Abstract
Transcript annotation in plant databases is incomplete and often inaccurate, leading to misinterpretation. As more and more RNA-seq data are generated, plant scientists need to be aware of potential pitfalls and understand the nature and impact of specific alternative splicing transcripts on protein production. A primary area of concern and the topic of this article is the (mis)annotation of open reading frames and premature termination codons. The basic message is that to adequately address expression and functions of transcript isoforms, it is necessary to be able to predict their fate in terms of whether protein isoforms are generated or specific transcripts are unproductive or degraded.
We are now in an era where alternative splicing (AS) in plants is widely recognized as an essential and important level of regulation of gene expression and of transcriptome and proteome diversity likely to contribute to plant adaptation and speciation (Syed et al., 2012; Carvalho et al., 2013; Reddy et al., 2013; Staiger and Brown, 2013). The number of plant AS articles published per annum has risen steadily over the last 15 years and has doubled in the last 5 years. Currently, the observed number of intron-containing genes showing AS in plants can be up to 70% (Chamala et al., 2015), including the model plant Arabidopsis thaliana with >61% of genes with AS (Marquez et al., 2012). RNA-seq is generating vast amounts of new information on transcript variants and AS events in a wide range of plant species, and newer technologies will help to define isoform variants by generating sequences of full-length transcripts. Exploitation of these data requires the accurate deciphering of AS transcripts ultimately to allow dynamic variation in transcript isoforms to be assessed during development and under different environmental conditions. The growing interest in AS and the potential pitfalls of using incorrect transcript annotation motivated us to write this short article.
Alternative splicing generates proteome diversity and affects protein abundance by regulating transcript levels via nonsense-mediated decay (NMD) (Schweingruber et al., 2013). A number of recent high-profile publications demonstrate the importance of AS and differential functions of AS variants in, for example, organ development (Zhang and Mount, 2009), flowering time control and the circadian clock (Sanchez et al., 2010; James et al., 2012; Posé et al., 2013; Li et al., 2015), light signaling (Shikata et al., 2014), dark-light retrograde signaling from chloroplast to nucleus (Petrillo et al., 2014), and zinc tolerance (Remy et al., 2014). AS of around 18% of Arabidopsis genes generates unproductive mRNA transcript isoforms that are degraded by NMD, which modulates transcript levels thereby regulating levels of protein produced from a gene (Kalyna et al., 2012; Drechsel et al., 2013). One recently described function for AS/NMD is in regulating plant-pathogen responses (Gloggnitzer et al., 2014; Wachter and Hartmann, 2014). AS therefore represents an important level of regulation of gene expression and must be considered by plant scientists in their goal of understanding gene function and plant biology.
