Significance
In plants, 12-oxo-phytodienoic acid (12-OPDA) and jasmonic acid are key 13-lipoxygenase-derived linolenate oxidation products termed jasmonates that regulate diverse processes in development and innate immunity. A less-studied metabolic pathway branch is generated by 9-lipoxygenase activity on linoleic acid, enabling the production of 10-oxo-11-phytoenoic acid (10-OPEA). In maize, fungal infection by southern leaf blight (Cochliobolus heterostrophus) results in the localized production of 10-OPEA, and a series of related 12- and 14-carbon cyclopente(a)nones, collectively termed “death acids” (DAs). DAs far exceed jasmonates in abundance within infected tissues, display direct phytoalexin activity against biotic agents, mediate defense gene expression, and can promote cytotoxicity resulting in cell death. Collectively DA activities are consistent with specialized local roles in plant defense.
Keywords: oxylipin, 9-lipoxygenase, 10-oxo-11-phytoenoic acid, maize, defense
Abstract
Plant damage promotes the interaction of lipoxygenases (LOXs) with fatty acids yielding 9-hydroperoxides, 13-hydroperoxides, and complex arrays of oxylipins. The action of 13-LOX on linolenic acid enables production of 12-oxo-phytodienoic acid (12-OPDA) and its downstream products, termed “jasmonates.” As signals, jasmonates have related yet distinct roles in the regulation of plant resistance against insect and pathogen attack. A similar pathway involving 9-LOX activity on linolenic and linoleic acid leads to the 12-OPDA positional isomer, 10-oxo-11-phytodienoic acid (10-OPDA) and 10-oxo-11-phytoenoic acid (10-OPEA), respectively; however, physiological roles for 9-LOX cyclopentenones have remained unclear. In developing maize (Zea mays) leaves, southern leaf blight (Cochliobolus heterostrophus) infection results in dying necrotic tissue and the localized accumulation of 10-OPEA, 10-OPDA, and a series of related 14- and 12-carbon metabolites, collectively termed “death acids.” 10-OPEA accumulation becomes wound inducible within fungal-infected tissues and at physiologically relevant concentrations acts as a phytoalexin by suppressing the growth of fungi and herbivores including Aspergillus flavus, Fusarium verticillioides, and Helicoverpa zea. Unlike previously established maize phytoalexins, 10-OPEA and 10-OPDA display significant phytotoxicity. Both 12-OPDA and 10-OPEA promote the transcription of defense genes encoding glutathione S transferases, cytochrome P450s, and pathogenesis-related proteins. In contrast, 10-OPEA only weakly promotes the accumulation of multiple protease inhibitor transcripts. Consistent with a role in dying tissue, 10-OPEA application promotes cysteine protease activation and cell death, which is inhibited by overexpression of the cysteine protease inhibitor maize cystatin-9. Unlike jasmonates, functions for 10-OPEA and associated death acids are consistent with specialized roles in local defense reactions.
In plants, cellular damage results in the enzymatic and nonenzymatic peroxidation of fatty acids (FAs) termed “oxylipins.” Enzymatic biosynthesis can be initiated by lipase-based cleavage of linoleic acid (18:2) or α-linolenic acid (18:3) from membrane lipids and subsequent dioxygenation by lipoxygenases (LOXs) with regiospecificity at carbons 9 or 13. Specific oxylipins function as direct antimicrobial defenses and plant signaling molecules that regulate diverse processes including development, reproduction, stress acclimation, and innate immune responses against pests and pathogens (1–3).
The most studied 13-LOX 18:3-derived plant oxylipins are 12-oxo-phytodienoic acid (12-OPDA) and jasmonic acid (JA). Biosynthesis requires conversion of 13-hydroperoxides to unstable epoxides via allene oxide synthase (AOS), cyclization by allene oxide cyclase (AOC) to form 12-OPDA, reduction of the cyclopentenone ring by 12-oxo-phytodienoic acid reductase (OPR), and subsequent β-oxidation steps to produce (+)-7-iso-JA (1, 4) (Fig. 1F). JA is further conjugated by jasmonate resistant 1 (JAR1) to its bioactive form, JA-Ile, which mediates formation of the Coronatine Insensitive (COI1)-Jasmonate Zim domain (JAZ) family complex to promote gene expression (5, 6). Diverse roles for JA and 12-OPDA as signals include developmental processes and inducible defenses against biotic threats (1–3, 7). Genetic evidence in maize (Zea mays) supports a role for JA biosynthesis in the survival of biotic stress, regulation of senescence, and cell death processes mediating male sex determination (8, 9). As a precursor, 12-OPDA can trigger developmental processes and defense signaling different from JA-Ile (10–12). During physiological stress, 12-OPDA binds cyclophilin 20–3 which facilitates recruitment of a cysteine synthase complex leading to elevated glutathione levels and cellular redox homeostasis (11). 12-OPDA signaling is partly dependent upon TGA transcription factors that govern detoxification responses such as OPRs, glutathione S transferases (GSTs) and cytochrome P-450s (CYPs) consistent with a role in cell protection and survival (13, 14). The activity of structurally related nonenzymatic cyclopentenone oxylipins, such as the phytoprostanes, overlap with 12-OPDA–regulated defense responses (13, 15, 16). In the context of nonenzymatic lipid signals, many reactive electrophile species can promote cell protection by inducing genes responsible for detoxification, cell cycle regulation, and chaperones (17).
