Abstract
EPEC (enteropathogenic Escherichia coli) and EHEC (enterohaemorrhagic Escherichia coli) are attaching and effacing pathogens frequently associated with infectious diarrhoea. EPEC and EHEC use a T3SS (type III secretion system) to translocate effectors that subvert different cellular processes to sustain colonization and multiplication. The eukaryotic proteins NHERF2 (Na+ /H+ exchanger regulatory factor 2) and AnxA2 (annexin A2), which are involved in regulation of intestinal ion channels, are recruited to the bacterial attachment sites. Using a stable HeLa-NHERF2 cell line, we found partial co-localization of AnxA2 and NHERF2; in EPEC-infected cells, AnxA2 and NHERF2 were extensively recruited to the site of bacterial attachment. We confirmed that NHERF2 dimerizes and found that NHERF2 interacts with AnxA2. Moreover, we found that AnxA2 also binds both the N- and C-terminal domains of the bacterial effector Tir through its C-terminal domain. Immunofluorescence of HeLa cells infected with EPEC showed that AnxA2 is recruited to the site of bacterial attachment in a Tir-dependent manner, but independently of Tir-induced actin polymerization. Our results suggest that AnxA2 and NHERF2 form a scaffold complex that links adjacent Tir molecules at the plasma membrane forming a lattice that could be involved in retention and dissemination of other effectors at the bacterial attachment site.
Keywords: annexin A2, enterohaemorrhagic Escherichia coli (EHEC), enteropathogenic Escherichia coli (EPEC), Na+ /H+ exchanger regulatory factor 2 (NHERF2), protein interaction, Tir
INTRODUCTION
Diarrhoeal diseases are the third leading cause of death in developing countries, mainly affecting young children [1]. Diarrhoeagenic Escherichia coli, which are among the major causative agents of infectious diarrhoea, are classified into six categories, including EPEC (enteropathogenic Escherichia coli) and EHEC (enterohaemorrhagic Escherichia coli) (reviewed in [2]). EPEC and EHEC colonize the intestinal mucosa via A/E (attaching and effacing) lesions [3,4], which are characterized by intimate bacterial attachment, effacement of the brush border microvilli and localized actin polymerization [3]. The ability to form A/E lesions is mediated by the LEE (locus of enterocyte effacement) pathogenicity island [5], which encodes a T3SS (type III secretion system), effector proteins, the outer membrane adhesin intimin and transcriptional regulators (reviewed in [6]). Intimate attachment and actin polymerization are dependent on the interaction between intimin and its translocated receptor Tir [7]. Following translocation, Tir is integrated, by as yet unknown mechanisms, into the plasma membrane in a hairpin loop topology where the N- and C-termini remain intracellular, whereas the central loop is exposed on the cell surface functioning as an intimin receptor [8]. Other translocated effectors are targeted to different cellular compartments, including the mitochondria, endoplasmic reticulum and the Golgi, subverting a wide range of cellular processes [6], including deregulation of ion channels [9].
TRPV (transient receptor potential vanilloid) 5/6 are Ca2+ -selective channels, belonging to the TRP (transient receptor potential) superfamily, the largest family of ion channels known [10]. TRPV5/6 are found in the apical brush-border membrane of the intestinal enterocytes, being responsible for the first step in Ca2+ absorption in the intestine. Five regulatory proteins that associate with TRPV5/6 have been identified: p11–AnxA2 (annexin A2), calmodulin, 80K-H, NHERF2 (Na+ /H+ exchange regulatory factor 2) and Rab11a [11]. Importantly, two of these regulatory proteins, p11–AnxA2 and NHERF2, which associate with the TRPV5/6 C-terminal tail, are recruited by A/E pathogens to the site of bacterial attachment [12-14].
AnxA2 is a 36 kDa protein which can be found as a monomer or a heterotetramer, where a dimer of AnxA2 subunits interacts with a dimer of p11, forming a stable (p11)2–(AnxA2)2 heterotetramer complex [15]. The monomeric form of AnxA2 is composed of an unfolded hypervariable N-terminal domain, amino acids 1–30, and a conserved C-terminal core, amino acids 31–339, bears the common features of annexins, including annexin repeats, Ca2+ - and F-actin (filamentous actin)-binding domains [15,16]. AnxA2 is localized at the apical plasma membranes [17,18], extracellular surfaces [19] and the luminal microvillar brush border of intestinal epithelial cells [17,20]. AnxA2 is recruited to sites of actin polymerization that are intimately associated with membrane surfaces, such as raft–cytoskeleton interfaces [21,22], playing a role in membrane organization, cell–cell adhesion and trafficking of ion channels to the plasma membrane [15]. Zobiack et al. [12] have shown that AnxA2, with its ligand p11, is recruited to actin assembly sites at the plasma membrane of HeLa cells infected with EPEC, in a T3SS-dependent but Tir-independent manner. The effector protein involved in AnxA2 recruitment in EPEC is not yet known. Miyahara et al. [13] showed that EHEC recruited AnxA2 to the bacterial adhesion sites. Moreover, AnxA2 recruited the effector protein EspL2 to adherent EHEC. EspL2, in combination with Tir, enhances AnxA2-induced F-actin aggregation. Knockdown of AnxA2 did not affect actin polymerization under attached EHEC bacteria, indicating that AnxA2 is not necessary for Tir-induced actin polymerization in vitro [13].
NHERF proteins, which are abundant in polarized epithelia [23], form multiprotein complexes that function in addressing, recycling and regulating receptors and ion channels [24,25]. NHERF1 and NHERF2 are composed of two PDZ [PSD-95 (postsynaptic density 95), Dlg (discs large) and ZO-1 (zonula occludens 1)] domains and a C-terminal EBD (ezrin/radixin/moesin-binding domain) [26]. Previously, we have shown that EPEC recruits NHERF1 [27] and NHERF2 [14] to the bacterial attachment sites. We found that the T3SS effectors Map, EspI and NleH1 bind the NHERFs, which affect their intracellular trafficking and activity [14]. Importantly, none of these effectors is involved in the recruitment of NHERFs. Therefore the mechanism by which EPEC recruits NHERFs remains unknown. The aim of the present study was to investigate the mechanism by which A/E pathogens recruit NHERF2 and AnxA2 to the bacterial attachment sites.
EXPERIMENTAL
Bacterial strains and growth conditions
E. coli strains used in the present study are listed in Table 1. Bacteria were cultured in LB (Luria–Bertani) medium or in DMEM (Dulbecco’s modified Eagle’s medium) supplemented with ampicillin (100 μg/ml) or kanamycin (50 μg/ml) as appropriate.
Table 1. Strains and plasmids employed in the present study.
Primer sequences are given in Supplementary Table S1 at http://www.BiochemJ.org/bj/445/bj4450383add.htm.
