PPD2 interacts with KIX8 and KIX9 proteins to regulate the transcription of its target genes, including the CYCD3s, and helps to control final leaf size in Arabidopsis.
Abstract
Cell number is an important determinant of final organ size. In the leaf, a large proportion of cells are derived from the stomatal lineage. Meristemoids, which are stem cell-like precursor cells, undergo asymmetric divisions, generating several pavement cells adjacent to the two guard cells. However, the mechanism controlling the asymmetric divisions of these stem cells prior to differentiation is not well understood. Here, we characterized PEAPOD (PPD) proteins, the only transcriptional regulators known to negatively regulate meristemoid division. PPD proteins interact with KIX8 and KIX9, which act as adaptor proteins for the corepressor TOPLESS. D3-type cyclin encoding genes were identified among direct targets of PPD2, being negatively regulated by PPDs and KIX8/9. Accordingly, kix8 kix9 mutants phenocopied PPD loss-of-function producing larger leaves resulting from increased meristemoid amplifying divisions. The identified conserved complex might be specific for leaf growth in the second dimension, since it is not present in Poaceae (grasses), which also lack the developmental program it controls.
INTRODUCTION
Variations in size and shape are one of the most apparent distinctions among different varieties of plants or animals. Plant organ size is highly heritable, and within a given variety, the sizes of leaves, flowers, and seeds are remarkably constant when grown in similar environments. Answering the question on how plant organ size is regulated is an important challenge from both a fundamental and a more applied perspective in view of the increasing demand for food, fiber, and other plant-derived products. However, the molecular mechanisms that control organ or organism size are still poorly understood.
In multicellular organisms, development of a functional organ requires differentiation mechanisms conferring cell and tissue identity, as well as growth regulatory mechanisms determining the number and size of cells that make up the organ. The two underlying processes, cell division and cell expansion, occur in different cell types, at different moments during development, and at different rates, implying a tight spatial and temporal genetic coordination. In addition, cells can divide either symmetrically to produce daughter cells of equivalent fates or asymmetrically to produce daughter cells with distinct developmental potentials (Jan and Jan, 1998; Scheres and Benfey, 1999).
In the model plant Arabidopsis thaliana, the leaf primordium, after emerging from the shoot apical meristem, grows through mainly symmetric division of undifferentiated protodermal cells (Donnelly et al., 1999). After a few days of intense cell division, cell expansion starts at the tip of the leaf and a front of cell cycle arrest moves from the tip to the base (Donnelly et al., 1999; Andriankaja et al., 2012). Simultaneously, meristemoids, transiently proliferating precursor cells with stem cell-like properties that are dispersed in the leaf epidermis, give rise to the stomatal lineage (White, 2006; Bergmann and Sack, 2007; Pillitteri and Torii, 2012). Consequently, final leaf size in Arabidopsis is determined by at least five parameters: the number of cells incorporated into the leaf primordia, the rate of cell division, the developmental window of cell proliferation, the extent of cell expansion, and the timing of meristemoid division (reviewed in Gonzalez et al., 2012). Particularly, the number of cells in the epidermis is significantly increased by the activity of early stomatal lineage cells that undergo several asymmetric divisions allowing self-renewal and the formation of neighboring pavement cells. In Arabidopsis, a meristemoid cell undergoes up to three sequential asymmetric divisions, thereby generating three pavement cells. At the mature stage, 67% of all pavement cells in cotyledons and 48% in leaves originate from the division of meristemoids (Geisler et al., 2000). The amplifying divisions of meristemoids are specific to dicots compared with grasses in which no self-renewing cells are produced in the stomatal lineage (Liu et al., 2009; Vatén and Bergmann, 2012).
Several studies have identified key factors involved in the regulation of the five parameters determining leaf growth in Arabidopsis (Gonzalez et al., 2009; Hepworth and Lenhard, 2014). A positive effect on leaf growth due to a prolonged period of division activity of the meristemoids in the epidermis has been described so far only for one mutant, Δppd, in which the two tandemly repeated PEAPOD (PPD) genes have been deleted (White, 2006). The Arabidopsis genome contains two PPD genes, PPD1 and PPD2, that encode proteins sharing 84% identity and belong to the plant-specific TIFY protein family, containing a conserved motif TIF[F/Y]XG, in which all characterized members are transcriptional regulators (Cuéllar Pérez et al., 2014). The Arabidopsis genome encodes 18 TIFY proteins divided into two classes based on the presence or absence of a C2C2-GATA protein domain, and are known as class I and II, respectively. PPD1 and PPD2 belong to class II, along with TIFY8 and 12 JASMONATE ZIM DOMAIN (JAZ) proteins (Vanholme et al., 2007; Bai et al., 2011; Pauwels and Goossens, 2011; Cuéllar Pérez et al., 2014). Diverse protein domains present in TIFY proteins are associated with different types of protein-protein interactions. First, the ZIM domain characterizing the family is known to mediate homo- and heterodimerization between different TIFY proteins and with other proteins, respectively, such as the transcriptional corepressor NOVEL INTERACTOR OF JAZ (NINJA) (Chini et al., 2009; Chung and Howe, 2009; Pauwels et al., 2010). Second, a C-terminal Jas domain in the JAZ proteins mediates the interaction with transcription factors such as the bHLH MYC2 and the F-box protein CORONATINE INSENSITIVE1 (Chini et al., 2007; Thines et al., 2007). The PPD proteins are distinct from the other TIFY proteins as they contain next to the ZIM domain, a divergent C-terminal Jas domain and a specific N-terminal PPD domain (Bai et al., 2011).
To gain insight into the mode of action of the PPD transcriptional regulators and their role in the regulation of final leaf size, we first characterized the leaf phenotype of Col-0 plants overexpressing an artificial microRNA targeting the PPD transcripts. We used genome-wide transcript profiling to analyze the effect of downregulating PPD expression at the molecular level. We also identified target genes of PPD2 by chromatin affinity purification and sequencing of PPD2 bound genomic fragments. Finally, we identified KIX8 and KIX9 as interacting protein partners of PPD2, enhancing its function of transcriptional repressor, and showed that their mutations phenocopy both the leaf and the molecular phenotype of plants in which PPD expression is decreased. This work provides important insights in the molecular mechanisms by which PPD proteins regulate meristemoid division and hence final leaf size.
RESULTS
Downregulation of PPD Increases the Amplifying Divisions of Meristemoids
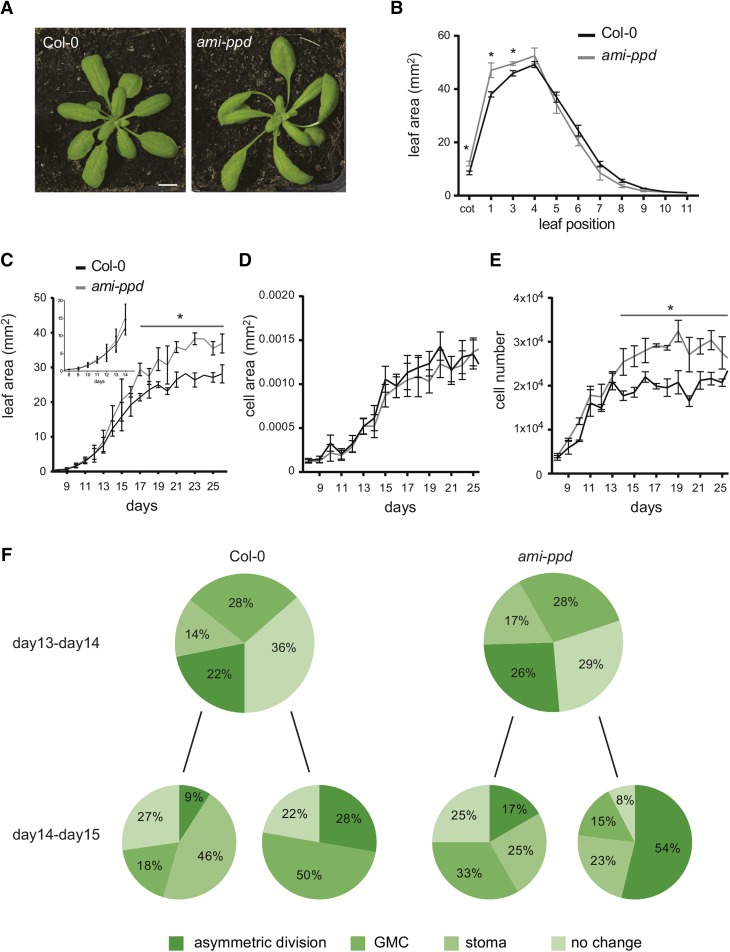
In the Landsberg erecta (Ler) background, a genomic deletion of the tandemly repeated PPD genes (Δppd) causes a change in leaf shape and an increase of cotyledon and leaf area (White, 2006). Because most work on leaf growth has used the Col-0 ecotype (Gonzalez et al., 2009), and to further analyze the effect of the down-regulation of the PPDs, we used a transgenic line (ami-ppd) overexpressing an artificial microRNA targeting PPD1 and PPD2 in this genetic background. The ami-ppd Col-0 plants had dome-shaped leaves (Gonzalez et al., 2010); Figure 1A), comparable to those in the Δppd Ler line. In plants grown in vitro for 21 DAS (days after stratification), the cotyledons and the first three leaves were significantly larger in the ami-ppd line than in the wild type (Figure 1B).
Figure 1.
ami-ppd Rosette, Leaf, and Cellular Phenotype.
(A) Wild-type (left) and ami-ppd (right) plants grown for 25 DAS in soil. Bar = 1 cm
(B) Area of individual leaves of wild-type and ami-ppd plants grown in vitro for 21 DAS (n = 3; error bars represent ±se; *, significant difference from the wild type at P < 0.05).
(C) Leaf area (the inset shows leaf area from 8 to 14 DAS) (n = 3; error bars represent ±se; *, significant difference from the wild type at P < 0.05).
(D) and (E) Cell area (D) and cell number (E) over time of ami-ppd leaves 1 and 2 compared with the wild type (error bars represent ±se from three to five leaves; *, significant difference from the wild type at P < 0.05).
(F) Proportion of cell types (meristemoid after asymmetric division, guard mother cell [GMC], stoma, or meristemoid) originating from meristemoids followed from 13 to 15 DAS by making daily dental resin imprints of the abaxial epidermis of leaves.
