A DCL1-dependent siRNA pathway bypasses the requirement for DCL4 in development, which can reduce selective constraints on DCL4, allowing it to diversify in response to viral suppressors.
Abstract
Dicer enzymes function at the core of RNA silencing to defend against exogenous RNA or to regulate endogenous genes. Plant DICER-LIKE4 (DCL4) performs dual functions, acting in antiviral defense and in development via the biogenesis of trans-acting short-interfering RNAs (siRNAs) termed tasiR-ARFs. These small RNAs play an essential role in the grasses, spatially defining the expression domain of AUXIN RESPONSE FACTOR3 (ARF3) transcription factors. However, contrary to tasiR-ARFs’ essential function in development, DCL4 proteins exhibit strong evidence of recurrent adaptation typical of host factors involved in antiviral immunity. Here, we address how DCL4 balances its role in development with pressures to diversify in response to viral attack. We show that, in contrast to other tasiR-ARF biogenesis mutants, dcl4 null alleles have an uncharacteristically mild phenotype, correlated with normal expression of select arf3 targets. Loss of DCL4 activity yields a class of 22-nucleotide tasiR-ARF variants associated with the processing of arf3 transcripts into 22-nucleotide secondary siRNAs by DCL1. Our findings reveal a DCL1-dependent siRNA pathway that bypasses the otherwise adverse developmental effects of mutations in DCL4. This pathway is predicted to have important implications for DCL4’s role in antiviral defense by reducing the selective constraints on DCL4 and allowing it to diversify in response to viral suppressors.
INTRODUCTION
RNA silencing is an evolutionarily conserved process with central roles in the regulation of endogenous gene expression and in the protection against foreign RNAs derived from viruses and transposable elements. These functions involve discrete genetic pathways resulting from diversification of the underlying core components. For example, most plant genomes encode for four DICER-LIKE (DCL) enzymes that have subfunctionalized to generate distinct small RNAs. The canonical microRNA (miRNA) pathway relies on DCL1, whereas DCL2, DCL3, and DCL4 mediate the processing of 22-, 24-, and 21-nucleotide short-interfering RNAs (siRNAs), respectively, from a diverse array of long double-stranded RNA (dsRNA) precursors (Axtell, 2013; Bologna and Voinnet, 2014). Although these three DCL enzymes show hierarchical activity with respect to processing of RNA templates (e.g., DCL2 can substitute for DCL4 in the processing of viral-derived dsRNAs), the small RNAs they generate are not necessarily functionally equivalent, in part owing to their distinct lengths. Specifically, DCL3 is required for the production of heterochromatic siRNAs, whereas DCL4 is the predominant DCL enzyme required for antiviral defense (Pumplin and Voinnet, 2013; Seo et al., 2013).
Consistent with this role, DCL4 proteins show strong evidence for recurrent adaptation that is not seen in other DCL enzymes (Mukherjee et al., 2013). Such sequence diversification is a hallmark of many defense proteins and implies a long-term evolutionary arms race between DCL4 and viral factors that evolved to evade or suppress the RNA silencing pathway. Evidence of recurrent selection is particularly strong at the essential PAZ domain and especially so in the monocots, where this RNA binding pocket has undergone a complete reversal of electrostatic charge relative to other known DCL enzymes in eudicots and animals (Mukherjee et al. 2013).
Distinct from its function in antiviral immunity, DCL4 also mediates the processing of trans-acting short-interfering RNAs (ta-siRNAs), including the developmentally relevant tasiR-ARFs (Gasciolli et al., 2005; Xie et al., 2005; Yoshikawa et al., 2005). Importantly, this role for DCL4 in development cannot be functionally substituted for by DCL2, contrary to its role in antiviral defense. The tasiR-ARFs are generated through the subspecialized and evolutionarily conserved TAS3 ta-siRNA pathway, in which miR390-loaded ARGONAUTE7 (AGO7) targets TAS3 transcripts and triggers their conversion into dsRNAs by the activity of RNA-DEPENDENT RNA POLYMERASE6 (RDR6) and SUPPRESSOR-OF-GENE-SILENCING3 (SGS3) (Allen et al., 2005; Montgomery et al., 2008; Fei et al., 2013). These dsRNA intermediates are subsequently processed by DCL4 into 21-nucleotide ta-siRNAs that are phased to yield discrete small RNA species. A subset of these, the tasiR-ARFs, are biologically active and target transcripts of the genes encoding the AUXIN RESPONSE FACTOR3 (ARF3) and ARF4 transcription factors (Allen et al., 2005; Nogueira et al., 2007; Chitwood et al., 2009; Yifhar et al., 2012; Zhou et al., 2013).
ARF3 and ARF4 function in the specification of adaxial-abaxial (top-bottom) leaf polarity, which underlies the flattened outgrowth of the leaf and directs the differentiation of distinct cell types within the leaf’s adaxial and abaxial domains (Waites and Hudson, 1995). Patterning of the adaxial-abaxial axis is driven by an intricate gene regulatory network (reviewed in Husbands et al., 2009). Integral to this network are two sets of conserved transcription factors that promote either adaxial or abaxial cell fate and are expressed in complementary domains on the upper or lower side of the developing leaf, respectively. In addition, small RNAs form central network components that provide positional information needed to separate the two domains. miR166, which accumulates in a gradient originating from the bottom side of the leaf, guides the cleavage of HD-ZIPIII transcripts, limiting expression of these adaxial determinants to the top side of primordia (Emery et al., 2003; Juarez et al., 2004a; Nogueira et al., 2009; Yao et al., 2009). Conversely, movement of tasiR-ARFs, whose biogenesis is confined to the two uppermost cell layers of developing leaf primordia, generates an abaxially dissipating gradient that restricts expression of the ARF3/4 abaxial determinants to a precisely defined domain on the leaf’s lower side (Chitwood et al. 2009).
In Arabidopsis thaliana, mutants that block ta-siRNA biogenesis, i.e., sgs3, rdr6, ago7, and dcl4, exhibit subtle phenotypes, developing downward curled leaves that are weakly abaxialized and undergo an accelerated transition from the juvenile to adult phase (Peragine et al., 2004; Xie et al., 2005; Yoshikawa et al., 2005). Disruption of this pathway in other eudicot species similarly results in viable plants with partially abaxialized or highly lobed leaves (Yan et al., 2010; Zhou et al., 2013). Even in tomato (Solanum lycopersicum), where the effects of ta-siRNA biogenesis mutants on leaf polarity are more exacerbated, such mutants develop to maturity and remain partially fertile (Yifhar et al., 2012; Brooks et al., 2014). In contrast, ta-siRNA pathway mutants in the grasses show severe developmental defects. Partial loss-of-function mutations in maize leafbladeless1 (lbl1/sgs3) and ragged seedling2 (rgd2/ago7) or the rice shootless genes give rise to severe adaxial-abaxial leaf polarity defects and strong mutant alleles in the pathway perturb stem cell activity in the shoot apical meristem, leading to embryonic or early seedling arrest (Nogueira et al., 2007; Nagasaki et al., 2007; Itoh et al., 2008; Douglas et al., 2010).
The essential developmental role of the ta-siRNA pathway in the grasses is expected to place unique constraints on the evolution of DCL4. However, such limitations are contradictory to what is observed and DCL4’s role in antiviral defense (Mukherjee et al., 2013). Here, we address how DCL4 might balance its two disparate functions and describe a novel DCL1-dependent small RNA pathway that bypasses the requirement for DCL4 in development. We show that in contrast to other ta-siRNA pathway mutants, loss of DCL4 function results in an uncharacteristically mild phenotype, correlated with wild-type expression of select tasiR-ARF targets (arf3a and arf3d). Deep sequencing of small RNAs shows that maize (Zea mays) dcl4 mutants generate a population of 22-nucleotide tasiR-ARF variants that are associated with the processing of arf3a and arf3d transcripts into a novel class of 22-nucleotide transitive siRNAs by DCL1. Loss of the 22-nucleotide arf3a- and arf3d-derived siRNAs in dcl1 dcl4 double mutants results in misexpression of these polarity determinants and a strongly enhanced phenotype that is equivalent to other ta-siRNA biogenesis mutants. These findings uncover a DCL1-dependent siRNA pathway that bypasses potential adverse developmental effects of mutations in DCL4. This novel DCL1 activity is predicted to have major implications for DCL4’s role in antiviral defense by reducing selective constraints on DCL4 and allowing it to diversify in response to viral suppressors.
RESULTS
The dcl4 Phenotype Is Suppressed in Comparison to That of Other ta-siRNA Pathway Mutants
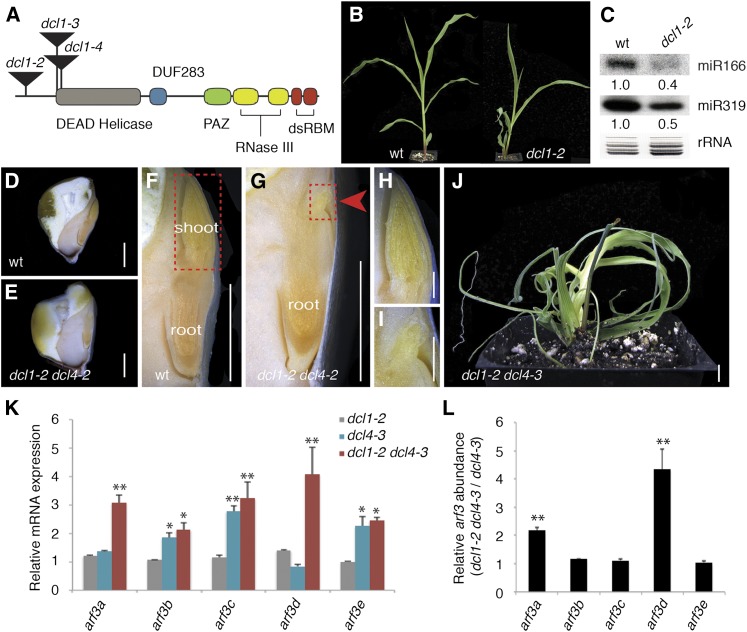
DCL4 proteins, particularly those in the grasses, show strong evidence for recurrent adaptation targeting the essential PAZ domain that is not seen in other DCL enzymes (Mukherjee et al., 2013). While such sequence diversification is consistent with DCL4’s role in antiviral defense, and is indeed a hallmark of many defense proteins, this is unexpected considering the essential role of tasiR-ARFs in development. To address how DCL4 might balance its role in development with pressures to diversify as an antiviral defense protein, we screened mutagenized maize populations for plants exhibiting adaxial-abaxial leaf polarity defects reminiscent of lbl1 and rgd2 that map near dcl4 on chromosome 10. Of the leaf polarity mutants identified, four resulted from mutations in dcl4. Sequence analysis revealed that the dcl4-1 allele contains a small rearrangement, which results in a net insertion of 21 nucleotides into the PAZ domain (Figure 1A; Supplemental Figure 1A), whereas the dcl4-3 allele carries a missense mutation that converts the highly conserved Ser-473 in the Helicase domain to Leu (Supplemental Figure 1B). Sequence analysis of the remaining dcl4-2 and dcl4-4 alleles showed that these result from nonsense mutations at amino acid positions 283 and 1109, respectively. The dcl4-4 allele is predicted to encode a truncated protein that lacks part of the enzymatic RNase III domain, as well as the dsRNA binding motifs, and is thus likely a complete loss-of-function allele. Likewise, the 5′ location of the stop codon in dcl4-2 predicts this to be a null allele, especially as alternatively spliced transcripts that could potentially excise the premature stop codon from dcl4-2 were not detected.
