A unifying peptide classification system is proposed, reflecting the huge diversity of plant peptides.
Abstract
Peptides fulfill a plethora of functions in plant growth, development, and stress responses. They act as key components of cell-to-cell communication, interfere with signaling and response pathways, or display antimicrobial activity. Strikingly, both the diversity and amount of plant peptides have been largely underestimated. Most characterized plant peptides to date acting as small signaling peptides or antimicrobial peptides are derived from nonfunctional precursor proteins. However, evidence is emerging on peptides derived from a functional protein, directly translated from small open reading frames (without the involvement of a precursor) or even encoded by primary transcripts of microRNAs. These novel types of peptides further add to the complexity of the plant peptidome, even though their number is still limited and functional characterization as well as translational evidence are often controversial. Here, we provide a comprehensive overview of the reported types of plant peptides, including their described functional and structural properties. We propose a novel, unifying peptide classification system to emphasize the enormous diversity in peptide synthesis and consequent complexity of the still expanding knowledge on the plant peptidome.
INTRODUCTION
Peptides (from the Greek πεπτος meaning “digestive”) are the smallest biological molecules of the plant proteome, often arbitrarily restricted to proteins of 2 to 100 amino acids. They fulfill diverse roles in plant growth, development, reproduction, symbiotic interactions, and stress responses (Albert, 2013; Czyzewicz et al., 2013; Matsubayashi, 2014). On the one hand, peptides can interact directly with pathogens through their antimicrobial properties (Goyal and Mattoo, 2014). On the other hand, they function by interfering with signaling cascades or by representing important messages in cell-to-cell communication (Murphy et al., 2012; Tintor et al., 2013; Uchida and Tasaka, 2013; Araya et al., 2014; Costa et al., 2014; Farkas et al., 2014; Haruta et al., 2014; Qu et al., 2015). Despite many studies on their occurrence in various plant species and involvement in different plant processes, peptides are still often overlooked in gene predictions and mass spectrometric analyses (Yang et al., 2011; Guillén et al., 2013; Niarchou et al., 2013). As a result, the total number of peptides in plants as well as the diversity of various peptide types is underestimated (Silverstein et al., 2007; De Coninck et al., 2013a). Here, we review the reported types of peptides starting from a more adapted classification to emphasize the enormous diversity in peptide synthesis, aiming at boosting research on the “shortest” members of the plant proteome.
A NEW, COMPREHENSIVE, AND UNIFYING PEPTIDE CLASSIFICATION SYSTEM
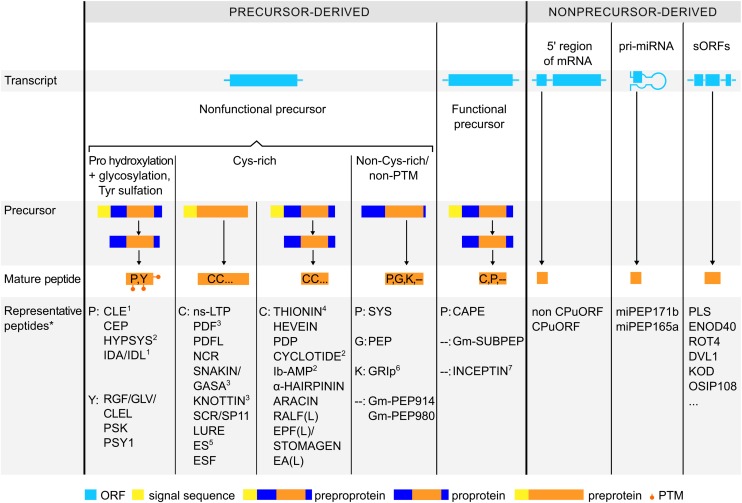
Peptides are often referred to as proteins smaller than 100 amino acids. Although this length restriction is rather artificial, in this Review, we maintain this upper limit and, moreover, only discuss peptides that are ribosomally synthesized. Up to now, so-called secreted small signaling peptides and antimicrobial peptides (AMPs) have mainly been studied (Goyal and Mattoo, 2014; Matsubayashi, 2014). Although they strikingly differ in amino acid sequence, length, 3D structure, and function, they are all derived from nonfunctional precursor proteins (Figure 1). The latter can be a preprotein that results in the mature peptide upon removal of an N-terminal signal sequence (NSS) (also referred to as N-terminal signal peptide), which directs the precursor via the endoplasmic reticulum to the secretory pathway. Alternatively, such a precursor can be a proprotein that is enzymatically modified to the mature peptide and is further termed preproprotein when also harboring an NSS.
Figure 1.
The Diversity of Plant Peptide Synthesis.
Plant peptides are synthesized from precursor proteins or directly translated from sORFs embedded in transcripts. The former type of peptides is derived from nonfunctional precursors or functional precursors. Such a precursor can be a preprotein that results in the mature peptide upon removal of an NSS (yellow rectangle). Alternatively, such a precursor can be a proprotein that contains a prodomain (blue rectangle) and is enzymatically modified to the mature peptide (orange rectangle). If the proprotein also harbors an NSS, it is termed preproprotein. So far, peptides derived from nonfunctional precursors can be posttranslationally modified (PTM), Cys-rich, or non-Cys-rich and non-posttranslationally modified (non-Cys-rich/non-PTM). Representative peptides of these three subgroups are classified by their dominant residues where appropriate (*), though sometimes a single dominant amino acid cannot be identified (‐‐). Nonprecursor-derived peptides are encoded by sORFs (<100 amino acids) that are located (1) upstream of the main ORF in the 5′ leader sequence of a gene, (2) in primary transcripts of miRNA (pri-miRNAs), or (3) in other transcripts not encoding longer (>100 amino acids) proteins. Most peptides in this group follow the presented scheme, but some exceptions may arise, including 1some members do not contain an NSS, 2multiple mature peptides are released from one single precursor, 3some members can contain a prodomain, 4an unprocessed THIONIN of 15 kD was reported, 5an internal signal sequence is present instead of an NSS, 6an NSS is present and proteolytic cleavage is executed by a metacaspase, and 7proteolytic cleavage is executed by herbivorous insect processing machinery.
Despite the majority of reported plant peptides being derived from such nonfunctional precursors, different studies also point to the existence of peptides originating from a functional protein but with an activity differing from that of the precursor (Schmelz et al., 2006; Pearce et al., 2010a; Chen et al., 2014) (Figure 1). This phenomenon raises the question whether many other well-known protein-encoding genes could have a double function, both as parent protein and as derived peptide. In addition, an increasing number of recent reports indicate that peptides can also be translated directly from a short open reading frame (sORF) (<100 codons) that is present in the 5′ leader sequence of an mRNA, in primary transcripts of microRNAs (miRNAs) or in other transcripts not encoding longer (>100 amino acids) proteins (Hanada et al., 2013; von Arnim et al., 2014; Lauressergues et al., 2015) (Figure 1). No precursor protein appears to be involved in their synthesis, but their maturation process has not yet been investigated in detail. It was shown that this type of peptides can influence plant morphogenesis or has a regulatory function (Hanada et al., 2013; von Arnim et al., 2014; Lauressergues et al., 2015). Additionally, one must note that features of the transcripts encoding this type of peptides overlap with long and short noncoding RNA transcripts, as they also contain one or more sORFs (Dinger et al., 2008). This controversial phenomenon leads to questioning the distinctive border between peptide/protein-coding and noncoding transcripts.
The above-mentioned examples of novel types of peptides could be added to the short list of exceptions, but we rather expect that they represent a glimpse of completely new classes of peptides that are underexplored so far. We perceive that the plant peptidome as a whole is composed of peptides that are (1) derived from nonfunctional precursors, (2) derived from functional precursors, or (3) not derived from a precursor protein (Figure 1). In the following sections, we will further discuss these groups and illustrate them and some key features with representative examples.
PEPTIDES DERIVED FROM A NONFUNCTIONAL PRECURSOR
Most peptides identified up to now are derived from a longer precursor that has no known biological function as a preprotein, proprotein, or preproprotein. Based on specific characteristics, peptides within this group can be divided into three subgroups: (1) peptides characterized by specific posttranslational modifications, such as Pro hydroxylation, Pro glycosylation, and Tyr sulfation; (2) Cys-rich peptides; and (3) non-Cys-rich peptides without posttranslational modifications (Figure 1) (Matsubayashi, 2011a; Meng et al., 2012a; Murphy et al., 2012). Peptides within this group carry specific amino acid residues with dominant roles, such as Cys, Pro, Tyr, Gly, or Lys, because these are often essential for the peptide’s activity.
Peptides with Specific Posttranslational Modifications
Posttranslationally modified (PTM) peptides typically consist of a maximum of 20 amino acids, have few or no Cys residues, contain modifications on their Pro and Tyr residues, and are released from preprotein precursors (Figure 1). Many peptide families involved in plant development, defense, cell identity, and cell-to-cell communication belong to this subgroup (Matsubayashi, 2011b). These peptides function as signaling molecules and are perceived by specific receptors. While progress has been made on molecular and physiological events downstream of PTM peptides, detailed modes of action are generally not known. In some cases, it has, for example, been demonstrated that PTM peptides can signal through a MITOGEN-ACTIVATED PROTEIN KINASE cascade, as such regulating transcription factor activity and/or other downstream events, that their activity ultimately leads to altered transcription factor expression patterns, that they affect apoplastic pH regulation, that they mediate the inhibition of proton transport, that they act as a chemo-attractant, etc. (Sparks et al., 2013; Haruta et al., 2014; Tabata et al., 2014; Butenko and Simon, 2015). To illustrate this subgroup of peptides, we selected some well-studied representatives (Figures 2 and 3) and refer to excellent, recent reviews for additional information (Fernandez et al., 2013b; Endo et al., 2014; Marmiroli and Maestri, 2014; Matsubayashi, 2014; Grienenberger and Fletcher, 2015; Sauter, 2015).
Figure 2.
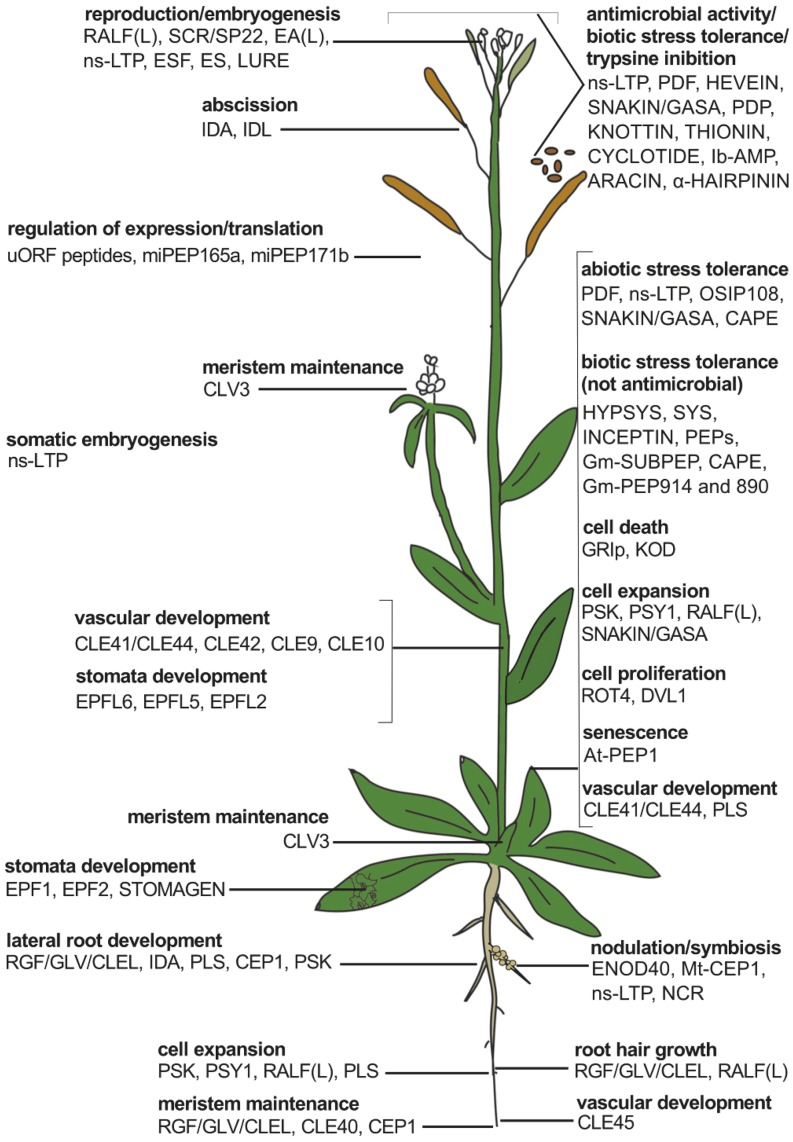
The Functional Diversity of Plant Peptides.
Plant peptides have a wide range of biological roles and act in different plant parts, as indicated in an illustration of a simplified plant (adapted from Czyzewicz et al. [2013]).
Figure 3.
Key Features of All Peptide Types Based on Representative Examples.
Peptides derived from nonfunctional precursors can be classified in three subgroups: (1) PTM peptides, (2) Cys-rich peptides, and (3) non-Cys-rich/non-PTM peptides. PTM peptides are characterized by specific posttranslational modifications, such as Pro hydroxylation (orange), glycosylation (asterisk), and Tyr sulfation (blue). Cys-rich peptides carry at least two Cys residues to form stable disulfide bridges (square brackets). Non-Cys-rich/non-PTM peptides are not characterized by specific PTM or two or more Cys. Peptides can also be formed from functional precursors or directly translated from sORFs (<100 amino acids) in case no protein precursor is involved in the maturation process. To illustrate the features of all peptide types, the amino acid sequence of a representative mature peptide and the main characteristics of its 3D structure are shown. Additionally, dominant residues (bold), such as Pro, Gly, Lys, Cys, or Tyr, and conserved motifs (underlined) are indicated in the amino acid sequences of these representative peptides. Arabidopsis thaliana (At), Glycine max (Gm), Helianthus annuus (Ha), Hevea brasiliensis (Hb), Impatiens balsamina (Ib), Mirabilis jalapa (Mj), Medicago truncatula (Mt), Oryza sativa (Os); Petunia hybrida (phyb), SUCROSE CONTROL-peptide (SC-peptide); Solanum lycopersicum (Sl), Stellaria media (Sm), Solanum tuberosum (St), Torenia fournieri (Tf), Vigna unguiculata (Vu), Zea mays (Zm). nd, not determined.
In the context of this Review, we focus on specific posttranslational modifications, such as Pro hydroxylation (ProHyp), ProHyp glycosylation (mainly arabinosylation), and Tyr sulfation (Matsubayashi, 2011a), which are often important for maturation of these peptides and play a role in peptide stability, activity, and interactions with their receptor (Ogawa et al., 2008; Matsubayashi, 2011a; Okamoto et al., 2013; Tabata and Sawa, 2014). We do not consider other posttranslational modifications, such as proteolytic cleavage or folding, as listed more extensively by Wang (2012).
Pro hydroxylation is catalyzed by PROLYL-4-HYDROXYLASE, a type II membrane protein with an N-terminal transmembrane domain that is localized in the endoplasmic reticulum and Golgi apparatus (Gorres and Raines, 2010). Hydroxylation as such is not always important for the conformation and bioactivity of peptides, but can function as a necessary bridge for the posttranslational arabinosylation (Ito et al., 2006; Kondo et al., 2008). ProHyp arabinosylation is catalyzed by HYDROXYPROLINE O-ARABINOSYLTRANSFERASE, a Golgi-localized transmembrane protein. This enzyme transfers l-arabinose to the hydroxyl group of ProHyp residues (Amano et al., 2007; Ohyama et al., 2009; Ogawa-Ohnishi et al., 2013; Ogawa-Ohnishi and Matsubayashi, 2015). ProHyp arabinosylation is important for maintenance of proper peptide structure and as a consequence affects peptide activity (Amano et al., 2007; Ohyama et al., 2009; Shinohara and Matsubayashi, 2013; Mohd-Radzman et al., 2015; C. Xu et al., 2015). Tyr sulfation is mediated by the Golgi-localized TYROSYLPROTEIN SULFOTRANSFERASE, which catalyzes the transfer of a sulfonate moiety from 3′-phosphoadenosine-5′-phosphosulfate to the hydroxyl group of a protein-bound Tyr residue to form a Tyr O-sulfate ester and 3′-phosphoadenosine-5′-phosphate (Moore, 2003). Tyr sulfation possibly regulates biological activity, proteolytic processing of bioactive peptides, and/or extracellular protein-protein interactions (Kehoe and Bertozzi, 2000; Matsubayashi, 2011a).
A well-studied example of such PTM peptides is the CLAVATA3 (CLV3)/EMBRYO SURROUNDING REGION (CLE) family. Mature CLE consist of 12 to 13 amino acids with one to three highly conserved Pro residues, which are part of the conserved CLE motif located at or near the C terminus of the preproprotein (Cock and McCormick, 2001) (Figure 3). The fourth and seventh Pro residues of these CLE peptides are often hydroxylated and the seventh ProHyp residue is also modified with three residues of l-arabinose (Ohyama et al., 2009; Matsubayashi, 2011b; Shinohara and Matsubayashi, 2013). Ala substitutions of CLV3 (Kondo et al., 2008) and experiments with clv3 mutants (Ohyama et al., 2009; Shinohara and Matsubayashi, 2013) indicated that hydroxylation and arabinosylation, more specifically arabinose chain length and its influence on peptide conformation, are essential for the biological activity of CLV3. In Arabidopsis thaliana, the CLV3 peptide acts as a regulator of the shoot apical meristem by maintaining the stem cell population within the shoot apical meristem (Clark et al., 1995; Schoof et al., 2000). Since the discovery of CLV3 (Clark et al., 1995; Ito et al., 2006; Kondo et al., 2006; Ohyama et al., 2009), (putative) CLE genes have been identified in dicots, monocots, mosses, and algae (Jun et al., 2008; Oelkers et al., 2008; Miwa et al., 2009; Zhang et al., 2014; Strabala et al., 2014), and they were shown to act on diverse developmental processes, including shoot and root meristem development, nodulation, embryo and endosperm development, regulation of root architecture in response to nutrients, and vascular development (Hobe et al., 2003; Fiers et al., 2005; Hirakawa et al., 2008; Stahl et al., 2009; Jun et al., 2010; Mortier et al., 2010; Lim et al., 2011; Reid et al., 2011; Fiume and Fletcher, 2012; Okamoto et al., 2013; Araya et al., 2014; Richards et al., 2015; T.-T. Xu et al., 2015). Strikingly, plant-parasitic cyst nematodes also secrete CLE-like effector proteins that are required for successful nematode infection (Wang et al., 2010, 2011; Replogle et al., 2011; Miyawaki et al., 2013). 3D structure modeling demonstrated that CLV3, CLE1, CLE2, and CLE6 shared a similar arch-shaped molecular structure with conserved residues at the C terminus and middle region, suggesting that these regions could be involved in receptor selection and binding (Meng and Feldman, 2010). Within the CLE family, the peptide-receptor relationships are diverse, since one receptor kinase can serve as the receptor for different peptides, but on the other hand, one peptide can be recognized by different receptors. To illustrate the former, CLV1 is a likely receptor for the CLV3 peptide during shoot meristem maintenance (Ogawa et al., 2008), for the CLE40 peptide during root meristem maintenance (Hobe et al., 2003) and for the CLE3 peptide during regulation of lateral root formation (Araya et al., 2014). To illustrate the latter, the CLE9 peptide interacts with the BARLEY ANY MERISTEM1 (BAM1) receptor, BAM2, BAM3, and CLV1, although with different affinity (Shinohara et al., 2012).
