Abstract
In order to explore the diversity and selective signatures of duplication and deletion human copy number variants (CNVs), we sequenced 236 individuals from 125 distinct human populations. We observed that duplications exhibit fundamentally different population genetic and selective signatures than deletions and are more likely to be stratified between human populations. Through reconstruction of the ancestral human genome, we identify megabases of DNA lost in different human lineages and pinpoint large duplications that introgressed from the extinct Denisova lineage now found at high frequency exclusively in Oceanic populations. We find that the proportion of CNV base pairs to single nucleotide variant base pairs is greater among non-Africans than it is among African populations, but we conclude that this difference is likely due to unique aspects of non-African population history as opposed to differences in CNV load.
In the past decade, genome sequencing has provided insights into demography and migration patterns of human populations (1–4), ancient DNA (5–7), de novo mutation rates (8–10), and the relative deleteriousness and frequency of coding mutations (11, 12). Global human diversity, however, has only been partially sampled and the genetic architecture of many populations remains uncharacterized. To date, the majority of human diversity studies have focused on single nucleotide variants (SNVs) although copy number variants (CNVs) have contributed significantly to hominid evolution (13, 14), adaptation and disease (15–18). Much of the research into CNV diversity has been performed with SNP microarray and array comparative genomic hybridization (aCGH) platforms (19–22), which provide limited resolution. In addition, comparisons of population CNV diversity with heterogeneous discovery platforms may lead to spurious population-specific trends in CNV diversity (22, 23). Although there are many other forms of structural variation (e.g., inversions or mobile element insertions) in this study, we focused on understanding the population genetics and normal pattern of copy number variation by deep sequencing a diverse panel of human genomes.
Results
CNV discovery
We sequenced to high coverage a panel of 236 human genomes representing 125 diverse human populations from across the globe (Fig. 1 and table S2). Sequencing was performed to a mean genome coverage of 41-fold from libraries prepared using a standard PCR-free protocol on the HiSeq 2000 Illumina sequencing platform (24). The panel includes representation from a broad swathe of human diversity, including individuals from across Siberia, the Indian subcontinent, and Oceania. We also analyzed the high-coverage archaic Neanderthal (25) and Denisova (26) as well as three ancient human genomes to refine the evolutionary origin and timing of CNV differences (24). We applied a read-depth-based digital comparative genomic hybridization (dCGH) approach (13, 24) to discover 14,467 autosomal CNVs and 545 X-linked CNVs among individuals relative to the reference genome (Table 1 and table S1), which we estimate provides breakpoint resolution to ~210 bp (24). CNV calls were validated with SNP microarrays and a custom aCGH microarray that targeted all CNVs identified in 20 randomly selected individuals (24).
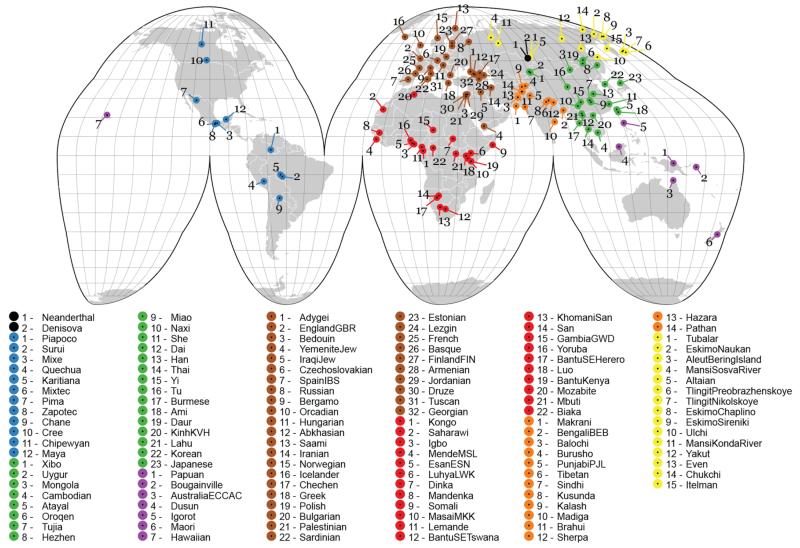
Fig. 1. Analysis of CNVs in several world populations.
The geographical locations of the 125 human populations, including two archaic genomes, assessed in this study. Populations are colored by their continental population groups, and archaic individuals are indicated in black.
Table 1. CNVs and SNVs broken down by their intersection with genomic region.
The number of Mbp of exonic and segmentally duplicated CNVs reflects the amount of exonic and segmental duplication sequence affected, respectively, not the total sum of the intersecting CNVs.
| Class | Autosomal (Mbp) | X chromosome (Mbp) |
Exonic (Mbp) | Segmentally duplicated (Mbp) |
|---|---|---|---|---|
| Deletions | 7,233 (78.99) | 278 (6.61) | 636 (0.32) | 331 (8.47) |
| Duplications | 7,234 (129.62) | 267 (6.46) | 2,093 (1.56) | 4,462 (96.93) |
| Subtotal | 14,467 (204.54) | 545 (12.61) | 2,729 (1.84) | 4,793 (99.84) |
| SNVs | 32,630,650 (32.63) | 1,175,170 (1.18) | 314,872 (0.31) | 1,559,158 (1.56) |
| All | 32,645,117 (237.17) | 1,175,715 (13.79) | 317,601 (2.15) | 1,563,951 (101.4) |
The median CNV size was 7,396 bp with 82.2% of events (n = 12,338) less than 25 kbp (24). CNVs mapping to segmental duplications were larger on average (median of 14.4 kbp), than CNVs mapping to the unique portions of the genome (median of 6.2 kbp). Almost one-half of CNV base pairs mapped within previously annotated segmental duplications (a 10-fold enrichment) (Table 1). In total, 217.1 Mbp (7.01%) of the human genome is variable due to CNVs in contrast to 33.8 Mbp (1.1%) due to single-nucleotide variation (Table 1). Deletions (loss of sequence) were less common (representing 85.6 Mbp or 2.77% of the genome) compared to duplications (gain of sequence, 136.1 Mbp or 4.4% of the genome). Furthermore, comparing our dataset with other studies of CNVs (21, 27) 67-73% of calls we report are unique to our study while we capture 68-77% of previously identified CNVs (24).
CNV diversity and selection
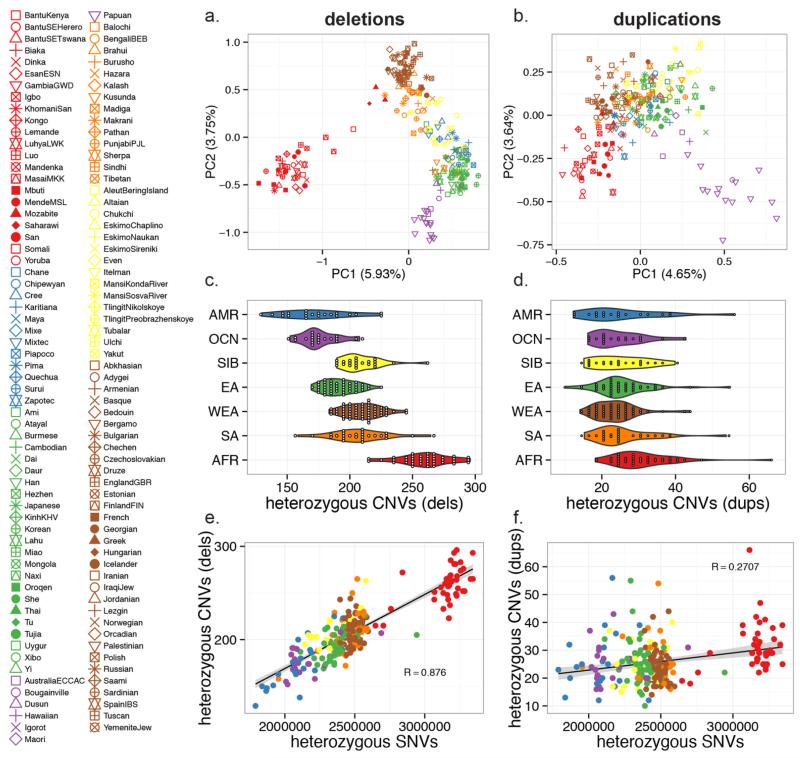
African populations are broadly distinguished from non-African populations by a principal component analysis (PCA) for either deletions (Fig. 2A and fig. S20) (24) or duplications (Fig. 2B). In this analysis, we limited the variants to bi-allelic deletions or bi-allelic duplications (diploid genotypes of 2, 3 or 4) to eliminate difficulty of inferring phase from multicopy CNVs. For deletions, PC1 (6.8% of the variance) and PC2 (3.94%) distinguish Africans, West Eurasians, East Asians and Oceanic populations. PC3 and PC4, describing 2.8% and 2.0% of the total variance, cluster Papuans and populations of the Americas, respectively. Many other populations were predictably distributed along clines between these clusters (e.g., Northern Africans, Siberian, South Asian, Amerindian and indigenous peoples of Philippines and North Borneo). PCAs generated from SNVs showed similar patterns as those from deletions. Africans also show much greater heterozygosity (Fig. 2C and Table 2), for instance, ~25% more heterozygous bi-allelic deletions and more than a twofold difference when compared to Amerindians (θAfrican = 535 versus θAmericas = 209). The archaic Neanderthal and Denisova genomes form an out-group to all humans (24).
