Significance
RNA-binding proteins play central roles in posttranscriptional gene regulation. HuD is one of the first markers used for neuronal lineage; however, the function of HuD in neural stem cell differentiation is largely unexplored. In addition, although it has been shown that HuD mRNA levels increase during neuronal differentiation, to date few studies have examined the mechanism controlling the expression of HuD during neural differentiation. In this study, we investigated the role of HuD in neural stem cell differentiation and uncovered an underlying molecular mechanism. Our results unveil a novel positive feedback network between an RNA-binding protein and a transcription factor that plays critical regulatory roles during neuronal differentiation.
Keywords: HuD, neural stem cells, NeuroD1, neurogenesis, SATB1
Abstract
The mammalian embryonic lethal abnormal vision (ELAV)-like protein HuD is a neuronal RNA-binding protein implicated in neuronal development, plasticity, and diseases. Although HuD has long been associated with neuronal development, the functions of HuD in neural stem cell differentiation and the underlying mechanisms have gone largely unexplored. Here we show that HuD promotes neuronal differentiation of neural stem/progenitor cells (NSCs) in the adult subventricular zone by stabilizing the mRNA of special adenine–thymine (AT)-rich DNA-binding protein 1 (SATB1), a critical transcriptional regulator in neurodevelopment. We find that SATB1 deficiency impairs the neuronal differentiation of NSCs, whereas SATB1 overexpression rescues the neuronal differentiation phenotypes resulting from HuD deficiency. Interestingly, we also discover that SATB1 is a transcriptional activator of HuD during NSC neuronal differentiation. In addition, we demonstrate that NeuroD1, a neuronal master regulator, is a direct downstream target of SATB1. Therefore, HuD and SATB1 form a positive regulatory loop that enhances NeuroD1 transcription and subsequent neuronal differentiation. Our results here reveal a novel positive feedback network between an RNA-binding protein and a transcription factor that plays critical regulatory roles in neurogenesis.
Posttranscriptional regulation of messenger RNAs (mRNAs) is an essential mechanism for controlling gene expression, and RNA-binding proteins play key roles in this process (1). Hu antigen D (HuD), a neuron-enriched RNA-binding protein (RBP) expressed early in embryonic neurogenesis, is one of the first markers of neuronal differentiation (2, 3). This protein belongs to the highly conserved ELAV/Hu family of RBPs that consists of four family members—HuR, HuB, HuC, and HuD—which are the mammalian homologs of Drosophila embryonic lethal abnormal vision (ELAV) and encoded by the ELAVL1–4 genes, respectively. Much literature has implicated HuD in neurite outgrowth, neuronal dendritic maturation, and neuronal circuitry development (2, 3). Genetic mutations and functional deficiencies of HuD are associated with a number of neurologic disorders, including paraneoplastic encephalomyelitis, spinal muscular atrophy, Parkinson’s disease, schizophrenia, epilepsy, and neuroblastoma (3). HuD depletion in a rodent model results in brain development deficiencies and impaired motor performance (4).
Although several studies have suggested that HuD is important for the differentiation of immature cells into neurons, most of this work was done using the rat pheochromocytoma PC12 cell line or avian neural crest cultures (5–7). Akamatsu et al. (4) created the first HuD knockout (KO) mouse line and demonstrated that primary neural progenitor cells isolated from the cortex of embryonic HuD KO mice exhibit increased neurosphere formation and decreased neuronal differentiation, as well as greater cell death. The mechanisms underlying these neurogenic deficits remain unexplored, however. In mammals, neuronal production ceases after birth but persists throughout life in the subventricular zone (SVZ) of the lateral ventricles and the dentate gyrus of the hippocampus. Adult-born neurons undergo a neuronal development process that recapitulates the one during early development (8). Both embryonic and adult neurogenesis is tightly controlled at many levels by both extrinsic factors, such as physiological and pathological conditions, and intricate molecular networks, such as transcriptional or translational processes. Disruptions of these molecular pathways lead to neuronal development deficits that are characteristic of human disorders (9, 10). Akamatsu et al. (4) have shown that, after a prolonged exposure to BrdU, a thymidine analog incorporated into dividing cells, the SVZ of adult HuD KO mice have more BrdU+ cells compared with wild-type mice; however, they determined neither the identity nor the fate of these BrdU+ cells. Thus, the role of HuD in adult neural stem/progenitor cells (NSCs) remains unclear. Understanding the precise regulatory mechanisms that enable lifelong neurogenesis from stem cells in the adult mammalian brain is crucial to understanding both the development and plasticity of mammalian brains.
At the molecular level, HuD is known to interact with AU-rich instability conferring sequences or AU-rich elements (AREs) in the 3′ UTRs of target mRNAs and stabilizes these mRNAs (1, 2, 11, 12). The identification of molecular targets regulated by HuD is critical for understanding the mechanisms underlying its biological functions and associated diseases. A recent study has identified RNAs bound to a combination of all neuronal ELAV-like proteins (HuB, HuC, and HuD) in the brain (12). However, only one study to date has focused exclusively on HuD targets. Previously, we used RNA immunoprecipitation of HuD from a mouse line overexpressing myc-tagged HuD combined with GST-HuD target pull-down as well as novel bioinformatics analyses to identify ∼700 new HuD targets; this revealed novel HuD binding motifs in the 3′ UTR of mRNAs (11). Although many of these predicted targets are associated with neuronal development and functions, their roles in HuD-mediated NSC differentiation have yet to be assessed.
HuD mRNA levels increase during neural differentiation (2). Despite studies demonstrating the roles of HuD in regulating gene expression, only a few so far have examined the mechanism, particularly transcriptional regulation controlling the expression of HuD. Thyroid hormone is known to repress HuD transcription (13, 14). Recently, Ngn2 was identified as the first transcriptional activator of HuD during neuronal differentiation of P19 neuroblastoma cells (15). Therefore, the spatial and temporal induction of HuD is regulated at least in part via transcriptional regulation. However, how HuD is regulated during mammalian neural stem cell differentiation and neurogenesis is largely unknown.
In the present study, we investigated the role of HuD in neuronal differentiation in NSCs of the adult SVZ and explored the potential mechanism behind HuD regulation of neurogenesis. We discovered that HuD promotes adult NSC differentiation into neuronal lineage. We also found that the mRNA of special AT-rich DNA-binding protein 1 (SATB1) is a direct target of HuD in NSCs. HuD binds specific regions of the 3′UTR of Satb1 mRNA and enhances its stability during NSC neuronal differentiation. In addition, we found that SATB1 acts as a transcriptional activator for both HuD and NeuroD1, a neuronal master regulator. Therefore, HuD and SATB1 form a positive regulatory loop that activates NeuroD1 transcription and promotes neuronal differentiation. Our results have revealed a novel positive feedback network between an RNA-binding protein and a transcription factor that plays critical regulatory roles during neuronal differentiation.
Results
HuD Regulates Neuronal Differentiation in the SVZ.
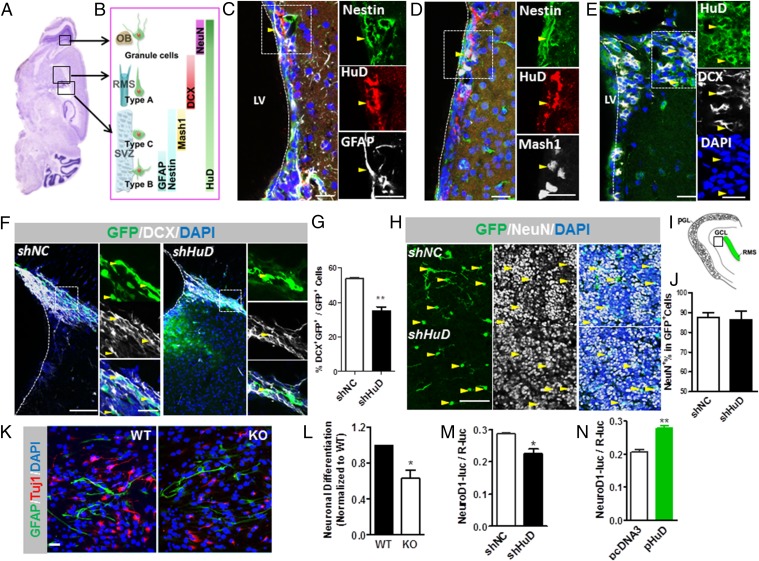
To determine the role of HuD in neural stem/progenitor cell (NSC) differentiation in the adult SVZ, we first assessed HuD expression patterns using cell lineage markers define SVZ neurogenesis (Fig. 1 A and B). HuD was localized in Nestin and GFAP double-positive (Nestin+GFAP+) radial glia-like (type B) cells (Fig. 1C) and Nestin and Marsh1 double-positive (Nestin+Marsh1+) transient amplifying (type C) cells (Fig. 1D) in the SVZ. In addition, consistent with our previous findings (16), HuD is present in doublecortin-positive (DCX+) immature neurons in both the SVZ and rostral migratory stream (RMS) where newly differentiated neurons are en route to their terminal destination in the olfactory bulb (OB) (Fig. 1E), as well as NeuN-positive (NeuN+) mature neurons in the OB (SI Appendix, Fig. S1 A and B). The expression patterns of HuD suggest it may play a potential regulatory role in adult SVZ neurogenesis.
