The phenotypes of Arabidopsis plants with altered aldehyde contents and guaiacyl-to-syringyl ratios provide insights into the effects of lignin composition on plant growth and cell wall degradability.
Abstract
Modifying lignin composition and structure is a key strategy to increase plant cell wall digestibility for biofuel production. Disruption of the genes encoding both cinnamyl alcohol dehydrogenases (CADs), including CADC and CADD, in Arabidopsis thaliana results in the atypical incorporation of hydroxycinnamaldehydes into lignin. Another strategy to change lignin composition is downregulation or overexpression of ferulate 5-hydroxylase (F5H), which results in lignins enriched in guaiacyl or syringyl units, respectively. Here, we combined these approaches to generate plants enriched in coniferaldehyde-derived lignin units or lignins derived primarily from sinapaldehyde. The cadc cadd and ferulic acid hydroxylase1 (fah1) cadc cadd plants are similar in growth to wild-type plants even though their lignin compositions are drastically altered. In contrast, disruption of CAD in the F5H-overexpressing background results in dwarfism. The dwarfed phenotype observed in these plants does not appear to be related to collapsed xylem, a hallmark of many other lignin-deficient dwarf mutants. cadc cadd, fah1 cadc cadd, and cadd F5H-overexpressing plants have increased enzyme-catalyzed cell wall digestibility. Given that these CAD-deficient plants have similar total lignin contents and only differ in the amounts of hydroxycinnamaldehyde monomer incorporation, these results suggest that hydroxycinnamaldehyde content is a more important determinant of digestibility than lignin content.
INTRODUCTION
Lignin is a complex polymer present in the secondary cell wall of all tracheophytes (Bonawitz and Chapple, 2010; Vanholme et al., 2010a). In angiosperms, these polymers are primarily composed of guaiacyl (G) and syringyl (S) subunits with minor amounts of p-hydroxyphenyl (H) subunits, derived from the hydroxycinnamyl alcohol monomers (or monolignols) coniferyl, sinapyl, and p-coumaryl alcohols (Vanholme et al., 2012). The hydroxycinnamaldehydes and hydroxycinnamic acids corresponding to some of these alcohols are also incorporated into lignin in smaller amounts (Baucher et al., 1996; Sibout et al., 2005; Ralph et al., 2008a). In addition to the predominant monolignols, other naturally occurring monomers, such as caffeyl alcohol, the flavonoid tricin, hydroxybenzaldehydes, dihydro-hydroxycinnamyl alcohols, and the variously acylated (by acetate, p-coumarate, p-hydroxybenzoate, and ferulate) monolignols, are incorporated into the polymer to varying degrees in certain species and in specific tissues (Boerjan et al., 2003; Del Río et al., 2007; Lu and Ralph, 2008; Chen et al., 2012; del Río et al., 2012; Rencoret et al., 2013; Wilkerson et al., 2014; Lu et al., 2015).
Lignin polymers interact with wall polysaccharides, adding to the structural integrity of the cell wall, and are responsible for rendering certain cell types impermeable to water (Boerjan et al., 2003); however, the interactions between lignin and cell wall polysaccharides and the enzymes that hydrolyze them greatly impede the conversion of these polymers for industrial and agricultural purposes (Pauly and Keegstra, 2010; Somerville et al., 2010; Carpita, 2012; Jung et al., 2012; Kim et al., 2015; Ko et al., 2015). To overcome this challenge, many strategies to reduce lignin content or alter lignin composition and structure have been implemented with the overall goal of increasing cell wall degradability (Vanholme et al., 2008; Van Acker et al., 2013).
Most of the enzymes that play a role in lignin biosynthesis have been identified and are well characterized (Fraser and Chapple, 2011). Disruptions of several lignin biosynthetic genes result in plants that accumulate atypical lignin polymers that are either not present or are not abundant in wild-type plants. For example, downregulation of p-coumaroylshikimate 3ʹ-hydroxylase or hydroxycinnamoyl-CoA:shikimate hydroxycinnamoyl transferase results in plants with elevated levels of H lignin (Hoffmann et al., 2004; Reddy et al., 2005; Besseau et al., 2007; Shadle et al., 2007; Coleman et al., 2008; Li et al., 2010a; Bonawitz et al., 2014). Downregulation of ferulate 5-hydroxylase (F5H) reduces or eliminates S subunits (Chapple et al., 1992; Reddy et al., 2005); conversely, its overexpression results in plants with a greatly elevated %S levels (Meyer et al., 1998; Franke et al., 2000; Huntley et al., 2003; Stewart et al., 2009). Reduction in caffeic acid-O-methyltransferase activity leads to the incorporation of 5-hydroxyconiferyl alcohol into lignin polymers, even though it is not detectable in the wild-type polymer (Ralph et al., 2001b; Vanholme et al., 2010b; Weng et al., 2010). These manipulations have not only demonstrated the plasticity of lignin biosynthesis but have also led to the development of plants with superior cell wall digestibility characteristics (Ralph et al., 2004; Li et al., 2008; Vanholme et al., 2012; Van Acker et al., 2014).
Downregulation of (hydroxy)cinnamyl alcohol dehydrogenase (CAD) genes is another promising strategy to increase cell wall digestibility (Halpin et al., 1994; Vailhé et al., 1998; Baucher et al., 1999; Chen et al., 2003; Jouanin et al., 2004; Saathoff et al., 2011). CAD is required for the reduction of hydroxycinnamaldehydes to hydroxycinnamyl alcohols, and downregulation of CAD genes generates unusual lignin polymers partially derived from hydroxycinnamaldehyde subunits (Kim et al., 2003; Kim et al., 2004). In Arabidopsis thaliana, two CADs (CADC and CADD) are those primarily involved in lignin monomer synthesis and, as in other systems, cadc cadd plants deposit aldehyde-enriched lignins (Sibout et al., 2005), although the structural details of the modified lignins remain elusive. The characterization of CAD-deficient plants has been conducted not only in model plants like Arabidopsis, but also in agriculturally relevant plants including poplar (Populus sp), alfalfa (Medicago sativa), tobacco (Nicotiana tabacum), maize (Zea mays), sorghum (Sorghum bicolor), rice (Oryza sativa), switchgrass (Panicum virgatum), and tall fescue (Festuca arundinacea). In addition to the increase in cell wall digestibility, CAD-deficient mutants and transgenics display red-pigmented stems and, in the case of Arabidopsis, a limp floral stem (Pillonel et al., 1991; Baucher et al., 1996; Sibout et al., 2005; Zhang et al., 2006). Although the disruption of CAD activity results in a major shift in lignin composition, this modification has a minimal effect on the overall plant yield under most environmental conditions (Kaur et al., 2012; Zhao et al., 2013). Although unaltered growth appears to be the general case for CAD-deficient plants, other manipulations of lignin biosynthesis have resulted in dwarfed and, in some severe cases, infertile plants (Franke et al., 2002; Mir Derikvand et al., 2008; Schilmiller et al., 2009; Huang et al., 2010; Li et al., 2010a).
Here, we used a combinatorial approach to generate plants containing two unusual lignin polymers enriched in aldehydes by disrupting two Arabidopsis CAD genes, CADC and CADD, in G/S-modified lignin backgrounds. NMR resolved the major shifts in the lignin composition and structures in the series of cad mutants. These data further support the observation that CADC and CADD are partially functionally redundant; however, these data also suggest that either CADD has an important role in catalyzing the reduction of sinapaldehyde or is the major form of CAD expressed in cells that deposit S lignin. In addition, these aldehyde lignin subunit compositional manipulations lead to increased enzymatic cell wall digestibility even though total lignin content was conserved. Taken together, these results show that lignin compositional changes can increase cell wall digestibility compared with manipulating lignin content alone and bolster the contention that viable plants can result from lignins derived from essentially no traditional monolignols.
RESULTS
Stacking Lignin Modification Strategies Generates Plants with Unexpected Dwarf Growth Phenotypes
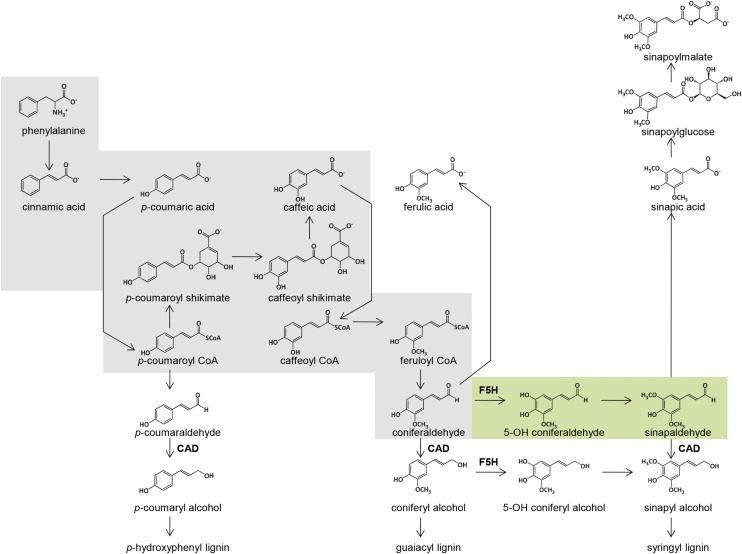
The biosynthesis of lignin, one of the main end-products of the phenylpropanoid pathway, is metabolically plastic and depends heavily on subunit availability to determine composition (Chapple et al., 1992; Meyer et al., 1998; Sederoff et al., 1999; Boerjan et al., 2003; Vanholme et al., 2008, 2012; Ralph et al., 2004, 2008b; Ralph, 2010; Anderson and Chapple, 2014; Wilkerson et al., 2014; Bonawitz et al., 2014). To generate plants enriched in units derived from either coniferaldehyde (Gʹ) or sinapaldehyde (Sʹ), we crossed the previously studied Arabidopsis cadc and cadd mutations (Sibout et al., 2003, 2005) into the F5H-deficient (high G) ferulic acid hydroxylase1 (fah1) mutant and a transgenic line in which the F5H gene is overexpressed under the control of the cinnamate 4-hydroxylase (C4H) promoter (high S) (C4H-F5H), respectively (Figure 1).
Figure 1.
The Angiosperm Phenylpropanoid Pathway.
The highlighted gray region outlines the strategy for lignin-incorporated coniferaldehyde enrichment that is achieved by blocking at F5H and CAD. The strategy for lignin-incorporated sinapaldehyde enrichment expands on the coniferaldehyde enrichment strategy (gray, and the additional area outlined in green) and includes the combination of F5H overexpression and CAD disruption.
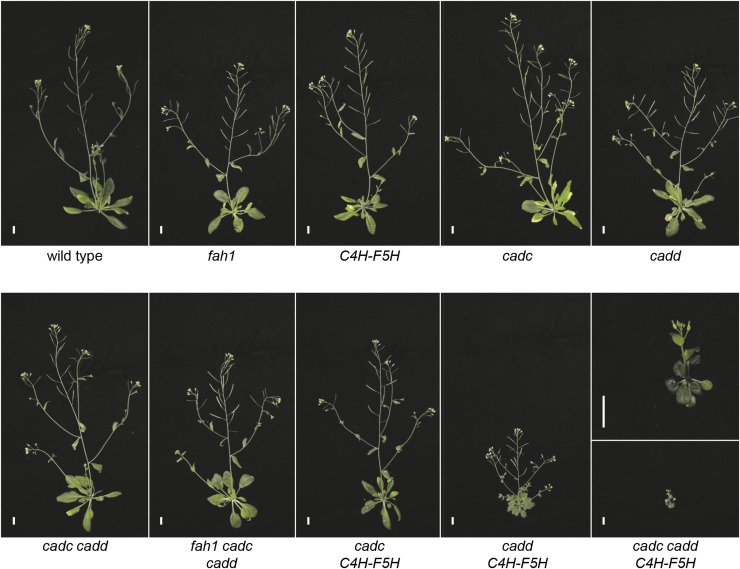
The fah1, cadc, and cadd single mutants as well as the C4H-F5H line display wild-type growth (Figures 2 and 3). Similarly, cadc cadd, fah1 cadc cadd, and cadc C4H-F5H plants obtain a wild-type height even though they display the limp floral stem phenotype previously reported for cadc cadd (Sibout et al., 2005). Surprisingly, cadd C4H-F5H is dwarfed, with a significant reduction in both rosette size and stem length. The most drastic change in morphology is observed in cadc cadd C4H-F5H, which bolts but is severely dwarfed and is infertile.
