Significance
The activation of kinases by Ca2+ represents a vital class of signaling interactions that regulates many biological processes. The mechanism of activation for these enzymes is conserved and characterized by removal of an inhibitory element from the kinase domain. We report a previously unidentified mechanism for the activation of essential apicomplexan calcium-dependent protein kinases (CDPKs). Using Toxoplasma CDPK1 as a representative, we demonstrate that the kinase domain is intrinsically inactive and requires stabilization for activity. This distinct mechanism of activation reveals a susceptibility in CDPKs, which we exploit to effectively inhibit them. When viewed in the context of the entire protein kinase family, our results emphasize the remarkable adaptability of the kinase fold to diverse forms of regulation.
Keywords: calcium-dependent protein kinase, kinase activation, VHH
Abstract
Calcium-dependent protein kinases (CDPKs) comprise the major group of Ca2+-regulated kinases in plants and protists. It has long been assumed that CDPKs are activated, like other Ca2+-regulated kinases, by derepression of the kinase domain (KD). However, we found that removal of the autoinhibitory domain from Toxoplasma gondii CDPK1 is not sufficient for kinase activation. From a library of heavy chain-only antibody fragments (VHHs), we isolated an antibody (1B7) that binds TgCDPK1 in a conformation-dependent manner and potently inhibits it. We uncovered the molecular basis for this inhibition by solving the crystal structure of the complex and simulating, through molecular dynamics, the effects of 1B7–kinase interactions. In contrast to other Ca2+-regulated kinases, the regulatory domain of TgCDPK1 plays a dual role, inhibiting or activating the kinase in response to changes in Ca2+ concentrations. We propose that the regulatory domain of TgCDPK1 acts as a molecular splint to stabilize the otherwise inactive KD. This dependence on allosteric stabilization reveals a novel susceptibility in this important class of parasite enzymes.
Calcium-dependent protein kinases (CDPKs) are major transducers of Ca2+ signaling in plants and protists. CDPKs regulate many essential processes in medically important parasites, including the apicomplexans responsible for malaria (Plasmodium spp.), toxoplasmosis (Toxoplasma gondii), and cryptosporidiosis (Cryptosporidium parvum). In these parasites, CDPKs control Ca2+-dependent secretion, parasite motility, gametogenesis, as well as invasion and egress from infected cells (reviewed in ref. 1). Their absence from mammalian genomes makes CDPKs attractive drug targets. Inhibition of TgCDPK1 (T. gondii CDPK1) phenocopies a conditional knock-down of the kinase by blocking Ca2+-regulated secretion (2). The closest paralog of TgCDPK1, TgCDPK3, regulates the initiation of T. gondii egress from host cells (3–5), and its homolog in Plasmodium falciparum, PfCDPK1, is essential (6).
CDPKs comprise an N-terminal kinase domain (KD), linked by a junction region to four EF hands that form a calmodulin-like domain (CaM-LD) and bind Ca2+. Studies of plant CDPKs suggest that the key mechanism of regulation is inhibition of the KD by the junction region preceding the EF hands, predicted to act as a pseudosubstrate (7, 8). The junction region interacts with the CaM-LD, and Ca2+ binding alters this interaction, presumably to relieve inhibition of the KD (9, 10). Structures of TgCDPK1 corroborated these predictions (11, 12) and showed the extreme Ca2+-dependent rearrangement of the CaM-LD, which warranted its distinction as a new regulatory domain, dubbed the CDPK activation domain (CAD) (11). In the absence of Ca2+, the junction domain and the first α-helix of EF hand I form a single long α-helix that extends from the base of the KD, occupies the substrate-binding site, and occludes the catalytic pocket (11). This structure is reminiscent of the autoinhibited state of mammalian Ca2+/CaM-dependent protein kinases (CaMKs) (13, 14), reinforcing the notion that autoinhibition is the major mechanism of CDPK regulation.
Here we show that removal of the regulatory domain of TgCDPK1 is not sufficient to activate the kinase. To identify new tools to probe the mechanism of kinase activation, we turned to single domain antibody fragments, or VHHs, derived from alpaca (Vicugna pacos) heavy chain-only antibodies. In screening phage display libraries of VHHs from alpacas naturally infected with T. gondii, we isolated a single domain antibody (VHH 1B7) against TgCDPK1. This antibody recognizes the kinase in a conformation-dependent manner and selectively inhibits the Ca2+-bound form. Cocrystallization of the antibody with TgCDPK1 has yielded new insights into the activation of these enzymes and revealed a novel mechanism for CDPK inhibition.
Results
Removal of the Regulatory Domain of TgCDPK1 Is Not Sufficient for Kinase Activation.
By analogy to other Ca2+-regulated kinases, it has been assumed that apicomplexan CDPKs are activated by de-repression of the KD upon Ca2+ binding to the regulatory domain (CAD) (11, 12, 15). We reasoned that expression of TgCDPK1 without the predicted repressive domains would yield a constitutively active kinase. We expressed two constructs that either removed the CAD completely (residues 1–314) or retained the autoinhibitory α-helix (residues 1–336). Like the full-length enzyme, both constructs expressed well as soluble proteins, but neither showed measurable catalytic activity (Fig. S1A).
Fig. S1.
TgCDPK1 truncations lacking the regulatory domain are catalytically inactive. (A) Two constructs were expressed and assayed for kinase activity: Truncation I lacked the CAD completely, whereas truncation II retained the autoinhibitory α-helix. The constructs’ sequence coverage of WT-enzyme amino acids is indicated in parentheses. Values are normalized to maximal activity. Mean ± SD, representative experiment. (B) The isolated TgCDPK1 KD cannot be activated by the CAD in trans. The purified KD yielded no measurable activity even at the 5 µM tested. Addition of purified CAD to the KD did not lead to any measurable kinase activity. Values are normalized to maximum WT activity. Mean ± SEM, n = 3 independent experiments.
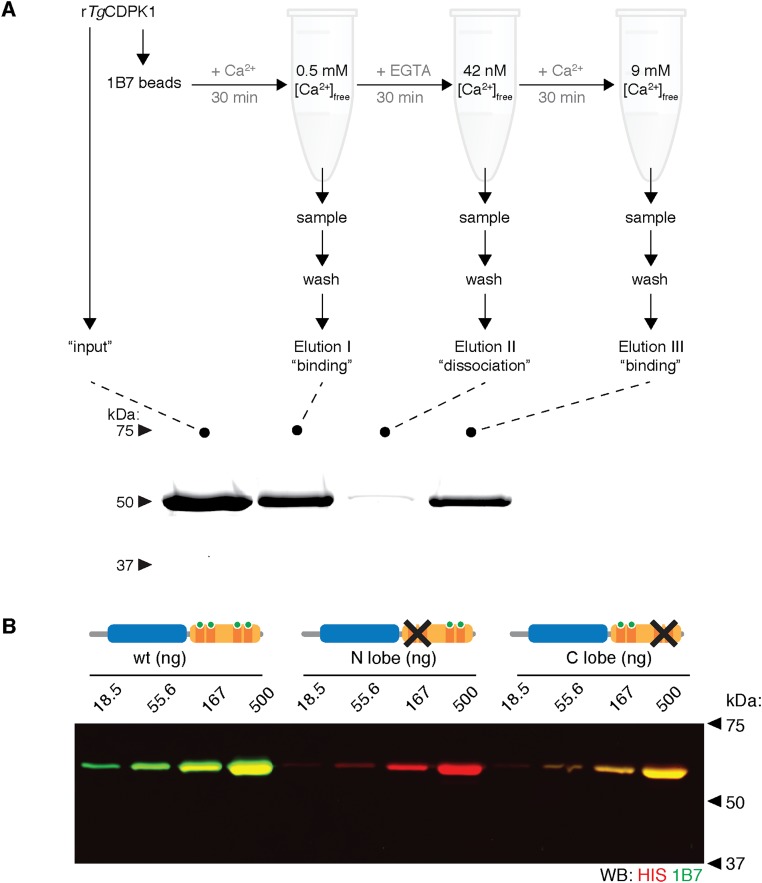
To examine the effect of severing the regulatory domain from the active kinase, we introduced the human rhinovirus 3C protease cleavage site at the junction of the KD and the CAD of TgCDPK1 to generate the kinase referred to as “3C.” Treatment of the mutant enzyme with the protease separated the two domains of TgCDPK1-3C but did not cleave the wild-type enzyme (Fig. 1A). Although introduction of the protease site decreased activity somewhat, cleaving the connection between the CAD and the KD abolished kinase activity (Fig. 1B). Separating the KD from the CAD by size exclusion chromatography (SEC) or inclusion of a fivefold molar excess of the CAD failed to yield appreciable kinase activity (Fig. S1B). We conclude that tethering the CAD to the KD is necessary for TgCDPK1 activity.
Fig. 1.
TgCDPK1 is not activated by removal of its regulatory domain. (A) Introduction of a 3C protease cleavage site between the CAD and KD of TgCDPK1, in the kinase referred to as 3C, enables separation of the two domains upon protease treatment. The wild-type (WT) enzyme was unaffected by the protease treatment. The CAD and KD could be further isolated from the digested 3C. (B) Kinase activity of WT and 3C, before (–) and after (+) protease treatment. Mean ± SEM, n = 3 independent experiments.
Identification of Single Domain Antibodies to Probe TgCDPK1 Structure.
Single domain antibodies derived from the heavy chain-only antibodies of alpacas (VHHs) are useful crystallization chaperones that often stabilize transient conformations (16). We identified VHHs that bind TgCDPK1 from alpacas in our cohort that had antibodies against T. gondii proteins (Fig. S2A). VHH phage display libraries from the seropositive animals were pooled and panned against recombinant TgCDPK1. VHH clones that showed reactivity against TgCDPK1 by ELISA were enriched for the sequence represented by clone 1B7 (Fig. S2B). 1B7 was subcloned into a bacterial cytoplasmic expression vector, produced in Escherichia coli, and purified to homogeneity (Fig. S2 C and D). By immunoblot, 1B7 recognized both endogenous and recombinant versions of TgCDPK1 and its closest paralog TgCDPK3 (Fig. 2A), with a limit of detection below 15 ng (Fig. 2A and Fig. S3). Measured against a standard curve of recombinant proteins, these kinases are expressed at ∼4–7 femtograms per cell, equivalent to 40,000–80,000 copies.
Fig. S2.
Identification of heavy chain-only antibodies to study the structure of TgCDPK1. (A) Individual or pooled serum from alpacas was tested for reactivity against T. gondii lysate by immunoblot. The secondary antibody showed no reactivity on its own. (B) Panning of a combined VHH phage display library from alpacas 1–4 and 8 against immobilized TgCDPK1 enriched for a group of nearly identical sequences from which 1B7 was selected. Sequences from 96 clones before enrichment are shown for comparison. (C) Sequence of 1B7 highlighting the variable loops (red), the sortase substrate motif (green), and the 6xHis-tag (blue). (D) Expression and purification of 1B7. Following the induction of 1B7 expression by IPTG, a lysate was prepared. The lysate was collected on a Ni-NTA column, and a sample of the flow-through was collected. The column was washed and 1B7 was eluted with buffer containing 500 mM imidazole. The eluted material was further enriched by SEC, and peak fractions were collected and concentrated. Samples collected during the purification were resolved by SDS/PAGE and stained with Coomassie for total protein.
Fig. 2.
1B7 recognizes a calcium-dependent conformation of TgCDPK1. (A) IRDye800-labeled 1B7 was used to probe decreasing concentrations of total T. gondii lysate or recombinant kinase (Top). Samples were separately probed for parasite actin (ACT1; red) or the 6xHis-tag on recombinant proteins (HIS; green). (B) Incubation of parasite lysates with immobilized 1B7 depletes TgCDPK1/TgCDPK3 in the presence of CaCl2 but not in EGTA. The immunoprecipitated material could be eluted from the beads by subsequent incubation with EGTA. Blot was probed with 1B7 (green), and ACT1 (red) is included as a control. (C) SEC of 1B7-TAMRA and TgCDPK1, alone or incubated at equimolar concentrations, in buffer containing 1 mM CaCl2 or EGTA. Absorbance was recorded at 280 nm for total protein (blue) and 550 nm for 1B7-TAMRA (red). (D) IP of recombinant kinases by 1B7 in the presence of 1 mM CaCl2 or EGTA. The domain structure of each kinase, indicating the KD and each of the EF hands (I–IV), is depicted for wild-type (WT), D368A/D415A (N-lobe mutant), and D451A/D485A (C-lobe mutant). Means ± SEM, n = 3 independent experiments. Insert shows the results from a representative experiment. (E) Calcium dependency of TgCDPK1 activation or 1B7 binding. Values are normalized to maximal activity or binding. Means ± SEM, n = 3 independent experiments.
Fig. S3.
Extended dilution series of recombinant TgCDPKs. (A) Western blots using either IRdye800-labeled 1B7 or an antibody recognizing the 6xHis-tag of the recombinant proteins. (B) Densitometric analysis of Western blots showing similar recognition of both kinases by 1B7.