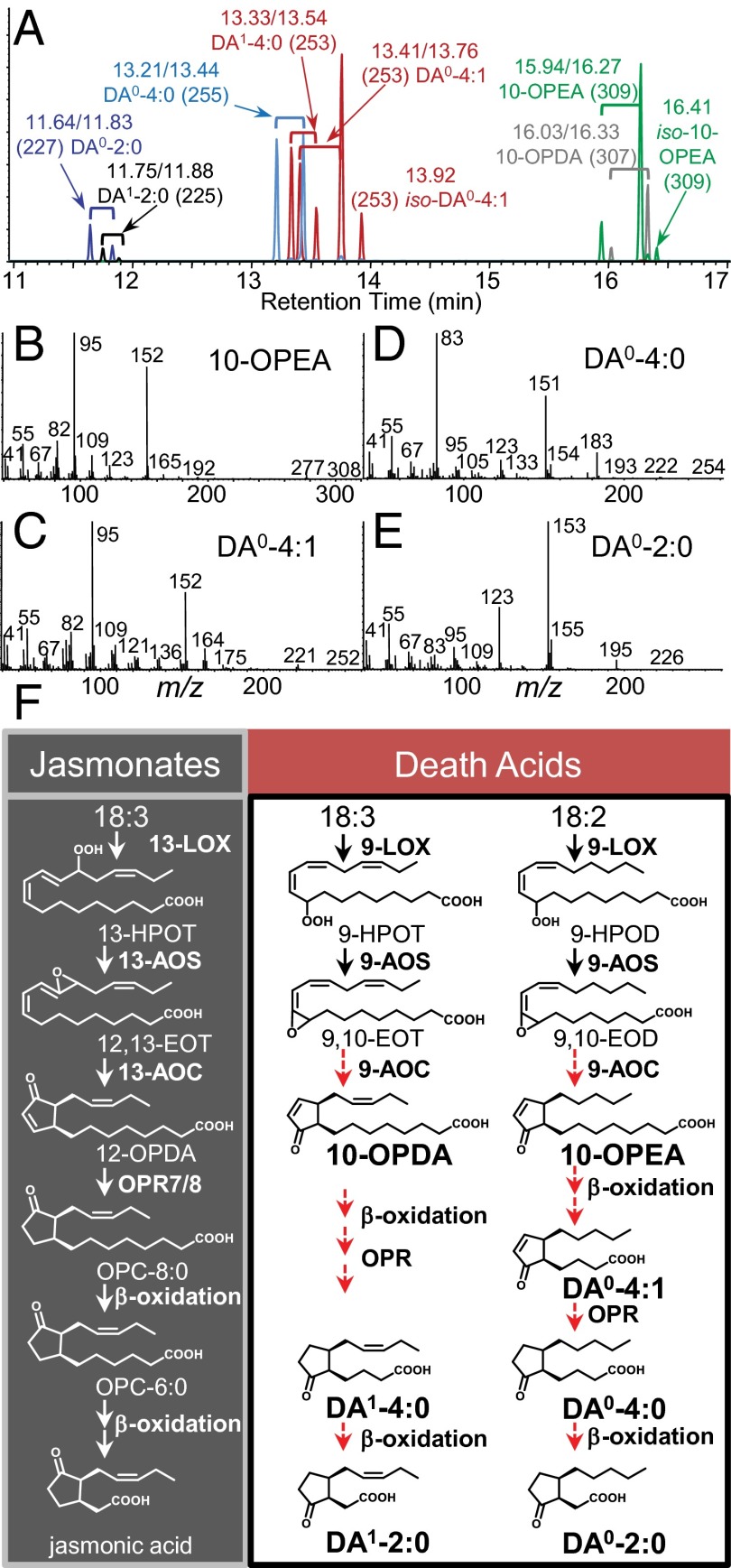
Fig. 1.
Identity, spectra, and conceptual working model for the biosynthetic pathway of death acids (DAs) derived from linoleic and linolenic acid. (A) Combined purified standards showing GC retention times (trans/cis pairs: min) and predominant positive-CI MS [M+H]+ ions of DA0-2:0, DA1-2:0, DA0-4:0, DA1-4:0, DA0-4:1, iso-DA0-4:1,10-OPEA, 10-OPDA, and iso-10-OPEA as methyl esters. Comparative EI spectra of purified linoleic acid-derived DAs as methyl esters: (B) 10-OPEA, (C) DA0-4:1, (D) DA0-4:0, and (E) DA0-2:0. (F) Proposed model of death acid biosynthesis with respect to the jasmonic acid pathway. Red dashed arrows indicate the presence of predicted enzyme activities requiring future confirmation. Abbreviations are as follows: lipoxygenase (LOX), allene oxide synthase (AOS), allene oxide cyclase (AOC), 12-oxo-phytodienoate reductases (OPR), linolenic acid (18:3), linoleic acid (18:2), 9-hydroperoxy-10,12-octadecadienoic acid (9-HPOD), 9-hydroperoxy-10,12,15-octadecatrienoic acid (9-HPOT), 13-hydroperoxy-10,12,15-octadecatrienoic acid (13-HPOT); 9,10-epoxyoctadecatrienoic acid (9,10-EOT); 9,10-epoxyoctadecadienoic acid (9,10-EOD); 12,13-epoxyoctadecatrienoic acid (12,13-EOT); 10-oxo-11,15-phytodienoic acid (10-OPDA); 10-oxo-11-phytoenoic acid (10-OPEA); 12-oxo-10,15-phytodienoic acid (12-OPDA); 3-oxo-2-(2′-pentenyl)-cyclopentane-1-octanoic acid (OPC-8:0), and hexanoic acid (OPC-6:0).
In addition to jasmonates, specific 9-LOX oxylipins also influence seed germination, root growth, and senescence as well as mediation of either susceptibility or resistance to pests and pathogens (18–22). As direct defenses, 9-LOX oxylipins can act as phytoalexins; for example, colneleic and colnelenic acids are pathogen inducible and inhibitory to Phytophthora infestans (23). As mediators, metabolites derived from the 9-LOX pathway in pepper (Capsicum annuum) and Arabidopsis positively regulate defense and cell death responses to diverse pathogens (21). Despite numerous studies linking the 9-LOX pathway to cell death processes (24–26), many oxylipin identities, activities, and links to enzyme activation remain unknown (27, 28). As a positional isomer of 12-OPDA, the 9-LOX oxylipin 10-oxo-11,15-phytodienoic acid (10-OPDA) and the 18:2-derived 10-oxo-11-phytoenoic acid (10-OPEA) are structurally similar to jasmonates (29). In potato (Solanum tuberosum), cis-10-OPEA is formed as a racemic product of 9-AOS–derived 9,10-epoxyoctadecadienoic acid (9,10-EOD) that is inefficiently cyclized in the absence of AOC (29, 30). Similarly, the 9-LOX/AOS pathway in tomato (Solanum lycopersicum) can act on 18:3 to yield 10-OPDA (31). These studies draw attention to the possibility of downstream 9-LOX derivatives of 10-OPEA and 10-OPDA; however, the existence of additional jasmonate-like metabolites has remained elusive.
While searching for defense-related metabolites present in southern leaf blight (SLB; Cochliobolus heterostrophus) infected maize, we detected high levels of 10-OPEA and a series of related cyclopente(a)none oxylipins. Localized within dying and necrotic tissues, we collectively refer to 10-OPEA, 10-OPDA, and derivatives as “death acids” (DAs). As direct defenses, levels of 10-OPEA produced in diseased tissues after wounding match those of known maize phytoalexins [>100 μg⋅g−1 fresh weight (FW)] and can impair growth in pathogens, insects, and plant cells. In contrast to 10-OPEA and 10-OPDA, previously established maize phytoalexins sharing reactive α,β-unsaturated carbonyls lack significant phytotoxicity in maize. Exogenous application of 10-OPEA to plant tissues strongly induces defense genes, cysteine protease activity, and common cell death symptoms including lesions, ion leakage, and DNA fragmentation. Unlike JA and 12-OPDA, 10-OPEA and related DAs elicit significantly lower transcriptional up-regulation of multiple protease inhibitors (PIs) including the maize cystatin-9 (ZmCC9), a negative regulator of apoplastic mediated cell death (27). Overexpression of ZmCC9 reduces the extent of 10-OPEA–induced lesions and provides a mechanistic link to the cytotoxic action in maize.
Results and Discussion
Identity of Cyclopente(a)none Death Acids in Maize.
