Abstract
Dysfunction of the intramembrane protease γ-secretase is thought to cause Alzheimer’s disease (AD), with most AD-derived mutations mapping to the catalytic subunit presenilin 1 (PS1). Here we report an atomic structure of human γ-secretase at 3.4 Å resolution, determined by single-particle cryo-electron microscopy. AD-derived mutations in PS1 affect residues at two hotspots, each located at the center of a distinct four transmembrane segment (TM) bundle. TM2, and to a lesser extent TM6, exhibit considerable flexibility, yielding a plastic active site and adaptable surrounding elements. The active site of PS1 is accessible from the convex side of the TM horseshoe, suggesting considerable conformational changes in nicastrin extracellular domain (ECD) following substrate recruitment. Aph-1 serves as a scaffold, anchoring the lone TM from nicastrin and supporting the flexible conformation of PS1. Ordered phospholipids stabilize the complex inside the membrane. Our structure serves as a molecular basis for mechanistic understanding of γ-secretase function.
A hallmark of Alzheimer’s disease (AD) is accumulation of β-amyloid plaque in patient brain1. The intramembrane protease γ-secretase is thought to contribute to AD development by generating β-amyloid peptides, particularly those that are prone to aggregation such as Aβ422-5. A mature γ-secretase consists of four components: presenilin, Pen-2, nicastrin, and Aph-16. Among these components, presenilin is responsible for the Aβ-producing proteolytic activity7,8.
Presenilin comprises 9 TMs, with the signature motifs YD on TM6 and GxGD on TM78. During assembly of γ-secretase, presenilin undergoes an autocatalytic cleavage to produce an N-terminal fragment (NTF, comprising TMs 1-6) and a C-terminal fragment (CTF, comprising TMs 7-9)9. Among the 300 or so mutations derived from patients with familial AD (FAD), more than two thirds are mapped to PS1, and about three dozen each are derived from PS2 and amyloid precursor protein (APP). Nicastrin contains a large ECD and a single TM; its ECD is heavily glycosylated and thought to recognize the N-terminus of substrate protein10-13. Pen-2 directly binds PS1 and is required for its autocatalytic maturation and protease activity14,15. Aph-1 contains 7 TMs and is indispensable for γ-secretase assembly16,17.
Our previous cryo-EM structure of human γ-secretase at 4.5 Å resolution led to identification of 19 TMs and construction of a partial atomic model for the ECD18. Analysis of the crystal structures of the archaeal presenilin homologue PSH19 and the nicastrin homologue from Dictyostelium purpureum (DpNCT)20 yielded a tantalizing clue on TM assignment and an improved atomic model for the ECD. A subsequent cryo-EM structure of human γ-secretase at 4.3 Å resolution allowed assignment of all 20 TMs to the four components21. In this study, we report the first atomic structure of an intact human γ-secretase, which allows visualization of the atomic structures of all four components and determination of the specific interactions that underlie γ-secretase assembly.
Structure determination of human γ-secretase
We performed cryo-EM single-particle analysis on the same sample in amphipols that was used to calculate our previous map18. With the aim of reaching higher resolution, we used zero-loss energy-filtered imaging, a higher magnification, and a lower dose rate on the single-electron counting detector (see Methods). We also collected a larger data set. An initial set of 412,272 particle images yielded a 3.5-Å map. Subsequent three-dimensional (3D) classification that was focused on the TMs resulted in a more structurally homogeneous subset of 159,549 particles. This subset was used to calculate the final map at 3.4 Å resolution (Extended Data Figs. 1 & 2). Our map displays excellent main-chain connectivity and side-chain densities for almost all residues (Extended Data Fig. 2a).
Twenty TMs were identified in the transmembrane region, including a highly mobile TM (TM2 of PS1) that is only visible as rod-shaped density in a 7 Å low-pass filtered map (Extended Data Fig. 2b). This low-pass filtered map also shows a second rod-shaped density in the PS1 cavity between TM3, TM5, and TM2, which we were unable to identify. As previously reported20,21, PS1 and Aph-1 are located at the center of the TM horseshoe. All seven TMs of Aph-1 have exceptional density, with most side chains clearly identifiable (Extended Data Fig. 2c). Except TM2 and TM6, all other TMs of PS1 exhibit excellent density, with aromatic and bulky residues easily recognizable (Extended Data Fig. 2d). The three TMs of Pen-2 on the thin end of the TM horseshoe and the lone TM of nicastrin on the thick end both display discernible side chain features (Extended Data Figs. 2e, f). A number of N-linked glycans and lipid molecules are also defined by clear EM density (Extended Data Figs. 2g, h).
On the basis of these unambiguous EM densities, we built and refined a near-complete atomic model for human γ-secretase (Fig. 1, Extended Data Table 1), which includes 598 residues in the TMs and 632 residues in the ECD. Assignment of specific residues in the TMs was aided by a large number of aromatic amino acids (Extended Data Fig. 2c-f). The density map for PS1-TM2 is inadequate for model building; nonetheless, we generated an atomic model for this TM based on its sequence homology to the archaeal homologue PSH19. This is the first structure of human γ-secretase at a near-atomic resolution (Fig. 1; Extended Data Fig. 3).
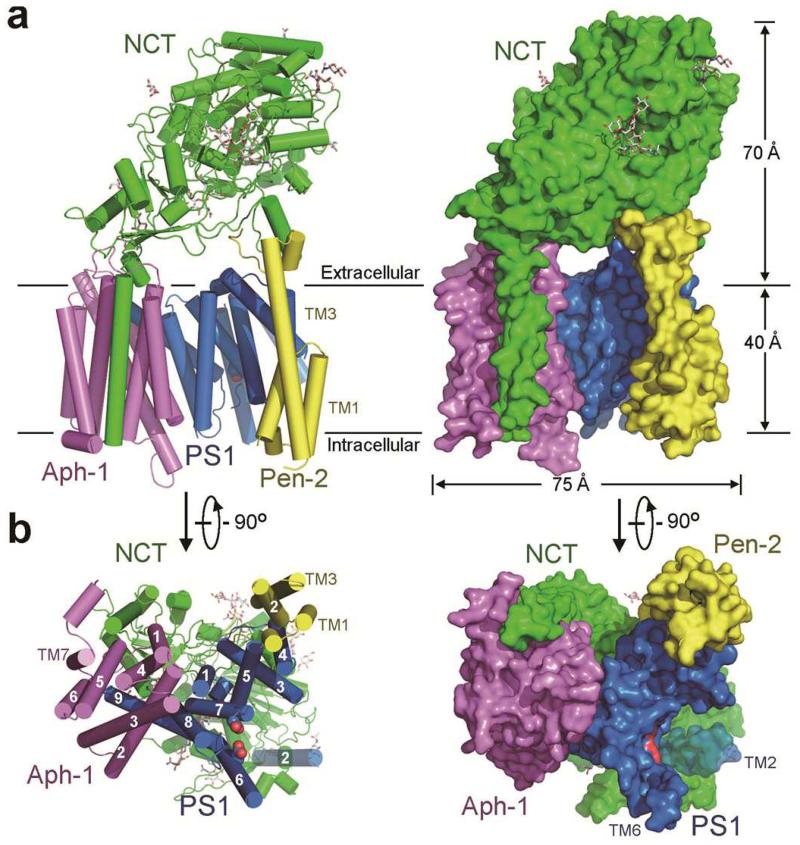
Figure 1. Atomic structure of human γ-secretase.
a, The γ-secretase structure is shown in cartoon representation (left panel) and surface view (right panel). Eleven N-linked glycans are displayed in stick. b, The γ-secretase structure is viewed perpendicular to the lipid membrane from the intracellular side. TM2 of PS1 is most flexible and shown in a semi-transparent fashion. The catalytic residues Asp257 and Asp385 are located on the convex side of the TM horseshoe. All structural figures were prepared using UCSF Chimera35 or PyMol36.