The C-TERMINALLY ENCODED PEPTIDE (CEP) family gives rise to peptides of 15 amino acids with two to four Pro residues that may be hydroxylated, thereby affecting biological activity (Ohyama et al., 2008; Imin et al., 2013; Roberts et al., 2013; Ogilvie et al., 2014) (Figure 3). In silico analysis showed that CEP-containing genes occur only in seed plants and are absent in mosses and algae (Ohyama et al., 2008; Delay et al., 2013; Imin et al., 2013; Roberts et al., 2013; Li et al., 2014; Ogilvie et al., 2014). Overexpression of CEP1 in Arabidopsis results in the arrest of root growth (Ohyama et al., 2008), and, in Medicago truncatula, overexpression of CEP1 or application of synthetic CEP1 results in inhibition of lateral root formation, enhancement of nodulation, and the induction of periodic circumferential root swellings (Imin et al., 2013; Mohd-Radzman et al., 2015). In addition, altering the levels of CEP5 affects aboveground Arabidopsis growth and development (Roberts et al., 2013). Structural analysis showed that Mt-CEP1 contains a β-turn-like conformation and that ProHyp increases conformational plasticity and alters surface area of the CEP peptide, which plays an important role in efficient physical interaction between peptide and receptor (Bobay et al., 2013). Remarkably, in an Mt-CEP1 overexpression line, a vast majority of hydroxylated peptides was isolated, as well as only a minority of triarabinosylated peptides (Mohd-Radzman et al., 2015). Recently, two receptors for several members of the CEP family, CEP RECEPTOR1 (CEPR1) and CEPR2, were described in Arabidopsis (Tabata et al., 2014). Interestingly, plant-parasitic root-knot nematodes, such as Meloidogyne hapla, secrete peptides with high sequence similarity to CEPs (Bobay et al., 2013), and these nematode peptides possibly play a role in the formation of the novel plant cell types from which nematodes feed.
HYDROXYPROLINE-RICH GLYCOPEPTIDE SYSTEMINS (HYPSYS) peptides are 15- to 20-amino acid glycopeptides with four to seven Pro residues possibly hydroxylated that are mainly found in members of the Solanaceae family (Pearce et al., 2001a, 2009; Pearce and Ryan, 2003; Chen et al., 2008; Bhattacharya et al., 2013) (Figure 3). Mature HypSys peptides are formed from large preproproteins. Remarkably, one single protein precursor contains multiple mature HYPSYS peptides. The latter contain either a characteristic ProHypProHypXProHyp-motif (where X is Ala, Ser, Thr, or ProHyp) or a slightly different ProHypXProHypProHyp motif (where X is Thr or Ser) in the case of potato (Solanum tuberosum) HYPSYS I and black nightshade (Solanum nigrum) HYPSYS I (Chen et al., 2008; Bhattacharya et al., 2013). Mass spectrometry analysis of HYPSYS isolated from tomato (Solanum lycopersicum) leaf extracts revealed the presence of 3 to 17 pentose residues attached to the ProHyp residues. Chemically synthesized tobacco (Nicotiana tabacum) HYPSYS I and II, both devoid of pentose residues, were found to be 10,000 times less active than native peptides in an alkalinization assay (Pearce and Ryan, 2003). In general, HYPSYS peptides adopt a polyproline helix stabilized by pentose appendages attached to ProHyp (Taylor et al., 2012). HYPSYS has a role in defense against insect herbivores, activates protease inhibitors, and can, for example, enhance the resistance against Helicoverpa armigera larvae when overexpressed in tobacco (Ren and Lu, 2006; Pearce, 2011).
The mature peptide structure of INFLORESCENCE DEFICIENT IN ABSCISSION (IDA) and IDA-LIKE (IDL) peptides has not yet been determined, but they are believed to be derived from a preproprotein carrying a C-terminal conserved motif (EPIP) (Butenko et al., 2003). The EPIP motif (FGYLPKGVPIPPSAPSKRHN) of Arabidopsis IDA and IDL1 can replace or partially substitute IDA function in vivo, and synthetic IDA and IDL1 EPIP peptides rescue ida mutants and induce early floral abscission in wild-type flowers (Stenvik et al., 2008). In addition to fulfilling a role in (floral) abscission, IDA also regulates cell separation during lateral root emergence (Butenko et al., 2003; Stenvik et al., 2006; Kumpf et al., 2013). Several IDL transcripts were predicted in dicots and monocots (Butenko et al., 2003) and the EPIP domain, which is well conserved between the IDL peptides of Arabidopsis, is also present in IDA and IDL sequences of bean (Phaseolus vulgaris), tomato, and soybean (Glycine max) (Tucker and Yang, 2012). IDA/IDL peptides act through the leucine-rich repeat receptor-like kinases HAESA (HAE) and HAE-LIKE2 (HSL2) (Stenvik et al., 2008; Kumpf et al., 2013; Butenko et al., 2014). It has been shown that the PIP motif (PIPPSAPSKRHN) within the EPIP domain is significantly more effective for HSL2 than EPIP and induces an oxidative burst (used as a marker for receptor activation). However, no response to the PIP peptide was observed for the HAE receptor (Butenko et al., 2014). Substitution of ProHyp for Pro-1, Pro-3, Pro-7, but not Pro-4, revealed the importance of hydroxylation in the PIP peptide (Butenko et al., 2014) (Figure 3).
ROOT GROWTH FACTOR/CLE-LIKE/GOLVEN (RGF/CLEL/GLV) peptides are 13 to 18 amino acids long and contain ProHyp residues as well as sulfated Tyr residues (Matsuzaki et al., 2010; Meng et al., 2012b; Whitford et al., 2012) (Figure 3). The RGF/CLEL/GLV peptide family was discovered in Arabidopsis and is conserved throughout different plants, such as rice (Oryza sativa), quaking aspen (Populus tremuloides), and conifers (Whitford et al., 2012; Strabala et al., 2014). Mature RGF/CLEL/GLV peptides are produced by proteolytic cleavage of a preproprotein with a C-terminal conserved motif (Matsuzaki et al., 2010; Meng et al., 2012a; Whitford et al., 2012). To date, the RGF/CLEL/GLV family contains 12 members in Arabidopsis, including CLE18, previously classified as a member of the CLE peptide family (Meng et al., 2012b). These peptides have a role in root gravitropism, root apical meristem maintenance, root hair development, and lateral root and shoot development (Matsuzaki et al., 2010; Whitford et al., 2012; Fernandez et al., 2013a). So far, receptors for this peptide family are unknown. Intriguingly, the putative Arabidopsis CLEL protein in the nucleus (CLELn) is possibly processed to a peptide that functions in intracellular signaling in plants, but this still requires further study (Meng et al., 2012a).
PHYTOSULFOKINE (PSK) is a 5-amino acid peptide containing two sulfated Tyr residues that has been detected in conditioned medium of plant cell cultures (Matsubayashi and Sakagami, 1996) (Figure 3). PSK is produced from a preprotein with a conserved characteristic motif in the C terminus, namely, YIYTQ in Arabidopsis and YIYSQ in rice (Lorbiecke and Sauter, 2002). PSK homologs are present in both monocots and dicots as small gene families (Lorbiecke and Sauter, 2002). In Arabidopsis, exogenous application of PSK caused enhanced root growth that was mainly a result of root cell elongation (Kutschmar et al., 2009). Posttranslational Tyr sulfation of PSK is required for its activity (Matsubayashi and Sakagami, 1996). The sulfate group of Tyr-1 is more important for PSK activity than that of Tyr-3 (Matsubayashi et al., 1996). PSK acts through PHYTOSULFOKINE RECEPTORs (PSKRs) with kinase activity, and these were identified in Arabidopsis (At-PSKR2 and At-PSKR1) and in carrot (Daucus carota) (Dc-PSKR1) (Matsubayashi et al., 2006). Overexpression of Dc-PSKR1 caused enhanced callus growth in response to PSK (Matsubayashi et al., 2002).
PLANT PEPTIDE CONTAINING SULFATED TYROSINE1 (PSY1) is an 18-amino acid glycopeptide containing two ProHyp residues and one sulfated Tyr residue (Amano et al., 2007) (Figure 3). This peptide is glycosylated with three l-arabinose units attached to the ProHyp-16 residue (Amano et al., 2007). In addition, as for PSK, Tyr sulfation is required for PSY1 activity (Matsubayashi and Sakagami, 1996). PSY1, as well as PSK, significantly promotes cellular proliferation and expansion at nanomolar concentrations. The Arabidopsis genome contains two genes encoding PSY1 precursor homologs with significant similarity within the conserved PSY1 domain and flanked by basic amino acid residues possibly involved in proteolytic processing (Amano et al., 2007). PSY RECEPTOR1 (PSYR1) was identified as a receptor for PSY, and recent studies revealed that both PSKR1 and PSY1R are involved in the plant defense response by regulating salicylate- and jasmonate-dependent defense pathways (Matsubayashi et al., 2002; Mosher and Kemmerling, 2013).
Cysteine-Rich Peptides
A second family of preprotein- or preproprotein-derived peptides are the Cys-rich peptides, characterized by a Cys-rich domain with 2 to 16 Cys residues, but considerably varying in both their length and primary sequences between peptide families and across plant species (Figures 1 and 3). Each Cys-rich peptide class has a characteristic number and linear arrangement of Cys residues. The latter form intramolecular disulfide bonds that are essential for proper class-specific secondary folding and activity (Marshall et al., 2011; Haag et al., 2012; Vriens et al., 2014). The majority of known Cys-rich plant peptides are thought to function as AMPs during plant-microbe interactions, and they have been isolated from roots, leaves, stems, flowers, and seeds (Nawrot et al., 2014). Most AMPs are cationic, allowing them to interact with the negatively charged membranes of pathogens. However, anionic antimicrobial peptides, belonging to known AMP families (e.g., hevein and cyclotides) have been reported (Prabhu et al., 2013). Additionally, it has been shown that several Cys-rich peptides also have a role in plant development, pollen recognition and guidance, and seed development (Marshall et al., 2011; Qu et al., 2015) (Figure 2). In this section, we highlight some typical representatives of those Cys peptides, but refer to excellent recent reviews for a more complete overview on Cys-rich peptides (Marshall et al., 2011; Beale and Johnson, 2013; Richardson and Torii, 2013; van der Weerden et al., 2013; de Souza Cândido et al., 2014; Murphy and De Smet, 2014; Nawrot et al., 2014).
Probably the best studied Cys-rich peptides are PLANT DEFENSINs (PDFs) (Terras et al., 1995). This family of ∼5-kD peptides is characterized by an α-helix and a triple-stranded β-sheet, stabilized by four disulfide bonds (Figure 3). Though PDFs are derived from a preprotein, there are some reports on PDFs derived from preproproteins containing a C-terminal prodomain necessary for vacuolar targeting and preventing phytotoxicity (De Coninck et al., 2013b; Lay et al., 2014). PDFs are widely distributed over monocots and dicots and belong to the pathogenesis-related protein family 12 (PR12 family) (van Loon et al., 2006). They mainly have antifungal activity, although some of them have reported antibacterial activity. Heterologous overexpression of various PDFs leads to increased resistance of both model plants and crops against different fungi and bacteria (Carvalho Ade and Gomes, 2011; De Coninck et al., 2013b; Gaspar et al., 2014). The modes of action of several PDFs, including radish (Raphanus sativus) AFP2, Nicotiana alata D1, alfalfa (Medicago sativa) Def1, M. truncatula Def4, and pea (Pisum sativum) d1, have been well studied and point toward specific interactions with various fungal sphingolipids and phospholipids (Thevissen et al., 2004, 2012; Aerts et al., 2007; Lobo et al., 2007; Ramamoorthy et al., 2007; van der Weerden et al., 2008, 2010; Sagaram et al., 2011, 2013; Muñoz et al., 2014). Upon interaction, PDFs are either internalized by the fungal cell and interact with intracellular targets, or they stay outside the cell and induce cell death through induction of a signaling cascade (Vriens et al., 2014). As such, they represent a typical example of host defense peptides acting in a more targeted way than through the aspecific lipid bilayer peturbation classically proposed for AMPs (Wilmes et al., 2011). Other activities have also been reported for PDFs, including trypsin, α-amylase, and protein synthesis inhibition and blocking of sodium channels. Additionally, some PDFs appear to play a role in pollen recognition and abiotic stress tolerance and inhibit root growth (van der Weerden and Anderson, 2013).
Initially, 15 PDFs were identified in Arabidopsis (Thomma et al., 2002), but motif model-based searches uncovered 317 defensin-like genes (DEFLs) in the Arabidopsis genome (Silverstein et al., 2007), 70% for which evidence for transcription has been provided (Tesfaye et al., 2013). Most Arabidopsis DEFLs have eight Cys but some contain only four to six Cys (Figure 3). This is also the case for a special group of DEFLs, i.e., the nodule-specific Cys-rich (NCR) peptides abundantly formed during the interaction between rhizobia soil bacteria and legumes. In M. truncatula, for example, ∼500 genes encode putative NCRs that are targeted to the nitrogen-fixing bacteroids and via a recent proteomics study, 138 NCR peptides could be detected (Figure 3). Interestingly, in contrast to most AMP family members, a majority of these peptides are anionic or neutral (Durgo et al., 2015), and they were suggested to neutralize the toxic activity of cationic NCRs. However, although cationic NCRs display in vitro antimicrobial activity, it is doubtful that they exert this activity in vivo, as peptide concentrations in nodules are lower than those used in in vitro assays (Van de Velde et al., 2010; Tiricz et al., 2013; Maróti and Kondorosi, 2014). Instead, they are essential for bacteroid differentiation and for maintenance of their bacterial endosymbionts (Van de Velde et al., 2010; Farkas et al., 2014; Penterman et al., 2014). Intriguingly, it seems that these NCRs interact with bacterial proteins (Farkas et al., 2014) and target intracellular regulatory pathways of the bacteroids (Penterman et al., 2014). These data, together with their specific expression in nodules and their lack of induced expression during pathogenic interactions, suggest that NCRs do not have a role as AMPs (Guefrachi et al., 2014).
Nonspecific LIPID TRANSFER PROTEINS (nsLTPs) (Kader, 1996) are grouped into two subfamilies, based on their molecular masses of ∼10 kD (LTP1) and ∼7 kD (LTP2) (Douliez et al., 2000). The LTP mature peptide, derived from a preprotein, shows a structure of four α-helices, stabilized by four disulfide bridges (Figure 3). This structure allows the formation of a hydrophobic cavity accommodating binding to lipids, hence, their reported in vitro ability to bind and transport various lipids (Carvalho Ade and Gomes, 2007; Tousheh et al., 2013). Almost 600 nsLTPs from dicots, monocots, and gymnosperms have been identified (Wang et al., 2012), but their mode of action is still poorly understood. Emphasis lies on their role in plant-microorganism interactions, although they have reported roles in cutin synthesis, somatic embryogenesis, reproduction, as well as responses to abiotic stress. nsLTPs, belonging to the PR14 family (van Loon et al., 2006), can have direct antimicrobial activity or play a role as signaling molecules during both pathogenic as well as symbiotic interactions (Terras et al., 1992; Cammue et al., 1995; Blein et al., 2002; Pii et al., 2012; Champigny et al., 2013; Nawrot et al., 2014).
THIONINs are peptides of ∼5 kD, with a common structure of two antiparallel α-helices and a double-stranded β-sheet, stabilized by three to four disulfide bonds (Figure 3). It is generally accepted that THIONINs are derived from a preproprotein containing an acidic C-terminal prodomain, hypothesized to neutralize the basic mature peptide (Stec, 2006). Recently, however, an unprocessed 15-kD THIONIN has also been detected in Arabidopsis (Asano et al., 2013). THIONINs are found in both monocots and dicots and are grouped within the PR13 family (van Loon et al., 2006). They display a broad range of toxicity against bacteria, fungi, yeast, insect larvae, and mammalian cells (Stec, 2006), and this is attributed to their interaction with anionic phospholipids leading to membrane permeabilization (Stec et al., 2004; Majewski and Stec, 2010). Moreover, overexpression of THIONIN genes in several plants led to decreased susceptibility against various bacteria, fungi as well as nematodes (Epple et al., 1997; Shirasawa-Seo et al., 2002; Muramoto et al., 2012; Ji et al., 2015).
KNOTTIN-type peptides of 3 to 4 kD are characterized by a triple-stranded β-sheet and three disulfide bonds, one of which crosses the macrocycle formed by the two other disulfide bridges (Figure 3). This knotted structure, also referred to as cystine knot, provides an extraordinary stability to peptides and has also been described for CYCLOTIDES (see below). KNOTTIN-type peptides, found in Mirabilis jalapa (Mj-AMP1 and Mj-AMP2) and Phytolacca americana (PAFP-S), are derived from a preprotein and display mainly antifungal activity (Cammue et al., 1992; Gao et al., 2001; de Souza Cândido et al., 2014). Interestingly, PA1b, a KNOTTIN from pea, is embedded in albumin PA1. Upon processing, this preproprotein yields two peptides, including PA1a, an uncharacterized peptide, and PA1b, displaying insecticidal activity (Chouabe et al., 2011).
A special type of knotted peptides is represented by the CYCLOTIDES, characterized by ∼30-amino acid peptides with a head-to-tail cyclized backbone resulting in ultrastable peptides (Figure 3). They have been detected in plants of the Rubiaceae, Violaceae, Fabaceae, Solanaceae, and Cucurbitaceae. CYCLOTIDES are embedded in preproproteins with a C-terminal prodomain and often containing several CYCLOTIDE peptides (Craik and Malik, 2013). A wide range of activities has been reported for CYCLOTIDES, including inhibitory activity on fungi, bacteria, viruses, insects, molluscs, barnacles, and nematodes, as well as inhibition of trypsin (Daly et al., 2009; Craik et al., 2010). Within the CYCLOTIDES, a special subclass is formed by the PawS-derived peptide (PDP) family. These cyclic Asteraceae peptides consist of 12 to 18 amino acids and two Cys residues (Elliott et al., 2014). PDPs are embedded in precursors for albumin seed storage protein named PawS (Mylne et al., 2011). The latter encodes a preproprotein comprising a central PDP domain and two C-terminal regions for the small and large subunit of albumin. Sunflower (Helianthus annuus) Trypsin Inhibitor 1 (SFTI-1) and Trypsin inhibitor 1-Like (SFT-L1) were the first members of this family to be isolated from sunflower seeds (Mylne et al., 2011). So far, only SFT-1 was shown to inhibit trypsin (Elliott et al., 2014).
SNAKIN/GIBBERELLIC ACID STIMULATED-LIKE (GASA) peptides are 6-kD peptides with 12 Cys residues, detected in monocots and dicots (Figure 3). The structure of the SNAKIN/GASA peptides has not yet been determined (Harris et al., 2014) but through a combination of ab initio and comparative molecular modeling and disulfide bond prediction, it was suggested that they consist of two long α-helices (Porto and Franco, 2013). The mature peptide is derived from a preprotein, in the case of S. tuberosum St-GSL1 (previously known as St-SN1), or a preproprotein, in the case of St-GSL2 (St-SN2) harboring a 15-amino acid region between the NSS and the mature St-GSL2. Both St-GSL peptides, as well as the alfalfa SNAKIN Ms-SN1, display in vitro broad antimicrobial activity (Segura et al., 1999; Berrocal-Lobo et al., 2002; García et al., 2014) and overexpression of their corresponding genes in several plants resulted in increased resistance against fungal and bacterial pathogens (Almasia et al., 2008; Rong et al., 2013; García et al., 2014). Additionally, SNAKIN/GASA peptides have been suggested to have an important role in plant development. For example, silencing of St-GSL1 resulted in plants with smaller leaves and affected cell division, metabolism, and cell wall composition of leaves (Nahirñak et al., 2012).
