Abstract
Faithful chromosome segregation during mitosis is essential for genome integrity and is mediated by the bi-oriented attachment of replicated chromosomes to spindle microtubules through kinetochores. Errors in kinetochore–microtubule (k–MT) attachment that could cause chromosome mis-segregation are frequent and are corrected by the dynamic turnover of k–MT attachments. Thus, regulating the rate of spindle microtubule attachment and detachment to kinetochores is crucial for mitotic fidelity and is frequently disrupted in cancer cells displaying chromosomal instability. A model based on homeostatic principles involving receptors, a core control network, effectors and feedback control may explain the precise regulation of k–MT attachment stability during mitotic progression to ensure error-free mitosis.
Accurate chromosome segregation during cell division ensures that each daughter cell inherits a complete and identical copy of the genome, which is essential for cell and organismal viability. Chromosome missegregation leads to aneuploidy, a cellular state defined by an abnormal number of chromosomes. Aneuploidy causes human conditions such as Down syndrome and is associated with cancer, as illustrated by the observation that most solid tumours exhibit aneuploid karyotypes1–5.
Faithful chromosome segregation is ensured by the bi-oriented (amphitelic) attachment of chromosomes to the spindle through the end-on attachment of microtubules to kinetochores6 (FIG. 1). Replicated chromosomes have two kinetochores and bi-orientation is achieved when one kinetochore binds microtubules oriented towards one spindle pole and the other kinetochore binds microtubules oriented towards the opposite spindle pole. The initial capture of microtubules by kinetochores is asynchronous and stochastic7–10. Consequently, kinetochore–microtubule (k–MT) attachments that are not bi-oriented frequently form during prometaphase. Some of these are normal transient attachments that eventually lead to the formation of bi-oriented attachments by anaphase onset. Such normal intermediates include monotelic attachments (in which only one kinetochore binds to spindle microtubules) and lateral attachments (in which kinetochores bind to the side wall of a microtubule); lateral attachments have a major role in chromosome alignment at the spindle equator10–14. However, erroneous k–MT attachments can also form, which include merotelic attachments (in which the same kinetochore binds to microtubules oriented towards both spindle poles) and syntelic attachments (in which microtubules from the same spindle pole bind to both kinetochores). These erroneous attachments must be converted to bi-oriented attachments to ensure faithful chromosome segregation1,10,15 (FIG. 1). The number of these errors is determined by both their rate of formation and their rate of correction. The efficiency of correction depends on the rate of turnover of kinetochore microtubules (defined by the attachment and detachment of microtubules from kinetochores) because erroneous k–MT attachments must be released from the kinetochore to enable the formation of new, correct attachments. As microtubule capture is stochastic, the error correction rate must be sufficient to correct initial errors and exceed the rate of new error formation15.
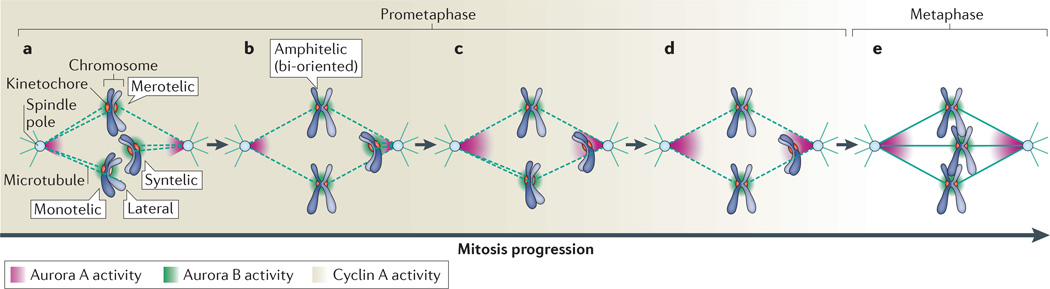
Figure 1. Kinetochore–microtubule attachments in mitosis.
Different types of kinetochore–microtubule (k–MT) attachments occur in prometaphase (a–d). These include transient intermediates such as monotelic attachments (in which only one of the sister kinetochores is attached to microtubules from one spindle pole) and lateral attachments (in which kinetochores are bound to the side wall of microtubules). In addition, errors in attachment exist, including syntelic attachments (in which both sister kinetochores are attached to microtubules from the same spindle pole) and merotelic attachments (in which a single kinetochore is attached to microtubules from both spindle poles). As cells progress through mitosis, the erroneous attachments are corrected, leading to end-on, bi-oriented attachments, in which sister kinetochores are attached to microtubules from opposite spindle poles to support faithful chromosome segregation. A core control network regulates the stability of k–MT attachments to promote efficient error correction and ensure faithful chromosome segregation. Note that, for simplicity, only cyclin A, Aurora A kinase and Aurora B kinase — which are key components of the core control network — are shown in the figure. Cyclin A forms a temporal gradient as its abundance declines during prometaphase (a–d), whereas Aurora A and Aurora B kinases form spatial gradients at spindle poles and at centromeres, respectively. Correction of syntely involves the recognition and targeted destabilization of k–MT attachments through the combined activities of Aurora A and Aurora B kinases as chromosomes are pulled towards the spindle poles (a-d). The release of microtubules from kinetochores permits the chromosome to move to the spindle midzone through lateral k–MT attachments to re-establish bi-oriented attachments (c,d). Correction of merotely involves the indiscriminate destabilization of k–MT attachments on aligned chromosomes during prometaphase (a to b, c to d). The high detachment rate of k–MT attachments in prometaphase (dashed lines) that is ensured by cyclin A activity combines with the back-to-back geometry of sister kinetochores to facilitate bi-oriented attachments. In metaphase (e), k–MT attachments switch to more stable attachments (solid lines) as cyclin A levels fall below a critical threshold.
Thus, mitotic fidelity relies on the dynamic association and dissociation rates of microtubules from kinetochores, and the combination of these two rates determines k–MT stability. Different models have been proposed for how these dynamics are regulated during mitosis. One model posits that tension generated as a consequence of the bi-oriented attachment of chromosomes to spindle microtubules determines the stability of k–MT attachments and the efficiency of error correction. However, new data suggest that a refined model that relies on the principles of homeostatic control more accurately describes how the stability of k–MT attachments is regulated during mitosis to ensure efficient error correction. In this Opinion article, we discuss the data that support a homeostatic model and the implications of this model for efficient error correction during mitosis to preserve genome integrity.
Importance of k–MT dynamics
Detachment of microtubules from kinetochores is likely to be the rate-limiting step in correcting erroneous attachments16,17. As microtubules detach from kinetochores, the opportunity arises for new, correctly oriented attachments to be created, thereby correcting previously formed erroneous attachments. Thus, unstable k–MT attachments, in which microtubules detach frequently, should improve the efficiency of error correction.
This relationship between k–MT attachment stability and chromosome segregation fidelity was highlighted through studies of chromosomal instability (CIN) in human cancer cells. CIN results from the persistence of whole-chromosome mis-segregation that leads to random losses and/or gains of whole chromosomes18–20. By measuring the stability of k–MT attachments in live cells using fluorescence dissipation after photoactivation21, it was shown that cells with CIN have hyperstable k–MT attachments relative to chromosomally stable diploid cells22,23. These hyperstable k–MT attachments reduce the efficiency of error correction20, leading to the persistence of erroneous attachments into anaphase and increased chromosome mis-segregation rates19. Consistent with these observations, targeted destabilization of k–MT attachments restored faithful chromosome segregation to cancer cells with CIN23–25, whereas increasing k–MT attachment stability in normal cells decreased the efficiency of error correction and elevated the frequency of chromosome mis-segregation22,24. Technical challenges have precluded the direct measurement of k–MT turnover in primary tumours; however, primary tumour cells display lagging chromosomes at anaphase and heterogeneous karyotypes similar to cancer cell lines in which k–MT attachments are hyperstable, suggesting that CIN in primary tumours may be dependent on k–MT attachment stability26,27.