Nicastrin ECD, which constitutes the bulk of the extracellular region of human γ-secretase, contains 11 glycans and directly interacts with two ends of the TM horseshoe (Fig. 1a). The lone TM of nicastrin closely stacks against TM1/TM5/TM7 of Aph-1 through predominantly hydrophobic interactions at the thick end. TM2 and TM4 of Aph-1 associate with TM8 and TM9 of PS1 at the center of the TM horseshoe, with the C-terminal three residues Phe465-Tyr466-Ile467 of PS1 inserting into a greasy central cavity on the extracellular side of Aph-1 (Fig. 1a). The three TMs of Pen-2 bind to TM4 of PS1 at the thin end. Notably, this arrangement places the two catalytic residues, Asp257 and Asp385 of PS1, on the convex side of the TM horseshoe (Fig. 1b).
Atomic structure of presenilin
PS1 exhibits an extended organization, with empty spaces between a few adjacent TMs (Fig. 2a; Extended Data Fig. 4). The N-terminal 77 residues have no EM density and are presumably disordered, likely due to their intrinsic flexibility. Among the seven surface loops that connect neighboring TMs, four exhibit clear and contiguous density. The extended sequences between TM6 and TM7 harbor the site of autocatalytic cleavage; but these sequences are hydrophilic and mostly disordered (Fig. 2b). The nine TMs exhibit a large variation in length, with TM9 containing 30 residues and TM7 only 18.
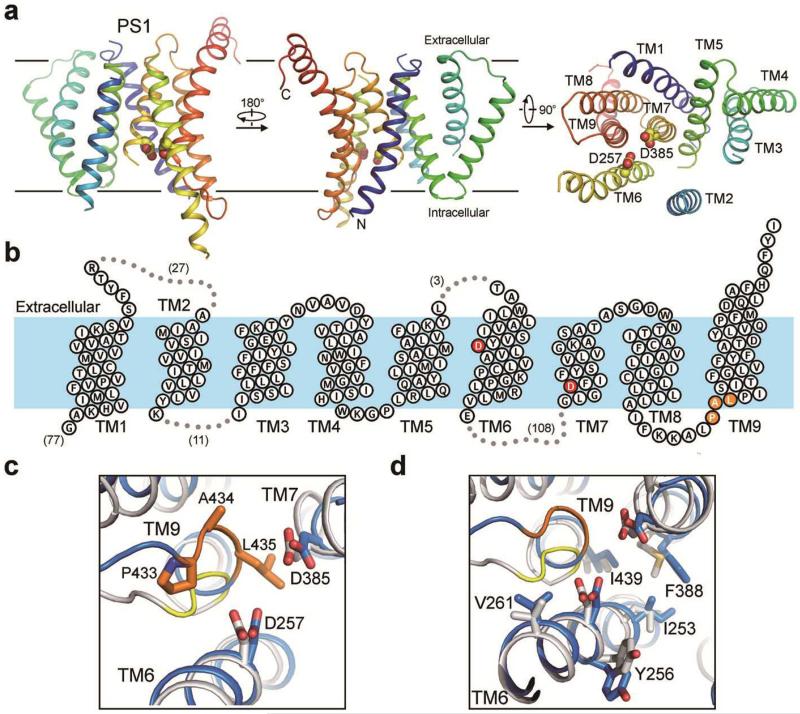
Figure 2. Atomic structure of PS1.
a, PS1 has a loosely organized structure and exhibits considerable flexibility. The cartoon representation of PS1 is rainbow-colored. TM2 is visible only at low resolutions, and the density map contains no features for side chains. Nonetheless, an atomic model for TM2 was built based on sequence and structural homology between PS1 and PSH19,21. b, A membrane topology diagram of PS1. The two catalytic aspartate residues are colored red. c, The two catalytic aspartate residues of PS1 are in nearly perfect registry with those in PSH19. The PAL sequence motif implicated in substrate recognition is shown. d, PS1 and PSH share similar features at their active sites.
Among the two catalytic residues, Asp257 is located in the middle of TM6 slightly to the extracellular side, whereas Asp385 maps to the cytoplasmic side of TM7 (Fig. 2b). The distance between the Cα atoms of Asp257 and Asp385 measures approximately 10.6 Å (Fig. 2c), considerably longer than that in an activated aspartate protease such as pepsin22. Importantly, however, these catalytic residues are placed next to the PAL signature motif on TM9 that is thought to play a role in substrate recognition23-25 (Fig. 2c). We speculate that substrate binding may trigger alignment of these two aspartate residues and consequent catalysis.
Despite a relatively low sequence identity of 19 percent between PS1 and the archaeal intramembrane protease PSH, their overall structures are similar to each other19,21. In particular, the catalytic residues are in nearly perfect registry between PS1 and PSH (Fig. 2c). The amino acids that surround the catalytic residues, including the PAL motif, are also highly conserved. Relative to Asp257, three residues of PS1 (Ile253, Tyr256, and Val261) are located on the same side of TM6; these residues are replaced by Leu158, Tyr161, and Val166, respectively, in PSH (Fig. 2d). Similarly, Gly382 and Phe388 of PS1 correspond to Gly219 and Met223 of PSH, respectively. Ile439 on TM9 of PS1 and Ile283 of PSH nearly coincide with each other (Fig. 2d). The observed structural conservation may underlie the finding that PSH exhibits similar cleavage preferences towards APP C99 as human γ-secretase26.
Mutational hotspots in PS1 Structure
The PS1 structure at a near-atomic resolution allows mapping and analysis of disease-derived mutations in PS1. On the basis of public information (http://www.alzforum.org/mutations), the 212 AD-derived missense mutations in PS1 affect 135 amino acids, of which 53 are targeted for two or more mutations. Presumably these residues affect the specific function of PS1 that is directly related to AD development. Among these 53 residues, 35 are identified in the TMs of our current PS1 structure, and the rest are located on surface elements that have no EM density. Notably, there are 8 such residues in TM5, 7 in TM6, but 0 in TM1. Together, these 35 amino acids account for a total of 101 AD-derived missense mutations.
