LEUNIG_HOMOLOG interacts with the transcription factor PHYTOCHROME-INTERACTING FACTOR1 and acts as a transcriptional coregulator to inhibit light-dependent seed germination in Arabidopsis.
Abstract
PHYTOCHROME-INTERACTING FACTOR1 (PIF1) is a basic helix-loop-helix transcription factor that inhibits light-dependent seed germination in Arabidopsis thaliana. However, it remains unclear whether PIF1 requires other factors to regulate its direct targets. Here, we demonstrate that LEUNIG_HOMOLOG (LUH), a Groucho family transcriptional corepressor, binds to PIF1 and coregulates its targets. Not only are the transcriptional profiles of the luh and pif1 mutants remarkably similar, more than 80% of the seeds of both genotypes germinate in the dark. We show by chromatin immunoprecipitation that LUH binds a subset of PIF1 targets in a partially PIF1-dependent manner. Unexpectedly, we found LUH binds and coregulates not only PIF1-activated targets but also PIF1-repressed targets. Together, our results indicate LUH functions with PIF1 as a transcriptional coregulator to inhibit seed germination.
INTRODUCTION
Phytochrome-interacting factors (PIFs) are basic helix-loop-helix transcription factors that interact with phytochromes in a light-dependent manner (Ni et al., 1998; Huq and Quail, 2002; Khanna et al., 2004; Oh et al., 2004; Leivar et al., 2008). The seven PIFs in Arabidopsis thaliana (PIF1, PIF3, PIF4, PIF5, PIF6, PIF7, and PIF8) regulate light responses ranging from seed germination (Oh et al., 2004) and hypocotyl negative gravitropism (Oh et al., 2004; Shin et al., 2009; Kim et al., 2011) to seedling photomorphogenesis (Ni et al., 1998; Huq and Quail, 2002; Kim et al., 2003; Fujimori et al., 2004; Huq et al., 2004), plant shade avoidance (Lorrain et al., 2008; Leivar et al., 2012; Li et al., 2012), and circadian gating of shade avoidance (Salter et al., 2003). PIF1 inhibits the germination of imbibed seeds in the dark when phytochromes are inactive by repressing several genes required for cell wall loosening (Oh et al., 2009). These include six EXPANSIN (EXP) genes and seven XYLOGLUCAN ENDO-TRANSGLYCOSYLASE/HYDROLASE (XTH) genes.
PIF1 also inhibits seed germination in the dark by regulating the abscisic acid (ABA), gibberellin (GA), brassinosteroid, and jasmonate hormone signaling pathways (Oh et al., 2006, 2007, 2009). PIF1’s coordinate regulation of plant hormone signaling pathways is exemplified by the mechanisms of its activation of ABA signaling and simultaneous inhibition of GA signaling. Not only does PIF1 regulate the expression of the genes required for ABA and GA biosynthesis, it also regulates the expression of key ABA and GA signaling components. Specifically, PIF1 increases ABA levels in imbibed seeds by activating the expression of ABA biosynthetic genes (ABA1, NCED6, and NCED9) and repressing the expression of ABA catabolic genes (CYP707A2) (Oh et al., 2007). PIF1 reduces GA levels by repressing GA biosynthetic genes (GA3ox1 and GA3ox2) and activating GA catabolic genes (GA2ox2) (Oh et al., 2006). PIF1 also enhances ABA signaling by activating the expression of positive components of the ABA signaling pathway (ABI3 and ABI5) (Oh et al., 2009). Conversely, PIF1 inhibits GA signaling by activating the expression of negative components of the GA signaling pathway (GAI and RGA) (Oh et al., 2006). This reciprocal pattern of regulation for both biosynthetic and signaling genes drives strong ABA signaling and limits GA signaling, a hormonal balance that inhibits the germination of imbibed seeds in the dark.
Of the PIF1-regulated genes mentioned above, only two of the six EXP genes, one of the seven XTH genes, and all four signaling pathway components (ABI3, ABI5, GAI, and RGA) are PIF1 targets (Oh et al., 2009). This implies that the rest of PIF1’s influence on cell wall loosening and hormone biosynthesis occurs indirectly via genetic interactions with the PIF1 targets. SOMNUS (SOM) and DOF AFFECTING GERMINATION1 (DAG1) function downstream of PIF1 to regulate subsets of the ABA and GA biosynthetic genes. SOM encodes a zinc finger protein that represses GA3ox1 and GA3ox2 partly by decreasing the expression of two histone arginine demethylase genes (Jumonji20 [JMJ20] and JMJ22) (Cho et al., 2012). DAG1 directly represses GA3ox1 by binding its promoter (Gabriele et al., 2010).
Whole processes reverse in the light when phytochromes become active and enter the nucleus (Sakamoto and Nagatani, 1996; Kircher et al., 1999; Yamaguchi et al., 1999). Activated phytochromes bind PIF1 (Huq et al., 2004; Oh et al., 2004), causing it to release DNA (Park et al., 2012). This subsequently leads to PIF1 protein degradation (Oh et al., 2004; Shen et al., 2008). Loss of PIF1 reverses the mechanisms described above, resulting in enhanced GA signaling, reduced ABA signaling, cell wall loosening, and seed germination.
PIF1 has a transcription activation domain that can activate reporter gene expression in yeast and in transient expression systems (Huq et al., 2004; Shen et al., 2008), but PIF1 shows both activator and repressor activity in plants. Of the PIF1 targets mentioned above, PIF1 activates ABI3, ABI5, GAI, and RGA and represses EXP8, EXP10, and XTH28 (Oh et al., 2009). In a genome-wide binding analysis performed on imbibed seeds, we identified 748 PIF1 binding sites closely associated with 842 genes. PIF1 activates 116 and represses 72 of these for a total of 188 PIF1 binding, PIF1-regulated targets in imbibed seeds. Other PIFs show similar dual activator/repressor activity profiles. In seedlings, PIF3 activates 20 and represses 2 targets (Zhang et al., 2013), PIF4 activates 893 and represses 540 targets (Oh et al., 2012), and PIF5 activates 115 and represses 3 targets (Hornitschek et al., 2012). The mechanism by which PIFs achieve this differential regulation of their direct targets remains unclear.
Groucho (Gro), which was discovered in Drosophila melanogaster, is the founding member of the highly conserved Gro family of transcriptional corepressors with WD repeats and Q-rich domains (Chen and Courey, 2000; Jennings and Ish-Horowicz, 2008). Gro family members target specific promoters by interacting with DNA binding transcription factors or other cofactors and they repress transcription by recruiting either HDAC (Chen et al., 1999) or mediator complexes (Kuchin and Carlson, 1998; Conlan et al., 1999). In Drosophila, Gro interacts with Hairy and E(Spl) to repress members of the proneural achaete-scute complex (AS-C) and suppress the formation of extra bristles around the developing eye (Heitzler et al., 1996).
In Arabidopsis, the 13 members of the Gro family are divided into the LEUNIG (LUG) and TOPLESS (TPL) subclades (Liu and Karmarkar, 2008; Lee and Golz, 2012), but both subclades act as transcriptional corepressors to regulate plant development and hormonal signaling (Liu and Meyerowitz, 1995; Conner and Liu, 2000; Long et al., 2006; Szemenyei et al., 2008; Stahle et al., 2009; Grigorova et al., 2011; Oh et al., 2012). The first function for LUG in plants was found in flower development. AGAMOUS (AG), a C class floral homeotic gene normally expressed in the inner two floral whorls, is derepressed in the outer two whorls of lug mutants, causing homeotic conversion of sepals to carpels and petals to stamens (Liu and Meyerowitz, 1995). In addition, lug mutants also show disrupted leaf (Stahle et al., 2009), gynoecium (Chen et al., 2000), and pollen development (Schneitz et al., 1997; Liu et al., 2000). Unlike lug mutants, leunig_homolog (luh) mutants do not show any floral homeotic conversion. Instead, luh mutants produce seed coats with pectin containing an altered rhamnogalacturonan I (RG I) (Bui et al., 2011; Huang et al., 2011; Walker et al., 2011). The decreased hydration capacity of this altered RG I prevents luh mutant seeds from extruding mucilage upon imbibition. This luh mutant mucilage phenotype is likely caused by reduced expression of MUM2, which encodes a β-galactosidase that hydrolyzes a galactose residue from RG I (Huang et al., 2011; Walker et al., 2011). Although the developmental processes regulated by LUG and LUH are mainly distinct, the synthetic lethality of the lug luh double mutant (Sitaraman et al., 2008) and the partial rescue of the luh mutant phenotype by LUG (Walker et al., 2011) suggest partial functional redundancy.
Like other Gro family members, LUG is a transcriptional corepressor. LUG binds DNA indirectly via transcription factors like APETALA1 (AP1), SEPALLATA3 (SEP3), SHORT VEGETATIVE PHASE (SVP), AGL24, and YABBY to repress targets like AG (Sridhar et al., 2006; Stahle et al., 2009; Gregis et al., 2013). LUG binds these transcription factors either directly (YABBY) or indirectly (AP1, SEP3, SVP, and AGL24) through an adaptor protein called SEUSS (SEU) (Gregis et al., 2006; Sridhar et al., 2006). The LUG-SEU complex then represses gene expression by recruiting HISTONE DEACETYLASE19 or by interacting with the mediator complex (Gonzalez et al., 2007). LUH also interacts with SEU and SEU-LIKEs (SLKs) (Sitaraman et al., 2008; Stahle et al., 2009; Shrestha et al., 2014), but LUH’s transcription factor partners remain unclear.
Here, we asked whether LUG or LUH act as corepressors of PIF1 in imbibed seeds. We report that the transcriptional profile of luh mutant seeds is similar to that of pif1 mutant seeds. In addition, more than 80% of both pif1 mutant seeds and luh mutant seeds germinate under phyBoff conditions, in which phytochrome B (phyB) is inactivated by a far-red light pulse. LUH interacts with PIF1 at the protein level and binds the promoters of PIF1 targets. It is clear that LUH binding to the promoters of PIF1 targets requires PIF1 because LUH binding is significantly reduced in both red light-irradiated wild-type seeds and pif1 mutant seeds. Consistent with its known function as a corepressor, we found that LUH represses many PIF1-repressed targets. Unexpectedly, LUH also activates many PIF1-activated targets. Thus, rather than acting as a simple corepressor, our results indicate LUH interacts with PIF1 to coregulate a subset of its targets in imbibed seeds.
RESULTS
LUH Inhibits Light-Dependent Seed Germination
Although PIF1 has a transcription activation domain (Huq et al., 2004; Shen et al., 2008), it functions as both a transcriptional activator and a repressor in vivo (Oh et al., 2009). This raises the possibility that PIF1 regulates its target genes in a context-dependent manner by associating with other transcriptional regulators. We asked whether LUG or its homolog LUH, two related Gro family transcriptional corepressors, assist PIF1 to regulate its direct targets in imbibed seeds.