Figure 2.
Representative Pictures of F5H- and CAD-Manipulated Plants at 2 Months of Age.
Bars = 1 cm.
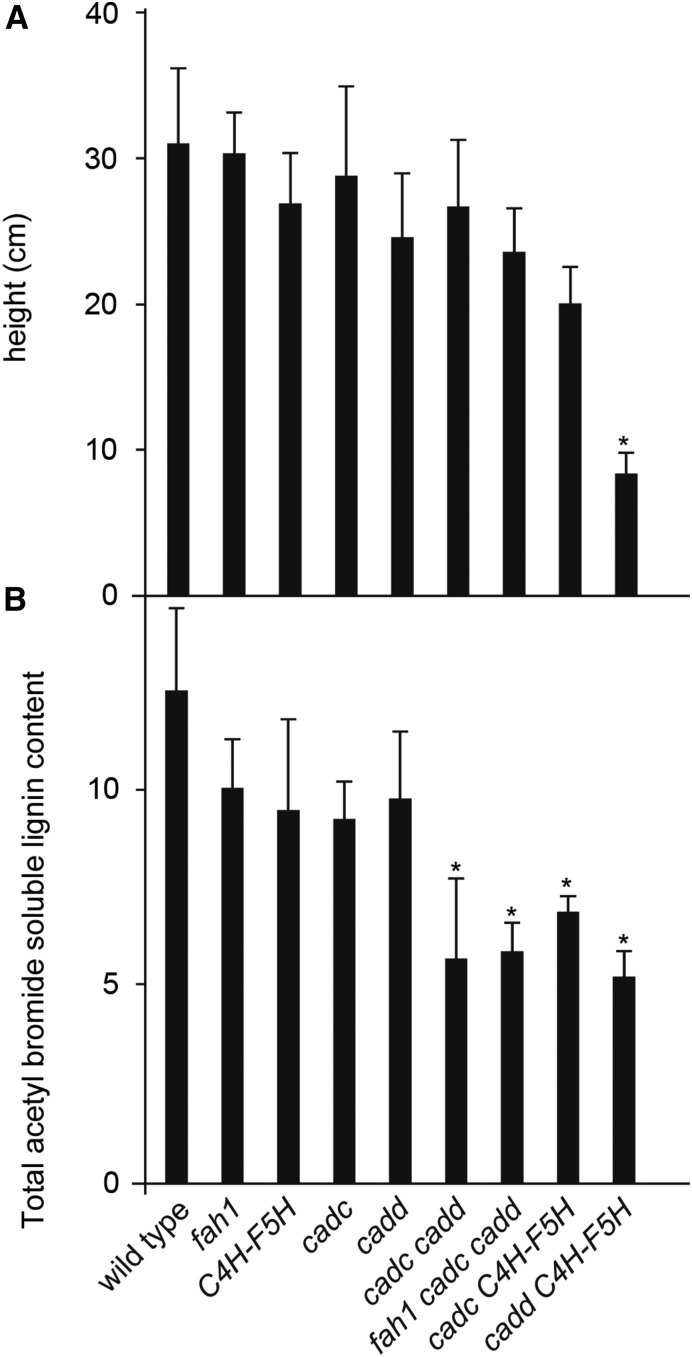
Figure 3.
The Dwarf Phenotype of cadd C4H-F5H Does Not Appear to Be Caused Solely by a Decrease in Total Lignin Content.
(A) Height of the primary inflorescence was measured in 2-month-old plants.
(B) Lignin content of 8-week-old cell wall-extracted tissue from inflorescence stems was determined by the acetyl bromide method.
Error bars represent the sd of biological triplicates. Asterisks indicate the significant difference between the plants with modified lignin composition compared with the wild type (*P < 0.05).
Dwarfism in plants with disrupted lignin biosynthesis is not uncommon (Hoffmann et al., 2004; Mir Derikvand et al., 2008; Stout et al., 2008; Schilmiller et al., 2009), but the mechanisms that lead to dwarfism are still in contention and may be multifaceted in nature (Patten et al., 2005; Laskar et al., 2006; Bonawitz and Chapple, 2013; Bonawitz et al., 2014). To test if the dwarfism we observed is associated with a deficiency in lignin, we quantified lignin using the acetyl bromide method (Chang et al., 2008). In our analysis, we found a 30% total lignin decrease in cadc cadd compared with the wild type and plants disrupted in either CADC or CADD alone (Figure 3), as has been described previously (Sibout et al., 2003, 2005), and not surprisingly also in fah1 cadc cadd. A similar reduction in lignin content was unexpectedly observed for cadc C4H-F5H and for the dwarfed cadd C4H-F5H plants (Figure 3). These results suggest that the dwarfism observed in cadd C4H-F5H plants is not directly or solely the result of reduced lignin content given that they deposit levels of lignin comparable to other CAD-disrupted plants that are wild-type in growth.
Hydroxycinnamaldehydes Can Be Incorporated into Lignin Polymers
To determine how disruption of monolignol biosynthesis alters lignin structure, we performed NMR analysis on cell walls from the series of plants described above (Figure 4). Gel-state NMR spectra were acquired with samples prepared by directly swelling the whole stem cell wall fractions, after fine milling, in DMSO-d6/pyridine-d5 (4:1, v/v) (Kim and Ralph, 2010; Mansfield et al., 2012a). Although we were able to generate the cadc cadd C4H-F5H line, we were not able to obtain sufficient material to conduct the NMR analysis as it displays such a severe dwarf phenotype (Figure 2).
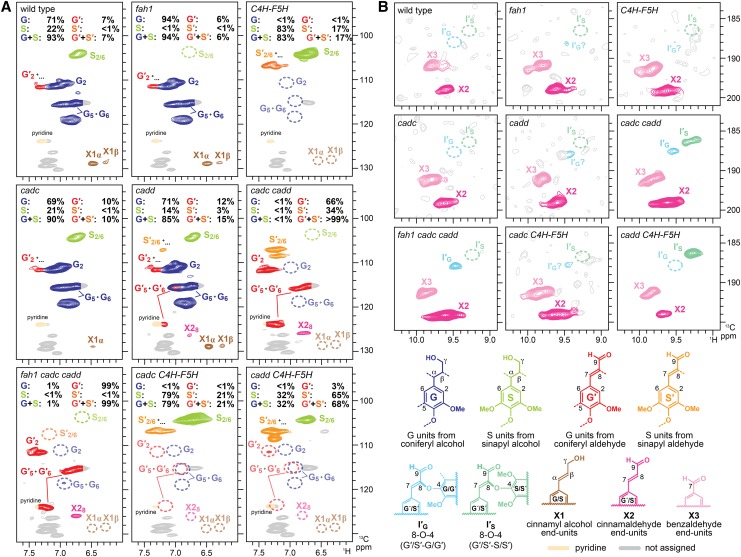
Figure 4.
HSQC Spectra Showing the Impact That CAD and F5H Manipulation Have on Lignin Subunit Composition.
Expanded aromatic (A) and aldehyde (B) subregions of HSQC NMR spectra of ball-milled whole cell walls from Arabidopsis manipulated lines.
Changes in lignin monomer composition are most readily visualized from the aromatic regions of the two-dimensional 1H–13C correlation (heteronuclear single quantum coherence [HSQC]) spectra (Figure 4). The spectra of the wild type displayed signals from G-rich G/S-type lignins derived from polymerization of both coniferyl and sinapyl alcohols, as is typical for most angiosperm lignins. The lignin alterations seen in the fah1 and C4H-F5H lines are consistent with the outcomes of the gene manipulations described in the earlier studies (Chapple et al., 1992; Meyer et al., 1998), with the former showing G lignins derived exclusively from polymerization of coniferyl alcohol and the latter S lignins from sinapyl alcohol. The natural presence of the aldehyde units (G' and S') in wild-type and fah1 plants was minimal and typical; G'2 and S'2/6 signals are overestimates of hydroxycinnamaldehyde incorporation as they also occur due to the presence of benzaldehyde units and oxidized β-aryl ether units with α-carbonyl carbons, some of which, particularly the syringyl units S', are produced during ball-milling (Kim and Ralph, 2010; Zhao et al., 2013). On the other hand, as expected, levels of aldehyde-derived units are significantly elevated in the CAD mutants when both CADC and CADD genes are downregulated. Our NMR data clearly show that the lignins produced in the cadc cadd line are striking G'/S' lignins derived exclusively from polymerization of coniferaldehyde and sinapaldehyde. In addition, the fah1 cadc cadd triple mutant produced G' lignins solely from coniferaldehyde. The typical alcohol-derived lignin units (G and S) are below the level of detection in these CAD-deficient mutants. Loss of function in either only CADC or CADD did not eliminate lignin units derived from coniferyl and/or sinapyl alcohols. The CADC mutation hardly affects the monomer composition of the lignins, which is consistent with the previous observation in these Arabidopsis mutants (Sibout et al., 2005). Also consistent with previous observations, our NMR data showed that disruption of CADD could only modestly elevate S' lignin unit levels in the cell walls (Sibout et al., 2003). Strikingly, cadd C4H-F5H produced considerably S'-rich S/S' lignins, mainly derived from polymerization of sinapaldehyde (∼70% based on the HSQC signal intensities). As noted above, and regrettably, we were unable to conduct the NMR analysis on the cadc cadd C4H-F5H line that displays a severe dwarf phenotype (Figure 2); we anticipated that the lignin in this line would be entirely sinapaldehyde-derived.
The aldehyde regions of the HSQC spectra (Figure 4) determine the nature of the various aldehyde units in the polymers, whereas aliphatic regions (Supplemental Figure 1) show the distributions of the intermonomeric linkages in the lignins. Consistent with the above aromatic compositional data, the characteristic and aldehyde-derived 8–O–4-units (I′G and I′S) that are diagnostic for the (cross-)coupling of hydroxycinnamaldehydes onto a growing lignin chain (Kim et al., 2000, 2003; Zhao et al., 2013) are clearly seen in the mutant lines producing aldehyde-rich lignins (cadc cadd, fah1 cadc cadd, and cadd C4H-F5H), whereas these aldehyde signals, except for the naturally occurring cinnamaldehyde (X2) and benzaldehyde (X3) end-units, are not detectable in the spectra from the wild type and other mutant lines producing typical alcohol-derived lignins (fah1, cadc, cadd, and cadc C4H-F5H) (Figure 4). In addition, typical lignin units, such as β-aryl ethers (IG and IS), phenylcoumarans (II), resinols (III), and cinnamyl alcohol end-groups (X1), are practically absent or significantly depleted in the mutant lines (cadc cadd, fah1 cadc cadd, and cadd C4H-F5H) producing lignins mainly from hydroxycinnamaldehydes, in comparison to the wild type and other mutant lines (fah1, cadc, cadd, and cadc C4H-F5H) producing normal lignins from p-hydroxycinnamyl alcohols (Supplemental Figure 1). We also analyzed cell wall polysaccharide unit profiles by HSQC NMR (Kim and Ralph, 2010; Kim and Ralph, 2014). The signal distribution patterns in sugar anomeric regions appear similar among the wild type and all of the mutants analyzed here (Supplemental Figure 2), suggesting that alteration of lignin biosynthesis in these plants does not dramatically affect the composition of cell wall polysaccharides.
