The moss Physcomitrella patens has over 1000 small RNA loci that produce 23- to 24-nucleotide heterochromatic short interfering RNAs, some of which are dependent upon a minimal Dicer-Like gene.
Abstract
Many plant small RNAs are sequence-specific negative regulators of target mRNAs and/or chromatin. In angiosperms, the two most abundant endogenous small RNA populations are usually 21-nucleotide microRNAs (miRNAs) and 24-nucleotide heterochromatic short interfering RNAs (siRNAs). Heterochromatic siRNAs are derived from repetitive regions and reinforce DNA methylation at targeted loci. The existence and extent of heterochromatic siRNAs in other land plant lineages has been unclear. Using small RNA-sequencing (RNA-seq) of the moss Physcomitrella patens, we identified 1090 loci that produce mostly 23- to 24-nucleotide siRNAs. These loci are mostly in intergenic regions with dense DNA methylation. Accumulation of siRNAs from these loci depends upon P. patens homologs of DICER-LIKE3 (DCL3), RNA-DEPENDENT RNA POLYMERASE2, and the largest subunit of DNA-DEPENDENT RNA POLYMERASE IV, with the largest subunit of a Pol V homolog contributing to expression at a smaller subset of the loci. A MINIMAL DICER-LIKE (mDCL) gene, which lacks the N-terminal helicase domain typical of DCL proteins, is specifically required for 23-nucleotide siRNA accumulation. We conclude that heterochromatic siRNAs, and their biogenesis pathways, are largely identical between angiosperms and P. patens, with the notable exception of the P. patens-specific use of mDCL to produce 23-nucleotide siRNAs.
INTRODUCTION
Small noncoding RNAs regulate gene expression to modulate growth, development, differentiation, genome silencing, and stress responses in eukaryotic organisms (Matzke and Mosher, 2014). There are two main categories of endogenous small RNAs in plants: microRNAs (miRNAs) and short interfering RNAs (siRNAs). Although the silencing pathways utilizing small RNAs have much in common, there are some fundamental distinctions between the two classes of small RNAs, particularly in regard to their biogenesis, evolutionary conservation, targets, and modes of action (Axtell, 2013a). Most importantly, miRNAs and siRNAs differ in their precursors: While siRNA precursors are the products of RNA-dependent RNA polymerase synthesized double-stranded RNA duplexes, miRNAs are derived from single RNA molecules that fold back to form self-complementary hairpin RNAs. Endogenous siRNAs are the dominant small RNA type in many plant species, while miRNAs have received more attention, particularly in regard to annotations of specific loci (Coruh et al., 2014).
Heterochromatin, which contains repetitive sequences and transposable elements, is silenced by conserved epigenetic modifications of histones and DNA. Epigenetic silencing is believed to prevent abnormal chromosomal rearrangements and activation of transposons, which can cause mutations if they are integrated into genes (Lippman and Martienssen, 2004). In flowering plants, siRNAs are known to induce DNA methylation at targeted genomic regions (Matzke and Mosher, 2014). Repressive histone modifications and siRNA-directed DNA methylation form a positive feedback loop to reinforce transcriptional silencing. This pathway is particularly well understood in Arabidopsis thaliana, where the presence of H3K9 methylation leads the SAWADEE HOMEODOMAIN HOMOLOG1/DNA BINDING TRANSCRIPTION FACTOR1 protein to recruit an alternative DNA-dependent RNA polymerase (Pol IV) to chromatin (Law et al., 2013; H. Zhang et al., 2013). Pol IV transcribes precursors of heterochromatic siRNAs, which are promptly converted into double-stranded RNAs by RNA-DEPENDENT RNA POLYMERASE2 (RDR2), and then processed by DICER-LIKE3 (DCL3) to generate 24-nucleotide siRNAs (Xie et al., 2004; Daxinger et al., 2009). The 24-nucleotide siRNAs are then bound to ARGONAUTE4 (AGO4) or a related AGO protein and seek nascent transcripts produced by another alternative DNA-dependent RNA polymerase, Pol V (Wierzbicki et al., 2008, 2009). Binding of an AGO4-siRNA complex to Pol V nascent transcripts is thought to recruit DNA and histone methyltransferases to the vicinity of the target chromatin.
The 24-nucleotide heterochromatic siRNAs dominate endogenous small RNA populations in most tissues of most angiosperms, but their presence in other land plants has been less clear. All of the early small RNA-sequencing (RNA-seq) efforts from the mosses Physcomitrella patens (Arazi et al., 2005) and Polytrichum juniperinum (Axtell and Bartel, 2005), several gymnosperm species (Dolgosheina et al., 2008), and the lycophyte Selaginella moellendorffii (Banks et al., 2011) found a conspicuous absence of endogenous 24-nucleotide RNAs. It has also been suggested that conifers lack homologs of DCL3 (Dolgosheina et al., 2008). However, there are several hints suggesting that the heterochromatic siRNA pathway may indeed be present outside of angiosperms. Significant amounts of 24-nucleotide RNAs have been observed in conifers in a highly tissue-specific manner (Nystedt et al., 2013; J. Zhang et al., 2013). Nonconiferous gymnosperms (Cycas and Gingko) contain clear 24-nucleotide RNA populations, as does the fern Marsilea quadrifolia (Chávez Montes et al., 2014). The Selaginella genome contains DCL3, RDR2, AGO4, and Pol IV/V largest subunit homologs (Banks et al., 2011), suggesting that the absence of 24-nucleotide RNAs in initial small RNA-seq libraries may be due to tissue-restricted expression. Phylogenetic analysis clearly identifies DCL3, AGO4, and Pol V-related homologs in basal plants (Huang et al., 2015). Finally, our previous analysis demonstrated that the P. patens DCL3 homolog is required for the accumulation of 22-, 23-, and 24-nucleotide RNAs from a handful of siRNA hot spots (Cho et al., 2008). Nonetheless, direct experimental proof of a bona fide heterochromatic siRNA system in plants basal to the angiosperms is currently lacking. In this study, we used extensive small RNA-seq analysis in wild-type and several P. patens mutants (two RDRs, Pol IV, Pol V, two canonical DCLs, and a minimal DCL gene) to rigorously test the hypothesis that heterochromatic siRNAs are expressed in this basal land plant.
RESULTS
Most DCL-Derived Small RNA Loci Produce Mixtures of 23- to 24-Nucleotide Small RNAs in P. patens
Several previous studies have annotated P. patens miRNAs and endogenous siRNAs using small RNA-seq (Arazi et al., 2005; Axtell et al., 2006; Fattash et al., 2007; Cho et al., 2008; Arif et al., 2012). However, these previous small RNA-seq efforts all had low sequencing depth by current standards (<0.5 million mapped reads per library in all cases). Reliability of annotating and quantifying small RNA producing loci is largely dependent on the sequencing depth. Therefore, to create a more comprehensive annotation of P. patens small RNA genes, we obtained 10 small RNA-seq libraries (from six biological replicates; four samples were run twice) from 10-d-old wild-type protonemata totaling more than 100 million mapped reads (Supplemental Table 1). The majority of the small RNAs aligned to the nuclear genome, while a substantial minority aligned to the plastid genome. In order to identify a reference set of small RNA genes, we first de novo annotated potential small RNA clusters separately from six wild-type small RNA libraries using ShortStack (Axtell, 2013b). Then, genomic regions that were covered by cluster annotations in at least four of the six wild-type replicates were used as our reference set of P. patens small RNA genes. This stringent strategy allowed us to identify 1462 small RNA-producing loci (Supplemental Data Set 1). For each locus, the fraction of included reads 20 to 24 nucleotides in length was calculated, and a cutoff of 0.8 was used to discriminate non-DCL-derived loci from DCL-derived loci. Non-DCL-derived small RNAs are most often breakdown products of abundant long RNAs, such as rRNA, tRNA, and mRNAs. Only 17 loci were found to have a predominant RNA size (which we refer to as the “DicerCall”) outside of the DCL size range. Therefore, we focused on the 1445 DCL loci for the remainder of the study (Supplemental Data Set 1). These annotations, and all underlying data, can be interactively explored via a genome browser and other tools at http://plantsmallrnagenes.psu.edu/physcomitrella_patens/.
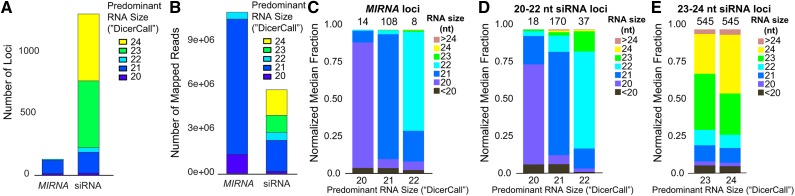
All annotated small RNA loci were classified into two categories: MIRNA loci and siRNA loci. Most DCL-derived small RNA loci were classified as siRNA loci (Figure 1). The majority were siRNA loci with DicerCalls of 23 or 24 (Figure 1A). However, when analyzed by total abundance, 21-nucleotide RNAs dominated both MIRNA and siRNA loci (Figure 1B). Thus, we conclude that a relatively small number of MIRNA and siRNA loci produce large amounts of 21-nucleotide RNAs, while a much larger number of mostly siRNA loci produce more modest amounts of 23- and 24-nucleotide RNAs.
Figure 1.
Properties of P. patens Small RNA Genes.
(A) and (B) Classification of DCL-derived small RNA loci, counted either by number of loci (A) or by total small RNA abundance (B).
(C) to (E) Small RNA size distributions within each class of DicerCall at MIRNA loci (C), 20- to 22-nucleotide siRNA loci (D), and 23- to 24-nucleotide siRNA loci (E).