Figure 1.
In Contrast to Other tasiR-ARF Biogenesis Components, Null Alleles of dcl4 Have an Uncharacteristically Mild Leaf Polarity Phenotype.
(A) Structure of DCL4 with conserved protein domains and amino acid substitutions in dcl4-2, dcl4-3, and dcl4-4 indicated. The dcl4-1 allele results from a net insertion of 21-nucleotide in the critical PAZ domain.
(B) Compared with the wild type, dcl4-2 seedlings show a mild leaf polarity defect. The arrow marks an occasional radial, thread-like leaf.
(C) Diagrammatic representations of the LBL1/SGS3, RGD2/AGO7, and RDR6 proteins with conserved domains and mutations indicated.
(D) to (F) When introgressed into B73, lbl1-rgd1 (D) causes most seedling leaves to become radial and strongly abaxialized, whereas rgd2-Ds1 (E) and rdr6-1 (F) give rise to seedlings with reduced fully abaxialized organs that arrest shortly after germination. Bars = 1 cm.
Introgression of the dcl4 alleles into the B73 inbred background yielded plants that, despite the difference in mutation, display comparable phenotypes. The dcl4 mutants develop irregular leaves associated with a variable loss of adaxial identity (Figure 1B; Supplemental Figure 2). Even in a single plant, the phenotype of individual leaves varies widely, ranging from having a near wild-type morphology, to occasionally appearing thread-like (Supplemental Figures 3A to 3E). As seen for lbl1 and rgd2 (Timmermans et al., 1998; Dotto et al., 2014), the thread-like dcl4 leaves lack adaxial features, such as macrohairs and bulliform cells, and show a radial symmetry in cross sections with photosynthetic and epidermal cells surrounding a central vascular bundle (Supplemental Figures 3F and 3I). However, this retains adaxial-abaxial polarity, as xylem forms at the adaxial pole and phloem at the abaxial pole. More often, dcl4 leaves show a mild polarity defect and develop pairs of ectopic blade outgrowths surrounding regions of abaxial identity on the upper leaf surface (Supplemental Figures 3C and 3G). In addition, the dcl4 phenotype includes a subtle stomatal defect that is prominent in the first juvenile leaves (Supplemental Figure 4). A defect in stomatal morphology has not been noted previously for other ta-siRNA biogenesis mutants. Moreover, the adaxial-abaxial leaf polarity defects of dcl4 plants, even those carrying a dcl4 null allele, appear attenuated in comparison to the phenotypes of mutants affecting earlier steps in ta-siRNA biogenesis.
The lbl1-rgd1 allele, which results from an amino acid substitution at a critical residue in the Zn-finger domain, causes most seedling leaves to become radial and strongly abaxialized when introgressed into B73 (Figures 1C and 1D; Nogueira et al., 2007). Such mutants are also severely stunted and fail to flower. The polarity phenotype conditioned by the rgd2-Ds1 allele, which carries a Ds transposon insertion in the essential PIWI domain, is even more severe and leads to early seedling lethality or occasionally a shootless phenotype (Figures 1C and 1E; Dotto et al., 2014). The less severe phenotype of lbl1-rgd1 mutants may be attributed to the fact that this allele still retains some activity. Supporting this notion, double mutants between lbl1-rgd1 and dcl4-2 exhibit an enhanced phenotype that resembles the phenotype of rgd2-Ds1 (Supplemental Figure 5). In addition, we isolated two independent null alleles for the single maize ortholog of RDR6, which together with LBL1/SGS3 converts TAS3 cleavage products into dsRNA templates for processing by DCL4 (Fei et al., 2013). The rdr6-1 allele results from a nonsense mutation in the conserved RNA-dependent RNA polymerase domain (Figure 1C), whereas the rdr6-2 allele derives from a Mu transposon insertion in the coding sequence, 1005 bp downstream from the start codon. Like rgd2-Ds1, rdr6-1 (Figure 1F) and rdr6-2 in the B73 inbred background give rise to seedlings with reduced and fully abaxialized organs that arrest shortly after germination.
In eudicots, the developmental defects of dcl4 loss-of-function mutants are mostly indistinguishable from those of mutations in other ta-siRNA pathway components (Gasciolli et al., 2005; Xie et al., 2005; Yoshikawa et al., 2005; Yifhar et al., 2012). It is therefore noteworthy that null alleles of maize dcl4, present as a single copy in the maize genome, give rise to a relatively weak phenotype compared with the developmental defects of mutations blocking earlier steps in ta-siRNA biogenesis.
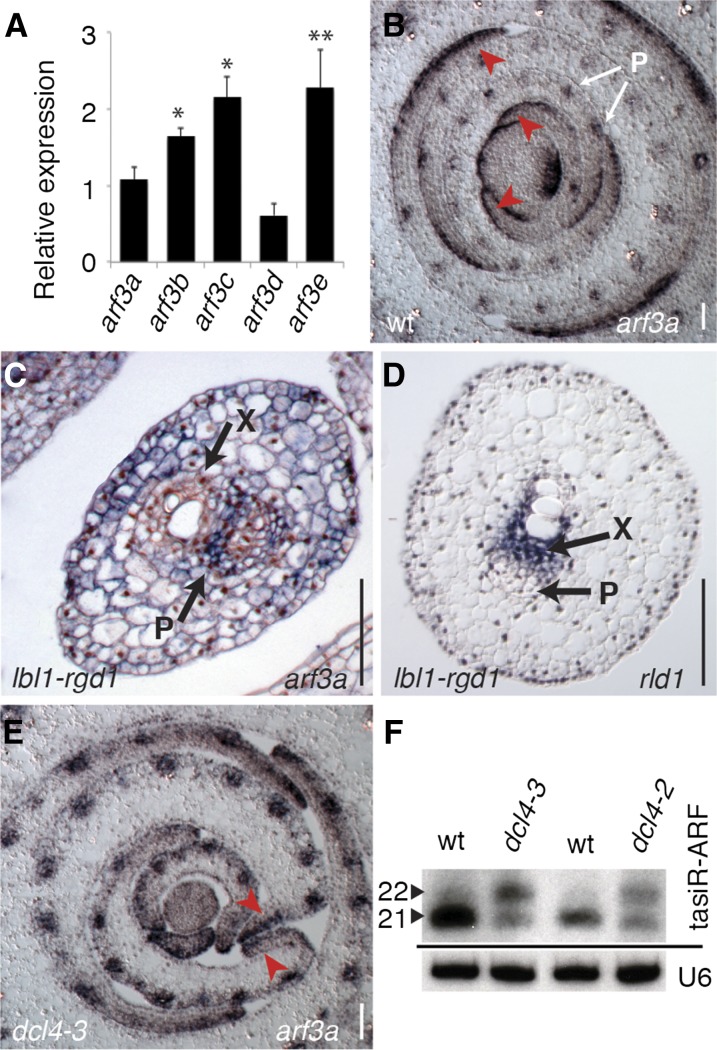
dcl4 and lbl1 Differentially Affect Expression of the arf3 tasiR-ARF Targets
The mild effect loss of DCL4 has on leaf polarity suggests its impact on ta-siRNA target expression is suppressed in comparison to lbl1, rgd2, and rdr6. A recent study revealed that the five maize ARF3 homologs (arf3a-e) are the only ta-siRNA targets with a role in shoot development (Dotto et al., 2014). A positive correlation between ARF3 dosage and phenotype severity has been observed in Arabidopsis and tomato (Fahlgren et al., 2006; Hunter et al., 2006; Yifhar et al., 2012), further supporting the idea that in comparison to other ta-siRNA biogenesis mutants, dcl4 affects expression of the arf3 targets to a lesser extent. To test this possibility directly, we examined the transcript levels for arf3a-e in dcl4 shoot apices. Considering that the arf3 targets are cleaved at one or both of their tasiR-ARF target sites (Dotto et al., 2014), their expression is predicted to increase upon loss of tasiR-ARF function. Indeed, arf3 transcript levels are increased in lbl1-rgd1 and rgd2-Ds1 shoot apices; likewise, arf3b, arf3c, and arf3e transcripts are upregulated approximately 2-fold in dcl4-3 mutants (Figure 2A). However, expression levels for arf3a and arf3d are not significantly changed in dcl4. This is particularly intriguing, as transcript levels for arf3a and arf3d increased 2- to 3-fold in lbl1-rgd1 and rgd2-Ds1 mutants, and expression of a tasiR-ARF-insensitive allele of arf3a largely recapitulates the phenotypic defects seen in these ta-siRNA biogenesis mutants (Dotto et al., 2014).
Figure 2.
Expression of the arf3 tasiR-ARF Targets Is Affected Differently in dcl4 and lbl1.
(A) RT-qPCR analysis shows transcript levels for arf3b, arf3c, and arf3e are significantly increased in dcl4-3 shoot apices. Expression values (means ± se) normalized to the wild type were calculated based on at least three independent biological replicates (*P < 0.05 and **P < 0.01).
(B) arf3a is expressed on the abaxial side of wild-type leaf primordia (red arrows) and in phloem precursors of vascular bundles.
(C) In radial lbl1-rgd1 leaves, arf3a is expressed throughout the ground tissue, although expression in the vascular bundle remains limited to phloem precursor cells.
(D) Expression of rld1 is lost from the adaxial side of radial lbl1-rgd1 leaves but persists in pro-xylem cells.
(E) In dcl4-3, arf3a shows a mostly abaxial pattern of expression, including in leaves with abnormal margins (red arrows).
(F) Small RNA gel blots show that 21-nucleotide tasiR-ARFs persist in dcl4-3 and dcl4-2 shoot apices. In addition, these mutants accumulate 22-nucleotide tasiR-ARF variants. X, xylem; P, phloem. Bars = 100 μm.