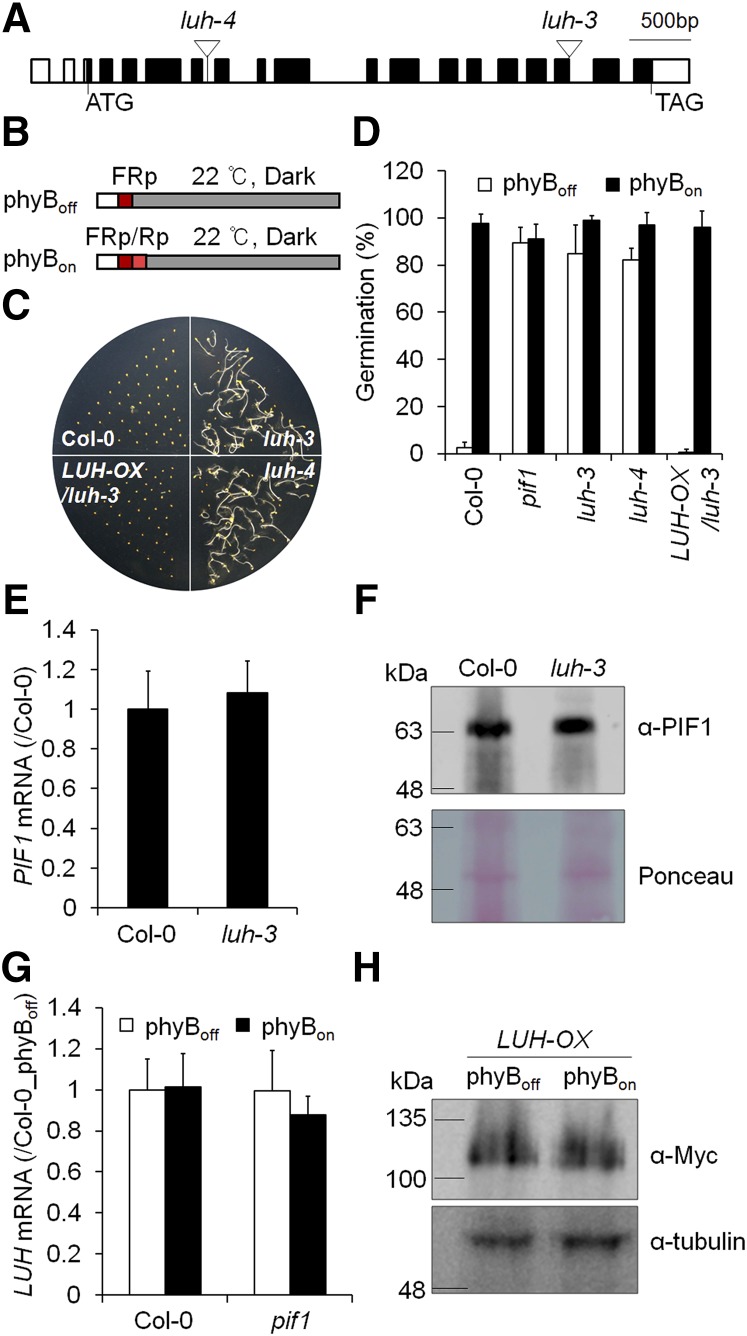
First, we tested whether lug or luh mutant seeds germinate independent of light like pif1 mutant seeds (Figure 1). We obtained two independent luh mutant alleles from the Salk T-DNA collection (luh-3 [Sitaraman et al., 2008] and luh-4 [Stahle et al., 2009]). We then examined the germination of these two independent luh mutant alleles (Figure 1A) under phyBoff and phyBon conditions (Figure 1B). Under the phyBoff condition, imbibed seeds were subjected to a protocol with a far-red light pulse that converts phyB to its inactive Pr form. Under the phyBon condition, seeds were subjected to an additional red light pulse that converts phyB to its active Pfr form. We determined the resulting germination frequencies by counting the number of seeds with protruding radicles after 4 d dark incubation following the light treatment. Wild-type seeds germinated well under the phyBon condition but not the phyBoff condition, whereas pif1 mutant seeds germinated well regardless of light exposure (Figures 1C and 1D). Like the pif1 mutant seeds, more than 80% of luh mutant seeds germinated under the phyBoff condition (Figures 1C and 1D). Expression of Myc-tagged LUH under the 35S promoter in the luh-3 background (LUH-OX/luh-3) rescued the light-independent germination phenotype of the luh mutant, confirming that the luh mutation is responsible for the light-independent seed germination of luh mutant seeds. The germination kinetics of the pif1 luh double mutant were identical to those of the pif1 mutant but faster than those of the luh mutant (Supplemental Figure 1), indicating epistasis.
Figure 1.
LUH Inhibits Light-Dependent Seed Germination.
(A) The T-DNA insertion sites for two luh mutants are indicated by inverted triangles. Black boxes indicate the LUH exons, white boxes indicate the untranslated regions, and the translation start and termination sites are indicated by ATG and TAG, respectively.
(B) The phyBoff and phyBon light treatment schemes. Dry seeds were sterilized and irradiated with a far-red light pulse (5 min, 2.59 μmol·m−2·s−1) for the phyBoff condition or with a far-red light pulse followed by a red light pulse (5 min, 11.5 μmol·m−2·s−1) for the phyBon condition. Irradiated seeds were then incubated in the dark for various periods. Germination assays were performed on seeds incubated in the dark for 96 h and 4 d, whereas gene expression analysis was performed on seeds incubated in the dark for 12 h.
(C) and (D) The two luh mutants germinate in the phyBoff condition. This is rescued by the expression of Myc-tagged LUH under the 35S promoter (LUH-OX/luh-3) (sd, n = 3 biological replicates).
(E) and (F) Wild-type and luh-3 seeds show comparable levels of PIF1 mRNA (E) (sd, n = 3 biological replicates) and protein (F) under the phyBoff condition.
(G) and (H) Red light neither alters LUH mRNA levels (G) (sd, n = 3 biological replicates) nor LUH protein stability (H).
As reported, homozygous lug-444 (Stahle et al., 2009) mutants are infertile and cannot be maintained. Thus, we examined the germination of seeds harvested from LUG+/− heterozygotes, a quarter of which should be homozygous lug mutants. Unlike luh mutant seeds, lug mutant seeds did not germinate under the phyBoff condition (Supplemental Figure 2). Together, our results implicate LUH as a major transcriptional coregulator of phytochrome-dependent seed germination.
We next asked whether the light-independent germination phenotype of luh mutant seeds is caused by their reduced seed mucilage extrusion. According to previous studies (Bui et al., 2011; Huang et al., 2011; Walker et al., 2011), LUH, which is also known as MUM1 (MUCILAGE MODIFIED1), is required for seed coat mucilage extrusion during imbibition. The reduced mucilage extrusion of luh mutant seeds has been attributed to their reduced expression of the β-galactosidase MUM2 (Huang et al., 2011; Walker et al., 2011). To determine whether the defect in mucilage extrusion causes light-independent seed germination, we examined the germination of mucilage extrusion-defective mum2 mutant seeds (Supplemental Figure 2). Unlike luh mutant seeds, mum2 mutant seeds germinated only under phyBon conditions, not phyBoff conditions. This indicates that mucilage extrusion is unrelated to light-dependent seed germination. The light-independent germination of luh mutant seeds was not a consequence of reduced PIF1 mRNA or PIF1 protein level (Figures 1E and 1F), nor did red light change LUH expression or protein stability (Figures 1G and 1H). Since corepressors generally function with other DNA binding transcription factors, our results suggest LUH works either directly with PIF1 or indirectly with other transcription factors that promote PIF1 activity to inhibit the germination of imbibed seeds.
LUH Interacts with PIF1
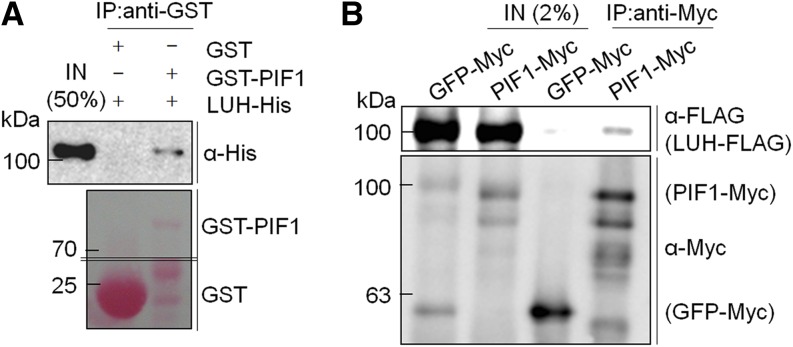
If LUH inhibits seed germination as a transcriptional corepressor of PIF1, LUH may physically interact with PIF1. Thus, we performed an in vitro binding assay using recombinant His-tagged LUH (LUH-His) and GST-tagged PIF1 (GST-PIF1) proteins. GST-PIF1, but not GST alone, was capable of pulling down LUH protein when precipitated by glutathione sepharose resin (Figure 2A). This confirmed an in vitro interaction between PIF1 and LUH. We next performed an in vivo binding assay using transgenic lines stably expressing either Myc-tagged PIF1 or GFP (PIF1-Myc or GFP-Myc) and transiently expressing similar levels of FLAG-tagged LUH protein (LUH-FLAG) via Agrobacterium tumefaciens infiltration. We immunoprecipitated PIF1-Myc and GFP-Myc with an anti-Myc antibody from infiltrated seedling extracts and detected any coimmunoprecipitated LUH-FLAG with an anti-FLAG antibody. PIF1 coimmunoprecipitated LUH, but GFP did not (Figure 2B). This confirmed that LUH interacts with PIF1.
Figure 2.
LUH Interacts with PIF1.
(A) GST pull-down assay showing the interaction between recombinant His-tagged LUH (LUH-His) and GST-tagged PIF1 (GST-PIF1). Resin-bound GST or GST-PIF1 proteins were mixed with LUH-His and precipitated. Precipitated proteins were then analyzed by immunoblot using an anti-His antibody. The lower panel shows precipitated GST and GST-PIF1 proteins stained with Ponceau S.
(B) In vivo binding assay between FLAG-tagged LUH (LUH-FLAG) and Myc-tagged PIF1 (PIF1-Myc). Seven-day-old, light-grown, stable, transgenic lines expressing Myc-tagged GFP (GFP-Myc) or PIF1-Myc were treated to transiently express LUH-FLAG by Agrobacterium infiltration. Myc-tagged proteins were immunoprecipitated with an anti-Myc antibody and coimmunoprecipitated LUH-FLAG was detected with an anti-FLAG antibody. IN, input samples; IP, immunoprecipitated samples. Both in vitro and in vivo binding assays were performed at least three times, and representative data are shown.