Previously, it was shown that the increased leaf area in the Δppd mutant results from a prolonged division of meristemoids in the epidermis (White, 2006). We identified the time during leaf development when these cellular changes triggered the formation of larger leaves in the ami-ppd plants. Leaves 1 and 2 were harvested daily to measure leaf area and quantify epidermal cell area and number. At early time points, leaves of ami-ppd and the wild type were similar in size, but became larger in the mutant 11 to 13 DAS, and this difference became significant from 17 DAS onward (Figure 1C). At maturity (25 DAS), the average cell area was not different from the wild type (Figure 1D), showing that the increase in leaf area was due to an increase in cell number (Figure 1E). This increase, observed early during development, became significant at 14 DAS. Although the average cell area was similar in the ami-ppd and wild-type plants, the proportion of small cells (between 0.25 to 0.75 10−5 mm2) was increased in ami-ppd leaves compared with wild-type leaves from 10 DAS onward (Supplemental Figure 1A).
To explain this early increase in cell number, we followed the fate of meristemoid cells in wild-type and ami-ppd plants over time by making dental resin imprints of the abaxial side of the leaf epidermis from 13 to 15 DAS. We evaluated whether, over time, a meristemoid became a guard mother cell, a stoma, whether it divided asymmetrically or whether it did not change (Supplemental Figure 1B). We found that more meristemoids were dividing asymmetrically in the ami-ppd line than in the wild type both between 13 and 14 DAS (26% versus 22%) and between 14 and 15 DAS (54% versus 28%; Figure 1F). In addition, in the mutant line, more meristemoids that had already divided asymmetrically showed an extra round of asymmetric division. These data demonstrated that, in the ami-ppd line, there were more amplifying divisions of the meristemoid cells. We also observed that the density of mesophyll cells in 17-d-old ami-ppd leaves is slightly increased compared with the wild type, indicating that more small mesophyll cells are produced and that cell division is also increased for this internal leaf cell type (Supplemental Figure 1C).
In summary, the data show that downregulation of PPD genes in the Col-0 background leads to an increase in leaf area resulting from an increase in cell number observed early during development that originated from an increase in amplifying divisions of meristemoid cells.
PPD Downregulation Results in an Increased Expression of a Coregulated Gene Set
To gain more insight into the molecular changes associated with PPD downregulation, RNA was extracted from the first leaf pair of ami-ppd and wild-type plants at 13 DAS (the time point at which differences in leaf area start to be visible; Figure 1C) and subjected to microarray transcript profiling. Excluding the target PPD2, 49 genes were differentially expressed in ami-ppd compared with wild-type plants (P value < 0.05). Thirty-six genes were upregulated and 13 genes were downregulated (Table 1).
Table 1. List of Differentially Expressed Genes in the ami-ppd Line.
| AGI Codes | Annotation | LinFC | P Value |
|---|---|---|---|
| AT2G12210 | Transposable element gene | 3.76 | 0.0040 |
| AT5G54510 | DFL1, DWARF IN LIGHT1 | 3.00 | 0.0020 |
| AT2G42840 | PDF1, PROTODERMAL FACTOR1 | 2.78 | 0.0003 |
| AT3G05600 | Epoxide hydrolase, putative | 2.45 | 0.0020 |
| AT5G49700 | DNA binding protein-related | 2.42 | 0.0001 |
| AT1G14900 | HMGA (HIGH MOBILITY GROUP A) | 2.38 | 0.0003 |
| AT2G37030 | SAUR46, SAUR-like auxin-responsive protein family | 2.31 | 0.0040 |
| AT5G62280 | Unknown protein | 2.18 | 0.0080 |
| AT1G23870 | ATTPS9, TREHALOSE-6-PHOSPHATE SYNTHASE S9 | 2.06 | 0.0070 |
| AT5G20740 | Invertase/pectin methylesterase inhibitor family protein | 2.04 | 0.0070 |
| AT3G04290 | LTL1 (LI-TOLERANT LIPASE1) | 1.97 | 0.0080 |
| AT1G27030 | Unknown protein | 1.96 | 0.0050 |
| AT3G09260 | PYK10 | 1.89 | 0.0020 |
| AT3G54990 | SMZ,SCHLAFMUTZE | 1.88 | 0.0070 |
| AT5G20630 | GER3, GERMIN 3 | 1.84 | 0.0280 |
| AT4G34250 | KCS16, 3-KETOACYL-COA SYNTHASE16 | 1.78 | 0.0070 |
| AT1G52000 | Jacalin lectin family protein | 1.77 | 0.0180 |
| AT5G67110 | ALC, ALCATRAZ | 1.75 | 0.0360 |
| AT5G04530 | KCS19, 3-KETOACYL-COA SYNTHASE19 | 1.73 | 0.0020 |
| AT5G53210 | SPCH, SPEECHLESS | 1.73 | 0.0200 |
| AT3G54400 | Aspartyl protease family protein | 1.72 | 0.0370 |
| AT5G04190 | PKS4,phytochrome kinase substrate 4 | 1.70 | 0.0390 |
| AT3G08770 | LTP6; lipid binding | 1.66 | 0.0250 |
| AT3G50570 | Hydroxyproline-rich glycoprotein family protein | 1.62 | 0.0050 |
| AT4G29020 | Glycine-rich protein | 1.61 | 0.0040 |
| AT1G06080 | ADS1, DELTA 9 DESATURASE1 | 1.61 | 0.0460 |
| AT3G12700 | Aspartyl protease family protein | 1.61 | 0.0090 |
| AT3G50070 | CYCD3;3, CYCLIN D3;3 | 1.60 | 0.0090 |
| AT2G23170 | GH3.3; indole-3-acetic acid amido synthetase | 1.56 | 0.0090 |
| AT1G26210 | Unknown protein | 1.55 | 0.0070 |
| AT2G37300 | Unknown protein | 1.53 | 0.0070 |
| AT5G43020 | Leucine-rich repeat transmembrane protein kinase, putative | 1.43 | 0.0290 |
| AT5G51750 | ATSBT1.3, ARABIDOPSIS THALIANA SUBTILASE 1.3 | 1.39 | 0.0350 |
| AT2G37630 | AS1, ASYMMETRIC LEAVES1 | 1.38 | 0.0250 |
| AT5G59540 | Oxidoreductase, 2OG-Fe(II) oxygenase family protein | 1.38 | 0.0490 |
| AT5G67260 | CYCD3;2, CYCLIN D3;2 | 1.34 | 0.0310 |
| AT5G56870 | BGAL4, β-galactosidase 4 | −1.33 | 0.0390 |
| AT1G70810 | C2 domain-containing protein | −1.46 | 0.0120 |
| AT2G15042 | Protein binding | −1.48 | 0.0300 |
| AT4G00880 | SAUR31 | −1.50 | 0.0460 |
| AT1G76240 | Unknown protein | −1.57 | 0.0200 |
| AT3G63110 | ATIPT3, Arabidopsis thaliana isopentenyltransferase 3 | −1.62 | 0.0460 |
| AT1G71140 | MATE efflux family protein | −1.69 | 0.0310 |
| AT4G14720 | PPD2 | −1.70 | 0.0200 |
| AT1G32450 | NRT1.5, NITRATE TRANSPORTER 1.5 | −1.79 | 0.0030 |
| AT5G09220 | AAP2, AMINO ACID PERMEASE2 | −1.91 | 0.0050 |
| AT3G04720 | PR4, PATHOGENESIS-RELATED4 | −1.97 | 0.0460 |
| AT2G32160 | Unknown protein | −2.14 | 0.0450 |
| AT1G19960 | Unknown protein | −2.16 | 0.0470 |
| AT1G67865 | Unknown protein | −2.17 | 0.0230 |
Wild-type and ami-ppd plants were grown for 13 DAS, and the first pair of leaves was harvested for RNA extraction and genome transcript profiling. LinFC, linear fold change.
PPDs regulate stomatal meristemoid division (White, 2006). Among the genes involved in stomatal development (Pillitteri and Dong, 2013), SPEECHLESS (SPCH) expression was significantly increased in the ami-ppd line (Table 1). Also, CYCD3;2, ATSBT1.3, and AT4G29020, which are upregulated in the meristemoid-enriched scrm-D;mute background (Pillitteri et al., 2011), were also upregulated in ami-ppd leaves (Table 1). Due to the potential role of PPD in the regulation of cell division, we also compared the differentially expressed genes in ami-ppd leaves with a data set of proliferation-specific genes (Beemster et al., 2005). A second gene encoding a D3-type cyclin, CYCD3;3, was found to be upregulated in the ami-ppd line as was AT5G43020, which encodes a leucine-rich repeat transmembrane protein kinase that is also specifically expressed in proliferating tissues. We used qRT-PCR to analyze the expression of several of these genes and stomatal lineage-related genes that showed a slightly but not significant increased expression in the microarray data. We performed a time-course experiment in which the first leaf pair from ami-ppd and wild-type plants was harvested from 11 to 16 DAS. For most of the genes tested, we confirmed the increased expression in ami-ppd leaves during early leaf development (Supplemental Figure 2A).
We also performed coexpression analysis with predefined sets of microarray expression data. We obtained two main networks containing 23 and 7 of the 49 differentially expressed genes, connected with 56 and 8 edges, respectively (Supplemental Figure 3). Among the genes of the large network, we found the two CYCD3s, SPCH and AT4G29020, but also HMGA and ATSBT1.3, were highly connected with the others, having 14 and 10 edges, respectively.
In conclusion, we found a specific set of differentially expressed genes, mainly upregulated, in the first pair of ami-ppd leaves. These genes were highly connected in terms of coexpression and were related to cell division, meristemoid cells and stomatal development.