1B7 Dynamically Senses TgCDPK1 Conformation.
To determine whether 1B7 could sense Ca2+-dependent conformational changes (11, 12), we performed immunoprecipitations (IPs) in the presence of either CaCl2 or the cation chelator EGTA. 1B7 required Ca2+ to deplete lysates of its cognate antigens (Fig. 2B). Chelation of Ca2+ with EGTA readily eluted the bound CDPKs from the 1B7 beads, which could be reversed by further Ca2+ addition, demonstrating the reversibility of the interaction (Fig. S4A). To identify parasite proteins retrieved by 1B7, the eluted samples were analyzed by mass spectrometry. The two paralogs, TgCDPK1 and TgCDPK3, were prominently represented with 26 unique peptides each (52% and 41% sequence coverage, respectively) and were the only CDPKs found (Table S1). 1B7 thus specifically recognizes the Ca2+-bound form of TgCDPK1 and TgCDPK3.
Fig. S4.
Ca2+ dependency of 1B7 binding. (A) 1B7 binds TgCDPK1 reversibly. Precipitation of recombinant TgCDPK1 by covalently immobilized 1B7 is shown. The experimental setup is diagrammed on top. Kinase was incubated with 1B7 beads, and the free [Ca2+] was modulated by adding EGTA or CaCl2, as indicated. Samples were taken after each free [Ca2+] change to determine whether complex formation had occurred, as measured by the presence of TgCDPK1 in the eluate. (B) Immunoblot comparing the detection of different kinases by IRDye800-labeled 1B7 (green). The domain structure of each kinase, indicating the KD and each of the EF hands (I–IV), is depicted for wild type (WT), D368A/D415A (N-lobe mutant), and D451A/D485A (C-lobe mutant). Detection of recombinant kinases by their His-tags (red) is included as a loading control.
Table S1.
Mass spectrometric analysis of 1B7 IP
| Gene ID | Product description | MW, kDa | Coverage | Unique peptides | Total spectra |
| TGGT1_301440 | TgCDPK1 | 57 | 52% | 26 | 70 |
| TGGT1_305860 | TgCDPK3 | 60 | 41% | 26 | 80 |
| TGGT1_247550 | HSP60 | 61 | 36% | 15 | 20 |
| TGGT1_225050 | Adenosylhomocysteinase | 52 | 31% | 14 | 30 |
| TGGT1_211680 | Protein disulfide isomerase | 53 | 25% | 9 | 14 |
| TGGT1_266960 | Beta-tubulin | 50 | 20% | 8 | 10 |
| TGGT1_270510 | Asparaginyl-tRNA synthetase | 75 | 15% | 8 | 12 |
| TGGT1_209030 | Actin | 42 | 28% | 8 | 9 |
| TGGT1_311240 | DnaJ family chaperone | 56 | 16% | 7 | 7 |
| TGGT1_411430 | ROP5 | 61 | 13% | 5 | 5 |
| TGGT1_253430 | Asparagine synthetase | 66 | 9.20% | 5 | 8 |
| TGGT1_305770 | ABC transporter | 40 | 5.80% | 3 | 3 |
| TGGT1_233110 | IMPDH | 101 | 4.50% | 3 | 3 |
| TGGT1_290670 | Leucyl aminopeptidase | 83 | 4.70% | 3 | 3 |
| TGGT1_219590 | RuvB family 1 protein | 53 | 7.70% | 3 | 3 |
| TGGT1_215775 | ROP8 | 62 | 4.60% | 2 | 2 |
| TGGT1_228490 | Hypothetical protein | 43 | 8.90% | 2 | 2 |
| TGGT1_291960 | ROP40 | 58 | 5.40% | 2 | 2 |
| TGGT1_290200 | NAD/NADP dehydrogenase | 48 | 6.00% | 2 | 4 |
| TGGT1_230450 | Bifunctional GMP synthase | 62 | 4.00% | 2 | 2 |
| TGGT1_294800B | eIF-1α | 49 | 5.40% | 2 | 3 |
| TGGT1_268890 | Citrate synthase I | 60 | 4.10% | 2 | 2 |
| TGVEG_050040 | ROP4/7 | 64 | 5.20% | 2 | 2 |
| TGGT1_220950 | Hypothetical protein | 48 | 5.60% | 2 | 2 |
| TGGT1_215590 | Subunit of succinate dehydrogenase | 73 | 3.30% | 2 | 2 |
We next determined whether 1B7 could form a stable complex with TgCDPK1. Using 1B7 labeled with a carboxytetramethylrhodamine (TAMRA) fluorophore at its C terminus, we tracked 1B7 binding to recombinant TgCDPK1 by SEC. When preincubated with TgCDPK1 in the presence of Ca2+, 1B7-TAMRA coeluted with the kinase, well-resolved from 1B7-TAMRA alone (Fig. 2C). Thus, 1B7 binds a conformation of TgCDPK1 that appears in the presence of Ca2+ and releases the kinase upon Ca2+ chelation.
1B7 Recognizes an Intermediate Conformation of TgCDPK1.
To explore the conformational changes imposed by Ca2+ on TgCDPK1 and their influence on 1B7 binding, we mutated the Ca2+-binding EF hands of the kinase. The four EF hands of canonical CDPKs are segregated into two lobes, proposed to have differing Ca2+ affinities (10). To isolate the contribution of each lobe, we mutated the first invariant Asp in each EF hand loop in either lobe (N-lobe Asp368Ala/Asp415Ala and C-lobe Asp451Ala/Asp485Ala). Both the C-lobe mutant and wild-type kinase bound immobilized 1B7 in a Ca2+-dependent manner with indistinguishable affinities, whereas binding was abolished by mutation of the N lobe (Fig. 2D). The N-lobe mutations also prevented 1B7 recognition by immunoblot (Fig. S4B). The N-lobe EF hands are thus crucial for 1B7 binding.
We next compared the precise [Ca2+] necessary for 1B7 binding and kinase activation. Using buffers with defined free [Ca2+], we measured the amount of kinase bound by 1B7 and, separately, the phosphorylation of a heterologous substrate. Binding of TgCDPK1 by 1B7 occurred at a low free [Ca2+], with an EC50 of 300–350 nM (95% confidence interval), fivefold lower than that required for activation (Fig. 2E). The discrepancy between the [Ca2+] required for binding and that required for activation suggests that 1B7 binds a TgCDPK1 conformation that is distinct from the active state of the kinase.
1B7 Is a Specific Inhibitor of TgCDPK1 and Related Kinases.
We wondered whether 1B7-bound CDPKs would retain activity in the presence of Ca2+. We measured kinase activity in increasing concentrations of 1B7, or an irrelevant VHH. 1B7 inhibited both kinases—TgCDPK1 and TgCDPK3—with an IC50 of ∼40 nM and achieved complete inhibition at approximately equimolar concentrations of antibody and enzyme (Fig. 3A).
Fig. 3.
1B7 inhibits TgCDPK1 and related kinases. (A) In vitro kinase assay with increasing concentrations of 1B7. Mean ± SEM, n = 3 independent experiments. (B) TgCDPK1-dependent (6-Fu-ATPγS) and total (ATPγS) thiophosphorylation in parasite lysates incubated with varying 1B7 concentrations. Thiophosphorylation is visualized by Western blot with rabbit mAb 51-8 (red); 1B7 staining is shown as a loading control (green). (C) Immunoblot of P. falciparum lysate from strain 3D7 or W2mef probed with 1B7-IRdye800 (green) or an antibody recognizing parasite actin (ACT1; red).
Although ATPγS is a substrate of many parasite kinases, 6-Fu-ATPγS is used exclusively by TgCDPK1 in parasite lysates (17). Specificity is conferred by the unusually large ATP-binding pocket of TgCDPK1 that harbors a Gly at a key position typically occupied by a bulky residue, which prevents other kinases—like TgCDPK3—from using 6-Fu-ATPγS as a substrate. Thiophosphorylation can be detected following alkylation using the monoclonal antibody 51-8 (17). We measured thiophosphorylation in total cell extracts treated with increasing concentrations of 1B7. The presence of 1B7 robustly inhibited TgCDPK1-mediated thiophosphorylation but did not affect overall kinase activity (Fig. 3B).
We also probed lysates from erythrocytic stages of two different P. falciparum strains. A single band of the correct molecular weight for the homologs of TgCDPK1 and TgCDPK3 was detected in each lysate (Fig. 3C). Accordingly, 1B7 also inhibits recombinant P. falciparum CDPK1 (Fig. 3A). 1B7 is thus a potent and specific inhibitor of TgCDPK1 and related apicomplexan kinases.
1B7 Interacts Directly with the Regulatory Domain of TgCDPK1.
To understand the Ca2+ dependency of 1B7 binding and kinase inhibition, we obtained crystals of the TgCDPK1:1B7 complex that diffracted to 2.94 Å (Table 1). We solved the structure by molecular replacement (MR) and single anomalous dispersion (SAD) phasing using selenomethionine-derivatized TgCDPK1. The structure was refined to a final Rwork of 22.6% and an Rfree of 26.2%. We used the structures of TgCDPK1 previously solved in the Ca2+-bound [Protein Data Bank (PDB) code 3HX4] and Ca2+-depleted (PDB ID codes 3KU2 and 3V51) states (11, 12) for comparison.
Table 1.
Data collection and refinement statistics
| Protein | TgCDPK1-1B7 native | TgCDPK1-1B7 SeMet derivative |
| Organism | T. gondii | T. gondii |
| PDB ID code | 4YGA | |
| Data collection | ||
| Space group | P1 | P21 |
| a, b, c, Å | 76.92, 91.07, 106.64 | 77.61, 91.92, 107.90 |
| α, β, γ, ° | 88.7, 108.3, 90.3 | 90.0, 108.5, 90.0 |
| Wavelength, Å | 0.9792 | 0.9792 |
| Resolution range, Å | 73.04–2.94 (3.05–2.94) | 102.3–4.09 (4.24–4.09) |
| Total reflections | 215,441 | 126,898 |
| Unique reflections | 53,985 | 21,729 |
| Completeness, % | 93.5 (94.8) | 98.2 (95.6) |
| Redundancy | 3.9 (3.8) | 5.8 (4.3) |
| Anomalous completeness, % | 96.4 | |
| Rsym, % | 11.0 (94.3) | 30.1 (74.4) |
| Rp.i.m., % | 6.1 (53.8) | 13.4 (37.7) |
| I/σ | 10.3 (1.4) | 6.0 (1.8) |
| CC1/2, % | 99.4 (69.6) | 99.1 (75.0) |
| Refinement | ||
| Resolution range, Å | 70.67–2.94 | |
| Rwork, % | 22.6 | |
| Rfree, % | 26.2 | |
| Coordinate error, Å | 0.46 | |
| Number of reflections | ||
| Total | 53,984 | |
| Rfree reflections | 1,715 | |
| Nonhydrogen atoms | 17,348 | |
| Protein atoms | 17,332 | |
| Calcium atoms | 16 | |
| rmsds | ||
| Bond lengths, Å | 0.003 | |
| Bond angles, ° | 0.78 | |
| Average B factors, Å2 | ||
| Protein | 113.3 | |
| Calcium | 84.4 | |
| Ramachandran, % | ||
| Favored, % | 94.9 | |
| Allowed, % | 4.3 | |
| Outlier, % | 0.8 | |
| Clashscore | 9.05 | |
| Molprobity percentile | 97th |
Values in parentheses are for the highest-resolution shell.