LUH Regulates a Subset of PIF1-Regulated Genes
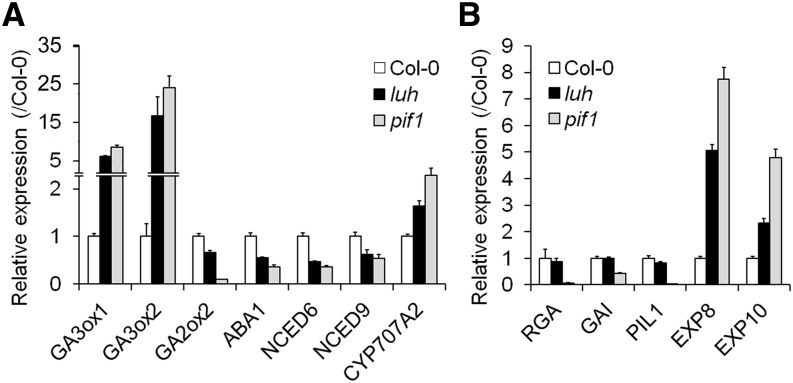
Since LUH interacts with PIF1, a subset of PIF1-regulated genes are likely affected by the luh mutation. We previously showed that PIF1 represses two GA biosynthetic genes (GA3ox1 and GA3ox2) and activates a GA catabolic gene (GA2ox2). Conversely, PIF1 activates ABA synthetic genes (ABA1, NCED6, and NCED9) and represses an ABA catabolic gene (CYP707A2) (Oh et al., 2006, 2007, 2009). We thus examined whether LUH regulates the expression of the PIF1-regulated genes required for the biosynthesis and catabolism of GA and ABA (Figure 3A). To do so, we incubated phyBoff seeds for 12 h in darkness and sampled them for expression analysis. Compared with wild-type seeds, luh mutant seeds expressed higher levels of GA3ox1 and GA3ox2, lower levels of GA2ox2, lower levels of ABA1, NCED6, and NCED9, and higher levels of CYP707A2. These shifts in the expression of GA- and ABA-related genes in luh mutant seeds match those for pif1 mutant seeds. Since the above-mentioned GA and ABA biosynthetic genes are indirectly regulated by PIF1, we also examined the expression of five other PIF1-regulated targets (Figure 3B). We found that luh mutant seeds expressed higher levels of two PIF1-repressed target genes (EXP8 and EXP10), but similar levels of three PIF1-activated targets (RGA, GAI, and PIL1). These results indicated that LUH coregulates a subset of PIF1-regulated targets in imbibed seeds.
Figure 3.
LUH Regulates a Subset of PIF1-Regulated Genes in Imbibed Seeds.
(A) The expression levels of GA and ABA biosynthetic genes in wild-type, luh-3 mutant, and pif1 mutant seeds treated with a far-red light pulse for 5 min and incubated for 12 h in darkness (phyBoff condition) (sd, n = 3 biological replicates).
(B) The expression levels of PIF1-regulated targets under the phyBoff condition (sd, n = 3 biological replicates).
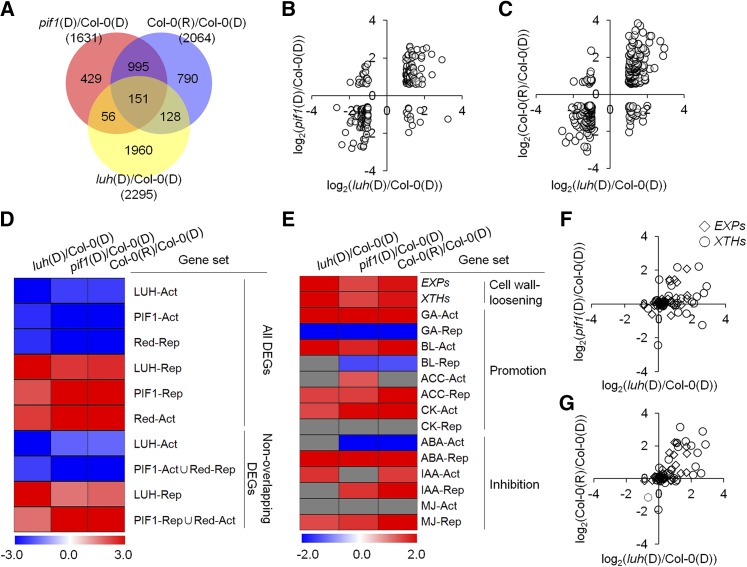
We expanded this expression analysis using microarray analysis and identified 2295 genes that were differentially expressed [luh(D)/Col-0(D), >2-fold, false discovery rate (FDR) <0.05] in 12-h dark-incubated phyBoff seeds from a wild-type strain [Col-0(D)] and from those of a luh mutant [luh(D)] (Figure 4A; Supplemental Data Set 1). We will refer to these 2295 differentially expressed genes (DEGs) as LUH-regulated genes. We next compared our list of LUH-regulated genes with a list of 1631 previously reported (Oh et al., 2009) PIF1-regulated genes [pif1(D)/Col-0(D), >1.5-fold, FDR <0.05] in imbibed seeds and found a significant overlap (207 genes, P < 2.25 × 10−12, hypergeometric test) (Figure 4A; Supplemental Data Set 1). Most of the 207 overlapping genes were regulated in the same direction by LUH and PIF1 (Figure 4B), consistent with the hypothesis that these genes are coregulated by LUH and PIF1. We also compared our list of LUH-regulated genes with a list of 2064 previously reported (Oh et al., 2009) red light-regulated genes [Col-0(R)/Col-0(D), >1.5-fold, FDR <0.05] and found a significant overlap (279 genes, P < 7.26 × 10−20, hypergeometric test) (Figure 4A; Supplemental Data Set 1). As with the comparison between LUH- and PIF1-regulated genes, most of these 279 genes were regulated in the same direction by the luh mutation and red light (Figure 4C). Similar expression patterns of LUH- and PIF1-regulated genes could be also visualized by hierarchical clustering (Supplemental Figure 3). Together, our transcriptome analysis is consistent with the hypothesis that LUH and PIF1 coregulate a subset of genes in imbibed seeds.
Figure 4.
LUH and PIF1 Coregulate Genes in Imbibed Seeds.
(A) A Venn diagram summarizing the overlap of differentially expressed genes from imbibed pif1, luh, and red light-treated seeds. X(D) and Y(R) indicate 12-h dark-incubated X seeds and 12-h red light-incubated Y seeds, respectively. X(D)/Z(D) indicates the transcriptome comparison between 12-h dark-incubated seeds of the X and Z genotypes. Numbers indicate the numbers of DEGs.
(B) and (C) Dot plots comparing the log2-transformed expression levels of 207 LUH and PIF1 coregulated genes (B) or 279 LUH and red light coregulated genes (C) in luh mutant, pif1 mutant, and red light-treated wild-type seeds.
(D) GSEA plotted as a heat map of NESs. All LUH-, PIF1-, or red light-regulated genes (All DEGs) were divided into activated (X-Act) and repressed (X-Rep) gene sets. Alternatively, only DEGs appearing uniquely in the list of LUH-regulated genes or in the union of PIF1- or red light-regulated genes (Non-overlapping DEGs) were divided into activated and repressed gene sets. An NES for each gene set was determined in the luh(D)/Col-0(D), pif1(D)/Col-0(D), and Col-0(R)/Col-0(D) transcriptome data sets and plotted on the heat map.
(E) GSEA plotted as a heat map of NESs. Various hormone-responsive genes were divided into hormone-activated (X-Act) and -repressed (X-Rep) gene sets. Cell wall loosening genes (EXPs and XTHs) were also used as gene sets. An NES for each gene set was determined in the luh(D)/Col-0(D), pif1(D)/Col-0(D), and Col-0(R)/Col-0(D) transcriptome data sets and plotted on the heat map. The gray cells have an FDR > 0.05 and are thus nonsignificant. Germination-promoting hormones are labeled “Promotion,” while germination-inhibiting hormones are labeled “Inhibition.” BL, brassinolide; ACC, 1-amino-cyclopropane-1-carboxylic acid; CK, zeatin; IAA, indole-3-acetic acid; MJ, methyl jasmonate.
(F) and (G) Dot plots comparing the log2-transformed expression levels of 36 EXP (open diamonds) and 33 XTH (open circles) genes in luh and pif1 mutant seeds (F) or in luh mutant and red light-treated wild-type seeds (G).
The transcriptome analysis above suggests LUH coregulates a statistically significant but relative small proportion (207 of 1631) of PIF1-regulated genes. However, the stringent cutoffs we used to determine differential expression may have excluded some legitimately coregulated genes. We investigated this possibility using gene set enrichment analysis (GSEA) (Subramanian et al., 2005). GSEA determines whether the members of a gene set tend to appear at the top or bottom of a transcriptome gene list ranked by expression fold changes. Sets whose members tend to appear at the top of the ranked transcriptome data show a positive normalized enrichment score (NES), while sets whose members appear at the bottom show a negative NES. To determine if LUH and PIF1 coregulate genes more extensively than the number of small overlapping DEGs suggests, we divided the entire list of LUH-, PIF1-, and red light-regulated genes into LUH-, PIF1-, and red light-activated or -repressed gene sets (Supplemental Data Set 2) and calculated NES values for each set against ranked luh(D)/Col-0(D), pif1(D)/Col-0(D), and Col-0(R)/Col-0(D) transcriptome data.
According to our GSEA, LUH-activated genes were biased toward the bottom of both the pif1(D)/Col-0(D) data and the Col-0(R)/Col-0(D) data, meaning LUH-activated genes tend to be repressed in pif1 mutant seeds and red light-treated wild-type seeds. Similarly, PIF1-activated genes were biased toward the bottom of both the luh(D)/Col-0(D) data and the Col-0(R)/Col-0(D) data, and red light-repressed genes were biased toward the bottom of both the luh(D)/Col-0(D) data and the pif1(D)/Col-0(D) data (Figure 4D, All DEGs). Reciprocally, LUH- and PIF1-repressed genes and red light-activated genes were biased toward the top of the indicated transcriptome data sets (Figure 4D, All DEGs). This implies an even more substantial overlap between LUH- and PIF1-regulated genes than the small number of overlapping DEGs in the Figure 4A Venn diagram suggests.
We also performed a second, similar GSEA, limiting the gene sets to those unique to the LUH-regulated DEGs or unique to the PIF1- and red light-regulated DEGs (Supplemental Data Set 2). By excluding the overlapping DEGs, we were asking if unique DEGs alone also showed the bias to the top or bottom of the ranked transcriptome data. The removal of the overlapping DEGs from the GSEA reduced the resulting NES values (Figure 4D, Non-overlapping DEGs), but still revealed a trend toward apparent coregulation. This indicated that the unique DEGs are also coregulated by LUH and PIF1, though less coherently than the overlapping DEGs.
PIF1 inhibits seed germination by repressing cell wall loosening and by coordinating hormone responses (Oh et al., 2009). We thus investigated whether LUH also regulates these processes by performing a GSEA using gene sets comprising either cell wall loosening genes (EXPs [Lee et al., 2001] and XTHs [Rose et al., 2002]) or hormone-related genes (Nemhauser et al., 2006; Goda et al., 2008) (Supplemental Data Set 3). Both the EXPs and the XTHs, two types of cell wall loosening genes, were biased toward the top of the luh(D)/Col-0(D), pif1(D)/Col-0(D), and Col-0(R)/Col-0(D) data sets (Figure 4E). This means the EXPs and XTHs tend to be activated in the luh mutant, the pif1 mutant, and in red light-treated wild-type seeds. Plotting the expression patterns of all 36 EXP genes and 33 XTH genes in dot plots made it clear that many were coinduced by the luh mutation, the pif1 mutation, and the red light treatment (Figures 4F and 4G). Together, these results indicated that LUH, PIF1, and darkness all repress cell wall loosening genes in imbibed seeds.