DicerCall is a somewhat crude indicator and could mask cases where loci actually tend to produce mixtures of different sized RNAs. For MIRNA and 20- to 22-nucleotide siRNA loci, the DicerCall generally reflected a strong majority of RNAs of that size (Figures 1C and 1D). However, siRNA loci with DicerCalls of 23 or 24 in fact often produce mixtures of 23 and 24 nucleotide RNAs (Figure 1E). For further analyses, we classified three different groupings of DCL loci, listed here in order from most to least numerous: 23- to 24-nucleotide siRNA loci, 20- to 22-nucleotide siRNA loci, and MIRNA loci (Figures 1C to 1E).
P. patens 23- to 24-Nucleotide siRNA Loci Are Associated with Intergenic Regions, Repeats, and Regions with Dense 5-Methyl Cytosine
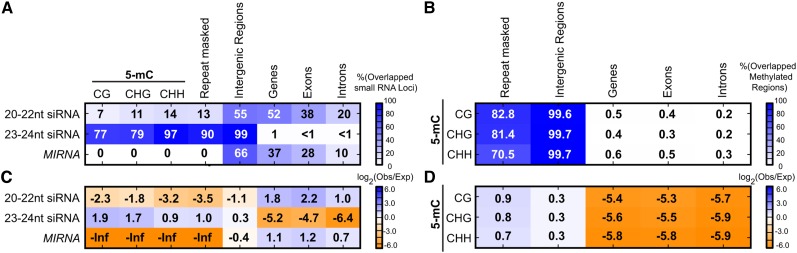
We performed co-occupancy analysis of the three groupings of DCL-derived small RNA loci against various genomic features. Two broad patterns were apparent. At one extreme, MIRNAs, and to a lesser extent 20- to 22-nucleotide siRNA loci, avoid regions with dense 5-methyl cytosine (5-mC) and repeats, but instead have some tendencies to overlap with genes (Figure 2). At the other extreme, 23- to 24-nucleotide siRNA loci are enriched for overlaps with 5-mC-dense regions of all contexts, intergenic regions, and repeats, but are severely depleted in overlaps with genes (Figures 2A and 2C). Consistent with a previous analysis (Zemach et al., 2010), P. patens loci with dense 5-mC are almost entirely confined to intergenic regions, enriched for association with repeats, and avoid genes (Figures 2B and 2D). It has been previously shown that Arabidopsis AGO4 preferentially binds to 24-nucleotide siRNAs with a 5′-bias toward A residues (Mi et al., 2008). Nucleotide frequency analysis revealed that small RNAs produced from 23- to 24-nucleotide siRNA loci in P. patens exhibit a weak 5′-A preference (Supplemental Figure 1). We conclude that P. patens 23- to 24-nucleotide siRNA loci are heterochromatic siRNAs, with grossly similar genomic arrangements as the heterochromatic siRNAs of angiosperms.
Figure 2.
Genomic Features of P. patens Small RNA-Producing Loci.
(A) Percentages of small RNA-producing loci that overlap various genomic features: % overlap = (# small RNA loci overlapping with one or more of the indicated genomic feature/# total small RNA loci) * 100.
(B) Percentages of regions of dense DNA methylation (5-mC) in three different contexts (CG, CHG, and CHH) relative to different genomic features.
(C) Heat map showing log2 (observed overlapped bases/expected overlapped bases) for each of the pairwise comparisons shown. Small RNA loci versus various genomic features are indicated. Cell values are shown in bold text.
(D) Heat map showing log2 (observed overlapped bases/expected overlapped bases) for each of the pairwise comparisons shown. Regions of dense DNA methylation (5-mC) in three different contexts (CG, CHG, and CHH) versus various genomic features are indicated. Repeat masked: repeats of various types as annotated by the Joint Genome Initiative genome annotation.
Improved P. patens MIRNA Annotations
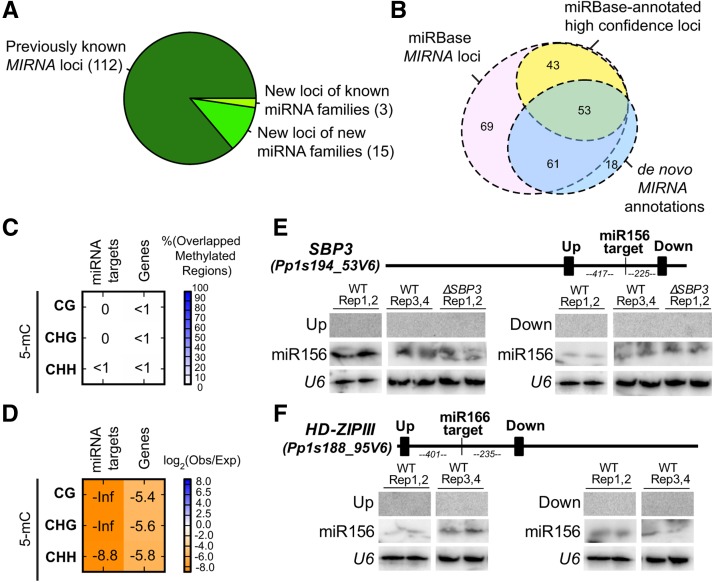
Our entirely de novo annotation of MIRNAs found 130 loci, of which 112 were already annotated in miRBase release 21 (Kozomara and Griffiths-Jones, 2014) (Figure 3; Supplemental Data Sets 2 and 3). We compared our mature miRNAs from novel loci with all mature miRNAs present in miRBase release 21. Three of the novel loci were found to be paralogs of known families (miR1027c, miR1049b, and miR1034b), and the remaining 15 new loci are not easily placed in previously known plant miRNA families (Figure 3A). We also found that most abundant miRNA reads have a 5′-U bias at P. patens MIRNA loci (Supplemental Figure 1), similar to what is observed for AGO1-bound miRNAs in Arabidopsis (Mi et al., 2008).
Figure 3.
Refinement of P. patens MIRNA Annotations and Functions.
(A) Classification of MIRNA loci confidently identified in this study.
(B) Euler diagram comparing our de novo MIRNA annotations with previously annotated P. patens MIRNA loci from miRBase 21.
(C) Percentage overlaps of regions of dense DNA methylation with miRNA targets and P. patens genes. Calculated as in Figure 2A.
(D) Enrichment/depletion analysis of methylated regions of genome with miRNA targets and P. patens genes. Calculated as in Figure 2C.
(E) RNA gel blot of small RNAs surrounding miR156 target site in Pp SBP3. Filled rectangles on the gene schematic indicate probe positions. Up, upstream of target site; down, downstream of target site.
(F) RNA gel blot of small RNAs surrounding miR166 target site in HD-ZIPIII. Filled rectangles on the gene schematic indicate probe positions. Numbers indicate distances (nucleotides) between target sites and probed regions. Up, upstream of target site; down, downstream of target site.
miRBase release 21 lists 229 P. patens MIRNA loci. We were unable to align three of these hairpins to the reference genome, leaving 226 loci to consider. Among these 226 loci, 96 are annotated in miRBase 21 as “high confidence” based on older and smaller RNA-seq data sets. Our deeper data set coupled with improved MIRNA annotation methods allowed us to further assess these prior annotations. Most P. patens miRBase loci (149 out of 226) were discovered as small RNA producing loci in our analysis (Supplemental Data Set 4). Only 114 of the prior miRBase annotations satisfied the strict structure and expression criteria we imposed (Figure 3B). Interestingly, the overlap between those 114 and the loci noted as “high confidence” loci in miRBase 21 (Kozomara and Griffiths-Jones, 2014) was not very high. Only 53 of the 96 miRBase 21 high confidence loci were accepted by our analysis (Figure 3B; Supplemental Data Set 4). We attribute this to the much greater sequencing depth, and consequent increased specificity, that our new small RNA-seq data allowed.
No Evidence for Widespread 5-mC or Secondary siRNA Accumulation from P. patens miRNA Targets
It has been proposed that high ratios of miRNA-to-target abundance promote 5-mC modification of target gene DNA in P. patens (Khraiwesh et al., 2010). However, bisulfite-seq data from Zemach et al. (2010) indicated that P. patens genes from a wild-type specimen are largely devoid of 5-mC in all sequence contexts in genes (Figures 3C and 3D). This lack of gene body 5-mC was even more strongly apparent in a set of 42 validated miRNA targets (Addo-Quaye et al., 2009) (Figures 3C and 3D; Supplemental Data Set 5). We conclude that either the earlier hypothesis is incorrect or that none of the natural miRNA-to-target ratios in wild-type 10-d-old protonemata are high enough to promote this effect. It is also possible that miRNA-directed target DNA methylation is only seen under certain conditions, such as at a miR1026 target after abscisic acid-induced expression (Khraiwesh et al., 2010).
It has been reported that protein-coding P. patens miRNA targets often spawn large amounts of ∼21-nucleotide secondary siRNAs both upstream and downstream of miRNA target sites (Khraiwesh et al., 2010). Despite the fact that about half of the 21- to 22-nucleotide siRNA loci we found overlapped a gene annotation (Figure 2A), only one overlapped with a validated miRNA target. This was the Pp TAS3a locus, a well known and deeply conserved producer of secondary siRNAs (Axtell et al., 2006) (Supplemental Data Set 5). RNA gel blots against two mRNAs that have validated as targets of conserved, highly abundant miRNAs (miR156 and miR166) failed to detect any secondary small RNA accumulation (Figures 3E and 3F). Our genome-wide analysis thus did not find evidence that P. patens miRNA targets generally spawn secondary siRNAs under our growth conditions.