Analysis of these 35 residues on the structure of PS1 led to identification of two mutational hotspots, each located at the inner core of a structural repeat (Fig. 3a). The first hotspot involves the inner core of TMs 2-5 (Fig. 3b). Among the 20 affected residues in these TMs, 18 have their side chains facing the inner core of TMs 2-5. Consequently, only one side of each TM helix is affected. For example, Leu219, Glu222, Leu226, Ser230, Met233, and Phe237 are placed on the same side of TM5 (Fig. 3b). Similarly, the second hotspot is located at the inner core of TMs 6-9, in the vicinity of the catalytic residues Asp257 and Asp385 (Fig. 3c).
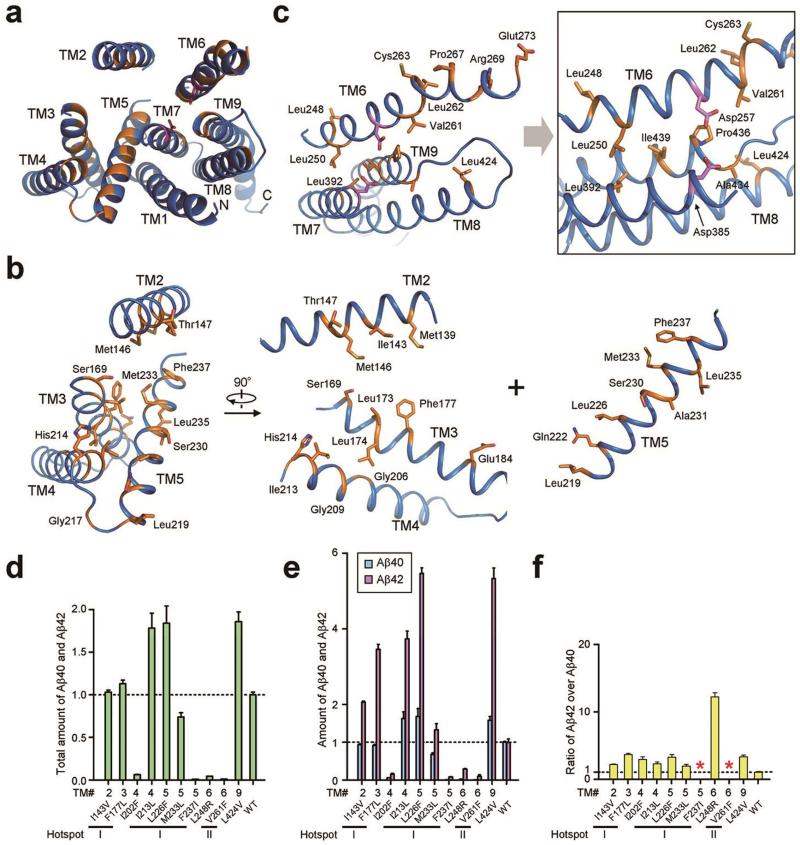
Figure 3. AD-derived mutations map to two hotspots in PS1.
a, An overall view of the PS1 residues targeted for mutations in AD patients. PS1 is viewed from the extracellular side. Mutated residues are colored orange. b, Close-up views of the mutation-targeted residues in TMs 2-5. The vast majority of these residues map to the center of this four-TM bundle. c, Close-up views of the mutation-targeted residues in TMs 6-9. The two catalytic residues Asp257 and Asp385 are shown. d, FAD-derived mutations in PS1 have varying degrees of effect on the combined Aβ40 and Aβ42 cleavage activity of γ-secretase. Shown here are results of 10 such γ-secretase mutants, each containing a specific mutation derived from FAD. The activity of WT γ-secretase is normalized to 1.0. e, FAD-derived mutations either suppressed the production of Aβ40 more than Aβ42 or increased the production of Aβ40 less than Aβ42. f, All but two FAD-derived mutations led to increased Aβ42/Aβ40 ratios. Two mutations F237I and V261F in PS1 abrogated Aβ40 cleavage altogether, disallowing calculation of the Aβ42/Aβ40 ratio. Each experiment was independently repeated three times. All error bars represent standard deviations.
To examine the functional consequence of disease-derived mutations, we generated 10 γ-secretase variants, each containing a distinct missense mutation in PS1. Among these mutations, seven map to the two mutational hotspots. These γ-secretase variants were individually purified to homogeneity and examined for their protease activities towards the APP C99 substrate in an in vitro cleavage assay. Compared to the wild-type (WT) γ-secretase, four mutations (I202F, F237I, L248R, and V261F) lead to severely compromised protease activity (Fig. 3d). Of these four mutations, F237I and V261F failed to generate any detectable amount of Aβ40 (Fig. 3e), disallowing calculation of the Aβ42/Aβ40 ratio (Fig. 3f). In contrast, three mutations (I143V, F177L, and M233L) had relatively little impact on the total cleavage activity of γ-secretase (Fig. 3d), whereas the remaining three mutations (I213L, L226F, and L424V) actually increased the total cleavage activity (Fig. 3d). These observations strongly suggest a disconnection between the total protease activity of γ-secretase and the development of FAD (where these mutations were isolated). All eight mutations for which the Aβ42/Aβ40 ratio can be calculated led to increased Aβ42/Aβ40 ratios compared to WT γ-secretase (Fig. 3f). The generally increased ratios of Aβ42 over Aβ40 may suggest a causal relationship for development of FAD, but could also be explained by other possibilities.
Atomic structure of nicastrin
Similar to DpNCT20, human nicastrin also contains a large lobe, a small lobe, and a lone TM (Fig. 4a; Extended Data Fig. 5). These two nicastrin structures can be aligned to each other with a root-mean-squared deviation (RMSD) of 2.2 Å. Of the 230-kDa molecular mass in mature γ-secretase, up to 70-kDa can be attributed to glycosylation of nicastrin27. Among the 16 predicted N-linked glycosylation sites in nicastrin, at least 11 are glycosylated as judged by the EM density. Because the extended glycans are flexible in solution, only a small portion close to each Asn residue was modeled (Fig. 4a).
Figure 4. Structural features of nicastrin.
a, Two perpendicular views of nicastrin. The Lid from the small lobe is highlighted in red. Surface glycans are shown. b, The Lid hovers above a hydrophilic pocket in the large lobe. Two large glycans on Asn55 and Asn435 sandwich the Lid and interact with surrounding residues. c, Glu333 and Tyr337 are surrounded by a number of charged and polar residues in the pocket. These structural features are consistent with the pocket being a binding site for substrate protein. d, Trp164 from the Lid makes van der Waals contacts to Pro424, Phe448, and the aliphatic side chain of Gln420. e, Phe287 from the large lobe may serve as the hydrophobic pivot. Phe287 interacts with four hydrophobic residues from the small lobe.
