Significance
Harnessing the inherent capability of stem cells to maintain and regenerate injured tissues is a prerequisite for their use in mending damage to the nervous system. In the olfactory epithelium stem cells accomplish neurogenesis and epithelial repair throughout life to an extent not seen elsewhere in the nervous system. Here we show that the transcription factor protein 63 (p63) is a master regulator of the transition from the reserve to the active stem cell pool in the epithelium. Loss of p63 expression in reserve basal cells is necessary and sufficient for activation, without compromising stem cell status. Identification of this central mechanism provides a target for stem cell activation in this uniquely accessible source of patient-specific, neurogenic stem cells.
Keywords: reserve stem cell, colony-forming unit, retroviral transduction, neural regeneration, lineage tracing
Abstract
Adult tissue stem cells can serve two broad functions: to participate actively in the maintenance and regeneration of a tissue or to wait in reserve and participate only when activated from a dormant state. The adult olfactory epithelium, a site for ongoing, life-long, robust neurogenesis, contains both of these functional stem cell types. Globose basal cells (GBCs) act as the active stem cell population and can give rise to all the differentiated cells found in the normal tissue. Horizontal basal cells (HBCs) act as reserve stem cells and remain dormant unless activated by tissue injury. Here we show that HBC activation following injury by the olfactotoxic gas methyl bromide is coincident with the down-regulation of protein 63 (p63) but anticipates HBC proliferation. Gain- and loss-of-function studies show that this down-regulation of p63 is necessary and sufficient for HBC activation. Moreover, activated HBCs give rise to GBCs that persist for months and continue to act as bona fide stem cells by participating in tissue maintenance and regeneration over the long term. Our analysis provides mechanistic insight into the dynamics between tissue stem cell subtypes and demonstrates that p63 regulates the reserve state but not the stem cell status of HBCs.
Stem cells harvested from adult tissues are a promising source of material for use in regenerative medicine if they can be appropriately identified and manipulated. Recent studies in a wide range of adult tissues, including the lining of the gastrointestinal tract, the adult and embryonic CNS, the hematopoietic elements of bone marrow, and the olfactory epithelium (OE), which is the neuroepithelium within the lining of the nasal cavity that subserves odorant transduction, have yielded a somewhat unexpected result: Multiple molecularly and morphologically distinct cell types within a tissue have the capacity to function as stem cells and satisfy even the most stringent criteria of stemness (1–7). Broadly, these populations can be functionally distinguished as active (participating in routine tissue maintenance) and reserve (dormant under normal circumstances but capable of activation by injury to participate in tissue regeneration) (8–10).
The OE provides a unique system to study the dynamics between active and reserve stem cells. The extensive neurogenic and regenerative capacity of the OE in both rodents and humans persists throughout adult life and is unmatched elsewhere in the nervous system (11–14). The two populations of stem cells that are present in the OE fit the active–quiescent dichotomy described above. The active population, globose basal cells (GBCs), constitutes a heterogeneous set of ostensibly lineage-committed and uncommitted progenitor cells that normally persist throughout adult life (6, 15, 16). GBCs are unique to the OE, identifiable at early stages of embryonic development, and are functionally and molecularly homologous to the embryonic olfactory placode progenitor cells (17–19). In contrast, the dormant population, horizontal basal cells (HBCs), which share molecular and morphological similarities with basal cells in other tissues, first appear perinatally at a time when the architecture and cellular constituents of the OE are fully formed (20, 21). HBCs very rarely contribute to cellular maintenance in undamaged tissue, even in the face of accelerated neuronal turnover, but can become activated by severe and direct injury to the epithelium and thereby give rise to all the cellular components of the OE during regeneration (7).
The transcription factor protein 63 (p63) is expressed in the basal stem cell populations of stratified epithelia such as the epidermis, and p63-knockout mice fail to form stratified epithelia altogether (22, 23). Cell-autonomous, conditional ablation of p63 causes cell death, senescence, spontaneous differentiation, or robust mitotic amplification depending on the cell type and mutated isoform (24–27). Because of the complexity of the p63 gene, which encodes six potential isoforms, and the large number of putative target genes, a wide array of functions has been attributed to this transcription factor including the generation, maintenance, self-renewal, proliferation, and differentiation of stem cells as well as both tumor-suppressor and oncogenic roles. Because many of these processes would be mutually exclusive within a given cell population, it is most likely that p63 operates in a highly context-dependent manner as a pivotal switch in a variety of processes including cell adhesion, cell-cycle regulation, and cell-signaling pathways known to regulate stem cell function.
During embryonic development, olfactory placodal-like progenitors and GBCs express p63 in the perinatal period before giving rise to HBCs. By adulthood, p63 (specifically, the isoform lacking the N-terminal trans-activation domain, ΔNp63) is expressed exclusively in mature HBCs. Genetic knockout of p63 results in an OE in which HBCs do not differentiate but that appears otherwise normal at the light and electron microscopic level (20). This effect is in stark contrast to the knockout of p63 in other stratified epithelial tissues, which do not form in mutant animals (22, 23). Moreover, HBC activation following injury is accompanied by p63 down-regulation (20), and conditional knockout (cKO) of p63 results in the spontaneous differentiation of HBCs (28). In one interpretation of these data, p63 is essential for the self-renewal of the stem cell population in the OE. However, this interpretation does not take into account several features unique to olfactory epithelial maintenance, namely, a subset of GBCs are likely to be stem cells (6, 15), activated HBCs must transition through a GBC stage during regeneration (7), and HBCs are largely dispensable in both the embryonic generation and in the adult maintenance of the OE (7, 20).
To define the role of p63 in the dynamics of the cellular context in the OE, a more thorough analysis of the transitions between active and reserve stem cells is necessary. Here, we use transplantation, gain and loss of function, and a variety of lesion models in vivo to define the timing and nature of HBC dynamics. Our data demonstrate that p63 preserves the pool of reserve HBCs in the OE but does not maintain stemness per se and also highlight the contribution of the GBC population to long-term epithelial homeostasis.
Results
p63 Down-Regulation Anticipates Proliferation of Activated HBCs.
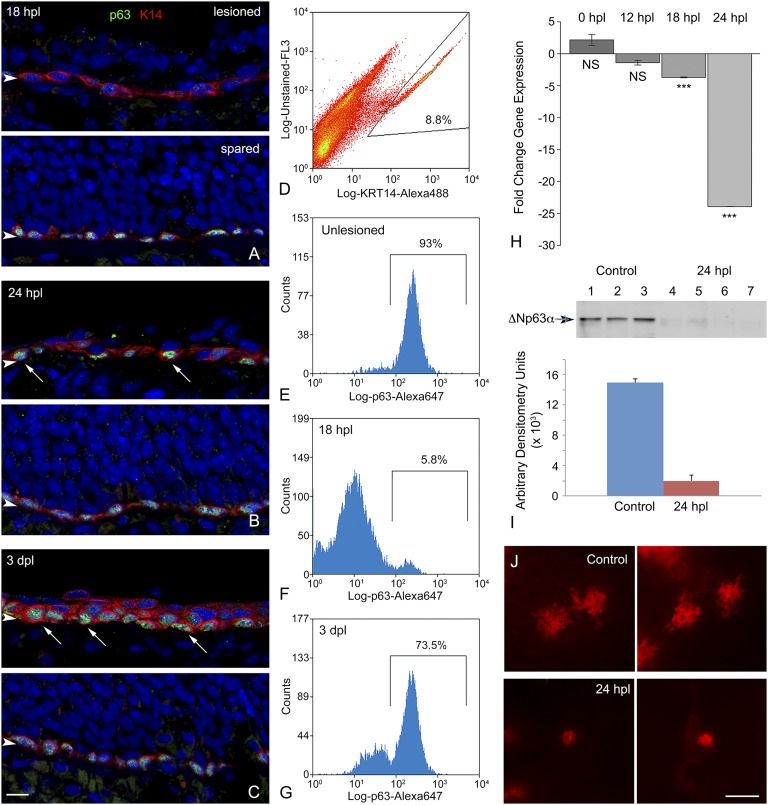
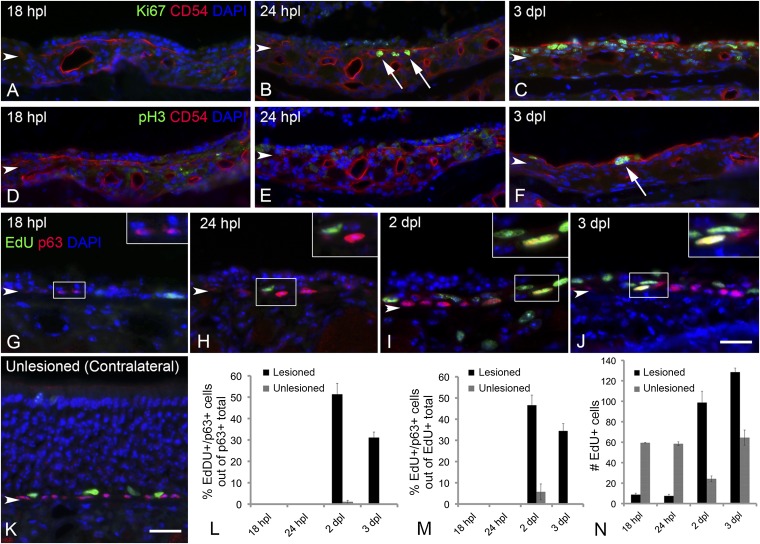
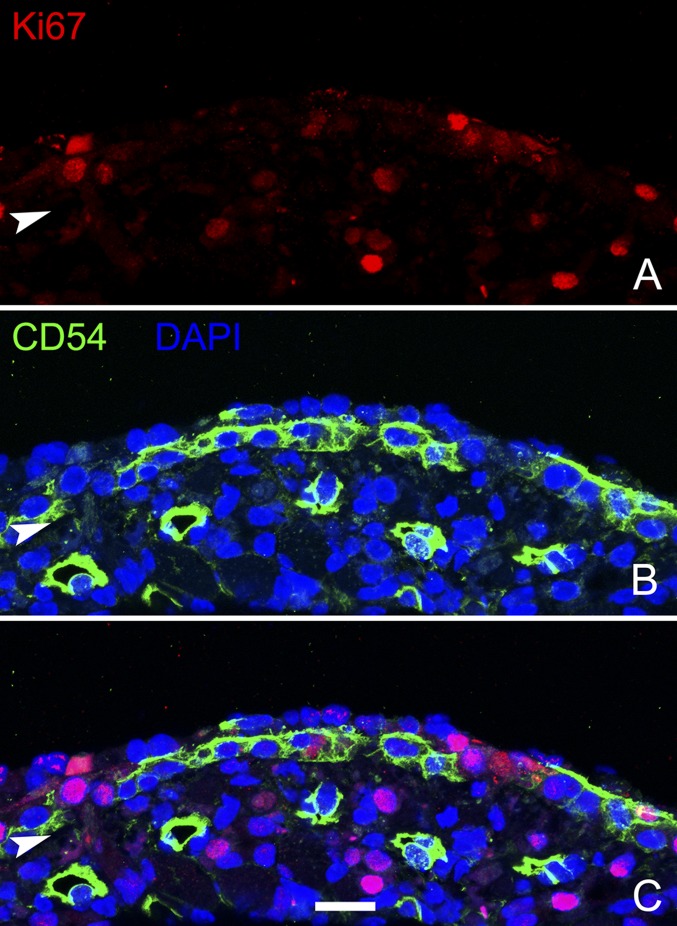
To analyze the role of p63 in the dynamics of HBC activation, we first sought to describe the timeline of p63 protein down-regulation after unilateral exposure to the olfactotoxic gas methyl bromide (MeBr), which causes activation of HBCs (7). By comparing the lesioned side and the unlesioned side, we found that p63 levels are maximally reduced in cytokeratin 5/cytokeratin 14 (K5/K14)-expressing HBCs at 18 h post lesion (hpl) both by immunohistochemistry and flow cytometric analysis (Fig. 1 A and D–F); it is worth noting that the spread of labeling intensities observed with flow cytometric analysis is broader at 18 hpl than in the unlesioned control (Fig. 1 E and F). By 24 hpl the K14+ population begins to bifurcate; in a few cells p63 levels have recovered to normal or nearly normal, but in others p63 has disappeared entirely (Fig. 1B). The segregation of K14+ cells into two populations of different p63 staining intensities is strikingly pronounced by 3 d post lesion (dpl) (Fig. 1 C and G). The overall decline in p63 expression over the first 24 hpl also is observed in both quantitative RT-PCR and Western blot assays (Fig. 1 H and I). The decline in protein levels revealed by immunohistochemistry or by flow cytometry precedes the drop in p63 transcript level that is revealed by the quantitative RT-PCR analysis. By 24 hpl the drop in p63 expression is associated with a conspicuous simplification of HBC morphology and elimination of the extensive processes that normally characterize them (Fig. 1 B and J). Down-regulation of p63 occurs significantly earlier than the proliferative burst of HBCs, which previously has been associated with activation (7). K14+ or CD54+ HBCs are first labeled by the expression of Ki67, a marker of active cell cycling, at 24 hpl (Fig. S1 A–C), and mitotic rates peak at 3 dpl [phospho-histone H3 (pH3) staining] (Fig. S1 D–F). Moreover, both p63+ and p63− cells are labeled by 5-ethynyl-2'-deoxyuridine (EdU) incorporation with a similar time course after lesion, indicating that the lack of p63 expression, in and of itself, does not correlate with active cell cycling (Fig. S1 G–N).