To explore the potential coregulation of the hormone signaling pathways that control seed germination, we divided the hormone-responsive gene sets (Nemhauser et al., 2006; Goda et al., 2008) into those that promote germination and those that inhibit germination (Figure 4E). Of the germination-promoting hormone gene sets, GA-activated genes were biased toward the top of the luh(D)/Col-0(D), pif1(D)/Col-0(D), and Col-0(R)/Col-0(D) data sets, while GA-repressed genes tended to occur at the bottom (Figure 4E). This means GA signaling is activated in luh mutant, pif1 mutant, and red light-treated wild-type seeds. Similarly, brassinolide-activated genes were biased toward the top of luh(D)/Col-0(D), pif1(D)/Col-0(D), and Col-0(R)/Col-0(D) data sets, while brassinolide-repressed genes were biased toward the bottom of the pif1(D)/Col-0(D) and Col-0(R)/Col-0(D) data sets but not the luh(D)/Col-0(D) data set. Of the germination-inhibiting hormone gene sets, ABA-repressed and methyl jasmonate-repressed genes were biased toward the top of the luh(D)/Col-0(D), pif1(D)/Col-0(D), and Col-0(R)/Col-0(D) data sets. ABA-activated genes were biased toward the bottom of the pif1(D)/Col-0(D) and Col-0(R)/Col-0(D) data sets but not the luh(D)/Col-0(D) data set. Thus, the expression patterns of hormone-related genes in luh mutant seeds and pif1 mutant or red light-treated wild-type seeds are similar but not identical.
LUH Regulates a Subset of Genes Both Directly Activated and Directly Repressed by PIF1
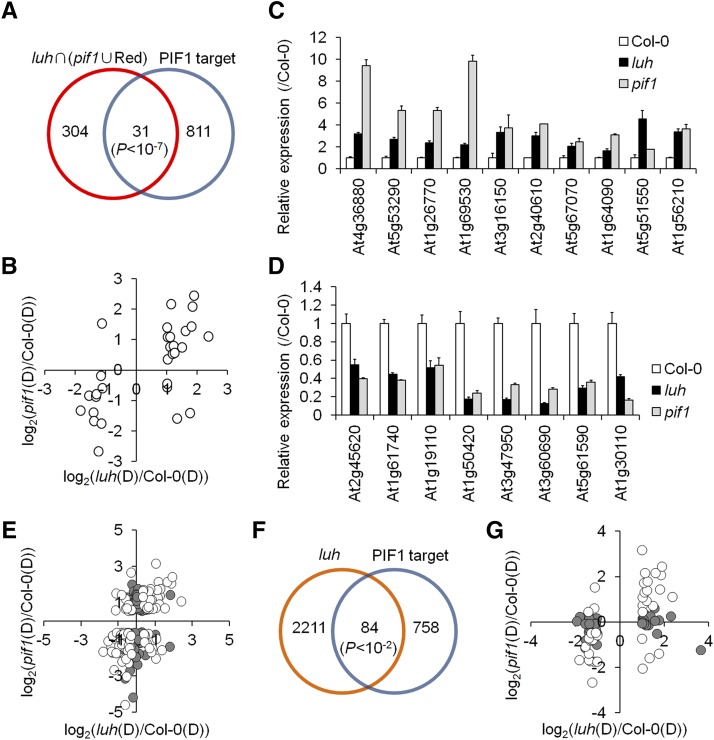
The PIF1-regulated genes identified in the microarray above included both direct and indirect PIF1 targets. By comparing PIF1 targets with the 335 overlapping DEGs from the comparison of the LUH-regulated genes with the combined PIF1- and red light-regulated genes, we identified 31 PIF1 targets strongly regulated by both LUH and PIF1 in imbibed seeds (Figure 5A; Supplemental Data Set 4). We next visualized the expression patterns of these 31 genes using a dot plot. The expression of the 15 genes in the upper-right quadrant was increased in both the luh and pif1 mutants, meaning these were LUH- and PIF1-repressed genes. The 11 genes in the lower-left quadrant were LUH- and PIF1-activated genes. The remaining five genes were regulated in opposite directions by LUH and PIF1 (Figure 5B). Using quantitative PCR, we were able to validate the direction of the changes in expression for 18 genes arbitrarily chosen from the 26 coregulated by LUH and PIF1. The expression levels of 10 genes were elevated in both the luh and the pif1 mutants (Figure 5C), and the levels of eight genes were reduced (Figure 5D). These results indicated that LUH regulates in the same direction genes both directly activated and directly repressed by PIF1.
Figure 5.
LUH Regulates a Subset of Genes Both Directly Activated and Directly Repressed by PIF1.
(A) Venn diagram identifying 31 PIF1 targets whose expression is regulated by LUH and PIF1. The significance of the overlap was determined with a hypergeometric test (P < 1.29 × 10−8).
(B) Dot plot comparing the log2-transformed expression levels of these 31 genes in pif1 and luh mutant seeds.
(C) and (D) Validation of the expression patterns for 10 PIF1-repressed targets (C) and 8 PIF1-activated targets (D) in luh and pif1 mutant seeds exposed to the phyBoff condition (sd, n = 3 biological replicates).
(E) Dot plot comparing the log2-transformed expression levels of 188 PIF1-regulated targets in luh and pif1 mutant seeds. Empty circles indicate genes significantly regulated (FDR < 0.05) by both PIF1 and LUH.
(F) Venn diagram identifying 84 PIF1 targets whose expression is also regulated by LUH (P = 0.004627, hypergeometric test).
(G) Dot plot comparing the log2-transformed expression levels in the luh and the pif1 mutant seeds of the 84 PIF1 targets whose expression is also regulated by LUH. Empty circles indicate genes significantly regulated (FDR < 0.05) by both PIF1 and LUH.
We next plotted all 188 PIF1-regulated targets to rule out any bias introduced by limiting our analysis to the 31 PIF1 targets whose expression was strongly regulated both by LUH and PIF1 (Figure 5E). Of the 72 PIF1-repressed targets falling in quadrants 1 and 2, 30 were significantly repressed (FDR < 0.05) by LUH (quadrant 1), while 12 were significantly activated (FDR < 0.05) by LUH (quadrant 2). Similarly, of the 116 PIF1-activated targets falling in quadrants 3 and 4, 55 were significantly activated (FDR < 0.05) by LUH (quadrant 3) and 11 genes were significantly repressed (FDR < 0.05) by LUH (quadrant 4). We also determined the expression patterns of 84 LUH-regulated PIF1 targets (Figures 5F and 5G; Supplemental Data Set 5). These 84 LUH-regulated PIF1 targets were PIF1 targets that showed a >2-fold change in expression in luh mutant seeds, regardless of their expression level in pif1 mutant seeds. Among them, 39 genes were repressed by LUH (quadrants 1 and 4) and 45 were activated by LUH (quadrants 2 and 3). Of the 39 LUH-repressed genes, 16 genes were significantly repressed (FDR < 0.05) by PIF1 (quadrant 1) and 4 were significantly activated (FDR < 0.05) by PIF1 (quadrant 4). Three of these genes were not included in the Affymetrix probe set. Similarly, of the 45 LUH-activated genes, 16 were significantly activated (FDR < 0.05) by PIF1 (quadrant 3) and 3 were significantly repressed (FDR < 0.05) by PIF1 (quadrant 2). Six of these genes were not included in the Affymetrix probe set. Taken together, these results indicated that LUH represses many PIF1-repressed targets and activates many PIF1-activated targets. In other words, LUH acts not as a simple corepressor, but in a way consistent with the notion of a coregulator that either activates or represses its targets depending on context.
LUH Binds a Subset of PIF1 Target Promoters
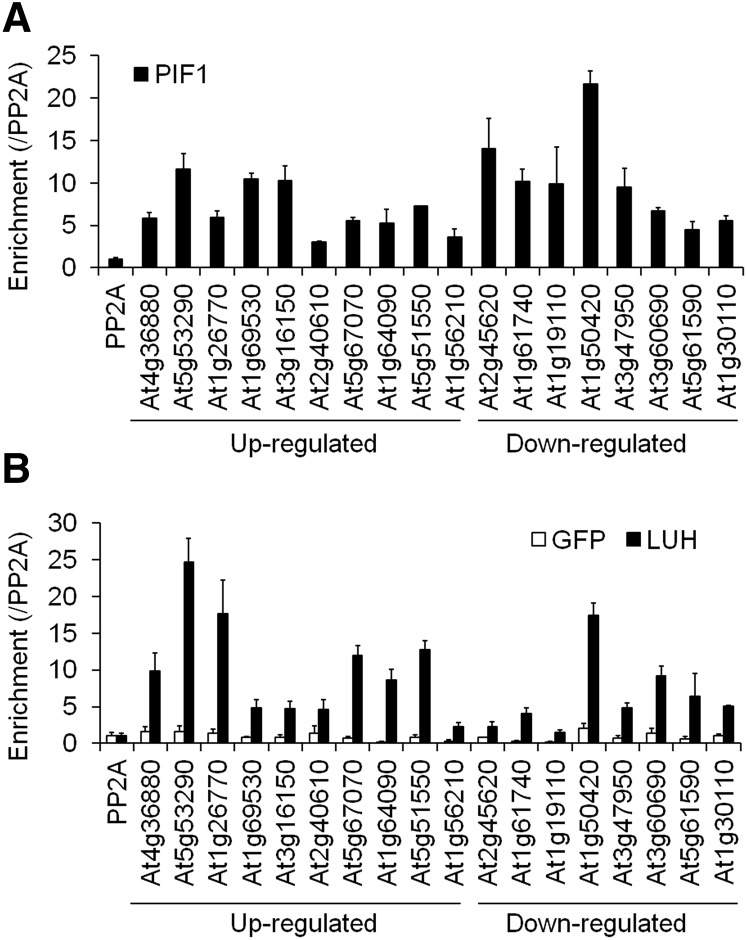
Although LUG lacks a DNA binding domain, it can bind target promoters and regulate their expression via physical interactions with other transcription factors. Thus, LUH likely binds the promoters of the targets it coregulates with PIF1 in imbibed seeds via its physical interaction with PIF1. The 18 genes in Figures 5C and 5D that were regulated in the same direction by both PIF1 and LUH are good candidates for cotargets. We thus performed a chromatin immunoprecipitation (ChIP) assay using transgenic lines expressing Myc-tagged LUH (LUH-OX), Myc-tagged PIF1 (PIF1-OX), or Myc-tagged GFP (GFP-OX) to determine if LUH also binds these 18 promoters. We irradiated seeds of each genotype with far-red light for 5 min and incubated them in the dark for 12 h before preparing the samples for the ChIP assay. As expected, PIF1 enriched promoter fragments of all 18 genes above that of the nonbinding PP2A control promoter (Figure 6A). Under the same conditions, LUH also enriched nearly all of these same promoter fragments above that of the nonbinding PP2A control promoter (Figure 6B). GFP did not enrich any of them. These results indicated both LUH and PIF1 bind these 18 promoters.
Figure 6.
LUH and PIF1 Bind an Overlapping Subset of Genes in Imbibed Seeds.
Enrichment of 18 PIF1-regulated target promoters by PIF1 (A) and LUH (B). ChIP assays were performed with an anti-Myc antibody using imbibed phyBoff GFP-OX (GFP), LUH-OX (LUH), and PIF1-OX (PIF1) seeds. The enrichment of each promoter was normalized both by the level of input DNA and by the ChIP enrichment value of the nonbinding PP2A promoter (sd, n = 3 biological replicates). Genes whose expression was either up- or downregulated in both luh and pif1 mutant seeds are indicated.