Importantly, there is an upper limit to the k–MT detachment rate, over which the detachment rate becomes detrimental rather than beneficial, because the attachments must be sufficiently stable to generate and maintain chromosome alignment at the metaphase plate and maintain a sufficient number of microtubules bound to kinetochores to satisfy the spindle assembly checkpoint (SAC). The SAC is a signalling network that ensures the proper timing of mitosis by eliciting a signal from kinetochores to prevent chromosome segregation at anaphase until all chromosomes are attached to spindle microtubules. Therefore, k–MT attachment stability is precisely regulated to be maintained within a narrow range, in which the SAC is satisfied but still allowing for error correction28.
Control of k–MT attachment stability
There has been substantial research aimed at understanding the processes that facilitate the bi-orientation of k–MT attachments before anaphase onset. Recent data suggest that the stability of k–MT attachments is regulated by multiple pathways.
Centromere tension controls k–MT stability
It was proposed that bi-oriented k–MT attachments are preferentially stable relative to other k–MT attachments and that tension provides a mechanism to discriminate between these two states17. This concept was expanded through molecular studies based on the activity of Aurora B kinase, which is the enzymatic component of a conserved chromosome passenger complex that localizes to inner centromeres29,30. Aurora B kinase activity, which destabilizes k–MT attachments to enhance error correction31–33, is highest at the inner centromere and progressively decreases towards the outer kinetochore34,35. Tension is generated across the centromere in bi-oriented attachments because sister kinetochores are being pulled towards opposite spindle poles. This tension increases the physical distance between the inner centromere and kinetochore. Therefore, as kinetochore substrates are pulled away from Aurora B kinase residing at the inner centromere, these substrates display reduced Aurora B kinase-dependent phosphorylation and are also subject to more frequent dephosphorylation by phosphatases localized to kinetochores. By contrast, erroneous k–MT attachments — notably, syntelic attachments — fail to generate extensive centromere tension and kinetochore substrates experience more phosphorylation owing to their closer physical proximity to Aurora B kinase36. Importantly, in this tension-dependent model for regulating k–MT attachment stability, each chromosome acts autonomously and independent of all other chromosomes.
However, recent data demonstrate that k–MT attachments remain relatively unstable on all chromosomes in prometaphase, including bi-orientated chromosomes, and that the physical location of Aurora B kinase to inner centromeres is not essential for efficient error correction24,37,38. Furthermore, changes in k–MT attachment stability are regulated in a coordinated manner among all chromosomes during phase transitions in mitosis21,24.
These data suggest that the regulation of k–MT attachment stability that is crucial to error correction and mitotic fidelity cannot be explained by models that rely on centromere tension alone and that such models are overly deterministic. For example, tension-dependent models posit that k– MT attachments on stretched centromeres should always be more stable relative to those on unstretched centromeres. However, live cell imaging demonstrates that inter-kinetochore distances vary dramatically as bi-oriented chromosomes undergo oscillatory motions between spindle poles39,40. Moreover, there is no evidence to indicate that centromere ‘breathing’ associated with these chromosome oscillations alters k–MT attachment stability as centromere distance transiently lengthens and shortens.
Homeostatic control of k–MT stability
An expanded model of k–MT attachment regulation is that k–MT attachment stability is governed by input from multiple sources in addition to centromere tension, including cell cycle regulators, and that feedback networks integrate these inputs to adjust k–MT attachment stability to different set points. This model describes a homeostatic control system that uses feedback networks to precisely adjust k–MT attachment stability during each specific stage of mitosis.
Homeostatic systems require three elements: a receptor that monitors and responds to the environmental conditions; a core control network that sets the appropriate response based on input from the receptor and engages with effectors to execute changes; and negative feedback pathways to modulate the core control network as well as effectors to adjust the system. Accordingly, we propose the following: microtubule-binding proteins of the kinetochore serve as the primary receptor that responds to microtubule attachment stability; the core control network is composed of proteins of the SAC41, cyclin–cyclin-dependent kinase (CDK) complexes, and Aurora and polo-like kinases (PLKs)42, and the effectors are microtubule stabilizers and destabilizers at kinetochores; and the protein phosphatases PP1 and PP2A provide negative feedback to the core control centre (FIG. 2).
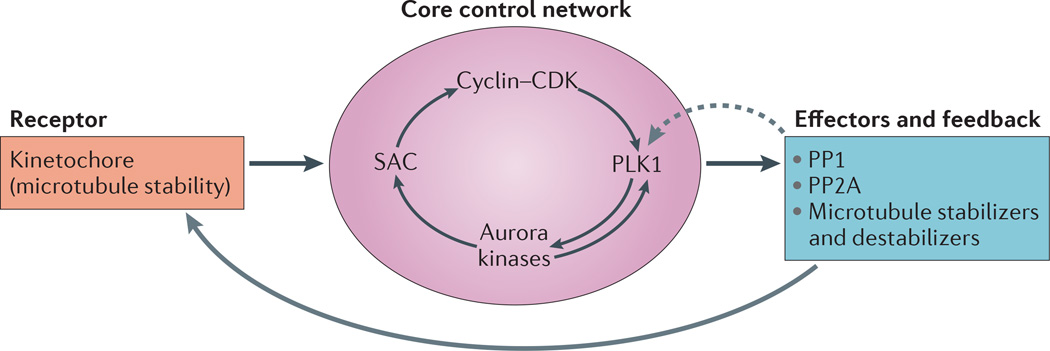
Figure 2. Homeostatic control circuit for regulating kinetochore–microtubule attachment stability in mitosis.
A receptor, a core control network, effector modules and feedback regulatory mechanisms (grey arrows) of a homeostatic control system regulate kinetochore–microtubule (k–MT) attachment stability. Conserved kinetochore proteins directly bind microtubules, thus forming the receptor. These probably include the proteins of the KMN network (BOX 1). Kinetochore proteins respond to microtubule attachment stability to send signals to the interactive core control network, which is composed of the spindle assembly checkpoint (SAC) and cyclin–cyclin-dependent kinase (CDK) complexes, polo-like kinase 1 (PLK1) and Aurora kinases (and possibly others, including the inner centromere protein shugoshin (SGO1) and haspin kinase). Arrows inside the core control network reflect some of the known functional interactions among these components. This network integrates input from the cell cycle regulatory machinery and the microtubule occupancy status of kinetochores to regulate the activity of effector molecules to increase or decrease the stability of k–MT attachments accordingly (solid grey arrow). The system is regulated by negative feedback from protein phosphatases PP1 and PP2A (dashed grey arrow) to enable control in the context of changing environmental conditions; that is, mitotic phase transitions.
Evidence for a homeostatic model
Quantitative measurement of k–MT attachment stability in live cells reveals a broad spectrum of proteins encompassing all three elements that are required for a homeostatic system to regulate the stability of k–MT attachments (TABLE 1). For simplicity, we have placed each protein in only one category, but we acknowledge that some proteins may have dual roles and either stabilize or destabilize k–MT attachments depending on the stage of mitosis and/or the state of protein phosphorylation43–45.
