The inability of the cytokinin perception mutant cre1 to form symbiotic nodules in Medicago truncatula was linked to a deficiency in auxin transport regulation mediated by endogenous flavonoids.
Abstract
Initiation of symbiotic nodules in legumes requires cytokinin signaling, but its mechanism of action is largely unknown. Here, we tested whether the failure to initiate nodules in the Medicago truncatula cytokinin perception mutant cre1 (cytokinin response1) is due to its altered ability to regulate auxin transport, auxin accumulation, and induction of flavonoids. We found that in the cre1 mutant, symbiotic rhizobia cannot locally alter acro- and basipetal auxin transport during nodule initiation and that these mutants show reduced auxin (indole-3-acetic acid) accumulation and auxin responses compared with the wild type. Quantification of flavonoids, which can act as endogenous auxin transport inhibitors, showed a deficiency in the induction of free naringenin, isoliquiritigenin, quercetin, and hesperetin in cre1 roots compared with wild-type roots 24 h after inoculation with rhizobia. Coinoculation of roots with rhizobia and the flavonoids naringenin, isoliquiritigenin, and kaempferol, or with the synthetic auxin transport inhibitor 2,3,5,-triiodobenzoic acid, rescued nodulation efficiency in cre1 mutants and allowed auxin transport control in response to rhizobia. Our results suggest that CRE1-dependent cytokinin signaling leads to nodule initiation through the regulation of flavonoid accumulation required for local alteration of polar auxin transport and subsequent auxin accumulation in cortical cells during the early stages of nodulation.
INTRODUCTION
Symbiotic bacteria collectively called rhizobia can initiate the formation of nitrogen-fixing root nodules in many species of legumes. Nodule initiation involves the reinitiation of cell divisions in the cortex, endodermis, and pericycle of the root, followed by differentiation of the nodule primordium into a mature organ, similar to the process of lateral root formation (Hirsch and LaRue, 1997, Mathesius, 2008; Herrbach et al., 2014; Xiao et al., 2014). Legume species differ in the types of nodules formed. Typically in temperate legumes, including the model legume Medicago truncatula, alfalfa (Medicago sativa), or clover (Trifolium sp), indeterminate nodules are initiated from pericycle and inner cortical cell divisions, and nodules maintain a persistent meristem. In many tropical legumes, however, including common bean (Phaseolus vulgaris) and soybean (Glycine max), determinate nodules form from outer cortical cell divisions, and the resulting nodules contain a temporary meristem that later differentiates (Hirsch, 1992; van Spronsen et al., 2001)
The mechanism of nodule initiation is only partially elucidated. Nod factors produced by rhizobia are in most cases necessary and in some legumes sufficient to induce nodules (Truchet et al., 1991). The signaling cascade mediating Nod factor action leads to the activation of cytokinin signaling in the cortex of the root (Crespi and Frugier, 2008; Oldroyd et al., 2011), which is accompanied by activation of the gene encoding the cytokinin biosynthesis enzyme LOG1, a cytokinin riboside 5-monophosphate phosphoribohydrolase (Mortier et al., 2014), and an increase in cytokinin concentration at the nodule initiation site in M. truncatula (van Zeijl et al., 2015). Several studies have implicated cytokinin as a central regulator in nodule development (Frugier et al., 2008). The expression of a cytokinin synthesis gene in Sinorhizobium meliloti nodulation mutants was sufficient to induce cortical cell divisions and expression of nodulation genes in alfalfa (Cooper and Long, 1994). Application of cytokinin can induce cortical cell divisions and expression of nodulation genes in alfalfa (Hirsch et al., 1997; Fang and Hirsch, 1998), white clover (Trifolium repens) (Mathesius et al., 2000), and Lotus japonicus (Heckmann et al., 2011), and pseudonodules on the actinorhizal plant Alnus glutinosa (Rodriguez-Barrueco and Bermudez de Castro, 1973). Legume mutants impaired in cytokinin perception typically fail to initiate nodules (Murray et al., 2007; Plet et al., 2011), while constitutive cytokinin signaling was found to cause spontaneous nodule formation in the absence of rhizobia (Tirichine et al., 2007). In M. truncatula, the cytokinin receptor CRE1 (CYTOKININ RESPONSE1) is necessary for perceiving exogenous cytokinin in roots, and either mutation or local RNAi-induced silencing of CRE1 in roots leads to the inhibition of nodule formation (Gonzalez-Rizzo et al., 2006; Plet et al., 2011). In response to rhizobia and cytokinins, the cre1 mutant fails to induce several cytokinin primary response genes, such as RESPONSE REGULATOR4 and NSP2 (NODULATION SIGNALING PATHWAY2) (Ariel et al., 2012). A recent transcriptome comparison of gene expression changes in wild-type and cre1 mutant roots in response to a 3-h Nod factor treatment in M. truncatula demonstrated that cytokinin signaling is necessary for the majority (∼75% or ∼600 transcripts) of Nod factor-induced transcriptional changes (van Zeijl et al., 2015). However, the mechanism of how cytokinin controls nodule development is still not defined.
Previous studies have suggested that cytokinin signaling may regulate plant developmental processes through its action on auxin transport. Auxin transport is essential for establishing auxin gradients in the plant, leading to cell-type specification and induction of meristematic cell divisions (Benková et al., 2003). Auxin transport regulation has been most closely studied for indole-3-acetic acid (IAA), which enters the cell partially by diffusion and by facilitated import via AUX1/LAX (AUXIN RESISTANT1/LIKE AUX1) and ABCB (ATP Binding Cassette subfamily B)-related transporters. Auxin export is strictly regulated through PIN (PIN-FORMED) and ABCB-type transporters that are polarly inserted into the plasma membrane on specific sides of the cell to control the direction of auxin transport in acropetal, basipetal, or lateral direction (Petrásek and Friml, 2009). In the Arabidopsis thaliana root, auxin transport in the acropetal direction, from the root base to the root tip, is mainly mediated by PIN1, whereas basipetal auxin transport, from the root tip to the elongation zone, is mediated by PIN2 (Rashotte et al., 2000; Michniewicz et al., 2007).
Certain flavonoids act as modulators of auxin transport by affecting the expression (Peer et al., 2004) and localization of PIN proteins (Santelia et al., 2008), the cycling of PIN proteins to endosomal vesicles (Geldner et al., 2001), as well as modifying the activity of ABCB-type auxin transporters (Di Pietro et al., 2002; Peer and Murphy, 2007; Bailly et al., 2008). Flavonoids bind to two types of protein complexes, a low-affinity binding complex containing an aminopeptidase and a high-affinity complex containing ABCB-type transporters (Noh et al., 2001; Murphy et al., 2002). External application of certain flavonoids, in particular flavonols, can inhibit auxin export (Jacobs and Rubery, 1988), and auxin transport in flavonoid-deficient mutants or transgenic plants is altered (Brown et al., 2001; Buer and Muday, 2004; Peer et al., 2004; Wasson et al., 2006; Laffont et al., 2010). In addition, flavonoids are required to control auxin transport during nodule initiation in legumes forming indeterminate nodules (Wasson et al., 2006), and flavonols are the most likely subclass of flavonoids responsible for this auxin transport control (Zhang et al., 2009).
Several studies indeed demonstrated that a local auxin transport inhibition is required for indeterminate nodule initiation. Early studies showed that application of synthetic auxin transport inhibitors can induce pseudonodules in some legumes that are characterized by a peripheral vasculature, which does not extend into the distal part of the nodule, an uninfected central zone, and a diffuse meristem (Allen et al., 1953; Hirsch et al., 1989). During nodulation, acropetal root auxin transport is temporarily inhibited by rhizobia-inducing indeterminate nodules, and this is followed by a local increase in auxin response in the pericycle and inner cortex at the site of nodule initiation (Mathesius et al., 1998; Boot et al., 1999; van Noorden et al., 2007). In the cre1 mutant, rhizobia are unable to inhibit acropetal auxin transport and this is associated with a misregulation of some PIN auxin carrier-encoding gene expression and protein accumulation (Plet et al., 2011). The hypothesis that cytokinin acts upstream of auxin transport regulation is supported by studies from Arabidopsis showing that cytokinin regulates PIN expression, accumulation, selective degradation, and subsequently polar auxin transport (Pernisová et al., 2009; Ruzicka et al., 2009; Marhavý et al., 2011, 2014). This is thought to regulate auxin accumulation during lateral root formation (Laplaze et al., 2007; Marhavý et al., 2014). The finding that auxin transport inhibitors can cause pseudonodule formation in Nod factor signaling defective mutants acting downstream of CRE1, such as nsp2 and nin, suggests that auxin transport control of nodulation acts downstream of cytokinin signaling (Rightmyer and Long, 2011). Furthermore, cytokinin application induces auxin responses in pseudonodule primordia in white clover (Mathesius et al., 2000), and spontaneous nodules formed in the L. japonicus constitutively activated cytokinin signaling mutant snf2 also show activation of auxin responses in dividing cortical cells during nodule primordium formation (Suzaki et al., 2012).
Collectively, these findings suggest that a crucial role of cytokinins may be to regulate auxin transport and/or response during nodule initiation. To investigate this hypothesis, we performed a detailed study of auxin transport, auxin response, and auxin metabolite accumulation in the M. truncatula cre1 mutant. We found that cre1 mutant roots did not mediate rhizobia-induced changes in acropetal or basipetal auxin transport and had a limited activation of auxin response in the inner cortex. Following inoculation with rhizobia, cre1 roots also showed reduced accumulation of the auxin IAA and of several flavonoids. Nodule initiation was rescued by the application of synthetic auxin transport inhibitors, as well as certain flavonoids that also rescued auxin transport inhibition and auxin responses in rhizobia-infected cre1 roots. These results suggest that cytokinins act through induction of flavonoids, which would alter auxin transport and accumulation during the initiation of indeterminate nodules.
RESULTS
The cre1 Mutant Is Defective in Acropetal and Basipetal Auxin Transport Regulation and Auxin Accumulation following S. meliloti Inoculation
To investigate auxin transport control in the cre1 mutant, we quantified acropetal and basipetal auxin transport using radiolabeled IAA applied either above the site of inoculation or at the root tip, respectively. To exclude the possibility that defects in flavonoids in the cre1 mutant prevented proper nod gene activation in the symbiont (Zhang et al., 2009), we used, instead of a wild-type strain, a genotype of S. meliloti that constitutively expresses nodD3, i.e., that produces Nod factors in the absence of nod gene-inducing flavonoids from the legume host (Barnett et al., 2004); hereafter, this strain is referred to as E65. This strain formed a significantly higher number of nodules on wild-type roots compared with the reference strain S. meliloti 1021 (Supplemental Figure 1). Under our growth conditions in agar plates, both strains usually did not lead to nodule formation in the cre1 mutant (Supplemental Figure 1), and when nodules formed, this was at a significantly lower level than in the wild type, as reported previously for the cre1 mutant (Plet et al., 2011) and other cytokinin receptor mutants (Murray et al., 2007; Held et al., 2014).
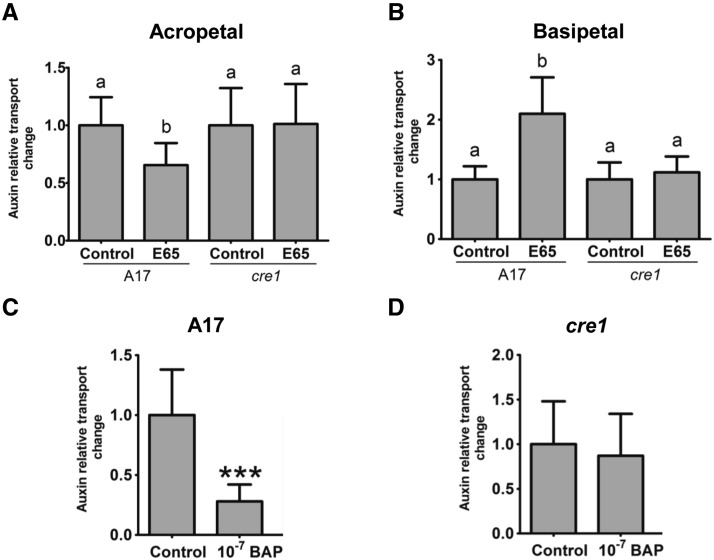
Acropetal (from root base to root tip) auxin transport capacity was measured just below the site of inoculation with E65, where previous studies detected a reduction in auxin transport capacity following inoculation with strain 1021 (Wasson et al., 2006; Plet et al., 2011; Supplemental Figure 2). Consistent with those studies, the E65 S. meliloti strain led in wild-type roots to a significant reduction in acropetal auxin transport into a root segment below the inoculation site within 24 h. In the cre1 mutant, inoculation with E65 failed to inhibit acropetal auxin transport (Figure 1A). Near the root tip, auxin transport also occurs in a basipetal (from the root tip upwards) direction, and we therefore tested whether this basipetal auxin flow was also affected by rhizobia. We measured basipetal auxin transport capacity of the root segment just above the inoculation site (Supplemental Figure 2). In wild-type roots, E65 inoculation significantly increased basipetal auxin transport capacity at 24 h postinoculation (hpi), while in cre1 mutant roots, there was no significant difference between inoculated and control roots (Figure 1B).
Figure 1.
Relative Auxin Transport Changes in Wild-Type (A17) and cre1 Mutant Roots.