Genome-Wide Determination of PPD2 Target Sites
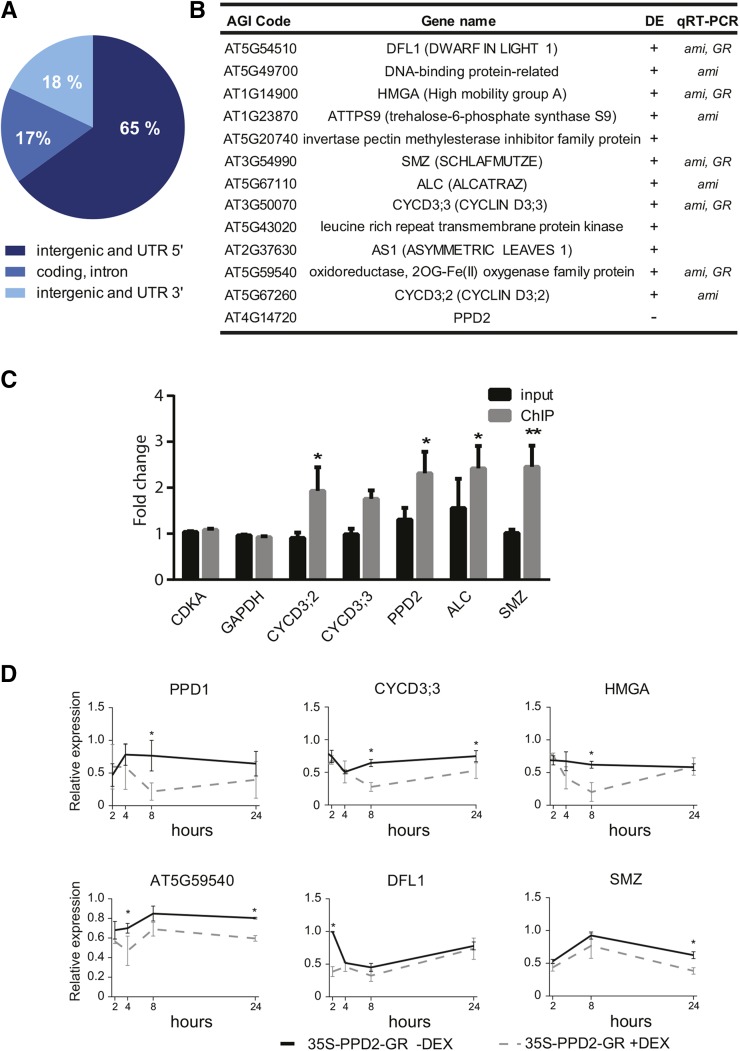
To identify the direct target genes of PPD2, we performed tandem chromatin affinity purification (TChAP; Verkest et al., 2014) using an Arabidopsis cell suspension culture overexpressing an HBH-tagged PPD2.
After sequencing of the purified DNA, 19.61 and 16.57 million reads were obtained for PPD2-HBH and the wild-type control, respectively. A total of 2042 peaks representing nonredundant reads and reads mapping uniquely to the genome, and corresponding to 1919 genes, were identified to be specific to the PPD2-HBH sample (Supplemental Data Set 1 and Supplemental Figure 4A). This specificity was also confirmed by comparing the peaks found for the PPD2-HBH sample to the TChAP-sequencing (TChAP-seq) data from ERF115, an unrelated transcription factor, which was sequenced in the same experiment using an ERF115-HBH cell culture (Heyman et al., 2013; Supplemental Figure 4A). Among the genes bound by PPD2, two Page-Man categories (Usadel et al., 2006) were overrepresented: “regulation of transcription” and “hormone metabolism” (Supplemental Figure 4B). The analysis of the location of these peak sequences, having an average length of ∼300 bp (Supplemental Figure 4C), showed that 83% were found in the intergenic/untranslated (UTR) regions (65 and 18% in the 5′ and 3′ intergenic/UTR region, respectively) and only 17% in the coding and intron regions (Figure 2A). In addition, ∼40% of the peaks had their summit located between −300 and 100 bp from the translation start site with a maximum between −200 and 0 bp (Supplemental Figure 4D). Among the 2042 peak sequences identified, we found that two specific motifs were highly represented. The first motif, GmCACGTGkC, containing an ABF (abscisic acid-responsive element binding factor) binding site sequence (CACGTGGC) or less specific a G-box sequence (CACGTG), was preferentially located near the peak summit and is present in 726 peak sequences (Supplemental Figure 4E). The second motif, yctCACGCGCyt, is also related to a G-box sequence and was found in 275 peak sequences.
Figure 2.
Expression of PPD2 Target Genes Identified by TChAP-Seq Is Altered in Growing ami-ppd Leaves.
(A) Genome-wide distribution of PPD2 DNA binding sites (peaks identified with MACS; Zhang et al., 2008) in the intergenic and UTR 5′ regions, the coding region and introns, and the intergenic and UTR 3′regions.
(B) Overlap between the genes identified by TChAP-Seq and the genes differentially expressed in the growing ami-ppd leaves (13 DAS). DE, differential expression of the target gene in the ami-ppd line as determined by microarray analysis. +, upregulation; −, downregulation. qRT-PCR: change in expression of the target gene, analyzed by qRT-PCR, in at least one time point, in the ami-ppd (ami) and/or in the 35S_PPD2-GR (GR) lines.
(C) Validation of PPD2 target genes by ChIP-qPCR in planta. ChIP was done with anti-GFP antibody on 21-d-old plants expressing GFP-tagged PPD2, and enrichment was determined with qPCR by comparing the input and anti-GFP purified samples of the 35S-PPD2-GFP to the control 35S-GFP line. ChIP: DNA purified using the anti-GFP antibody. Asterisks indicate significant difference to the input (n = 5, error bars represent ±se, *P < 0.05 and **P < 0.01).
(D) Time-course analysis of PPD2 target gene expression after induction of PPD2 in the 35S-PPD2-GR line treated with DEX (n = 3, error bars represent ± se; *, significant difference from the 35S-PPD2-GR without DEX at P < 0.05).
The PPD2-HBH TChAP-Seq data set was compared with the set of genes differentially expressed in the ami-ppd line. Of the 49 differentially expressed genes, 13 (including PPD2) were bound by PPD2 (Figure 2B). CYCD3;2, CYCD3;3, and HMGA were part of these direct targets of PPD2. To validate the target genes identified by TChAP-seq, we performed chromatin immunoprecipitation-quantitative PCR (ChIP-qPCR) using both an Arabidopsis cell culture expressing a PPD2-GSyellow fusion protein under the control of the 35S promoter and an Arabidopsis transgenic line in which a PPD2-GFP fusion protein is expressed from the 35S promoter. In both systems, a significant enrichment of DNA corresponding to the five tested PPD2 target genes (CYCD3;2, CYCD3;3, PPD2, ALC, and SMZ) was found in the PPD2-tagged pulled-down DNA relative to the reference genes, input, and negative control sample (Figure 2C; Supplemental Figure 4F). Subsequently, we analyzed the time-dependent expression of the direct target genes from 11 to 16 DAS in the first leaf pair of ami-ppd and wild-type plants by qRT-PCR. The increase in expression of SMZ, DFL1, ALC, HMGA, ATTPS9, AT5G59540, and AT5G49700 was confirmed at 13 DAS (Supplemental Figure 2A).
In conclusion, we identified PPD2 direct target genes, among which several are differentially expressed in ami-ppd.
PPD2 Negatively Regulates the Expression of Its Target Genes
The observation that target genes of PPD2 identified by TChAP-seq were upregulated in ami-ppd leaves suggests that PPD2 acts as a negative regulator for these targets. To test this hypothesis, we generated transgenic plants constitutively expressing a glucocorticoid-inducible PPD2 gain-of-function construct, 35S-PPD2-GR (PPD2-GR), using the CaMV 35S promoter and the rat glucocorticoid receptor (GR). When grown in vitro or in soil, the PPD2-GR plants do not appear different from control plants (Supplemental Figure 2B). When germinated on medium supplemented with dexamethasone (DEX), a glucocorticoid hormone causing the translocation of PPD2-GR to the nucleus, we saw only a slight decrease in leaf area (Supplemental Figure 2B). This mild decrease in leaf area was also seen when plants were grown in soil and sprayed with DEX (Supplemental Figure 2B). At 9 DAS, PPD2-GR plants grown on Murashige and Skoog medium were transferred to medium with or without DEX because at this time point, the cell cycle arrest front has disappeared in the first two leaves and, therefore, the majority of the cells dividing in the epidermis are meristemoids. RNA was extracted from the first leaf pair harvested 2, 4, 8, and 24 h after transfer and candidate target gene expression was analyzed by qRT-PCR (Figures 2D; Supplemental Figure 2C). Upon activation of PPD2 after DEX treatment, the mRNA level of some target genes tested decreased, albeit at various time points (Figure 2D), whereas, for some genes, no change in expression was observed (Supplemental Figure 2C).
In conclusion, we identified at least 13 direct target genes of PPD2, including the D-type cyclins, CYCD3;2 and CYCD3;3. In addition, PPD2 was shown to be a repressor of the expression of most of the target genes tested, such as CYCD3;3 and HMGA.
PPD Proteins Contain a Functional ZIM Domain
PPD proteins belong to the class II of the TIFY protein family along with TIFY8 and the 12 JAZ proteins (Cuéllar Pérez et al., 2014). In TIFY proteins, diverse domains are associated with different protein-protein interaction abilities (Pauwels et al., 2010). To determine the nature of the protein complex associated with PPD2, we performed tandem affinity purification (TAP) (Van Leene et al., 2015), using an Arabidopsis cell culture overexpressing PPD2 with a GS tag at the C-terminal region. PPD2 interacted with the TIFY proteins JAZ3, JAZ10, and TIFY8, confirming that heterodimerization within the TIFY family is not restricted to the JAZ proteins (Supplemental Data Set 2 and Supplemental Figure 5A). To evaluate this hypothesis, we tested all 12 JAZ proteins, PPD1, and PPD2 for interaction with PPD2 using yeast two-hybrid (Y2H) assays. We found direct interaction of PPD2 with JAZ3 and JAZ9 but not JAZ10. We also found evidence of homodimerization of PPD2 and heterodimerization with PPD1 (Supplemental Figure 5B).
Dimerization of JAZ proteins requires the ZIM (zinc-finger expressed in inflorescence meristem) domain, which is also present in PPD proteins (Chini et al., 2009; Chung and Howe, 2009). We designed truncated versions of the PPD2 protein comprising different combinations of its N-terminal PPD, central ZIM, and C-terminal Jas-like domain and tested these fragments for interaction with JAZ3 in a Y2H. These results showed that the ZIM domain is necessary and sufficient for interaction with JAZ3 and demonstrated that the PPD2 ZIM domain is a functional protein-protein interaction domain (Supplemental Figure 5C).
In conclusion, this analysis showed that PPD2 interacts with some of the class II TIFY proteins through its ZIM domain.
KIX8 and KIX9 Proteins Directly Interact with the PPD Domain and Are TPL Adaptors
Besides the TIFY proteins identified by TAP as part of PPD2 protein complexes, we found the TOPLESS-adaptor protein NINJA (Pauwels et al., 2010) and two proteins containing a KIX (kinase-inducible domain interacting) domain, encoded by AT3G24150 and AT4G32295 and named KIX8 and KIX9 (Supplemental Figure 5A; Thakur et al., 2013). The KIX protein domain is well characterized in several non-plant species and is present in transcriptional coactivator proteins such as the HISTONE ACETYLTRANFERASE (HAT), CREB BINDING PROTEIN (CBP), p300 proteins (Radhakrishnan et al., 1997), and the Mediator subunit ARC105/MED15/Gal11 (Yang et al., 2006; Jedidi et al., 2010). The KIX domain in these proteins mediates the interaction with activation domains of transcription factors (Jedidi et al., 2010).