Table 1.
Modifiers of kinetochore–microtubule attachment stability in mitosis
| Protein | Localization | Activity | Refs |
|---|---|---|---|
| Destabilizers | |||
| Aurora B | Centromere | Kinase | 32 |
| CENPE | Outer kinetochore | Chromosome alignment | 43 |
| CLASP1 | Outer kinetochore | Non-motor microtubule-associated protein | 43 |
| CLASP2 | Outer kinetochore | Microtubule plus-end tracking protein | 44 |
| Cyclin A | Cytoplasm | Cyclin | 24 |
| KIF2B | Outer kinetochore | Error correction | 23 |
| MCAK | Centromere | Error correction | 23 |
| SGO1 | Centromere | Cohesion | 81 |
| Stabilizers | |||
| APC | Cytoplasm | Non-motor microtubule-associated protein | 22 |
| Astrin | Outer kinetochore | Non-motor microtubule-associated protein | 45 |
| BUB3 | Outer kinetochore | Spindle assembly checkpoint | 82 |
| BUBR1 | Outer kinetochore | Spindle assembly checkpoint | 83 |
| CDC20 | Outer kinetochore | Spindle assembly checkpoint | 83 |
| CENPH | Inner kinetochore | Centromere-associated protein | 84 |
| CENPL | Inner kinetochore | Centromere-associated protein | 85 |
| HEC1 (amino terminus) | Outer kinetochore | KMN network | 46 |
| HURP | Spindle | Microtubule-associated protein | 86 |
| KIF18A | Spindle | Chromosome alignment | 45 |
| MAD2 | Outer kinetochore | Spindle assembly checkpoint | 83 |
| MIS13 | Outer kinetochore | Microtubule binding | 87 |
| PLK1 | Centromere | Kinase | 52 |
| SKAP | Outer kinetochore | Kinetochore-associated protein | 87 |
| No effect | |||
| EG5 | Spindle | Plus-end directed kinesin | 24 |
| MAD1 | Outer kinetochore | Spindle assembly checkpoint | 83 |
| Loss of kinetochore–microtubule attachments | |||
| HEC1 | Outer kinetochore | KMN network | 88 |
| HDAC3 | Spindle | Histone deacetylase | 89 |
| NUF2 | Outer kinetochore | KMN network | 88* |
| SKA1 and SKA3 | Outer kinetochore | Microtubule-associated protein | 90 |
L.K., unpublished observations.
APC, adenomatous polyposis coli; BUB3, BUB3 mitotic checkpoint protein; BUBR1, BUB1-related kinase 1; CDC20, cell division cycle 20; CENP, centromere protein; CLASP, cytoplasmic linker-associated protein; EG5, also known as KIF11; HDAC3, histone deacetylase 3; HURP, hepatoma upregulated protein (also known as DAP5); KIF, kinesin family member; KMN, kinetochore null protein 1 (KNL1)–mis-segregation 12 (MIS12)–nuclear division cycle 80 (NDC80); MAD, mitotic spindle checkpoint protein; MCAK, mitotic centromere-associated kinesin; NUF2, nuclear filament-containing protein; PLK1, polo-like kinase 1; SGO1, shugoshin; SKA, spindle- and kinetochore-associated complex subunit; SKAP, SRC kinase-associated phosphoprotein.
The conserved kinetochore machinery referred to as the KMN — kinetochore null protein 1 (KNL1)–mis-segregation 12 (MIS12)–nuclear division cycle 80 (NDC80) — network is essential for the attachment of microtubule plus ends to kinetochores and is appropriate to act as a receptor for k–MT attachments46,47 (BOX 1). Loss-of-function experiments demonstrate that end-on k–MT attachments are almost eliminated without this conserved complex, indicating that it provides an all-or-nothing attachment for microtubule end binding. Evidence also shows that phosphorylation can modify the microtubule-binding affinity of components of this network36,38,48, suggesting that the role of the KMN network in binding microtubules can be modulated in response to environmental cues.
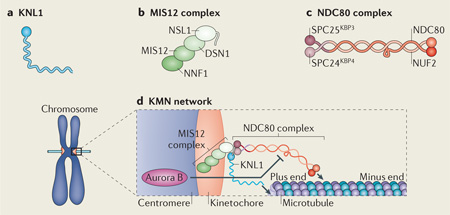
Box 1. The KMN network.
The KMN network is an association of conserved kinetochore protein complexes. It is composed of kinetochore null protein 1 (KNL1; see the figure, part a), the mis-segregation 12 (MIS12) complex (see the figure, part b) and the nuclear division cycle 80 (NDC80) complex (see the figure, part c). Multiple subunits from different complexes in the KMN network bind microtubules directly (as indicated by the arrows; see the figure, part d) and some components are direct targets of Aurora B kinase phosphorylation as indicated (DSN1 and KNL1). Phosphorylation of DSN1 and KNL1 by Aurora B reduces the microtubule-binding affinity of the KMN network in vitro. In the homeostatic model discussed in this article, the KMN network provides the essential function of binding to microtubule ends and signals to the core control network to ensure that the stability of the kinetochore–microtubule attachments is subsequently properly regulated. NUF2, nuclear filament-containing protein; SPC, spindle pole component.
In addition to this direct microtubule-binding machinery, core control and effector proteins promote the stabilization or destabilization of k–MT attachments to ensure that k–MT attachment stability remains within a defined range. The core control proteins include the SAC proteins and the protein kinases PLK1, Aurora A and B kinases, and cyclin–CDK complexes (and possibly others, including the inner centromere protein shugoshin (SGO1) and haspin kinase). This core control network regulates the function of the KMN machinery and the activity of effector proteins. The effector proteins are not part of the KMN machinery and include k–MT stabilizing and destabilizing proteins — for example, astrin and mitotic centromere-associated kinesin (MCAK; also known as KIF2C) — and the phosphatases PP1 and PP2A, which provide negative feedback to the core control network (FIG. 2).
The core control network
Here, we discuss the mechanistic details of how the SAC, PLK1, Aurora A and B kinases, and cyclin–CDK complexes act as a core control network to regulate k–MT attachment stability.
PLK1
One example of a core control protein is PLK1 (FIG. 3). PLK1 localizes to kinetochores during prometaphase and is most abundant on unaligned chromosomes49,50. Paradoxically, PLK1 stimulates the activity of both k–MT stabilizers and destabilizers at kinetochores. PLK1 activity is necessary for the initial formation of stable k–MT attachments, probably by phosphorylating the k–MT stabilizing protein BUB1-related kinase 1 (BUBR1; also known as BUB1B)50–53. Conversely, PLK1 phosphorylates and stimulates the microtubule depolymerase activity of kinesin family member 2B (KIF2B), which localizes to kinetochores in prometaphase and is required to destabilize k–MT attachments and efficiently correct errors54,55. Therefore, PLK1 functions to maintain k–MT attachment stability within a range that enables microtubules to bind to kinetochores but to remain sufficiently unstable to facilitate error correction.