(A) and (B) Acropetal (A) and basipetal (B) auxin transport measurements 24 h after mock or E65 inoculation in a segment just below (for acropetal) and above (for basipetal) the inoculation site, respectively.
(C) and (D) Acropetal auxin transport measurements in A17 (C) and cre1 mutant (D) roots 24 h after mock or BAP treatment (10−7 M) in a segment just below the mock or BAP application sites. Control treatments were set to “1” in each case.
A two-way ANOVA with a Tukey-Kramer multiple comparison post-test was used for statistical analyses in (A) and (B) (P < 0.05, n = 15 to 20). Different lowercase letters indicate significant differences in relative auxin transport rate. A Student’s t test was used for statistical analyses in (C) and (D) (P < 0.05, n = 20), where asterisks in (C) indicate an extremely very significant difference in relative auxin transport rate (P < 0.001). Graphs show mean and sd.
To test whether cytokinin signaling may directly lead to auxin transport inhibition, we measured acropetal auxin transport in wild-type and cre1 mutant roots 24 h after treatment with the synthetic cytokinin benzyl amino purine (BAP) at 10−7 M. This cytokinin application led to a significant inhibition of acropetal auxin transport in the wild type (Figure 1C), but not in cre1 mutant roots (Figure 1D).
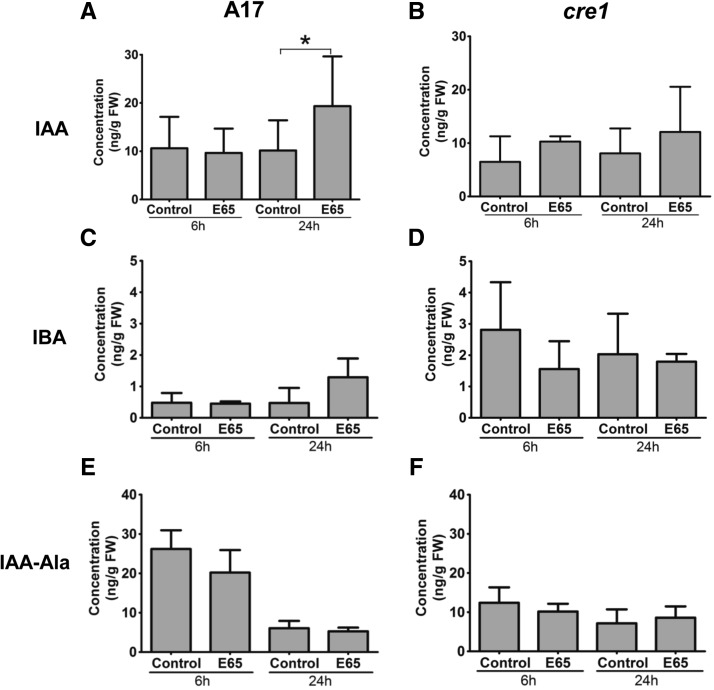
We predicted that a reduction of auxin export below the inoculation site would likely increase auxin concentrations at the S. meliloti inoculation site in the wild type. Thus, we quantified the auxin content in a 4-mm-long root segment comprising the inoculation site (2 mm above and 2 mm below the inoculation site), harvested at 6 and 24 hpi. The most abundant nonconjugated auxin measured in the roots of both wild-type and cre1 mutants was IAA (Figure 2). The IAA concentration in wild-type root segments inoculated with rhizobia did not change at 6 hpi but significantly increased at 24 hpi (Figure 2A). In cre1 roots, no significant increase was measured at 6 or 24 hpi (Figure 2B). Concentrations of indole-3-butyric acid (IBA) were not significantly changed in response to inoculation in wild-type or cre1 mutants, but cre1 roots showed elevated IBA concentrations compared with the wild type (Figures 2C and 2D). The conjugated auxin IAA-alanine showed no significant changes in response to E65 inoculation in either genotype, but higher concentrations were observed in the wild type at 6 hpi compared with cre1 roots (Figures 2E and 2F). Overall, combined auxin concentrations were higher in the wild type than in cre1 roots under both control and inoculated conditions (Supplemental Figure 3). We could not detect 4-chloro-IAA, phenylacetic acid, and the auxin conjugates IAA-phenylalanine, IAA-tryptophan, IAA-leucine, IAA-isoleucine, IAA-valine, or IAA-aspartate in any of the root segments. These auxin metabolites were targeted for quantification because a previous study suggested their presence in M. truncatula based on enzyme feeding assays (Campanella et al., 2008). The detection limits for these auxins are shown in Supplemental Table 1 and are below the concentrations measured for some of these auxins in other studies (Schneider et al., 1985; Kowalczyk and Sandberg, 2001; Matsuda et al., 2005).
Figure 2.
Auxin Concentration in Wild-Type (A17) and cre1 Mutant Roots at 6 and 24 h after Mock or E65 Inoculation.
(A) and (B) IAA concentration.
(C) and (D) IBA concentration.
(E) and (F) IAA-Ala concentration.
A three-way ANOVA with a Bonferroni post-test was used for statistical analyses. Asterisk in (A) indicates a significant difference in IAA concentration with a Bonferroni post-test (P < 0.05; n = 5 to 6). Each biological replicate consists of at least 30 root segments. Graphs show mean and sd.
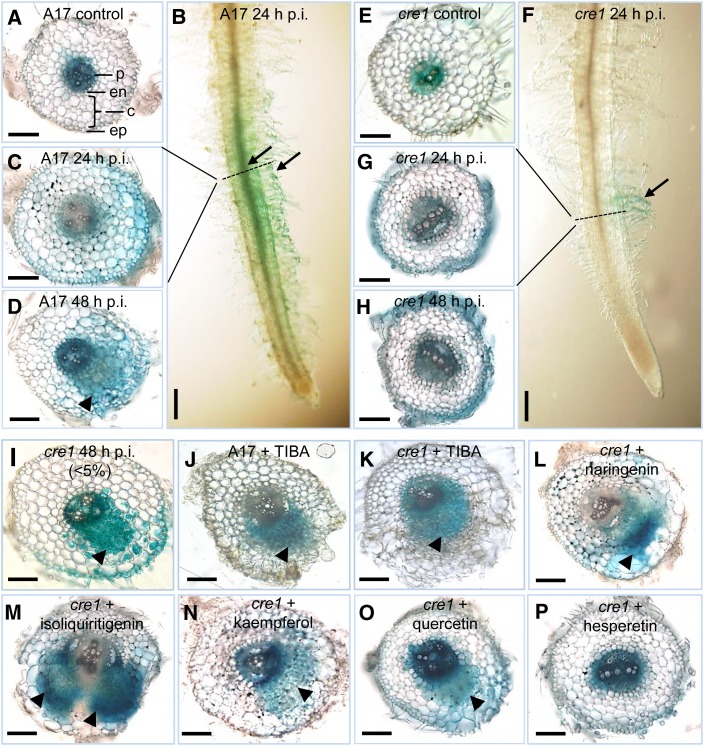
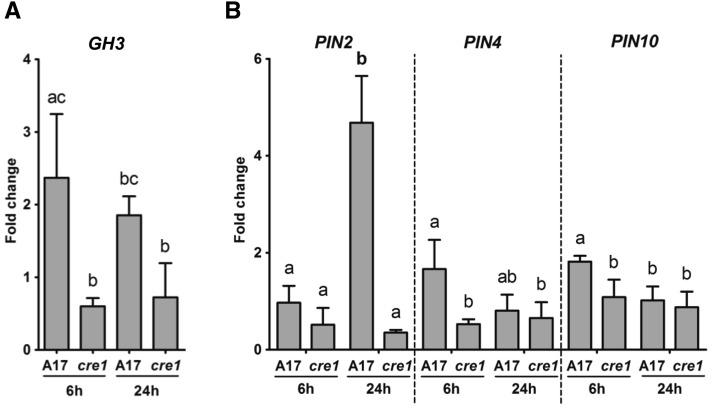
Next, we visualized changes in auxin response in roots transformed with the auxin reporter GH3:GUS (Hagen et al., 1991; Figure 3). In uninfected wild-type roots, GH3:GUS expression was detected in the vascular bundle and pericycle (Figure 3A), similar to the expression in uninfected cre1 roots (Figure 3E). At 24 hpi, an auxin response was activated in the cortex and root hairs of wild-type plants before the initiation of cortical cell divisions (Figures 3B and 3C). The extent of GH3:GUS induction in the cortex varied between roots from partial to complete staining of the cortex. In cre1 mutants, only a weak GH3:GUS response was seen in root hairs and cortical GH3:GUS expression was absent (Figures 3F and 3G). At 48 hpi, dividing cortical and pericycle cells were detected in wild-type roots inoculated with E65, and these dividing cells, as well as cortical and epidermal cells surrounding the nodule primordium, showed a strong auxin response (Figure 3D). In cre1 roots, no activation of auxin response was observed in the cortex, which typically fails to induce cell divisions in response to E65 (Figure 3H). GH3:GUS expression was seen in epidermal cells of cre1 mutants in the absence of cortical cell divisions (Figure 3H). However, in a few cases, the cre1 mutant did form small and delayed nodules, and in these cases, GH3:GUS expression was found in dividing cells of the nodule primordia, as well as in overlying outer cortical and epidermal cells (Figure 3I), similar to the wild type. RT-qPCR showed significantly higher expression of GH3 inoculated in wild-type roots compared with cre1 roots at 6 hpi (Figure 4A).
Figure 3.
Auxin Response (GH3:GUS Expression) Is Localized to Dividing Cells during the Early Stages of Nodule and Pseudonodule Development.
(A) Auxin response in a mock-treated wild-type (A17) root.
(B) Auxin response is localized to the root hairs and underlying cortex directly below the root hairs of an A17 root spot-inoculated with E65 at 24 hpi.
(C) Cross section of (B).
(D) Auxin response in the dividing pericycle, endodermal, and cortical cells during early symbiotic stages in an A17 root inoculated with E65 at 48 hpi.
(E) Auxin response in a mock-treated cre1 mutant root.
(F) Auxin response is absent in the root cortex but present in the root hairs of a cre1 mutant root spot-inoculated with E65 at 24 hpi.
(G) Cross section of (F).
(H) In most of cre1 mutant roots, no cell divisions occur in response to E65 inoculation at 48 hpi.
(I) Dividing cells in cre1 mutants are associated with an enhanced auxin response and are observed in <5% of cre1 mutant roots inoculated with E65.
(J) and (K) Auxin response in the dividing cells of A17 (J) and cre1 (K) roots in response to TIBA treatment.
(L) to (O) Auxin response in a cre1 nodule primordium rescued with naringenin (L), isoliquiritigenin (M), kaempferol (N), and quercetin (O).
(P) No enhanced auxin response or cell divisions were observed in hesperetin-treated roots.
Arrowheads indicate nodule primordia. Arrows indicate auxin response in the root hairs and/or the root cortex. At least 30 individual samples were observed for each treatment. Horizontal and vertical scale bars represent 100 µm and 1 mm, respectively. ep, epidermis; c, cortex; en, endodermis; p, pericycle.
Figure 4.
RT-qPCR Showing Transcript Abundance in Root Segments of the Wild Type (A17) and cre1 Mutants Inoculated for 6 and 24 h with E65 Relative to Mock-Treated Roots.
Expression was normalized to the GAPDh reference gene. The auxin response gene GH3 (A) and the IAA exporter-encoding genes PIN2, PIN4, and PIN10 (B) were analyzed. A two-way ANOVA with a Tukey-Kramer multiple comparison post-test was used for statistical analyses (P < 0.05, n = 3). Different lowercase letters indicate significant differences in transcript abundance within each gene. Each biological replicate consists of at least 50 root segments. Graphs show mean and sd.
To test whether changes in auxin transport and auxin accumulation were accompanied by changes in the expression of auxin transporter-encoding genes, we monitored the relative expression of all 10 known PIN genes and of the five known LAX genes (Schnabel and Frugoli, 2004) using RT-qPCR. A significantly higher PIN4 and PIN10 expression at 6 hpi and PIN2 expression at 24 hpi was found in E65-inoculated wild-type roots compared with cre1 roots (Figure 4B). None of the other PIN genes showed a significantly differential expression between wild-type and cre1 roots in the first 24 h, while some minor changes in expression occurred at later time points (Supplemental Figure 4). None of the LAX genes showed altered expression levels between genotypes or in response to inoculation with E65 (Supplemental Figure 5).