To elucidate pathogen-induced defense metabolites in maize, we conducted metabolic profiling of SLB-infected tissues. Among the analytes were the rarely encountered 9-LOX derived cyclopentenones, 10-OPEA, 10-OPDA, and seven other related yet unknown cyclopente(a)nones (Fig. 1A) (29, 31). To establish identities, we performed a large-scale purification of these analytes and used 1H and 13C NMR spectroscopy (SI Appendix, Table S1). Chemical and electron ionization (CI/EI) mass spectrometry of the corresponding methyl esters provides useful diagnostic spectra of these 10-OPEA and 10-OPDA derivatives (Fig. 1 B–E and SI Appendix, Fig. S1). Additional related isomerization products of 10-OPEA, including iso-10-OPEA were also identified (SI Appendix, Table S1) (29). Given multiple FA precursors and additional anticipated metabolites, we assigned each DA according to the number of carbons in the carboxylic acid side chain (e.g., DA-X), the presence/absence of a double bond in the cyclopente(a)none ring (e.g., DA-X:1 or DA-X:0), and its FA origin (e.g., 18:2 = DA0; 18:3 = DA1). In our proposed model, DA biosynthesis initiates with 9-LOX activity on 18:2 and 18:3 to form 9-hydroperoxides followed by 9-AOS–mediated allene oxide formation (30, 31). Analysis of ZmLOX and ZmAOS gene expression from SLB-infected leaves indicated that ZmLOX3/4/5 and ZmAOS1/3 are candidate genes for the initial steps in pathogen-induced DA biosynthesis (SI Appendix, Fig. S2). SLB-infection screening of available single and double LOX mutants, including Zmlox3, Zmlox4, Zmlox5, Zmlox3/5, and Zmlox4/5, showed no significant reduction in 10-OPEA production (SI Appendix, Fig. S2). This suggests that multiple LOXs provide substrates for DA biosynthesis, similar to the complexity observed for JA biosynthesis (32). In contrast to the AOC-mediated enzymatic cyclization of 12,13-epoxyoctadecatrienoic acid yielding pure (9S,13S)-12-OPDA [i.e., cis(+)-12-OPDA] in potato and tomato, cis-10-OPEA and cis-10-OPDA are formed as minor racemic (9S,13S and 9R,13R) cyclization products of AOS-derived 9-allene oxides, which are predominantly hydrolyzed to α-ketols (29, 30, 33). Analysis of these relationships in maize unexpectedly revealed that inducible levels of cis-10-OPEA exceeded those of the predicted dominant α-ketol (9-hydroxy-10-oxo-12(Z)-octadecenoic acid) product by more than 15-fold (SI Appendix, Fig. S3). Chiral phase high performance liquid chromatography (CP-HPLC) analysis of maize cis-10-OPEA was then performed and resulted in a single chromatographic peak, whereas a standard of racemic (9S,13S and 9R,13R) cis-10-OPEA produced two separate peaks (SI Appendix, Fig. S3). Our results are consistent with recently published findings demonstrating that maize synthesis of predominantly (9S,13S)-10-OPEA is enzymatic and is the result of an undiscovered AOC acting on 9-LOX derived allene oxide(s) (34). Curiously, a portion of the 10-OPEA cyclopentenone appears to undergo two β-oxidation–like steps to form DA0-4:1 before an OPR-like mediated cyclopentenone reduction to DA0-4:0 (Fig. 1F and SI Appendix, Fig. S4). This differs from the JA biosynthetic pathway where OPR activity first reduces cis-12-OPDA to a cyclopentanone. Additional processing of DA0-4:0 and DA1-4:0 by a β-oxidation step is envisioned to produce the 9-LOX positional isomers of dihydro-JA and JA, respectively denoted as DA0-2:0 and DA1-2:0. To investigate the capacity of healthy control maize plants to synthesize processed DA pathway metabolites, we applied cis-10-OPEA to leaves and observed the accumulation of C14 and C12 DAs within 2 h (SI Appendix, Fig. S4). Although not excluding the potential for multiple origins in nature, these results establish the capacity of maize to produce cyclopente(a)none DAs.
10-OPEA Acts as a Cytotoxic Phytoalexin.
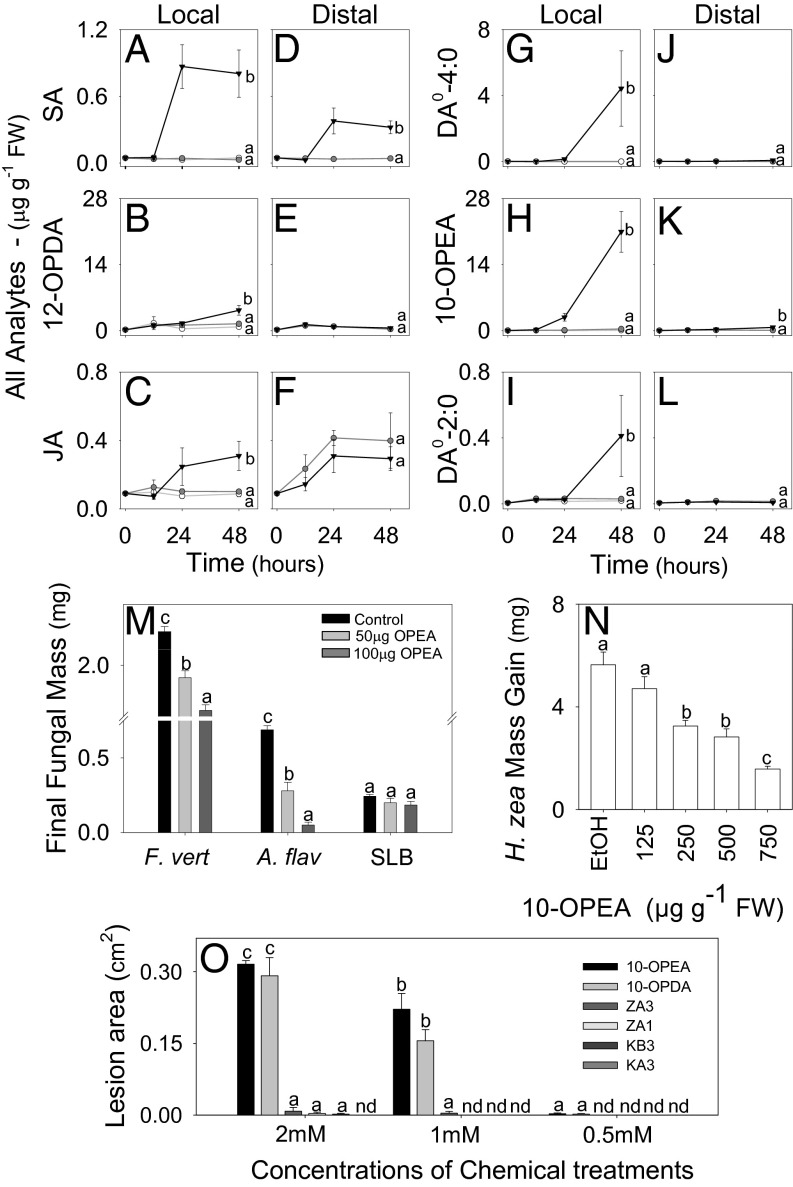
To compare the dynamics of DA accumulation to structurally related oxylipins and defense signals, both SLB-infected (local) and adjacent tissues (distal, 1–2 mm from the site of visible necrosis) were analyzed over 48 h. Within 24 h, local infected tissues displayed 8-fold increases in salicylic acid (SA) and 10-OPEA accumulation, whereas 12-OPDA and JA concentrations only moderately increased (Fig. 2 A–C and H). By 48 h, 10-OPEA concentrations within local infected tissues predominated over SA, JA, and 12-OPDA by >5-fold. Accumulation of DA0-4:0 and DA0-2:0 also significantly increased by 48 h, suggesting active processing of 10-OPEA in plants during disease progression (Fig. 2 G and I). Interestingly, the 18:3 derivatives 10-OPDA and DA1-4:0 were 7- and 2-fold lower than 10-OPEA and DA0-4:0 in SLB-infected tissues, respectively, consistent with a tissue bias in 18:2 precursor present (SI Appendix, Fig. S5). In the tissue distal to SLB infection, SA accumulation was 6-fold higher than mechanically damaged controls (Fig. 2D); however, there was little or no distal accumulation of 12-OPDA or 10-OPEA family DAs at 48 h (Fig. 2 E and J–L). Thus, DA production is localized and confined to diseased tissue. In an attempt to mimic the collapse of plant cells under pathogen attack, tissues were subsequently crush damaged either 2 or 4 d after SLB inoculation. Damaged plants with subsequent crush damage produced high levels of 18:2 and 18:3, yet 10-OPEA, 10-OPDA, DA0-4:0, DA1-4:0, and DA0-2:0 levels remained very low (Fig. 2 and SI Appendix, Figs. S5 and S6). In contrast, SLB-infected tissue subjected to subsequent crush damage resulted in 10-OPEA accumulation exceeding 150 µg·g−1 FW, which surpassed 12-OPDA and JA production by >7-fold (SI Appendix, Figs. S5 and S6). On average, diseased tissues also produced higher levels of DA0-4:0 and DA0-2:0 in response to crush damage, surpassing JA levels by 15- and 3-fold, respectively.