Fig. 2. Population structure and CNV diversity.
Principal component analysis (PCA) of individuals assessed in this study plotted for bi-allelic deletions (A) and duplications (B) with colors and shapes representing continental and specific populations, respectively. Individuals are projected along the PC1 and PC2 axes. The deletion (C) and duplication (D) heterozygosity plotted and grouped by continental population. The relationship between SNV heterozygosity and deletion (E) or duplication (F) heterozygosity is compared.
Table 2. Summary statistics of bi-allelic CNV deletions versus SNVs by continental population group.
| Continental population group |
n | Segregating SNVs |
Segregating CNVs |
CNVs / individual (median) |
Heterozygous CNVs / individual (median) |
Continental population group- specific CNVs (allele count ≥2) |
θCNV / genome |
|---|---|---|---|---|---|---|---|
| West Eurasian (WEA) | 58 | 13610715 | 1728 | 279.0 | 209.0 | 688 (89) | 324.42 |
| Oceanic (OCN) | 21 | 9467426 | 1022 | 263.0 | 173.0 | 353 (84) | 237.51 |
| East Asian (EA) | 45 | 17452049 | 1463 | 271.0 | 191.0 | 525 (59) | 288.48 |
| Siberian (SIB) | 23 | 9644914 | 1102 | 285.0 | 205.0 | 214 (30) | 250.74 |
| South Asian (SA) | 27 | 11308883 | 1405 | 279.0 | 208.0 | 418 (43) | 308.32 |
| Americas (AMR) | 21 | 8127639 | 899 | 266.0 | 169.0 | 208 (25) | 208.93 |
| African (AFR) | 41 | 21698517 | 2663 | 319.0 | 261.0 | 1772 (702) | 534.97 |
Duplication heterozygosity and PCA in general show similar trends (Fig. 2D); albeit with far less definition. Interestingly, Oceanic populations, especially those from Papua New Guinea, Australia, and Bougainville showed the greatest separation on PC1 by duplication. Bi-allelic duplications appear somewhat less informative markers of human ancestry in contrast to SNVs, which provide the greatest resolution (e.g., SNV PCs 1-4 describe 5.8, 3.4, 2.6 and 1.7% of the variance, respectively). This difference is also seen when comparing SNV and CNV heterozygosity (Fig. 2, E and F). While heterozygous bi-allelic deletions were strongly correlated (R = 0.88) with SNV heterozygosity, the correlation between SNVs and duplications was much weaker (R = 0.27). We compared this correlation for duplications located adjacent to segmental duplications (within or proximal 150 kbp) in contrast to those occurring in unique regions of the genome and therefore less likely to be subject to recurrent mutation. Heterozygous duplications occurring in unique regions were better correlated with heterozygous SNVs (r = 0.29) than those adjacent or within segmental duplications (r = 0.17), though the difference was not significant (two-sided Williams’ test P < 0.1).
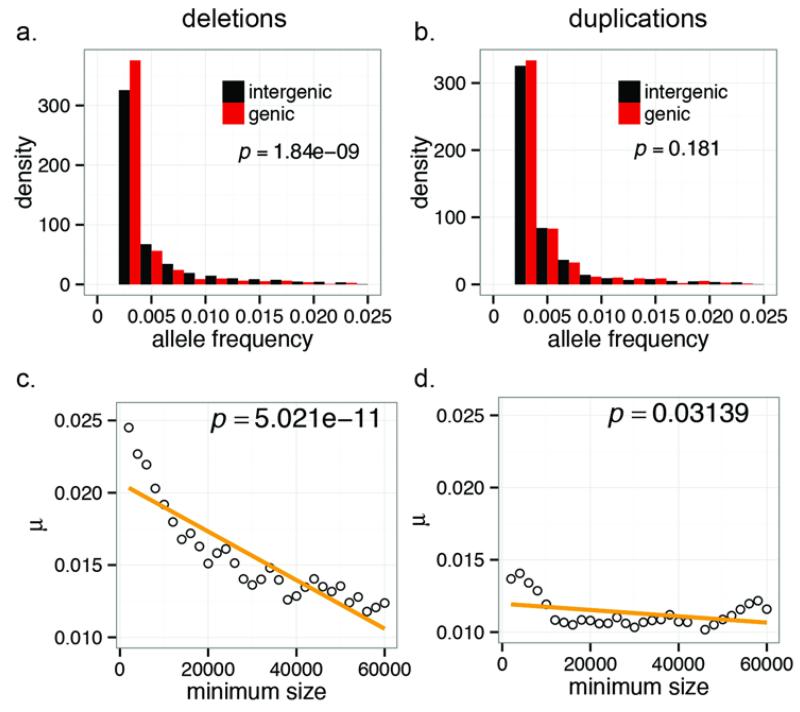
Studies of larger (>100 kbp) deletion and duplication events indicate that deletions are more deleterious than duplications (28). We reasoned that this may be reflected in the allele frequency spectrum (AFS) of normal genetic variation and compared the AFS of genic versus intergenic deletions and duplications for smaller events (Fig. 3, A and B). Genic deletions were significantly rarer than intergenic deletions (Wilcoxon rank sum test, P = 1.84e-9), but genic duplications showed no such skew (Wilcoxon rank sum test, P = 0.181). Size also had a significant impact on the AFS of CNVs. Deletions increased in rarity as a function of size (F-test, P = 5.02e-11) (Fig. 3C), but only a nominally significant trend was observed for duplications (P = 0.031) (Fig. 3D). These data suggest that selection has shaped the extant diversity of deletions and duplications differently during human evolution.
Fig. 3. Selection on CNVs.
Folded allele frequency spectra of exon-intersecting deletions (A) and duplications (B). While deletions intersecting exons are significantly rarer than intergenic deletions, exon-intersecting duplications show no difference compared to intergenic duplications. The mean frequency of CNVs beyond a minimum size threshold is plotted for deletions (C) and duplications (D). A strong negative correlation between size and allele frequency is observed for deletions but less so for duplications.
Population stratification
As population stratification can be indicative of loci under adaptive selection, we calculated Vst statistics for each CNV among all pairs of continental population groups, a metric analogous to Fst (the fixation index) (29). Vst and Fst statistics compare the variance in allele frequencies between populations with Vst allowing comparison of multi-allelic or multicopy CNVs. We identified 1,036 stratified copy number variable loci (CNVRs with maximum population Vst > 0.2, ~10% of the total), 295 of which intersected the exons of genes and 199 that exhibited extreme stratification (Vst > 0.5) (table S3). After correcting for copy number, duplicated loci were 1.8-fold more likely to be stratified than deletions. This finding is more remarkable in light of the fact that duplications were less discriminatory by PCA suggesting that a subset of multi-allelic duplicated CNVs show large allele frequency differences between different populations (see discussion below). The Vst of stratified duplicated CNVs was weakly correlated with the Fst of flanking SNVs (R2 = 0.03, P = 3.27e-12) in contrast to deletions (R2 = 0.2, p < 2e-16). Stratified duplication loci, thus, are far less likely to be tagged by adjacent SNPs through linkage disequilibrium.