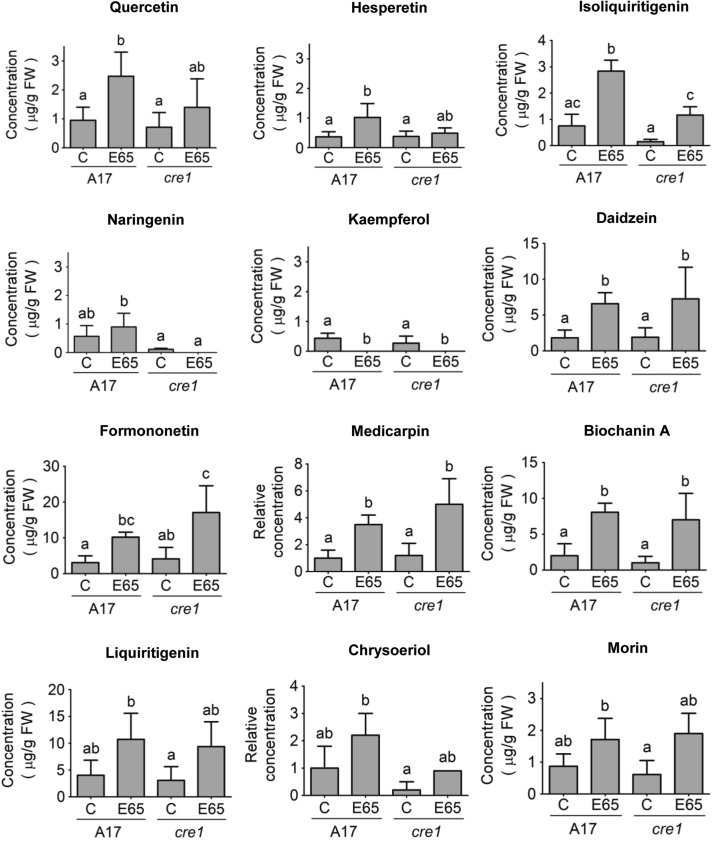
The cre1 Mutant Shows an Altered Flavonoid Profile
Because flavonoids can act as modulators of auxin transport (Peer and Murphy, 2007) and are required for acropetal auxin transport control during nodulation of M. truncatula (Wasson et al., 2006), we studied the flavonoid profile of wild-type and cre1 mutant roots. We quantified the abundance of flavonoids in wild-type and cre1 mutant roots in mock-inoculated and E65-inoculated roots comprising 2 cm of roots from the root tip upwards and including the inoculation site at 24 hpi using liquid chromatography-tandem mass spectrometry. We performed two assays: first, a quantification of free aglycones (Figure 5); and, second, a quantification of the total flavonoid pool following acid hydrolysis of root extracts, which converts flavonoid glycosides into free aglycones (Supplemental Figure 6). Quantification of the total flavonoid pool could reconcile physiological changes mediated by flavonoid glycosides in response to rhizobia infection, which might not be detected with the quantification of free flavonoid aglycones alone. In uninoculated roots, cre1 mutants showed no significant changes in concentrations for any of the measured free aglycones compared with wild-type roots (Figure 5). After inoculation with E65, wild-type, but not cre1, mutant roots showed a significant increase in concentrations of the flavonol quercetin and the flavone hesperetin (Figure 5). Isoliquiritigenin was significantly increased in response to E65 inoculation in both genotypes, but the concentration in inoculated wild-type roots was significantly higher than in cre1 mutant roots. Naringenin concentration was also significantly higher in inoculated wild-type roots in comparison to inoculated cre1 mutant roots. The concentration of kaempferol decreased to levels below the detection limit in both genotypes following inoculation. Inoculation significantly elevated the concentrations of the isoflavonoids daidzein, formononetin, medicarpin, and biochanin A, with similar responses in both genotypes. Concentrations of liquiritigenin, chrysoeriole, and morin were not significantly altered by genotype or inoculation. A summary of differences in flavonoid aglycone abundance between wild-type and cre1 roots in response to E65 inoculation is depicted in Supplemental Figure 7 to correlate the observed defects with the flavonoid metabolic pathway.
Figure 5.
Concentrations of Major Flavonoids in Wild-Type (A17) and cre1 Mutant Root Segments 24 h after Mock or E65 Inoculation.
Flavonoids analyzed include the flavanones (naringenin and hesperetin), flavonols (quercetin, kaempferol, and morin), isoflavonoids (isoliquiritigenin, liquiritigenin, medicarpin, formononetin, daidzein, and biochanin A), and a flavone (chrysoeriole). Relative quantification was performed on chrysoeriol and medicarpin, where commercial standards were not available. A two-way ANOVA with a Tukey-Kramer multiple comparison post-test was used for statistical analyses (P < 0.05, n = 3 to 5). Different lowercase letters indicate statistically significant differences between treatments. A total of 15 root segments were harvested for each biological replicate. Graphs show mean and sd.
Quantification of flavonoid aglycones after acid hydrolysis, i.e., after release from glycosides (Supplemental Figure 6), showed no significant differences between genotypes or depending on E65 inoculation, except for a significant induction of liquiritigenin, formononetin, and medicarpin concentrations in cre1 mutants after inoculation, which was not observed in the wild type. This suggests that the cre1 mutant is compromised in its ability to regulate flavonoid accumulation in response to rhizobia either through cytokinin-mediated changes in biosynthesis or conversion of flavonoids during nodule initiation. A previous study showed that a large number of flavonoid metabolic genes were transcriptionally regulated by cytokinin in M. truncatula roots (Ariel et al., 2012; genes listed in Supplemental Table 2).
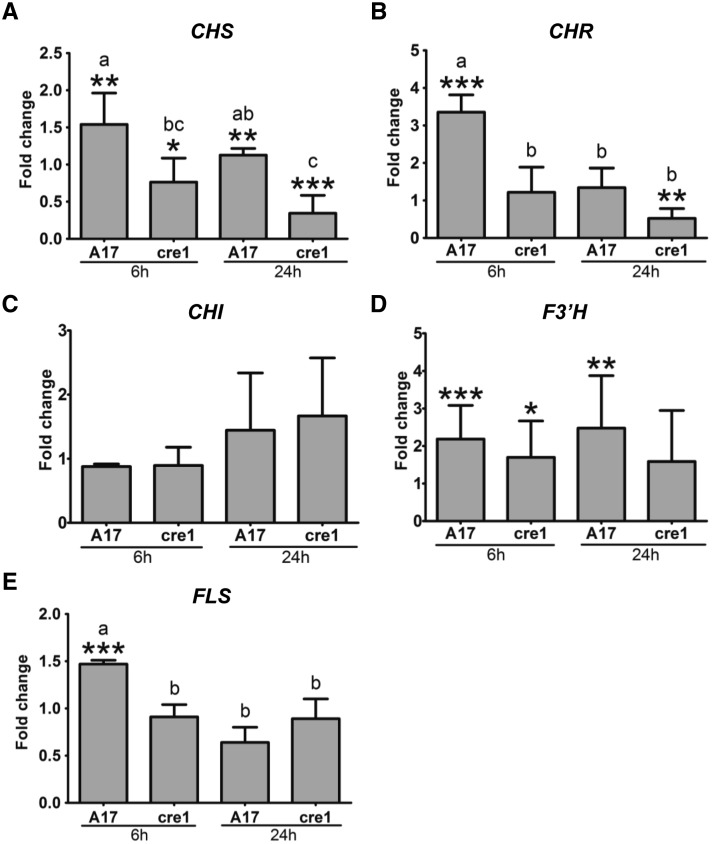
To test whether the observed changes in flavonoid content, in particular those with differential responses in wild-type and cre1 mutants, i.e., naringenin, isoliquiritigenin, quercetin, and hesperetin (highlighted in bold red in Supplemental Figure 7), are accompanied by altered expression of the respective flavonoid synthesis genes, we monitored the expression of CHALCONE SYNTHASE (CHS), CHALCONE ISOMERASE (CHI), CHALCONE REDUCTASE (CHR), FLAVONOID-3′-HYDROXYLASE (F3′H), and FLAVONOL SYNTHASE (FLS) by RT-qPCR. Expression of these flavonoid synthesis genes in response to a short-term cytokinin treatment (10−7 M BAP for 30 min) revealed significant induction of CHR, F3′H, and FLS expression in BAP-treated wild-type but not cre1 mutant roots compared with mock-treated roots (Supplemental Figure 8), indicating that these genes are cytokinin inducible. A comparison between uninoculated roots of the two genotypes revealed significantly higher expression of CHS, CHR, F3′H, and FLS in cre1 mutants roots compared with the wild type at 24 h and of CHR and FLS also at 6 h (Supplemental Figure 9). In response to inoculation with E65, we found a significant induction of CHS, CHR, and FLS in the wild type but not cre1 roots at 6 hpi and of CHS and F3′H also at 24 hpi (Figure 6; shown in bold blue in Supplemental Figure 7). On the contrary, expression of CHS and CHR was significantly reduced by E65 inoculation in the cre1 mutant at 24 hpi for both genes and at 6 hpi for CHS (Figure 6). F3′H was significantly induced in wild-type and cre1 roots after E65 inoculation at 6 h, but this increase was only significant in the wild type at 24 hpi (Figure 6). CHI was neither induced by BAP nor by rhizobia. These gene expression assays support the hypothesis that the observed differential accumulation of isoliquiritigenin (requiring CHS and CHR activities) and quercetin (requiring CHS, F3′H, and FLS activities) in wild-type roots in response to rhizobia is mediated through cytokinin signaling. For naringenin, which only requires CHS activity, there was a discrepancy between induction of CHS by rhizobia in the wild type (but not cre1 mutants), while CHS was not significantly induced by BAP. It is possible that this is due to the different timing of BAP application and rhizobia inoculation or biological variation of the RT-qPCR experiment.
Figure 6.
RT-qPCR Showing Transcript Abundance of Flavonoid-Related Genes in Root Segments.
Transcript abundance of flavonoid-related genes in the wild type (A17) and the cre1 mutant inoculated for 6 and 24 h with E65 relative to mock-treated roots. Expression was normalized to the GAPDh reference gene. CHS (A), CHR (B), CHI (C), F3′H (D), and FLS (E) genes were analyzed. A Student’s t test was used for statistical analyses between roots inoculated with E65 relative to mock-treated roots (fold change) (P < 0.05, n = 3). Asterisks indicate significant differences in induction/repression in roots inoculated with E65 relative to mock-treated roots within individual treatments. A two-way ANOVA with a Tukey-Kramer multiple comparison post-test was used for statistical analyses between genotypes (P < 0.05, n = 3). Different lowercase letters indicate significant differences in induction/repression between A17 and cre1 mutants. Graphs show mean and sd.
Nodulation in cre1 Mutants Can Be Rescued by Auxin Transport Inhibitors
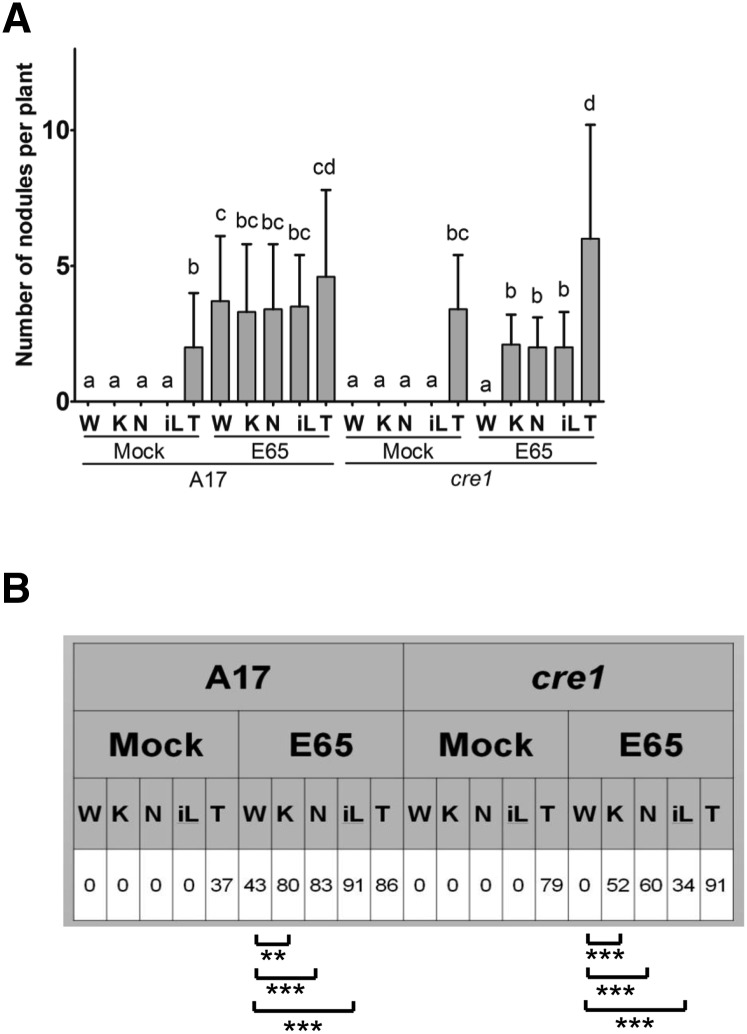
As the cre1 mutant is deficient in the acropetal auxin transport inhibition preceding nodule initiation, we tested whether the failure to initiate nodules in this mutant could be rescued by application of synthetic or natural auxin transport inhibitors, including flavonoids. We used flooding application of the synthetic auxin transport inhibitors 1-N-naphthylphthalamic acid (NPA) and 2,3,5-triiodobenzoic acid (TIBA), which were previously shown to induce the formation of pseudonodules (Rightmyer and Long, 2011). Pseudonodules were indeed observed in the absence of rhizobia, both in wild-type and cre1 mutants (Figure 7A). We observed that NPA caused pleiotropic effects on roots, such as inhibiting root growth or causing root curling, while TIBA did not induce these phenotypes at the concentrations used in our experiments. We therefore only used TIBA for subsequent tests as a synthetic auxin transport inhibitor.
Figure 7.
Complementation of Nodulation in cre1 Mutants Using Flavonoids or the Synthetic Auxin Transport Inhibitor TIBA.
Nodules were quantified 3 weeks after treatment.
