Significance
Recently the preoptic area (POa) has been shown to be a source of GABAergic neurons in the medial amygdala and cerebral cortex, where they are thought to play a pivotal role in emotions and intelligence, respectively. However, it is unknown how the POa-derived neurons migrate and selectively segregate into either the amygdala or cortex. By using focal in utero labeling of the POa, we show that switching on/off the transcription factor COUP-TFII (Chicken ovalbumin upstream promoter transcription factor II) and the receptor Neuropilin-2 (Nrp2) directs the POa-derived neurons toward either the amygdala or cortex. Our study revealed an essential role of COUP-TFII/Nrp2 expression dynamics in the development of the amygdala and cortex.
Keywords: preoptic area, COUP-TFII/Neuropilin-2, caudal migratory stream, cortex, amygdala
Abstract
The preoptic area (POa) of the rostral diencephalon supplies the neocortex and the amygdala with GABAergic neurons in the developing mouse brain. However, the molecular mechanisms that determine the pathway and destinations of POa-derived neurons have not yet been identified. Here we show that Chicken ovalbumin upstream promoter transcription factor II (COUP-TFII)–induced expression of Neuropilin-2 (Nrp2) and its down-regulation control the destination of POa-derived GABAergic neurons. Initially, a majority of the POa-derived migrating neurons express COUP-TFII and form a caudal migratory stream toward the caudal subpallium. When a subpopulation of cells steers toward the neocortex, they exhibit decreased expression of COUP-TFII and Nrp2. The present findings show that suppression of COUP-TFII/Nrp2 changed the destination of the cells into the neocortex, whereas overexpression of COUP-TFII/Nrp2 caused cells to end up in the medial part of the amygdala. Taken together, these results reveal that COUP-TFII/Nrp2 is a molecular switch determining the pathway and destination of migrating GABAergic neurons born in the POa.
Neuronal migration is the fundamental process that determines the final allocation of neurons in the nervous system. During cerebral cortical development, most GABAergic inhibitory neurons are generated in the medial ganglionic eminence (MGE) and the caudal ganglionic eminence (CGE) of the ventral telencephalon, from where they migrate tangentially into the dorsal telencephalon (1). Several independent reports indicate that defects in the migration of GABAergic neurons are involved in brain malformations and psychiatric disorders (1).
The ventral telencephalon is characterized by distinct expression patterns of transcription factors, including NK2 homeobox 1 (NKx2.1) in the MGE (2) and Chicken ovalbumin upstream promoter transcription factor II (COUP-TFII) in the CGE (3). Such segregation of transcription factor expression is relevant to the separate migratory profiles of the cortical GABAergic neurons derived from each region (4). Although MGE-derived cells express LIM homeobox protein 6 (Lhx6), a downstream target of Nkx2.1, and uniformly migrate toward the entire neocortex (5–7), most CGE-derived cells express COUP-TFII and migrate caudally to the neocortex (3, 7). Thus, the migratory profiles of the cortical GABAergic neurons born at each subregion of the ganglionic eminence (GE) differ from each other, however the molecular mechanism of the migration profiles, especially the caudal migration of cortical GABAergic neurons, remains to be elucidated.
The GE is the main source of cortical GABAergic neurons, and there is increasing evidence showing that GABAergic neurons are also derived from regions outside the ventral telencephalon. Recent studies using Nkx5.1-Cre and Dbx1-Cre mice show the preoptic area (POa) as a new source of cortical GABAergic neurons containing an Lhx6-expressing population (8, 9). Other investigators report that the POa supplies GABAergic neurons to the medial nucleus of amygdala, in a study using Dbx1-Cre knock-in mice (10). The neurons migrate caudally to the amygdala via the ventral GE. However, the profile of the migration of POa-derived cortical GABAergic neurons remains largely unknown.
Although Lhx6 and COUP-TFII expression are mutually exclusive in the dorsal part of the GE in that they determine the MGE and CGE properties, respectively, their expression profiles in the ventral region of the GE are not clearly segregated from each other. Previous studies showed that in addition to MGE, there is a large Lhx6-expressing domain covering the caudal telencephalon (11). This distinct Lhx6 expression converges into the ventro-caudal edge of the MGE, a region termed the anterior peduncular area (AEP) (11, 12), which partially overlaps with the COUP-TFII–positive region. Although our previous study demonstrated that AEP-derived cells migrate caudally (3), the origin of this Lhx6-positive cell population and the physiological significance of the dual expression of Lhx6 and COUP-TFII remains unknown.
In this study, we examined COUP-TFII and Lhx6 expression in the migrating neurons of the AEP and found that the Lhx6 and COUP-TFII double-expressing cells originated from the POa and that these GABAergic neurons migrated through the AEP to the amygdala and the cortex. We also found that COUP-TFII expression in the POa-derived neurons varied both spatially and temporally and that the dynamic expression profiles of COUP-TFII, and its downstream molecule Neuropilin-2 (Nrp2), were crucial for the appropriate selection of two major migratory routes toward the amygdala or the neocortex. Thus, these results demonstrate a novel molecular mechanism for correctly sorting GABAergic neurons in the developing brain.