Many of the population-differentiated loci were multi-allelic and mapped to segmental duplications including the repeat domain of ANKRD36, and the DUF1220 domain of NBPF (24) (Table 3). Several of these population differences involve genes of medical consequence, such as the multi-allelic duplication of CLPS, a pancreatic colipase involved in dietary metabolism of long chain triglyceride fatty acids (Fig. 4A). Increased expression in mouse models of this gene is negatively correlated with blood glucose levels (30). A duplication of the haptoglobin and haptoglobin-related (HP and HPR) genes expanded exclusively in Africa. The duplication has recently been associated with a possible protective effect against trypanosomiasis in Africa, though only copy 3 and 4 alleles were reported (31). We find this locus has further expanded to five and six copies in Esan, Gambian, Igbo, Mandenka, and Yoruban individuals (Fig. 4A). We also compared the location of our CNVs with disease loci identified by GWAS (32) and sites of potential positive selection (33). Although only a small fraction of our CNVs (1-6%) overlapped such functional annotation, we note that 21% of putative adaptive loci intersected with a CNV when compared to 6% of disease GWAS loci (table S4). Because many of the intervals are large, further refinement and investigation are needed to determine the significance of such overlaps.
Table 3. CNVs differentiated between human populations.
CNVs intersecting genes that show dramatic difference in copy number (as measured by Vst) between human populations (see Fig. 1 for definition of populations).
| Locus | Genes | Vst | Copy range |
Description |
|---|---|---|---|---|
| chr2:97849921-97899292 | ANKRD36 | 0.49 (OCN-WEA) | 30-41 | Repeat domain expanded to 45 copies in Papuans. |
| chr1:144146792-144224420 | NBPF | 0.32 (AFR-EA) | 185-271 | Expansion of the DUF1220 repeat domain in Africans and Amerindians. Copy number associated with cognitive function and autism severity (47). |
| chr6:35749042-35767153 | CLPS | 0.29 (AMR-SA) | 2-6 | Pancreatic colipase involved in dietary metabolism of long chain triglyceride fatty acids. Increased expression is negatively correlated with blood glucose in mice (30). |
| chr16:72088031-72119241 | HP, HPR | 0.25 (AFR-WEA) | 1-6 | Haptoglobin and haptoglobin-related genes are expanded exclusively in Africa and associated with a possible protective effect against trypanosomiasis (31). |
| chr12:64011854-64015265 | DPY19L2 | 0.32 (OCN-SA) | 5-7 | DPY genes are required for sperm head elongation and acrosome formation during spermatogenesis and DPY19L2 homozygous deletions have been identified as a major cause of globozoospermia (48). |
| chr1:74648583-74664195 | LRRIQ3 | 0.23 (AMR-WEA) | 2-3 | LRRIQ3 is duplicated exclusively in Siberian and Amerindian populations. |
| chr17:43692284-43708692 | CRHR1 | 0.25 (EA-WEA) | 4-7 | Deletions of corticotropin-releasing hormone receptor 1 result in reduced anxiety and neurotransmission impairments in mice (49). |
| chr5:150201231-150223428 | IRGM promoter | 0.25 (AFR-WEA) | 0-2 | The IRGM promoter CNV is a Crohn’s disease risk factor (50). |
| chr3:195771149-195776591 | TFRC promoter | 0.57 (AFR-EA) | 0-2 | Transferrin receptor is a cellular receptor for New World haemorrhagic fever arenaviruses (51). |
Fig. 4. Population-stratified CNVs and archaic introgression.
(A) Four specific examples of population-stratified CNVs intersecting genes are shown, including LRRIQ3, the pancreatic collipase CLPS, the sperm head an acrosome formation gene DPY19L2, and the haptoglobin and haptoglobin-related genes HP and HPR. Dot-plots indicating the copy of the locus in each individual and pie charts with colors depicting the continental population distribution per copy number (see text for details and Figs. 1 and 2 and dot plots for color scheme). (B) Predicted copy number on the basis of read-depth for a 73.5 kbp duplication on chromosome 16. It is observed in the archaic Denisovan genome and at 0.84 allele frequency in Papuan and Bougainville populations, yet absent from all other assessed populations. The duplication intersects two microRNAs. The orange arrow corresponds to the position and orientation of this duplication as further highlighted in (C) and (D). (C) A heatmap representation of a ~1 Mbp region of chromosome 16p12 (chr16:21518638-22805719). Each row of the heatmap represents the estimated copy number in 1 kbp windows of a single individual across this locus. Genes, annotated segmental duplications, and arrows highlighting the size and orientation in the reference of the Denisova/Papuan-specific duplication locus (locus D) and three other duplicated loci (A, B, and C) of interest are shown below. (D) The structure of duplications A, B, C and D (as shown in 4C over the same locus) in the reference genome and the discordant paired-end read placements used to characterize two duplication structures. Structure A/C is found in all individuals, though not present in the reference genome, while structure B/D is only found in Papuan and Bougainville individuals indicating a large complex, duplication (~225 kbp) composed of different segmental duplications. Both the A/C and B/D duplication architectures exhibit inverted orientations compared to the reference. The number of reads in all Oceanic and non-Oceanic individuals supporting each structure are indicated. (E) Maximum likelihood tree of the 16p12 duplication locus (duplication D in 4B, 4C, and 4D) constructed from the locus in Orangutan, Denisova, the human reference and the inferred sequence of the Papuan duplication (24). All bootstrap values are 100%.
Denisovan CNVs are retained and expanded in Oceanic populations
We further searched for highly stratified population-specific CNVs sharing alleles with the archaic Neanderthal and Denisovan individuals assessed in our study. While no Neanderthal-shared population-specific CNVs were identified, five Oceanic-specific CNVs were identified that shared the Denisova allele at high frequency (24). Papuan genomes have previously been reported to harbor 3-6% Denisovan admixture (6, 26). CNVs of putative Denisovan ancestry were at remarkably high frequency in Papuan individuals (all >0.2 allele frequency), with one ~9 kbp deletion lying 2 kbp upstream of the long noncoding RNA LINC00501, another 5 kbp duplication lying 8 kbp upstream of the METTL9 methyltransferase gene, and a 73.5 kbp duplication intersecting the MIR548D2 and MIR548AA2 microRNAs (Fig. 4B).
We determined that the latter two are part of a larger composite segmental duplication that appears to have almost fixed among human Papuan–Bougainville genomes (AF = 0.84) but has not been observed in any other extant human population (Fig. 4, B and C). We noted three additional duplications proximal to this locus exhibiting strikingly correlated copy number, despite being separated by >1 Mbp in the reference genome (Fig. 4C) (24). We suggest that these constitute a single, larger (~225 kbp) complex duplication composed of different segmental duplications. Using discordantly mapping paired end reads, we resolved the organization of two duplication architectures not represented in the human reference (Fig. 4D). The first of which (architecture A/C) is present in all individuals assessed in this study (5,625 discordant paired-end reads supporting) but not in the human reference genome. The second (B/D) corresponds to the Denisova–Papuan-specific duplication and is only present in these individuals and the Denisova genome. 70 paralogous sequence variants (markers distinct to paralogous locus (34, 35)) distinguish the Papuan duplication of which 65/70 (92.9%) were shared with the archaic Denisova genome. On the basis of single-nucleotide divergence we estimate that the duplication emerged ~440 kya and rose to high frequency in Papuan (>0.80 AF) but not Australian genomes probably over the last 40,000 years after introgression from Denisova (Fig. 4E). This duplication polymorphism represents the largest introgressed archaic hominin locus in modern humans.
The ancestral human genome
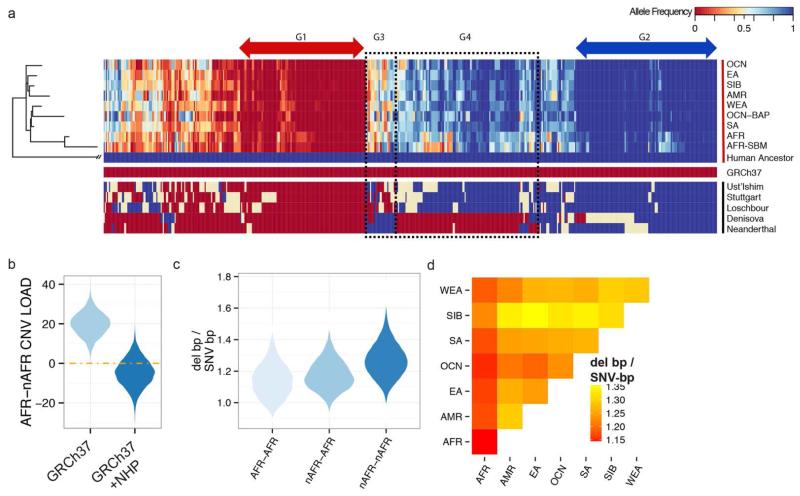
The breadth of the dataset allowed us to reconstruct the structure and content of the ancestral human genome prior to human migration and subsequent gene loss. To identify ancestral sequences potentially lost by deletion, we identified a set of sequences present in chimpanzee and orangutan reference genomes but absent from the human reference genome (20,373 nonredundant loci corresponding to 40.7 Mbp of sequence). Of these, 9,666 (27.6 Mbp) were unique (i.e., not composed of common repeats). Due to the inability to accurately genotype copy number for unique segments less than 500 bp by read-depth analysis, we limited our ancestral reconstruction to nonrepetitive sequences greater than this length threshold. While the majority represented deletions specifically lost in the human lineage since divergence from great apes (6,341 loci) or else reference genome artifacts (2,026 loci fixed-copy 2 in all individuals assessed, 6.2 Mbp), a small subset of these (n = 571 or 1.55 Mbp) segregate as bi-allelic polymorphisms in human populations (Fig. 5A). As expected, Africans were more likely to show evidence of these ancestral sequences compared to non-African populations, as the latter have experienced more population bottlenecks and thus retained less of the ancestral human diversity. A comparison to archaic genomes allowed us to identify sequences (50 loci or 104 kbp) that were present in Denisova or Neanderthal but lost in all contemporary humans as well as ancestral sequences present in all humans but not found in Denisova or Neanderthal (17 loci or 33.3 kbp).