Nicastrin is thought to rely on Glu333 and Tyr337 for substrate recruitment10-13. In the structure of human nicastrin, Glu333 and Tyr337 are buried in a hydrophilic pocket that is covered by an extended surface loop known as the Lid20 (Fig. 4b). The Lid, containing five aromatic residues, is sandwiched by two prominent glycans on Asn55 and Asn435 (Fig. 4b). A number of charged and polar amino acids are located in the pocket, including four arginine residues Arg281, Arg285, Arg429, and Arg432 (Fig. 4c). With the potential to mediate specific interactions such as hydrogen bonds, buried charged and/or polar residues often serve a functional role. These structural features support the notion that this pocket is responsible for substrate recruitment, with these residues directly contributing to recognition.
The closed conformation of the Lid is sustained by specific interactions between residues from the Lid and residues from surrounding structural elements. For example, the indole ring of Trp164 makes a number of van der Waals contacts to the side chains of Pro424 and Phe448 and the aliphatic portion of Gln420 (Fig. 4d). Substrate binding requires opening of the Lid and disruption of these interactions. Because the Lid comes from the small lobe, a rotation of the large lobe around a central pivot – Phe287 – is proposed to be both necessary and sufficient for Lid opening20. Such a rotation around Phe287 would be greatly facilitated by hydrophobic interactions, which are more adaptive to conformational changes than hydrogen bonds. Consistent with this analysis, Phe287 is nestled in a greasy pocket formed by Phe103, Leu171, Phe176, and Ile180 from the small lobe (Fig. 4e).
Inter-component interactions
Assembly of the four components into a functional human γ-secretase involves specific interactions mainly in the membrane-spanning region, resulting in the burial of approximately 3,794 Å2 otherwise membrane-exposed surface areas (Fig. 5a). Binding of the ECD onto the TMs of γ-secretase involves an additional buried surface area of 1,320 Å2. The intra-membrane interactions comprise predominantly van der Waals contacts among hydrophobic amino acids, exemplified by the insertion of three residues at the C-terminus of PS1 into a greasy pocket formed by TM2/3/4/6/7 of Aph-1 (Fig. 5b). Phe465 from PS1 contacts Leu72 and Phe205 in Aph-1, whereas Tyr466 from PS1 stacks against Leu163 and Ile167 in Aph-1. The aliphatic side chain of Ile467 from PS1 interacts with Leu46, Leu86, and Phe125 at the bottom of the cavity in Aph-1. The C-terminus of PS1 is in close proximity of the highly conserved His171 and His197 in Aph-1, which were found to be important for Aph-1 binding to γ-secretase28. Interestingly, a peptide comprising the C-terminal 8 residues of PS1 was shown to inhibit Aβ production29, presumably by weakening or disrupting normal assembly of γ-secretase.
Figure 5. Assembly interfaces among the four components of γ-secretase in the transmembrane region.
a, An overall view of the packing interfaces in the transmembrane region. The three boxed interfaces are detailed in panels b-d. b, The C-terminal three residues Phe465-Tyr466-Ile467 of PS1 insert into a cavity formed by TMs in Aph-1. c, Nicastrin interacts with Pen-2 through van der Waals contacts on a flat interface. d, Pen-2 binds to PS1 through mostly van der Waals contacts. In particular, Phe94 of Pen-2 is nestled in the greasy pocket of PS1, formed by Phe179, Leu182, Phe186, Val193, Tyr195, and Va198. e, Two phospholipids appear to stabilize the inter-component interfaces in γ-secretase. One lipid is bound at the interface between PS1 and Aph-1, whereas the other is intercalated between the lone TM of nicastrin and TMs 1/4/5/7 of Aph-1 (left panel). The aliphatic tails of the latter phospholipid may interact with a number of hydrophobic residues whereas its phosphate group likely hydrogen bonds to Arg115 and Gln116 (right panel).
On the thin end of the TM horseshoe, an α-helix and its preceding and ensuing loops in nicastrin stack against an extended loop at the carboxyl-terminal end of Pen-2 through multiple van der Waals contacts (Fig. 5c). At the nearby interface between PS1 and Pen-2, Phe94 from Pen-2 is nestled in a hydrophobic pocket formed by six residues in PS1 (Fig. 5d). Hydrophobic residues from TM3 of Pen-2, exemplified by Leu71, Ile75, and Phe78, also interact with residues in PS1. Notably, although Pen-2 has only 3 TMs and interacts with both PS1 and nicastrin, it actually contains three hydrophobic structural cores (Extended Data Fig. 6).
At 3.4-Å resolution, at least two phospholipid molecules were identified, each with two aliphatic tails linked to a small head group (Extended Data Fig. 2h). One lipid binds to the interface between PS1 and Aph-1, making close contacts with residues in TM1/TM8 of PS1 and TM4 of Aph-1 (Fig. 5e). The other lipid molecule is intercalated into the interface between Aph-1 and the lone TM of nicastrin (Fig. 5e, right panel). The aliphatic tails of this phospholipid interact with a number of surrounding hydrophobic residues, whereas the phosphate group may directly hydrogen bond to the side chains of Arg115 and Gln116 from Aph-1. This lipid likely stabilizes the nicastrin-Aph-1 interface within the membrane.
To corroborate the observed inter-component interfaces, we generated eight mutant γ-secretase complexes, each containing two specific cysteine mutations on two neighboring components to facilitate formation of designed disulfide bonds. Three such γ-secretase mutants target the interface between Aph-1 and nicastrin (Extended Data Fig. 7a). The distance between the Cα atoms of Val147 on Aph-1 and Ile40 on nicastrin is 4.1 Å, ideal for formation of a disulfide bond if these two residues were replaced by cysteine. Indeed, in the absence of the reducing agent DTT, Aph-1 V147C was crosslinked to nicastrin I40C within the mutant γ-secretase, resulting in a high molecular weight complex on denaturing SDS-PAGE (Extended Data Fig. 7a, lane 1). This crosslinked complex disappeared in the presence of DTT (lane 2) and was absent for the WT γ-secretase regardless of DTT (lanes 7-8). Similarly, two additional γ-secretase mutants, one containing Aph-1 V146C and nicastrin A664C and the other containing Aph-1 A4C and nicastrin L673C, allowed specific crosslinking only in the absence of DTT (lanes 3-6). The same strategy was successfully applied to verify the specific interactions at the interfaces between PS1 and Pen-2 (Extended Data Fig. 7b), between Pen-2 and nicastrin (Extended Data Fig. 7c), and between Aph-1 and PS1 (Extended Data Fig. 7d).
Discussion
In this manuscript, we report the cryo-EM structure of human γ-secretase at an overall resolution of 3.4 Å. The qualitative improvement in resolution over earlier studies allowed us to derive an atomic model for all four components of γ-secretase. Except nicastrin18,20, such atomic models are being reported for the first time.