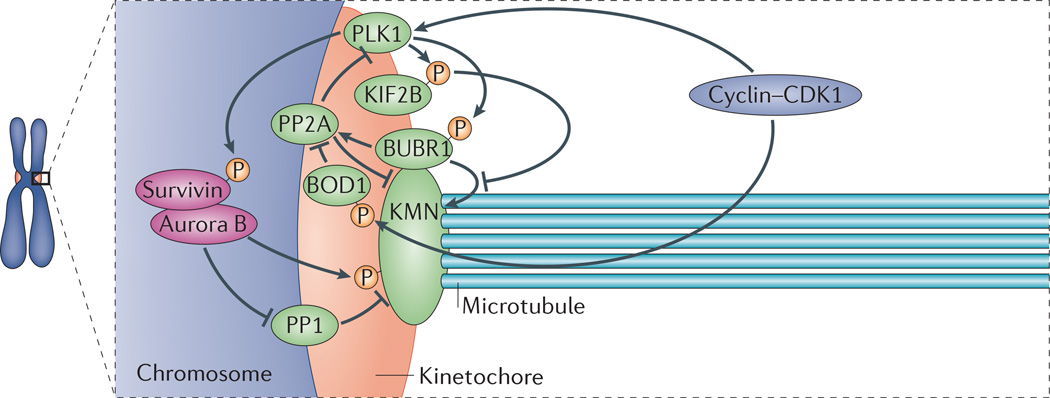
Figure 3. The network of regulatory components at kinetochores.
The complexity of the signalling pathways acting to regulate kinetochore–microtubule (k–MT) attachment stability is displayed in a nonspecific stage of mitosis (not necessarily prometaphase or metaphase). The KMN network (BOX 1) provides the primary microtubule-binding element in the kinetochore. The core control network — which is composed of polo-like kinase 1 (PLK1), Aurora B kinase and cyclin–cyclin-dependent kinases (CDKs) (and possibly other components, including the inner centromere protein shugoshin (SGO1) and haspin kinase) — acts to regulate the stability of k–MT attachments. The phosphorylation status of their substrates (such as kinesin family member 2B (KIF2B), BUB1-related kinase 1 (BUBR1), bi-orientation of chromosomes in cell division 1 (BOD1) and survivin) is determined by feedback from the phosphatases PP1 and PP2A. The combined activities of the core control proteins and phosphatases determines the relative activities of effector proteins such as the k–MT stabilizer BUBR1 and the k–MT destabilizer KIF2B, which collectively modulate k–MT attachment stability.
It remains unknown how PLK1 coordinates the regulation of these opposing activities. Temporal control of PLK1 activity could be crucial because artificially tethering PLK1 to kinetochores so that its levels remain unduly high into metaphase inappropriately stabilizes k–MT attachments, and this increases the frequency of merotelic attachment errors and causes lagging chromosomes in anaphase52. Alternatively, substrate access at kinetochores may determine whether the effect of PLK1 is to stabilize or destabilize k–MT attachments. Importantly, a fluorescence resonance energy transfer (FRET)-based biosensor for PLK1 activity suggests there is no gradient of substrate phosphorylation generated by PLK1 akin to the type of gradient that has been observed with Aurora B kinase52. Thus, the data obtained using biosensors indicate that PLK1-dependent phosphorylation of k–MT stabilizers and destabilizers is not directly controlled by centromere tension.
A key component of a homeostatic system is negative feedback to the core control network, and an important negative regulator of PLK1 activity is the protein phosphatase PP2A (FIG. 3). As mentioned above, BUBR1 is a target of PLK1 that serves to recruit protein phosphatase PP2A to kinetochores through binding of the B56 regulatory subunit of PP2A56,57. Depletion of the B56 regulatory subunit increases PLK1 levels at centromeres, suggesting that PP2A both dephosphorylates PLK1 substrates and regulates the targeting of PLK1 to kinetochores58. PP2A also dephosphorylates Aurora B kinase substrates at kinetochores.
Aurora B kinase
Aurora B kinase also has an essential and conserved role in destabilizing k–MT attachments to promote error correction, and participates in regulatory feedback loops as part of the core control network (FIG. 3). Aurora B kinase has numerous substrates in mitosis, including kinetochore proteins, and the extent of phosphorylation of kinetochore substrates varies according to distance from the inner centromere34,35. Aurora B is enriched at centromeres during early mitosis, including centromeres of mis-aligned chromosomes59. This enrichment is promoted by PLK1 activity, in part through phosphorylation of the chromosome passenger complex subunit survivin, creating a positive-feedback loop between the two kinases56,60,61. This feedback loop includes PLK1 priming phosphorylation on some substrates for Aurora B kinase and Aurora B kinase priming phosphorylation on some substrates for PLK1 (REFS 56,62). The feedback loop between PLK1 and Aurora B kinase provides an example of a regulatory loop occurring within the core control network to modulate k–MT stability.
Aurora B activity also directly influences the microtubule receptors at the kinetochore. For example, the microtubule-binding affinity of the KMN network proteins DSN1 and KNL1 (BOX 1) is reduced by Aurora B kinase phosphorylation36, and the protein phosphatase PP2A has been shown to counteract this activity through targeting via the B56 regulatory subunit58. This process provides a direct and reversible mechanism to tune the microtubule-binding affinity of these kinetochore proteins.
Finally, it has been shown that the protein phosphatase PP1 has a major role in dephosphorylating Aurora B kinase substrates in mitosis, and PP1 abundance at kinetochores is inversely proportional to Aurora B activity, thereby creating a negative-feedback loop63–65. The KMN network protein KNL1 (BOX 1) serves as the docking site for PP1 at kinetochores65. Aurora B kinase phosphorylates KNL1 and inhibits PP1 binding to KNL1, which prevents PP1 from dephosphorylating Aurora B kinase substrates. In this context, the tension generated on bi-oriented chromosomes stretches KNL1 beyond the reach of Aurora B kinase. Consequently, PP1 is recruited to kinetochores to antagonize Aurora B kinase activity and to stabilize k–MT attachments. This creates a negative-feedback loop between the kinetochore (that is, the receptor) and the core control network to regulate Aurora B kinase activity and influence the stability of microtubule binding.
CDKs
Another core control component controlling k–MT attachment stability involves cyclin–CDK activities (FIG. 3). There is a coordinated switch of k–MT attachment stability on all chromosomes during the transitions from prometaphase to metaphase and from metaphase to anaphase. In prometaphase, k– MT attachment stability on bi-oriented chromosomes is similar between a cell with all chromosomes aligned at the metaphase plate except for one and an early prometaphase cell with many unaligned chromosomes24. These data demonstrate that k–MT attachments on bi-oriented chromosomes in prometaphase are relatively unstable regardless of the duration of prometaphase. Cyclin A–CDK activity is responsible for maintaining unstable k–MT attachments on bi-oriented chromosomes during prometaphase. This function is important in promoting the correction of k–MT attachment errors as demonstrated by an increased rate of segregation errors in anaphase in cyclin A-depleted cells24. Moreover, k–MT attachments become even more stable as cells transit from metaphase to anaphase21.
The transition from metaphase to anaphase coincides with the loss of cyclin B– CDK activity, and the destruction of cyclin B might be responsible for the increase in k–MT attachment stability at this transition. Data have shown varied responses of k–MT attachments to cyclin B levels. For example, the expression of a non-degradable cyclin B in Drosophila melanogaster embryos increases merotelic k–MT attachments, the depletion of cyclin B in mammalian cells increases lagging chromosomes at anaphase, and the prevention of cyclin B1 destruction at the time of sister chromatid disjunction destabilizes k–MT attachments in human cells66–68. These seemingly contradictory results may reflect alterations in the error correction or error formation rate that is dependent on the absence or presence of cyclin B. Cyclin B degradation coincides with an increase in k–MT attachment stability; therefore, premature degradation may lead to hyperstable k–MT attachments and a decrease in the error correction efficiency. By contrast, overexpression of non-degradable cyclin B in D. melanogaster embryos and mammalian cells causes inner centromere protein (INCENP) and Aurora B kinase to remain at kinetochores in anaphase, resulting in destabilized k–MT attachments and potentially increasing the rate of new error formation66,68.