Fig. 5. The ancestral human genome and CNV burden.
(A) A heatmap of the allele frequency of 571 (1.55 Mbp) nonrepetitive sequences absent from the human reference genome yet segregating in at least one human population ordered in humans by a maximum likelihood tree (49). Four groups of interest are highlighted: G1 – ancestral sequences that have almost been completely lost from the human lineage, G2 – ancestral sequences that are largely fixed but rarely deleted (also absent in human reference), G3 – ancestral sequences that have become copy number variable since the divergence of humans and Neanderthals/Denisovans ~700 kya, and G4 – sequences potentially lost in Neanderthals and Denisovans since their divergence from humans. (B) The resulting distributions of 10,000 block-bootstrapped estimates of the difference in load between African (AFR) and non-African (nAFR) populations considering only the reference genome (GRCh37) and supplemented by sequence absent from the human reference genome (GRCh37 + NHP) included (see text for details). (C) Violin plots of the distribution of the ratio of deletion base pairs to SNV base pairs differing between every pair of African individuals (AFR-AFR), all pairs of non-African individuals (nAFR-nAFR) and every non-African, African pair (nAFR-AFR). (D) Heatmap representation of the mean ratio of deletion to SNV base pairs differing between individuals from pairs of populations.
No difference in the CNV load between Africans and non-Africans
The high coverage and uniformity allowed us to contrast putatively deleterious, exon-removing CNVs among human populations, of interest in disease studies (36–38). In our callset we identified 2,437 CNVRs intersecting exons. The distribution of allele counts of these tended toward lower frequency events with, again, deletions more rare than duplications (Wilcoxon rank sum test, P = 1.25e-5). Collectively, individuals harbor a mean of 19.2 exon-intersecting deletions per genome (22.8 per diploid genome), with African individuals exhibiting, on average, a mean of 22.4 deletions compared to 18.6 in non-Africans (26.1 and 22.1 per diploid genome, respectively), consistent with the increased diversity of African populations and consistent with data observed for loss-of-function SNVs ((12, 39), ~122 LoF SNVs in Africans versus ~104 in non-Africans).
While non-African individuals exhibited more homozygous deletion variants compared to Africans, among exon-intersecting deletions no such pattern was observed. Exon-intersecting duplications were much more balanced with African populations showing only a slight excess when compared to non-Africans (98.4 versus 95.2 events per genome). Studies of SNVs have not found consistent evidence of difference in load between African compared to non-African populations (40–42). We compared the difference in load between African and non-African populations for deletions and duplications, respectively. Here, we defined the difference in load as the difference in the sum of derived allele frequencies between African and non-African populations, where PAfr(i) is the derived allele frequency of a variant i. Prima facie Africans exhibited an apparent higher deletion load than non-African populations (Fig. 5B) (P = 0.0003, block bootstrap test), though only a nominal difference in the load of exonic deletions (P = 0.0482). Duplications showed no such effect.
We reasoned that this striking difference might potentially be driven by high-frequency derived alleles, absent from the human reference genome, which was enriched for clone libraries of non-African ancestry (5). Approaches that rely on identifying CNVs based on read placements to the reference genome would necessarily miss these CNVs, decreasing the number of variants identified in individuals more closely resembling the reference, i.e., non-Africans. To test this hypothesis we incorporated the bi-allelic 571 non-repetitive human CNV loci described above. Copy numbers were estimated for these sequences in each of the individuals assessed by remapping raw reads against an ancestral human reference genome. As expected, the deletion allele of this sequence was at a high frequency (mean derived allele frequency, DAF = 0.58). After including these sequences we observed no difference in the CNV load between Africans and non-Africans (95% confidence interval −18.4 to 8.8 load difference as defined above) (Fig. 5B) underscoring the importance of an unbiased human reference for such population genetic assessments.
Although we found no CNV or SNV load differences between populations, we examined whether the relative proportion of base pairs differing among individuals derived from CNVs versus SNVs showed any population-specific trends. We calculated the number of base pairs varying between all pairs of individuals assessed in our study contributed either from SNVs or from deletions calculating the DEL-bp/SNV-bp ratio. As expected, the number of base pairs differing between individuals by deletions or by SNVs independently was always higher among African individuals when compared to other populations. Surprisingly, the ratio of deletion-bp to SNV-bp was substantially higher within non-African populations (mean 1.27 compared to 1.14, Fig. 5, C and D). This relative increase in deleted base pairs was most pronounced among non-African populations, which have experienced more recent genetic bottlenecks (e.g., Siberian and Amerindian). Given the absence of a significant difference in the deletion load comparing African and non-African populations, there is no reason to believe that this finding is due to differences in the effectiveness of selection against deletions since the populations separated. However, selection places a downward pressure on the allele frequencies of both deletions and SNVs, with the pressure being stronger for deletions because the selection coefficients are stronger on average. As has been previously shown for SNVs, different allele frequency spectra for deletions in contrast to SNVs has the potential to interact with the differences in demographic history across populations—even without differences in the effectiveness of selection after population separation—to contribute to observed differences in the apportionment of genetic variation among human populations (41).
Discussion
While the mutational properties and selective signatures of SNVs have been explored extensively, similar analyses of CNVs have lagged behind. As a class, duplications show generally poor correlations with SNV density, have poor linkage disequilibrium to SNVs (43, 44), and are less informative as phylogenetic markers but are more likely to be stratified than deletions among human populations. This observation may be explained by the fact that directly orientated duplications show a gradient of elevated mutation rates due to non-allelic homologous recombination and, as such, can change their copy number state more dynamically over short periods of time. This property also makes this class of variation, similar to highly mutable loci such as minisatellites (45), particularly susceptible to homoplasy— i.e., identity by state as opposed identity by descent. Deletions, in contrast, recapitulate most properties of SNVs because they are more likely to exhibit identity by descent as a result of single ancestral mutation event.
We have provided here sequencing data for the study of human diversity and utilize this resource to explore patterns of human CNV diversity at a fine scale of resolution (>1 kbp). As expected, human genomes differ more with respect to CNVs than SNVs and almost one-half of these CNV differences map to regions of segmental duplication. Both deletion and duplication analyses consistently distinguish African, Oceanic, and Amerindian human populations. Africans show the greatest deletion and duplication diversity and have the lowest rate of fixed deletions with respect to ancestral human insertion sequences. Oceanic and Amerindian, in contrast, show greater CNV differentiation likely as a result of longer periods of genetic isolation and founder effects (46). Among the Oceanic, the Papuan–Bougainville group stands out in sharing more derived CNV alleles in common with Denisova, including a massive interspersed duplication that rose to high frequency over a short period of time.
We find that duplications and deletions exhibit fundamentally different population-genetic properties. Duplications are subjected to weaker selective constraint and are four times more likely to affect genes than deletions (Table 1) indicating that they provide a larger target for adaptive selection. After controlling for reference genome biases, we find no difference in CNV load between human populations when measured on a per-genome basis which is what matters to disease risk assuming that CNVs act additively. However, we find that the proportion of human variation that can be ascribed to CNVs rather than to SNVs is greater among non-Africans than among Africans. The biological significance of this difference should be interpreted cautiously and will require association studies to determine its relevance to disease and other phenotypic differences.
