Equal–outFilterIntronMotifs RemoveNoncanonicalUnannotated–clip3pAdapterSeq TCGTATGCCGTCTTCTGCTTG–outSAMstrandField intronMotif–outStd SAM) (78). Considering all replicates of a particular genotype, differential expression statistics were computed between genotypes of interest using cuffdiff v. 2.1.1 (options–library-type fr-unstranded -c 100 -b mm10.fa -u–max-bundle-frags 100000000) (79), using mouse transcript models of protein-coding genes annotated in Ensembl release 72. Before all subsequent analyses, we filtered away genes annotated by cuffdiff as “NOTEST” in all genotypes, indicating the genes were too lowly expressed to accuracy quantify their abundances. To evaluate functional gene categories that were statistically enriched, we loaded differentially expressed genes (i.e., genes with Q < 0.05) into the Core Analysis function of Ingenuity Pathway Analysis software (Ingenuity Systems), testing gene categories related to development and function. All P values reported from this analysis were adjusted using the Benjamini–Hochberg method to control the false discovery rate.
miRNA Target Analysis.
To identify predicted miRNA targets, the 3′ UTR sequences of protein-coding genes were searched to identify 6mer, 7mer-A1, 7mer-m8, and 8mer miRNA binding sites cognate to the miR-196 seed (35, 80). A context+ score was computed for each target site within a given 3′ UTR, and scores were summed to produce a total context+ score for each gene, which was used for all miRNA-related analyses (35). TargetScanMouse 6.2 was further used to assess target site conservation or to include predicted miR-196 targets containing noncanonical 3′ compensatory sites, such as in the case of Hoxb8 (81).
Permutation Test for Significance Testing.
A permutation test was devised to evaluate the significance of differences in vertebral number. Briefly, given two groups of count-based data of size n and m, we randomly partitioned the counts (without replacement) from the union of the two groups to generate 100,000 pairs of data, again of size n and m. To compute an empirical one-sided P value, we then computed the proportion of pairs that satisfied the condition that the difference in the means of each pair exceeded the difference in means of the original two groups.
In Vitro Luciferase Assay.
A 3′ UTR sequence (300–700 nucleotides) of protein-coding genes of interest were commercially synthesized and cloned into psiCheck2 vector. For each, a mutant version containing four nucleotide substitutions within the miR-196 seed sequence was generated. Constructs were transfected into NIH 3T3 cells with or without 25 pmol mmu-miR-196b duplex. Transfection (Lipofectamine2000; Life Technologies) and luciferase analysis (Dual Luciferase Reporter Assay System; Promega) were performed as per the manufacturer’s instructions.
Chick Electroporation and in Vivo BatLuc Reporter Analysis.
Chicken embryos were harvested at Hamburger and Hamilton stage 5 (82) and electroporated ex ovo as previously described (30), with a DNA mix containing BATLuc (1 μg/μL final), CMV-Renilla (Promega; used as a control to normalize the differences of electroporation intensity between embryos; 0.2 μg/μL final), a control pCAGGS-Venus vector (gift from K. Hadjantonakis, Memorial Sloan Kettering Cancer Center, NY), or a Hox gene of interest (Hoxb1, a5, a7, b7, b8, or c8) cloned in pCAGGS-IRES2-Venus (5 μg/μL final). Electroporated embryos were cultured in a humidified incubator at 38 °C for 20 h. Embryos were analyzed using a fluorescent microscope, and only embryos showing restricted expression of Venus in the paraxial mesoderm were selected (90–100% of the electroporated embryos) for luciferase assay (between three and five embryos for each condition). The posterior region (from somite 1 to tailbud) of the selected embryos was dissected and lysed in passive lysis buffer (Promega) for 15 min at room temperature. Lysates were then distributed in a 96-well plate, and luciferase assays were performed using a Centro LB 960 luminometer (Berthold Technology) and the dual luciferase kit (Promega) following manufacturer’s instructions. Raw intensity values for Firefly luciferase signal were normalized with corresponding Renilla luciferase values (RLUs), and the control experiment was set to 1.