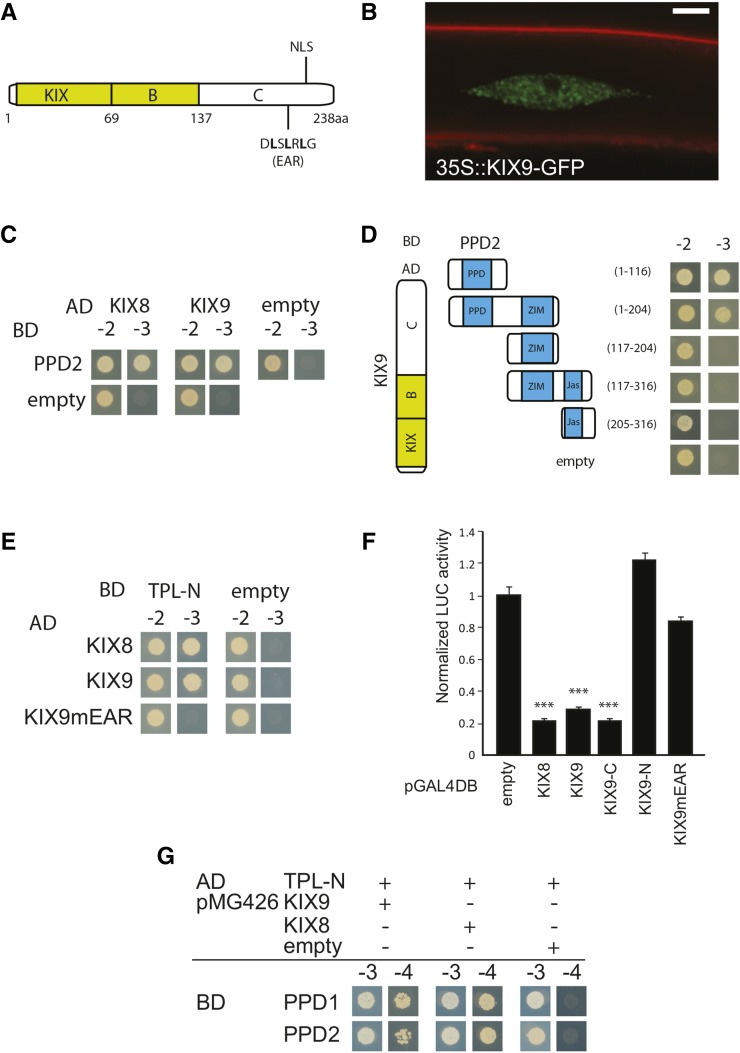
KIX8 and KIX9 are uncharacterized proteins that belong to the KIX protein family composed of 11 members in Arabidopsis. They contain a KIX domain in their N-terminal region (Thakur et al., 2013), show no similarity with HATs or MED15, and have a completely different domain structure compared with the other KIX proteins (Figure 3A). Using the PLAZA comparative genomics platform (Proost et al., 2015), we found that KIX8 and KIX9, similarly to PPD, are found in the majority of vascular plants, with the exception of Poaceae (grasses) (Supplemental Figure 5D). Based on the protein sequence alignment, we identified three conserved regions (Figure 3A; Supplemental Figure 6 and Supplemental Data Set 3): a conserved B domain (amino acids 70 to 137 in KIX9) next to the N terminus, a highly conserved KIX domain (amino acids 1 to 69), and a less conserved EAR motif (amino acids 212 to 220) in the C terminus. Predicted nuclear localization by using the Web tool WoLF PSORT was confirmed in Arabidopsis seedlings expressing KIX9-GFP (Figure 3B). Fluorescence was seen only in the nucleus, corresponding with the observed nuclear localization of PPD2 (Lacatus and Sunter, 2009).
Figure 3.
The PPD Domain Mediates the Interaction of PPD Proteins with the EAR Domain-Containing TPL Adaptor Proteins KIX8 and KIX9.
(A) Schematic overview of KIX9 structure.
(B) Representative confocal microscopy image of a root cell from an Arabidopsis transgenic line transformed with a 35S-KIX9-GFP construct that expresses KIX9-GFP in the nucleus. Bar = 5 µm.
(C) Y2H interaction analysis using PPD2 and KIX proteins. Transformants were selected on medium lacking Leu and Trp (-2) or medium lacking Leu, Trp, and His (-3) to test the interaction. AD, activating domain; BD, binding domain.
(D) Truncations of PPD2 that were tested to identify the KIX9 interaction domain.
(E) Direct interaction with TPL and the necessity of the EAR domain as tested by Y2H.
(F) GAL4DBD-fused KIX8 and KIX9 proteins were tested for transcriptional repression activity of the UAS-fLUC reporter in tobacco BY-2 protoplasts. Error bars represent ± se of eight biological replicates (***P < 0.001).
(G) Yeast three-hybrid experiment to test the bridging capacity of KIX for the PPD-TPL interaction. Transformants were selected on medium lacking Leu, Trp, and Ura (-3) or selective medium additionally lacking His (-4) to test the interaction.
To further analyze the KIX8/9-related protein complex, we performed a TAP experiment with KIX8 (GS tag fused to the C or N terminus) that confirmed the in vivo interaction with PPD2 (Supplemental Data Set 2 and Supplemental Figure 5A). Y2H assays further supported the direct interaction between PPD2 and the KIX proteins (Figure 3C) and identified the N-terminal PPD domain of PPD2 as being necessary and sufficient for the interaction with KIX9 (Figure 3D).
TAP using the N-terminal tagged KIX8 identified the transcriptional corepressor TOPLESS (TPL), as well as NINJA, as part of the KIX8 protein complex (Supplemental Figure 5A). The EAR motif, present in KIX proteins (Figure 3A), mediates binding with TPL (Kagale et al., 2010; Causier et al., 2012). To confirm the direct interaction between the two KIX proteins and TPL, we performed Y2H assays. KIX8 and KIX9 interacted directly with the N-terminal LisH-domain of TPL (TPL-N) (Figure 3E). Mutation of the Leu residues in the LxLxL core of the EAR motif to Ala abolished the TPL interaction (Figure 3E).
To test whether the KIX proteins have repressor activity, both of them were fused to the GAL4 DNA binding domain (GAL4DBD) and coexpressed with a construct expressing the firefly luciferase (fLUC) reporter gene under the control of GAL4 binding elements in tobacco (Nicotiana tabacum) Bright Yellow 2 (BY-2) protoplasts. Both KIXs were capable of strong repression mediated by the EAR motif (Figure 3F). Finally, we tested if KIX was capable of forming a molecular bridge between PPD2 and TPL. Due to the absence of an EAR motif within its sequence, PPD2 was unable to bind directly to TPL (Figure 3G). Co-expression with either KIX8 or KIX9 was sufficient for GAL4 reconstitution, providing evidence for PPD2/KIX/TPL ternary complexes (Figure 3G).
In conclusion, we showed that PPD2 interacted directly with KIX8 and KIX9 that act as adaptor proteins linking PPD proteins with their EAR motifs to the corepressor TPL.
Double kix8 kix9 Mutants Phenocopy the ami-ppd Leaf Phenotype
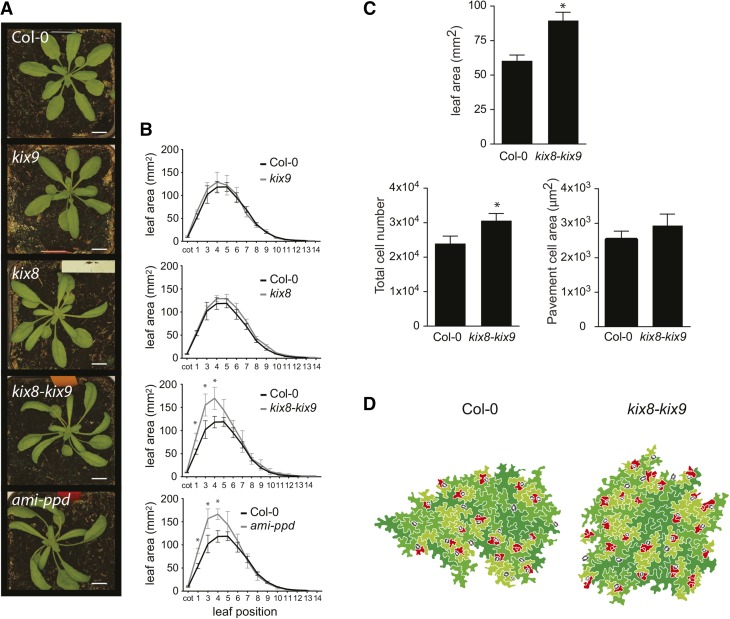
To study the potential role of the KIX proteins during leaf development, single T-DNA insertion lines for KIX8 (GABI_422H04) and KIX9 (SAIL_1168_G09) were obtained and kix8 kix9 double homozygous lines were generated. Plants were grown for 21 DAS in soil and leaf series were done on wild-type, ami-ppd, kix8, kix9, and kix8 kix9 lines. As shown in Figure 4A, the dome-shape phenotype previously described for the leaves of ami-ppd was also observed in the kix8 kix9 double mutant and was seen to a lesser extent in the kix8 single mutant, but not in the kix9 mutant. In addition, in ami-ppd as well as in the kix8 kix9 double mutant, we observed twisting of the rosette (“propeller” phenotype), which is not present in the wild-type plants (Figure 4A). From the leaf series analysis, a similar increase in leaf area was observed for the ami-ppd line and the kix8 kix9 double mutant, whereas no increase was found when only one of the two KIX genes was downregulated (Figure 4B).
Figure 4.
kix8, kix9, and kix8 kix9 Mutant Phenotypes.
(A) Plants grown for 25 DAS in soil. From top to bottom: wild type, kix9, kix8, kix8 kix9, and ami-ppd. Bar = 1 cm.
(B) Area of individual leaves of wild-type, kix9, kix8, kix8 kix9, and ami-ppd plants grown in soil for 21 DAS (n = 3; error bars represent ±se; *, significant difference from the wild type at P < 0.05).
(C) Leaf (first pair) area, cell area, and cell number of 25-d-old kix8 kix9 mutants compared with the wild type (n = 3; error bars represent ±se; *, significant difference from the wild type at P < 0.05).
(D) Representative drawing of cells from the first leaf pair of 14-d-old kix8 kix9 and wild-type plants. Cells are colored in relation to their area, from smaller cells (red) to larger (dark green). Red, cells smaller than 500 μm2; light green, cell area ranging from 500 to 1500 μm2; medium green, cell area ranging from 1500 to 3000 μm2; dark green, cells larger than 3000 μm2; stomata are marked in gray.