The available structural evidence supports the notion that Glu333 and Tyr337 of nicastrin, both buried in a hydrophilic pocket, may play a key role in substrate recruitment12. Under this scenario, displacement of the Lid, caused by a movement of the large lobe relative to the small lobe, is required prior to substrate binding20. This proposal is supported by the unique pattern of contacts between the large and small lobes and by the conversed interactions around the central pivot Phe287 in nicastrin (Fig. 4). In substrate-free γ-secretase, the Lid is positioned right above the concave side of the TM horseshoe whereas the active site is located on the convex side, resulting in a relatively long distance between the putative binding pocket for the N-terminus of substrate and the site of cleavage by PS1 (Extended Data Fig. 8). This distance may shorten, however, in response to conformational changes within γ-secretase. In particular, rotation of the large lobe relative to the small lobe, induced by substrate binding, may also reorient the substrate for cleavage, perhaps by aligning the pocket in nicastrin and the active site in PS1. Dynamic conformations of γ-secretase have been observed30, particularly in the ECD region31.
TM6 of PS1, which harbors the catalytic residue Asp257, exhibits relatively poor EM density, whereas TM2 is visible only at low resolutions. These structural observations are consistent with the notion that binding by inhibitors or modulators may induce pronounced conformational changes30-32. In particular, substrate binding may trigger a conformational change, rendering the active site suitable for catalysis32. The plasticity observed in our structure may expedite the conformational changes that are needed to bring the two catalytic aspartates within hydrogen bonding distance of each other, and may play a role in the relaxed substrate specificity of the complex. Activation of the active site also depends on the binding of Pen-2, which was observed to have an allosteric effect on TM633.
The prevailing β-amyloid hypothesis suggests an increased ratio of Aβ42 over Aβ40 as the major culprit for the accumulation of β-amyloid plaque and consequent development of AD2,3. Our finding that FAD-derived mutations have varying degrees of effect on the protease activity of γ-secretase argues against a causal relationship between the total protease activity of γ-secretase and the development of FAD. Our finding is consistent with the poor clinical performance of γ-secretase-inhibiting drugs34. Notably, the generally increased ratios of Aβ42 over Aβ40 only suggest, but fail to prove, a direct causal relationship for the development of FAD. For example, the assumption that γ-secretase has evolved to optimize production of Aβ40 over all other peptides would naturally lead to the prediction that any mutation, disease-causing or not, will increase the Aβ42/Aβ40 ratio.
Intriguingly, the AD-causing mutations map to two hotspots, each located at the inner core of a four-TM bundle (Fig. 3). The mutations on TMs 6-9 affect residues in the immediate neighborhood of the catalytic residues Asp257 and Asp385 and thus may cripple the protease activity of γ-secretase. The biochemical data seem to support this analysis (Fig. 3d). However, the mutations on TMs 2-5 defy such rationale – they either abrogate or increase the protease activity of γ-secretase. The generally inward-facing nature of the mutation-targeted residues on TMs 2-5 (Fig. 3b) may suggest a transport function of some sort, or a binding site for another transmembrane protein. These tantalizing clues await experimental examination.
Our structure represents a milestone for the rapidly emerging technique of high-resolution cryo-EM structure determination. Because the signal-to-noise ratio in cryo-EM images correlates with the size of the particles, determining the relative orientations of individual particles becomes a limitation for small complexes. With a protein mass of ~170 kDa, γ-secretase is much smaller than any other atomic-resolution cryo-EM structure determined to date, while its lack of symmetry further complicates structure determination. Whereas glycosylation adds another 30-70 kDa of mass to the complex, only a small proportion of the sugar moieties are ordered and their effect on alignment is limited. Although glycosylation may inhibit crystal growth, it did not seem to hamper cryo-EM structure determination. On the contrary, glycosylation may stabilize the protein, possibly by protecting the protein from the denaturing air-water interface of the thin cryo-EM sample.
Method
Protein preparation
To improve protein production, we co-expressed the four components of γ-secretase, each in its own pMLink plasmid18. This strategy allowed both convenient manipulation of individual components and optimization of protein expression by altering the relative molar ratios of the four plasmids. Specific mutations in PS1, Pen-2, Aph-1, or nicastrin were generated only in the corresponding pMLink vector using standard PCR-based approach. Pen-2 has an amino-terminal FLAG tag, Aph-1 contains a carboxyl-terminal HA tag, and nicastrin is tagged with a V5-His6 sequence at the carboxyl terminus. The ten γ-secretase variants, each carrying a FAD-derived mutation on PS1, and the eight γ-secretase complexes, each carrying two introduced cysteine mutations, were overexpressed similarly as the WT γ-secretase18. Purification of both WT and mutant human γ-secretases followed published protocol18.
Electron microscopy
Aliquots of 3 μl of purified γ-secretase in amphipols at a concentration of ~4 μM were placed on glow-discharged holey carbon grids (Quantifoil Au R1.2/1.3, 300 mesh), and flash frozen in liquid ethane using an FEI Vitrobot. Zero-energy-loss images were recorded manually on an FEI Titan Krios electron microscope at 300 kV, using a slit width of 20 eV on a GIF-Quantum energy filter. A Gatan K2-Summit detector was used in super-resolution counting mode at a calibrated magnification of 35,714× (yielding a pixel size of 1.4 Å), and a dose rate of ~2.5 electrons/Å2/s (~5 electrons/pixel/s). Exposures of 16 seconds were dose-fractionated into 20 movie frames. Defocus values in the final data set ranged from 0.7-3.2 μm.
Image processing
We used MOTIONCORR37 for whole-frame motion correction, CTFFIND338 for estimation of the contrast transfer function parameters, and RELION-1.339 for all other image processing steps. Templates for reference-based particle picking were obtained from 2D class averages that were calculated from a manually picked subset of the micrographs. Using low-pass filtered templates to 20 Å to limit reference bias, 1.8 million particles were picked automatically from a total of 2,925 micrographs. Because the picking procedure is prone to false positives40, we used reference-free 2D class averaging and an initial run of 3D classification to select 412,272 particles for a first 3D refinement. After per-particle motion correction and radiation-damage weighting (particle polishing41), these particles gave a reconstruction with a resolution of 3.5 Å. In a subsequent 3D classification, we applied a mask around the transmembrane domain and did not perform any alignments. This yielded a subset of 159,549 particles, for which a reconstruction with improved density in the trans-membrane domain was obtained. The resolution of this final map was 3.4 Å. Reported resolutions are based on the gold-standard FSC=0.143 criterion42, and FSC curves were corrected for the effects of a soft mask on the FSC curve using high-resolution noise substitution42. All 3D classifications and refinements were started from a 40 Å low-pass filtered initial model, the first of which was made from our previous 4.5 Å map. Prior to visualization, all density maps were corrected for the modulation transfer function (MTF) of the detector, and then sharpened by applying a negative B-factor that was estimated using automated procedures43. Local resolution variations were estimated using ResMap44.
Atomic modelling
The first atomic model was built from an intermediate, 3.9-Å EM map which was of sufficient quality to allow side chain assignment for all four components of γ-secretase. The initial models used for nicastrin ECD and presenilin 1 was generated from the crystal structures of DpNCT20 (PDB ID: 4R12) and PSH19 (PDB ID: 4HYG) by CHAINSAW45. The two generated structures were first refined in real-space by PHENIX46 with secondary structure and geometry restraints. Aph-1 and Pen-2 were built de novo from a poly-Ala model. Then the entire atomic model was manually improved in COOT47. Sequence assignment was guided mainly by bulky residues such as Phe, Tyr, Trp and Arg. Unique patterns of sequences were exploited for validation of residue assignment. Eleven glycosylation sites with obvious sugar densities and five pairs of disulfide bond in nicastrin ECD domain also facilitated sequence assignment. This initial model was largely confirmed and further improved by the 3.4-Å resolution EM map, with minor adjustment of a few residues and identification of two additional glycosylation sites and two lipid molecules. This model was refined in real-space by PHENIX46.