SI Materials and Methods
To enable quantitative PCR analysis of Hox gene expression (Fig. S6), WT and 196a2;b mutant embryos of various genotypes were collected at E9.5; yolk sac tissue was used for genotyping. The entire caudal embryo, encompassing the PSM, was isolated by cutting caudal to the last somite and immediately stored at −80 °C in RTL buffer (Qiagen). In situ hybridization was performed to detect Uncx4.1 in the embryonic trunk, and somite number was determined. Exact somite-matched embryos were processed to extract total RNA using RNeasy with added on column DNaseI treatment (Qiagen), and reverse transcription was performed using the RT2 HT First Strand Kit (Qiagen). A custom PCR panel was generated (RT2 Profiler PCR Array) to analyze the expression of 17 Hox genes. Quantitative PCR was performed using the Lightcycler 480 (Roche) in technical duplicate at a minimum. Raw Cp values were normalized to GAPDH control values. Hox expression within WT samples was set to 1, and somite-matched mutant samples were compared. Data from six biological replicates (across various somite stages) were combined, and statistical significance was assessed using a t test.
SI Discussion
For final numbers associated with vertebral defects displayed in Table S1 and Fig. S3, unilateral and bilateral defects were counted together. eGFP knock-in and straight KO alleles were used interchangeably, with the only discrepancy observed being a shift of the transitional vertebrae from T10 to T11 in 196a2GFP/GFP compared with 196a2−/− embryos. This defect did not segregate with rib fusion defects, and L1-to-T transformations were observed on both 196a2 lines and thus do not impact our analysis. The196a2GFP line was used for double and triple mutant generation. A reduction in the length of the 13th rib (T13), with or without anterior sacral displacement, was observed at low frequency in 196a1GFP/+, 196a1GFP/GFP and 196a1−/− animals (Table S1). Anterior displacement of the sacrum, resulting in variation of lumbar number between 5 and 6, is often observed in WT populations, although we only see this defect in 1 of 47 WT embryos on our >99.9% C57B6 background. It is of note, that in our RNAseq analysis, we observe an up-regulation of Hoxb9 specifically in 196a1GFP/GFP samples (Fig. 3A), and ectopic Hoxb9 was shown to cause anterior sacral displacement. Hoxb9 lies adjacent to miR-196a1, so this may represent a genomic alteration due to GFP insertion. However, anterior sacral displacement is observed in both 196a1 GFP knock-in and straight KO lines and may represent a true vertebral alteration.
Supplementary Material
Acknowledgments
We thank Xin Sun and Denis Duboule for supplying in situ probes and A. Dobin for help in understanding RNA-seq mapping parameters. We thank Allan Bradley for providing three KO mouse ES cell lines used in these studies. We thank Christophe Marcelle, Eran Hornstein, and Jan Manent for critical reading of the manuscript. This work was supported by a National Science Foundation Graduate Research fellowship (to V.A.), Wellcome Trust Grant 098051 (to support miRNA KO reagent production by H.M.P.), National Institutes of Health Grant GM067031 (to D.P.B.), National Institutes of Health Grant R37HD032443-19 (to C.J.T.), and National Health and Medical Research Council Project Grant APP1051792 (to E.M.). E.M. thanks Bioplatforms Australia for support. D.P.B. is a Howard Hughes Medical Institute Investigator. The Australian Regenerative Medicine Institute is supported by grants from the State Government of Victoria and the Australian Government.
Footnotes
The authors declare no conflict of interest.
Data deposition: The data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo (accession no. GSE53018).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1512655112/-/DCSupplemental.