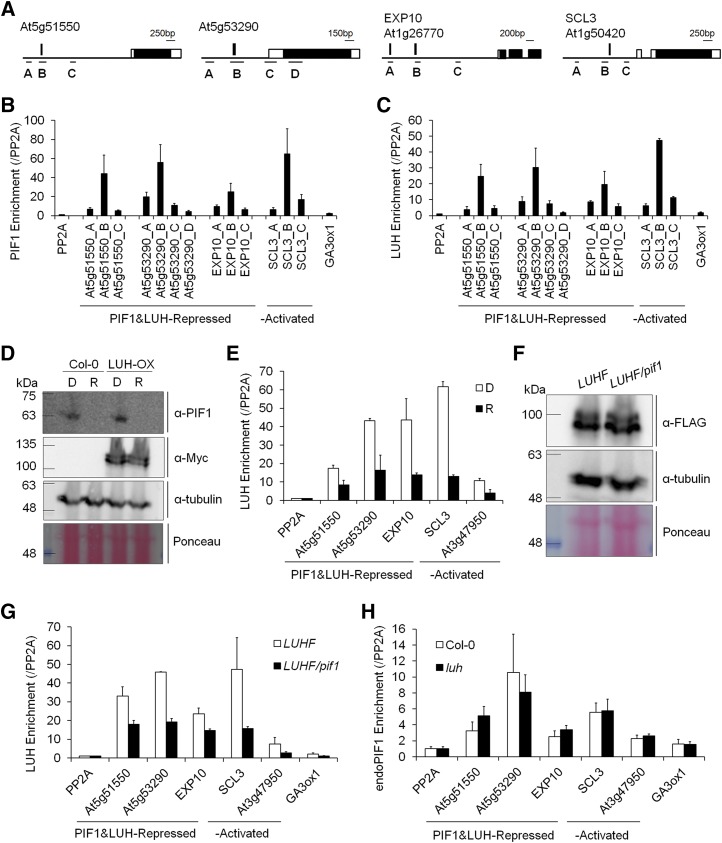
Binding of LUH to Its Target Promoters Is Partly PIF1 Dependent
To determine whether LUH is binding its targets via its interaction with PIF1, we selected four genes (At5g51550, At5g53290, EXP10, and SCL3) enriched in both the LUH-ChIP and PIF1-ChIP experiments for further analysis and designed primer sets to amplify different fragments of their promoters (Figure 7A). LUH activated one of these genes (SCL3) and repressed the other three (At5g51550, At5g53290, and EXP10). Consistent with our previous ChIP-chip data, PIF1 enriched the fragment of each target gene promoter that contains the PIF1 binding site (fragment B) more than fragments A, C, and D and much more than two nonbinding PP2A and GA3ox1 promoter fragments (Figure 7B). LUH also enriched the PIF1 binding site-containing fragments of each promoter (the B fragments) more than control fragments (Figure 7C).
Figure 7.
PIF1 Recruits LUH to Target Promoters.
(A) ChIP amplicons for the At5g53290, At5g51550, EXP10, and SCL3 loci. Black and white boxes indicate exons and untranslated regions, respectively, vertical bars indicate PIF1 binding sites, and the ChIP amplicons are labeled with capital letters.
(B) and (C) Enrichment profiles for each promoter fragment by PIF1 (B) and LUH (C). ChIP assays were performed using imbibed phyBoff LUH-OX (LUH) and PIF1-OX (PIF1) seeds. The enrichment of each promoter was normalized both by the level of input DNA and by the ChIP enrichment value of the nonbinding PP2A promoter (se, n = 3 biological replicates).
(D) Immunoblot showing endogenous PIF1 levels detected with an anti-PIF1 antibody (α-PIF1) and Myc-tagged LUH levels detected with an anti-Myc antibody (α-Myc) in the wild type (Col-0) and an Myc-tagged LUH overexpression (LUH-OX) line. Imbibed phyBoff seeds were incubated for 6 h in either the dark (D) or in red light (R). α-Tubulin detected with an antitubulin antibody and Ponceau S staining were used as loading controls.
(E) Reduced enrichment of target promoters by LUH under red light. The enrichment of each promoter was normalized both by the level of input DNA and by the ChIP enrichment value of the nonbinding PP2A promoter (se, n = 2 biological replicates).
(F) Immunoblot showing similar levels of FLAG-tagged LUH in LUHF and the LUHF/pif1 genotypes. The LUHF and LUHF/pif1 genotypes indicate stable transgenic lines expressing FLAG-tagged LUH in the wild-type and pif1 mutant backgrounds, respectively. Imbibed phyBoff seeds were incubated for 12 h in the dark. α-Tubulin detected with an antitubulin antibody and Ponceau S staining were used as loading controls.
(G) Reduced enrichment of target promoters by LUH in the absence of PIF1. The enrichment of each promoter was normalized both by the level of input DNA and by the ChIP enrichment value of the nonbinding PP2A promoter (se, n = 2 biological replicates).
(H) Comparable enrichment of target promoters by endogenous PIF1 in wild-type and luh mutant backgrounds. ChIP assays were performed with an anti-PIF1 antibody using imbibed phyBoff seeds. The enrichment of each promoter was normalized both by the level of input DNA and by the ChIP enrichment value of the nonbinding PP2A promoter (se, n = 2 biological replicates).
We next asked whether LUH binding is affected by red light. Since red light destabilizes PIF1, LUH binding to its target promoters should decrease if LUH is binding via its physical interaction with PIF1. We irradiated LUH-OX seeds with far-red light for 5 min and incubated them for 6 h in either the dark or continuous red light. As expected, PIF1 accumulated in dark-incubated seeds but not in red light-incubated seeds (Figure 7D). LUH levels, by contrast, were unaffected by red light (Figure 7D). According to the ChIP assay using fragment B, LUH enriched all four target promoters above that of the nonbinding PP2A promoter in both dark- and red light-incubated imbibed seeds (Figure 7E). However, the degree of enrichment was significantly higher in the dark-incubated seeds than the red light-incubated seeds (Figure 7E). This is consistent with LUH binding showing a strong, albeit incomplete PIF1 dependence.
Finally, we examined the ability of LUH to enrich the same target promoters in the pif1 mutant. To do so, we performed ChIP assays using dark-incubated imbibed seeds from transgenic plants expressing FLAG-tagged LUH in both the wild-type (LUHF) and pif1 mutant (LUHF/pif1) backgrounds. Both transgenic lines expressed similar levels of LUH protein (Figure 7F). Like the Myc-tagged LUH, FLAG-tagged LUH also enriched the same target promoters more than the nonbinding PP2A and GA3ox1 promoters (Figure 7G). This LUH enrichment was significantly reduced in the pif1 mutant background, consistent with substantial PIF1-dependent LUH binding. Interestingly, though, the pattern of LUH enrichment peaking at fragment B was still similar (Supplemental Figure 4), suggesting that LUH also targets similar promoters by binding to factors in addition to PIF1. Unlike LUH, PIF1 bound to its target promoters irrespective of the luh mutation (Figure 7H). Together, our results indicate LUH regulates gene expression in imbibed seeds partly via its interaction with PIF1.
DISCUSSION
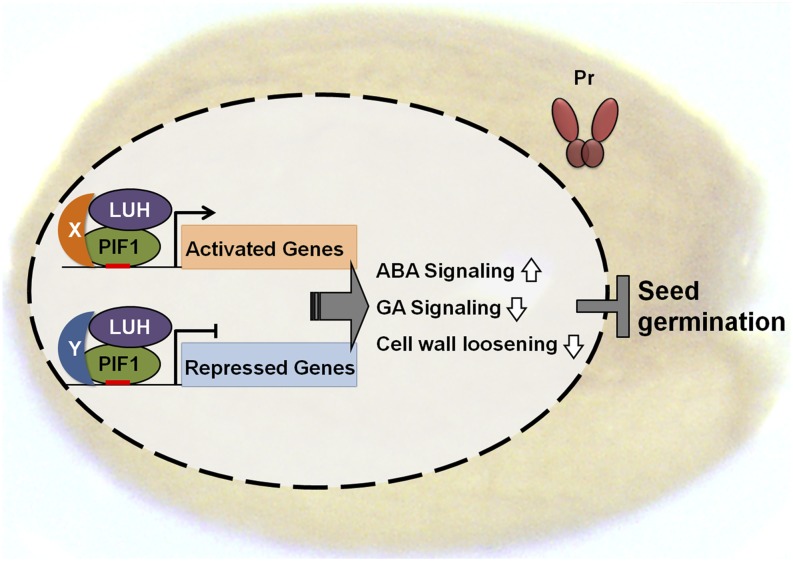
We showed previously that PIF1, a basic helix-loop-helix transcription factor, inhibits phytochrome-dependent seed germination by regulating various hormone- and cell wall-related genes in imbibed seeds. PIF1 has 842 targets across the Arabidopsis genome and regulates the expression of 188 genes in imbibed seeds, activating 116 and repressing 72 (Oh et al., 2009). In this study, we show that LUH, a Gro family corepressor, physically interacts with PIF1 to bind and either activate or repress subsets of PIF1-regulated targets (Figure 8). Both luh mutant seeds and pif1 mutant seeds share a remarkably similar gene expression profile characterized by activated GA signaling and increased expression of cell wall loosening genes. These changes promote germination to the point that, like pif1 mutant seeds, more than 80% of luh mutant seeds germinate under phyBoff conditions. Thus, our results implicate LUH as an important and previously undiscovered coregulator of seed germination working with PIF1.
Figure 8.
A Model Depicting How LUH Inhibits Light-Dependent Seed Germination.
In the dark, when phytochromes are in the cytosol in their inactive Pr form, PIF1 binds various promoters and recruits LUH in a context-dependent manner to either activate or repress its target genes in imbibed seeds. This increases ABA signaling, decreases GA signaling, and decreases cell wall loosening, ultimately inhibiting seed germination. Although showing a slight bias toward corepression, LUH is capable of both coactivating PIF1-activated targets and corepressing PIF1-repressed targets. This model also suggests the existence of one or more unidentified factors (X and Y) that permit the context-dependent switching of the PIF1-LUH complex from an activator to a repressor. Genes independently regulated by either PIF1 or LUH are not included in the model. In the light, phytochromes in their active Pfr form enter the nucleus and inhibit PIF1 both by dissociating it from DNA and promoting its degradation.
LUH and the closely related LUG both belong to the Gro family of proteins. Studies in yeast and protoplasts support their functional classification as a family of transcriptional corepressors (Sridhar et al., 2004, 2006; Gonzalez et al., 2007; Shrestha et al., 2014). Our data, rather, suggest LUH acts as a transcriptional coregulator, apparently by both activating and repressing its targets in imbibed seeds. Of its 188 known PIF1-regulated targets in imbibed seeds, PIF1 activates 1.6 times more genes than it represses (i.e., 116 genes activated and 72 genes repressed). This ratio represents a significant bias toward activation (P = 0.001712, χ2 test). Of these 188 genes, 85 are significantly regulated by both LUH and PIF1 in the same direction. According to our microarray data, of these 85 genes, 55 are coactivated by LUH and PIF1 and 30 are corepressed (Figure 5E). This ratio is not significantly different from the ratio of 116 PIF1-activated and 72 PIF1-repressed genes (P = 0.6469, χ 2 test), implying no particular bias of the interactions of LUH with PIF1 toward corepression.
