Small RNA derived from the virulence modulating region of the Potato spindle tuber viroid downregulates callose synthase by RNA silencing in tomato.
Abstract
The tomato (Solanum lycopersicum) callose synthase genes CalS11-like and CalS12-like encode proteins that are essential for the formation of callose, a major component of pollen mother cell walls; these enzymes also function in callose formation during pathogen infection. This article describes the targeting of these callose synthase mRNAs by a small RNA derived from the virulence modulating region of two Potato spindle tuber viroid variants. More specifically, viroid infection of tomato plants resulted in the suppression of the target mRNAs up to 1.5-fold, depending on the viroid variant used and the gene targeted. The targeting of these mRNAs by RNA silencing was validated by artificial microRNA experiments in a transient expression system and by RNA ligase-mediated rapid amplification of cDNA ends. Viroid mutants incapable of targeting callose synthase mRNAs failed to induce typical infection phenotypes, whereas a chimeric viroid obtained by swapping the virulence modulating regions of a mild and a severe variant of Potato spindle tuber viroid greatly affected the accumulation of viroids and the severity of disease symptoms. These data provide evidence of the silencing of multiple genes by a single small RNA derived from a viroid.
INTRODUCTION
In plant pathology, viroids are the most enigmatic pathogens, due to their small (246 to 401 nucleotides), single-stranded, highly structured RNA genome. Viroids do not encode any proteins and rely entirely on host factors for their replication (Ding, 2009). Viroid infection is often accompanied by a wide array of symptoms, including stunting, leaf epinasty, leaf distortion, flower distortion, and reduced numbers of flowers. How exactly these small, noncoding RNAs can invade plants and induce host responses has long been a subject of interest (Owens et al., 2012).
The current viroid classification scheme consists of 32 species, broadly divided into two families (Di Serio et al., 2014). The Avsunviroidae family, whose type species is the Avocado sunblotch viroid, includes four members that replicate in chloroplasts through a symmetric rolling-circle mechanism and exhibit self-cleavage activity. Members of the Pospiviroidae family, whose type species is the Potato spindle tuber viroid (PSTVd), replicate in the nucleus and contain five structural/functional domains: the terminal left, pathogenicity, central conserved region, variable, and terminal right regions. The importance of the viroid pathogenicity domain in symptom severity is well established (Keese and Symons, 1985), but the mechanism responsible for symptom development upon the infection of plants has remained elusive.
Due to their highly base-paired structures and RNA-RNA mode of replication, viroids act as both inducers and targets of RNA silencing (reviewed in Pallás et al., 2012). RNA silencing, also known as RNA interference (RNAi), provides a multilayer defense system, which protects plants from invasion by exogenous RNA replicons such as viruses and viroids (Ding, 2010). Silencing is triggered by the conversion of long double-stranded RNAs into small RNA duplexes of 21 and 24 nucleotides (Hamilton et al., 2002). The accumulation of such viroid-derived small RNAs (vd-sRNAs) has been extensively studied in different viroid-host combinations (Bolduc et al., 2010; Ivanova et al., 2014; Tsushima et al., 2015), and multiple plant Argonaute proteins bind these vd-sRNAs (Minoia et al., 2015). These observations support the hypotheses that viroids infection trigger RNA silencing and that RNAi might play a role in symptom induction. Studies on the Peach latent mosaic viroid, a chloroplast replicating viroid, showed that vd-sRNA could target heat shock protein 90 in its natural host, Prunus persica, further supporting the hypothesis that vd-sRNA directed RNAi induces symptoms in viroid-infected plants (Navarro et al., 2012).
Callose is a polysaccharide (β-1,3-glucan) that contains some β-1,6-branches (Chen and Kim, 2009). It functions in multiple stages of pollen development as a structural component (Enns et al., 2005). In addition, callose is also deposited at plasmodesmata (PD) to regulate the passive diffusion of small, soluble molecules but restrict the passage of macromolecular structures, such as viruses and viroids (Chen and Kim, 2009). Viruses have evolved mechanisms to overcome this barrier, thus facilitating their spread. For example, many virus-encoded movement proteins induce an increase in the PD exclusion limit, thus allowing the cell-to-cell movement of viruses (Benitez-Alfonso et al., 2010). PSTVd moves through PD and can be transmitted at high frequency through contaminated seeds (Ding et al., 1997; Matsushita et al., 2011). Previously, RT-PCR assays showed that PSTVd was distributed in all floral tissues, including sepals, petals, stamens, and pistils of infected tomato (Solanum lycopersicum) plants (Singh, 2006). In fact, in situ hybridization of flowers from PSTVd-infected tomato plants revealed viroids in the outer integuments, but not in the inner integuments around the embryo sac, suggesting that PSTVd may enter the seed through infected pollen (Matsushita et al., 2011). Nevertheless, the key mechanistic question remains how does a highly structured, noncoding RNA overcome the plant’s defenses, including callose, to achieve cell-to-cell trafficking and entry into the pollen.
PSTVd is a good model for studying viroids given the previous demonstration of its cell-to-cell trafficking through PD and its systemic movement through phloem (Ding et al., 1997). Recently, the structure of PSTVd in solution was resolved by high-throughput selective 2'-hydroxyl acylation analyzed by primer extension (SHAPE). PSTVd-infected tomato plants have also been used to both identify small RNAs (sRNAs) derived from PSTVd upon infection, as well as to monitor tomato gene regulation by microarray analysis (Owens et al., 2012; Giguère et al., 2014). Furthermore, different PSTVd variants induce an array of symptoms in S. lycopersicum cv Rutgers, ranging from mild to severe (Tsushima et al., 2015). In addition, the availability of the tomato genome allows for prediction of sRNA target genes (Tomato Genome Consortium, 2012). In this study, we used bioinformatic and molecular approaches to analyze the effect of vd-sRNA derived from PSTVd variants on two callose synthase genes in tomato. The results provide conclusive molecular and biological evidence that vd-sRNAs target and silence at least two host callose synthase genes.
RESULTS
PSTVd Variants Fold into Different Secondary Structures in Solution
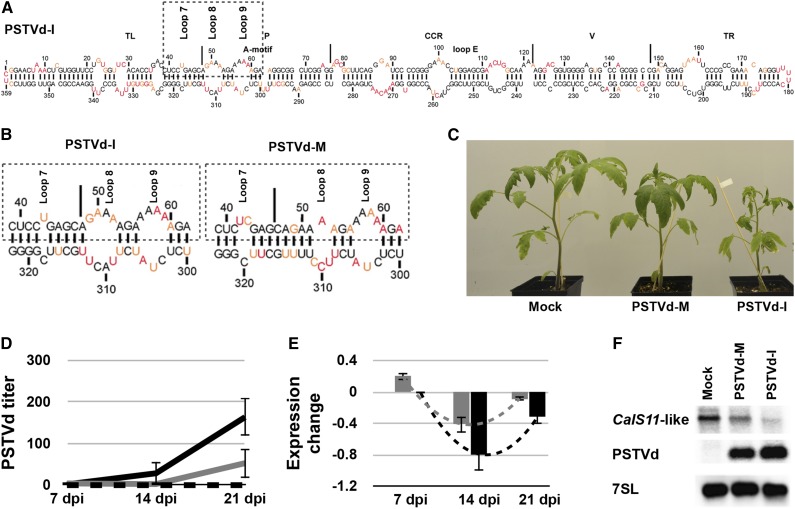
PSTVd-intermediate (PSTVd-I) and PSTVd-mild (PSTVd-M synonyms: PSTVd-Dahlia and PSTVd-D) induce different symptoms in S. lycopersicum cv Rutgers. Sequence analysis has shown that these variants differ in two nucleotides in the VMR, the stretch of sequence comprising the terminal left and pathogenicity domains of PSTVd, which is essential for disease symptom development (Keese and Symons, 1985; Tsushima et al., 2011). Secondary structure predictions revealed that these two changes affect the sizes of loops 7 and 8, which are required for the initiation of systemic trafficking and cell-to-cell movement through PD of PSTVd (Supplemental Figure 1; Zhong et al., 2008). Recently, new methods, such as high-throughput SHAPE, have been used on viroids in solution, in combination with prediction algorithms, to elucidate viroid structure (Giguère et al., 2014). We used SHAPE to verify and compare the structure of two PSTVd variants in solution. As the secondary structure of PSTVd-M in solution had been previously described (Giguère et al., 2014), transcripts of PSTVd-I were prepared for benzoyl cyanide treatment followed by primer extension, as previously described (Giguère et al., 2014). All resulting mapping data were inputted into the RNAstructure software to determine the secondary structures (Figure 1A; Reuter and Mathews, 2010). Comparing the PSTVd-I structure obtained with that of PSTVd-M revealed considerable changes in the VMRs (Figure 1B). More specifically, loop 7 of PSTVd-M is bigger than that of PSTVd-I by one nucleotide, while loop 8 is considerably smaller. Sequence variation within this region is often associated with dramatic differences in symptom induction; however, the mechanism behind this is not completely understood (Eamens et al., 2014). As this VMR is known to be involved in PSTVd movement, it was suspected that the vd-sRNA produced from this region might play a role in PD-related gene regulation.
Figure 1.
Effect of the PSTVd Variants on the CalS11-like mRNA.
(A) The most stable predicted secondary structure for PSTVd-I generated using in solution mapping data obtained by SHAPE is shown. Low-reactivity nucleotides are indicated in black, and highly reactive nucleotides are shown in red. Intermediately reactive nucleotides are shown in orange. The structural/functional motifs of PSTVd-I are delimited by the vertical solid lines and are named accordingly. The A-motif and loop E are indicated.
(B) Enlarged structure at the VMR of both PSTVd-I and PSTVd-M. The boxed regions indicate the VMRs, which are often associated with the induction of symptoms in the bioassay. On the structures, loops 7, 8, and 9 are indicated.
(C) PSTVd variants exhibiting differences in the VMR were inoculated into tomato plants. At 14 dpi, PSTVd-M-inoculated plants showed slight stunting, compared with mock-inoculated plants, while PSTVd-I-inoculated plants showed both leaf curling and severe stunting.
(D) and (E) RNA extracted from tomato plants at 7, 14, and 21 dpi was used to monitor both the titer of PSTVd (D) and the expression of the CalS11-like mRNA (E) at different time intervals. The expression of the CalS11-like mRNA is presented in a log2 scale. In both graphs, the dotted black lines represent the mock-inoculated plants, while the gray and the black solid lines indicate the PSTVd-M- and PSTVd-I-inoculated plants, respectively. The changes in the expression levels of the CalS11-like mRNA between the time points are shown with dotted lines. Data represent the mean of three independent experiments, each performed in triplicate. Error bars indicate sd.
(F) Total RNA from plants infected as in (C) was extracted at 14 dpi and subjected to RNA gel blotting using probes against the CalS11-like mRNA and PSTVd, as well as the 7SL RNA as an internal control.