We then harvested the first leaf pair of Col-0 and kix8 kix9 plants grown in soil for 14 and 25 d and quantified epidermal cell area and number. At 25 DAS, the 48% increase in leaf area in the double kix8 kix9 mutant (Figure 4C) resulted mainly from an increase in total cell number, whereas the average pavement cell area was not significantly different from control plants (Figure 4C). At 14 DAS, we also observed an increase in total cell number (Supplemental Figure 7A) and the presence of a larger amount of smaller cells surrounding the stomata in the kix8 kix9 double mutants compared with the wild-type plants (Figure 4D; Supplemental Figure 7).
In conclusion, this analysis showed that the double mutant kix8 kix9 phenocopied the ami-ppd line, whereas the single lines did not show changes in leaf size, implying that KIX8 and KIX9 may be partially redundant.
KIX8 and KIX9 Are Required for PPD2 Target Gene Expression
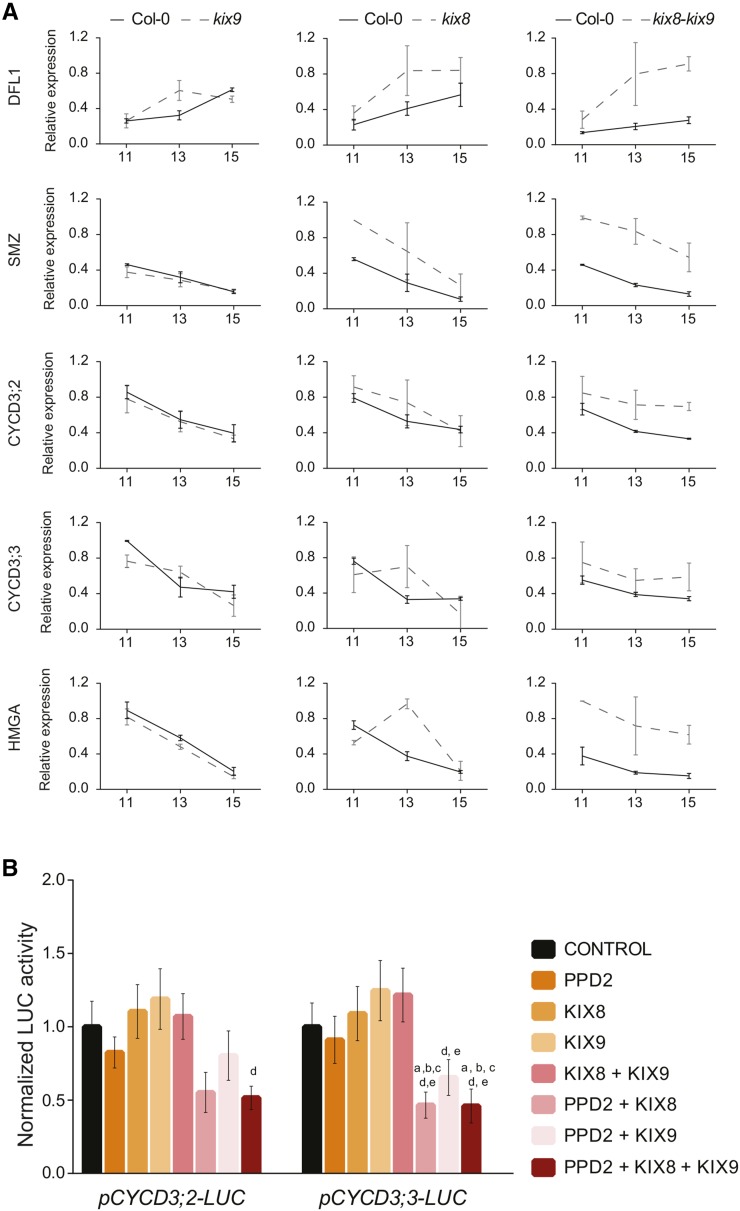
To study the involvement of the KIX proteins in the regulation of PPD2 target gene expression, wild type, single kix8 and kix9, and double kix8 kix9 mutants were grown in vitro and leaves 1 and 2 were harvested at 11, 13, and 15 DAS for RNA extraction. We quantified mRNA levels of several PPD2 target genes (DFL1, SMZ, CYCD3;2, CYCD3;3, and HMGA) by qRT-PCR.
In kix9 leaves, none of the genes tested showed an obvious change in expression compared with the wild type, except for DFL1 having higher expression levels at 13 DAS (Figure 5A). In kix8 leaves, the expression of all genes tested, except CYCD3;2, was significantly increased compared with the wild type, although not at all time points. Similar to what was observed in the ami-ppd line, the expression of DFL1, SMZ, CYCD3;2, CYCD3;3, and HMGA was significantly increased in the double kix8 kix9 mutant (Figure 5A).
Figure 5.
Regulation of PPD2 Target Gene Expression by KIX8 and KIX9.
(A) Time-course analysis of PPD2 target gene expression in wild-type, kix9, kix8, and kix8 kix9 leaves 1 and 2 (n = 3, error bars represent ±se; *, significant difference from the wild type at P < 0.05).
(B) PPD2-, KIX8-, and KIX9-dependent activation of the promoters of CYCD3;2 and CYCD3;3 in the transient expression assay. Indicated values are average relative luciferase activity compared with the control (35S-GUS) for four biological repeats [a, b, c, d, and e represent significantly different values compared with control (a), PPD2 (b), KIX8 (c), KIX9 (d), and KIX8+KIX9 (e)]. P value < 0.05; error bars indicate ±se.
To analyze the contribution of KIX8 and KIX9 in the function of PPD2, we performed transient expression assays in protoplasts with promoter-luciferase reporter constructs, in which promoters of PPD2 target genes were cloned upstream of the fLUC gene (encoding the firefly luciferase enzyme, pCYCD3;2-LUC and pCYCD3;3-LUC) and coexpressed in BY-2 protoplasts with either a 35S-PPD2, 35S-KIX8, or 35S-KIX9 construct alone or in combination. We found that when pCYCD3;2-LUC and pCYCD3;3-LUC constructs were cotransformed with the 35S-PPD2 construct, only a modest repression of the luciferase activity was observed compared with the control (Figure 5B). When these constructs were cotransformed with only the 35S-KIX8 or 35S-KIX9 constructs, no change or a slight increase of the luciferase signal was observed. On the other hand, when PPD2 was coexpressed with KIX8 or KIX9 in the protoplasts, the luciferase signal for pCYCD3;2-LUC and pCYCD3;3-LUC decreased significantly compared with the control (45 and 50% for PPD2+KIX8 and 46 and 44% for PPD2+KIX9, respectively). Finally, when PPD2 was coexpressed with both KIX8 and KIX9, the luciferase signal for pCYCD3;2-LUC and pCYCD3;3-LUC was equivalent than when PPD2 was coexpressed only with KIX8 (Figure 5B). These results confirmed the functional redundancy of KIX8 and KIX9 in the repression of PPD2 target genes observed in loss-of-function lines.
Taken together, these data suggest the KIX proteins are important for the regulation of leaf growth by PPD2 and the repression of its target genes.
DISCUSSION
PPD2 Represses the Expression of D3-Type Cyclins
Downregulation of PPD expression leads to the formation of larger, dome-shaped leaves in both the Arabidopsis Ler (White, 2006) and the Col-0 genetic background. This increase in leaf size results from an enhanced division of meristemoid cells, which are proliferating precursor cells dispersed in the leaf epidermis that give rise to the stomatal lineage (White, 2006; Pillitteri and Dong, 2013). Therefore, PPD might act by limiting the division of meristemoids during leaf development. As measured here, the amplifying divisions of meristemoids were increased in the ami-ppd line, but increased division of the stomatal lineage ground cells cannot be excluded. Here, we show that several genes involved in stomatal development (Pillitteri and Dong, 2013), such as SPCH, MUTE, TMM, and POLAR, have an increased expression in plants with a reduced expression of PPD genes. However, this may be an indirect effect derived from the PPD-mediated increased number of meristemoid cells, given that none of the stomatal related genes were identified as direct target of PPD2.
We also demonstrated that PPD2 bound to the promoter region of the two cell cycle-related genes CYCD3;2 and CYCD3;3 and that their expression increased when the expression level of both PPD genes was downregulated. In contrast, PPD2 overexpression in protoplasts can downregulate the promoter activity of these two D3-type cyclin encoding genes in the presence of KIX8 and KIX9. Arabidopsis encodes three D3-type cyclins and the activity of these three proteins has been shown to be important for determining cell number in developing leaves (Dewitte et al., 2007). In a triple cycD3;1-3 mutant, leaves contain less cells due to an early arrest of the mitotic phase (Dewitte et al., 2007). Remarkably, it has been shown that in the triple cycD3;1-3 mutant, the duration of epidermal meristemoid activity and the frequency of the formation of satellite meristemoids originating from the asymmetric division of a meristemoid-derived sister cell, appear to be reduced compared with the wild type (Elsner et al., 2012). In addition, it was recently shown that CYCD3;2 is a direct target of SPCH, a positive regulator of meristemoid division, and is upregulated upon induction of SPCH (Lau et al., 2014). Beside CYCD3;2, among the 13 PPD2 bound and regulated genes, only one additional SPCH target was found: AT5G59540. Taken together, it is likely that PPD2 regulates in part meristemoid activity by repressing the expression of CYCD3 genes.
We also found that PPD2 inhibits the expression of HMGA, encoding a member of the chromatin-associated high mobility group (HMG) proteins. Moreover, HMGA was identified as an important hub in the transcriptional network identified in PPD-regulated genes. In animals, HMGA proteins act as architectural factors, facilitating various nuclear processes, including chromatin organization and transcription, and are therefore involved in a wide variety of biological processes including growth, cell proliferation, differentiation, and death (Reeves, 2001). In mice, a null mutation of the HMG I-C/HMGA2 gene results in a decreased rate of cell proliferation (Zhou et al., 1995). By inhibiting HMGA expression, PPD2 could therefore negatively influence cell proliferation through a global change in chromatin organization.
Additional potential target genes of PPD2 were identified by TChAP-seq and their expression was increased in the ami-ppd line, but for some, no downregulation was found in the PPD2-GR line after DEX induction. Several reasons could account for the differences between the TChAP-seq data and the expression analyses. First, the use of a cell culture for the identification of the DNA regions bound by PPD2 might have led to altered binding to target regions (either false positive or negative) due to the potential absence of important protein partners. Nonetheless, in the case of PPD2, we identified KIX and others proteins as interacting partners in the cell cultures, indicating that at least some of the partners necessary for PPD2 function are present in the cell culture and that several targets identified in the cell culture were confirmed by ChIP-qPCR in planta. Second, because PPD2 is presumably acting in a cell type- and developmental stage-specific manner, the alteration of the expression of the target genes might be dependent on the time point analyzed. Such developmental dependency can therefore complicate interpretation of the activating or repressing action of PPD2. Third, the expression analysis of the ami-ppd line and of PPD2-GR upon nuclear translocation at later time points reflects a steady state of the molecular phenotype accompanying the developmental effects and identified genes may therefore include indirect targets of PPD2 signaling. The role of putative PPD2 targets will need further investigation.