LUH’s dual role as both a coactivator and a corepressor may extend to LUG and other Gro family members. By comparing the transcriptome of lug mutant flowers (Gonzalez et al., 2007) with a ChIP-seq data set for SEP3 (Kaufmann et al., 2009), a transcription factor that recruits LUG (Sridhar et al., 2006) to its targets, we identified 131 SEP3 targets whose expression is significantly altered in the absence of LUG (Supplemental Figure 5). Of these, 79 genes are activated by LUG and 52 are repressed, displaying no bias toward corepression. In addition, TLE1, a mammalian Gro family member that interacts with the estrogen receptor is required for RNA polymerase II recruitment to a subset of estrogen receptor target promoters (Holmes et al., 2012). This also suggests that TLE1 is not a simple corepressor, but a transcriptional coregulator that can either activate or repress its targets.
This study still has unresolved issues that will have to be addressed in future studies. Both LUH and LUG can interact with transcription factors either directly or indirectly through SEU or SLKs. Specifically, LUG interacts with AP1 and SEP3 only through an SEU adaptor protein (Sridhar et al., 2006). However, YABBY family transcription factors interact with LUH and LUG both directly and indirectly via SEU and SLKs (Stahle et al., 2009). Similarly, TPR/TPL, another Gro family corepressor, also interacts with transcription factors both directly (Kieffer et al., 2006; Long et al., 2006) and indirectly (Pauwels et al., 2010; Causier et al., 2012). Although we show that PIF1 interacts directly with LUH in vitro in the absence of both SEU and SLK adaptor proteins (Figure 2), it remains possible that SEU and/or SLK adaptor proteins are involved in vivo.
The precise tissues in the seed where LUH exerts its influence on germination are still unclear. It is clear that PIF1 suppresses GA signaling and activates ABA signaling by regulating the expression of both biosynthetic and signaling genes in each pathway (Oh et al., 2006, 2007, 2009). In the light, activated phytochromes promote seed germination by inhibiting PIF1 and thus increasing GA signaling and decreasing ABA signaling. PIF1 is known to function in both the embryo and the endosperm. Although all phytochromes function in the embryo to promote seed germination, presumably by inhibiting PIF1, phyB but not phyA inhibits PIF1 in the endosperm to reduce the amount of ABA entering the embryo (Lee et al., 2010, 2012). Both PIF1 and LUH mRNAs are expressed in the embryo and the endosperm, but whereas PIF1 expression is even in each tissue, LUH levels are higher in the endosperm (Penfield et al., 2006; Walker et al., 2011). This suggests LUH, like PIF1, may function in both tissues. Consistent with this hypothesis, the overlapping DEGs among LUH- and PIF1-regulated genes are expressed in both the embryo and the endosperm (Supplemental Figure 6). Nonoverlapping LUH- and PIF1-regulated genes are also expressed similarly in both tissues. These expression patterns suggest LUH functions with PIF1 in both the embryo and the endosperm. Further transcriptome analyses using separated embryos and endosperms of pif1 and luh mutants coupled with seed coat bedding assays will help to further clarify the precise tissue requirements for LUH.
Finally, context-dependent switching of the mode by which coregulators function is found not only in PIFs but also in other plant transcription factors. For example, SVP, a MADS box transcription factor that inhibits flowering, directly activates 77 genes in the vegetative stage of Arabidopsis development and directly represses another 31 (Gregis et al., 2013). During soybean (Glycine max) seedling development, NAC activates 21 genes and represses 10, while YABBY activates 27 genes and represses 19 (Shamimuzzaman and Vodkin, 2013). Similar context-dependent switching has also been observed with animal transcription factors. In Drosophila, for example, Suppressor of hairless [Su(H)] represses the expression of Enhancer of split [E(Spl)] in the absence of Notch signaling but activates E(Spl) upon Notch activation (Cinnamon and Paroush, 2008). This context-dependent switching of Su(H) likely depends upon its binding to either the Hairy/Gro repressor complex (Barolo et al., 2002; Nagel et al., 2005) or the Notch-ICD activator complex (Bailey and Posakony, 1995; Artavanis-Tsakonas et al., 1999). Another good example from Drosophila is Dorsal, a Rel family transcription factor that directly activates twi, sna, and miR-1 in the presumptive mesoderm, but directly represses Dpp in the mesoderm and neuroectoderm via an association with the Gro corepressor (Zeitlinger et al., 2007). To account for the apparently context-dependent switch in LUH’s mode of action from corepressor to coactivator, the data we present here suggest the presence of one or more partners for LUH, PIF1, or both (Figure 8). Such factors have not yet been identified.
METHODS
Plant Materials and Growth Conditions
Arabidopsis thaliana plants were grown in a growth room with a 16-h-light/8-h-dark cycle at 22 to 24°C for general growth and seed harvesting. All mutants in this study are T-DNA insertion lines obtained from the ABRC: luh-3 (Salk_107245), luh-4 (Salk_097509), pif1 (Salk_131872, pil5-2; Penfield et al., 2005), mum2-sk25 (Salk_015025), mum2-sk46 (Salk_022846), mum2-sk21 (Salk_060221), and lug-444 (Salk_126444). To generate the LUH-OX transgenic lines expressing Myc-tagged LUH, a full-length LUH cDNA was amplified (for primers, see Supplemental Data Set 6), cloned into a modified pBI121 vector, and introduced into the wild-type strain Col-0. The LUH-OX/luh-3 transgenic line expressing Myc-tagged LUH was generated by cloning the full-length LUH cDNA into a pCAMBIA1300 vector and introducing it into the luh-3 background. For the two transgenic lines expressing FLAG-tagged LUH, the full-length LUH cDNA was cloned into a pCAMBIA1300 vector modified for expressing FLAG-tagged proteins and introduced into the wild-type or pif1 backgrounds. All tags are attached to the C terminus of LUH. Myc-tagged PIF1- or GFP-expressing PIF1-OX and GFP-OX transgenic lines correspond to the previously described PIL5-OX3 and GFP-Myc (Oh et al., 2007). All plants used in these experiments were of the Col-0 ecotype background.
phyB-Mediated Seed Germination Assays
Seeds were surface sterilized, plated on Murashige and Skoog (MS) medium containing 0.6% phytoagar without sucrose and imbibed for 1 h at 22°C under white light (15 µmol·m−2·s−1). The plates were irradiated with far-red light (2.59 μmol·m−2·s−1) for 5 min (phyBoff) or far-red light followed by red light (11.5 μmol·m−2·s−1) for 5 min (phyBon). After incubation for 4 d in the dark at 22°C, germination frequencies were determined by counting the numbers of seeds with protruding radicles.
RNA Isolation and Gene Expression Quantification
For gene expression analysis, seeds were irradiated with far-red light (2.59 μmol·m−2·s−1) for 5 min (phyBoff) and then incubated for 12 h in the dark. Total RNAs were isolated using the Spectrum plant total RNA kit (Sigma-Aldrich) according to the manufacturer’s protocol and converted to cDNA using MMLV-RTase (Promega). The transcript levels were determined by real-time PCR using specific primer sets (Supplemental Data Set 6) and normalized with respect to the expression levels of PP2A.
Protein Analysis
For protein analysis, seeds were irradiated with far-red light (2.59 μmol·m−2·s−1) for 5 min (phyBoff) and imbibed for 12 h in the dark, except for Figure 7D. The seeds were then frozen in liquid nitrogen, ground to a fine powder, dissolved in urea-based denaturing buffer (100 mM NaH2PO4, 8 M urea, and 10 mM Tris-HCl, pH 8.0), and cleared by centrifugation at 20,000g for 10 min at 4°C. The extracted total proteins were then boiled for 5 min at 100°C, and protein levels were determined by immunoblot analysis using an anti-c-Myc antibody (sc-789, lot number D1715; Santa Cruz Biotechnology), an anti-FLAG antibody (F7425, lot number 093M4798; Sigma-Aldrich), an anti-His antibody (sc-803, lot number J3111; Santa Cruz Biotechnology), a goat anti-rabbit antibody (sc-2004, lot number K0414; Santa Cruz Biotechnology), and an antibody against endogenous PIF1 (Park et al., 2012).
ChIP
ChIP was performed as described previously (Oh et al., 2007). Briefly, 60 mg dry seed was imbibed for 1 h, irradiated with far-red light for 5 min (phyBoff), incubated for 12 h in the dark, and then cross-linked in 1% formaldehyde solution under vacuum for 1 h in the dark. Chromatin was isolated and sonicated with a Bioruptor 12 times (30 s on/30 s off cycles and high-power output) to obtain 200- to 600-bp DNA fragments. The sonicated chromatin was precipitated with antibodies against Myc (2276S, lot number 24; Cell Signaling) or FLAG (F7425, lot number 093M4798; Sigma-Aldrich). Reverse cross-linking and proteinase K treatment using an elution buffer containing 20 mM Tris-HCl, pH 7.5, 5 mM EDTA, 50 mM NaCl, 1% SDS, and 50 mg/mL proteinase K were performed for at least 6 h at 68°C, and the immunoprecipitated DNA was purified by phenol/chloroform extraction. The enriched DNA levels were quantified by real-time PCR using specific primer sets (Supplemental Data Set 6).
In Vitro Binding Assay
To express recombinant His-tagged LUH, a full-length LUH cDNA was cloned into a pET22b-CPD vector. For the in vitro binding assay, glutathione-agarose bead-bound GST or GST-fused PIF1 protein expressed from a modified pET50 vector was mixed with His-tagged LUH in a binding buffer (50 mM Tris-Cl, pH 8.0, 100 mM NaCl, 0.1% [v/v] Triton X-100, 0.5 mM EDTA, 10% glycerol, 1 mM DTT, and 1× complete protease inhibitor cocktail) for 4 h at 4°C in the dark. After incubation, glutathione-agarose beads were collected by centrifugation at 340g for 1 min and washed three times with 500 μL binding buffer. Precipitated proteins were detected by immunoblot analysis.
In Vivo Binding Assay
Seven-day-old light grown Myc-tagged GFP (GFP-OX) or Myc-tagged PIF1 (PIF1-OX) transgenic seedlings were exposed to Agrobacterium tumefaciens-based infiltration to induce transient expression of LUH-FLAG. Briefly, GV3101 cells carrying the binary LUH-FLAG construct were suspended in half-strength MS solution (0.5× MS, 5% sucrose, and 0.05% MES, pH 5.7) and used to infiltrate the seedlings under a vacuum (2 min of vacuum, break, 2 min of vacuum). Infiltrated seedlings were grown further for 2 d under white light and transferred to the dark for 6 h followed by a 5 min far-red pulse treatment before sampling. For the in vivo binding assay, 2 g of seedlings were homogenized in 1 mL lysis buffer (50 mM Tris-HCl, pH 7.5, 100 mM NaCl, 0.1% Nonidet P-40, 50 μM MG132, and 1× complete protease inhibitor cocktail). Samples were centrifuged at 20,000g in 4°C for 10 min, and then the supernatant was collected in a new tube. Three microliters of anti-Myc antibody (2276S, lot number 24; Cell Signaling) was added and incubated at 4°C for 4 h in the dark. Then, 40 μL protein A-agarose beads (20333; Pierce) was added and further incubated for 2 h to immunoprecipitate Myc-tagged GFP or PIF1. After incubation, the beads were washed three times with 1 mL lysis buffer and then boiled.