HEVEIN-like peptides are chitin binding peptides of ∼4 kD with 6 to 10 Cys residues and are derived from a preproprotein (Andreev et al., 2012; R Shukurov et al., 2012). The structure is represented by an α-helix and an antiparallel three- or four-stranded β-sheet (Andersen et al., 1993; Xiang et al., 2004; Dubovskii et al., 2011) (Figure 3). Up to now, only a few HEVEIN-like peptides have been detected in both monocots and dicots (Porto et al., 2012). Based on their chitin binding capacity, they were proposed to play a role in plant defense. Indeed, HEVEIN-like peptides display antimicrobial activity against various fungi (Broekaert et al., 1992; de Souza Cândido et al., 2014), although the exact mode of action has not yet been elucidated. Strikingly, some HEVEIN-like peptides also affect microorganisms without chitin in their cell wall, such as oomycetes and bacteria (de Souza Cândido et al., 2014). Overexpression of HEVEIN-like peptides resulted in increased plant resistance against various fungal and oomycete pathogens (Lee et al., 2003; R Shukurov et al., 2012).
Impatiens balsamina ANTIMICROBIAL PEPTIDES (Ib-AMPs), isolated from seeds (Tailor et al., 1997), represent four closely related basic peptides of 20 amino acid, each defined by two disulfide bonds which form a distinctive loop structure (Patel et al., 1998) (Figure 3). They display antifungal and antibacterial activity. Ib-AMP1 was shown to bind the cell surface and penetrate into the cell membrane of fungi (Lee et al., 1999), albeit not through interaction with proteinaceous receptors nor with glucosylceramides reported to act as specific binding sites for PDFs (Thevissen et al., 2005). The Ib-AMPs appear unique as no homologs proteins could be identified in protein databases. Moreover, Ib-AMPs are all embedded in a single preproprotein and are processed from a multipeptide precursor.
Recently, a novel class of ∼4-kD Cys-rich peptides, referred to as α-HAIRPININS, has been identified in seeds of several monocots and dicots (Oparin et al., 2012; Utkina et al., 2013; Slavokhotova et al., 2014). α-HAIRPININS are defined by four cysteines and two α-helices (Figure 3). They are derived from a preproprotein containing 5 to 12 α-HAIRPININS (Utkina et al., 2013; Slavokhotova et al., 2014). Various peptides display broad antimicrobial and/or trypsin-inhibitory activity (Oparin et al., 2012; Utkina et al., 2013; de Souza Cândido et al., 2014; Slavokhotova et al., 2014).
Finally, two novel cationic and hydrophobic peptides, named ARACINs, were recently identified in Arabidopsis and appear specific for the Brassicaceae family. The mature peptides of 39 to 40 amino acids are derived from a preproprotein and contain two Cys residues (Figure 3). Chemically synthesized ARACINs displayed in vitro antifungal activity while overexpression of the ARACIN1 precursor in Arabidopsis resulted in increased resistance against Botrytis cinerea and Alternaria brassicicola (Neukermans et al., 2015).
All above-mentioned Cys-rich peptides have been mainly studied for their antimicrobial activity, but based on the examples given, it seems that several of those peptides acquired novel functions in plant development. This phenomenon, called protein promiscuity, has been reported for several peptides in the plant defense response and postulated as being essential for peptide evolution (Franco, 2011). Although less abundantly documented, some Cys-rich peptide families lack reported antimicrobial activity, and those will be exemplified below.
RAPID ALKALINIZATION FACTOR (RALF) peptides of ∼5 kD, derived from the C-terminal end of a preproprotein, have been detected using mass spectrometry techniques on tobacco leaves (Pearce et al., 2001b). RALF and RALF-like peptides were found in Arabidopsis (Pearce et al., 2001b), rice (Cao and Shi, 2012), poplar (Populus tremula) (Haruta and Constabel, 2003), tomato (Pearce et al., 2001b), sugarcane (Saccharum officinarum) (Mingossi et al., 2010), M. truncatula (Combier et al., 2008), and maize (Zea mays) (Cao and Shi, 2012). RALF and RALF-like peptides mainly contain four Cys residues forming two disulfide bridges (Pearce et al., 2001b) (Figure 3). They are reportedly involved in plant development through arresting primary root growth, inhibiting root hair growth and cell elongation in hypocotyl and root cells, decreasing nodulation, and regulating pollen tube growth (Pearce et al., 2001b; Wu et al., 2007; Combier et al., 2008; Covey et al., 2010). Using a phosphoproteomics approach, FERONIA (FER), a member of the Catharanthus roseus RLK1-LIKE KINASE (Cr-RLK1L) family of receptors, could be identified as a RALF receptor (Haruta et al., 2014); subsequently, a model for the signaling transduction pathway was presented (Wolf and Höfte, 2014). For more information on the RALF peptide family, we refer to a recent review (Murphy and De Smet, 2014).
EPIDERMAL PATTERNING FACTOR (EPF) and EPF-LIKE (EPFL) peptides contain six or eight conserved Cys residues in the C-terminal mature peptide region (Figure 3). They are 45 to 76 amino acids long and are derived from a preproprotein. In Arabidopsis, the EPF family consists of two EPF peptides (EPF1 and EPF2) and nine EPF-like peptides (EPFL1 to EPFL9). EPF/EPFL genes are present in mosses, monocots, and dicots (Takata et al., 2013). Overexpression of EPF1, EPF2, EPFL4, and EPFL5 inhibits stomatal development when ectopically overexpressed (Hara et al., 2009). In contrast, overexpression of EPFL9 (STOMAGEN) results in increased stomatal density and clustering (Hunt et al., 2010). EPFL9 was recently shown to be involved in the light response of the stomatal pathway (Hronková et al., 2015). NMR-derived structural information showed that STOMAGEN represents two antiparallel β-strands connected by a loop (Ohki et al., 2011) (Figure 3). Additional 3D structure modeling revealed that EPF1/EPF2-LIKE (EPFL) peptides form a disulfide bond in the loop region absent in EPFL9/STOMAGEN-LIKE peptides (Takata et al., 2013). Recent studies demonstrated direct binding of EPF peptides to ERECTA family receptor kinases (Lee et al., 2012).
Based on their Cys arrangement and predicted Cys stabilized structure, several peptides, including S-LOCUS CYSTEINE-RICH PROTEIN/S-LOCUS PROTEIN11 (SCR/SP11), LURE, and maize EMBRYO SAC4 (ES4), have been classified as DEFLs. SCR/SP11 peptides, identified in Brassicaceae, consist of ∼50 amino acids with eight Cys residues linked by disulfide bridges and are derived from a preprotein. SCR/SP11 functions as the male determinant in the pollen coat and is perceived by the female determinant, the S-locus receptor kinase. Recognition leads to inhibition of self-pollen germination and pollen tube growth (Takayama et al., 2000, 2001; Higashiyama, 2010). LURE peptides are 60- to 70-amino acid peptides abundantly expressed in synergid cells (an egg-accompanying haploid cell) of the ornamental plant torenia (Torenia fournieri) and Arabidopsis. They have six conserved Cys residues and are derived from a preprotein (Figure 3). Tf-LUREs were shown to attract pollen tubes in vitro, while silencing of At-LURE1 resulted in impaired pollen tube guidance (Okuda et al., 2009; Takeuchi and Higashiyama, 2012). ES4 is abundantly expressed in the synergid cell and is involved in pollen tube growth arrest, burst, and explosive sperm release in maize (Amien et al., 2010). ES1-4 peptides have antifungal activity, since they inhibit the germination and growth of the maize pathogens Fusarium graminearum and Ustilago maydis (Woriedh et al., 2015). The ES family of maize encodes 61- to 68-amino acid peptides with eight Cys residues, and they are highly conserved with little variation at the C terminus. Secondary structure analysis of mature ES peptides predicts an α-helix and a triple-stranded antiparallel β-sheet (Cordts et al., 2001; Woriedh et al., 2015) (Figure 3).
EMBRYO SURROUNDING FACTOR1 (ESF1) peptides were recently discovered in Arabidopsis as regulators of suspensor development and auxin distribution in early developing embryos (Costa et al., 2014). Mature ESF1 peptide, containing 68 amino acids, is characterized by an α-helix and three β-sheets supported by four disulfide bonds. It was shown that ESF1 peptides protein structure was essential for its biological activity, as ESF1 peptide application induced suspensor cell elongation at nanomolar concentrations only when it was folded correctly (Costa et al., 2014). In addition, removal of any of the disulfide bonds or both tryptophan residues also resulted in the loss of peptide activity (Costa et al., 2014).
Maize EGG APPARATUS1 (EA1) is a 49-amino acid peptide with two Cys residues and is released from an Ala-rich precursor containing an internal signal sequence (Figure 3). This peptide is secreted from the egg apparatus to the micropylar region of the ovule integument, required for micropylar pollen tube guidance in maize, the last step of the pollen tube journey during the double fertilization process in flowering plants. EA1 attracts maize pollen tubes in vitro and arrests their growth at higher concentrations (Márton et al., 2012). In-depth in silico analysis of peptide sequence revealed that all peptides in this family shared a highly conserved C-terminal domain and the EA box (Zm-EA1-like motif) found in both monocot and dicot species (Gray-Mitsumune and Matton, 2006). The EA box is 27 to 29 amino acids in length with the conserved residues near the C terminus (Gray-Mitsumune and Matton, 2006). EAL transcripts were found in both reproductive and vegetative tissues (Gray-Mitsumune and Matton, 2006).
Non-Cysteine-Rich Peptides without Specific Posttranslational Modifications
A diverse group of plant peptides is not characterized by specific PTMs, such as Pro hydroxylation and glycosylation and/or Tyr sulfation, or the presence of two or more Cys, though we do not exclude the possibility that any of these PTMs could be detected in the future (Pearce et al., 1991; Huffaker et al., 2006; Yamaguchi et al., 2011; Wrzaczek et al., 2015). These “non-Cys-rich/non-PTM peptides” can contain functionally important amino acids in their primary structure such as Pro, Gly, and Lys and are mainly released from a proprotein (Figures 1 and 3) (Constabel et al., 1995; Huffaker et al., 2006; Yamaguchi et al., 2011; Wrzaczek et al., 2015). They are 8 to 36 amino acids in length and have reported roles mainly in the defense response of plants (Figure 2).
SYSTEMINS (SYS) are 18-amino acid peptides characterized by four to six Pro residues and a conserved C terminus (PPKMQTD) (Figure 3). They are specifically found in the Solanoideae subfamily of Solanaceae, including tomato, bell pepper (Capsicum anuum), potato, and black nightshade (S. nigrum) and are not posttranslationally modified in contrast to HYPSYS (Pearce et al., 1991; Constabel et al., 1998). SYS is a primary, mobile signal that is released into the vascular system of tomato plants at sites of herbivore attacks and further transported to distal tissues. SYS also induces the production of volatiles to attract natural enemies of insect herbivores and the synthesis of two wound-inducible proteinase inhibitor proteins (Pearce et al., 1991; Pearce, 2011). Ala-scan studies revealed that Pro-13 or Thr-17 of SYS are essential residues for the latter activity (Pearce et al., 1993). In addition to a role in defense against herbivores, it was demonstrated that SYS mediates the jasmonate signaling pathway required for resistance against the necrotrophic fungus B. cinerea in tomato (El Oirdi et al., 2011). Although a SYS receptor was not detected yet, SYS was found to adopt a poly(L-Pro) II type helix known to be important for receptor recognition events in animals (Toumadje and Johnson, 1995).
PLANT ELICITOR PEPTIDES (PEPs) are 23- to 36-amino acid peptides carrying Gly as dominant residue in their motif (S/G)(S)Gxx(G/P)xx(N) and are conserved across monocots and dicots (Figure 3). All PEPs reveal a strict conservation of Gly-17, and this residue as well as Ser-15 was proven to be important for the alkalinizing activity of At-PEPs (Pearce et al., 2008). PEPs are likely to function as enhancers of immunity, damage-signaling peptides, and elicitors of systemic defense responses against pathogens and/or herbivores (Huffaker et al., 2011, 2013; Albert, 2013; Bartels et al., 2013; Klauser et al., 2015). Recently, it was shown that At-PEPs also have a role in dark/starvation-induced senescence via an early induction of chlorophyll degradation and autophagy (Gully et al., 2015). Although PEPs are released from precursors without NSS, PEPs must be exported from the cells to interact with cell surface leucine-rich repeat receptor kinases PEP RECEPTOR1 (PEPR1) and PEPR2 (Krol et al., 2010; Yamaguchi et al., 2010). Recently, analysis of the crystal structure of the At-PEP1-PEPR1 complex revealed that At-PEP1 adopts a fully extended conformation and binds to the inner surface of the superhelical extracellular LRR domain of PEPR1 (Tang et al., 2015). The LRR domains of PEPRs coevolved with the PEPs, leading to distinct PEP motifs in each plant family and interfamily incompatibility of recognition, in contrast to the highly conserved downstream signaling (Lori et al., 2015). In general, the PEP immune signaling pathway is associated with an early cytosolic Ca2+ influx that activates protein kinases, reactive oxygen species, and nitrogen oxide (Ma et al., 2013). Moreover, this pathway coactivates the jasmonate- and salicylate-mediated immune branches to link local and systemic immunity (Ross et al., 2014).
Recently, GRIM REAPER PEPTIDE (GRIp) was identified in Arabidopsis as a novel, basic Lys-rich 11-amino acid peptide with homologs in crucifers (Wrzaczek et al., 2015) (Figure 3). The formation of GRIp from the preproprotein GRI is unique because this is the first report in plants of a specific type of protease, METACASPASE-9, involved in the release of a mature functional peptide. Since METACASPASE-9 also cleaves GRI at other sites, it cannot be excluded that other peptides are formed. GRIp is released in the extracellular space and binds to the extracellular domain of the plasma membrane-localized atypical leucine-rich repeat receptor-like kinase POLLEN-SPECIFIC RECEPTOR-LIKE KINASE5 (PRK5). Upon binding with its receptor, GRIp triggers oxidative stress and reactive oxygen species-dependent cell death in Arabidopsis (Wrzaczek et al., 2015).
In soybean, the PLANT ELICITOR PEPTIDEs 914 and 890 (PEP914 and PEP890) and homologs in Fabales and Cucurbitales are 8-amino acid peptides that share a highly similar sequence [DxPRG(G/H)NY] (Yamaguchi et al., 2011) (Figure 3). Although no single, dominant residue occurs in Gm-PEP914, the C-terminal Asp-7 or Tyr-8 residues are important for its alkalinizing activity. Moreover, synthetic Gm-PEP914 and Gm-PEP890 induce the expression of defense related genes in soybean cell cultures (Yamaguchi and Huffaker, 2011).
PEPTIDES DERIVED FROM A FUNCTIONAL PRECURSOR
Although most known peptides are derived from nonfunctional precursors (see above), some studies reported the existence of unique peptides derived from functional precursor proteins (Schmelz et al., 2006; Pearce et al., 2010a; Chen et al., 2014) (Figure 1). In some cases, these functional precursors have a different primary function from that of the peptides buried within them. Such peptides, released from the precursor by proteolytic enzyme activity, are called cryptides (Samir and Link, 2011). In humans, ∼35 cryptides have been reported (Pimenta and Lebrun, 2007), but in plants, only three have been reportedly identified (Figures 2 and 3) (Schmelz et al., 2006; Pearce et al., 2010a; Chen et al., 2014).
The soybean SUBTILASE PEPTIDE (Gm-SUBPEP) is a unique 12-amino acid peptide abundant in Pro and Arg residues (Pearce et al., 2010a) (Figure 3). An Ala-scan revealed that Arg-10 and His-12 are important for Gm-SUBPEP alkalinizing activity and putative receptor interaction (Pearce et al., 2010b). Gm-SUBPEP has a high basic net charge and was purified from soybean leaves as a non-posttranslationally modified peptide. Gm-SUBPEP is embedded in the C-terminal end of a preproprotein with an independent metabolic role as subtilase, an extracellular protease, and member of the subtilisin-like protease family. Gm-SUBPEP induces the same defense-related genes as Gm-PEP914 and Gm-PEP890, without sharing sequence homology (Pearce et al., 2010a; Yamaguchi and Huffaker, 2011).
Recently, a targeted and quantitative peptidomics strategy was employed to discover novel defense signaling peptides in tomato (Chen et al., 2014). CYSTEINE-RICH SECRETORY PROTEINS, ANTIGEN 5, AND PATHOGENESIS-RELATED 1 PROTEINS derived peptide 1 (CAPE1) was successfully identified as an 11-amino acid peptide derived from the preproprotein PATHOGENESIS-RELATED PROTEIN 1b (PR1b), a marker gene for the salicylic acid signaling pathway and systemic acquired resistance. For many years it was thought that tomato PR-1b encodes a 14-kD protein with antifungal activity (Niderman et al., 1995). Now it appears that it also encodes the CAPE1 peptide with antibacterial activity and antiherbivory activity and a role in the regulation of salt stress responses (Chen et al., 2014; Chien et al., 2015). At least 30 novel putative CAPE1-like peptides carrying the characteristic Pro-rich PxGNxxxxxPY motif were found in Solanoideae, Nicotianoideae, Viticeae, Brassicaceae, Fabaceae, and Poaceae (Chen et al., 2014) (Figure 3). This could mean that CAPE1 and its homologs form a new, highly conserved peptide family functioning in the defense response of plants. Tomato plants presprayed with CAPE1 exhibited increased resistance to the bacterial pathogen Pseudomonas syringae pv tomato DC3000 and reduced Spodoptera litura larval growth and weight. CAPE1 was significantly upregulated after wounding and/or methyl jasmonate treatment, similar to systemin (Chen et al., 2014). It is suggested that CAPE1 may be a novel DAMP (damage-associated molecular pattern) for induction of immunity, similar to HYPSYS, RALF, and At-Pep1 (Pearce et al., 2001a, 2001b; Huffaker et al., 2006; Albert, 2013; Trivilin et al., 2014). Recently, it was shown that At-CAPE1 functions as a negative regulator of salt stress (Chien et al., 2015).
INCEPTINS are 11- to 13-amino acid long peptides embedded in a functional plant protein, i.e., chloroplastic ATP synthase. Remarkably, after uptake by herbivorous insects, the latter undergoes nonspecific proteolysis and releases the INCEPTIN back into the plant where it functions as an elicitor of herbivore defense (Schmelz et al., 2006; Schmelz et al., 2012). These acidic INCEPTINS carry a core motif consisting of one disulfide bridge (Figure 3). However, the latter is nonessential for INCEPTIN’s reported antiherbivore activity, in contrast to Asp-3, Asp-10, Cys-8, and the C-terminal Ala (Schmelz et al., 2007). INCEPTINS are conserved in cowpea (Vigna unguiculata), bean, Arabidopsis, maize, and rice (Schmelz et al., 2006). This example shows that diversity in plant peptide synthesis is not only created by the plant processing machinery, but also by that of species interacting with plants, thereby increasing even more the potential of the plant proteome.
Evidence is emerging on bioactive peptides formed from functional protein precursors that are suggested to serve (themselves) as important functional components of plant metabolism (Fesenko et al., 2015). For example, in gametophore, protonema, and protoplast cells of the moss Physcomitrella patens, more than 20,000 unique endogenous peptides ranging from 5 to 78 amino acids were detected with mass spectrometry analysis (Fesenko et al., 2015). These specific peptide pools are hydrolysis products of functional proteins, although the exact mechanisms of endogenous peptide formation have not been elucidated yet. In both the protonema and protoplast states of the moss, plastid proteins served as the main source of peptides. In gametophores, stress-related proteins were among the most productive precursors (Fesenko et al., 2015). The functional characterization of endogenous peptide pools is ongoing, though 117 peptides already were predicted to have antimicrobial potential (Torrent et al., 2012; Fesenko et al., 2015). It is hypothesized that the formation of these specific endogenous peptide pools or “peptide burst” is a form of biotic stress response based on the production of AMPs from existing functional proteins (Fesenko et al., 2015). Nevertheless, experimental evidence on the bioactivity of these endogenous peptides is essential to consider them as functional components of plant metabolism and not as degradation products of the proteome.