(A) Number of nodules forming on wild-type (A17) and cre1 mutant roots, with or without E65 inoculation and various treatments. Note that nodules formed with TIBA in the absence of rhizobia were uninfected pseudonodules. A three-way ANOVA with a Tukey-Kramer multiple comparison post-test was used for statistical analysis (P < 0.05, n = 35). Different lowercase letters indicate statistically significant difference in nodule numbers per plant between treatments. Graph shows mean and sd. W, water; K, 3 µM kaempferol; N, 3 µM naringenin; iL, 3 µM isoliquiritigenin; T, 50 µM TIBA.
(B) Percentage of plants forming nodules in A17 and cre1 mutants. A two-sample t test was used for statistical analyses between treatments (**P < 0.01 and ***P < 0.001; n = 35).
We selected those flavonoid aglycones that were differentially altered by rhizobia in wild-type and cre1 mutants, as well as kaempferol, which responded similarly to rhizobia in both genotypes but is a likely auxin transport inhibitor acting in nodulation, or a precursor thereof (Zhang et al., 2009). The flavonoid concentration used for these rescue experiments (3 μM) was selected as it is in the range of the measured tissue concentrations for free aglycones (Figure 5) and roots were flooded with the flavonoids for 10 s at the start of the experiment, identical to the NPA and TIBA treatments. This treatment of seedlings with the flavonoids kaempferol, naringenin, and isoliquiritigenin at a 3 μM concentration did not lead to pseudonodule formation in either genotype in the absence of rhizobia (Figure 7A). Similarly, root growth was not affected by these flavonoids at the 3 μM concentration (Supplemental Figure 10). We also tested kaempferol application at concentrations up to 100 μM, but this did not cause pseudonodule formation either. However, when wild-type or cre1 mutant roots were inoculated with E65 in the presence of TIBA, kaempferol, naringenin, or isoliquiritigenin, nodules formed on both genotypes with a similar efficiency (Figures 7A and 7B). Addition of quercetin increased percentage of nodulated cre1 roots and increased nodule numbers in cre1 mutants such that there was no significant difference in nodule numbers on E65-inoculated roots between mock-treated wild-type and cre1 mutants treated with quercetin (Supplemental Figures 11A and 11B), suggesting a partial rescue effect of quercetin. In contrast, hesperetin was not able to rescue nodulation of cre1 mutants to a wild-type level (Supplemental Figures 11A and 11B). Overall, naringenin, kaempferol, and isoliquiritigenin were able to rescue the cre1 nodulation phenotype, while quercetin partially rescued nodulation and hesperetin did not rescue nodulation under our conditions.
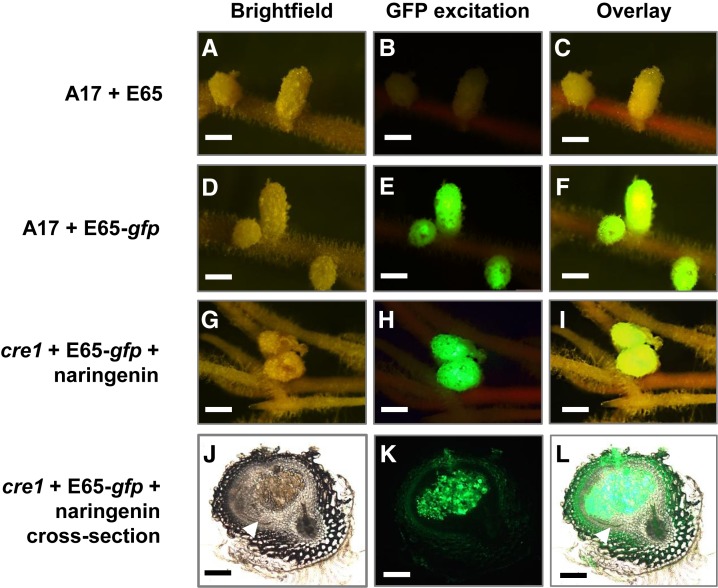
Subsequently, we tested whether application of natural or synthetic auxin transport inhibitors also enabled the induction of GH3:GUS expression in nodule primordia. In the absence of rhizobia, pseudonodule primordia that formed in the presence of TIBA expressed GH3:GUS in dividing root pericycle, endodermal, and cortical cells similarly in the wild-type and the cre1 mutant (Figures 3J and 3K). Likewise, nodules formed in cre1 mutants in the presence of E65 and naringenin (Figure 3L) showed an auxin response in developing nodule primordia. Similar GH3:GUS expression was seen in nodule primordia induced in cre1 roots after addition of either isoliquiritigenin, kaempferol, or quercetin (Figures 3M to 3O) but not after addition of hesperetin (Figure 3P). To test whether nodules formed in cre1 mutants in the presence of auxin transport inhibitors were infected, we used a GFP-labeled strain of S. meliloti transformed with the E65 plasmid. Nodules that formed on cre1 mutant roots supplemented with naringenin looked similar to the wild type and were infected with GFP-labeled rhizobia (Figure 8). Similarly, supplementation with TIBA, isoliquiritigenin, kaempferol, or quercetin also led to the formation of infected nodules in cre1 mutants (Supplemental Figure 12).
Figure 8.
Nodules Restored on cre1 Mutant Roots Treated with Selected Flavonoids Are Infected by Rhizobia.
Nodules were quantified 3 weeks after treatment.
(A) to (C) Wild-type (A17) roots inoculated with E65 without gfp to show background autofluorescence under GFP excitation.
(D) to (F) A17 roots inoculated with a gfp-expressing Sm1021 pE65 strain.
(G) to (I) cre1 mutant roots inoculated with a gfp-expressing Sm1021 pE65 strain and treated with naringenin (3 µM).
(J) to (L) Cross section of a cre1 naringenin-rescued nodule shown in (G) to (I), with gfp-expressing Sm1021 pE65 in infected cells of the nodule.
(A), (D), (G), and (J) Bright-field images,
(B), (E), (H), and (K) Images taken under GFP excitation (maximum excitation 470 nm; 515-nm long-pass filter) of the same nodules as in (A), (D), (G), and (J).
(C), (F), (I), and (L) Overlay of bright-field and fluorescence images from the same row. More than 20 nodule-forming roots were observed under fluorescence for each treatment.
White arrowheads in (J) and (L) indicate the nodule peripheral vasculature. Bars = 1 mm in (A) to (I) and 200 µm in (J) to (L).
To determine whether supplementation with TIBA or flavonoids rescued acropetal auxin transport regulation in cre1 mutants, we measured auxin transport 24 h after the flood treatment. TIBA alone, which is sufficient to induce pseudonodules, inhibited acropetal auxin transport significantly in wild-type and cre1 mutants (Figure 9A). Flood treatment with naringenin alone also significantly inhibited acropetal auxin transport in wild-type and cre1 roots (Figure 9B). Cotreatment with E65 and naringenin, which is sufficient to rescue nodulation in cre1 mutants, significantly reduced acropetal auxin transport in the wild type and cre1 (Figure 9B). A comparable result was found with isoliquiritigenin, kaempferol, and quercetin (Supplemental Figures 13A and 13B), but not with hesperetin (Supplemental Figures 13C and 13D), which also did not rescue nodulation in cre1 mutants.
Figure 9.
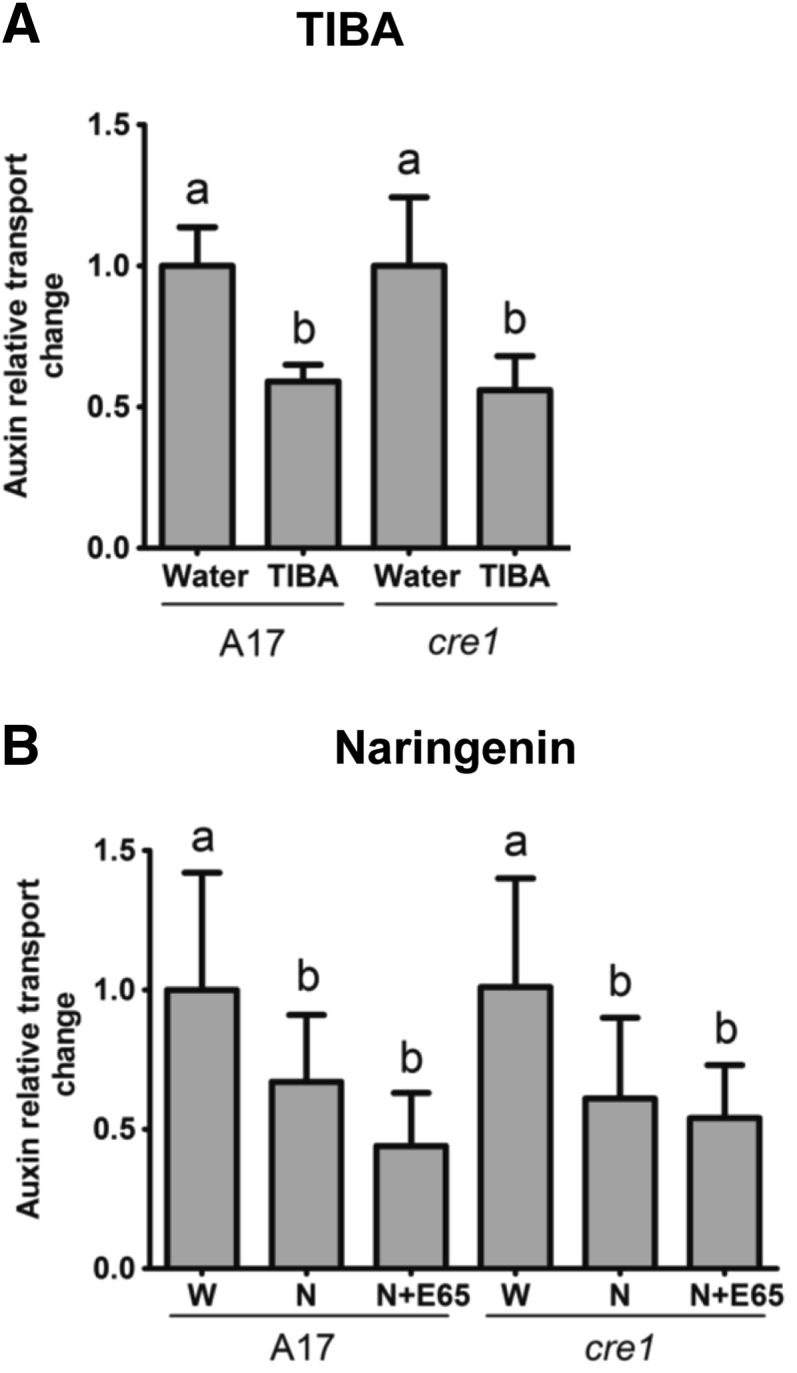
Acropetal Auxin Transport Measurements in Roots of the Wild Type (A17) and cre1 Mutants.
Auxin relative transport rate changes at 24 h after treatment are shown. Seedlings were treated with TIBA (A) or naringenin (B), either in the presence or absence of E65. A two-way ANOVA with a Tukey-Kramer multiple comparison post-test was used for statistical analyses (P < 0.05, n = 15 to 25). Different lowercase letters indicate significant differences in relative auxin transport rates. Graphs show mean and sd. W, water; N, naringenin.
In contrast to acropetal auxin transport, basipetal auxin transport was not inhibited by application of TIBA in wild-type or in cre1 mutants (Supplemental Figure 14A). Flood treatment of roots with a number of flavonoids also failed to alter basipetal auxin transport (Supplemental Figure 14B). Cotreatment of roots with E65 and quercetin reduced basipetal auxin transport in the wild type but not in cre1 mutants (Supplemental Figure 14C). Thus, none of the tested auxin transport inhibitors mimicked the increased basipetal auxin transport that was observed in the wild type after E65 inoculation (Figure 1B), even though they were sufficient to rescue nodulation. This suggests that the acropetal, but not the basipetal, auxin transport changes following S. meliloti infection are crucial for successful nodule initiation.