Fig. 1.
HuD regulates neural stem/progenitor cell neuronal differentiation in the adult SVZ. (A) Schematic of a sagittal section of adult mouse brain showing the regions from which the images in C–I were taken. (B) Schematic of HuD expression in different lineages of cells during SVZ adult neurogenesis. (C–E) Adult brain sections containing the SVZ and RMS were stained with antibodies against HuD and lineage markers for neurogenesis. (C) HuD (red) is expressed in Nestin (green)- and GFAP (gray)-expressing radial glia-like (type B) cells. Blue, DAPI. (D) HuD (red) is expressed in Nestin (green)- and Mash1 (gray)-expressing transient (type C) amplifying cells. Blue, DAPI. (E) HuD is expressed in DCX (gray)-positive neuroblasts (type A) in the SVZ and the RMS. In C–E, the right panels are high-magnification images of the boxed regions in the left panels. Arrows indicate the colocalized cells. (Scale bars: 20 µm.) (F) Sample confocal images showing that retrovirus-infected cells in the adult SVZ differentiated into DCX+ immature neurons at 1 wk after virus injection. DCX, gray; GFP, green; DAPI, blue. (Scale bar: 50 μm.) (Right) High-magnification image of the boxed region in the left panel. (Scale bar: 20 µm.) (G) Quantitative analysis showing that shHuD-infected cells differentiated into fewer DCX+ neurons compared with control shNC virus-infected cells (n = 3). (H) Sample confocal images showing that retrovirus-infected cells in the GCL of the OB differentiated into NeuN+ neurons at 4 wk after virus injection. GFP, green; NeuN, gray; DAPI, blue. Arrows indicate cells that are positive for both shRNA (GFP) and NeuN. (Scale bar: 50 μm.) (I) Schematic showing different layers in the adult OB. The boxed region indicates where the images were taken and quantitative analyses were performed. (J) Acute knockdown of HuD in adult SVZ NSCs has no effect on the terminal differentiated into NeuN+ neurons (n = 5). (K) Sample confocal images of differentiated adult SVZ NSCs derived from WT and HuD KO mice. Tuj1, red; GFAP, green; DAPI, blue. (Scale bar: 20 μm.) (L) Quantitative analysis showing that HuD KO NSCs differentiated into fewer Tuj1+ neurons compared with control WT NSCs (n = 5). (M) Quantitative analysis showing that acute knockdown of HuD in NSCs (shHuD) led to reduced neuronal differentiation compared with control NSCs (shNC), as assessed by NeuroD1 promoter activities (n = 3). (N) Quantitative analysis showing that overexpression of HuD (pcHuD) in NSCs led to increased neuronal differentiation compared with controls (pCDNA3), as assessed by NeuroD1 promoter activities (n = 3). Data are expressed as mean ± SEM. *P < 0.05; **P < 0.01, Student’s t test.
Given that HuD KO mice exhibit altered embryonic brain development (4), to determine the role of HuD in adult neurogenesis without the confound of developmental impact, we acutely deleted HuD in NSCs in the adult SVZ using retrovirus expressing a small hairpin inhibitory RNA against HuD (shHuD) (17) as well as GFP (SI Appendix, Fig. S2 A–C). Recombinant retroviruses that are only capable of infecting dividing cells selectively transduce NSCs, allowing for fate tracking at single-cell levels in the adult germinal zone (18–20). One group of mice also received BrdU injections and analyzed at 12 h after BrdU injection for assessing cell proliferation. We found that NSCs infected with retrovirus expressing shHuD (retro-shHuD) incorporated more BrdU compared with NSCs infected with control retrovirus (retro-shNC) (SI Appendix, Fig. S3, BrdU+GFP+/ GFP+). At 1 wk after viral injection, many of the retrovirus-labeled NSCs (eGFP+) would be expected to have differentiated into neurons expressing the early neuronal marker DCX (Fig. 1F and SI Appendix, Fig. S2B). We found that retro-shHuD–infected NSCs differentiated into fewer DCX+ neurons compared with retro-shNC–infected NSCs (Fig. 1G; DCX+GFP+/GFP+). We then determined the impact of HuD deficiency on NSC terminal differentiation in the OB at 4 wk after viral injection. We found no difference in the percentage of NeuN+ mature neurons between retro-shHuD– and control retro-shNC–infected cells (Fig. 1 H–J). To validate the shRNA results, we injected HuD mutant (KO) mice with BrdU and analyzed terminal differentiation of BrdU-labeled cells 4 wk later (SI Appendix, Fig. S4 A and B). We analyzed NeuN+ total neurons as well as Calretinin (CR)+, Calbindin (CB)+, and tyrosine hydroxylase (TH)+ interneurons known to be produced by adult SVZ neurogenesis (21, 22). Quantitative analyses showed that although the total number of BrdU+ cells were lower in KO mice compared with WT mice (SI Appendix, Fig. S4C), the percentage differentiation into NeuN+, CR+, CB+, or TH+ interneurons was not different between KO and WT mice (SI Appendix, Fig. S4 D–F). Because HuD KO mice had significantly smaller volume in both the granule cell layer (GCL) and periglomerular layer (PGL) of the OB (SI Appendix, Fig. S4G), the total numbers of NeuN+, CR+, and CB+, but not TH+ were lower in KO mice compared with WT mice (SI Appendix, Fig. S4 H–K). Therefore, HuD deficiency in adult NSCs impairs early neuronal differentiation rather than terminal differentiation.
To further confirm the effect of HuD on NSCs, we isolated NSCs from the SVZ of adult HuD KO mice and wild-type (WT) littermate controls. In WT NSCs, HuD mRNA expression levels increased during neuronal differentiation (SI Appendix, Fig. S5 A and B), consistent with previous findings in cell lines (15, 23). We then found that HuD KO NSCs differentiated into fewer Tuj1+ neurons (Fig. 1 K and L) with no significant alteration in proliferation or astrocyte differentiation compared with WT controls (SI Appendix, Fig. S5 C and D). To validate our immunocytochemical data, we further assessed the differentiation of NSCs by measuring the promoter activities of a neuronal transcription factor, Neurogenic differentiation 1 (NeuroD1), and the promoter activities of astrocyte lineage marker GFAP (24, 25). Acute knockdown of HuD in NSCs using lentivirus expressing shHuD led to decreased NeuroD1 promoter activity (Fig. 1M) without a significant impact on Gfap promoter activities (SI Appendix, Fig. S5E). Next, we performed a gain-of-function assay by overexpressing HuD in NSCs. Overexpression of HuD led to increased NeuroD1 promoter activities (Fig. 1N), again with no significant effect on the Gfap promoter (SI Appendix, Fig. S5F). Taken together, our results provide further evidence that HuD plays an important role in promoting the neuronal differentiation of NSCs in the adult SVZ.
HuD Regulates SATB1 Expression in NSCs.
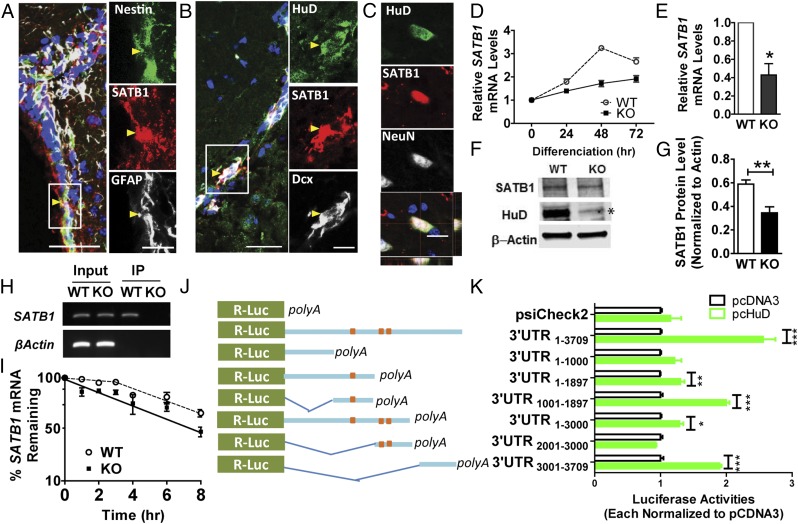
We next searched for downstream targets of HuD that might mediate its regulation of neurogenesis. HuD is known to bind and regulate the stability or translation of a large number of mRNAs (2, 11). We previously used RNA immunoprecipitation coupled with microarray (RIP-ChIP) and novel bioinformatics methods to identify approximately 700 novel HuD targets in the mouse brain and discovered three new HuD-binding motifs (11). Interestingly, many of these mRNAs encoded RBPs and transcription factors, suggesting that HuD is part of a complex transcriptional-translational gene regulatory network (11). Given our observation that HuD mRNA levels were up-regulated during neuronal differentiation (SI Appendix, Fig. S5B), we decided to investigate transcription factors among predicted HuD targets to identify a novel regulatory network between HuD and transcriptional regulators in neural differentiation. Among the transcription factors on the HuD target list, we focused on SATB1 because of its known function in regulating cell linage-specific gene expression during both T-cell development (26) and cortical neuron maturation (27, 28). We first assessed the expression of SATB1 and confirmed that it was localized in Nestin+GFAP+ type B cells in the SVZ (Fig. 2A) and colocalized with HuD in DCX+ cells in the adult SVZ (Fig. 2B) and NeuN+ cells in the OB (Fig. 2C). In addition, similar to HuD, the mRNA levels of SATB1 also increased during adult NSC neuronal differentiation, and such an increase was partially diminished in HuD KO NSCs (Fig. 2D), further supporting a positive correlation between HuD and SATB1. Indeed, both Satb1 mRNA (Fig. 2E) and protein (Fig. 2 F and G) levels were lower in proliferating HuD KO NSCs compared with WT NSCs. Furthermore, the fluorescent intensity of an anti-SATB1 antibody staining was lower in HuD KO SVZ NSCs (SI Appendix, Fig. S6). Therefore, HuD is likely a positive regulator for Satb1 mRNA levels.