Supplementary Material
ACKNOWLEDGMENTS
We are grateful to the volunteers who donated the DNA samples used in this study. This work was supported, in part, by U.S. National Institutes of Health (NIH) grant 2R01HG002385 and a grant (11631) from The Paul G. Allen Family Foundation to E.E.E. The sequencing for this study was supported by a grant from the Simons Foundation to D.R. (SFARI 280376) and by a HOMINID grant from The National Science Foundation to D.R. (BCS-1032255). T.K. is supported by ERC Starting Investigator grant FP7 - 261213. R.S. and S.D. received support from The Ministry of Education and Science, Russian Federation (14.Z50.31.0010). H.S., E.M., R.V., and M.M. are supported by Institutional Research Funding from the Estonian Research Council IUT24-1 and by the European Regional Development Fund (European Union) through the Centre of Excellence in Genomics to Estonian Biocentre and University of Tartu. S.A.T. is supported by grants 5DP1ES022577 05, 1R01DK104339-01, and 1R01GM113657-01. C.T.S. is supported by The Wellcome Trust grant 098051. C.M.B. is supported by the National Science Foundation (award numbers 0924726 and 1153911). E.E.E. and D.R. are investigators of the Howard Hughes Medical Institute. Data are deposited into ENA, and variant calls are deposited in dbVar (PRJEB9586, PRJNA285786). E.E.E. is on the scientific advisory board (SAB) of DNAnexus, Inc. and is a consultant for Kunming University of Science and Technology (KUST) as part of the 1000 China Talent Program.
Footnotes
www.sciencemag.org/cgi/content/full/science.aab3761/DC1
Supplementary Text
Figs. S1 to S48
Tables S1 to S18
References (52–57)
REFERENCES AND NOTES
- 1.Schuster SC, Miller W, Ratan A, Tomsho LP, Giardine B, Kasson LR, Harris RS, Petersen DC, Zhao F, Qi J, Alkan C, Kidd JM, Sun Y, Drautz DI, Bouffard P, Muzny DM, Reid JG, Nazareth LV, Wang Q, Burhans R, Riemer C, Wittekindt NE, Moorjani P, Tindall EA, Danko CG, Teo WS, Buboltz AM, Zhang Z, Ma Q, Oosthuysen A, Steenkamp AW, Oostuisen H, Venter P, Gajewski J, Zhang Y, Pugh BF, Makova KD, Nekrutenko A, Mardis ER, Patterson N, Pringle TH, Chiaromonte F, Mullikin JC, Eichler EE, Hardison RC, Gibbs RA, Harkins TT, Hayes VM. Complete Khoisan and Bantu genomes from southern Africa. Nature. 2010;463:943–947. doi: 10.1038/nature08795. Medline doi:10.1038/nature08795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Steinberg KM, Antonacci F, Sudmant PH, Kidd JM, Campbell CD, Vives L, Malig M, Scheinfeldt L, Beggs W, Ibrahim M, Lema G, Nyambo TB, Omar SA, Bodo JM, Froment A, Donnelly MP, Kidd KK, Tishkoff SA, Eichler EE. Structural diversity and African origin of the 17q21.31 inversion polymorphism. Nat. Genet. 2012;44:872–880. doi: 10.1038/ng.2335. Medline doi:10.1038/ng.2335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gravel S, Zakharia F, Moreno-Estrada A, Byrnes JK, Muzzio M, Rodriguez-Flores JL, Kenny EE, Gignoux CR, Maples BK, Guiblet W, Dutil J, Via M, Sandoval K, Bedoya G, Oleksyk TK, Ruiz-Linares A, Burchard EG, Martinez-Cruzado JC, Bustamante CD. 1000 Genomes Project, Reconstructing Native American migrations from whole-genome and whole-exome data. PLOS Genet. 2013;9:e1004023. doi: 10.1371/journal.pgen.1004023. Medline. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Raghavan M, DeGiorgio M, Albrechtsen A, Moltke I, Skoglund P, Korneliussen TS, Grønnow B, Appelt M, Gulløv HC, Friesen TM, Fitzhugh W, Malmström H, Rasmussen S, Olsen J, Melchior L, Fuller BT, Fahrni SM, Stafford T, Jr., Grimes V, Renouf MA, Cybulski J, Lynnerup N, Lahr MM, Britton K, Knecht R, Arneborg J, Metspalu M, Cornejo OE, Malaspinas AS, Wang Y, Rasmussen M, Raghavan V, Hansen TV, Khusnutdinova E, Pierre T, Dneprovsky K, Andreasen C, Lange H, Hayes MG, Coltrain J, Spitsyn VA, Götherström A, Orlando L, Kivisild T, Villems R, Crawford MH, Nielsen FC, Dissing J, Heinemeier J, Meldgaard M, Bustamante C, O’Rourke DH, Jakobsson M, Gilbert MT, Nielsen R, Willerslev E. The genetic prehistory of the New World Arctic. Science. 2014;345:1255832. doi: 10.1126/science.1255832. Medline doi:10.1126/science.1255832. [DOI] [PubMed] [Google Scholar]
- 5.Green RE, Krause J, Briggs AW, Maricic T, Stenzel U, Kircher M, Patterson N, Li H, Zhai W, Fritz MH, Hansen NF, Durand EY, Malaspinas AS, Jensen JD, Marques-Bonet T, Alkan C, Prüfer K, Meyer M, Burbano HA, Good JM, Schultz R, Aximu-Petri A, Butthof A, Höber B, Höffner B, Siegemund M, Weihmann A, Nusbaum C, Lander ES, Russ C, Novod N, Affourtit J, Egholm M, Verna C, Rudan P, Brajkovic D, Kucan Z, Gusic I, Doronichev VB, Golovanova LV, Lalueza-Fox C, de la Rasilla M, Fortea J, Rosas A, Schmitz RW, Johnson PL, Eichler EE, Falush D, Birney E, Mullikin JC, Slatkin M, Nielsen R, Kelso J, Lachmann M, Reich D, Pääbo S. A draft sequence of the Neandertal genome. Science. 2010;328:710–722. doi: 10.1126/science.1188021. Medline doi:10.1126/science.1188021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Reich D, Green RE, Kircher M, Krause J, Patterson N, Durand EY, Viola B, Briggs AW, Stenzel U, Johnson PL, Maricic T, Good JM, Marques-Bonet T, Alkan C, Fu Q, Mallick S, Li H, Meyer M, Eichler EE, Stoneking M, Richards M, Talamo S, Shunkov MV, Derevianko AP, Hublin JJ, Kelso J, Slatkin M, Pääbo S. Genetic history of an archaic hominin group from Denisova Cave in Siberia. Nature. 2010;468:1053–1060. doi: 10.1038/nature09710. Medline doi:10.1038/nature09710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rasmussen M, Li Y, Lindgreen S, Pedersen JS, Albrechtsen A, Moltke I, Metspalu M, Metspalu E, Kivisild T, Gupta R, Bertalan M, Nielsen K, Gilbert MT, Wang Y, Raghavan M, Campos PF, Kamp HM, Wilson AS, Gledhill A, Tridico S, Bunce M, Lorenzen ED, Binladen J, Guo X, Zhao J, Zhang X, Zhang H, Li Z, Chen M, Orlando L, Kristiansen K, Bak M, Tommerup N, Bendixen C, Pierre TL, Grønnow B, Meldgaard M, Andreasen C, Fedorova SA, Osipova LP, Higham TF, Ramsey CB, Hansen TV, Nielsen FC, Crawford MH, Brunak S, Sicheritz-Pontén T, Villems R, Nielsen R, Krogh A, Wang J, Willerslev E. Ancient human genome sequence of an extinct Palaeo-Eskimo. Nature. 2010;463:757–762. doi: 10.1038/nature08835. Medline. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Conrad DF, Keebler JE, DePristo MA, Lindsay SJ, Zhang Y, Casals F, Idaghdour Y, Hartl CL, Torroja C, Garimella KV, Zilversmit M, Cartwright R, Rouleau GA, Daly M, Stone EA, Hurles ME, Awadalla P. 1000 Genomes Project, Variation in genome-wide mutation rates within and between human families. Nat. Genet. 