To further assess the possibility that the relatively mild phenotype of dcl4 mutants results from divergent expression of arf3 family members, we compared the spatial pattern of arf3a expression in lbl1-rgd1 and dcl4-3 leaf primordia. In wild-type plants, arf3a exhibits a precise polar expression pattern, with transcripts accumulating specifically on the abaxial side of developing leaf primordia and in immature phloem cells (Figure 2B). However, in lbl1-rgd1 mutants, arf3a accumulates throughout the ground tissue of young radialized leaves, yet maintains its polar expression in the vasculature (Figure 2C). Concomitant with the misexpression of arf3a, expression of the HD-ZIPIII family member, rolled leaf1 (rld1), which is normally expressed on the adaxial side of leaf primordia (Juarez et al., 2004a), is lost from the ground tissue of radialized lbl1-rgd1 leaves (Figure 2D). These data corroborate a role for the ARF3 transcription factors in promoting abaxial cell fate and highlight the importance of their regulation by tasiR-ARFs in establishing adaxial-abaxial polarity in the incipient leaf. However, expression of rld1 in the xylem precursors of the central vasculature does persist in lbl1-rgd1, suggesting a less critical role for the ta-siRNA pathway in patterning this tissue.
Examination of dcl4 leaf primordia shows that arf3a expression remains mostly polarized with transcripts localizing preferentially to the abaxial side, even in leaf margins that do not resemble those of the wild type (Figure 2E). However, unlike the wild type, expression in the abaxial domain is more diffuse and slightly expanded, perhaps indicative of less efficient tasiR-ARF regulation. Thus, while arf3a is misexpressed in lbl1-rgd1 leaf primordia, its spatial distribution is largely unaffected in dcl4. Together with their distinct effects on arf3a and arf3d transcript levels (Figure 2A; Dotto et al., 2014), this finding supports the premise that the tasiR-ARF-mediated regulation of arf3 targets is affected differently depending on whether DCL4 or LBL1 function is perturbed.
21-Nucleotide tasiR-ARF Biogenesis Persists in the Absence of DCL4
While differential expression of arf3a and arf3d provides a plausible basis for the distinct severities of the dcl4 and lbl1 phenotypes, the reason why dcl4 and lbl1 affect these targets differently is not immediately apparent. One possibility is that the hierarchical activity of DCL enzymes allows a population of functional tasiR-ARFs to persist in dcl4. Indeed, small RNA gel blot analysis showed that a population of 21-nucleotide tasiR-ARF-related small RNAs remains in dcl4, even in the dcl4-2 mutant (Figure 2F). In addition, a fraction of tasiR-ARFs is shifted in size to form a class of 22-nucleotide variants. A shift in ta-siRNA size has also been observed in Arabidopsis and tomato dcl4 mutants; however, in these species, the 21-nucleotide ta-siRNA class is completely lost (Gasciolli et al., 2005; Xie et al., 2005; Dunoyer et al., 2007; Howell et al., 2007; Yifhar et al., 2012). Moreover, as processing of the TAS3 precursors into 22-nucleotide siRNAs offsets the normal phasing of ta-siRNAs, the biologically active tasiR-ARFs are lost in Arabidopsis and tomato dcl4 mutants, such that their developmental phenotypes resemble those of mutants blocking earlier steps in ta-siRNA biogenesis (Gasciolli et al., 2005; Xie et al., 2005; Yoshikawa et al., 2005; Yifhar et al., 2012). In some plant species, such as the moss Physcomitrella patens, dcl4 mutants give rise to a strongly enhanced developmental phenotype due to detrimental “off target” effects of the novel siRNAs generated by the hierarchically acting DCL enzymes (Arif et al., 2012). Thus, it is noteworthy that in maize, a 21-nucleotide class of tasiR-ARFs remains, even in dcl4-2 mutants. Although this finding does not immediately explain why arf3a and arf3d specifically are differentially regulated between lbl1 and dcl4 mutants, the 21-nucleotide tasiR-ARFs remaining in dcl4 may contribute to its comparatively mild phenotype. Indeed, tasiR-ARF levels are severely reduced in lbl1 and rgd2, both of which exhibit dramatically enhanced phenotypes (Nogueira et al., 2007; Douglas et al., 2010).
dcl1 and dcl4 Mutations Show a Synergistic Interaction
A persistence of 21-nucleotide tasiR-ARFs in dcl4-2 suggests the presence of functional redundancy or hierarchy among the maize DCL enzymes that allows for the correct processing of double-stranded TAS3 transcripts in the absence of DCL4. The maize genome encodes five DCL enzymes (Nogueira et al., 2009). In addition to dcl4, single gene copies exist of dcl1 and dcl2, whereas dcl3 underwent a duplication event to give rise to two genes, dcl3a and dcl3b (aka dcl5), which are expressed primarily during the vegetative and reproductive stages of development, respectively (Supplemental Figure 5). Considering that DCL1 is the only other DICER enzyme shown to generate 21-nucleotide small RNAs (Axtell, 2013; Bologna and Voinnet, 2014), we hypothesized it to be the most likely family member to partially substitute for DCL4 function in the production of 21-nucleotide ta-siRNAs. To test this hypothesis, we screened transposon-mutagenized populations for insertions into dcl1 (Meeley and Briggs, 1995). When introgressed into the B73 background, two of the three Mu-insertion alleles recovered result in early embryonic lethality; however, dcl1-2 is viable and conditions a partial loss-of-function phenotype (Figures 3A and 3B). The dcl1-2 allele contains a Mu insertion in the coding sequence 140 bp downstream from the start codon, but upstream of all evolutionary conserved protein domains. Growth of dcl1-2 plants is stunted and mutant leaves display a variable loss in adaxial-abaxial leaf polarity (Figure 3B; Supplemental Figures 7A and 7B). However, in contrast to mutants defective for ta-siRNA biogenesis, dcl1-2 leaves curl upward and are partially adaxialized with adaxial leaf characters, including the ligule, macrohairs, and bulliform cells, present also on the abaxial side (Supplemental Figures 7E to 7K). In addition, dcl1-2 plants show defects in inflorescence development reminiscent of the recently described dcl1-fzt mutant and are both male and female sterile (Supplemental Figures 7C and 7D; Thompson et al., 2014). Importantly, small RNA gel blot analysis shows that levels of miR166 and miR319 are reduced in dcl1-2 shoot apices relative to their wild-type siblings (Figure 3C), providing evidence that the Mu insertion negatively impacts DCL1 activity.
Figure 3.
Mutations in dcl1 and dcl4 Show a Synergistic Interaction.
(A) Diagrammatic representation of DCL1 with conserved domains and positions of transposon insertions in the dcl1 alleles indicated.
(B) Compared with the wild type, dcl1-2 mutants develop upward curling leaves that are partially adaxialized.
(C) Small RNA gel blot showing miR166 and miR319 levels are reduced in dcl1-2 shoot apices.
(D) to (I) Relative to the wild type ([D], [F], and [H]), the embryonic shoot of dcl1-2 dcl4-2 mutants ([E], [G], and [I]) is dramatically reduced (arrow in [G]). Red boxes in (F) and (G) mark shoot regions enlarged in (H) and (I), respectively.
(J) dcl1-2 dcl4-3 seedlings similarly reveal a synergistic interaction and resemble mutants blocked at early steps in ta-siRNA biogenesis.
(K) and (L) The severity of the dcl1-2 dcl4-3 phenotype is marked by elevated expression of arf3a and arf3d specifically. arf3 transcript levels in dcl1-2, dcl4-3, and dcl1-2 dcl4-3 shoot apices (means ± se) normalized to the wild type (K) were calculated based on three independent biological replicates. arf3 expression values (means ± se) in dcl1-2 dcl4-3 normalized to dcl4-3 (L) show arf3a and arf3d transcript levels specifically are increased in dcl1-2 dcl4-3 (*P < 0.05 and **P < 0.01).
Bars in (D) to (J) = 5 mm.
To address whether DCL1 acts hierarchically to DCL4 in the processing of TAS3 precursor transcripts, we generated double mutants between the weak dcl1-2 allele and the dcl4-2 and dcl4-3 alleles. In contrast to the single mutants, dcl1-2 dcl4-2 double mutants fail to germinate (Figures 3D to 3I). Inspection of mature dcl1-2 dcl4-2 embryos reveals that while a root primordium is present, the embryonic shoot is dramatically reduced (Figures 3G and 3I). Likewise, dcl1-2 dcl4-3 double mutants, while viable, exhibit a dramatically enhanced seedling phenotype compared with either single mutant (Figure 3J). The synergistic interaction between dcl1-2 and the dcl4 alleles might be explained by the converging roles of small RNA targets onto select developmental processes, but it is also consistent with overlapping or hierarchical functions for the DCL enzymes themselves in small RNA biogenesis.
Importantly, the phenotype of dcl1-2 dcl4-3 double mutants more closely resembles the phenotype of partial loss-of-function mutations like lbl1-rgd1, affecting early steps in ta-siRNA biogenesis (Nogueira et al., 2007; Douglas et al., 2010). Correspondingly, reverse transcription-quantitative PCR (RT-qPCR) analysis shows a highly significant increase especially in arf3a and arf3d transcript levels in dcl1-2 dcl4-3 double versus dcl4-3 single mutants (Figures 3K and 3L). Expression levels of arf3a are increased ∼2-fold in dcl1-2 dcl4-3 versus dcl4-3, whereas transcript levels for arf3d are up over 4-fold in the double mutant. The remaining arf3 genes (arf3b, c, and e) show an increase in expression over the wild type that is comparable between dcl1-2 dcl4-3 and dcl4-3, and expression of all five arf3 genes remains unchanged in dcl1-2. Together, these findings reveal a role for DCL1 in maintaining normal arf3a and arf3d transcript levels specifically in the dcl4 mutant background and suggest that the synergistic interaction between dcl1-2 and dcl4 in part reflects a convergence of these DCL enzymes on the regulation of arf3 targets, possibly via control of the tasiR-ARFs.