| (a) Strains | |||
|---|---|---|---|
|
| |||
| Strain | Description | Reference | |
| E2348/69 | Wild-type EPEC O127:H6 | [41] | |
| ICC192 | E2348/69 ΔescN | [36] | |
| ICC225 | E2348/69 Δtir | [42] | |
| E2348/69TirY474S | E2348/69tirY474S | [38] | |
| EDL933 | Prototypic O157:H7 EHEC strain | [43] | |
| ICC169 | Prototypic Citrobacter rodentium, nalr | [44] | |
| (b) Plasmids | |||
|---|---|---|---|
|
| |||
| Plasmid | Description | Reference | Primers used |
| pGFP-AnxA2 | pEGFP-C1 encoding AnxA2 fused to GFP | [32] | |
| pICC587 | pRK5-anxA2 encoding Myc-tagged AnxA2 | The present study | Fw-BamH1-AnxA2 and Rv-AnxA2-BamHI |
| pVenus | pCDNA3 encoding full-length YFPVenus protein | [37] | |
| pBiFC-VN173 | N-terminal fragment of the YFPVenus protein (VN, residues 1–173) | [29] | |
| pBiFC-VC155 | C-terminal fragment of the YFPVenus protein (VC, residues 155–238) | [29] | |
| pICC588 | pBiFC-VN173 derivative encoding AnxA2 Myc-tagged fused to VN | The present study | SacI-Myc-VN173-For and VN-Anx-BamH1-Rev |
| pICC589 | pBiFC-VN173 derivative encoding NHERF2 FLAG-tagged fused to VN | The present study | Fw-EcoRI-Nherf2-VN and Rv-Nherf2-KpnI-VN |
| pICC590 | pBiFC-VC155 derivative encoding NHERF2 HA-tagged fused to VC | The present study | Fw-EcoRI-Nherf2-VC and Rv-Nherf2-KpnI-VC |
| pICC591 | pBiFC-VN173 derivative encoding TccP FLAG-tagged fused to VN | The present study | Fw-EcoRI-TccP-VN and Rv-TccP-KpnI-VN |
| pICC592 | pBiFC-VC155 derivative encoding TccP HA-tagged fused to VC | The present study | Fw-TccP-EcoR1 and Rv-TccP-VC-Kpn1 |
| pICC593 | pBiFC-VC155 derivative encoding Tir-EHEC HA-tagged fused to VC | The present study | Fw-Tir-EHEC-BglII and Rv-Tir-EHEC-KpnI |
| pICC594 | pBiFC-VC155 derivative encoding Tir-EPEC HA-tagged fused to VC | The present study | Fw-Tir-EPEC-EcoRI and Rv-TirVC-EPEC-KpnI |
| pICC514 | pET28a-nherf2 encoding His6–NHERF2 | [14] | |
| pICC595 | pET28a-int280γ encoding His6–Int280γ | The present study | NcoI-Int280γ and EcoRI-Int280g-Rv |
| pICC596 | pET28a-anxA2 encoding AnxA2 (residues 1–339) His6-tagged | The present study | XbaI-AnxA2 and AnxA2-XhoI |
| pICC597 | pET28a-anxA2 encoding AnxA2 (residues 1–339) HA- and His6-tagged | The present study | XbaI-AnxA2 and AnxA2-C-HA-Xho1 |
| pICC598 | pET28a-anxA2-N encoding N-terminal region of AnxA2 (residues 1–190) His6-tagged | The present study | XbaI-AnxA2 and AnxA2-N-HA-XhoI |
| pICC599 | pET28a-anxA2-C encoding C-terminal region of AnxA2 (residues 191–339) His6-tagged | The present study | XbaI-AnxA2-Cterm and AnxA2-C-HA-Xho1 |
| pICC602 | pET28a-tir-EHEC encoding Tir-EHEC (residues 1–558) His6-tagged | The present study | XbaI-Tir-Fw and XhoI-Tir-Rv |
| pICC603 | pET28a-tir-EPEC encoding Tir-EPEC (residues 1–550) His6-tagged | The present study | Fw-Xba-Tir-EPEC and TirC-EPEC-Not1-b |
| pICC604 | pET28a-tir-Citrobacter encoding Tir-Citrobacter (residues 1–547) His6-tagged | The present study | Fw-Xba-Tir-Citrob and Rv-Tir-Citrob-NotI |
| pICC605 | pET28a-tirN-EHEC encoding TirN-EHEC (residues 1–237) His6-tagged | The present study | XbaI-TirN and TirN-XhoI |
| pICC606 | pET28a-tirN-EPEC encoding TirN-EPEC (residues 1–240) His6-tagged | The present study | Fw-Xba-Tir-EPEC and Rv-TirN-EPEC-NotI |
| pICC607 | pET28a-tirN-Citrobacter encoding TirN-Citrobacter (residues 1–238) His6-tagged | The present study | Fw-Xba-Tir-Citrob and Rv-TirN-Citrob-NotI |
| pICC608 | pET28a-tirC-EHEC encoding TirC-EHEC (residues 366–558) His6-tagged | The present study | XbaI-TirC and TirC-XhoI |
| pICC609 | pET28a-tirC-EPEC encoding TirC-EPEC (residues 369–550) His6-tagged | The present study | XbaI-TirC-EPEC and TirC-EPEC-Not1-b |
| pICC610 | pET28a-tirC-Citrobacter encoding TirC-Citrobacter (residues 367–547) His6-tagged | The present study | XbaI-TirC-Citrob and TirC-Citrob-Not1-b |
| pICC481 | pSA10 derivative expressing Tir HA-tagged | [45] | |
Plasmids
Plasmids and primers used in the present study are listed in Table 1 and Supplementary Table S1 at http://www.BiochemJ.org/bj/445/bj4450383add.htm respectively. All PCRs were carried out using Pfu polymerase. For cloning into the eukaryotic pRK5 expression vector, human ANXA2 was amplified from pBSAxA2 plasmid [28] using the primer pairs indicated. For BiFC (bimolecular fluorescence complementation) experiments, we amplified human ANXA2 from pBSAxA2 plasmid, nherf2 from pICC514 plasmid [14], tccP/espFU from EDL933 genomic DNA and tir from the corresponding EPEC or EHEC genomic DNA with the indicated primer pairs. PCR products were cloned into pBiFC-VN173 or pBiFC-VC155 vectors [29] with the indicated enzymes generating fusions to the N-terminal (VN, residues 1–173) or the C-terminal (VC, residues 155–238) fragment of the Venus protein, a YFP (yellow fluorescent protein) variant that is optimized for better folding at 37°C. For generation of His6-tagged proteins, ANXA2 was amplified from pBSAxA2 plasmid, Int280γ was amplified from EDL933 genomic DNA, and tir from EPEC, EHEC and the mouse A/E pathogen Citrobacter rodentium were amplified from the corresponding genomic DNA. PCR products were cloned into pET28a(+) (Novagen). All constructs were verified by DNA sequencing.
Cell culture and transfections
Simian kidney fibroblast COS-7 and HeLa cell lines (A.T.C.C.. Manassas, VA, U.S.A.) were routinely maintained in DMEM containing 10% (v/v) heat-inactivated FBS (fetal bovine serum) and 1 mM l-glutamine (Gibco) in a humidified atmosphere of 5% CO2 at 37°C. For maintenance of HeLa-NHERF2 stable cell line, the medium was supplemented with 100 μg/ml geneticin (G418, Gibco), as described previously [14]. For BiFC experiments, COS-7 cells were used, as it is the standard cell line used for these assays. COS-7 cells were seeded in 24-well plates at a density of 2.5×105 cells per well and 500 ng of DNA was transfected using FuGene™ 6 transfection reagent (Roche Applied Science) according to the manufacturer’s recommendations. The cells were incubated at 37°C in a humidified incubator with 5% CO2 for 48 h. Transfection of HeLa and HeLa-NHERF2 cells with pRK5 and pGFP vectors encoding ANXA2 was performed using Lipofectamine™ 2000 (Invitrogen) according to the manufacturer’s recommendations. The cells were incubated at 37°C in a humidified incubator with 5% CO2 for 5 h, washed twice in PBS before having their medium replaced with DMEM and incubated for further 19 h. When indicated, transfected cells were infected with the appropriate strain as described below.
Infection of HeLa/HeLa-NHERF2 cells
At 48 h before infection, HeLa and HeLa-NHERF2 cells were seeded on to glass coverslips at a density allowing to reach 50% confluence on the day of infection and maintained in the medium described above at 37°C in 5% CO2. At 3 h before infection, cells were washed three times with PBS and the medium was replaced with fresh DMEM without FBS. Overnight cultures of the appropriate bacteria were inoculated 1:50 into DMEM and primed as described previously [30]. Bacteria were diluted to reach an approximate MOI (multiplicity of infection) of 100 and added to the cell culture medium. All infections were carried out during 2 h at 37°C in a 5% CO2 incubator.
Immunofluorescence staining and microscopy
Coverslips were washed three times in PBS and fixed with 3% (w/v) paraformaldehyde for 20 min before washing three more times in PBS. For immunostaining, the fixed cells were quenched for 20 min with PBS containing 50 mM NH4Cl, permeabilized for 4 min in PBS containing 0.1% Triton X-100 and washed three times in PBS. The coverslips were then blocked for 10 min with PBS/1% (w/v) BSA before incubation with primary and secondary antibodies. HeLa-NHERF2 cells were incubated with mouse anti-HA (haemagglutinin) primary antibody (Covance) followed by secondary antibody donkey anti-(mouse IgG) conjugated to a Cy3 (indocarbocyanine) fluorophore (Jackson ImmunoResearch Laboratories). HeLa cells were incubated with primary rabbit anti-Tir antibody followed by donkey anti-(rabbit IgG) conjugated to a Cy3 fluorophore (Jackson ImmunoResearch Laboratories). Dilutions of primary and secondary antibodies were at 1:500 and 1:200 respectively. Actin was stained using Alexa Fluor® 647–phalloidin (Invitrogen) at 1:100 dilution and DNA was stained with DAPI (4′,6-diamidino-2-phenylindole) at 1:1000 dilution. All dilutions were carried out in PBS/0.1% BSA. Coverslips were incubated at room temperature (21°C) with the primary antibody for 1 h, washed three times in PBS and incubated with the secondary antibody for 1 h. Coverslips were mounted on slides using ProLong Gold anti-fade reagent (Invitrogen) and visualized using a Zeiss Axioimager immunofluorescence microscope. All images were analysed using the Axiovision Rel 4.5 software and trimmed to 5 cm2 (300 pixels) using Adobe Photoshop.