Results
The COUP-TFII–Expressing Stream of Migrating Cells and the POa Share Molecular Properties with Both the MGE and the CGE.
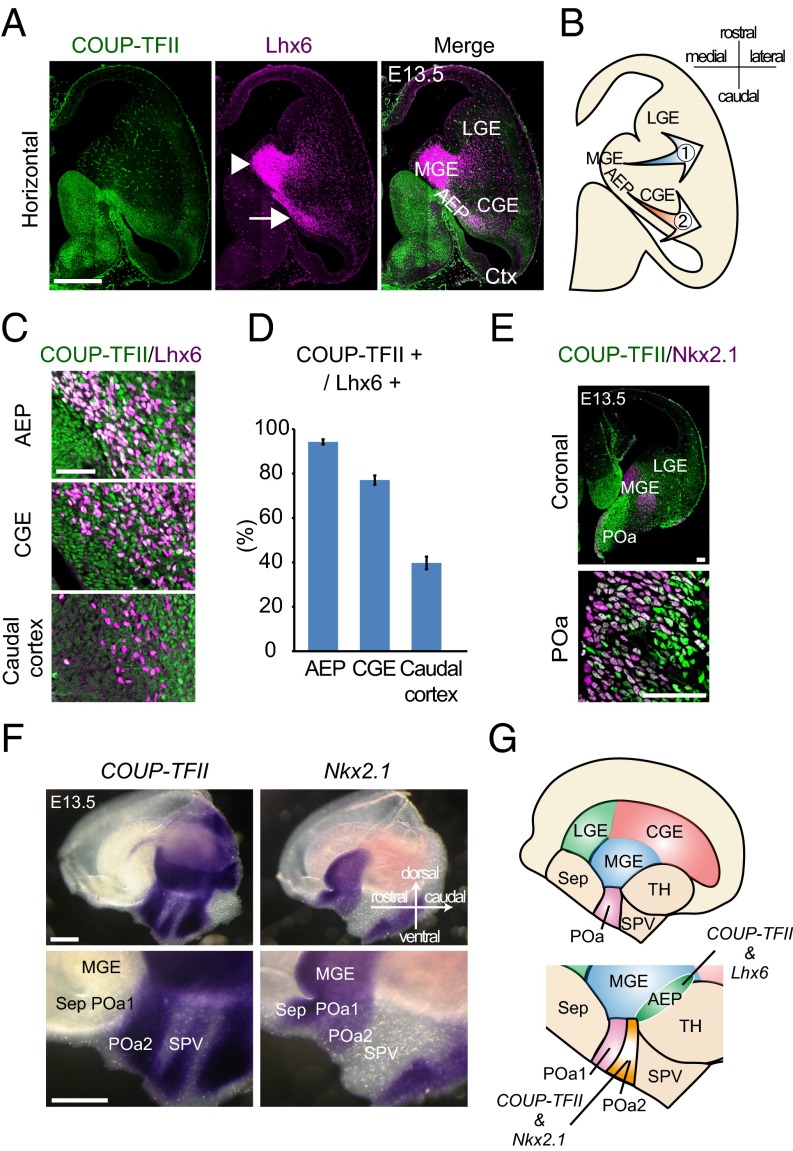
Because the molecular mechanisms underlying the migration of the cell population through/from the AEP are poorly understood, we performed immunohistochemistry using antibodies specific for Lhx6 and COUP-TFII in mice at embryonic day 13.5 (E13.5) and found that AEP partially overlaps with the previously reported COUP-TFII–expressing region (3) (Fig. 1A). A large number of Lhx6-expressing cells formed a migratory stream from the AEP toward the caudal cortex (Fig. 1A, arrow). Quantifying the number of caudally migrating Lhx6-positive cells in the AEP revealed that 94.2 ± 1.1% cells (n = 395) from five brains (n = 5) were coexpressing COUP-TFII (Fig. 1D). The temporal and spatial expression of COUP-TFII was then studied. In caudally migrating Lhx6-positive cells, the expression of COUP-TFII gradually decreased as the cells reached the CGE (77.1 ± 2.1%, n = 1,018 cells, five brains) and the cortex (39.7 ± 2.9%, n = 410 cells, five brains) (Fig. 1 C and D).
Fig. 1.