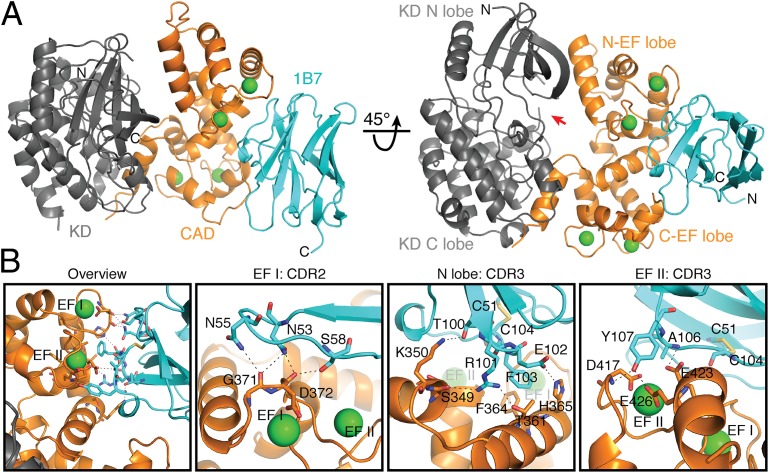
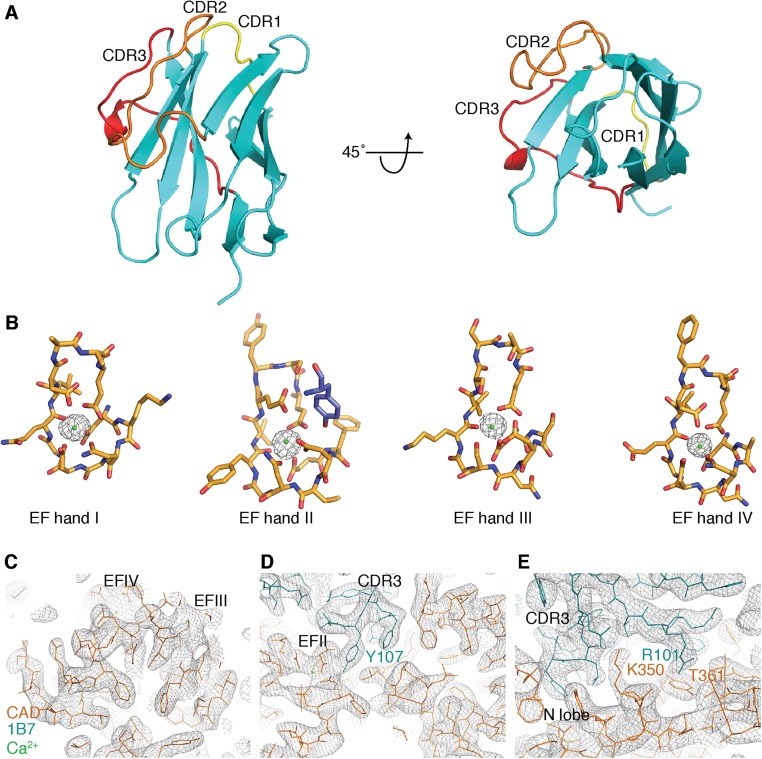
The antibody engages the N lobe of the CAD (Fig. 4A and Fig. S5A), consistent with loss of antibody binding to the N-lobe mutant (Fig. 2D and Fig. S4B). All four EF hands are occupied by Ca2+ in the Ca2+/1B7-bound structure (Fig. S5B). The Ca2+ dependency of the interaction is explained by 1B7’s engagement of residues that participate in Ca2+ coordination and form the Ca2+-binding regions of EF hands I and II. The Ca2+-binding loop of EF hand II is ordered in the presence of 1B7, in contrast to the structures derived in the absence of Ca2+ (11, 12). Asn53, Asn55, and Ser58 of 1B7 CDR2 form a hydrogen-bonding network with the carbonyl oxygens of Gly371 and Asp372 in the loop segment of EF hand I. CDR3 of 1B7 contacts α2 of the N lobe via Thr100–Lys350 and Arg101–Ser349 hydrogen bonds and α3 (the first helix of EF hand I) via an Arg101–Thr361 hydrogen bond, a Glu102–His365 salt bridge, and a Phe103–Phe364 aromatic ring stack. CDR3 also contacts the loop segment and second helix of EF hand II (α6) through Tyr107, which forms a bidentate hydrogen bond to Asp417 and Glu426 on the CAD and a hydrogen bond between the amide of Ala106 and the Glu423 side chain (Fig. 4B; see Fig. S5 C–E for density maps). There are additional Van der Waals contacts that result in a total buried surface area of 790 Å2 between 1B7 and TgCDPK1.
Fig. 4.
1B7 stabilizes a novel conformation of TgCDPK1. (A) Cartoon depicting the structure of the complex with the catalytic KD (gray), the CaM-like CAD (orange), bound calcium ions (green spheres), and 1B7 (blue). The position of the active site pocket is indicated (red arrow). (B) Specific interactions within the CAD:1B7 binding interface. Hydrogen bonds and salt bridges are depicted as black dotted lines. Interacting residues are shown as sticks for the CAD and 1B7. Cys51, in the beta strand immediately preceding CDR2, and Cys104, in CDR3, form a stabilizing, intrachain disulfide in 1B7. CDR1 on 1B7 makes no contacts with TgCDPK1.
Fig. S5.
Details of the Ca2+/1B7-bound TgCDPK1 structure. (A) CDRs of 1B7. CDR1 (yellow), CDR2 (orange), and CDR3 (red) of 1B7 are colored. The orientation of 1B7 shown is the same as in Fig. 4A. (B) Ca2+ occupancy of the TgCDPK1 EF hands in the presence of 1B7. Stick representation of the four EF hands that comprise the CAD is shown. The built Ca2+ ions are depicted as green spheres. The electron density shown is from a Sigma-A weighted difference (mFo-DFc) omit map for Ca2+ contoured at 6 σ. (C–E) Representative images of the final 2mFo-DFc electron density map contoured at 1 σ. 1B7 (teal) and the CAD (orange) are shown as sticks, and calcium ions (green) are depicted as dots. (C) View of the hydrophobic core of the CAD C lobe. (D) Interaction site between CDR3 of 1B7 and EF hand II of the CAD. (E) Interaction site between CDR3 and the N-lobe of the CAD.
Having defined the interactions between TgCDPK1 and 1B7, we explored the basis of 1B7’s specificity. We generated a sequence alignment of known 1B7 binders (TgCDPK1, TgCDPK3, and PfCDPK1) together with CDPKs from T. gondii, P. falciparum, and C. parvum. Of the six residues involved in sequence-specific interactions, Lys350 and Thr361 are conserved in the 1B7-binding CDPKs and variable in the others (Fig. S6).
Fig. S6.
Sequence comparison of Apicomplexan CDPKs. (A) Multiple sequence alignment of the 1B7-binding region of the CAD of 30 CDPKs from T. gondii, P. falciparum, and C. parvum. Residues known to interact with 1B7 at the six sequence-specific CAD positions are highlighted in red and numbered according to their position in TgCDPK1. Conservative mutations expected to still interact with 1B7 are highlighted in green. The remaining residues are highlighted white to blue according to sequence identity. Nonconserved insertions are omitted for clarity. Phe364 and Glu426 are conserved in nearly all CDPKs. His365 is only present in TgCDPK1. Glu423 or the conservative aspartate mutant are present in most CDPKs. Positions 350 and 361 are considered the important sequence determinants. Although non–1B7-binding CDPKs may have a conservative mutation (i.e., K350R or T361S) at an individual position, none of these homologs have the correct identity at both of the sites. (B) Phylogenetic tree of TgCDPK1 homologs. (C) Table of sequences used.
Allosteric Inhibition of TgCDPK1 by 1B7.
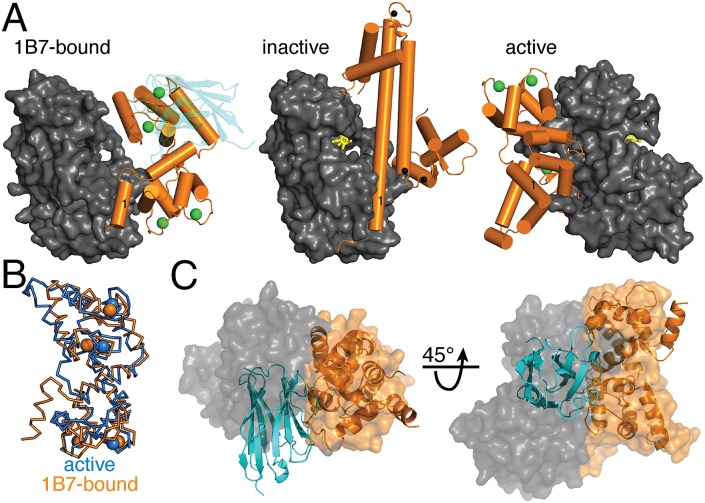
We aligned the three solved TgCDPK1 structures by their KDs to determine the mechanism through which 1B7 inhibits the kinase. The CADs from the Ca2+-depleted and Ca2+/1B7-bound conformations occupy the opposite face of the KD from where the CAD lies in the Ca2+-bound state (Fig. 5A). The first helix of the CAD (CH1) sits in the same cleft of the KD in the Ca2+/1B7-bound and Ca2+-depleted structures, although the helix terminates earlier in the Ca2+/1B7-bound structure. Superposition of the CAD domains shows that the Ca2+/1B7-bound CAD closely resembles the Ca2+-bound conformer (Fig. 5B).
Fig. 5.
1B7 prevents reorganization of the CAD to its activation-associated conformation. (A) Comparison of 1B7-bound TgCDPK1 to the inactive (3KU2) and active (3HX4) conformations. The three TgCDPK1 conformations are aligned by their KDs (gray), depicting the CAD (orange), 1B7 (blue), bound calcium (green), and unknown cations (black). ANP (phosphoaminophosphonic acid-adenylate ester) is built as yellow sticks to indicate the active site. The first helix of the CAD (CH1) is labeled with “1.” (B) Aligned carbon trace of the CADs from the active (blue) and 1B7-bound structures (orange). (C) Superposition of 1B7 and the bound CAD as ribbon diagrams over a space-filling model of the active structure.
The Ca2+-depleted conformation of TgCDPK1 has been proposed to be inhibited by an autoinhibitory triad between Lys338 in CH1 and the catalytic Glu135 and Asp138 and steric occlusion of the substrate binding site (11, 12). In contrast, Lys338 in our Ca2+/1B7-bound structure participates in interactions within the CAD. We observed the formation of a disulfide bond linking Cys247 in the KD to Cys505 in the CAD in our Ca2+/1B7-bound structure (Fig. S7A). Addition of a reducing agent did not affect 1B7 inhibition of kinase activity (Fig. S7B), although crystals were not obtained in the presence of 1 mM DTT. It is therefore likely that the Ca2+/1B7-bound CAD is sampling many conformations in solution, including the state captured by the oxidized crystal structure, making steric occlusion of the substrate-binding pocket an unlikely mechanism for 1B7-mediated inhibition of TgCDPK1. The Ca2+/1B7-bound kinase may be inactive because 1B7 and the KD bind to the same face of the CAD in the active conformation (Fig. 5C). This would imply an unprecedented role for the interaction of the CAD with the KD during activation.
Fig. S7.
A disulfide bond stabilizes the structure of Ca2+/1B7-bound TgCDPK1. (A) Disulfide bond (yellow) between Cys247 of the KD (gray) and Cys505 of the CAD (orange) rendered as sticks in the Ca2+/1B7-bound TgCDPK1 structure. (B) 1B7 inhibition of TgCDPK1 is not affected by reducing conditions. Addition of up to 10 mM DTT did not affect kinase activity or inhibition by 1B7. Kinase and 1B7 were assayed at 100 nM and 150 nM, respectively. Values are normalized to maximal WT activity. Mean ± SD, representative experiment.
Molecular Dynamics Show That the KD of TgCDPK1 Is Intrinsically Unstable.
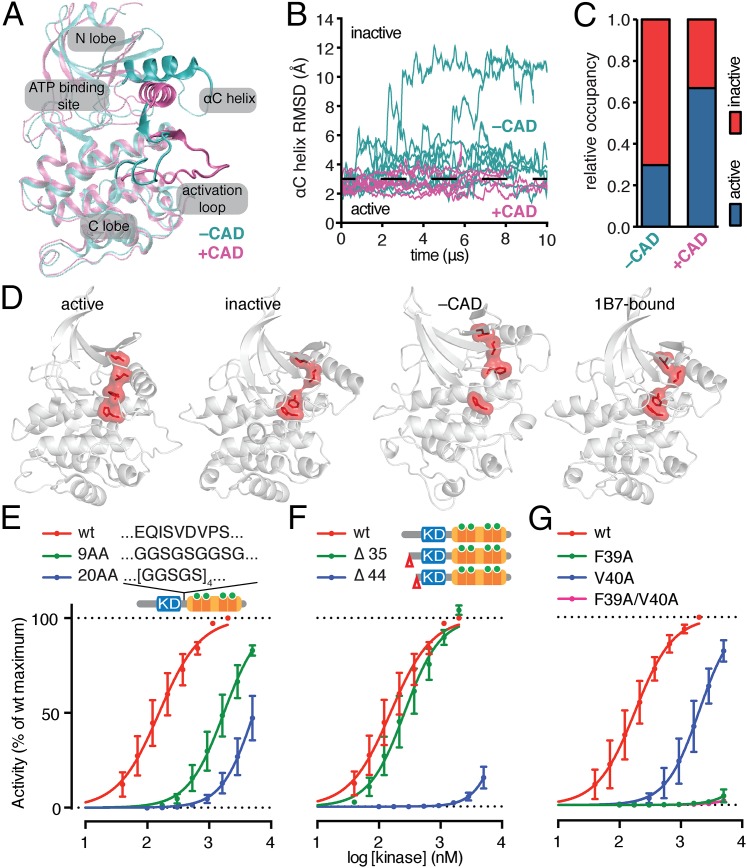
Long-timescale molecular dynamics (MD) simulations have been used to assess the intrinsic stability of various tyrosine kinases and inform their mechanisms of activation (18). To model the interaction of the CAD and the KD, we performed MD simulations using the Ca2+-bound conformation of TgCDPK1 as the starting point. Simulations performed in the absence of the CAD showed that the KD diverged from that of the Ca2+-bound, active structure (Fig. 6A). The major changes were in the rearrangement of the activation loop and in an approximate 30° turn in the αC helix of the KD to a so-called “αC-out” conformation that dissolves the catalytically important salt bridge between Lys80 and Glu99 and marks the inactive state of many protein kinases. We noted a clear preference for the inactive state when the CAD was removed from the KD (Fig. 6 B and C).