References
- 1.Bénazéraf B, Pourquié O. Formation and segmentation of the vertebrate body axis. Annu Rev Cell Dev Biol. 2013;29:1–26. doi: 10.1146/annurev-cellbio-101011-155703. [DOI] [PubMed] [Google Scholar]
- 2.Cambray N, Wilson V. Axial progenitors with extensive potency are localised to the mouse chordoneural hinge. Development. 2002;129(20):4855–4866. doi: 10.1242/dev.129.20.4855. [DOI] [PubMed] [Google Scholar]
- 3.Cambray N, Wilson V. Two distinct sources for a population of maturing axial progenitors. Development. 2007;134(15):2829–2840. doi: 10.1242/dev.02877. [DOI] [PubMed] [Google Scholar]
- 4.McGrew MJ, et al. Localised axial progenitor cell populations in the avian tail bud are not committed to a posterior Hox identity. Development. 2008;135(13):2289–2299. doi: 10.1242/dev.022020. [DOI] [PubMed] [Google Scholar]
- 5.Psychoyos D, Stern CD. Fates and migratory routes of primitive streak cells in the chick embryo. Development. 1996;122(5):1523–1534. doi: 10.1242/dev.122.5.1523. [DOI] [PubMed] [Google Scholar]
- 6.Iimura T, Yang X, Weijer CJ, Pourquié O. Dual mode of paraxial mesoderm formation during chick gastrulation. Proc Natl Acad Sci USA. 2007;104(8):2744–2749. doi: 10.1073/pnas.0610997104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Neijts R, Simmini S, Giuliani F, van Rooijen C, Deschamps J. Region-specific regulation of posterior axial elongation during vertebrate embryogenesis. Dev Dyn. 2014;243(1):88–98. doi: 10.1002/dvdy.24027. [DOI] [PubMed] [Google Scholar]
- 8.Gomez C, et al. Control of segment number in vertebrate embryos. Nature. 2008;454(7202):335–339. doi: 10.1038/nature07020. [DOI] [PubMed] [Google Scholar]
- 9.Wellik DM. Hox patterning of the vertebrate axial skeleton. Dev Dyn. 2007;236(9):2454–2463. doi: 10.1002/dvdy.21286. [DOI] [PubMed] [Google Scholar]
- 10.Duboule D, Dollé P. The structural and functional organization of the murine HOX gene family resembles that of Drosophila homeotic genes. EMBO J. 1989;8(5):1497–1505. doi: 10.1002/j.1460-2075.1989.tb03534.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Graham A, Papalopulu N, Krumlauf R. The murine and Drosophila homeobox gene complexes have common features of organization and expression. Cell. 1989;57(3):367–378. doi: 10.1016/0092-8674(89)90912-4. [DOI] [PubMed] [Google Scholar]
- 12.Wellik DM, Capecchi MR. Hox10 and Hox11 genes are required to globally pattern the mammalian skeleton. Science. 2003;301(5631):363–367. doi: 10.1126/science.1085672. [DOI] [PubMed] [Google Scholar]
- 13.Favier B, Le Meur M, Chambon P, Dollé P. Axial skeleton homeosis and forelimb malformations in Hoxd-11 mutant mice. Proc Natl Acad Sci USA. 1995;92(1):310–314. doi: 10.1073/pnas.92.1.310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Favier B, et al. Functional cooperation between the non-paralogous genes Hoxa-10 and Hoxd-11 in the developing forelimb and axial skeleton. Development. 1996;122(2):449–460. doi: 10.1242/dev.122.2.449. [DOI] [PubMed] [Google Scholar]
- 15.Heimberg A, McGlinn E. Building a robust a-p axis. Curr Genomics. 2012;13(4):278–288. doi: 10.2174/138920212800793348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bartel DP. MicroRNAs: Target recognition and regulatory functions. Cell. 2009;136(2):215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yekta S, Shih IH, Bartel DP. MicroRNA-directed cleavage of HOXB8 mRNA. Science. 2004;304(5670):594–596. doi: 10.1126/science.1097434. [DOI] [PubMed] [Google Scholar]