Quantification of the BiFC signal by flow cytometry
COS-7 cells were transfected as described above and incubated in a humidified atmosphere of 5% CO2 at 37°C for 48 h to enable maturation of Venus protein. Cells were washed three times with PBS, detached by incubation for 5 min at 37°C with trypsin/EDTA and neutralized with DMEM/10% (v/v) FBS. The BiFC signal was measured using a FACSCalibur analyser (Becton Dickson) with an appropriate filter set for Venus protein. For each sample, 10000 cells were counted in three independent experiments and used to calculate the mean ± S.D. BiFC signal intensity (n = 3). Results were expressed as the mean fluorescence index, calculated as geometric mean fluorescence multiplied by the percentage of positive cells. Full-length Venus fluorescence signal (obtained by transfecting cells with 500 ng of corresponding plasmid cDNA) was used as the interassay reference value (100%) for maximal fluorescence in each experiment, against which the BiFC signal intensities were normalized.
Protein expression and purification
Genes cloned into pET28a vector were expressed in E. coli K-12 strain BL21 Star™ (DE3) strain (Invitrogen) in the presence of 1 mM IPTG (isopropyl β-d-thiogalactopyranoside) at 30°C for 5 h. The bacteria were pelleted, lysed using a French press and His6-tagged proteins were purified using Ni2+ resin (Novagen) according to the manufacturer’s recommendations. Purified proteins were dialysed to remove imidazole using 10 kDa MWCO (molecular-mass cut-off) Slide-A-Lyzer Dialysis Cassettes (Thermo Fisher). The yield and purity of the resulting proteins were assayed by BCA (bicinchoninic acid) Protein Assay Kit (Pierce) and SDS/PAGE.
ELISA
Purified His6-tagged proteins or BSA as control were coated on to 96-well microtitre plates (Maxisorb, Nunc) overnight at 4°C at a concentration of 100 μg/ml in PBS. The next day, the plates were blocked with PBS/0.1% Tween 20 containing 3% (w/v) dried skimmed milk powder for 2 h at room temperature. Different concentrations of purified AnxA2–HA or NHERF2 in PBS were incubated in the wells for 2 h. Subsequently, the plates were washed three times with PBS/0.1% Tween 20. Binding of AnxA2 was detected with horseradish-peroxidase-conjugated anti-HA antibodies (Sigma), whereas binding of NHERF2 was detected with rabbit anti-NHERF2 (Covalab) followed by horseradish-peroxidase-conjugated goat anti-(rabbit IgG) antibodies (Sigma). After washing the plates as above, the ELISAs were developed using OPD (o-phenylenediamine) (Sigma) and the absorbance was measured at 490 nm. Data points have been adjusted using a fit by regression to a square hyperbola.
Overlay assays and Western blots
Protein samples for overlays were prepared under denaturing (1% SDS, 100°C, 10 min) conditions and 15 μl of each protein at 10 μM was separated on SDS (0.1%)-containing polyacrylamide (12%) gels using standard methods [31]. The gels were stained with Coomassie Blue or transferred on to a PVDF membrane using a semi-dry electrophoresis transfer apparatus (Bio-Rad Laboratories) and blocked overnight with PBS/0.1% Tween 20 containing 5% (w/v) dried skimmed milk powder. The PVDF membranes were washed in PBS/0.1% Tween 20 for 5 min and allowed to react with 125 μg of the purified proteins in 5 ml of PBS/0.1% Tween 20 for 3 h at room temperature. After three washes in PBS/0.1% Tween 20, binding of purified NHERF2 and Tir was detected with polyclonal rabbit anti-NHERF2 (Covalab) and rabbit anti-Tir antibodies followed by horseradish-peroxidase-conjugated goat anti-(rabbit IgG) antibodies (Sigma). Binding of AnxA2–HA was detected with horseradish-peroxidase-conjugated anti-HA antibodies (Sigma). The membranes were washed as above and developed using ECL (enhanced chemiluminescence) (GE Healthcare). Western blots were performed using standard methods [31] with horseradish-peroxidase-conjugated anti-HA, rabbit anti-AnxA2 (Abcam), rabbit anti-Tir or mouse anti-tubulin (Abcam) antibodies. Horseradish-peroxidase-conjugated goat anti-(rabbit IgG) or rabbit anti-(mouse IgG) (Sigma) was used as the secondary antibody.
Co-IP (co-immunoprecipitation)
HeLa and HeLa-NHERF2 cells were grown to confluence in 150 cm2 cell culture plates and transfected with pRK5-ANXA2. After 24 h, cells were washed three times with PBS and lysed in 500 μl of co-IP buffer [50 mM Tris/HCl (pH 7.5), 100 mM NaCl, 1% Nonidet P40, 0.5% sodium deoxycholate, 10% glycerol and Complete™ protease inhibitors (Roche)], scraped, vortex-mixed and incubated in a spinning wheel for 15 min at 4°C. The lysate was centrifuged at 9200 g for 10 min at 4°C, the supernatant was transferred to a fresh pre-chilled Eppendorf tube containing 30 μl of pre-equilibrated anti-HA–agarose beads (Sigma) and incubated on a spinning wheel for 2 h at 4°C. The suspension was washed three times in co-IP buffer followed by a brief centrifugation at 12000 g at 4°C after each wash. Samples were boiled with Laemmli buffer and analysed by Western blotting as described before.
RESULTS
Co-localization of NHERF2 and AnxA2 at the EPEC attachment site
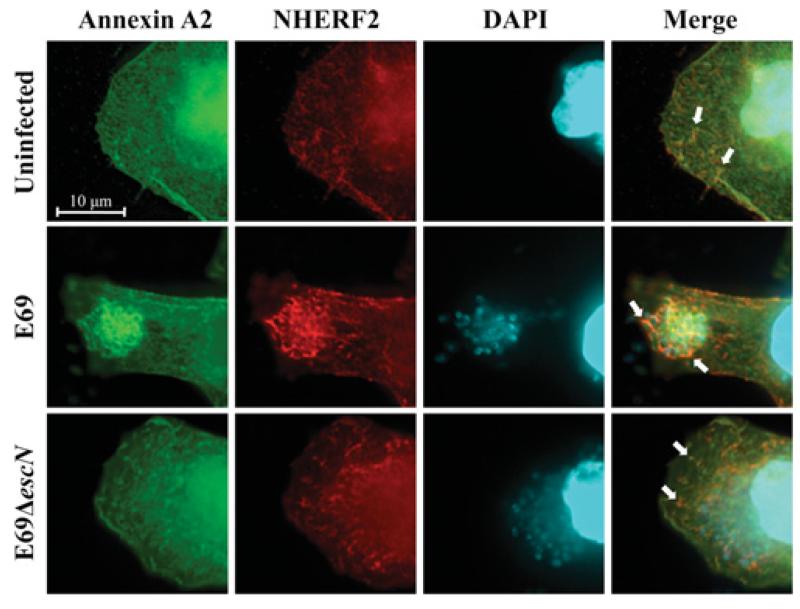
In order to investigate the mechanism involved in the recruitment of NHERF2 and AnxA2 by EPEC, HeLa cells stably expressing HA–NHERF2 (HeLa-NHERF2 [14]) were transfected with GFP (green fluorescent protein)–AnxA2 [32]. Immunofluorescence using anti-HA antibodies combined with the GFP signal revealed partial co-localization of NHERF2 and AnxA2 in 70–80% of examined uninfected cells (Figure 1; uninfected). In order to determine the localization of NHERF2 and AnxA2 during EPEC infection, HeLa-NHERF2 cells ectopically expressing AnxA2 were infected with wild-type EPEC strain E2348/69 and E2348/69ΔescN (encoding the ATPase) mutant, defective in T3SS, as a control. This has shown that both NHERF2 and AnxA2 are recruited by wild-type EPEC and co-localized at the bacterial attachment sites in 90–100% of examined cells. In contrast, the localization of AnxA2 and NHERF2 in cells infected with E2348/69ΔescN was similar to that of the uninfected control, suggesting that NHERF2 and AnxA2 are recruited to EPEC in a T3SS-dependent manner.
Figure 1. AnxA2 and NHERF2 co-localize in human cultured cells.
Fluorescence microscopy of HeLa-NHERF2 cells transfected with GFP–AnxA2, uninfected or infected with E2348/69 (E69) or E2348/69ΔescN strains. A total of 100 transfected cells were examined in triplicate. The green GFP signal corresponds to AnxA2, NHERF2 was stained with mouse anti-HA followed by Cy3-conjugated anti-mouse antibodies (red) and DNA from bacteria and eukaryotic cells was stained with DAPI (cyan). White arrows indicate partial co-localization of AnxA2 and NHERF2 in uninfected or E2348/69ΔescN-infected cells and intensified co-localization upon infection of E2348/69.