Although the detailed molecular pathways that the cyclin–CDK complexes control to modulate k–MT stability are currently unknown, one potential pathway involves the core control protein PLK1. PP2A is a negative regulator of PLK1 that is antagonized by the bi-orientation of chromosomes in cell division 1 (BOD1)69,70(FIG. 3). Cells lacking BOD1 function display defects in efficiently establishing bi-oriented chromosomes on the mitotic spindle consistent with disruption of k–MT attachments. Evidence indicates that BOD1 is regulated by cyclin– CDK activity, creating a network for regulation of PLK1 activity at kinetochores that includes input from both upstream kinases and phosphatases. Additionally, cyclin–CDK complexes frequently provide priming phosphorylation to PLK1 substrates, providing important downstream regulation of PLK1 function.
The SAC
Cyclin A and cyclin B levels are differentially regulated by components of the SAC71. Degradation of cyclin A is initiated in early prometaphase and is regulated by competition with the mitotic checkpoint complex for binding to cell division cycle 20 (CDC20)71–73. Degradation of cyclin B is initiated once the SAC is satisfied at kinetochores and levels of the mitotic checkpoint complex diminish to permit recognition of cyclin B by the anaphase-promoting complex (APC/C; also known as the cyclosome)74. Thus, the switches that increase k–MT attachment stability as cells transit from prometaphase to metaphase and from metaphase to anaphase are sensitive to checkpoint signalling. Moreover, evidence indicates that intra-kinetochore stretch regulates the timely satisfaction of the SAC75,76. Thus, intra-kinetochore stretch may indirectly influence the stability of k–MT attachments through regulation of the quantities of cyclin A and/or cyclin B. Importantly, the signal emerging from checkpoint activity at individual kinetochores affects cyclin levels that subsequently diffuse throughout the cell to influence the stability of k–MT attachments on all chromosomes.
Integrated homeostatic model
The complexity of the interactions of the regulatory components within this signalling network (FIG. 3) fits into a broader homeostatic model for controlling the stability of k–MT attachments (FIG. 2). As cells transit through different phases of mitosis, the core control network integrates input from multiple sources to adjust the set point for the stability of k–MT attachments to ensure the efficient formation of bi-oriented attachments and facilitate robust error correction.
In this context, the core control network receives negative feedback from PP1 and PP2A in early prometaphase to promote the initial formation of k–MT attachments. However, the countervailing activity of microtubule destabilizers maintains the attachments in a relatively unstable state to promote the efficient correction of errors that are common in early phases of mitosis. Later in mitosis, as kinetochores are bound by a higher number of microtubules and all chromosomes become bi-oriented, increased centromere tension and reduced levels of cyclin A facilitate the rapid, switch-like transition to more stable k–MT attachments as cells enter metaphase.
Importantly, homeostatic systems are designed to regulate systems around specific set points that are far from extremes. Here, this is evident by the fact that kinetochore proteins are not fully saturated with respect to phosphorylation36,48,58,77. Moreover, inhibition of Aurora B kinase or expression of mutant versions of the kinetochore protein NDC80 lacking all the phosphorylation sites (that is, mimicking a saturated condition) disrupts feedback control, leading to extreme stabilization of k–MT attachments32,38,48.
Correcting k–MT attachment errors
Cells use a homeostatic system to execute correction of k–MT attachment errors. Principally, cells must correct both merotelic and syntelic attachments before anaphase onset to ensure faithful chromosome segregation and to preserve genome integrity. Cumulative data from analyses of these regulatory networks reveals that two (non-exclusive) strategies are used by cells to execute error correction.
Targeted k–MT destabilization
One strategy used by cells to correct k–MT attachment errors involves the detection and systematic destabilization of incorrectly oriented k–MT attachments. This may be the predominant strategy used by cells to correct syntelic attachments because the distortion of the centromere induced by syntely enables efficient recognition of erroneous attachments (FIG. 1). Moreover, syntelic chromosomes might move closer to their attached pole owing to the unbalanced force between the spindle poles. The centromere distortion created by syntely would disrupt the phosphorylation gradient generated by Aurora B kinase at centromeres and promote destabilization of k–MT attachments.
Also, Aurora A kinase is abundant at spindle poles and has been shown to phosphorylate the NDC80 component of the KMN network microtubule-binding machinery78. The combination of Aurora A and Aurora B kinase activities would push the core control network to an extreme position, leading to the destabilization of k–MT attachments on that chromosome, and perhaps leading to complete detachment of both kinetochores from spindle microtubules. The chromosome would then utilize lateral k–MT attachments to move to the spindle midzone to subsequently form bi-oriented attachments.
This model suggests that the core control network can act locally on individual chromosomes when the conditions require extreme changes in k–MT attachment stability, and indicates that Aurora A and Aurora B kinases cooperate to destabilize k–MT attachments on syntelic chromosomes near spindle poles. Moreover this model predicts that k–MT attachments on chromosomes located near spindle poles are exceptionally unstable. Unfortunately, technical limitations currently preclude the direct measurement of k–MT attachment stability when chromosomes are adjacent to spindle poles.
Indiscriminate k–MT turnover
The other strategy used by cells to correct k–MT attachment errors is to destabilize k–MT attachments without discriminating between microtubules that are oriented towards the correct pole and those oriented towards the incorrect pole. This probably has an important role in the correction of merotelic attachments that are not detected by the SAC and would rely on the geometric arrangement of sister kinetochores on either side of the centromeres79,80(FIG. 1). The back-to-back arrangement of sister kinetochores would favour microtubule attachment towards opposite spindle poles as microtubules undergo repeated rounds of detachment and attachment to kinetochores. Thus, unstable k–MT attachments would improve the efficiency of correction of merotely.
Consistent with this idea, it was recently demonstrated that k–MT attachments on bi-oriented chromosomes continue to undergo rapid turnover during prometaphase. This is governed by cyclin A abundance, which, by diffusive mechanisms, affects k–MT attachment stability on all chromosomes equally24. This subjugates the influence of other components of the core control network such as Aurora B kinase that respond to centromere tension on individual chromosomes. The dominance of cyclin A control of k–MT attachment stability on bi-oriented chromosomes during prometaphase creates conditions in which kinetochores are primed to switch to more stable attachments that are typical of metaphase once cyclin A is degraded. Once the influence of cyclin A falls below a critical threshold, centromere tension may become a dominant input into the core control network because all chromosomes would be bi-oriented.
The switch to increased k–MT attachment stability observed between prometaphase and metaphase represents a clear example of how the core control network can change the set point for k–MT stability. One implication of the rapid and irreversible stabilization of k–MT attachments as cells enter metaphase, owing to cyclin A destruction, is that it promotes microtubule occupancy at kinetochores to satisfy the spindle checkpoint and usher the transition to anaphase. Another prediction of this model is that there is no difference in the stability of k–MT attachments on merotelic kinetochores versus bi-oriented kinetochores. Testing this prediction will require technical advances to both reliably identify merotelic kinetochores in live cells and measure k– MT stability associated with individual kinetochores.