Fig. 1.
HBCs respond to MeBr exposure with declines in p63 protein- and gene-expression levels and morphological changes. (A–C) K14 and p63 staining over an acute time course after unilateral MeBr exposure. The lesioned side is the top image in each pair, and the spared side of the same animal is on the bottom. (A) At 18 hpl the mature cells of the OE on the lesioned side have been largely shed, and the K14+ HBCs are still arrayed as a monolayer. However, the level of p63 staining has declined precipitously in all the cells in this field, by comparison with a comparable area on the unlesioned side. (B) At 24 hpl, the level of p63 staining on the lesions side has bifurcated. For some K14+ cells, particularly those adjacent to the basal lamina, staining for p63 has rebounded to be indistinguishable from that on the unlesioned side (arrows). (C) By 3 dpl, K14+ cells are disposed in multiple layers, with the intensely p63+ cells adjacent to the basal lamina (arrows). (Scale bar: 10 µm for A–C.) (D–G) The immunohistochemical pattern observed in A–C is confirmed when the septal OE is dissected and dissociated and the constituent cells are fixed, jointly stained with anti-K14 and anti-p63 antibodies, sorted on the basis of K14 positivity, and analyzed for p63-staining. (D) Example of the sorting strategy highlighting the population of K14+ cells subjected to analysis of p63 staining. (E) Unlesioned OE. The overwhelming majority of K14+ cells are strongly stained for p63. (F) At 18 hpl, in contrast to the unlesioned OE, the great majority of the K14+ cells are much more weakly stained for p63 and fall below the analytical gate. In addition, the levels of staining are more heterogeneous, extending over two orders of fluorescent intensity. (G) At 3 dpl there has been substantial recovery in the percentage of K14+ cells in which p63 staining exceeds the lower end of the analytic gate, although a substantial population is more lightly stained for p63. The FACS pattern fits well with the immunohistochemical analysis. (H) Quantitative PCR (qPCR) analysis for ΔNp63 isoforms as a function of time after the period of MeBr inhalation; 0 hpl signifies animals killed immediately at the end of the 8-h exposure. The fall-off in gene expression slightly lags the precipitous decline in immunohistochemical staining for p63.Statistical comparison with unlesioned control mice by Student’s t test; ***P < 0.001; NS, not significant. (I, Upper) Western blot analysis of p63 protein levels in normal OE (lanes 1–3) vs. 24 hpl (lanes 4–7). The arrowhead designates the position of the unmodified ΔNp63 band, which is markedly reduced at 24 hpl as shown by densitometry analysis of the blots (Lower). (J) By 24 hpl the HBCs have undergone substantial simplification of their morphology, as shown in CLARITY-prepared whole mounts of K5.CreERT2 > R26R(TdTomato) mice. In the septal OE of normal mice, the HBCs display elaborate extensions that presumably indicate hemidesmosomal attachments to the basal lamina, based on comparisons with published electron microscopic examinations. After lesion, the cells lose most of the extensions, rendering them simpler in outline. (Scale bar: 20 μm.)
Fig. S1.
Proliferation of HBCs does not correlate with p63 expression. The decline in p63 levels anticipates the onset of proliferation by a substantial period; when proliferation accelerates, mitotic activity increases p63+ basal cells as well. At 18 hpl there are virtually no proliferating CD54+ HBCs as labeled by the cell-cycle marker Ki67 (A), by the mitotic marker pH3 (D), or by the incorporation of EdU (G); the nonproliferation of HBCs is typical of the normal OE (K and M). Rare CD54+ HBCs have entered the cell cycle at 24 hpl, as shown by Ki-67 expression (arrows in B), but they are very infrequent and generally have not progressed to incorporate EdU (H) and do not enter mitosis (E). At 2 dpl a large number of p63+ basal cells incorporate EdU (I). Likewise, at 3 dpl substantial numbers of CD54+ basal cells are labeled for Ki67 (C) and pH3 (arrow in F); many of the p63+ cells incorporate EdU at this time (J). Arrowheads mark the basal lamina. (Scale bar: 20 µm for all images.) (L) Quantitative assessment of the percentage of p63+ cells that incorporate EdU confirms the finding that HBCs are proliferating at 2 and 3 dpl, whereas p63+ basal cells reemerge at 24 hpl (see Fig. 1); for each type of count, the unexposed side of the same animal is plotted for comparison. (M) Likewise, a large proportion of the EdU+ proliferating cells are labeled for p63. (N) The recruitment of p63+ basal cells into the cell cycle parallels the overall time course of proliferation in the epithelium, independent of cell type. Arrowheads mark the basal lamina.
An in Vivo cfu Assay Identifies Activated HBCs and Correlates with p63 Down-Regulation.
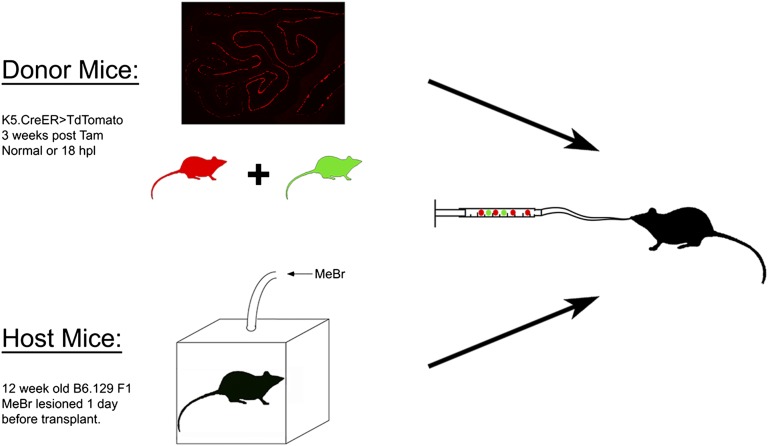
We have demonstrated previously that, in contrast to GBCs, HBCs isolated from normal, intact OE do not engraft and do not participate in epitheliopoiesis after transplantation into MeBr-lesioned OE (6). In accordance with the expectation that multipotency after transplantation is a hallmark of active, but not of dormant, stem cells, we transplanted an equal number of labeled HBCs harvested from uninjured mice or mice killed 18 h after MeBr exposure. HBCs were labeled by injecting mice bearing the K5.CreERT2 driver and the R26R(TdTomato) reporter (abbreviated KT) with 300 mg/kg tamoxifen 2 wk before harvesting. Tamoxifen treatment resulted in TdTomato expression in 75 ± 17% of HBCs and their progeny. Because the uninjured OE contains one or more multipotent stem cell populations capable of engrafting and participating in epithelial regeneration, we mixed the KT donor cells with cells from the olfactory mucosa of an unlesioned β-Actin.GFP (BACT.GFP) mouse (in which GFP is constitutively expressed by a chicken β-actin/CMV promoter–driven transgene) (6) to serve as a positive control (Fig. S2).
Fig. S2.
Experimental design of transplantation experiments. OE (normal, postlesion, or p63fl/fl) was dissociated from donor animals at the indicated interval after tamoxifen (Tam) injection. As a positive control, OE cells from one B6.GFP mouse, which expresses GFP in all cells, were mixed with the labeled donor cells. Cells then were infused into the nasal cavity of a single host animal, which had been lesioned 24 h before transplantation. Each condition was repeated in three host animals.
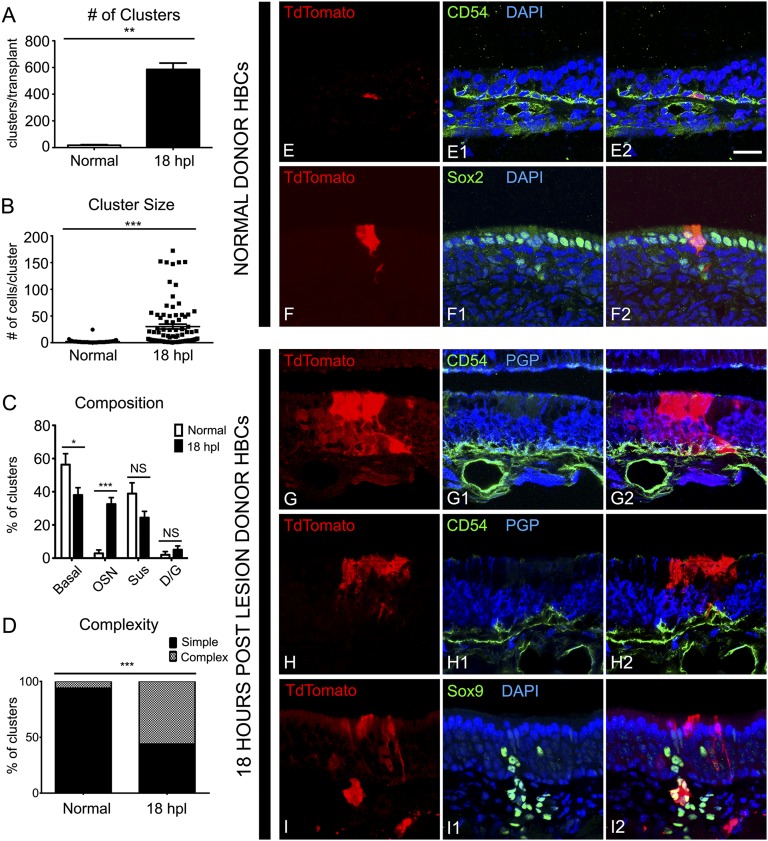
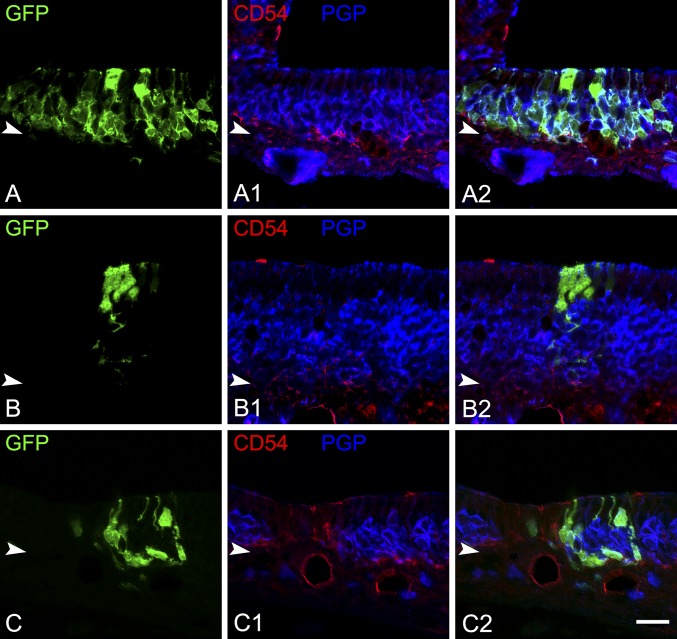
After transplantation of TdTomato-labeled HBCs from unlesioned OE, we observed only 17 ± 4 engrafted clusters per host animal (n = 3) (Fig. 2A). These clusters were relatively small with an average of 2.2 ± 0.5 cells per cluster (Fig. 2B). Of the 52 clusters, 23 (44%) contained only HBCs as identified by morphology and K14 or CD54 staining (Fig. 2 C and E); 15 clusters (29%) contained only apical supporting cells, as identified by morphology and intense sex-determining region Y-box 2 (Sox2) staining (Fig. 2 C and F); and six clusters (12%) contained basal cells and supporting cells (Fig. 2C). Only three clusters (6%) contained neurons, and of those only one cluster (2%) was found to contain olfactory sensory neurons (OSNs) and supporting cells (Fig. 2C). These results are in contrast to the BACT.GFP+ control cells, which engrafted and gave rise to many simple (consisting of one cell type) and complex (containing more than one cell type) clusters (Fig. S3).