AnxA2 binds NHERF2
Owing to their co-localization in uninfected cells, we have used the BiFC approach [33] to determine whether AnxA2 and NHERF2 interact. BiFC technology is based on the assembly of a fluorescent protein (e.g. YFP) through the association of its non-fluorescent N- and C-terminal fragments when they are brought together through interacting proteins. For BiFC constructs, we fused AnxA2 and NHERF2 to the N-terminal (VN, residues 1–173) or the C-terminal (VC, residues 155–238) fragment of the Venus protein, a YFP variant that is optimized for folding at 37°C [34], generating AnxA2–VN and NHERF2–VC (Table 1). As a positive control, we generated NHERF2–VN, which was co-transfected with NHERF2–VC in order to detect NHERF2–NHERF2 interaction, as this interaction has been described previously [35]. As a negative control, we used TccP/EspFU [36] fused to VN (TccP–VN). BiFC constructs were transfected into HeLa cells, and expression of the fusion proteins was confirmed by immunofluorescence with antibodies that recognize protein tags that are present in BiFC constructs (Supplementary Figure S1 at http://www.BiochemJ.org/bj/445/bj4450383add.htm). The YFP fluorescence (BiFC signal) of 10000 cells was quantified by FACS as described previously [37] and expressed as the percentage of Venus fluorescence. Transfections of cells with single constructs did not produce any detectable BiFC signal (Figure 2A). However, when cells were co-transfected with NHERF2–VC and AnxA2–VN or NHERF2–VN (positive control), we obtained fluorescence equivalent to 75% of Venus protein (Figure 2A), indicating a direct interaction between AnxA2 and NHERF2. When NHERF2–VC was co-transfected with the negative control TccP–VN, we recorded 8% of Venus fluorescence.
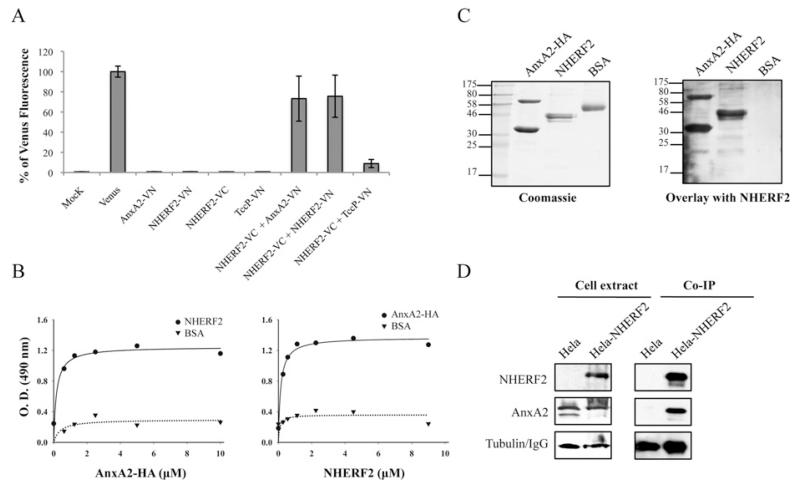
Figure 2. AnxA2 binds NHERF2.
(A) Measured BiFC signal intensities by FACS in COS-7 cells. Results are means ± S.D. for three independent experiments (10000 cells/experiment) expressed as percentages of the signal intensity obtained with an equivalent amount (500 ng) of the pVenus plasmid. (B) ELISA quantification of binding of AnxA2 and NHERF2. A range of concentrations of purified AnxA2–HA were added to microtitre plate wells coated with purified NHERF2 or BSA. Binding was detected with horseradish-peroxidase-conjugated anti-HA antibodies (left-hand panel). A range of concentrations of purified NHERF2 were added to microtitre plate wells coated with purified AnxA2–HA or BSA. Binding was detected with rabbit polyclonal anti-NHERF2 antibodies followed by horseradish-peroxidase-conjugated anti-rabbit antibodies (right-hand panel). O.D. = absorbance. (C) Protein overlay assay showing binding of NHERF2 to AnxA2. Purified proteins AnxA2–HA, NHERF2 and BSA were separated by SDS/PAGE and stained with Coomassie Blue (left-hand panel) or immobilized on to PVDF membranes and incubated with purified NHERF2 (right-hand panel). Bound NHERF2 was detected by immunostaining with rabbit polyclonal anti-NHERF2 antibodies followed by horseradish-peroxidase-conjugated anti-rabbit antibodies, showing specific interaction of NHERF2 with AnxA2 either as monomer or as stable dimer. Molecular masses are indicated in kDa. (D) NHERF2 co-immunoprecipitates with AnxA2. HA–NHERF2 was immunoprecipitated with mouse anti-HA antibody from HeLa or HeLa-NHERF2 cells. Western blots revealed that AnxA2 was immunoprecipitated with NHERF2 in HeLa-NHERF2 cells, but not in HeLa cells, whereas equivalent levels of AnxA2 were found in the whole-cell extracts. Equivalent protein loading was confirmed by Western blotting with anti-tubulin antibody in cell extracts and anti-(mouse IgG) antibody in co-IP samples.
In order to confirm the interaction of AnxA2 with NHERF2, we performed an ELISA in which microtitre plates coated with purified His6–NHERF2 [14] or BSA, as a negative control, were incubated with serial dilutions of purified His6–AnxA2 fused to an HA tag (AnxA2–HA). Detection of bound protein with HA antiserum revealed that NHERF2 interacts with AnxA2 (Figure 2B, left-hand panel). In a reciprocal experiment, AnxA2–HA-coated microtitre plates were incubated with His6–NHERF2, and the bound proteins were detected with NHERF2 rabbit polyclonal antiserum followed by rabbit antiserum, confirming the interaction between NHERF2 and AnxA2 (Figure 2B, right).
The interaction between NHERF2 and AnxA2 was confirmed further in gel overlay-based assays. Purified AnxA2–HA, NHERF2 and BSA, as a negative control, were immobilized on a membrane and overlaid with purified His6–NHERF2. Detection of bound protein with NHERF2 rabbit polyclonal antiserum followed by rabbit antiserum confirmed further the interaction of NHERF2 with 36 and 64 kDa AnxA2 proteins, which correspond to monomeric and dimeric AnxA2 (Figure 2C). No signal was observed in a control overlay in the absence of purified His6–NHERF2 (results not shown).
Finally, we investigated the interaction of AnxA2 and NHERF2 in eukaryotic cells by co-IP. Towards this end, we ectopically expressed AnxA2 in HeLa and HeLa-NHERF2 cells. Probing cell extracts with HA and AnxA2 antisera revealed that whereas NHERF2 was found only in HeLa-NHERF2 cells, as expected, AnxA2 was found in equal amounts in both samples (Figure 2D). Following cell lysis, the supernatants were subjected to co-IP with anti-HA antibodies. AnxA2 co-immunoprecipitated with NHERF2 in HeLa-NHERF2 cell, whereas it did not co-immunoprecipitate from the control HeLa cells (Figure 2D).
Taken together, these results show, for the first time, an interaction between NHERF2 and Anx2A.