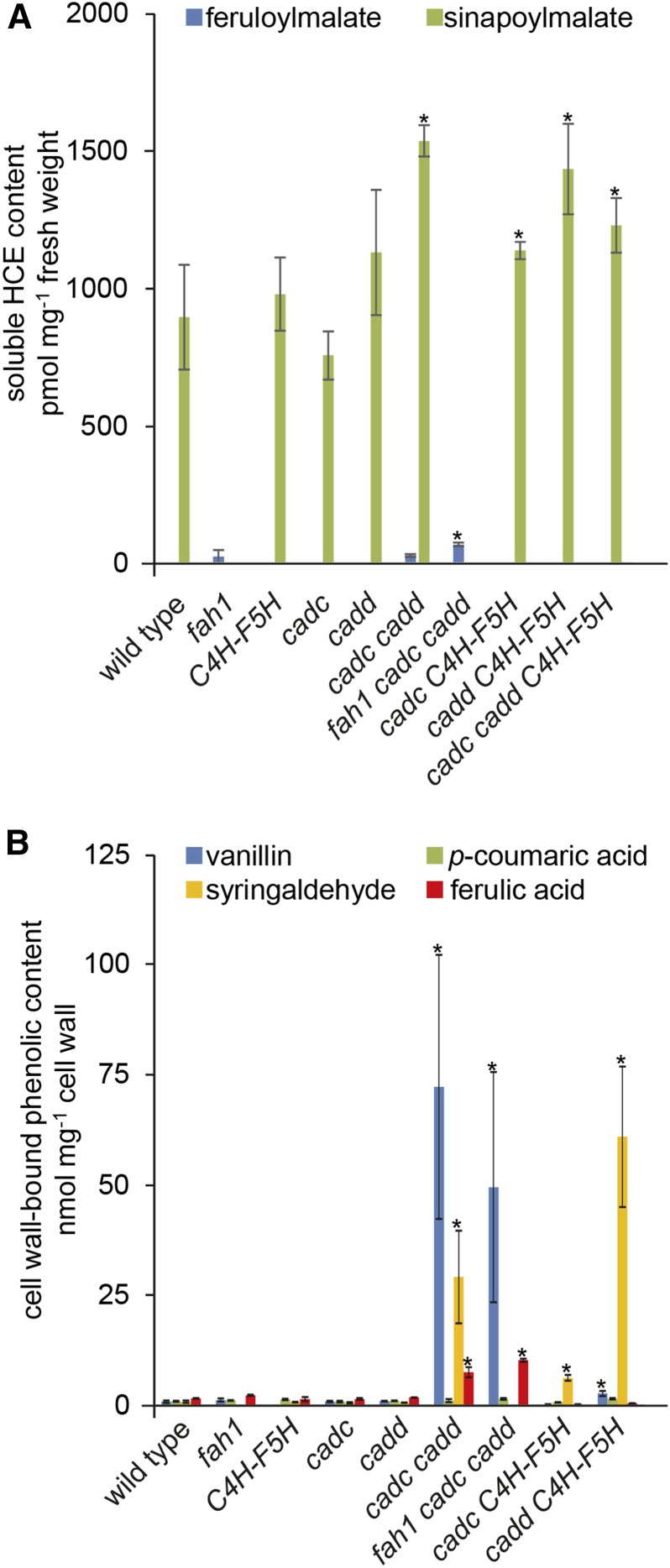
Major Soluble and Wall-Bound Metabolites Are Only Modestly Affected by CAD Disruption
In addition to lignin, the phenylpropanoid pathway is required for the synthesis of cell wall-bound hydroxycinnamic acids and soluble hydroxycinnamate esters (HCEs). To assess how the pathway manipulations described above affect these metabolites, we analyzed leaf HCE content (Figure 5) and the phenolics released after alkaline hydrolysis of inflorescence cell wall tissue (Figure 5). In these experiments, we observed a modest yet significant increase in total HCEs (measured primarily as sinapoylmalate) in cadc cadd, fah1 cadc cadd, cadc C4H-F5H, cadd C4H-F5H, and cadc cadd C4H-F5H (Figure 5). These data suggest that, in HCE-producing cells, reduced CAD activity permits enhanced conversion of coniferaldehyde and sinapaldehyde, presumably by the hydroxycinnamaldehyde dehydrogenase encoded by REDUCED EPIDERMAL FLUORESCENCE1 (REF1), to ferulic acid and sinapic acid, respectively, which are subsequently available for enhanced HCE synthesis.
Figure 5.
Soluble Metabolites Are Modestly Affected by CAD Disruption, Whereas Cell Wall-Bound Metabolite Levels Are Increased.
Soluble (A) and cell wall-bound (B) metabolites of 3-week-old whole rosettes and 2-month-old inflorescence stems, respectively. Error bars represent the sd of biological triplicates. Asterisks indicate the significant difference between the plants with modified lignin composition compared with either the wild type or fah1 (*P < 0.05).
Unlike the relatively minor changes in soluble leaf metabolite biosynthesis, in all CAD-disrupted lines, dramatic changes are observed in phenolic compounds released from inflorescence tissue by alkaline hydrolysis (Figure 5). We observed about a 10-fold increase in cell wall-bound ferulic acid in cadc cadd and fah1 cadc cadd compared with the wild type and fah1, respectively, but no increase in p-coumaric acid. In addition, we observed a 30-fold increase in vanillin release from both cadc cadd and fah1 cadc cadd, which is most likely the result of degradation of aldehyde-rich lignins during the saponification treatment. Another likely lignin degradation product, syringaldehyde, increased 6-fold in cadc C4H-F5H samples and 60-fold in cadd C4H-F5H hydrolysates compared with C4H-F5H. These data indicate that, as in leaves, reductions in CAD activity results in redirection of carbon flux toward hydroxycinnamic acid biosynthesis and enhanced esterification of these metabolites by cell wall components.
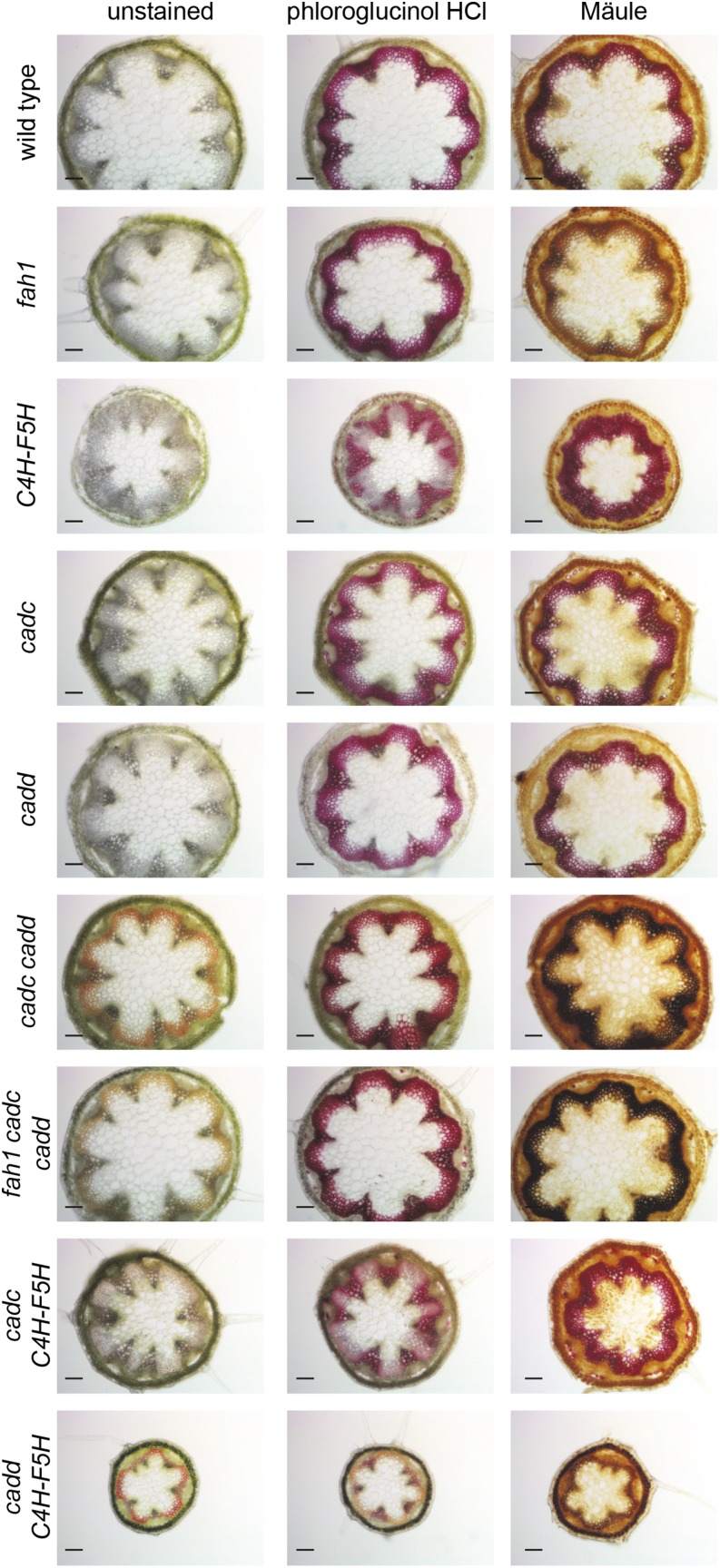
CAD-Manipulated Plants Have Typical, Noncollapsed Vasculature Despite Having Altered Cell Wall Composition and Structure
To assess the effects lignin compositional changes have on vascular anatomy, we first conducted several histochemical analyses on sectioned inflorescence tissue (Figure 6). Previous characterization of CAD-deficient plants revealed a reddish-orange coloration in the lignifying cells (Ralph et al., 1997; Sibout et al., 2005; Sirisha et al., 2012). For wild-type and fah1 plants, disruption of both CADC and CADD were required to observe this phenotype; however, lignified cells in cadc C4H-F5H are pigmented, and this coloration is even more intense in cadd C4H-F5H. These results indicate that disruption of only one CAD is required to generate this phenotype in a high S-lignin background, consistent with the NMR results that indicated enhanced aldehyde content in cadd C4H-F5H. Phloroglucinol-HCl is known to interact with coniferaldehyde and sinapaldehyde end-groups and is frequently used to stain lignifying cell walls (Black et al., 1953; Pomar et al., 2002). All lines stained positively for phloroglucinol-HCl; however, the staining of cadc cadd and fah1 cadc cadd is a deeper red than that observed in other plants, which correlates with the increased aldehyde content in the lignin in these plants. In addition, plants overexpressing F5H exhibit phloroglucinol staining primarily in the xylem with less staining in the interfascicular fibers, a pattern that has been observed previously (Franke et al., 2000; Weng et al., 2010). It is tempting to overinterpret the reduction in fiber staining to suggest that the observed increased aldehyde content (Figure 4) is restricted to the xylem and that lignin in the cell walls of fibers is not decorated with aldehyde end-units in plants overexpressing F5H. However, as has been shown previously, the staining cannot be used as a reliable indicator of hydroxycinnamaldehyde incorporation as the fully incorporated units, i.e., those units incorporated into the chain of the polymer as opposed to the (terminal) end-groups, are not colored and do not stain (Kim et al., 2002). We also conducted Mäule staining, which stains cell walls high in S lignin red and walls high in G lignin tan. Staining of the wild type, fah1, and C4H-F5H gave results consistent with these expectations as has been observed previously (Weng et al., 2010). In contrast, the cell walls of plants with the highest aldehyde content, cadc cadd, fah1 cadc cadd, and cadd C4H-F5H, stained black, which is similar to what has been reported previously for CAD-disrupted mutants (Jourdes et al., 2007). This difference in staining obviously reflects the structural differences in the lignins between these lines, but as the molecular mechanism behind this stain is poorly understood, any further interpretation would be only speculative.
Figure 6.
Histochemical Staining of 2-Month-Old Inflorescence Stem Sections from F5H- and CAD-Disrupted Plants.
Sections were either left unstained or stained with phloroglucinol-HCl or Mäule. Bars = 100 μm.