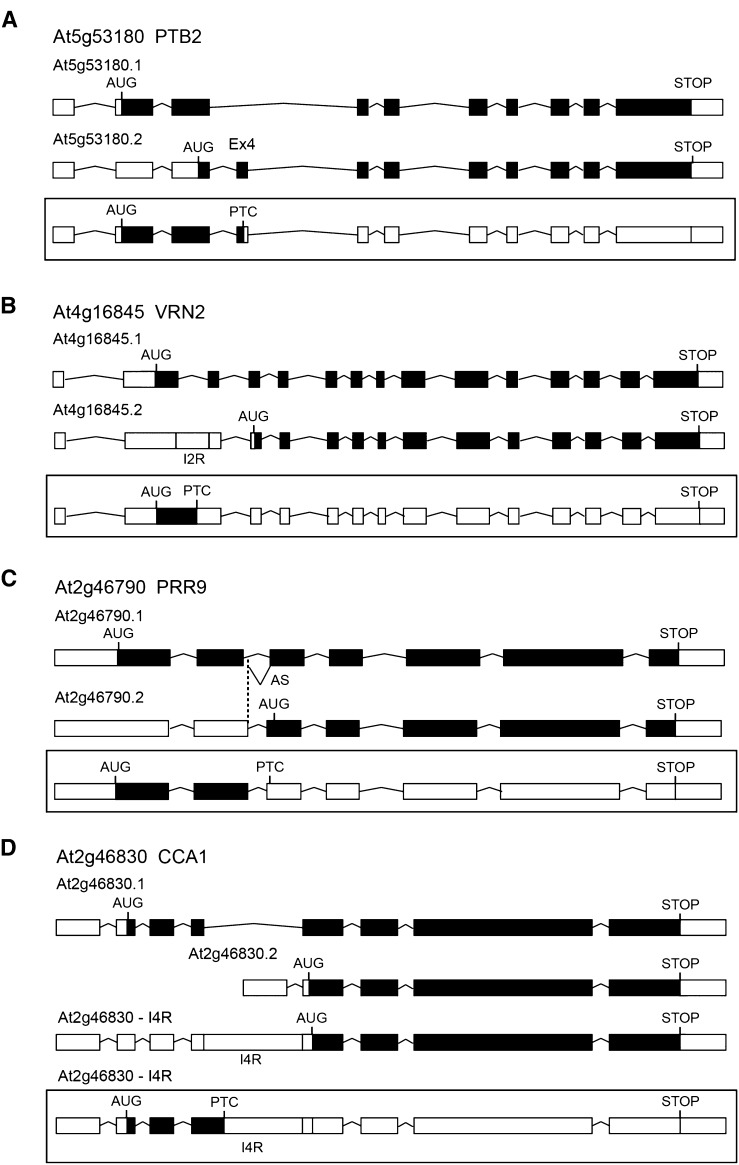
We believe that awareness needs to be raised about the annotation of protein coding potential of some AS transcripts. TAIR transcript models are presented based on the gene exon-intron structure and with open reading frame (ORF) information. However, the program that generates the translational models identifies and illustrates the longest open reading frame. This is most likely due to automated genome annotation programs often dismissing shorter ORFs (less than approximately 100 amino acids) so as not to predict false-positive ORFs and thereby leading to annotation of an AUG downstream of the authentic translation start site. We use “authentic” here to denote the AUG that is used in the translation of the transcript from the gene that gives the expected protein and that, if present in alternatively spliced transcripts, will be used for translation. The consequence is that in numerous cases where translation from the authentic translation start site would encounter a premature termination codon (PTC) and generate a short ORF, instead a downstream AUG is suggested (by annotation software) as the translation start site. Often, this creates a transcript model that contains multiple exons/introns upstream of the suggested translation start site and an extended and unlikely 5′ untranslated region (UTR) (Figures 1A to 1D). In addition, not only is the authentic translation start site ignored but often other AUG and stop codons in the three reading frames are discounted. For example, POLYPYRIMIDINE TRACT BINDING PROTEIN2 (PTB2) is known to autoregulate its transcript levels by AS/NMD through the inclusion of exon 4 (which contains a PTC) (Stauffer et al., 2010; Rühl et al., 2012). The TAIR model of this transcript (AT5G53180.2) shows an AUG in exon 3 that recreates the ORF; however, translation from the authentic start site generates the PTC in exon 4 (Figure 1A), which targets the transcript for degradation by NMD, consistent with experimental data (Kalyna et al., 2012; Rühl et al., 2012). Similarly, VRN2 has a transcript (TAIR model AT4G16845.2) that retains intron 2 (I2R) and has an annotated AUG in exon 4 that recreates the ORF (Figure 1B). However, translation from the authentic translation start site would generate a PTC within the retained intron 2 sequence. The clock gene PRR9 has an alternative 5′ splice site in intron 2 that adds eight nucleotides, thereby changing the reading frame and generating a PTC in exon 3 triggering NMD (Sanchez et al., 2010; Kalyna et al., 2012). However, the TAIR model (AT2G46790.2) has an annotated AUG in exon 3 that recreates the reading frame, while translation from the authentic translation start site generates a PTC in exon 3 (Figure 1C). This problem goes beyond TAIR as new variants are discovered and new assemblies are generated. For example, a transcript where intron 4 is retained has been identified for the clock gene CCA1 (Figure 1D). Translation from the authentic start site would stop at a PTC in intron 4; however, erroneous annotation has suggested that an AUG in exon 5, which recreates the reading frame for the C-terminal half of the CCA1, is the translation initiation start codon (Figure 1D).