Fig. 2.
DAs predominate in localized diseased tissue and display antimicrobial, antiinsect, and phytotoxic activity. Average (n = 4, ± SEM) concentrations (µg⋅g−1 FW) of (A and D) SA, (B and E) 12-OPDA, (C and F) JA, (G and J) DA0-4:0, (H and K) 10-OPEA, and (I and L) DA0-2:0 in maize interior whorl tissue following no treatment (open circle), mechanical incision damage (gray circle), and SLB inoculation (solid triangle). Two segments consisting of the infected site (local; A–C and G–I) and visually asymptomatic tissue 1–2 mm adjacent to the treatment site (distal; D–F and J–L) were collected for analysis at indicated times. (M) Average (n = 8, ± SEM) southern leaf blight (SLB), Fusarium verticillioides (F.v.), and Aspergillus flavus (A.f.) hyphae mass (in milligrams) after 72 h in media containing 0, 50, or 100 µg·ml−1 10-OPEA. (N) Average mass gain (in milligrams) (n = 20, ± SEM) in H. zea larval growth after 24 h on maize leaf diet containing 10-OPEA (125–750 µg·g−1 FW). (O) Average (n = 4, ± SEM) maize leaf lesion area (in square centimeters) at 24 h for phytoalexin toxicity analysis. Fourth leaves were treated with two 10-µL droplets of 10-OPEA, 10-OPDA, zealexins (Z; ZA1 and ZA3) and kauralexins (K; KA3 and KB3) at concentrations of 0.5, 1, and 2 mM dissolved in 95:5:0.1 H2O:DMSO:Tween 20 (vol/vol/vol). Within plots, different letters (a–d) represent significant differences at indicated time points (nd, not detected; all ANOVA P < 0.05; Tukey test for corrections for multiple comparisons: P < 0.05).
Collectively, wound-induced accumulation of 10-OPDA and 10-OPEA in SLB-infected tissues and damaged silks can range between 180 and 340 µg·g−1 FW (SI Appendix, Figs. S5–S7) concentrations similar to fungal induced levels of terpenoid phytoalexins (35, 36). Given that oxylipins can function as phytoalexins, we investigated the potential for 10-OPEA to act as a direct defense (23, 37). 10-OPEA displayed minor inhibitory effects against SLB growth in vitro, suggesting appreciable fungal tolerance or detoxification (Fig. 2M). Conversely, growth of Fusarium verticillioides and Aspergillus flavus was significantly suppressed by 10-OPEA at 100 µg·mL−1 (Fig. 2M) and is consistent with the variation in fungal species tolerance observed in oxylipin antimicrobial assays (38). Given the abundance of 10-OPEA wounded silks and modest accumulation in stems following 6 d of corn earworm (CEW; Helicoverpa zea) herbivory (SI Appendix, Fig. S7), we examined a role for 10-OPEA as an insect growth inhibitor. Over a range of 125–750 µg·g−1 FW, 10-OPEA promoted a concentration-dependent inhibition of insect growth (Fig. 2N). These results indicate that 10-OPEA can function as a broadly active phytoalexin that shares similar antibiotic potencies to maize zealexins and kauralexins (35, 36). Because phytoalexins can also harm plant cells (39), we tested the cytotoxic properties of 10-OPEA in comparison with four different maize phytoalexins (zealexin A1/A3, kauralexin A3/B3). Comparatively, 10-OPEA and 10-OPDA were equally cytotoxic and resulted in 35-fold greater lesion areas than acidic terpenoid phytoalexins tested (Fig. 2O).
10-OPEA and DA0-4:0 Mediate Defense Gene Expression.
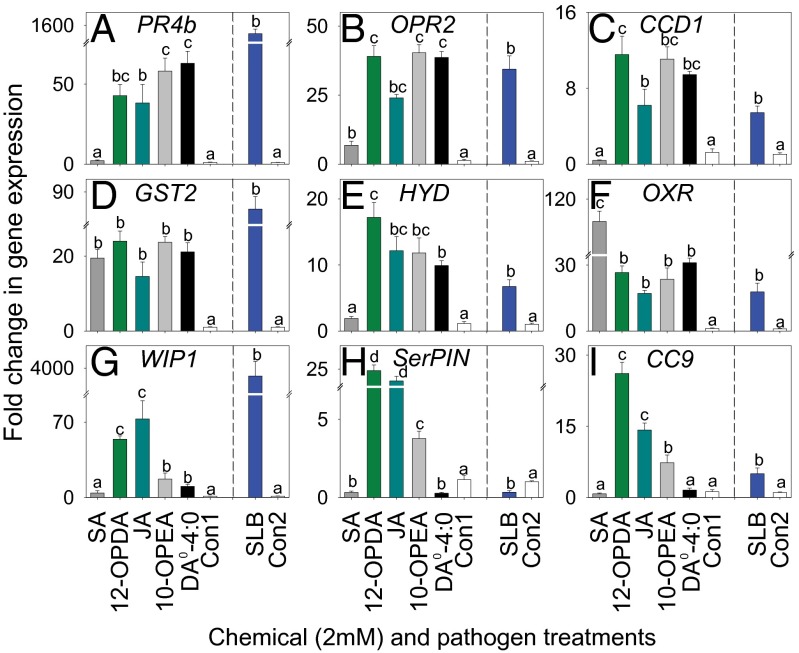
Given that cyclopentenones and cyclopentanones can have distinct signaling properties, we first investigated the potential for 10-OPEA–mediated transcriptional regulation using the Affymetrix GeneChip maize genome array (7, 12, 16). Treatment with 10-OPEA after 90 min resulted in 55 genes with a twofold or greater change in probe intensity at 95% confidence. Among the strongest induced transcripts were genes associated with OPR activity, pathogen defense, cell death, calcium signaling, and redox homeostasis (SI Appendix, Dataset S1). To validate the microarray results and to compare 10-OPEA activity to other meaningful metabolites, we treated plants with either SA, 12-OPDA, JA, 10-OPEA, DA0-4:0, or SLB and compared the expression levels of pathogen- and insect-related genes at 3 h using qRT-PCR. Defense marker genes included ZmOPR2, the maize OPR most strongly up-regulated in response to SLB (40); pathogenesis-related 4b (ZmPR4b) and EF-hand Ca2+-binding protein (ZmCCD1), genes involved in the promotion of cell death and pathogen-elicited defense responses (41–43); GST 2 (ZmGST2) and hydrolase (ZmHYD), previously shown to be induced by both insect- and pathogen-related treatments (35, 44); a stress-inducible oxidoreductase (ZmOXR); and maize cystatin 9 (ZmCC9), wound inducible protein 1 (ZmWIP1), and serine protease inhibitor (ZmSerPIN), that encode PIs inducible by biotic attack (27, 45, 46). The defense genes ZmPR4b, ZmOPR2, and ZmCCD1 were strongly induced by 10-OPEA and DA0-4:0 treatments with expression levels either statistically equivalent to or higher than those of 12-OPDA, JA, and SA (Fig. 3 A–C). The insect- and pathogen-inducible transcripts encoding ZmGST2 and ZmHYD were also significantly up-regulated by 10-OPEA and DA0-4:0 (Fig. 3 D and