An independent effort of atomic model building used stereochemical refinement tools in Coot47 and REFMAC48 that were originally designed for X-ray crystallography, but later adapted for cryo-EM49. The structures of DpNCT20 (PDB ID: 4R12) and PSH19 (PDB ID: 4HYC) were used as a guide, while Aph-1 and Pen-2 were built de novo in Coot47, starting from idealized α-helices. This model was refined in REFMAC48, with secondary structure restraints generated by ProSMART50. Superposition of the two independently built models revealed that both models were in excellent agreement. Refinement of the final, consensus model was performed in REFMAC, and overfitting of the atomic model was monitored by refining the model in one of the two independent maps from the gold-standard refinement approach, and testing this model against the other map51. At no point in our procedures did we use the atomic model to modify the cryo-EM map.
γ-secretase activity assay
The assay was performed as described in the AlphaLISA Kit (PerkinElmer). Briefly, 2 μl reaction products were incubated with 8 μl AlphaLISA Aβ1-40/42 Acceptor beads at 23°C for 1 hour. After another 30-minute incubation with 10 μl AlphaLISA Aβ1-40/42 Donor beads in the dark at 23°C, the samples were read using Envision-Alpha Reader (PerkinElmer). The readings were expressed in arbitrary unit.
Crosslinking assay
For the crosslinking assay, copper sulfate (CuSO4) was added to the purified protein at a final concentration of 1 mM, and the sample was incubated at 4 °C for 1 hour. Then, the protein was applied to gel filtration (Superose-6, GE Healthcare). The peak fractions were divided into two aliquots. One aliquot was treated with the reducing agent, DL-dithiothreitol (DTT), at a final concentration of 10 mM and incubated at room temperature for 30 minutes. Samples from the two aliquots, treated with or without DTT, were analyzed by Western blotting using monoclonal antibodies against the HA tag on Aph-1 or FLAG tag on Pen-2. The antibodies were purchased from Beijing ComWin Biotech.
Extended Data
Extended Data Figure 1 . Cryo-EM, single-particle analysis of human γ-secretase.
a, Representative raw particles from an original micrograph. b, Representative reference-free 2-D class averages of the γ-secretase particles. Two classes identified by a red rectangle box (lower right corner) may contain some density for the extended cytosolic loop sequences between TM6 and TM7 of PS1, which are disordered in the final maps. c, Resolution estimation of the EM structure. The overall resolution is calculated to be 3.4 Å on the basis of gold-standard FSC curve42. d, Color-coded resolution variations in the γ-secretase structure as estimated by ResMap44. e, FSC curves of the final, Refmac-refined model versus the map it was refined against (in black); of a model refined in the first of the two independent maps used for the gold-standard FSC versus that same map (in red); and of a model refined in the first of the two independent maps versus the second independent map (in green). The small difference between the red and green curves indicates that the refinement of the atomic coordinates did not suffer from severe overfitting.
Extended Data Figure 2 . An atomic model of human γ-secretase.
a, The γ-secretase structure is viewed parallel to the lipid membrane. Shown here is EM density for the entire γ-secretase complex. EM density is colored blue for PS1, yellow for Pen-2, magenta for Aph-1, and green for nicastrin. b, The density map for TM2 of PS1. Among the 20 TMs, TM2 of PS1 shows the highest degree of flexibility and only becomes visible at as rod-shaped density in a 7 Å low-pass filtered map. At this resolution, another rod-shaped density is visible next to TM2 and remains unaccounted for. c, EM density map and the atomic model are shown for all 7 TMs of Aph-1. 2-3 bulky residues are indicated for each TM. d, EM density map and the atomic model are shown for 7 TMs of hPS1. TM6 exhibits relatively poor EM density, likely due to its intrinsic flexibility. e, EM density map and the atomic model are shown for the 3 TMs of Pen-2. f, EM density map and the atomic model are shown for the only TM and select regions of nicastrin. g, EM density map and the atomic model for three representative glycans are shown. h, EM density map and putative assignment are shown for two lipid molecules.
Extended Data Figure 3 . Overall structure of human γ-secretase.
a, Structure of human γ-secretase is shown in cartoon representation (top panels) and surface view (bottom panels) in four successively perpendicular views. The γ-secretase structure is viewed parallel to the lipid membrane. The coloring scheme is the same as in Fig. 1. Two lipid molecules are shown. Eleven glycosylated Asn residues and their glycans are displayed in stick. b, The γ-secretase structure is represented by electrostatic surface potential.
Extended Data Figure 4 . Electrostatic surface potential of PS1.
PS1 exhibits a loosely folded structure, with a number of large cavities and empty spaces between adjacent TMs.
Extended Data Figure 5 . Structural comparison between human nicastrin and nicastrin from Dictyostelium purpureum (DpNCT).
Individual structures of human nicastrin and DpNCT20 are shown in the left and middle panels, respectively. The overlay is shown in the right panel, with an RMSD of 2.2 Å. Two perpendicular views for each structure are displayed here.
Extended Data Figure 6 . Pen-2 contains three small hydrophobic cores in its three TMs.
Unlike previous prediction14,52, Pen-2 contains three, not two, TMs. Pen-2 contains a small hydrophobic core in the extracellular side and two in the transmembrane region. These three regions are boxed and shown in close-up views.
Extended Data Figure 7 . Results of crosslinking experiments corroborate the atomic model of γ-secretase.
a, Crosslinking results for the interface between Aph-1 and nicastrin (NCT). Three mutant γ-secretase complexes were examined: Aph-1-V147C/NCT-I40C, Aph-1-V146C/NCT-A664C, and Aph-1-A4C/NCT-L673C. Shown in the upper panel is a SDS-PAGE gel blotted by a monoclonal antibody against the HA tag on Aph-1. Only in the absence of DTT, crosslinking led to high molecular weight complexes for the mutant γ-secretase, but not for the WT γ-secretase. The two bands likely represent Aph-1 crosslinked to mature NCT (mNCT) and immature NCT (iNCT). The structural basis is shown in the lower panel. The distances between the Cα atoms of the two residues targeted for cysteine mutation range 4.1-6.1 Å, which would facilitate convenient crosslinking reactions. b, Crosslinking results for the interface between PS1 and Pen-2. The mutant γ-secretase contains Pen-2-P97C and PS1-N190C. Shown in the upper panel is a SDS-PAGE gel blotted by a monoclonal antibody against the FLAG tag on Pen-2. c, Crosslinking results for the interface between Pen-2 and nicastrin. Two γ-secretase mutants were examined: Pen-2-T100C/NCT-V224C and Pen-2-L98C/NCT-H222C. d, Crosslinking results for the interface between Aph-1 and PS1. Two γ-secretase mutants were examined: Aph-1-T204C/PS1-F465C and Aph-1-A76C/PS1-I467C.