Fig. 6.
The KD of TgCDPK1 is intrinsically inactive and stabilized by the Ca2+-bound CAD. (A) Superposition of the active conformation of the TgCDPK1 KD with an inactive conformation obtained from a simulation without the CAD, in which the KD readily departed from the (starting) active conformation. The superposition is obtained using the C-lobe backbone atoms. (B) The rmsd trace of the αC helix relative to the (starting) active conformation in simulations of the TgCDPK1 KD with or without the CAD. Conformations with an rmsd less than 3 Å are considered active. (C) The relative occupancy of the catalytically active conformation in simulations of the TgCDPK1 KD with or without the CAD. As shown, the presence of the CAD stabilizes the active conformation. (D) Structure of the KD highlighting the integrity of the R-spine (red) in the Ca2+-bound (active) conformation, and its disruption in the Ca2+-depleted (inactive), the simulation without the CAD (–CAD), and the Ca2+/1B7-bound (1B7-bound) conformations. (E) Kinase activity of TgCDPK1 mutants carrying different flexible linkers between the KD and CAD domains. (F) Kinase activity of different N-terminal truncations removing the first 35 (Δ35) or 44 (Δ44) amino acids preceding the KD. (G) Kinase activity of point mutants in the N-terminal extension, alone or in combination. All kinase experiments plotted as means ± SEM, n = 3 independent experiments.
The “regulatory” spine (R-spine) is a series of noncontiguous hydrophobic residues (Leu103, Leu114, His172, and Phe196 in TgCDPK1) that are aligned in the active conformations of all protein kinases (19). Misalignment of the R-spine residues is a key feature of the inactive states found in the MD simulation in the absence of the CAD and the Ca2+/1B7-bound structure (Fig. 6D). The R-spine was similarly disrupted in the Ca2+-depleted structure, in contrast to its perfect alignment in the Ca2+-bound conformation (Fig. 6D). The Ca2+-bound CAD appears to impart an active conformation on the KD by stabilizing the position of the αC helix required for alignment of the R-spine and catalysis.
The CAD Acts as a Molecular Splint to Stabilize the KD of TgCDPK1.
During activation, the CAD imposes a 15° rotation in the relative orientations of the N and C lobes of the KD, suggesting that points of attachment to both kinase lobes are needed to generate torque upon Ca2+ binding. Having established that integrity of the tether between the KD and the CAD is necessary (Fig. 1B), we investigated whether the flexibility or length of the tether would affect kinase activity. We replaced nine residues from the tether with a flexible linker of equal length (9AA) or one 11 residues longer (20AA). We observed a loss of kinase activity proportional to the length and presumed flexibility of the tether (Fig. 6E). The greater loop entropy associated with the longer linkers presumably disfavors proper positioning of CAD and KD (20).
Interaction between the CAD and an N-terminal extension of the KD, also visible in the Ca2+-bound structure (11), could provide the proposed tether to the N lobe of the KD. Superposition of the Ca2+-bound and Ca2+/1B7-bound structures showed that the same groove in the Ca2+-bound CAD was engaged by both the N-terminal extension and CDR3 of 1B7 (Fig. S8). We therefore asked whether the N-terminal extension of TgCDPK1 participates in kinase activation. Of the 50 residues in the N-terminal extension, only the final 20 are observed in the Ca2+-bound structure, and only the final six in both inactive structures. Although truncation of the first 35 residues minimally affected kinase activity, elimination of 44 residues from the N-terminal extension led to a complete loss of function (Fig. 6F).
Fig. S8.
Superposition of the Ca2+/1B7-bound and Ca2+-bound structures. (A) The two structures are shown individually and superimposed. For clarity, the KD of the Ca2+/1B7-bound structure is not shown in the superimposed image. (B) Detailed view of the predicted clash between the N-terminal extension (red) in the active, Ca2+-bound structure and CDR3 of the VHH (blue). The KD (gray) and CAD (yellow) from the active structure are shown.
We identified two key residues important for this interaction: Phe39 and Val40. We mutated Phe39 and Val40 individually and in combination to Ala and measured their effects. Val40Ala shifted the EC50 of the kinase by ∼10-fold, whereas Phe39Ala strongly inhibited kinase activity, as did the double mutation (Fig. 6G). The N-terminal extension is therefore crucial for the activation of TgCDPK1.
Allosteric Activation Is Required for TgCDPK1 Function in Vivo.
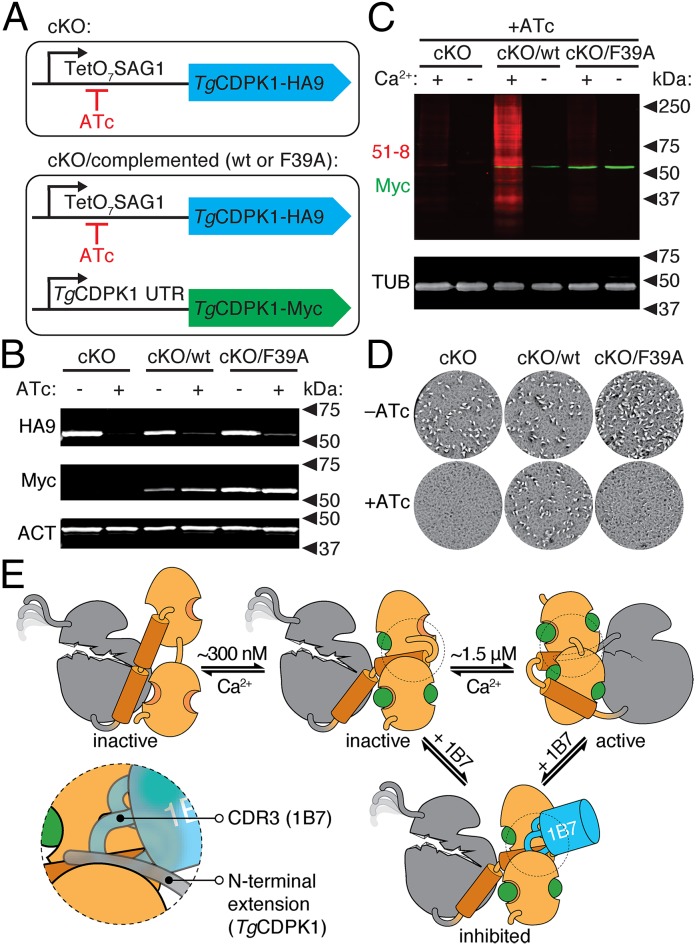
To examine the role of the N-terminal extension in vivo, we used a conditional knockout (cKO) of TgCDPK1 (2). This strain uses a tetracycline transactivator to express an HA9-tagged allele of TgCDPK1, in the absence of the endogenous gene (Fig. 7A). Growth of the cKO strain in the presence of anhydrotetracycline (ATc) blocks expression of TgCDPK1, which was used to demonstrate the essentiality of the kinase in the T. gondii lytic cycle (2). The cKO can be complemented with an allele of the kinase constitutively expressed under the endogenous promoter of TgCDPK1 (Fig. 7A). We generated strains complemented either with wild type or with the Phe39Ala mutant incapable of allosteric activation. We confirmed regulation of the HA9-tagged allele and constitutive expression of the complementing, Myc-tagged alleles by immunoblot (Fig. 7B). To determine whether the expressed enzymes were active, we performed thiophosphorylation reactions, using 6-Fu-ATPγS as a substrate, with lysates from parasites grown for 48 h in the presence of ATc. We observed robust Ca2+-dependent thiophosphorylation in the wild-type complemented strain. However, consistent with our analysis of the recombinant enzymes, no TgCDPK1 activity was observed in the Phe39Ala mutant strain (Fig. 7C).
Fig. 7.
Allosteric activation is required for TgCDPK1 activity in vivo. (A) Strategy depicting the TgCDPK1 cKO carrying an HA9-tagged allele that can be shut down by addition of ATc. The complemented strains additionally carry a constitutive Myc-tagged allele (WT or F39A), expressed under the endogenous TgCDPK1 promoter. (B) Immunoblot of regulatable (HA9) or constitutive (Myc) TgCDPK1 alleles in the different strains grown in the presence or absence of ATc for 48 h. Parasite actin (ACT) is included as a loading control. (C) TgCDPK1-dependent thiophosphorylation (51-8; red) in lysates from the various parasite strains following growth in the presence of ATc. Reactions were performed in the presence or absence of Ca2+. Tubulin is included as a loading control. (D) Plaque formation by the various strains in the presence or absence of ATc. (E) Model for the activation of TgCDPK1 and its inhibition by 1B7. The KD (gray) is intrinsically inactive and its catalytic site is occluded by the CAD (orange). At ∼300 nM Ca2+, the CAD is partially occupied by Ca2+ and can bind 1B7 (blue). Higher Ca2+ concentrations are required for full Ca2+ occupancy of the CAD and stabilization of the KD, which leads to activation. The region of the CAD competitively bound by 1B7 and the N-terminal extension is circled and highlighted in the Inset.
We next measured plaque formation to assess functional complementation of the cKO. All strains formed plaques of similar size when grown in the absence of ATc (Fig. 7D). However, plaque formation was impaired when the cKO was grown in the presence of ATc (2). This defect was complemented by constitutive expression of the wild-type allele, but not the Phe39Ala mutant (Fig. 7D). Taken together, these data demonstrate that the Phe39 residue of TgCDPK1, which mediates allosteric activation, is necessary for the function of the kinase in vivo.
Discussion
Unlike many other Ca2+-regulated kinases, the KD of T. gondii CDPK1 is intrinsically inactive. We identified an alpaca-derived single domain antibody (1B7) that binds to and inhibits TgCDPK1 in a reversible, Ca2+-dependent manner. Solving the structure of TgCDPK1 in complex with 1B7 showed a common binding site on the Ca2+-bound CAD for both the antibody and the TgCDPK1 N-terminal extension. 1B7 binding prevents the stabilization of the KD by blocking the interaction of the N-terminal extension and the CAD. We refer to this mode of stabilization as a molecular splint because anchoring of the KD to the CAD at both the N and C termini is required for activation.
Ca2+-regulated kinases are activated by removal of an inhibitory element from the KD (7, 8, 13, 14). Animal Ca2+/CaMKs possess intrinsically active KDs, kept inactive by an autoinhibitory region (13, 14). A similar autoinhibitory region is part of the CAD of CDPKs (11). Peptide mimetics of this region inhibit both plant (7, 8) and parasite kinases (6, 21). Based on observations of plant CDPK truncations that appeared constitutively active and Ca2+-independent, CDPK KDs—like those of CaMKs—were thought to be intrinsically active (7, 21). These truncations consisted of N-terminal fusions with GST, which can dimerize and stabilize the conformation of its fusion partners. For phosphatidylinositol 3′-kinase and the insulin receptor KD, fusion to GST is sufficient for activation (22, 23). In contrast, the KD of TgCDPK1 is inactive on its own. Removal of the CAD, either genetically or proteolytically, fails to activate TgCDPK1. MD simulations showed that the KD adopts an inactive conformation in the absence of the CAD. The KDs of different CDPKs might well differ in stability, but our work suggests that instability is a conserved feature of the CDPKs most closely related to TgCDPK1.
Ca2+ binding to the CAD imposes rigidity to its fold. When connected to the N and C termini of the KD, the CAD acts as a molecular splint that supports and stabilizes the KD in its active conformation (Fig. 7E). The CAD therefore exerts a far greater degree of regulation over the KD than assumed previously. Perturbation of either point of contact between the CAD and KD impairs kinase activity. Disruption of the N-terminal extension (Fig. 6 F and G) strongly inhibited kinase activity, congruent with studies of PfCDPK1 (24). We identified Phe39 and Val40 in the N-terminal extension as crucial for kinase activation. The presence of Phe39 is required for TgCDPK1 function in vivo, because the Phe39Ala mutant, when expressed at wild-type levels, fails to complement a cKO of the kinase (Fig. 7). The similarity of these anchor residues among diverse CDPKs suggests that the corresponding regions may function like the N-terminal extension of TgCDPK1.
KDs have evolved diverse modes of positive and negative regulation, often through acquisition of additional protein domains or noncovalent interacting partners. The diversity of these regulatory systems often converges on similar themes, including regulation of the αC helix position via a hydrophobic patch in the N lobe of the KD (25). Regulatory elements can interact directly with this hydrophobic patch, as is the case with the nonreceptor tyrosine kinases Fes and Abl (26). In the case of Abl, the same regulatory domain can also act to lock the kinase in an inactive conformation (27). Only a few domains are known to exert dual functions in the regulation of kinase activity, by serving both as repressors and activators. In a remarkable convergence, the CAD has adopted such a role in the regulation of TgCDPK1.