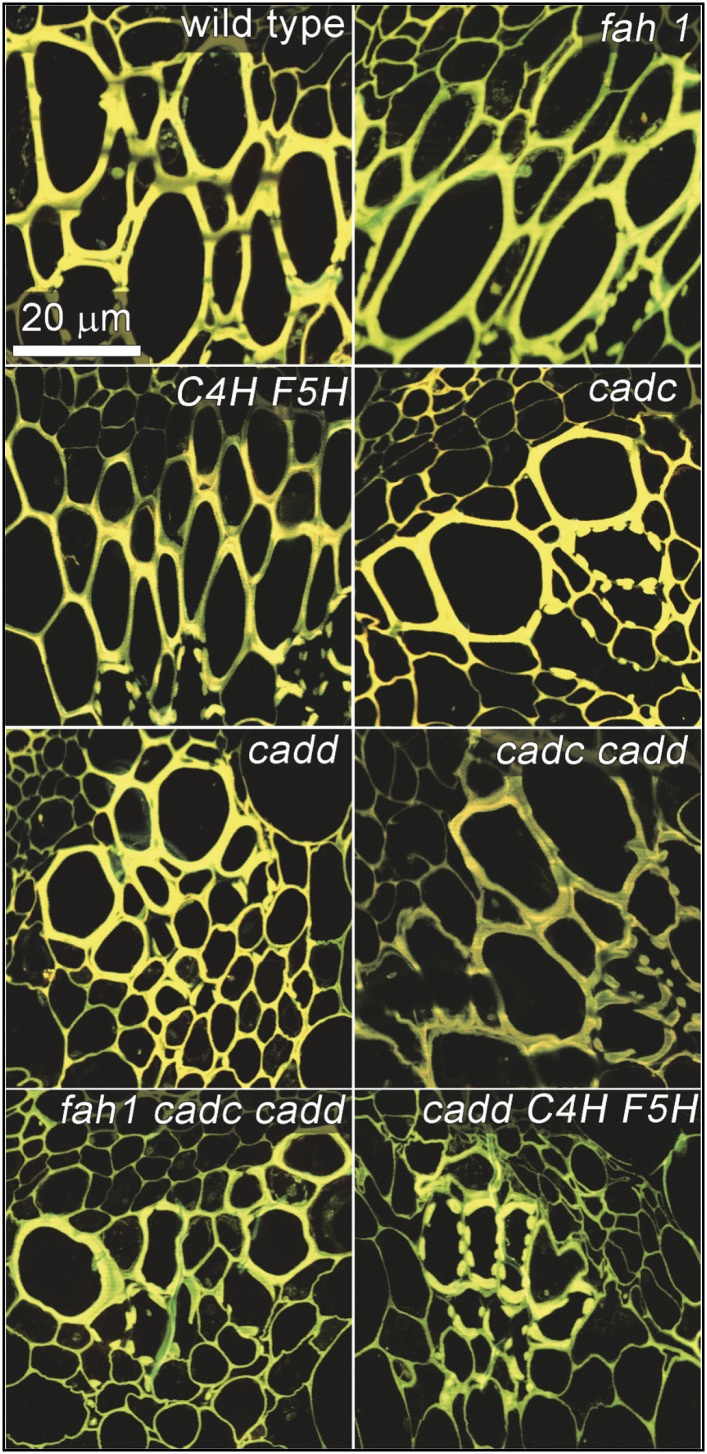
The structure of the vascular tissue of these plants was next examined by confocal laser scanning microscopy (Figure 7). Vascular bundles in the fah1 and C4H-F5H transgenics appear largely similar to those of the wild type. The xylem tissue from the CAD-disrupted plants display fewer tracheary elements compared with the wild type and the fah1 and C4H-F5H lines. This phenotype was most evident in the cadd C4H-F5H tissue, in which the semicontinuous secondary cell walls typical of metaxylem cells are virtually absent from the vascular bundles. Instead, the xylem tissues of the cadd C4H-F5H samples display secondary cell wall thickenings typical of the sparse helical morphology found in protoxylem. This pattern of sparse and incomplete secondary cell walls is markedly different from the collapsed and occluded vasculature recently reported in the lignin-modified dwarf reduced epidermal fluorescence8 mutant (Bonawitz et al., 2014; Kim et al., 2014), but is likely still an impediment to effective transport of water and nutrients throughout the plant. A less continuous lignified secondary wall is likely to allow more lateral water transport at the expense of efficient axial transport.
Figure 7.
Confocal Laser Scanning Microscopy of Vascular Bundles of 2-Month-Old Inflorescence Sections Stained Using the Fluorescent Dye Acriflavine.
Bars = 20 μm.
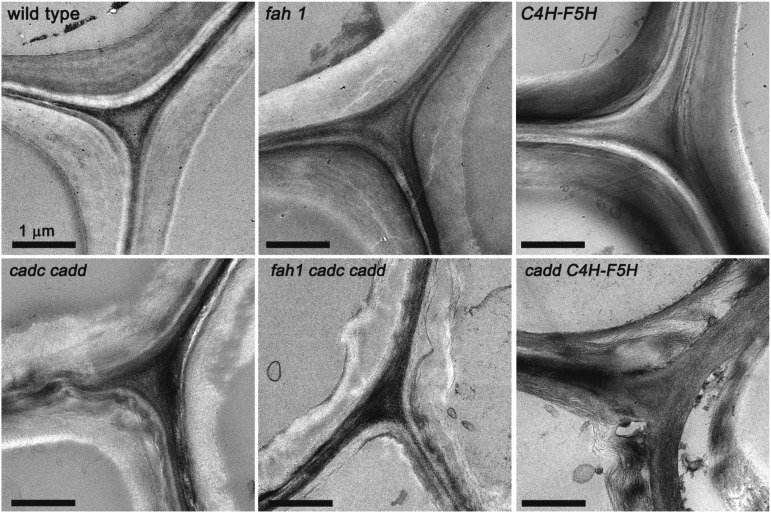
The effects of CAD disruption on cell wall ultrastructure were investigated by transmission electron microscopy of fiber cells adjacent to the xylem tissue (Figure 8). Compared with cell walls of wild-type, fah1, and C4H-F5H plants, the secondary cell walls of the cadc cadd, fah1 cadc cadd, and cadd C4H-F5H variants appear loose, less dense, and disorganized. In the extreme case of the cadd C4H-F5H plant, secondary cell walls are barely identifiable as coherent organelles and instead appear as loose assemblies of cellulose microfibrils. Such a deficiency in properly formed secondary cell walls will weaken the mechanical properties of the vascular tissue and provide little structural support to the plant, which may contribute to the dwarfism of the cadd C4H-F5H variant and the limp floral stem phenotype of cadc cadd mutants.
Figure 8.
Transmission Electron Microscopy Imaging of 2-Month-Old Inflorescence Tissue Showing Cell Wall Ultrastructure.
Bars = 1 μm.
Aldehyde Enrichment in Lignin Increases Cell Wall Digestibility
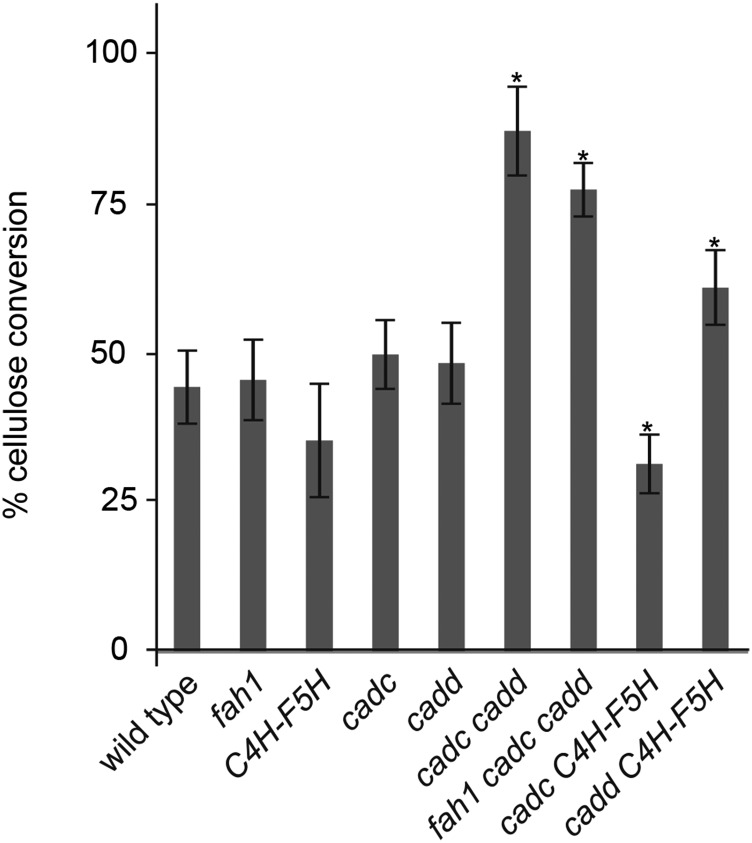
To assess how changes in cell wall composition affect cell wall digestibility, we analyzed enzymatic cellulose conversion on untreated samples. Previous studies in Arabidopsis showed that modifications of cell wall composition can have a dramatic effect on cellulose conversion (Li et al., 2010b; Weng et al., 2010; Bonawitz et al., 2014). We observed for the wild type, fah1, and C4H-F5H that there was no significant difference in cellulose conversion, with ∼50% conversion of available cellulose to glucose after a 48-h incubation. In contrast, we found that cadc cadd and fah1 cadc cadd yielded almost twice as much glucose under the same conditions (Figure 9). This increase was observed to a lesser extent for cadd C4H-F5H and not observed (or diminished) for cadc C4H-F5H. The increased aldehyde lignin content in cadc cadd and fah1 cadc cadd is most likely responsible for the increase in enzymatic cellulose conversion compared with cadc C4H-F5H and cadd C4H-F5H, which have lower aldehyde contents.
Figure 9.
Cellulose Conversion of Ground Inflorescence Tissue from Aldehyde-Enriched Plants.
Error bars represent the sd of biological triplicates. Asterisks indicate the significant difference between the plants with modified cellulose conversion compared with the wild type (*P < 0.05).
DISCUSSION
Lignin content and composition are two factors that contribute to the recalcitrance of the cell wall (Vanholme et al., 2012). Genetic and transgenic approaches to manipulate these parameters have resulted in varying degrees of success with regard to improving cell wall digestibility (Chen and Dixon, 2007; Van Acker et al., 2013). On the one hand, several plants with disruptions in lignin content exhibit an increase in cell wall digestibility, although the reduced lignin has been correlated with decreased yield (Bonawitz and Chapple, 2013). On the other hand, plants with perturbations that affect lignin composition show improvements in cell wall degradability; however, this increase is sometimes only detectable after pretreatment (Li et al., 2010b; Ciesielski et al., 2014). Lignin composition is plastic and several genetic manipulations have generated plants with nearly homogeneous lignin subunit profiles or lignins derived from noncanonical monomers. These include the alteration of syringyl monomer content by manipulation of F5H expression and the perturbation of alcohol and aldehyde subunit content through the disruption of CAD (Meyer et al., 1998; Sibout et al., 2005). We stacked these two manipulation strategies to generate two unusual lignin compositions in anticipation that this approach might deliver unique lignins and provide insights into the impact of lignin content and composition on plant growth and approaches to overcome cell wall recalcitrance to enzyme hydrolysis.
CADD Has the Predominant Role in Catalyzing the Reduction of Sinapaldehyde
It has been established that both CADC and CADD need to be disrupted in a wild-type background to increase aldehyde lignin subunits (Figure 4; Sibout et al., 2005; Jourdes et al., 2007). Here, we show that both CADC and CADD also need to be disrupted in a fah1 background to synthesize a lignin polymer composed almost exclusively from coniferaldehyde. These results indicate that, in this genetic background, CADC and CADD function redundantly in the conversion of hydroxycinnamaldehydes to hydroxycinnamyl alcohols in lignin biosynthesis.
The production of monolignols in many species including Arabidopsis involves the activity of several CAD isoenzymes (Feuillet et al., 1995; Barakat et al., 2009; Saballos et al., 2009). In Arabidopsis, CADC and CADD are thought to be the result of a recent gene duplication event (Guo et al., 2010). Although they both have similar catalytic efficiencies toward most hydroxycinnamaldehydes (Kim et al., 2004), there has been some dispute over whether or not angiosperms have evolved CADs that specifically utilize sinapaldehyde and whether this neofunctionalization explains the occurrence of S lignin biosynthesis in flowering plants (Li et al., 2001; Peter and Neale, 2004; Bomati and Noel, 2005; Chiang, 2006; Barakate et al., 2011). Although Li et al. (2001) identified a CAD homolog, which they termed sinapyl alcohol dehydrogenase (SAD), from Populus tremuloides that has a high catalytic efficiency toward sinapaldehyde and found that this enzyme is missing in gymnosperms, which are deficient in S lignin, Barakate et al. (2011) thoroughly demonstrated that S lignin is dependent on CAD activity and not SAD. Even though our data clearly show a differential contribution of the two CAD isoforms to S lignin biosynthesis in Arabidopsis, neither CADC nor CADD can be considered an ortholog of the SAD protein discussed by Li et al. (2001).