Fig. 2.
A cfu assay demonstrates HBC activation at 18 hpl. (A) As shown by the number of clusters, HBCs transplanted from HBCs 18 h after MeBr lesion (18 hpl) more efficiently than HBCs from unlesioned donors; **P < 0.001, Mann–Whitney test. (B) Clusters derived from 18 hpl HBCs are significantly larger than clusters derived from unlesioned donors; ***P < 0.0001, Mann–Whitney test. (C) Summed across all clusters, cells derived from 18 hpl HBCs differentiate into all the mature cell types of the OE including OSNs and Bowman’s D/Gs, whereas cells derived from normal HBCs preferentially differentiate into cytokeratin-expressing basal cells and Sus cells; *P < 0.05, ***P < 0.0001; NS, not significant; ANOVA with Bonferroni correction. (D) Clusters from 18 hpl HBC are more likely to be complex, containing more than one cell type, than clusters from unlesioned donors; ***P < 0.0001, χ2 with Yates’ correction performed on raw data. (E and F) Examples of clusters derived from unlesioned donor HBCs. Unlesioned donors gave rise to predominantly CD54+ HBCs (E) and Sox2+ Sus cells (F). (G–I) Examples of clusters derived from18 hpl donor HBCs. At 18 hpl HBCs gave rise to complex clusters consisting of CD54+ HBCs, PGP9.5+ OSNs, and morphologically distinct Sus cells (G), simple clusters consisting of morphologically distinct Sus cells apical to the PGP+ layer of OSNs (H), and entire, morphologically distinct, Sox9+ Bowman’s D/G units (I). (Scale bar: 20 µm.)
Fig. S3.
Transplantation of B6.GFP+ cells from unlesioned donor mice results in the expected range of cluster size and complexity in contrast to the results obtained following HBC transplantation. (A–A2) A large, complex cluster containing basal cells, OSNs, and Sus cells derived from the transplantation of an unsorted, dissociated preparation of cells from the OE of unlesioned mice. (B–B2) A small, simple cluster consisting of Sus cells only. (C–C2) An example of a cluster with OSNs and basal cells (white arrow). Arrowheads mark the basal lamina. (Scale bar: 20 µm.)
The relative paucity of graft-derived clusters following transplantation of HBCs from the intact OE of unlesioned mice stands in marked contrast to the results obtained when labeled HBCs were transplanted from the OE of mice killed 18 hpl. In the latter case, transplant efficiency, as measured by engraftment, exhibited a 30-fold increase, with 585 ± 48 TdTomato+ clusters observed per host animal (n = 3) compared with 17 ± 4 clusters per animal from unlesioned HBCs (Fig. 2A). The size of these clusters is more than an order of magnitude larger on average than the normal HBC grafts (27.4 ± 8.3 cells per cluster vs. 2.2 ± 0.5 cells per cluster) (Fig. 2B). The 18-hpl–derived clusters included simple colonies consisting of only neurons or only supporting cells as well as significantly more complex colonies, which included combinations of neurons, HBCs, GBCs, supporting cells, and/or Bowman’s duct/gland cells (D/Gs) (P < 0.0001, χ2 with Yates’ correction) (Fig. 2 C, D, and G–I). Thus, activated HBCs can be identified by their ability to engraft and their multipotency coincident with the maximal down-regulation of p63 in HBCs.
Down-Regulation of p63 Is Necessary for HBC Activation.
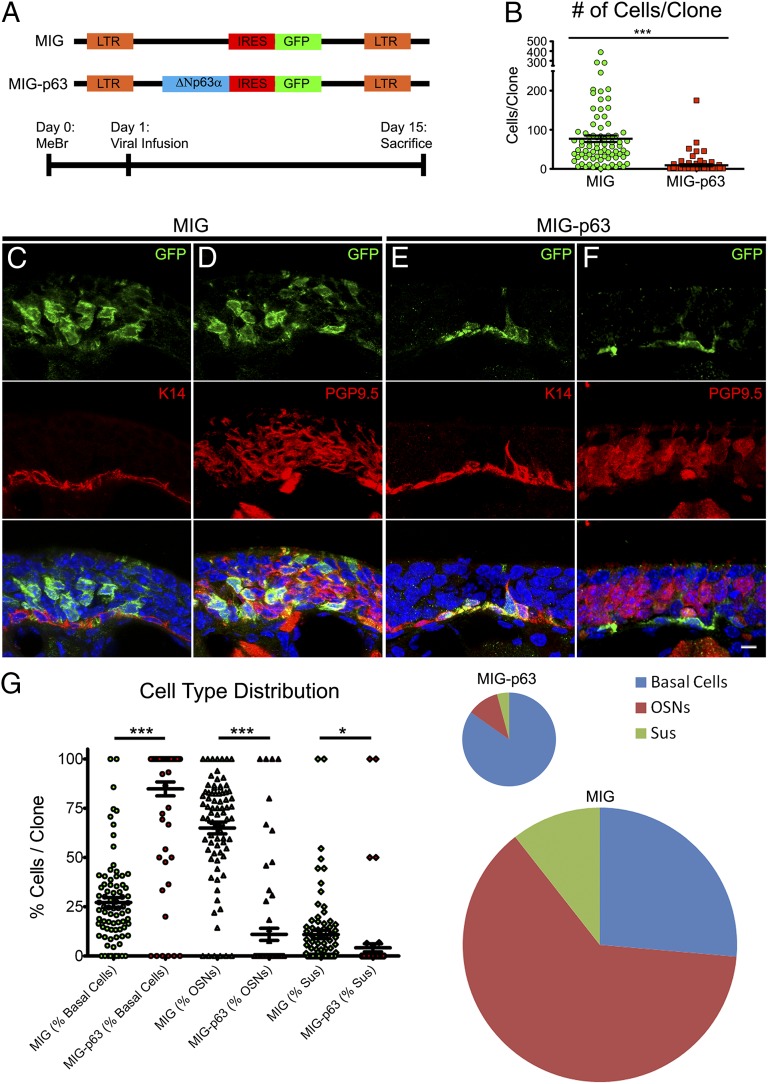
To test the functional significance of p63 down-regulation after injury, we expressed ΔNp63α in the dividing cells of the OE 24 hpl by intranasal infusion of the murine stem cell virus (MSCV)-ΔNp63α-internal ribosome entry site (IRES)-GFP (MIG-p63) retroviral vector to transduce infected cells and their progeny (Fig. 3A). At this time point (24 hpl), the majority (81%) of proliferating, Ki67+ cells are CD54−, and they are more superficial and accessible to viruses following intranasal infusion, suggesting that most, but not all, infected cells are CD54− GBCs (Fig. S4). MIG-p63–transduced progenitor cells gave rise to much smaller clones compared with the MSCV-IRES-GFP (MIG) control (9.3 ± 2.6 vs. 77 ± 8.3 cells per clone, P < 0.0001, Mann–Whitney u test = 472.5, two tailed, n = 3) (Fig. 3B). Of the MIG-p63 clones, 74% were composed of basal cells only. Upon closer examination, we found that all the analyzed basal cells were immunopositive for HBC markers such as K14 and CD54 and were protein gene product 9.5-negative (PGP9.5−) (Fig. 3 C and D). On the other hand, MIG-transduced clones were heterogeneous and included both simple and complex clones (Fig. 3 E and F). When all cell types were grouped independently of clone of origin, a similar pattern emerged: MIG-p63–infected clones rarely differentiate into sensory neurons (11% of progeny compared with 65% in MIG-derived cells) or sustentacular (Sus) cells (4% vs. 11%), whereas MIG-transduced clones reflect the full plastic heterogeneity of proliferating cells at 24 hpl, consistent with past data using retroviral lineage tracing after MeBr lesion in rats (Fig. 3G) (29). These findings demonstrate that p63 expression inhibits HBC activation and promotes maintenance of an HBC phenotype and/or its adoption by GBCs, indicating that p63 down-regulation is necessary for HBC activation.
Fig. 3.
Down-regulation of p63 is necessary for HBC activation. (A, Upper) Schematics of retroviral constructs. (Lower) Experimental timeline. (B) Infection with MIG-p63 significantly attenuated the size of the resultant clones compared with empty vector, MIG-infected controls; ***P < 0.0001, Mann–Whitney test. (C and D) Representative clones resulting from MIG infection. Infection results in large clones including both simple clones, consisting of only one cell type (C), and complex clones, consisting of more than one differentiated cell type (D). (E and F) Representative clones resulting from MIG-p63 infection. MIG-p63 infection results in smaller, simple clones consisting of K14+ HBCs (E) and, rarely, smaller clones consisting of K14+ HBCs with very few PGP9.5+ OSNs (F). (Scale bar: 10 µm.) (G, Left) Quantitative analysis of cell type distribution by percentage of the contribution of each differentiated cell type to each individual clone. *P < 0.05, ***P < 0.0001; Kruskal–Wallis test. (Right) Pooled data of cell type contribution normalized to average clone size as indicated by pie chart size.
Fig. S4.
Proliferation at the time of retroviral transduction (24 hpl). Many of the Ki67+ dividing cells 24 h after MeBr lesion are CD54− GBCs situated superficial to the layer of CD54+ HBCs. (A–B) Ki67+ and CD54 staining in individual panels. (C) Overlay of all three channels to show lack of staining colocalization. Arrowheads mark the basal lamina. (Scale bar: 10 µm.)
Down-Regulation of p63 Is Sufficient for HBC Activation.