Microarray Analysis
For the transcriptome analyses, total RNA samples were isolated as biological triplicates from Col-0 and luh mutant seeds as described above. The Agilent Arabidopsis Genome 44k chip was used, and the data were analyzed in R with packages from Bioconductor. Background correction and normalization between and within arrays were performed using the LIMMA package. Then, lmFit and eBayes were used to fit a linear model for each probe set and to compute statistics. DEGs were defined as genes with a FDR < 0.05 and with at least a 2-fold difference in expression level. For PIF1-regulated and red light-regulated DEGs, raw data from the NCBI Gene Expression Omnibus were obtained and processed as described.
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL data libraries under the following accession numbers: LUH (At2g32700), LUG (At4g32551), MUM2 (At5g63800), PIF1 (At2g20180), GA3ox1 (At1g15550), GA3ox2 (At1g80340), GA2ox2 (At1g30040), ABA1 (At5g67030), NCED6 (At3g24220), NCED9 (At1g78390), CYP707A2 (At2g29090), RGA (At2g01570), GAI (At1g14920), PIL1 (At2g46970), EXP8 (At2g40610), EXP10 (At1g26770), SCL3 (At1g50420), and PP2A (At1g13320). Microarray data have been deposited in the NCBI Gene Expression Omnibus database (www.ncbi.nlm.nih.gov/geo; accession number GSE70406).
Supplemental Data
Supplemental Figure 1. pif1 mutants and pif1 luh double mutants have comparable germination kinetics under the phyBoff condition.
Supplemental Figure 2. Comparable germination frequencies for wild-type seeds and seeds harvested from the LUG+/− or mum2 mutants.
Supplemental Figure 3. Heat map showing a hierarchical clustering analysis of LUH- and PIF1-regulated genes.
Supplemental Figure 4. LUH-mediated enrichment of promoter fragments in the wild type and pif1 mutants.
Supplemental Figure 5. LUG regulates SEP3 targets either positively or negatively.
Supplemental Figure 6. Expression patterns of LUH- and PIF1-regulated genes in the embryo and endosperm.
Supplemental Data Set 1. LUH- and PIF1-regulated genes.
Supplemental Data Set 2. LUH- and PIF1-activated and -repressed genes.
Supplemental Data Set 3. Cell wall loosening enzyme genes and hormone-responsive genes.
Supplemental Data Set 4. LUH- and PIF1-regulated PIF1 target genes.
Supplemental Data Set 5. LUH-regulated PIF1 target genes.
Supplemental Data Set 6. List of primers used in this study.
Supplementary Material
Acknowledgments
We thank TAIR and NASC for providing information and mutant seeds. This work was supported in part by grants from the National Research Foundation of Korea (2015R1A2A1A05001091 and 2011-0031955) and the Rural Development Administration (SSAC-PJ011073) to G.C.
AUTHOR CONTRIBUTIONS
N.L., J.P., K.K., and G.C. designed experiments. N.L., J.P., and K.K. performed experiments. N.L. and G.C. wrote the article.
Glossary
- ABA
abscisic acid
- GA
gibberellin
- FDR
false discovery rate
- DEG
differentially expressed gene
- GSEA
gene set enrichment analysis
- NES
normalized enrichment score
- ChIP
chromatin immunoprecipitation
- MS
Murashige and Skoog
References
- Artavanis-Tsakonas S., Rand M.D., Lake R.J. (1999). Notch signaling: cell fate control and signal integration in development. Science 284: 770–776. [DOI] [PubMed] [Google Scholar]
- Bailey A.M., Posakony J.W. (1995). Suppressor of hairless directly activates transcription of enhancer of split complex genes in response to Notch receptor activity. Genes Dev. 9: 2609–2622. [DOI] [PubMed] [Google Scholar]
- Barolo S., Stone T., Bang A.G., Posakony J.W. (2002). Default repression and Notch signaling: Hairless acts as an adaptor to recruit the corepressors Groucho and dCtBP to Suppressor of Hairless. Genes Dev. 16: 1964–1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bui M., Lim N., Sijacic P., Liu Z. (2011). LEUNIG_HOMOLOG and LEUNIG regulate seed mucilage extrusion in Arabidopsis. J. Integr. Plant Biol. 53: 399–408. [DOI] [PubMed] [Google Scholar]
- Causier B., Ashworth M., Guo W., Davies B. (2012). The TOPLESS interactome: a framework for gene repression in Arabidopsis. Plant Physiol. 158: 423–438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C., Wang S., Huang H. (2000). LEUNIG has multiple functions in gynoecium development in Arabidopsis. Genesis 26: 42–54. [DOI] [PubMed] [Google Scholar]
- Chen G., Courey A.J. (2000). Groucho/TLE family proteins and transcriptional repression. Gene 249: 1–16. [DOI] [PubMed] [Google Scholar]
- Chen G., Fernandez J., Mische S., Courey A.J. (1999). A functional interaction between the histone deacetylase Rpd3 and the corepressor groucho in Drosophila development. Genes Dev. 13: 2218–2230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho J.N., Ryu J.Y., Jeong Y.M., Park J., Song J.J., Amasino R.M., Noh B., Noh Y.S. (2012). Control of seed germination by light-induced histone arginine demethylation activity. Dev. Cell 22: 736–748. [DOI] [PubMed] [Google Scholar]
- Cinnamon E., Paroush Z. (2008). Context-dependent regulation of Groucho/TLE-mediated repression. Curr. Opin. Genet. Dev. 18: 435–440. [DOI] [PubMed] [Google Scholar]
- Conlan R.S., Gounalaki N., Hatzis P., Tzamarias D. (1999). The Tup1-Cyc8 protein complex can shift from a transcriptional co-repressor to a transcriptional co-activator. J. Biol. Chem. 274: 205–210. [DOI] [PubMed] [Google Scholar]
- Conner J., Liu Z. (2000). LEUNIG, a putative transcriptional corepressor that regulates AGAMOUS expression during flower development. Proc. Natl. Acad. Sci. USA 97: 12902–12907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujimori T., Yamashino T., Kato T., Mizuno T. (2004). Circadian-controlled basic/helix-loop-helix factor, PIL6, implicated in light-signal transduction in Arabidopsis thaliana. Plant Cell Physiol. 45: 1078–1086. [DOI] [PubMed] [Google Scholar]
- Gabriele S., Rizza A., Martone J., Circelli P., Costantino P., Vittorioso P. (2010). The Dof protein DAG1 mediates PIL5 activity on seed germination by negatively regulating GA biosynthetic gene AtGA3ox1. Plant J. 61: 312–323. [DOI] [PubMed] [Google Scholar]
- Goda H., et al. (2008). The AtGenExpress hormone and chemical treatment data set: experimental design, data evaluation, model data analysis and data access. Plant J. 55: 526–542. [DOI] [PubMed] [Google Scholar]
- Gonzalez D., Bowen A.J., Carroll T.S., Conlan R.S. (2007). The transcription corepressor LEUNIG interacts with the histone deacetylase HDA19 and mediator components MED14 (SWP) and CDK8 (HEN3) to repress transcription. Mol. Cell. Biol. 27: 5306–5315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregis V., Sessa A., Colombo L., Kater M.M. (2006). AGL24, SHORT VEGETATIVE PHASE, and APETALA1 redundantly control AGAMOUS during early stages of flower development in Arabidopsis. Plant Cell 18: 1373–1382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregis V., et al. (2013). Identification of pathways directly regulated by SHORT VEGETATIVE PHASE during vegetative and reproductive development in Arabidopsis. Genome Biol. 14: R56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grigorova B., Mara C., Hollender C., Sijacic P., Chen X., Liu Z. (2011). LEUNIG and SEUSS co-repressors regulate miR172 expression in Arabidopsis flowers. Development 138: 2451–2456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heitzler P., Bourouis M., Ruel L., Carteret C., Simpson P. (1996). Genes of the Enhancer of split and achaete-scute complexes are required for a regulatory loop between Notch and Delta during lateral signalling in Drosophila. Development 122: 161–171. [DOI] [PubMed] [Google Scholar]
- Holmes K.A., Hurtado A., Brown G.D., Launchbury R., Ross-Innes C.S., Hadfield J., Odom D.T., Carroll J.S. (2012). Transducin-like enhancer protein 1 mediates estrogen receptor binding and transcriptional activity in breast cancer cells. Proc. Natl. Acad. Sci. USA 109: 2748–2753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hornitschek P., Kohnen M.V., Lorrain S., Rougemont J., Ljung K., López-Vidriero I., Franco-Zorrilla J.M., Solano R., Trevisan M., Pradervand S., Xenarios I., Fankhauser C. (2012). Phytochrome interacting factors 4 and 5 control seedling growth in changing light conditions by directly controlling auxin signaling. Plant J. 71: 699–711. [DOI] [PubMed] [Google Scholar]
- Huang J., DeBowles D., Esfandiari E., Dean G., Carpita N.C., Haughn G.W. (2011). The Arabidopsis transcription factor LUH/MUM1 is required for extrusion of seed coat mucilage. Plant Physiol. 156: 491–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huq E., Quail P.H. (2002). PIF4, a phytochrome-interacting bHLH factor, functions as a negative regulator of phytochrome B signaling in Arabidopsis. EMBO J. 21: 2441–2450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huq E., Al-Sady B., Hudson M., Kim C., Apel K., Quail P.H. (2004). Phytochrome-interacting factor 1 is a critical bHLH regulator of chlorophyll biosynthesis. Science 305: 1937–1941. [DOI] [PubMed] [Google Scholar]
- Jennings B.H., Ish-Horowicz D. (2008). The Groucho/TLE/Grg family of transcriptional co-repressors. Genome Biol. 9: 205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaufmann K., Muiño J.M., Jauregui R., Airoldi C.A., Smaczniak C., Krajewski P., Angenent G.C. (2009). Target genes of the MADS transcription factor SEPALLATA3: integration of developmental and hormonal pathways in the Arabidopsis flower. PLoS Biol. 7: e1000090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khanna R., Huq E., Kikis E.A., Al-Sady B., Lanzatella C., Quail P.H. (2004). A novel molecular recognition motif necessary for targeting photoactivated phytochrome signaling to specific basic helix-loop-helix transcription factors. Plant Cell 16: 3033–3044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kieffer M., Stern Y., Cook H., Clerici E., Maulbetsch C., Laux T., Davies B. (2006). Analysis of the transcription factor WUSCHEL and its functional homologue in Antirrhinum reveals a potential mechanism for their roles in meristem maintenance. Plant Cell 18: 560–573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J., Yi H., Choi G., Shin B., Song P.S., Choi G. (2003). Functional characterization of phytochrome interacting factor 3 in phytochrome-mediated light signal transduction. Plant Cell 15: 2399–2407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim K., Shin J., Lee S.H., Kweon H.S., Maloof J.N., Choi G. (2011). Phytochromes inhibit hypocotyl negative gravitropism by regulating the development of endodermal amyloplasts through phytochrome-interacting factors. Proc. Natl. Acad. Sci. USA 108: 1729–1734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kircher S., Kozma-Bognar L., Kim L., Adam E., Harter K., Schafer E., Nagy F. (1999). Light quality-dependent nuclear import of the plant photoreceptors phytochrome A and B. Plant Cell 11: 1445–1456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuchin S., Carlson M. (1998). Functional relationships of Srb10-Srb11 kinase, carboxy-terminal domain kinase CTDK-I, and transcriptional corepressor Ssn6-Tup1. Mol. Cell. Biol. 18: 1163–1171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J.E., Golz J.F. (2012). Diverse roles of Groucho/Tup1 co-repressors in plant growth and development. Plant Signal. Behav. 7: 86–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee K.P., Piskurewicz U., Turecková V., Strnad M., Lopez-Molina L. (2010). A seed coat bedding assay shows that RGL2-dependent release of abscisic acid by the endosperm controls embryo growth in Arabidopsis dormant seeds. Proc. Natl. Acad. Sci. USA 107: 19108–19113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee K.P., Piskurewicz U., Turečková V., Carat S., Chappuis R., Strnad M., Fankhauser C., Lopez-Molina L. (2012). Spatially and genetically distinct control of seed germination by phytochromes A and B. Genes Dev. 26: 1984–1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y., Choi D., Kende H. (2001). Expansins: ever-expanding numbers and functions. Curr. Opin. Plant Biol. 4: 527–532. [DOI] [PubMed] [Google Scholar]