Discovery and Mutagenesis of a P. patens MINIMAL DICER-LIKE Gene
Next, we revisited annotation of the P. patens DCL gene family. Dicers emerged early in the eukaryotic lineage and independently diverged in plants and animals (Mukherjee et al., 2013). Plants contain four ancient clades of DCL genes, with members in each clade being sub-functionalized for different types of small RNAs (Margis et al., 2006; Huang et al., 2015). P. patens has been reported to have no members of the DCL2 clade, single members of both the DCL3 and DCL4 clades, and two members of the DCL1 clade (Pp DCL1a and Pp DCL1b) (Axtell et al., 2007). Mutants in all four genes have been described (Cho et al., 2008; Khraiwesh et al., 2010; Arif et al., 2012). Upon review of P. patens DCL genes, we noticed several discrepancies in the annotation of the Pp DCL1b locus. All of the three available mRNA models were strongly contradicted by RNA-seq data (Chen et al., 2012), and none were capable of producing a full-length DCL protein (Supplemental Figure 2). De novo mRNA assembly using RNA-seq data resulted in several transcript models, none of which possessed any open reading frames > 100 codons in length (Supplemental Figure 2; Supplemental Data Set 6). Given these ambiguities, we excluded DCL1b when constructing a phylogeny of DCL proteins. We suspect that DCL1b is an expressed, spliced pseudogene.
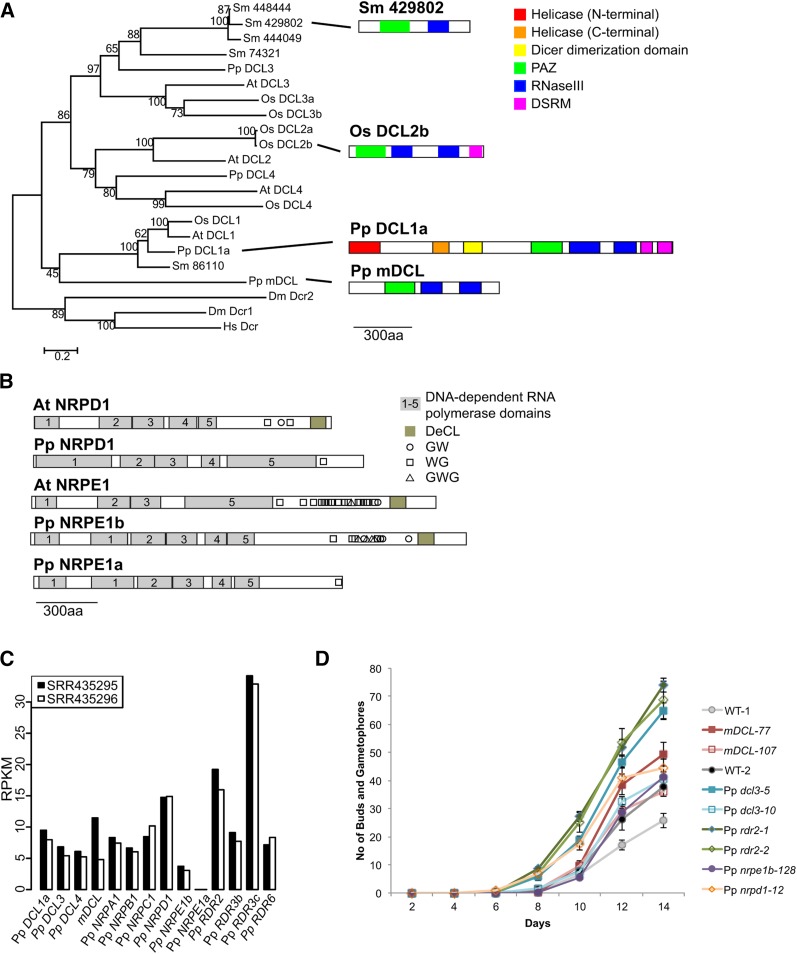
We also identified a P. patens MINIMAL DICER-LIKE (mDCL) gene encoding only the PAZ domain and two RNaseIII domains (Figure 4A). The mDCL locus is not located near Pp DCL1b, as they reside on different chromosomes. mDCL is not an obvious member of any of the canonical four clades of plant DCL proteins, although its phylogenetic position was difficult to resolve (Figure 4A). The protozoan parasite Giardia intestinalis has been shown to produce a functional Dicer with a similarly minimal domain composition (Macrae et al., 2006). In addition, the ciliated protozoan Tetrahymena thermophila also produces a Dicer protein that lacks an N-terminal helicase domain (although it also lacks a PAZ domain); T. thermophila DCL1 is required for accumulation of scan RNAs that direct programmed DNA deletion events (Malone et al., 2005; Mochizuki and Gorovsky, 2005). Rice (Oryza sativa) DCL2b has a similar truncated domain structure, and a S. moellendorffii DCL protein (429802) has a PAZ domain followed by a single RNaseIII domain (Figure 4A). Importantly, mDCL was basal to all four clades of plant DCLs (Figure 4A), while the Os DCL2b and Sm 429802 were clear recent duplicates of DCL2 and DCL3, respectively. We hypothesized that mDCL contributes to production of endogenous P. patens siRNAs. To test this hypothesis, we used homologous recombination to obtain two independent mdcl mutant lines (Supplemental Figure 3).
Figure 4.
Analysis of P. patens Small RNA Biogenesis Genes and Their Mutant Phenotypes.
(A) Phylogenetic analysis of DCL proteins. At, Arabidopsis thaliana; Os, Oryza sativa; Sm, Selaginella moellendorffii; Pp: P. patens; Dm, Drosophila melanogaster; Hs, Homo sapiens. Numbers are bootstrap percentages from 1000 replicates. Scale bar indicates substitutions per site. Sidebar shows example domain structures of mDCL and two other proteins with a similar minimal domain structure, and Pp DCL1a was included as an example of a full-length DCL protein.
(B) Domain structures of Arabidopsis (At) and P. patens (Pp) largest subunits of DNA-dependent RNA polymerases. DeCL, DEFECTIVE CHLOROPLASTS AND LEAVES domain.
(C) mRNA accumulation for the indicated genes in protonemata according to RNA-seq data from the indicated accessions. RPKM, reads per kilobase per million.
(D) Rates of bud and gametophore production. Seven-day-old protonemal tissues were inoculated on BCD media, and the total numbers of buds and gametophores were counted every 2 d. Points show means and whiskers show standard errors of the means from 12 replicates.
Heterochromatic siRNA Mutants and mdcl Mutants Have a Similar Accelerated Growth Phenotype
Previous analyses collectively indicated that P. patens has one homolog of the Arabidopsis Pol IV largest subunit (Pp NRPD1) and two homologs of the Pol V largest subunit (Pp NPRE1a and Pp NRPE1b) (Arif et al., 2013; Huang et al., 2015). A high density of multiple GW/WG/GWG motifs in the C-terminal domain is a characteristic of the Pol V, but not the Pol IV largest subunit (Haag and Pikaard, 2011). We found that Pp NRPE1b, but not Pp NRPE1a, had many C-terminal GW/WG/GWG motifs (Figure 4B). Pp NRPE1a is positioned around 600 kb away from Pp NRPE1b, suggestive of a recent tandem duplication. The Pp NRPE1a gene was previously suggested to be a Pol IV largest subunit homolog (Arif et al., 2013). Using homologous recombination, we obtained a single mutant line each for Pp nrpe1b and Pp nrpd1 (Supplemental Figures 4 and 5). Attempts to isolate a Pp nrpe1a mutant failed for unknown reasons.
P. patens contains RDR genes in the α and γ clades (Zong et al., 2009; Huang et al., 2015). Pp rdr6 mutants have an accelerated juvenile-to-adult gametophyte transition phenotype and lose the accumulation of trans-acting siRNAs (Talmor-Neiman et al., 2006; Cho et al., 2008). Huang et al. (2015) referred to the only other P. patens member of the α clade as Pp RDR1/2 because it is basal to the flowering plant RDR1 and RDR2 clades. Because our subsequent functional analysis demonstrated that the function of this gene is homologous to that of Arabidopsis RDR2, we refer to it here as simply Pp RDR2. It remains to be tested whether this locus also might perform RDR1-like functions. Two independent Pp rdr2 mutant lines were created using homologous recombination (Supplemental Figure 6).
Expression levels in protonemata, as estimated by RNA-seq data (Chen et al., 2012), were moderate for all of the P. patens genes we studied, with the exception of Pp NRPE1a, for which we were unable to obtain a mutant (Figure 4C). This suggests that the protonematal stage of growth is a valid time point to assay for effect of these mutations on small RNA populations.
We previously observed that Pp dcl3 mutants display an accelerated juvenile-to-adult transition in gametophyte growth (Cho et al., 2008). In flowering plants, DCL3, RDR2, Pol IV, and Pol V are known to collaborate in the heterochromatic siRNA pathway, so we hypothesized that Pp rdr2, Pp nrpd1, and Pp nrpe1b mutants would also display the same phenotype. We found that this was indeed the case (Figure 4D). We also found that mdcl plants had an accelerated juvenile-to-adult transition (Figure 4D), suggesting that mDCL also contributes to the heterochromatic siRNA pathway.
mDCL Promotes Accumulation of 23-Nucleotide RNAs from Heterochromatic siRNA Loci
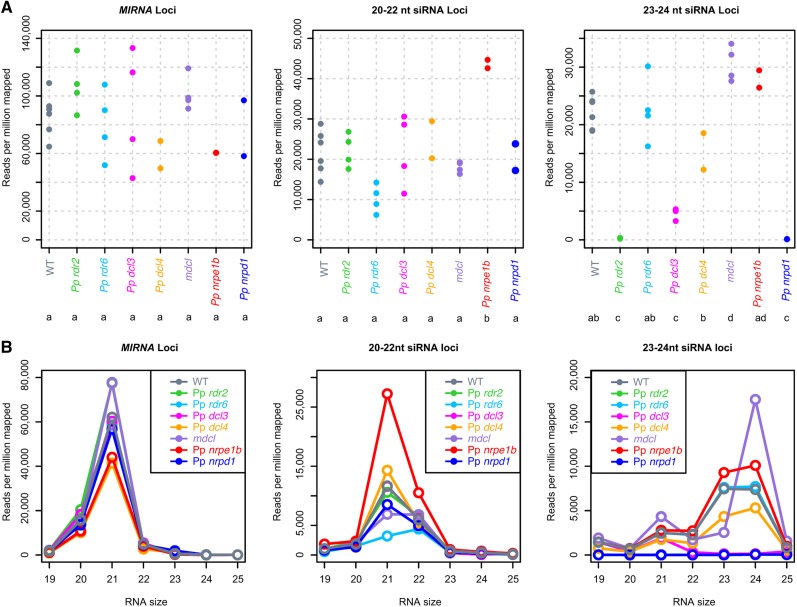
We tested the hypothesis that the Pp rdr2, Pp nrpd1, Pp nrpe1b, and mdcl mutants affected 23- to 24-nucleotide siRNA accumulation by constructing and sequencing multiple small RNA-seq libraries from 10-d-old protonemata (Supplemental Table 1). Also included were Pp dcl4 and Pp rdr6 mutants (known to affect secondary siRNAs; Talmor-Neiman et al., 2006; Arif et al., 2012), and Pp dcl3 mutants (which our previous analysis implicated in 23- to 24-nucleotide siRNA accumulation; Cho et al., 2008). All mutants were represented by two to four biological replicates (Figure 5A; Supplemental Table 1).