Conclusion
Biological systems use homeostatic control mechanisms because of their utility in maintaining system stability through responsiveness to environmental changes. The homeostatic model we describe here is designed to ensure high mitotic fidelity by tightly controlling the stability of k–MT attachments. However, this model for the regulation of k–MT attachment stability raises many questions and presents opportunities for additional experimentation. For example, the model predicts that systematic manipulation of the core control network should result in defined changes in the stability of k–MT attachments. It will therefore be important to develop strategies to induce quantitative changes to the level of activity of different core control components at defined times during mitosis to investigate the response in k–MT attachment stability. Moreover, the idea that the core control network may respond to the stability of k–MT attachments remains to be tested, and the underlying molecular mechanisms remain to be defined.
Acknowledgements
The authors thank members of the laboratory for their input and feedback on the manuscript. This work was supported by grants from the US National Institutes of Health (GM51542 to D.A.C and GM008704 to L.K.) and the American Cancer Society (PF-12-103-01-CCG to K.M.G.)
Footnotes
Competing interests statement
The authors declare no competing interests.
References
- 1.Cimini D, Degrassi F. Aneuploidy: a matter of bad connections. Trends Cell Biol. 2001;15:442–451. doi: 10.1016/j.tcb.2005.06.008. [DOI] [PubMed] [Google Scholar]
- 2.Cimini D. Merotelic kinetochore orientation, aneuploidy, and cancer. Biochim. Biophys. Acta. 2008;1786:32–40. doi: 10.1016/j.bbcan.2008.05.003. [DOI] [PubMed] [Google Scholar]
- 3.Weaver BA, Cleveland DW. Aneuploidy: instigator and inhibitor of tumorigenesis. Cancer Res. 2006;67:10103–10105. doi: 10.1158/0008-5472.CAN-07-2266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Siegel JJ, Amon A. New insights into the troubles of aneuploidy. Annu. Rev. Cell Devel. Biol. 2012;28:189–214. doi: 10.1146/annurev-cellbio-101011-155807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Duijf PH, Benezra R. The cancer biology of whole-chromosome instability. Oncogene. 2013;32:4727–4736. doi: 10.1038/onc.2012.616. [DOI] [PubMed] [Google Scholar]
- 6.Cheeseman I. The kinetochore. Cold Spring Harb. Perspect. Biol. 2014;6:a015826. doi: 10.1101/cshperspect.a015826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cimini D, Moree B, Canman JC, Salmon ED. Merotelic kinetochore orientation occurs frequently during early mitosis in mammalian tissue cells and error correction is achieved by two different mechanisms. J. Cell Sci. 2003;116:4213–4225. doi: 10.1242/jcs.00716. [DOI] [PubMed] [Google Scholar]
- 8.Salmon ED, Cimini D, Cameron LA, DeLuca JG. Merotelic kinetochores in mammalian tissue cells. Phil. Trans. R. Soc. 2005;360:553–568. doi: 10.1098/rstb.2004.1610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rieder CL. The formation, structure, and composition of the mammalian kinetochore and kinetochore fiber. Int. Rev. Cytol. 1982;79:1–58. doi: 10.1016/s0074-7696(08)61672-1. [DOI] [PubMed] [Google Scholar]
- 10.Magidson V, et al. The spatial arrangement of chromosomes during prometaphase facilitates spindle assembly. Cell. 2011;146:555–567. doi: 10.1016/j.cell.2011.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Maure JF, et al. The Ndc80 looop region facilitates formation of kinetochore attachment to the dynamic microtubule plus end. Curr. Biol. 2011;21:207–213. doi: 10.1016/j.cub.2010.12.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shrestha RL, Draviam VM. Lateral to end on conversion of chromosome-microtubule attachment requires kinesins CENP-E and MCAK. Curr. Biol. 2013;23:1514–1526. doi: 10.1016/j.cub.2013.06.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cai S, O’Connell CB, Khodjakov A, Walczak CE. Chromosome congression in the absence of kinetochore fibers. Nat. Cell Biol. 2009;11:832–838. doi: 10.1038/ncb1890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tanaka K, et al. Molecular mechanisms of kinetochore capture by spindle microtubules. Nature. 2005;434:987–994. doi: 10.1038/nature03483. [DOI] [PubMed] [Google Scholar]
- 15.Maiato H, DeLuca J, Salmon ED, Earnshaw WC. The dynamic kinetochore-microtubule interface. J. Cell Sci. 2004;117:5461–5477. doi: 10.1242/jcs.01536. [DOI] [PubMed] [Google Scholar]
- 16.Nicklas RB, Ward SC. Elements of error correction in mitosis: microtubule capture, release, and tension. J. Cell Biol. 1994;126:1241–1253. doi: 10.1083/jcb.126.5.1241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li S, Nicklas RB. Mitotic forces control a cell-cycle checkpoint. Nature. 1995;373:630–632. doi: 10.1038/373630a0. [DOI] [PubMed] [Google Scholar]
- 18.Lengauer C, Kinzler KW, Vogelstein B. Genetic instabilities in human cancers. Nature. 1998;396:643–649. doi: 10.1038/25292. [DOI] [PubMed] [Google Scholar]
- 19.Thompson SL, Compton DA. Examining the link between chromosomal instability and aneuploidy in human cells. J. Cell Biol. 2008;180:665–672. doi: 10.1083/jcb.200712029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Thompson SL, Bakhoum SF, Compton DA. Mechanisms of chromosomal instability. Curr. Biol. 2010;20:R285–R295. doi: 10.1016/j.cub.2010.01.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhai Y, Kronebusch PJ, Borisy GG. Kinetochore microtubule dynamics and the metaphase-anaphase transition. J. Cell Biol. 1995;131:721–734. doi: 10.1083/jcb.131.3.721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bakhoum SF, Genovese G, Compton DA. Deviant kinetochore microtubule dynamics underlie chromosomal instability. Curr. Biol. 2009;19:1937–1942. doi: 10.1016/j.cub.2009.09.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bakhoum SF, Thompson SL, Manning AL, Compton DA. Genome stability is ensured by temporal control of kinetochore-microtubule dynamics. Nat. Cell Biol. 2009;11:27–35. doi: 10.1038/ncb1809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kabeche L, Compton DA. Cyclin A regulates kinetochore microtubules to promote faithful chromosome segregation. Nature. 2013;502:110–113. doi: 10.1038/nature12507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stolz A, et al. The CHIF2 BRCA1 tumor suppressor pathway ensures chromosomal stability in human somatic cells. Nat. Cell Biol. 2010;12:492–499. doi: 10.1038/ncb2051. [DOI] [PubMed] [Google Scholar]
- 26.Bakhoum SF, Danilova OV, Kaur P, Levy NB, Compton DA. Chromosomal instability substantiates poor prognosis in patients with diffuse large B cell lymphoma. Clin. Cancer Res. 2011;17:7704–7711. doi: 10.1158/1078-0432.CCR-11-2049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hecht BK, et al. Cytogenetics of malignant gliomas: I. The autosomes with reference to rearrangements. Cancer Genet. Cytogenet. 1995;84:1–8. doi: 10.1016/0165-4608(95)00091-7. [DOI] [PubMed] [Google Scholar]