Fig. 2.
HuD regulates SABT1 mRNA stability. (A–C) A sagittal section of the adult mouse brain containing the SVZ, RMS, and the OB was costained with antibodies against SATB1, HuD, and different lineage markers for neurogenesis. (A) SATB1 (red) is localized in Nestin (green)- and GFAP (gray)-expressing (type B) cells. Blue, DAPI. (B) SATB1 (red) is colocalized with HuD (green) in DCX+ (gray) neuroblasts in the RMS. The right panels in A and B are high-magnification images of the boxed regions in the left panels. (Scale bars: Left, 50 µm; Right, 20 µm.) Arrows indicate colocalized cells. (C) Sample confocal images showing that SATB1 (red) is colocalized with HuD (green) in NeuN+ neurons in the OB. (Scale bar: 20 µm.) (D) qPCR analysis showing that Satb1 mRNA expression levels increase during adult NSC differentiation, but such an increase is significantly diminished in HuD KO NSCs (n = 3). Both WT and KO were normalized to their own proliferating conditions. P < 0.001, two-way ANOVA with Bonferroni posttest. (E) Satb1 mRNA levels were lower in HuD KO NSCs compared with WT (n = 3). P < 0.05, paired t test. (F and G) Sample Western blot analysis (F) and quantitative results of three Western blots (G) showing that SATB1 protein levels are lower in aNSCs isolated from the SVZ of adult HuD KO mice compared with those from WT mice. β-actin served as a loading control. n = 3. The asterisk indicates a cross-reactive band in HuD KO mice that has been observed previously in both HuD KO mice (4) and with other HuD-specific antibodies (49). (H) Sample reverse transcription PCR (RT-PCR) analysis of Satb1 mRNAs in input and HuD-antibody IP NSC samples. HuD KO NSCs served as negative controls for assessing HuD antibody specificity. β-actin mRNA served as an internal control for input. (I) Adult NSCs were treated with 10 μM actinomycin D to inhibit gene transcription, and the amounts of Satb1 mRNA in WT and HuD KO NSCs were quantified using RT-PCR. Regression analyses indicate that in WT NSCs, Satb1 mRNA degradation followed a two-rate exponential decay, with a slow phase (T1/2 = 18.9 h) during the first 3 h and a rapid phase (T1/2 = 5.6 h) during the last 5 h of the experiment. In contrast, in HuD KO NSCs, Satb1 mRNA degradation followed a single rate of decay with a significantly shortened half-life (T1/2 = 5.2 h). n = 3. Genotype F(1,4) = 41.30; P = 0.003, two-way ANOVA. For the first 3 h, the genotype–time interaction: F(3,16) = 4.076; P = 0.025, two-way ANOVA. (J) Schematics of Satb1 3′ UTR fragments used for reporter assays. The predicted HuD-binding motifs are marked in orange. (K) R-luc activities produced by various fragments of 3′ UTR constructs were normalized to control firefly luciferase (fLuc) activities in the same psiCheck2 vectors. Luciferase activities in pcHuD-transfected conditions were normalized to the pcDNA3-transfected condition. Statistical analyses were carried out between each pcHuD condition vs. pcDNA3 control conditions (before normalization) for each 3′ UTR fragment. n = 4. Data are presented as mean ± SEM. *P < 0.05; **P < 0.01; ***P < 0.001, Student’s t test.
We next explored whether SATB1 is a molecular target of HuD in adult NSCs. HuD is known to regulate gene expression by binding to the 3′UTR of targeted mRNAs and, in most cases, acts as a mRNA stabilizer (2, 11). Using a HuD antibody RNA-IP followed by real-time quantitative PCR (qPCR), we confirmed that HuD bound to Satb1 mRNA (Fig. 2H). Next, to determine whether the reduction of Satb1 mRNA levels in HuD KO NSCs might be due to decreased mRNA stability, we treated HuD KO and WT NSCs with actinomycin D to inhibit gene transcription and assessed the Satb1 mRNA levels over an 8-h period (Fig. 2I). Regression analyses indicated that, in WT NSCs, Satb1 mRNA degradation followed a two-rate exponential decay, with a slow phase (T1/2 = 18.9 h) during the first 3 h and a rapid phase (T1/2 = 5.6 h) during the last 5 h of the experiment. In contrast, in HuD KO NSCs, Satb1 mRNA degradation followed a single rate of decay, with a significantly shortened half-life (T1/2 = 5.2 h). Previously, we had observed a similar biphasic decay for another HuD target mRNA, GAP-43, and had shown that the 3-h delay in the onset of the fast decay was due to an interaction of HuD’s third recognition motif with long poly(A) tails (29). In the absence of HuD, Satb1 mRNA, like GAP-43 mRNA, decayed at a single fast rate. Therefore, HuD increases Satb1 mRNA levels by enhancing RNA stability in NSCs.
We next investigated which regions of Satb1 mRNA interacted with HuD. There are multiple putative HuD binding motifs in Satb1 3′UTR (SI Appendix, Table S1), as predicted by our motif-searching method (11). To identify the regions of Satb1 3′UTR that might be regulated by HuD, we cloned a 3.7-kb 3′ UTR of SATB1 containing all predicted HuD binding sites (3′ UTR1–3709, with 1 denoting the first base after the termination codon) into a pSicheck2 luciferase reporter vector (Fig. 2 J and K) so that the Renilla luciferase (R-luc) expression would be regulated via 3′ UTR sequence of SATB1. When the reporter was cotransfected with HuD expression plasmids (pcHuD) (30) into NSCs, the overexpression of HuD significantly increased luciferase activities compared with control vector (pCDNA3)-transfected conditions (Fig. 2K; 3′ UTR1–3709). We then cloned fragments containing partial sequences of Satb1 3′UTR (3′UTR1–1000, 3′ UTR1–1897, 3′ UTR1001–1897, 3′ UTR1–3000, 3′ UTR2001–3000, and 3′ UTR3001–3709) into reporter vectors. Each Satb1 3′UTR reporter was cotransfected with either pcHuD or pCDNA3 empty vector into adult NSCs. HuD significantly increased luciferase reporter activities through 3′ UTR1–1897, 3′ UTR1000–1897, 3′ UTR1–3000, and 3′ UTR3001–3709, but not through 3′ UTR1–1000, or 3′ UTR2001–3000 sequences (Fig. 2K). Therefore, HuD appears to bind Satb1 3′UTR at sites located within bases 1001–1897 and 3001–3709. Taken together, these data support HuD as a direct posttranscriptional mRNA stabilizer of SATB1 in NSCs.
SATB1 Deficiency Impairs Neuronal Differentiation of SVZ NSCs.