Figure 5.
mDCL Promotes 23-Nucleotide RNA Accumulation and Represses 24-Nucleotide RNA Accumulation at Heterochromatic siRNA Loci.
(A) Overall small RNA abundance at MIRNA, 20- to 22-nucleotide siRNA loci, and 23- to 24-nucleotide siRNA loci. Each dot represents a biological replicate small RNA-seq sample. Genotypes sharing a letter have means that are not significantly different (Tukey's honestly significant difference test, α = 0.05).
(B) Small RNA abundance by size within MIRNA loci, 20- to 22-nucleotide siRNA loci, and 23- to 24-nucleotide siRNA loci for the indicated genotypes. Reads for all biological replicates within each genotype were summed and converted to reads per million mapped.
None of the mutants examined had significant effects on the MIRNA population (Figure 5). At 20- to 22-nucleotide siRNA loci, the only significant difference in overall abundance was an increase in the Pp nrpe1b mutant (Figure 5A). However, when examining size distributions, Pp rdr6 mutants clearly lost 21-nucleotide RNAs, but not 22-nucleotide RNAs, from the 20- to 22-nucleotide loci (Figure 5B). We noted that none of the Dicer-like mutants tested affected the levels of small RNAs from 20- to 22-nucleotide siRNA loci. Pp DCL1a was the only P. patens dcl mutant not examined in our study, and we suspect that many of these loci are Pp DCL1a dependent. Several of the mutants had major effects on small RNAs from 23- to 24-nucleotide siRNA loci. Pp rdr2, Pp nrpd1, and Pp dcl3 mutants had the most severe effects; siRNAs of all sizes were essentially absent from 23- to 24-nucleotide siRNA loci in the Pp rdr2 and Pp nrpd1 plants, while only 21-nucleotide RNAs remained in Pp dcl3 (Figure 5B). The specific loss of 23- to 24-nucleotide RNAs, but not 21-nucleotide RNAs, in the Pp dcl3 background is consistent with our earlier smaller scale observations (Cho et al., 2008). Pp nrpe1b and Pp rdr6 mutants did not have strong changes in overall RNA accumulation levels from 23- to 24-nucleotide siRNA loci (Figure 5). mdcl mutants had unique alterations in their small RNA profiles at 23- to 24-nucleotide siRNA loci; the levels of 23-nucleotide siRNAs were decreased, but the levels of 21-nucleotide, and especially 24-nucleotide, siRNAs were increased (Figure 5B). We conclude that mDCL affects the heterochromatic siRNA pathway by promoting the production of 23-nucleotide siRNAs at the expense of 21- and 24-nucleotide siRNAs.
Differential Expression Analysis Reveals Distinct Subgroups of Heterochromatic siRNA Loci
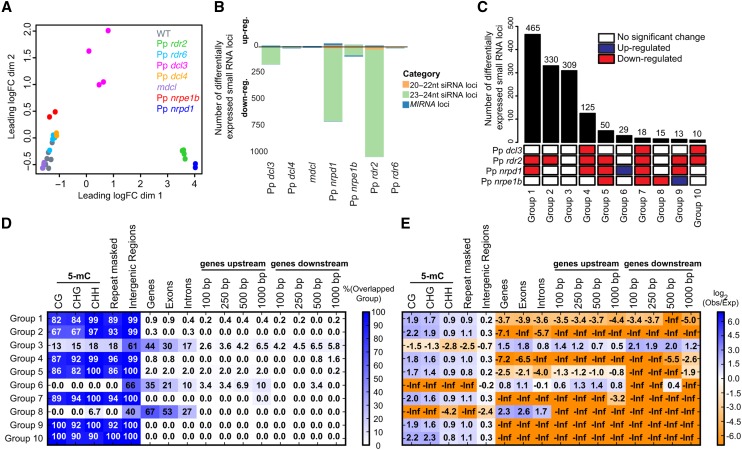
A differential expression analysis was performed by tallying small RNA alignments of all sizes from each library within each of our annotated DCL-derived small RNA loci. A multidimensional scaling plot of these data was prepared to illustrate overall differences in small RNA accumulation between each of the samples (Figure 6). Biological replicates for each genotype were generally consistent with each other, indicated by their tight groupings on the multidimensional scaling plot (Figure 6A). Pp rdr6, Pp dcl4, mdcl, and Pp nrpe1b mutants clustered closely with the wild type, while Pp rdr2 and Pp nrpd1 formed a second cluster of libraries distinct from the wild-type and from all of the other mutants (Figure 6A). A third, looser cluster was formed by the Pp dcl3 mutants.
Figure 6.
Differential Expression Analysis of P. patens Small RNAs in Mutants.
(A) Multidimensional scaling plot showing the overall relationship between each mutant and wild-type biological replicate small RNA-seq library. Leading fold change (FC) is the (root mean square) average of the largest absolute log2 fold changes between each pair of samples.
(B) Numbers of differentially downregulated or upregulated small RNA loci in each of the indicated mutants compared with the wild type. Differential expression for a locus is defined as with at least 2-fold-change with a false discovery rate of <0.01.
(C) Numbers of differentially expressed loci (bar chart; upper panel) represented by different mutant combinations (heat map; lower panel).
(D) Observed overlap/expected overlap ratios for different mutant groups defined in (C) relative to various genomic features. Calculated as in Figure 2C.
(E) Overlap percentage for mutant groups defined in (C) relative to various genomic features. Calculated as in Figure 2A. Repeat-masked: repeats of various types as annotated by the Joint Genome Initiative genome annotation.
Loci were considered differentially expressed (DE) in a particular mutant if they had at least a 2-fold change compared with the wild type at a false discovery rate of <0.01 (Supplemental Data Set 7). Very few DE loci were found in Pp dcl4 and Pp rdr6 mutants, indicating that the secondary siRNA pathway does not make a major contribution to most of the endogenous small RNA loci under study (Figure 6B). Large numbers of downregulated 23- to 24-nucleotide siRNA loci were found in Pp rdr2 and Pp nrpd1 mutants (Figure 6B). A much smaller number of downregulated 23- to 24-nucleotide siRNA loci were apparent in Pp dcl3 and Pp nrpe1b mutants (Figure 6B). The modest numbers of upregulated loci observed in Pp dcl3, Pp nrpd1, and Pp rdr2 mutants were mostly MIRNAs (Figure 6B). It is possible that the heterochromatic siRNAs dependent on Pp dcl3, Pp nrpd1, and Pp rdr2 compete with miRNA accumulation. Alternatively, because small RNA-seq quantification is proportional rather than absolute, small RNAs from these loci may appear upregulated only because of the gross absence of 23- to 24-nucleotide siRNAs in these samples. Interestingly, relatively small numbers of 23- to 24-nucleotide siRNA loci were upregulated in the Pp nrpe1b mutant (Figure 6B), suggesting the existence of distinct subsets of heterochromatic siRNA loci. This is consistent with the effect of Arabidopsis Pol V on only a subset of heterochromatic siRNAs (Mosher et al. 2008). Finally, 23- to 24-nucleotide siRNA loci did not appear as DE in mdcl mutants because decreased accumulation of 23 nucleotide RNAs were compensated for by the increased accumulation of 24-nucleotide RNAs, resulting in similar total small RNA accumulations to that of the wild type (Figures 5B and 6B).
We next integrated DE calls for loci between the various mutants. We plotted and analyzed the 10 most common patterns of loci for the four mutants of major effect (Figure 6C). Pp rdr6, Pp dcl4, and mdcl were not involved in any of the top 10 patterns, so were omitted from the figure. Loci downregulated in both Pp rdr2 and Pp nrpd1 mutants were most numerous (Group 1, Figure 6C). Group 2 loci were downregulated only in the Pp rdr2 mutant, while group 4 was composed of loci downregulated in Pp dcl3, Pp rdr2, and Pp nrpd1 (Figure 6C). Group 5 loci were downregulated in Pp rdr2, Pp nrpd1, and Pp nrpe1b. Except for groups 6 and 9, the remainder of the most common patterns were loci that were downregulated in one or more of the Pp dcl3, Pp rdr2, Pp nrpd1, and Pp nrpe1b mutants. To sum up, the DE patterns of mutants whose direct homologs in Arabidopsis are involved in the canonical Pol IV RdDM pathway tend to be similar.