- 18.Yekta S, Tabin CJ, Bartel DP. MicroRNAs in the Hox network: An apparent link to posterior prevalence. Nat Rev Genet. 2008;9(10):789–796. doi: 10.1038/nrg2400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vonk FJ, et al. The king cobra genome reveals dynamic gene evolution and adaptation in the snake venom system. Proc Natl Acad Sci USA. 2013;110(51):20651–20656. doi: 10.1073/pnas.1314702110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Asli NS, Kessel M. Spatiotemporally restricted regulation of generic motor neuron programs by miR-196-mediated repression of Hoxb8. Dev Biol. 2010;344(2):857–868. doi: 10.1016/j.ydbio.2010.06.003. [DOI] [PubMed] [Google Scholar]
- 21.Hornstein E, et al. The microRNA miR-196 acts upstream of Hoxb8 and Shh in limb development. Nature. 2005;438(7068):671–674. doi: 10.1038/nature04138. [DOI] [PubMed] [Google Scholar]
- 22.McGlinn E, et al. In ovo application of antagomiRs indicates a role for miR-196 in patterning the chick axial skeleton through Hox gene regulation. Proc Natl Acad Sci USA. 2009;106(44):18610–18615. doi: 10.1073/pnas.0910374106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mansfield JH, et al. MicroRNA-responsive ‘sensor’ transgenes uncover Hox-like and other developmentally regulated patterns of vertebrate microRNA expression. Nat Genet. 2004;36(10):1079–1083. doi: 10.1038/ng1421. [DOI] [PubMed] [Google Scholar]
- 24.He X, et al. miR-196 regulates axial patterning and pectoral appendage initiation. Dev Biol. 2011;357(2):463–477. doi: 10.1016/j.ydbio.2011.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schröter C, Oates AC. Segment number and axial identity in a segmentation clock period mutant. Curr Biol. 2010;20(14):1254–1258. doi: 10.1016/j.cub.2010.05.071. [DOI] [PubMed] [Google Scholar]
- 26.Harima Y, Takashima Y, Ueda Y, Ohtsuka T, Kageyama R. Accelerating the tempo of the segmentation clock by reducing the number of introns in the Hes7 gene. Cell Reports. 2013;3(1):1–7. doi: 10.1016/j.celrep.2012.11.012. [DOI] [PubMed] [Google Scholar]
- 27.Dubrulle J, McGrew MJ, Pourquié O. FGF signaling controls somite boundary position and regulates segmentation clock control of spatiotemporal Hox gene activation. Cell. 2001;106(2):219–232. doi: 10.1016/s0092-8674(01)00437-8. [DOI] [PubMed] [Google Scholar]
- 28.Economides KD, Zeltser L, Capecchi MR. Hoxb13 mutations cause overgrowth of caudal spinal cord and tail vertebrae. Dev Biol. 2003;256(2):317–330. doi: 10.1016/s0012-1606(02)00137-9. [DOI] [PubMed] [Google Scholar]
- 29.Young T, et al. Cdx and Hox genes differentially regulate posterior axial growth in mammalian embryos. Dev Cell. 2009;17(4):516–526. doi: 10.1016/j.devcel.2009.08.010. [DOI] [PubMed] [Google Scholar]
- 30.Denans N, Iimura T, Pourquié O. Hox genes control vertebrate body elongation by collinear Wnt repression. eLife. 2015;4:4. doi: 10.7554/eLife.04379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pollock RA, Jay G, Bieberich CJ. Altering the boundaries of Hox3.1 expression: Evidence for antipodal gene regulation. Cell. 1992;71(6):911–923. doi: 10.1016/0092-8674(92)90388-s. [DOI] [PubMed] [Google Scholar]
- 32.Pollock RA, Sreenath T, Ngo L, Bieberich CJ. Gain of function mutations for paralogous Hox genes: Implications for the evolution of Hox gene function. Proc Natl Acad Sci USA. 1995;92(10):4492–4496. doi: 10.1073/pnas.92.10.4492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chen F, Capecchi MR. Targeted mutations in hoxa-9 and hoxb-9 reveal synergistic interactions. Dev Biol. 1997;181(2):186–196. doi: 10.1006/dbio.1996.8440. [DOI] [PubMed] [Google Scholar]
- 34.Burke AC, Nelson CE, Morgan BA, Tabin C. Hox genes and the evolution of vertebrate axial morphology. Development. 1995;121(2):333–346. doi: 10.1242/dev.121.2.333. [DOI] [PubMed] [Google Scholar]