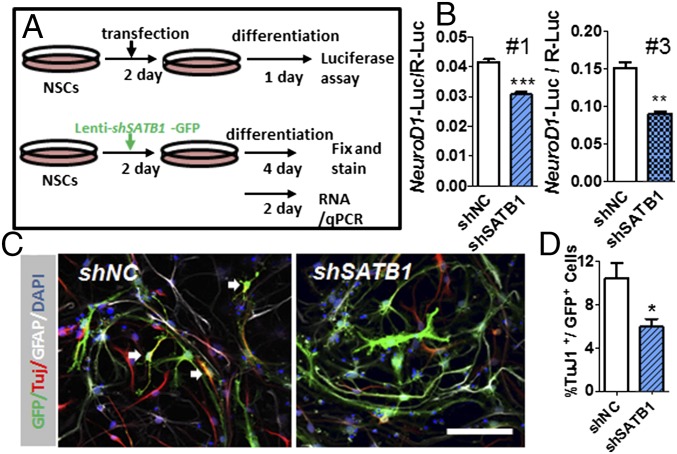
SATB1 is known to play a critical role in the execution of T-cell-specific gene expression programs (26, 31) and can also be induced by neuronal activity and control the transition of tangentially migrating immature interneurons into terminally differentiated somatostatin neurons (28); however, its role in adult NSCs is unknown. Because HuD deficiency leads to reduced SATB1 levels, we reasoned that SATB1 might promote neuronal differentiation, similar to HuD. We used lentivirus expressing two different shRNAs against SATB1 (1 and 3) to acutely knock down SATB1 in SVZ NSCs and subjected the NSCs to differentiation (SI Appendix, Fig. S7 A–D). We found that both shSATB1-1 and 3 led to reduced NeuroD1 promoter activities in differentiating NSCs (Fig. 3 A and B). To validate the above results, we subjected lenti-shSATB1 infected NSCs to differentiation, followed by high-content imaging analysis of cell lineage-specific markers. We found that both lentivirus-shSATB1-1– and -3–infected NSCs differentiated into significantly fewer Tuj1+ neurons compared with lentivirus-shNC-infected NSCs (SI Appendix, Fig. S7E). Acute knockdown of SATB1 also led to a mild reduction in NSC differentiation into GFAP+ astrocytes but had no significant effect on NSC proliferation (SI Appendix, Fig. S7 F–H). To validate these data, we quantified cell lineage markers using our established unbiased stereology method (32, 33). Since both shSATB1-1 and 3 can knockdown SATB1 efficiently and had similar effects on NSC differentiation, we decided to focus on the shSATB1-#1 first (SI Appendix, Fig. S7 A–D). Again, we found that knockdown of shSATB1 led to reduced differentiation into Tuj1+ neurons (Fig. 3 C and D), but without affecting terminal differentiation into NeuN+ mature neurons in the OB (SI Appendix, Fig. S8). Therefore, SATB1 is an important activator for SVZ NSC differentiation and its deficiency impairs NSC differentiation into the neuronal lineage.
Fig. 3.
SATB1 deficiency impairs neuronal differentiation. (A) Experimental scheme for assessing the function of SATB1 in adult NSC differentiation in vitro using both luciferase reporter assays and cell lineage-specific gene expression analyses. (B) Acute knockdown of SATB1 in adult NSCs using two different shRNAs (shSATB1-#1 and -#3) leads to decreased NeuroD1 promoter activities. A cotransfected R-luc plasmid served as a transfection control. n = 3. **P < 0.01; ***P < 0.001, Student’s t test. (C) Sample images showing Lenti-shNC-GFP– and Lenti-shSATB1-GFP–infected NSCs differentiated into Tuj1+ neurons (red) and GFAP+ astrocytes (gray). GFP, green; DAPI, blue. Arrows indicate GFP+Tuj1+ cells. (Scale bar: 50 μm.) (D) Quantitative analysis indicates that shSATB1-infected cells differentiated into fewer Tuj1+ (red) cells compared with shNC-infected cells. n = 4. Data are expressed as mean ± SEM. *P < 0.05; **P < 0.01, Student’s t test.
SATB1 Rescues Decreased Neuronal Differentiation Caused by HuD Deficiency.
We next tested whether SATB1 could rescue the impaired neuronal differentiation of HuD KO NSCs. Using luciferase reporter assays, we found that transfected SATB1 expression plasmid could enhance NeuroD1 promoter activities in both WT and HuD KO cells (Fig. 4A). We then confirmed these results by infecting WT and HuD KO NSCs with lentivirus expressing a Flag-tagged SATB1 as well as mCherry (SI Appendix, Fig. S9 A–C) and assessed the effects on NSC differentiation. Lenti-SATB1 infection enhanced neuronal differentiation in WT NSCs, and more importantly, it rescued the differentiation of HuD KO NSCs into Tuj1+ neurons and restored it to a level comparable to WT NSCs (Fig. 4 B and C).
Fig. 4.
SATB1 rescues the decreased neuronal differentiation caused by HuD deficiency. (A) Lenti-SATB1 transfection led to increased NeuroD1 promoter activities in both WT and HuD KO NSCs. A cotransfected R-luc plasmid served as a transfection control. fLuc activities produced by NeuroD1 promoter were normalized to R-luc activities. n = 3. One-way ANOVA followed by Tukey post hoc analysis. (B) Both control (Lenti-mCherry)- and Lenti-FLAG-SATB1 virus-infected NSCs differentiated into Tuj1+ neurons (green). Red, mCherry (Upper) or FLAG-SATB1 (Lower), detected by an anti-FLAG antibody; blue, DAPI. Arrows indicate virus-infected cells expressing Tuj1. (Scale bar: 50 μm.) (C) Quantitative analysis showing that lenti-SATB1 infection led to increased neuronal differentiation of HuD KO NSCs without a significant effect on WT NSCs. n = 3. One-way ANOVA followed by Tukey’s post hoc analysis. (D) Sample confocal images of double virus-infected cells in the SVZ used for quantification of percentage of DCX+ cells. (Right) High-magnification pictures of the white boxed area in the left panel. DCX, gray; GFP, green; Flag-SATB1, red; DAPI, blue. Arrows indicate cells that were positive for Retro-GFP (green) and negative for Lenti-SATB1 (red). Stars indicate cells that were positive for both GFP (green) and SATB1 (red). (Scale bars: Left, 50 μm; Right, 20 μm.) (E) Quantification of infected cells differentiated into DCX+ neurons (n = 4). (F) Sample confocal images of double virus-infected cells in the GCL of the OB used for quantification of percentage of NeuN+ cells. NeuN, gray; GFP, green; Flag-SATB1, red; DAPI, blue. Arrows indicate cells positive for retroviral labeling (green) and negative for Lenti-SATB1 (red). Asterisks indicate cells positive for both retroviral labeling (green) and Lenti-SATB1 infection (red). (Scale bars: 50 μm.) (G) Quantification of virus-infected cells differentiated into NeuN+ neurons (n = 5). Data are expressed as mean ± SEM. *P < 0.05; **P < 0.01; ***P < 0.001, one-way ANOVA followed by Tukey’s post hoc test.
We next determined whether SATB1 could rescue the HuD deficiency-induced neuronal differentiation deficit in vivo. We stereotaxically injected lentivirus expressing Flag-tagged SATB1 (also expressing mCherry) together with a retrovirus expressing either shHuD or control shNC (also GFP) into the adult SVZ (SI Appendix, Fig. S9D). At 1 wk postinjection, we collected brain tissues and assessed neuronal differentiation of GFP+ only (HuD knockdown or control, but without SATB1 overexpression) or GFP+SATB1+ double-positive cells (HuD knockdown with SATB1 rescue). We found that exogenous SATB1 enhanced NSC differentiation into DCX+ neurons in both shNC- and shHuD-infected cells and, more importantly, rescued the neuronal differentiation deficits in HuD-deficient cells (Fig. 4 D and E). There was no effect on terminal differentiation into NeuN+ mature neurons at 4 wk after viral injection (Fig. 4 F and G). Therefore, both in vitro and in vivo data indicated that SATB1 is a molecular target and mediator of HuD regulation of NSC neuronal differentiation.
SATB1 Is a Transcriptional Activator of HuD.
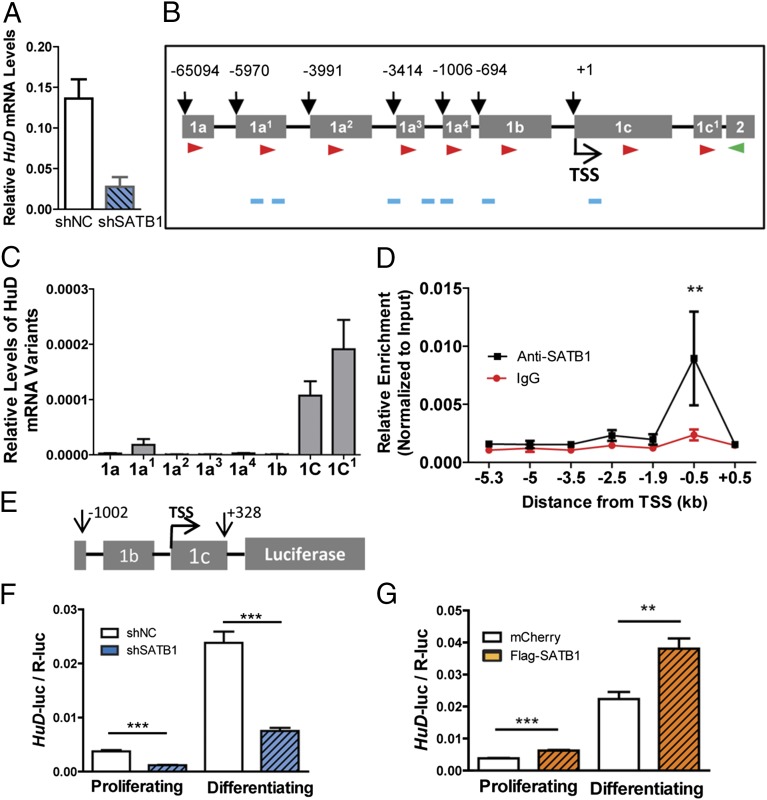
Although an extensive literature focuses on HuD regulation of its downstream targets, much less is known about how HuD expression is regulated. Recent evidence suggests that the levels of HuD mRNAs in neurons are determined predominantly by transcription (2, 3, 15). In our study, both HuD and Satb1 mRNA levels increased during NSC neuronal differentiation, suggesting that a positive regulatory loop may exist between these two proteins. We hypothesized that SATB1 may act as a transcriptional activator for HuD expression. We first assessed HuD mRNA levels in NSCs with acute SATB1 knockdown. Indeed, Lenti-shSATB1-infected NSCs exhibited significantly reduced HuD mRNA levels (Fig. 5A). The HuD signal detected by an anti-HuD antibody staining was lower in SATB1 knockdown cells in OB (SI Appendix, Fig. S9E). We next determined whether SATB1 activates HuD transcription through the HuD promoter. The mammalian HuD gene is known to contain eight conserved leader exons (E1a to E1c), and each of them, except for E1c1, is individually spliced into a common exon 2 (E2), which results in HuD transcript variants with alternative 5′ ends (15). We first examined the expression levels of all eight E1 mRNA variants in NSCs using variant-specific forward primers together with a reverse primer derived from the common E2 (Fig. 5 B and C). We found that E1c and E1c1 are the most abundant variants, with E1a1 as a minor variant, in adult NSCs (Fig. 5C). The other variants were expressed at minimal levels. Our data are consistent with the previous finding that the E1c variant is enriched in neurons and is up-regulated during neuronal differentiation (15).