AnxA2 is recruited in EPEC infection in a Tir-dependent and actin-independent manner
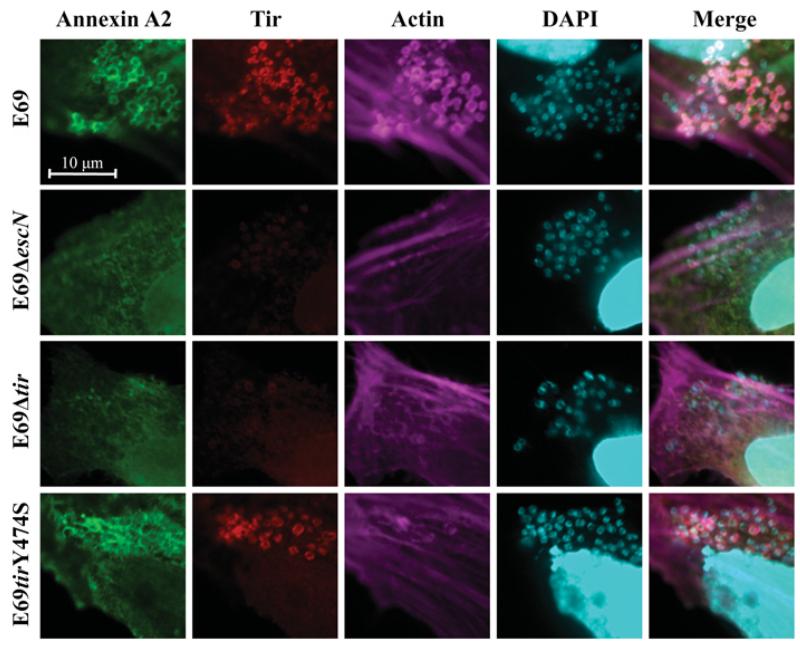
As NHERF2 and AnxA2 form a complex and are recruited to the bacterial attachment sites in a T3SS-dependent manner, we investigated whether recruitment of AnxA2 upon EPEC infection is dependent on the membrane-associated bacterial effector Tir. HeLa cells ectopically expressing GFP–AnxA2 were infected for 2 h with wild-type E2348/69 (E69) and E69Δtir strains. E69ΔescN was used as a control. Immunofluorescence revealed that, whereas AnxA2 was recruited to wild-type EPEC in 90–100% of examined cells, it was not recruited to the attachment sites of EPEC Δtir or ΔescN in any of the cells examined (Figure 3); recruitment was restored when the Δtir mutant was complemented (Supplementary Figure S2 at http://www.BiochemJ.org/bj/445/bj4450383add.htm), indicating that Tir plays a direct role in AnxA2 recruitment. As AnxA2 binds actin [16] that is absent from the sites of EPEC Δtir [7], we wanted to determine whether AnxA2 recruitment was dependent on Tir-induced actin polymerization. To test this, we infected HeLa cells with EPEC E69tirY474S [38], a chromosomal tir mutant unable to trigger actin polymerization. This revealed that AnxA2 was still recruited following infection with E69tirY474S, in 80–90% of the examined cells (Figure 3), indicating that AnxA2 recruitment depends on Tir and is independent of Tir-induced actin polymerization.
Figure 3. AnxA2 is recruited during EPEC infection in a Tir-dependent and actin-independent manner.
Fluorescence microscopy of HeLa cells transfected with GFP–AnxA2 and infected with EPEC E2348/69 (E69) or its derivative strains. A total of 100 transfected cells were examined in triplicate. The green GFP signal corresponds to AnxA2, Tir was stained with rabbit anti-Tir followed by Cy3-conjugated anti-rabbit antibodies (red), actin was stained with Alexa Fluor® 647–phalloidin (magenta), and DNA was stained with DAPI (cyan). AnxA2 is recruited to the sites of bacterial attachment during E2348/69 and E2348/69 tirY474S infection, but not during E2348/69ΔescN or E2348/69Δtir infection.
AnxA2 binds Tir-EPEC and Tir-EHEC through its C-terminal domain
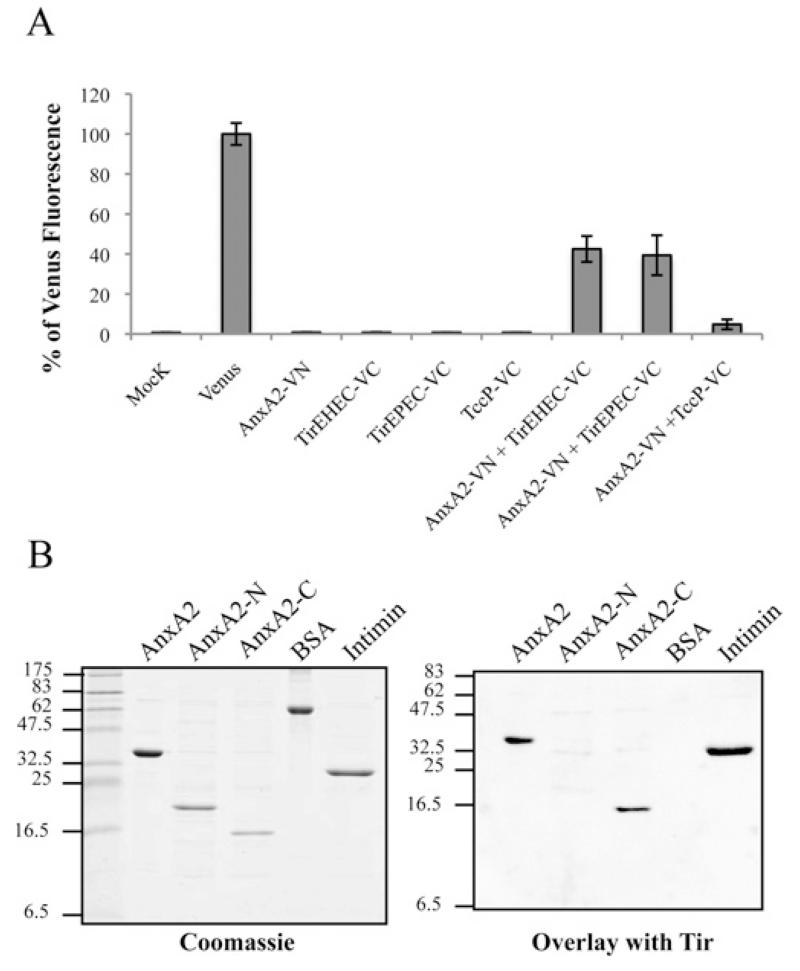
We investigated whether recruitment of Anx2A to EPEC is mediated by a direct Tir–Anx2A and/or Tir–NHERF2 interaction. Although we did not detect any interaction of Tir with NHERF2 by any of the techniques we tried (results not shown), using the BiFC approach described above, we found that AnxA2 binds Tir. For this experiment, COS-7 cells were co-transfected with AnxA2–VN together with Tir-EPEC fused to the C-terminal fragment of the Venus protein (TirEPEC–VC) or TccP–VC as a negative control. When AnxA2–VN was co-transfected with TirEPEC–VC, the BiFC signal showed a >40% signal relative to the fluorescence of Venus protein (Figure 4A). Neither single construct transfections nor co-transfection of AnxA2–VN with TccP–VC produced any significant BiFC signal, thus indicating a direct interaction between AnxA2 and Tir-EPEC (Figure 4A). In order to determine whether this finding could be generalized to other A/E pathogens, we repeated the experiment using Tir-EHEC. This has shown that, similarly to Tir-EPEC, Tir-EHEC can also interact with AnxA2 (Figure 4A). Expression of all BiFC constructs was confirmed by immunofluorescence of transfected cells with antibodies that recognize protein tags that are present in the constructs (Supplementary Figure S1).
Figure 4. AnxA2 binds Tir-EPEC and Tir-EHEC through its C-terminal domain.
(A) Measured BiFC signal intensities determined by FACS in COS-7 cells. Results are means ± S.D. for three independent experiments (10000 cells/experiment) expressed as percentages of the signal intensity obtained with an equivalent amount (500 ng) of the pVenus plasmid. (B) Identification of Tir-binding sites on AnxA2. Protein overlay assay showing binding of Tir-EHEC to AnxA2. Purified full-length human AnxA2, AnxA2-N (amino acids 1–190), AnxA2-C (amino acids 191–339), BSA and intimin (Int280γ ) proteins were separated by SDS/PAGE and stained with Coomassie Blue (left-hand panel) or immobilized on to PVDF membranes and incubated with purified His6–Tir-EHEC (right-hand panel). Bound Tir-EHEC was detected by immunostaining with rabbit polyclonal anti-Tir antibodies followed by alkaline phosphatase-linked anti-(rabbit IgG) antibody, showing specific interaction of Tir with the intact C-terminal AnxA2 region. Molecular masses are indicated in kDa.
In order to identify which domain of AnxA2 is involved in the interaction with Tir, we constructed and purified His6-tagged full-length AnxA2 (amino acids 1–339), AnxA2-N (amino acids 1–190) and AnxA2-C (amino acids 191–339) and tested their interaction with purified Tir in protein overlay assays. The AnxA2 variants, together with BSA and intimin (Int280γ) as negative and positive controls respectively, were separated by SDS/PAGE and stained with Coomassie Blue (Figure 4B, left-hand panel) or blotted and then incubated with purified Tir (Figure 4B, right-hand panel). Tir specifically bound to the C-terminus domain of AnxA2 and the intimin control. Similar results were seen when the experiments were performed with either Tir-EPEC or Tir-EHEC. No signal was observed in the BSA control or in an overlay in the absence of purified Tir (results not shown).