- 35.Garcia DM, et al. Weak seed-pairing stability and high target-site abundance decrease the proficiency of lsy-6 and other microRNAs. Nat Struct Mol Biol. 2011;18(10):1139–1146. doi: 10.1038/nsmb.2115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Carapuço M, Nóvoa A, Bobola N, Mallo M. Hox genes specify vertebral types in the presomitic mesoderm. Genes Dev. 2005;19(18):2116–2121. doi: 10.1101/gad.338705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Davis AP, Capecchi MR. Axial homeosis and appendicular skeleton defects in mice with a targeted disruption of hoxd-11. Development. 1994;120(8):2187–2198. doi: 10.1242/dev.120.8.2187. [DOI] [PubMed] [Google Scholar]
- 38.Spitz F, et al. Large scale transgenic and cluster deletion analysis of the HoxD complex separate an ancestral regulatory module from evolutionary innovations. Genes Dev. 2001;15(17):2209–2214. doi: 10.1101/gad.205701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ito K, Nakamura H, Watanabe Y. Protogenin mediates cell adhesion for ingression and re-epithelialization of paraxial mesodermal cells. Dev Biol. 2011;351(1):13–24. doi: 10.1016/j.ydbio.2010.11.024. [DOI] [PubMed] [Google Scholar]
- 40.Chung AC, et al. Loss of orphan receptor germ cell nuclear factor function results in ectopic development of the tail bud and a novel posterior truncation. Mol Cell Biol. 2001;21(2):663–677. doi: 10.1128/MCB.21.2.663-677.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mikawa S, et al. Fine mapping of a swine quantitative trait locus for number of vertebrae and analysis of an orphan nuclear receptor, germ cell nuclear factor (NR6A1) Genome Res. 2007;17(5):586–593. doi: 10.1101/gr.6085507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Li Z, et al. miR-196b directly targets both HOXA9/MEIS1 oncogenes and FAS tumour suppressor in MLL-rearranged leukaemia. Nat Commun. 2012;3:688. doi: 10.1038/ncomms1681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Velu CS, et al. Therapeutic antagonists of microRNAs deplete leukemia-initiating cell activity. J Clin Invest. 2014;124(1):222–236. doi: 10.1172/JCI66005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chamorro MN, et al. FGF-20 and DKK1 are transcriptional targets of beta-catenin and FGF-20 is implicated in cancer and development. EMBO J. 2005;24(1):73–84. doi: 10.1038/sj.emboj.7600460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dequéant ML, et al. A complex oscillating network of signaling genes underlies the mouse segmentation clock. Science. 2006;314(5805):1595–1598. doi: 10.1126/science.1133141. [DOI] [PubMed] [Google Scholar]
- 46.Lickert H, et al. Dissecting Wnt/beta-catenin signaling during gastrulation using RNA interference in mouse embryos. Development. 2005;132(11):2599–2609. doi: 10.1242/dev.01842. [DOI] [PubMed] [Google Scholar]
- 47.Weidinger G, Thorpe CJ, Wuennenberg-Stapleton K, Ngai J, Moon RT. The Sp1-related transcription factors sp5 and sp5-like act downstream of Wnt/beta-catenin signaling in mesoderm and neuroectoderm patterning. Curr Biol. 2005;15(6):489–500. doi: 10.1016/j.cub.2005.01.041. [DOI] [PubMed] [Google Scholar]
- 48.Buttitta L, Tanaka TS, Chen AE, Ko MS, Fan CM. Microarray analysis of somitogenesis reveals novel targets of different WNT signaling pathways in the somitic mesoderm. Dev Biol. 2003;258(1):91–104. doi: 10.1016/s0012-1606(03)00116-7. [DOI] [PubMed] [Google Scholar]
- 49.Takahashi M, et al. Isolation of a novel human gene, APCDD1, as a direct target of the beta-Catenin/T-cell factor 4 complex with probable involvement in colorectal carcinogenesis. Cancer Res. 2002;62(20):5651–5656. [PubMed] [Google Scholar]