Fig. 5.
SATB1 is a transcriptional activator of HuD. (A) Acute knockdown of SATB1 led to reduced HuD mRNA levels in NSCs as assessed by qPCR analysis. Gapdh mRNA served as a control. n = 3. P < 0.05, t test. (B) Schematic representation of the genomic loci of eight different HuD E1 variants (adapted from ref. 15). Red and green arrows indicate the forward and backward primers, respectively, used for qPCR. Primers used for ChIP assays are marked in blue. (C) qPCR analysis of mRNA levels for different HuD E1 variants in adult NSCs. Gapdh mRNA served as a control. n = 3. (D) SATB1-specific ChIP followed by qPCR showing enrichment of SATB1 at a genomic sequence 0.5 kb upstream of the HuD TSS in NSCs, as assessed by SATB1-specific ChIP. IgG rabbit in WT NSCs served as negative controls. The enrichment was normalized to input. Quantities of DNA were calculated using standard curves generated from input DNA. n = 3. **P < 0.01, two-way ANOVA with Bonferroni’s posttest. (E) Schematic representation of the HuD promoter luciferase plasmid (pLuc1.0) used in F and G (adapted from ref. 15). (F) SATB1 knockdown led to decreased HuD promoter luciferase activity, both in proliferating and differentiating NSCs. n = 4. (G) SATB1 overexpression led to increased HuD promoter luciferase activities in both proliferating and differentiating SVZ NSCs. n = 4. Data are expressed as mean ± SEM. **P < 0.01; ***P < 0.001 vs. shNC or mCherry, Student’s t test.
We next investigated whether SATB1 interacts directly with genomic regions proximal to HuD E1c by using SATB1-specific chromatin immunoprecipitation (ChIP) followed by qPCR across a ∼6-kb region from 5.3 kb upstream (−5.3 kb) to 0.5 kb downstream (+0.5 kb) relative to the transcription start site (TSS) of HuD E1c. We found that SATB1 antibody was enriched approximately threefold relative to IgG in a genomic region 0.5 kb upstream (−0.5 kb) of the E1c1 TSS (Fig. 5D). This result is consistent with the earlier finding that a ∼400-bp sequence between −1002 and −606 is responsible for transcriptional activation of the HuD E1c variant during neuronal differentiation (15).
To determine whether SATB1 can regulate the transcriptional activity of HuD E1c through the genomic region it binds in adult NSCs, we used a published luciferase reporter construct (pLuc1.0) harboring the ∼1-kb (−1002 to +328) regulatory region of HuD-E1c1 (15)). We first cotransfected pLuc1.0 together with either shSATB1 or control shNC into NSCs (Fig. 5 E and F) and analyzed luciferase activities in both proliferating and differentiating NSCs. The pLuc1.0 and control shNC cotransfected NSCs exhibited dramatically increased luciferase activities in differentiating compared with proliferating NSCs, demonstrating increased HuD E1c1 promoter activities on differentiation. Although SATB1-deficient NSCs (shSATB1) also showed increased pLuc1.0 luciferase activities upon differentiation, HuD 1c1 promoter activities were much diminished in both proliferating and differentiating conditions (Fig. 5F). We next cotransfected pLuc1.0 with either SATB1 expression vector or control mCherry expression vector into NSCs and found that overexpression of SATB1 led to increased HuD promoter activities in both proliferating and differentiating NSCs compared with controls (Fig. 5G). Collectively, these data support our hypothesis that SATB1 binds the promoter of HuD and promotes HuD transcription during NSC differentiation. Therefore, SATB1 and HuD form a positive transcription and posttranscription regulatory loop in NSCs and during neuronal differentiation.
HuD and SATB1 Form a Positive Regulatory Loop That Regulates Neuronal Differentiation Through NeuroD1.
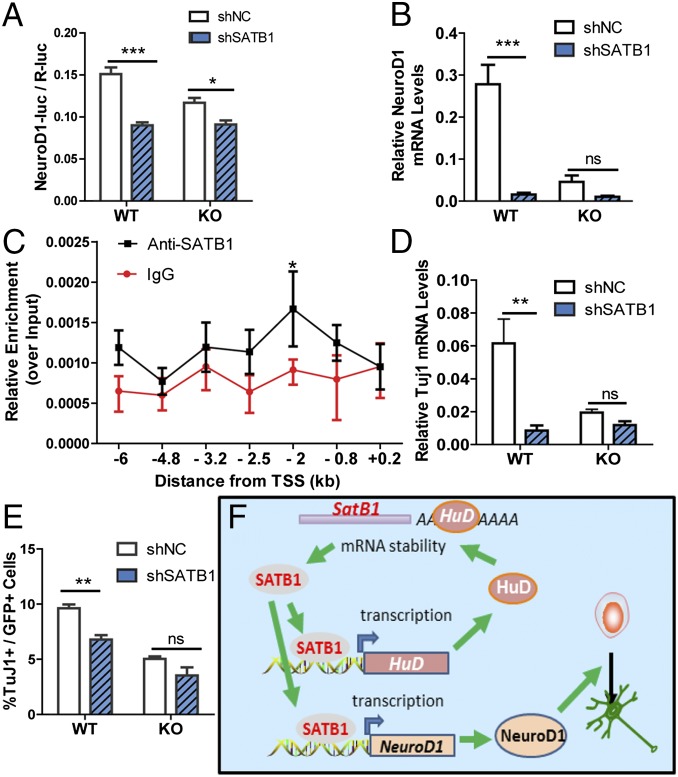
We next searched for downstream effectors controlled by the HuD and SATB1 regulatory loop during neuronal differentiation. We acutely knocked down SATB1 in WT and HuD KO NSCs with lentivirus expressing either shSATB1 or shNC, and analyzed changes in gene expression profiles using neurogenesis pathway arrays. We first searched genes and pathways that exhibited similar changes in NSCs infected with lentivirus expressing two different shSATB1 (#1 and #3). We found that both Lenti-shSATB1-#1- and -#3-infected NSCs showed similar up- or down-regulated genes, confirming the specificities of these two shSATB1s (SI Appendix, Tables S2 and S3). Among them, 31 genes exhibited more than a 1.5-fold change in shSATB1-infected NSCs (21 genes up-regulated and 10 genes down-regulated) (SI Appendix, Table S2 and S3). We next compared neuronal lineage-specific genes that were down-regulated in shSATB1-infected NSCs with those genes down-regulated in HuD KO-NSCs compared with WT controls. We identified NeuroD1 as a gene down-regulated in both SATB1-deficient and HuD KO NSCs. NeuroD1 is a known neuronal master activator, and NeuroD1 promoter activities were reduced in both HuD KO (Fig. 1) and SATB1-deficient NSCs (Fig. 3) during neuronal differentiation.
These findings led us to assess whether NeuroD1 is a downstream effector of the HuD and SATB1 regulatory loop. We found that SATB1 knockdown led to a ∼40.0% reduction in NeuroD1 promoter activities in WT NSCs, but interestingly, the reduction in HuD KO NSCs was much smaller (22.1%) (Fig. 6A). We then validated these data by analyzing NeuroD1 gene expression and found that acute SATB1 knockdown led to dramatically decreased NeuroD1 mRNA levels in WT NSCs, but there was no significant effect in HuD KO NSCs (Fig. 6B). Therefore, HuD and SATB1 may regulate NeuroD1 through a common pathway.
Fig. 6.