21-Nucleotide ta-siRNA Levels Are Unchanged between dcl4 and dcl1 dcl4
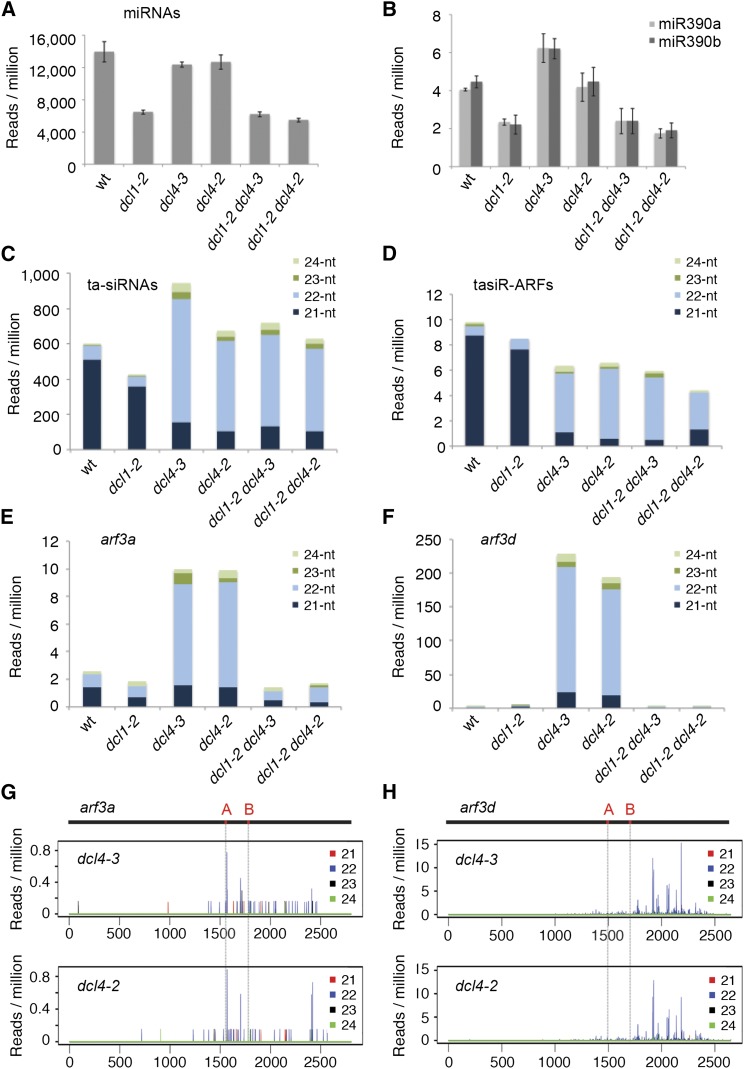
A synergistic interaction between dcl1-2 and dcl4 with respect to ta-siRNA biogenesis can be explained by functional overlap of these enzymes specifically at the ta-siRNA processing step. Alternatively, enhancement of the dcl4 phenotype by dcl1-2 is also expected if dcl1 additively reduces ta-siRNA levels by impacting miR390 biogenesis, rather than ta-siRNA processing directly. To assess the contribution of DCL1 to ta-siRNA biogenesis in the dcl4 mutants, we used a deep sequencing approach to evaluate ta-siRNA and miRNA levels in imbibed embryos, which also permits analysis of the dcl1-2 dcl4-2 double mutant. Duplicate small RNA libraries were examined for dcl1-2 dcl4-2, dcl1-2 dcl4-3, their single mutants, and wild-type controls (Supplemental Table 1).
Although the overall small RNA profiles are minimally changed in each of the mutant backgrounds (Supplemental Figure 8A), levels of the known maize miRNAs are decreased by ∼50% in dcl1-2 (Figure 4A; Supplemental Table 2), consistent with a reduction in miR166 and miR319 levels detected by small RNA gel blot analysis (Figure 3C). Levels of miR390a and miR390b, specifically, are reduced similarly (Figure 4B). Neither known miRNA levels nor the accumulation of miR390a and miR390b is significantly affected in the dcl4 single mutants (Supplemental Table 3), indicating that DCL4 is not required for the biogenesis of miRNAs. Accordingly, both overall miRNA levels and the abundance of miR390 specifically, are indistinguishable between dcl1-2 and its double mutants with dcl4-2 and dcl4-3 (Supplemental Table 3).
Figure 4.
DCL1-Dependent arf3a and arf3d-Derived Secondary siRNAs Accumulate in dcl4 Embryos.
(A) miRNA levels are unchanged in dcl4 but are significantly reduced in dcl1 mutants. miRNA levels in dcl1 dcl4 are comparable to dcl1, indicating that dcl1 is epistatic to dcl4 with respect to miRNA biogenesis.
(B) Similarly, miR390a and miR390b are only reduced in dcl1 and dcl1 dcl4 mutants.
(C) The 21-nucleotide ta-siRNA levels in dcl1, dcl4, and dcl1 dcl4 are significantly reduced relative to the wild type; however, 21-nucleotide ta-siRNA levels between dcl4 and dcl1 dcl4 embryos are comparable. These mutants also accumulate an abundant class of 22-nucleotide ta-siRNAs.
(D) Similarly, the biologically active tasiR-ARFs are significantly reduced in dcl4 and dcl1 dcl4, and these mutants accumulate a novel class of 22-nucleotide tasiR-ARFs that precisely match the canonical 21-nucleotide small RNA in sequence but are extended by one nucleotide. The 21- and 22-nucleotide tasiR-ARF levels are unchanged between dcl4 and dcl1 dcl4.
(E) and (F) dcl4 imbibed embryos accumulate 22-nucleotide arf3a- (E) and arf3d-derived (F) siRNAs that are abolished in the dcl1-2 dcl4 mutants.
(G) and (H) arf3a- (G) and arf3d-derived (H) siRNAs in dcl4-3 and dcl4-2 originate from transcribed regions downstream of the tasiR-ARF target sites.
tasiR-ARF target sites ([A] and [B]) are indicated in red on the arf3 transcripts, and vertical gray lines mark their position relative to siRNA accumulation. Note: P values for individual small RNA level comparisons are listed in Supplemental Tables 3 and 4.
Genome-wide analysis of LBL1-regulated phased small RNAs functioning in the maize vegetative apex identified nine TAS loci, all belonging to the TAS3 family (Dotto et al., 2014). These loci yield 194 potential ta-siRNAs, including 11 tasiR-ARFs with varying complementarity to the five arf3 targets. Analysis of all TAS3-derived small RNAs indicates that ta-siRNA levels are minimally affected in dcl1-2 compared with the wild type (Figure 4C). Much greater changes in the ta-siRNA profile are evident in dcl4. In both mutants, the 21-nucleotide ta-siRNA class is significantly reduced by ∼3- to 5-fold (Supplemental Table 4). A highly abundant 22-nucleotide class of ta-siRNAs is now the dominant size species, although smaller, novel 23- and 24-nucleotide ta-siRNAs are also present. A similar trend is evident for the biologically active tasiR-ARFs (Figure 4D). Although their levels are strongly reduced, 21-nucleotide tasiR-ARFs persist, even in the dcl4-2 null mutant. In addition, a class of 22-nucleotide small RNAs that exactly match the tasiR-ARFs but are extended by a single nucleotide are detected in dcl4 mutants. Furthermore, an abundant class of 22-nucleotide tasiR-ARF variants that are offset by one to two nucleotides are also detected in dcl4. Thus, processing of the TAS3 precursors into 22-nucleotide siRNAs in maize dcl4 does not prevent the production of tasiR-ARFs, a finding that is in contrast to eudicot species (Howell et al., 2007; Yifhar et al., 2012).
Importantly, levels of the 21-nucleotide ta-siRNAs, and the 21-nucleotide tasiR-ARFs specifically, are not significantly changed in dcl1-2 dcl4-2 and dcl1-2 dcl4-3 double mutants compared with the respective dcl4 single mutants (Supplemental Table 4). This finding is contrary to what would be expected if DCL1 compensates for loss of DCL4 activity in the biogenesis of 21-nucleotide ta-siRNAs and suggests that either DCL2 or DCL3 substitutes for loss of DCL4 function in this process. Moreover, the data indicate that partial loss of DCL1 function does not dramatically impact overall ta-siRNA levels, particularly in dcl4, signifying that the level of miR390 is not rate limiting for TAS3 ta-siRNA biogenesis.
DCL1 Is Required for the Biogenesis of arf3-Derived 22-Nucleotide siRNAs in dcl4 Mutants
Considering that the 21-nucleotide tasiR-ARF levels are not changed significantly between dcl4 single and dcl1 dcl4 double mutants, we sought an alternative explanation for the enhanced phenotype of the latter and the requirement for DCL1 to maintain arf3a and arf3d expression in dcl4. Because arf3 transcripts are the primary ta-siRNA targets and their misregulation is responsible for the developmental defects of ta-siRNA pathway mutants (Dotto et al., 2014), we next examined for the accumulation of arf3-derived small RNAs. Few small RNA reads are associated with arf3b, arf3c, and arf3e in mature embryos of both the wild type and the various dcl mutants (Supplemental Figures 9A to 9C). In contrast, arf3a and particularly arf3d accumulate substantially more small RNAs, but only in dcl4 single mutants (Figures 4E to 4H). These small RNAs are phased, primarily 22-nucleotide in length and originate from both strands of the transcribed sequences immediately downstream of the tasiR-ARF target sites (Supplemental Figures 10 and 11). These characteristics are reminiscent of ta-siRNAs generated via the “one-hit” model. In this scenario, a 22-nucleotide miRNA directs cleavage of its mRNA target at a single site and triggers the production of phased siRNAs downstream of this cleavage site, although these are typically 21-nucleotide in length (Fei et al., 2013). In contrast, the TAS3 ta-siRNA pathway operates via the “two-hit” model that requires dual targeting by the miR390-AGO7 complex to trigger the processing of ta-siRNAs upstream of the 3′ miRNA cleavage site. Processing of the arf3a and arf3d transcripts into 22-nucleotide siRNAs provides an explanation as to why transcript levels of these targets are unchanged in dcl4 and a basis for the relatively mild phenotype of dcl4 mutants. Importantly, production of the 22-nucleotide arf3a- and arf3d-derived siRNAs is reduced to wild-type levels in dcl1 dcl4 double mutants (Figures 4E and 4F). This finding is consistent with the synergistic interaction between these dcl mutants and moreover implies a novel role for DCL1 in the biogenesis of the 22-nucleotide arf3a- and arf3d-derived siRNAs.