2011;43:712–714. doi: 10.1038/ng.862. Medline doi:10.1038/ng.862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Campbell CD, Chong JX, Malig M, Ko A, Dumont BL, Han L, Vives L, O’Roak BJ, Sudmant PH, Shendure J, Abney M, Ober C, Eichler EE. Estimating the human mutation rate using autozygosity in a founder population. Nat. Genet. 2012;44:1277–1281. doi: 10.1038/ng.2418. Medline doi:10.1038/ng.2418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kong A, Frigge ML, Masson G, Besenbacher S, Sulem P, Magnusson G, Gudjonsson SA, Sigurdsson A, Jonasdottir A, Jonasdottir A, Wong WS, Sigurdsson G, Walters GB, Steinberg S, Helgason H, Thorleifsson G, Gudbjartsson DF, Helgason A, Magnusson OT, Thorsteinsdottir U, Stefansson K. Rate of de novo mutations and the importance of father’s age to disease risk. Nature. 2012;488:471–475. doi: 10.1038/nature11396. Medline doi:10.1038/nature11396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fu W, O’Connor TD, Jun G, Kang HM, Abecasis G, Leal SM, Gabriel S, Rieder MJ, Altshuler D, Shendure J, Nickerson DA, Bamshad MJ, Akey JM. NHLBI Exome Sequencing Project, Analysis of 6,515 exomes reveals the recent origin of most human protein-coding variants. Nature. 2013;493:216–220. doi: 10.1038/nature11690. Medline doi:10.1038/nature11690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tennessen JA, Bigham AW, O’Connor TD, Fu W, Kenny EE, Gravel S, McGee S, Do R, Liu X, Jun G, Kang HM, Jordan D, Leal SM, Gabriel S, Rieder MJ, Abecasis G, Altshuler D, Nickerson DA, Boerwinkle E, Sunyaev S, Bustamante CD, Bamshad MJ, Akey JM. Broad GO, Seattle GO, NHLBI Exome Sequencing Project, Evolution and functional impact of rare coding variation from deep sequencing of human exomes. Science. 2012;337:64–69. doi: 10.1126/science.1219240. Medline doi:10.1126/science.1219240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sudmant PH, Huddleston J, Catacchio CR, Malig M, Hillier LW, Baker C, Mohajeri K, Kondova I, Bontrop RE, Persengiev S, Antonacci F, Ventura M, Prado-Martinez J, Marques-Bonet T, Eichler EE. Great Ape Genome Project, Evolution and diversity of copy number variation in the great ape lineage. Genome Res. 2013;23:1373–1382. doi: 10.1101/gr.158543.113. Medline doi:10.1101/gr.158543.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Marques-Bonet T, Kidd JM, Ventura M, Graves TA, Cheng Z, Hillier LW, Jiang Z, Baker C, Malfavon-Borja R, Fulton LA, Alkan C, Aksay G, Girirajan S, Siswara P, Chen L, Cardone MF, Navarro A, Mardis ER, Wilson RK, Eichler EE. A burst of segmental duplications in the genome of the African great ape ancestor. Nature. 2009;457:877–881. doi: 10.1038/nature07744. Medline. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kumar RA, KaraMohamed S, Sudi J, Conrad DF, Brune C, Badner JA, Gilliam TC, Nowak NJ, Cook EH, Jr., Dobyns WB, Christian SL. Recurrent 16p11.2 microdeletions in autism. Hum. Mol. Genet. 2008;17:628–638. doi: 10.1093/hmg/ddm376. Medline doi:10.1093/hmg/ddm376. [DOI] [PubMed] [Google Scholar]
- 16.Sharp AJ, Mefford HC, Li K, Baker C, Skinner C, Stevenson RE, Schroer RJ, Novara F, De Gregori M, Ciccone R, Broomer A, Casuga I, Wang Y, Xiao C, Barbacioru C, Gimelli G, Bernardina BD, Torniero C, Giorda R, Regan R, Murday V, Mansour S, Fichera M, Castiglia L, Failla P, Ventura M, Jiang Z, Cooper GM, Knight SJ, Romano C, Zuffardi O, Chen C, Schwartz CE, Eichler EE. A recurrent 15q13.3 microdeletion syndrome associated with mental retardation and seizures. Nat. Genet. 2008;40:322–328. doi: 10.1038/ng.93. Medline. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sebat J, Lakshmi B, Troge J, Alexander J, Young J, Lundin P, Månér S, Massa H, Walker M, Chi M, Navin N, Lucito R, Healy J, Hicks J, Ye K, Reiner A, Gilliam TC, Trask B, Patterson N, Zetterberg A, Wigler M. Large-scale copy number polymorphism in the human genome. Science. 2004;305:525–528. doi: 10.1126/science.1098918. Medline doi:10.1126/science.1098918. [DOI] [PubMed] [Google Scholar]
- 18.Weiss LA, Shen Y, Korn JM, Arking DE, Miller DT, Fossdal R, Saemundsen E, Stefansson H, Ferreira MA, Green T, Platt OS, Ruderfer DM, Walsh CA, Altshuler D, Chakravarti A, Tanzi RE, Stefansson K, Santangelo SL, Gusella JF, Sklar P, Wu BL, Daly MJ. Autism Consortium, Association between microdeletion and microduplication at 16p11.2 and autism. N. Engl. J. Med. 2008;358:667–675. doi: 10.1056/NEJMoa075974. Medline doi:10.1056/NEJMoa075974. [DOI] [PubMed] [Google Scholar]
- 19.McCarroll SA, Hadnott TN, Perry GH, Sabeti PC, Zody MC, Barrett JC, Dallaire S, Gabriel SB, Lee C, Daly MJ, Altshuler DM. International HapMap Consortium, Common deletion polymorphisms in the human genome. Nat. Genet. 2006;38:86–92. doi: 10.1038/ng1696. Medline doi:10.1038/ng1696. [DOI] [PubMed] [Google Scholar]
- 20.McCarroll SA, Kuruvilla FG, Korn JM, Cawley S, Nemesh J, Wysoker A, Shapero MH, de Bakker PI, Maller JB, Kirby A, Elliott AL, Parkin M, Hubbell E, Webster T, Mei R, Veitch J, Collins PJ, Handsaker R, Lincoln S, Nizzari M, Blume J, Jones KW, Rava R, Daly MJ, Gabriel SB, Altshuler D. Integrated detection and population-genetic analysis of SNPs and copy number variation. Nat. Genet. 2008;40:1166–1174. doi: 10.1038/ng.238. Medline doi:10.1038/ng.238. [DOI] [PubMed] [Google Scholar]
- 21.Conrad DF, Pinto D, Redon R, Feuk L, Gokcumen O, Zhang Y, Aerts J, Andrews TD, Barnes C, Campbell P, Fitzgerald T, Hu M, Ihm CH, Kristiansson K, Macarthur DG, Macdonald JR, Onyiah I, Pang AW, Robson S, Stirrups K, Valsesia A, Walter K, Wei J, Tyler-Smith C, Carter NP, Lee C, Scherer SW, Hurles ME. Wellcome Trust Case Control Consortium, Origins and functional impact of copy number variation in the human genome. Nature. 2010;464:704–712. doi: 10.1038/nature08516. Medline doi:10.1038/nature08516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jakobsson M, Scholz SW, Scheet P, Gibbs JR, VanLiere JM, Fung HC, Szpiech ZA, Degnan JH, Wang K, Guerreiro R, Bras JM, Schymick JC, Hernandez DG, Traynor BJ, Simon-Sanchez J, Matarin M, Britton A, van de Leemput J, Rafferty I, Bucan M, Cann HM, Hardy JA, Rosenberg NA, Singleton AB. Genotype, haplotype and copy-number variation in worldwide human populations. Nature. 2008;451:998–1003. doi: 10.1038/nature06742. Medline. [DOI] [PubMed] [Google Scholar]
- 23.Itsara A, Cooper GM, Baker C, Girirajan S, Li J, Absher D, Krauss RM, Myers RM, Ridker PM, Chasman DI, Mefford H, Ying P, Nickerson DA, Eichler EE. Population analysis of large copy number variants and hotspots of human genetic disease. Am. J. Hum. Genet. 2009;84:148–161. doi: 10.1016/j.ajhg.2008.12.014. Medline doi:10.1016/j.ajhg.2008.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Materials and methods are available as supplementary materials at Science Online.