PPD Proteins Are Associated with the JA Signaling Module
We identified several protein partners of PPD2, including NINJA and the TIFY proteins JAZ3, JAZ9, JAZ10, PPD1, and TIFY8. Earlier TAP profiling of NINJA, JAZ10, and TIFY8 had already identified PPD2 as an interactor, confirming our current findings (Pauwels et al., 2010; Moreno et al., 2013; Cuéllar Pérez et al., 2014). These protein-protein interaction data therefore reinforce the concept that group II TIFY proteins can form hetero- and homodimers through the ZIM domain (Chini et al., 2009; Chung and Howe, 2009). NINJA and JAZ proteins are negative regulators of jasmonate signaling (Pauwels et al., 2010). Although the hormone JA-Ile is known to be involved in the repression of cell proliferation and cell size during leaf growth (Noir et al., 2013), the relation of JA-Ile to PPD function is currently unclear. Interestingly, JAZ10 was also found to be upregulated in the meristemoid-enriched scrm-D;mute mutant and is expressed in the stomatal lineage (Pillitteri et al., 2011).
KIX8 and KIX9 were also identified in Arabidopsis TIFY8 protein complexes and TIFY8 heterodimerized with PPD1 and PPD2, but not with any JAZ protein (Cuéllar Pérez et al., 2014). Although a TIFY8 ortholog can be found in the moss Physcomitrella patens, hinting at a more ancient evolutionary origin compared with PPD1/2 and KIX8/9, TIFY8 is also lost in grasses (Cuéllar Pérez et al., 2014). Together, these results suggest a function for TIFY8 in the PPD/KIX repressor complex in Arabidopsis.
KIX8 and KIX9 Are TPL Adaptor Proteins
Transcription factors can interact directly with constituents of the transcriptional machinery to recruit them at promoters or indirectly through cofactors. In addition to the interaction of various TIFY proteins with PPD2, binding was found, through the PPD-specific domain, with two KIX domain-containing proteins, KIX8 and KIX9, which are important for the repression activity of PPD2. In animals and yeast, KIX protein domains are present mainly in transcriptional coactivator proteins such as HAT or MEDIATOR (MED) subunit proteins (Radhakrishnan et al., 1997; Yang et al., 2006) and mediate the interaction with activation domains of transcription factors (Jedidi et al., 2010). In Arabidopsis, 11 proteins containing KIX-like domains have been described, including not only CBP/p300-like proteins (KIX3, KIX4, KIX5, and KIX7) and MED15-like proteins (KIX1 and KIX6) but also proteins such as KIX8 or KIX9 that have no characterized domain other than a KIX domain (Thakur et al., 2013, 2014). KIX8 and KIX9 do not have similarity with MED15 or HATs outside the KIX domain. In this study, we showed that KIX8 and KIX9 possessed strong transcriptional repression activity. This repression activity was mediated by the EAR motif which is present in numerous transcriptional repressors, such as Aux/IAA, NINJA, and some JAZ proteins (Pauwels and Goossens, 2011). In addition, we have shown that KIX8, like NINJA, is capable of recruiting the corepressor protein TPL (Causier et al., 2012) through the EAR motif.
It is likely that the KIX-fold, through which transcriptional coactivators such as HATs and MED15 interact with the activation domains of transcription factors, functions in the KIX proteins to link transcription factors with the transcriptional corepressor TPL. It has become clear that recruitment of TPL to transcription factors functions as a general repression mechanism employed to steer a variety of plant processes. TPL proteins are the functional orthologs of the Tup1 and Groucho/TLE corepressors in yeast and metazoans (Lee and Golz, 2012). Similar to Groucho (Winkler et al., 2010), TPL function is associated with histone deacetylase interaction (Krogan et al., 2012; Wang et al., 2013). Nevertheless, it is proposed that the Cyc8-Tup1 complex in yeast acts by shielding activation domains and preventing transcription factors of recruiting coactivators (Wong and Struhl, 2011). In this complex, Cyc8 functions as a Tup1 adaptor (Tzamarias and Struhl, 1994). In plants, many transcription factors can bind TPL directly (Causier et al., 2012), but others, as in Cyc8-Tup1, recruit TPL via molecular bridges such as NINJA, JAZ, Aux/IAA, and TIE1, which links TEOSINTE BRANCHED1/CYCLOIDEA/PCF (TCP) transcription factors to TPL (Tao et al., 2013), including the KIX proteins presented here.
KIX8 and KIX9: New Leaf Growth Regulators
In Arabidopsis, the role of the KIX proteins during development is poorly known. Only two KIX domain-containing proteins, CBP/p300-like and MED15-like, have been described and are involved in the regulation of flowering time and the response to salicylic acid, respectively (Han et al., 2007; Canet et al., 2012). Here, we report another function for the KIX proteins during leaf development, having shown that the double mutant kix8 kix9 produces larger and dome-shaped leaves like the ami-ppd line. This phenotype was only observed when both genes are downregulated but we also observed that downregulation of the expression of KIX8 alone leads to the formation of dome-shaped leaves. In addition, we observed in the ami-ppd line as well as in the double mutant kix8 kix9 that the rosette leaves are twisted. Finally, at cellular level, the kix8 kix9 double mutants produce more small cells located around the stomata, suggesting that similar processes are altered in ami-ppd and kix8 kix9 lines.
The PPD2-KIX8/9 Complex, a New Signaling Module Regulating Leaf Growth, Is Lost in Grasses
Final cell number is an important parameter influencing leaf size, and meristemoid cells have been shown to generate, through asymmetric division, half of the total number of pavement cells (Geisler et al., 2000), therefore contributing significantly to the determination of final leaf size. PPD proteins, by regulating meristemoid division, are therefore important for regulating leaf growth (White, 2006). In this study, we identified the molecular mechanisms downstream of PPD2 and its protein partners and propose two possible, but not mutually exclusive, models by which PPD2 could act to regulate leaf growth.
Using the PLAZA comparative genomics platform (Proost et al., 2015), we found that the smallest common phylogenetic clade in which PPD2 and KIX8/9 proteins are found is the class of angiosperms. In the monocot lineage, PPD2 and KIX8/9 proteins are present in the genus Musa, but may have been subject to gene loss in Poaceae (grasses). PPD2 is a negative regulator of meristemoid division (White, 2006). Here, we demonstrated that this regulation occurs during the amplifying division phase of these cells. This step, during which meristemoid cells undergo several asymmetric divisions allowing self-renewal and the formation of neighboring pavement cells, is specific for dicots. In grasses, no self-renewing cells are produced in the stomatal lineage (Liu et al., 2009; Vatén and Bergmann, 2012). The absence of the PPD2-KIX8/9 complex in grasses might therefore reflect the absence of meristemoid amplifying divisions observed in leaves having a two-dimensional growth.
METHODS
Plant Material and Growth Conditions
Seeds from all transgenics and mutants were in the Col-0 background. The 35S-ami-ppd line was previously described by Gonzalez et al. (2010). The T-DNA insertion lines kix8 (GABI_422H04) and kix9 (SAIL_1168_G09) were obtained from GABI-KAT and SAIL, respectively (Sessions et al., 2002; Kleinboelting et al., 2012) and genotyped by PCR to verify the homozygosity. Through Gateway cloning, the 35S-PPD2-GR and the 35S-KIX9-GFP constructs were made and introduced into pK7m34GW and pK7FWG2, respectively (Karimi et al., 2002). The expression vectors were used for transformation of Arabidopsis thaliana Col-0 background plants using the floral dip method using Agrobacterium tumefaciens (C58C1, pMP90) (Clough and Bent, 1998). Transformants were selected on media supplied with the corresponding antibiotic and homozygous T3 plant lines were used in the assays. The expression of PPD1, PPD2, KIX8, and KIX9 in the different mutants was verified by qRT-PCR from RNA extracted from seedlings or leaves (Supplemental Figure 8). PPD1 and PPD2 expression were reduced by 36 and 75%, respectively, in the 35S-ami-ppd line; a truncated version of KIX8 mRNA was expressed in the kix8 and kix8 kix9 mutants, and KIX9 expression was reduced by more than 90% in kix9 and kix8 kix9 mutants.
Experiments were conducted with wild-type and transgenic seeds harvested from plants grown side-by-side on the same tray. Plants were grown in vitro in half-strength Murashige and Skoog medium (Murashige and Skoog, 1962) supplemented with 1% sucrose at 21°C under a 16-h-day/8-h-night regime. Plants were also grown in soil under the same day/night regime.
Leaf Growth Parameter Analysis
Individual rosette leaf area and abaxial epidermal cell number and size were measured with ImageJ software (http://rsb.info.nih.gov/ij/) and the fate of meristemoid cells was determined over time. The developmental stages harvested and the protocol followed are described in Supplemental Methods.
Gene Expression Analysis
Total RNA from three biological replicates was extracted with an RNeasy plant mini kit (Qiagen) from the first pair of leaves. For the genome-wide expression analysis, RNAs were hybridized to the ATH1 array (Affymetrix), and expression data were analyzed as previously described (Gonzalez et al., 2010). The developmental stages harvested for RNA extraction, the protocol for the qRT-PCR analysis, and the primers used are described in Supplemental Methods.
Transient expression assays were performed as described previously (Cuéllar Pérez et al., 2014) using protoplasts prepared from a BY-2 tobacco cell culture. The vectors and a detailed protocol used to investigate transcriptional repression potential and transient promoter activation/repression activity are presented in Supplemental Methods.
TChAP-Seq and ChIP-qPCR
TChAP was done according to Verkest et al. (2014) using an Arabidopsis cell suspension culture overexpressing a HBH-tagged PPD2, with a wild-type cell suspension culture as control. Pulled-down DNA libraries were prepared according to the protocol of Illumina and sequenced on a Genome II Analyzer (http://www.illumina.com/applications/sequencing.html). DNA sequencing data processing and peak identification are described in Supplemental Methods. Chromatin affinity purification was done according to Kaufmann et al. (2010) using Arabidopsis transgenic plants overexpressing a GFP-tagged PPD2, with 35S-GFP transgenic plants as control. Chromatin affinity purification was done according to a protocol adapted from Morohashi et al. (2012) using cell cultures overexpressing a GSyellow-tagged PPD2, with a 35S-GSyellow cell culture as control. This TAP tag is a derivative of the TAP tag GSrhino (Van Leene et al., 2015), encoding a fragment of protein-G (G) peptide and a streptavidin binding peptide (S) and replacing the IgG binding domains in the latter by YFP. Pulled-down DNA was analyzed by qPCR as described in Supplemental Methods.