The N- and C-terminal domains of Tir bind AnxA2
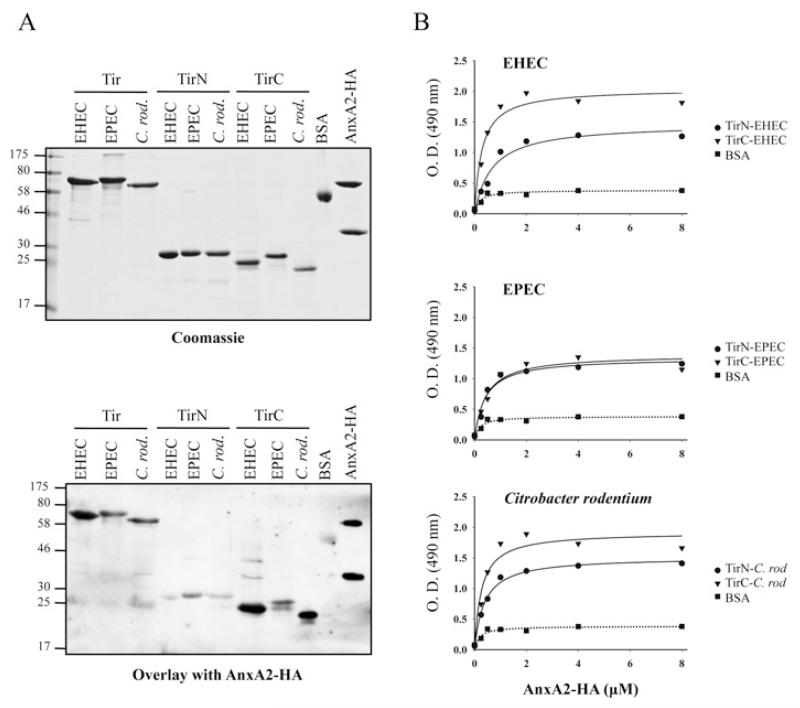
Next, we investigated which domain/s of Tir are involved in AnxA2 interaction. We purified His6-tagged TirN (N-terminal Tir domain) and TirC (C-terminal Tir domain), as well as full-length Tir from EPEC, EHEC and from the mouse A/E pathogen C. rodentium [39]. Purified His6-tagged Tir variants were separated by SDS/PAGE and stained with Coomassie Blue (Figure 5A, upper panel) or blotted and then incubated with purified AnxA2–HA. This revealed that Tir of all three pathogens could bind AnxA2 (Figure 5A, lower panel). Specific interaction of AnxA2 with TirN and TirC showed that TirC of EHEC and C. rodentium interacted with AnxA2 more strongly than their corresponding TirN domains, which showed a reduced level of binding to AnxA2. However, in EPEC, the interaction of AnxA2 with TirN and TirC revealed equivalent binding levels. In order to confirm these results, we performed ELISAs in which purified TirN and TirC from EPEC, EHEC and C. rodentium or BSA, as a negative control, were used to coat microtitre plates which were then incubated with serial dilutions of purified AnxA2–HA. As Tir harbours two transmembrane domains that could aggregate the protein in solution, we did not include full-length Tir in this assay. Detection of bound protein with HA antiserum showed that AnxA2 interacts with both the TirN and TirC domains of A/E pathogens, revealing stronger interactions of TirN from EHEC and C. rodentium than expected, according to the overlay assay. This difference could be due to the native nature of the proteins used in the ELISA compared with the denatured proteins used in the overlay. The strongest interactions were detected with TirC-EHEC and TirC-C. rodentium, confirming the results obtained with the overlay assay.
Figure 5. TirN and TirC from EHEC, EPEC and C. rodentium bind AnxA2 with different affinities.
(A) Protein overlay assay showing binding of AnxA2 to Tir-EHEC, Tir-EPEC and Tir-C. rodentium (C. rod.) derivatives. Purified full-length Tir, TirN and TirC from EHEC, EPEC and C. rodentium were separated by SDS/PAGE and stained with Coomassie Blue (upper panel) or immobilized on to PVDF membranes and incubated with purified AnxA2–HA (lower panel). Bound AnxA2 was detected with horseradish-peroxidase-conjugated anti-HA antibodies. Molecular masses are indicated in kDa. (B) ELISA quantification of binding of AnxA2 to EHEC, EPEC and C. rodentium TirN and TirC fragments. A range of concentrations of purified AnxA2–HA were added to microtitre plate wells coated with EHEC (top panel), EPEC (middle panel) or C. rodentium (C. rod) (bottom panel) purified TirN, TirC or BSA proteins. Binding was detected with horseradish-peroxidase-conjugated anti-HA antibodies. O.D. = absorbance.
DISCUSSION
In the present paper, we report that NHERF2 and AnxA2 are partially co-localized in HeLa cells and that this co-localization is intensified upon infection with EPEC. Consistently, we found that AnxA2 binds directly to NHERF2. We demonstrated this interaction using BiFC technology and in vitro assays with recombinant proteins. To our knowledge, this is the first report showing such an interaction. Moreover, we found that NHERF2 and AnxA2 co-immunoprecipitated from HeLa-NHERF2 cells. AnxA2 binds to the periphery of membranes containing acidic phospholipids, playing a role as membrane scaffold proteins and being recruited at sites of actin assembly at cellular membranes [40]. NHERF2 is a eukaryotic protein usually linked to membranes. Previous studies have shown that AnxA2 [12,13] and NHERFs [14,27] are recruited to the EPEC/EHEC-attachment sites.
Although EPEC-induced diarrhoea is arguably the most important aspect of infection, our understanding of the mechanisms involved remains incomplete. We know that diarrhoea caused by A/E pathogens is a multifactorial process involving an array of virulence factors and host signalling pathways. The eventual loss of the electrolyte homoeostasis in the intestine is usually linked to an impaired regulation of the intestinal ion channels. In this sense, the regulatory proteins AnxA2 (in complex with p11) and NHERF2, critical for Ca2+ homoeostasis through their association with TRPV5/6 are recruited to the EPEC- and EHEC-attachment sites [12-14]. It is therefore possible that EPEC and EHEC modify the microenvironment around the sites of bacterial adhesion, exploiting and sequestering cellular factors such as AnxA2 and NHERF2 to disrupt ion channel homoeostasis and establishing the infection that leads to diarrhoea.
As TRPV5/6 binds both AnxA2 and NHERF2 at cellular membranes, we investigated whether the membrane bacterial effector Tir is involved in their recruitment to the bacterial attachment sites. This revealed that, upon EPEC infection, AnxA2 accumulated at the site of bacterial attachment in a Tir-dependent manner. Importantly, recruitment of AnxA2 was independent of Tir Tyr474, which is involved in strong and localized actin polymerization. This result disagrees with Zobiack et al. [12] who reported that an EPEC Δtir mutant is still capable of recruiting AnxA2 to the bacterial attachment sites. The reason for this discrepancy could be that Zobiack et al. [12] employed a C-terminally truncated Tir protein, still harbouring a functional TirN terminal domain, which, as we have shown by overlay- and ELISA-based assays, is able to bind AnxA2 and possibly recruit it during EPEC infection.
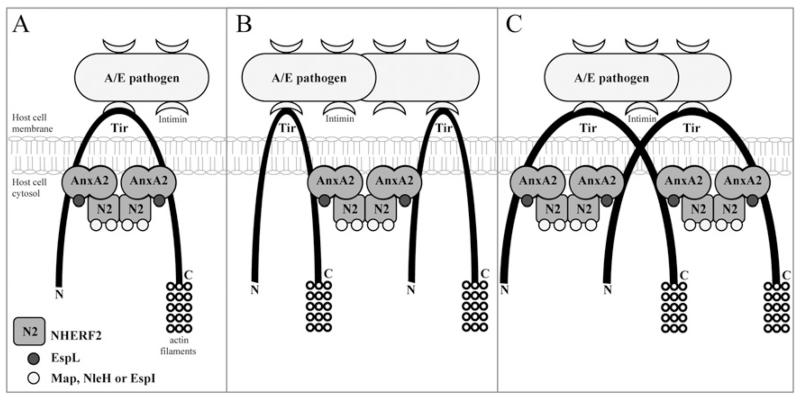
The finding that Tir is involved in the recruitment of AnxA2 to the bacterial attachment site led us to investigate whether Tir binds AnxA2 and/or NHERF2. Although we did not find evidence that Tir binds NHERF2, we found that it binds AnxA2, specifically the AnxA2 C-terminal region (amino acids 191–339). Importantly, the AnxA2 C-terminal binds both the N- and C-termini of Tir. We propose a model by which AnxA2 is recruited to both the N- and C-termini of Tir. NHERF2 is recruited either to Tir-bound AnxA2 or in complex with it. Via NHERF2–NHERF2 interaction, the NHERF2–AnxA2 complex can then bridge between adjacent Tir molecules that could either space the N- and C-termini of a single Tir molecule (Figure 6A), and/or bridge between two neighbouring Tir molecules (Figure 6B). Alternatively, as Tir can also form membrane dimers, the AnxA2–NHERF2 linker can bridge between two N- or C-termini of different Tir molecules (Figure 6C). This cross-linking is likely to play a role in stabilizing and spacing Tir in the membrane, which might be important for its ability to bind intimin extracellularly or host cell proteins intracellularly. In addition, the Tir–AnxA2–NHERF2 complex can provide a localized signalling scaffold/hub at the plasma membrane. Indeed, a number of EPEC and EHEC effectors bind to NHERF2 [14] and Anx2A [13], and our model provides a possible mechanism for their recruitment to the cytoplasmic face of the plasma membrane which is needed to execute their activity (e.g. EspL and Map are involved in modulating localized actin dynamics) or before dissemination to different organelles (e.g. NleH and EspI target the endoplasmic reticulum and Golgi respectively). Further studies are needed to determine whether the Tir–AnxA2–NHERF2 complex recruits additional bacterial effectors or host cell proteins that are involved in A/E lesion formation or disruption of channels and diarrhoea, both being the hallmarks of EPEC infection.