A population of the caudally migrating cells and the POa share molecular properties with the MGE and the CGE. (A) Immunohistochemical staining for COUP-TFII (green) and Lhx6 (magenta) in an E13.5 horizontal section. Multiple images were tiled and combined to show the whole brain slice. Lhx6-expressing cells form two migratory streams from the MGE (arrowhead) and the AEP (arrow). (B) Schematic diagram of the two migratory pathways. (C) Magnified views of the COUP-TFII/Lhx6–positive stream in the AEP, CGE, and caudal cortex at E13.5. (D) Quantification of the proportion of Lhx6-expressing cells that also express COUP-TFII. (E) Immunohistochemical staining for COUP-TFII (green) and Nkx2.1 (magenta) in an E13.5 coronal section, including the POa. Multiple images were tiled and combined to show the whole brain slice. High-magnification views show that COUP-TFII and Nkx2.1 are coexpressed in the ventricular zone of the POa. (F) Whole mount in situ hybridization of COUP-TFII and Nkx2.1 in the E13.5 hemisphere. COUP-TFII and Nkx2.1 are coexpressed in the POa2. (G) Schematic diagram of the expression of COUP-TFII, Nkx2.1, and Lhx6 in the POa and the AEP. Ctx, cortex; LGE, lateral ganglionic eminence; Sep, septum; SPV, supraoptic paraventricular region; TH, thalamus. [Scale bars, (A and F) 500 µm, (E) 100 µm, and (C) 50 µm.]
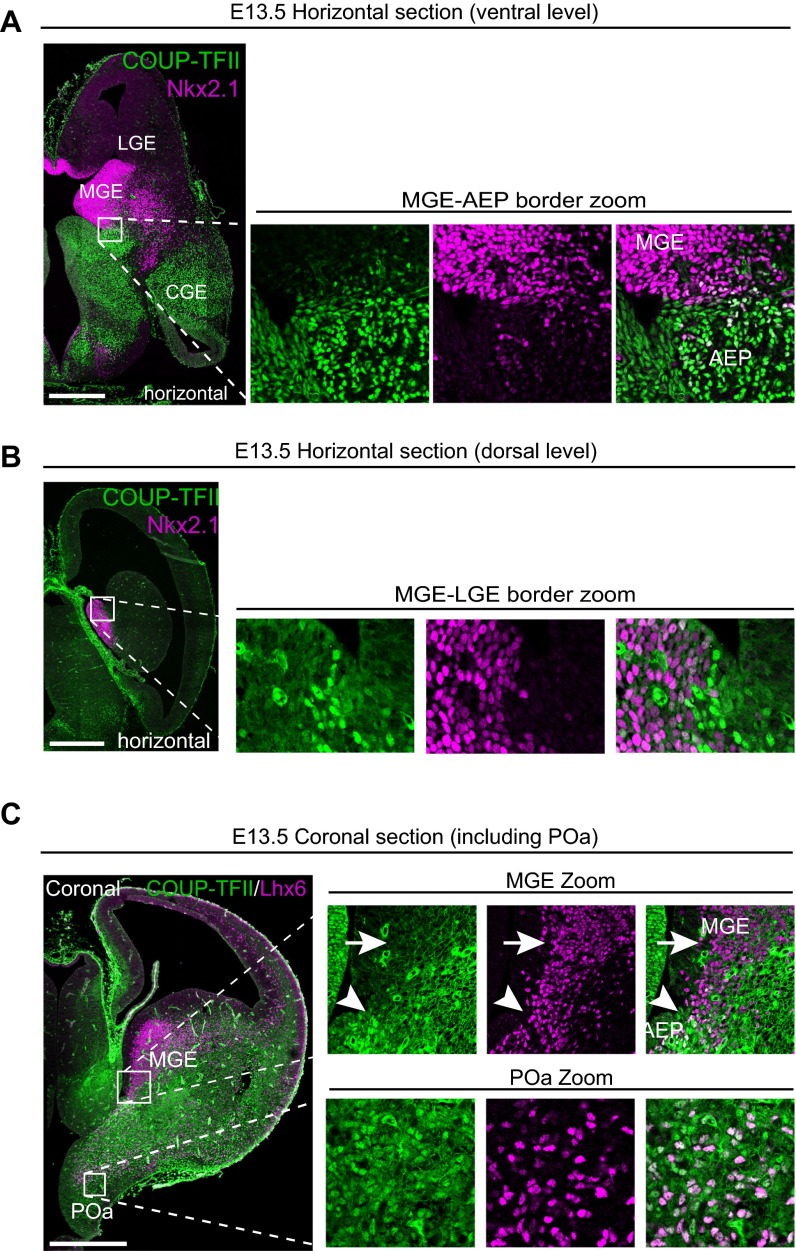
To examine the origin of the Lhx6/COUP-TFII double-positive cells, we performed immunostaining for Nkx2.1, a transcription factor regulating the generation of Lhx6-expressing GABAergic neurons (2, 6, 13, 14). The expression of Nkx2.1 and COUP-TFII overlapped in the ventricular zone of the POa at E13.5 (Fig. 1E) but not in the GE (Fig. S1 A and B). Nkx2.1 expression in the AEP was weak and not uniform, suggesting that the AEP contributes very little, if at all, to the Lhx6/COUP-TFII–positive population (Fig. S1A).
Fig. S1.