DISCUSSION
Cytokinin Signaling Is Required for Increased Auxin Accumulation at the Infection Site
Our results suggest that cytokinin signaling through CRE1 is required for the accumulation of auxin in pericycle, endodermal, and cortical cells targeted to divide to form a nodule. Indeed, cre1 mutants did not show increased IAA concentrations following S. meliloti inoculation, and the auxin reporter GH3:GUS was only induced in dividing cells leading to nodule formation in the wild type or in the few cases of successful nodulation in cre1 mutants. Our evidence points to a correlation between IAA concentrations and GH3:GUS expression level. However, we cannot exclude the importance of other auxins that were not detectable or not measured in our assays. While we concentrated on auxin amino acids conjugates, which were previously suggested to be present in M. truncatula (Campanella et al., 2008), a number of other auxin conjugates exist in plants that could contribute to the dynamics of auxin breakdown, storage, and activity (Korasick et al., 2013). In addition, it is likely that the GH3:GUS reporter construct does not reflect the exact localization and activity of all different auxins in the root. Nevertheless, auxin activity analyzed through GH3:GUS expression agrees with the induction of the DR5 promoter in nodule primordia of L. japonicus (Suzaki et al., 2012) and soybean (Turner et al., 2013) and in the cortex of M. truncatula (Breakspear et al., 2014). Interestingly, an auxin response in root hairs at the infection site was visible after 24 hpi in both genotypes. A similar auxin response was identified in M. truncatula recently and was shown to be involved in the infection process (Breakspear et al., 2014). This suggests that the cortical auxin response is associated with nodule primordium formation, depending on CRE1, while the epidermal auxin response associated with infection still occurs in the cre1 mutant. In addition, supply of cre1 mutants with auxin transport inhibitors restored the induction of cortical GH3:GUS expression in nodule primordia and the development of nodules. These results are consistent with the findings that cytokinin application induced expression of GH3:GUS in dividing cortical cells in white clover (Mathesius et al., 2000) and that spontaneous cortical cell divisions in the L. japonicus snf2 mutant are associated with DR5:GFP expression (Suzaki et al., 2012). These data are reminiscent of studies showing that, in Arabidopsis, cytokinin signaling regulates auxin accumulation during lateral root initiation (Laplaze et al., 2007). In Arabidopsis, cytokinin was previously shown to control the auxin pool via alteration of PIN protein expression and localization, for example, during lateral root initiation (Marhavý et al., 2011, 2014; Pernisová et al., 2009; Ruzicka et al., 2009). In our study, we did not find any clear evidence that the decrease in acropetal auxin transport was accompanied by a reduction in PIN or LAX gene expression. Indeed, in response to S. meliloti inoculation in the wild type, we found an increased M. truncatula PIN4 and PIN10 expression, the closest homolog of Arabidopsis PIN1, which encodes a protein that transports auxin acropetally in Arabidopsis (Petrásek and Friml, 2009). PIN4 and PIN10 transcript levels also increased within the first 6 hpi with Nod factors in the wild type but not cre1 mutant roots (Plet et al., 2011). Therefore, it is most likely that PIN expression levels are not a sufficient predictor of actual auxin transport capacity. Instead, additional posttranscriptional regulatory mechanisms likely modulate PIN protein activity. Alternatively, M. truncatula PIN4 and PIN10 may not be the main transporters mediating acropetal auxin transport, but might be involved in lateral auxin transport.
As both synthetic auxin transport inhibitors as well as certain flavonoids inhibit auxin export (Jacobs and Rubery, 1988), it is expected that the accumulation of auxin in nodule primordia is related to a decreased auxin export from these cells. Reduced acropetal auxin transport was measured below the nodule initiation site in M. truncatula in this study, as well as in Plet et al. (2011) and Wasson et al. (2006). Accordingly, in Vicia sativa roots, Nod factor application reduced acropetal auxin transport (Boot et al., 1999). Our evidence thus supports the in silico modeling of auxin maximum generation during nodule initiation, which suggested that auxin accumulation in nodule primordia is most likely explained by a reduced auxin export (Deinum et al., 2012). While BAP application to white clover roots did not lead to reduced GH3:GUS expression below the application site in white clover (Mathesius et al., 2000), our finding that BAP application to roots significantly reduced acropetal auxin transport in M. truncatula supports the hypothesis that the induction of cytokinin signaling during nodulation acts upstream of auxin transport control, in accordance with results from Plet et al. (2011).
We also measured basipetal auxin transport and found increased basipetal auxin transport in response to rhizobia in the wild type at 24 hpi, which was not detected in cre1 mutants. This was accompanied in the wild type with increased PIN2 expression; M. truncatula PIN2 is a homolog of the Arabidopsis PIN2 transporter responsible for basipetal auxin transport from the root tip to the elongation zone (Schnabel and Frugoli, 2004). However, the increased basipetal auxin transport was not mimicked in cre1 mutants rescued with auxin transport inhibitors; therefore, we cannot confirm at this stage whether the increased basipetal auxin transport is necessary for nodule initiation.
Collectively, our results show that the cre1 mutant is defective in the inhibition of acropetal auxin transport in response to rhizobia. As TIBA and specific flavonoids were able to inhibit acropetal auxin transport similarly in inoculated cre1 mutants as in wild-type roots, this suggests that cre1 mutants are defective in the induction of an endogenous auxin transport inhibitor.
Flavonoids Rescue Nodulation in the cre1 Mutant
Our results showed that mRNA levels of CHS, CHR, and FLS were induced significantly by rhizobia in wild-type but not in cre1 mutant roots and that cre1 mutant roots did not show increased free naringenin, quercetin, and hesperetin concentrations in E65-inoculated roots, while the concentration of isoliquiritigenin was increased in cre1 mutants after inoculation with E65, but was significantly lower than in inoculated wild-type roots. CHS activity is required for the synthesis of all flavonoids, while CHR leads to isoliquiritigenin synthesis and FLS leads to kaempferol and quercetin synthesis (Supplemental Figure 7). Supplementation of cre1 mutant roots with rhizobia and naringenin, isoliquiritigenin, or kaempferol rescued nodulation to a wild-type level, while hesperetin did not, and quercetin showed a partial rescue. These results strongly suggest that the transcriptional induction of flavonoids during nodule initiation is a (direct or indirect) target of cytokinin signaling. Similarly, results by van Zeijl et al. (2015) showed that a number of flavonoid synthesis genes are induced by Nod factors after 3 h in wild-type but not in cre1 roots, including a flavonoid hydroxylase, a dihydroflavonol reductase-like protein, a flavonoid glucosyl transferase, five isoflavone methyltransferases, and a malonyl-CoA:isoflavone 7-O-glucoside malonyl transferase, while two copies of flavonol synthase/flavanone-3-hydroxylase were significantly reduced in wild-type but not cre1 roots. In addition, our study showed induction of CHR, F3′H, and FLS expression by the cytokinin BAP, while a previous transcriptomic analysis of M. truncatula root apices response to cytokinins demonstrated that flavonoid-metabolic genes constituted the most significantly enriched functional category (Ariel et al., 2012; Supplemental Table 2). Previous results showing that a local BAP treatment of white clover roots led to cortical cell divisions, CHS induction, and flavonoid accumulation in the dividing cells also support this model (Mathesius et al., 2000). In addition, in Arabidopsis, expression of some flavonoid genes was also found to be under the control of cytokinins (Bhargava et al., 2013). However, it is likely that BAP treatment does not correctly mimic the induction of specific endogenous cytokinins during nodule initiation (van Zeijl et al., 2015), since BAP also induced changes in flavonoid gene expression in the cre1 mutant. This suggests that the BAP-induced changes are mediated by CRE1, as well as other cytokinin receptors.
So far, it is unknown which flavonoid(s) is (are) responsible for auxin transport regulation during nodulation. Silencing of the flavonoid pathway in M. truncatula indirectly suggested that flavonols, most likely kaempferol, are required for auxin transport control because kaempferol addition to CHS-silenced roots, in combination with supplying rhizobia with nod gene inducing flavones, rescued nodulation (Zhang et al., 2009). Our results show that even though nodulation in the cre1 mutant could be rescued by application of kaempferol, the cre1 mutant roots were not deficient in free or total kaempferol concentrations compared with the wild type. However, we detected in cre1 a deficiency in the induction of free quercetin by rhizobia, which was observed in the wild type at 24 hpi with E65. Kaempferol can be converted to quercetin (Supplemental Figure 7); thus, it is possible that experiments with kaempferol addition may lead to an increase of the quercetin pool. However, quercetin supplementation only partially rescued nodule formation in cre1 mutants. Therefore, it is unlikely that flavonol aglycones are sufficient candidates for acropetal auxin transport control during nodulation in M. truncatula. We could also rescue nodulation in cre1 mutants with naringenin and isoliquiritigenin, which accumulated to significantly higher concentrations in inoculated wild-type compared with inoculated cre1 roots. In our assays, naringenin and isoliquiritigenin could rescue auxin transport inhibition in cre1 roots and are therefore also good candidates for auxin transport inhibitors during nodulation. A summary of the characteristics of the different flavonoids tested and their relative abundance in wild-type and cre1 roots in response to S. meliloti inoculation is shown in Supplemental Table 3 and Supplemental Figure 15. From this comparison, the most likely candidates for auxin transport control regulated by the CRE1 cytokinin pathway are naringenin and isoliquiritigenin, or derivatives thereof, formed in the root after supplementation.
Recent studies have suggested that in addition to flavonoid aglycones, flavonoid glycosides might act as auxin transport inhibitors. For example, an Arabidopsis flavonoid 3-O-glycosyltransferase mutant, which overaccumulated kaempferol 3-O-rhamnoside-7-O-rhamnoside, showed reduced polar auxin transport (Yin et al., 2014). Similarly, auxin transport phenotypes in several Arabidopsis flavonoid mutants have been linked to altered accumulation of flavonoid glycosides as they contain undetectable levels of free kaempferol or quercetin aglycones (Buer et al., 2013). In addition, overexpression of the transcription factor WRKY23 in Arabidopsis led to increased levels of quercetin-3-O-rhamnoside, accompanied by a reduced auxin transport in seedling roots (Grunewald et al., 2012). Among the 18 cytokinin-inducible flavonoid-related genes, 14 were putative (iso)flavonoid glycosyl transferases (Ariel et al., 2012; Supplemental Table 2). Thus, future studies could be focused on detailed analysis of flavonoid glycosides during nodulation.
In addition to their activity as endogenous auxin transport inhibitors, flavonoids also play a role as nod gene inducers in rhizobia. As the cre1 mutant was partially defective in the induction of isoliquiritigenin, which can act as a nod gene inducer in S. meliloti (Zuanazzi et al., 1998), this may affect the ability of rhizobia to induce nodules in the cre1 mutant. However, in our study, we used the A2102 strain harboring the pE65 plasmid, which expresses the NodD3 gene from a constitutive promoter (Barnett et al., 2004). Therefore, it is unlikely that a deficiency in nod gene inducing flavonoids was the reason for the lack of nodulation in the cre1 mutant inoculated with this S. meliloti strain.
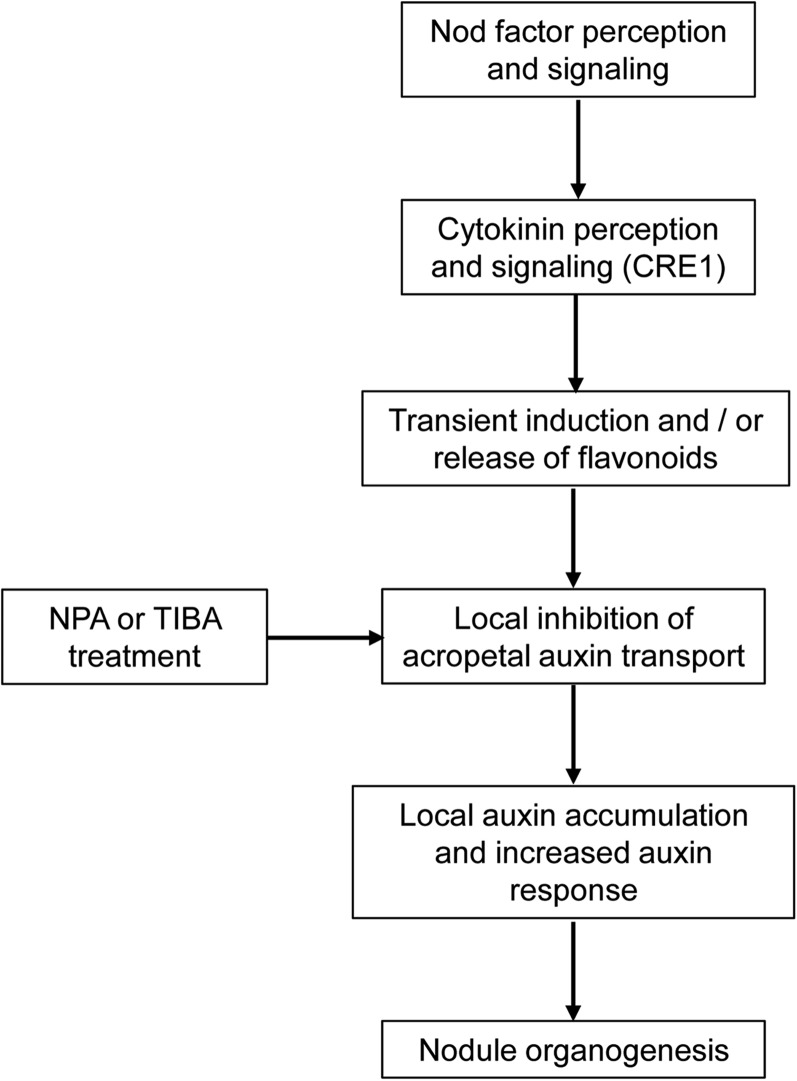
To conclude, our results suggest a model in which cytokinin signaling via the CRE1 receptor leads to a transient induction of flavonoid synthesis or release, which is required for local, acropetal auxin transport inhibition and subsequent auxin accumulation in the initiating nodule (Figure 10). Our current evidence points to naringenin and isoliquiritigenin as the most likely candidates for flavonoids acting as auxin transport inhibitors and IAA as the most likely active auxin induced during early nodule development. Since the cre1 mutant is defective in the induction of a large number of transcripts following Nod factor application (van Zeijl et al., 2015), it will be interesting to determine in the future whether these gene expression changes are also dependent on changes in flavonoid accumulation.
Figure 10.
Proposed Model for the Action of Cytokinin on Auxin Transport and Accumulation during Nodule Initiation in M. truncatula.
Our data suggest a model in which cytokinin signaling mediated by the CRE1 receptor transiently activates or releases certain flavonoids (most likely naringenin and/or isoliquiritigenin) in the root, which then act as auxin export inhibitors that cause auxin (IAA) accumulation and subsequently enhance auxin response in cells that will divide to form a nodule primordium. Both transient application of synthetic auxin transport inhibitors like NPA and TIBA, or flavonoids, induce (pseudo)nodules infected by rhizobia.