Figure 6. Model for AnxA2 and NHERF2 recruitment to the bacterial attachment sites during infection with attaching and effacing pathogens.
AnxA2 and NHERF2, which are partially co-localized at the luminal microvillar brush border of intestinal epithelial cells, are recruited to adherent bacteria that are attached to the host by the intimate intimin–Tir interaction. TirN and TirC recruit AnxA2 through an interaction with its C-terminal domain. NHERF2 then binds Tir-associated AnxA2, forming a bridge that can span the N- and C- termini of a single Tir molecule (A), span two adjacent Tir molecules (B), or span two TirN regions or TirC regions of a Tir dimer (C). The scaffold formed by the Tir–AnxA2–NHERF2 complex serves to relay cellular information of the docking bacterial effectors that function at the membrane, e.g. EspL and Map, or are targeted to deeper organelles, e.g. NleH and EspI.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Dr Sakari Kellokumpu (University of Oulu, Oulu, Finland) for the gift of pVenus plasmid. The excellent technician work of Leah Ensell is much appreciated.
FUNDING
This work was supported by grants from the Wellcome Trust. D.M. holds a fellowship from the Ministerio de Educación y Ciencia (MEC) (Spain).
Abbreviations used
- A/E
attaching and effacing
- AnxA2
annexin A2
- BiFC
bimolecular fluorescence complementation
- co-IP
co-immunoprecipitation
- Cy3
indocarbocyanine
- DAPI
4′,6-diamidino-2-phenylindole
- DMEM
Dulbecco’s modified Eagle’s medium
- EHEC
enterohaemorrhagic Escherichia coli
- EPEC
enteropathogenic Escherichia coli
- F-actin
filamentous actin
- FBS
fetal bovine serum
- GFP
green fluorescent protein
- HA
haemagglutinin
- NHERF2
Na+ /H+ exchange regulatory factor 2
- T3SS
type III secretion system
- TirC
C-terminal Tir domain
- TirN
N-terminal Tir domain
- TRPV
transient receptor potential vanilloid
- VC
C-terminal fragment of Venus protein
- VN
N-terminal fragment of Venus protein
- YFP
yellow fluorescent protein
REFERENCES
- 1.World Health Organization . The World Health Report 2004: Changing History. WHO; Geneva: 2004. [Google Scholar]
- 2.Croxen MA, Finlay BB. Molecular mechanisms of Escherichia coli pathogenicity. Nat. Rev. Microbiol. 2010;8:26–38. doi: 10.1038/nrmicro2265. [DOI] [PubMed] [Google Scholar]
- 3.Frankel G, Phillips AD, Rosenshine I, Dougan G, Kaper JB, Knutton S. Enteropathogenic and enterohemorrhagic Escherichia coli: more subversive elements. Mol. Microbiol. 1998;30:911–921. doi: 10.1046/j.1365-2958.1998.01144.x. [DOI] [PubMed] [Google Scholar]
- 4.Wong AR, Pearson JS, Bright MD, Munera D, Robinson KS, Lee SF, Frankel G, Hartland EL. Enteropathogenic and enterohaemorrhagic Escherichia coli: even more subversive elements. Mol. Microbiol. 2011;80:1420–1438. doi: 10.1111/j.1365-2958.2011.07661.x. [DOI] [PubMed] [Google Scholar]
- 5.McDaniel TK, Jarvis KG, Donnenberg MS, Kaper JB. A genetic locus of enterocyte effacement conserved among diverse enterobacterial pathogens. Proc. Natl. Acad. Sci. U.S.A. 1995;92:1664–1668. doi: 10.1073/pnas.92.5.1664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Garmendia J, Frankel G, Crepin VF. Enteropathogenic and enterohemorrhagic Escherichia coli infections: translocation, translocation, translocation. Infect. Immun. 2005;73:2573–2585. doi: 10.1128/IAI.73.5.2573-2585.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kenny B, DeVinney R, Stein M, Reinscheid DJ, Frey EA, Finlay BB. Enteropathogenic Escherichia coli (EPEC) transfers its receptor for intimate adherence into mammalian cells. Cell. 1997;91:511–520. doi: 10.1016/s0092-8674(00)80437-7. [DOI] [PubMed] [Google Scholar]
- 8.Hartland EL, Batchelor M, Delahay RM, Hale C, Matthews S, Dougan G, Knutton S, Connerton I, Frankel G. Binding of intimin from enteropathogenic Escherichia coli to Tir and to host cells. Mol. Microbiol. 1999;32:151–158. doi: 10.1046/j.1365-2958.1999.01338.x. [DOI] [PubMed] [Google Scholar]
- 9.Laohachai KN, Bahadi R, Hardo MB, Hardo PG, Kourie JI. The role of bacterial and non-bacterial toxins in the induction of changes in membrane transport: implications for diarrhea. Toxicon. 2003;42:687–707. doi: 10.1016/j.toxicon.2003.08.010. [DOI] [PubMed] [Google Scholar]
- 10.Vennekens R, Owsianik G, Nilius B. Vanilloid transient receptor potential cation channels: an overview. Curr. Pharm. Des. 2008;14:18–31. doi: 10.2174/138161208783330763. [DOI] [PubMed] [Google Scholar]
- 11.van de Graaf SF, Hoenderop JG, Bindels RJ. Regulation of TRPV5 and TRPV6 by associated proteins. Am. J. Physiol. Renal Physiol. 2006;290:F1295–F1302. doi: 10.1152/ajprenal.00443.2005. [DOI] [PubMed] [Google Scholar]
- 12.Zobiack N, Rescher U, Laarmann S, Michgehl S, Schmidt MA, Gerke V. Cell-surface attachment of pedestal-forming enteropathogenic E. coli induces a clustering of raft components and a recruitment of annexin 2. J. Cell Sci. 2002;115:91–98. doi: 10.1242/jcs.115.1.91. [DOI] [PubMed] [Google Scholar]
- 13.Miyahara A, Nakanishi N, Ooka T, Hayashi T, Sugimoto N, Tobe T. Enterohemorrhagic Escherichia coli effector EspL2 induces actin microfilament aggregation through annexin 2 activation. Cell. Microbiol. 2009;11:337–350. doi: 10.1111/j.1462-5822.2008.01256.x. [DOI] [PubMed] [Google Scholar]
- 14.Martinez E, Schroeder GN, Berger CN, Lee SF, Robinson KS, Badea L, Simpson N, Hall RA, Hartland EL, Crepin VF, Frankel G. Binding to Na+ /H+ exchanger regulatory factor 2 (NHERF2) affects trafficking and function of the enteropathogenic Escherichia coli type III secretion system effectors Map, EspI and NleH. Cell. Microbiol. 2010;12:1718–1731. doi: 10.1111/j.1462-5822.2010.01503.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gerke V, Moss SE. Annexins: from structure to function. Physiol. Rev. 2002;82:331–371. doi: 10.1152/physrev.00030.2001. [DOI] [PubMed] [Google Scholar]
- 16.Filipenko NR, Waisman DM. The C terminus of annexin II mediates binding to F-actin. J. Biol. Chem. 2001;276:5310–5315. doi: 10.1074/jbc.M009710200. [DOI] [PubMed] [Google Scholar]
- 17.Kassam G, Le BH, Choi KS, Kang HM, Fitzpatrick SL, Louie P, Waisman DM. The p11 subunit of the annexin II tetramer plays a key role in the stimulation of t-PA-dependent plasminogen activation. Biochemistry. 1998;37:16958–16966. doi: 10.1021/bi981713l. [DOI] [PubMed] [Google Scholar]
- 18.Hajjar KA, Jacovina AT, Chacko J. An endothelial cell receptor for plasminogen/tissue plasminogen activator. I. Identity with annexin II. J. Biol. Chem. 1994;269:21191–21197. [PubMed] [Google Scholar]
- 19.Deora AB, Kreitzer G, Jacovina AT, Hajjar KA. An annexin 2 phosphorylation switch mediates p11-dependent translocation of annexin 2 to the cell surface. J. Biol. Chem. 2004;279:43411–43418. doi: 10.1074/jbc.M408078200. [DOI] [PubMed] [Google Scholar]