- 50.Naiche LA, Holder N, Lewandoski M. FGF4 and FGF8 comprise the wavefront activity that controls somitogenesis. Proc Natl Acad Sci USA. 2011;108(10):4018–4023. doi: 10.1073/pnas.1007417108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Aulehla A, et al. A beta-catenin gradient links the clock and wavefront systems in mouse embryo segmentation. Nat Cell Biol. 2008;10(2):186–193. doi: 10.1038/ncb1679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Dunty WC, Jr, et al. Wnt3a/beta-catenin signaling controls posterior body development by coordinating mesoderm formation and segmentation. Development. 2008;135(1):85–94. doi: 10.1242/dev.009266. [DOI] [PubMed] [Google Scholar]
- 53.Galceran J, Sustmann C, Hsu SC, Folberth S, Grosschedl R. LEF1-mediated regulation of Delta-like1 links Wnt and Notch signaling in somitogenesis. Genes Dev. 2004;18(22):2718–2723. doi: 10.1101/gad.1249504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hofmann M, et al. WNT signaling, in synergy with T/TBX6, controls Notch signaling by regulating Dll1 expression in the presomitic mesoderm of mouse embryos. Genes Dev. 2004;18(22):2712–2717. doi: 10.1101/gad.1248604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhang Z, et al. The microRNA-processing enzyme Dicer is dispensable for somite segmentation but essential for limb bud positioning. Dev Biol. 2011;351(2):254–265. doi: 10.1016/j.ydbio.2011.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.McPherron AC, Lawler AM, Lee SJ. Regulation of anterior/posterior patterning of the axial skeleton by growth/differentiation factor 11. Nat Genet. 1999;22(3):260–264. doi: 10.1038/10320. [DOI] [PubMed] [Google Scholar]
- 57.Jurberg AD, Aires R, Varela-Lasheras I, Nóvoa A, Mallo M. Switching axial progenitors from producing trunk to tail tissues in vertebrate embryos. Dev Cell. 2013;25(5):451–462. doi: 10.1016/j.devcel.2013.05.009. [DOI] [PubMed] [Google Scholar]
- 58.Takada S, et al. Wnt-3a regulates somite and tailbud formation in the mouse embryo. Genes Dev. 1994;8(2):174–189. doi: 10.1101/gad.8.2.174. [DOI] [PubMed] [Google Scholar]
- 59.Jurberg AD, Aires R, Nóvoa A, Rowland JE, Mallo M. Compartment-dependent activities of Wnt3a/β-catenin signaling during vertebrate axial extension. Dev Biol. 2014;394(2):253–263. doi: 10.1016/j.ydbio.2014.08.012. [DOI] [PubMed] [Google Scholar]
- 60.Takemoto T, et al. Tbx6-dependent Sox2 regulation determines neural or mesodermal fate in axial stem cells. Nature. 2011;470(7334):394–398. doi: 10.1038/nature09729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Nowotschin S, Ferrer-Vaquer A, Concepcion D, Papaioannou VE, Hadjantonakis AK. Interaction of Wnt3a, Msgn1 and Tbx6 in neural versus paraxial mesoderm lineage commitment and paraxial mesoderm differentiation in the mouse embryo. Dev Biol. 2012;367(1):1–14. doi: 10.1016/j.ydbio.2012.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.van den Akker E, et al. Axial skeletal patterning in mice lacking all paralogous group 8 Hox genes. Development. 2001;128(10):1911–1921. doi: 10.1242/dev.128.10.1911. [DOI] [PubMed] [Google Scholar]
- 63.Davis AP, Witte DP, Hsieh-Li HM, Potter SS, Capecchi MR. Absence of radius and ulna in mice lacking hoxa-11 and hoxd-11. Nature. 1995;375(6534):791–795. doi: 10.1038/375791a0. [DOI] [PubMed] [Google Scholar]
- 64.Zákány J, Gérard M, Favier B, Duboule D. Deletion of a HoxD enhancer induces transcriptional heterochrony leading to transposition of the sacrum. EMBO J. 1997;16(14):4393–4402. doi: 10.1093/emboj/16.14.4393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lee YJ, et al. Growth differentiation factor 11 signaling controls retinoic acid activity for axial vertebral development. Dev Biol. 2010;347(1):195–203. doi: 10.1016/j.ydbio.2010.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Sawada A, et al. Fgf/MAPK signalling is a crucial positional cue in somite boundary formation. Development. 2001;128(23):4873–4880. doi: 10.1242/dev.128.23.4873. [DOI] [PubMed] [Google Scholar]