To determine whether p63 down-regulation is sufficient for HBC activation in the absence of injury, we used mice homozygous for the p63fl/fl allele and bearing the K5.CreERT2 driver (K5.CreERT2 > p63fl/fl) to accomplish cKO of p63 in HBCs of the unlesioned OE. Recombination events were traced using either LacZ [R26R(LacZ)] or TdTomato [R26R(TdT)] reporter transgenic strains, depending on technical convenience for immunostaining purposes. In accordance with prior findings (28), we found that 91 ± 2.1% of the reporter-labeled p63fl/fl cells no longer had HBC morphology and were K14−, consistent with HBC activation (Fig. 4 A–C). All the labeled p63fl/fl cells that were K14+ (a small minority at 9%) also were immunopositive for p63, indicating incomplete recombination of all three alleles in HBCs following tamoxifen treatment (Fig. 4I). Consistent with past studies, the cells that derived from p63fl/fl HBCs included differentiated sensory neurons, GBCs, and rare Sus cells (Fig. 4 D–H). We also observed other HBC-derived cells that have not been reported previously. A significant proportion of X-Gal+ cells (2.7 ± 0.9%) were constituents of morphologically identifiable Bowman’s D/G units, indicating that HBCs can readily differentiate along this lineage to contribute to Bowman’s D/G units in the context of an otherwise unmanipulated OE (Fig. 4G). We also investigated the identity of the apical, nonneuronal, X-Gal+ cells, and found that many of them stain positive for the microvillar cell marker TrpM5 (transient receptor potential cation channel, subfamily M, member 5) (Fig. 4H). It is noteworthy that the cell types generated following HBC activation include nonneuronal cells, in contrast to the descendants of NeuroD1+ GBCs (29). Thus, it is likely that the persistent GBCs generated by activated HBCs belong to a more upstream GBC subpopulation, such as the Paired box 6 (Pax6)/Sox2+ and/or Ascl1+ cells, which have been shown to be multipotent by retroviral and transplantation analysis (6, 17, 18, 30–32).
Fig. 4.
Down-regulation of p63 is sufficient for HBC activation. (A and B) Comparison of HBC lineage trace in WT K5.CreERT2 > LacZ mice (A and A′) and p63 cKO in K5.CreERT2 > p63fl/fl;R26R(LacZ) mice (B and B′) 2 wk after 50 mg/kg tamoxifen induction shows robust HBC activation in cKO vs. WT tissue. (C) Quantification of the proportion of non-HBCs derived from K5+ cells in WT [K5.CreERT2 > R26R(LacZ)], heterozygote [HET, K5.CreERT2 > p63fl/+;R26R(LacZ)], and homozygote [HOMO, K5.CreERT2 > p63fl/fl;R26R(LacZ)] tissue, showing a relationship between p63 gene dosage and activation efficiency; *P < 0.0001; ANOVA with Bonferroni correction. (D–H) At 4 wk after tamoxifen (50 mg/kg) administration, HBCs activated by p63 cKO give rise to all the differentiated cell types of the OE, including K14−/PGP9.5− GBCs situated between the HBC monolayer and the sensory neurons (arrows in D); Sox2+ columnar Sus cells situated at the apex of the epithelium (arrow in E); mature OMP+ OSNs (arrows in F); Bowman’s D/G units (arrow in G); and TrpM5+ microvillar cells (arrow in H). (I) In the K5.CreERT2 > p63fl/fl;R26R(LacZ) mice some LacZ+ HBCs (identified by their flattened shape) abutting the basal lamina retain expression of p63 protein, indicating that they have not undergone complete recombination of all three alleles. (Scale bars: 50 µm in A and B; 20 µm in A′, B′, and D–I.)
Finally, in addition to the findings in the p63fl/fl animals, we found that after tamoxifen administration the incidence of marker-positive/K14− non-HBCs was significantly higher in p63fl/+ animals than in WT p63+/+ mice (14.3 ± 3.6% vs. 0.88 ± 0.5%, P < 0.0001, Kruskal–Wallis K = 23.47, n = 3) (Fig. 4C). The haploinsufficiency seen with p63 heterozygosity is further evidence that the effect of p63 in the OE is dose dependent, as was previously demonstrated in the developing HBCs of the perinatal OE (20). Taken together, the results of p63 cKO indicate that cell-autonomous loss of p63 is sufficient for HBC activation in a dose-dependent manner even in the absence of tissue injury.
Activated HBCs Give Rise to Persistent, Multipotent GBCs.
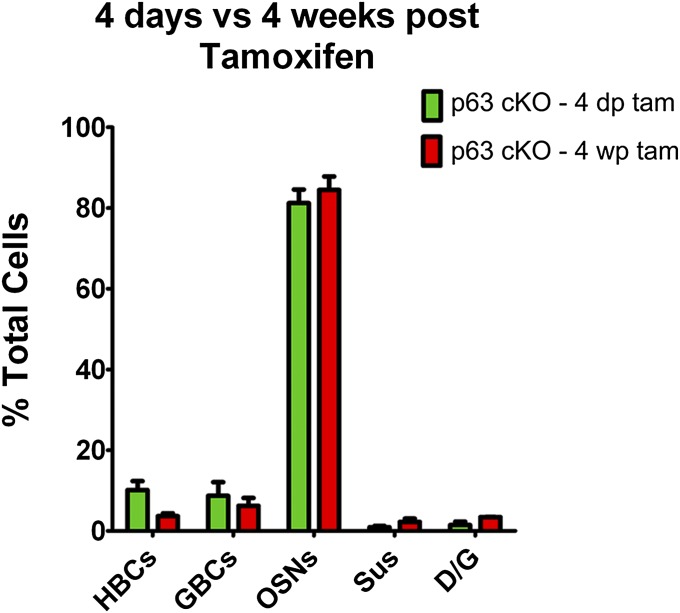
Past studies have demonstrated that activated HBCs give rise to GBCs when they participate in tissue regeneration, but it is unclear whether the HBC-derived GBCs function as transient progenitors or assume a persistently active stem cell state (7, 28). Published data indicate that GBCs of the normal OE constitute a heterogeneous population, which includes relatively quiescent, long-lived stem cells (GBCQ), dividing, Sox2/Pax6+ multipotent progenitors (GBCMPP), rapidly dividing transit-amplifying cells (GBCTA), and immediate neuronal progenitors (GBCINP) (17, 18, 31–33). The similarity of the composition of the HBC-derived clones at 4 d and 4 wk after recombination and p63 deletion suggests that the GBCs function similarly over that time period (Fig. S5).
Fig. S5.
The behavior of HBC-derived progenitor cells is equivalent early and late after their formation. K5.CreERT2 > p63fl/fl;R26R(TdT) mice were injected with tamoxifen to delete p63 by recombination and thereby activate the HBCs (and prevent their redifferentiation into HBCs). The distribution of cells generated by the HBC-derived progenitors was determined 4 d and 4 wk postinjection. There is no statistically significant difference in the cell types generated by the progenitors derived from activated HBCs at 4 d (4 d tam) and 4 wk (4 wk tam) after tamoxifen injection (two-way ANOVA). The similarity in outcomes suggests that the progenitors are functionally equivalent at the two time points.
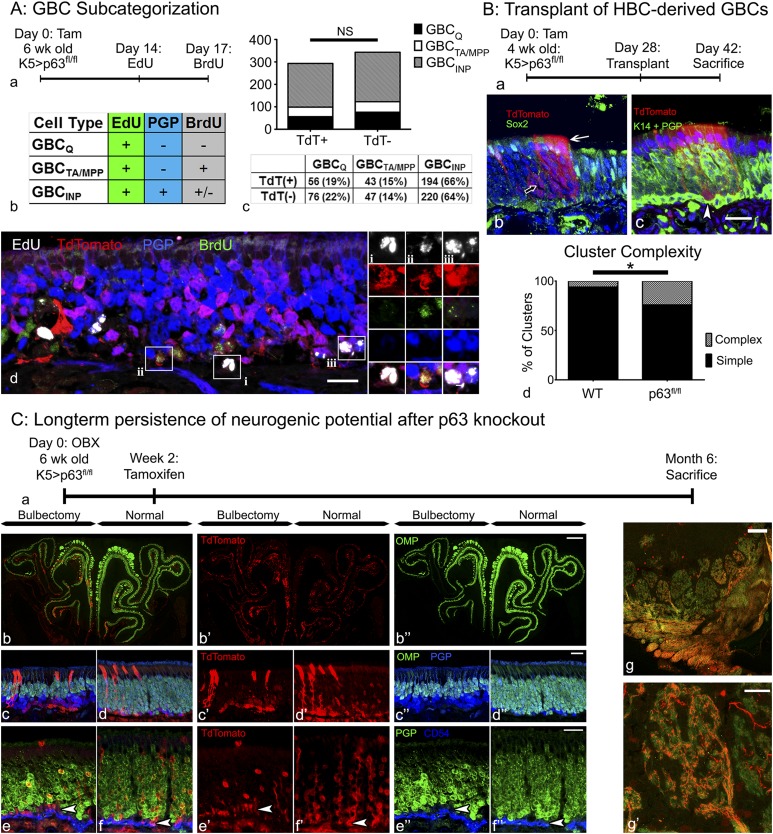
To analyze the several GBC subtypes, we labeled proliferating GBCs by injecting EdU 2 wk after tamoxifen induction of K5.CreERT2 > p63fl/fl;R26R(TdT) HBCs; a follow-up BrdU pulse was administered 2 h before the animal was killed to label rapidly cycling cells (Fig. 5 A, a); PGP9.5 labeling was used to identify immature and mature neurons. By operational definition, GBCQs remained EdU+/BrdU−, whereas GBCMPPs and GBCTAs were double-labeled (EdU+/BrdU+). Finally, the descendants of GBCINPs could be identified as EdU+/PGP9.5+ (Fig. 5 A, b).
Fig. 5.
Activated HBCs generate GBCs, which integrate into the tissue and retain stem cell potential comparable to endogenous GBCs. (A) GBC subcategorization: analysis of the proliferation dynamics of endogenous TdTomato− (TdT−), and HBC-derived TdTomato+ (TdT+) GBCs. (a and b) Timeline (a) and experimental design (b) for assessing the different GBC subtypes in K5.CreERT2 > p63fl/fl;R26R(TdT) animals as described in the table. Note that the population of GBCINPs is defined by their differentiation into PGP+ sensory neurons. (c) Quantification and comparison of TdT+ (i.e., HBC-derived) and TdT− GBCs, showing no significant (NS) difference between these two GBC populations in terms of subtype distribution. (d) Representative section (included in the quantitative analysis in c) containing each of the three classifications of GBCs: (i) GBCQs: TdT+/EdU+/BrdU−/PGP−; (ii) GBCTA/MPPs: TdT+/EdU+/BrdU+/PGP−; (iii) GBCINPs: TdT+/EdU+/PGP+. (Scale bars: 10 µm in A, d; 2 µm in A, d, i–iii;) (B) Transplantation of HBC-derived cells: Transplantation of cells from K5.CreERT2 > p63fl/fl;R26R(TdT) mice [Kp63fl/flT] 4 wk after tamoxifen injection; note that the tamoxifen-treated donor animals were unlesioned at the time of harvest. (a) Timeline of experimental design. (b) An example of a complex clone consisting of morphologically distinct OSNs (open arrow) and Sox2+ Sus cells (white arrow). (c) An example of a complex clone containing a row of labeled Sus cells (arrow) and K14−/PGP− GBCs (white arrowhead). (Scale bar: 10 µm in B, b and c.) (d) Quantification of HBC-derived clones from Kp63fl/flT donors vs. WT controls (i.e., tamoxifen-treated Kp63+/+T from Fig. 1). *P < 0.05; χ2 with Yates’ correction, performed on raw data. (C) Long-term persistence of neurogenic potential after p63 knockout: Analysis of HBC-derived cells in KpT animals 6 mo after OBX. (a) Timeline of experimental design. (b–d) Mosaic image encompassing the whole section (b) and representative high-magnification images (c and d) of KpT tissue 6 mo after OBX demonstrating significant thinning of the OMP+ neuronal layer on the bulbectomized side (c) compared with the unoperated contralateral control side (d). HBC-derived TdTomato+ GBCs remain neurogenic and make a substantial contribution to the population of PGP9.5+ sensory neurons. (e and f) Representative high-magnification images from the bulbectomized side (e) and the unoperated contralateral control side (f) demonstrating the persistence of PGP9.5−/CD54− GBCs (white arrowheads). (g) On the unlesioned side, TdTomato+ OSNs (i.e., derived from recombined HBCs) actively contribute OMP+ axons to the olfactory sensory nerve, fasciculate, and extensively innervate the olfactory bulb glomeruli (g′). (Scale bars: 100 µm in C, b and g; 20 µm in C, c–f and g′.)