The interaction of 1B7 with its target is characteristic of VHHs, which often present large convex paratopes that can access buried epitopes less accessible to conventional antibodies (28). The 1B7 CDR3 protrudes into a groove that forms upon Ca2+ binding to the CAD, displacing the N-terminal extension in the active conformation of the kinase (11). Features associated with autoinhibition are notably absent from the Ca2+/1B7-bound structure. 1B7 may bind an intermediate state in the activation of TgCDPK1 as inferred from the lower [Ca2+] required for 1B7 binding, versus kinase activation (Fig. 2E). Preventing the stabilization of the KD is the primary mechanism for the inhibition of TgCDPK1 by 1B7, a trait generalizable at least to the clade of CDPKs to which TgCDPK1 belongs.
CDPKs are obvious therapeutic targets. Development of small molecule inhibitors against them has focused exclusively on ATP analogs, which often suffer from off-target effects against other host and parasite kinases. Targeting the atypical ATP-binding pocket of TgCDPK1 and related kinases has afforded some measure of specificity. However, a single mutation suffices to confer resistance to these compounds (3). We identify a vulnerability in this family of parasite kinases, exposed by their need for stabilization. Through the use of 1B7, we provide proof of principle that this feature can be exploited to inhibit TgCDPK1 and related kinases, including PfCDPK1, a kinase essential for the erythrocytic stages of the malaria parasite (6). Conservation of the residues that mediate allosteric activation suggests that this mechanism may extend to Cryptosporidium CDPK1 and Plasmodium CDPK4. Inhibition of these kinases has already shown promising activity in murine infection models, in the case of CpCDPK1 (29), and in blocking the transmission of malaria, in the case of PbCDPK4 (30). We expect that the binding of 1B7 to TgCDPK1 can be used to guide the design of small molecules with the therapeutic potential to allosterically inhibit these essential parasite kinases.
Methods
Protein Purification.
Vectors encoding TgCDPK1-6xHis and TgCDPK3-6xHis were generated as previously described (3). Mutations in the EF hands or the N-terminal extension were introduced using the QuikChange II Site-Directed Mutagenesis Kit (Agilent Technologies), with specific primers designed according to the manufacturer’s instructions. N-terminally truncated versions of TgCDPK1 were generated using primers P1 and P2 (Δ35) or P1 and P3 (Δ44). Mutant kinases with insertions in the junction between KD and CAD were ordered as gBlocks from Integrated DNA Technologies (B1 for 9AA, B2 for 20AA, and B3 for 3C) and cloned into pET22b+ (Novagen). Primer and gBlock sequences are listed in Table S2.
Table S2.
Primer and gBlock sequences
| ID | Sequence |
| P1 | 5′GCGCTCGAGGTTTCCGCAGAGCTTCAAGAGC3′ |
| P2 | 5′GCGCATATGATGGGGCCCGGGATGTTCGTTCAGCA3′ |
| P3 | 5′GCGCATATGATGGGGGCGATCTTCTCCGACCGGTA3′ |
| P4 | 5′GGGTTTCATATGCAGGTGCAGCTCGTGGAGACG3′ |
| P5 | 5′CACCCCCTCGAGACCACCGGTTTCCGGCAGCCCACCTCCTGAGGAGACGGTGACCTGGGTCCC3′ |
| B1 | 5′caTATGATGGGGCAGCAGGAAAGCACTCTTGGGGGTGCGGCCGGGGAGCCTCGCTCGCGCGGTCATGCGGCGGGGACCAGCGGTGGACCAGGAGACCATCTCCACGCGACGCCCGGGATGTTCGTTCAGCATTCGACTGCGATCTTCTCCGACCGGTACAAGGGACAGCGGGTGTTGGGGAAGGGGTCTTTTGGTGAGGTGATTCTGTGCAAAGACAAGATCACCGGTCAGGAGTGTGCGGTGAAGGTCATTAGCAAGCGCCAAGTGAAGCAGAAGACGGACAAGGAGTCTCTGCTTCGCGAGGTGCAGTTGCTGAAGCAGCTGGACCACCCCAACATCATGAAGCTGTATGAATTCTTCGAGGACAAAGGCTACTTCTACCTCGTCGGCGAAGTGTACACGGGAGGCGAGTTGTTCGACGAGATCATTTCCCGCAAGCGCTTCAGCGAAGTCGATGCGGCGCGGATCATCCGCCAAGTCCTCAGCGGCATCACGTACATGCACAAAAATAAAATCGTACATCGGGACCTCAAACCGGAAAACCTGCTCCTGGAAAGCAAAAGCAAGGACGCGAACATCCGCATCATCGACTTTGGCCTCAGCACTCACTTCGAAGCGAGCAAGAAGATGAAGGACAAAATCGGAACTGCATACTACATCGCACCCGAGGTCCTCCACGGCACTTACGACGAGAAATGCGACGTATGGTCGACCGGTGTTATTCTCTACATCCTTCTTTCCGGATGCCCACCCTTCAATGGAGCAAACGAGTACGACATCCTGAAGAAGGTTGAGAAAGGCAAGTACACCTTTGAACTGCCTCAGTGGAAGAAAGTGTCAGAGAGCGCAAAAGATTTGATTCGCAAAATGCTGACCTACGTCCCCAGCATGAGAATCAGCGCGCGAGATGCTCTGGACCACGAGTGGATCCAGACCTACACGAAGGAGCAGATCAGCCTGGAAGTTCTGTTCCAGGGGCCCGTGGACGTTCCGTCTCTGGACAACGCCATTCTCAACATCCGACAGTTCCAAGGCACTCAGAAGCTCGCGCAAGCTGCGCTGCTCTACATGGGCTCGAAACTGACAAGCCAAGACGAAACGAAGGAACTGACGGCCATCTTCCATAAGATGGACAAGAATGGAGACGGGCAACTGGACCGTGCAGAGCTCATCGAAGGGTACAAGGAGTTGATGCGGATGAAGGGCCAAGATGCGAGCATGCTCGACGCGAGCGCTGTTGAACACGAAGTTGACCAGGTCTTGGACGCAGTCGACTTCGACAAAAACGGCTACATCGAGTACTCTGAGTTCGTCACCGTGGCGATGGACAGAAAGACGCTGCTTTCGCGAGAACGCCTGGAGCGCGCCTTCCGGATGTTTGACTCCGACAACTCAGGAAAGATTTCTTCCACTGAGCTGGCCACCATCTTTGGCGTCTCCGACGTGGACAGCGAAACATGGAAGAGCGTGCTGTCTGAGGTCGACAAGAATAACGACGGCGAAGTCGACTTTGACGAGTTTCAACAGATGCTCTTGAAGCTCTGCGGAAACCtCGAG3′ |
| B2 | 5′CATATGATGGGGCAGCAGGAAAGCACTCTTGGGGGTGCGGCCGGGGAGCCTCGCTCGCGCGGTCATGCGGCGGGGACCAGCGGTGGACCAGGAGACCATCTCCACGCGACGCCCGGGATGTTCGTTCAGCATTCGACTGCGATCTTCTCCGACCGGTACAAGGGACAGCGGGTGTTGGGGAAGGGGTCTTTTGGTGAGGTGATTCTGTGCAAAGACAAGATCACCGGTCAGGAGTGTGCGGTGAAGGTCATTAGCAAGCGCCAAGTGAAGCAGAAGACGGACAAGGAGTCTCTGCTTCGCGAGGTGCAGTTGCTGAAGCAGCTGGACCACCCCAACATCATGAAGCTGTATGAATTCTTCGAGGACAAAGGCTACTTCTACCTCGTCGGCGAAGTGTACACGGGAGGCGAGTTGTTCGACGAGATCATTTCCCGCAAGCGCTTCAGCGAAGTCGATGCGGCGCGGATCATCCGCCAAGTCCTCAGCGGCATCACGTACATGCACAAAAATAAAATCGTACATCGGGACCTCAAACCGGAAAACCTGCTCCTGGAAAGCAAAAGCAAGGACGCGAACATCCGCATCATCGACTTTGGCCTCAGCACTCACTTCGAAGCGAGCAAGAAGATGAAGGACAAAATCGGAACTGCATACTACATCGCACCCGAGGTCCTCCACGGCACTTACGACGAGAAATGCGACGTATGGTCGACCGGTGTTATTCTCTACATCCTTCTTTCCGGATGCCCACCCTTCAATGGAGCAAACGAGTACGACATCCTGAAGAAGGTTGAGAAAGGCAAGTACACCTTTGAACTGCCTCAGTGGAAGAAAGTGTCAGAGAGCGCAAAAGATTTGATTCGCAAAATGCTGACCTACGTCCCCAGCATGAGAATCAGCGCGCGAGATGCTCTGGACCACGAGTGGATCCAGACCTACACGAAGGGCGGTTCTGGCAGCGGCGGTTCTGGCCTGGACAACGCCATTCTCAACATCCGACAGTTCCAAGGCACTCAGAAGCTCGCGCAAGCTGCGCTGCTCTACATGGGCTCGAAACTGACAAGCCAAGACGAAACGAAGGAACTGACGGCCATCTTCCATAAGATGGACAAGAATGGAGACGGGCAACTGGACCGTGCAGAGCTCATCGAAGGGTACAAGGAGTTGATGCGGATGAAGGGCCAAGATGCGAGCATGCTCGACGCGAGCGCTGTTGAACACGAAGTTGACCAGGTCTTGGACGCAGTCGACTTCGACAAAAACGGCTACATCGAGTACTCTGAGTTCGTCACCGTGGCGATGGACAGAAAGACGCTGCTTTCGCGAGAACGCCTGGAGCGCGCCTTCCGGATGTTTGACTCCGACAACTCAGGAAAGATTTCTTCCACTGAGCTGGCCACCATCTTTGGCGTCTCCGACGTGGACAGCGAAACATGGAAGAGCGTGCTGTCTGAGGTCGACAAGAATAACGACGGCGAAGTCGACTTTGACGAGTTTCAACAGATGCTCTTGAAGCTCTGCGGAAACCTCGAG3′ |
| B3 | 5′CATATGATGGGGCAGCAGGAAAGCACTCTTGGGGGTGCGGCCGGGGAGCCTCGCTCGCGCGGTCATGCGGCGGGGACCAGCGGTGGACCAGGAGACCATCTCCACGCGACGCCCGGGATGTTCGTTCAGCATTCGACTGCGATCTTCTCCGACCGGTACAAGGGACAGCGGGTGTTGGGGAAGGGGTCTTTTGGTGAGGTGATTCTGTGCAAAGACAAGATCACCGGTCAGGAGTGTGCGGTGAAGGTCATTAGCAAGCGCCAAGTGAAGCAGAAGACGGACAAGGAGTCTCTGCTTCGCGAGGTGCAGTTGCTGAAGCAGCTGGACCACCCCAACATCATGAAGCTGTATGAATTCTTCGAGGACAAAGGCTACTTCTACCTCGTCGGCGAAGTGTACACGGGAGGCGAGTTGTTCGACGAGATCATTTCCCGCAAGCGCTTCAGCGAAGTCGATGCGGCGCGGATCATCCGCCAAGTCCTCAGCGGCATCACGTACATGCACAAAAATAAAATCGTACATCGGGACCTCAAACCGGAAAACCTGCTCCTGGAAAGCAAAAGCAAGGACGCGAACATCCGCATCATCGACTTTGGCCTCAGCACTCACTTCGAAGCGAGCAAGAAGATGAAGGACAAAATCGGAACTGCATACTACATCGCACCCGAGGTCCTCCACGGCACTTACGACGAGAAATGCGACGTATGGTCGACCGGTGTTATTCTCTACATCCTTCTTTCCGGATGCCCACCCTTCAATGGAGCAAACGAGTACGACATCCTGAAGAAGGTTGAGAAAGGCAAGTACACCTTTGAACTGCCTCAGTGGAAGAAAGTGTCAGAGAGCGCAAAAGATTTGATTCGCAAAATGCTGACCTACGTCCCCAGCATGAGAATCAGCGCGCGAGATGCTCTGGACCACGAGTGGATCCAGACCTACACGAAGGGCGGTTCTGGCAGCGGtGGcagtGGCAGCGGCGGTTCaGGCAGCGGCGGTTCTGGtAGCCTGGACAACGCCATTCTCAACATCCGACAGTTCCAAGGCACTCAGAAGCTCGCGCAAGCTGCGCTGCTCTACATGGGCTCGAAACTGACAAGCCAAGACGAAACGAAGGAACTGACGGCCATCTTCCATAAGATGGACAAGAATGGAGACGGGCAACTGGACCGTGCAGAGCTCATCGAAGGGTACAAGGAGTTGATGCGGATGAAGGGCCAAGATGCGAGCATGCTCGACGCGAGCGCTGTTGAACACGAAGTTGACCAGGTCTTGGACGCAGTCGACTTCGACAAAAACGGCTACATCGAGTACTCTGAGTTCGTCACCGTGGCGATGGACAGAAAGACGCTGCTTTCGCGAGAACGCCTGGAGCGCGCCTTCCGGATGTTTGACTCCGACAACTCAGGAAAGATTTCTTCCACTGAGCTGGCCACCATCTTTGGCGTCTCCGACGTGGACAGCGAAACATGGAAGAGCGTGCTGTCTGAGGTCGACAAGAATAACGACGGCGAAGTCGACTTTGACGAGTTTCAACAGATGCTCTTGAAGCTCTGCGGAAACCTCGAG3′ |
Kinase constructs were transformed into BL21(DE3)V2RpAcYc-LIC+LamP E. coli, which express the LamP phosphatase as previously described (11). Cultures were grown to 0.6 OD600 at 37 °C and induced with 1 mM isopropyl-β-D-thiogalactopyranoside (IPTG) for 2.5 h. Pelleted cells were lysed in CelLytic B buffer (Sigma) containing 0.1 mg⋅mL−1 lysozyme (Sigma), 650 units benzonase (Sigma), and HALT Protease Inhibitor Mixture (Thermo-Scientific). Lysates were clarified by centrifugation at 24,000 × g for 30 min at 4 °C. The soluble fraction was added to prewashed Ni-NTA agarose (Qiagen) and allowed to bind 1 h at 4 °C, rotating. The resin was washed with 50 mM sodium phosphate pH 8.0, 500 mM NaCl, 15 mM imidazole, and HALT Protease Inhibitor Mixture, and bound proteins were eluted by increasing imidazole to 500 mM. Eluates were concentrated using Amicon Centrifugal Filters (EMD Millipore) and loaded onto a Superdex 200 26/60 SEC column equilibrated with 50 mM Tris, pH 7.5, and 150 mM NaCl. Peak elution fractions were pooled and concentrated. Protein purity was assessed by SDS/PAGE, and concentrations were determined using the DC Protein Assay (Bio-Rad). Recombinant CDPKs were stored at –80 °C after addition of 25% (vol/vol) glycerol and 1 mM DTT.