- Leivar P., Tepperman J.M., Cohn M.M., Monte E., Al-Sady B., Erickson E., Quail P.H. (2012). Dynamic antagonism between phytochromes and PIF family basic helix-loop-helix factors induces selective reciprocal responses to light and shade in a rapidly responsive transcriptional network in Arabidopsis. Plant Cell 24: 1398–1419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leivar P., Monte E., Al-Sady B., Carle C., Storer A., Alonso J.M., Ecker J.R., Quail P.H. (2008). The Arabidopsis phytochrome-interacting factor PIF7, together with PIF3 and PIF4, regulates responses to prolonged red light by modulating phyB levels. Plant Cell 20: 337–352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L., et al. (2012). Linking photoreceptor excitation to changes in plant architecture. Genes Dev. 26: 785–790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z., Meyerowitz E.M. (1995). LEUNIG regulates AGAMOUS expression in Arabidopsis flowers. Development 121: 975–991. [DOI] [PubMed] [Google Scholar]
- Liu Z., Karmarkar V. (2008). Groucho/Tup1 family co-repressors in plant development. Trends Plant Sci. 13: 137–144. [DOI] [PubMed] [Google Scholar]
- Liu Z., Franks R.G., Klink V.P. (2000). Regulation of gynoecium marginal tissue formation by LEUNIG and AINTEGUMENTA. Plant Cell 12: 1879–1892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long J.A., Ohno C., Smith Z.R., Meyerowitz E.M. (2006). TOPLESS regulates apical embryonic fate in Arabidopsis. Science 312: 1520–1523. [DOI] [PubMed] [Google Scholar]
- Lorrain S., Allen T., Duek P.D., Whitelam G.C., Fankhauser C. (2008). Phytochrome-mediated inhibition of shade avoidance involves degradation of growth-promoting bHLH transcription factors. Plant J. 53: 312–323. [DOI] [PubMed] [Google Scholar]
- Nagel A.C., Krejci A., Tenin G., Bravo-Patiño A., Bray S., Maier D., Preiss A. (2005). Hairless-mediated repression of notch target genes requires the combined activity of Groucho and CtBP corepressors. Mol. Cell. Biol. 25: 10433–10441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nemhauser J.L., Hong F., Chory J. (2006). Different plant hormones regulate similar processes through largely nonoverlapping transcriptional responses. Cell 126: 467–475. [DOI] [PubMed] [Google Scholar]
- Ni M., Tepperman J.M., Quail P.H. (1998). PIF3, a phytochrome-interacting factor necessary for normal photoinduced signal transduction, is a novel basic helix-loop-helix protein. Cell 95: 657–667. [DOI] [PubMed] [Google Scholar]
- Oh E., Kim J., Park E., Kim J.I., Kang C., Choi G. (2004). PIL5, a phytochrome-interacting basic helix-loop-helix protein, is a key negative regulator of seed germination in Arabidopsis thaliana. Plant Cell 16: 3045–3058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh E., Kang H., Yamaguchi S., Park J., Lee D., Kamiya Y., Choi G. (2009). Genome-wide analysis of genes targeted by PHYTOCHROME INTERACTING FACTOR 3-LIKE5 during seed germination in Arabidopsis. Plant Cell 21: 403–419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh E., Yamaguchi S., Hu J., Yusuke J., Jung B., Paik I., Lee H.S., Sun T.P., Kamiya Y., Choi G. (2007). PIL5, a phytochrome-interacting bHLH protein, regulates gibberellin responsiveness by binding directly to the GAI and RGA promoters in Arabidopsis seeds. Plant Cell 19: 1192–1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh E., Yamaguchi S., Kamiya Y., Bae G., Chung W.I., Choi G. (2006). Light activates the degradation of PIL5 protein to promote seed germination through gibberellin in Arabidopsis. Plant J. 47: 124–139. [DOI] [PubMed] [Google Scholar]
- Oh E., Zhu J.Y., Wang Z.Y. (2012). Interaction between BZR1 and PIF4 integrates brassinosteroid and environmental responses. Nat. Cell Biol. 14: 802–809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park E., Park J., Kim J., Nagatani A., Lagarias J.C., Choi G. (2012). Phytochrome B inhibits binding of phytochrome-interacting factors to their target promoters. Plant J. 72: 537–546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pauwels L., et al. (2010). NINJA connects the co-repressor TOPLESS to jasmonate signalling. Nature 464: 788–791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penfield S., Li Y., Gilday A.D., Graham S., Graham I.A. (2006). Arabidopsis ABA INSENSITIVE4 regulates lipid mobilization in the embryo and reveals repression of seed germination by the endosperm. Plant Cell 18: 1887–1899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penfield S., Josse E.M., Kannangara R., Gilday A.D., Halliday K.J., Graham I.A. (2005). Cold and light control seed germination through the bHLH transcription factor SPATULA. Curr. Biol. 15: 1998–2006. [DOI] [PubMed] [Google Scholar]
- Rose, J.K., Braam, J., Fry, S.C., and Nishitani, K. (2002). The XTH family of enzymes involved in xyloglucan endotransglucosylation and endohydrolysis: current perspectives and a new unifying nomenclature. Plant Cell Physiol. 43: 1421–1435. [DOI] [PubMed] [Google Scholar]
- Sakamoto K., Nagatani A. (1996). Nuclear localization activity of phytochrome B. Plant J. 10: 859–868. [DOI] [PubMed] [Google Scholar]
- Salter M.G., Franklin K.A., Whitelam G.C. (2003). Gating of the rapid shade-avoidance response by the circadian clock in plants. Nature 426: 680–683. [DOI] [PubMed] [Google Scholar]
- Schneitz K., Hülskamp M., Kopczak S.D., Pruitt R.E. (1997). Dissection of sexual organ ontogenesis: a genetic analysis of ovule development in Arabidopsis thaliana. Development 124: 1367–1376. [DOI] [PubMed] [Google Scholar]
- Shamimuzzaman M., Vodkin L. (2013). Genome-wide identification of binding sites for NAC and YABBY transcription factors and co-regulated genes during soybean seedling development by ChIP-Seq and RNA-Seq. BMC Genomics 14: 477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen H., Zhu L., Castillon A., Majee M., Downie B., Huq E. (2008). Light-induced phosphorylation and degradation of the negative regulator PHYTOCHROME-INTERACTING FACTOR1 from Arabidopsis depend upon its direct physical interactions with photoactivated phytochromes. Plant Cell 20: 1586–1602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin J., Kim K., Kang H., Zulfugarov I.S., Bae G., Lee C.H., Lee D., Choi G. (2009). Phytochromes promote seedling light responses by inhibiting four negatively-acting phytochrome-interacting factors. Proc. Natl. Acad. Sci. USA 106: 7660–7665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shrestha B., Guragain B., Sridhar V.V. (2014). Involvement of co-repressor LUH and the adapter proteins SLK1 and SLK2 in the regulation of abiotic stress response genes in Arabidopsis. BMC Plant Biol. 14: 54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sitaraman J., Bui M., Liu Z. (2008). LEUNIG_HOMOLOG and LEUNIG perform partially redundant functions during Arabidopsis embryo and floral development. Plant Physiol. 147: 672–681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sridhar V.V., Surendrarao A., Liu Z. (2006). APETALA1 and SEPALLATA3 interact with SEUSS to mediate transcription repression during flower development. Development 133: 3159–3166. [DOI] [PubMed] [Google Scholar]
- Sridhar V.V., Surendrarao A., Gonzalez D., Conlan R.S., Liu Z. (2004). Transcriptional repression of target genes by LEUNIG and SEUSS, two interacting regulatory proteins for Arabidopsis flower development. Proc. Natl. Acad. Sci. USA 101: 11494–11499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stahle M.I., Kuehlich J., Staron L., von Arnim A.G., Golz J.F. (2009). YABBYs and the transcriptional corepressors LEUNIG and LEUNIG_HOMOLOG maintain leaf polarity and meristem activity in Arabidopsis. Plant Cell 21: 3105–3118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian A., Tamayo P., Mootha V.K., Mukherjee S., Ebert B.L., Gillette M.A., Paulovich A., Pomeroy S.L., Golub T.R., Lander E.S., Mesirov J.P. (2005). Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA 102: 15545–15550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szemenyei H., Hannon M., Long J.A. (2008). TOPLESS mediates auxin-dependent transcriptional repression during Arabidopsis embryogenesis. Science 319: 1384–1386. [DOI] [PubMed] [Google Scholar]
- Walker M., Tehseen M., Doblin M.S., Pettolino F.A., Wilson S.M., Bacic A., Golz J.F. (2011). The transcriptional regulator LEUNIG_HOMOLOG regulates mucilage release from the Arabidopsis testa. Plant Physiol. 156: 46–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi R., Nakamura M., Mochizuki N., Kay S.A., Nagatani A. (1999). Light-dependent translocation of a phytochrome B-GFP fusion protein to the nucleus in transgenic Arabidopsis. J. Cell Biol. 145: 437–445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeitlinger J., Zinzen R.P., Stark A., Kellis M., Zhang H., Young R.A., Levine M. (2007). Whole-genome ChIP-chip analysis of Dorsal, Twist, and Snail suggests integration of diverse patterning processes in the Drosophila embryo. Genes Dev. 21: 385–390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Mayba O., Pfeiffer A., Shi H., Tepperman J.M., Speed T.P., Quail P.H. (2013). A quartet of PIF bHLH factors provides a transcriptionally centered signaling hub that regulates seedling morphogenesis through differential expression-patterning of shared target genes in Arabidopsis. PLoS Genet. 9: e1003244. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.