Surprisingly, we found that disruption of CADD alone in the high-S background leads to sinapaldehyde incorporation into lignin (Figure 4), a phenotype not observed in CADC mutants in the same background. This observation indicates that CADD has a predominant role in catalyzing the reduction of sinapaldehyde, which is consistent with the in vitro evidence that shows that CADD has a higher catalytic efficiency toward sinapaldehyde compared with CADC (Kim et al., 2004) and with the finding that only CADD mutants display a modest increase in Sʹ lignin (Figure 4; Sibout et al., 2003). This final observation may also indicate that CADD is more highly expressed than CADC in interfascicular fibers, in which S lignin is most abundantly deposited in wild-type stems, although previous promoter-reporter gene fusion data do not strongly support this hypothesis (Sibout et al., 2003).
Previous radiolabeled substrate feeding studies and the in vitro characterization of F5H and caffeic acid-O-methyltransferase suggested that sinapyl alcohol can be synthesized by hydroxylation and subsequent O-methylation of coniferyl alcohol, a pathway that obviates the requirement for SAD activity (Chen et al., 1999; Humphreys et al., 1999; Tsuji et al., 2004). If this metabolic route permits plants to bypass the reduction of sinapaldehyde completely, then it should not be possible to generate plants that accumulate Sʹ lignin in the relative absence of Gʹ lignin by disrupting CAD. The accumulation of Sʹ lignin in C4H-F5H, cadd, cadc cadd, cadc C4H-F5H, and cadd C4H-F5H suggests, at least for Arabidopsis, that some phenylpropanoid flux proceeds through sinapaldehyde in S lignin biosynthesis. However, we cannot differentiate whether or not the residual S lignin in cadd C4H-F5H is derived directly from the partial redundancy of CADC or indirectly derived from bypassed flux coming from the hydroxylation and O-methylation of coniferyl alcohol.
Hydroxycinnamaldehyde Monomers Can Serve as Lignin Precursors
Coniferaldehyde and sinapaldehyde incorporation into lignin resulting in lignin end-groups (on the starting end of a chain) has been well established and is the basis for phloroglucinol-HCl staining (Black et al., 1953; Pomar et al., 2002). In fah1 cadc cadd, coniferaldehyde not only acts as an end-group but also as the major internal subunit component of the lignin polymer through 8–O–4-linkages (Figure 4; Kim et al., 2002, 2003; Sibout et al., 2005; Jourdes et al., 2007). Unfortunately, we were only able to document an enrichment of sinapaldehyde levels to ∼70% in cadd C4H-F5H, as estimated by NMR, as a copolymer with sinapyl alcohol because we were unable to analyze cadc cadd C4H-F5H plants. We would anticipate that their sinapaldehyde levels would be still higher, perhaps even close to 100%. Although it remains unclear what impact the incorporation of sinapaldehyde units has on the lignin polymer, it may compromise the function of this polymer when the content becomes too high. A similar phenomenon has been observed when lignins greatly enriched in 5-hydroxyconiferyl alcohol monomers were engineered into Arabidopsis (Weng et al., 2010; Vanholme et al., 2010b). Unlike conventional monolignols and hydroxycinnamaldehydes, which mostly form rotatable β–O–4 and analogous 8–O–4 bonds, lignification via 5-hydroxyconiferyl alcohol forms cyclic benzodioxane units (from β–O–4-coupling of a monolignol with 5-hydroxyguaiacyl phenolic end-units) resulting in a linkage that is rigid (Marita et al., 2001; Ralph et al., 2001a, 2001b; Lu et al., 2010). Both Weng et al. (2010) and Vanholme et al. (2010b) speculate that the enrichment of this unusual linkage affects the physical properties of the polymer. In moderation, benzodioxane linkages are not problematic; however, when 5-hydroxy units rise to the level of G subunits, dwarfism of the plant is observed. However, that such benzodioxane-only homopolymers can function in certain roles is demonstrated by the recent observation of caffeyl alcohol-only and 5-hydroxyconiferyl alcohol-only lignins in seed coats of some Cactaceae (Chen et al., 2013). Although this hypothesis remains untested, lignin polymers derived from sinapaldehyde alone may also have undesirable physical or architectural properties that negatively affect plant growth. From our results, it appears that plants can maintain wild-type growth when Sʹ levels are lower than those of G, S, or Gʹ units; however, severe changes in plant morphology arise when Sʹ units rival the combined levels of the other monomers.
Viable Plants Can Generate Lignins Highly Enriched in Hydroxycinnamaldehyde-Derived Units
To the casual observer, it might appear that the polymers generated in the most extreme cases here resemble the normal phenolic polymer. However, closer inspection shows that, because of the differences in functional groups, reactivity, coupling propensities, and in the altered way in which intermediate quinone methides rearomatize, the lignins are in fact entirely different materials that bear little structural resemblance to the “normal” lignins derived from hydroxycinnamyl alcohols. As noted above, and as most clearly seen in the aromatic regions of the HSQC spectra in Figure 4, the fah1 cadc cadd mutant is derived from close to 100% coniferaldehyde. As such it has none of the traditional structural units normally present in lignins (Supplemental Figure 1), although it is still derived from analogous radical coupling reactions. Similarly, the cadc cadd mutant is almost solely (>99%) derived from the two hydroxycinnamaldehydes, coniferaldehyde and sinapaldehyde. These two mutants, both of which form viable and fertile plants, are developmentally relatively normal, a finding that demonstrates that plants can contend with major structural alterations in a crucial and abundant cell wall polymer. These mutants are even more extreme than the recently reported CAD1 mutant of Medicago truncatula, which has only ∼5% normal monolignols (Zhao et al., 2013). Why plants high in sinapaldehyde-derived lignin are developmentally compromised, and whether their dwarfism is even attributable to a poorly functional lignin polymer, remain unclear at this time.
Dwarfism in Lignin Biosynthetic Mutants Is Not the Result of Collapsed Xylem
Previous studies have highlighted the correlation between dwarfism and metabolic blocks in lignin biosynthesis that lead to decreased lignin content (Meyer et al., 1998; Besseau et al., 2007; Mir Derikvand et al., 2008; Li et al., 2010a); however, the mechanism by which growth is impeded in these plants remains unknown (Bonawitz and Chapple, 2013). There are examples of plants with reduced total lignin content that maintain wild-type growth, including the cadc cadd mutant itself (Rohde et al., 2004; Sibout et al., 2005), but many dwarfed low-lignin plants have a collapsed xylem phenotype that has been suggested to lead to reduced water transport that is the proximal cause of growth inhibition. These include plants defective in, or downregulated for C4H, p-coumaroylshikimate 3ʹ-hydroxylase, hydroxycinnamoyl-CoA:shikimate hydroxycinnamoyl transferase, and cinnamoyl-CoA reductase (Franke et al., 2002; Mir Derikvand et al., 2008; Schilmiller et al., 2009; Li et al., 2010a; Vanholme et al., 2010b). Although cadd C4H-F5H plants are similar in height, or even shorter than many of these low-lignin plants, we observed no evidence of collapsed xylem, suggesting that dwarfing arises via some other mechanism(s) in these plants and is even further exacerbated in cadc cadd C4H-F5H plants. These mechanisms could include deficiencies in the production of dehydrodiconiferyl alcohol glucoside, CAD-dependent compounds suggested to have a role in cell expansion and growth (Binns et al., 1987; Orr and Lynn, 1992; Tamagnone et al., 1998a, 1998b), or changes in transcription related to pathway flux perturbations that were recently shown to be dependent upon the Med5 subunit of the Mediator complex (Bonawitz et al., 2014).
Increasing Aldehyde Content Correlates with Increased Potential for Enzymatic Hydrolysis of Cellulose
Reduction in lignin content is known to improve sugar release from biomass upon treatment with cocktails of polysaccharide hydrolases (Studer et al., 2011), and similar beneficial effects have recently been reported to arise from modifications to lignin composition (Li et al., 2010b; Mansfield et al., 2012b; Ciesielski et al., 2014). We observed that of the four variants examined with low lignin content, cadc cadd and fah1 cadc cadd have the highest enzymatic cellulosic conversion for nonpretreated Arabidopsis inflorescence tissue. This increase in cell wall digestibility in CAD-disrupted plants has been observed before (Chen et al., 2003; Fu et al., 2011; Muguerza et al., 2014). There is some disagreement as to whether this increase in digestibility is the result of modified lignin content or, as our study suggests, modified lignin composition (Baucher et al., 1996; Reddy et al., 2005). The effect may result from a significant alteration in the intermolecular forces between biopolymers within the cell wall caused by the increased aldehyde content. More specifically, the strong dipoles contributed by the aldehyde groups may disrupt typical hydrogen bonding, electrostatic, and dispersive forces that facilitate integration of the lignin polymer with other cell wall biopolymers, thereby facilitating enhanced penetration of enzymes and increasing accessibility of the carbohydrate components. These data suggest that increased aldehyde content is an important determinant of cell wall digestibility and that synergistic increases in yield can be achieved by combining changes in both lignin content and composition. Based on thioacidolysis experiments following exhaustive methylation (Jouanin et al., 2004), researchers have noted that aldehyde units might be more prone to terminate a chain, resulting in lower molecular mass lignins that would indeed be expected to protect polysaccharides to a lesser degree than larger polymers (Lapierre et al., 2004).
METHODS
Plant Material and Growth Conditions
Arabidopsis thaliana Col-0 plants were grown under a 16-h-light/8-h-dark photoperiod at 100 μE m−2 s−1 in Redi-Earth Plug and Seedling Mixture (Sun Gro Horticulture) supplemented with Scotts Osmocote Plus controlled release fertilizer (Hummert International) at 22°C. The cadc and cadd mutants (Sibout et al., 2003, 2005) were crossed to either the fah1 mutant (Chapple et al., 1992) or the transgenic line in which F5H is overexpressed (Meyer et al., 1998). The putative triple mutants and transgenic double mutants were identified and confirmed by genomic PCR analysis.
Lignin Analysis
Eight-week-old inflorescence tissues stripped of leaves and siliques were pooled, ground in liquid N2, washed with 80% ethanol four or five times to remove soluble metabolites, then washed with acetone and dried, as performed by Meyer et al. (1998). Acetyl bromide analysis was conducted according to Chang et al. (2008). Briefly, samples were exposed to a 4:1 (v/v) mixture of acetic acid/acetyl bromide and incubated at 70°C for 2 h. Samples were cooled to room temperature and then transferred to 50-mL volumetric flasks containing 2 M NaOH, acetic acid, and 7.5 M hydroxylamine hydrochloride. Acetic acid was used to bring each sample up to volume and then each sample was measured for absorbance at 280 nm. Absorbance values were converted to mass using the extinction coefficient of 23.29 g−1 L cm−1.
NMR Spectroscopy
Preparation of cell walls samples for NMR was conducted as described previously (Kim and Ralph, 2010; Mansfield et al., 2012a). In brief, preground dried stems were extracted with 80% aqueous ethanol (sonication 3 × 20 min). Isolated cell wall fractions (∼200 mg) were ball-milled (5 × 5 min milling and 5-min cooling cycles) using a Fritsch Planetary Micro Pulverisette 7 ball mill at 600 rpm with ZrO2 vessels containing ZrO2 ball bearings. Aliquots of the ball-milled whole cell walls (∼60 mg) were transferred into NMR sample tubes and swollen in DMSO-d6/pyridine-d5 (4:1, v/v, 600 μL). NMR spectra were acquired on a Bruker Biospin Avance 700-MHz spectrometer fitted with a cryogenically cooled 5-mm TXI gradient probe with inverse geometry (proton coils closest to the sample). The central DMSO solvent peak was used as an internal reference (δC, 49.5; δH, 3.49 ppm). Adiabatic HSQC experiments (hsqcetgpsisp2.2) were performed using the parameters described previously (Kim and Ralph, 2010; Mansfield et al., 2012a). Processing used typical matched Gaussian apodization in F2 (LB = −0.5, GB = 0.001) and squared cosine-bell apodization and one level of linear prediction (32 coefficients) in F1. Volume integration of contours in HSQC spectra (processed using no linear prediction) used Bruker’s TopSpin 3.1 (Mac) software with no correction factors; i.e., the data represent volume integrals only. For quantification of lignin aromatic distributions, only the carbon/proton-2 correlations from G and G' units and the carbon/proton-2/6 correlations from S and S' units were used, and the G and G' integrals were logically doubled.