NONPRECURSOR-DERIVED PEPTIDES
So far, the majority of characterized plant peptides are derived from precursor proteins (Tabata and Sawa, 2014). Recently, some sORFs (usually <100 codons) were proposed to represent a potential additional source of functional peptides, called “short peptides encoded by sORFs” or sPEPs (Andrews and Rothnagel, 2014). This type of peptide is directly translated from an sORF. The involvement of an intermediate precursor or further processing is not required for the synthesis of these peptides (Figure 1). Therefore, we define this type of peptides as “nonprecursor-derived peptides.” In contrast to previous literature, we do not employ the terminology “micropeptides” for sPEPs for two reasons (Crappé et al., 2013). First, posttranslationally modified peptides are 20 amino acids or less and thus are smaller than micropeptides. Second, the prefix “micro” was used to point to a regulatory function, as in microproteins (Staudt and Wenkel, 2011) rather than commonly implemented as a small-size indicator. sPEPs have been demonstrated to fulfill various biological roles ranging from plant development to regulation of gene expression, and we will illustrate this novel group with representative examples (Figures 2 and 3).
Up to now, nonprecursor-derived peptides from plants could be subdivided in three subgroups, depending on the genomic location of the sORF(s) encoding the peptide(s). These sORFs can be located (1) upstream of the main ORF in the 5′ leader sequence of a gene, (2) in primary transcripts of miRNA, or (3) in other transcripts not encoding longer (>100 amino acid) proteins. The latter class includes annotated protein-encoding transcripts with one or more ORFs encoding peptides <100 amino acids, transcripts from intergenic regions, and some transcripts previously annotated as long noncoding RNA (lncRNA) (Figure 1).
Peptides Encoded by Upstream ORFs
Upstream ORFs (uORFs) are putative protein-coding regions located in the 5′ leader sequence of a main protein-coding region or overlapping with such a main ORF (Andrews and Rothnagel, 2014; von Arnim et al., 2014) (Figure 4). Currently, more than 20,000 uORFs are annotated in Arabidopsis (von Arnim et al., 2014). Only a minor fraction of these uORFs show conservation of their primary sequences (conserved peptide uORFs or CPuORFs), whereas the majority does not (nonCPuORFs) (Jorgensen and Dorantes-Acosta, 2012; von Arnim et al., 2014). The length of uORFs with a reported function varies from 1 to 92 codons (von Arnim et al., 2014). For an overview of all plant uORFs, we refer to uORFdb, a database on eukaryotic uORF biology (http://cbdm.mdc-berlin.de/tools/uorfdb; Wethmar et al., 2014).
Figure 4.
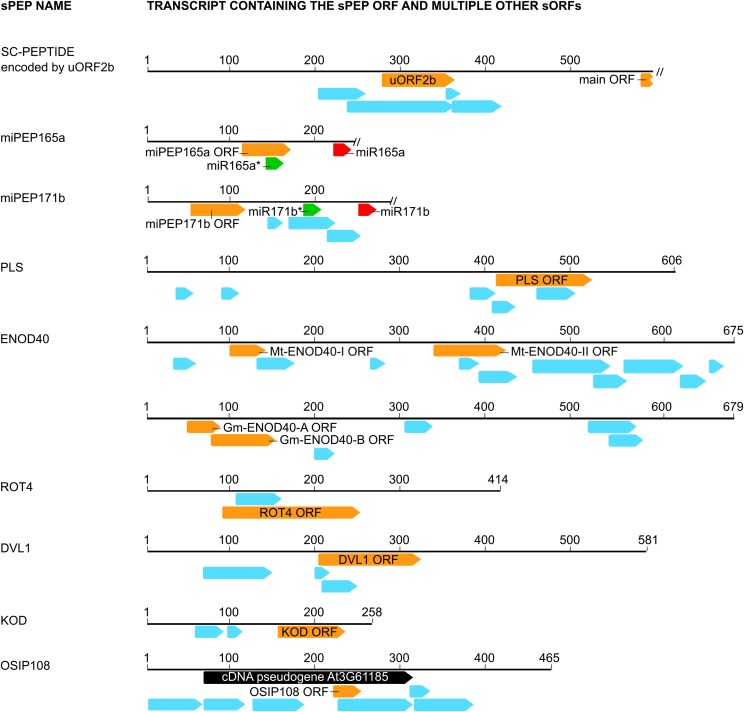
A Wide Variety of sPEP Transcripts.
Transcripts of sPEPs harbor many sORFs (<100 amino acids; blue arrows), though only a few sORFs have been experimentally characterized (orange arrows). All sORFs starting with AUG and consisting of at least six codons in sense direction are displayed. Interior ORFs located within the same frame of a longer ORF are omitted, except for the SC-PEPTIDE. Only the 5′ part of the SC-PEPTIDE transcript and the miPEP transcripts is displayed (indicated by a double slash). A red arrow represents mature miRNA found from one arm of a hairpin; a green arrow represents mature miRNA (with asterisk) formed from the opposite arm of a hairpin.
In several case studies, uORFs function as regulators of metabolism or mediators of developmental gene regulation by influencing the expression and/or translation of the main ORF. Often, this posttranscriptional regulation occurs in response to compounds such as sucrose, polyamines, phosphocholine, and ascorbate (Jorgensen and Dorantes-Acosta, 2012; von Arnim et al., 2014; Ebina et al., 2015; Laing et al., 2015). A genome-wide ribosome footprinting study in Arabidopsis revealed that uORFs capture ribosomes and thereby limit the translation of the downstream main ORF, meaning that uORFs can act as a barrier for translation (Juntawong et al., 2014).
Translational evidence for uORFs is supported by in vitro translation, mutational analysis in planta, and ribosome footprinting data, though not all uORFs are necessarily translated (Andrews and Rothnagel, 2014; Juntawong et al., 2014; von Arnim et al., 2014). In contrast to human cell lines, uORF peptides have not yet been detected by mass spectrometry in plants (Kim et al., 2014). One possible explanation could be that uORF peptides do not accumulate because short peptides may typically be turned over rapidly in the cell (von Arnim et al., 2014). Remarkably, in a recent report, a functional uORF initiated by a noncanonical codon (a non-AUG initiation codon) has been described, meaning that noncanonically translated uORFs too should be taken into account in future research (Laing et al., 2015). Leaky scanning and reinitiation are two major mechanisms via which translation of uORFs and the main ORF are coordinated. For more information on the latter, we refer to excellent recent reviews (Andrews and Rothnagel, 2014; von Arnim et al., 2014).
Peptides Encoded by sORFs in Primary Transcripts of miRNAs
Recently, it was shown that primary transcripts of microRNAs (pri-miRNAs) contain sORFs encoding regulatory peptides termed miRNA-ENCODED PEPTIDES (miPEPs) (Lauressergues et al., 2015). For each of the 50 pri-miRNAs in Arabidopsis, a putative peptide with a canonical initiation codon was predicted. These 50 miPEPs are 3 to 59 amino acids long and share no common signature, suggesting that each miPEP is specific for its corresponding miRNA. The M. truncatula miPEP171b and Arabidopsis miPEP165a were studied in detail and function as transcriptional activators. Overproduction of miPEP171b and miPEP165a in roots of M. truncatula and Arabidopsis, respectively, enhanced the accumulation of their respective endogenous miRNAs, resulting in downregulation of target genes involved in root development. miPEPs were not yet detected by mass spectrometry analysis in planta, though translational evidence for miPEP171b and miPEP165a was supported by a combination of in vivo translational GUS fusions, overexpression experiments, immunolocalization studies, and ribosome footprinting (Juntawong et al., 2014; Lauressergues et al., 2015). Altogether, these findings strongly indicate that the first ORF in pri-miR171b and pri-miR165a functions as a peptide rather than as RNA (Lauressergues et al., 2015) (Figure 4). As such, these peptides derived from pri-miRNAs form a novel subclass with important regulatory features in plants and probably in other eukaryotes as well.
Peptides Encoded by sORFs in Transcripts Not Encoding Longer (>100 Amino Acids) Proteins
sORFs (<100 codons) are not only found in the 5′ leader sequence of a main protein-coding region or in primary transcripts of miRNA, but also in other transcripts not encoding longer (>100 amino acid) proteins. A striking overlap exists between features of some of the latter coding transcripts and lncRNAs, since some lncRNAs also contain one or more ORFs <100 codons (Mercer et al., 2009). An unambiguous definition of sPEPs encoded by these sORFs and a strict distinction with lncRNA is impeded at the moment because functional studies and translational evidence for these peptides are limited (Andrews and Rothnagel, 2014). Therefore, we carefully discuss the indications for functionality and translatability of these sPEPs to discriminate between functional peptides and functional transcripts.
Currently, six functionally characterized plant sPEPs encoded by transcripts not encoding longer proteins have been reported in the literature, namely, POLARIS (PLS; 36 amino acids), EARLY NODULIN GENE 40 (ENOD40; 12, 13, 24, or 27 amino acids), ROTUNDIFOLIA FOUR (ROT4; 53 amino acids), KISS OF DEATH (KOD; 25 amino acids), BRICK1 (BRK1; 84 amino acids), Zm-908p11 (97 amino acids), and Zm-401p10 (89 amino acids) (Ma et al., 2008; Dong et al., 2013; Crappé et al., 2013; Andrews and Rothnagel, 2014). However, based on the TAIR10 annotation of the Arabidopsis genome, 131 functionally annotated peptides (<100 amino acids) without an NSS can be identified. This group of peptides, previously not regarded as sPEPs, includes 18 DEVIL (DVL)/ROT peptides, 6 methallothioneins, 2 NUCLEAR RNA POLYMERASE (NRPD) peptides, 10 members of the SMALL AUXIN-UP RNA (SAUR)-like auxin-responsive protein family, and many other Arabidopsis peptides encoded by sORFs in transcripts not encoding longer (>100 amino acids) proteins (Wen et al., 2004; Ream et al., 2009; Grennan, 2011; Spartz et al., 2012) (Supplemental Data Set 1). Therefore, we emphasize that many more peptides than previously thought are covered by the definition of this type of sPEPs. Here, we discuss the functionality and translatability of the five best-characterized sPEPs (PLS, ENOD40, ROT4, DVL1, and KOD) in plants and of novel, potential sPEPs in Arabidopsis.
The 36-amino acid peptide PLS is required for correct root growth and leaf vascular patterning (Casson et al., 2002; Chilley et al., 2006). This peptide is encoded by the longest and fifth of six sORFs in the PLS transcript (Figure 4). Overexpression of the PLS transcript in a pls knockout mutant resulted in partial rescue of the short root mutant phenotype, whereas overexpression of a modified PLS transcript (with mutated initiation codon) did not. These complementation assays demonstrate that the PLS initiation codon is required for activity, supporting a functionality as sPEP (Casson et al., 2002). Nevertheless, it remains unclear how the PLS peptide is translated, since the PLS-ORF is preceded by three other sORFs. It was hypothesized that these sORFs are uORFs and the long PLS-ORF is the main ORF.
M. truncatula ENOD40 is involved in symbiotic nodule development, and its activity can be mediated by the peptides ENOD40-I (13 amino acids) and ENOD40-II (27 amino acids), as well as by the ENOD40 transcript (Sousa et al., 2001; Campalans et al., 2004). This transcript contains 12 sORFs, though only ENOD40-I and -II span two nucleotide sequences that are conserved in all legume ENOD40 genes (Wan et al., 2007). Local overexpression of ENOD40 and derived variants into M. truncatula roots demonstrated the involvement of ENOD40-I and -II peptides as well as of a structured RNA signal in cortical cell division (Sousa et al., 2001). The translatability of ENOD40-I (second sORF) and -II (fifth sORF) has been demonstrated by in vivo translational GUS fusions in M. truncatula roots, even if they were preceded by other sORFs in the transcript (Sousa et al., 2001) (Figure 4). These results suggest that sORFs do not arrest 5′-to-3′ ribosome scanning, since reinitiation can occur along the ENOD40-1 transcript and the initiation codons of ENOD40-I and -II can be recognized by ribosomes for translation (Sousa et al., 2001). However, in vitro translation studies of all ENOD40 sORFs in wheat germ extract could not detect the corresponding peptides, possibly due to instability of such small peptides (<39 amino acids) under artificial conditions (Sousa et al., 2001). Strikingly, the ENOD40 transcript as well as a modified variant (with mutated initiation codons of ENOD40-I and -II) relocalized an RNA BINDING PROTEIN1 (RBP1) from the cytoplasm to the nucleus, proving that the ENOD40 transcript itself, and not the ENOD40 encoded peptides, is responsible for this specific activity (Campalans et al., 2004). Overall, it can be hypothesized that ENOD40 encodes a bifunctional or dual RNA, since both the ENOD40 transcript and peptides are functional in M. truncatula and exert different roles (Bardou et al., 2011).
The soybean ENOD40-A and -B peptides, 12 and 24 amino acids long, respectively, are involved in the regulation of sucrose use in nitrogen-fixing nodules and specifically bind to NODULIN 100, a subunit of sucrose synthase (Röhrig et al., 2002). The sORFs encoding these peptides are embedded in a transcript containing six sORFs (Figure 4). However, only Gm-ENOD40-A and -B, similar to Mt-ENOD40-I, span a nucleotide sequence that is conserved in all legume ENOD40 genes. The translatability of Gm-ENOD40-A and -B is supported by in vitro translation studies and mutational assays (Röhrig et al., 2002). Moreover, the Gm-ENOD40-B peptide was detected in nodules via protein gel blotting (Röhrig et al., 2002). Since Gm-ENOD40-A and -B are located out of frame, it is remarkable that both sORFs are translated and function as peptides in the same biological conditions. The translation mechanism of Gm-ENOD40-A and -B could be explained by leaky scanning and ribosome reinitiation, as was also reported in the context of uORFs (Andrews and Rothnagel, 2014).
The 53-amino acid ROT4/DVL16 peptide regulates polar cell proliferation in lateral organs and leaf morphogenesis in Arabidopsis (Narita et al., 2004). The ROT4 transcript contains a longer sORF encoding ROT4 and a shorter sORF (Figure 4). A dominant mutant, rot4-1D, overexpressing ROT4 possesses shortened, rounded leaves and short floral organs. Moreover, overexpression of only the conserved ROT4-LIKE (RTFL) domain (RKCVVKEQRARLYIIRRCVLMLLCWHD) embedded in ROT4 and its homologs was sufficient to confer the rot4-1D phenotype (Figure 3). Loss-of-function mutations in ROT4 and several ROT4-LIKE genes were aphenotypic, suggesting that there may be some functional redundancy between family members (Narita et al., 2004; Guo et al., 2015). Translatability of ROT4 is supported by in vivo translational GFP fusion experiments (Narita et al., 2004). In contrast to ENOD40 and PLS, ROT4 could be translated via a classical translation mechanism as this peptide is encoded by the first and longest sORF in the transcript.
ROT18 (or DVL1) is a 51-peptide member of the RTFL or DEVIL (DVL) family in Arabidopsis, which consists of many proteins that are widely conserved among land plants and function mainly in plant organogenesis (Wen et al., 2004; Valdivia et al., 2012; Guo et al., 2015). ROT18/DVL1 is encoded by the longest ORF in the ROT18/DVL1 transcript, which contains four sORFs in total (Figure 4). Downregulation and overexpression of the ROT18/DVL1 ORF resulted in phenotypic changes similar to corresponding mutants of ROT4 (Wen et al., 2004). Overexpression of modified ROT18/DVL1 (with frame-shift or point mutations inhibiting ROT18/DVL1 translation) and complementation assays suggest a role for the ROT18/DVL1 peptide in plant development (Wen et al., 2004). However, it remains unclear how this peptide is translated, since the ROT18/DVL1-ORF is preceded by two other sORFs, similar to PLS.
KOD is a 25-amino acid peptide unique in Arabidopsis and involved in programmed cell death regulation (Blanvillain et al., 2011). More specifically, it modulates suspensor elimination during embryogenesis and root hair programmed cell death. KOD is encoded by the longest and last ORF in a short KOD-transcript containing three sORFs in sense. Experiments using knockout mutants, carrying a T-DNA insertion and a point mutation (Pro-9 to Ser), respectively, and transient assays in onion (Allium cepa) using mutated and truncated variants of KOD indicate that KOD functions as a peptide rather than as RNA (Blanvillain et al., 2011). In vivo translational GFP fusions showed that the initiation codon of KOD can be recognized for translation in planta, though it remains unclear how the KOD peptide is translated, similar as for PLS (Figure 4). Nevertheless, NMR studies on synthetic KOD peptide pointed out that it adopts an α-helix (from Pro-9 to Arg-21), whereas the N-terminal region is disordered (Blanvillain et al., 2011).
Based on a recent transcriptome study of Arabidopsis leaves upon oxidative stress and subsequent functional analysis in the eukaryotic model yeast Saccharomyces cerevisiae, different putative oxidative stress-induced peptides (OSIPs) could be identified (De Coninck et al., 2013a). The sORF encoding the decapeptide OSIP108 is contained within a pseudogene harboring seven sORFs and induced by oxidative stress. Both overexpression and infiltration of synthetic OSIP108 in Arabidopsis leaves resulted in increased oxidative stress tolerance. Moreover, treatment of other eukaryotes with OSIP108 was demonstrated to prevent apoptosis triggered by different oxidative stress inducers (Spincemaille et al., 2014a, 2014b). Although these findings support a function for OSIP108 as a peptide, the latter could not yet be identified as such in planta and its translation can be questioned.
Recently, 7901 sORFs potentially encoding sPEPs of 30 to 100 amino acids were identified in intergenic regions of the Arabidopsis genome (Hanada et al., 2007, 2013). A subset of these sORFs (473), showing high homology with sORFs in other plants and unbiased expression evidence, was overexpressed in Arabidopsis. Altered morphogenesis of Arabidopsis plants was shown for ∼10% of these sORFs, a proportion that is approximately 7 times higher than that of randomly chosen known genes (Hanada et al., 2013). Translational evidence for the latter 49 functional sORFs could not be found in reported data sets of ribosome footprinting experiments and mass spectrometry analysis (Castellana et al., 2008; Juntawong et al., 2014). On the other hand, other sORFs predicted in the initial pool but without reported phenotypes are associated with ribosomes and possibly translated (Juntawong et al., 2014) or detected by mass spectrometry (Castellana et al., 2008).