Prediction of mRNA Targets of sRNAs Derived from the PSTVd VMR
Upon infection, both PSTVd variants produce vd-sRNAs of 21, 22, 23, and 24 nucleotides from throughout their genome (Tsushima et al., 2015). To understand the effect of the VMR on disease symptom induction, we dissected the VMRs of PSTVd variants in silico into 21- to 24-nucleotide fragments, which we then used to interrogate publicly available tomato transcriptome data sets using the WMD3 Web-based tool (http://wmd3.weigelworld.org/cgi-bin/webapp.cgi). The resulting putative target sequences were analyzed by BLAST and mRNAs coding for proteins with known functions were selected for further analysis. The free energy, represented as ∆G, for each vd-sRNA/target duplex was calculated using PairFold (Andronescu et al., 2003; Supplemental Table 1). Interestingly, vd-sRNAs derived from this region of PSTVd-I, but not of PSTVd-M, were predicted to target the open reading frame of a callose synthase gene (CalS), specifically, Callose synthase 11-like mRNA (CalS11-like; GenBank accession number XM_004232828). CalS11-like (homologous to GSL1 of Arabidopsis thaliana) is required for the formation of the callose wall that separates the tetraspores (interstitial wall). Callose is a major component of the pollen mother cell walls, the pollen tubes, and in PD (Enns et al., 2005; http://www.uniprot.org/uniprot/Q9S9U0). At the same time, a manual alignment of the vd-sRNAs derived from the VMR of PSTVd-M against the CalS11-like gene indicated the presence of a number of mismatches in the seed region of the vd-sRNA/target duplex (Supplemental Table 1). This resulted in increased predicted ∆G values and in decreased base-pairing percentage compared with the PSTVd-I vd-sRNA/target duplex. Due to the presence of two mismatches in the seed region, specifically at positions 4 and 5 of the 5′ end of the vd-sRNA, WMD3 failed to predict a match between PSTVd-M and the CalS11-like mRNA. As such, the vd-sRNAs derived from PSTVd-M still likely target the CalS11-like mRNA, but less efficiently than the vd-sRNAs derived from PSTVd-I.
Effect of the PSTVd Variants on the CalS11-like mRNA Levels
To understand the effect of PSTVd infection on CalS11-like mRNA levels, we performed time-course studies by inoculating tomato plants with the PSTVd variants. Plants inoculated with PSTVd-I exhibited initial disease symptoms at 10 to 12 d postinoculation (dpi) and displayed leaf curling and stunted growth, while PSTVd-M inoculated plants were almost identical to mock-inoculated plants, except for a slight stunting at 14 dpi (Figure 1C). To monitor both viroid accumulation and CalS11-like mRNA levels, leaf samples were collected at 7, 14, and 21 dpi. Viroid levels at different time intervals were quantified by subjecting total RNA extracted from these samples to RT-qPCR as described previously (Boonham et al., 2004). PSTVd-I RNA levels increased rapidly in comparison to those of PSTVd-M: At 7 dpi, the levels of PSTVd-I and PSTVd-M RNAs were very similar, but by 14 dpi, the PSTVd-I RNA levels were at least 10-fold higher than those of PSTVd-M (Figure 1D). To determine the effect of the different PSTVd variants on the expression of CalS11-like at different time intervals, we used CalS11-like gene-specific primers (Supplemental Table 2) to perform RT-qPCR analysis on the same RNA samples used to quantify viroid accumulation. Expression levels were normalized to those of the mock-inoculated plants. As shown in Figure 1E, the greatest degree of repression of CalS11-like was seen at 14 dpi with both PSTVd variants. At all time points, PSTVd-I induced a stronger downregulation than did PSTVd-M. The lower expression level of CalS11-like mRNA is consistent with PSTVd infection. Interestingly, in both cases, at 21 dpi, the degree of CalS11-like repression decreased by more than 50%. The decrease in the downregulation of the CalS11-like mRNA level at 21 dpi compared with 14 dpi can be attributed to the hypersensitive reaction of the plant’s defense system against the invading pathogen (Li et al., 2012). To confirm the decreased expression of the CalS11-like mRNA, we performed RNA gel blot assays using probes specific to the CalS11-like mRNA and to PSTVd. The data obtained demonstrated the repression of the CalS11-like mRNA in the viroid-infected plants, regardless of the variant used. The downregulation of CalS11-like mRNA was higher in the PSTVd-I-infected plants than in plants infected with PSTVd-M, a fact that might be due to the higher accumulation of PSTVd-I compared with PSTVd-M (Figure 1F; Supplemental Figure 2). These data are in agreement with the RT-qPCR results, thus further supporting the hypothesis that PSTVd infection downregulates the accumulation of the CalS11-like mRNA in tomato plants.
PSTVd-sRNA Derived from the VMR Favors RNAi-Mediated Cleavage of the CalS11-Like Target Sequence
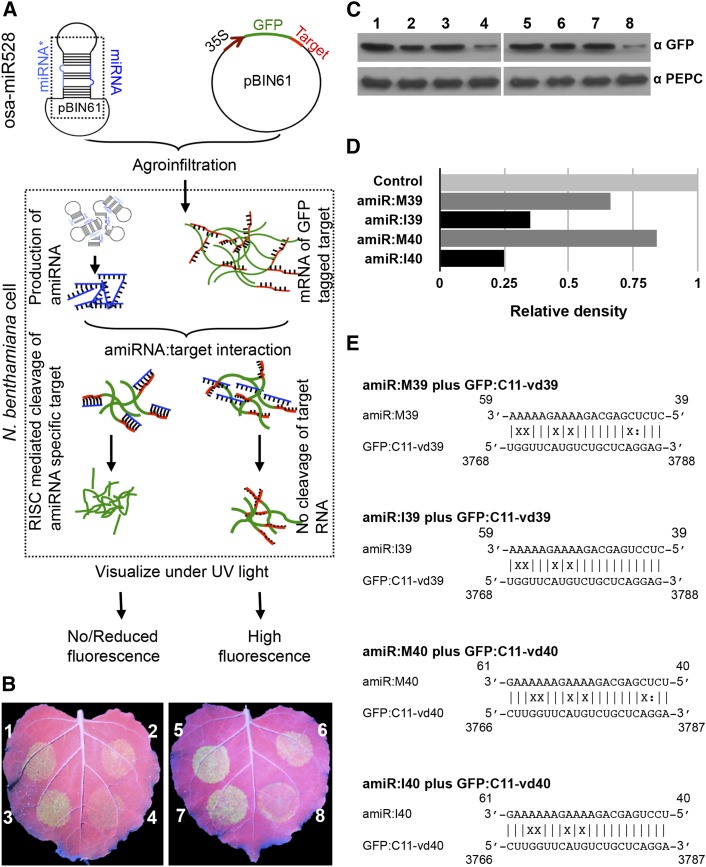
To demonstrate RNA-induced silencing complex (RISC)-mediated targeting of the CalS11-like mRNA, we developed an artificial microRNA (amiRNA) system for the transient expression of the vd-sRNA derived from the VMR of both PSTVd variants in Nicotiana benthamiana leaves. Since 21- and 22-nucleotide vd-sRNA were the most abundant vd-sRNA species recovered from PSTVd-infected tomato plants (Tsushima et al., 2015), only these two vd-sRNA species were used to design the amiRNAs amiR:I39, amiR:I40, amiR:M39, and amiR:M40 (Table 1). We used oligonucleotides corresponding to the sequences of the vd-sRNAs to create amiRNA constructs based on the osa-MIR528 backbone by PCR amplification (Yan et al., 2012). The PCR products were then ligated into the binary vector pBIN61. Additional constructs were prepared by ligating either 21- or 22-nucleotide predicted target sequences into the 3′-untranslated region (UTR) of the GFP gene under the control of the 35S promoter in pBIN61 (Supplemental Tables 3 and 4). If the vd-sRNA produced by the amiRNA binds the target, the fluorescence level will be decreased as binding blocks the translational process and/or stability of the target mRNA (Figure 2A). The GFP constructs with CalS11-like target sites were named GFP:C11-vd39, which is targeted by amiR:I39 and amiR:M39, and GFP:C11-vd40, which is targeted by both amiR:I40 and amiR:M40. The GFP constructs were coagroinfiltrated in N. benthamiana leaves along with their respective targeting amiRNAs or empty vector. To demonstrate the feasibility of this technique, a GFP construct with a fragment of the UPF1 gene in the 3′ UTR was expressed with either empty vector or with an amiRNA targeting it (Yan et al., 2012; Supplemental Figure 3). At 3 dpi, agroinfiltrated leaves were observed under UV illumination and GFP fluorescence was detected. All combinations of vd-sRNA:amiRNA plus GFP/target combinations showed less GFP fluorescence than the same GFP/targets expressed with empty vector, suggesting that the GFP-expressing mRNAs were targeted by the amiRNAs (Figure 2B). An immunoblot was performed using anti-GFP antibody to verify the amount of GFP produced in the various vd-sRNA:amiRNA plus GFP/target combinations (Figure 2C). The degree of GFP downregulation by each vd-sRNA:amiRNA plus GFP/target combination was quantified and expressed as relative density (Figure 2D) using phosphoenolpyruvate carboxylase (PEPC) protein levels for normalization. All of the vd-sRNA/target complexes predicted to be formed by the interaction between the amiRNAs and the putative CalS11-like mRNA target sequences are shown in Figure 2E. To rule out an effect of the vd-sRNA:amiRNAs on GFP, an additional control experiment was performed by coexpressing the vd-sRNA:amiRNAs with pBIN-GFP. None of the vd-sRNA:amiRNAs showed any repressive effect on GFP levels expressed from pBIN-GFP, compared with empty vector (Supplemental Figure 4). Comparing the data obtained with the different vd-sRNAs, the vd-sRNAs derived from PSTVd-I were more effective in downregulating GFP than those derived from PSTVd-M. This can be attributed to the presence of the mismatches located in the seed region formed by the PSTVd-M vd-sRNA/target complex, as well as to higher hybridization energy for the base pairing compared with that of PSTVd-I vd-sRNA. These data demonstrate that the vd-sRNA derived from the VMR of PSTVd has the potential to downregulate CalS11-like mRNA.
Table 1. Predicted Duplexes Formed between the Tomato CalS11-like mRNA Target and vd-sRNAs Derived from the VMRs of PSTVd Variants.
| PSTVd Variant | CalS11-like mRNA/vd-sRNA | ∆Ga | % Pairing |
|---|---|---|---|
| PSTVd-I | 3768 GG A G 3788 | −27.96 | 80.9 |
| 5′-U UUC U UCUGCUCAGGAG-3′ | |||
| 3′-A AAG A AGACGAGUCCUC-5′ | |||
| 59 AA A A 39 | |||
| PSTVd-I | 3766 GG A G 3787 | −27.87 | 81.8 |
| 5′-CUU UUC U UCUGCUCAGGA-3′ | |||
| 3′-GAA AAG A AGACGAGUCCU-5′ | |||
| 61 AA A A 40 | |||
| PSTVd-M | 3768 GG A G A 3788 | −22.11 | 71.4 |
| 5′-U UUC U UCUGCUC GGAG-3′ | |||
| 3′-A AAG A AGACGAG UCUC-5′ | |||
| 59 AA A A C39 | |||
| PSTVd-M | 3766 GG A G A 3787 | −22.02 | 72.7 |
| 5′-CUU UUC U UCUGCUC GGA-3′ | |||
| 3′-GAA AAG A AGACGAG UCU-5′ | |||
| 61 AA A A C 40 |
The PairFold online tool was used to predict the minimum secondary structure free energy of pairs of RNA sequences.
Underlined nucleotides represent G:U wobble base pairs.
Figure 2.
Predicted Targeting of the CalS11-like mRNA by vd-sRNA Validated by amiRNA.
(A) Flow chart illustrating the details of the amiRNA experiment. The vd-sRNA, expressed as an amiRNA, and the GFP construct with target sequence in the 3′ UTR, both in the vector pBIN61, were agroinfiltrated into N. benthamiana leaves. Specific interaction of miRNA with the target sequence will lead to either RISC-mediated cleavage or translational repression, which in turn results in decreased GFP production.