Figure 1.
Gene and Transcript Structures Illustrating Erroneous Open Reading Frame Identification.
PTB2 (A), VRN2 (B), PRR9 (C), and CCA1 (D). In all cases, the top two transcripts are redrawn from TAIR. In (A) to (C), the third transcript (boxed) shows the consequence of translation beginning at the authentic translation start site AUG and terminating at a premature termination codon. All three transcripts have been shown to be NMD-sensitive (Kalyna et al., 2012). In (D), the second TAIR transcript is truncated, beginning in intron 4. The third and fourth transcripts show intron retention of intron 4 (I4R), the third is redrawn from Seo et al. (2012), and the fourth transcript (boxed) shows the more likely outcome when translation begins at the authentic translation start site. White boxes, UTRs; black boxes, coding exons; lines, introns; InR, retention of intron number n. The positions of authentic and predicted start and stop codons are indicated by AUG and STOP. The positions of the first PTC following the AUG are indicated by PTC.
It is also important to note that while many transcripts with PTCs are targets of NMD, in plants, transcripts with a retained intron are not NMD-sensitive (e.g., Figure 1D, CCA1 I4R) (James et al., 2012; Kalyna et al., 2012; Marquez et al., 2012; Leviatan et al., 2013). This is due to such transcripts being retained in the nucleus (Göhring et al., 2014) and, therefore, not encountering translation or the NMD machinery (Kalyna et al., 2012; Leviatan et al., 2013). Indeed, gene expression in eukaryotes can be stalled by intron retention to control developmental transitions or stress responses (Yap et al., 2012; Boothby et al., 2013; Shalgi et al., 2014; Boutz et al., 2015). Transcripts containing intron sequences are recognized as incompletely processed and remain in the nucleus until introns are removed posttranscriptionally when, for example, a stress condition is removed or at a particular developmental stage (Boothby et al., 2013; Shalgi et al., 2014; Boutz et al., 2015). In these cases, clearly, annotation of protein coding potential of intron retention transcripts is actually the same as for fully spliced isoforms.
Assuming that translation of some AS isoforms starts at a downstream AUG instead of the authentic AUG can lead to erroneous hypotheses, experimental design, results, and conclusions. To avoid such misinterpretation, it is necessary to apply basic molecular and biochemical knowledge to understand the likely fates of different transcripts. We should remember the rules of translation and that ribosomes do not refer to genome annotation programs (and databases) before translating a message (Figure 2). In eukaryotes, translation initiation is usually cap dependent with a cap binding complex recruiting the mRNA and the 40S ribosomal subunit scanning to the AUG translation start site. The AUG must be in the proper sequence context to be used as the initiation codon by the translation machinery (Kozak, 1999, 2002; Lukaszewicz et al., 2000). On encountering a stop codon, translation terminates with the release of the polypeptide, mRNA, and dissociation of the ribosomal subunits. Therefore, in the majority of cases, translation will start at the authentic AUG and terminate when a PTC is encountered. Exceptions involve reinitiation of translation, use of internal ribosome entry sites, or leaky scanning when the first AUG is in a weak context, and a further potentially confounding factor is the use of noncanonical translation start sites. The potential for reinitiation of translation at a downstream AUG parallels the situation with genes containing upstream open reading frames (uORFs) in their 5′UTRs. If a uORF is recognized and translated, it can affect gene expression by a variety of mechanisms: coding for an active peptide, affecting translational efficiency, or reducing transcript levels by triggering NMD (Morris and Geballe, 2000; Meijer and Thomas, 2002; Kalyna et al., 2012; Liu et al., 2013; Remy et al., 2014). Although in some genes reinitiation after uORF translation can occur, this process is generally thought to be inefficient (Meijer and Thomas, 2002; Kochetov et al., 2008). Indeed, Arabidopsis genome-wide ribosome profiling detected the use of only 35 potential downstream AUGs in a total of 31 genes (Liu et al., 2013). Similarly, the use of internal ribosome entry sites in cellular mRNAs is thought to be inefficient and rare (Jackson, 2013). In general, ribosome profiling data could be used to determine translation start codon usage, but currently these data are scarce.