For crystallization, the following changes were made. Proteins were expressed in BL21(DE3)-LOBSTR-RIL E. coli, which has been optimized for low background in Ni-affinity purifications (31). Cultures were shifted to 18 °C before induction with 0.2 mM IPTG for 16 h. Cells were lysed using a cell disruptor (Constant Systems) in 50 mM potassium phosphate, pH 8.0, 500 mM NaCl, 30 mM imidazole, 3 mM β-mercaptoethanol, 1 mM PMSF, and 250 units of Turbonuclease (Eton Bioscience). The soluble fraction was added to prewashed Ni-Sepharose fast-flow beads (GE Healthcare) and agitated 20 min at 4 °C. Protein was eluted with 250 mM imidazole, pH 8.0, 150 mM NaCl, and 3 mM β-mercaptoethanol. SEC was performed in 10 mM Tris, pH 7.4, 150 mM NaCl, 1 mM DTT, and 1 mM CaCl2. SeMet-derivatized TgCDPK1 was purified identically, with the exception that 10 mM β-mercaptoethanol and 5 mM DTT were used in purification steps.
Upon VHH selection by phage display and ELISA, the 1B7 coding sequence was subcloned for expression. First, VHH coding sequences were amplified with primers P4 and P5, which include a sortase A recognition motif. The resulting product was subcloned into the pET30b+ (Novagen) expression vector using NdeI and XhoI sites for inclusion of a C-terminal 6xHis tag. BL21(DE3) E. coli containing the plasmid were grown to midlog phase at 37 °C in 2YT plus kanamycin and induced with 1 mM IPTG overnight at 30 °C. Total soluble protein was collected by French press and cleared at 39,000 × g for 30 min, before loading onto Ni-NTA (Qiagen) in 50 mM Tris, pH 7.5, 150 mM NaCl, and 10 mM imidazole. Protein was eluted in 50 mM Tris, pH 7.5, 150 mM NaCl, 500 mM imidazole, and 10% (vol/vol) glycerol, and then loaded onto a Superdex 75 10/300 column in 50 mM Tris, pH 7.5, 150 mM NaCl, and 10% (vol/vol) glycerol. Peak fractions were pooled and concentrated. Purity was assessed by SDS/PAGE.
The TgCDPK1 variant carrying a human rhinovirus 3C protease cleavage site was digested with 3C protease for 36 h at 4 °C to achieve near-complete digestion. Wild-type kinase underwent the same treatment, to be used as a control. KD and CAD were separated from digested samples by SEC through a Superdex 75 16/60 column equilibrated with 50 mM Tris, pH 7.5, 150 mM NaCl, 1 mM CaCl2, and 1 mM DTT. Fractions for each peak were pooled and concentrated as described above.
Growth of Host Cells and Parasite Strains.
T. gondii tachyzoites were maintained by growth in monolayers of human foreskin fibroblasts cultured in DMEM containing 10% (vol/vol) tetracycline-free FBS (HyClone), 2 mM glutamine, 10 mM Hepes pH 7.5, and 20 µg⋅mL−1 gentamicin, as described (2). The TgCDPK1 cKO was generated as previously described (2). The wild-type complementation plasmid was previously generated and contains a Myc-tagged version of TgCDPK1 downstream of the endogenous promoter sequence of the kinase (2). Phe39 was mutated to Ala in this construct using the QuikChange II Site-Directed Mutagenesis Kit (Agilent Technologies), according to the manufacturer’s instructions. Mutagenesis and integrity of the entire ORF was confirmed by sequencing. The complementing plasmids were cotransfected with pDHFR-TS (32), and stable integration was selected for by growth in 3 µM pyrimethamine (Sigma). Clones were isolated by limiting dilution, and expression of the transgenes was confirmed by immunoblot. We added 1.5 µg⋅mL−1 ATc (Clontech) as indicated to suppress expression of the regulatable, HA9-tagged wild-type allele.
Plaque Assays.
Assays were performed as previously described (2). Confluent monolayers of human foreskin fibroblasts in six-well plates were infected with 400–1,000 parasites per well in media with or without 1.5 µg⋅mL−1 ATc. At 24 h after infection, additional medium was added to decrease the concentration of ATc to 1 µg⋅mL−1. Monolayers were fixed 7 d after infection and stained with crystal violet.
Immunoblots.
Parasites from fully lysed monolayers were lysed directly in SDS/PAGE loading buffer, or in a 50 mM Tris, pH 7.5, 150 mM NaCl, 1% Triton X-100, and HALT Protease Inhibitor Mixture, before measuring protein concentration. Serum was collected from each alpaca before immunization and stored at –80 °C (33). T. gondii lysates were run on SDS/PAGE and transferred to nitrocellulose. Membranes were blocked overnight at 4 °C in PBS, 5% (wt/vol) nonfat dry milk. Serum was diluted 1:5,000 in blocking solution and incubated 1 h with the respective section of the blot. Horseradish peroxidase-conjugated goat anti-llama IgG (Bethyl) was used as a secondary antibody at a 1:20,000 dilution in blocking solution. To compare T. gondii lysates with recombinant proteins, lysates corresponding to different numbers of parasites were loaded besides dilutions of the recombinant proteins. 1B7 was labeled using the LI-COR IRDye 800CW labeling kit according to the manufacturer’s instructions and used at a concentration of 1 µg⋅mL−1 for immunoblots. Expression of the regulatable and complementing alleles of TgCDPK1 was similarly assessed by immunoblot following growth of the parasites in the presence or absence of ATc. Immunoblots were probed, as indicated, with Rabbit anti-TgACT1 (34) diluted 1:10,000, mouse anti-penta HIS (Qiagen) diluted 1:5,000, rabbit anti-HA9 (Invitrogen) diluted 1:10,000, mouse anti-Myc (9E10; Sigma) diluted 1:5,000, and mouse anti-αTubulin (12G10; Developmental Studies Hybridoma Bank) diluted 1:10,000.
VHH Library Generation.
Alpacas (V. pacos) were purchased locally, maintained in pasture, and immunized following a protocol authorized by the Tufts University Cummings Veterinary School Institutional Animal Care and Use Committee. For each alpaca, total RNA was isolated from ∼106 fresh peripheral blood lymphocytes using the RNeasy Plus Mini Kit (Qiagen), following the manufacturer’s instructions. First strand cDNA synthesis was performed using SuperScript III reverse transcriptase (Life Technologies) and a combination of oligo dT, random hexamer, or Ig-specific primers, AlCH2 and AlCH2.2, as previously described (33). Subsequent PCR amplification of VHH sequences and phage library generation followed an established procedure (33). This included the use of alpaca-specific primers for VHH gene amplification and the use of a phagemid vector adapted for VHH expression as a pIII fusion. Following transformation into E. coli TG1 cells (Agilent), the total number of independent clones for each library was estimated to range from 1 × 106–107. The resulting phagemid libraries were stored at –80 °C.
Generation of M13 Phage Displaying Anti-T. gondii VHH Library.
Several independent libraries were generated from animals immunized with different mixtures of antigens. None of the animals were deliberately immunized with T. gondii. Libraries were pooled and used to inoculate 100 mL SOC Medium with 50 µg⋅mL−1 ampicillin. The culture was grown to midlog phase and infected with 100 µL 1014 pfu⋅mL−1 VCSM13 helper phage. Following a 2-h incubation at 37 °C, the cells were harvested by centrifugation and resuspended in 100 mL 2YT, 0.1% glucose, 50 µg⋅mL−1 kanamycin, and 50 µg⋅mL−1 ampicillin. Cultures were incubated overnight at 30 °C, then centrifuged for 20 min at 7,700 × g, followed by phage precipitation from the resulting supernatant with 1% PEG-6000, 500 mM NaCl at 4 °C, and resuspension in PBS.
Selection of VHHs by Phage Display.
VHHs were selected by panning against recombinant TgCDPK1. We biotinylated 100 µg recombinant TgCDPK1 by coupling Chromalink NHS-biotin reagent (Solulink) to primary amines for 90 min in 100 mM sodium phosphate, pH 7.4, 150 mM NaCl. Unreacted biotin was removed using a 10 kDa molecular weight cut-off concentrator (Millipore). Biotin incorporation was monitored, following the manufacturer’s guidelines. We blocked 100 µL MyOne Streptavidin-T1 Dynabeads (Life Technologies) in PBS, 2% (wt/vol) BSA (Sigma) for 2 h at 37 °C. We added 20 µg biotinlyated TgCDPK1 in PBS to the blocked beads and incubated it for 30 min at 25 °C, with agitation. The beads were then washed three times in PBS, and 200 µL of 1014 pfu·mL−1 M13 phage displaying the VHH library were added in PBS, 2% (wt/vol) BSA for 1 h at room temperature. The beads were then washed 15 times with PBS, 0.1% Tween-20. Phage was eluted by the addition of E. coli ER2738 (NEB) for 15 min at 37 °C, followed by elution with 200 mM glycine, pH 2.2, for 10 min at 25 °C. The eluate was neutralized; pooled with the E. coli culture; plated onto 2YT agar plates supplemented with 2% (wt/vol) glucose, 5 µg·mL−1 tetracycline, and 10 µg·mL−1 ampicillin; and grown overnight at 37 °C. A second round of panning was performed with the following modifications: 2 µg of biotinylated TgCDPK1 was used as bait and incubated with 2 µL 1014 pfu/mL M13 phage displaying the first-round VHH library for 15 min at 37 °C, followed by extended washes in PBS, 0.1% Tween-20.
ELISA.
Following two rounds of phage panning, 96 colonies were isolated in 96-well round-bottom plates and grown to midlog phase at 37 °C in 200 µL 2YT, 10 µg·mL−1 ampicillin, and 5 µg·mL−1 tetracycline, induced with 3 mM IPTG and grown overnight at 30 °C. Plates were centrifuged at 12,000 × g for 10 min, and 100 µL of supernatant was mixed with an equal volume of PBS, 5% (wt/vol) nonfat dry milk. This mixture was added to an ELISA plate coated with 1µg·mL−1 TgCDPK1. Following four washes in PBS, 1% Tween-20, anti-llama–HRP antibody (Bethyl) was added at a 1:10,000 dilution in PBS, 5% (wt/vol) nonfat dry milk for 1 h at 25 °C. The plate was developed with fast kinetic 3,3′,5,5′-tetramethylbenzidine (Sigma) and quenched with 1 M HCl. Absorbance at 450 nm was determined in a plate reader (Spectramax; Molecular Devices).
IP of TgCDPK1 and TgCDPK3.