MGE and CGE markers are coexpressed in the POa, but not in the GE. (A and B) Immunohistochemical staining for COUP-TFII (green) and Nkx2.1 (magenta) in an E13.5 horizontal section at the ventral (A) and dorsal level (B). Higher magnification views of the caudal region of the MGE in the boxed areas show that COUP-TFII and Nkx2.1 expression is mutually segregated in the GE at both the ventral and dorsal level. (C) Immunohistochemical staining for COUP-TFII (green) and Lhx6 (magenta) in an E13.5 coronal section, including the POa. Whole section image and higher magnification views of the boxed areas are shown. Lhx6-expressing cells in the MGE do not express COUP-TFII (arrows in MGE zoom). Arrowhead shows the border of the MGE region and Lhx6-expressing cells at the more ventral level expressing COUP-TFII. Multiple images were tiled and combined to show the whole brain slice. [Scale bars, (A–C) 500 µm.]
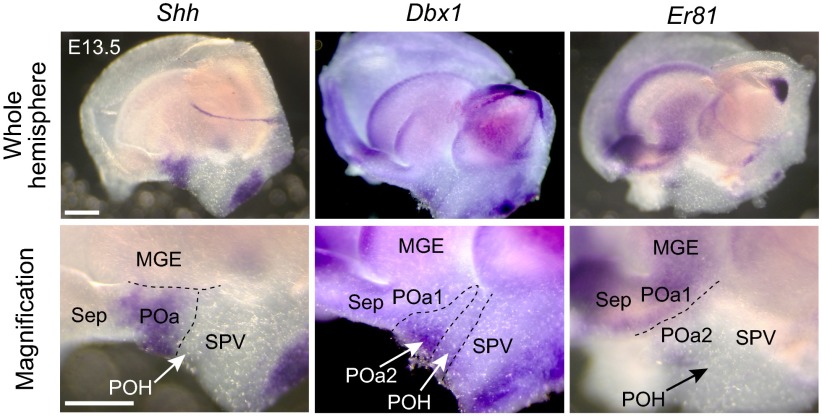
To study the expression of COUP-TFII and Nkx2.1 in the POa, we then carried out whole mount in situ hybridization at E13.5 using FANTOM clones (15). The POa and its subdomains [the dorsal POa (POa1) and ventro-caudal POa (POa2)] were characterized by expression of Shh (POa1 and 2), Er81 (POa1), and Dbx1 (POa2), respectively (Fig. 1F and Fig. S2), as previously shown (16). COUP-TFII was detected in the ventro-caudal side of the POa (POa2), a domain in which Nkx2.1 was also detected (Fig. 1F). Most cells in the POa expressed both COUP-TFII and Lhx6 (Fig. S1C), suggesting that the POa and the caudally migrating cells in the AEP share molecular properties with both the MGE and the CGE cells.
Fig. S2.
Whole mount identification of the expression patterns of regional markers in the POa. Whole mount in situ hybridization of the regional markers Shh, Dbx1, and Er81 at E13.5 with higher magnification views in the POa (Bottom). LGE, lateral ganglionic eminence; POH, the preoptic–hypothalamic border region; Sep, septum; SPV, supraoptic paraventricular region; TH, thalamus. The image of each coronal section was prepared by tiling and combining multiple images to show the whole brain slice. (Scale bars, 500 µm.)
POa-Derived Cells Show a Novel Two-Step Migratory Profile.
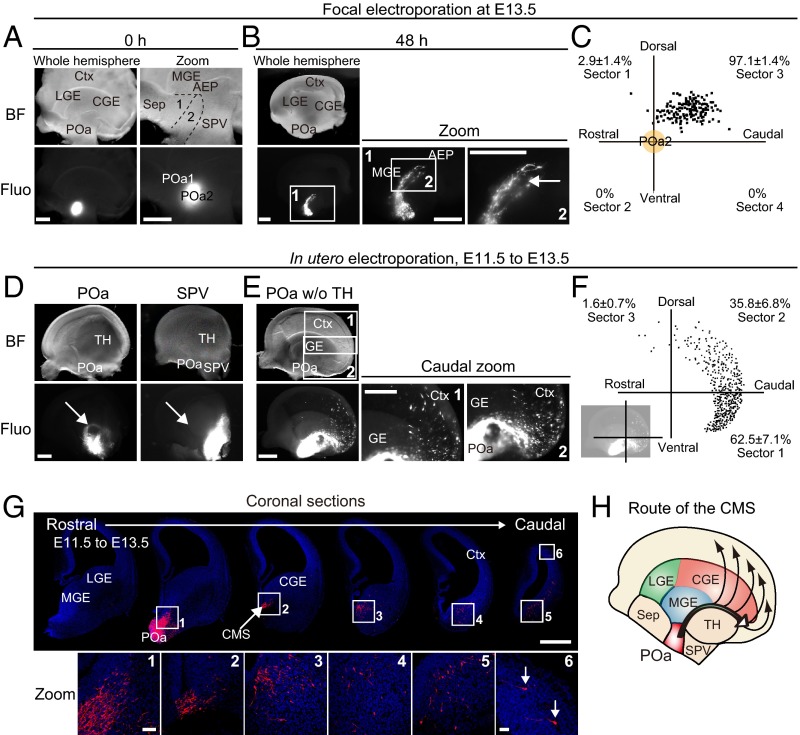
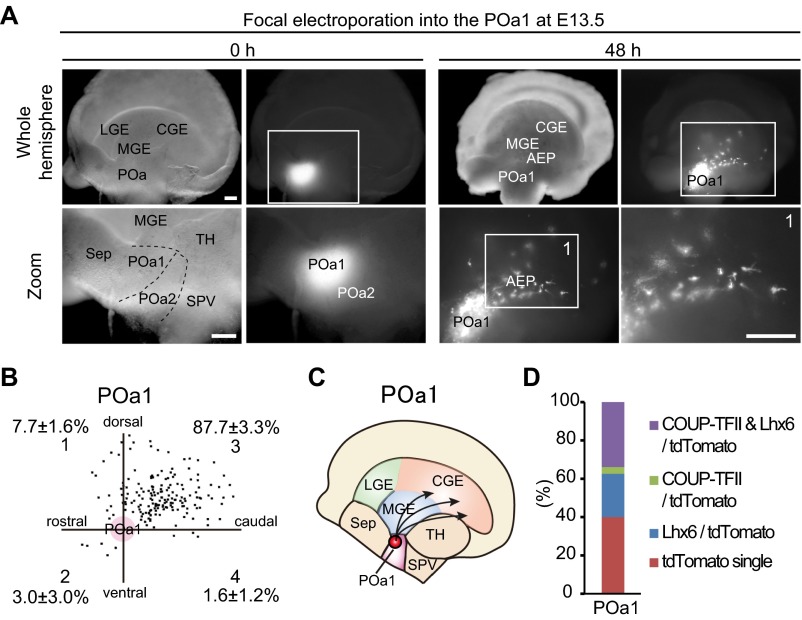