HuD and SATB1 form a positive regulatory loop that regulates neuronal differentiation through NeuroD1. (A) SATB1 knockdown led to decreased NeuroD1 transcription as assessed by NeuroD1 promoter-luciferase activity in differentiating NSCs. n = 3. P < 0.05 (interaction), two-way ANOVA with Bonferroni’s posttest. *P < 0.05; ***P < 0.001, post hoc analysis. (B) SATB1 knockdown led to decreased NeuroD1 mRNA levels. n = 3. P < 0.05, two-way ANOVA with Bonferroni’s posttest. ***P < 0.001, post hoc analysis. (C) SATB1-specific ChIP assay demonstrates enrichment of SATB1 protein at a genomic sequence 2 kb upstream of the NeuD1 TSS in NSCs. ChIPs with IgG rabbit served as negative controls. The enrichment was normalized to the input. Quantities of DNA were calculated using a standard curve generated from an input DNA. n = 3. *P < 0.05 (post hoc value), two-way ANOVA with Bonferroni’s posttest. (D and E) SATB1 knockdown led to reduced neuronal differentiation in WT NSCs with no further effect on HuD KO NSCs, as demonstrated by both Tuj1 mRNA level (D; n = 3, interaction P < 0.05) and immunostaining using neuronal marker Tuj1 (E; n = 3, interaction P > 0.05, two-way ANOVA with Bonferroni’s posttest.) **P < 0.01; ***P < 0.001, post hoc analysis. Data are expressed as mean ± SEM. (F) Model for the HuD and SATB1 regulatory network for the regulation of adult NSC neuronal differentiation through NeuroD1.
We then used SATB1-specific ChIP to determine whether SATB1 binds to NeuroD1 promoter. We performed SATB1-ChIP followed by qPCR across a ∼7-kb region encompassing 6 kb upstream (−6 kb) to 0.2 kb downstream (+0.2 kb) relative to the TSS of NeuroD1. We found that SATB1-specific antibody was enriched in a region ∼2 kb upstream (−2 kb) of NeuroD1 TSS relative to IgG control (Fig. 6C). Therefore, SATB1 directly interact with NeuroD1 promoter in NSCs and NeuroD1 is likely a transcriptional target of SATB1.
It is possible that HuD can also regulate NeuroD1 through RNA metabolism; however, we found no predicted binding motif of HuD in NeuroD1 3′ UTR (SI Appendix, Table S4), suggesting that NeuroD1 might not be a direct target for HuD. To validate this hypothesis, we assessed the mRNA stability of NeuroD1 and found that NeuroD1 mRNA stability exhibited no significant difference in HuD KO NSCs compared with WT NSCs in both proliferating and differentiating NSCs (SI Appendix, Fig. S10 A and B). Therefore, NeuroD1 mRNA might not be a direct target of HuD. Thus, our data suggest that NeuroD1 is a direct transcriptional target of SATB1 and is regulated by the positive feedback loop between HuD and SATB1 during neuronal differentiation.
Since NeuroD1 plays an instructional role in neuronal fate determination in both embryonic and adult NSCs (34, 35), reduced NeuroD1 expression levels in SATB1-deficient cells would have a direct impact on neuronal differentiation. We found that Tuj1 mRNA levels were reduced in shSATB1-infected WT NSCs but were not further reduced in HuD KO cells (Fig. 6D); whereas mRNA levels of astrocyte Gfap and Aquaporin4 exhibited no significant change in either HuD KO or SATB1 knockdown cells compared with controls (SI Appendix, Fig. S10 C and D). Consistent with these gene expression changes, acute knockdown of SATB1 using Lenti-shSATB1 infection led to reduced differentiation into Tuj1+ neurons in WT NSCs, but had no significant effect in HuD KO NSCs (Fig. 6E). Therefore, the reduced neuronal differentiation mirrors the reduced NeuroD1 expression levels in shSATB1 or HuD KO conditions. Hence NeuroD1 may serve as a functional effector for the HuD and SATB1 regulatory network during NSC neuronal differentiation (Fig. 6F).
Discussion
Despite intense interest, we still do not fully understand how neurogenesis is regulated (9, 10). In this study we revealed how HuD regulates neural stem cell differentiation and discovered a novel feedback loop between an RNA-binding protein, HuD, and a transcriptional regulator, SATB1, in the regulation of stem cell fate and neurogenesis.
Hu Proteins in Neurogenesis.
Drosophila elav is required for both development of young neurons and the maintenance of mature neurons (36). HuD is expressed by embryonic day 10 during mouse brain development and is one of the earliest markers of neuronal differentiation (37). Although many literature focuses on how HuD regulates neurite extension (38, 39) and neuronal maturation (40), only a limited number of studies have addressed HuD function in neural differentiation. Overexpression of HuD leads to neuronal morphology and neuronal marker expression in cultured avian neural crest cells (6) and stalled proliferation of immortalized rat neural progenitor cell line (37); down-regulation of HuD blocks neurite induction in mouse embryonic carcinoma cells (7). The function of HuD in mammalian NSC differentiation was first addressed by Akamatsu et al. (4) using a HuD knockout (KO) mouse line they created. They demonstrated that neural progenitor cells isolated from the cortex of embryonic HuD KO mice exhibit increased neurosphere formation; however, the percentage of neurospheres differentiated into Tuj1+ neurons was reduced. Akamatsu et al. also showed that HuD KO mice had more BrdU+ cells in the SVZ compared with WT mice after a prolonged (4 wk) exposure to BrdU. However, we found that at 4 wk after a BrdU pulse, not continuous, labeling, the number of BrdU+ cells in the OB was reduced in HuD KO compared with WT mice. Since BrdU labels all proliferating cells in SVZ, RMS, and OB, we further showed that acute knockdown of HuD in the adult SVZ NSCs led to increased proliferation. Since SVZ NSCs isolated from adult HuD KO mice exhibited no difference in proliferation compared with controls, HuD may not regulate NSC proliferation through intrinsic mechanisms. One clear observation is that HuD deficiency reduces neuronal fate specification. Akamatsu et al. (4) did not determine either the identity of BrdU-positive cells in the SVZ. Therefore, we focused on neuronal differentiation and provided the first assessment for the role of HuD in adult NSC neuronal differentiation and underlying mechanism. Interestingly, HuD deficiency had no effect on the percentage of mature neurons in the OB. Therefore, HuD plays a significant role in early neuronal specification rather than terminal differentiation of SVZ NSCs.
HuD Targets.
Several studies, including ours, have shown that HuD preferentially interacts with AU-rich elements, known as AREs, in the 3′ UTR of target mRNAs and stabilizes these mRNAs (2, 11, 41). We have previously identified novel HuD-binding motifs in the 3′UTR of target mRNAs (11). Using our bioinformatics algorithm, we identified three predicted HuD target sites in the Satb1 3′ UTR, two of which match the consensus of a typical AU-rich motif. Surprisingly, we found that only one of the ARE-like target sites, the one at position 1578 from the stop codon, resulted in HuD-enhanced expression in our luciferase reporter assay. Interestingly, the last 700 bp of sequence near the polyA (3001–3709) containing an ARE-like sequence with two mismatched nucleotides at position 3583 was also sensitive to HuD overexpression. This may be related to HuD’s function of binding polyA tails (3). It is likely that other factors, in addition to sequence motifs, can significantly modulate the binding of HuD and other RBPs to mRNAs, including interactions with other RNA-binding proteins.
Regulation of HuD Expression.
Despite extensive studies on the roles of HuD in regulating gene expression, far less is known about the mechanisms controlling the expression of HuD. Several studies have shown that HuD can be regulated at posttranscriptional levels by RNA-binding proteins and microRNAs (3). Still, little is known about HuD regulation at the transcriptional level. Thyroid hormone was shown to repress HuD transcription (13, 14). However, the significant increase of HuD expression during cellular differentiation suggests that transcriptional activation is likely a major player. Recently, Neurogenin 2 was identified as the first transcriptional activator of HuD by binding to the proximal promoter of the HuD gene in P19 neuroblastoma cells and activating transcription during neuronal differentiation (15). Therefore, the spatial and temporal induction of HuD is regulated at least in part via transcriptional regulation. This study also identified several exon 1 (E1) variants and demonstrated that E1c and E1b are the most abundant variants in adult murine brains. We found that E1c and E1c1 are the most abundant variants in NSCs from adult SVZ. It is possible that HuD variants represent its molecular diversity in subtypes of cells in the mammalian brain. Future studies dissecting the functional significance of this molecular diversity would help us understand the regulation and function of this protein.
Posttranscriptional Regulation of SATB1.
Among the most highly enriched mRNAs of the 700 putative HuD targets that we have identified (11), SATB1, a specific T-lineage–enriched transcription regulator, stood out because there were a number of predicted HuD binding sites in its 3′ UTR. SATB1 is well known to orchestrate the temporal and spatial expression of genes during T-cell proliferation and differentiation, thereby ensuring the proper development of this lineage (26, 31). SATB1 is differentially expressed in various subsets of neuronal cells and regulates a large number of genes involved in development and differentiation (27, 28). Recent studies found that SATB1 is the major SATB family protein in postnatal brains and acts as a ‘‘docking site’’ to recruit chromatin modifiers to gene promoters. SATB1 binds to genomic loci of multiple immediate early genes (IEGs) and is required for the proper temporal expression of these genes during postnatal development. SATB1 is also induced by neuronal activity and promotes interneuron maturation (27, 28). However, the role of SATB1 in neural stem cell differentiation during embryonic or adult neurogenesis has not been uncovered and our study provides the first evidence for the role of SATB1 in adult neurogenesis. Previously, studies have shown that both SATB1-null or HuD-null mice exhibit an abnormal hind limb-clasping reflex, which is seen in mutant mice with cortical and basal ganglia defects (4, 26), suggesting that both HuD and SATB1 are critical for mice motor-sensory circuit development and might have an overlapping signaling pathway regulating cortical development. Our results show that HuD and SATB1 form a positive regulatory loop in NSCs that regulates NeuroD1 transcription and neuronal differentiation. Our observation that SATB1 knockdown had a significant effect on WT NSCs but not on HuD KO NSCs suggests that these two proteins may largely share pathways for regulating neuronal differentiation.