Pp nrpe1b mutants had an interesting pattern of DE loci. Fifteen loci were downregulated solely in this mutant while not significantly changed in the other three mutants (Group 8, Figure 6C). Thirteen loci were upregulated in Pp nrpe1b mutants but downregulated in Pp rdr2 and Pp nrpd1 mutants (Group 9, Figure 6C). Thus, there appear to be distinct subsets of heterochromatic siRNA loci uniquely affected by Pp NRPE1b. Pp nrpd1 mutants were also interesting, with 29 loci upregulated only in Pp nrpd1 mutants but not significantly changed in the other three mutants (Group 6, Figure 6C). Pp NRPD1 therefore negatively regulates a unique subset of heterochromatic siRNA loci in terms of overall small RNA abundance. Co-occupancy analysis of the loci in these groups of interest relative to general genomic features revealed distinct patterns (Figures 6D and 6E). First, loci that were not DE in any of the mutants (Group 3) seemed to overlap with genes and gene-proximal regions and were depleted in regions characteristic of heterochromatic siRNAs. Second, loci that were upregulated only in Pp nrpd1 (Group 6) and loci that were solely downregulated in Pp nrpe1b (Group 8) mutants were depleted for overlaps with 5-mC, repeats, and intergenic regions. Pp nrpe1b-dependent loci (Group 8) were highly enriched in genes, while Pp nrpd1-dependent loci (Group 6) were associated with both genes and upstream of genes (Figures 6D and 6E). All of the other groups were enriched for overlaps with 5-mC and repeats and depleted for overlaps with genes and gene-proximal regions (Figures 6D and 6E).
P. patens Heterochromatic siRNAs Suppress Expression of LTR Retrotransposon-Related Sequences Despite Minimal Changes in DNA Methylation
To test whether loss of P. patens 23- to 24-nucleotide siRNAs affects 5-mC patterns, whole-genome shotgun sequencing of bisulfite converted DNA was performed from one wild-type specimen and one specimen each from both Pp rdr2-1 and Pp rdr2-2 mutant plants. Numerous PCR duplicates were observed in these libraries (Supplemental Table 2). After removing them, genomic coverage was very sparse (over 80% of Cs in the nuclear genome had no coverage) and biased toward repetitive regions (Supplemental Figures 7A and 7B). Bulk DNA methylation patterns in the nuclear genome were very similar between the wild type and the two Pp RDR2 mutant specimens in all three sequence contexts (Supplemental Figure 7C). Mutation of rdr2 caused no obvious changes in DNA methylation in the population of 23- to 24-nucleotide siRNA loci nor in any of the groups of differentially expressed loci from Figure 6 (Supplemental Figure 8). However, because of low coverage, groups 6 and 8 were not able to be analyzed.
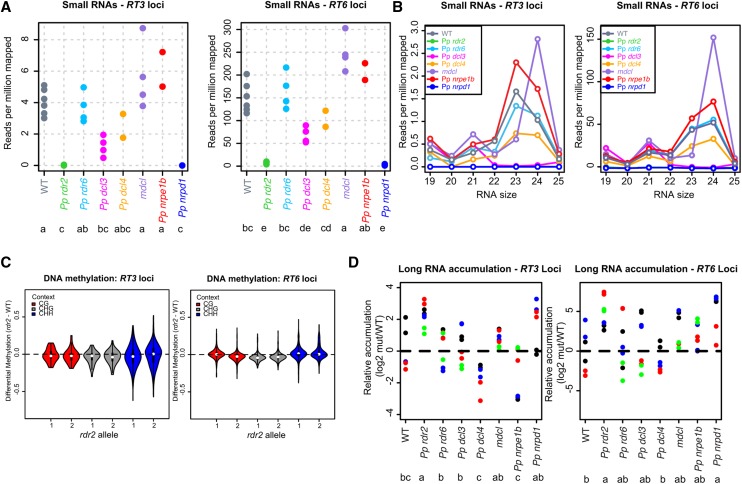
Previously, we described two families of reverse transcriptase genes, RT3 and RT6, from P. patens high-copy LTR retrotransposons that were silent in the wild type but expressed in the Pp dcl3 mutant (Cho et al., 2008). The overall patterns of small RNA accumulation at these loci in the various mutants mirrored those of the 23- to 24-nucleotide siRNA loci (Figures 7A and 7B). At RT3 loci, no strong changes in DNA methylation were observed in the Pp rdr2 mutants (Figure 7C). However, at more than 75% of the RT6 loci, there was a decrease in 5-mC in Pp rdr2 mutants specifically in the CHG context (Figure 7C), suggesting that Pp RDR2 is required to maintain wild-type levels of CHG methylation at many of these loci. Pp rdr2 mutants have significantly increased accumulation of long RNAs from both RT3 and RT6 loci (Figure 7D). Pp nrpd1 mutants have increased accumulation of long RNAs from RT6, but not RT3 loci (Figure 7D). These results indicate that Pp RDR2 and Pp NRPD1 play a role in suppressing long RNA accumulation from these LTR retrotransposon-related loci. Overall, however, this analysis did not find strong evidence that Pp RDR2-dependent siRNAs are required to maintain most existing patterns of 5-mC.
Figure 7.
Effects of the P. patens Heterochromatic siRNA Pathway on the LTR Retrotransposon-Related Sequences RT3 and RT6.
(A) Overall small RNA abundance at RT3 and RT6 loci. Each dot represents a biological replicate small RNA-seq sample. Genotypes sharing a letter have means that are not significantly different (Tukey's honestly significant difference test, α = 0.05).
(B) Small RNA abundance by size within RT3 loci and RT6 loci for the indicated genotypes. Reads for all biological replicates within each genotype were summed and converted to reads per million mapped.
(C) Differential DNA methylation between the indicated Pp rdr2 alleles and the wild type at RT3 loci (left) and RT6 loci (right). Violin plots show a Tukey box plot (line = median, box edges = 1st and 3rd quartiles, whiskers = 1.5 IQR) surrounded by a twinned kernel density plot to visually show the distribution of the data.
(D) Long RNA accumulation based on quantitative RT-PCR from RT3 loci (left) and RT6 loci (right). Dots of the same color represent different technical replicates of the same biological replicate. Genotypes sharing a letter have means that are not significantly different (Tukey's honestly significant difference test, α = 0.05).
DISCUSSION
We analyzed more than 100 million mapped small RNA-seq reads from wild-type P. patens and used these data to produce a comprehensive set of small RNA gene annotations. Setting aside degradation products that are unlikely to be part of the DCL/AGO regulatory system, most P. patens small RNA genes produce 23- to 24-nucleotide siRNAs. These loci are enriched for overlaps with repeats and regions of dense 5-mC and nearly always avoid protein-coding genes. The P. patens 23- to 24-nucleotide siRNA loci are also strongly dependent on Pp RDR2, Pp NRPD1 (the presumed largest subunit of a Pol IV complex), and Pp DCL3 for small RNA production. Altogether, these data lead us to conclude that P. patens uses a heterochromatic siRNA pathway fundamentally similar to that of flowering plants. Therefore, the potential absence of heterochromatic siRNAs in conifers (Dolgosheina et al., 2008; Morin et al., 2008) and lycophytes (Banks et al., 2011) could reflect secondary loss of the pathway in those specific lineages. However, more recent data show that, for conifers, endogenous 24-nucleotide siRNAs can be found, albeit sometimes in tissue-specific patterns (Nystedt et al., 2013; J. Zhang et al., 2013) or in unique lineages such as cycads and Gingko (Chávez Montes et al., 2014). Thus, we favor the hypothesis that, like miRNAs, heterochromatic siRNAs are a universal feature of land plant transcriptomes.
However, P. patens heterochromatic siRNAs do have some atypical features compared with those in flowering plants. Small RNA-seq samples from most wild-type tissues of most flowering plants are dominated by 24-nucleotide heterochromatic siRNAs. In contrast, heterochromatic siRNAs are weakly expressed in P. patens protonemata, where 21-nucleotide miRNA expression dominates the small RNA profile in terms of abundance. Also in contrast to flowering plants, whose heterochromatic siRNAs are mostly 24 nucleotides, P. patens heterochromatic siRNA loci produce a mixture of 23- and 24-nucleotide RNAs at nearly equal levels, with much lower levels of 21- and 22-nucleotide RNAs. Our genetic analysis indicates that the mDCL gene is responsible specifically for 23-nucleotide siRNA accumulation from these loci; in mdcl mutants, 23-nucleotide RNAs are strongly reduced, while 24-nucleotide RNAs are strongly increased at heterochromatic siRNA loci. At the same loci, loss of Pp DCL3 function eliminates all sizes of small RNA accumulation. We speculate that mDCL is dependent upon Pp DCL3 due to its lack of an N-terminal helicase domain. We also speculate that mDCL competes with Pp DCL3 for small RNA precursors produced by Pol IV and Pp RDR2. In mdcl mutants, Pp DCL3 processes the excess precursors to make mostly 24-nucleotide RNAs. In Pp dcl3 mutants, mDCL cannot function, leading to the loss of both the mDCL-dependent 23-nucleotide and Pp DCL3-dependent 24-nucleotide RNAs. Further investigation is required to test this hypothesis. There are precedents for competition in small RNA size classes in plants: Mutation of maize (Zea mays) mop1, an RDR2 homolog, reduces 24-nucleotide siRNA accumulation but enhances 22-nucleotide siRNA accumulation (Nobuta et al., 2008). Also, there are known Arabidopsis loci where multiple DCLs engage the same template, such as the inverted repeat locus IR71 (Dunoyer et al., 2010). We find proteins with a mDCL-like arrangement of domains in S. moellendorffii and rice, but neither are clear mDCL homologs; instead, they are recent duplications of DCL3 clade and DCL2 clade genes, respectively (Figure 4A). This suggests that mDCL genes might arise frequently during plant evolution.
Our genetic analyses show that P. patens expresses heterochromatic siRNAs that have a largely similar biogenesis pathway as in flowering plants. Therefore, the most parsimonious scenario is that, as for miRNAs, the heterochromatic siRNA pathway is an ancestral trait that was present in the last common ancestor of bryophytes and all other subsequently diverged lineages of plants. The major differences are the relative levels of expression in vegetative tissue and the use of the novel mDCL gene to produce 23-nucleotide heterochromatic siRNAs in P. patens. We also suggest that P. patens miRNA functions may not be as unusual as has previously been suggested (Khraiwesh et al., 2010); we find no evidence that P. patens miRNAs spawn abundant secondary siRNAs from protein-coding target mRNAs nor direct 5-mC deposition at target chromatin. It remains possible that these miRNA functions occur under specific conditions not represented in our study. Finally, our publically available and browsable annotations of P. patens small RNA genes (http://plantsmallrnagenes.psu.edu/physcomitrella_patens) provide a comprehensive and useful resource for further study of all classes of small RNAs in this model organism.