(B) N. benthamiana leaves were agroinfiltrated with (1) empty pBIN61 vector (EV) plus GFP:C11-vd39, (2) amiR:M39 plus GFP:C11-vd39, (3) EV plus GFP:C11-vd39, (4) amiR:I39 plus GFP:C11-vd39, (5) EV plus GFP:C11-vd40, (6) amiR:M40 plus GFP:C11-vd40, (7) EV plus GFP:C11-vd40, and (8) amiR:I40 plus GFP:C11-vd40. At 3 dpi, the leaves were photographed under UV illumination.
(C) N. benthamiana leaves were agroinfiltrated in the same combinations as in (B). At 3 dpi, total protein extracts were subjected to immunoblotting with anti-GFP (top panel) and anti-PEPC (lower panel) antibodies.
(D) The immunoblot signals from (C) were quantified and expressed as a ratio of the GFP to the PEPC signal. For each set of experiments, the ratio of GFP to PEPC obtained with EV plus GFP:C11-vdXX (control) was set at a value of 1. The additional bars indicate the relative GFP/PEPC ratio for each amiRNA (as indicated) expressed with its respective GFP target. Each experiment was performed at least three times, and the indicated graph shows a representative quantification of one experiment.
(E) Duplexes predicted to be formed by complexes of amiRNAs and GFP reporter constructs containing CalS11-like mRNA target sequences.
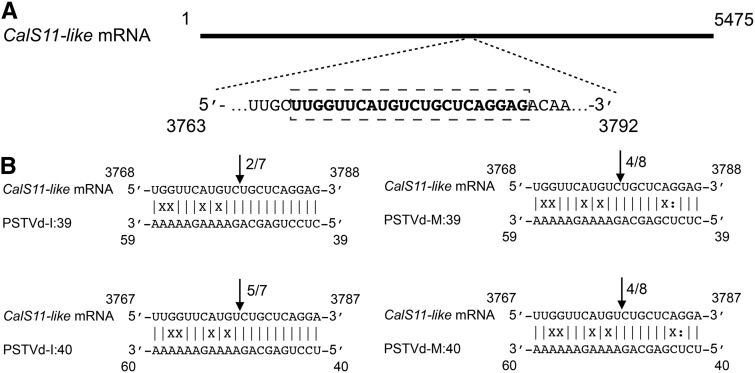
Direct cleavage of a target by a miRNA can be confirmed by 5′ RNA ligase-mediated rapid amplification of cDNA ends (5′ RLM-RACE) (Thomson et al., 2011). Briefly, in 5′ RLM-RACE, an RNA adapter is ligated to the free 5′ phosphate end of an uncapped mRNA produced from RISC-mediated cleavage. The ligation product is reverse transcribed using a gene-specific reverse primer, which is subsequently amplified by nested PCR, cloned, and sequenced. To verify RISC-mediated cleavage at the predicted site, a 5′ RLM-RACE experiment was performed on the 21-dpi RNA preparations using a CalS11-like gene-specific nested PCR (Supplemental Table 5). The nested PCR products were analyzed by agarose gel electrophoresis and the bands of the expected size were eluted, cloned, and sequenced. The cDNA clones from the CalS11-like transcripts were aligned to detect the 5′ termini. All of the CalS11-like transcripts had 5′ termini identical to that of the predicted cleavage site, i.e., between positions 10 and 11 from the 5′ termini (Figure 3). No PCR amplification was obtained when similar experiments were performed with RNA preparations obtained from mock-inoculated plants, indicating specific cleavage of the CalS11-like mRNA by the PSTVd-sRNA (Supplemental Figure 5). The data presented here validate the cleavage of the predicted CalS11-like mRNA sequence in PSTVd-infected plants.
Figure 3.
Analysis of RISC-Mediated Cleavage of the CalS11-like mRNA by 5′ RLM-RACE.
(A) Schematic representation of the CalS11-like mRNA. The bold letters inside the box indicate the predicted vd-sRNA binding site.
(B) CalS11-like mRNA/vd-sRNA duplexes predicted to be formed by the sRNA derived from the PSTVd-I and PSTVd-M variants. The arrows indicate the 5′ termini of CalS11-like mRNA fragments isolated from the PSTVd-infected plants, as identified by 5′ RLM-RACE products, with the frequency of clones shown (ex. 2/7, indicates that two cleavage products were found out of seven analyzed clones). Sequences are shown in the complementary polarity.
VMR vd-sRNA Targets More Than One Gene
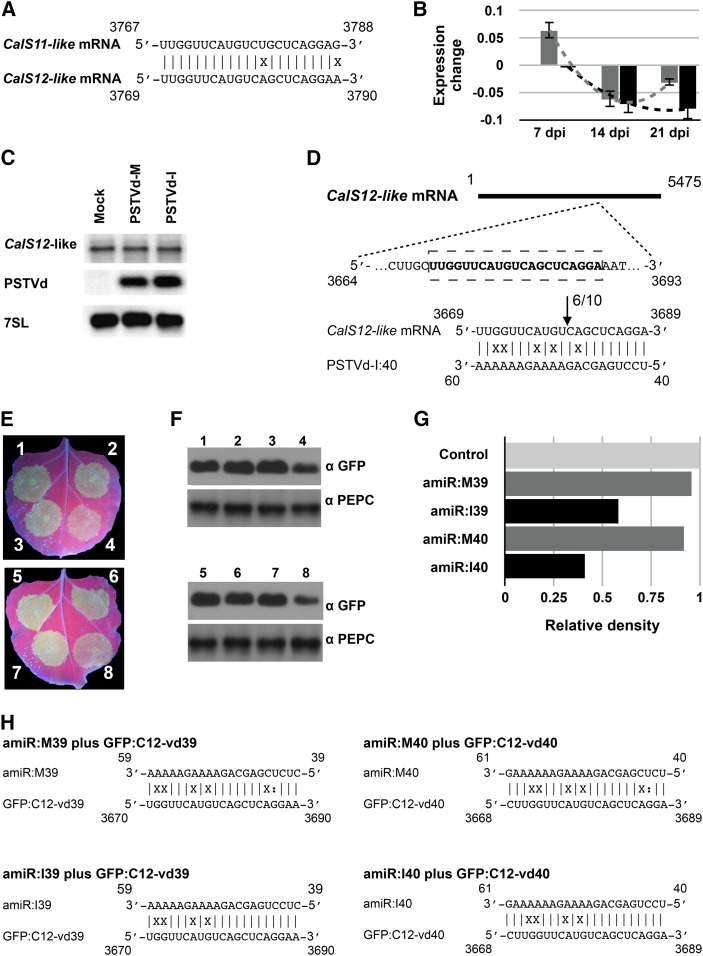
Callose synthase 12-like (CalS12-like; homologous to GSL5 of Arabidopsis) is redundant to CalS11-like (Enns et al., 2005). Alignment of the CalS11-like and CalS12-like mRNA sequences showed a high degree of sequence similarity at the predicted vd-sRNA binding site (Figure 4A). To investigate the possibility of vd-sRNA targeting, we manually aligned the CalS12-like mRNA to the CalS12-like coding region (Supplemental Table 6) and calculated hybridization energies and base-pairing percentages of the vd-sRNA/target interactions. Analysis of the vd-sRNA/CalS12-like duplex revealed the presence of a mismatch with the target that is located either at position 9 (for the vd-sRNA nucleotide covering positions 40 to 61) or 10 (for the vd-sRNA nucleotide covering positions 39 to 59) from the 5′ end of vd-sRNA, indicating a possible decrease in RNA silencing efficiency (Table 2). All four vd-sRNA interactions with CalS12-like exhibited higher ∆G values and lower percentage base pairings. The vd-sRNA derived from PSTVd-M has at least four mismatches in positions 1 to 12 of the 5′ end of vd-sRNA, which could further decrease the stability of the duplex. This also explains why we initially failed to predict this target along with CalS11-like.
Figure 4.
Effect of vd-sRNA on CalS12-like mRNA.
(A) Alignment of the tomato CalS11-like and CalS12-like mRNAs in the region predicted to be targeted by the PSTVd sRNA.
(B) RNA extracted from tomato plants inoculated with the two PSTVd variants at 7, 14, and 21 dpi was used to monitor expression of the CalS12-like mRNA over time. The values are presented in log2 scale. The gray and black color bars in the graph indicate the PSTVd-M- and the PSTVd-I-inoculated plants, respectively. Changes in the expression levels of the CalS12-like mRNA between the time points are shown by dotted lines. Data represent the mean of three independent experiments, each performed in triplicate. Error bars indicate sd.
(C) Total RNA from plants infected with PSTVd-M or PSTVd-I, or mock inoculated, was extracted at 14 dpi and subjected to RNA gel blotting using probes against the CalS11-like mRNA and PSTVd, as well as the 7SL RNA as an internal control (note that mRNA samples are from the same experiment as in Figure 1F and the same 7SL control blot is shown here).
(D) The upper panel shows a schematic representation of the CalS12-like mRNA. The bold letters inside the box indicate the predicted vd-sRNA binding site. The lower panel shows the predicted CalS12-like mRNA/vd-sRNA duplex. The arrows indicate the 5′ termini of CalS12-like mRNA fragments isolated from the PSTVd-I infected plants, as identified by 5′ RLM-RACE products, with the frequency of clones indicated. The sequences shown are those of the complementary polarity.
(E) N. benthamiana leaves were agroinfiltrated with (1) empty pBIN61 vector (EV) plus GFP:C12-vd39, (2) amiR:M39 plus GFP:C12-vd39, (3) EV plus GFP:C12-vd39, (4) amiR:I39 plus GFP:C12-vd39, (5) EV plus GFP:C12-vd40, (6) amiR:M40 plus GFP:C12-vd40, (7) EV plus GFP:C12-vd40, and (8) amiR:I40 plus GFP:C12-vd40. At 3 dpi, the leaves were photographed under UV illumination.
(F) N. benthamiana leaves were agroinfiltrated in the same combinations as in (E). At 3 dpi, the total protein extracts were subjected to immunoblotting with anti-GFP (top panel) and anti-PEPC (lower panel) antibodies.
(G) The immunoblot signals from (F) were quantified and expressed as a ratio of the GFP-to-PEPC signal. For each set of experiments, the ratio of GFP to PEPC obtained with EV plus GFP:C12-vdXX (control) was set at a value of 1. The additional bars indicate the relative GFP/PEPC ratio for each amiRNA (as indicated) expressed with its respective GFP target. Each experiment was performed at least three times, and the indicated graph shows a representative quantification of one experiment.
(H) Duplexes predicted to be formed by complexes of amiRNAs and GFP reporter constructs containing CalS11-like mRNA target sequences.
Table 2. Predicted Duplexes Formed between the Tomato CalS12-like mRNA Target and vd-sRNAs Derived from the VMRs of PSTVd Variants.
| PSTVd Variant | CalS12-like mRNA/vd-sRNA | ∆Ga | % Pairing |
|---|---|---|---|
| PSTVd-I | 3670 GG A G A A 3690 | −22.06 | 71.4 |
| 5′-U UUC U UC GCUCAGGA -3′ | |||
| 3′-A AAG A AG CGAGUCCU -5′ | |||
| 59 AA A A A C 39 | |||
| PSTVd-I | 3668 GG A G A 3689 | −22.95 | 77.3 |
| 5′-CUU UUC U UC GCUCAGGA-3′ | |||
| 3′-GAA AAG A AG CGAGUCCU-5′ | |||
| 61 AA A A A 40 | |||
| PSTVd-M | 3670 GG A G A A A 3690 | −16.21 | 66.7 |
| 5′-U UUC U UC GCTC GGA -3′ | |||
| 3′-A AAG A AG CGAG UCU -5′ | |||
| 59 AA A A A C C 39 | |||
| PSTVd-M | 3668 GG AG A A 3689 | −17.1 | 68.2 |
| 5′-CUU UUC U UC GCUC GGA-3′ | |||
| 3′-GAA AAG A AG CGAG UCU-5′ | |||
| 61 AA A A A C 40 |
The PairFold online tool was used to predict the minimum secondary structure free energy of pairs of RNA sequences.