METHODS
Plant Material, Bacteria Strains, and Growth and Inoculation Conditions
Wild-type Medicago truncatula cv Jemalong A17 (The South Australian Research and Development Institute) and the cre1-1 mutant (Plet et al., 2011) were used. The Sinorhizobium meliloti strain A2102, a triple nodD mutant for nodD1, nodD2, and nodD3 derived from Sm1021, containing the pE65 plasmid encoding a constitutively expressed copy of nodD3 (Barnett et al., 2004) was used for all inoculations. The E65 strain was maintained on Bergersen’s Modified Medium (BMM) (Rolfe et al., 1980) supplemented with 10 µg/mL tetracycline and 100 µg/mL streptomycin (Sigma Chemicals).
To generate a gfp-labeled E65 strain, the plasmid pTE3, containing a nodD3 expression cassette driven by the tcp promoter, was isolated from S. meliloti A2102 (Barnett et al., 2004). A 957-bp region from the plasmid pHC60 that constitutively expresses gfp (Cheng and Walker, 1998) was excised using BglII (New England Biolabs), ligated into the pTE3 vector containing nodD3 expression cassette using T4 DNA ligase (New England Biolabs), and electroporated into competent S. meliloti 1021. The colonies were cultured in BMM with 10 μg/mL tetracyclin (Sigma-Aldrich) and screened for GFP fluorescence to confirm insertion of the gfp fragment.
Plant transformation of A17 or cre1 mutants with the GH3:GUS construct, described by van Noorden et al. (2007), was performed using Agrobacterium rhizogenes “composite plants” (Boisson-Dernier et al., 2001).
For all plant assays, M. truncatula seeds were scarified with sandpaper, surface-sterilized in 6% (w/v) sodium hypochlorite for 10 min, washed with sterilized milliQ water five times, and then washed with 0.25 mg/mL Augmentin for 6 h. Sterilized seeds were plated onto F medium (Fåhraeus, 1957) containing 0.04% (w/v) Augmentin to prevent bacterial contamination. Seeds were incubated at 4°C in the dark for two nights. Germination of seeds was synchronized by incubating inverted plates at 25°C overnight. Seedlings with radical length of 5 to 10 mm were transferred onto F plates. Plates were semisealed with Parafilm and placed vertically in a container with a black cardboard interspersed between each plate to shield roots from direct light. Plates were incubated at 25°C, with a 16-h-light and 8-h-dark period at 150 µmol m−2 s−1 light intensity.
For plant inoculation, an overnight culture of S. meliloti strain E65 in BMM at 28°C was used. The optical density (OD600) of the culture was adjusted to 0.1 for spot-inoculation and 0.01 for flood-inoculation. Spot-inoculation was performed by placing ∼1 μL of S. meliloti culture or BMM 2 mm above the root tip, corresponding to the nodulation-susceptible zone (Bhuvaneswari et al., 1981). For analyzing GH3:GUS expression, spot-inoculation was performed with a glass capillary pulled into a fine tip over a flame and glued to a hypodermic needle. Flood inoculation was used for complementation assays and involved flooding each root with 500 μL of bacterial culture.
Auxin Transport Measurements
Plants were germinated as described above. Four-day-old seedlings were spot-inoculated with either BMM (mock treatment) or the E65 strain. To investigate the effects of flavonoids, of the synthetic auxin transport inhibitor TIBA , and of BAP on auxin transport, roots were pretreated with 10−7 M BAP or, in the case of cotreatments, flavonoids or TIBA prior to E65 or mock treatment. Plants were incubated vertically in a growth chamber at 25°C, with a 16-h-light and 8-h-dark period at 150 µmol m−2 s−1 light intensity for 24 h.
For auxin transport measurements, a tritium-labeled IAA (3H-IAA) solution (7.5 μL of 1 mCi/mL) (Amersham Biosciences) was diluted in 30 μL ethanol and mixed with 1.5 mL of melted and cooled 4% agarose at pH 4.8 in a Petri dish. For acropetal auxin transport measurements, roots were cut 8 mm from the inoculation spot (or an equivalent spot for flood-treated roots), the top segment discarded, and a small 3H-IAA block (2 mm × 2 mm × 2 mm) placed on the cut end of the bottom segment. A Parafilm strip was placed underneath the root segments to prevent diffusion of 3H-IAA from the growth media into parts of the root. Samples were incubated vertically for 6 h in the dark, and a 4-mm segment just below the inoculation site was excised and placed into scintillation vials containing 2 mL of scintillation fluid (Perkin-Elmer). For basipetal auxin transport measurements, 3H-IAA agar blocks were placed on Parafilm strips on top of the root tip of intact seedlings and incubated vertically for 6 h in the dark. A 2-mm-long segment just above the inoculation site was excised and placed into scintillation vials containing 2 mL of scintillation fluid. We chose a shorter segment than for the acropetal auxin transport assays because cre1 roots grew more slowly and the distance from the root tip to the inoculation site was shorter. A figure depicting the auxin transport assays is shown in Supplemental Figure 2. Vials were shaken overnight at room temperature, and radioactivity was measured in a scintillation counter (Tri-Carb Liquid Scintillation Analyzer B2810TR; Perkin-Elmer) for 1 min each. Between 15 and 20 samples were analyzed for each treatment.
Auxin Quantification by LC-ESI-QTOF Tandem Mass Spectrometry Analysis
Plant roots mock-inoculated or inoculated with rhizobia as described above were collected at 6 and 24 hpi. Root segments of 4 mm, from 2 mm below to 2 mm above the inoculation site, were collected and frozen immediately in liquid nitrogen. A total of 30 to 40 root segments were collected for each treatment for each biological replicate. Samples from six biological replicates for the wild type and five biological replicates for cre1 were analyzed. The frozen tissue samples were mechanically lysed with stainless steel beads in a Qiagen TissueLyser LT with a precooled tube holder. To each tube 20 μL of the internal standard (1 µg/mL of 3-[2H5]indolylacetic acid) followed by 1 mL extraction solvent (20% methanol:79% propanol:1% glacial acetic acid) were added and auxin extraction was performed in a sonicator bath for 15 min at 4°C. Samples were then centrifuged at 16,100g for 15 min. The supernatant was transferred to a fresh tube and dried in a SpeedVac centrifuge. Extraction was repeated and the supernatant combined with the first batch and dried again. Dried samples were resuspended in methanol/water (60:40, v/v) and filtered with a Nanosep MF GHP (hydrophilic polypropylene) 0.45-µm filter (Pall Life Sciences) prior to injection. Tandem mass spectrometry was performed using an Agilent 6530 Accurate Mass LC-MS Q-TOF. Samples were subjected to ESI in the jet stream interface in both ion positive and negative polarities. Based on optimized LC-ESI-QTOF parameters using auxin standards, the auxins IAA, IBA, and IAA-Ala had better sensitivity in the positive mode. The other auxin species were better detected in the negative mode. Optimized conditions in the positive mode were as follows: gas temperature 250°C, drying gas 5 liters min−1, nebulizer 30 psig, sheath gas temperature 350°C, and flow rate of 11 liters min−1, capillary voltage 2500 V, nozzle voltage 500 V and fragmentor voltage 138 V. Conditions in the negative mode were as follow: gas temperature 300°C, drying gas 9 liters min−1, nebulizer 25 psig, sheath gas temperature 350°C, flow rate of 11 liters min−1, capillary voltage 3000 V, nozzle voltage 500 V, and fragmentor voltage 140 V. Samples were injected (7 µL) onto an Agilent Zorbax Eclipse 1.8 µm XDB-C18 2.1 × 50 mm column. Solvent A consisted of 0.1% aqueous formic acid and solvent B consisted of 90% methanol/water with 0.1% formic acid. Free auxins and conjugates were eluted with a linear gradient from 10 to 50% solvent B over 8 min, 50 to 70% solvent B from 8 to 12 min (then held at 70% from 12 to 20 min) at a flow rate of 200 μL min−1. The QTOF was run in targeted MS/MS mode using collision-induced dissociation (N2 collision gas supplied at 18 p.s.i. with a m/z 1.3 isolation window) where the MS extended dynamic range (2 Hz) was m/z 100 to 1000 with an acquisition rate of 3 spectra s−1 and MS/MS at m/z 50 to 1000 at 3 spectra s−1. Data were analyzed using the Agilent Technologies MassHunter software (version B.5.0). Auxin standards and deuterated internal standards were used to determine elution times, collision energies, limits of detection, and limits of quantification in order for subsequent quantification of the endogenous auxins. Auxin standards were obtained from OlChemim (IAA-phenylalanine, IAA-leucine, IAA-valine, IAA-tryptophan, and 4-Cl-IAA), Sigma-Aldrich (IAA-aspartate, IAA-alanine, IAA-isoleucine, IAA, IBA, and phenylacetic acid), and Cambridge Isotope Laboratories (indole-2,4,5,6,7-d5-3-acetic). HPLC-grade methanol was from Acros Organics. Detection limits for the different auxins are shown in Supplemental Table 1.
LC-ESI-QTOF Tandem Mass Spectrometry for Flavonoid Quantification
Plant roots mock-inoculated and inoculated with rhizobia as described above were collected at 24 hpi. Root segments of 2 cm length starting from the root tip (including the inoculation site) were collected and frozen immediately in liquid nitrogen. To determine the total content (free form and glycosides) of flavonoids, 30 root segments were collected for each treatment in each biological replicate and flavonoid concentration from three biological replicates was analyzed. The frozen tissue samples were mechanically lysed with stainless steel beads in a Qiagen TissueLyser LT with a precooled tube holder. Extraction of total flavonoids was based on Farag et al. (2007), with some modifications. As an internal standard, 20 ng luteolin (Indofine Chemicals) was added to each tube and 1 mL 80% methanol/water was used as the extraction solvent. Flavonoids were extracted overnight on a rotating wheel at 4°C in the dark. Tubes were centrifuged at 16,100g for 30 min and the supernatant transferred to a fresh tube and evaporated in a SpeedVac centrifuge. The residue was redissolved in 100 μL of 2 n HCl and heated at 80°C for 90 min to deglycosylate flavonoids. The acid hydrolyzed sample was mixed with 200 μL of ethyl acetate and vortexed, and the ethyl acetate fraction (containing flavonoid aglycones) was transferred to a separate tube before being dried in a SpeedVac centrifuge. Samples were resuspended in 50 μL of 45% methanol, passed through a Nanosep MF GHP (hydrophilic polypropylene) 0.45-µm filter (Pall Life Sciences), and analyzed by targeted LC-ESI-MS/MS.
To determine the concentration of selected flavonoid free aglycones, 15 root segments were collected for each treatment in each biological replicate and flavonoid concentration from five biological replicates was analyzed. Extraction was similar to total flavonoids without the acid hydrolysis and ethyl acetate addition step, i.e., following overnight extraction, samples were dried and resuspended for targeted LC-ESI-MS/MS analysis.
LC-ESI-MS/MS analysis was performed as described above in the negative mode with collision energies optimized for the targeted flavonoids. Samples were injected (7 µL) onto an Ascentis Express 2.7 µm C18 2.1 × 50 mm column (Supelco). Mobile phase A consisted of 0.1% aqueous formic acid and mobile phase B comprised 90% acetonitrile/water with 0.1% formic acid. The applied gradient was as described above and data analysis was performed using the Agilent Technologies MassHunter software. Flavonoid standards were obtained from Sigma-Aldrich (kaempferol, naringenin, quercetin, morin, and hesperetin), Indofine Chemical Company (isoliquiritigenin, liquiritigenin, formononetin, daidzein, and biochanin A), Fluka (apigenin), and ICN Biomedicals (genistein). For identification and relative quantification (peak area ratio of the endogenous analyte/internal standard) of flavonoid compounds where standards were not available to us (medicarpin and chrysoeriole), the mass spectra of flavonoid compounds detected in samples were compared with the mass bank database (Horai et al., 2010) and the literature (Guo et al., 2008; Li et al., 2013).
Complementation of Nodulation by Flood Treatment with Auxin Transport Inhibitors
Seeds were germinated as described above. At 1 week postgermination, seedlings were treated by flooding, as described by Rightmyer and Long (2011). Diluted solutions of NPA (100 µM), TIBA (50 µM), kaempferol (3 µM), quercetin (3 µM), naringenin (3 µM), and isoliquiritigenin (3 µM) were made in sterile 50-mL tubes. Control treatments contained equivalent dilutions of methanol used as a solvent for stock solutions. The concentrations of auxin transport inhibitors and flavonoids were chosen because they were previously demonstrated to induce pseudonodule formation and to complement nodule formation, respectively, in M. truncatula (Zhang et al. 2009; Rightmyer and Long, 2011). Seedlings were flooded for 10 s with 20 to 30 mL of diluted auxin transport inhibitors or flavonoids, and then the solution was decanted. Following flooding, the entire root of individual seedlings was inoculated with BMM (mock-inoculation) or strain E65 (OD600 = 0.01).
Histochemistry and Microscopy
GUS staining was performed as described by van Noorden et al. (2007). Sections were made using a Vibratome 1000 (Vibratome Company) and viewed under bright field using a DMLB microscope (Leica Microsystems), and images were collected with a CCD camera (RT Slider; Diagnostic Instruments).