METHODS
Small RNA-seq and Reference Annotation of Wild-Type P. patens Small RNA Genes
Total RNA was extracted using the miRNeasy Mini kit (Qiagen) per the manufacturer's instructions from 10-d-old Physcomitrella patens protonemata grown on cellophane-overlaid BCD medium (Ashton and Cove, 1977) supplemented with 5 mM ammonium tartrate and cultured at 25°C, 16 h day/8 h night. Small RNA libraries were constructed using the TruSeq Small RNA kit (Illumina) per the manufacturer’s instructions and sequenced on a HiSequation 2500 (Illumina) instrument using 50-nucleotide single-end runs. Small RNA-seq data from the wild-type libraries (Supplemental Table 1) were analyzed with ShortStack version 2.0.0 (Axtell, 2013b). First, each library was adapter trimmed and aligned to the reference genome (version 3.0 nuclear assembly from Phytozome v10.1; Physcomitrella patens v3.0, DOE-JGI, http://phytozome.jgi.doe.gov/pz/portal.html#!info?alias=Org_Ppatens; Rensing et al., 2008; Goodstein et al., 2012) combined with the plastid and mitochondrial genomes) using default butter version 0.0.3 settings with –adapter TGGAATTC. The alignments were then run with ShortStack mindepth 4 to find abundance of small RNAs at the de novo ShortStack-identified small RNA loci. Bedtools (Quinlan and Hall, 2010) was used to find shared clusters in six different wild-type libraries and then the output was filtered so that only regions that were present in at least 4 (out of 6) libraries were used to serve as the final reference small RNA locus boundaries. ShortStack-count mode under default settings was then used to find relative small RNA abundance on this reference list of de novo-identified small RNA loci for each wild-type and mutant library. Full results are listed in Supplemental Data Set 1, and the annotations are also hosted at http://plantsmallrnagenes.psu.edu/physcomitrella_patens. Raw wild-type small RNA-seq data, processed data, and alignments were deposited to the NCBI Gene Expression Omnibus (GEO) (GSE44900) and are also available at http://plantsmallrnagenes.psu.edu/physcomitrella_patens.
Co-Occupancy Analyses
Regions of dense 5-mC occupancy in the CG, CHG, and CHH contexts were calculated in 50-nucleotide intervals based on protonematal bisulfite-seq data from NCBI GEO accession GSM497264 (Zemach et al., 2010). A given 50-nucleotide interval was considered densely methylated in a particular context if there were more than six reads of all Cs in that context and more than a threshold percentage of those were unconverted (60% threshold for CG and CHG contexts; 20% threshold for CHH contexts). Repeat-masked regions were obtained from version 3.0 of the nuclear assembly from Phytozome v10.1 (Physcomitrella patens v3.0, DOE-JGI, http://phytozome.jgi.doe.gov/pz/portal.html#!info?alias=Org_Ppatens; Rensing et al., 2008; Goodstein et al., 2012). Intergenic, genic, exonic, intronic, gene upstream, and gene downstream locations were calculated based on version 3.0 of the transcriptome assembly gff3 file obtained from Phytozome 10.1. rRNA gene locations were based on regions of significant similarity (BLASTn e-value of ≤ 1E-10) to the rRNA consensus sequences. tRNA gene locations were based on genome-wide analysis with tRNAscan-SE version 1.3.1 under default parameters (Lowe and Eddy, 1997). Raw data for all of these annotations are retrievable and browsable at plantsmallrnagenes.psu.edu/physcomitrella_patens/jbrowse/.
The absolute numbers of overlapping loci and the total of nonredundant overlapping nucleotides for each pairwise comparison of feature types were calculated. Enrichment/depletion was calculated based on the ratio of the observed to the expected number of overlapping nucleotides. The expected number of overlapping nucleotides for any pairwise feature comparison is given by E = (x/g) * (y/g) * g, where E is the expected number of overlapping nucleotides under the null hypothesis of random location, x is the total number of nonredundant nucleotides for feature type 1, y is the total number of nonredundant nucleotides for feature type 2, and g is the total genome size.
miRNA and miRNA Target Analyses
MIRNA hairpin sequences and mature miRNA sequences identified by our de novo annotation effort were compared with the prior annotations in miRBase 21. The 18 loci that had not been previously annotated were registered with miRBase. We also used miRBase’s “confidence” community annotation system (Kozomara and Griffiths-Jones, 2014) to up-vote and comment on the existing annotations. A set of 42 high-confidence miRNA target genes was curated from Addo-Quaye et al. (2009) (Supplemental Data Set 5) and compared with the 5-mC data (see above for processing methods) from Zemach et al. (2010).
Small RNA Gel Blots
Small RNA gel blots were performed as described (Cho et al., 2012) with modification. Total RNAs from 10-d-old samples were extracted using Tri-Reagent (Sigma-Aldrich), and small RNAs were fractionated as described (Pall and Hamilton, 2008). Twenty micrograms of total RNAs was separated on 20% PAGE gel, transblotted onto the Hybond X membrane (GE Healthcare), and cross-linked using 1-ethyl-3-(3-dimethylamonipropyl) carbodiimide (Pall and Hamilton, 2008). Probes were independently labeled with T4 polynucleotide kinase (New England Biolabs) and mixed before hybridization. Hybridization, washing, and detection were performed as described (Cho et al., 2012). The probe sequences are listed in Supplemental Table 3.
Phylogenetic Analysis
DCL protein sequences were trimmed to include only the C-terminal portions, beginning 50 residues before the PAZ domain. Sequence alignments of trimmed DCL proteins were generated using MUSCLE in the MEGA5 software (Tamura et al., 2011) with default parameters (Supplemental Data Set 8) and used for phylogenetic analysis (MEGA5) using the maximum likelihood method based on the JTT matrix-based model with uniform rates. All positions with alignment gaps were used. The tree with the highest log likelihood was shown, and topology reliability was checked using bootstrap analysis with 1000 replicates.
Construction of Vectors
For the construction of knockout vectors, two ∼1-kb regions 5′ and 3′ from the open reading frame of desired genes were amplified using specific primer sets (Supplemental Table 3) and inserted into the pUQ vector (Cho et al., 2008) as previously described (Cho et al., 2012).
DNA Gel Blot Analysis
Genomic DNAs were extracted using a Phytopure DNA Extraction kit (GE Healthcare). For DNA gel blot analysis, the BglII-digested genomic DNAs of mdcl and Pp rdr2 were blotted onto a Hybond NX nylon membrane (GE Healthcare) and hybridized following a standard protocol (Sambrook and Russell, 2001). For a probe, PCR-amplified hptII fragment was radiolabeled with [α-32P]dCTP using an NEblot Kit (New England Biolabs) per the manufacturer's instructions.
RT-PCR
Total RNAs were extracted from 10-d-old protonemata using the miRNeasy Mini kit (Qiagen). RT-PCR reactions were performed as previously described (Cho et al., 2012). Primer sequences are listed in Supplemental Table 3.
Differential Expression Analysis
Small RNA-seq samples from the various mutants (Supplemental Table 1) were trimmed (–adapter TGGAATTC), aligned to the version 3.0 genome (including plastid and mitochondrial genomes), and analyzed using ShortStack version 2.0.0 in count mode using the wild-type de novo small RNA gene annotations as the–count file. Counts from separate sequencing runs of the same libraries were combined (Supplemental Table 1) and used for differential expression analysis with the R package edgeR (Robinson et al., 2010). Libraries were normalized with the “calcNormFactors” function and analyzed with the “exactTest” function analysis for each mutant in comparison with the wild type. DE genes at a 1% false discovery rate were retrieved using the “decideTestsDGE” function and further filtered to retain only those with 2-fold or greater deviation from the wild type. Supplemental Data Set 7 contains the full details and results of these analyses.
Whole-Genome Bisulfite Sequencing
Library preparation was essentially as described by Hsieh et al. (2009). In brief, total genomic DNA was isolated from 10-d-old protonemata cultivated on BCDA media from three samples: the wild type (Gransden 2004 isolate), Pp rdr2-1, and Pp rdr2-2 plants. DNA was isolated using DNeasy Plant Mini Kit (Qiagen) sheared by sonication to fragments of 350 to 850 bp, repaired using NEBNext End Repair Module protocol, purified using QIAquick PCR purification kit, and then ligated to custom Illumina paired-end adapters (sequencing primer A and sequencing primer B) that were synthesized with 5-methyl modifications on each cytosine position (Supplemental Table 3). The library was then treated with bisulfite with the Qiagen EpiTect Bisulfite kit, followed by 26 cycles of PCR amplification using Illumina PE PCR Primer A and Illumina PE PCR Primer B (Supplemental Table 3) and paired-end sequencing (100 × 100 nucleotides) on an Illumina GAIIx sequencing instrument.