We used the above labeling scheme to compare the various categories of GBCs in the GBC population derived from TdTomato+ HBCs vs. the population derived from TdTomato− GBCs (which did not differentiate from HBCs activated by excision of p63) within the same fields of view. The experiment was performed in four different animals, and 8–12 nonconsecutive sections were analyzed per animal. We found that the majority of both the TdTomato− and TdTomato+ (i.e., HBC-derived) GBCs were GBCINPs (64% and 66% respectively); GBCQs constituted 22% and 19%, respectively, and GBCTA/MPPs constituted 14% and 15%, respectively (Fig. 5 A, c and d). These slight differences in distribution were not statistically significant (P = 0.63, χ2 = 0.92), indicating that activated HBCs give rise to GBCs that integrate into the normal OE and function as endogenous GBCs, some of which are persistent, quiescent progenitors.
We next analyzed the functional capacity of HBC-derived GBCs by harvesting and transplanting OE cells from K5.CreERT2 > p63fl/fl;R26R (TdTomato) mice 28 d after tamoxifen administration and HBC activation (Fig. 5 B, a). At 14 d after transplantation (42 d after activation), there were numerous TdTomato+ colonies including neurons, Sus cells, and GBCs, but, as expected, there were no HBCs (Fig. 5 B, b). The p63fl/fl-derived clusters also were significantly more complex than the clusters derived from WT HBCs described earlier, with 17 of 71 (24%) clusters containing cells of more than one type, compared with three of 52 (5.5%) (P < 0.05, χ2 with Yates’ correction) (Fig. 5 B, c). The persistence of HBC-derived GBCs indicates that at least some GBCs generated in this manner remain multipotent, continue to cycle after the loss of p63, and do not progress into GBCs with more limited differentiative capacity and/or differentiate terminally into neurons. Thus, HBC-derived GBCs apparently persist as stem cells capable of engraftment and multipotency over the relatively long term.
Finally, we investigated the long-term persistence of HBC-derived, functional GBC stem cells in situ and their ability to contribute to neurogenesis in a neuron-specific model of injury and repair. In this model we performed a unilateral olfactory bulbectomy (OBX) by surgically removing the olfactory bulb, which is the postsynaptic target for the OSNs (Fig. 5 C, a). It is known that the neurons depend on the presence of the olfactory bulb for trophic support, and hence the lifespan of all OSNs born after OBX is truncated at about 2 wk or less (34). The continuous loss of neurons is accompanied by a significant increase in GBC division and neuronal production, without significant concomitant HBC activation (7, 34–36). To assess whether HBC-derived GBCs are able to contribute to such ongoing neurogenesis alongside endogenous GBCs, we administered tamoxifen to K5.CreERT2 > p63fl/fl;R26R (TdTomato) mice 2 wk after OBX, a time point after the initial wave of neuronal death. The animals were left to recover for 6 mo before analysis; as a consequence, all TdTomato+ neurons on the lesioned side must have been born 5 mo or more after GBC differentiated from genetically activated HBCs. In these animals both the lesioned and unlesioned sides contain abundant TdTomato+ GBCs and neurons, as well as microvillar and Sus supporting cells (Fig. 5 C, b–f). Thus, GBCs derived from activated HBCs persist for an extended time despite the significantly enhanced demand for neurogenesis when neuronal lifespan is cut short. Of note, on the unlesioned side the TdTomato+ sensory neurons are capable of maturing into olfactory marker protein-positive (OMP+) cells, which innervate glomeruli of the olfactory bulb (Fig. 5 C, f), indicating that the HBC-derived GBCs not only are multipotent and persistent but also contribute to the maintenance of tissue and generate mature OSNs that reach their postsynaptic target.
Discussion
Our findings illustrate the contextual requirements for the activation vs. a return to quiescence of a population of reserve stem cells in the intact and regenerating OE. We have shown that activation of HBCs can be tested efficiently by transplantation into lesioned hosts. Using this approach, we were able to demonstrate that HBC activation correlates with p63 down-regulation and anticipates enhanced mitotic cycling. Down-regulation of p63 is necessary and sufficient for the activation of dormant HBCs. However, HBC activation does not necessarily lead to a global loss of stem cell capacity, because GBCs derived from HBCs are functionally similar to endogenous GBCs, retain engraftment and multipotency potential, and remain capable of ongoing neurogenesis for 5 mo or more even in the face of accelerated production of OSNs postbulbectomy. Taken together, the data indicate that p63 is a master regulator of the reserve status and dormancy of HBCs but is not responsible for stemness in the OE, per se.
The Role of p63 in the Maintenance and Repair of the OE.
Understanding the role of p63 in stem cell regulation has been complicated by conflicting data from various stem cell systems and conditions. For example, in the thymus, p63 is necessary to maintain the proliferative capacity of the stem cell population (37). On the other hand, in HaCaT cells (a keratinocyte-derived cell line) p63 promotes quiescence by repressing transcription of a number of positive cell-cycle regulators such as cyclin B2, cdc2, and topoisomerase II and activating transcription of the negative cell-cycle regulator p57kip2 (38, 39). These data underscore the context-dependency of p63 function.
Our study in the OE defines the contexts in which p63 is associated with cell proliferation vs. quiescence in this tissue. The data presented here uncouple the p63-dependent process of functional activation from the proliferation of activated cells and fit with previous data in which HBC proliferation is elicited apparently independent of activation (40, 41) and other results demonstrating the proliferation of K5/K14+ basal cells in respiratory epithelium (42). Our finding that retroviral-driven expression of p63 attenuates clone size and maintains the low mitotic index of mature, p63+ HBCs suggests that p63 exerts a net inhibitory effect on the proliferation of these reserve stem cells. Nevertheless, this net negative effect is not absolute, as demonstrated here by the finding that the p63+ cells actively cycle early in the regeneration of the lesioned OE.
The observation that p63 down-regulation and HBC activation occur only in response to severe and direct injury to the OE and do not happen with neuronal turnover alone strongly implicates exogenous, i.e., non–cell-autonomous, mechanisms in the governance of HBC activation. The enhancement of HBC activation by a halving of p63 gene dosage as demonstrated by analysis of the p63fl/+ animals suggests that the overall balance of activating and homeostatic environmental influences and their respective signaling pathways likely serves to regulate p63 transcript and/or protein levels. A number of pathways known to be active in the OE, including Wnt, Notch, and EGF, have been shown to affect p63 expression levels in other tissues (21, 43–45). Moreover, the environment at the time of activation can direct different outcomes, because very severe injury to the OE, worse than the damage occasioned by MeBr exposure here, causes spared HBCs to give rise to metaplastic respiratory epithelium rather than regenerating GBCs and restoring the OE (46). Further elucidation of the role of activating and quiescence-maintaining pathways and more detailed analysis of HBC expression profiles with and without epithelial injury will be necessary to use these cells in regenerative applications.
General Implications for the Dynamics of Stem Cell Transitions.
The existence of multiple functional classes of stem cells, whose respective contributions to epithelial homeostasis and regeneration are contextualized by epithelial status, is not unique to the OE. Such heterogeneity has been identified in diverse tissues including intestine, skin, the hematopoietic system, and the CNS (8, 47–49). In these tissues, injury above and beyond the normal cellular turnover also serves to activate the reserve stem cells. For example, in the context of the irradiated small intestine, Bmi-1+ reserve cells in the +4 position of the crypts can regenerate entire villi, including the active Lgr5+ stem cell population. However, in this and other cases it is not completely understood which cell-autonomous and nonautonomous mechanisms maintain the reserve phenotype or activate reserve stem cells (47–49).
Here, we demonstrate that p63 is the central hub of a cell-autonomous network that regulates HBC maintenance and activation, so that even partial down-regulation of p63, as seen in the p63fl/+ cells, is sufficient to promote activation. The observation that HBC activation gives rise to long-term repopulating cells of the GBC type indicates that p63 serves to maintain a reserve phenotype rather than to regulate self-renewal of a stem cell phenotype, as has been suggested previously (28). Interestingly, the role of p63 in the OE is distinct from its role in the epidermis, where p63 regulates and maintains active stem cells. Such differences likely reflect the context dependency of different cellular and molecular environments, as discussed above.
Finally, the notions that stem cell capacity is hardier and more distributed than previously suspected and that transitions between stem cell subtypes can be governed by targeting a single conserved molecule are promising for the future use of adult stem cells in regenerative medicine. Despite the inherent challenges of harnessing these molecular mechanisms, the OE provides an easily accessible source of neurocompetent stem cells, and future studies of the contexts upstream and downstream of p63 in the OE have the potential to move toward the development of cell-based regenerative therapies for neurodegenerative disease.
Materials and Methods
C57/B6, 129S1/Sv1M, BACT.GFP, R26R(TdTomato), and R26R(LacZ) mice were purchased from Jackson Laboratories. K5.CreERT2 mice were generously provided by P. Chambon, Institut d'Études Avancées de l'Université de Strasbourg, Strasbourg, France and R. Reed, Johns Hopkins University, School of Medicine, Baltimore (50). Floxed p63 mice (p63fl/fl) were kindly provided by A. Mills, Cold Spring Harbor Laboratory, Cold Spring Harbor, NY (51). All animals were housed in the American Association for Laboratory Animal Care (AALAC)-accredited vivarium at Tufts University School of Medicine. All vertebrate animal protocols were approved by the Committee for the Humane Use of Animals at Tufts University School of Medicine. MeBr exposure, OBX, transplantation, retroviral infection, and immunohistochemistry were performed as previously described (6, 29, 37, 52). Primary antibodies used, their dilutions, and detection methods are listed in Table S1. Images were obtained on a Zeiss 510 confocal microscope or on a Nikon 800E epifluorescent microscope. Image analysis was performed using ImageJ. Image preparation and figure assembly were performed in Adobe Photoshop CS2. In all photographs, only balance, contrast, and evenness of the illumination were altered. For a full description of the experimental procedures and staining conditions used in this study, please see SI Materials and Methods.
Table S1.