In general, although various approaches have been employed for sPEPs to discriminate between a functional peptide and a functional transcript, direct evidence demonstrating the presence of sPEPs in planta is lacking as well as an in-depth explanation of the translation mechanism for certain sORFs. Generally applied to assess potential translation of an ORF are ribosome footprinting techniques that determine whether ribosomes are associated with a certain region in the transcript. While these could support the translatability of, e.g., miPEPs, no such conclusions can yet be made for PLS, ROT4, DVL1, KOD, and OSIP108 ORFs due to insufficient reads present in recent ribosome footprinting data from Arabidopsis (Juntawong et al., 2014). However, this approach still provides indirect evidence for the presence of a specific sPEP, since the observed interaction of an ORF with ribosomes does not necessarily imply translation of that ORF, but it can also function in regulating the translation of other ORFs. Cutting-edge techniques such as puromycin-associated nascent chain proteomics (PUNCH-P) allowing intact ribosome-nascent polypeptide chains to be analyzed by mass spectrometry (Aviner et al., 2014) could overcome this problem, but are not yet been optimized for plants. The most direct evidence for the presence of peptides in planta can be obtained from mass spectrometry analyses. The latter allowed successful detection of several sPEPs in other higher eukaryotes, but many of these were noncanonical sPEPs (Slavoff et al., 2013; Prabakaran et al., 2014). Moreover, since sPEPs often occur at low abundance (Hanada et al., 2013; Andrews and Rothnagel, 2014) and classical proteomic analyses involving data-dependent acquisition mass spectrometry have a bias for highly abundant peptides, they have limited potential to detect sPEPs. Targeted proteomic techniques as data-independent acquisition mass spectrometry and selected/multiple reaction monitoring could be promising in this regard for detection of specific sPEPs though less suitable for discovery-based applications (Samir and Link, 2011; Chen et al., 2014; Doerr, 2014; Taylor et al., 2014). Finally, it should be noted that in planta identification of a peptide does not necessarily imply that it fulfills any biological role but the peptide can instead be part of suggested translational noise (Guttman and Rinn, 2012).
CONCLUDING REMARKS
Peptides (<100 amino acids) form a subset of the plant proteome that has been receiving a lot of attention over the past several years. In particular, the different types and the number of peptides identified in different plant species have been expanding. In addition to peptides derived from a nonfunctional precursor, such as well-known AMPs and signaling peptides, evidence is emerging on peptides derived from functional protein precursors as well as peptides directly translated from sORFs (<100 codons). These sORFs can be located in the 5′ leader sequence of protein-encoding genes, in primary transcripts of miRNA, or in other transcripts not encoding longer (>100 amino acids) proteins. Moreover, plant peptides display diverse functions. This functional diversity ranges from roles in plant development, growth, fertilization, senescence, cell death, cell-to-cell communication, nodulation, and defense response to regulation of expression and translation. Peptides exert these functions via direct interactions with pathogens or through receptor binding and downstream signaling. In addition to this diversity in synthesis and function, a peptide varies in length, 3D structure, amino acid composition, and possible posttranslational modifications. Thus, it can be concluded that diversity is key in the peptide world.
Nevertheless, this diversity still remains largely unexplored, mainly because of technical limitations. First, identification of ligand-receptor pairs remains challenging and even less is known about the downstream signaling. Various approaches are available to study physical ligand-receptor interactions, including photoaffinity labeling of peptide-receptor interactions (Tabata et al., 2014), phosphoproteomics revealing the phosphorylation status of plasma membrane proteins in planta upon treatment with synthetic peptides (Haruta et al., 2014), and a cellular bioassay using the oxidative burst as readout for receptor activation by synthetic peptides (Butenko et al., 2014). For more details, we refer to a recent discussion on these approaches (Stes et al., 2015). Second, it is still controversial and unclear whether the plant peptidome harbors a wide array of functional peptides encoded by sORFs. Future experiments should focus on cutting-edge ribosome footprinting, mass spectrometry techniques and in-depth functional analyses of nonprecursor-derived peptides in planta to further explore this dark matter of the peptidome. Expanding the peptidome brings us close to the field of noncoding RNA, since it was recently hypothesized that lncRNA transcripts too could form a repository for new peptides (Ruiz-Orera et al., 2014). Furthermore, transcripts could be bifunctional, meaning that the transcript as well as one or more peptides from one transcript play a role, as was shown for Mt-ENOD40 (Bardou et al., 2011). Therefore, we want to put forward the notion that it is difficult to define an exact border, since there is rather a continuum between coding and noncoding sequences. In conclusion, bringing together biological questions and emerging trends as well as cutting-edge techniques in the field of peptide research will deepen our understanding of the complex peptidome as a fascinating and essential part of the plant proteome.
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL libraries under the following accession numbers: CEP1 (AT1G47485), CEP5 (AT5G66815), GSL1 (FJ195646), EPF1 (AT2G20875), EPF2 (AT1G34245), EPFL4 (AT4G14723), EPFL5 (AT3G22820), EPFL9 (AT4G12970), LURE1 (AT5G43285), ES4 (NM_001112150.1), PR-1b (AT2G14610), PLS (AT4G39403), Mt-ENOD40 (X80264.1), Gm-ENOD40 (X69154), ROT4 (AT2G36985), DVL1 (AT5G16023), and KOD (JF411843).
Supplemental Data
Supplemental Data Set 1. List of potential Arabidopsis peptides encoded by short ORFs (<100 amino acids) in transcripts not encoding longer (>100 amino acids) proteins.
Supplementary Material
Acknowledgments
This work was supported by a BBSRC David Phillips Fellowship (BB_BB/H022457/1) to I.D.S., a PhD grant from ‘Agentschap voor Innovatie door Wetenschap en Techniek (IWT)-Vlaanderen’ (SB-111657) to P.T., a postdoctoral fellowship from ‘Fonds Wetenschappelijk Onderzoek (FWO)-Vlaanderen’ to B.D.C. (FWO/12A7213N), and a ‘FWO-Vlaanderen’ project for B.P.A.C. (G0D5113N).
AUTHOR CONTRIBUTIONS
All authors contributed to writing the article.
Footnotes
Articles can be viewed online without a subscription.
References
- Aerts A.M., François I.E.J.A., Meert E.M.K., Li Q.T., Cammue B.P.A., Thevissen K. (2007). The antifungal activity of RsAFP2, a plant defensin from raphanus sativus, involves the induction of reactive oxygen species in Candida albicans. J. Mol. Microbiol. Biotechnol. 13: 243–247. [DOI] [PubMed] [Google Scholar]
- Albert M. (2013). Peptides as triggers of plant defence. J. Exp. Bot. 64: 5269–5279. [DOI] [PubMed] [Google Scholar]
- Almasia N.I., Bazzini A.A., Hopp H.E., Vazquez-Rovere C. (2008). Overexpression of snakin-1 gene enhances resistance to Rhizoctonia solani and Erwinia carotovora in transgenic potato plants. Mol. Plant Pathol. 9: 329–338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amano Y., Tsubouchi H., Shinohara H., Ogawa M., Matsubayashi Y. (2007). Tyrosine-sulfated glycopeptide involved in cellular proliferation and expansion in Arabidopsis. Proc. Natl. Acad. Sci. USA 104: 18333–18338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amien S., Kliwer I., Márton M.L., Debener T., Geiger D., Becker D., Dresselhaus T. (2010). Defensin-like ZmES4 mediates pollen tube burst in maize via opening of the potassium channel KZM1. PLoS Biol. 8: e1000388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersen N.H., Cao B., Rodríguez-Romero A., Arreguin B. (1993). Hevein: NMR assignment and assessment of solution-state folding for the agglutinin-toxin motif. Biochemistry 32: 1407–1422. [DOI] [PubMed] [Google Scholar]
- Andreev Y.A., Korostyleva T.V., Slavokhotova A.A., Rogozhin E.A., Utkina L.L., Vassilevski A.A., Grishin E.V., Egorov T.A., Odintsova T.I. (2012). Genes encoding hevein-like defense peptides in wheat: distribution, evolution, and role in stress response. Biochimie 94: 1009–1016. [DOI] [PubMed] [Google Scholar]
- Andrews S.J., Rothnagel J.A. (2014). Emerging evidence for functional peptides encoded by short open reading frames. Nat. Rev. Genet. 15: 193–204. [DOI] [PubMed] [Google Scholar]
- Araya T., Miyamoto M., Wibowo J., Suzuki A., Kojima S., Tsuchiya Y.N., Sawa S., Fukuda H., von Wirén N., Takahashi H. (2014). CLE-CLAVATA1 peptide-receptor signaling module regulates the expansion of plant root systems in a nitrogen-dependent manner. Proc. Natl. Acad. Sci. USA 111: 2029–2034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asano T., Miwa A., Maeda K., Kimura M., Nishiuchi T. (2013). The secreted antifungal protein thionin 2.4 in Arabidopsis thaliana suppresses the toxicity of a fungal fruit body lectin from Fusarium graminearum. PLoS Pathog. 9: e1003581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aviner R., Geiger T., Elroy-Stein O. (2014). Genome-wide identification and quantification of protein synthesis in cultured cells and whole tissues by puromycin-associated nascent chain proteomics (PUNCH-P). Nat. Protoc. 9: 751–760. [DOI] [PubMed] [Google Scholar]
- Bardou F., Merchan F., Ariel F., Crespi M. (2011). Dual RNAs in plants. Biochimie 93: 1950–1954. [DOI] [PubMed] [Google Scholar]
- Bartels S., Lori M., Mbengue M., van Verk M., Klauser D., Hander T., Böni R., Robatzek S., Boller T. (2013). The family of Peps and their precursors in Arabidopsis: differential expression and localization but similar induction of pattern-triggered immune responses. J. Exp. Bot. 64: 5309–5321. [DOI] [PubMed] [Google Scholar]
- Beale K.M., Johnson M.A. (2013). Speed dating, rejection, and finding the perfect mate: advice from flowering plants. Curr. Opin. Plant Biol. 16: 590–597. [DOI] [PubMed] [Google Scholar]
- Berrocal-Lobo M., Segura A., Moreno M., López G., García-Olmedo F., Molina A. (2002). Snakin-2, an antimicrobial peptide from potato whose gene is locally induced by wounding and responds to pathogen infection. Plant Physiol. 128: 951–961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharya R., Koramutla M.K., Negi M., Pearce G., Ryan C.A. (2013). Hydroxyproline-rich glycopeptide signals in potato elicit signalling associated with defense against insects and pathogens. Plant Sci. 207: 88–97. [DOI] [PubMed] [Google Scholar]
- Blanvillain R., Young B., Cai Y.M., Hecht V., Varoquaux F., Delorme V., Lancelin J.-M., Delseny M., Gallois P. (2011). The Arabidopsis peptide kiss of death is an inducer of programmed cell death. EMBO J. 30: 1173–1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blein J.P., Coutos-Thévenot P., Marion D., Ponchet M. (2002). From elicitins to lipid-transfer proteins: a new insight in cell signalling involved in plant defence mechanisms. Trends Plant Sci. 7: 293–296. [DOI] [PubMed] [Google Scholar]
- Bobay B.G., DiGennaro P., Scholl E., Imin N., Djordjevic M.A., Mck Bird D. (2013). Solution NMR studies of the plant peptide hormone CEP inform function. FEBS Lett. 587: 3979–3985. [DOI] [PubMed] [Google Scholar]
- Broekaert W.F., et al. (1992). Antimicrobial peptides from Amaranthus caudatus seeds with sequence homology to the cysteine/glycine-rich domain of chitin-binding proteins. Biochemistry 31: 4308–4314. [DOI] [PubMed] [Google Scholar]
- Butenko M.A., Patterson S.E., Grini P.E., Stenvik G.E., Amundsen S.S., Mandal A., Aalen R.B. (2003). Inflorescence deficient in abscission controls floral organ abscission in Arabidopsis and identifies a novel family of putative ligands in plants. Plant Cell 15: 2296–2307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butenko M.A., Wildhagen M., Albert M., Jehle A., Kalbacher H., Aalen R.B., Felix G. (2014). Tools and strategies to match peptide-ligand receptor pairs. Plant Cell 26: 1838–1847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butenko M.A., Simon R. (2015). Beyond the meristems: similarities in the CLAVATA3 and INFLORESCENCE DEFICIENT IN ABSCISSION peptide mediated signalling pathways. J. Exp. Bot. pii: erv310. [DOI] [PubMed] [Google Scholar]
- Cammue B.P., De Bolle M.F., Terras F.R., Proost P., Van Damme J., Rees S.B., Vanderleyden J., Broekaert W.F. (1992). Isolation and characterization of a novel class of plant antimicrobial peptides form Mirabilis jalapa L. seeds. J. Biol. Chem. 267: 2228–2233. [PubMed] [Google Scholar]
- Cammue B.P., et al. (1995). A potent antimicrobial protein from onion seeds showing sequence homology to plant lipid transfer proteins. Plant Physiol. 109: 445–455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campalans A., Kondorosi A., Crespi M. (2004). Enod40, a short open reading frame-containing mRNA, induces cytoplasmic localization of a nuclear RNA binding protein in Medicago truncatula. Plant Cell 16: 1047–1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao J., Shi F. (2012). Evolution of the RALF gene family in plants: gene duplication and selection patterns. Evol. Bioinform. Online 8: 271–292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carvalho Ade O., Gomes V.M. (2007). Role of plant lipid transfer proteins in plant cell physiology-a concise review. Peptides 28: 1144–1153. [DOI] [PubMed] [Google Scholar]
- Carvalho Ade O., Gomes V.M. (2011). Plant defensins and defensin-like peptides - biological activities and biotechnological applications. Curr. Pharm. Des. 17: 4270–4293. [DOI] [PubMed] [Google Scholar]
- Casson S.A., Chilley P.M., Topping J.F., Evans I.M., Souter M.A., Lindsey K. (2002). The POLARIS gene of Arabidopsis encodes a predicted peptide required for correct root growth and leaf vascular patterning. Plant Cell 14: 1705–1721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castellana N.E., Payne S.H., Shen Z., Stanke M., Bafna V., Briggs S.P. (2008). Discovery and revision of Arabidopsis genes by proteogenomics. Proc. Natl. Acad. Sci. USA 105: 21034–21038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Champigny M.J., Isaacs M., Carella P., Faubert J., Fobert P.R., Cameron R.K. (2013). Long distance movement of DIR1 and investigation of the role of DIR1-like during systemic acquired resistance in Arabidopsis. Front. Plant Sci. 4: 230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y.C., Siems W.F., Pearce G., Ryan C.A. (2008). Six peptide wound signals derived from a single precursor protein in Ipomoea batatas leaves activate the expression of the defense gene sporamin. J. Biol. Chem. 283: 11469–11476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y.-L., Lee C.-Y., Cheng K.-T., Chang W.-H., Huang R.-N., Nam H.G., Chen Y.-R. (2014). Quantitative peptidomics study reveals that a wound-induced peptide from PR-1 regulates immune signaling in tomato. Plant Cell 26: 4135–4148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chien P.-S., Nam H.G., Chen Y.-R. (2015). A salt-regulated peptide derived from the CAP superfamily protein negatively regulates salt-stress tolerance in Arabidopsis. J. Exp. Bot. pii: erv263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chilley P.M., Casson S.A., Tarkowski P., Hawkins N., Wang K.L.-C., Hussey P.J., Beale M., Ecker J.R., Sandberg G.K., Lindsey K. (2006). The POLARIS peptide of Arabidopsis regulates auxin transport and root growth via effects on ethylene signaling. Plant Cell 18: 3058–3072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chouabe C., Eyraud V., Da Silva P., Rahioui I., Royer C., Soulage C., Bonvallet R., Huss M., Gressent F. (2011). New mode of action for a knottin protein bioinsecticide: pea albumin 1 subunit b (PA1b) is the first peptidic inhibitor of V-ATPase. J. Biol. Chem. 286: 36291–36296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark S.E., Running M.P., Meyerowitz E.M. (1995). CLAVATA3 is a specific regulator of shoot and floral meristem development affecting the same processes as CLAVATA1. Development 121: 2057–2067. [Google Scholar]
- Cock J.M., McCormick S. (2001). A large family of genes that share homology with CLAVATA3. Plant Physiol. 126: 939–942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Combier J.-P., Küster H., Journet E.-P., Hohnjec N., Gamas P., Niebel A. (2008). Evidence for the involvement in nodulation of the two small putative regulatory peptide-encoding genes MtRALFL1 and MtDVL1. Mol. Plant Microbe Interact. 21: 1118–1127. [DOI] [PubMed] [Google Scholar]
- Constabel C.P., Bergey D.R., Ryan C.A. (1995). Systemin activates synthesis of wound-inducible tomato leaf polyphenol oxidase via the octadecanoid defense signaling pathway. Proc. Natl. Acad. Sci. USA 92: 407–411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Constabel C.P., Yip L., Ryan C.A. (1998). Prosystemin from potato, black nightshade, and bell pepper: primary structure and biological activity of predicted systemin polypeptides. Plant Mol. Biol. 36: 55–62. [DOI] [PubMed] [Google Scholar]
- Cordts S., Bantin J., Wittich P.E., Kranz E., Lörz H., Dresselhaus T. (2001). ZmES genes encode peptides with structural homology to defensins and are specifically expressed in the female gametophyte of maize. Plant J. 25: 103–114. [DOI] [PubMed] [Google Scholar]
- Costa L.M., et al. (2014). Central cell-derived peptides regulate early embryo patterning in flowering plants. Science 344: 168–172. [DOI] [PubMed] [Google Scholar]
- Covey P.A., Subbaiah C.C., Parsons R.L., Pearce G., Lay F.T., Anderson M.A., Ryan C.A., Bedinger P.A. (2010). A pollen-specific RALF from tomato that regulates pollen tube elongation. Plant Physiol. 153: 703–715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craik D.J., Mylne J.S., Daly N.L. (2010). Cyclotides: macrocyclic peptides with applications in drug design and agriculture. Cell. Mol. Life Sci. 67: 9–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craik D.J., Malik U. (2013). Cyclotide biosynthesis. Curr. Opin. Chem. Biol. 17: 546–554. [DOI] [PubMed] [Google Scholar]
- Crappé J., Van Criekinge W., Trooskens G., Hayakawa E., Luyten W., Baggerman G., Menschaert G. (2013). Combining in silico prediction and ribosome profiling in a genome-wide search for novel putatively coding sORFs. BMC Genomics 14: 648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czyzewicz N., Yue K., Beeckman T., De Smet I. (2013). Message in a bottle: small signalling peptide outputs during growth and development. J. Exp. Bot. 64: 5281–5296. [DOI] [PubMed] [Google Scholar]
- Daly N.L., Rosengren K.J., Craik D.J. (2009). Discovery, structure and biological activities of cyclotides. Adv. Drug Deliv. Rev. 61: 918–930. [DOI] [PubMed] [Google Scholar]