To examine whether the POa2 is the origin of the caudally migrating cells observed in the AEP, focal electroporation was applied on a whole mount hemisphere culture system (7) (Fig. 2 A and B). A CAG-tdTomato vector was electroporated into the POa2 at E13.5 with Fast Green or fluorescein isothiocyanate (FITC)-oligonucleotide DNA as a fluorescent marker (Fig. 2A). After 48 h, virtually all of the POa2-derived cells (97.1 ± 1.4%, n = 148 and n = 5) had migrated into the AEP (Fig. 2C), where they formed a bundle-like stream (Fig. 2B, arrow). The migratory profile of the dorsal POa (POa1) cells tended to migrate caudally and were scattered in the MGE and the CGE (Fig. S3). Most POa1-derived cells did not form a bundle-like stream, suggesting that the POa2 is the major source of the caudally migrating cell population in the AEP.
Fig. 2.
POa-derived cells migrate to the dorsal cortex in a two-step manner. (A and B) The CAG-tdTomato vector was focally electroporated into the POa2 at E13.5 with FITC-oligo DNA as a fluorescent marker or Fast Green, and the hemisphere was cultured for 48 h. Bright-field and fluorescence micrographs show the site of electroporation specifically labeled by FITC-oligo DNA in the POa2 (A). A sharp and thin stream originating from the POa2 was observed 48 h later (arrow in B). (C) The distribution of POa2-derived cells that migrate in the AEP. The percentages shown in each sector (mean ± SEM) were quantified (n = 148 cells, five brain hemispheres). (D) Targeted in utero electroporation into the POa or SPV. Representative micrographs of an E13.5 brain electroporated into the POa or SPV at E11.5. Median views of E13.5 hemispheres show that a clearly delineated migratory stream originated from the POa, not SPV (arrows). (E) Median views of dissected E13.5 hemispheres electroporated into the POa after removal of the thalamus and medial cortex. A clear migratory stream from the POa (CMS) and subsequent scattered cell migration were observed (caudal zoom 1 and 2). (F) Representative distributions of POa-derived cells at E13.5, electroporated at E11.5. The percentages shown in each sector (mean ± SEM) were quantified (n = 696 cells, seven brain hemispheres). Representation of cell quantification is shown in the bottom left. (G) A series of coronal sections of an E13.5 hemisphere electroporated at E11.5. The image of each section was prepared by tiling and combining multiple images to show the whole brain slice. Magnified views in each section show migrating cells from the POa to the caudal cortex (arrows in zoom 6). (H) Schematic diagram of the migratory route of the CMS from the POa. BF, bright field; Ctx, cortex; Fluo, fluorescence; LGE, lateral ganglionic eminence; Sep, septum; SPV, supraoptic paraventricular region; TH, thalamus. [Scale bars, (A, B, D, E, and G) 500 µm and (G zoom) 50 µm.]
Fig. S3.