Extended Data Figure 8 . Implication on substrate access to γ-secretase.
Structure of γ-secretase is displayed in three relevant views. The middle panel shows the overall structure, with key features labeled. The left panel displays electrostatic surface potential from the convex side of γ-secretase. The right panel suggests a putative path for substrate access to the active site of γ-secretase.
Extended Data Table 1 . Refinement and model statistics.
| Data Collection | |
| Particles | 159,549 |
| Pixel size (Å) | 1.4 |
| Defocus range (μm) | 0.7-3.2 |
| Voltage (kV) | 300 |
| Electron dose (e-Å−2) | 40 |
| Model composition | |
| Non-hydrogen atoms | 10,016 |
| Protein residues | 1,251 |
| Phospholipids (PC) | 2 |
| Sugars | 15 |
| Refinement | |
| Resolution (Å) | 3.4 |
| Map sharpening B-factor (Å2) | −109 |
| Fourier Shell Correlation | 0.8503 |
| R factor | 0.3494 |
| Rms deviations | |
| Bonds (Å) | 0.0077 |
| Angles (°) | 1.4224 |
| Validation (proteins) | |
| Molprobity score | 2.65 |
| Clashscore, all atoms | 8.22 |
| Good rotamers (%) | 93.5% |
| Ramachandran plot | |
| Favored (%) | 88.5 |
| Allowed (%) | 8.6 |
| Outliers (%) | 2.9 |
Acknowledgments
We thank Shaoxia Chen and Christos Savva for support with electron microscopy and Jake Grimmett and Toby Darling for support with high-performance computing. This work was supported by funds from the Ministry of Science and Technology (2014ZX09507003006 to YS), the National Natural Science Foundation of China (31130002 and 31321062 to YS), a European Union Marie Curie Fellowship (to XCB), and the UK Medical Research Council (MC_UP_A025_1013, to SHWS).
Footnotes
Author information
The atomic coordinates have been deposited in the Protein Data Bank with the accession code 5A63, and the EM maps have been deposited in EMDB with the accession code EMD-3061.
The authors declare no competing financial interests.
References
- 1.Alzheimer A. About a peculiar disease of the cerebral cortex. Centralblatt für Nervenheilkunde Psychiatrie. 1907;30:177–179. [Google Scholar]
- 2.Hardy JA, Higgins GA. Alzheimer’s disease: the amyloid cascade hypothesis. Science. 1992;256:184–5. doi: 10.1126/science.1566067. [DOI] [PubMed] [Google Scholar]
- 3.De Strooper B, Iwatsubo T, Wolfe MS. Presenilins and gamma-Secretase: Structure, Function, and Role in Alzheimer Disease. Cold Spring Harbor perspectives in medicine. 2012;2:a006304. doi: 10.1101/cshperspect.a006304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Goate A, Hardy J. Twenty years of Alzheimer’s disease-causing mutations. J Neurochem. 2012;120(Suppl 1):3–8. doi: 10.1111/j.1471-4159.2011.07575.x. [DOI] [PubMed] [Google Scholar]
- 5.Tanzi RE, Bertram L. Twenty years of the Alzheimer’s disease amyloid hypothesis: a genetic perspective. Cell. 2005;120:545–55. doi: 10.1016/j.cell.2005.02.008. [DOI] [PubMed] [Google Scholar]
- 6.Kimberly WT, et al. Gamma-secretase is a membrane protein complex comprised of presenilin, nicastrin, Aph-1, and Pen-2. Proc Natl Acad Sci U S A. 2003;100:6382–7. doi: 10.1073/pnas.1037392100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.De Strooper B, et al. Deficiency of presenilin-1 inhibits the normal cleavage of amyloid precursor protein. Nature. 1998;391:387–90. doi: 10.1038/34910. [DOI] [PubMed] [Google Scholar]
- 8.Wolfe MS, et al. Two transmembrane aspartates in presenilin-1 required for presenilin endoproteolysis and gamma-secretase activity. Nature. 1999;398:513–7. doi: 10.1038/19077. [DOI] [PubMed] [Google Scholar]
- 9.Thinakaran G, et al. Endoproteolysis of presenilin 1 and accumulation of processed derivatives in vivo. Neuron. 1996;17:181–90. doi: 10.1016/s0896-6273(00)80291-3. [DOI] [PubMed] [Google Scholar]
- 10.Yu G, et al. Nicastrin modulates presenilin-mediated notch/glp-1 signal transduction and betaAPP processing. Nature. 2000;407:48–54. doi: 10.1038/35024009. [DOI] [PubMed] [Google Scholar]
- 11.Shah S, et al. Nicastrin functions as a gamma-secretase-substrate receptor. Cell. 2005;122:435–47. doi: 10.1016/j.cell.2005.05.022. [DOI] [PubMed] [Google Scholar]
- 12.Dries DR, et al. Glu-333 of nicastrin directly participates in gamma-secretase activity. J Biol Chem. 2009;284:29714–24. doi: 10.1074/jbc.M109.038737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Goo JS, et al. Nicastrin overexpression in transgenic mice induces aberrant behavior and APP processing. Mol Neurobiol. 2013;48:232–43. doi: 10.1007/s12035-013-8453-3. [DOI] [PubMed] [Google Scholar]
- 14.Francis R, et al. aph-1 and pen-2 are required for Notch pathway signaling, gamma-secretase cleavage of betaAPP, and presenilin protein accumulation. Dev Cell. 2002;3:85–97. doi: 10.1016/s1534-5807(02)00189-2. [DOI] [PubMed] [Google Scholar]
- 15.Takasugi N, et al. The role of presenilin cofactors in the gamma-secretase complex. Nature. 2003;422:438–441. doi: 10.1038/nature01506. [DOI] [PubMed] [Google Scholar]
- 16.Goutte C, Tsunozaki M, Hale VA, Priess JR. APH-1 is a multipass membrane protein essential for the Notch signaling pathway in Caenorhabditis elegans embryos. Proc Natl Acad Sci U S A. 2002;99:775–9. doi: 10.1073/pnas.022523499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gu Y, et al. APH-1 interacts with mature and immature forms of presenilins and nicastrin and may play a role in maturation of presenilin.nicastrin complexes. J Biol Chem. 2003;278:7374–80. doi: 10.1074/jbc.M209499200. [DOI] [PubMed] [Google Scholar]
- 18.Lu P, et al. Three-dimensional structure of human gamma-secretase. Nature. 2014;512:166–170. doi: 10.1038/nature13567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li X, et al. Structure of a presenilin family intramembrane aspartate protease. Nature. 2013;493:56–61. doi: 10.1038/nature11801. [DOI] [PubMed] [Google Scholar]
- 20.Xie T, et al. Crystal structure of the gamma-secretase component nicastrin. Proc Natl Acad Sci U S A. 2014;111:13349–54. doi: 10.1073/pnas.1414837111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sun L, et al. Structural basis of human gamma-secretase assembly. Proc Natl Acad Sci U S A. 2015 doi: 10.1073/pnas.1506242112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cooper JB, Khan G, Taylor G, Tickle IJ, Blundell TL. X-ray analyses of aspartic proteinases. II. Three-dimensional structure of the hexagonal crystal form of porcine pepsin at 2.3 A resolution. J Mol Biol. 1990;214:199–222. doi: 10.1016/0022-2836(90)90156-G. [DOI] [PubMed] [Google Scholar]