Raw FASTQ data were processed to remove 3′ adapters and for quality using cutadapt version 1.8.1 (Martin, 2011) using nondefault settings: -O 1,–max-n 2, -q 20, -m 30, -a AGATCGGAAGAGCG, -A AGATCGGAAGAGCG. These trimmed FASTQ files were further processed with a perl script (Supplemental Data Set 9, script 1) that converts all Cs in the first mate set to Ts and all of the Gs in the second mate set to As, in both bases tracking the conversions by modifying the names of the reads. After this conversion, any mate-pair where one or more of the mates had a homopolymeric stretch of 20 or more was discarded, and the converted mates output to FASTA-formatted files. The reference genome (consisting of the plastid genome, the mitochondrial genome, and the version 3.0 nuclear genome from Phytozome 10; http://phytozome.jgi.doe.gov/pz/portal.html#!info?alias=Org_Ppatens; Rensing et al., 2008; Goodstein et al., 2012) was processed with a perl script (Supplemental Data Set 9, script 2) that makes two copies of each chromosome: one representing a fully converted top strand in which all Cs are converted to Ts, and a second representing the reverse complement of the bottom strand in which all Gs and converted to As. In the process, a large table documenting the positions of every C on both strands of the genome is created. The converted genome was then indexed using bowtie2-build (version 2.2.4; Langmead and Salzberg, 2012) under default parameters. The converted mate-paired read sets were then aligned to the converted genome using bowtie2 (version 2.2.5; Langmead and Salzberg, 2012) with nondefault settings -f, -X 800,–no-discordant,–no-mixed, -p 6 -D 30, -R 6, and –non-deterministic. The resulting SAM streams were piped through sequential samtools (version 1.2; Li et al., 2009) view and sort commands to create coordinate-sorted BAM alignment files. Fragments resulting from PCR duplication were defined as those sharing a common left end and flagged by setting bit 1024 in the SAM FLAG field using a perl script (Supplemental Data Set 9, script 3). The BAM files were then filtered using samtools view (version 1.2; Li et al., 2009) with options -b, -f 3, -F 1028, which outputs only alignments from concordantly mapped mate-pairs that are not flagged as PCR duplicates.
Using another perl script (Supplemental Data Set 8, script 4), along with the large table of genome-wide C positions (see above), the frequencies of converted and nonconverted Cs at each position in the genome were calculated and output into large tables. This analysis avoided “double-counting” information for positions that were covered by both mates of a mate-pair. All subsequent summaries and analyses of the results were based on analysis of these tables. Analyses of specific loci or 100-nucleotide genomic bins required a minimum number of reads covering Cs of 10 in each of the three libraries. Violin plots were produced in R (version 3.2.0) using vioplot (package version 0.2). The plots show a Tukey box plot (line = median, box edges = 1st and 3rd quartiles, whiskers = 1.5 IQR) surrounded by a twinned kernel density plot to visually show the distribution of the data.
Analyses of RT3 and RT6 Loci
RT3 and RT6 loci were defined based on degenerate oligos first described by Cho et al. (2008) (Supplemental Table 3). The locations of RT3 and RT6 loci in the version 3.0 nuclear genome were determined by searching for all cases where exact matches to any permutation of the forward and any permutation of the reverse oligo were within 500 nucleotides of each in the opposite orientation. Lists of these loci are in Supplemental Data Sets 10 and 11. These lists were then compared with small RNA and bisulfite-seq data. Quantitative RT-PCR of these loci was performed from RNA samples extracted from 10-d-old protonemata (using the Qiagen miRNeasy Mini kit). cDNA was synthesized using Applied Biosystems High Capacity cDNA kit [mixture of oligo(dT) and random primers], amplified with Quantitect SYBR-Green master mix (Qiagen), and analyzed on an Applied Biosystems StepOne Plus real-time PCR system. Accumulation, relative to the control amplicon Pp EF1-α, was calculated and normalized for PCR efficiencies. The mean of the wild-type samples was set to one. Oligos are listed in Supplemental Table 3.
Accession Numbers
cDNA sequences for mDCL, Pp NRPE1a, and Pp NRPE1b have been deposited to NCBI under accession numbers KF179046, KF908783, and KF908782, respectively. Small RNA-seq data have been deposited to NCBI GEO under accession numbers GSE44900 (wild type) and GSE51419 (mutants). Bisulfite-seq data have been deposited at the Sequence Read Archive under accession numbers SRR2013850 (wild type), SRR2013877 (Pp rdr2-1), and SRR2013879 (Pp rdr2-2). The full set of P. patens small RNA gene annotations and associated data are also available and browsable at http://plantsmallrnagenes.psu.edu/physcomitrella_patens. Other accession numbers are as follows: At DCL1 (At1g01040), At DCL2 (At3g03300), At DCL3 (At3g43920), At DCL4 (At5g20320), Pp DCL1a (ABV31244.1), Pp DCL1b (DQ675601), Pp DCL3 (ABV31245), Pp DCL4 (EF670438), mDCL (KF179046), Os DCL1 (Os03g02970), Os DCL2a (Os03g38740), Os DCL2b (Os09g14610), Os DCL3a (Os01g68120), Os DCL3b/5 (Os10g34430), Os DCL4 (Os04g43050), Hs Dcr (Uniprot Q9UPY3), Dm Dcr1 (FlyBase FBgn0039016), and Dm Dcr2 (FlyBase FBgn0034246). Selaginella moellendorffii accession numbers are given in Figure 4A.
Supplemental Data
Supplemental Figure 1. Nucleotide frequencies of Physcomitrella patens small RNAs for each position.
Supplemental Figure 2. Discrepancies in Pp DCL1b annotations.
Supplemental Figure 3. Targeted knockout of mDCL.
Supplemental Figure 4. Targeted knockout of Pp NRPE1b.
Supplemental Figure 5. Targeted knockout of Pp NRPD1.
Supplemental Figure 6. Targeted knockout of Pp RDR2.
Supplemental Figure 7. Overview of bisulfite-seq libraries.
Supplemental Figure 8. Analysis of DNA methylation at 23- to 24-nucleotide siRNA loci in Pp rdr2 mutants.
Supplemental Table 1. Physcomitrella patens small RNA-seq libraries.
Supplemental Table 2. Summary of bisulfite-seq libraries.
Supplemental Table 3. Oligonucleotide sequences used in this study.
Supplemental Data Set 1. P. patens small RNA-producing loci.
Supplemental Data Set 2. Text-based alignments of 130 P. patens MIRNA loci.
Supplemental Data Set 3. Summary of ShortStack-annotated miRNAs.
Supplemental Data Set 4. All miRBase loci and overlapping ShortStack loci.
Supplemental Data Set 5. Degradome-validated P. patens miRNA target genes and overlapping ShortStack small RNA loci.
Supplemental Data Set 6. Inferred mRNA sequence of Pp DCL1b from RNA-seq data.
Supplemental Data Set 7. Differential expression analysis details.
Supplemental Data Set 8. Alignment of DCL proteins used for phylogenetic analysis.
Supplemental Data Set 9. Perl scripts used for processing of bisulfite-seq data.
Supplemental Data Set 10. List of Pp RT3 loci (version 3.0 nuclear genome).
Supplemental Data Set 11. List of Pp RT6 loci (version 3.0 nuclear genome).
Supplementary Material
Acknowledgments
We thank Tzahi Arazi for the gift of Pp rdr6 mutant plants, Wolfgang Frank for the gift of Pp dcl4 mutant plants, Brian Gregory for helpful discussions on this work and pilot small RNA-seq experiments, and Craig Praul and his colleagues at the Huck Institutes Genomics Core Facility for small RNA sequencing. Purchase of the Core Facility Illumina HiSeq 2500 instrument was supported by National Science Foundation Award 1229046 to M.J.A. This work was supported by grants from the Searle Scholars Program and NIH-NIGMS (R01-GM084051) to M.J.A.
AUTHOR CONTRIBUTIONS
C.C. and M.J.A. analyzed small RNA-seq data and wrote the article with input from all authors. S.H.C. generated P. patens mutants, characterized their phenotypes, and performed phylogenetic analyses. S.S. analyzed MIRNA annotations and functions. S.H.C. and Q.L. created small RNA-seq libraries. A.W. identified and cloned cDNAs for P. patens Pol IV and Pol V largest subunit homologs. M.J.A. conceived and supervised the project.
Glossary
- miRNA
microRNA
- siRNA
short interfering RNA
- 5-mC
5-methyl cytosine
- DE
differentially expressed
- GEO
Gene Expression Omnibus
Footnotes
Articles can be viewed online without a subscription.