Figure 2.
Lost in Translation.
In translating an alternatively spliced transcript containing a PTC, the ribosome encounters the authentic AUG and would begin translation. Some transcript misannotations suggest that that the ribosome would ignore the authentic translation start site and continue downstream to an AUG, which would generate an open reading frame.
In conclusion, before planning experiments, it is necessary to look closely at the transcript variants of a gene and predict, as far as possible, likely fates of specific transcripts. Transcript variants should be translated in silico using the authentic translation initiation AUG or at least the AUG common to most transcripts of a gene. This will allow the detection of PTCs that potentially trigger NMD and thereby unproductive AS isoforms. Predicted NMD transcripts can be experimentally validated by testing for NMD sensitivity. Publicly available NMD data (for example, by Drechsel et al. [2013], at http://gbrowse.cbio.mskcc.org/gb/gbrowse/NMD2013/) can be also used to check whether an AS event triggers NMD. It is also important to bear in mind that many plant intron retention transcripts, despite containing PTCs, avoid NMD by nuclear retention and may be spliced posttranscriptionally to yield fully spliced transcripts and protein. Similarly, in silico translation will clearly define in-frame alternative splicing events and will also identify transcripts that potentially generate proteins with altered C-terminal sequences. This allows more accurate identification of potential functional changes in protein isoform sequence and structure. Studies designed to experimentally address the coding potential of AS variants should ideally use the original gene sequences including introns, the 5′ and 3′ UTR such that alternatively spliced transcripts contain their authentic complement of RNA binding proteins (e.g., the exon-junction complex), any regulatory elements in UTRs, and the translation start codons are in their authentic context. N- and C-terminally tagged versions of the gene in question can be used to test experimentally for the production of protein isoforms from alternatively spliced transcripts by, for example, immunoblotting. With the growing use of RNA-seq, it is important to understand the potential problems caused by misannotation of AS transcripts. Time invested in understanding and validating the possible fates of transcript variants is time well spent, and we can look forward to exploiting the power of RNA-seq and opening up new and exciting plant discoveries.
ACKNOWLEDGMENTS
This research was supported by funding from the Biotechnology and Biological Sciences Research Council (BB/K006568/1 to J.W.S.B.), by the Scottish Government Rural and Environment Science and Analytical Services division, and by the Austrian Science Fund (P26333 to M.K. and DK W1207 SFB RNAreg F43-P10 to A.B.).
AUTHOR CONTRIBUTIONS
J.W.S.B., C.G.S., Y.M., A.B., and M.K. wrote the article. G.M.G. and J.W.S.B. designed Figure 2, and G.M.G. drew Figure 2.