Underlined nucleotides represent G:U wobble base pairs.
To investigate the effect of PSTVd infection on CalS12-like expression levels, we performed RT-qPCR on RNA from PSTVd-I- and PSTVd-M-inoculated tomato plants at 7, 14, and 21 dpi. Normalization to mock-inoculated plants revealed a slight, almost negligible, decrease in CalS12-like mRNA levels in the PSTVd-infected plants (Figure 4B). Next, we used RNA gel blot assays with a radiolabeled CalS12-like mRNA-specific probe to validate the repression of the CalS12-like mRNA in the PSTVd-infected plants. No observable changes were detected in the repression of the CalS12-like mRNA in both the PSTVd-I- and PSTVd-M-infected tomato plants compared with the control (Figure 4C). In order to confirm the cleavage of the CalS12-like mRNA by vd-sRNA, 5′ RLM-RACE experiments were performed as described above using CalS12-like gene-specific primers (Supplemental Table 7). Only the PSTVd-I inoculated plants yielded amplicons of the expected size upon nested PCR (Supplemental Figure 6). The obtained amplicons were eluted, cloned, and sequenced as above. Sequence data from 10 clones were aligned with the CalS12-like mRNA to detect the 5′ termini. Six resulting sequences had identical 5′ termini that were consistent with the predicted cleavage site, i.e., between positions 10 and 11 from the 5′ termini (Figure 4D; Supplemental Figure 7).
To investigate a possible effect of vd-sRNAs on translational repression of CalS12-like mRNA, we performed an amiRNA experiment. A binary vector expressing GFP with the predicted 21-nucleotide target sequence from CalS12-like in the 3′ UTR (GFP:C12-vd39) was coexpressed with either empty vector or the same amiRNA constructs (amiR:I39 and amiR:M39) designed to validate the CalS11-like targets. Similar experiments were performed with a second construct (GFP:C12-vd40), predicted to be targeted by amiR:I40 and amiR:M40 (Figure 4E). Three days postinoculation, GFP levels were assessed by both visual inspection and by immunoblot (Figure 4F). The relative GFP densities for each of the amiRNA/target combinations were calculated as before (Figure 4G). All of the vd-sRNA/target complexes predicted to be formed by the interaction between the amiRNAs, and the putative CalS12-like mRNA target sequences are shown in Figure 4H. As anticipated, no significant reduction in GFP was induced by amiRNAs derived from PSTVd-M (amiR:M39 and amiR:M40). This can be explained by the fact that the PSTVd-M vd-sRNA/CalS12-like interaction has more mismatches located in the 5′ seed region than does the predicted PSTVd-I vd-sRNA/CalS12-like duplex. The amiR:I40 construct decreased GFP to a greater extent than did amiR:I39, consistent with the fact that the latter vd-sRNA/mRNA duplex has a mismatch at position 10 from the 5′ of the vd-sRNA (Table 2). Taken together, these data suggest that the vd-sRNA derived from the VMR of PSTVd-I, but not PSTVd-M, has the potential to downregulate the CalS12-like mRNA.
Infection with Both PSTVd-M and PSTVd-I Leads to Accumulation of CalS-Targeting sRNA in Infected Tomato Plants
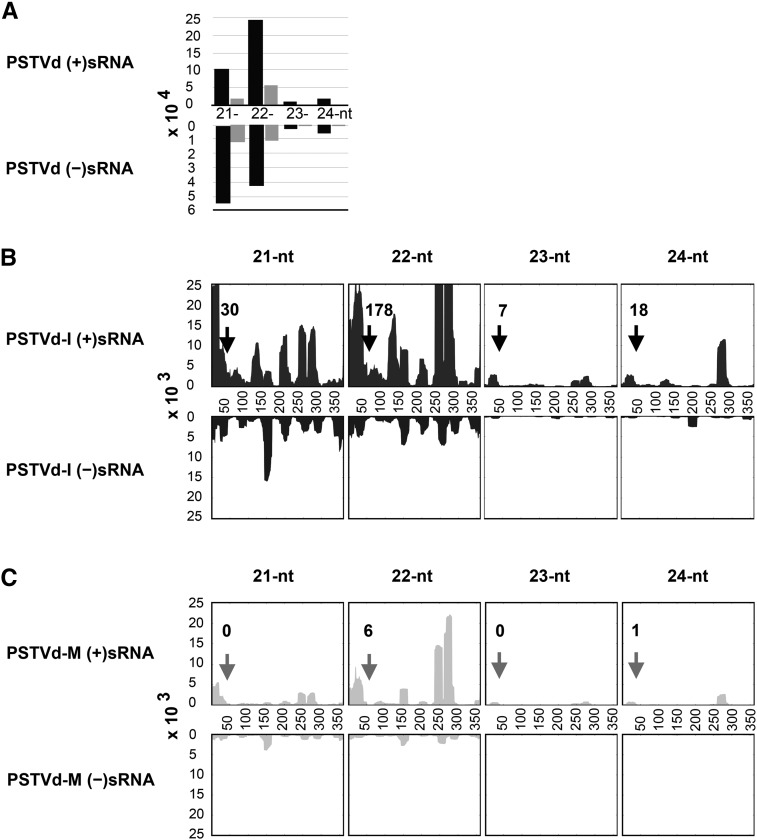
To validate the production of sRNA by the PSTVd variants upon infection and to investigate the differences in the accumulation levels of the CalS gene targeting sRNAs, total RNA extracted from samples at 21 dpi was subjected to high-throughput sequencing. This produced ∼4.3 million reads of sRNA, ranging in length from 21 to 24 nucleotides, from leaf samples of mock-, PSTVd-I-, and PSTVd-M-inoculated plants (Table 3). Sequence analysis of over four million sRNAs from mock-inoculated plants identified only 106 and 108 vd-sRNA of PSTVd-I and PSTVd-M type, respectively, that were 100% identical to either the genomic (+) and antigenomic (−) strands of PSTVd-I and PSTVd-M. Similar analysis with the sRNA recovered from the PSTVd-I- and PSTVd-M-inoculated plants revealed that PSTVd-I-inoculated plants had more numbers of vd-sRNA than PSTVd-M-inoculated plants (Table 3). The 21- to 24-nucleotide-long vd-sRNAs identified from both PSTVd-I- and PSTVd-M-inoculated plants were analyzed for their size and distribution on the viroid genomes. As shown in Figure 5A, comparison of the vd-sRNA populations between the PSTVd variants showed a higher accumulation of vd-sRNA in PSTVd-I-inoculated plants compared with PSTVd-M-inoculated plants. The lower recovery of vd-sRNA in the PSTVd-M-inoculated plants is presumably due to the lower total accumulation of this variant. In both viroid infections, more vd-sRNAs corresponding to the genomic strand (+) were recovered, compared with antigenomic strand (−) derived sRNA. This may be due to the higher accumulation of the genomic strand of PSTVd in the infected plants (Hutchins et al., 1985). Interestingly, analysis of the vd-sRNA revealed that the genomic strand produced more 22-nucleotide sRNAs, while the antigenomic strand produced more 21-nucleotide sRNAs, irrespective of the PSTVd variant. All vd-sRNAs were individually profiled on both the genomic and the antigenomic strands of the respective PSTVd variants in order to understand the production of the vd-sRNAs in the VMR region of both viroids (Figures 5B and 5C). Analysis of the total number of vd-sRNA targeting the CalS gene (as identified in Supplemental Table 1) revealed that PSTVd-I produced a total of 233, while PSTVd-M produced only 7 PSTVd-sRNA. The data presented here demonstrate that infection with both PSTVd variants does indeed result in the production of the vd-sRNAs predicted above, which can target CalS mRNAs.
Table 3. Summary of sRNAs Identified by High-Throughput Sequencing.
| sRNA Population | Mocka | PSTVd-Ib | PSTVd-Mc | ||||||
|---|---|---|---|---|---|---|---|---|---|
| sRNAd | PSTVd-I sRNAe | PSTVd-M sRNAf | sRNAd | vd-sRNA | % vd-sRNAg | sRNAd | vd-sRNA | % vd-sRNA | |
| 21 nucleotides | 585,563 | 39 | 41 | 700,131 | 160,598 | 22.9 | 560,960 | 29,273 | 5.2 |
| 22 nucleotides | 548,009 | 57 | 58 | 839,033 | 288,180 | 34.3 | 560,365 | 66,438 | 11.8 |
| 23 nucleotides | 721,920 | 2 | 2 | 622,373 | 13,218 | 2.1 | 791,306 | 3,121 | 0.4 |
| 24 nucleotides | 2,424,949 | 8 | 7 | 2,155,006 | 26,150 | 1.2 | 2,627,483 | 5,101 | 0.2 |
| 21 to 24 nucleotides | 4,280,441 | 106 | 108 | 4,316,543 | 488,146 | 11.3 | 4,540,114 | 103,933 | 2.3 |
sRNA isolated from a mock-inoculated plant.
sRNA isolated from a PSTVd-I-inoculated plant.
sRNA isolated from a PSTVd-M-inoculated plant.
Total number of individual sRNAs.
sRNAs identical to the genomic (+) and antigenomic (−) strands of PSTVd-I .
sRNA identical to the genomic (+) and antigenomic (−) strands of PSTVd-M.
Figure 5.
Sequence Profile of the PSTVd-sRNAs Recovered from Infected Tomato Leaves.
(A) Size distribution of PSTVd-sRNAs. The horizontal axis indicates the lengths of the sRNA, and the vertical axis indicates the total number of sRNA reads. The black bars indicate PSTVd-I and the gray bars PSTVd-M.
(B) and (C) Profiles of the PSTVd-sRNA populations recovered from leaf tissues of tomato plants infected with PSTVd-I (B) and PSTVd-M (C). The upper panel shows the profile of the PSTVd-sRNAs derived from the genomic strand (+), and the lower panel presents those from the antigenomic strand (−). The regions of the vd-sRNA targeting the CalS genes are indicated with an arrow, along with the number of reads for sRNAs mapping to this region.
Mutations in the VMR Affect Viroid Titer
To establish a systemic infection, PSTVd inoculated into leaves must be capable of both replication and movement not only from cell to cell, but also through the vascular system to invade new tissues. For efficient RNAi-mediated silencing of the target mRNA, a strong sequence complementarity is required in both the 5′ seed region of the targeting miRNA and across the cleavage site (Figure 6A; Ossowski et al., 2008). To identify the effect of vd-sRNA targeting the CalS genes, two types of PSTVd mutants were created by altering the sequence of the VMR: (1) disruption of the predicted vd-sRNA cleavage site and (2) swapping the 5′ seed region of the vd-sRNA/target duplexes (Figure 6B). In the former case, nucleotides 47 to 50 (CAGA to GUCU) of the viroid, which encompass the predicted cleavage site of the CalS11-like targeting vd-sRNA, were mutated. The mutants created in the PSTVd-I and PSTVd-M backgrounds were named PSTVd-Imut and PSTVd-Mmut, respectively. The PSTVd-I mutant in which the VMR has been changed to that of PSTVd-M (nucleotides 42 and 43; CU to UC) was denoted as PSTVd-I→M, while the PSTVd-M having the VMR of PSTVd-I (nucleotides 42 and 43; UC to CU) was denoted as PSTVd-M→I. The predicted duplexes formed from all of the mutant vd-sRNA/target pairs significantly affected the predicted hybridization energies (Table 4). Upon infection, both PSTVd-Imut and PSTVd-Mmut should produce a mutated vd-sRNA possessing a high ∆G compared with that of the wild type. Similarly, PSTVd-I→M should produce a vd-sRNA similar to PSTVd-M, which has higher ∆G as compared with its wild type (PSTVd-I). On the other hand, PSTVd-M→I will produce a PSTV-I-type vd-sRNA, which is predicted to be more efficient in downregulating CalS11-like mRNA compared with the wild type (PSTVd-M; Table 3).