- De Coninck B., Carron D., Tavormina P., Willem L., Craik D.J., Vos C., Thevissen K., Mathys J., Cammue B.P.A. (2013a). Mining the genome of Arabidopsis thaliana as a basis for the identification of novel bioactive peptides involved in oxidative stress tolerance. J. Exp. Bot. 64: 5297–5307. [DOI] [PubMed] [Google Scholar]
- De Coninck B., Cammue B.P.A., Thevissen K. (2013b). Modes of antifungal action and in planta functions of plant defensins and defensin-like peptides. Fungal Biol. Rev. 26: 109–120. [Google Scholar]
- Delay C., Imin N., Djordjevic M.A. (2013). CEP genes regulate root and shoot development in response to environmental cues and are specific to seed plants. J. Exp. Bot. 64: 5383–5394. [DOI] [PubMed] [Google Scholar]
- de Souza Cândido E., e Silva Cardoso M.H., Sousa D.A., Viana J.C., de Oliveira-Júnior N.G., Miranda V., Franco O.L. (2014). The use of versatile plant antimicrobial peptides in agribusiness and human health. Peptides 55: 65–78. [DOI] [PubMed] [Google Scholar]
- Dinger M.E., Pang K.C., Mercer T.R., Mattick J.S. (2008). Differentiating protein-coding and noncoding RNA: challenges and ambiguities. PLOS Comput. Biol. 4: e1000176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doerr A. (2014). DIA mass spectrometry. Nat. Methods 12: 35. [Google Scholar]
- Dong X., Wang D., Liu P., Li C., Zhao Q., Zhu D., Yu J. (2013). Zm908p11, encoded by a short open reading frame (sORF) gene, functions in pollen tube growth as a profilin ligand in maize. J. Exp. Bot. 64: 2359–2372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douliez J.P., Michon T., Elmorjani K., Marion D. (2000). Structure, biological and technological functions of lipid transfer proteins and indolines, the major lipid binding proteins from cereal kernels. J. Cereal Sci. 32: 1–20. [Google Scholar]
- Dubovskii P.V., Vassilevski A.A., Slavokhotova A.A., Odintsova T.I., Grishin E.V., Egorov T.A., Arseniev A.S. (2011). Solution structure of a defense peptide from wheat with a 10-cysteine motif. Biochem. Biophys. Res. Commun. 411: 14–18. [DOI] [PubMed] [Google Scholar]
- Durgo H., Klement E., Hunyadi-Gulyas E., Szucs A., Kereszt A., Medzihradszky K.F., Kondorosi E. (2015). Identification of nodule-specific cysteine-rich plant peptides in endosymbiotic bacteria. Proteomics 15: 2291–2295. [DOI] [PubMed] [Google Scholar]
- Ebina I., et al. (2015). Identification of novel Arabidopsis thaliana upstream open reading frames that control expression of the main coding sequences in a peptide sequence-dependent manner. Nucleic Acids Res. 43: 1562–1576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elliott A.G., et al. (2014). Evolutionary origins of a bioactive peptide buried within preproalbumin. Plant Cell 26: 981–995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El Oirdi M., El Rahman T.A., Rigano L., El Hadrami A., Rodriguez M.C., Daayf F., Vojnov A., Bouarab K. (2011). Botrytis cinerea manipulates the antagonistic effects between immune pathways to promote disease development in tomato. Plant Cell 23: 2405–2421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endo S., Betsuyaku S., Fukuda H. (2014). Endogenous peptide ligand-receptor systems for diverse signaling networks in plants. Curr. Opin. Plant Biol. 21: 140–146. [DOI] [PubMed] [Google Scholar]
- Epple P., Apel K., Bohlmann H. (1997). Overexpression of an endogenous thionin enhances resistance of Arabidopsis against Fusarium oxysporum. Plant Cell 9: 509–520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farkas A., Maróti G., Durgő H., Györgypál Z., Lima R.M., Medzihradszky K.F., Kereszt A., Mergaert P., Kondorosi É. (2014). Medicago truncatula symbiotic peptide NCR247 contributes to bacteroid differentiation through multiple mechanisms. Proc. Natl. Acad. Sci. USA 111: 5183–5188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez A., Drozdzecki A., Hoogewijs K., Nguyen A., Beeckman T., Madder A., Hilson P. (2013a). Transcriptional and functional classification of the GOLVEN/ROOT GROWTH FACTOR/CLE-like signaling peptides reveals their role in lateral root and hair formation. Plant Physiol. 161: 954–970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez A., Hilson P., Beeckman T. (2013b). GOLVEN peptides as important regulatory signalling molecules of plant development. J. Exp. Bot. 64: 5263–5268. [DOI] [PubMed] [Google Scholar]
- Fesenko I.A., et al. (2015). Specific pools of endogenous peptides are present in gametophore, protonema, and protoplast cells of the moss Physcomitrella patens. BMC Plant Biol. 15: 87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiers M., Golemiec E., Xu J., van der Geest L., Heidstra R., Stiekema W., Liu C.-M. (2005). The 14-amino acid CLV3, CLE19, and CLE40 peptides trigger consumption of the root meristem in Arabidopsis through a CLAVATA2-dependent pathway. Plant Cell 17: 2542–2553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiume E., Fletcher J.C. (2012). Regulation of Arabidopsis embryo and endosperm development by the polypeptide signaling molecule CLE8. Plant Cell 24: 1000–1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franco O.L. (2011). Peptide promiscuity: an evolutionary concept for plant defense. FEBS Lett. 585: 995–1000. [DOI] [PubMed] [Google Scholar]
- Gao G.H., Liu W., Dai J.X., Wang J.F., Hu Z., Zhang Y., Wang D.C. (2001). Solution structure of PAFP-S: a new knottin-type antifungal peptide from the seeds of Phytolacca americana. Biochemistry 40: 10973–10978. [DOI] [PubMed] [Google Scholar]
- García A.N., Ayub N.D., Fox A.R., Gómez M.C., Diéguez M.J., Pagano E.M., Berini C.A., Muschietti J.P., Soto G. (2014). Alfalfa snakin-1 prevents fungal colonization and probably coevolved with rhizobia. BMC Plant Biol. 14: 248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaspar Y.M., McKenna J.A., McGinness B.S., Hinch J., Poon S., Connelly A.A., Anderson M.A., Heath R.L. (2014). Field resistance to Fusarium oxysporum and Verticillium dahliae in transgenic cotton expressing the plant defensin NaD1. J. Exp. Bot. 65: 1541–1550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorres K.L., Raines R.T. (2010). Prolyl 4-hydroxylase. Crit. Rev. Biochem. Mol. Biol. 45: 106–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goyal R.K., Mattoo A.K. (2014). Multitasking antimicrobial peptides in plant development and host defense against biotic/abiotic stress. Plant Sci. 228: 135–149. [DOI] [PubMed] [Google Scholar]
- Gray-Mitsumune M., Matton D.P. (2006). The Egg apparatus 1 gene from maize is a member of a large gene family found in both monocots and dicots. Planta 223: 618–625. [DOI] [PubMed] [Google Scholar]
- Grennan A.K. (2011). Metallothioneins, a diverse protein family. Plant Physiol. 155: 1750–1751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grienenberger E., Fletcher J.C. (2015). Polypeptide signaling molecules in plant development. Curr. Opin. Plant Biol. 23: 8–14. [DOI] [PubMed] [Google Scholar]
- Guefrachi I., Nagymihaly M., Pislariu C.I., Van de Velde W., Ratet P., Mars M., Udvardi M.K., Kondorosi E., Mergaert P., Alunni B. (2014). Extreme specificity of NCR gene expression in Medicago truncatula. BMC Genomics 15: 712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guillén G., Díaz-Camino C., Loyola-Torres C.A., Aparicio-Fabre R., Hernández-López A., Díaz-Sánchez M., Sanchez F. (2013). Detailed analysis of putative genes encoding small proteins in legume genomes. Front. Plant Sci. 4: 208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gully K., Hander T., Boller T., Bartels S. (2015). Perception of Arabidopsis AtPep peptides, but not bacterial elicitors, accelerates starvation-induced senescence. Front. Plant Sci. 6: 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo P., Yoshimura A., Ishikawa N., Yamaguchi T., Guo Y., Tsukaya H. (2015). Comparative analysis of the RTFL peptide family on the control of plant organogenesis. J. Plant Res. 128: 497–510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guttman M., Rinn J.L. (2012). Modular regulatory principles of large non-coding RNAs. Nature 482: 339–346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haag A.F., Kerscher B., Dall’Angelo S., Sani M., Longhi R., Baloban M., Wilson H.M., Mergaert P., Zanda M., Ferguson G.P. (2012). Role of cysteine residues and disulfide bonds in the activity of a legume root nodule-specific, cysteine-rich peptide. J. Biol. Chem. 287: 10791–10798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanada K., Zhang X., Borevitz J.O., Li W.-H., Shiu S.-H. (2007). A large number of novel coding small open reading frames in the intergenic regions of the Arabidopsis thaliana genome are transcribed and/or under purifying selection. Genome Res. 17: 632–640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanada K., et al. (2013). Small open reading frames associated with morphogenesis are hidden in plant genomes. Proc. Natl. Acad. Sci. USA 110: 2395–2400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hara K., Yokoo T., Kajita R., Onishi T., Yahata S., Peterson K.M., Torii K.U., Kakimoto T. (2009). Epidermal cell density is autoregulated via a secretory peptide, EPIDERMAL PATTERNING FACTOR 2 in Arabidopsis leaves. Plant Cell Physiol. 50: 1019–1031. [DOI] [PubMed] [Google Scholar]
- Harris P.W.R., Yang S.-H., Molina A., López G., Middleditch M., Brimble M.A. (2014). Plant antimicrobial peptides snakin-1 and snakin-2: chemical synthesis and insights into the disulfide connectivity. Chemistry 20: 5102–5110. [DOI] [PubMed] [Google Scholar]
- Haruta M., Constabel C.P. (2003). Rapid alkalinization factors in poplar cell cultures. Peptide isolation, cDNA cloning, and differential expression in leaves and methyl jasmonate-treated cells. Plant Physiol. 131: 814–823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haruta M., Sabat G., Stecker K., Minkoff B.B., Sussman M.R. (2014). A peptide hormone and its receptor protein kinase regulate plant cell expansion. Science 343: 408–411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higashiyama T. (2010). Peptide signaling in pollen-pistil interactions. Plant Cell Physiol. 51: 177–189. [DOI] [PubMed] [Google Scholar]
- Hirakawa Y., Shinohara H., Kondo Y., Inoue A., Nakanomyo I., Ogawa M., Sawa S., Ohashi-Ito K., Matsubayashi Y., Fukuda H. (2008). Non-cell-autonomous control of vascular stem cell fate by a CLE peptide/receptor system. Proc. Natl. Acad. Sci. USA 105: 15208–15213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hobe M., Müller R., Grünewald M., Brand U., Simon R. (2003). Loss of CLE40, a protein functionally equivalent to the stem cell restricting signal CLV3, enhances root waving in Arabidopsis. Dev. Genes Evol. 213: 371–381. [DOI] [PubMed] [Google Scholar]
- Hronková M., Wiesnerová D., Šimková M., Skůpa P., Dewitte W., Vráblová M., Zažímalová E., Šantrůček J. (2015). Light-induced STOMAGEN-mediated stomatal development in Arabidopsis leaves. J. Exp. Bot. 66: 4621–4630. [DOI] [PubMed] [Google Scholar]
- Huffaker A., Dafoe N.J., Schmelz E.A. (2011). ZmPep1, an ortholog of Arabidopsis elicitor peptide 1, regulates maize innate immunity and enhances disease resistance. Plant Physiol. 155: 1325–1338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huffaker A., Pearce G., Ryan C.A. (2006). An endogenous peptide signal in Arabidopsis activates components of the innate immune response. Proc. Natl. Acad. Sci. USA 103: 10098–10103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huffaker A., Pearce G., Veyrat N., Erb M., Turlings T.C.J., Sartor R., Shen Z., Briggs S.P., Vaughan M.M., Alborn H.T., Teal P.E., Schmelz E.A. (2013). Plant elicitor peptides are conserved signals regulating direct and indirect antiherbivore defense. Proc. Natl. Acad. Sci. USA 110: 5707–5712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunt L., Bailey K.J., Gray J.E. (2010). The signalling peptide EPFL9 is a positive regulator of stomatal development. New Phytol. 186: 609–614. [DOI] [PubMed] [Google Scholar]
- Imin N., Mohd-Radzman N.A., Ogilvie H.A., Djordjevic M.A. (2013). The peptide-encoding CEP1 gene modulates lateral root and nodule numbers in Medicago truncatula. J. Exp. Bot. 64: 5395–5409. [DOI] [PubMed] [Google Scholar]
- Ito Y., Nakanomyo I., Motose H., Iwamoto K., Sawa S., Dohmae N., Fukuda H. (2006). Dodeca-CLE peptides as suppressors of plant stem cell differentiation. Science 313: 842–845. [DOI] [PubMed] [Google Scholar]
- Ji H., Gheysen G., Ullah C., Verbeek R., Shang C., De Vleesschauwer D., Höfte M., Kyndt T. (2015). The role of thionins in rice defence against root pathogens. Mol. Plant Pathol., in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jorgensen R.A., Dorantes-Acosta A.E. (2012). Conserved peptide upstream open reading frames are associated with regulatory genes in angiosperms. Front. Plant Sci. 3: 191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jun J., Fiume E., Roeder A.H.K., Meng L., Sharma V.K., Osmont K.S., Baker C., Ha C.M., Meyerowitz E.M., Feldman L.J., Fletcher J.C. (2010). Comprehensive analysis of CLE polypeptide signaling gene expression and overexpression activity in Arabidopsis. Plant Physiol. 154: 1721–1736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jun J.H., Fiume E., Fletcher J.C. (2008). The CLE family of plant polypeptide signaling molecules. Cell. Mol. Life Sci. 65: 743–755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juntawong P., Girke T., Bazin J., Bailey-Serres J. (2014). Translational dynamics revealed by genome-wide profiling of ribosome footprints in Arabidopsis. Proc. Natl. Acad. Sci. USA 111: E203–E212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kader J.C. (1996). Lipid-transfer proteins in plants. Annu. Rev. Plant Physiol. Plant Mol. Biol. 47: 627–654. [DOI] [PubMed] [Google Scholar]
- Kehoe J.W., Bertozzi C.R. (2000). Tyrosine sulfation: a modulator of extracellular protein-protein interactions. Chem. Biol. 7: R57–R61. [DOI] [PubMed] [Google Scholar]
- Kim M.-S., et al. (2014). A draft map of the human proteome. Nature 509: 575–581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klauser D., Desurmont G.A., Glauser G., Vallat A., Flury P., Boller T., Turlings T.C.J., Bartels S. (2015). The Arabidopsis Pep-PEPR system is induced by herbivore feeding and contributes to JA-mediated plant defence against herbivory. J. Exp. Bot. 66: 5327–5336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondo T., Sawa S., Kinoshita A., Mizuno S., Kakimoto T., Fukuda H., Sakagami Y. (2006). A plant peptide encoded by CLV3 identified by in situ MALDI-TOF MS analysis. Science 313: 845–848. [DOI] [PubMed] [Google Scholar]
- Kondo T., Nakamura T., Yokomine K., Sakagami Y. (2008). Dual assay for MCLV3 activity reveals structure-activity relationship of CLE peptides. Biochem. Biophys. Res. Commun. 377: 312–316. [DOI] [PubMed] [Google Scholar]
- Krol E., Mentzel T., Chinchilla D., Boller T., Felix G., Kemmerling B., Postel S., Arents M., Jeworutzki E., Al-Rasheid K.A.S., Becker D., Hedrich R. (2010). Perception of the Arabidopsis danger signal peptide 1 involves the pattern recognition receptor AtPEPR1 and its close homologue AtPEPR2. J. Biol. Chem. 285: 13471–13479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumpf R.P., Shi C.-L., Larrieu A., Stø I.M., Butenko M.A., Péret B., Riiser E.S., Bennett M.J., Aalen R.B. (2013). Floral organ abscission peptide IDA and its HAE/HSL2 receptors control cell separation during lateral root emergence. Proc. Natl. Acad. Sci. USA 110: 5235–5240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kutschmar A., Rzewuski G., Stührwohldt N., Beemster G.T.S., Inzé D., Sauter M. (2009). PSK-α promotes root growth in Arabidopsis. New Phytol. 181: 820–831. [DOI] [PubMed] [Google Scholar]
- Laing W.A., Martínez-Sánchez M., Wright M.A., Bulley S.M., Brewster D., Dare A.P., Rassam M., Wang D., Storey R., Macknight R.C., Hellens R.P. (2015). An upstream open reading frame is essential for feedback regulation of ascorbate biosynthesis in Arabidopsis. Plant Cell 27: 772–786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauressergues D., Couzigou J.-M., Clemente H.S., Martinez Y., Dunand C., Bécard G., Combier J.-P. (2015). Primary transcripts of microRNAs encode regulatory peptides. Nature 520: 90–93. [DOI] [PubMed] [Google Scholar]
- Lay F.T., Poon S., McKenna J.A., Connelly A.A., Barbeta B.L., McGinness B.S., Fox J.L., Daly N.L., Craik D.J., Heath R.L., Anderson M.A. (2014). The C-terminal propeptide of a plant defensin confers cytoprotective and subcellular targeting functions. BMC Plant Biol. 14: 41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee D.G., Shin S.Y., Kim D.H., Seo M.Y., Kang J.H., Lee Y., Kim K.L., Hahm K.S. (1999). Antifungal mechanism of a cysteine-rich antimicrobial peptide, Ib-AMP1, from Impatiens balsamina against Candida albicans. Biotechnol. Lett. 21: 1047–1050. [Google Scholar]
- Lee J.S., Kuroha T., Hnilova M., Khatayevich D., Kanaoka M.M., McAbee J.M., Sarikaya M., Tamerler C., Torii K.U. (2012). Direct interaction of ligand-receptor pairs specifying stomatal patterning. Genes Dev. 26: 126–136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee O.S., et al. (2003). Pn-AMPs, the hevein-like proteins from Pharbitis nil confers disease resistance against phytopathogenic fungi in tomato, Lycopersicum esculentum. Phytochemistry 62: 1073–1079. [DOI] [PubMed] [Google Scholar]
- Li Y.L., Dai X.R., Yue X., Gao X.-Q., Zhang X.S. (2014). Identification of small secreted peptides (SSPs) in maize and expression analysis of partial SSP genes in reproductive tissues. Planta 240: 713–728. [DOI] [PubMed] [Google Scholar]
- Lim C.W., Lee Y.W., Hwang C.H. (2011). Soybean nodule-enhanced CLE peptides in roots act as signals in GmNARK-mediated nodulation suppression. Plant Cell Physiol. 52: 1613–1627. [DOI] [PubMed] [Google Scholar]
- Lobo D.S., Pereira I.B., Fragel-Madeira L., Medeiros L.N., Cabral L.M., Faria J., Bellio M., Campos R.C., Linden R., Kurtenbach E. (2007). Antifungal Pisum sativum defensin 1 interacts with Neurospora crassa cyclin F related to the cell cycle. Biochemistry 46: 987–996. [DOI] [PubMed] [Google Scholar]
- Lorbiecke R., Sauter M. (2002). Comparative analysis of PSK peptide growth factor precursor homologs. Plant Sci. 163: 321–332. [Google Scholar]
- Lori M., van Verk M.C., Hander T., Schatowitz H., Klauser D., Flury P., Gehring C.A., Boller T., Bartels S. (2015). Evolutionary divergence of the plant elicitor peptides (Peps) and their receptors: interfamily incompatibility of perception but compatibility of downstream signalling. J. Exp. Bot. pii: erv236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma J., Yan B., Qu Y., Qin F., Yang Y., Hao X., Yu J., Zhao Q., Zhu D., Ao G. (2008). Zm401, a short-open reading-frame mRNA or noncoding RNA, is essential for tapetum and microspore development and can regulate the floret formation in maize. J. Cell. Biochem. 105: 136–146. [DOI] [PubMed] [Google Scholar]