Methanol-Soluble Secondary Metabolite Analysis
Hydroxycinnamoylmalate esters from 3-week-old whole rosette leaves were analyzed using methods previously described by Hemm et al. (2003). Briefly, tissue extracts were prepared at a concentration of 100 mg fresh weight mL−1 50% methanol, extracted for 2 h at 65°C (n = 3). Compounds were quantified using ferulic and sinapic acid as standards and normalized per fresh plant tissue.
Cell Wall-Bound Hydroxycinnamic Acid Analysis
Ground, 8-week-old inflorescence cell wall tissue was saponified using 1 M NaOH and incubated for 24 h at 37°C with constant agitation. The solution was acidified with 1 M HCl and extracted using ethyl acetate. The organic phase was then dried, redissolved in 50% methanol, and analyzed by HPLC. Compounds were quantified using p-coumaric and ferulic acid standards obtained from Sigma-Aldrich.
Histochemical Staining
Eight-week-old inflorescence stems were sectioned using a Leica VT1000 S vibrating blade microtome and left either unstained or stained with phloroglucinol-HCl or Mäule reagent as described (Chapple et al., 1992; Franke et al., 2000).
Transmission Electron Microscopy
Sections from the inflorescence stem were taken just below the fifth elongating silique of the inflorescence and were high-pressure frozen with a Leica EMPact2 high-pressure freezer in 0.2-mm-deep planchets (Leica Microsystems) using 0.15 M sucrose as a cryoprotectant. Next, freeze-substitution was performed in a Leica AFS2 automated freeze substitution unit in 1% osmium tetroxide (EMS). The samples were dehydrated by treating by solution exchange with increasing concentrations of acetone (15%, 30%, 60%, 90%, and 3 × 100% acetone). The samples were then infiltrated with Eponate 812 (EMS) by incubating at room temperature for several hours to overnight in increasing concentrations of resin (15%, 30%, 60%, 90%, 3 × 100% resin, diluted in acetone). The samples were transferred to capsules and the resin polymerized in an oven at 60°C overnight. Resin-embedded samples were sectioned to ∼50 nm with a Diatome diamond knife on a Leica EM UTC ultramicrotome. Sections were collected on 200 mesh copper transmission electron microscopy grids (SPI Supplies) and were poststained for 30 s with 1% aqueous KMnO4. Images were taken with a 4-megapixel Gatan UltraScan 1000 camera on an FEI Tecnai G2 20 Twin 200-kV LaB6 transmission electron microscope.
Confocal Microscopy
Samples were preserved and embedded as described above. The embedded samples were sectioned to 250 nm and positioned on glass microscope slides and stained with 0.1% acriflavine. Images were captured in fluorescence mode with a Nikon C1 Plus microscope, equipped with the Nikon C1 confocal system with a 488-nm excitation laser.
Cellulose Conversion
Cellulose conversion was conducted as described by Li et al. (2010b) on 8-week-old hand-ground inflorescence tissue that had been stripped of leaves and siliques. Cellulose conversion was conducted without pretreatment and evaluated colorimetrically using the Megazyme D-Glucose (glucose oxidase/peroxidase) assay kit after 48-h incubation.
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL libraries under accession numbers C4H (At2g30490), F5H (At4g36220), CADC (At3g19450), CADD (At4g34239), and REF1 (At3g24503).
Supplemental Data
Supplemental Figure 1. Aliphatic subregions of HSQC NMR spectra of ball-milled whole cell walls from Arabidopsis mutant lines.
Supplemental Figure 2. Polysaccharide anomeric subregions of HSQC NMR spectra of ball-milled whole cell walls from Arabidopsis manipulated lines.
Supplementary Material
Acknowledgments
This work was supported as part of the Center for Direct Catalytic Conversion of Biomass to Biofuels, an Energy Frontier Research Center funded by the U.S. Department of Energy, Office of Science, Basic Energy Sciences under Award DE-SC0000997. This material is also based upon work supported by the U.S. Department of Energy, Office of Science, Office of Basic Energy Sciences, Chemical Sciences, Geosciences, and Biosciences Division under Award DE-FG02-07ER15905. J.R. and Y.T. were funded by Stanford University’s Global Climate and Energy Project and the Great Lakes Bioenergy Research Center by the U.S. DOE’s Office of Science (DE-FC02-07ER64494). Y.T. also acknowledges a support from The Ministry of Education, Culture, Sports, Science, and Technology (MEXT), Japan (Grant 26892014). E.X. and M.L. were supported by the U.S. DOE through Grant DE-FG02-06ER64301 to M.L. and C.C. and by the Purdue University Office of Agricultural Research Programs.
AUTHOR CONTRIBUTIONS
N.A.A. and C.C. conceived of the study. N.A.A., Y.T., P.N.C., and E.X. performed the research and along with the other authors wrote the article.
Glossary
- CAD
(hydroxy)cinnamyl alcohol dehydrogenase
- HCE
hydroxycinnamate ester
- SAD
sinapyl alcohol dehydrogenase
- HSQC
heteronuclear single quantum coherence
References
- Anderson N.A., Chapple C. (2014). Perturbing lignin biosynthesis: metabolic changes in response to manipulation of the phenylpropanoid pathway. In Recent Advances in Polyphenol Research, Vol. 4, Romani A., Lattanzio V., Quideau S., eds (Chichester, UK: John Wiley & Sons; ), pp. 39–59. [Google Scholar]
- Barakat A., Bagniewska-Zadworna A., Choi A., Plakkat U., DiLoreto D.S., Yellanki P., Carlson J.E. (2009). The cinnamyl alcohol dehydrogenase gene family in Populus: phylogeny, organization, and expression. BMC Plant Biol. 9: 26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barakate A., Stephens J., Goldie A., Hunter W.N., Marshall D., Hancock R.D., Lapierre C., Morreel K., Boerjan W., Halpin C. (2011). Syringyl lignin is unaltered by severe sinapyl alcohol dehydrogenase suppression in tobacco. Plant Cell 23: 4492–4506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baucher M., Bernard-Vailhé M.A., Chabbert B., Besle J.M., Opsomer C., Van Montagu M., Botterman J. (1999). Down-regulation of cinnamyl alcohol dehydrogenase in transgenic alfalfa (Medicago sativa L.) and the effect on lignin composition and digestibility. Plant Mol. Biol. 39: 437–447. [DOI] [PubMed] [Google Scholar]
- Baucher M., Chabbert B., Pilate G., Van Doorsselaere J., Tollier M.T., Petit-Conil M., Cornu D., Monties B., Van Montagu M., Inze D., Jouanin L., Boerjan W. (1996). Red xylem and higher lignin extractability by down-regulating a cinnamyl alcohol dehydrogenase in poplar. Plant Physiol. 112: 1479–1490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Besseau S., Hoffmann L., Geoffroy P., Lapierre C., Pollet B., Legrand M. (2007). Flavonoid accumulation in Arabidopsis repressed in lignin synthesis affects auxin transport and plant growth. Plant Cell 19: 148–162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Binns A.N., Chen R.H., Wood H.N., Lynn D.G. (1987). Cell division promoting activity of naturally occurring dehydrodiconiferyl glucosides: do cell wall components control cell division? Proc. Natl. Acad. Sci. USA 84: 980–984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Black R.A., Rosen A.A., Adams S.L. (1953). The chromatographic separation of hardwood extractive components giving color reactions with phloroglucinol. J. Am. Chem. Soc. 75: 5344–5346. [Google Scholar]
- Boerjan W., Ralph J., Baucher M. (2003). Lignin biosynthesis. Annu. Rev. Plant Biol. 54: 519–546. [DOI] [PubMed] [Google Scholar]
- Bomati E.K., Noel J.P. (2005). Structural and kinetic basis for substrate selectivity in Populus tremuloides sinapyl alcohol dehydrogenase. Plant Cell 17: 1598–1611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonawitz N.D., Chapple C. (2010). The genetics of lignin biosynthesis: connecting genotype to phenotype. Annu. Rev. Genet. 44: 337–363. [DOI] [PubMed] [Google Scholar]
- Bonawitz N.D., Chapple C. (2013). Can genetic engineering of lignin deposition be accomplished without an unacceptable yield penalty? Curr. Opin. Biotechnol. 24: 336–343. [DOI] [PubMed] [Google Scholar]
- Bonawitz N.D., Kim J.I., Tobimatsu Y., Ciesielski P.N., Anderson N.A., Ximenes E., Maeda J., Ralph J., Donohoe B.S., Ladisch M., Chapple C. (2014). Disruption of Mediator rescues the stunted growth of a lignin-deficient Arabidopsis mutant. Nature 509: 376–380. [DOI] [PubMed] [Google Scholar]
- Carpita N.C. (2012). Progress in the biological synthesis of the plant cell wall: new ideas for improving biomass for bioenergy. Curr. Opin. Biotechnol. 23: 330–337. [DOI] [PubMed] [Google Scholar]
- Chang X.F., Chandra R., Berleth T., Beatson R.P. (2008). Rapid, microscale, acetyl bromide-based method for high-throughput determination of lignin content in Arabidopsis thaliana. J. Agric. Food Chem. 56: 6825–6834. [DOI] [PubMed] [Google Scholar]
- Chapple C.C., Vogt T., Ellis B.E., Somerville C.R. (1992). An Arabidopsis mutant defective in the general phenylpropanoid pathway. Plant Cell 4: 1413–1424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiang V.L. (2006). Monolignol biosynthesis and genetic engineering of lignin in trees, a review. Environ. Chem. Lett. 4: 143–146. [Google Scholar]
- Chen F., Dixon R.A. (2007). Lignin modification improves fermentable sugar yields for biofuel production. Nat. Biotechnol. 25: 759–761. [DOI] [PubMed] [Google Scholar]
- Chen F., Tobimatsu Y., Havkin-Frenkel D., Dixon R.A., Ralph J. (2012). A polymer of caffeyl alcohol in plant seeds. Proc. Natl. Acad. Sci. USA 109: 1772–1777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen F., Tobimatsu Y., Jackson L., Nakashima J., Ralph J., Dixon R.A. (2013). Novel seed coat lignins in the Cactaceae: structure, distribution and implications for the evolution of lignin diversity. Plant J. 73: 201–211. [DOI] [PubMed] [Google Scholar]
- Chen F., Yasuda S., Fukushima K. (1999). Evidence for a novel biosynthetic pathway that regulates the ratio of syringyl to guaiacyl residues in lignin in the differentiating xylem of Magnolia kobus DC. Planta 207: 597–603. [Google Scholar]
- Chen L., Auh C.K., Dowling P., Bell J., Chen F., Hopkins A., Dixon R.A., Wang Z.Y. (2003). Improved forage digestibility of tall fescue (Festuca arundinacea) by transgenic down-regulation of cinnamyl alcohol dehydrogenase. Plant Biotechnol. J. 1: 437–449. [DOI] [PubMed] [Google Scholar]
- Ciesielski P.N., et al. (2014). Engineering plant cell walls: Tuning lignin monomer composition for efficient biofuels feedstocks or resilient biomaterials. Green Chem. 16: 2627–2635. [Google Scholar]