For complementation of nodulation by flood treatment, whole roots were viewed under a Leica M205 FA stereomicroscope. GFP fluorescence was visualized using an ET Blue LP filter system (maximum excitation at 470 nm with a 515-nm long-pass filter). Nodule hand-sections were viewed under a Leica DM4000-6000 epifluorescence microscope. GFP fluorescence was visualized using a +L5 filter cube (maximum excitation at 480 nm, band-pass filter at 527 ± 30 nm). Photos were taken with a Leica DFC550 high-speed digital camera.
RNA Extraction, cDNA Synthesis, and RT-qPCR
Frozen root samples were ground in liquid nitrogen, and total RNA extraction was performed with an RNeasy Plant Mini Kit (Qiagen) or a Spectrum Plant Total RNA kit (Sigma-Aldrich). RNA quality and quantity were analyzed with a NanoDrop ND-1000 (Labtech International). First-strand cDNA synthesis was performed using a SuperScript first-strand synthesis system for RT-PCR kit (Invitrogen). RNA quantity from each sample in each biological replicate was standardized prior to first-strand cDNA synthesis. Primers were designed using Clone Manager for Windows version 9.0 (GE Healthcare Life Sciences) or Primer 3. Gene primers are listed in Supplemental Table 4. Standard criteria for RT-qPCR primer design were used, based on Udvardi et al. (2008). Sample mix for qRT-PCR was prepared in 384-well plates using standard reaction mixture from Power SYBR Green (Applied Biosystems) and analyzed with an Applied Biosystems-7900HT. Raw data were analyzed in Excel using a relative quantification method based on Pfaffl (2001), with glyceraldehyde-3-phosphate dehydrogenase (GAPDh) or RNA binding protein (RBP1) as reference genes, as indicated in the figures. Correspondence between various Medicago resources (MtGI-TIGR, 16K+ array IDs and the Medicago genome v4.0) was established using the Legoo “nickname” tool (http://www.legoo.org/).
Statistical Analysis
Statistical analyses were performed with Genstat 15th Edition (VSN International) and Instat version 3.06 (Graphpad Software).
Accession Numbers
All IMGAG (International Medicago Genome Annotation Group; http://jcvi.org/medicago/) accession numbers are listed in Supplemental Table 4.
Supplemental Data
Supplemental Figure 1. Nodulation efficiency on wild-type and cre1 mutant roots spot-inoculated with Sinorhizobium meliloti strains Rm1021 and E65.
Supplemental Figure 2. Cartoon showing auxin transport measurements performed in this study.
Supplemental Figure 3. Combined auxin concentrations (sum of IAA, IBA, and IAA-Ala) in wild-type (A17) and cre1 mutant roots mock-treated or inoculated with E65 rhizobia.
Supplemental Figure 4. Quantitative RT-PCR of Mt-PIN genes.
Supplemental Figure 5. Quantitative RT-PCR of Mt-LAX genes.
Supplemental Figure 6. Concentrations of total flavonoid aglycones following acid hydrolysis of flavonoid glycosides in wild-type and cre1 roots.
Supplemental Figure 7. Schematic overview of the flavonoid biosynthesis pathway in Medicago truncatula.
Supplemental Figure 8. Quantitative RT-PCR showing relative transcript abundance of flavonoid-related genes in roots treated with cytokinin (benzylaminopurine, BAP).
Supplemental Figure 9. Quantitative RT-PCR showing transcript abundance of flavonoid-related genes in root segments.
Supplemental Figure 10. Root growth on Medicago truncatula wild-type plants with or without flavonoids and E65.
Supplemental Figure 11. Nodulation on wild-type and cre1 mutant roots treated with or without quercetin or hesperetin, in the presence of E65.
Supplemental Figure 12. Nodules formed on cre1 mutant roots with addition of auxin transport inhibitors are infected by rhizobia.
Supplemental Figure 13. Acropetal auxin transport in roots treated with or without flavonoids, in the presence of E65.
Supplemental Figure 14. Basipetal auxin transport in wild-type and cre1 mutant roots in response to auxin transport inhibitors at 24 hpi.
Supplemental Figure 15. Relative flavonoid abundance (free aglycones) in wild-type and cre1 mutant roots in control or E65-inoculated roots.
Supplemental Table 1. Quality parameters for auxin detection using LC-MS/MS in our study.
Supplemental Table 2. Induction of flavonoid-related genes by the cytokinin BAP, extracted and modified from the publication by Ariel et al. (2012).
Supplemental Table 3. Summary table showing the roles of different flavonoid aglycones identified in this study.
Supplemental Table 4. Primer sequences of genes used in this study wild-type and their gene IDs (v4.0).
Supplementary Material
Acknowledgments
We thank Melanie Barnett and Sharon Long (Stanford University, Stanford, CA) for the S. meliloti E65 strain; Anton Wasson (CSIRO Canberra Australia) and Giel van Noorden (Australian National University) for valuable discussion; Britta Winterberg (Australian National University) for primers (PIN2/4 and LAX2/4); and the anonymous reviewers for constructive comments. Research in the U.M. laboratory was supported by a Future Fellowship from the Australian Research Council to U.M. (FT100100669) and by Saclay Plant Science “Labex” in the F.F. laboratory.
AUTHOR CONTRIBUTIONS
J.L.P.N., F.F., and U.M. designed the research. J.L.P.N., S.H., C.L., and U.M. performed research. T.T.T. and C.H.H. contributed new analytic tools for mass spectrometry. J.L.P.N. and S.H. analyzed data. J.L.P.N., S.H., F.F., and U.M. wrote the article.
Glossary
- IAA
indole-3-acetic acid
- hpi
hours postinoculation
- BAP
benzyl amino purine
- IBA
indole-3-butyric acid
- NPA
1-N-naphthylphthalamic acid
- TIBA
2,3,5-triiodobenzoic acid
- BMM
Bergersen’s Modified Medium
References
- Allen E.K., Allen O.N., Newman A.S. (1953). Pseudonodulation of leguminous plants induced by 2-bromo-3,5-dichlorobenzoic acid. Am. J. Bot. 40: 429–435. [Google Scholar]
- Ariel F., et al. (2012). Two direct targets of cytokinin signaling regulate symbiotic nodulation in Medicago truncatula. Plant Cell 24: 3838–3852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailly A., Sovero V., Vincenzetti V., Santelia D., Bartnik D., Koenig B.W., Mancuso S., Martinoia E., Geisler M. (2008). Modulation of P-glycoproteins by auxin transport inhibitors is mediated by interaction with immunophilins. J. Biol. Chem. 283: 21817–21826. [DOI] [PubMed] [Google Scholar]
- Barnett M.J., Toman C.J., Fisher R.F., Long S.R. (2004). A dual-genome Symbiosis Chip for coordinate study of signal exchange and development in a prokaryote-host interaction. Proc. Natl. Acad. Sci. USA 101: 16636–16641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benková E., Michniewicz M., Sauer M., Teichmann T., Seifertová D., Jürgens G., Friml J. (2003). Local, efflux-dependent auxin gradients as a common module for plant organ formation. Cell 115: 591–602. [DOI] [PubMed] [Google Scholar]
- Bhargava A., Clabaugh I., To J.P., Maxwell B.B., Chiang Y.H., Schaller G.E., Loraine A., Kieber J.J. (2013). Identification of cytokinin-responsive genes using microarray meta-analysis and RNA-Seq in Arabidopsis. Plant Physiol. 162: 272–294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhuvaneswari T.V., Bhagwat A.A., Bauer W.D. (1981). Transient susceptibility of root cells in four common legumes to nodulation by rhizobia. Plant Physiol. 68: 1144–1149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boisson-Dernier A., Chabaud M., Garcia F., Bécard G., Rosenberg C., Barker D.G. (2001). Agrobacterium rhizogenes-transformed roots of Medicago truncatula for the study of nitrogen-fixing and endomycorrhizal symbiotic associations. Mol. Plant Microbe Interact. 14: 695–700. [DOI] [PubMed] [Google Scholar]
- Boot K.J.M., van Brussel A.A.N., Tak T., Spaink H.P., Kijne J.W. (1999). Lipochitin oligosaccharides from Rhizobium leguminosarum bv. viciae reduce auxin transport capacity in Vicia sativa subsp nigra roots. Mol. Plant Microbe Interact. 12: 839–844. [Google Scholar]
- Breakspear A., Liu C., Roy S., Stacey N., Rogers C., Trick M., Morieri G., Mysore K.S., Wen J., Oldroyd G.E.D., Downie J.A., Murray J.D. (2014). The root hair “infectome” of Medicago truncatula uncovers changes in cell cycle genes and reveals a requirement for Auxin signaling in rhizobial infection. Plant Cell 26: 4680–4701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown D.E., Rashotte A.M., Murphy A.S., Normanly J., Tague B.W., Peer W.A., Taiz L., Muday G.K. (2001). Flavonoids act as negative regulators of auxin transport in vivo in arabidopsis. Plant Physiol. 126: 524–535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buer C.S., Kordbacheh F., Truong T.T., Hocart C.H., Djordjevic M.A. (2013). Alteration of flavonoid accumulation patterns in transparent testa mutants disturbs auxin transport, gravity responses, and imparts long-term effects on root and shoot architecture. Planta 238: 171–189. [DOI] [PubMed] [Google Scholar]
- Buer C.S., Muday G.K. (2004). The transparent testa4 mutation prevents flavonoid synthesis and alters auxin transport and the response of Arabidopsis roots to gravity and light. Plant Cell 16: 1191–1205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campanella J., Smith S., Leibu D., Wexler S., Ludwig-Müller L. (2008). The auxin conjugate hydrolase family of Medicago truncatula and their expression during the interaction with two symbionts. J. Plant Growth Regul. 27: 26–38. [Google Scholar]
- Cheng H.P., Walker G.C. (1998). Succinoglycan is required for initiation and elongation of infection threads during nodulation of alfalfa by Rhizobium meliloti. J. Bacteriol. 180: 5183–5191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper J.B., Long S.R. (1994). Morphogenetic rescue of Rhizobium meliloti nodulation mutants by trans-zeatin secretion. Plant Cell 6: 215–225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crespi M., Frugier F. (2008). De novo organ formation from differentiated cells: root nodule organogenesis. Sci. Signal. 1: re11. [DOI] [PubMed] [Google Scholar]
- Deinum E.E., Geurts R., Bisseling T., Mulder B.M. (2012). Modeling a cortical auxin aximum for nodulation: different signaures of potential strategies. Front. Plant Sci. 3: 1–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Pietro A., et al. (2002). Modulation by flavonoids of cell multidrug resistance mediated by P-glycoprotein and related ABC transporters. Cell. Mol. Life Sci. 59: 307–322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fåhraeus G. (1957). The infection of clover root hairs by nodule bacteria studied by a simple glass slide technique. J. Gen. Microbiol. 16: 374–381. [DOI] [PubMed] [Google Scholar]
- Fang Y., Hirsch A.M. (1998). Studying early nodulin gene ENOD40 expression and induction by nodulation factor and cytokinin in transgenic alfalfa. Plant Physiol. 116: 53–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farag M.A., Huhman D.V., Lei Z., Sumner L.W. (2007). Metabolic profiling and systematic identification of flavonoids and isoflavonoids in roots and cell suspension cultures of Medicago truncatula using HPLC-UV-ESI-MS and GC-MS. Phytochemistry 68: 342–354. [DOI] [PubMed] [Google Scholar]
- Frugier F., Kosuta S., Murray J.D., Crespi M., Szczyglowski K. (2008). Cytokinin: secret agent of symbiosis. Trends Plant Sci. 13: 115–120. [DOI] [PubMed] [Google Scholar]
- Geldner N., Friml J., Stierhof Y.D., Jürgens G., Palme K. (2001). Auxin transport inhibitors block PIN1 cycling and vesicle trafficking. Nature 413: 425–428. [DOI] [PubMed] [Google Scholar]
- Gonzalez-Rizzo S., Crespi M., Frugier F. (2006). The Medicago truncatula CRE1 cytokinin receptor regulates lateral root development and early symbiotic interaction with Sinorhizobium meliloti. Plant Cell 18: 2680–2693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grunewald W., et al. (2012). Transcription factor WRKY23 assists auxin distribution patterns during Arabidopsis root development through local control on flavonol biosynthesis. Proc. Natl. Acad. Sci. USA 109: 1554–1559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo J., Liu D., Nikolic D., Zhu D., Pezzuto J.M., van Breemen R.B. (2008). In vitro metabolism of isoliquiritigenin by human liver microsomes. Drug Metab. Dispos. 36: 461–468. [DOI] [PubMed] [Google Scholar]
- Hagen G., Martin G., Li Y., Guilfoyle T.J. (1991). Auxin-induced expression of the soybean GH3 promoter in transgenic tobacco plants. Plant Mol. Biol. 17: 567–579. [DOI] [PubMed] [Google Scholar]