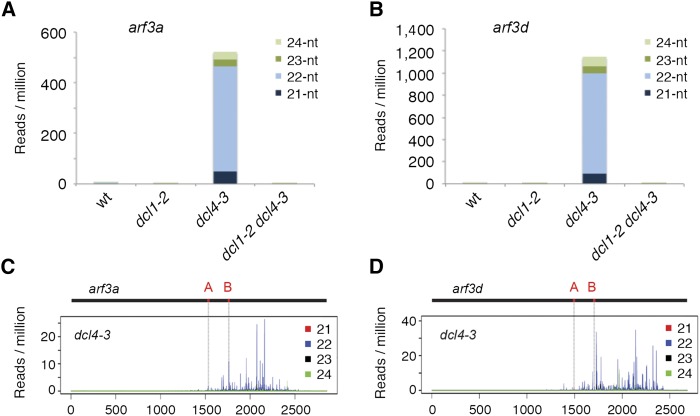
Finally, we examined whether the 22-nucleotide arf3a and arf3d-derived siRNAs persist in postembryonic dcl4 mutants, as would be expected if these small RNAs underlie the relatively mild phenotype of dcl4 seedlings. Small RNA libraries were analyzed from vegetative shoot apices of 2-week-old wild-type, dcl1-2, dcl4-3, and dcl1-2 dcl4-3 double mutants (Supplemental Table 1). As in the embryo, dcl4 acts epistatically to dcl1 in the processing of ta-siRNAs and tasiR-ARFs specifically (Supplemental Figures 8B to 8D). While a small subset of 21-nucleotide ta-siRNAs persist in dcl4-3, a large fraction of these small RNAs shift from being 21- to 22-nucleotide in length. Moreover, abundant levels of 22-nucleotide siRNAs are only detected at the arf3a and arf3d loci downstream of the tasiR-ARF target sites, specifically in dcl4-3 (Figure 5; Supplemental Figures 9D to 9F). In fact, these arf3-derived siRNAs are more abundant in the seedling apex than in the mature embryo. However, as in the embryo, the 22-nucleotide siRNAs are lost in the dcl1-2 dcl4-3 double mutant, indicating a unique requirement for DCL1 in the biogenesis of this 22-nucleotide siRNA class.
Figure 5.
DCL1-Dependent arf3a- and arf3d-Derived Secondary siRNAs Accumulate in dcl4 Shoot Apices.
(A) and (B) The 22-nucleotide arf3a- (A) and arf3d-derived (B) siRNAs accumulate in dcl4-3 but not in dcl1-2 dcl4-3 mutant apices. Levels of these small RNAs are substantially higher in dcl4-3 vegetative apices than in imbibed embryos.
(C) and (D) arf3a- (C) and arf3d-derived (D) siRNAs in dcl4-3 originate from transcribed regions downstream of the tasiR-ARF target sites.
tasiR-ARF target sites ([A] and [B]) are indicated in red on the arf3 transcripts, and vertical gray lines mark their position relative to siRNA accumulation.
Taken together, these data reveal that maize dcl4 mutants, in contrast to similar mutants in eudicot species, retain a population of 21-nucleotide tasiR-ARFs and produce a class of 22-nucleotide tasiR-ARF variants. These are associated with the processing of arf3a and arf3d tasiR-ARF target transcripts into a novel class of 22-nucleotide siRNAs by DCL1. As a result, transcript levels for both ARF3 abaxial determinants remain largely unaffected in dcl4. This novel DCL1-dependent siRNA pathway thereby mitigates the otherwise detrimental effects of mutations in DCL4, providing a mechanism to relieve the selective constraints the essential developmental role of tasiR-ARFs imposes on DCL4.
DISCUSSION
tasiR-ARF-Directed Regulation of arf3 Is Essential for Development
The ta-siRNA pathway is critical for the regulation of arf3 transcripts. In maize, a defining feature of mutations that affect ta-siRNA biogenesis is that they condition adaxial-abaxial leaf polarity defects, although the severity of these defects is variable depending on which step in the pathway is perturbed. While dcl4 mutants exhibit a relatively mild phenotype, the loss of upstream components in tasiR-ARF biogenesis, such as in rgd2-Ds1 and rdr6-1, results in severe leaf polarity defects that lead to shoot meristem arrest. In the absence of tasiR-ARF activity, expression of arf3 targets persists on the adaxial side of the incipient leaf leading to reduced expression of HD-ZIPIII adaxial determinants and an abaxialization of the primordium. As lateral blade outgrowth requires the juxtaposition of adaxial and abaxial domains (Waites and Hudson, 1995), such leaves develop as radial, thread-like structures. These findings argue for a conserved role of the maize ARF3 transcription factors in promoting abaxial cell fate and highlight the importance of their regulation by tasiR-ARFs in setting up the leaf’s adaxial domain. However, polarity of the central vasculature, along with the polarized expression of arf3a and rld1, remains unchanged in radialized organs of ta-siRNA biogenesis mutants. This suggests that specification of adaxial-abaxial polarity during vascular and primordium development is governed by divergent gene networks, distinguished in part by the respective contributions of the ta-siRNA pathway, a prospect that seems particularly intriguing considering the independent origins of the central vasculature and flattened leaf blade (Gifford and Foster, 1989).
The tasiR-ARF-ARF3 regulatory module is conserved across land plants (Fei et al., 2013). Yet, the developmental effects of mutants defective for ta-siRNA biogenesis differ widely across species. In contrast to the essential role of this pathway in maize and rice development (Nogueira et al., 2007; Nagasaki et al., 2007; Itoh et al., 2008; Dotto et al., 2014), ta-siRNA biogenesis mutants in the eudicots exhibit relatively subtle defects. In Arabidopsis, Medicago, and Lotus, such mutants give rise to downward-curled, lobed, or narrow leaves that are weakly abaxialized (Peragine et al., 2004; Xie et al., 2005; Yan et al., 2010; Zhou et al., 2013). Even in tomato, where ta-siRNA biogenesis mutants show a more severe effect on leaf polarity, these mutants are viable and male fertile (Yifhar et al., 2012; Brooks et al., 2014). The divergent phenotypes of ta-siRNA pathway mutants seem to result in part from expression variation in key pathway components that alters the spatiotemporal pattern of tasiR-ARF activity. While tasiR-ARFs in maize act to polarize arf3 expression in the incipient leaf, tasiR-ARF biogenesis in Arabidopsis and Medicago is delayed until later in primordium development (Chitwood et al., 2009; Husbands et al., 2009; Zhou et al., 2013). In addition, the polarity network in Arabidopsis and maize appears to be organized differently, as reflected in the distinct contributions of the YABBY genes to organ polarity in these species (Siegfried et al., 1999; Juarez et al., 2004b; Kidner and Timmermans, 2007). Thus, although the tasiR-ARF-ARF3 module itself is highly conserved, differences in the spatiotemporal pattern in which this module functions, or diversity in the gene networks it regulates, could lead to variation in phenotypes.
22-Nucleotide arf3-Derived Transitive siRNAs Suppress the Maize dcl4 Phenotype
Strikingly, loss of DCL4 activity in maize results in a distinctively mild developmental phenotype that is greatly suppressed in comparison to the phenotypes of mutations in lbl1, rgd2, and rdr6. This is surprising considering that morphological defects of dcl4 null mutants in eudicot species are indistinguishable from mutants affecting earlier steps in ta-siRNA biogenesis (Gasciolli et al., 2005; Xie et al., 2005; Yoshikawa et al., 2005; Yifhar et al., 2012). Deep sequencing revealed two important effects on small RNA biogenesis that distinguish loss-of-function dcl4 alleles in maize from other plant species. Maize dcl4 mutants retain a population of 21- and 22-nucleotide tasiR-ARF variants and, in addition, generate a novel class of DCL1-dependent 22-nucleotide siRNAs from arf3a and arf3d transcripts. The persistence of 21- and 22-nucleotide tasiR-ARF-related small RNAs indicates direct redundancy among the maize DCL enzymes. Hierarchy among DCL proteins has been noted in other plants. For example, in Arabidopsis, tomato, and moss, mutation of DCL4 allows the processing of TAS precursor transcripts into 22- to 24-nucleotide siRNAs by DCL2 and/or DCL3 (Xie et al., 2005; Arif et al., 2012; Yifhar et al., 2012). However, the increased length of these ta-siRNAs offsets their normal phasing pattern such that functional tasiR-ARFs are no longer produced. Considering the role of DCL1 in generating 21-nucleotide miRNAs, this seemed a likely candidate to substitute for DCL4 in the production of the 21-nucleotide ta-siRNAs, and tasiR-ARFs in particular. However, dcl4 was found to be epistatic to dcl1-2 in ta-siRNA processing, indicating that DCL2 and/or DCL3 are responsible for the production of these 21- and 22-nucleotide ta-siRNAs (Figure 6).
Figure 6.
DCL1-Dependent 22-Nucleotide siRNAs Bypass the Loss of DCL4.
(A) In the wild type, tasiR-ARFs are generated via the two-hit TAS3 ta-siRNA pathway. Biogenesis of these small RNAs is confined to the adaxial side of leaf primordia via the localized accumulation of miR390. tasiR-ARFs target arf3 transcripts, limiting expression of these abaxial determinants to the lower side of developing leaves.
(B) In dcl4, targeting of TAS3 transcripts by miR390-AGO7 complexes proceeds on the adaxial side of leaf primordia as in the wild type. However, processing of the double-stranded TAS3 precursors by DCL2/3 leads to the accumulation of 22-nucleotide tasiR-ARF variants. These trigger a unique one-hit mechanism of secondary siRNA biogenesis at the arf3a and arf3d transcripts specifically, leading to the processing of arf3a/d transcripts into 22-nucleotide siRNAs by DCL1. As a result, expression for both ARF3 abaxial determinants remains largely unaffected. See text for further details.
The persistence of tasiR-ARFs is consistent with the mild phenotype of dcl4 mutants. However, the mechanism by which these small RNAs maintain regulation of the arf3 targets in dcl4 is distinct and more complex than observed in the wild type. Specifically, the pattern and wild-type level of arf3a and arf3d expression is maintained in dcl4, whereas transcript abundance for the remaining arf3 genes is increased, as in other mutants defective for ta-siRNA biogenesis (Dotto et al., 2014). That a distinct mechanism of arf3 regulation is at play in dcl4 is also supported by the finding that the combined reduction of DCL1 and DCL4 activity has little or no effect on tasiR-ARF levels but leads to increased arf3a and arf3d expression and a strongly enhanced mutant phenotype.
Importantly, processing of the TAS3 precursors by DCL2 and/or DCL3 yields a population of 22-nucleotide ta-siRNAs that match the sequence of tasiR-ARFs, as well as 22-nucleotide variants that are shifted by one to two nucleotides. It is well established that 22-nucleotide small RNAs can function as triggers for secondary siRNA production via the one-hit mechanism of ta-siRNA biogenesis (Chen et al., 2010; Cuperus et al., 2010; Manavella et al., 2012; Fei et al., 2013). Our observation that the arf3a and arf3d transcripts in dcl4 generate phased siRNAs immediately downstream of the tasiR-ARF recognition sites implies a similar mechanism for their biogenesis, although these siRNAs are 22 rather than 21 nucleotides in length. Critically, a one-hit mechanism of secondary siRNA biogenesis can also explain why arf3a and arf3d specifically accumulate 22-nucleotide siRNAs in dcl4 mutants. Although the precise requirements for triggering ta-siRNA biogenesis are not completely understood, perfect complementarity between the target and the 3′ end of the small RNA trigger is a prerequisite (Zhang et al., 2012; Yoshikawa et al., 2013). A single nucleotide polymorphism at the 5′ end of the tasiR-ARF recognition sites distinguishes arf3a and arf3d from the other arf3 genes (Supplemental Figure 12). These polymorphisms create perfect complementarity between the arf3a and arf3d transcripts and the 3′ end of certain tasiR-ARFs, thereby biasing secondary siRNAs production to these arf3 targets. However, perfect complementarity is not essential for canonical tasiR-ARF-directed transcript cleavage, providing an explanation as to why all arf3 transcripts are subject to 21-nucleotide tasiR-ARF regulation in the wild type (Dotto et al., 2014).