Figure 6.
Effect of the vd-sRNA on Viroid Symptoms and Titer.
(A) Diagram showing the predicted target mRNA/miRNA duplex.
(B) Diagram showing the VMRs of the wild type and the mutated PSTVd variants. The black and gray bold underlined letters indicate the PSTVd-I- and PSTVd-M-type nucleotides, respectively. The underlined nucleotides denote the introduced mutations.
(C) At 14 dpi, plants inoculated with the PSTVd-I-derived mutants PSTVd-Imut and PSTVd-I→M showed morphological changes.
(D) Quantification of viroid titer at 14 dpi in inoculated plants by RT-qPCR. Lane 1, mock inoculated; lane 2, PSTVd-I inoculated; lane 3, PSTVd-Imut inoculated; lane 4, PSTVd-I→M inoculated plants. Data represent the mean of three independent experiments, each performed in triplicate. Error bars indicate sd.
Table 4. Mutation of the PSTVd Variants in a Predicted CalS11-like Targeting vd-sRNA.
| PSTVd Variant | Mutated vd-sRNA | CalS11-like mRNA/Mutant vd-sRNA | ∆Ga | Mutant |
|---|---|---|---|---|
| PSTVd-I | CUCCUGAGGUCUAAAGAAAAAb | 3768 GG A GUCUG 3788 | −13.90 | PSTVd-Imut |
| 5′-U UUC U CUCAGGAG-3′ | ||||
| 3′-A AAG A GAGUCCUC-5′ | ||||
| 59 AA A AUCUG 39 | ||||
| PSTVd-I | CUCUCGAGCAGAAAAGAAAAAc | 3768 GG A G AG 3788 | −18.20 | PSTVd-I→M |
| 5′-U UUC U UCUGCUC GAG-3′ | ||||
| 3′-A AAG A AGACGAG CUC-5′ | ||||
| 59 AA A A CU 39 | ||||
| PSTVd-M | CUCUCGAGGUCUAAAGAAAAAb | 3768 GG A GUCUG AG 3788 | −7.50 | PSTVd-Mmut |
| 5′-U UUC U CUC GAG-3′ | ||||
| 3′-A AAG A GAG CUC-5′ | ||||
| 59 AA A AUCUG CU 39 | ||||
| PSTVd-M | CUCCUGAGCAGAAAAGAAAAAd | 3768 GG A G 3788 | −24.60 | PSTVd-M→I |
| 5′-U UUC U UCUGCUCAGGAG-3′ | ||||
| 3′-A AAG A AGACGAGUCCUC-5′ | ||||
| 59 AA A G 39 |
As PSTVd-I induces more severe symptoms than PSTVd-M, it was expected that the mutants having a PSTVd-I backbone would cause distinguishable phenotypic changes compared with those possessing PSTVd-M backbone, for several reasons. First, as PSTVd-Imut is not capable of producing a vd-sRNA effectively targeting CalS11-like, there should be a decrease in symptom severity due to the defective vd-sRNA. Second, although PSTVd-I→M is capable of producing a vd-sRNA predicted to target CalS11-like, this vd-sRNA is not as efficient as that produced by PSTVd-I due to the presence of mismatches located in 5′ seed region. Thus, it was expected that this chimeric mutant would cause intermediate symptoms compared with those observed in PSTVd-I- and PSTVd-Imut-inoculated plants. At 14 dpi, PSTVd-I and its mutant derivatives induced distinguishable symptoms in inoculated tomato plants (Figure 6C). As predicted, the PSTVd-I→M-inoculated plants exhibited very slight stunting, whereas plants inoculated with PSTVd-Imut were essentially asymptomatic. Surprisingly, at 21 dpi, PSTVd-Imut-inoculated plants exhibited disease symptoms with leaf curling and stunting, but little change was observed in PSTVd-I→M-infected plants (Supplemental Figure 8). Initially, it was predicted that PSTVd-M→I infection should result in an increased amount of disease symptoms compared with those caused by the wild type if, indeed, the predicted vd-sRNA is involved in the suppression of CalS, because the chimeric PSTVd-M→I can produce a more efficient vd-sRNA than its wild type (PSTVd-M). Surprisingly, however, the wild-type viroid induced slightly stronger symptoms than did PSTVd-M→I at 14 dpi (Supplemental Figure 9).
Previously, mutagenic studies conducted on the VMR resulted in the reversion of PSTVd mutants to the wild type (Zhong et al., 2008). Consequently, we investigated whether the sudden increase in symptom expression in PSTVd-Imut-inoculated plants might be due to reversion mutations; to this end, we extracted RNA from 14- and 21-dpi samples followed by RT-PCR amplification, cloning, and sequencing of at least 20 clones from two individual plants for each mutant. The plants inoculated with PSTVd-I→M showed sequences 100% identical to that of the original PSTVd-I→M. However, 19 out of the 20 clones obtained from PSTVd-Imut-inoculated plants had reverted to the wild type, PSTVd-I. Surprisingly, detailed analysis of VMR revealed that almost all the clones obtained at 21 dpi were identical in sequence to that of PSTVd-M (i.e., at nucleotide positions 42 and 43). Furthermore, full-length sequence alignment of these clones revealed that all of the clones were longer than the original PSTVd-I by one nucleotide, due to the insertion of a nucleotide at position 63/64. We performed RT-qPCR on RNA samples obtained at 14 dpi to evaluate the viroid RNA levels (Figure 6D). The PSTVd RNA level was almost negligible in PSTVd-Imut inoculated tomato plants, while plants inoculated with PSTVd-I→M showed approximately half as much viroid RNA as did PSTVd-I-inoculated plants. Taken together, the mutation of the nucleotides of the vd-sRNA/target cleavage site attenuated the symptoms and decreased the viroid titer, whereas the changing of the 5′ seed region of PSTVd-I resulted in mild symptoms, indicating the importance of this vd-sRNA in determining viroid titer and disease severity.
Polymorphism in the vd-sRNA Binding Region of the CalS11-like mRNA Affects Viroid Accumulation in Different Tomato Cultivars
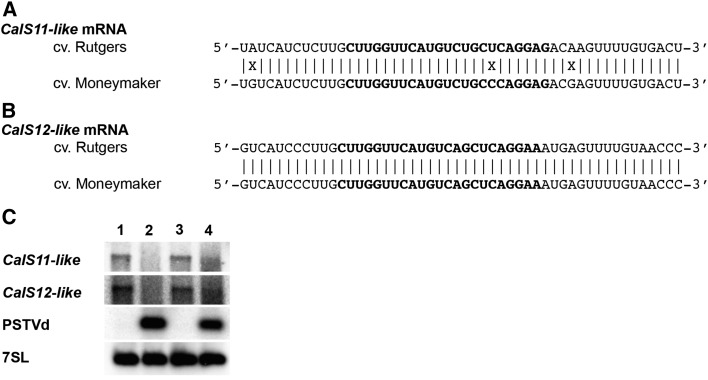
Infection of tomatoes of the cultivar ‘Moneymaker’ (tomato cv Moneymaker) results in only very mild symptoms upon PSTVd infection (Owens et al., 2012). To identify any possible polymorphisms located in the vd-sRNA binding site of the CalS genes of symptomatic versus asymptomatic cultivars, the relevant portions of the CalS genes from both cv Rutgers and cv Moneymaker were amplified and sequenced. BLAST analysis of the CalS11-like mRNA of the tomato cv Moneymaker revealed 99% sequence identity with the Nicotiana sylvestris CalS11-like mRNA (N. sylvestris; GenBank accession number XM_009775011) and 95% sequence identity with the tomato CalS11-like mRNA (GenBank accession number XM_010318449). Similarly, BLAST analysis of the CalS12-like mRNA of tomato cv Moneymaker revealed 99% sequence identity with the N. sylvestris CalS12-like mRNA (N. sylvestris; GenBank accession number XM_009784675) and 94% sequence identity with the tomato CalS12-like mRNA (GenBank accession number XM_010325437). Alignment of the Rutgers CalS11-like sequence with that of Moneymaker revealed a sequence polymorphism in the vd-sRNA binding site (Figure 7A), while the CalS12-like mRNAs from both cultivars were 100% identical within the predicted vd-sRNA binding site (Figure 7B). The change in the CalS11-like sequence from Moneymaker resulted in a mismatch in the seed region of the vd-sRNA/CalS11-like mRNA duplex, thus increasing the ∆G and decreasing the percentage of base pairing compared Rutgers (Table 5). In order to verify the effect of this observed mismatch on the PSTVd accumulation level both Rutgers and Moneymaker plants were inoculated with PSTVd-I. Total RNA was extracted at 14 dpi and analyzed by RNA gel blot. As shown in Figure 7D, PSTVd-I accumulated more in Rutgers than in Moneymaker. Although PSTVd-I infection repressed the CalS genes in the cultivars to somewhat different degrees, this difference was not very significant. Taken together, these data suggest a direct link between the sequence of the vd-sRNA binding site in the CalS11-like mRNA and the accumulation of PSTVd in different tomato cultivars.
Figure 7.
Polymorphisms in the PSTVd-sRNA Binding Site in the CalS11-like mRNA Correlate with Viroid Accumulation.
(A) and (B) Sequence alignment of the tomato cv Rutgers and Moneymaker CalS11-like (A) and CalS12-like (B) mRNAs in the region predicted to be targeted by the PSTVd sRNA. The bold letters indicate the predicted PSTVd-sRNA binding region.
(C) RNA extracted from tomato plants at 14 dpi was used monitor both the accumulation of PSTVd and the repression of both the CalS11-like and the CalS12-like mRNAs by RNA gel blot assay. Lane 1, mock-inoculated cv Rutgers; lane 2, PSTVd-I-inoculated cv Rutgers; lane 3, mock-inoculated cv Moneymaker; lane 4, PSTVd-I-inoculated cv Moneymaker. The 7SL mRNA was used as the internal control.
Table 5. Predicted Duplexes Formed between vd-sRNA Derived from the VMRs of PSTVd Variants and the CalS11-like mRNA from Tomato cv Moneymaker.
| PSTVd Variant | CalS11-like mRNA/vd-sRNA | ∆Ga | % Pairing |
|---|---|---|---|
| PSTVd-I | 170 GG A G C 190 | −19.58 | 76.2 |
| 5′-U UUCU UCUGC CAGGAG-3′ | |||
| 3′-A AAGA AGACG GUCCUC-5′ | |||
| 59 AA A A A 39 | |||
| PSTVd-I | 168 GG A G C 189 | −18.98 | 77.3 |
| 5′-CUU UUC U UCUGC CAGGA-3′ | |||
| 3′-GAA AAG A AGACG GUCCU-5′ | |||
| 61 AA A A A 40 | |||
| PSTVd-M | 170 GG AG C A 189 | −13.18 | 71.4 |
| 5′-U UUC U UCUGC C GGAG-3′ | |||
| 3′-A AAG A AGACG G UCUC-5′ | |||
| 59 AA A A A C 39 | |||
| PSTVd-M | 168 GG A G C A 189 | −12.58 | 72.7 |
| 5′-CUU UUC U UCUGC C GGA-3′ | |||
| 3′-GAA AAG A AGACG G UCU-5′ | |||
| 61 AA A A A C 40 |
The PairFold online tool was used to predict the minimum free energy secondary structure of a pair of RNA sequences.