- Ma Y., Zhao Y., Walker R.K., Berkowitz G.A. (2013). Molecular steps in the immune signaling pathway evoked by plant elicitor peptides (Peps): CPKs, NO and ROS are downstream from the early Ca2+ signal. Plant Physiol. 163: 1459–1471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majewski J., Stec B. (2010). X-ray scattering studies of model lipid membrane interacting with purothionin provide support for a previously proposed mechanism of membrane lysis. Eur. Biophys. J. 39: 1155–1165. [DOI] [PubMed] [Google Scholar]
- Marmiroli N., Maestri E. (2014). Plant peptides in defense and signaling. Peptides 56: 30–44. [DOI] [PubMed] [Google Scholar]
- Maróti G., Kondorosi E. (2014). Nitrogen-fixing Rhizobium-legume symbiosis: are polyploidy and host peptide-governed symbiont differentiation general principles of endosymbiosis? Front. Microbiol. 5: 326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall E., Costa L.M., Gutierrez-Marcos J. (2011). Cysteine-rich peptides (CRPs) mediate diverse aspects of cell-cell communication in plant reproduction and development. J. Exp. Bot. 62: 1677–1686. [DOI] [PubMed] [Google Scholar]
- Márton M.L., Fastner A., Uebler S., Dresselhaus T. (2012). Overcoming hybridization barriers by the secretion of the maize pollen tube attractant ZmEA1 from Arabidopsis ovules. Curr. Biol. 22: 1194–1198. [DOI] [PubMed] [Google Scholar]
- Matsubayashi Y. (2011a). Post-translational modifications in secreted peptide hormones in plants. Plant Cell Physiol. 52: 5–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsubayashi Y. (2011b). Small post-translationally modified Peptide signals in Arabidopsis. Arabidopsis Book 9: e0150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsubayashi Y. (2014). Posttranslationally modified small-peptide signals in plants. Annu. Rev. Plant Biol. 65: 385–413. [DOI] [PubMed] [Google Scholar]
- Matsubayashi Y., Hanai H., Hara O., Sakagami Y. (1996). Active fragments and analogs of the plant growth factor, phytosulfokine: structure-activity relationships. Biochem. Biophys. Res. Commun. 225: 209–214. [DOI] [PubMed] [Google Scholar]
- Matsubayashi Y., Ogawa M., Morita A., Sakagami Y. (2002). An LRR receptor kinase involved in perception of a peptide plant hormone, phytosulfokine. Science 296: 1470–1472. [DOI] [PubMed] [Google Scholar]
- Matsubayashi Y., Ogawa M., Kihara H., Niwa M., Sakagami Y. (2006). Disruption and overexpression of Arabidopsis phytosulfokine receptor gene affects cellular longevity and potential for growth. Plant Physiol. 142: 45–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsubayashi Y., Sakagami Y. (1996). Phytosulfokine, sulfated peptides that induce the proliferation of single mesophyll cells of Asparagus officinalis L. Proc. Natl. Acad. Sci. USA 93: 7623–7627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuzaki Y., Ogawa-Ohnishi M., Mori A., Matsubayashi Y. (2010). Secreted peptide signals required for maintenance of root stem cell niche in Arabidopsis. Science 329: 1065–1067. [DOI] [PubMed] [Google Scholar]
- Meng L., Buchanan B.B., Feldman L.J., Luan S. (2012a). A putative nuclear CLE-like (CLEL) peptide precursor regulates root growth in Arabidopsis. Mol. Plant 5: 955–957. [DOI] [PubMed] [Google Scholar]
- Meng L., Buchanan B.B., Feldman L.J., Luan S. (2012b). CLE-like (CLEL) peptides control the pattern of root growth and lateral root development in Arabidopsis. Proc. Natl. Acad. Sci. USA 109: 1760–1765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng L., Feldman L.J. (2010). CLE14/CLE20 peptides may interact with CLAVATA2/CORYNE receptor-like kinases to irreversibly inhibit cell division in the root meristem of Arabidopsis. Planta 232: 1061–1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercer T.R., Dinger M.E., Mattick J.S. (2009). Long non-coding RNAs: insights into functions. Nat. Rev. Genet. 10: 155–159. [DOI] [PubMed] [Google Scholar]
- Mingossi F.B., Matos J.L., Rizzato A.P., Medeiros A.H., Falco M.C., Silva-Filho M.C., Moura D.S. (2010). SacRALF1, a peptide signal from the grass sugarcane (Saccharum spp.), is potentially involved in the regulation of tissue expansion. Plant Mol. Biol. 73: 271–281. [DOI] [PubMed] [Google Scholar]
- Miwa H., Tamaki T., Fukuda H., Sawa S. (2009). Evolution of CLE signaling: origins of the CLV1 and SOL2/CRN receptor diversity. Plant Signal. Behav. 4: 477–481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyawaki K., Tabata R., Sawa S. (2013). Evolutionarily conserved CLE peptide signaling in plant development, symbiosis, and parasitism. Curr. Opin. Plant Biol. 16: 598–606. [DOI] [PubMed] [Google Scholar]
- Mohd-Radzman N.A., Binos S., Truong T.T., Imin N., Mariani M., Djordjevic M.A. (2015). Novel MtCEP1 peptides produced in vivo differentially regulate root development in Medicago truncatula. J. Exp. Bot. pii: erv008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore K.L. (2003). The biology and enzymology of protein tyrosine O-sulfation. J. Biol. Chem. 278: 24243–24246. [DOI] [PubMed] [Google Scholar]
- Mortier V., Den Herder G., Whitford R., Van de Velde W., Rombauts S., D’Haeseleer K., Holsters M., Goormachtig S. (2010). CLE peptides control Medicago truncatula nodulation locally and systemically. Plant Physiol. 153: 222–237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosher S., Kemmerling B. (2013). PSKR1 and PSY1R-mediated regulation of plant defense responses. Plant Signal. Behav. 8: e24119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muñoz A., Chu M., Marris P.I., Sagaram U.S., Kaur J., Shah D.M., Read N.D. (2014). Specific domains of plant defensins differentially disrupt colony initiation, cell fusion and calcium homeostasis in Neurospora crassa. Mol. Microbiol. 92: 1357–1374. [DOI] [PubMed] [Google Scholar]
- Muramoto N., Tanaka T., Shimamura T., Mitsukawa N., Hori E., Koda K., Otani M., Hirai M., Nakamura K., Imaeda T. (2012). Transgenic sweet potato expressing thionin from barley gives resistance to black rot disease caused by Ceratocystis fimbriata in leaves and storage roots. Plant Cell Rep. 31: 987–997. [DOI] [PubMed] [Google Scholar]
- Murphy E., De Smet I. (2014). Understanding the RALF family: a tale of many species. Trends Plant Sci. 19: 664–671. [DOI] [PubMed] [Google Scholar]
- Murphy E., Smith S., De Smet I. (2012). Small signaling peptides in Arabidopsis development: how cells communicate over a short distance. Plant Cell 24: 3198–3217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mylne J.S., Colgrave M.L., Daly N.L., Chanson A.H., Elliott A.G., McCallum E.J., Jones A., Craik D.J. (2011). Albumins and their processing machinery are hijacked for cyclic peptides in sunflower. Nat. Chem. Biol. 7: 257–259. [DOI] [PubMed] [Google Scholar]
- Nahirñak V., Almasia N.I., Hopp H.E., Vazquez-Rovere C. (2012). Snakin/GASA proteins: involvement in hormone crosstalk and redox homeostasis. Plant Signal. Behav. 7: 1004–1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narita N.N., Moore S., Horiguchi G., Kubo M., Demura T., Fukuda H., Goodrich J., Tsukaya H. (2004). Overexpression of a novel small peptide ROTUNDIFOLIA4 decreases cell proliferation and alters leaf shape in Arabidopsis thaliana. Plant J. 38: 699–713. [DOI] [PubMed] [Google Scholar]
- Nawrot R., Barylski J., Nowicki G., Broniarczyk J., Buchwald W., Goździcka-Józefiak A. (2014). Plant antimicrobial peptides. Folia Microbiol. (Praha) 59: 181–196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neukermans J., Inzé A., Mathys J., De Coninck B., van de Cotte B., Cammue B.P.A., Van Breusegem F. (2015). ARACINs, Brassicaceae-specific peptides exhibiting antifungal activities against necrotrophic pathogens in Arabidopsis. Plant Physiol. 167: 1017–1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niarchou A., Alexandridou A., Athanasiadis E., Spyrou G. (2013). C-PAmP: large scale analysis and database construction containing high scoring computationally predicted antimicrobial peptides for all the available plant species. PLoS One 8: e79728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niderman T., Genetet I., Bruyère T., Gees R., Stintzi A., Legrand M., Fritig B., Mösinger E. (1995). Pathogenesis-related PR-1 proteins are antifungal. Isolation and characterization of three 14-kilodalton proteins of tomato and of a basic PR-1 of tobacco with inhibitory activity against Phytophthora infestans. Plant Physiol. 108: 17–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oelkers K., Goffard N., Weiller G.F., Gresshoff P.M., Mathesius U., Frickey T. (2008). Bioinformatic analysis of the CLE signaling peptide family. BMC Plant Biol. 8: 1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogawa M., Shinohara H., Sakagami Y., Matsubayashi Y. (2008). Arabidopsis CLV3 peptide directly binds CLV1 ectodomain. Science 319: 294. [DOI] [PubMed] [Google Scholar]
- Ogawa-Ohnishi M., Matsubayashi Y. (2015). Identification of three potent hydroxyproline O-galactosyltransferases in Arabidopsis. Plant J. 81: 736–746. [DOI] [PubMed] [Google Scholar]
- Ogawa-Ohnishi M., Matsushita W., Matsubayashi Y. (2013). Identification of three hydroxyproline O-arabinosyltransferases in Arabidopsis thaliana. Nat. Chem. Biol. 9: 726–730. [DOI] [PubMed] [Google Scholar]
- Ogilvie H.A., Imin N., Djordjevic M.A. (2014). Diversification of the C-TERMINALLY ENCODED PEPTIDE (CEP) gene family in angiosperms, and evolution of plant-family specific CEP genes. BMC Genomics 15: 870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohki S., Takeuchi M., Mori M. (2011). The NMR structure of stomagen reveals the basis of stomatal density regulation by plant peptide hormones. Nat. Commun. 2: 512. [DOI] [PubMed] [Google Scholar]
- Ohyama K., Ogawa M., Matsubayashi Y. (2008). Identification of a biologically active, small, secreted peptide in Arabidopsis by in silico gene screening, followed by LC-MS-based structure analysis. Plant J. 55: 152–160. [DOI] [PubMed] [Google Scholar]
- Ohyama K., Shinohara H., Ogawa-Ohnishi M., Matsubayashi Y. (2009). A glycopeptide regulating stem cell fate in Arabidopsis thaliana. Nat. Chem. Biol. 5: 578–580. [DOI] [PubMed] [Google Scholar]
- Okamoto S., Shinohara H., Mori T., Matsubayashi Y., Kawaguchi M. (2013). Root-derived CLE glycopeptides control nodulation by direct binding to HAR1 receptor kinase. Nat. Commun. 4: 2191. [DOI] [PubMed] [Google Scholar]
- Okuda S., et al. (2009). Defensin-like polypeptide LUREs are pollen tube attractants secreted from synergid cells. Nature 458: 357–361. [DOI] [PubMed] [Google Scholar]
- Oparin P.B., Mineev K.S., Dunaevsky Y.E., Arseniev A.S., Belozersky M.A., Grishin E.V., Egorov T.A., Vassilevski A.A. (2012). Buckwheat trypsin inhibitor with helical hairpin structure belongs to a new family of plant defence peptides. Biochem. J. 446: 69–77. [DOI] [PubMed] [Google Scholar]
- Patel S.U., Osborn R., Rees S., Thornton J.M. (1998). Structural studies of Impatiens balsamina antimicrobial protein (Ib-AMP1). Biochemistry 37: 983–990. [DOI] [PubMed] [Google Scholar]
- Pearce G. (2011). Systemin, hydroxyproline-rich systemin and the induction of protease inhibitors. Curr. Protein Pept. Sci. 12: 399–408. [DOI] [PubMed] [Google Scholar]
- Pearce G., Johnson S., Ryan C.A. (1993). Structure-activity of deleted and substituted systemin, an 18-amino acid polypeptide inducer of plant defensive genes. J. Biol. Chem. 268: 212–216. [PubMed] [Google Scholar]
- Pearce G., Moura D.S., Stratmann J., Ryan C.A. (2001a). Production of multiple plant hormones from a single polyprotein precursor. Nature 411: 817–820. [DOI] [PubMed] [Google Scholar]
- Pearce G., Moura D.S., Stratmann J., Ryan C.A. Jr (2001b). RALF, a 5-kDa ubiquitous polypeptide in plants, arrests root growth and development. Proc. Natl. Acad. Sci. USA 98: 12843–12847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearce G., Munske G., Yamaguchi Y., Ryan C.A. (2010b). Structure-activity studies of GmSubPep, a soybean peptide defense signal derived from an extracellular protease. Peptides 31: 2159–2164. [DOI] [PubMed] [Google Scholar]
- Pearce G., Ryan C.A. (2003). Systemic signaling in tomato plants for defense against herbivores. Isolation and characterization of three novel defense-signaling glycopeptide hormones coded in a single precursor gene. J. Biol. Chem. 278: 30044–30050. [DOI] [PubMed] [Google Scholar]
- Pearce G., Strydom D., Johnson S., Ryan C.A. (1991). A polypeptide from tomato leaves induces wound-inducible proteinase inhibitor proteins. Science 253: 895–897. [DOI] [PubMed] [Google Scholar]
- Pearce G., Yamaguchi Y., Munske G., Ryan C.A. (2008). Structure-activity studies of AtPep1, a plant peptide signal involved in the innate immune response. Peptides 29: 2083–2089. [DOI] [PubMed] [Google Scholar]
- Pearce G., Bhattacharya R., Chen Y.-C., Barona G., Yamaguchi Y., Ryan C.A. (2009). Isolation and characterization of hydroxyproline-rich glycopeptide signals in black nightshade leaves. Plant Physiol. 150: 1422–1433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearce G., Yamaguchi Y., Barona G., Ryan C.A. (2010a). A subtilisin-like protein from soybean contains an embedded, cryptic signal that activates defense-related genes. Proc. Natl. Acad. Sci. USA 107: 14921–14925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penterman J., Abo R.P., De Nisco N.J., Arnold M.F.F., Longhi R., Zanda M., Walker G.C. (2014). Host plant peptides elicit a transcriptional response to control the Sinorhizobium meliloti cell cycle during symbiosis. Proc. Natl. Acad. Sci. USA 111: 3561–3566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pii Y., Molesini B., Masiero S., Pandolfini T. (2012). The non-specific lipid transfer protein N5 of Medicago truncatula is implicated in epidermal stages of rhizobium-host interaction. BMC Plant Biol. 12: 233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pimenta D.C., Lebrun I. (2007). Cryptides: Buried secrets in proteins. Peptides 28: 2403–2410. [DOI] [PubMed] [Google Scholar]
- Porto W.F., Souza V.A., Nolasco D.O., Franco O.L. (2012). In silico identification of novel hevein-like peptide precursors. Peptides 38: 127–136. [DOI] [PubMed] [Google Scholar]
- Porto W.F., Franco O.L. (2013). Theoretical structural insights into the snakin/GASA family. Peptides 44: 163–167. [DOI] [PubMed] [Google Scholar]
- Prabakaran S., Hemberg M., Chauhan R., Winter D., Tweedie-Cullen R.Y., Dittrich C., Hong E., Gunawardena J., Steen H., Kreiman G., Steen J.A. (2014). Quantitative profiling of peptides from RNAs classified as noncoding. Nat. Commun. 5: 5429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prabhu S., Dennison S.R., Lea B., Snape T.J., Nicholl I.D., Radecka I., Harris F. (2013). Anionic antimicrobial and anticancer peptides from plants. Crit. Rev. Plant Sci. 32: 303–320. [Google Scholar]
- Qu L.-J., Li L., Lan Z., Dresselhaus T. (2015). Peptide signalling during the pollen tube journey and double fertilization. J. Exp. Bot. pii: erv275. [DOI] [PubMed] [Google Scholar]
- Ramamoorthy V., Cahoon E.B., Li J., Thokala M., Minto R.E., Shah D.M. (2007). Glucosylceramide synthase is essential for alfalfa defensin-mediated growth inhibition but not for pathogenicity of Fusarium graminearum. Mol. Microbiol. 66: 771–786. [DOI] [PubMed] [Google Scholar]
- Ream T.S., Haag J.R., Wierzbicki A.T., Nicora C.D., Norbeck A.D., Zhu J.K., Hagen G., Guilfoyle T.J., Pasa-Tolić L., Pikaard C.S. (2009). Subunit compositions of the RNA-silencing enzymes Pol IV and Pol V reveal their origins as specialized forms of RNA polymerase II. Mol. Cell 33: 192–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reid D.E., Ferguson B.J., Gresshoff P.M. (2011). Inoculation- and nitrate-induced CLE peptides of soybean control NARK-dependent nodule formation. Mol. Plant Microbe Interact. 24: 606–618. [DOI] [PubMed] [Google Scholar]
- Ren F., Lu Y.T. (2006). Overexpression of tobacco hydroxyproline-rich glycopeptide systemin precursor A gene in transgenic tobacco enhances resistance against Helicoverpa armigera larvae. Plant Sci. 171: 286–292. [Google Scholar]
- Replogle A., Wang J., Bleckmann A., Hussey R.S., Baum T.J., Sawa S., Davis E.L., Wang X., Simon R., Mitchum M.G. (2011). Nematode CLE signaling in Arabidopsis requires CLAVATA2 and CORYNE. Plant J. 65: 430–440. [DOI] [PubMed] [Google Scholar]
- Richards S., Wink R.H., Simon R. (2015). Mathematical modelling of WOX5- and CLE40-mediated columella stem cell homeostasis in Arabidopsis. J. Exp. Bot. pii: erv257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson L.G.L., Torii K.U. (2013). Take a deep breath: peptide signalling in stomatal patterning and differentiation. J. Exp. Bot. 64: 5243–5251. [DOI] [PubMed] [Google Scholar]
- Roberts I., Smith S., De Rybel B., Van Den Broeke J., Smet W., De Cokere S., Mispelaere M., De Smet I., Beeckman T. (2013). The CEP family in land plants: evolutionary analyses, expression studies, and role in Arabidopsis shoot development. J. Exp. Bot. 64: 5371–5381. [DOI] [PubMed] [Google Scholar]
- Röhrig H., Schmidt J., Miklashevichs E., Schell J., John M. (2002). Soybean ENOD40 encodes two peptides that bind to sucrose synthase. Proc. Natl. Acad. Sci. USA 99: 1915–1920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rong W., Qi L., Wang J., Du L., Xu H., Wang A., Zhang Z. (2013). Expression of a potato antimicrobial peptide SN1 increases resistance to take-all pathogen Gaeumannomyces graminis var. tritici in transgenic wheat. Funct. Integr. Genomics 13: 403–409. [DOI] [PubMed] [Google Scholar]
- Ross A., Yamada K., Hiruma K., Yamashita-Yamada M., Lu X., Takano Y., Tsuda K., Saijo Y. (2014). The Arabidopsis PEPR pathway couples local and systemic plant immunity. EMBO J. 33: 62–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Shukurov R., D Voblikova V., Nikonorova A.K., Komakhin R.A., V Komakhina V., A Egorov T., V Grishin E., V Babakov A. (2012). Transformation of tobacco and Arabidopsis plants with Stellaria media genes encoding novel hevein-like peptides increases their resistance to fungal pathogens. Transgenic Res. 21: 313–325. [DOI] [PubMed] [Google Scholar]
- Ruiz-Orera J., Messeguer X., Subirana J.A., Alba M.M. (2014). Long non-coding RNAs as a source of new peptides. eLife 3: e03523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sagaram U.S., El-Mounadi K., Buchko G.W., Berg H.R., Kaur J., Pandurangi R.S., Smith T.J., Shah D.M. (2013). Structural and functional studies of a phosphatidic acid-binding antifungal plant defensin MtDef4: identification of an RGFRRR motif governing fungal cell entry. PLoS One 8: e82485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sagaram U.S., Pandurangi R., Kaur J., Smith T.J., Shah D.M. (2011). Structure-activity determinants in antifungal plant defensins MsDef1 and MtDef4 with different modes of action against Fusarium graminearum. PLoS One 6: e18550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samir P., Link A.J. (2011). Analyzing the cryptome: uncovering secret sequences. AAPS J. 13: 152–158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauter M. (2015). Phytosulfokine peptide signalling. J. Exp. Bot. pii: erv071. [DOI] [PubMed] [Google Scholar]