Antibodies and staining protocols used in this study
| Primary antibody | Source/vendor | Protocol | Cell types marked (reference) |
| Rat α-BrdU | Abcam | Pre-Tx: 6N HCl for 10 min or steam for 10 min (1:200) → bDαRat → fluor-SA | Dividing cells labeled with a BrdU pulse |
| Rb α-CK14 | Labvision | (1:500) → fluor-DαRb | HBCs (21) |
| Gt α-mouse CD54 | R&D | (1:100) → fluor-DαGt or (1:400) → bDαGt → fluor-SA | HBCs (58) |
| Rb α-dsRed | Rockland Inc. | (1:400) → fluor-DαRb | All TdTomato+ cells |
| Rb α-Ki67 | Epitomics | (1:100) → fluor- DαRb | Cells in the cell cycle (33, 59) |
| Gt α-mCherry | Acris | (1:150) →fluor-DαGt | All TdTomato+ cells |
| Gt α-OMP | Wako (Frank Margolis, University of Maryland, Baltimore) | (1:400) → DAB or (1:100) → fluor-DαGt | Mature OSNs (34) |
| Mo α-p63 | Santa Cruz | Pre-Tx: Steam (1:150) → bDαMo →fluor-SA |
All HBCs, except early activated HBCs (20) |
| Rb α-pH3(Ser10) | Millipore | (1:300) →fluor-DαRb | Cells in mitosis (59) |
| Ch α-PGP9.5 | Rockland Inc. | Pre-Tx: 6N HCl for 10 min (1:200) → fluor-DαCh | All neurons (29) |
| Rb α-PGP9.5 | Ultraclone | (1:1,200) → fluor-DαRb or (1:5,000) → DAB | All neurons (29) |
| Gt α-Sox2 | Santa Cruz | Pre-Tx: DNaseI + Steam or 30 mins 65 °C (1:25) → bDαGt → fluor-SA or DAB | Sus cells, HBCs, GBCs (17) |
| Rb α-Sox9 | Millipore | Pre-Tx: Steam+EtOH (1:300) → bDαGt → fluor-SA | Duct/gland, microvillar cells (60) |
| GP α-TrpM5 | Gift from E. Liman (University of Southern California, Los Angeles) | (1:400) → DAB | microvillar cells (61) |
Antibody hosts: Ch, chicken; D, donkey; GP, guinea pig; Gt, goat; Mo, mouse; Rb, rabbit. Antibody visualization: DAB: the HRP substrate diaminobenzidine was used for chromogenic staining; DNaseI: sections were treated with 500 U/mL DNaseI in buffer (40 mM Tris⋅HCl, 10 mM NaCl, 6 mM MgCl2, 10 mM CaCl2, pH 7.9) for 10 min; EtOH: slides were dehydrated and rehydrated through graded EtOH (70% →95% →100% →95% → 70%), 1 min each; fluor: a variety of Alexa fluorophores, including Alexa 405, 488, 546, 594, and 647(Jackson Immunological) were used at 1:100; tertiary reagent amplification: biotinylated secondary antibody (bDαX) was followed by incubation in fluor-streptavidin (fluor-SA). Pretreatments (Pretx): 30 min 65 °C: slides were incubated in 0.01 M citrate, pH6, at 65 °C for 30 min; 6N HCl: sections were preincubated in 6N HCl for 10 min; steam: sections wee covered with 0.01 M citrate, pH6, and steamed in a commercial food steamer for 10 min.
SI Materials and Methods
Animals and Breeding.
WT F1 mice used in lesion, transplantation, and viral infection studies were bred from C57/B6J and 129S1/Sv1MJ mice in house or were ordered from Jackson Laboratories when necessary (stock #101043). B6.GFP mice were purchased from Jackson Laboratories [C57BL/6-Tg(ACTB-EGFP)1Osb/J; stock # 003291]. K5CreERT2 mice have been described elsewhere (50) and were generously provided by P. Chambon via R. Reed. The floxed p63 mice (p63fl/fl) were kindly provided by A. Mills (51). The Cre reporter strains R26R(LacZ) [B6.129S4-Gt(Rosa)26Sortm1Sor/J; stock #003474] and R26R(TdTomato) [B6.Cg-Gt(ROSA)26Sortm9(CAG-TdTomato)Hze/J; stock #007909] were purchased from Jackson Laboratories and then were bred in house (52). All mice were maintained on ad libitum rodent chow and water. All animals were housed in a heat- and humidity-controlled, AALAC-accredited vivarium operating under a 12:12-h light:dark cycle. All protocols for the use of vertebrate animals were approved by the Committee for the Humane Use of Animals at Tufts University School of Medicine, where the animals were housed and the experiments were conducted.
MeBr and OBX Lesions.
Animals were passively exposed to MeBr gas for 8 h as previously described (53). Optimal lesion conditions in terms of age, dose, extent of injury, and regenerative capacity were empirically determined for each strain individually. C57/B6.129 F1 mice were exposed to 180 ppm MeBr in pure air at 12 wk of age. All the transgenic mouse lines were exposed to 175 ppm MeBr in pure air at 8 wk of age. Unilateral lesions were performed by plugging the right naris with a 5-mm piece of PE10 tubing, and the lumen of the tube was obstructed with a knotted 7.0 suture thread and superglue.
OBX was performed as described previously (34). Eight-week-old K5CreER;p63(fl/fl);R26(TdTomato) mice were anesthetized with an induction mixture (37.5 mg/kg ketamine, 7.5 mg/kg xylazine, and 1.25 mg/kg acepromazine) supplemented with a maintenance mixture (47.5 mg/kg ketamine, 0.9 mg/kg acepromazine) as needed to maintain an anesthetic plane. Overlying frontal bone was removed to expose the bulb, which was removed by aspiration. Oxycel, purchased from Becton Dickinson (375846), was placed within the ablation cavity to achieve hemostasis, and the overlying skin was sutured.
Drug Preparation and Administration.
Tamoxifen purchased from Sigma (#T5648) was dissolved in sterile corn oil at 30 mg/mL by vortexing for 20 min at 37 °C. This solution was injected i.p. at 50–300 mg/kg as indicated. EdU purchased from Invitrogen (#A10044) was dissolved in sterile PBS at 5 mg/mL and injected s.c. under the back skin at 25–50 mg/kg as indicated. BrdU was purchased from Sigma (B5002), dissolved in sterile PBS at 10 mg/mL, and injected s.c. at 50 mg/kg as indicated.
Tissue Processing.
At the indicated time points mice were anesthetized by an i.p. injection of a triple mixture of ketamine (37.5 mg/kg), xylazine (7.5 mg/kg), and acepromazine (1.25 mg/kg). Anesthetized animals were transcardially flushed with PBS and perfused with 1% PLP (1% paraformaldehyde, 0.01 M monobasic and dibasic phosphates, 90 mM lysine, 0.1 M sodium periodate). After dissection the tissue was postfixed in 1% PLP under vacuum for 2 h, washed in PBS, and decalcified in saturated EDTA overnight. All tissue was cryoprotected in 30% (wt/vol) sucrose in PBS, embedded in optimal cutting temperature (OCT) compound (Miles Inc.), and frozen in liquid nitrogen. Coronal sections (10 µm) were generated on a Leica cryostat, mounted on “Plus” slides (Fischer Scientific), and stored at −20 °C until needed.
Tissue Dissociation.
Donor animals were anesthetized with an i.p. injection of a triple mixture of ketamine (37.5 mg/kg), xylazine (7.5 mg/kg), and acepromazine (1.25 mg/kg) and perfused with cold low Ca2+ Ringer’s solution (140 mM NaCl, 5 mM KCl, 10 mM Hepes, 1 mM EDTA, 10 mM glucose, 1 mM sodium pyruvate, pH 7.2). The nose was dissected and placed in cold low Ca2+ Ringer’s solution on ice. After fine mincing the tissue was incubated in 0.05% trypsin-EDTA for 15 min at 37 °C until it formed a sticky ball. The trypsin-EDTA solution was discarded and replaced with an enzyme mixture containing 100 U/mL collagenase, 250 U/mL hyaluronidase, 75 U/mL DNase I, 0.1 mg/mL trypsin inhibitor, 2.5 U/mL Dispase II, and 5 U/mL papain (Worthington Biochemical, Roche, and Sigma) in lactated Ringer’s solution (140 mM NaCl, 5 mM KCl, 10 mM Hepes, 1 mM EDTA, 10 mM glucose, 1 mM sodium pyruvate, 1 mM CaCl2, 1 mM MgCl2, pH 7.2). The tissue was incubated in enzyme mixture at 37 °C for 30 min with light vortexing every 10 min followed by filtration through a 125-µm mesh, cell pelleting, and filtration through a 35-µm mesh. The cells were resuspended in DMEM + 2% FBS + 1% penicillin/streptomycin for transplantation.
FACS Analysis of p63 Expression Level Postlesion.
The OE was dissociated as described above, with the following modifications: Minced tissue was incubated in 0.05% trypsin-EDTA for 10 min, Dispase II was removed from the enzyme mixture, and cells were finally resuspended in 1× HBSS containing 25 mM Hepes, 10 mM EDTA, and 0.5% BSA. The single-cell suspension then was fixed and permeabilized using the Foxp3 transcription factor staining kit (Affymetrix, catalog no. 00-5523-00) with modifications. Briefly, cells in HBSS buffer were pelleted and resuspended in 50 µL of residual buffer. Then 1 mL of fresh fixation/permeabilization working solution was added to the cells, mixed briefly, and incubated on ice for 30 min. Two milliliters of 1× permeabilization buffer was added at the end of the incubation, mixed briefly, and spun down at 500 × g. Cells were resuspended in 1× permeabilization buffer and incubated with 1:100 mouse anti-KRT14 (Vector Laboratories, catalog no. VP-C410) and 1:200 rabbit anti–p63-delta (Novus, catalog no. NBP2-29467) for 1 h at room temperature, followed by two washes with permeabilization buffer. Cells then were resuspended and incubated with 1:400 Alexa 488-donkey anti-mouse IgG and 1:400 Alexa 647-donkey anti-rabbit IgG (Jackson ImmunoResearch, catalog nos. 715-095-151 and 711-605-152) for 30 min at room temperature followed by two more washes with permeabilization buffer. Finally, cells were resuspended in 100 µL of permeabilization buffer and transferred for analytical flow cytometer analysis on a BD Biosciences FACSCalibur using a 670-nm long-pass filter with a 635-nm red diode laser and a 515- to 545-nm band-pass filter with a 488-nm argon laser. Detector voltages, gains, and collector modes were held constant among samples after being calibrated on positive and negative controls. Samples then were analyzed using Summit V6.2.2 software.
qPCR Analysis of p63 Expression.
Twelve-week-old male F1 mice were exposed to MeBr at 175 ppm for 8 h and were killed at 0, 12, 18, or 24 h after MeBr exposure. Littermate controls were not exposed to MeBr gas. The OE was dissociated as described above. After filtration through a 35-µm mesh, cells were resuspended in 1× HBSS containing 10 mM EDTA. Cells were incubated with goat anti-mouse CD54 primary antibody (R&D) at a concentration of 1:100 for 20 min on ice. The cells were washed, pelleted, resuspended, and incubated with biotinylated donkey anti-goat IgG secondary antibody at a concentration of 1:400 for 20 min on ice. The cells were washed, pelleted, resuspended, and incubated with Alexa 647-strepavidin at a concentration of 1:400 for 20 min on ice. The cells were washed, pelleted, and resuspended a final time, and the stained HBCs were FACS-purified. Detector voltages, gains, and collector modes were held constant among samples after being calibrated on positive and negative controls. RNA was isolated from cells using the Zymo Research DNA-free RNA purification kit. cDNA was generated using SuperScript III reverse transcriptase (Invitrogen). cDNA was subjected to qPCR using primers and conditions as described (54). The fold-change in gene expression at various time points after MeBr lesion was calculated using noninjured F1 littermate control FACS-purified HBCs as a reference.
Western Blot Analysis of p63 Expression.
Equal 2-mm2 areas of OE tissue from 12-wk-old male C57/B6.129 F1 MeBr-lesioned mice killed 18 h after exposure and littermate controls were dissected and flash frozen in liquid nitrogen. Samples were resuspended in ice-cold lysis buffer [20 mM Tris (pH 7.5), 1 mM EDTA (pH 8.0), 1% Triton X-100, 150 mM NaCl, HALT Protease/Phosphatase Inhibitor (Roche)] and pulsed twice for 2 s at 2 kHz on ice using a Branson 250 Sonifier (Branson Ultrasonics Corporation). The protein concentration of lysates was determined using the bicinchoninic acid assay method. Control tissue was diluted to a concentration of 1 µg/µL. The loss of cells and decline in total protein as a consequence of injury led us to normalize the samples from the MeBr-lesioned mice on the basis of equivalent surface area compared with control tissue. Accordingly, tissue from the lesioned mice was diluted with a volume of lysis buffer equal to that added to controls to compare closely equivalent numbers of basal epithelial cells across control and lesioned samples. Bolt LDS sample buffer and reducing agent (Life Technologies) were added to a final concentration of 1× according to the manufacturer’s protocol, and samples were denatured at 95 °C for 10 min. Then 30 µg total lysate from controls and equal volumes from lesioned samples were electrophoresed in a 4–20% gradient Bis-Tris Plus gel using the Bolt Electrophoresis system (Life Technologies). Gels then were transferred to PVDF membrane, blocked in TBS blocking buffer containing 0.1% Tween-20 and 5% (wt/vol) milk, and probed with anti-p63 (NBP2-29467; Novus Biologicals) overnight at 4 °C. Membranes were washed three times with blocking buffer, incubated for 1 h in HRP-conjugated anti-rabbit secondary antibody (Jackson ImmunoResearch) at room temperature, and washed twice with blocking buffer. Immune complexes were detected using SuperSignal West Pico Luminescent Substrate (Thermo Scientific).