- 25.Prüfer K, Racimo F, Patterson N, Jay F, Sankararaman S, Sawyer S, Heinze A, Renaud G, Sudmant PH, de Filippo C, Li H, Mallick S, Dannemann M, Fu Q, Kircher M, Kuhlwilm M, Lachmann M, Meyer M, Ongyerth M, Siebauer M, Theunert C, Tandon A, Moorjani P, Pickrell J, Mullikin JC, Vohr SH, Green RE, Hellmann I, Johnson PL, Blanche H, Cann H, Kitzman JO, Shendure J, Eichler EE, Lein ES, Bakken TE, Golovanova LV, Doronichev VB, Shunkov MV, Derevianko AP, Viola B, Slatkin M, Reich D, Kelso J, Pääbo S. The complete genome sequence of a Neanderthal from the Altai Mountains. Nature. 2014;505:43–49. doi: 10.1038/nature12886. Medline doi:10.1038/nature12886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Meyer M, Kircher M, Gansauge MT, Li H, Racimo F, Mallick S, Schraiber JG, Jay F, Prüfer K, de Filippo C, Sudmant PH, Alkan C, Fu Q, Do R, Rohland N, Tandon A, Siebauer M, Green RE, Bryc K, Briggs AW, Stenzel U, Dabney J, Shendure J, Kitzman J, Hammer MF, Shunkov MV, Derevianko AP, Patterson N, Andrés AM, Eichler EE, Slatkin M, Reich D, Kelso J, Pääbo S. A high-coverage genome sequence from an archaic Denisovan individual. Science. 2012;338:222–226. doi: 10.1126/science.1224344. Medline. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mills RE, Walter K, Stewart C, Handsaker RE, Chen K, Alkan C, Abyzov A, Yoon SC, Ye K, Cheetham RK, Chinwalla A, Conrad DF, Fu Y, Grubert F, Hajirasouliha I, Hormozdiari F, Iakoucheva LM, Iqbal Z, Kang S, Kidd JM, Konkel MK, Korn J, Khurana E, Kural D, Lam HY, Leng J, Li R, Li Y, Lin CY, Luo R, Mu XJ, Nemesh J, Peckham HE, Rausch T, Scally A, Shi X, Stromberg MP, Stütz AM, Urban AE, Walker JA, Wu J, Zhang Y, Zhang ZD, Batzer MA, Ding L, Marth GT, McVean G, Sebat J, Snyder M, Wang J, Ye K, Eichler EE, Gerstein MB, Hurles ME, Lee C, McCarroll SA, Korbel JO. 1000 Genomes Project, Mapping copy number variation by population-scale genome sequencing. Nature. 2011;470:59–65. doi: 10.1038/nature09708. Medline doi:10.1038/nature09708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cooper GM, Coe BP, Girirajan S, Rosenfeld JA, Vu TH, Baker C, Williams C, Stalker H, Hamid R, Hannig V, Abdel-Hamid H, Bader P, McCracken E, Niyazov D, Leppig K, Thiese H, Hummel M, Alexander N, Gorski J, Kussmann J, Shashi V, Johnson K, Rehder C, Ballif BC, Shaffer LG, Eichler EE. A copy number variation morbidity map of developmental delay. Nat. Genet. 2011;43:838–846. doi: 10.1038/ng.909. Medline doi:10.1038/ng.909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Redon R, Ishikawa S, Fitch KR, Feuk L, Perry GH, Andrews TD, Fiegler H, Shapero MH, Carson AR, Chen W, Cho EK, Dallaire S, Freeman JL, González JR, Gratacòs M, Huang J, Kalaitzopoulos D, Komura D, MacDonald JR, Marshall CR, Mei R, Montgomery L, Nishimura K, Okamura K, Shen F, Somerville MJ, Tchinda J, Valsesia A, Woodwark C, Yang F, Zhang J, Zerjal T, Zhang J, Armengol L, Conrad DF, Estivill X, Tyler-Smith C, Carter NP, Aburatani H, Lee C, Jones KW, Scherer SW, Hurles ME. Global variation in copy number in the human genome. Nature. 2006;444:444–454. doi: 10.1038/nature05329. Medline doi:10.1038/nature05329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang J, Kaasik K, Blackburn MR, Lee CC. Constant darkness is a circadian metabolic signal in mammals. Nature. 2006;439:340–343. doi: 10.1038/nature04368. Medline doi:10.1038/nature04368. [DOI] [PubMed] [Google Scholar]
- 31.Hardwick RJ, Ménard A, Sironi M, Milet J, Garcia A, Sese C, Yang F, Fu B, Courtin D, Hollox EJ. Haptoglobin (HP) and Haptoglobin-related protein (HPR) copy number variation, natural selection, and trypanosomiasis. Hum. Genet. 2014;133:69–83. doi: 10.1007/s00439-013-1352-x. Medline doi:10.1007/s00439-013-1352-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hindorff LA, Sethupathy P, Junkins HA, Ramos EM, Mehta JP, Collins FS, Manolio TA. Potential etiologic and functional implications of genome-wide association loci for human diseases and traits. Proc. Natl. Acad. Sci. U.S.A. 2009;106:9362–9367. doi: 10.1073/pnas.0903103106. Medline doi:10.1073/pnas.0903103106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Grossman SR, Andersen KG, Shlyakhter I, Tabrizi S, Winnicki S, Yen A, Park DJ, Griesemer D, Karlsson EK, Wong SH, Cabili M, Adegbola RA, Bamezai RN, Hill AV, Vannberg FO, Rinn JL, Lander ES, Schaffner SF, Sabeti PC. 1000 Genomes Project, Identifying recent adaptations in large-scale genomic data. Cell. 2013;152:703–713. doi: 10.1016/j.cell.2013.01.035. Medline doi:10.1016/j.cell.2013.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Horvath JE, Schwartz S, Eichler EE. The mosaic structure of human pericentromeric DNA: A strategy for characterizing complex regions of the human genome. Genome Res. 2000;10:839–852. doi: 10.1101/gr.10.6.839. Medline doi:10.1101/gr.10.6.839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cheung J, Estivill X, Khaja R, MacDonald JR, Lau K, Tsui LC, Scherer SW. Genome-wide detection of segmental duplications and potential assembly errors in the human genome sequence. Genome Biol. 2003;4:R25. doi: 10.1186/gb-2003-4-4-r25. Medline doi:10.1186/gb-2003-4-4-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Krumm N, Sudmant PH, Ko A, O’Roak BJ, Malig M, Coe BP, Quinlan AR, Nickerson DA, Eichler EE. NHLBI Exome Sequencing Project, Copy number variation detection and genotyping from exome sequence data. Genome Res. 2012;22:1525–1532. doi: 10.1101/gr.138115.112. Medline doi:10.1101/gr.138115.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fromer M, Moran JL, Chambert K, Banks E, Bergen SE, Ruderfer DM, Handsaker RE, McCarroll SA, O’Donovan MC, Owen MJ, Kirov G, Sullivan PF, Hultman CM, Sklar P, Purcell SM. Discovery and statistical genotyping of copy-number variation from whole-exome sequencing depth. Am. J. Hum. Genet. 2012;91:597–607. doi: 10.1016/j.ajhg.2012.08.005. Medline doi:10.1016/j.ajhg.2012.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Deciphering Developmental Disorders Study, Large-scale discovery of novel genetic causes of developmental disorders. Nature. 2015;519:223–228. doi: 10.1038/nature14135. Medline. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.MacArthur DG, Balasubramanian S, Frankish A, Huang N, Morris J, Walter K, Jostins L, Habegger L, Pickrell JK, Montgomery SB, Albers CA, Zhang ZD, Conrad DF, Lunter G, Zheng H, Ayub Q, DePristo MA, Banks E, Hu M, Handsaker RE, Rosenfeld JA, Fromer M, Jin M, Mu XJ, Khurana E, Ye K, Kay M, Saunders GI, Suner MM, Hunt T, Barnes IH, Amid C, Carvalho-Silva DR, Bignell AH, Snow C, Yngvadottir B, Bumpstead S, Cooper DN, Xue Y, Romero IG, Wang J, Li Y, Gibbs RA, McCarroll SA, Dermitzakis ET, Pritchard JK, Barrett JC, Harrow J, Hurles ME, Gerstein MB, Tyler-Smith C. 1000 Genomes Project Consortium, A systematic survey of loss-of-function variants in human protein-coding genes. Science. 2012;335:823–828. doi: 10.1126/science.1215040. Medline doi:10.1126/science.1215040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fu W, Gittelman RM, Bamshad MJ, Akey JM. Characteristics of neutral and deleterious protein-coding variation among individuals and populations. Am. J. Hum. Genet. 2014;95:421–436. doi: 10.1016/j.ajhg.2014.09.006. Medline doi:10.1016/j.ajhg.2014.