A Novel Role for DCL1 in Phased 22-Nucleotide siRNA Biogenesis
Notably, the arf3a- and arf3d-derived 22-nucleotide siRNAs are completely abolished in dcl1 dcl4 double mutants, indicating an essential role for DCL1 in their biogenesis. Considering a one-hit mechanism of secondary siRNA biogenesis triggered in response to tasiR-ARF activity, DCL1 could conceivably act at multiple steps in the pathway. However, as DCL1 is dispensable for the biogenesis of 22-nucleotide TAS3-derived ta-siRNAs in dcl4 (Figure 4), the most parsimonious model for our findings is that DCL1 mediates the processing of the tasiR-ARF-directed arf3a and arf3d cleavage products into 22-nucleotide phased siRNAs (Figure 6). This represents a unique function for DCL1, which normally has a negligible role in the processing of siRNAs and secondary siRNAs specifically (Henderson et al., 2006; Dunoyer et al., 2007; Kasschau et al., 2007). DCL1 is known to function in cis-natural antisense siRNA biogenesis. However, DCL1’s contribution to their biogenesis appears variable, and in cases where phased secondary siRNAs are produced, these are 21 nucleotides in length (Borsani et al., 2005; Katiyar-Agarwal et al., 2006; Ron et al., 2010). In contrast, arf3a- and arf3d-derived siRNAs in dcl4 are 22 nucleotides in length. The reason why secondary siRNAs at arf3a and arf3d are 22 nucleotides in length, whereas DCL1-generated miRNAs are typically 21 nucleotides long is not currently understood. One possibility is that small RNA length is defined in part by distinctive properties of the precursors and the protein complexes they recruit.
The finding that the arf3-derived 22-nucleotide siRNAs are completely abolished in a dcl1 dcl4 background further indicates that DCL2 and DCL3 are unable to substitute for DCL1 in their biogenesis. This is particularly noteworthy, considering that DCL2 and DCL3 act hierarchically to DCL4 in the production of TAS3-derived ta-siRNAs, a process for which DCL1 is dispensable. The critical features that direct some siRNA precursors to be processed by DCL2/3 and others by DCL1 is an area for future research. However, one notable difference between these siRNAs is that, in contrast to the DCL1-dependent secondary siRNAs at arf3a and arf3d, the TAS3 ta-siRNAs are generated following a two-hit mechanism involving a specialized miR390-AGO7 effector complex (Montgomery et al., 2008). Our findings thus highlight the presence of additional diversity and complexity in small RNA networks and reveal a novel role for DCL1 in the production of 22-nucleotide secondary siRNAs.
Plasticity in Small RNA Regulation Bypasses Selective Constraints on DCL4
Taken together, the data reveal the presence of a DCL1-dependent siRNA pathway that bypasses the otherwise essential role for DCL4 in development. But how might processing of arf3a and arf3d transcripts into 22-nucleotide siRNAs maintain leaf polarity in dcl4? tasiR-ARFs normally act preferentially on the adaxial side, and this polarized activity limits arf3 expression to the abaxial side of leaf primordia (Nogueira et al., 2007; Chitwood et al., 2009). In Arabidopsis, the polarized activity of tasiR-ARF reflects a threshold-based readout of a tasiR-ARF gradient generated through the cell-to-cell movement of tasiR-ARFs away from their adaxial source of biogenesis (Chitwood et al., 2009; Skopelitis et al., 2012). Whether this is the case in maize remains to be formally shown. In maize, tasiR-ARF biogenesis is confined to the adaxial side of incipient and developing leaf primordia by the localized accumulation of miR390 (Nogueira et al., 2009; Douglas et al., 2010). As such, the 22-nucleotide tasiR-ARFs generated in dcl4 also likely derive from the adaxial domain of the leaf. This in turn would trigger the DCL1-dependent processing of arf3a and arf3d transcripts into 22-nucleotide secondary siRNAs preferentially on the adaxial side of dcl4 primordia (Figure 6). Indeed, biogenesis of these small RNAs in dcl4 is accompanied by a negligible change in arf3a and arf3d transcript levels, and the abaxial expression of arf3a is maintained.
The presence of a small RNA pathway that bypasses the requirement for DCL4 in development is predicted to have major implications for DCL4’s other function in antiviral defense. The essential role of the tasiR-ARFs in maize development is expected to place unique constraints on the evolution of DCL4 and negatively impact its ability to evade the possible inhibitory effects of viral suppressors, a situation that is at odds with DCL4’s role in antiviral defense. The existence of a DCL1-dependent siRNA pathway that buffers for reduced DCL4 activity in development could reduce selective constraints on DCL4 and allow it to diversify in response to viral suppressors. Such adaptations are unlikely to favor null alleles, as depending on the level of redundancy with DCL2, at least some DCL4 activity would be required for effective antiviral defense (Pumplin and Voinnet, 2013; Seo et al., 2013). Without the DCL1 bypass pathway, however, even subtle adaptive mutations can have dramatic effects on maize development, as illustrated by the severe seedling defects conditioned by weak alleles of other ta-siRNA biogenesis components and perhaps the dcl1-2 dcl4-3 double mutant (Nogueira et al., 2007; Douglas et al., 2010). Moreover, the existence of the DCL1 bypass pathway may favor selection of mutations that separate DCL4’s contributions in development from those in antiviral defense, i.e., by compromising ta-siRNA biogenesis while minimally impacting viral-induced siRNA processing. In Arabidopsis, partial loss-of-function alleles of dcl4 have been identified that genetically uncouple DCL4’s contributions to development from its role in antiviral immunity (Dunoyer et al., 2007). Intriguingly, mutations in two out of the three such dcl4 alleles identified are located in the PAZ domain, the domain that was identified as being subject to long-term recurrent adaptation (Mukherjee et al., 2013). While the PAZ domain is primarily known to anchor the 3′ overhang of dsRNA templates, the ability of single point mutations to uncouple DCL4’s disparate functions hints that this domain also provides an interface for protein-protein interactions that lend specificity to DCL4 in its role in either development or antiviral defense. This possibility is consistent with observations that DCL enzymes can distinguish between dsRNA templates based in part on the biosynthetic pathway that generates them (see above; Bologna and Voinnet, 2014). Furthermore, some viral suppressors are known to target this domain, indicating importance with respect to viral defense strategies (Zhang et al., 2006; Baumberger et al., 2007; Singh et al., 2009; Hamera et al., 2012). Thus, the ability to buffer for reduced DCL4 activity via a DCL1-dependent siRNA pathway presents a mechanism to release DCL4 from selective constraints and allow it to adapt to challenges in antiviral defense. Conceivably, this buffering capacity may further explain why monocots, like maize and rice, appear to have undergone more extreme adaptive changes to their PAZ domain, despite the critical role of the ta-siRNA pathway for shoot meristem function and organogenesis in these species.
METHODS
Plant Materials
The dcl4-1 allele originated in an Ac transposon population (Pilu et al., 2002). The dcl4-2 and dcl4-4 mutations arose in independent ethyl methanesulfonate-mutagenized populations, whereas the dcl4-3 allele corresponds to the classical rgd3 (aka rgd-N766B) mutation. The lbl1-rgd1 and the rgd2-Ds1 alleles have been described previously (Nogueira et al., 2007; Douglas et al., 2010). The rdr6-1 allele was acquired from the Maize TILLING Project (http://popcorn.maizegdb.org/search/project_search/project_search.php?record=1233073; tilling line ID: 03IN-W22CW-0612), whereas the rdr6-2 allele was identified in a Mu transposon population. DuPont Pioneer kindly screened for the dcl1 Mu insertion alleles in the maize (Zea mays) TUSC collection (Meeley and Briggs, 1995). All mutants used in this study were introgressed for four to six generations into the B73 inbred backgrounds prior to further genetic and phenotypic analyses. The dcl4-2 and dcl4-3 alleles were additionally introgressed into the A619 inbred background for five or six generations, respectively, before phenotypic analysis.
Histology
Leaf tissues from 2-week old seedlings were fixed and embedded as described (Javelle et al., 2011). Paraplast blocks were sectioned at a thickness of up to 10 μM and stained with Safranin-O and Fast Green according to Johansen’s method. For Toluidine blue O staining of the leaf epidermis, 1- to 2-cm-long leaf sections from fully expanded leaves 1 to 8 were fixed at room temperature in ethanol and acetic acid (3:1) and incubated in Toluidine blue staining solution (0.05% Toluidine blue in 0.1 M sodium acetate, pH 4.4, diluted in water [1:15]). The percentage of abnormal subsidiary cells was calculated by examining at least 150 subsidiary cells from each leaf sample, and three plants per genotype were collected per leaf stage.
In Situ Hybridization
Vegetative shoot apices from 2-week old maize seedlings were fixed and processed as described (Javelle et al., 2011). DIG-labeled probes comprising nucleotides 973 to 2438 of the arf3a (GRMZM2G030710_T01) and nucleotides 619 to 1674 of the rld1 (GRMZM2G109987_T02) cDNAs were prepared by in vitro transcription (Roche), according to the manufacturer’s protocol.
Small RNA Gel Blot Hybridization
Total RNA was isolated from vegetative apices, including the SAM and five leaf primordia, using TRIzol reagent (Invitrogen). At least five apices were collected and pooled for each sample and 15 μg of total RNA per pool was used for gel electrophoresis and blotting. Hybridization with an end-labeled 21-nucleotide miR166, miR390, or 16-nucleotide LNA-modified oligonucleotide probe (Exiqon) complementary to the tasiR-ARF sequence (5′-CTTACAAGGTCAAGAA-3′) was performed as previously described (Nogueira et al., 2007). A U6 probe (5′-TCATCCTTGCGCAGGGGCCA-3′) was used to establish equal loading.
RT-qPCR
For RT-qPCR, shoot apices consisting of the shoot apical meristem and five leaf primordia were collected from 2-week old seedlings. Expression of stomatal patterning genes was analyzed on RNA isolated from the proximal 1 cm of immature juvenile leaves of 11-d-old seedlings. Total RNA was isolated using TRIzol reagent (Invitrogen) and treated with DNase I (Promega), and 2 to 4 μg was primed with oligo(dT) and reverse transcribed with SuperScript III first-strand synthesis system (Invitrogen) according to the manufacturer’s protocol. RT-qPCR was performed on the CFX96 real-time PCR detection system with iQ SYBR Green Supermix (Bio-Rad). The specificity of all amplification products was determined using dissociation curve analyses. Relative quantification values were calculated based on at least three biological replicates using the 2−∆Ct method, with the ∆Ct of glyceraldehyde-3-phosphate dehydrogenase (gapc) as normalization control. Primer sequences are listed in Supplemental Table 5.