Our discovery of this positive feedback loop between a RNA-binding protein and a transcription factor in NSC differentiation indicates that neuronal development is regulated by a complex network of RNA- and DNA-binding proteins. Among the list of HuD targets are several additional transcription factors and RNA-binding proteins. Future studies of the extent and dimension of the posttranscriptional and transcriptional interactome in neural stem cell differentiation will provide critical insight into the developmental regulation and disease mechanisms associated with this protein.
Materials and Methods
Additional and more detailed descriptions of the methodology of this study are available in the SI Appendix.
Mice.
All animal procedures were performed according to protocols approved by the University of Wisconsin–Madison’s Institutional Animal Care and Use Committee. The HuD KO mice were described previously (4). Mice were group-housed up to four per cage with the same sex and maintained on a 14/10-h light/dark cycle with food and water available ad libitum.
Production of Lentivirus and Retrovirus and in Vivo Grafting of Virus.
Recombinant viral production and in vivo viral grafting using stereotaxic surgery were performed as described previously (33, 42, 43). In brief, 7- to 8-wk-old C57B/L6 male mice were anesthetized with isofluorane and placed in a stereotactic instrument (KOPF). Virus (1 μL with titer >1 × 108/mL) was stereotaxically injected into the SVZ using the following coordinates relative to bregma: caudal, +1.0 mm; lateral, ±1.0 mm; ventral, −2.2 mm, and caudal, +0 mm; lateral, ±1.4 mm; ventral, −1.9 mm. At indicated time points after viral grafting, mice were deeply anesthetized with pentobarbital and perfused with saline, followed by 4% (wt/vol) paraformaldehyde.
Immunohistology and neurogenesis analysis were performed as we described previously (24, 43, 44).
For isolation and analyses of adult NSCs, SVZ NSCs were isolated from 6- to 8-wk-old HuD KO mice and WT littermates as described previously (45). Cell proliferation and differentiation analyses were carried out as described previously (24, 32, 46). RNA-IP was performed as described previously (24, 42).
For luciferase reporter assays, the 3′ UTR sequence of SATB1 was PCR amplified directly from purified mouse cortical genomic DNA and cloned into psiCHECK-2 dual luciferase vector (Promega; C8021) using In-Fusion HD Cloning Kit (Takara; 011614). The 4-kb and 1.3-kb HuD promoter-reporter plasmids cloned into the MCS of the PGL4.14 vector were kindly provided by B. J. Jasmin (University of Ottawa) (15). Transfection of NSCs was carried out using Fugene HD (Roche; 04709713001) based on the manufacturer’s protocol with modifications.
For the mRNA stability assay, cultured hippocampus neurons were treated with 10 μg/mL actinomycin D (Sigma-Aldrich; A1410) to inhibit gene transcription (33) and SVZ NSCs were collected at various time intervals for RNA isolation and qPCR analysis. SATB1 and NeuroD1 mRNA levels were normalized to Gapdh. RNA decay kinetics and half-life were analyzed using published methods (29, 47, 48). In brief, we used the exponential function M = M0e−λt, where M is the amount of mRNA at time t, M0 is the amount of mRNA at time 0, and λ = (ln 2)/T1/2, where T1/2 is the half-life of the mRNA.
RT-PCR, qPCR, and pathway array analyses were performed using standard methods as described previously (33, 43, 46). The first-strand cDNA was generated by reverse transcription with random primers using a Transcriptor First-Strand cDNA Synthesis Kit (Roche; 04896866001). Standard RT-PCR was performed using GoTaq DNA polymerase (Promega; M3005).
ChIP.
ChIP was performed according to published methods (46).
Statistical Analysis.
The results were assessed by Student’s t test to compare two groups or by one-way ANOVA with Bonferroni post hoc test or two-way ANOVA with Bonferroni’s post hoc test for multiple comparisons, using GraphPad Prism. Statistical comparisons between two genotypes within the same treatment group and between different treatment groups within the same genotype were carried out for each experiment. In all tables and figures, data are expressed as mean ± SEM. *P < 0.05; **P < 0.01; ***P < 0.001.
Supplementary Material
Acknowledgments
We thank Cheryl T. Strauss for editing, Yina Xing for technical assistance, and members of the Zhao laboratory for helpful discussions. We also thank Drs. Hideyuki Okano, Hirotaka J. Okano, and Robert Darnell for providing HuD KO mice and Dr. Bernard J. Jasmin for providing the HuD-luciferase plasmids. This work was supported by grants from the National Institutes of Health (R01 MH080434 and R01 MH078972, to X.Z.; P30HD03352, to the Waisman Center; and R01 NS30255 and R21DA034452, to N.P.B.), the International Rett Syndrome Foundation (3007, to X.Z.), Chinese Academy of Sciences-Institute of Genetics and Developmental Biology start-up funds (O7600O941, Y4655021941, and Y472011941, to W.G.). W.G. was funded by a postdoctoral fellowship from the University of Wisconsin Center for Stem Cells and Regenerative Medicine and the Recruitment Program of the Global Youth Experts of China, 2015. E.D.P. was funded by a University of Wisconsin–Hilldale fellowship for undergraduate research.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1513780112/-/DCSupplemental.
References
- 1.Darnell JC, Richter JD. Cytoplasmic RNA-binding proteins and the control of complex brain function. Cold Spring Harb Perspect Biol. 2012;4(8):a012344. doi: 10.1101/cshperspect.a012344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bronicki LM, Jasmin BJ. Emerging complexity of the HuD/ELAVl4 gene: Implications for neuronal development, function, and dysfunction. RNA. 2013;19(8):1019–1037. doi: 10.1261/rna.039164.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Perrone-Bizzozero N, Bird CW. Role of HuD in nervous system function and pathology. Front Biosci (Schol Ed) 2013;5:554–563. doi: 10.2741/s389. [DOI] [PubMed] [Google Scholar]
- 4.Akamatsu W, et al. The RNA-binding protein HuD regulates neuronal cell identity and maturation. Proc Natl Acad Sci USA. 2005;102(12):4625–4630. doi: 10.1073/pnas.0407523102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Marusich MF, Furneaux HM, Henion PD, Weston JA. Hu neuronal proteins are expressed in proliferating neurogenic cells. J Neurobiol. 1994;25(2):143–155. doi: 10.1002/neu.480250206. [DOI] [PubMed] [Google Scholar]
- 6.Wakamatsu Y, Weston JA. Sequential expression and role of Hu RNA-binding proteins during neurogenesis. Development. 1997;124(17):3449–3460. doi: 10.1242/dev.124.17.3449. [DOI] [PubMed] [Google Scholar]
- 7.Kasashima K, et al. Cytoplasmic localization is required for the mammalian ELAV-like protein HuD to induce neuronal differentiation. Genes Cells. 1999;4(11):667–683. doi: 10.1046/j.1365-2443.1999.00292.x. [DOI] [PubMed] [Google Scholar]
- 8.Ge S, Pradhan DA, Ming GL, Song H. GABA sets the tempo for activity-dependent adult neurogenesis. Trends Neurosci. 2007;30(1):1–8. doi: 10.1016/j.tins.2006.11.001. [DOI] [PubMed] [Google Scholar]
- 9.Song J, Christian KM, Ming GL, Song H. Modification of hippocampal circuitry by adult neurogenesis. Dev Neurobiol. 2012;72(7):1032–1043. doi: 10.1002/dneu.22014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Eisch AJ, Petrik D. Depression and hippocampal neurogenesis: A road to remission? Science. 2012;338(6103):72–75. doi: 10.1126/science.1222941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bolognani F, Contente-Cuomo T, Perrone-Bizzozero NI. Novel recognition motifs and biological functions of the RNA-binding protein HuD revealed by genome-wide identification of its targets. Nucleic Acids Res. 2010;38(1):117–130. doi: 10.1093/nar/gkp863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ince-Dunn G, et al. Neuronal Elav-like (Hu) proteins regulate RNA splicing and abundance to control glutamate levels and neuronal excitability. Neuron. 2012;75(6):1067–1080. doi: 10.1016/j.neuron.2012.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cuadrado A, Navarro-Yubero C, Furneaux H, Muñoz A. Neuronal HuD gene encoding a mRNA stability regulator is transcriptionally repressed by thyroid hormone. J Neurochem. 2003;86(3):763–773. doi: 10.1046/j.1471-4159.2003.01877.x. [DOI] [PubMed] [Google Scholar]