- 67.Aulehla A, et al. Wnt3a plays a major role in the segmentation clock controlling somitogenesis. Dev Cell. 2003;4(3):395–406. doi: 10.1016/s1534-5807(03)00055-8. [DOI] [PubMed] [Google Scholar]
- 68.Bajard L, et al. Wnt-regulated dynamics of positional information in zebrafish somitogenesis. Development. 2014;141(6):1381–1391. doi: 10.1242/dev.093435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Iimura T, Pourquié O. Collinear activation of Hoxb genes during gastrulation is linked to mesoderm cell ingression. Nature. 2006;442(7102):568–571. doi: 10.1038/nature04838. [DOI] [PubMed] [Google Scholar]
- 70.Tsakiridis A, et al. Distinct Wnt-driven primitive streak-like populations reflect in vivo lineage precursors. Development. 2014;141(6):1209–1221. doi: 10.1242/dev.101014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Ryan JF, Baxevanis AD. Hox, Wnt, and the evolution of the primary body axis: Insights from the early-divergent phyla. Biol Direct. 2007;2:37. doi: 10.1186/1745-6150-2-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Moran Y, et al. Cnidarian microRNAs frequently regulate targets by cleavage. Genome Res. 2014;24(4):651–663. doi: 10.1101/gr.162503.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Lagos-Quintana M, Rauhut R, Meyer J, Borkhardt A, Tuschl T. New microRNAs from mouse and human. RNA. 2003;9(2):175–179. doi: 10.1261/rna.2146903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Prosser HM, Koike-Yusa H, Cooper JD, Law FC, Bradley A. A resource of vectors and ES cells for targeted deletion of microRNAs in mice. Nat Biotechnol. 2011;29(9):840–845. doi: 10.1038/nbt.1929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.McLeod MJ. Differential staining of cartilage and bone in whole mouse fetuses by alcian blue and alizarin red S. Teratology. 1980;22(3):299–301. doi: 10.1002/tera.1420220306. [DOI] [PubMed] [Google Scholar]
- 76.McGlinn E, Mansfield JH. Detection of gene expression in mouse embryos and tissue sections. Methods Mol Biol. 2011;770:259–292. doi: 10.1007/978-1-61779-210-6_10. [DOI] [PubMed] [Google Scholar]
- 77.Denzler R, Agarwal V, Stefano J, Bartel DP, Stoffel M. Assessing the ceRNA hypothesis with quantitative measurements of miRNA and target abundance. Mol Cell. 2014;54(5):766–776. doi: 10.1016/j.molcel.2014.03.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Dobin A, et al. STAR: Ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29(1):15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Trapnell C, et al. Differential analysis of gene regulation at transcript resolution with RNA-seq. Nat Biotechnol. 2013;31(1):46–53. doi: 10.1038/nbt.2450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Grimson A, et al. MicroRNA targeting specificity in mammals: Determinants beyond seed pairing. Mol Cell. 2007;27(1):91–105. doi: 10.1016/j.molcel.2007.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Friedman RC, Farh KK, Burge CB, Bartel DP. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 2009;19(1):92–105. doi: 10.1101/gr.082701.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Hamburger V, Hamilton HL. A series of normal stages in the development of the chick embryo. J Morphol. 1951;88(1):49–92. [PubMed] [Google Scholar]
- 83.Krämer A, Green J, Pollard J, Jr, Tugendreich S. Causal analysis approaches in Ingenuity Pathway Analysis. Bioinformatics. 2014;30(4):523–530. doi: 10.1093/bioinformatics/btt703. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.