E), suggesting functional overlap with maize responses to biotic stress. Curiously, ZmOXR transcript accumulation was more responsive to SA than all other treatments (Fig. 3F). Whereas similar expression patterns between DAs and established signals were observed in other defense genes (SI Appendix, Fig. S8), there was a consistent difference between DAs and jasmonates in the expression of multiple PIs. As expected, transcript accumulation of pathogen- and insect-inducible PIs was strongly regulated by JA and 12-OPDA; however, 10-OPEA and DA0-4:0 resulted in significantly lower PI transcript accumulation (Fig. 3 G–I). Compared with jasmonates, weak DA-mediated PI transcriptional regulation may leave respective protease activities unaltered. To identify additional responses, we subsequently performed whole transcriptome analyses using RNA-Seq. In this experiment, 10-OPEA and 12-OPDA treatments resulted in over 5,000 differentially expressed candidate genes with 60% being either equally elicited or suppressed by both cyclopentenones at 3 h (SI Appendix, Dataset S2). Enrichment analyses of transcripts displaying comparatively stronger elicitation to 10-OPEA than 12-OPDA were dominated by heat shock proteins (HSPs) and GSTs (SI Appendix, Table S2). Transcript accumulation specific to 10-OPEA treatments encodes proteins that function in transport and detoxification (e.g., P450s). Conversely, enrichment of transcripts displaying comparatively stronger elicitation to 12-OPDA than 10-OPEA were dominated by genes associated with aromatic amino acid biosynthesis, secondary metabolism, and jasmonate regulation. Importantly, 12-OPDA application results in partial conversion to JA and thus the combination of two distinct jasmonate pathway responses (12, 13). Compared with 10-OPEA, 12-OPDA specific transcripts are enriched for encoded proteins predicted to function in cell wall synthesis. Collectively, these analyses confirm a broad coregulation of the 12-OPDA and 10-OPEA transcriptome (Fig. 3) with narrow subsets of gene ontology enrichments consistent with defense and cell fortification for 12-OPDA and detoxification, molecular chaperones (HSPs), and transport for 10-OPEA (SI Appendix, Dataset S2 and Table S2).
Fig. 3.
10-OPEA and DA0-4:0 induce defense gene expression in maize. Average (n = 4, ± SEM) qRT-PCR fold change in transcript levels of genes encoding pathogen and insect defense proteins including (A) ZmPR4b, (B) ZmOPR2, (C) ZmCCD1, (D) ZmGST2, (E) ZmHYD, (F) ZmOXR, (G) ZmWIP1, (H) ZmSerPIN, and (I) ZmCC9, 3 h posttreatment (all 2 mM) with SA (dark gray), 12-OPDA (light green), JA (dark green), 10-OPEA (light gray), DA0-4:0 (black), or control (white; Con1: 95:5:0.1 H20:DMSO:Tween 20, vol/vol/v), or 24 h postinfection with SLB spores (blue bar) and H20 with 0.1% Tween 20 (white; Con2). Within chemical- and SLB-infection treatment plots, different letters (a–d) represent significant differences (all ANOVA P < 0.05; Tukey test corrections for multiple comparisons: P < 0.05).
10-OPEA Cytotoxicity Is Mediated by a Cysteine Protease Inhibitor.
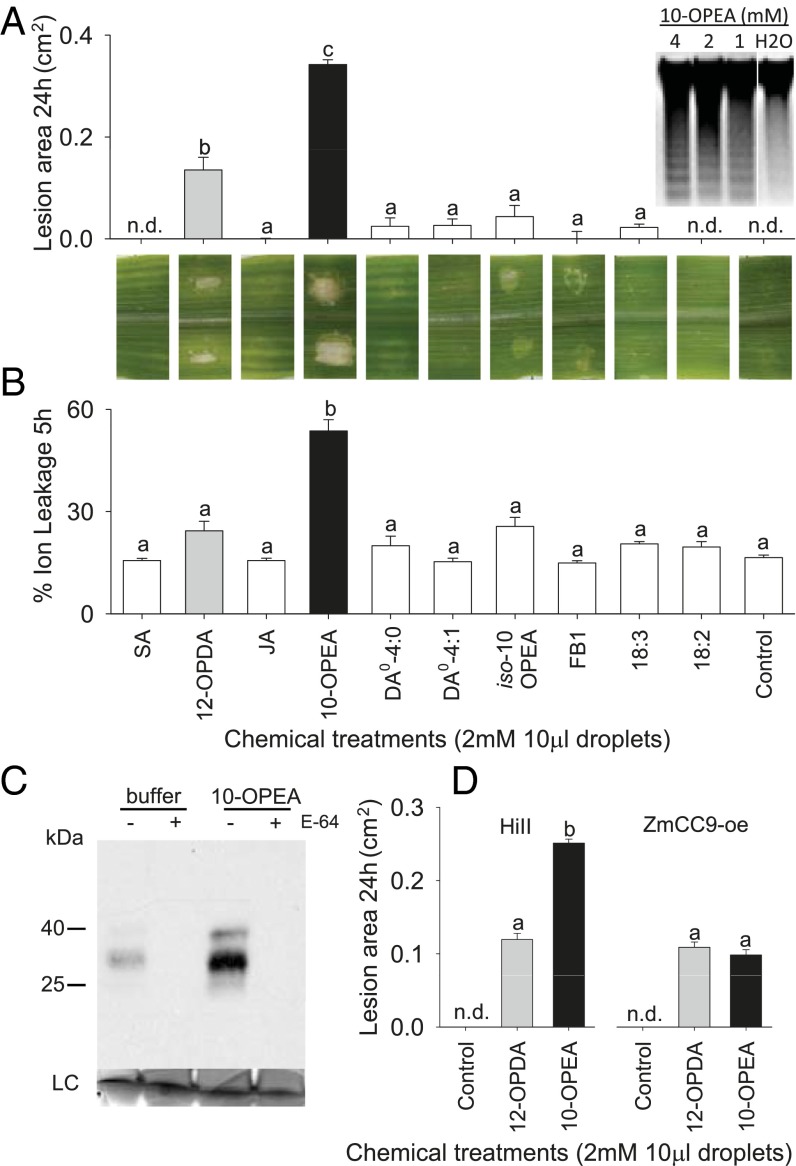
Given DA accumulation in necrotic tissue, comparatively high cytotoxicity, and weak induction of both ZmCC9 and ZmSerPIN, established inhibitors of cysteine protease-mediated cell death (27, 45), we hypothesized that high concentrations of 10-OPEA may function as a positive mediator of cell death. To compare lesion-inducing activity, we examined leaves treated with 10-OPEA, DA0-4:0, DA0-4:1, iso-10-OPEA, 18:2, 18:3, and the phytohormones JA, 12-OPDA, and SA (1, 47). Fumonisin B1 (FB1), a cell death inducing mycotoxin, was also included (48). Little or no visual evidence for cell death was found in either SA, JA, FB1, or solvent control treated plants at 24 h (Fig. 4A). However, significant cell death was observed in 10-OPEA–treated leaves with chlorotic lesion areas at least twofold larger than all other treatments (Fig. 4A). Interestingly, DA0-4:1 contains an α,β-unsaturated carbonyl similar to 10-OPEA and 12-OPDA yet lesion promotion was greatly reduced. This suggests that the presence of a Michael addition site alone does not control the observed toxicity (17). Similarly, within 5 h, electrolyte leakage in 10-OPEA–treated leaves was at least twofold greater than all other treatments and remained significantly higher over a 15-h period (Fig. 4B and SI Appendix, Fig. S9). DNA fragmentation was also observed in tissues treated with 1–4 mM 10-OPEA, a response consistent with cell death (Fig. 4A) (49, 50). As cell death processes are often mediated by cysteine proteases (27, 28), we examined the capacity of 10-OPEA to induce cysteine protease activity using DCG-04, a biotinylated substrate that reacts with the catalytic cysteine residue of papain-like cysteine proteases. Immunoblot detection of DCG-04–labeled proteases from leaf tissue collected 8 h posttreatment with 10-OPEA showed stable induction of protease activity, indicated by two bands with the expected masses of 30 and 40 kDa (Fig. 