The migratory profiles of POa1-derived cells differ from those of POa2-derived cells. (A) Focal electroporation was performed into the POa1 of an E13.5 brain hemisphere and cultured for 48 h. Micrographs of the telencephalic hemisphere and of the site of electroporation, indicated by FITC-oligo DNA, are shown. POa1-derived cells tend to migrate caudally into the GE, but their migratory profile is more scattered than that of POa2-derived cells. (B) The distribution of POa1 cells. POa1 cells from five brains and 179 cells were dotted in the four sectors. The percentages shown in each sector are mean ± SEM. (C) Schematic diagram of the migratory profile of cells from the POa1. (D) Total proportions of cells expressing COUP-TFII and/or Lhx6. The percentage of cells expressing COUP-TFII is lower in the POa1 than in the POa2. [Scale bars, (A) 250 µm.]
Examining whether caudally migrating cells in the AEP and CGE at E13.5 derive from the POa in vivo, performing in utero electroporation at E11.5 and fixation at 13.5 revealed a narrow migratory stream from the POa (Fig. 2D). In utero labeling of the supraoptic paraventricular region (SPV), which is adjacent to the POa, failed to detect the caudal migratory stream (CMS) (Fig. 2D). After removing the thalamus and the medial cortex, where the narrow stream was not observed, from the electroporated hemispheres, a larger fraction of labeled cells was found to migrate to the ventral CGE, anatomically corresponding to the medial amygdala, and the caudal cortex (Fig. 2E). The shape of the POa-derived stream was narrow until it reached the middle of the CGE and widened in the caudal cortex. Coronal sections taken at E13.5 (Fig. 2G) revealed that most of the migrating POa-derived cells initially formed a bundle-like stream in the AEP and then migrated to the ventral CGE. Thereafter, they gradually spread within the CGE and the ventro-caudal cortex and migrated into the dorsal cortex. The first part of this two-step migratory profile, in which the POa-derived cells caudally migrated through the AEP and the ventral CGE, should be included in the CMS, as previously reported for the CGE cells (3, 7). Because the AEP and the CGE are located caudally to the POa, direct labeling of the CGE cells in our previous study must have also visualized the POa-derived caudally migrating cells in the CGE. Therefore, the CMS contains POa-derived cells migrating through the ventral CGE (Fig. 2H), in addition to the cells born in the CGE. The two-step migration from the POa is distinct from the single tangential migratory profile in the ventral telencephalon. These results support our hypothesis that the POa domain within the diencephalon provides the most significant source of caudally migrating cells forming the CMS.
The CMS Consists of POa-Derived GABAergic Neurons with Dynamic COUP-TFII Expression.
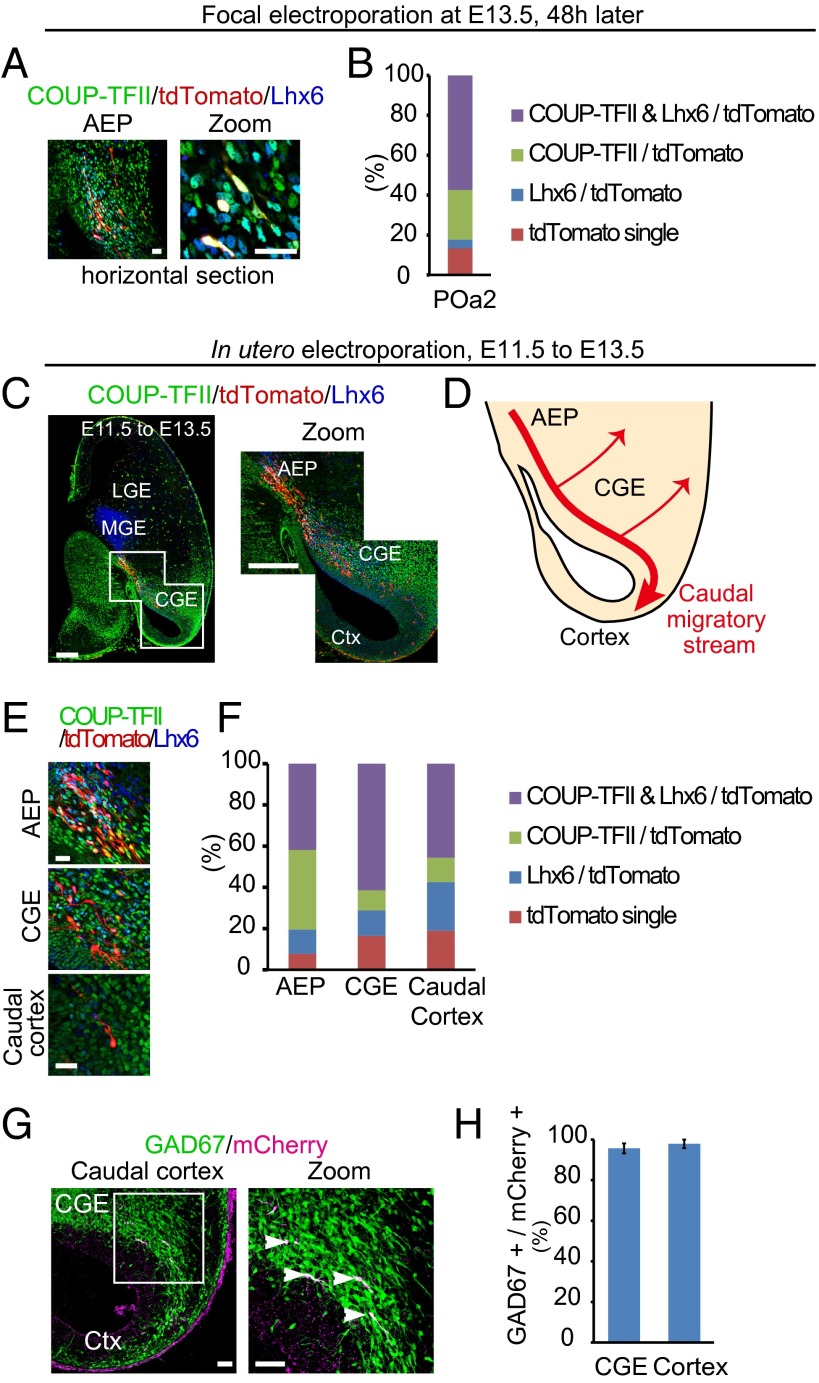
To further examine the molecular properties of caudally migrating cells from the POa, cultured hemispheres electroporated into the POa2 were horizontally sectioned and immunostained for COUP-TFII and Lhx6 (Fig. 3A). Quantification showed that 82.3 ± 2.8% (n = 280 and n = 6) of the POa2-derived cells migrating in the stream bundle also expressed high levels of COUP-TFII (Fig. 3B). Immunohistochemical analysis showed that only 37.4 ± 9.4% (n = 97 and n = 5) of the POa1-derived cells expressed COUP-TFII (Fig. S3D), supporting the hypothesis that the POa2 region is the major source of COUP-TFII–positive migrating cells in the CMS.