The underlined nucleotides represent G:U wobble base pairs.
DISCUSSION
In order to cause systemic infections and to be transmitted through seeds, viruses and viroids move through the PD and enter seeds. Callose deposition at the PD is a critical factor in restricting the cell-to-cell viral movement of Soybean mosaic virus (Li et al., 2012). Previous studies of the thermodynamic prediction of the PSTVd structure underscored the importance of some of its RNA motifs such as loops 7 and 8 of the VMR that are critical to both the viroid’s replication and its systemic trafficking (Zhong et al., 2008). Comparison of the structures obtained by SHAPE for two PSTVd variants inducing different symptoms in tomato plants revealed significant changes in these two loops (Figure 1B). Visual analysis of these structures showed that the sequence changes in the VMRs alter the structures of these loops, thus indicating a possible involvement of vd-sRNAs produced from this region in viroid trafficking. In order to understand the importance of the sequence of the VMR, the VMRs of two PSTVd variants was divided into 21- to 24-nucleotide-long sRNAs and these were used to search for possible RNA/RNA duplex formation with the tomato transcriptome data set. This search revealed the possibility of vd-sRNA/mRNA duplex formation with the CalS11-like mRNA. Interestingly, CalS11-like and CalS12-like are functionally redundant. They are required for the formation of the callose wall separating the tetraspores. In addition, during plant growth and development, callose is a major component of both the pollen mother cell wall and the pollen tubes. It is also found in the PD canal (Enns et al., 2005). Hence, the possibility of the downregulation of CalS11-like mRNA by PSTVd vd-sRNA, which could in turn enable viroid movement through PD, was hypothesized. Previously, the disruption of a loop in this VMR sequence by mutating two nucleotides (A50G and A51U) greatly impaired viroid trafficking, and the mutants reverted to the wild type in the next generation (Zhong et al., 2008). These mutations are predicted to disrupt the cleavage site of the vd-sRNA/CalS11-like mRNA duplex.
Time-course experiments conducted to evaluate the effect of both PSTVd variants on their titers revealed that PSTVd-M accumulates at a slower pace than does PSTVd-I at all points up to 21 dpi. Additionally, PSTVd-I downregulates CalS11-like to a higher degree than does PSTVd-M. This may be due to the greater accumulation of PSTVd-I, which in turn produces more vd-sRNA, or to the less efficient systemic movement of PSTVd-M, which eventually results in lower accumulation of vd-sRNAs. This hypothesis was validated by comparing the data obtained from the large-scale sequencing of vd-sRNAs obtained from both PSTVd-I- and PSTVd-M-inoculated plants. The results are in agreement with data obtained from experiments in which tomato plants were inoculated with similar PSTVd variants (Tsushima et al., 2015). Validation of duplex formation between the predicted vd-sRNA/CalS11-like mRNA sequence by an amiRNA experiment and the subsequent quantification of GFP revealed that the vd-sRNA derived from PSTVd-I is more effective in the downregulation of its target than is the PSTVd-M sRNA.
The efficient RISC-mediated cleavage of a target by miRNA in both plants and animals requires an absence of mismatches in the 5′ seed regions as well as in the miRNA cleavage site. That said, some mismatches are allowed at the 3′ compensatory end of the miRNA (Schwab et al., 2006). Surprisingly, in the case of the PSTVd-M sRNA that targets CalS11-like, the RNA/RNA duplex contains a total of six mismatches, out of 21 nucleotides, including two mismatches in the 5′ seed region. Even so, it is still capable of targeting an mRNA, albeit with a lower efficiency (Table 1, Figure 2B). This fact is further supported by recent studies where an amiRNA targeting the soluble inorganic pyrophosphatase gene was able to cleave its target by RNAi even though the miRNA/target duplex possessed six mismatches, including one located in the seed region (Eamens et al., 2014). Often, the predicted RNA/RNA duplex formed between the miRNA and the target results in translational repression, but does not lead to the cleavage of the mRNA (Huntzinger and Izaurralde, 2011). In this scenario, the repression of GFP observed in the amiRNA experiments might be due in part to such a phenomenon. The absence of successful nested PCR reactions using 5′ RLM-RACE on RNA preparations from mock-inoculated plants helped to confirm the cleavage of the CalS11-like mRNA that is associated with PSTVd infection. Sequence analysis of the cDNA clones of CalS11-like transcripts obtained from PSTVd-I- and PSTVd-M-inoculated plants confirm the RISC-mediated cleavage sites in the CalS11-like mRNA. Taken together, these two in vivo experiments prove the long-standing hypothesis that viroids cause disease symptoms by RNA silencing and that 100% sequence complementarity in the seed region is not required for vd-sRNAs to target host genes via the RNA silencing machinery.
Sequence alignment between the CalS11-like and CalS12-like mRNAs showed a high degree of similarity at the vd-sRNA binding site, suggesting the possibility of the RNA silencing of CalS12-like by this vd-sRNA. Quantitative RT-PCR performed to monitor the expression levels of CalS12-like mRNA revealed that its downregulation was not very strong, compared with that of the CalS11-like mRNA. In 5′ RLM-RACE experiments, neither mock-inoculated nor PSTVd-M-inoculated plants showed any amplification of CalS12-like cleavage products. However, both amiRNAs based on PSTVd-I sRNAs resulted in a lower accumulation of GFP produced from a construct with the CalS12-like binding site (Figure 4). These data imply that the predicted vd-sRNA/target is less efficient in inducing the RISC-mediated cleavage of the CalS12-like mRNA, but that it still represses protein production, at least in case of PSTVd-I. To the best of our knowledge, this is the first report on the identification of a single vd-sRNA silencing of more than one gene, thus illustrating the versatile nature of vd-sRNAs.
We performed two types of mutagenic experiments to study the importance of the CalS targeting sRNA on the systemic accumulation of PSTVd: one disrupting the RISC mediated cleavage site and the other disrupting the 5′ seed region. In the first case, plants inoculated with the mutants should be devoid of viroid or should show a decreased viroid titer compared with plants inoculated with the wild type, as the mutants are unable to move through the PD because the RISC-mediated cleavage site is disrupted. This hypothesis appeared to be validated up to 14 dpi, as mutant (PSTVd-Imut) inoculated plants phenotypically resembled mock-inoculated plants. However, at 21 dpi, plants exhibited disease symptoms. Sequence analysis of viroids recovered at 14 and 21 dpi revealed that all of the mutants had reverted to the wild type during the course of the infection. These findings are in agreement with previous mutagenic studies, in which A50G/A51U mutants reverted to the wild type (Zhong et al., 2008). Interestingly, almost all of the sequences obtained at 21 dpi possessed changes C42U and U43C and the insertion of a U at position 63/64 in a manner, which is very much similar to PSTVd-M. Although PSTVd-M infects and induces mild symptoms in tomato plants, this variant was first isolated from dahlia (Dahlia × hybrid) and is thought to help the viroid to adapt under unfavorable conditions (Tsushima et al., 2011).
PSTVd-I and PSTVd-M differ by two nucleotides in the vd-sRNA targeting the CalS mRNA, out of a total of nine nucleotide mismatches. In order to check the effect of these mismatches on PSTVd accumulation, the region was swapped between the two PSTVd variants. In the case of PSTVd-I→M, it is logical to expect decreased symptom severity as the swap decreases the efficiency of the CalS targeting vd-sRNA due to the fact that the sRNA derived from PSTVd-M contains mismatches in the 5′ seed region. In contrast, PSTVd-M→I should result in an increased accumulation if PSTVd-M replicates and accumulates as well as PSTVd-I does, as the swap leads to the production of more efficient vd-sRNA targeting the CalS mRNA. As expected, at 14 dpi, the plants inoculated with PSTVd-I→M exhibited mild disease symptoms compared with those inoculated with the wild-type viroid (PSTVd-I; Figure 4B). However, in the case of PSTVd-M→I, few phenotypic changes were observed compared with when the wild-type viroid was used (PSTVd-M). This can be attributed to the fact that PSTVd-M accumulates more slowly and hence produces fewer sRNAs, as described previously (Tsushima et al., 2015). In contrast, although PSTVd-I→M accumulates faster, the vd-sRNA produced is less effective than that of PSTVd-I. This affects its overall accumulation and eventually results in a decrease in disease symptoms. The importance of the vd-sRNA binding sequence in the induction of disease symptoms is further supported by the results obtained for both the PSTVd-I accumulation and the CalS11-like mRNA repression observed in tomato cv Moneymaker.
Potato (Solanum tuberosum) plants are the natural host of PSTVd, but for convenience, tomato plants are widely used for experimental purposes. As both of these species belong to the same genus, we expect a high degree of sequence similarity between their CalS genes. Indeed, sequence alignment of the CalS11-like and CalS12-like mRNAs from tomato cv Rutgers and potato (CalS11-like, GenBank accession number XM_006346978; CalS12-like, GenBank accession number XM_006357419) revealed 100% identity in the vd-sRNA binding site (Supplemental Figure 10), indicating that the vd-sRNA described in this study could play a crucial role in the downregulation of the CalS genes in both species, thus ensuring PSTVd trafficking upon infection.
The data presented here clearly show that, although vd-sRNAs are not very efficient in cleaving their target mRNAs, they are capable of inducing RNA silencing to establish themselves. If there is any alteration in the sequence that is vital to the viroid’s life cycle, it is capable of reverting to the wild type to establish a systemic infection. Furthermore, validation of the vd-sRNA targeting the CalS11-like and CalS12-like mRNA, by amiRNA experiments, showed that a single vd-sRNA is capable of silencing more than one target. The data presented here should open new avenues in development of transgenic viroid-resistant plants, based on non-host resistance such as the engineering of plants harboring a CalS gene from viroid resistant plant.
METHODS
PSTVd cDNA Construction, Transcription, and SHAPE
The dimeric construct of PSTVd-Dahlia (GenBank accession number AB623143: the term PSTVd-M was used as this variant induces mild symptoms in tomato plants), which was previously used to study structure in solution, was used to synthesize infectious dimeric transcripts (Giguère et al., 2014). In the case of the PSTVd-I variant, the head-to-tail dimers were constructed using monomeric PSTVd-I (GenBank accession number AY937179) obtained from gBlock Gene Fragments (Integrated DNA Technologies) in the pBluescript KS+ vector (Stratagene) as described previously (Giguère et al., 2014). Monomeric mutants of PSTVd were generated by PCR amplification using PSTVd-81F primers combined with appropriate reverse primer as listed in Supplemental Table 8. All introduced mutations were verified by DNA sequencing. These monomers were then dimerized to prepare infectious clones as described (Giguère et al., 2014).
Recombinant plasmids were linearized with the appropriate restriction enzymes and transcribed with either T7 or T3 RNA polymerase. The resulting products were analyzed by electrophoresis in 1.5% agarose gels containing 1× TAE buffer (40 mM Tris, 20 mM acetic acid, pH 8.0, and 1 mM EDTA). The transcripts obtained from both PSTVd-M and PSTVd-I were subjected to the SHAPE reaction. The QuSHAPE software using the default parameters was used to analyze the obtained electropherograms. The normalized reactivity values were then loaded into the RNAstructure 5.5 software Fold tool as described previously (Giguère et al., 2014). The structures with the lowest Gibbs free energies are presented here.