We coupled 4 mg of 1B7 to 500 mg of cyanogen bromide-activated Sepharose (Sigma) according to the manufacturer’s instructions. Coupling efficiency was monitored by SDS/PAGE. 1B7-Sepharose was stored in 50 mM Tris, pH 7.5, 150 mM NaCl at 4 °C. Approximately 109 parasites were harvested, resuspended in lysis buffer [50 mM Hepes, pH 7.4, 100 mM KCl, 1 mM MgCl2, 10% (vol/vol) glycerol, and 0.05% Triton X-100], and disrupted with a Dounce homogenizer. We prepared 0.25 g 1B7-Sepharose for each sample by washing with lysis buffer supplemented with either 1 mM EGTA or CaCl2. Half of the parasite lysate was diluted to 10 mL containing 1 mM EGTA or CaCl2 and incubated with the respective beads for 1 h at 4 °C, with agitation. The resin was then washed with 200 mL of the appropriate lysis buffer supplemented with EGTA or CaCl2 and eluted with three consecutive 500 µL volumes of lysis buffer containing 1 mM EGTA. The eluted material was analyzed by SDS/PAGE followed by silver staining. Bands were excised, reduced, alkylated, and digested with trypsin at 37 °C overnight. The resulting peptides were extracted, concentrated, and injected by standard reverse-phase chromatography into a Thermo Orbitrap Elite mass spectrometer operated in a data-dependent manner. The resulting fragmentation spectra were correlated against the known database using SEQUEST. Scaffold Q+S (Proteome Software) was used to provide consensus reports for the identified proteins.
The IP of recombinant, wild-type, and double EF hand mutant kinases was performed similarly. We diluted 5 µg of kinase in 1 mL of the respective binding buffer (50 mM Tris pH 7.5, 150 mM NaCl, 1% Triton X-100, supplemented with either 1 mM EGTA or 1 mM CaCl2, respectively) for 1 h. For each binding condition, 1B7-Sepharose beads were washed twice with the respective binding buffer, followed by incubation of ∼100 µg 1B7-Sepharose with 1 mL of diluted kinase for 30 min, with agitation. Following incubation, the beads were loaded onto 200 µL filter tips and washed with three column volumes of the respective buffer, before eluting with 40 µL 200 mM glycine, pH 2.2, followed by neutralization with 10 µL 1 M Tris, pH 9.0. The amount of immunoprecipitated kinase was determined by SDS/PAGE, SYPRO Ruby protein staining (Life Technologies), and fluorescent gel imaging using a Typhoon FLA 9500 laser scanner (GE Healthcare Life Sciences). Densitometry was conducted using ImageQuant TL software (GE Healthcare Life Sciences).
The IP of wild-type TgCDPK1 in the presence of specific [Ca2+]free concentrations was performed correspondingly, but with distinct buffer compositions. These binding buffers were made using the Calcium Calibration Buffer Kit (Life Technologies), adjusting the supplied buffer composition to 30 mM Mops, pH 7.2, 100 mM KCl, 4 mM CaEGTA/K2EGTA, 1 mM MgCl2, and 0.05% Tween-20 to more closely mimic physiological conditions and to assure consistency between the conditions used to measure kinase activity and 1B7 binding. The [Ca2+]free were determined using WEBMAX STANDARD (35). Before IP, wild-type TgCDPK1 was dialyzed against chelex-100 (Bio Rad)-treated buffer, to minimize contaminating the Ca2+ present in the buffers.
SEC.
Sortase was used to label 1B7 by incubating 1 mg of purified 1B7 with 1 mg Staphylococcus aureus sortase A and 1 mM GGGK-TAMRA in 50 mM Tris, pH 7.5, 150 mM NaCl, and 10 mM CaCl2. The reaction proceeded overnight at 25 °C, and unreacted VHH and sortase A were removed by adsorption onto Ni-NTA agarose (Qiagen). Labeled 1B7 was isolated by SEC, concentrated, and its purity determined as described above. TgCDPK1 and purified TAMRA-labeled 1B7 were incubated at a 1:1 molar ratio in 50 mM Tris, pH 7.5, and 150 mM NaCl, supplemented with 1 mM CaCl2 or 1 mM EGTA for 2 h at 25 °C. Samples were then loaded onto a Superdex 75 10/30 column in 50 mM Tris, pH 7.5, and 150 mM NaCl, supplemented with 1 mM CaCl2 or 1 mM EGTA, and absorbance was monitored at both 280 nm and 550 nm.
Kinase Assays.
Kinase assays were conducted using a peptide-based ELISA, as previously described (3). We coated 96-well plates with Syntide-2 peptide (Calbiochem) diluted to 100 µg·mL−1 in carbonate coating buffer (pH 9.6) at 4 °C overnight. Following incubation, plates were rinsed with wash buffer (50 mM Tris, pH 7.5, and 0.2% Tween-20) and blocked with 3% (wt/vol) BSA in wash buffer for 2 h at 25 °C. Subsequent washes were performed with wash buffer. Reactions were conducted at 30 °C for 30 min in kinase buffer (20 mM Hepes, pH 7.0, 10 mM MgCl2, 1 mM CaCl2, 0.005% Tween-20, and 1 mM ATP). Phosphorylation was detected with mAb MS-6E6 (MBL), followed by HRP-conjugated goat–anti-mouse IgG, developed with the substrate o-phenylenediamine dihydrochloride (Thermo Scientific), and detected by absorbance at 492 nm. Where appropriate, each kinase preparation was individually tested to determine its half maximal effective concentration (EC50). For VHH inhibition assays and Ca2+ titration experiments, TgCDPK1 was used at its EC50. For the inhibition assays, 1B7 or a control VHH (2A7) was titrated into the reaction, before the addition of 1 mM ATP. Ca2+ titration experiments used reaction buffers with specific [Ca2+]free, prepared as in the IP experiments, with 1 mM ATP added. EC50 and IC50 values were calculated from a dose–response curve plotted in Prism (GraphPad). Duplicate or triplicate experiments were run for all assays.
Thiophosphorylation Assay.
Thiophosphorylation was measured as previously described (17). Briefly, T. gondii parasites were lysed on ice in the reaction buffer (20 mM Hepes, pH 7.4, 137 mM KCl, and 10 mM MgCl2), supplemented with 1% Triton X-100. Individual aliquots of the lysate were combined with an equal volume of the reaction buffer supplemented with 8 mM CaEGTA alone or in combination with different amounts of 1B7. To start the reactions, an equal volume of reaction buffer supplemented with 1 mM GTP, 0.1 mM ATP, and 50 µM of either ATPγS or 6-Fu-ATPγS (BioLog) was added to each sample. Reactions were allowed to proceed for 30 min at 30 °C, before alkylating with 2 mM p-nitrobenzyl mesylate (Epitomics) for 1 h. Samples were analyzed by Western blot using a rabbit–anti-thiophosphate ester (51-8; Epitomics) at a dilution of 1:10,000 and an appropriate loading control. Goat–anti-rabbit conjugated to IRdye 680 (Li-Cor) was used to visualize the signal from the thiophosphate ester antibody. To compare TgCDPK1-dependent thiophosphorylation in the different T. gondii strains, lysates were prepared from strains grown in 1.5 µg·mL−1 for 48 h. Reactions were initiated as described above, except CaEGTA was substituted for K2EGTA in the samples without Ca2+.
Crystallization.
Recombinant TgCDPK1 and 1B7 were combined in a 1:1.5 molar ratio at room temperature, rotating for 1 h. The mixture was loaded on a Superdex 200 16/60 SEC column to separate complex from excess 1B7. The complex peak was pooled, concentrated, and dialyzed into 50 mM Tris, pH 7.4, and 150 mM NaCl for 16–20 h before crystal-tray setup. Initial crystals of TgCDPK1-1B7 complex grew via vapor diffusion at 4 mg·mL−1 in 25% (wt/vol) PEG 3350, 0.2 M (NH4)2SO4, and 0.1 M Bis-Tris, pH 6.5 at 18 °C, after 1 d of incubation in a 96-well, sitting drop tray (IndexHT, Hampton Research). Protein and mother liquor were mixed in a 1:1 ratio. The use of 4% (vol/vol) 1-propanol, as an additive, produced optically superior crystals. Scaling-up to a 24-well, hanging-drop format and immediate microseeding of the crystallization drop produced diffraction-quality crystals that grew as thin, mechanically separable plate clusters. SeMet-derivatized protein crystals were obtained following this protocol, albeit they grew singular, were smaller, and took several days longer to grow. Individual crystals were serially cryoprotected in mother liquor supplemented with 20% (vol/vol) glycerol or 35% (wt/vol) PEG 3350 before cooling in liquid nitrogen.
Data Processing and Structure Determination.
Complete native and SeMet datasets were collected at the Advanced Photon Source user end station 24-IDC. Data reduction was performed in HKL2000 (36). MR and SAD phasing was performed using phaser in the PHENIX suite (37), available through SBGrid (38). An initial MR solution was obtained in the SeMet dataset using the Ca2+-depleted KD of TgCDPK1 (PDB ID code 3KU2, residues 44–313) and a VHH with the complementarity determining regions (CDRs) removed (PDB ID code 1BZQ) as search models in phaser-MR. No solution was obtained with the active KD (PDB ID code 3HX4) or the CAD from either Ca2+-bound or Ca2+-depleted TgCDPK1. Even with weak anomalous data, an MR–SAD search strategy using the MR solution in phaser-EP yielded seven selenium sites of the possible 30 (2 copies per asymmetric unit in P21). Only one of these sites was in the placed KDs. We took advantage of the 14 known methionine positions in the two placed KDs by inserting selenium atoms at the side chain sulfur positions. Inputting these sites into the subsequent phaser-EP run resulted in the deletion of only a single site, the XYZ, B-factor, and occupancy refinement of all other input sites, and the identification of five new sites in the CAD. Application of the known noncrystallographic symmetry operator to the selenium sites resulted in a total of 23 sites. After solvent flattening and noncrystallographic symmetry map averaging in parrot, available through the CCP4 suite (39), clear helical density could be seen corresponding to the missing CAD. Iterative rounds of model building in Coot (40), phaser-EP map calculation, and parrot density modification were performed until the map figure of merit was greater than 0.5. Phenix.refine (37) was used for model refinement in the SeMet dataset until it was clear that experimental phase restraints were hindering further model improvement. The partially built and refined model was placed into the native P1 unit cell (4 copies per asymmetric unit) using phaser-MR. Sequence assignment was aided by the use of Se sites as positional markers and structural homology to the high-resolution, active CAD structure (rmsd, 3.2 Å; 137 Cαs aligned). Iterative building in Coot and refinement in Phenix.refine were performed in the native dataset to obtain the final, fully refined structure.
Structure Analysis.
The CDPK1:1B7 interface was analyzed using PDBePISA (41). A multiple sequence alignment of TgCDPK1 homologs was created using ClustalW in JalView (42).
MD Simulations.
Simulated systems were modeled from the X-ray crystal structure of active, Ca2+-bound TgCDPK1 (PDB ID code 3HX4) (11). TgCDPK1 with the Ca2+-bound CAD present (+CAD, residues 30–503) or removed (–CAD, residues 30–314) was solvated with 150 mM NaCl in cubic simulation boxes of 93 Å or 75 Å per side, respectively. There were ∼81,000 atoms in each +CAD simulation and 43,000 atoms in each –CAD simulation. The systems were parameterized using the Amber ff99SB-ILDN (43–45) force field, combined with the ff99SB* backbone correction (46) for proteins, and a TIP3P water model (47). After parameterization, the systems were energy-minimized and then equilibrated in a 10-ns simulation with an ensemble of constant number of atoms and constant temperature and pressure (NPT) at 310 °K and 1 bar. In the equilibration, 1 fs time step was used, and harmonic positional restraints of a force constant of 1 kcal/mol/Å2 were initially applied to the protein backbone atoms. Production simulations were performed starting from the last frame of equilibration simulations with an NPT ensemble at 310 °K and 1 bar using the Nosé–Hoover thermostat (48) with a time constant of 0.05 ps. All hydrogen-containing bonds were constrained using an implementation (49) of the M-SHAKE algorithm (50). A reversible reference system propagator algorithm integrator (51) was used. Bonded, Van der Waals, and short-range electrostatic interactions were computed every 2 fs, whereas long-range electrostatic interactions were computed every 6 fs. The short-range electrostatic interactions were calculated at a cutoff of 9.0 Å, and the long-range electrostatic interactions were computed using Gaussian-split Ewald (52). All simulations were carried out on either Anton or Anton 2, special purpose supercomputers designed for MD (53).
Acknowledgments
We thank Majida El Bakkouri and Raymond Hui at the Structural Genomics Consortium for providing us with the PfCDPK1 protein and helpful advice, Manoj T. Duraisingh for providing malaria lysates, Charles Shoemaker and Jacqueline Tremblay at Tufts Cummings School of Veterinary Medicine for their assistance with alpaca immunizations and library construction, and Eric Spooner at the Whitehead Institute Proteomics Facility for technical support. The X-ray diffraction data were collected at the Advanced Photon Source Northeastern Collaborative Access Team beamlines supported by the National Institute of General Medical Sciences, NIH (GM103403). Use of the APS is supported by the US Department of Energy, Office of Basic Energy Sciences (Contract DE-AC02-06CH11357). K.E.K. received fellowship support from NIH Grant T32GM007287 and National Science Foundation Graduate Research Fellowship Grant 1122374. B.M.M. received stipend support from the German Academic Exchange Service (Programm zur Steigerung der Mobilität von deutschen Studierenden) under the sponsorship of Christoph Borner at the Institute for Molecular Medicine and Cell Research and with support from Damaris Braun, both at the University of Freiburg. This work was supported in part by NIH Director’s Early Independence Award Grant 1DP5OD017892 (to S.L.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The atomic coordinates have been deposited in the Protein Data Bank, www.pdb.org (PDB ID code 4YGA).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1505914112/-/DCSupplemental.