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Do R, Balick D, Li H, Adzhubei I, Sunyaev S, Reich D. No evidence that selection has been less effective at removing deleterious mutations in Europeans than in Africans. Nat. Genet. 2015;47:126–131. doi: 10.1038/ng.3186. Medline doi:10.1038/ng.3186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Simons YB, Turchin MC, Pritchard JK, Sella G. The deleterious mutation load is insensitive to recent population history. Nat. Genet. 2014;46:220–224. doi: 10.1038/ng.2896. Medline doi:10.1038/ng.2896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Campbell CD, Sampas N, Tsalenko A, Sudmant PH, Kidd JM, Malig M, Vu TH, Vives L, Tsang P, Bruhn L, Eichler EE. Population-genetic properties of differentiated human copy-number polymorphisms. Am. J. Hum. Genet. 2011;88:317–332. doi: 10.1016/j.ajhg.2011.02.004. Medline doi:10.1016/j.ajhg.2011.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Locke DP, Sharp AJ, McCarroll SA, McGrath SD, Newman TL, Cheng Z, Schwartz S, Albertson DG, Pinkel D, Altshuler DM, Eichler EE. Linkage disequilibrium and heritability of copy-number polymorphisms within duplicated regions of the human genome. Am. J. Hum. Genet. 2006;79:275–290. doi: 10.1086/505653. Medline doi:10.1086/505653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jeffreys AJ, Wilson V, Thein SL. Hypervariable ‘minisatellite’ regions in human DNA. Nature. 1985;314:67–73. doi: 10.1038/314067a0. Medline doi:10.1038/314067a0. [DOI] [PubMed] [Google Scholar]
- 46.Duggan AT, Evans B, Friedlaender FR, Friedlaender JS, Koki G, Merriwether DA, Kayser M, Stoneking M. Maternal history of Oceania from complete mtDNA genomes: Contrasting ancient diversity with recent homogenization due to the Austronesian expansion. Am. J. Hum. Genet. 2014;94:721–733. doi: 10.1016/j.ajhg.2014.03.014. Medline doi:10.1016/j.ajhg.2014.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Davis JM, Searles Quick VB, Sikela JM. Replicated linear association between DUF1220 copy number and severity of social impairment in autism. Hum. Genet. 2015;134:569–575. doi: 10.1007/s00439-015-1537-6. Medline doi:10.1007/s00439-015-1537-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Koscinski I, Elinati E, Fossard C, Redin C, Muller J, Velez de la Calle J, Schmitt F, Ben Khelifa M, Ray PF, Kilani Z, Barratt CL, Viville S. DPY19L2 deletion as a major cause of globozoospermia. Am. J. Hum. Genet. 2011;88:344–350. doi: 10.1016/j.ajhg.2011.01.018. Medline. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Refojo D, Schweizer M, Kuehne C, Ehrenberg S, Thoeringer C, Vogl AM, Dedic N, Schumacher M, von Wolff G, Avrabos C, Touma C, Engblom D, Schütz G, Nave KA, Eder M, Wotjak CT, Sillaber I, Holsboer F, Wurst W, Deussing JM. Glutamatergic and dopaminergic neurons mediate anxiogenic and anxiolytic effects of CRHR1. Science. 2011;333:1903–1907. doi: 10.1126/science.1202107. Medline. [DOI] [PubMed] [Google Scholar]
- 50.McCarroll SA, Huett A, Kuballa P, Chilewski SD, Landry A, Goyette P, Zody MC, Hall JL, Brant SR, Cho JH, Duerr RH, Silverberg MS, Taylor KD, Rioux JD, Altshuler D, Daly MJ, Xavier RJ. Deletion polymorphism upstream of IRGM associated with altered IRGM expression and Crohn’s disease. Nat. Genet. 2008;40:1107–1112. doi: 10.1038/ng.215. Medline doi:10.1038/ng.215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Radoshitzky SR, Abraham J, Spiropoulou CF, Kuhn JH, Nguyen D, Li W, Nagel J, Schmidt PJ, Nunberg JH, Andrews NC, Farzan M, Choe H. Transferrin receptor 1 is a cellular receptor for New World haemorrhagic fever arenaviruses. Nature. 2007;446:92–96. doi: 10.1038/nature05539. Medline doi:10.1038/nature05539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rosenberg NA, Pritchard JK, Weber JL, Cann HM, Kidd KK, Zhivotovsky LA, Feldman MW. Genetic structure of human populations. Science. 2002;298:2381–2385. doi: 10.1126/science.1078311. Medline doi:10.1126/science.1078311. [DOI] [PubMed] [Google Scholar]
- 53.Coe BP, Witherspoon K, Rosenfeld JA, van Bon BW, Vulto-van Silfhout AT, Bosco P, Friend KL, Baker C, Buono S, Vissers LE, Schuurs-Hoeijmakers JH, Hoischen A, Pfundt R, Krumm N, Carvill GL, Li D, Amaral D, Brown N, Lockhart PJ, Scheffer IE, Alberti A, Shaw M, Pettinato R, Tervo R, de Leeuw N, Reijnders MR, Torchia BS, Peeters H, O’Roak BJ, Fichera M, Hehir-Kwa JY, Shendure J, Mefford HC, Haan E, Gécz J, de Vries BB, Romano C, Eichler EE. Refining analyses of copy number variation identifies specific genes associated with developmental delay. Nat. Genet. 2014;46:1063–1071. doi: 10.1038/ng.3092. Medline doi:10.1038/ng.3092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sudmant PH, Kitzman JO, Antonacci F, Alkan C, Malig M, Tsalenko A, Sampas N, Bruhn L, Shendure J, Eichler EE. 1000 Genomes Project, Diversity of human copy number variation and multicopy genes. Science. 2010;330:641–646. doi: 10.1126/science.1197005. Medline doi:10.1126/science.1197005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lazaridis I, Patterson N, Mittnik A, Renaud G, Mallick S, Kirsanow K, Sudmant PH, Schraiber JG, Castellano S, Lipson M, Berger B, Economou C, Bollongino R, Fu Q, Bos KI, Nordenfelt S, Li H, de Filippo C, Prüfer K, Sawyer S, Posth C, Haak W, Hallgren F, Fornander E, Rohland N, Delsate D, Francken M, Guinet JM, Wahl J, Ayodo G, Babiker HA, Bailliet G, Balanovska E, Balanovsky O, Barrantes R, Bedoya G, Ben-Ami H, Bene J, Berrada F, Bravi CM, Brisighelli F, Busby GB, Cali F, Churnosov M, Cole DE, Corach D, Damba L, van Driem G, Dryomov S, Dugoujon JM, Fedorova SA, Gallego Romero I, Gubina M, Hammer M, Henn BM, Hervig T, Hodoglugil U, Jha AR, Karachanak-Yankova S, Khusainova R, Khusnutdinova E, Kittles R, Kivisild T, Klitz W, Kučinskas V, Kushniarevich A, Laredj L, Litvinov S, Loukidis T, Mahley RW, Melegh B, Metspalu E, Molina J, Mountain J, Näkkäläjärvi K, Nesheva D, Nyambo T, Osipova L, Parik J, Platonov F, Posukh O, Romano V, Rothhammer F, Rudan I, Ruizbakiev R, Sahakyan H, Sajantila A, Salas A, Starikovskaya EB, Tarekegn A, Toncheva D, Turdikulova S, Uktveryte I, Utevska O, Vasquez R, Villena M, Voevoda M, Winkler CA, Yepiskoposyan L, Zalloua P, Zemunik T, Cooper A, Capelli C, Thomas MG, Ruiz-Linares A, Tishkoff SA, Singh L, Thangaraj K, Villems R, Comas D, Sukernik R, Metspalu M, Meyer M, Eichler EE, Burger J, Slatkin M, Pääbo S, Kelso J, Reich D, Krause J. Ancient human genomes suggest three ancestral populations for present-day Europeans. Nature. 2014;513:409–413. doi: 10.1038/nature13673. Medline doi:10.1038/nature13673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Fu Q, Li H, Moorjani P, Jay F, Slepchenko SM, Bondarev AA, Johnson PL, Aximu-Petri A, Prüfer K, de Filippo C, Meyer M, Zwyns N, Salazar-García DC, Kuzmin YV, Keates SG, Kosintsev PA, Razhev DI, Richards MP, Peristov NV, Lachmann M, Douka K, Higham TF, Slatkin M, Hublin JJ, Reich D, Kelso J, Viola TB, Pääbo S. Genome sequence of a 45,000-year-old modern human from western Siberia. Nature. 2014;514:445–449. doi: 10.1038/nature13810. Medline doi:10.1038/nature13810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zook JM, Chapman B, Wang J, Mittelman D, Hofmann O, Hide W, Salit M. Integrating human sequence data sets provides a resource of benchmark SNP and indel genotype calls. Nat. Biotechnol. 2014;32:246–251. doi: 10.1038/nbt.2835. Medline doi:10.1038/nbt.2835. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.