- Heckmann A.B., Sandal N., Bek A.S., Madsen L.H., Jurkiewicz A., Nielsen M.W., Tirichine L., Stougaard J. (2011). Cytokinin induction of root nodule primordia in Lotus japonicus is regulated by a mechanism operating in the root cortex. Mol. Plant Microbe Interact. 24: 1385–1395. [DOI] [PubMed] [Google Scholar]
- Held M., Hou H., Miri M., Huynh C., Ross L., Hossain M.S., Sato S., Tabata S., Perry J., Wang T.L., Szczyglowski K. (2014). Lotus japonicus cytokinin receptors work partially redundantly to mediate nodule formation. Plant Cell 26: 678–694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrbach V., Remblière C., Gough C., Bensmihen S. (2014). Lateral root formation and patterning in Medicago truncatula. J. Plant Physiol. 171: 301–310. [DOI] [PubMed] [Google Scholar]
- Hirsch A.M. (1992). Developmental biology of legume nodulation. New Phytol. 122: 211–237. [DOI] [PubMed] [Google Scholar]
- Hirsch A.M., Bhuvaneswari T.V., Torrey J.G., Bisseling T. (1989). Early nodulin genes are induced in alfalfa root outgrowths elicited by auxin transport inhibitors. Proc. Natl. Acad. Sci. USA 86: 1244–1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirsch A.M., Fang Y., Asad S., Kapulnik Y. (1997). The role of phytohormones in plant-microbe symbioses. Plant Soil 194: 171–184. [Google Scholar]
- Hirsch A.M., LaRue T. (1997). Is the legume nodule a modified root, stem or an organ sui generis? Crit. Rev. Plant Sci. 16: 361–392. [Google Scholar]
- Horai H., et al. (2010). MassBank: a public repository for sharing mass spectral data for life sciences. J. Mass Spectrom. 45: 703–714. [DOI] [PubMed] [Google Scholar]
- Jacobs M., Rubery P.H. (1988). Naturally occurring auxin transport regulators. Science 241: 346–349. [DOI] [PubMed] [Google Scholar]
- Korasick D.A., Enders T.A., Strader L.C. (2013). Auxin biosynthesis and storage forms. J. Exp. Bot. 64: 2541–2555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kowalczyk M., Sandberg G. (2001). Quantitative analysis of indole-3-acetic acid metabolites in Arabidopsis. Plant Physiol. 127: 1845–1853. [PMC free article] [PubMed] [Google Scholar]
- Laffont C., Blanchet S., Lapierre C., Brocard L., Ratet P., Crespi M., Mathesius U., Frugier F. (2010). The compact root architecture1 gene regulates lignification, flavonoid production, and polar auxin transport in Medicago truncatula. Plant Physiol. 153: 1597–1607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laplaze L., et al. (2007). Cytokinins act directly on lateral root founder cells to inhibit root initiation. Plant Cell 19: 3889–3900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li T., Yan Z., Zhou C., Sun J., Jiang C., Yang X. (2013). Simultaneous quantification of paeoniflorin, nobiletin, tangeretin, liquiritigenin, isoliquiritigenin, liquiritin and formononetin from Si-Ni-San extract in rat plasma and tissues by liquid chromatography-tandem mass spectrometry. Biomed. Chromatogr. 27: 1041–1053. [DOI] [PubMed] [Google Scholar]
- Marhavý P., Bielach A., Abas L., Abuzeineh A., Duclercq J., Tanaka H., Pařezová M., Petrášek J., Friml J., Kleine-Vehn J., Benková E. (2011). Cytokinin modulates endocytic trafficking of PIN1 auxin efflux carrier to control plant organogenesis. Dev. Cell 21: 796–804. [DOI] [PubMed] [Google Scholar]
- Marhavý P., Duclercq J., Weller B., Feraru E., Bielach A., Offringa R., Friml J., Schwechheimer C., Murphy A., Benková E. (2014). Cytokinin controls polarity of PIN1-dependent auxin transport during lateral root organogenesis. Curr. Biol. 24: 1031–1037. [DOI] [PubMed] [Google Scholar]
- Mathesius U. (2008). Auxin: at the root of nodule development? Funct. Plant Biol. 35: 651–668. [DOI] [PubMed] [Google Scholar]
- Mathesius U., Charon C., Rolfe B.G., Kondorosi A., Crespi M. (2000). Temporal and spatial order of events during the induction of cortical cell divisions in white clover by Rhizobium leguminosarum bv. trifolii inoculation or localized cytokinin addition. Mol. Plant Microbe Interact. 13: 617–628. [DOI] [PubMed] [Google Scholar]
- Mathesius U., Schlaman H.R.M., Spaink H.P., Of Sautter C., Rolfe B.G., Djordjevic M.A. (1998). Auxin transport inhibition precedes root nodule formation in white clover roots and is regulated by flavonoids and derivatives of chitin oligosaccharides. Plant J. 14: 23–34. [DOI] [PubMed] [Google Scholar]
- Matsuda F., Miyazawa H., Wakasa K., Miyagawa H. (2005). Quantification of indole-3-acetic acid and amino acid conjugates in rice by liquid chromatography-electrospray ionization-tandem mass spectrometry. Biosci. Biotechnol. Biochem. 69: 778–783. [DOI] [PubMed] [Google Scholar]
- Michniewicz, M., Brewer, P., and Friml, J. (2007). Polar auxin transport and asymmetric auxin distribution. The Arabidopsis Book 5: e0108, doi/10.1199/tab.0108. [DOI] [PMC free article] [PubMed]
- Mortier V., Wasson A., Jaworek P., De Keyser A., Decroos M., Holsters M., Tarkowski P., Mathesius U., Goormachtig S. (2014). Role of LONELY GUY genes in indeterminate nodulation on Medicago truncatula. New Phytol. 202: 582–593. [DOI] [PubMed] [Google Scholar]
- Murphy A.S., Hoogner K.R., Peer W.A., Taiz L. (2002). Identification, purification, and molecular cloning of N-1-naphthylphthalmic acid-binding plasma membrane-associated aminopeptidases from Arabidopsis. Plant Physiol. 128: 935–950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray J.D., Karas B.J., Sato S., Tabata S., Amyot L., Szczyglowski K. (2007). A cytokinin perception mutant colonized by Rhizobium in the absence of nodule organogenesis. Science 315: 101–104. [DOI] [PubMed] [Google Scholar]
- Noh B., Murphy A.S., Spalding E.P. (2001). Multidrug resistance-like genes of Arabidopsis required for auxin transport and auxin-mediated development. Plant Cell 13: 2441–2454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oldroyd G.E.D., Murray J.D., Poole P.S., Downie J.A. (2011). The rules of engagement in the legume-rhizobial symbiosis. Annu. Rev. Genet. 45: 119–144. [DOI] [PubMed] [Google Scholar]
- Peer W.A., Bandyopadhyay A., Blakeslee J.J., Makam S.N., Chen R.J., Masson P.H., Murphy A.S. (2004). Variation in expression and protein localization of the PIN family of auxin efflux facilitator proteins in flavonoid mutants with altered auxin transport in Arabidopsis thaliana. Plant Cell 16: 1898–1911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peer W.A., Murphy A.S. (2007). Flavonoids and auxin transport: modulators or regulators? Trends Plant Sci. 12: 556–563. [DOI] [PubMed] [Google Scholar]
- Pernisová M., Klíma P., Horák J., Válková M., Malbeck J., Souček P., Reichman P., Hoyerová K., Dubová J., Friml J., Zazímalová E., Hejátko J. (2009). Cytokinins modulate auxin-induced organogenesis in plants via regulation of the auxin efflux. Proc. Natl. Acad. Sci. USA 106: 3609–3614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrásek J., Friml J. (2009). Auxin transport routes in plant development. Development 136: 2675–2688. [DOI] [PubMed] [Google Scholar]
- Pfaffl M.W. (2001). A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 29: e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plet J., Wasson A., Ariel F., Le Signor C., Baker D., Mathesius U., Crespi M., Frugier F. (2011). MtCRE1-dependent cytokinin signaling integrates bacterial and plant cues to coordinate symbiotic nodule organogenesis in Medicago truncatula. Plant J. 65: 622–633. [DOI] [PubMed] [Google Scholar]
- Rashotte A.M., Brady S.R., Reed R.C., Ante S.J., Muday G.K. (2000). Basipetal auxin transport is required for gravitropism in roots of Arabidopsis. Plant Physiol. 122: 481–490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rightmyer A.P., Long S.R. (2011). Pseudonodule formation by wild-type and symbiotic mutant Medicago truncatula in response to auxin transport inhibitors. Mol. Plant Microbe Interact. 24: 1372–1384. [DOI] [PubMed] [Google Scholar]
- Rodriguez-Barrueco C., Bermudez de Castro F. (1973). Cytokinin-induced pseudonodules on Alnus glutinosa. Physiol. Plant. 29: 277–280. [Google Scholar]
- Rolfe B.G., Gresshoff P.M., Shine J. (1980). Rapid screening for symbiotic mutants of Rhizobium and white clover. Plant Sci. Lett. 19: 277–284. [Google Scholar]
- Ruzicka K., Simásková M., Duclercq J., Petrásek J., Zazímalová E., Simon S., Friml J., Van Montagu M.C.E., Benková E. (2009). Cytokinin regulates root meristem activity via modulation of the polar auxin transport. Proc. Natl. Acad. Sci. USA 106: 4284–4289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santelia D., Henrichs S., Vincenzetti V., Sauer M., Bigler L., Klein M., Bailly A., Lee Y., Friml J., Geisler M., Martinoia E. (2008). Flavonoids redirect PIN-mediated polar auxin fluxes during root gravitropic responses. J. Biol. Chem. 283: 31218–31226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnabel E.L., Frugoli J. (2004). The PIN and LAX families of auxin transport genes in Medicago truncatula. Mol. Genet. Genomics 272: 420–432. [DOI] [PubMed] [Google Scholar]
- Schneider E.A., Kazakoff C.W., Wightman F. (1985). Gas chromatography-mass spectrometry evidence for several endogenous auxins in pea seedling organs. Planta 165: 232–241. [DOI] [PubMed] [Google Scholar]
- Suzaki T., Yano K., Ito M., Umehara Y., Suganuma N., Kawaguchi M. (2012). Positive and negative regulation of cortical cell division during root nodule development in Lotus japonicus is accompanied by auxin response. Development 139: 3997–4006. [DOI] [PubMed] [Google Scholar]
- Tirichine L., Sandal N., Madsen L.H., Radutoiu S., Albrektsen A.S., Sato S., Asamizu E., Tabata S., Stougaard J. (2007). A gain-of-function mutation in a cytokinin receptor triggers spontaneous root nodule organogenesis. Science 315: 104–107. [DOI] [PubMed] [Google Scholar]
- Truchet G., Roche P., Lerouge P., Vasse J., Camut S., De Billy F., Promé J.C., Dénarié J. (1991). Sulphated lipo-oligosaccharide signals of Rhizobium meliloti elicit root nodule organogenesis in alfalfa. Nature 351: 670–673. [Google Scholar]
- Turner M., Nizampatnam N.R., Baron M., Coppin S., Damodaran S., Adhikari S., Arunachalam S.P., Yu O., Subramanian S. (2013). Ectopic expression of miR160 results in auxin hypersensitivity, cytokinin hyposensitivity, and inhibition of symbiotic nodule development in soybean. Plant Physiol. 162: 2042–2055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Udvardi M.K., Czechowski T., Scheible W.-R. (2008). Eleven golden rules of quantitative RT-PCR. Plant Cell 20: 1736–1737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Noorden G.E., Kerim T., Goffard N., Wiblin R., Pellerone F.I., Rolfe B.G., Mathesius U. (2007). Overlap of proteome changes in Medicago truncatula in response to auxin and Sinorhizobium meliloti. Plant Physiol. 144: 1115–1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Spronsen P.C., Grønlund M., Pacios Bras C., Spaink H.P., Kijne J.W. (2001). Cell biological changes of outer cortical root cells in early determinate nodulation. Mol. Plant Microbe Interact. 14: 839–847. [DOI] [PubMed] [Google Scholar]
- van Zeijl A., Op den Camp R.H.M., Deinum E.E., Charnikhova T., Franssen H., Op den Camp H.J.M., Bouwmeester H., Kohlen W., Bisseling T., Geurts R. (2015). Rhizobium lipo-chitooligosaccharide signaling triggers accumulation of cytokinins in Medicago truncatula roots. Mol. Plant http://dx.doi.org/10.1016/j.molp.2015.03.010. [DOI] [PubMed] [Google Scholar]
- Wasson A.P., Pellerone F.I., Mathesius U. (2006). Silencing the flavonoid pathway in Medicago truncatula inhibits root nodule formation and prevents auxin transport regulation by rhizobia. Plant Cell 18: 1617–1629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao T.T., Schilderink S., Moling S., Deinum E.E., Kondorosi E., Franssen H., Kulikova O., Niebel A., Bisseling T. (2014). Fate map of Medicago truncatula root nodules. Development 141: 3517–3528. [DOI] [PubMed] [Google Scholar]
- Yin R., Han K., Heller W., Albert A., Dobrev P.I., Zažímalová E., Schäffner A.R. (2014). Kaempferol 3-O-rhamnoside-7-O-rhamnoside is an endogenous flavonol inhibitor of polar auxin transport in Arabidopsis shoots. New Phytol. 201: 466–475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J., Subramanian S., Stacey G., Yu O. (2009). Flavones and flavonols play distinct critical roles during nodulation of Medicago truncatula by Sinorhizobium meliloti. Plant J. 57: 171–183. [DOI] [PubMed] [Google Scholar]
- Zuanazzi J.A.S., Clergeot P.H., Quirion J.C., Husson H.P., Kondorosi A., Ratet P. (1998). Production of Sinorhizobium meliloti nod gene activator and repressor flavonoids from Medicago sativa roots. Mol. Plant Microbe Interact. 11: 784–794. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.