References
- 1.Lourido S, Moreno SNJ. The calcium signaling toolkit of the Apicomplexan parasites Toxoplasma gondii and Plasmodium spp. Cell Calcium. 2015;57(3):186–193. doi: 10.1016/j.ceca.2014.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lourido S, et al. Calcium-dependent protein kinase 1 is an essential regulator of exocytosis in Toxoplasma. Nature. 2010;465(7296):359–362. doi: 10.1038/nature09022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lourido S, Tang K, Sibley LD. Distinct signalling pathways control Toxoplasma egress and host-cell invasion. EMBO J. 2012;31(24):4524–4534. doi: 10.1038/emboj.2012.299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Garrison E, et al. A forward genetic screen reveals that calcium-dependent protein kinase 3 regulates egress in Toxoplasma. PLoS Pathog. 2012;8(11):e1003049. doi: 10.1371/journal.ppat.1003049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.McCoy JM, Whitehead L, van Dooren GG, Tonkin CJ. TgCDPK3 regulates calcium-dependent egress of Toxoplasma gondii from host cells. PLoS Pathog. 2012;8(12):e1003066. doi: 10.1371/journal.ppat.1003066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Azevedo MF, et al. Inhibition of Plasmodium falciparum CDPK1 by conditional expression of its J-domain demonstrates a key role in schizont development. Biochem J. 2013;452(3):433–441. doi: 10.1042/BJ20130124. [DOI] [PubMed] [Google Scholar]
- 7.Harper JF, Huang JF, Lloyd SJ. Genetic identification of an autoinhibitor in CDPK, a protein kinase with a calmodulin-like domain. Biochemistry. 1994;33(23):7267–7277. doi: 10.1021/bi00189a031. [DOI] [PubMed] [Google Scholar]
- 8.Harmon AC, Yoo BC, McCaffery C. Pseudosubstrate inhibition of CDPK, a protein kinase with a calmodulin-like domain. Biochemistry. 1994;33(23):7278–7287. doi: 10.1021/bi00189a032. [DOI] [PubMed] [Google Scholar]
- 9.Chandran V, et al. Structure of the regulatory apparatus of a calcium-dependent protein kinase (CDPK): A novel mode of calmodulin-target recognition. J Mol Biol. 2006;357(2):400–410. doi: 10.1016/j.jmb.2005.11.093. [DOI] [PubMed] [Google Scholar]
- 10.Christodoulou J, Malmendal A, Harper JF, Chazin WJ. Evidence for differing roles for each lobe of the calmodulin-like domain in a calcium-dependent protein kinase. J Biol Chem. 2004;279(28):29092–29100. doi: 10.1074/jbc.M401297200. [DOI] [PubMed] [Google Scholar]
- 11.Wernimont AK, et al. Structures of apicomplexan calcium-dependent protein kinases reveal mechanism of activation by calcium. Nat Struct Mol Biol. 2010;17(5):596–601. doi: 10.1038/nsmb.1795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ojo KK, et al. Toxoplasma gondii calcium-dependent protein kinase 1 is a target for selective kinase inhibitors. Nat Struct Mol Biol. 2010;17(5):602–607. doi: 10.1038/nsmb.1818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rosenberg OS, Deindl S, Sung R-J, Nairn AC, Kuriyan J. Structure of the autoinhibited kinase domain of CaMKII and SAXS analysis of the holoenzyme. Cell. 2005;123(5):849–860. doi: 10.1016/j.cell.2005.10.029. [DOI] [PubMed] [Google Scholar]
- 14.Rellos P, et al. Structure of the CaMKIIdelta/calmodulin complex reveals the molecular mechanism of CaMKII kinase activation. PLoS Biol. 2010;8(7):e1000426. doi: 10.1371/journal.pbio.1000426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhao Y, et al. Calcium-binding properties of a calcium-dependent protein kinase from Plasmodium falciparum and the significance of individual calcium-binding sites for kinase activation. Biochemistry. 1994;33(12):3714–3721. doi: 10.1021/bi00178a031. [DOI] [PubMed] [Google Scholar]
- 16.De Meyer T, Muyldermans S, Depicker A. Nanobody-based products as research and diagnostic tools. Trends Biotechnol. 2014;32(5):263–270. doi: 10.1016/j.tibtech.2014.03.001. [DOI] [PubMed] [Google Scholar]
- 17.Lourido S, Jeschke GR, Turk BE, Sibley LD. Exploiting the unique ATP-binding pocket of toxoplasma calcium-dependent protein kinase 1 to identify its substrates. ACS Chem Biol. 2013;8(6):1155–1162. doi: 10.1021/cb400115y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shan Y, et al. Oncogenic mutations counteract intrinsic disorder in the EGFR kinase and promote receptor dimerization. Cell. 2012;149(4):860–870. doi: 10.1016/j.cell.2012.02.063. [DOI] [PubMed] [Google Scholar]
- 19.Taylor SS, Keshwani MM, Steichen JM, Kornev AP. Evolution of the eukaryotic protein kinases as dynamic molecular switches. Philos Trans R Soc Lond B Biol Sci. 2012;367(1602):2517–2528. doi: 10.1098/rstb.2012.0054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhou H-X. Loops, linkages, rings, catenanes, cages, and crowders: Entropy-based strategies for stabilizing proteins. Acc Chem Res. 2004;37(2):123–130. doi: 10.1021/ar0302282. [DOI] [PubMed] [Google Scholar]
- 21.Ranjan R, Ahmed A, Gourinath S, Sharma P. Dissection of mechanisms involved in the regulation of Plasmodium falciparum calcium-dependent protein kinase 4. J Biol Chem. 2009;284(22):15267–15276. doi: 10.1074/jbc.M900656200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yu J, et al. Regulation of the p85/p110 phosphatidylinositol 3′-kinase: Stabilization and inhibition of the p110alpha catalytic subunit by the p85 regulatory subunit. Mol Cell Biol. 1998;18(3):1379–1387. doi: 10.1128/mcb.18.3.1379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Baer K, et al. Dimerization-induced activation of soluble insulin/IGF-1 receptor kinases: An alternative mechanism of activation. Biochemistry. 2001;40(47):14268–14278. doi: 10.1021/bi015588g. [DOI] [PubMed] [Google Scholar]
- 24.Ahmed A, et al. Novel insights into the regulation of malarial calcium-dependent protein kinase 1. FASEB J. 2012;26(8):3212–3221. doi: 10.1096/fj.12-203877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jura N, et al. Catalytic control in the EGF receptor and its connection to general kinase regulatory mechanisms. Mol Cell. 2011;42(1):9–22. doi: 10.1016/j.molcel.2011.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Filippakopoulos P, et al. Structural coupling of SH2-kinase domains links Fes and Abl substrate recognition and kinase activation. Cell. 2008;134(5):793–803. doi: 10.1016/j.cell.2008.07.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nagar B, et al. Structural basis for the autoinhibition of c-Abl tyrosine kinase. Cell. 2003;112(6):859–871. doi: 10.1016/s0092-8674(03)00194-6. [DOI] [PubMed] [Google Scholar]
- 28.De Genst E, et al. Molecular basis for the preferential cleft recognition by dromedary heavy-chain antibodies. Proc Natl Acad Sci USA. 2006;103(12):4586–4591. doi: 10.1073/pnas.0505379103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Castellanos-Gonzalez A, et al. A novel calcium-dependent protein kinase inhibitor as a lead compound for treating cryptosporidiosis. J Infect Dis. 2013;208(8):1342–1348. doi: 10.1093/infdis/jit327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ojo KK, et al. Transmission of malaria to mosquitoes blocked by bumped kinase inhibitors. J Clin Invest. 2012;122(6):2301–2305. doi: 10.1172/JCI61822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Andersen KR, Leksa NC, Schwartz TU. Optimized E. coli expression strain LOBSTR eliminates common contaminants from His-tag purification. Proteins. 2013;81(11):1857–1861. doi: 10.1002/prot.24364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Donald RG, Roos DS. Stable molecular transformation of Toxoplasma gondii: A selectable dihydrofolate reductase-thymidylate synthase marker based on drug-resistance mutations in malaria. Proc Natl Acad Sci USA. 1993;90(24):11703–11707. doi: 10.1073/pnas.90.24.11703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Maass DR, Sepulveda J, Pernthaner A, Shoemaker CB. Alpaca (Lama pacos) as a convenient source of recombinant camelid heavy chain antibodies (VHHs) J Immunol Methods. 2007;324(1-2):13–25. doi: 10.1016/j.jim.2007.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dobrowolski JM, Carruthers VB, Sibley LD. Participation of myosin in gliding motility and host cell invasion by Toxoplasma gondii. Mol Microbiol. 1997;26(1):163–173. doi: 10.1046/j.1365-2958.1997.5671913.x. [DOI] [PubMed] [Google Scholar]
- 35.Bers DM, Patton CW, Nuccitelli R. A practical guide to the preparation of Ca(2+) buffers. Methods Cell Biol. 2010;99:1–26. doi: 10.1016/B978-0-12-374841-6.00001-3. [DOI] [PubMed] [Google Scholar]
- 36.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. In: Carter CJ, Sweet RM, editors. Methods in Enzymology. Elsevier; New York: 1997. pp. 307–326. [DOI] [PubMed] [Google Scholar]
- 37.Adams PD, et al. PHENIX: A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66(Pt 2):213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Morin A, et al. Collaboration gets the most out of software. eLife. 2013;2:e01456. doi: 10.7554/eLife.01456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Winn MD, et al. Overview of the CCP4 suite and current developments. Acta Crystallogr D Biol Crystallogr. 2011;67(Pt 4):235–242. doi: 10.1107/S0907444910045749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr D Biol Crystallogr. 2010;66(Pt 4):486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Krissinel E, Henrick K. Inference of macromolecular assemblies from crystalline state. J Mol Biol. 2007;372(3):774–797. doi: 10.1016/j.jmb.2007.05.022. [DOI] [PubMed] [Google Scholar]
- 42.Waterhouse AM, Procter JB, Martin DMA, Clamp M, Barton GJ. Jalview Version 2—A multiple sequence alignment editor and analysis workbench. Bioinformatics. 2009;25(9):1189–1191. doi: 10.1093/bioinformatics/btp033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cornell WD, et al. A second generation force field for the simulation of proteins, nucleic acids, and organic molecules. J Am Chem Soc. 1995;117(19):5179–5197. [Google Scholar]
- 44.Hornak V, et al. Comparison of multiple Amber force fields and development of improved protein backbone parameters. Proteins. 2006;65(3):712–725. doi: 10.1002/prot.21123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lindorff-Larsen K, et al. Improved side-chain torsion potentials for the Amber ff99SB protein force field. Proteins. 2010;78(8):1950–1958. doi: 10.1002/prot.22711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Best RB, Hummer G. Optimized molecular dynamics force fields applied to the helix-coil transition of polypeptides. J Phys Chem B. 2009;113(26):9004–9015. doi: 10.1021/jp901540t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jorgensen WL, Chandrasekhar J, Madura JD, Impey RW, Klein ML. Comparison of simple potential functions for simulating liquid water. J Chem Phys. 1983;79(2):926–935. [Google Scholar]
- 48.Hoover WG. Canonical dynamics: Equilibrium phase-space distributions. Phys Rev A. 1985;31(3):1695–1697. doi: 10.1103/physreva.31.1695. [DOI] [PubMed] [Google Scholar]
- 49.Lippert RA, et al. A common, avoidable source of error in molecular dynamics integrators. J Chem Phys. 2007;126(4):046101. doi: 10.1063/1.2431176. [DOI] [PubMed] [Google Scholar]
- 50.Kräutler V, van Gunsteren WF, Hünenberger PH. A fast SHAKE algorithm to solve distance constraint equations for small molecules in molecular dynamics simulations. J Comput Chem. 2001;22(5):501–508. [Google Scholar]
- 51.Tuckerman M, Berne BJ, Martyna GJ. Reversible multiple time scale molecular dynamics. J Chem Phys. 1992;97(3):1990–2001. [Google Scholar]
- 52.Shan Y, Klepeis JL, Eastwood MP, Dror RO, Shaw DE. Gaussian split Ewald: A fast Ewald mesh method for molecular simulation. J Chem Phys. 2005;122(5):54101. doi: 10.1063/1.1839571. [DOI] [PubMed] [Google Scholar]
- 53.Shaw DE, et al. Anton 2: Raising the Bar for Performance and Programmability in a Special-Purpose Molecular Dynamics Supercomputer. IEEE Press; Piscataway, NJ: 2014. [Google Scholar]