- Coleman H.D., Park J.Y., Nair R., Chapple C., Mansfield S.D. (2008). RNAi-mediated suppression of p-coumaroyl-CoA 3′-hydroxylase in hybrid poplar impacts lignin deposition and soluble secondary metabolism. Proc. Natl. Acad. Sci. USA 105: 4501–4506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Del Río J.C., Marques G., Rencoret J., Martínez A.T., Gutiérrez A. (2007). Occurrence of naturally acetylated lignin units. J. Agric. Food Chem. 55: 5461–5468. [DOI] [PubMed] [Google Scholar]
- del Río J.C., Prinsen P., Rencoret J., Nieto L., Jiménez-Barbero J., Ralph J., Martínez A.T., Gutiérrez A. (2012). Structural characterization of the lignin in the cortex and pith of elephant grass (Pennisetum purpureum) stems. J. Agric. Food Chem. 60: 3619–3634. [DOI] [PubMed] [Google Scholar]
- Feuillet C., Lauvergeat V., Deswarte C., Pilate G., Boudet A., Grima-Pettenati J. (1995). Tissue- and cell-specific expression of a cinnamyl alcohol dehydrogenase promoter in transgenic poplar plants. Plant Mol. Biol. 27: 651–667. [DOI] [PubMed] [Google Scholar]
- Franke R., Humphreys J.M., Hemm M.R., Denault J.W., Ruegger M.O., Cusumano J.C., Chapple C. (2002). The Arabidopsis REF8 gene encodes the 3-hydroxylase of phenylpropanoid metabolism. Plant J. 30: 33–45. [DOI] [PubMed] [Google Scholar]
- Franke R., McMichael C.M., Meyer K., Shirley A.M., Cusumano J.C., Chapple C. (2000). Modified lignin in tobacco and poplar plants over-expressing the Arabidopsis gene encoding ferulate 5-hydroxylase. Plant J. 22: 223–234. [DOI] [PubMed] [Google Scholar]
- Fraser, C.M., Chapple, C. (2011). The phenylpropanoid pathway in Arabidopsis The Arabidopsis Book 9: e0152, doi/10.1199/tab.0152. [DOI] [PMC free article] [PubMed]
- Fu C., Xiao X., Xi Y., Ge Y., Chen F., Bouton J., Dixon R., Wang Z. (2011). Downregulation of cinnamyl alcohol dehydrogenase (CAD) leads to improved saccharification efficiency in switchgrass. Bioenerg. Res. 4: 153–164. [Google Scholar]
- Guo D.M., Ran J.H., Wang X.Q. (2010). Evolution of the Cinnamyl/Sinapyl Alcohol Dehydrogenase (CAD/SAD) gene family: the emergence of real lignin is associated with the origin of bona fide CAD. J. Mol. Evol. 71: 202–218. [DOI] [PubMed] [Google Scholar]
- Halpin C., Knight M.E., Foxon G.A., Campbell M.M., Boudet A.M., Boon J.J., Chabbert B., Tollier M.T., Schuch W. (1994). Manipulation of lignin quality by down-regulation of cinnamyl alcohol-dehydrogenase. Plant J. 6: 339–350. [Google Scholar]
- Hemm M.R., Ruegger M.O., Chapple C. (2003). The Arabidopsis ref2 mutant is defective in the gene encoding CYP83A1 and shows both phenylpropanoid and glucosinolate phenotypes. Plant Cell 15: 179–194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann L., Besseau S., Geoffroy P., Ritzenthaler C., Meyer D., Lapierre C., Pollet B., Legrand M. (2004). Silencing of hydroxycinnamoyl-coenzyme A shikimate/quinate hydroxycinnamoyltransferase affects phenylpropanoid biosynthesis. Plant Cell 16: 1446–1465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J., Gu M., Lai Z., Fan B., Shi K., Zhou Y.H., Yu J.Q., Chen Z. (2010). Functional analysis of the Arabidopsis PAL gene family in plant growth, development, and response to environmental stress. Plant Physiol. 153: 1526–1538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humphreys J.M., Hemm M.R., Chapple C. (1999). New routes for lignin biosynthesis defined by biochemical characterization of recombinant ferulate 5-hydroxylase, a multifunctional cytochrome P450-dependent monooxygenase. Proc. Natl. Acad. Sci. USA 96: 10045–10050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huntley S.K., Ellis D., Gilbert M., Chapple C., Mansfield S.D. (2003). Significant increases in pulping efficiency in C4H-F5H-transformed poplars: improved chemical savings and reduced environmental toxins. J. Agric. Food Chem. 51: 6178–6183. [DOI] [PubMed] [Google Scholar]
- Jouanin L., Gujon T., Sibout R., Pollet B., Mila I., Leplé J.C., Pilate G., Petit-Conil M., Ralph J., Lapierre C. (2004). Comparison of the consequences on lignin content and structure of COMT and CAD down regulation in poplar and Arabidopsis thaliana. In Plantation Forest Biotechnology in the 21st Century, Walter C., Carson M., eds (Kerala, India: India Research Signpost; ), pp. 219–229. [Google Scholar]
- Jourdes M., Cardenas C.L., Laskar D.D., Moinuddin S.G.A., Davin L.B., Lewis N.G. (2007). Plant cell walls are enfeebled when attempting to preserve native lignin configuration with poly-p-hydroxycinnamaldehydes: evolutionary implications. Phytochemistry 68: 1932–1956. [DOI] [PubMed] [Google Scholar]
- Jung H.J., Samac D.A., Sarath G. (2012). Modifying crops to increase cell wall digestibility. Plant Sci. 185-186: 65–77. [DOI] [PubMed] [Google Scholar]
- Kaur H., Shaker K., Heinzel N., Ralph J., Gális I., Baldwin I.T. (2012). Environmental stresses of field growth allow cinnamyl alcohol dehydrogenase-deficient Nicotiana attenuata plants to compensate for their structural deficiencies. Plant Physiol. 159: 1545–1570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H., Ralph J. (2010). Solution-state 2D NMR of ball-milled plant cell wall gels in DMSO-d(6)/pyridine-d(5). Org. Biomol. Chem. 8: 576–591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H., Ralph J. (2014). A gel-state 2D-NMR method for plant cell wall profiling and analysis: a model study with the amorphous cellulose and xylan from ball-milled cotton linters. RSC Advances 4: 7549–7560. [Google Scholar]
- Kim H., Ralph J., Lu F., Pilate G., Leplé J.C., Pollet B., Lapierre C. (2002). Identification of the structure and origin of thioacidolysis marker compounds for cinnamyl alcohol dehydrogenase deficiency in angiosperms. J. Biol. Chem. 277: 47412–47419. [DOI] [PubMed] [Google Scholar]
- Kim H., Ralph J., Lu F., Ralph S.A., Boudet A.M., MacKay J.J., Sederoff R.R., Ito T., Kawai S., Ohashi H., Higuchi T. (2003). NMR analysis of lignins in CAD-deficient plants. Part 1. Incorporation of hydroxycinnamaldehydes and hydroxybenzaldehydes into lignins. Org. Biomol. Chem. 1: 268–281. [DOI] [PubMed] [Google Scholar]
- Kim H., Ralph J., Yahiaoui N., Pean M., Boudet A.M. (2000). Cross-coupling of hydroxycinnamyl aldehydes into lignins. Org. Lett. 2: 2197–2200. [DOI] [PubMed] [Google Scholar]
- Kim J.I., Ciesielski P.N., Donohoe B.S., Chapple C., Li X. (2014). Chemically induced conditional rescue of the reduced epidermal fluorescence8 mutant of Arabidopsis reveals rapid restoration of growth and selective turnover of secondary metabolite pools. Plant Physiol. 164: 584–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S.J., Kim M.R., Bedgar D.L., Moinuddin S.G.A., Cardenas C.L., Davin L.B., Kang C., Lewis N.G. (2004). Functional reclassification of the putative cinnamyl alcohol dehydrogenase multigene family in Arabidopsis. Proc. Natl. Acad. Sci. USA 101: 1455–1460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y., Kreke T., Ko J.-K., Ladisch M.R. (2015). Hydrolysis-determining substrate characteristics in liquid hot water pretreated hardwood. Biotechnol. Bioeng. 112: 677–687. [DOI] [PubMed] [Google Scholar]
- Ko J.-K., Ximenes E., Kim Y., Ladisch M.R. (2015). Adsorption of enzyme onto lignins of liquid hot water pretreated hardwoods. Biotechnol. Bioeng. 112: 447–456. [DOI] [PubMed] [Google Scholar]
- Lapierre C., Pilate G., Pollet B., Mila I., Leplé J.C., Jouanin L., Kim H., Ralph J. (2004). Signatures of cinnamyl alcohol dehydrogenase deficiency in poplar lignins. Phytochemistry 65: 313–321. [DOI] [PubMed] [Google Scholar]
- Laskar D.D., Jourdes M., Patten A.M., Helms G.L., Davin L.B., Lewis N.G. (2006). The Arabidopsis cinnamoyl CoA reductase irx4 mutant has a delayed but coherent (normal) program of lignification. Plant J. 48: 674–686. [DOI] [PubMed] [Google Scholar]
- Li L., Cheng X.F., Leshkevich J., Umezawa T., Harding S.A., Chiang V.L. (2001). The last step of syringyl monolignol biosynthesis in angiosperms is regulated by a novel gene encoding sinapyl alcohol dehydrogenase. Plant Cell 13: 1567–1586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X., Bonawitz N.D., Weng J.K., Chapple C. (2010a). The growth reduction associated with repressed lignin biosynthesis in Arabidopsis thaliana is independent of flavonoids. Plant Cell 22: 1620–1632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X., Weng J.K., Chapple C. (2008). Improvement of biomass through lignin modification. Plant J. 54: 569–581. [DOI] [PubMed] [Google Scholar]
- Li X., Ximenes E., Kim Y., Slininger M., Meilan R., Ladisch M., Chapple C. (2010b). Lignin monomer composition affects Arabidopsis cell-wall degradability after liquid hot water pretreatment. Biotechnol. Biofuels 3: 27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu F., et al. (2015). Naturally p-hydroxybenzoylated lignins in palms. BioEnergy Res., http://dx.doi.org/10.1007/s12155-015-9583-4. [Google Scholar]
- Lu F., Marita J.M., Lapierre C., Jouanin L., Morreel K., Boerjan W., Ralph J. (2010). Sequencing around 5-hydroxyconiferyl alcohol-derived units in caffeic acid O-methyltransferase-deficient poplar lignins. Plant Physiol. 153: 569–579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu F., Ralph J. (2008). Novel tetrahydrofuran structures derived from β-β-coupling reactions involving sinapyl acetate in Kenaf lignins. Org. Biomol. Chem. 6: 3681–3694. [DOI] [PubMed] [Google Scholar]
- Mansfield S.D., Kang K.Y., Chapple C. (2012b). Designed for deconstruction--poplar trees altered in cell wall lignification improve the efficacy of bioethanol production. New Phytol. 194: 91–101. [DOI] [PubMed] [Google Scholar]
- Mansfield S.D., Kim H., Lu F., Ralph J. (2012a). Whole plant cell wall characterization using solution-state 2D NMR. Nat. Protoc. 7: 1579–1589. [DOI] [PubMed] [Google Scholar]
- Marita J.M., Ralph J., Lapierre C., Jouanin L., Boerjan W. (2001). NMR characterization of lignins from transgenic poplars with suppressed caffeic acid O-methyltransferase activity. J. Chem. Soc. Perkin Trans. 1: 2939–2945. [Google Scholar]
- Meyer K., Shirley A.M., Cusumano J.C., Bell-Lelong D.A., Chapple C. (1998). Lignin monomer composition is determined by the expression of a cytochrome P450-dependent monooxygenase in Arabidopsis. Proc. Natl. Acad. Sci. USA 95: 6619–6623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mir Derikvand M., Sierra J.B., Ruel K., Pollet B., Do C.T., Thévenin J., Buffard D., Jouanin L., Lapierre C. (2008). Redirection of the phenylpropanoid pathway to feruloyl malate in Arabidopsis mutants deficient for cinnamoyl-CoA reductase 1. Planta 227: 943–956. [DOI] [PubMed] [Google Scholar]
- Muguerza M., Gondo T., Ishigaki G., Akashi R. (2014). Lignin content and digestibility in transgenic bahiagrass (Paspalum notatum Flugge) obtained by genetic manipulation of cinnamyl alcohol dehydrogenase gene. Asian J. Plant Sci. 13: 8–17. [Google Scholar]