Plant Materials and Growing Conditions
Tomato seeds (Solanum lycopersicum cv Rutgers; Livingston Seed Co.; S. lycopersicum cv Moneymaker; Nojaus Sèklos, Lithuania) were used for the bioassays. All plants were grown in a growth chamber at a temperature of 25°C with 16 h of light and 8 h of darkness. After the development of primary leaves, carborundum was used as an abrasive in order to inoculate plants with 1 µg of the RNA transcripts of the respective viroid variants. Leaf samples were collected at 7, 14, and 21 dpi for RT-qPCR assays. For the amiRNA experiments, Nicotiana benthamiana plants were grown at 23°C with 16 h of light and 8 h of darkness.
RNA Extraction and RT-qPCR
Total RNA from infected leaf samples was extracted using the mirVana miRNA isolation kit (Ambion), with modifications. Briefly, 100 mg of leaf sample was ground with 400 μL of lysis/binding buffer. Then, 60 μL of microRNA homogenate was added and the resulting solution was mixed by vortexing and then placed on ice for 10 min. Acid phenol-chloroform (5:1) extraction was then performed and was followed by reextraction of the supernatant with an equal volume of chloroform. The RNA was precipitated by adding 2.5 volumes of absolute alcohol. The RNA was further purified by DNase I (Promega) treatment followed by chloroform extraction. RNA integrity was examined in a 2100 Bioanalyzer (Agilent Technologies).
cDNA was prepared by reverse transcribing 1 µg of the total RNA using SuperScript III reverse transcriptase in the presence of random primer according to the manufacturer’s instructions (Invitrogen). For the evaluation of both the PSTVd titer and of the gene expression levels, 10 ng of cDNA was used for qPCR with the appropriate primer combinations (Supplemental Table 2). RT-qPCR data were obtained for the three housekeeping genes; specifically, the Ubiquitin-conjugating enzyme (UBC), Transducing/WD40 repeat family protein, ARF-like GTPase family protein (ASAR1) were used for normalization (Dekkers et al., 2012). Each experiment was performed at least three times with true biological replicates. Every quantitative PCR run included no-template primer-pair control samples. Relative expression levels were calculated using the qBASE framework as described previously (Hellemans et al., 2007). All of the RT-qPCR analyses were performed commercially at the Laboratoire de Génomique Fonctionnelle de l’Université de Sherbrooke (http://palace.lgfus.ca).
Bioinformatic Analysis, Construction of PSTVd-Specific amiRNA, and Immunoblot Analysis
The pathogenicity domain of PSTVd-I was dissected into 21-, 22-, 23-, and 24-nucleotide-long sRNA, which were then used to interrogate publicly available tomato transcriptome databases using the WMD3 Web-based tool. The free energy (∆G) for each predicted vd-sRNA/target duplex was calculated using the PairFold online tool as described previously (Andronescu et al., 2003). In order to validate the vd-sRNA/target duplex formation, amiRNAs for the different vd-sRNA sequences were synthesized on an osa-MIR528 backbone using the appropriate oligonucleotide pairs in the PCR reaction. The details on the primers used for the PCR amplification of the different constructs are presented in Supplemental Tables 3 and 4. The amiRNA/GFP-tagged target complex is shown in Supplemental Figure 11. Additionally, the positive control construct targeting the UPF1 gene of N. benthamiana was constructed as described previously (Yan et al., 2012). PCR products were digested with the restriction endonucleases XbaI and BamHI and then ligated into the same sites of the binary vector pBIN61. Target construct was prepared by ligating either 21- or 22-nucleotide predicted target sequences to the 3′ UTR of the GFP gene under the control of the 35S promoter in the pBIN61 vector. The resulting binary vectors were then transformed into an Agrobacterium tumefaciens strain C58C1 carrying the virulence plasmid pCH32 and were used for agroinfiltration as previously described (Moffett, 2011).
For GFP expression analysis, two leaf discs were ground in liquid nitrogen and resuspended in 60 μL of 1.5× of Laemmli sample buffer (10% SDS-glycerol, 1 M Tris-HCl, pH 6.8, and 0.02% bromophenol blue) and then heated at 95°C for 5 min. The proteins were separated by SDS-PAGE on 10.5% gels and then were transferred to polyvinylidene difluoride membranes (Bio-Rad). Anti-GFP-HRP antibodies were used to detect GFP according to the manufacturer’s instructions (Santa Cruz Biotechnology). Anti-PEPC rabbit antibodies (Rockland Immunochemicals) were used to detect PEPC at a 1:10,000 dilution, followed by a subsequent incubation with 1:10,000 donkey anti-IgG rabbit-HRP polyclonal antibodies (BioLegend). The proteins were subsequently visualized using ECL substrate (Bio-Rad) per the manufacturer’s instructions. The amount of GFP produced was quantified using ImageJ software (http://imagej.nih.gov/ij). The data obtained for the housekeeping gene (PEPC) were used for the normalization.
5′ RLM-RACE
An RNA adaptor described previously was used to ligate RNA with minor modifications (Navarro et al., 2012). Briefly, 10 µg of total RNA was mixed with an RNA adaptor and incubated for 6 h at 37°C in the presence of T4 RNA ligase I (New England Biolabs). The resulting product was reverse transcribed using gene-specific outer primers (CalS11 RACE-R and CalS12 RACE-R), which are located 252 and 317 nucleotides downstream of the predicted cleavage site of the CalS11-like and CalS12-like genes, respectively. The products were subjected to nested PCR using specific primers (Supplemental Tables 5 and 7). The nested PCR products were separated by 2.0% agarose gel electrophoresis, and the band corresponding to the expected amplicon was eluted using the QAIquick gel extraction kit (Qiagen). The purified products were cloned into the pGEM-T easy vector (Promega). The resulting clones were commercially sequenced (http://www.sequences.crchul.ulaval.ca) and the sequences analyzed using CLC Free Workbench version 4.6 software (http://www.clcbio.com/index.php?id=28).
High-Throughput Sequencing of sRNAs and Data Analysis
For high-throughput sequencing of sRNAs, sRNAs of 15 to 50 nucleotides in length were purified after denaturing total RNA extracted from plants at 21 dpi and subjected to small RNA library preparation commercially at Hokkaido System Science Co. using a Genetic Analyzer IIx (Illumina). The samples were quantified using an Agilent 2100 Bioanalyzer (Agilent Technologies) and were processed simultaneously in the Illumina system using an index sequence. Adapter sequences were removed from the ends of the resulting raw data, and sequences of 21 to 24 nucleotides were grouped (Adkar-Purushothama et al., 2015). These short reads were then mapped to either the genomic or antigenomic strand of both PSTVd-M and PSTVd-I using the standard pattern matching algorithm.
Amplification of Viroid cDNA and Host Genes
Total RNA extracted from tomato plants cv Rutgers and cv Moneymaker were subjected to cDNA synthesis using SuperScript III reverse transcriptase (Invitrogen) in the presence of either oligo(dT) or Vid-RE as primer (Verhoeven et al., 2004). In order to amplify the CalS genes, CalS11-like and CalS12-like gene-specific primers were used (Supplemental Table 9), while the primer pair Vid-RE and Vid-FW was used to amplify PSTVd (Verhoeven et al., 2004). All PCRs were performed using purified Pfu polymerase, and the resulting products were analyzed by 1.0% agarose gel electrophoresis. Amplicons of the expected size were cloned and sequenced as before. The resulting sequences were assembled and aligned using CLC Free Workbench version 4.6 software (http://www.clcbio.com/index.php?id=28) and were manually checked for variation in the sequence.
Probe Preparation and RNA Gel Blot Assay
In order to detect CalS11-like and CalS12-like mRNAs, the riboprobes were prepared using the previously amplified genes from tomato cv Rutgers, which was cloned in the pBluescript KS+ vector. The expression level of 7S signal recognition particle RNA (7SL) was used as the loading control for RNA gel blot assay. Hence, the portion of the 7SL RNA of tomato cv Rutgers was amplified using previously described primer pair and was then cloned into the pBluescript KS+ vector (Matousek et al., 2008). To prepare PSTVd riboprobes, the pBluescript KS+ vector with the dimeric PSTVd-I construct was used as the template. In order to prepare 7SL riboprobes, the T7 MAXIscript kit (Ambion) was used after linearizing the plasmid with KpnI restriction endonuclease as explained above.
For the RNA gel blot hybridizations, 2.0 µg of the total RNA samples extracted from 14-dpi plants was denatured at 65°C for 15 min using 3 volumes of sample buffer (50% formamide, 2.2 M formaldehyde [37%], and 1× MOPS) and was then separated on 0.8% agarose-formaldehyde gels containing 1× MOPS buffer. The RNAs were then transferred to a Hybond-XL nylon membrane (Amersham, GE Healthcare Life Sciences) and hybridized with radiolabeled probes to detect the CalS11-like mRNA, the CalS12-like mRNA, PSTVd, and the 7SL RNA. All the probes were hybridized overnight at 60°C and then the membranes were washed twice for 20 min at 62°C with washing buffer (1× SSC and 0.1× SDS). The RNA gel blot hybridization signals were visualized using a Typhoon imaging system (GE Healthcare Life Sciences) after exposing to the phosphor imager screen.
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers KR706381 (partial sequence of CalS11-like mRNA of tomato cv Rutgers), KR706382 (partial sequence of CalS12-like mRNA of tomato cv Rutgers), KR706379 (partial sequence of CalS11-like mRNA of tomato cv Moneymaker), and KR706380 (partial sequence of CalS12-like mRNA of tomato cv Moneymaker). The deep sequence data used in this study is deposited in the Gene Expression Omnibus under accession number GSE69225.
Supplemental Data
Supplemental Figure 1. Thermodynamically predicted secondary structure of PSTVd variants.
Supplemental Figure 2. RNA gel blot analysis of CalS11-like mRNA in PSTVd-infected tomato plants.
Supplemental Figure 3. Demonstration of feasibility of amiRNA technique.
Supplemental Figure 4. Evaluation of effect of vd-sRNA:amiRNAs on GFP.
Supplemental Figure 5. 5′ RLM-RACE products obtained with primers targeting the CalS11-like mRNA.
Supplemental Figure 6. 5′ RLM-RACE products obtained with primers targeting the CalS12-like mRNA
Supplemental Figure 7. Alignment of the CalS12-like mRNA with the portion of PSTVd-I located in the vicinity of the predicted mRNA/vd-sRNA duplexes.
Supplemental Figure 8. Effect of PSTVd-I mutants on tomato plants at 21 dpi.
Supplemental Figure 9. Effect of PSTVd-M mutants on tomato plants at 14 and 21 dpi.
Supplemental Figure 10. Alignment of the PSTVd-sRNA binding sites of the tomato CalS genes against the potato CalS genes.
Supplemental Figure 11. Duplexes predicted to be formed by complexes of amiRNAs and GFP-expressing reporter constructs.
Supplemental Table 1. Predicted duplex formation between tomato CalS11-like mRNA targets and all possible vd-sRNA derived from the VMR of PSTVd variants.
Supplemental Table 2. Details of the primers used in the RT-qPCR reactions for the evaluation of both the PSTVd titer and the callose synthase gene expression.
Supplemental Table 3. Details of the primers used for constructing amiRNAs on the osa-MIR528 backbone.
Supplemental Table 4. Oligonucleotides of the predicted target sequence of the CalS11-like mRNA used as inserts in to the binary vector pBIN61-GFP.
Supplemental Table 5. Details of the primers used for the 5′ RLM-RACE experiment used to prove RISC-mediated cleavage site of the CalS11-like mRNA.
Supplemental Table 6. Predicted duplexes formed between the tomato CalS12-like mRNA targets and all possible vd-sRNAs derived from the VMR of PSTVd variants.
Supplemental Table 7. Details of the primers used for the 5′ RLM-RACE experiment used to prove the RISC-mediated cleavage site of the CalS12-like mRNA.