Assessment of HBC Postlesion Morphology.
To visualize the morphology of HBCs, we adopted the Clarity tissue-clearing protocol and more specifically the passive Clarity technique with modifications for olfactory epithelial whole mounts (55, 56). Briefly, K5CreER:R26(TdTomato) mice were injected with tamoxifen as described above and were exposed to MeBr 2 wk later. One day after MeBr exposure the mice were perfused with hydrogel monomer solution [4% (wt/vol) acrylamide, 0.025% bis-acrylamide, 4% (wt/vol) paraformaldehyde, 0.25% VA-044 (polymerization thermal initiator), and 1× PBS]. The nasal septum was embedded and polymerized at 37 °C. The embedded tissue then was cleared in clearing solution [4% (wt/vol) SDS, 200 mM boric acid, pH 8.5] in a histological tissue cassette for 2 d at 50 °C with stirring, followed by washing with 0.1% Triton-X in PBS. The tissue was stained with goat anti-mCherry antibody to label TdTomato+ cells more robustly. After staining, the septum was mounted in 87% glycerol (vol/vol) before imaging. To image HBCs in isolation, areas of the epithelium with a lesser extent of recombination were imaged. Tamoxifen-injected but nonlesioned mice were used for comparison.
Transplantation and Retroviral Infection.
Transplantation and in vivo retroviral infection were performed as previously described (6, 20). Donor cells for transplantation were prepared as described in the text and Fig. S1. Briefly, host and infected animals were anesthetized with the induction and maintenance mixtures used for transplantation as above. The anterior neck was shaved and disinfected with iodine and ethanol. A tracheotomy was performed inferior to the thyroid gland to maintain a patent airway. The palate was raised with a 3-cm piece of PE-100 tubing to close the nasopharyngeal passage. Resuspended cells (75 µL) or concentrated virus were introduced into one naris through a cannula made from PE-10 tubing until the solution could be seen exiting the contralateral naris. The mice were positioned in a lateral decubitus position at a 45° angle, alternating sides every 45 min, to allow drainage of cells or virus into both lateral turbinates. After 3 h the solution was removed from the nasal cavity, the tracheotomy was sutured, and the mice were placed on a warm pad overnight. After 2 wk the animals were killed as described above.
Retroviral Constructs and Production.
MSCV (Clontech) was used as a backbone to generate the constructs (57). MSCV-IRES-GFP (MIG) was generated by replacing the PGK.puro selection cassette with an IRES-GFP cassette. The IRES-GFP template was obtained from pLIA-IRES-GFP provided by C. Cepko, Harvard Medical School, Boston. The template was amplified using the following primers that contained homology to the sequences flanking the EcoRI and ClaI restriction sites in MSCV.puro: F: 5′-CGAGGTTAACGAATTCCTAACGTTACTGGCCGAAGC-3′ and R: 5′- TCTTTTATTTTATCGATACTGTGCTGGCGCTTTACTT-3′. The amplified fragment was inserted into MSCV.puro using Infusion Advantage enzyme from Clontech to generate the MIG backbone. MIG-p63 was generated from this MIG backbone by cloning ΔNp63α into the BglII and XhoI restriction sites upstream of the IRES-GFP sequence. Full-length ΔNp63α cDNA in the pCMV-HA plasmid was provided by S. Sinha, University at Buffalo, Buffalo, NY. The cDNA was amplified using primers that contained homology to sequences flanking the BglII and XhoI restriction sites of MIG: F: 5′- CGCCGGAATTAGATCTCGCTCTTATGGCCATGGAGG-3′ and R: 5′-ATTCGTTAACCTCGAGATCATGTCTGGATCCCCGC-3′.
Viral particles were produced by cotransfecting HEK293T cells with the retroviral plasmid and the packaging vector pCL-Eco bearing the gag/pol/env genes (Addgene, #12371). Forty-eight hours after transfection the supernatant was collected and concentrated 100-fold using the Retro-Concentin reagent according to the manufacturer’s instructions (Systems Biosciences). Titers of 105–6 were confirmed by infecting 3T3 murine fibroblasts with limiting dilutions of concentrated supernatant.
Histochemistry and Immunohistochemistry.
To visualize β-galactosidase expression, tissue sections were incubated in a staining solution containing X-Gal (Sigma) overnight as described previously (30). After X-Gal staining the slides were subjected to the appropriate pretreatments and processed for immunostaining with 3,3′-diaminobenzidine (DAB). EdU staining with azide-fluor reagents was performed according to the manufacturer’s instructions (Invitrogen).
The primary antibodies used, their dilutions, and the details of their working conditions and detection are listed in Table S1. Before immunostaining, tissue sections were rinsed in PBS to remove OCT and subjected to antibody-specific pretreatments. The pretreatments include dehydration and rehydration in ethanol (70% > 95% > 100% > 95% > 70%, 1 min each), steaming in 0.01 M citrate buffer (pH 6.0) for 10 min in a commercial food steamer, incubation in 0.005% trypsin-EDTA for 5 min, and/or incubation in 3% (vol/vol) hydrogen peroxide in MeOH for 5 min. Sections were blocked with 10% (wt/vol) donkey serum/5% (wt/vol) nonfat dry milk/4% (wt/vol) BSA/0.1% TritonX-100 in PBS and incubated overnight in primary antibody. The following day the staining was visualized using an array of methods as indicated in Table S1. Unless otherwise indicated in the figure legends, blue represents the nuclear counterstain DAPI.
Imaging and Quantification.
Stained sections were imaged on a Zeiss 510 confocal microscope in multitracking mode or on a Nikon 800E epifluorescent microscope with a Spot RT2 digital camera. Image preparation and assembly were performed in Photoshop CS2 (Adobe). Image analysis and quantification were performed in ImageJ. In all photographs, only balance, contrast, and evenness of the illumination were altered.
The proliferation status of HBCs and other cells in the OE postlesion was assessed as follows. Twelve-week-old male F1 mice were unilaterally exposed to MeBr and killed at 18 h, 24 h, 2 d, or 3 d after MeBr; 2 h before they were killed, all animals received 50 mg/kg EdU s.c. EdU was detected by click-chemistry in prepared tissue sections, which subsequently were stained for p63. EdU and p63 colocalization were counted by direct observation of total EdU+ and total p63+ cells.
Cluster size, composition, and cell types were counted by direct observation with the epifluorescent microscope. GBC subtypes after p63 knockout were stained with Rbα-dsRed, EdU, BrdU, and ChαPGP using the appropriate pretreatments. Nonsequential sections were analyzed for TdTomato+ regions, which were imaged under epifluorescence using the 40× objectives. TdTomato+ and TdTomato− cells in the same image were quantified with respect to GBC subtype composition. Eight to 12 nonsequential sections per animal were analyzed, and data were pooled from four animals. For all experiments at least three animals were counted per condition examined, and the data were analyzed using GraphPad Prism software, with the appropriate statistical test as indicated in the text. In all graphs mean values and SEM are reported.
Acknowledgments
This work was supported by NIH Grants R01 DC002167 (to J.E.S.), F30 DC011241 (to N.S.), F30 DC013962 (to D.B.H.), F31 DC 014637 (to B.L.), and F31 DC014398 (to J.H.C.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1512272112/-/DCSupplemental.
References
- 1.Iscove NN, Nawa K. Hematopoietic stem cells expand during serial transplantation in vivo without apparent exhaustion. Curr Biol. 1997;7(10):805–808. doi: 10.1016/s0960-9822(06)00341-1. [DOI] [PubMed] [Google Scholar]
- 2.Price J, Turner D, Cepko C. Lineage analysis in the vertebrate nervous system by retrovirus-mediated gene transfer. Proc Natl Acad Sci USA. 1987;84(1):156–160. doi: 10.1073/pnas.84.1.156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barker N, et al. Crypt stem cells as the cells-of-origin of intestinal cancer. Nature. 2009;457(7229):608–611. doi: 10.1038/nature07602. [DOI] [PubMed] [Google Scholar]
- 4.Barker N, et al. Identification of stem cells in small intestine and colon by marker gene Lgr5. Nature. 2007;449(7165):1003–1007. doi: 10.1038/nature06196. [DOI] [PubMed] [Google Scholar]
- 5.Doetsch F, Caillé I, Lim DA, García-Verdugo JM, Alvarez-Buylla A. Subventricular zone astrocytes are neural stem cells in the adult mammalian brain. Cell. 1999;97(6):703–716. doi: 10.1016/s0092-8674(00)80783-7. [DOI] [PubMed] [Google Scholar]
- 6.Chen X, Fang H, Schwob JE. Multipotency of purified, transplanted globose basal cells in olfactory epithelium. J Comp Neurol. 2004;469(4):457–474. doi: 10.1002/cne.11031. [DOI] [PubMed] [Google Scholar]
- 7.Leung CT, Coulombe PA, Reed RR. Contribution of olfactory neural stem cells to tissue maintenance and regeneration. Nat Neurosci. 2007;10(6):720–726. doi: 10.1038/nn1882. [DOI] [PubMed] [Google Scholar]
- 8.Li L, Clevers H. Coexistence of quiescent and active adult stem cells in mammals. Science. 2010;327(5965):542–545. doi: 10.1126/science.1180794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Grompe M. Tissue stem cells: New tools and functional diversity. Cell Stem Cell. 2012;10(6):685–689. doi: 10.1016/j.stem.2012.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Göritz C, Frisén J. Neural stem cells and neurogenesis in the adult. Cell Stem Cell. 2012;10(6):657–659. doi: 10.1016/j.stem.2012.04.005. [DOI] [PubMed] [Google Scholar]
- 11.Graziadei PP, Graziadei GA. Neurogenesis and neuron regeneration in the olfactory system of mammals. I. Morphological aspects of differentiation and structural organization of the olfactory sensory neurons. J Neurocytol. 1979;8(1):1–18. doi: 10.1007/BF01206454. [DOI] [PubMed] [Google Scholar]
- 12.Graziadei GA, Graziadei PP. Neurogenesis and neuron regeneration in the olfactory system of mammals. II. Degeneration and reconstitution of the olfactory sensory neurons after axotomy. J Neurocytol. 1979;8(2):197–213. doi: 10.1007/BF01175561. [DOI] [PubMed] [Google Scholar]
- 13.Schwob JE. Neural regeneration and the peripheral olfactory system. Anat Rec. 2002;269(1):33–49. doi: 10.1002/ar.10047. [DOI] [PubMed] [Google Scholar]
- 14.Schwob JE, Jang W, Holbrook EH. Stem cells of the olfactory epithelium. In: Rao MS, editor. Neural Development and Stem Cells. 3rd Ed. Springer Science & Business Media; New York: 2012. pp. 201–222. [Google Scholar]
- 15.Goldstein BJ, Fang H, Youngentob SL, Schwob JE. Transplantation of multipotent progenitors from the adult olfactory epithelium. Neuroreport. 1998;9(7):1611–1617. doi: 10.1097/00001756-199805110-00065. [DOI] [PubMed] [Google Scholar]