Small RNA Library Construction and Data Analysis
Total RNA was extracted from embryos of 24 h imbibed kernels or shoot apices using TRIzol reagent (Invitrogen) followed by treatment with DNase I (Promega). Small RNA libraries from embryos were prepared from 1 μg of total RNA using the NEBNext Multiplex Small RNA Library Prep Set for Illumina (New England Biolabs). Shoot apex libraries were constructed from 1.2 μg of total RNA using the TruSeq Small RNA sample preparation kit (Illumina). Libraries were quantified with the KAPA Illumina Library Quantification Kit (KAPABIOSYSTEMS) and sequenced on the Illumina HiSequation 2000 platform at the CSHL Genome Center. Trimmed reads 18 to 26 nucleotides in length were aligned to the maize B73 RefGen_v2 genome (release 5a.57) using Bowtie v1.0.0 (Langmead et al., 2009), allowing no mismatches and up to 20 potential alignments per read. Reads matching known structural RNAs (rRNAs, tRNAs, sn-RNAs, and sno-RNAs) from Rfam 10.0 (Griffiths-Jones et al., 2005) were removed from further analysis. Read counts were normalized by millions of mapped reads in each library (reads per million) for comparisons across samples. Small RNAs were annotated using BEDtools (Quinlan and Hall, 2010) to miRNA (miRBase v20; http://www.mirbase.org), TAS3 precursor loci (Dotto et al., 2014), and maize genes (release 5b, January 2010) for subsequent analyses. Small RNA differential expression was calculated using the DESeq package (Anders and Huber, 2010) to identify statistically significant differential accumulation. To assess read distribution along the arf3 loci, small RNAs were remapped to the arf3 cDNA sequence, and the density of 5′ ends were plotted along the cDNA nucleotide position. Phasing of small RNAs was determined as described by Dotto et al. (2014).
Accession Numbers
All high-throughput sequencing data, both raw and processed files, are available through the Gene Expression Omnibus under number GSE66986. The sequences of genes used in this study can be found at MaizeGDB and in the GenBank data library under the following accession numbers: dcl4, GenBank accession KR230386 or GRMZM2G024466, GRMZM2G160473, GRMZM2G050882, and GRMZM2G050869; dcl1, GenBank accession KR230388 and GRMZM2G040762; lbl1, GRMZM2G020187; rgd2, GRMZM5G892991; rdr6, GenBank accession KR230387 or GRMZM2G145201 and GRMZM2G082437; arf3a, GRMZM2G030710; arf3b, GRMZM2G441325; arf3c, GRMZM2G056120; arf3d, GRMZM2G437460; and arf3e, GRMZM5G874163.
Supplemental Data
Supplemental Figure 1. Amino acid substitutions in mutant alleles of dcl4.
Supplemental Figure 2. The different dcl4 alleles condition a comparably mild phenotype.
Supplemental Figure 3. dcl4 mutants exhibit defects in adaxial-abaxial leaf polarity.
Supplemental Figure 4. dcl4 affects the asymmetric divisions of subsidiary cells within the stomatal complex.
Supplemental Figure 5. lbl1-rgd1 retains partial tasiR-ARF activity.
Supplemental Figure 6. Global expression profiles of the five maize dicer-like genes show that while dcl1, dcl2, dcl3a, and dcl4 are expressed broadly throughout plant development, dcl3b/dcl5 is expressed primarily during reproductive development and in the immature embryo.
Supplemental Figure 7. dcl1-2 conditions a weakly adaxialized leaf polarity phenotype.
Supplemental Figure 8. dcl1 and dcl4 mutants show defined changes in small RNA content.
Supplemental Figure 9. arf3b-, c-, and e-derived small RNAs are present at low levels in dcl4 imbibed embryos and shoot apices.
Supplemental Figure 10. In dcl4 mutants, arf3a transcripts are processed into 22-nucleotide phased siRNAs.
Supplemental Figure 11. arf3d transcripts are similarly processed into 22-nucleotide phased siRNAs in dcl4 mutants.
Supplemental Figure 12. SNPs in the tasiR-ARF recognition sites distinguish arf3a and arf3d from the remaining arf3 genes.
Supplemental Table 1. Summary analysis for small RNA libraries.
Supplemental Table 2. miRNA abundance (RPM) in imbibed embryos.
Supplemental Table 3. Two-tailed t test comparisons for miRNA levels in imbibed embryos of various dcl mutants.
Supplemental Table 4. Two-tailed t test comparisons for 21-nucleotide ta-siRNA and tasiR-ARF levels in imbibed embryos.
Supplemental Table 5. Primers used for RT-qPCR.
Supplementary Material
Acknowledgments
We thank members of the Timmermans laboratory and of the maize SAM project who have contributed ideas and thoughtful comments to the article as well as Tim Mulligan for plant care. O.H.T. is supported by a fellowship of the Human Frontier Science Program. This work was supported by grants from the National Science Foundation (IOS-1238142 and MCB-1159098).
AUTHOR CONTRIBUTIONS
K.P. and M.C.P.T. designed the project and experiments. K.P., P.S.M., and G.C. performed the phenotypic analyses and cloned the dcl4 alleles. R.M. provided the dcl1 alleles. K.P., O.H.T., M.H., and M.C.P.T. performed the small RNA data analysis. K.P. and M.C.P.T. wrote the article.
Glossary
- siRNA
small interfering RNA
- dsRNA
double-stranded RNA
- ta-siRNA
trans-acting short-interfering RNA
- RT-qPCR
reverse transcription-quantitative PCR
- miRNA
microRNA
Footnotes
Articles can be viewed online without a subscription.
References
- Allen E., Xie Z., Gustafson A.M., Carrington J.C. (2005). MicroRNA-directed phasing during trans-acting siRNA biogenesis in plants. Cell 121: 207–221. [DOI] [PubMed] [Google Scholar]
- Anders S., Huber W. (2010). Differential expression analysis for sequence count data. Genome Biol. 11: R106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arif M.A., Fattash I., Ma Z., Cho S.H., Beike A.K., Reski R., Axtell M.J., Frank W. (2012). DICER-LIKE3 activity in Physcomitrella patens DICER-LIKE4 mutants causes severe developmental dysfunction and sterility. Mol. Plant 5: 1281–1294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Axtell M.J. (2013). Classification and comparison of small RNAs from plants. Annu. Rev. Plant Biol. 64: 137–159. [DOI] [PubMed] [Google Scholar]
- Baumberger N., Tsai C.-H., Lie M., Havecker E., Baulcombe D.C. (2007). The Polerovirus silencing suppressor P0 targets ARGONAUTE proteins for degradation. Curr. Biol. 17: 1609–1614. [DOI] [PubMed] [Google Scholar]
- Bologna N.G., Voinnet O. (2014). The diversity, biogenesis, and activities of endogenous silencing small RNAs in Arabidopsis. Annu. Rev. Plant Biol. 65: 473–503. [DOI] [PubMed] [Google Scholar]
- Borsani O., Zhu J., Verslues P.E., Sunkar R., Zhu J.-K. (2005). Endogenous siRNAs derived from a pair of natural cis-antisense transcripts regulate salt tolerance in Arabidopsis. Cell 123: 1279–1291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brooks C., Nekrasov V., Lippman Z.B., Van Eck J. (2014). Efficient gene editing in tomato in the first generation using the clustered regularly interspaced short palindromic repeats/CRISPR-associated9 system. Plant Physiol. 166: 1292–1297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H.-M., Chen L.-T., Patel K., Li Y.-H., Baulcombe D.C., Wu S.-H. (2010). 22-Nucleotide RNAs trigger secondary siRNA biogenesis in plants. Proc. Natl. Acad. Sci. USA 107: 15269–15274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chitwood D.H., Nogueira F.T.S., Howell M.D., Montgomery T.A., Carrington J.C., Timmermans M.C.P. (2009). Pattern formation via small RNA mobility. Genes Dev. 23: 549–554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuperus J.T., Carbonell A., Fahlgren N., Garcia-Ruiz H., Burke R.T., Takeda A., Sullivan C.M., Gilbert S.D., Montgomery T.A., Carrington J.C. (2010). Unique functionality of 22-nt miRNAs in triggering RDR6-dependent siRNA biogenesis from target transcripts in Arabidopsis. Nat. Struct. Mol. Biol. 17: 997–1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dotto M.C., Petsch K.A., Aukerman M.J., Beatty M., Hammell M., Timmermans M.C.P. (2014). Genome-wide analysis of leafbladeless1-regulated and phased small RNAs underscores the importance of the TAS3 ta-siRNA pathway to maize development. PLoS Genet. 10: e1004826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douglas R.N., Wiley D., Sarkar A., Springer N., Timmermans M.C.P., Scanlon M.J. (2010). ragged seedling2 encodes an ARGONAUTE7-like protein required for mediolateral expansion, but not dorsiventrality, of maize leaves. Plant Cell 22: 1441–1451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunoyer P., Himber C., Ruiz-Ferrer V., Alioua A., Voinnet O. (2007). Intra- and intercellular RNA interference in Arabidopsis thaliana requires components of the microRNA and heterochromatic silencing pathways. Nat. Genet. 39: 848–856. [DOI] [PubMed] [Google Scholar]
- Emery J.F., Floyd S.K., Alvarez J., Eshed Y., Hawker N.P., Izhaki A., Baum S.F., Bowman J.L. (2003). Radial patterning of Arabidopsis shoots by class III HD-ZIP and KANADI genes. Curr. Biol. 13: 1768–1774. [DOI] [PubMed] [Google Scholar]
- Fahlgren N., Montgomery T.A., Howell M.D., Allen E., Dvorak S.K., Alexander A.L., Carrington J.C. (2006). Regulation of AUXIN RESPONSE FACTOR3 by TAS3 ta-siRNA affects developmental timing and patterning in Arabidopsis. Curr. Biol. 16: 939–944. [DOI] [PubMed] [Google Scholar]
- Fei Q., Xia R., Meyers B.C. (2013). Phased, secondary, small interfering RNAs in posttranscriptional regulatory networks. Plant Cell 25: 2400–2415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasciolli V., Mallory A.C., Bartel D.P., Vaucheret H. (2005). Partially redundant functions of Arabidopsis DICER-like enzymes and a role for DCL4 in producing trans-acting siRNAs. Curr. Biol. 15: 1494–1500. [DOI] [PubMed] [Google Scholar]
- Gifford E.M., Foster A.S. (1989). Morphology and Evolution of Vascular Plants. San Francisco, CA, W.H. Freeman & Company; ). [Google Scholar]