- 28.Bakhoum SF, Compton DA. Kinetochores and disease: keeping microtubule dynamics in check! Curr. Opin. Cell Biol. 2012;24:64–70. doi: 10.1016/j.ceb.2011.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Carmena M, Wheelock M, Funabiki H, Earnshaw WC. The chromosomal passenger complex (CPC): from easy rider to the godfather of mitosis. Nat. Rev. Mol. Cell. Biol. 2012;13:789–803. doi: 10.1038/nrm3474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vader G, Medema RH, Lens SM. The chromosomal passenger complex: guiding Aurora B through mitosis. J. Cell Biol. 2006;173:833–837. doi: 10.1083/jcb.200604032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lampson MA, Cheeseman IM. Sensing centromere tension: Aurora B and the regulation of kinetochore function. Trends Cell Biol. 2011;21:133–140. doi: 10.1016/j.tcb.2010.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cimini D, Wan X, Hirel CB, Salmon ED. Aurora kinase promotes turnover of kinetochore microtubules to reduce chromosome segregation errors. Curr. Biol. 2006;16:1711–1718. doi: 10.1016/j.cub.2006.07.022. [DOI] [PubMed] [Google Scholar]
- 33.Shannon KB, Salmon ED. Chromosome dynamics: new light on Aurora B kinase function. Curr. Biol. 2002;12:R458–R460. doi: 10.1016/s0960-9822(02)00945-4. [DOI] [PubMed] [Google Scholar]
- 34.Liu D, Vader G, Vromans MJ, Lampson MA, Lens SM. Sensing chromosome bi orientation by spatial separation of aurora B kinase from kinetochore substrates. Science. 2009;323:1350–1353. doi: 10.1126/science.1167000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang E, Ballister ER, Lampson MA. Aurora B dynamics at centromeres create a diffusion-based phosphorylation gradient. J. Cell Biol. 2011;194:539–549. doi: 10.1083/jcb.201103044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Welburn JP, et al. Aurora B phosphorylates spatially distinct targets to differentially regulate the kinetochore-microtubule interface. Mol. Cell. 2010;38:383–392. doi: 10.1016/j.molcel.2010.02.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Campbell CS, Desai A. Tension sensing by Aurora B kinase is independent of survivin-based centromere localization. Nature. 2013;497:118–121. doi: 10.1038/nature12057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.DeLuca KF, Lens SM, DeLuca JG. Temporal changes in Hec1 phosphorylation control kinetochore-microtubule attachment stability during mitosis. J. Cell Sci. 2011;124:622–634. doi: 10.1242/jcs.072629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Skibbens RV, Skeen VP, Salmon ED. Directional instability of kinetochore motility during chromosome congression and segregation in mitotic newt lung cells: a push-pull mechanism. J. Cell Biol. 1993;122:859–875. doi: 10.1083/jcb.122.4.859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jaqaman K, et al. Kinetochore alignment within the metaphase plate is regulated by centromere stiffness and microtubule depolymerases. J. Cell Biol. 2010;188:665–679. doi: 10.1083/jcb.200909005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lara-Gonzalez P, Westhorpe FG, Taylor SS. The spindle assembly checkpoint. Curr. Biol. 2012;22:R966–R980. doi: 10.1016/j.cub.2012.10.006. [DOI] [PubMed] [Google Scholar]
- 42.Bruinsma W, Baaijmakers JA, Medema RH. Switching Polo-like kinase-1 on and off in time and space. Trends Biochem. Sci. 2012;37:534–542. doi: 10.1016/j.tibs.2012.09.005. [DOI] [PubMed] [Google Scholar]
- 43.Maffini S, et al. Motor-independent targeting of CLASPs to kinetochores by CENP-E promotes microtubule turnover and poleward flux. Curr. Biol. 2009;19:1566–1572. doi: 10.1016/j.cub.2009.07.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Maia AR, et al. Cdk1 and Plk1 mediate a CLASP2 phospho-switch that stabilizes kinetochore-microtubule attachments. J. Cell Biol. 2012;199:285–301. doi: 10.1083/jcb.201203091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Manning AL, Bakhoum SF, Maffini S, Correia-Melo C, Maiato H, Compton DA. CLASP1, astrin and Kif2b form a molecular switch that regulates kinetochore-microtubule dynamics to promote mitotic progression and fidelity. EMBO J. 2010;29:3531–3543. doi: 10.1038/emboj.2010.230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.DeLuca JG, et al. Kinetochore microtubule dynamics and attachment stability are regulated by Hec1. Cell. 2006;127:969–982. doi: 10.1016/j.cell.2006.09.047. [DOI] [PubMed] [Google Scholar]
- 47.Cheeseman IM, Chappie JS, Wilson Kubalek EM, Desai A. The conserved KMN network constitutes the core microtubule-binding site of the kinetochore. Cell. 2006;127:983–997. doi: 10.1016/j.cell.2006.09.039. [DOI] [PubMed] [Google Scholar]
- 48.Zaytsev AV, Sundin LJ, DeLuca KF, Grishchuk EL, DeLuca JG. Accurate phosphoregulation of kinetochore-microtubule affinity requires unconstrained molecular interactions. J. Cell Biol. 2014;206:45–59. doi: 10.1083/jcb.201312107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ahonen LF, et al. Polo-like kinase 1 creates the tension-sensing 3F3/2 phosphoepitope and modulates the association of spindle-checkpoint proteins at kinetochores. Curr. Biol. 2005;15:1078–1089. doi: 10.1016/j.cub.2005.05.026. [DOI] [PubMed] [Google Scholar]
- 50.Lenart P, et al. The small-molecule inhibitor BI2536 reveals novel insights into mitotic roles of polo-like kinase 1. Curr. Biol. 2007;17:304–315. doi: 10.1016/j.cub.2006.12.046. [DOI] [PubMed] [Google Scholar]
- 51.Elowe S, Hummer S, Uldschmid A, Li X, Nigg EA. Tension-sensitive Plk1 phosphorylation on BubR1 regulates the stability of kinetochore microtubule interactions. Genes Dev. 2007;21:2205–2219. doi: 10.1101/gad.436007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Liu D, Davydenko O, Lampson MA. Polo-like kinase 1 regulates kinetocohore-microtubule dynamics and spindle checkpoint silencing. J. Cell Biol. 2012;198:491–499. doi: 10.1083/jcb.201205090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lampson MA, Kapoor M. The human mitotic checkpoint protein BubR1 regulates chromosome-spindle attachments. Nat. Cell Biol. 2005;7:93–98. doi: 10.1038/ncb1208. [DOI] [PubMed] [Google Scholar]
- 54.Manning AL, et al. The kinesin-13 proteins Kif2a, Kif2b, and Kif2c/MCAK have distinct roles during mitosis in human cells. Mol. Biol. Cell. 2007;18:2970–2979. doi: 10.1091/mbc.E07-02-0110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hood EA, Kettenbach AN, Gerber SA, Compton DA. Plk1 regulates the kinesin-13 protein Kif2b to promote faithful chromosome segregation. Mol. Biol. Cell. 2012;23:2264–2274. doi: 10.1091/mbc.E11-12-1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Carmena M, et al. The chromosomal passenger complex activated Polo kinase at centromeres. PLoS Biol. 2012;10:e1001250. doi: 10.1371/journal.pbio.1001250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kruse T, et al. Direct binding between BubR1 and B56-PP2A phosphatase complexes regulate mitotic progression. J. Cell Sci. 2013;126:1086–1092. doi: 10.1242/jcs.122481. [DOI] [PubMed] [Google Scholar]
- 58.Foley EA, Maldonado M, Kapoor TM. Formation of stable attachments between kinetochores and microtubules depends on the B56-PP2A phosphatase. Nat. Cell Biol. 2011;13:1265–1271. doi: 10.1038/ncb2327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Knowlton AL, Lan W, Stukenberg PT. Aurora B is enriched at merotelic attachment sites, where it regulates MCAK. Curr. Biol. 2006;16:1705–1710. doi: 10.1016/j.cub.2006.07.057. [DOI] [PubMed] [Google Scholar]