- Schmelz E.A., Carroll M.J., LeClere S., Phipps S.M., Meredith J., Chourey P.S., Alborn H.T., Teal P.E.A. (2006). Fragments of ATP synthase mediate plant perception of insect attack. Proc. Natl. Acad. Sci. USA 103: 8894–8899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmelz E.A., Huffaker A., Carroll M.J., Alborn H.T., Ali J.G., Teal P.E.A. (2012). An amino acid substitution inhibits specialist herbivore production of an antagonist effector and recovers insect-induced plant defenses. Plant Physiol. 160: 1468–1478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmelz E.A., LeClere S., Carroll M.J., Alborn H.T., Teal P.E.A. (2007). Cowpea chloroplastic ATP synthase is the source of multiple plant defense elicitors during insect herbivory. Plant Physiol. 144: 793–805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoof H., Lenhard M., Haecker A., Mayer K.F.X., Jürgens G., Laux T. (2000). The stem cell population of Arabidopsis shoot meristems in maintained by a regulatory loop between the CLAVATA and WUSCHEL genes. Cell 100: 635–644. [DOI] [PubMed] [Google Scholar]
- Segura A., Moreno M., Madueño F., Molina A., García-Olmedo F. (1999). Snakin-1, a peptide from potato that is active against plant pathogens. Mol. Plant Microbe Interact. 12: 16–23. [DOI] [PubMed] [Google Scholar]
- Shinohara H., Moriyama Y., Ohyama K., Matsubayashi Y. (2012). Biochemical mapping of a ligand-binding domain within Arabidopsis BAM1 reveals diversified ligand recognition mechanisms of plant LRR-RKs. Plant J. 70: 845–854. [DOI] [PubMed] [Google Scholar]
- Shinohara H., Matsubayashi Y. (2013). Chemical synthesis of Arabidopsis CLV3 glycopeptide reveals the impact of hydroxyproline arabinosylation on peptide conformation and activity. Plant Cell Physiol. 54: 369–374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirasawa-Seo N., Nakamura S., Ukai N., Honkura R., Takayoshi I., Ohashi Y. (2002). Ectopic expression of an oat thionin gene in carnation plants confers enhanced resistance to bacterial wilt disease. Plant Biotechnol. 19: 311–317. [Google Scholar]
- Silverstein K.A.T., Moskal W.A. Jr., Wu H.C., Underwood B.A., Graham M.A., Town C.D., VandenBosch K.A. (2007). Small cysteine-rich peptides resembling antimicrobial peptides have been under-predicted in plants. Plant J. 51: 262–280. [DOI] [PubMed] [Google Scholar]
- Slavoff S.A., Mitchell A.J., Schwaid A.G., Cabili M.N., Ma J., Levin J.Z., Karger A.D., Budnik B.A., Rinn J.L., Saghatelian A. (2013). Peptidomic discovery of short open reading frame-encoded peptides in human cells. Nat. Chem. Biol. 9: 59–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slavokhotova A.A., Rogozhin E.A., Musolyamov A.K., Andreev Y.A., Oparin P.B., Berkut A.A., Vassilevski A.A., Egorov T.A., Grishin E.V., Odintsova T.I. (2014). Novel antifungal α-hairpinin peptide from Stellaria media seeds: structure, biosynthesis, gene structure and evolution. Plant Mol. Biol. 84: 189–202. [DOI] [PubMed] [Google Scholar]
- Sousa C., Johansson C., Charon C., Manyani H., Sautter C., Kondorosi A., Crespi M. (2001). Translational and structural requirements of the early nodulin gene enod40, a short-open reading frame-containing RNA, for elicitation of a cell-specific growth response in the alfalfa root cortex. Mol. Cell. Biol. 21: 354–366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sparks E., Wachsman G., Benfey P.N. (2013). Spatiotemporal signalling in plant development. Nat. Rev. Genet. 14: 631–644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spartz A.K., Lee S.H., Wenger J.P., Gonzalez N., Itoh H., Inzé D., Peer W.A., Murphy A.S., Overvoorde P.J., Gray W.M. (2012). The SAUR19 subfamily of SMALL AUXIN UP RNA genes promote cell expansion. Plant J. 70: 978–990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spincemaille P., Chandhok G., Newcomb B., Verbeek J., Vriens K., Zibert A., Schmidt H., Hannun Y.A., van Pelt J., Cassiman D., Cammue B.P., Thevissen K. (2014a). The plant decapeptide OSIP108 prevents copper-induced apoptosis in yeast and human cells. Biochim. Biophys. Acta 1843: 1207–1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spincemaille P., Pham D.H., Chandhok G., Verbeek J., Zibert A., Libbrecht L., Schmidt H., Esguerra C.V., de Witte P.A.M., Cammue B.P.A., Cassiman D., Thevissen K. (2014b). The plant decapeptide OSIP108 prevents copper-induced toxicity in various models for Wilson disease. Toxicol. Appl. Pharmacol. 280: 345–351. [DOI] [PubMed] [Google Scholar]
- Stahl Y., Wink R.H., Ingram G.C., Simon R. (2009). A signaling module controlling the stem cell niche in Arabidopsis root meristems. Curr. Biol. 19: 909–914. [DOI] [PubMed] [Google Scholar]
- Staudt A.C., Wenkel S. (2011). Regulation of protein function by ‘microProteins’. EMBO Rep. 12: 35–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stec B. (2006). Plant thionins--the structural perspective. Cell. Mol. Life Sci. 63: 1370–1385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stec B., Markman O., Rao U., Heffron G., Henderson S., Vernon L.P., Brumfeld V., Teeter M.M. (2004). Proposal for molecular mechanism of thionins deduced from physico-chemical studies of plant toxins. J. Pept. Res. 64: 210–224. [DOI] [PubMed] [Google Scholar]
- Stenvik G.E., Butenko M.A., Urbanowicz B.R., Rose J.K.C., Aalen R.B. (2006). Overexpression of INFLORESCENCE DEFICIENT IN ABSCISSION activates cell separation in vestigial abscission zones in Arabidopsis. Plant Cell 18: 1467–1476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stenvik G.-E., Tandstad N.M., Guo Y., Shi C.-L., Kristiansen W., Holmgren A., Clark S.E., Aalen R.B., Butenko M.A. (2008). The EPIP peptide of INFLORESCENCE DEFICIENT IN ABSCISSION is sufficient to induce abscission in arabidopsis through the receptor-like kinases HAESA and HAESA-LIKE2. Plant Cell 20: 1805–1817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stes E., Gevaert K., De Smet I. (2015). Phosphoproteomics-based peptide ligand-receptor kinase pairing. Commentary on: “A peptide hormone and its receptor protein kinase regulate plant cell expansion”. Front. Plant Sci. 6: 224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strabala T.J., Phillips L., West M., Stanbra L. (2014). Bioinformatic and phylogenetic analysis of the CLAVATA3/EMBRYO-SURROUNDING REGION (CLE) and the CLE-LIKE signal peptide genes in the Pinophyta. BMC Plant Biol. 14: 47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabata R., Sawa S. (2014). Maturation processes and structures of small secreted peptides in plants. Front. Plant Sci. 5: 311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabata R., Sumida K., Yoshii T., Ohyama K., Shinohara H., Matsubayashi Y. (2014). Perception of root-derived peptides by shoot LRR-RKs mediates systemic N-demand signaling. Science 346: 343–346. [DOI] [PubMed] [Google Scholar]
- Tailor R.H., Acland D.P., Attenborough S., Cammue B.P.A., Evans I.J., Osborn R.W., Ray J.A., Rees S.B., Broekaert W.F. (1997). A novel family of small cysteine-rich antimicrobial peptides from seed of Impatiens balsamina is derived from a single precursor protein. J. Biol. Chem. 272: 24480–24487. [DOI] [PubMed] [Google Scholar]
- Takata N., Yokota K., Ohki S., Mori M., Taniguchi T., Kurita M. (2013). Evolutionary relationship and structural characterization of the EPF/EPFL gene family. PLoS One 8: e65183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takayama S., Shiba H., Iwano M., Shimosato H., Che F.S., Kai N., Watanabe M., Suzuki G., Hinata K., Isogai A. (2000). The pollen determinant of self-incompatibility in Brassica campestris. Proc. Natl. Acad. Sci. USA 97: 1920–1925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takayama S., Shimosato H., Shiba H., Funato M., Che F.-S., Watanabe M., Iwano M., Isogai A. (2001). Direct ligand-receptor complex interaction controls Brassica self-incompatibility. Nature 413: 534–538. [DOI] [PubMed] [Google Scholar]
- Takeuchi H., Higashiyama T. (2012). A species-specific cluster of defensin-like genes encodes diffusible pollen tube attractants in Arabidopsis. PLoS Biol. 10: e1001449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang J., Han Z., Sun Y., Zhang H., Gong X., Chai J. (2015). Structural basis for recognition of an endogenous peptide by the plant receptor kinase PEPR1. Cell Res. 25: 110–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor C.M., Karunaratne C.V., Xie N. (2012). Glycosides of hydroxyproline: some recent, unusual discoveries. Glycobiology 22: 757–767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor N.L., Fenske R., Castleden I., Tomaz T., Nelson C.J., Millar A.H. (2014). Selected reaction monitoring to determine protein abundance in Arabidopsis using the Arabidopsis proteotypic predictor. Plant Physiol. 164: 525–536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terras F.R., Goderis I.J., Van Leuven F., Vanderleyden J., Cammue B.P., Broekaert W.F. (1992). In vitro antifungal activity of a radish (Raphanus sativus L.) seed protein homologous to nonspecific lipid transfer proteins. Plant Physiol. 100: 1055–1058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terras F.R., et al. (1995). Small cysteine-rich antifungal proteins from radish: their role in host defense. Plant Cell 7: 573–588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tesfaye M., et al. (2013). Spatio-temporal expression patterns of Arabidopsis thaliana and Medicago truncatula defensin-like genes. PLoS One 8: e58992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thevissen K., François I.E.J.A., Sijtsma L., van Amerongen A., Schaaper W.M.M., Meloen R., Posthuma-Trumpie T., Broekaert W.F., Cammue B.P.A. (2005). Antifungal activity of synthetic peptides derived from Impatiens balsamina antimicrobial peptides Ib-AMP1 and Ib-AMP4. Peptides 26: 1113–1119. [DOI] [PubMed] [Google Scholar]
- Thevissen K., Warnecke D.C., François I.E.J.A., Leipelt M., Heinz E., Ott C., Zähringer U., Thomma B.P.H.J., Ferket K.K.A., Cammue B.P.A. (2004). Defensins from insects and plants interact with fungal glucosylceramides. J. Biol. Chem. 279: 3900–3905. [DOI] [PubMed] [Google Scholar]
- Thomma B.P.H.J., Cammue B.P.A., Thevissen K. (2002). Plant defensins. Planta 216: 193–202. [DOI] [PubMed] [Google Scholar]
- Tintor N., Ross A., Kanehara K., Yamada K., Fan L., Kemmerling B., Nürnberger T., Tsuda K., Saijo Y. (2013). Layered pattern receptor signaling via ethylene and endogenous elicitor peptides during Arabidopsis immunity to bacterial infection. Proc. Natl. Acad. Sci. USA 110: 6211–6216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiricz H., Szücs A., Farkas A., Pap B., Lima R.M., Maróti G., Kondorosi É., Kereszt A. (2013). Antimicrobial nodule-specific cysteine-rich peptides induce membrane depolarization-associated changes in the transcriptome of Sinorhizobium meliloti. Appl. Environ. Microbiol. 79: 6737–6746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torrent M., Di Tommaso P., Pulido D., Nogués M.V., Notredame C., Boix E., Andreu D. (2012). AMPA: an automated web server for prediction of protein antimicrobial regions. Bioinformatics 28: 130–131. [DOI] [PubMed] [Google Scholar]
- Toumadje A., Johnson W.C. Jr. (1995). Systemin has the characteristics of a poly (L-proline) II type helix. J. Am. Chem. Soc. 117: 7023–7024. [Google Scholar]
- Tousheh M., Miroliaei M., Asghar Rastegari A., Ghaedi K., Esmaeili A., Matkowski A. (2013). Computational evaluation on the binding affinity of non-specific lipid-transfer protein-2 with fatty acids. Comput. Biol. Med. 43: 1732–1738. [DOI] [PubMed] [Google Scholar]
- Trivilin A.P., Hartke S., Moraes M.G. (2014). Components of different signalling pathways regulated by a new orthologue of AtPROPEP1 in tomato following infection by pathogens. Plant Pathol. 2: 1110–1118. [Google Scholar]
- Tucker M.L., Yang R. (2012). IDA-like gene expression in soybean and tomato leaf abscission and requirement for a diffusible stelar abscission signal. AoB Plants 2012: pls035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uchida N., Tasaka M. (2013). Regulation of plant vascular stem cells by endodermis-derived EPFL-family peptide hormones and phloem-expressed ERECTA-family receptor kinases. J. Exp. Bot. 64: 5335–5343. [DOI] [PubMed] [Google Scholar]
- Utkina L.L., Andreev Y.A., Rogozhin E.A., Korostyleva T.V., Slavokhotova A.A., Oparin P.B., Vassilevski A.A., Grishin E.V., Egorov T.A., Odintsova T.I. (2013). Genes encoding 4-Cys antimicrobial peptides in wheat Triticum kiharae Dorof. et Migush.: multimodular structural organization, instraspecific variability, distribution and role in defence. FEBS J. 280: 3594–3608. [DOI] [PubMed] [Google Scholar]
- Valdivia E.R., Chevalier D., Sampedro J., Taylor I., Niederhuth C.E., Walker J.C. (2012). DVL genes play a role in the coordination of socket cell recruitment and differentiation. J. Exp. Bot. 63: 1405–1412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Weerden N.L., Anderson M.A. (2013). Plant defensins: Common fold, multiple functions. Fungal Biol. Rev. 26: 121–131. [Google Scholar]
- van der Weerden N.L., Lay F.T., Anderson M.A. (2008). The plant defensin, NaD1, enters the cytoplasm of Fusarium oxysporum hyphae. J. Biol. Chem. 283: 14445–14452. [DOI] [PubMed] [Google Scholar]
- van der Weerden N.L., Hancock R.E., Anderson M.A. (2010). Permeabilization of fungal hyphae by the plant defensin NaD1 occurs through a cell wall-dependent process. J. Biol. Chem. 285: 37513–37520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Weerden N.L., Bleackley M.R., Anderson M.A. (2013). Properties and mechanisms of action of naturally occurring antifungal peptides. Cell. Mol. Life Sci. 70: 3545–3570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van de Velde W., et al. (2010). Plant peptides govern terminal differentiation of bacteria in symbiosis. Science 327: 1122–1126. [DOI] [PubMed] [Google Scholar]
- van Loon L.C., Rep M., Pieterse C.M.J. (2006). Significance of inducible defense-related proteins in infected plants. Annu. Rev. Phytopathol. 44: 135–162. [DOI] [PubMed] [Google Scholar]
- von Arnim A.G., Jia Q., Vaughn J.N. (2014). Regulation of plant translation by upstream open reading frames. Plant Sci. 214: 1–12. [DOI] [PubMed] [Google Scholar]
- Vriens K., Cammue B.P., Thevissen K. (2014). Antifungal plant defensins: mechanisms of action and production. Molecules 19: 12280–12303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan X., Hontelez J., Lillo A., Guarnerio C., van de Peut D., Fedorova E., Bisseling T., Franssen H. (2007). Medicago truncatula ENOD40-1 and ENOD40-2 are both involved in nodule initiation and bacteroid development. J. Exp. Bot. 58: 2033–2041. [DOI] [PubMed] [Google Scholar]
- Wang G. (2012). Post-translational modifications of natural antimicrobial peptides and strategies for peptide engineering. Curr. Biotechnol. 1: 72–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J., Lee C., Replogle A., Joshi S., Korkin D., Hussey R., Baum T.J., Davis E.L., Wang X., Mitchum M.G. (2010). Dual roles for the variable domain in protein trafficking and host-specific recognition of Heterodera glycines CLE effector proteins. New Phytol. 187: 1003–1017. [DOI] [PubMed] [Google Scholar]
- Wang J., Replogle A., Hussey R., Baum T., Wang X., Davis E.L., Mitchum M.G. (2011). Identification of potential host plant mimics of CLAVATA3/ESR (CLE)-like peptides from the plant-parasitic nematode Heterodera schachtii. Mol. Plant Pathol. 12: 177–186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang N.-J., Lee C.-C., Cheng C.-S., Lo W.-C., Yang Y.-F., Chen M.-N., Lyu P.-C. (2012). Construction and analysis of a plant non-specific lipid transfer protein database (nsLTPDB). BMC Genomics 13 (suppl. 1): S9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen J., Lease K.A., Walker J.C. (2004). DVL, a novel class of small polypeptides: overexpression alters Arabidopsis development. Plant J. 37: 668–677. [DOI] [PubMed] [Google Scholar]
- Wethmar K., Barbosa-Silva A., Andrade-Navarro M.A., Leutz A. (2014). UORFdb - A comprehensive literature database on eukaryotic uORF biology. Nucleic Acids Res. 42: 60–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitford R., et al. (2012). GOLVEN secretory peptides regulate auxin carrier turnover during plant gravitropic responses. Dev. Cell 22: 678–685. [DOI] [PubMed] [Google Scholar]
- Wilmes M., Cammue B.P.A., Sahl H.G., Thevissen K. (2011). Antibiotic activities of host defense peptides: more to it than lipid bilayer perturbation. Nat. Prod. Rep. 28: 1350–1358. [DOI] [PubMed] [Google Scholar]
- Wolf S., Höfte H. (2014). Growth control: a saga of cell walls, ROS, and peptide receptors. Plant Cell 26: 1848–1856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woriedh M., Merkl R., Dresselhaus T. (2015). Maize EMBRYO SAC family peptides interact differentially with pollen tubes and fungal cells. J. Exp. Bot. pii: erv268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wrzaczek M., et al. (2015). GRIM REAPER peptide binds to receptor kinase PRK5 to trigger cell death in Arabidopsis. EMBO J. 34: 55–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J., Kurten E.L., Monshausen G., Hummel G.M., Gilroy S., Baldwin I.T. (2007). NaRALF, a peptide signal essential for the regulation of root hair tip apoplastic pH in Nicotiana attenuata, is required for root hair development and plant growth in native soils. Plant J. 52: 877–890. [DOI] [PubMed] [Google Scholar]
- Xiang Y., Huang R.H., Liu X.Z., Zhang Y., Wang D.C. (2004). Crystal structure of a novel antifungal protein distinct with five disulfide bridges from Eucommia ulmoides Oliver at an atomic resolution. J. Struct. Biol. 148: 86–97. [DOI] [PubMed] [Google Scholar]
- Xu C., et al. (2015). A cascade of arabinosyltransferases controls shoot meristem size in tomato. Nat. Genet. 47: 784–792. [DOI] [PubMed] [Google Scholar]
- Xu T.-T., Ren S.-C., Song X.-F., Liu C.-M. (2015). CLE19 expressed in the embryo regulates both cotyledon establishment and endosperm development in Arabidopsis. J. Exp. Bot. pii: erv293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi Y., Huffaker A., Bryan A.C., Tax F.E., Ryan C.A. (2010). PEPR2 is a second receptor for the Pep1 and Pep2 peptides and contributes to defense responses in Arabidopsis. Plant Cell 22: 508–522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi Y., Barona G., Ryan C.A., Pearce G. (2011). GmPep914, an eight-amino acid peptide isolated from soybean leaves, activates defense-related genes. Plant Physiol. 156: 932–942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi Y., Huffaker A. (2011). Endogenous peptide elicitors in higher plants. Curr. Opin. Plant Biol. 14: 351–357. [DOI] [PubMed] [Google Scholar]
- Yang X., et al. (2011). Discovery and annotation of small proteins using genomics, proteomics, and computational approaches. Genome Res. 21: 634–641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Yang S., Song Y., , J. (2014). Genome-wide characterization, expression and functional analysis of CLV3/ESR gene family in tomato. BMC Genomics 15: 827. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.