- 16.Caggiano M, Kauer JS, Hunter DD. Globose basal cells are neuronal progenitors in the olfactory epithelium: A lineage analysis using a replication-incompetent retrovirus. Neuron. 1994;13(2):339–352. doi: 10.1016/0896-6273(94)90351-4. [DOI] [PubMed] [Google Scholar]
- 17.Guo Z, et al. Expression of pax6 and sox2 in adult olfactory epithelium. J Comp Neurol. 2010;518(21):4395–4418. doi: 10.1002/cne.22463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Manglapus GL, Youngentob SL, Schwob JE. Expression patterns of basic helix-loop-helix transcription factors define subsets of olfactory progenitor cells. J Comp Neurol. 2004;479(2):216–233. doi: 10.1002/cne.20316. [DOI] [PubMed] [Google Scholar]
- 19.Tucker ES, et al. Proliferative and transcriptional identity of distinct classes of neural precursors in the mammalian olfactory epithelium. Development. 2010;137(15):2471–2481. doi: 10.1242/dev.049718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Packard A, Schnittke N, Romano RA, Sinha S, Schwob JE. DeltaNp63 regulates stem cell dynamics in the mammalian olfactory epithelium. J Neurosci. 2011;31(24):8748–8759. doi: 10.1523/JNEUROSCI.0681-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Holbrook EH, Szumowski KE, Schwob JE. An immunochemical, ultrastructural, and developmental characterization of the horizontal basal cells of rat olfactory epithelium. J Comp Neurol. 1995;363(1):129–146. doi: 10.1002/cne.903630111. [DOI] [PubMed] [Google Scholar]
- 22.Mills AA, et al. p63 is a p53 homologue required for limb and epidermal morphogenesis. Nature. 1999;398(6729):708–713. doi: 10.1038/19531. [DOI] [PubMed] [Google Scholar]
- 23.Yang A, et al. p63 is essential for regenerative proliferation in limb, craniofacial and epithelial development. Nature. 1999;398(6729):714–718. doi: 10.1038/19539. [DOI] [PubMed] [Google Scholar]
- 24.Carroll DK, et al. p63 regulates an adhesion programme and cell survival in epithelial cells. Nat Cell Biol. 2006;8(6):551–561. doi: 10.1038/ncb1420. [DOI] [PubMed] [Google Scholar]
- 25.Keyes WM, et al. p63 deficiency activates a program of cellular senescence and leads to accelerated aging. Genes Dev. 2005;19(17):1986–1999. doi: 10.1101/gad.342305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.King KE, et al. deltaNp63alpha functions as both a positive and a negative transcriptional regulator and blocks in vitro differentiation of murine keratinocytes. Oncogene. 2003;22(23):3635–3644. doi: 10.1038/sj.onc.1206536. [DOI] [PubMed] [Google Scholar]
- 27.Crum CP, McKeon FD. p63 in epithelial survival, germ cell surveillance, and neoplasia. Annu Rev Pathol. 2010;5:349–371. doi: 10.1146/annurev-pathol-121808-102117. [DOI] [PubMed] [Google Scholar]
- 28.Fletcher RB, et al. p63 regulates olfactory stem cell self-renewal and differentiation. Neuron. 2011;72(5):748–759. doi: 10.1016/j.neuron.2011.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Packard A, Giel-Moloney M, Leiter A, Schwob JE. Progenitor cell capacity of NeuroD1-expressing globose basal cells in the mouse olfactory epithelium. J Comp Neurol. 2011;519(17):3580–3596. doi: 10.1002/cne.22726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Huard JM, Youngentob SL, Goldstein BJ, Luskin MB, Schwob JE. Adult olfactory epithelium contains multipotent progenitors that give rise to neurons and non-neural cells. J Comp Neurol. 1998;400(4):469–486. [PubMed] [Google Scholar]
- 31.Cau E, Gradwohl G, Fode C, Guillemot F. Mash1 activates a cascade of bHLH regulators in olfactory neuron progenitors. Development. 1997;124(8):1611–1621. doi: 10.1242/dev.124.8.1611. [DOI] [PubMed] [Google Scholar]
- 32.Cau E, Casarosa S, Guillemot F. Mash1 and Ngn1 control distinct steps of determination and differentiation in the olfactory sensory neuron lineage. Development. 2002;129(8):1871–1880. doi: 10.1242/dev.129.8.1871. [DOI] [PubMed] [Google Scholar]
- 33.Jang W, Chen X, Flis D, Harris M, Schwob JE. Label-retaining, quiescent globose basal cells are found in the olfactory epithelium. J Comp Neurol. 2014;522(4):731–749. doi: 10.1002/cne.23470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schwob JE, Szumowski KE, Stasky AA. Olfactory sensory neurons are trophically dependent on the olfactory bulb for their prolonged survival. J Neurosci. 1992;12(10):3896–3919. doi: 10.1523/JNEUROSCI.12-10-03896.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schwartz Levey M, Chikaraishi DM, Kauer JS. Characterization of potential precursor populations in the mouse olfactory epithelium using immunocytochemistry and autoradiography. J Neurosci. 1991;11(11):3556–3564. doi: 10.1523/JNEUROSCI.11-11-03556.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Carr VM, Farbman AI. Ablation of the olfactory bulb up-regulates the rate of neurogenesis and induces precocious cell death in olfactory epithelium. Exp Neurol. 1992;115(1):55–59. doi: 10.1016/0014-4886(92)90221-b. [DOI] [PubMed] [Google Scholar]
- 37.Senoo M, Pinto F, Crum CP, McKeon F. p63 Is essential for the proliferative potential of stem cells in stratified epithelia. Cell. 2007;129(3):523–536. doi: 10.1016/j.cell.2007.02.045. [DOI] [PubMed] [Google Scholar]
- 38.Testoni B, Mantovani R. Mechanisms of transcriptional repression of cell-cycle G2/M promoters by p63. Nucleic Acids Res. 2006;34(3):928–938. doi: 10.1093/nar/gkj477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Beretta C, Chiarelli A, Testoni B, Mantovani R, Guerrini L. Regulation of the cyclin-dependent kinase inhibitor p57Kip2 expression by p63. Cell Cycle. 2005;4(11):1625–1631. doi: 10.4161/cc.4.11.2135. [DOI] [PubMed] [Google Scholar]
- 40.Farbman AI, Ezeh PI. TGF-alpha and olfactory marker protein enhance mitosis in rat olfactory epithelium in vivo. Neuroreport. 2000;11(16):3655–3658. doi: 10.1097/00001756-200011090-00051. [DOI] [PubMed] [Google Scholar]
- 41.Getchell TV, Narla RK, Little S, Hyde JF, Getchell ML. Horizontal basal cell proliferation in the olfactory epithelium of transforming growth factor-alpha transgenic mice. Cell Tissue Res. 2000;299(2):185–192. doi: 10.1007/s004419900149. [DOI] [PubMed] [Google Scholar]
- 42.Yu XM, et al. Reduced growth and proliferation dynamics of nasal epithelial stem/progenitor cells in nasal polyps in vitro. Sci Rep. 2014;4:4619. doi: 10.1038/srep04619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Barbieri CE, Barton CE, Pietenpol JA. Delta Np63 alpha expression is regulated by the phosphoinositide 3-kinase pathway. J Biol Chem. 2003;278(51):51408–51414. doi: 10.1074/jbc.M309943200. [DOI] [PubMed] [Google Scholar]
- 44.Nguyen BC, et al. Cross-regulation between Notch and p63 in keratinocyte commitment to differentiation. Genes Dev. 2006;20(8):1028–1042. doi: 10.1101/gad.1406006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wang F, et al. Loss of TACSTD2 contributed to squamous cell carcinoma progression through attenuating TAp63-dependent apoptosis. Cell Death Dis. 2014;5:e1133. doi: 10.1038/cddis.2014.96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Xie F, Fang C, Schnittke N, Schwob JE, Ding X. Mechanisms of permanent loss of olfactory receptor neurons induced by the herbicide 2,6-dichlorobenzonitrile: Effects on stem cells and noninvolvement of acute induction of the inflammatory cytokine IL-6. Toxicol Appl Pharmacol. 2013;272(3):598–607. doi: 10.1016/j.taap.2013.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tian H, et al. A reserve stem cell population in small intestine renders Lgr5-positive cells dispensable. Nature. 2011;478(7368):255–259. doi: 10.1038/nature10408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yan KS, et al. The intestinal stem cell markers Bmi1 and Lgr5 identify two functionally distinct populations. Proc Natl Acad Sci USA. 2012;109(2):466–471. doi: 10.1073/pnas.1118857109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Carlén M, et al. Forebrain ependymal cells are Notch-dependent and generate neuroblasts and astrocytes after stroke. Nat Neurosci. 2009;12(3):259–267. doi: 10.1038/nn.2268. [DOI] [PubMed] [Google Scholar]
- 50.Indra AK, et al. Temporally-controlled site-specific mutagenesis in the basal layer of the epidermis: Comparison of the recombinase activity of the tamoxifen-inducible Cre-ER(T) and Cre-ER(T2) recombinases. Nucleic Acids Res. 1999;27(22):4324–4327. doi: 10.1093/nar/27.22.4324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mills AA, Qi Y, Bradley A. Conditional inactivation of p63 by Cre-mediated excision. Genesis. 2002;32(2):138–141. doi: 10.1002/gene.10067. [DOI] [PubMed] [Google Scholar]
- 52.Soriano P. Generalized lacZ expression with the ROSA26 Cre reporter strain. Nat Genet. 1999;21(1):70–71. doi: 10.1038/5007. [DOI] [PubMed] [Google Scholar]
- 53.Schwob JE, Youngentob SL, Mezza RC. Reconstitution of the rat olfactory epithelium after methyl bromide-induced lesion. J Comp Neurol. 1995;359(1):15–37. doi: 10.1002/cne.903590103. [DOI] [PubMed] [Google Scholar]
- 54.Nakamuta N, Kobayashi S. Expression of p63 in the mouse ovary. J Reprod Dev. 2007;53(3):691–697. doi: 10.1262/jrd.18181. [DOI] [PubMed] [Google Scholar]
- 55.Chung K, Deisseroth K. CLARITY for mapping the nervous system. Nat Methods. 2013;10(6):508–513. doi: 10.1038/nmeth.2481. [DOI] [PubMed] [Google Scholar]
- 56.Tomer R, Ye L, Hsueh B, Deisseroth K. Advanced CLARITY for rapid and high-resolution imaging of intact tissues. Nat Protoc. 2014;9(7):1682–1697. doi: 10.1038/nprot.2014.123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Grez M, Akgün E, Hilberg F, Ostertag W. Embryonic stem cell virus, a recombinant murine retrovirus with expression in embryonic stem cells. Proc Natl Acad Sci USA. 1990;87(23):9202–9206. doi: 10.1073/pnas.87.23.9202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Carter LA, MacDonald JL, Roskams AJ. Olfactory horizontal basal cells demonstrate a conserved multipotent progenitor phenotype. J Neurosci. 2004;24(25):5670–5683. doi: 10.1523/JNEUROSCI.0330-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Cunha C, Hort Y, Shine J, Doyle KL. Morphological and behavioural changes occur following the X-ray irradiation of the adult mouse olfactory neuroepithelium. BMC Neurosci. 2012;13:134–148. doi: 10.1186/1471-2202-13-134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Holbrook EH, Wu E, Curry WT, Lin DT, Schwob JE. Immunohistochemical characterization of human olfactory tissue. Laryngoscope. 2011;121(8):1687–1701. doi: 10.1002/lary.21856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lin W, Ezekwe EA, Jr, Zhao Z, Liman ER, Restrepo D. TRPM5-expressing microvillous cells in the main olfactory epithelium. BMC Neurosci. 2008;9:114–127. doi: 10.1186/1471-2202-9-114. [DOI] [PMC free article] [PubMed] [Google Scholar]