- 60.Chu Y, et al. Aurora B kinase activation requires survivin priming phosphorylation by PLK1. J. Mol. Cell. Biol. 2011;3:260–267. doi: 10.1093/jmcb/mjq037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Salimian KJ, et al. Feedback control in sensing chromosome biorientation by the Aurora B kinase. Curr. Biol. 2011;21:1158–1165. doi: 10.1016/j.cub.2011.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Goto H, et al. Complex formation of Plk1 and INCENP required for metaphase-anaphase transition. Nature Cell Biol. 2006;8:180–187. doi: 10.1038/ncb1350. [DOI] [PubMed] [Google Scholar]
- 63.Trinkle-Mulcahy L, et al. Time-lapse imaging reveals dynamic relocalization of PP1γ throughout the mammalian cell cycle. Mol. Biol. Cell. 2003;14:107–117. doi: 10.1091/mbc.E02-07-0376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.De Wulf P, Montani F, Visintin R. Protein phosphatases take the mitotic stage. Curr. Opin. Cell Biol. 2009;21:806–815. doi: 10.1016/j.ceb.2009.08.003. [DOI] [PubMed] [Google Scholar]
- 65.Liu D, et al. Regulated targeting of protein phosphatase 1 to the outer kinetochore by KNL1 opposes Aurora B kinase. J. Cell Biol. 2010;188:809–820. doi: 10.1083/jcb.201001006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Parry DH, Hickson GR, O’Farrell PH. Cyclin B destruction triggers changes in kinetochore behavior essential for successful anaphase. Curr. Biol. 2003;13:647–653. doi: 10.1016/s0960-9822(03)00242-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Soni DV, Sramkoski RM, Lam M, Stegan T, Jacobberger JW. Cyclin B1 is rate limiting but not essential for mitotic entry and progression in mammalian somatic cells. Cell Cycle. 2008;7:1285–1300. doi: 10.4161/cc.7.9.5711. [DOI] [PubMed] [Google Scholar]
- 68.Vazquez-Novelle MD, et al. Cdk1 inactivation terminates mitotic checkpoint surveillance and stabilizes kinetochore attachments in anaphase. Curr. Biol. 2014;24:638–645. doi: 10.1016/j.cub.2014.01.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Porter IM, et al. Bod1, a novel kinetochore protein required for chromosome biorientation. J. Cell Biol. 2007;179:187–197. doi: 10.1083/jcb.200704098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Porter IM, Schleicher K, Porter M, Swedlow JR. Bod1 regulates protein phosphatase 2A at mitotic kinetochores. Nat. Commun. 2013;4:2677. doi: 10.1038/ncomms3677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Collin P, Nashchenkina O, Walker R, Pines J. The spindle assembly checkpoint works like a rheostat rather than a toggle switch. Nat. Cell Biol. 2013;15:1378–1385. doi: 10.1038/ncb2855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.DiFiore B, Pines J. How cyclin A destruction escapes the spindle assembly checkpoint. J. Cell Biol. 2010;190:501–509. doi: 10.1083/jcb.201001083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.den Elzen N, Pines J. Cyclin A is destroyed in prometaphase and can delay chromosome alignment and anaphase. J. Cell Biol. 2001;153:121–136. doi: 10.1083/jcb.153.1.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Musacchio A, Salmon ED. The spindle-assembly checkpoint in space and time. Nat. Rev. Mol. Cell. Biol. 2007;8:379–393. doi: 10.1038/nrm2163. [DOI] [PubMed] [Google Scholar]
- 75.Maresca TJ, Salmon ED. Intrakinetochore stretch is associated with changes in kinetochore phosphorylation and spindle assembly checkpoint activity. J. Cell Biol. 2009;184:373–381. doi: 10.1083/jcb.200808130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Uchida KSK, et al. Kinetochore stretching inactivates the spindle assembly checkpoint. J. Cell Biol. 2009;184:383–390. doi: 10.1083/jcb.200811028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Foley EA, Kapoor TM. Microtubule attachment and spindle assembly checkpoint signaling at the kinetochore. Nat. Rev. Mol. Cell. Biol. 2013;14:25–37. doi: 10.1038/nrm3494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Kettenbach AN, et al. Quantitative phosphoproteomics identifies substrates and functional modules of Aurora and Polo-like kinase activities in mitotic cells. Sci. Signal. 2011;4:rs5. doi: 10.1126/scisignal.2001497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Indjeian VB, Murray AW. Budding yeast mitotic chromosomes have an intrinsic bias to biorient on the spindle. Curr. Biol. 2007;17:1837–1846. doi: 10.1016/j.cub.2007.09.056. [DOI] [PubMed] [Google Scholar]
- 80.Loncarek J, et al. The centromere geometry essential for keeping mitosis error free is controlled by spindle forces. Nature. 2007;450:745–749. doi: 10.1038/nature06344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Salic A, Waters JC, Mitchison TJ. Vertebrate shugoshin links sister centromere cohesion and kinetochore microtubule stability in mitosis. Cell. 2004;118:567–578. doi: 10.1016/j.cell.2004.08.016. [DOI] [PubMed] [Google Scholar]
- 82.Logarinho E, Resende T, Torres C, Bousbaa H. The human spindle assembly checkpoint protein Bub3 is required for the establishment of efficient kinetochore-microtubule attachments. Mol. Biol. Cell. 2008;19:1798–1813. doi: 10.1091/mbc.E07-07-0633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Kabeche L, Compton DA. Checkpoint-independent stabilization of kinetochore-microtubule attachments by Mad2 in human cells. Curr. Biol. 2012;22:638–644. doi: 10.1016/j.cub.2012.02.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Amaro AC, et al. Molecular control of kinetochore-microtubule dynamics and chromosome oscillations. Nature Cell Biol. 2010;12:319–329. doi: 10.1038/ncb2033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.McHedlishvili N, et al. Kinetochores accelerate centrosome separation to ensure faithful chromosome segregation. J. Cell Sci. 2012;125:906–918. doi: 10.1242/jcs.091967. [DOI] [PubMed] [Google Scholar]
- 86.Ye F, et al. HURP regulates chromosome congression by modulating kinesin Kif18A function. Curr. Biol. 2011;21:1584–1591. doi: 10.1016/j.cub.2011.08.024. [DOI] [PubMed] [Google Scholar]
- 87.Wang X, et al. Mitotic regulator SKAP forms a link between kinetochore core complex KMN and dynamic spindle microtubules. J. Biol. Chem. 2012;287:39380–39390. doi: 10.1074/jbc.M112.406652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.DeLuca JG, et al. Hec1 and Nuf2 are core components of the kinetochore outer plate essential for organizing microtubule attachment sites. Mol. Biol. Cell. 2005;16:519–531. doi: 10.1091/mbc.E04-09-0852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Ishii S, Kurasawa Y, Wong J, Yu-Lee LY. Histone deacetylase 3 localizes to the mitotic spindle and is required for kinetochore-microtubule attachment. Proc. Natl Acad. Sci. USA. 2008;105:4179–4184. doi: 10.1073/pnas.0710140105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Gaitanos TN, et al. Stable kinetochore-microtubule interactions depend on the Ska complex and its new component Ska3/C13Orf3. EMBO J. 2009;28:1442–1452. doi: 10.1038/emboj.2009.96. [DOI] [PMC free article] [PubMed] [Google Scholar]