Protein-Protein Interaction Assays
Tandem affinity purification experiments used Arabidopsis cell cultures overexpressing PPD2 tagged with a GS tag at the C-terminal region or KIX8 tagged with a GS tag at the C-terminal region and N-terminal region as previously described (Van Leene et al., 2015). For more details, see Supplemental Methods.
Yeast two- and three-hybrid assays were performed as described by Cuéllar Pérez et al. (2014). Description of the vectors and the media used to test the interactions can be found in Supplemental Methods.
Accession Numbers
Sequence data from this article can be found in the EMBL/GenBank data libraries under the following accession numbers: AT5G59540 [oxidoreductase, 2OG-Fe(II) oxygenase family protein], AT5G67110 (ALCATRAZ [ALC]), AT2G37630 (ASYMMETRIC LEAVES1 [AS1]), AT1G23870 (TREHALOSE-6-PHOSPHATE SYNTHASE S9 [ATTPS9]), AT5G67260 (CYCLIN D3;2 [CYCD3;2]), AT3G50070 (CYCLIN D3;3 [CYCD3;3]), AT5G54510(DWARF IN LIGHT1 [DFL1]), AT5G49700 (DNA binding protein-related), AT2G20875 (kinase-inducible domain interacting 9 [KIX9]), AT1G34245 (EPIDERMAL PATTERNING FACTOR2 [EPF2]), AT5G07180 (ERECTA-LIKE2 [ERL2]), AT1G14900 (HIGH MOBILITY GROUP A [HMGA]), AT5G20740 (invertase/pectin methylesterase inhibitor family protein), AT5G43020 (leucine-rich repeat transmembrane protein kinase, putative), AT3G06120 (MUTE), AT4G31805 (POLAR LOCALIZATION DURING ASYMMETRIC DIVISION AND REDISTRIBUTION [POLAR]), AT4G14713 (PEAPOD1 [PPD1]), AT4G14720 (PEAPOD2 [PPD2]), AT3G54990 (SCHLAFMUTZE [SMZ]), AT5G53210 (SPEECHLESS [SPCH]), AT1G80080 (TOO MANY MOUTHS [TMM]), AT4G14720 (kinase-inducible domain interacting 8 [KIX8]), AT3G24150, AT4G32295, AT5G44200 (CAP BINDING PROTEIN20 [CBP20]), AT3G48750 (CYCLIN-DEPENDENT KINASEA [CDKA1]), M15077 (P. pyralis [firefly] luciferase), and Nr3c1 (rat glucocorticoid receptor). Raw microarray and DNA-sequencing data can be accessed at ArrayExpress (http://www.ebi.ac.uk/arrayexpress/) under the accession numbers E-MTAB-3045 and E-MTAB-3046, respectively.
Supplemental Data
Supplemental Figure 1. Pavement, meristemoid, and mesophyll cell characteristics in ami-ppd leaves.
Supplemental Figure 2. Expression of known stomatal development and patterning genes, cell cycle-related genes, and PPD2 target genes in growing ami-ppd leaves and expression of PPD2 target genes in the 35S-PPD2-GR line treated with or without dexamethasone.
Supplemental Figure 3. Coexpression networks generated with the differentially expressed genes in ami-ppd using CORNET.
Supplemental Figure 4. Genome-wide determination of PPD2 binding sites by TChAP-Seq.
Supplemental Figure 5. Proteins purified by tandem affinity purification using PPD2 and KIX8 as bait and heterodimerization of PPD with JAZ proteins.
Supplemental Figure 6. Alignment of different KIX protein orthologs from eudicots.
Supplemental Figure 7. Cellular phenotype of kix8 kix9 double mutant at 14 DAS.
Supplemental Figure 8. Expression of PPD1 and PPD2 in the ami-ppd line and expression of KIX8 and KIX9 in the T-DNA insertion lines for KIX8 (GABI_422H04) and KIX9 (SAIL_1168_G09) in the double mutant kix8 kix9.
Supplemental Data Set 1. List of genes bound by PPD2 identified by tandem chromatin affinity purification.
Supplemental Data Set 2. Protein identification details obtained with the LTQ Orbitrap after TAP.
Supplemental Data Set 3. Text file of the sequences for the alignment used for the phylogenetic analysis shown in Supplemental Figure 6.
Supplementary Material
Acknowledgments
We thank Paul Wintermans and Janine Colling for help with experiments and Annick Bleys for help in preparing the article. This research received funding from the European Union’s Seventh Framework Program FP7/2007-2013 under Grant 222716-SMARTCELL, the Research Foundation Flanders (FWO) under Project G005212N, the Bijzonder Onderzoeksfonds Methusalem Project (BOF08/01M00408), and the Interuniversity Attraction Poles Programme (IUAP P7/29 “MARS”) initiated by the Belgian Science Policy Office. L.P. and J.V.L. are postdoctoral fellows and A.B. a predoctoral fellow of the FWO. K.S.H. is indebted to the Agency for Innovation by Science and Technology in Flanders. This work was also supported by Ghent University: Multidisciplinary Research Partnership “Bioinformatics: from nucleotides to networks” (Project 01MR0410W) and “Biotechnology for a Sustainable Economy” (Project 01MRB510W).
AUTHOR CONTRIBUTIONS
N.G., L.P., A.B., and A.C.P. designed and performed the experiments. A.N.D. performed TAP experiments. D.E. analyzed mass spectrometry data. J.V.L., E.V.D.S., and N.B. performed TChAP experiments. K.S.H. analyzed TChAP-Seq data. L.D.M., R.D.C., and R.V.B. assisted experiments. D.I., A.G., G.D.J., K.V., and K.G. supervised the research. N.G. and L.P. wrote the article with critical input of the remaining authors.
Glossary
- Ler
Landsberg erecta
- DAS
days after stratification
- TChAP
tandem chromatin affinity purification
- UTR
untranslated region
- ChIP-qPCR
chromatin immunoprecipitation-quantitative PCR
- DEX
dexamethasone
- TAP
tandem affinity purification
- Y2H
yeast two-hybrid
- BY-2
Bright Yellow 2
References
- Andriankaja M., Dhondt S., De Bodt S., Vanhaeren H., Coppens F., De Milde L., Mühlenbock P., Skirycz A., Gonzalez N., Beemster G.T.S., Inzé D. (2012). Exit from proliferation during leaf development in Arabidopsis thaliana: a not-so-gradual process. Dev. Cell 22: 64–78. [DOI] [PubMed] [Google Scholar]
- Bai Y., Meng Y., Huang D., Qi Y., Chen M. (2011). Origin and evolutionary analysis of the plant-specific TIFY transcription factor family. Genomics 98: 128–136. [DOI] [PubMed] [Google Scholar]
- Beemster G.T.S., De Veylder L., Vercruysse S., West G., Rombaut D., Van Hummelen P., Galichet A., Gruissem W., Inzé D., Vuylsteke M. (2005). Genome-wide analysis of gene expression profiles associated with cell cycle transitions in growing organs of Arabidopsis. Plant Physiol. 138: 734–743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergmann D.C., Sack F.D. (2007). Stomatal development. Annu. Rev. Plant Biol. 58: 163–181. [DOI] [PubMed] [Google Scholar]
- Canet J.V., Dobón A., Tornero P. (2012). Non-recognition-of-BTH4, an Arabidopsis mediator subunit homolog, is necessary for development and response to salicylic acid. Plant Cell 24: 4220–4235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Causier B., Ashworth M., Guo W., Davies B. (2012). The TOPLESS interactome: a framework for gene repression in Arabidopsis. Plant Physiol. 158: 423–438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chini A., Fonseca S., Chico J.M., Fernández-Calvo P., Solano R. (2009). The ZIM domain mediates homo- and heteromeric interactions between Arabidopsis JAZ proteins. Plant J. 59: 77–87. [DOI] [PubMed] [Google Scholar]
- Chini A., Fonseca S., Fernández G., Adie B., Chico J.M., Lorenzo O., García-Casado G., López-Vidriero I., Lozano F.M., Ponce M.R., Micol J.L., Solano R. (2007). The JAZ family of repressors is the missing link in jasmonate signalling. Nature 448: 666–671. [DOI] [PubMed] [Google Scholar]
- Chung H.S., Howe G.A. (2009). A critical role for the TIFY motif in repression of jasmonate signaling by a stabilized splice variant of the JASMONATE ZIM-domain protein JAZ10 in Arabidopsis. Plant Cell 21: 131–145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough, S.J., and Bent, A.F. (1998). Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana Plant J. 16: 735–743. [DOI] [PubMed]
- Cuéllar Pérez A., Nagels Durand A., Vanden Bossche R., De Clercq R., Persiau G., Van Wees S.C.M., Pieterse C.M.J., Gevaert K., De Jaeger G., Goossens A., Pauwels L. (2014). The non-JAZ TIFY protein TIFY8 from Arabidopsis thaliana is a transcriptional repressor. PLoS One 9: e84891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dewitte W., Scofield S., Alcasabas A.A., Maughan S.C., Menges M., Braun N., Collins C., Nieuwland J., Prinsen E., Sundaresan V., Murray J.A.H. (2007). Arabidopsis CYCD3 D-type cyclins link cell proliferation and endocycles and are rate-limiting for cytokinin responses. Proc. Natl. Acad. Sci. USA 104: 14537–14542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donnelly P.M., Bonetta D., Tsukaya H., Dengler R.E., Dengler N.G. (1999). Cell cycling and cell enlargement in developing leaves of Arabidopsis. Dev. Biol. 215: 407–419. [DOI] [PubMed] [Google Scholar]
- Elsner J., Michalski M., Kwiatkowska D. (2012). Spatiotemporal variation of leaf epidermal cell growth: a quantitative analysis of Arabidopsis thaliana wild-type and triple cyclinD3 mutant plants. Ann. Bot. (Lond.) 109: 897–910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geisler M., Nadeau J., Sack F.D. (2000). Oriented asymmetric divisions that generate the stomatal spacing pattern in Arabidopsis are disrupted by the too many mouths mutation. Plant Cell 12: 2075–2086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez N., Beemster G.T.S., Inzé D. (2009). David and Goliath: what can the tiny weed Arabidopsis teach us to improve biomass production in crops? Curr. Opin. Plant Biol. 12: 157–164. [DOI] [PubMed] [Google Scholar]
- Gonzalez N., Vanhaeren H., Inzé D. (2012). Leaf size control: complex coordination of cell division and expansion. Trends Plant Sci. 17: 332–340. [DOI] [PubMed] [Google Scholar]
- Gonzalez N., et al. (2010). Increased leaf size: different means to an end. Plant Physiol. 153: 1261–1279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han S.-K., Song J.-D., Noh Y.-S., Noh B. (2007). Role of plant CBP/p300-like genes in the regulation of flowering time. Plant J. 49: 103–114. [DOI] [PubMed] [Google Scholar]
- Hepworth J., Lenhard M. (2014). Regulation of plant lateral-organ growth by modulating cell number and size. Curr. Opin. Plant Biol. 17: 36–42. [DOI] [PubMed] [Google Scholar]