- Orr J.D., Lynn D.G. (1992). Biosynthesis of dehydrodiconiferyl alcohol glucosides: implications for the control of tobacco cell growth. Plant Physiol. 98: 343–352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patten A.M., Cardenas C.L., Cochrane F.C., Laskar D.D., Bedgar D.L., Davin L.B., Lewis N.G. (2005). Reassessment of effects on lignification and vascular development in the irx4 Arabidopsis mutant. Phytochemistry 66: 2092–2107. [DOI] [PubMed] [Google Scholar]
- Pauly M., Keegstra K. (2010). Plant cell wall polymers as precursors for biofuels. Curr. Opin. Plant Biol. 13: 305–312. [DOI] [PubMed] [Google Scholar]
- Peter G., Neale D. (2004). Molecular basis for the evolution of xylem lignification. Curr. Opin. Plant Biol. 7: 737–742. [DOI] [PubMed] [Google Scholar]
- Pillonel C., Mulder M.M., Boon J.J., Forster B., Binder A. (1991). Involvement of cinnamyl-alcohol dehydrogenase in the control of lignin formation in Sorghum bicolor L. Moench. Planta 185: 538–544. [DOI] [PubMed] [Google Scholar]
- Pomar F., Merino F., Barceló A.R. (2002). O-4-Linked coniferyl and sinapyl aldehydes in lignifying cell walls are the main targets of the Wiesner (phloroglucinol-HCl) reaction. Protoplasma 220: 17–28. [DOI] [PubMed] [Google Scholar]
- Ralph J. (2010). Hydroxycinnamates in lignification. Phytochem. Rev. 9: 65–83. [Google Scholar]
- Ralph, J., Brunow, G., Harris, P.J., Dixon, R.A., Schatz, P.F., and Boerjan, W. (2008b). Lignification: Are lignins biosynthesized via simple combinatorial chemistry or via proteinaceous control and template replication? In Recent Advances in Polyphenol Research, Vol. 1, F. Daayf, A. El Hadrami, L. Adam, and G.M. Balance, eds (Oxford, UK: Wiley-Blackwell Publishing), pp. 36–66. [Google Scholar]
- Ralph J., Kim H., Lu F., Grabber J.H., Leplé J.C., Berrio-Sierra J., Derikvand M.M., Jouanin L., Boerjan W., Lapierre C. (2008a). Identification of the structure and origin of a thioacidolysis marker compound for ferulic acid incorporation into angiosperm lignins (and an indicator for cinnamoyl CoA reductase deficiency). Plant J. 53: 368–379. [DOI] [PubMed] [Google Scholar]
- Ralph J., Lapierre C., Lu F., Marita J.M., Pilate G., Van Doorsselaere J., Boerjan W., Jouanin L. (2001a). NMR evidence for benzodioxane structures resulting from incorporation of 5-hydroxyconiferyl alcohol into Lignins of O-methyltransferase-deficient poplars. J. Agric. Food Chem. 49: 86–91. [DOI] [PubMed] [Google Scholar]
- Ralph J., Lundquist K., Brunow G., Lu F., Kim H., Schatz P.F., Marita J.M., Hatfield R.D., Ralph S.A., Christensen J.H., Boerjan W. (2004). Lignins: natural polymers from oxidative coupling of 4-hydroxyphenylpropanoids. Phytochem. Rev. 3: 29–60. [Google Scholar]
- Ralph J., MacKay J.J., Hatfield R.D., O’Malley D.M., Whetten R.W., Sederoff R.R. (1997). Abnormal lignin in a loblolly pine mutant. Science 277: 235–239. [DOI] [PubMed] [Google Scholar]
- Ralph J., et al. (2001b). Elucidation of new structures in lignins of CAD- and COMT-deficient plants by NMR. Phytochemistry 57: 993–1003. [DOI] [PubMed] [Google Scholar]
- Reddy M.S.S., Chen F., Shadle G., Jackson L., Aljoe H., Dixon R.A. (2005). Targeted down-regulation of cytochrome P450 enzymes for forage quality improvement in alfalfa (Medicago sativa L.). Proc. Natl. Acad. Sci. USA 102: 16573–16578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rencoret J., Ralph J., Marques G., Gutiérrez A., Martínez Á., del Río J.C. (2013). Structural characterization of lignin isolated from coconut (Cocos nucifera) coir fibers. J. Agric. Food Chem. 61: 2434–2445. [DOI] [PubMed] [Google Scholar]
- Rohde A., et al. (2004). Molecular phenotyping of the pal1 and pal2 mutants of Arabidopsis thaliana reveals far-reaching consequences on phenylpropanoid, amino acid, and carbohydrate metabolism. Plant Cell 16: 2749–2771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saathoff A.J., Sarath G., Chow E.K., Dien B.S., Tobias C.M. (2011). Downregulation of cinnamyl-alcohol dehydrogenase in switchgrass by RNA silencing results in enhanced glucose release after cellulase treatment. PLoS One 6: e16416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saballos A., Ejeta G., Sanchez E., Kang C., Vermerris W. (2009). A genomewide analysis of the cinnamyl alcohol dehydrogenase family in sorghum [Sorghum bicolor (L.) Moench] identifies SbCAD2 as the brown midrib6 gene. Genetics 181: 783–795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schilmiller A.L., Stout J., Weng J.K., Humphreys J., Ruegger M.O., Chapple C. (2009). Mutations in the cinnamate 4-hydroxylase gene impact metabolism, growth and development in Arabidopsis. Plant J. 60: 771–782. [DOI] [PubMed] [Google Scholar]
- Sederoff R.R., MacKay J.J., Ralph J., Hatfield R.D. (1999). Unexpected variation in lignin. Curr. Opin. Plant Biol. 2: 145–152. [DOI] [PubMed] [Google Scholar]
- Shadle G., Chen F., Srinivasa Reddy M.S., Jackson L., Nakashima J., Dixon R.A. (2007). Down-regulation of hydroxycinnamoyl CoA:shikimate hydroxycinnamoyl transferase in transgenic alfalfa affects lignification, development and forage quality. Phytochemistry 68: 1521–1529. [DOI] [PubMed] [Google Scholar]
- Sibout R., Eudes A., Mouille G., Pollet B., Lapierre C., Jouanin L., Séguin A. (2005). CINNAMYL ALCOHOL DEHYDROGENASE-C and -D are the primary genes involved in lignin biosynthesis in the floral stem of Arabidopsis. Plant Cell 17: 2059–2076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sibout R., Eudes A., Pollet B., Goujon T., Mila I., Granier F., Séguin A., Lapierre C., Jouanin L. (2003). Expression pattern of two paralogs encoding cinnamyl alcohol dehydrogenases in Arabidopsis. Isolation and characterization of the corresponding mutants. Plant Physiol. 132: 848–860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sirisha V.L., Prashant S., Kumar D.R., Pramod S., Jalaja N., Kumari P.H., Rao P.M., Rao S.N., Mishra P., Karumanchi S.R., Khan B.M., Kishor P.B.K. (2012). Cloning, characterization and impact of up- and down-regulating subabul cinnamyl alcohol dehydrogenase (CAD) gene on plant growth and lignin profiles in transgenic tobacco. Plant Growth Regul. 66: 239–253. [Google Scholar]
- Somerville C., Youngs H., Taylor C., Davis S.C., Long S.P. (2010). Feedstocks for lignocellulosic biofuels. Science 329: 790–792. [DOI] [PubMed] [Google Scholar]
- Stewart J.J., Akiyama T., Chapple C., Ralph J., Mansfield S.D. (2009). The effects on lignin structure of overexpression of ferulate 5-hydroxylase in hybrid poplar. Plant Physiol. 150: 621–635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stout J., Romero-Severson E., Ruegger M.O., Chapple C. (2008). Semidominant mutations in reduced epidermal fluorescence 4 reduce phenylpropanoid content in Arabidopsis. Genetics 178: 2237–2251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Studer M.H., DeMartini J.D., Davis M.F., Sykes R.W., Davison B., Keller M., Tuskan G.A., Wyman C.E. (2011). Lignin content in natural Populus variants affects sugar release. Proc. Natl. Acad. Sci. USA 108: 6300–6305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamagnone L., Merida A., Parr A., Mackay S., Culianez-Macia F.A., Roberts K., Martin C. (1998a). The AmMYB308 and AmMYB330 transcription factors from antirrhinum regulate phenylpropanoid and lignin biosynthesis in transgenic tobacco. Plant Cell 10: 135–154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamagnone L., Merida A., Stacey N., Plaskitt K., Parr A., Chang C.F., Lynn D., Dow J.M., Roberts K., Martin C. (1998b). Inhibition of phenolic acid metabolism results in precocious cell death and altered cell morphology in leaves of transgenic tobacco plants. Plant Cell 10: 1801–1816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuji Y., Chen F., Yasuda S., Fukushima K. (2004). The behavior of deuterium-labeled monolignol and monolignol glucosides in lignin biosynthesis in angiosperms. J. Agric. Food Chem. 52: 131–134. [DOI] [PubMed] [Google Scholar]
- Vailhé M.A.B., Besle J.M., Maillot M.P., Cornu A., Halpin C., Knight M. (1998). Effect of down-regulation of cinnamyl alcohol dehydrogenase on cell wall composition and on degradability of tobacco stems. J. Sci. Food Agric. 76: 505–514. [Google Scholar]
- Van Acker R., Vanholme R., Storme V., Mortimer J.C., Dupree P., Boerjan W. (2013). Lignin biosynthesis perturbations affect secondary cell wall composition and saccharification yield in Arabidopsis thaliana. Biotechnol. Biofuels 6: 46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Acker R., et al. (2014). Improved saccharification and ethanol yield from field-grown transgenic poplar deficient in cinnamoyl-CoA reductase. Proc. Natl. Acad. Sci. USA 111: 845–850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanholme R., Demedts B., Morreel K., Ralph J., Boerjan W. (2010a). Lignin biosynthesis and structure. Plant Physiol. 153: 895–905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanholme R., Morreel K., Darrah C., Oyarce P., Grabber J.H., Ralph J., Boerjan W. (2012). Metabolic engineering of novel lignin in biomass crops. New Phytol. 196: 978–1000. [DOI] [PubMed] [Google Scholar]
- Vanholme R., Morreel K., Ralph J., Boerjan W. (2008). Lignin engineering. Curr. Opin. Plant Biol. 11: 278–285. [DOI] [PubMed] [Google Scholar]
- Vanholme R., et al. (2010b). Engineering traditional monolignols out of lignin by concomitant up-regulation of F5H1 and down-regulation of COMT in Arabidopsis. Plant J. 64: 885–897. [DOI] [PubMed] [Google Scholar]
- Weng J.K., Mo H., Chapple C. (2010). Over-expression of F5H in COMT-deficient Arabidopsis leads to enrichment of an unusual lignin and disruption of pollen wall formation. Plant J. 64: 898–911. [DOI] [PubMed] [Google Scholar]
- Wilkerson C.G., Mansfield S.D., Lu F., Withers S., Park J.Y., Karlen S.D., Gonzales-Vigil E., Padmakshan D., Unda F., Rencoret J., Ralph J. (2014). Monolignol ferulate transferase introduces chemically labile linkages into the lignin backbone. Science 344: 90–93. [DOI] [PubMed] [Google Scholar]
- Zhang K., Qian Q., Huang Z., Wang Y., Li M., Hong L., Zeng D., Gu M., Chu C., Cheng Z. (2006). GOLD HULL AND INTERNODE2 encodes a primarily multifunctional cinnamyl-alcohol dehydrogenase in rice. Plant Physiol. 140: 972–983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Q., Tobimatsu Y., Zhou R., Pattathil S., Gallego-Giraldo L., Fu C., Jackson L.A., Hahn M.G., Kim H., Chen F., Ralph J., Dixon R.A. (2013). Loss of function of cinnamyl alcohol dehydrogenase 1 leads to unconventional lignin and a temperature-sensitive growth defect in Medicago truncatula. Proc. Natl. Acad. Sci. USA 110: 13660–13665. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.