- 20.Danielsen EM, van Deurs B, Hansen GH. “Nonclassical” secretion of annexin A2 to the lumenal side of the enterocyte brush border membrane. Biochemistry. 2003;42:14670–14676. doi: 10.1021/bi0355239. [DOI] [PubMed] [Google Scholar]
- 21.Babiychuk EB, Draeger A. Annexins in cell membrane dynamics: Ca2+ -regulated association of lipid microdomains. J. Cell Biol. 2000;150:1113–1124. doi: 10.1083/jcb.150.5.1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Oliferenko S, Paiha K, Harder T, Gerke V, Schwarzler C, Schwarz H, Beug H, Gunthert U, Huber LA. Analysis of CD44-containing lipid rafts: recruitment of annexin II and stabilization by the actin cytoskeleton. J. Cell Biol. 1999;146:843–854. doi: 10.1083/jcb.146.4.843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yun CH, Oh S, Zizak M, Steplock D, Tsao S, Tse CM, Weinman EJ, Donowitz M. cAMP-mediated inhibition of the epithelial brush border Na+ /H+ exchanger, NHE3, requires an associated regulatory protein. Proc. Natl. Acad. Sci. U.S.A. 1997;94:3010–3015. doi: 10.1073/pnas.94.7.3010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shenolikar S, Weinman EJ. NHERF: targeting and trafficking membrane proteins. Am. J. Physiol. Renal Physiol. 2001;280:F389–F395. doi: 10.1152/ajprenal.2001.280.3.F389. [DOI] [PubMed] [Google Scholar]
- 25.Cao TT, Deacon HW, Reczek D, Bretscher A, von Zastrow M. A kinase-regulated PDZ-domain interaction controls endocytic sorting of the β2-adrenergic receptor. Nature. 1999;401:286–290. doi: 10.1038/45816. [DOI] [PubMed] [Google Scholar]
- 26.Reczek D, Bretscher A. The carboxyl-terminal region of EBP50 binds to a site in the amino-terminal domain of ezrin that is masked in the dormant molecule. J. Biol. Chem. 1998;273:18452–18458. doi: 10.1074/jbc.273.29.18452. [DOI] [PubMed] [Google Scholar]
- 27.Simpson N, Shaw R, Crepin VF, Mundy R, FitzGerald AJ, Cummings N, Straatman-Iwanowska A, Connerton I, Knutton S, Frankel G. The enteropathogenic Escherichia coli type III secretion system effector Map binds EBP50/NHERF1: implication for cell signalling and diarrhoea. Mol. Microbiol. 2006;60:349–363. doi: 10.1111/j.1365-2958.2006.05109.x. [DOI] [PubMed] [Google Scholar]
- 28.Ayala-Sanmartin J, Gouache P, Henry JP. N-terminal domain of annexin 2 regulates Ca2+ -dependent membrane aggregation by the core domain: a site directed mutagenesis study. Biochemistry. 2000;39:15190–15198. doi: 10.1021/bi000764r. [DOI] [PubMed] [Google Scholar]
- 29.Shyu YJ, Liu H, Deng X, Hu CD. Identification of new fluorescent protein fragments for bimolecular fluorescence complementation analysis under physiological conditions. BioTechniques. 2006;40:61–66. doi: 10.2144/000112036. [DOI] [PubMed] [Google Scholar]
- 30.Collington GK, Booth IW, Knutton S. Rapid modulation of electrolyte transport in Caco-2 cell monolayers by enteropathogenic Escherichia coli (EPEC) infection. Gut. 1998;42:200–207. doi: 10.1136/gut.42.2.200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ausubel FM, Brent R, Kingston RE, Moore DD, Seidman JG, Smith JA, Struhl K. Short Protocols in Molecular Biology. John Wiley & Sons; New York: 1997. [Google Scholar]
- 32.Merrifield CJ, Rescher U, Almers W, Proust J, Gerke V, Sechi AS, Moss SE. Annexin 2 has an essential role in actin-based macropinocytic rocketing. Curr. Biol. 2001;11:1136–1141. doi: 10.1016/s0960-9822(01)00321-9. [DOI] [PubMed] [Google Scholar]
- 33.Hu CD, Chinenov Y, Kerppola TK. Visualization of interactions among bZIP and Rel family proteins in living cells using bimolecular fluorescence complementation. Mol. Cell. 2002;9:789–798. doi: 10.1016/s1097-2765(02)00496-3. [DOI] [PubMed] [Google Scholar]
- 34.Liu H, Deng X, Shyu YJ, Li JJ, Taparowsky EJ, Hu CD. Mutual regulation of c-Jun and ATF2 by transcriptional activation and subcellular localization. EMBO J. 2006;25:1058–1069. doi: 10.1038/sj.emboj.7601020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lau AG, Hall RA. Oligomerization of NHERF-1 and NHERF-2 PDZ domains: differential regulation by association with receptor carboxyl-termini and by phosphorylation. Biochemistry. 2001;40:8572–8580. doi: 10.1021/bi0103516. [DOI] [PubMed] [Google Scholar]
- 36.Garmendia J, Phillips AD, Carlier MF, Chong Y, Schuller S, Marches O, Dahan S, Oswald E, Shaw RK, Knutton S, Frankel G. TccP is an enterohaemorrhagic Escherichia coli O157:H7 type III effector protein that couples Tir to the actin-cytoskeleton. Cell. Microbiol. 2004;6:1167–1183. doi: 10.1111/j.1462-5822.2004.00459.x. [DOI] [PubMed] [Google Scholar]
- 37.Hassinen A, Rivinoja A, Kauppila A, Kellokumpu S. Golgi N-glycosyltransferases form both homo- and heterodimeric enzyme complexes in live cells. J. Biol. Chem. 2010;285:17771–17777. doi: 10.1074/jbc.M110.103184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schuller S, Chong Y, Lewin J, Kenny B, Frankel G, Phillips AD. Tir phosphorylation and Nck/N-WASP recruitment by enteropathogenic and enterohaemorrhagic Escherichia coli during ex vivo colonization of human intestinal mucosa is different to cell culture models. Cell. Microbiol. 2007;9:1352–1364. doi: 10.1111/j.1462-5822.2006.00879.x. [DOI] [PubMed] [Google Scholar]
- 39.Mundy R, MacDonald TT, Dougan G, Frankel G, Wiles S. Citrobacter rodentium of mice and man. Cell. Microbiol. 2005;7:1697–1706. doi: 10.1111/j.1462-5822.2005.00625.x. [DOI] [PubMed] [Google Scholar]
- 40.Rescher U, Gerke V. Annexins: unique membrane binding proteins with diverse functions. J. Cell Sci. 2004;117:2631–2639. doi: 10.1242/jcs.01245. [DOI] [PubMed] [Google Scholar]
- 41.Levine MM, Bergquist EJ, Nalin DR, Waterman DH, Hornick RB, Young CR, Sotman S. Escherichia coli strains that cause diarrhoea but do not produce heat-labile or heat-stable enterotoxins and are non-invasive. Lancet. 1978;311:1119–1122. doi: 10.1016/s0140-6736(78)90299-4. [DOI] [PubMed] [Google Scholar]
- 42.Berger CN, Crepin VF, Jepson MA, Arbeloa A, Frankel G. The mechanisms used by enteropathogenic Escherichia coli to control filopodia dynamics. Cell. Microbiol. 2008;11:309–322. doi: 10.1111/j.1462-5822.2008.01254.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.O’Brien AO, Lively TA, Chen ME, Rothman SW, Formal SB. Escherichia coli O157:H7 strains associated with haemorrhagic colitis in the United States produce a Shigella dysenteriae 1 (SHIGA) like cytotoxin. Lancet. 1983;321:702. doi: 10.1016/s0140-6736(83)91987-6. [DOI] [PubMed] [Google Scholar]
- 44.Ghaem-Maghami M, Simmons CP, Daniell S, Pizza M, Lewis D, Frankel G, Dougan G. Intimin-specific immune responses prevent bacterial colonization by the attaching-effacing pathogen Citrobacter rodentium. Infect. Immun. 2001;69:5597–5605. doi: 10.1128/IAI.69.9.5597-5605.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Munera D, Crepin VF, Marches O, Frankel G. N-terminal type III secretion signal of enteropathogenic Escherichia coli translocator proteins. J. Bacteriol. 2010;192:3534–3539. doi: 10.1128/JB.00046-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.