4C). Preincubation with the cysteine protease inhibitor E-64 blocked protease labeling, confirming specificity of the probe. To genetically test if inhibition of cysteine protease activity impairs 10-OPEA–induced lesions, wild-type (HiII) plants and those overexpressing (oe) the papain-like cysteine PI ZmCC9 (ZmCC9-oe) (27) were comparatively treated with 10-OPEA and 12-OPDA (Fig. 4D). At 24 h, 10-OPEA–induced lesion areas were twofold greater than 12-OPDA as expected (Fig. 4 A and D); however, on ZmCC9-oe plants there was no significant difference between 10-OPEA and 12-OPDA. Collectively, these results are consistent with 10-OPEA acting as a broadly toxic phytoalexin with the additional capacity to activate cysteine proteases and promote cell death, which is negatively regulated in part by ZmCC9. As pathogenic fungi are known to manipulate host lipid metabolism to facilitate pathogenicity (51), we used wild-type HiII and ZmCC9oe plants to investigate whether SLB uses 10-OPEA to promote necrotrophy via cysteine protease-mediated cell death. Three days post-SLB inoculation, ZmCC9oe plants showed no difference in lesion areas compared with HiII (SI Appendix, Fig. S10). Curiously, interior whorl SLB infection on HiII and ZmCC9oe plants revealed higher 10-OPEA levels in ZmCC9oe plants than HiII (SI Appendix, Fig. S10). Although speculative, these results hint at the existence of localized homeostatic compensation in 10-OPEA production to counter ZmCC9oe-mediated cell death inhibition ultimately returning the phenotype to wild-type HiII lesion levels. Alternatively, the lack of lesion area differences between SLB-infected HiII and ZmCC9oe plants suggests that SLB does not rely upon 10-OPEA cytotoxicity and cysteine protease-activated cell death for pathogenicity.
Fig. 4.
10-OPEA promotes lesions in maize. (A) Quantification (in square centimeters) of macroscopic lesions (cell death) at 24 h and (B) leaf disk percent (%) ion leakage at 5 h following treatment with two droplets (all 2 mM) of SA, 12-OPDA (gray), JA, 10-OPEA (black), DA0-4:0, DA0-4:1, iso-10-OPEA, fumonisin B1 (FB1), 18:3 (linolenic acid), 18:2 (linoleic acid), or control solution (95:5:0.1 H20:DMSO:Tween 20, vol/vol/v). Reflective surface deposits were examined and excluded if lacking lesions. (A, Insert) DNA fragmentation from leaf midrib tissue 4 h following treatment with 4, 2, and 1 mM 10-OPEA and H2O control. (C) Activity-based protein profiling demonstrates activation of cysteine proteases by 10-OPEA. Immunoblot detection of DCG-04–labeled leaf extracts 8 h posttreatment with 2 mM 10-OPEA. Extracts were pretreated with 5 µM E-64, or untreated, and thereafter labeled with DCG-04. (D) Inhibition of papain-like cysteine proteases significantly reduces 10-OPEA–mediated cell death at 24 h. Wild type (HiII) and ZmCC9-overexpression (oe) plants were treated with two 10-µL droplets of 2 mM 10-OPEA, 12-OPDA, or control solution. Within plots, different letters (a–c) represent significant differences (all ANOVA P < 0.05; Tukey test corrections for multiple comparisons: P < 0.05; nd, not detected).
The existence of a 9-LOX initiated pathway conceptually similar to the 13-LOX jasmonate pathway was postulated 15 y ago when 10-OPEA and 10-OPDA production was first observed in potato homogenates (29). Our investigation of pathogen-elicited maize oxylipins enabled the discovery of 9-LOX–derived cyclopente(a)none DAs and the characterization of 10-OPEA as a directly defensive phytoalexin with significant cytotoxicity. 10-OPEA is also the source of nontoxic cyclopente(a)nones both of which have transcriptional activity. The most critical immediate question left unanswered is “do maize plants lacking 10-OPEA biosynthesis display altered biotic or abiotic stress responses?” Given the current work and recently published findings, the most elegant way to address this issue will be the identification of the AOC(s) responsible for 10-OPEA biosynthesis and creation of null mutant plants (34). Additional remaining questions involve the occurrence and function of DAs in multiple grain crops, the role of individual DAs as transcriptional mediators, and mechanistic basis for differential activity in gene expression. Although not as commonly encountered as jasmonates, in maize, DAs have selective localized activities consistent with defense and stress response mediation.
Materials and Methods
The isolation and identification of acidic maize metabolites follows from Schmelz et al. (36) with modification (SI Appendix, SI Materials and Methods). Quantification of maize metabolites by gas chromatography-mass spectrometry, chemical treatment of plants and bioassays with C. heterostrophus, A. flavus, and F. verticillioides follows from Huffaker et al. (35) as detailed (SI Appendix, SI Materials and Methods). Unless otherwise noted, all experiments in this work consisted of at least four or more independent biological replicates. Additional details on experimental protocols and methods on can be found in SI Appendix.