- Heyman J., Cools T., Vandenbussche F., Heyndrickx K.S., Van Leene J., Vercauteren I., Vanderauwera S., Vandepoele K., De Jaeger G., Van Der Straeten D., De Veylder L. (2013). ERF115 controls root quiescent center cell division and stem cell replenishment. Science 342: 860–863. [DOI] [PubMed] [Google Scholar]
- Jan Y.N., Jan L.Y. (1998). Asymmetric cell division. Nature 392: 775–778. [DOI] [PubMed] [Google Scholar]
- Jedidi I., Zhang F., Qiu H., Stahl S.J., Palmer I., Kaufman J.D., Nadaud P.S., Mukherjee S., Wingfield P.T., Jaroniec C.P., Hinnebusch A.G. (2010). Activator Gcn4 employs multiple segments of Med15/Gal11, including the KIX domain, to recruit mediator to target genes in vivo. J. Biol. Chem. 285: 2438–2455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kagale S., Links M.G., Rozwadowski K. (2010). Genome-wide analysis of ethylene-responsive element binding factor-associated amphiphilic repression motif-containing transcriptional regulators in Arabidopsis. Plant Physiol. 152: 1109–1134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karimi M., Inzé D., Depicker A. (2002). GATEWAYTM vectors for Agrobacterium-mediated plant transformation. Trends Plant Sci. 7: 193–195. [DOI] [PubMed] [Google Scholar]
- Kaufmann K., Muiño J.M., Østerås M., Farinelli L., Krajewski P., Angenent G.C. (2010). Chromatin immunoprecipitation (ChIP) of plant transcription factors followed by sequencing (ChIP-SEQ) or hybridization to whole genome arrays (ChIP-CHIP). Nat. Protoc. 5: 457–472. [DOI] [PubMed] [Google Scholar]
- Kleinboelting N., Huep G., Kloetgen A., Viehoever P., Weisshaar B. (2012). GABI-Kat SimpleSearch: new features of the Arabidopsis thaliana T-DNA mutant database. Nucleic Acids Res. 40: D1211–D1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krogan N.T., Hogan K., Long J.A. (2012). APETALA2 negatively regulates multiple floral organ identity genes in Arabidopsis by recruiting the co-repressor TOPLESS and the histone deacetylase HDA19. Development 139: 4180–4190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lacatus G., Sunter G. (2009). The Arabidopsis PEAPOD2 transcription factor interacts with geminivirus AL2 protein and the coat protein promoter. Virology 392: 196–202. [DOI] [PubMed] [Google Scholar]
- Lau O.S., Davies K.A., Chang J., Adrian J., Rowe M.H., Ballenger C.E., Bergmann D.C. (2014). Direct roles of SPEECHLESS in the specification of stomatal self-renewing cells. Science 345: 1605–1609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J.E., Golz J.F. (2012). Diverse roles of Groucho/Tup1 co-repressors in plant growth and development. Plant Signal. Behav. 7: 86–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu T., Ohashi-Ito K., Bergmann D.C. (2009). Orthologs of Arabidopsis thaliana stomatal bHLH genes and regulation of stomatal development in grasses. Development 136: 2265–2276. [DOI] [PubMed] [Google Scholar]
- Moreno J.E., Shyu C., Campos M.L., Patel L.C., Chung H.S., Yao J., He S.Y., Howe G.A. (2013). Negative feedback control of jasmonate signaling by an alternative splice variant of JAZ10. Plant Physiol. 162: 1006–1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morohashi K., et al. (2012). A genome-wide regulatory framework identifies maize pericarp color1 controlled genes. Plant Cell 24: 2745–2764. Erratum. Plant Cell 24: 3853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murashige T., Skoog F. (1962). A revised medium for rapid growth and bio assays with tobacco tissue cultures. Physiol. Plant. 15: 473–497. [Google Scholar]
- Noir S., Bömer M., Takahashi N., Ishida T., Tsui T.-L., Balbi V., Shanahan H., Sugimoto K., Devoto A. (2013). Jasmonate controls leaf growth by repressing cell proliferation and the onset of endoreduplication while maintaining a potential stand-by mode. Plant Physiol. 161: 1930–1951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pauwels L., Goossens A. (2011). The JAZ proteins: a crucial interface in the jasmonate signaling cascade. Plant Cell 23: 3089–3100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pauwels L., et al. (2010). NINJA connects the co-repressor TOPLESS to jasmonate signalling. Nature 464: 788–791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pillitteri L.J., Dong J. (2013). Stomatal development in Arabidopsis. Arabidopsis Book 11: e0162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pillitteri L.J., Torii K.U. (2012). Mechanisms of stomatal development. Annu. Rev. Plant Biol. 63: 591–614. [DOI] [PubMed] [Google Scholar]
- Pillitteri L.J., Peterson K.M., Horst R.J., Torii K.U. (2011). Molecular profiling of stomatal meristemoids reveals new component of asymmetric cell division and commonalities among stem cell populations in Arabidopsis. Plant Cell 23: 3260–3275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Proost S., Van Bel M., Vaneechoutte D., Van de Peer Y., Inzé D., Mueller-Roeber B., Vandepoele K. (2015). PLAZA 3.0: an access point for plant comparative genomics. Nucleic Acids Res. 43: D974–D981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radhakrishnan I., Pérez-Alvarado G.C., Parker D., Dyson H.J., Montminy M.R., Wright P.E. (1997). Solution structure of the KIX domain of CBP bound to the transactivation domain of CREB: a model for activator:coactivator interactions. Cell 91: 741–752. [DOI] [PubMed] [Google Scholar]
- Reeves R. (2001). Molecular biology of HMGA proteins: hubs of nuclear function. Gene 277: 63–81. [DOI] [PubMed] [Google Scholar]
- Scheres B., Benfey P.N. (1999). Asymmetric cell division in plants. Annu. Rev. Plant Physiol. Plant Mol. Biol. 50: 505–537. [DOI] [PubMed] [Google Scholar]
- Sessions A., et al. (2002). A high-throughput Arabidopsis reverse genetics system. Plant Cell 14: 2985–2994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao Q., Guo D., Wei B., Zhang F., Pang C., Jiang H., Zhang J., Wei T., Gu H., Qu L.-J., Qin G. (2013). The TIE1 transcriptional repressor links TCP transcription factors with TOPLESS/TOPLESS-RELATED corepressors and modulates leaf development in Arabidopsis. Plant Cell 25: 421–437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thakur J.K., Yadav A., Yadav G. (2014). Molecular recognition by the KIX domain and its role in gene regulation. Nucleic Acids Res. 42: 2112–2125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thakur J.K., Agarwal P., Parida S., Bajaj D., Pasrija R. (2013). Sequence and expression analyses of KIX domain proteins suggest their importance in seed development and determination of seed size in rice, and genome stability in Arabidopsis. Mol. Genet. Genomics 288: 329–346. [DOI] [PubMed] [Google Scholar]
- Thines B., Katsir L., Melotto M., Niu Y., Mandaokar A., Liu G., Nomura K., He S.Y., Howe G.A., Browse J. (2007). JAZ repressor proteins are targets of the SCF(COI1) complex during jasmonate signalling. Nature 448: 661–665. [DOI] [PubMed] [Google Scholar]
- Tzamarias D., Struhl K. (1994). Functional dissection of the yeast Cyc8-Tup1 transcriptional co-repressor complex. Nature 369: 758–761. [DOI] [PubMed] [Google Scholar]
- Usadel B., Nagel A., Steinhauser D., Gibon Y., Bläsing O.E., Redestig H., Sreenivasulu N., Krall L., Hannah M.A., Poree F., Fernie A.R., Stitt M. (2006). PageMan: an interactive ontology tool to generate, display, and annotate overview graphs for profiling experiments. BMC Bioinformatics 7: 535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanholme B., Grunewald W., Bateman A., Kohchi T., Gheysen G. (2007). The tify family previously known as ZIM. Trends Plant Sci. 12: 239–244. [DOI] [PubMed] [Google Scholar]
- Van Leene J., et al. (2015). An improved toolbox to unravel the plant cellular machinery by tandem affinity purification of Arabidopsis protein complexes. Nat. Protoc. 10: 169–187. [DOI] [PubMed] [Google Scholar]
- Vatén A., Bergmann D.C. (2012). Mechanisms of stomatal development: an evolutionary view. Evodevo 3: 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verkest A., et al. (2014). A generic tool for transcription factor target gene discovery in Arabidopsis cell suspension cultures based on tandem chromatin affinity purification. Plant Physiol. 164: 1122–1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L., Kim J., Somers D.E. (2013). Transcriptional corepressor TOPLESS complexes with pseudoresponse regulator proteins and histone deacetylases to regulate circadian transcription. Proc. Natl. Acad. Sci. USA 110: 761–766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White D.W.R. (2006). PEAPOD regulates lamina size and curvature in Arabidopsis. Proc. Natl. Acad. Sci. USA 103: 13238–13243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winkler C.J., Ponce A., Courey A.J. (2010). Groucho-mediated repression may result from a histone deacetylase-dependent increase in nucleosome density. PLoS One 5: e10166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong K.H., Struhl K. (2011). The Cyc8-Tup1 complex inhibits transcription primarily by masking the activation domain of the recruiting protein. Genes Dev. 25: 2525–2539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang F., et al. (2006). An ARC/Mediator subunit required for SREBP control of cholesterol and lipid homeostasis. Nature 442: 700–704. [DOI] [PubMed] [Google Scholar]
- Zhang Y., Liu T., Meyer C.A., Eeckhoute J., Johnson D.S., Bernstein B.E., Nusbaum C., Myers R.M., Brown M., Li W., Liu X.S. (2008). Model-based analysis of ChIP-Seq (MACS). Genome Biol. 9: R137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X., Benson K.F., Ashar H.R., Chada K. (1995). Mutation responsible for the mouse pygmy phenotype in the developmentally regulated factor HMGI-C. Nature 376: 771–774. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.