Fig. 3.
The majority of caudally migrating POa-derived cells express COUP-TFII in the CMS. (A) Immunostaining of the cultured hemisphere electroporated into the POa2 at E13.5 for COUP-TFII (green) and Lhx6 (blue). Low magnification of the micrograph of the AEP and higher magnification show the POa2 cells migrating in the CMS. (B) The majority of POa2 cells (tdTomato positive) in the CMS express COUP-TFII (82.3 ± 2.8%, n = 280 and n = 6). (C) Immunohistochemical staining for COUP-TFII (green) and Lhx6 (blue) in E13.5 brains electroporated at E11.5 (horizontal section). Whole image of the representative horizontal section (left), which was prepared by tiling and combining multiple images to show the whole brain slice, and a magnified micrograph of the boxed CGE region (right). POa-derived cells (red) migrate along the CMS. (D) Schematic diagram of the migratory route of the CMS from the POa in the horizontal section. (E) Higher magnifications of the AEP, CGE, and caudal cortex in E13.5 brains electroporated at E11.5. (F) Quantification analysis shows that most POa-derived cells express COUP-TFII in the CMS, AEP (80.5 ± 3.4%, n = 328, five brains), CGE (71.2 ± 1.4%, n = 391, five brains), and caudal cortex (57.4 ± 2.5%, n = 247, five brains). (G) Micrographs of the E13.5 caudal cortex of GAD67-GFP mice electroporated in the POa at E11.5. All migrating POa-derived cells express GFP (arrowheads in magnified boxed area). (H) Quantification analysis shows that the majority of POa-derived cells are GABAergic neurons (CGE, 95.7 ± 2.5%, n = 239 cells; caudal cortex, 97.9 ± 2.1%, n = 67 cells, three brains). Ctx, cortex. [Scale bars, (A) 25 µm, (C) 250 µm, (E) 25 µm, and (G) 100 µm.]
The temporal and spatial expression of COUP-TFII in the CMS was then studied in vivo using the electroporated brain at E11.5. In the horizontal section at E13.5, POa-derived cells formed the caudally migrating stream identical with the one from the AEP observed as a Lhx6-positive stream (Figs. 1 A and B and 3 C and D). In caudally migrating Lhx6-positive cells, the expression of COUP-TFII gradually decreased as the cells reached the cortex (Fig. 1 C and D). The same tendency of the COUP-TFII expression levels to decline was observed in E13.5 POa-derived cells electroporated at E11.5 (Fig. 3 E and F), further supporting the hypothesis that the POa is a source of the CMS.
To examine whether caudally migrating POa-derived cells were differentiating into GABAergic neurons, we used GAD67-GFP mice (Fig. 3G) (17). Electroporation into the POa of GAD67-GFP mice with mCherry at E11.5 showed that the majority of cells migrating in the CGE and the caudal cortex represent GABAergic neurons (CGE, 95.7 ± 2.5%, n = 239 and n = 3; caudal cortex, 97.9 ± 2.1%, n = 67 and n = 3) (Fig. 3H). These results suggest that the CMS is a stream of GABAergic neurons dynamically changing expression of COUP-TFII.
A Dynamic Expression of COUP-TFII Regulates Both the Caudal Migration and the Distribution of POa-Derived Cells in the Cortex.