Supplementary Material
Acknowledgments
Special thanks to Wayne Davis for the kind donation of maize tissues and James R. Rocca for facilitating NMR experiments at the McKnight Brain Institute (National High Magnetic Field Laboratory, Advanced Magnetic Resonance Imaging and Spectroscopy Facility). NMR work was supported by the National Science Foundation (NSF) Division of Materials Research Award 1157490, the State of Florida, and NIH Award S10RR031637. Research was funded by US Department of Agriculture (USDA)–Agricultural Research Service Projects 6615-21000-010-00/-22000-027-00, NSF Division of Integrative Organismal Systems Competitive Award 1139329, and University of California, San Diego startup funds allocated to E.A.S.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo (accession nos. GSE68589 and GSE69659).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1511131112/-/DCSupplemental.
References
- 1.Wasternack C, Hause B. Jasmonates: Biosynthesis, perception, signal transduction and action in plant stress response, growth and development. An update to the 2007 review in Annals of Botany. Ann Bot (Lond) 2013;111(6):1021–1058. doi: 10.1093/aob/mct067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Farmer EE, Alméras E, Krishnamurthy V. Jasmonates and related oxylipins in plant responses to pathogenesis and herbivory. Curr Opin Plant Biol. 2003;6(4):372–378. doi: 10.1016/s1369-5266(03)00045-1. [DOI] [PubMed] [Google Scholar]
- 3.Howe GA, Jander G. Plant immunity to insect herbivores. Annu Rev Plant Biol. 2008;59:41–66. doi: 10.1146/annurev.arplant.59.032607.092825. [DOI] [PubMed] [Google Scholar]
- 4.Koo AJK, Chung HS, Kobayashi Y, Howe GA. Identification of a peroxisomal acyl-activating enzyme involved in the biosynthesis of jasmonic acid in Arabidopsis. J Biol Chem. 2006;281(44):33511–33520. doi: 10.1074/jbc.M607854200. [DOI] [PubMed] [Google Scholar]
- 5.Katsir L, Chung HS, Koo AJK, Howe GA. Jasmonate signaling: A conserved mechanism of hormone sensing. Curr Opin Plant Biol. 2008;11(4):428–435. doi: 10.1016/j.pbi.2008.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sheard LB, et al. Jasmonate perception by inositol-phosphate-potentiated COI1-JAZ co-receptor. Nature. 2010;468(7322):400–405. doi: 10.1038/nature09430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dave A, Graham IA. Oxylipin signaling: A distinct role for the jasmonic acid precursor cis-(+)-12-oxo-phytodienoic acid (cis-OPDA) Front Plant Sci. 2012;3:42. doi: 10.3389/fpls.2012.00042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Acosta IF, et al. tasselseed1 is a lipoxygenase affecting jasmonic acid signaling in sex determination of maize. Science. 2009;323(5911):262–265. doi: 10.1126/science.1164645. [DOI] [PubMed] [Google Scholar]
- 9.Yan Y, et al. Disruption of OPR7 and OPR8 reveals the versatile functions of jasmonic acid in maize development and defense. Plant Cell. 2012;24(4):1420–1436. doi: 10.1105/tpc.111.094151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Böttcher C, Pollmann S. Plant oxylipins: Plant responses to 12-oxo-phytodienoic acid are governed by its specific structural and functional properties. FEBS J. 2009;276(17):4693–4704. doi: 10.1111/j.1742-4658.2009.07195.x. [DOI] [PubMed] [Google Scholar]
- 11.Park S-W, et al. Cyclophilin 20-3 relays a 12-oxo-phytodienoic acid signal during stress responsive regulation of cellular redox homeostasis. Proc Natl Acad Sci USA. 2013;110(23):9559–9564. doi: 10.1073/pnas.1218872110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stintzi A, Weber H, Reymond P, Browse J, Farmer EE. Plant defense in the absence of jasmonic acid: The role of cyclopentenones. Proc Natl Acad Sci USA. 2001;98(22):12837–12842. doi: 10.1073/pnas.211311098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mueller S, et al. General detoxification and stress responses are mediated by oxidized lipids through TGA transcription factors in Arabidopsis. Plant Cell. 2008;20(3):768–785. doi: 10.1105/tpc.107.054809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Danon A, Miersch O, Felix G, Camp RG, Apel K. Concurrent activation of cell death-regulating signaling pathways by singlet oxygen in Arabidopsis thaliana. Plant J. 2005;41(1):68–80. doi: 10.1111/j.1365-313X.2004.02276.x. [DOI] [PubMed] [Google Scholar]
- 15.Thoma I, et al. Cyclopentenone isoprostanes induced by reactive oxygen species trigger defense gene activation and phytoalexin accumulation in plants. Plant J. 2003;34(3):363–375. doi: 10.1046/j.1365-313x.2003.01730.x. [DOI] [PubMed] [Google Scholar]
- 16.Mueller MJ. Archetype signals in plants: The phytoprostanes. Curr Opin Plant Biol. 2004;7(4):441–448. doi: 10.1016/j.pbi.2004.04.001. [DOI] [PubMed] [Google Scholar]
- 17.Farmer EE, Mueller MJ. ROS-mediated lipid peroxidation and RES-activated signaling. Annu Rev Plant Biol. 2013;64:429–450. doi: 10.1146/annurev-arplant-050312-120132. [DOI] [PubMed] [Google Scholar]
- 18.Vellosillo T, et al. Oxylipins produced by the 9-lipoxygenase pathway in Arabidopsis regulate lateral root development and defense responses through a specific signaling cascade. Plant Cell. 2007;19(3):831–846. doi: 10.1105/tpc.106.046052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nalam VJ, Keeretaweep J, Sarowar S, Shah J. Root-derived oxylipins promote green peach aphid performance on Arabidopsis foliage. Plant Cell. 2012;24(4):1643–1653. doi: 10.1105/tpc.111.094110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.López MA, et al. Antagonistic role of 9-lipoxygenase-derived oxylipins and ethylene in the control of oxidative stress, lipid peroxidation and plant defence. Plant J. 2011;67(3):447–458. doi: 10.1111/j.1365-313X.2011.04608.x. [DOI] [PubMed] [Google Scholar]
- 21.Hwang IS, Hwang BK. The pepper 9-lipoxygenase gene CaLOX1 functions in defense and cell death responses to microbial pathogens. Plant Physiol. 2010;152(2):948–967. doi: 10.1104/pp.109.147827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gao X, et al. Disruption of a maize 9-lipoxygenase results in increased resistance to fungal pathogens and reduced levels of contamination with mycotoxin fumonisin. Mol Plant Microbe Interact. 2007;20(8):922–933. doi: 10.1094/MPMI-20-8-0922. [DOI] [PubMed] [Google Scholar]