References
- Boothby T.C., Zipper R.S., van der Weele C.M., Wolniak S.M. (2013). Removal of retained introns regulates translation in the rapidly developing gametophyte of Marsilea vestita. Dev. Cell 24: 517–529. [DOI] [PubMed] [Google Scholar]
- Boutz P.L., Bhutkar A., Sharp P.A. (2015). Detained introns are a novel, widespread class of post-transcriptionally spliced introns. Genes Dev. 29: 63–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carvalho R.F., Feijão C.V., Duque P. (2013). On the physiological significance of alternative splicing events in higher plants. Protoplasma 250: 639–650. [DOI] [PubMed] [Google Scholar]
- Chamala S., Feng G., Chavarro C., Barbazuk W.B. (2015). Genome-wide identification of evolutionarily conserved alternative splicing events in flowering plants. Front. Bioeng. Biotechnol. 3: 33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drechsel G., Kahles A., Kesarwani A.K., Stauffer E., Behr J., Drewe P., Rätsch G., Wachter A. (2013). Nonsense-mediated decay of alternative precursor mRNA splicing variants is a major determinant of the Arabidopsis steady state transcriptome. Plant Cell 25: 3726–3742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gloggnitzer J., Akimcheva S., Srinivasan A., Kusenda B., Riehs N., Stampfl H., Bautor J., Dekrout B., Jonak C., Jiménez-Gómez J.M., Parker J.E., Riha K. (2014). Nonsense-mediated mRNA decay modulates immune receptor levels to regulate plant antibacterial defense. Cell Host Microbe 16: 376–390. [DOI] [PubMed] [Google Scholar]
- Göhring J., Jacak J., Barta A. (2014). Imaging of endogenous messenger RNA splice variants in living cells reveals nuclear retention of transcripts inaccessible to nonsense-mediated decay in Arabidopsis. Plant Cell 26: 754–764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson R.J. (2013). The current status of vertebrate cellular mRNA IRESs. Cold Spring Harb. Perspect. Biol. 5: a011569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- James A.B., Syed N.H., Bordage S., Marshall J., Nimmo G.A., Jenkins G.I., Herzyk P., Brown J.W.S., Nimmo H.G. (2012). Alternative splicing mediates responses of the Arabidopsis circadian clock to temperature changes. Plant Cell 24: 961–981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalyna M., et al. (2012). Alternative splicing and nonsense-mediated decay modulate expression of important regulatory genes in Arabidopsis. Nucleic Acids Res. 40: 2454–2469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kochetov A.V., Ahmad S., Ivanisenko V., Volkova O.A., Kolchanov N.A., Sarai A. (2008). uORFs, reinitiation and alternative translation start sites in human mRNAs. FEBS Lett. 582: 1293–1297. [DOI] [PubMed] [Google Scholar]
- Kozak M. (1999). Initiation of translation in prokaryotes and eukaryotes. Gene 234: 187–208. [DOI] [PubMed] [Google Scholar]
- Kozak M. (2002). Pushing the limits of the scanning mechanism for initiation of translation. Gene 299: 1–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leviatan N., Alkan N., Leshkowitz D., Fluhr R. (2013). Genome-wide survey of cold stress regulated alternative splicing in Arabidopsis thaliana with tiling microarray. PLoS One 8: e66511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li P., Tao Z., Dean C. (2015). Phenotypic evolution through variation in splicing of the noncoding RNA COOLAIR. Genes Dev. 29: 696–701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu M.-J., Wu S.-H., Wu J.-F., Lin W.D., Wu Y.C., Tsai T.Y., Tsai H.L., Wu S.H. (2013). Translational landscape of photomorphogenic Arabidopsis. Plant Cell 25: 3699–3710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukaszewicz M., Feuermann M. I, Jérouville B., Stas A., Boutry M. (2000). In vivo evaluation of the context sequence of the translation initiation codon in plants. Plant Sci. 154: 89–98. [DOI] [PubMed] [Google Scholar]