References
- Addo-Quaye C., Snyder J.A., Park Y.B., Li Y.-F., Sunkar R., Axtell M.J. (2009). Sliced microRNA targets and precise loop-first processing of MIR319 hairpins revealed by analysis of the Physcomitrella patens degradome. RNA 15: 2112–2121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arazi T., Talmor-Neiman, M., Stav, R., Riese, M., Huijser, P., and Baulcombe, D.C. (2005). Cloning and characterization of micro-RNAs from moss. Plant J. 43: 837–848. [DOI] [PubMed] [Google Scholar]
- Arif M.A., Fattash I., Ma Z., Cho S.H., Beike A.K., Reski R., Axtell M.J., Frank W. (2012). DICER-LIKE3 activity in Physcomitrella patens DICER-LIKE4 mutants causes severe developmental dysfunction and sterility. Mol. Plant 5: 1281–1294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arif M.A., Frank W., Khraiwesh B. (2013). Role of RNA interference (RNAi) in the moss Physcomitrella patens. Int. J. Mol. Sci. 14: 1516–1540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashton N.W., Cove D.J. (1977). The isolation and preliminary characterisation of auxotrophic and analogue resistant mutants of the moss, Physcomitrella patens. Mol. Gen. Genet. 154: 87–95. [Google Scholar]
- Axtell M.J. (2013a). Classification and comparison of small RNAs from plants. Annu. Rev. Plant Biol. 64: 137–159. [DOI] [PubMed] [Google Scholar]
- Axtell M.J. (2013b). ShortStack: comprehensive annotation and quantification of small RNA genes. RNA 19: 740–751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Axtell M.J., Bartel D.P. (2005). Antiquity of microRNAs and their targets in land plants. Plant Cell 17: 1658–1673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Axtell M.J., Jan C., Rajagopalan R., Bartel D.P. (2006). A two-hit trigger for siRNA biogenesis in plants. Cell 127: 565–577. [DOI] [PubMed] [Google Scholar]
- Axtell M.J., Snyder J.A., Bartel D.P. (2007). Common functions for diverse small RNAs of land plants. Plant Cell 19: 1750–1769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banks J.A., et al. (2011). The Selaginella genome identifies genetic changes associated with the evolution of vascular plants. Science 332: 960–963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chávez Montes R.A., de Fátima Rosas-Cárdenas F., De Paoli E., Accerbi M., Rymarquis L.A., Mahalingam G., Marsch-Martínez N., Meyers B.C., Green P.J., de Folter S. (2014). Sample sequencing of vascular plants demonstrates widespread conservation and divergence of microRNAs. Nat. Commun. 5: 3722. [DOI] [PubMed] [Google Scholar]
- Chen Y.-R., Su Y.S., Tu S.-L. (2012). Distinct phytochrome actions in nonvascular plants revealed by targeted inactivation of phytobilin biosynthesis. Proc. Natl. Acad. Sci. USA 109: 8310–8315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho S.H., Addo-Quaye C., Coruh C., Arif M.A., Ma Z., Frank W., Axtell M.J. (2008). Physcomitrella patens DCL3 is required for 22-24 nt siRNA accumulation, suppression of retrotransposon-derived transcripts, and normal development. PLoS Genet. 4: e1000314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho S.H., Coruh C., Axtell M.J. (2012). miR156 and miR390 regulate tasiRNA accumulation and developmental timing in Physcomitrella patens. Plant Cell 24: 4837–4849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coruh C., Shahid S., Axtell M.J. (2014). Seeing the forest for the trees: annotating small RNA producing genes in plants. Curr. Opin. Plant Biol. 18: 87–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daxinger L., Kanno T., Bucher E., van der Winden J., Naumann U., Matzke A.J.M., Matzke M. (2009). A stepwise pathway for biogenesis of 24-nt secondary siRNAs and spreading of DNA methylation. EMBO J. 28: 48–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolgosheina E.V., Morin R.D., Aksay G., Sahinalp S.C., Magrini V., Mardis E.R., Mattsson J., Unrau P.J. (2008). Conifers have a unique small RNA silencing signature. RNA 14: 1508–1515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunoyer P., Brosnan C.A., Schott G., Wang Y., Jay F., Alioua A., Himber C., Voinnet O. (2010). An endogenous, systemic RNAi pathway in plants. EMBO J. 29: 1699–1712. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Fattash I., Voss B., Reski R., Hess W.R., Frank W. (2007). Evidence for the rapid expansion of microRNA-mediated regulation in early land plant evolution. BMC Plant Biol. 7: 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodstein D.M., Shu S., Howson R., Neupane R., Hayes R.D., Fazo J., Mitros T., Dirks W., Hellsten U., Putnam N., Rokhsar D.S. (2012). Phytozome: a comparative platform for green plant genomics. Nucleic Acids Res. 40: D1178–D1186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haag J.R., Pikaard C.S. (2011). Multisubunit RNA polymerases IV and V: purveyors of non-coding RNA for plant gene silencing. Nat. Rev. Mol. Cell Biol. 12: 483–492. [DOI] [PubMed] [Google Scholar]
- Hsieh T.-F., Ibarra C.A., Silva P., Zemach A., Eshed-Williams L., Fischer R.L., Zilberman D. (2009). Genome-wide demethylation of Arabidopsis endosperm. Science 324: 1451–1454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y., Kendall T., Forsythe E.S., Dorantes-Acosta A., Li S., Caballero-Pérez J., Chen X., Arteaga-Vázquez M., Beilstein M.A., Mosher R.A. (2015). Ancient origin and recent innovations of RNA polymerase IV and V. Mol. Biol. Evol. 32: 1788–1799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khraiwesh B., Arif M.A., Seumel G.I., Ossowski S., Weigel D., Reski R., Frank W. (2010). Transcriptional control of gene expression by microRNAs. Cell 140: 111–122. [DOI] [PubMed] [Google Scholar]
- Kozomara A., Griffiths-Jones S. (2014). miRBase: annotating high confidence microRNAs using deep sequencing data. Nucleic Acids Res. 42: D68–D73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B., Salzberg S.L. (2012). Fast gapped-read alignment with Bowtie 2. Nat. Methods 9: 357–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Law J.A., Du J., Hale C.J., Feng S., Krajewski K., Palanca A.M.S., Strahl B.D., Patel D.J., Jacobsen S.E. (2013). Polymerase IV occupancy at RNA-directed DNA methylation sites requires SHH1. Nature 498: 385–389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., Homer N., Marth G., Abecasis G., Durbin R.; 1000 Genome Project Data Processing Subgroup (2009). The sequence alignment/map format and SAMtools. Bioinformatics 25: 2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lippman Z., Martienssen R. (2004). The role of RNA interference in heterochromatic silencing. Nature 431: 364–370. [DOI] [PubMed] [Google Scholar]
- Lowe T.M., Eddy S.R. (1997). tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 25: 955–964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macrae I.J., Zhou K., Li F., Repic A., Brooks A.N., Cande W.Z., Adams P.D., Doudna J.A. (2006). Structural basis for double-stranded RNA processing by Dicer. Science 311: 195–198. [DOI] [PubMed] [Google Scholar]
- Malone C.D., Anderson A.M., Motl J.A., Rexer C.H., Chalker D.L. (2005). Germ line transcripts are processed by a Dicer-like protein that is essential for developmentally programmed genome rearrangements of Tetrahymena thermophila. Mol. Cell. Biol. 25: 9151–9164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margis R., Fusaro A.F., Smith N.A., Curtin S.J., Watson J.M., Finnegan E.J., Waterhouse P.M. (2006). The evolution and diversification of Dicers in plants. FEBS Lett. 580: 2442–2450. [DOI] [PubMed] [Google Scholar]
- Martin M. (2011). Cutadapt removes adapter sequences from high-throughput sequencing. EMBnet Journal 17: 10–12. [Google Scholar]
- Matzke M.A., Mosher R.A. (2014). RNA-directed DNA methylation: an epigenetic pathway of increasing complexity. Nat. Rev. Genet. 15: 394–408. [DOI] [PubMed] [Google Scholar]
- Mi S., et al. (2008). Sorting of small RNAs into Arabidopsis argonaute complexes is directed by the 5′ terminal nucleotide. Cell 133: 116–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mochizuki K., Gorovsky M.A. (2005). A Dicer-like protein in Tetrahymena has distinct functions in genome rearrangement, chromosome segregation, and meiotic prophase. Genes Dev. 19: 77–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morin R.D., Aksay G., Dolgosheina E., Ebhardt H.A., Magrini V., Mardis E.R., Sahinalp S.C., Unrau P.J. (2008). Comparative analysis of the small RNA transcriptomes of Pinus contorta and Oryza sativa. Genome Res. 18: 571–584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosher R.A., Schwach F., Studholme D., Baulcombe D.C. (2008). PolIVb influences RNA-directed DNA methylation independently of its role in siRNA biogenesis. Proc. Natl. Acad. Sci. USA 105: 3145–3150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukherjee K., Campos H., Kolaczkowski B. (2013). Evolution of animal and plant dicers: early parallel duplications and recurrent adaptation of antiviral RNA binding in plants. Mol. Biol. Evol. 30: 627–641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nobuta K., et al. (2008). Distinct size distribution of endogeneous siRNAs in maize: Evidence from deep sequencing in the mop1-1 mutant. Proc. Natl. Acad. Sci. USA 105: 14958–14963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nystedt B., et al. (2013). The Norway spruce genome sequence and conifer genome evolution. Nature 497: 579–584. [DOI] [PubMed] [Google Scholar]
- Pall, G.S., and Hamilton, A.J. (2008). Improved northern blot method for enhanced detection of small RNA. Nat. Protoc. 3: 1077–1084. [DOI] [PubMed] [Google Scholar]
- Quinlan A.R., Hall I.M. (2010). BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26: 841–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rensing, S.A., et al. (2008). The Physcomitrella genome reveals evolutionary insights into the conquest of land by plants. Science 319: 64–69. [DOI] [PubMed] [Google Scholar]
- Robinson M.D., McCarthy D.J., Smyth G.K. (2010). edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26: 139–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook, J., and Russell, D.W. (2001). Molecular Cloning: A Laboratory Manual, 3rd ed. (Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press). [Google Scholar]
- Talmor-Neiman M., Stav R., Klipcan L., Buxdorf K., Baulcombe D.C., Arazi T. (2006). Identification of trans-acting siRNAs in moss and an RNA-dependent RNA polymerase required for their biogenesis. Plant J. 48: 511–521. [DOI] [PubMed] [Google Scholar]
- Tamura K., Peterson D., Peterson N., Stecher G., Nei M., Kumar S. (2011). MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 28: 2731–2739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wierzbicki A.T., Haag J.R., Pikaard C.S. (2008). Noncoding transcription by RNA polymerase Pol IVb/Pol V mediates transcriptional silencing of overlapping and adjacent genes. Cell 135: 635–648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wierzbicki A.T., Ream T.S., Haag J.R., Pikaard C.S. (2009). RNA polymerase V transcription guides ARGONAUTE4 to chromatin. Nat. Genet. 41: 630–634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Z., Johansen L.K., Gustafson A.M., Kasschau K.D., Lellis A.D., Zilberman D., Jacobsen S.E., Carrington J.C. (2004). Genetic and functional diversification of small RNA pathways in plants. PLoS Biol. 2: E104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zemach A., McDaniel I.E., Silva P., Zilberman D. (2010). Genome-wide evolutionary analysis of eukaryotic DNA methylation. Science 328: 916–919. [DOI] [PubMed] [Google Scholar]
- Zhang H., et al. (2013). DTF1 is a core component of RNA-directed DNA methylation and may assist in the recruitment of Pol IV. Proc. Natl. Acad. Sci. USA 110: 8290–8295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J., Zhang S., Han S., Li X., Tong Z., Qi L. (2013). Deciphering small noncoding RNAs during the transition from dormant embryo to germinated embryo in Larches (Larix leptolepis). PLoS One 8: e81452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zong J., Yao X., Yin J., Zhang D., Ma H. (2009). Evolution of the RNA-dependent RNA polymerase (RdRP) genes: duplications and possible losses before and after the divergence of major eukaryotic groups. Gene 447: 29–39. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.