- 23.Weber H, Chételat A, Caldelari D, Farmer EE. Divinyl ether fatty acid synthesis in late blight-diseased potato leaves. Plant Cell. 1999;11(3):485–494. doi: 10.1105/tpc.11.3.485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sun L, et al. Cotton cytochrome P450 CYP82D regulates systemic cell death by modulating the octadecanoid pathway. Nat Commun. 2014;5:5372. doi: 10.1038/ncomms6372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rustérucci C, et al. Involvement of lipoxygenase-dependent production of fatty acid hydroperoxides in the development of the hypersensitive cell death induced by cryptogein on tobacco leaves. J Biol Chem. 1999;274(51):36446–36455. doi: 10.1074/jbc.274.51.36446. [DOI] [PubMed] [Google Scholar]
- 26.Montillet JL, et al. Fatty acid hydroperoxides and H2O2 in the execution of hypersensitive cell death in tobacco leaves. Plant Physiol. 2005;138(3):1516–1526. doi: 10.1104/pp.105.059907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.van der Linde K, et al. A maize cystatin suppresses host immunity by inhibiting apoplastic cysteine proteases. Plant Cell. 2012;24(3):1285–1300. doi: 10.1105/tpc.111.093732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Solomon M, Belenghi B, Delledonne M, Menachem E, Levine A. The involvement of cysteine proteases and protease inhibitor genes in the regulation of programmed cell death in plants. Plant Cell. 1999;11(3):431–444. doi: 10.1105/tpc.11.3.431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hamberg M. New cyclopentenone fatty acids formed from linoleic and linolenic acids in potato. Lipids. 2000;35(4):353–363. doi: 10.1007/s11745-000-532-z. [DOI] [PubMed] [Google Scholar]
- 30.Grechkin AN, et al. Tomato CYP74C3 is a multifunctional enzyme not only synthesizing allene oxide but also catalyzing its hydrolysis and cyclization. ChemBioChem. 2008;9(15):2498–2505. doi: 10.1002/cbic.200800331. [DOI] [PubMed] [Google Scholar]
- 31.Itoh A, Schilmiller AL, McCaig BC, Howe GA. Identification of a jasmonate-regulated allene oxide synthase that metabolizes 9-hydroperoxides of linoleic and linolenic acids. J Biol Chem. 2002;277(48):46051–46058. doi: 10.1074/jbc.M207234200. [DOI] [PubMed] [Google Scholar]
- 32.Chauvin A, Caldelari D, Wolfender J-L, Farmer EE. Four 13-lipoxygenases contribute to rapid jasmonate synthesis in wounded Arabidopsis thaliana leaves: A role for lipoxygenase 6 in responses to long-distance wound signals. New Phytol. 2013;197(2):566–575. doi: 10.1111/nph.12029. [DOI] [PubMed] [Google Scholar]
- 33.Laudert D, et al. Analysis of 12-oxo-phytodienoic acid enantiomers in biological samples by capillary gas chromatography-mass spectrometry using cyclodextrin stationary phases. Anal Biochem. 1997;246(2):211–217. doi: 10.1006/abio.1997.2012. [DOI] [PubMed] [Google Scholar]
- 34.Ogorodnikova AV, et al. Stereospecific biosynthesis of (9S,13S)-10-oxo-phytoenoic acid in young maize roots. Biochim Biophys Acta. 2015;1851(9):1262–1270. doi: 10.1016/j.bbalip.2015.05.004. [DOI] [PubMed] [Google Scholar]
- 35.Huffaker A, et al. Novel acidic sesquiterpenoids constitute a dominant class of pathogen-induced phytoalexins in maize. Plant Physiol. 2011;156(4):2082–2097. doi: 10.1104/pp.111.179457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schmelz EA, et al. Identity, regulation, and activity of inducible diterpenoid phytoalexins in maize. Proc Natl Acad Sci USA. 2011;108(13):5455–5460. doi: 10.1073/pnas.1014714108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shimada TL, et al. Leaf oil body functions as a subcellular factory for the production of a phytoalexin in Arabidopsis. Plant Physiol. 2014;164(1):105–118. doi: 10.1104/pp.113.230185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Prost I, et al. Evaluation of the antimicrobial activities of plant oxylipins supports their involvement in defense against pathogens. Plant Physiol. 2005;139(4):1902–1913. doi: 10.1104/pp.105.066274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bhandal IS, Paxton JD, Widholm JM. Phytophthora megasperma culture filtrate and cell-wall preparation stimulate glyceollin production and reduce cell viability in suspension-cultures of soybean. Phytochemistry. 1987;26(10):2691–2694. [Google Scholar]
- 40.Zhang J, et al. Genomic analysis of the 12-oxo-phytodienoic acid reductase gene family of Zea mays. Plant Mol Biol. 2005;59(2):323–343. doi: 10.1007/s11103-005-8883-z. [DOI] [PubMed] [Google Scholar]
- 41.Hemetsberger C, Herrberger C, Zechmann B, Hillmer M, Doehlemann G. The Ustilago maydis effector Pep1 suppresses plant immunity by inhibition of host peroxidase activity. PLoS Pathog. 2012;8(5):e1002684. doi: 10.1371/journal.ppat.1002684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hwang IS, Choi S, Kim NH, Kim DS, Hwang BK. Pathogenesis-related protein 4b interacts with leucine-rich repeat protein 1 to suppress PR4b-triggered cell death and defense response in pepper. Plant J. 2014;77(4):521–533. doi: 10.1111/tpj.12400. [DOI] [PubMed] [Google Scholar]
- 43.Takezawa D. A rapid induction by elicitors of the mRNA encoding CCD-1, a 14 kDa Ca2+ -binding protein in wheat cultured cells. Plant Mol Biol. 2000;42(6):807–817. doi: 10.1023/a:1006431724090. [DOI] [PubMed] [Google Scholar]
- 44.Huffaker A, et al. Plant elicitor peptides are conserved signals regulating direct and indirect antiherbivore defense. Proc Natl Acad Sci USA. 2013;110(14):5707–5712. doi: 10.1073/pnas.1214668110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lampl N, Alkan N, Davydov O, Fluhr R. Set-point control of RD21 protease activity by AtSerpin1 controls cell death in Arabidopsis. Plant J. 2013;74(3):498–510. doi: 10.1111/tpj.12141. [DOI] [PubMed] [Google Scholar]
- 46.Rohrmeier T, Lehle L. WIP1, a wound-inducible gene from maize with homology to Bowman-Birk proteinase inhibitors. Plant Mol Biol. 1993;22(5):783–792. doi: 10.1007/BF00027365. [DOI] [PubMed] [Google Scholar]
- 47.Shah J. The salicylic acid loop in plant defense. Curr Opin Plant Biol. 2003;6(4):365–371. doi: 10.1016/s1369-5266(03)00058-x. [DOI] [PubMed] [Google Scholar]
- 48.Asai T, et al. Fumonisin B1-induced cell death in arabidopsis protoplasts requires jasmonate-, ethylene-, and salicylate-dependent signaling pathways. Plant Cell. 2000;12(10):1823–1836. doi: 10.1105/tpc.12.10.1823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Doukhanina EV, et al. Identification and functional characterization of the BAG protein family in Arabidopsis thaliana. J Biol Chem. 2006;281(27):18793–18801. doi: 10.1074/jbc.M511794200. [DOI] [PubMed] [Google Scholar]
- 50.Pennell RI, Lamb C. Programmed cell death in plants. Plant Cell. 1997;9(7):1157–1168. doi: 10.1105/tpc.9.7.1157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Christensen SA, Kolomiets MV. The lipid language of plant-fungal interactions. Fungal Genet Biol. 2011;48(1):4–14. doi: 10.1016/j.fgb.2010.05.005. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.