- 23.Wang J, Brunkan AL, Hecimovic S, Walker E, Goate A. Conserved “PAL” sequence in presenilins is essential for gamma-secretase activity, but not required for formation or stabilization of gamma-secretase complexes. Neurobiol Dis. 2004;15:654–66. doi: 10.1016/j.nbd.2003.12.008. [DOI] [PubMed] [Google Scholar]
- 24.Sato C, Takagi S, Tomita T, Iwatsubo T. The C-terminal PAL motif and transmembrane domain 9 of presenilin 1 are involved in the formation of the catalytic pore of the gamma-secretase. Journal of Neuroscience. 2008;28:6264–71. doi: 10.1523/JNEUROSCI.1163-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang J, et al. C-terminal PAL motif of presenilin and presenilin homologues required for normal active site conformation. J Neurochem. 2006;96:218–27. doi: 10.1111/j.1471-4159.2005.03548.x. [DOI] [PubMed] [Google Scholar]
- 26.Dang S, et al. Cleavage of amyloid precursor protein by an archaeal presenilin homologue PSH. Proc Natl Acad Sci U S A. 2015;112:3344–9. doi: 10.1073/pnas.1502150112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schedin-Weiss S, Winblad B, Tjernberg LO. The role of protein glycosylation in Alzheimer disease. FEBS J. 2014;281:46–62. doi: 10.1111/febs.12590. [DOI] [PubMed] [Google Scholar]
- 28.Pardossi-Piquard R, et al. APH1 polar transmembrane residues regulate the assembly and activity of presenilin complexes. J Biol Chem. 2009;284:16298–307. doi: 10.1074/jbc.M109.000067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Esselens C, et al. Peptides based on the presenilin-APP binding domain inhibit APP processing and Abeta production through interfering with the APP transmembrane domain. FASEB J. 2012;26:3765–78. doi: 10.1096/fj.11-201368. [DOI] [PubMed] [Google Scholar]
- 30.Elad N, et al. The dynamic conformational landscape of gamma-secretase. J Cell Sci. 2014 doi: 10.1242/jcs.164384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li Y, et al. Structural Interactions between Inhibitor and Substrate Docking Sites Give Insight into Mechanisms of Human PS1 Complexes. Structure. 2014;22:125–35. doi: 10.1016/j.str.2013.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Takagi-Niidome S, et al. Cooperative roles of hydrophilic loop 1 and the C-terminus of presenilin 1 in the substrate-gating mechanism of gamma-secretase. J Neurosci. 2015;35:2646–56. doi: 10.1523/JNEUROSCI.3164-14.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Takeo K, Watanabe N, Tomita T, Iwatsubo T. Contribution of the gamma-secretase subunits to the formation of catalytic pore of presenilin 1 protein. J Biol Chem. 2012;287:25834–43. doi: 10.1074/jbc.M111.336347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.De Strooper B. Lessons from a failed γ-secretase Alzheimer trial. Cell. 2014;159:721–726. doi: 10.1016/j.cell.2014.10.016. [DOI] [PubMed] [Google Scholar]
- 35.Pettersen EF, et al. UCSF Chimera--a visualization system for exploratory research and analysis. J Comput Chem. 2004;25:1605–12. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 36.DeLano WL. The PyMOL Molecular Graphics System. on World Wide Web. 2002 http://www.pymol.org.
- 37.Li X, et al. Electron counting and beam-induced motion correction enable near-atomic-resolution single-particle cryo-EM. Nat Methods. 2013;10:584–90. doi: 10.1038/nmeth.2472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mindell JA, Grigorieff N. Accurate determination of local defocus and specimen tilt in electron microscopy. J Struct Biol. 2003;142:334–47. doi: 10.1016/s1047-8477(03)00069-8. [DOI] [PubMed] [Google Scholar]
- 39.Scheres SH. RELION: Implementation of a Bayesian approach to cryo-EM structure determination. J Struct Biol. 2012;180:519–30. doi: 10.1016/j.jsb.2012.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Scheres SH. Semi-automated selection of cryo-EM particles in RELION-1.3. J Struct Biol. 2015;189:114–22. doi: 10.1016/j.jsb.2014.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Scheres SH. Beam-induced motion correction for sub-megadalton cryo-EM particles. Elife. 2014;3:e03665. doi: 10.7554/eLife.03665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chen S, et al. High-resolution noise substitution to measure overfitting and validate resolution in 3D structure determination by single particle electron cryomicroscopy. Ultramicroscopy. 2013;135:24–35. doi: 10.1016/j.ultramic.2013.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rosenthal PB, Henderson R. Optimal determination of particle orientation, absolute hand, and contrast loss in single-particle electron cryomicroscopy. J Mol Biol. 2003;333:721–45. doi: 10.1016/j.jmb.2003.07.013. [DOI] [PubMed] [Google Scholar]
- 44.Kucukelbir A, Sigworth FJ, Tagare HD. Quantifying the local resolution of cryo-EM density maps. Nat Methods. 2014;11:63–5. doi: 10.1038/nmeth.2727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Stein N. CHAINSAW: a program for mutating pdb files used as templates in molecular replacement. Journal of Applied Crystallography. 2008;41:641–643. [Google Scholar]
- 46.Adams PD, et al. PHENIX: building new software for automated crystallographic structure determination. Acta Crystallographica Section D-Biological Crystallography. 2002;58:1948–1954. doi: 10.1107/s0907444902016657. [DOI] [PubMed] [Google Scholar]
- 47.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 48.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D Biol Crystallogr. 1997;53:240–55. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 49.Brown A, et al. Tools for macromolecular model building and refinement into electron cryo-microscopy reconstructions. Acta Crystallogr D Biol Crystallogr. 2015;71:136–53. doi: 10.1107/S1399004714021683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Nicholls RA, Fischer M, McNicholas S, Murshudov GN. Conformation-independent structural comparison of macromolecules with ProSMART. Acta Crystallogr D Biol Crystallogr. 2014;70:2487–99. doi: 10.1107/S1399004714016241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Amunts A, et al. Structure of the yeast mitochondrial large ribosomal subunit. Science. 2014;343:1485–9. doi: 10.1126/science.1249410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Crystal AS, et al. Membrane topology of gamma-secretase component PEN-2. J Biol Chem. 2003;278:20117–23. doi: 10.1074/jbc.M213107200. [DOI] [PubMed] [Google Scholar]