Supplemental Table 8. Details of the oligonucleotides used to generate, by PCR, both PSTVd mutants and chimeric constructs.
Supplemental Table 9. Details of the oligonucleotides used to amplify, by PCR, the CalS11-like and CalS12-like mRNA of tomato.
Supplementary Material
Acknowledgments
This work was supported by grants from the Natural Sciences and Engineering Research Council of Canada (Grants 155219-12 and 386223-2010) to J.-P.P. and P.M. T.S. received a grant from the Japan Society for the Promotion of Sciences KAKENHI (Grant 24380026). The RNA group is supported by grants from the Université de Sherbrooke. J.-P.P. holds the Research Chair of Université de Sherbrooke in RNA Structure and Genomics and is a member of the Centre de Recherche du CHUS. C.R.A.-P. received a postdoctoral scholarship from the Fonds Recherche Québec-Nature et Technologies. T.G. has a scholarship from the Centre de Recherche Médicale de l’Université de Sherbrooke. The funders had no role in study design, data collection and analysis, decision to publish, or in the preparation of the article.
AUTHOR CONTRIBUTIONS
C.R.A.-P., P.M., and J.-P.P. conceived and designed the experiments. C.R.A.-P., C.B., T.G., and T.S. performed the experiments. C.R.A.-P., C.B., and T.G. analyzed the data. T.S., P.M., and J.-P.P. contributed reagents/materials/analysis tools. C.R.A.-.P, C.B., T.G., T.S., P.M., and J.-P.P. contributed to the writing the article.
Glossary
- PSTVd
Potato spindle tuber viroid
- RNAi
RNA interference
- vd-sRNA
viroid-derived small RNA
- PD
plasmodesmata
- SHAPE
selective 2'-hydroxyl acylation analyzed by primer extension
- sRNA
small RNA
- dpi
days postinoculation
- RISC
RNA-induced silencing complex
- amiRNA
artificial microRNA
- UTR
untranslated region
- PEPC
phosphoenolpyruvate carboxylase
- 5′ RLM-RACE
5′ RNA ligase-mediated rapid amplification of cDNA ends
Footnotes
Articles can be viewed online without a subscription.
References
- Adkar-Purushothama C.R., Zhang Z., Li S., Sano T. (2015). Analysis and application of viroid-specific small RNAs generated by viroid-inducing RNA silencing. Methods Mol. Biol. 1236: 135–170. [DOI] [PubMed] [Google Scholar]
- Andronescu M., Aguirre-Hernández R., Condon A., Hoos H.H. (2003). RNAsoft: A suite of RNA secondary structure prediction and design software tools. Nucleic Acids Res. 31: 3416–3422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benitez-Alfonso Y., Faulkner C., Ritzenthaler C., Maule A.J. (2010). Plasmodesmata: gateways to local and systemic virus infection. Mol. Plant Microbe Interact. 23: 1403–1412. [DOI] [PubMed] [Google Scholar]
- Bolduc F., Hoareau C., St-Pierre P., Perreault J.-P. (2010). In-depth sequencing of the siRNAs associated with peach latent mosaic viroid infection. BMC Mol. Biol. 11: 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boonham N., Pérez L.G., Mendez M.S., Peralta E.L., Blockley A., Walsh K., Barker I., Mumford R.A. (2004). Development of a real-time RT-PCR assay for the detection of potato spindle tuber viroid. J. Virol. Methods 116: 139–146. [DOI] [PubMed] [Google Scholar]
- Chen X.Y., Kim J.Y. (2009). Callose synthesis in higher plants. Plant Signal. Behav. 4: 489–492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dekkers B.J., Willems L., Bassel G.W., van Bolderen-Veldkamp R.P., Ligterink W., Hilhorst H.W., Bentsink L. (2012). Identification of reference genes for RT-qPCR expression analysis in Arabidopsis and tomato seeds. Plant Cell Physiol. 53: 28–37. [DOI] [PubMed] [Google Scholar]
- Ding B. (2009). The biology of viroid-host interactions. Annu. Rev. Phytopathol. 47: 105–131. [DOI] [PubMed] [Google Scholar]
- Ding B., Kwon M.O., Hammond R., Owens R. (1997). Cell-to-cell movement of potato spindle tuber viroid. Plant J. 12: 931–936. [DOI] [PubMed] [Google Scholar]
- Ding S.W. (2010). RNA-based antiviral immunity. Nat. Rev. Immunol. 10: 632–644. [DOI] [PubMed] [Google Scholar]
- Di Serio F., Flores R., Verhoeven J.Th.J., Li S.-F., Pallás V., Randles J.W., Sano T., Vidalakis G., Owens R.A. (2014). Current status of viroid taxonomy. Arch. Virol. 159: 3467–3478. [DOI] [PubMed] [Google Scholar]
- Eamens, A.L., Smith, N.A., Dennis, E.S., Wassenegger, M., and Wang, M. (2014). In Nicotiana species, an artificial microRNA corresponding to the virulence modulating region of Potato spindle tuber viroid directs RNA silencing of a soluble inorganic pyrophosphatase gene and the development of abnormal phenotypes. Virology 450-451: 266–277. [DOI] [PubMed] [Google Scholar]
- Enns L.C., Kanaoka M.M., Torii K.U., Comai L., Okada K., Cleland R.E. (2005). Two callose synthases, GSL1 and GSL5, play an essential and redundant role in plant and pollen development and in fertility. Plant Mol. Biol. 58: 333–349. [DOI] [PubMed] [Google Scholar]
- Giguère T., Adkar-Purushothama C.R., Perreault J.-P. (2014). Comprehensive secondary structure elucidation of four genera of the family Pospiviroidae. PLoS One 9: e98655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamilton A., Voinnet O., Chappell L., Baulcombe D. (2002). Two classes of short interfering RNA in RNA silencing. EMBO J. 21: 4671–4679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hellemans J., Mortier G., De Paepe A., Speleman F., Vandesompele J. (2007). qBase relative quantification framework and software for management and automated analysis of real-time quantitative PCR data. Genome Biol. 8: R19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huntzinger E., Izaurralde E. (2011). Gene silencing by microRNAs: contributions of translational repression and mRNA decay. Nat. Rev. Genet. 12: 99–110. [DOI] [PubMed] [Google Scholar]
- Hutchins C.J., Keese P., Visvader J.E., Rathjen P.D., McInnes J.L., Symons R.H. (1985). Comparison of multimeric plus and minus forms of viroids and virusoids. Plant Mol. Biol. 4: 293–304. [DOI] [PubMed] [Google Scholar]
- Ivanova D., Milev I., Vachev T., Baev V., Yahubyan G., Minkov G., Gozmanova M. (2014). Small RNA analysis of Potato spindle tuber viroid infected Phelipanche ramosa. Plant Physiol. Biochem. 74: 276–282. [DOI] [PubMed] [Google Scholar]
- Keese P., Symons R.H. (1985). Domains in viroids: evidence of intermolecular RNA rearrangements and their contribution to viroid evolution. Proc. Natl. Acad. Sci. USA 82: 4582–4586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W., Zhao Y., Liu C., Yao G., Wu S., Hou C., Zhang M., Wang D. (2012). Callose deposition at plasmodesmata is a critical factor in restricting the cell-to-cell movement of Soybean mosaic virus. Plant Cell Rep. 31: 905–916. [DOI] [PubMed] [Google Scholar]
- Matousek J., Orctová L., Skopek J., Pesina K., Steger G. (2008). Elimination of hop latent viroid upon developmental activation of pollen nucleases. Biol. Chem. 389: 905–918. [DOI] [PubMed] [Google Scholar]
- Matsushita Y., Usugi R., Tsuda S. (2011). Distribution of tomato chlorotic dwarf viroid in floral organs of tomato. Eur. J. Plant Pathol. 130: 441–447. [Google Scholar]
- Minoia S., Navarro B., Delgado S., Serio F.D., Flores R. (2015). Viroid RNA turnover: characterization of the subgenomic RNAs of potato spindle tuber viroid accumulating in infected tissues provides insights into decay pathways operating in vivo. Nucleic Acids Res. 43: 2313–2325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moffett P. (2011). Fragment complementation and co-immunoprecipitation assays for understanding R protein structure and function. Methods Mol. Biol. 712: 9–20. [DOI] [PubMed] [Google Scholar]
- Navarro B., Gisel A., Rodio M.E., Delgado S., Flores R., Di Serio F. (2012). Small RNAs containing the pathogenic determinant of a chloroplast-replicating viroid guide the degradation of a host mRNA as predicted by RNA silencing. Plant J. 70: 991–1003. [DOI] [PubMed] [Google Scholar]
- Ossowski S., Schwab R., Weigel D. (2008). Gene silencing in plants using artificial microRNAs and other small RNAs. Plant J. 53: 674–690. [DOI] [PubMed] [Google Scholar]
- Owens R.A., Tech K.B., Shao J.Y., Sano T., Baker C.J. (2012). Global analysis of tomato gene expression during Potato spindle tuber viroid infection reveals a complex array of changes affecting hormone signaling. Mol. Plant Microbe Interact. 25: 582–598. [DOI] [PubMed] [Google Scholar]
- Pallás V., Martínez G., Gómez G. (2012). The interaction between plant viroid-induced symptoms and RNA silencing. Methods Mol. Biol. 894: 323–343. [DOI] [PubMed] [Google Scholar]
- Reuter J.S., Mathews D.H. (2010). RNAstructure: software for RNA secondary structure prediction and analysis. BMC Bioinformatics 11: 129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwab R., Ossowski S., Riester M., Warthmann N., Weigel D. (2006). Highly specific gene silencing by artificial microRNAs in Arabidopsis. Plant Cell 18: 1121–1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh R.P. (2006). Reassessment of the presence of viroid species of the genus Pospiviroid in infected floral parts, using reverse transcription-polymerase chain reaction and infectivity assays. Can. J. Plant Pathol. 28: 242–249. [Google Scholar]
- Tomato Genome Consortium (2012). The tomato genome sequence provides insights into fleshy fruit evolution. Nature 485: 635–641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomson D.W., Bracken C.P., Goodall G.J. (2011). Experimental strategies for microRNA target identification. Nucleic Acids Res. 39: 6845–6853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsushima D., Adkar-Purushothama C.R., Taneda A., Sano T. (2015). Changes in relative expression levels of viroid-specific small RNAs and microRNAs in tomato plants infected with severe and mild isolates of Potato spindle tuber viroid. J. Gen. Plant Pathol. 81: 49–62. [Google Scholar]
- Tsushima T., Murakami S., Ito H., He Y.-H., Adkar-Purushothama C.R., Sano T. (2011). Molecular characterization of Potato spindle tuber viroid in Dahlia. J. Gen. Plant Pathol. 4: 253–256. [Google Scholar]
- Verhoeven J.Th.J., Jansen C.C.C., Willemen T.M., Kox L.F.F., Owens R.A., Roenhorst J.W. (2004). Natural infections of tomato by Citrus exocortis viroid, Columnea latent viroid, Potato spindle tuber viroid and Tomato chlorotic dwarf viroid. Eur. J. Plant Pathol. 110: 823–831. [Google Scholar]
- Yan F., Lu Y., Wu G., Peng J., Zheng H., Lin L., Chen J. (2012). A simplified method for constructing artificial microRNAs based on the osa-MIR528 precursor. J. Biotechnol. 160: 146–150. [DOI] [PubMed] [Google Scholar]
- Zhong X., Archual A.J., Amin A.A., Ding B. (2008). A genomic map of viroid RNA motifs critical for replication and systemic trafficking. Plant Cell 20: 35–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.