- 14.Lee EK, et al. RNA-binding protein HuD controls insulin translation. Mol Cell. 2012;45(6):826–835. doi: 10.1016/j.molcel.2012.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bronicki LM, Bélanger G, Jasmin BJ. Characterization of multiple exon 1 variants in mammalian HuD mRNA and neuron-specific transcriptional control via neurogenin 2. J Neurosci. 2012;32(33):11164–11175. doi: 10.1523/JNEUROSCI.2247-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bolognani F, et al. Coordinated expression of HuD and GAP-43 in hippocampal dentate granule cells during developmental and adult plasticity. Neurochem Res. 2007;32(12):2142–2151. doi: 10.1007/s11064-007-9388-8. [DOI] [PubMed] [Google Scholar]
- 17.Allen M, et al. HuD promotes BDNF expression in brain neurons via selective stabilization of the BDNF long 3' UTR mRNA. PLoS One. 2013;8(1):e55718. doi: 10.1371/journal.pone.0055718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Smrt RD, et al. Mecp2 deficiency leads to delayed maturation and altered gene expression in hippocampal neurons. Neurobiol Dis. 2007;27(1):77–89. doi: 10.1016/j.nbd.2007.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.van Praag H, et al. Functional neurogenesis in the adult hippocampus. Nature. 2002;415(6875):1030–1034. doi: 10.1038/4151030a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ge S, et al. GABA regulates synaptic integration of newly generated neurons in the adult brain. Nature. 2006;439(7076):589–593. doi: 10.1038/nature04404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Batista-Brito R, Close J, Machold R, Fishell G. The distinct temporal origins of olfactory bulb interneuron subtypes. J Neurosci. 2008;28(15):3966–3975. doi: 10.1523/JNEUROSCI.5625-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hack MA, et al. Neuronal fate determinants of adult olfactory bulb neurogenesis. Nat Neurosci. 2005;8(7):865–872. doi: 10.1038/nn1479. [DOI] [PubMed] [Google Scholar]
- 23.Akamatsu W, et al. Mammalian ELAV-like neuronal RNA-binding proteins HuB and HuC promote neuronal development in both the central and the peripheral nervous systems. Proc Natl Acad Sci USA. 1999;96(17):9885–9890. doi: 10.1073/pnas.96.17.9885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Luo Y, et al. Fragile X mental retardation protein regulates proliferation and differentiation of adult neural stem/progenitor cells. PLoS Genet. 2010;6(4):e1000898. doi: 10.1371/journal.pgen.1000898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Barkho BZ, et al. Identification of astrocyte-expressed factors that modulate neural stem/progenitor cell differentiation. Stem Cells Dev. 2006;15(3):407–421. doi: 10.1089/scd.2006.15.407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Alvarez JD, et al. The MAR-binding protein SATB1 orchestrates temporal and spatial expression of multiple genes during T-cell development. Genes Dev. 2000;14(5):521–535. [PMC free article] [PubMed] [Google Scholar]
- 27.Close J, et al. Satb1 is an activity-modulated transcription factor required for the terminal differentiation and connectivity of medial ganglionic eminence-derived cortical interneurons. J Neurosci. 2012;32(49):17690–17705. doi: 10.1523/JNEUROSCI.3583-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Denaxa M, et al. Maturation-promoting activity of SATB1 in MGE-derived cortical interneurons. Cell Reports. 2012;2(5):1351–1362. doi: 10.1016/j.celrep.2012.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Beckel-Mitchener AC, Miera A, Keller R, Perrone-Bizzozero NI. Poly(A) tail length-dependent stabilization of GAP-43 mRNA by the RNA-binding protein HuD. J Biol Chem. 2002;277(31):27996–28002. doi: 10.1074/jbc.M201982200. [DOI] [PubMed] [Google Scholar]
- 30.Anderson KD, et al. Overexpression of HuD, but not of its truncated form HuD I+II, promotes GAP-43 gene expression and neurite outgrowth in PC12 cells in the absence of nerve growth factor. J Neurochem. 2000;75(3):1103–1114. doi: 10.1046/j.1471-4159.2000.0751103.x. [DOI] [PubMed] [Google Scholar]
- 31.Cai S, Lee CC, Kohwi-Shigematsu T. SATB1 packages densely looped, transcriptionally active chromatin for coordinated expression of cytokine genes. Nat Genet. 2006;38(11):1278–1288. doi: 10.1038/ng1913. [DOI] [PubMed] [Google Scholar]
- 32.Barkho BZ, et al. Endogenous matrix metalloproteinase (MMP)-3 and MMP-9 promote the differentiation and migration of adult neural progenitor cells in response to chemokines. Stem Cells. 2008;26(12):3139–3149. doi: 10.1634/stemcells.2008-0519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Guo W, et al. RNA-binding protein FXR2 regulates adult hippocampal neurogenesis by reducing Noggin expression. Neuron. 2011;70(5):924–938. doi: 10.1016/j.neuron.2011.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gao Z, et al. Neurod1 is essential for the survival and maturation of adult-born neurons. Nat Neurosci. 2009;12(9):1090–1092. doi: 10.1038/nn.2385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kuwabara T, et al. Wnt-mediated activation of NeuroD1 and retro-elements during adult neurogenesis. Nat Neurosci. 2009;12(9):1097–1105. doi: 10.1038/nn.2360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Robinow S, Campos AR, Yao KM, White K. The elav gene product of Drosophila, required in neurons, has three RNP consensus motifs. Science. 1988;242(4885):1570–1572. doi: 10.1126/science.3144044. [DOI] [PubMed] [Google Scholar]
- 37.Hambardzumyan D, et al. AUF1 and Hu proteins in the developing rat brain: Implication in the proliferation and differentiation of neural progenitors. J Neurosci Res. 2009;87(6):1296–1309. doi: 10.1002/jnr.21957. [DOI] [PubMed] [Google Scholar]
- 38.Anderson KD, et al. Overexpression of HuD accelerates neurite outgrowth and increases GAP-43 mRNA expression in cortical neurons and retinoic acid-induced embryonic stem cells in vitro. Exp Neurol. 2001;168(2):250–258. doi: 10.1006/exnr.2000.7599. [DOI] [PubMed] [Google Scholar]
- 39.Perrone-Bizzozero NI, Tanner DC, Mounce J, Bolognani F. Increased expression of axogenesis-related genes and mossy fibre length in dentate granule cells from adult HuD overexpressor mice. ASN Neuro. 2011;3(5):259–270. doi: 10.1042/AN20110015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.DeBoer EM, et al. Prenatal deletion of the RNA-binding protein HuD disrupts postnatal cortical circuit maturation and behavior. J Neurosci. 2014;34(10):3674–3686. doi: 10.1523/JNEUROSCI.3703-13.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bolognani F, Perrone-Bizzozero NI. RNA–protein interactions and control of mRNA stability in neurons. J Neurosci Res. 2008;86(3):481–489. doi: 10.1002/jnr.21473. [DOI] [PubMed] [Google Scholar]
- 42.Guo W, et al. Fragile X proteins FMRP and FXR2P control synaptic GluA1 expression and neuronal maturation via distinct mechanisms. Cell Reports. 2015;11(10):1651–1666. doi: 10.1016/j.celrep.2015.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Smrt RD, et al. MicroRNA miR-137 regulates neuronal maturation by targeting ubiquitin ligase mind bomb-1. Stem Cells. 2010;28(6):1060–1070. doi: 10.1002/stem.431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Guo W, et al. Ablation of Fmrp in adult neural stem cells disrupts hippocampus-dependent learning. Nat Med. 2011;17(5):559–565. doi: 10.1038/nm.2336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Guo W, et al. Inhibition of GSK3β improves hippocampus-dependent learning and rescues neurogenesis in a mouse model of fragile X syndrome. Hum Mol Genet. 2012;21(3):681–691. doi: 10.1093/hmg/ddr501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Liu C, et al. Epigenetic regulation of miR-184 by MBD1 governs neural stem cell proliferation and differentiation. Cell Stem Cell. 2010;6(5):433–444. doi: 10.1016/j.stem.2010.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bolognani F, et al. In vivo post-transcriptional regulation of GAP-43 mRNA by overexpression of the RNA-binding protein HuD. J Neurochem. 2006;96(3):790–801. doi: 10.1111/j.1471-4159.2005.03607.x. [DOI] [PubMed] [Google Scholar]
- 48.Perrone-Bizzozero NI, Cansino VV, Kohn DT. Posttranscriptional regulation of GAP-43 gene expression in PC12 cells through protein kinase C-dependent stabilization of the mRNA. J Cell Biol. 1993;120(5):1263–1270. doi: 10.1083/jcb.120.5.1263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mobarak CD, et al. The RNA-binding protein HuD is required for GAP-43 mRNA stability, GAP-43 gene expression, and PKC-dependent neurite outgrowth in PC12 cells. Mol Biol Cell. 2000;11(9):3191–3203. doi: 10.1091/mbc.11.9.3191. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.