- Marquez Y., Brown J.W.S., Simpson C., Barta A., Kalyna M. (2012). Transcriptome survey reveals increased complexity of the alternative splicing landscape in Arabidopsis. Genome Res. 22: 1184–1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meijer H.A., Thomas A.A. (2002). Control of eukaryotic protein synthesis by upstream open reading frames in the 5′-untranslated region of an mRNA. Biochem. J. 367: 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris D.R., Geballe A.P. (2000). Upstream open reading frames as regulators of mRNA translation. Mol. Cell. Biol. 20: 8635–8642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrillo E., Godoy Herz M.A., Fuchs A., Reifer D., Fuller J., Yanovsky M.J., Simpson C., Brown J.W.S., Barta A., Kalyna M., Kornblihtt A.R. (2014). A chloroplast retrograde signal regulates nuclear alternative splicing. Science 344: 427–430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Posé D., Verhage L., Ott F., Yant L., Mathieu J., Angenent G.C., Immink R.G.H., Schmid M. (2013). Temperature-dependent regulation of flowering by antagonistic FLM variants. Nature 503: 414–417. [DOI] [PubMed] [Google Scholar]
- Reddy A.S.N., Marquez Y., Kalyna M., Barta A. (2013). Complexity of the alternative splicing landscape in plants. Plant Cell 25: 3657–3683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Remy E., Cabrito T.R., Batista R.A., Hussein M.A.M., Teixeira M.C., Athanasiadis A., Sá-Correia I., Duque P. (2014). Intron retention in the 5'UTR of the novel ZIF2 transporter enhances translation to promote zinc tolerance in arabidopsis. PLoS Genet. 10: e1004375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rühl C., Stauffer E., Kahles A., Wagner G., Drechsel G., Rätsch G., Wachter A. (2012). Polypyrimidine tract binding protein homologs from Arabidopsis are key regulators of alternative splicing with implications in fundamental developmental processes. Plant Cell 24: 4360–4375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez S.E., et al. (2010). A methyl transferase links the circadian clock to the regulation of alternative splicing. Nature 468: 112–116. [DOI] [PubMed] [Google Scholar]
- Schweingruber C., Rufener S.C., Zünd D., Yamashita A., Mühlemann O. (2013). Nonsense-mediated mRNA decay - mechanisms of substrate mRNA recognition and degradation in mammalian cells. Biochim. Biophys. Acta 1829: 612–623. [DOI] [PubMed] [Google Scholar]
- Seo P.J., Park M.-J., Lim M.-H., Kim S.-G., Lee M., Baldwin I.T., Park C.-M. (2012). A self-regulatory circuit of CIRCADIAN CLOCK-ASSOCIATED1 underlies the circadian clock regulation of temperature responses in Arabidopsis. Plant Cell 24: 2427–2442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shalgi R., Hurt J.A., Lindquist S., Burge C.B. (2014). Widespread inhibition of posttranscriptional splicing shapes the cellular transcriptome following heat shock. Cell Reports 7: 1362–1370. [DOI] [PubMed] [Google Scholar]
- Shikata H., Hanada K., Ushijima T., Nakashima M., Suzuki Y., Matsushita T. (2014). Phytochrome controls alternative splicing to mediate light responses in Arabidopsis. Proc. Natl. Acad. Sci. USA 111: 18781–18786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staiger D., Brown J.W.S. (2013). Alternative splicing at the intersection of biological timing, development, and stress responses. Plant Cell 25: 3640–3656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stauffer E., Westermann A., Wagner G., Wachter A. (2010). Polypyrimidine tract-binding protein homologues from Arabidopsis underlie regulatory circuits based on alternative splicing and downstream control. Plant J. 64: 243–255. [DOI] [PubMed] [Google Scholar]
- Syed N.H., Kalyna M., Marquez Y., Barta A., Brown J.W.S. (2012). Alternative splicing in plants--coming of age. Trends Plant Sci. 17: 616–623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wachter A., Hartmann L. (2014). NMD: nonsense-mediated defense. Cell Host Microbe 16: 273–275. [DOI] [PubMed] [Google Scholar]
- Yap K., Lim Z.Q., Khandelia P., Friedman B., Makeyev E.V. (2012). Coordinated regulation of neuronal mRNA steady-state levels through developmentally controlled intron retention. Genes Dev. 26: 1209–1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X.N., Mount S.M. (2009). Two alternatively spliced isoforms of the Arabidopsis SR45 protein have distinct roles during normal plant development. Plant Physiol. 150: 1450–1458. [DOI] [PMC free article] [PubMed] [Google Scholar]