- Griffiths-Jones S., Moxon S., Marshall M., Khanna A., Eddy S.R., Bateman A. (2005). Rfam: annotating non-coding RNAs in complete genomes. Nucleic Acids Res. 33: D121–D124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamera S., Song X., Su L., Chen X., Fang R. (2012). Cucumber mosaic virus suppressor 2b binds to AGO4-related small RNAs and impairs AGO4 activities. Plant J. 69: 104–115. [DOI] [PubMed] [Google Scholar]
- Henderson I.R., Zhang X., Lu C., Johnson L., Meyers B.C., Green P.J., Jacobsen S.E. (2006). Dissecting Arabidopsis thaliana DICER function in small RNA processing, gene silencing and DNA methylation patterning. Nat. Genet. 38: 721–725. [DOI] [PubMed] [Google Scholar]
- Howell M.D., Fahlgren N., Chapman E.J., Cumbie J.S., Sullivan C.M., Givan S.A., Kasschau K.D., Carrington J.C. (2007). Genome-wide analysis of the RNA-DEPENDENT RNA POLYMERASE6/DICER-LIKE4 pathway in Arabidopsis reveals dependency on miRNA- and tasiRNA-directed targeting. Plant Cell 19: 926–942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter C., Willmann M.R., Wu G., Yoshikawa M., de la Luz Gutiérrez-Nava M., Poethig S.R. (2006). Trans-acting siRNA-mediated repression of ETTIN and ARF4 regulates heteroblasty in Arabidopsis. Development 133: 2973–2981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Husbands A.Y., Chitwood D.H., Plavskin Y., Timmermans M.C.P. (2009). Signals and prepatterns: new insights into organ polarity in plants. Genes Dev. 23: 1986–1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itoh J., Sato Y., Nagato Y. (2008). The SHOOT ORGANIZATION2 gene coordinates leaf domain development along the central-marginal axis in rice. Plant Cell Physiol. 49: 1226–1236. [DOI] [PubMed] [Google Scholar]
- Javelle M., Marco C.F., Timmermans M. (2011). In situ hybridization for the precise localization of transcripts in plants. J. Vis. Exp. 23: e3328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juarez M.T., Kui J.S., Thomas J., Heller B.A., Timmermans M.C.P. (2004a). MicroRNA-mediated repression of rolled leaf1 specifies maize leaf polarity. Nature 428: 84–88. [DOI] [PubMed] [Google Scholar]
- Juarez M.T., Twigg R.W., Timmermans M.C.P. (2004b). Specification of adaxial cell fate during maize leaf development. Development 131: 4533–4544. [DOI] [PubMed] [Google Scholar]
- Kasschau K.D., Fahlgren N., Chapman E.J., Sullivan C.M., Cumbie J.S., Givan S.A., Carrington J.C. (2007). Genome-wide profiling and analysis of Arabidopsis siRNAs. PLoS Biol. 5: e57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katiyar-Agarwal S., Morgan R., Dahlbeck D., Borsani O., Villegas A. Jr., Zhu J.-K., Staskawicz B.J., Jin H. (2006). A pathogen-inducible endogenous siRNA in plant immunity. Proc. Natl. Acad. Sci. USA 103: 18002–18007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kidner C.A., Timmermans M.C.P. (2007). Mixing and matching pathways in leaf polarity. Curr. Opin. Plant Biol. 10: 13–20. [DOI] [PubMed] [Google Scholar]
- Langmead B., Trapnell C., Pop M., Salzberg S.L. (2009). Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 10: R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manavella P.A., Koenig D., Weigel D. (2012). Plant secondary siRNA production determined by microRNA-duplex structure. Proc. Natl. Acad. Sci. USA 109: 2461–2466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meeley B., Briggs S.P. (1995). Reverse genetics for maize. Maize Genet. Coop. Newsl. 69: 67. [Google Scholar]
- Montgomery T.A., Howell M.D., Cuperus J.T., Li D., Hansen J.E., Alexander A.L., Chapman E.J., Fahlgren N., Allen E., Carrington J.C. (2008). Specificity of ARGONAUTE7-miR390 interaction and dual functionality in TAS3 trans-acting siRNA formation. Cell 133: 128–141. [DOI] [PubMed] [Google Scholar]
- Mukherjee K., Campos H., Kolaczkowski B. (2013). Evolution of animal and plant dicers: early parallel duplications and recurrent adaptation of antiviral RNA binding in plants. Mol. Biol. Evol. 30: 627–641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagasaki H., Itoh J., Hayashi K., Hibara K., Satoh-Nagasawa N., Nosaka M., Mukouhata M., Ashikari M., Kitano H., Matsuoka M., Nagato Y., Sato Y. (2007). The small interfering RNA production pathway is required for shoot meristem initiation in rice. Proc. Natl. Acad. Sci. USA 104: 14867–14871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nogueira F.T.S., Chitwood D.H., Madi S., Ohtsu K., Schnable P.S., Scanlon M.J., Timmermans M.C.P. (2009). Regulation of small RNA accumulation in the maize shoot apex. PLoS Genet. 5: e1000320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nogueira F.T.S., Madi S., Chitwood D.H., Juarez M.T., Timmermans M.C.P. (2007). Two small regulatory RNAs establish opposing fates of a developmental axis. Genes Dev. 21: 750–755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peragine A., Yoshikawa M., Wu G., Albrecht H.L., Poethig R.S. (2004). SGS3 and SGS2/SDE1/RDR6 are required for juvenile development and the production of trans-acting siRNAs in Arabidopsis. Genes Dev. 18: 2368–2379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pilu R., Consonni G., Busti E., MacCabe A.P., Giulini A., Dolfini S., Gavazzi G. (2002). Mutations in two independent genes lead to suppression of the shoot apical meristem in maize. Plant Physiol. 128: 502–511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pumplin N., Voinnet O. (2013). RNA silencing suppression by plant pathogens: defence, counter-defence and counter-counter-defence. Nat. Rev. Microbiol. 11: 745–760. [DOI] [PubMed] [Google Scholar]
- Quinlan A.R., Hall I.M. (2010). BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26: 841–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ron M., Alandete Saez M., Eshed Williams L., Fletcher J.C., McCormick S. (2010). Proper regulation of a sperm-specific cis-nat-siRNA is essential for double fertilization in Arabidopsis. Genes Dev. 24: 1010–1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo J.-K., Wu J., Lii Y., Li Y., Jin H. (2013). Contribution of small RNA pathway components in plant immunity. Mol. Plant Microbe Interact. 26: 617–625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegfried K.R., Eshed Y., Baum S.F., Otsuga D., Drews G.N., Bowman J.L. (1999). Members of the YABBY gene family specify abaxial cell fate in Arabidopsis. Development 126: 4117–4128. [DOI] [PubMed] [Google Scholar]
- Singh G., Popli S., Hari Y., Malhotra P., Mukherjee S., Bhatnagar R.K. (2009). Suppression of RNA silencing by Flock house virus B2 protein is mediated through its interaction with the PAZ domain of Dicer. FASEB J. 23: 1845–1857. [DOI] [PubMed] [Google Scholar]
- Skopelitis D.S., Husbands A.Y., Timmermans M.C. (2012). Plant small RNAs as morphogens. Curr. Opin. Cell Biol. 24: 217–224. [DOI] [PubMed] [Google Scholar]
- Thompson B.E., Basham C., Hammond R., Ding Q., Kakrana A., Lee T.F., Simon S.A., Meeley R., Meyers B.C., Hake S. (2014). The dicer-like1 homolog fuzzy tassel is required for the regulation of meristem determinacy in the inflorescence and vegetative growth in maize. Plant Cell 26: 4702–4717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timmermans M.C., Schultes N.P., Jankovsky J.P., Nelson T. (1998). Leafbladeless1 is required for dorsoventrality of lateral organs in maize. Development 125: 2813–2823. [DOI] [PubMed] [Google Scholar]
- Waites R., Hudson A. (1995). phantastica: a gene required for dorsoventrality of leaves in Antirrhinum majus. Development 121: 2143–2154. [Google Scholar]
- Xie Z., Allen E., Wilken A., Carrington J.C. (2005). DICER-LIKE 4 functions in trans-acting small interfering RNA biogenesis and vegetative phase change in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 102: 12984–12989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan J., Cai X., Luo J., Sato S., Jiang Q., Yang J., Cao X., Hu X., Tabata S., Gresshoff P.M., Luo D. (2010). The REDUCED LEAFLET genes encode key components of the trans-acting small interfering RNA pathway and regulate compound leaf and flower development in Lotus japonicus. Plant Physiol. 152: 797–807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao X., Wang H., Li H., Yuan Z., Li F., Yang L., Huang H. (2009). Two types of cis-acting elements control the abaxial epidermis-specific transcription of the MIR165a and MIR166a genes. FEBS Lett. 583: 3711–3717. [DOI] [PubMed] [Google Scholar]
- Yifhar T., Pekker I., Peled D., Friedlander G., Pistunov A., Sabban M., Wachsman G., Alvarez J.P., Amsellem Z., Eshed Y. (2012). Failure of the tomato trans-acting short interfering RNA program to regulate AUXIN RESPONSE FACTOR3 and ARF4 underlies the wiry leaf syndrome. Plant Cell 24: 3575–3589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshikawa M., Iki T., Tsutsui Y., Miyashita K., Poethig R.S., Habu Y., Ishikawa M. (2013). 3′ Fragment of miR173-programmed RISC-cleaved RNA is protected from degradation in a complex with RISC and SGS3. Proc. Natl. Acad. Sci. USA 110: 4117–4122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshikawa M., Peragine A., Park M.-Y., Poethig R.S. (2005). A pathway for the biogenesis of trans-acting siRNAs in Arabidopsis. Genes Dev. 19: 2164–2175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C., Ng D.W.-K., Lu J., Chen Z.J. (2012). Roles of target site location and sequence complementarity in trans-acting siRNA formation in Arabidopsis. Plant J. 69: 217–226. [DOI] [PubMed] [Google Scholar]
- Zhang X., Yuan Y.R., Pei Y., Lin S.S., Tuschl T., Patel D.J., Chua N.H. (2006). Cucumber mosaic virus-encoded 2b suppressor inhibits Arabidopsis Argonaute1 cleavage activity to counter plant defense. Genes Dev. 20: 3255–3268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou C., et al. (2013). The trans-acting short interfering RNA3 pathway and no apical meristem antagonistically regulate leaf margin development and lateral organ separation, as revealed by analysis of an argonaute7/lobed leaflet1 mutant in Medicago truncatula. Plant Cell 25: 4845–4862. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.