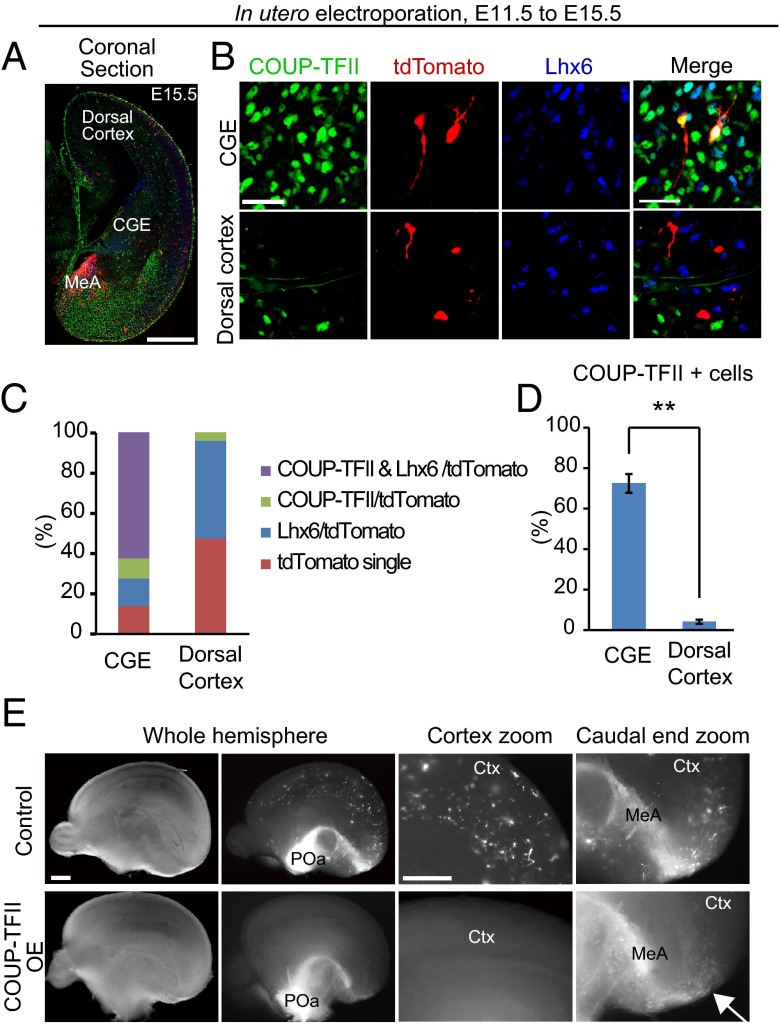
To examine COUP-TFII expression at the later developmental stages, we performed immunohistochemistry for COUP-TFII and Lhx6 using E15.5 brains that had been electroporated at E11.5 (Fig. 4 A and B). Similar to E13.5, the majority of electroporated cells (72.4 ± 4.6%, n = 338 and n = 5) in the ventral CGE expressed COUP-TFII at E15.5 (Fig. 4 B and C). Interestingly, only a few of the electroporated cells (4.1 ± 1.0%, n = 208 and n = 8) that had reached the dorsal cortex expressed COUP-TFII (Fig. 4 B–D). By contrast, approximately half (48.5 ± 3.6%, n = 208 and n = 8) of the POa-derived cells in the dorsal cortex expressed Lhx6 (Fig. 4 B and C). These results suggest that COUP-TFII is down-regulated in POa-derived cells before they enter the dorsal cortex.
Fig. 4.
Down-regulation of COUP-TFII expression is required for POa-derived cells to distribute in the dorsal cortex. (A and B) Immunohistochemical staining for COUP-TFII (green) and Lhx6 (blue) in an E15.5 brain electroporated at E11.5. (A) Whole image of the coronal section, including the CGE region. Multiple images were tiled and combined to show the whole brain slice. (B) High magnification of the CGE and dorsal cortex in the coronal section. Most POa cells (red) in the CGE express COUP-TFII (Upper); however, virtually no POa cells express COUP-TFII in the dorsal cortex (Bottom). (C) Quantification of COUP-TFII and Lhx6 expression in electroporated POa-derived cells. (D) Statistics test shows that populations of COUP-TFII–expressing cells were significantly decreased in the dorsal cortex (**P < 0.01, P = 0.002; Mann–Whitney rank sum test). (E) In utero electroporation of COUP-TFII overexpression in the POa at E11.5 and observed at E15.5. Medial views of the control (Upper) and COUP-TFII–overexpressing (Bottom) E15.5 hemisphere electroporated at E11.5. Bright-field (whole hemisphere) and fluorescence micrographs (whole hemisphere and zoom) are shown. POa-derived cells in the control are distributed within the entire cortex, whereas when COUP-TFII is OE, POa-derived cells accumulate around the medial amygdala and the caudal end of the hemisphere (arrow), and no cells are observed in the dorsal cortex (n = 4). Ctx, cortex; MeA, medial amygdala. [Scale bars, (A) 500 µm, (B) 30 µm, and (E) 500 µm.]
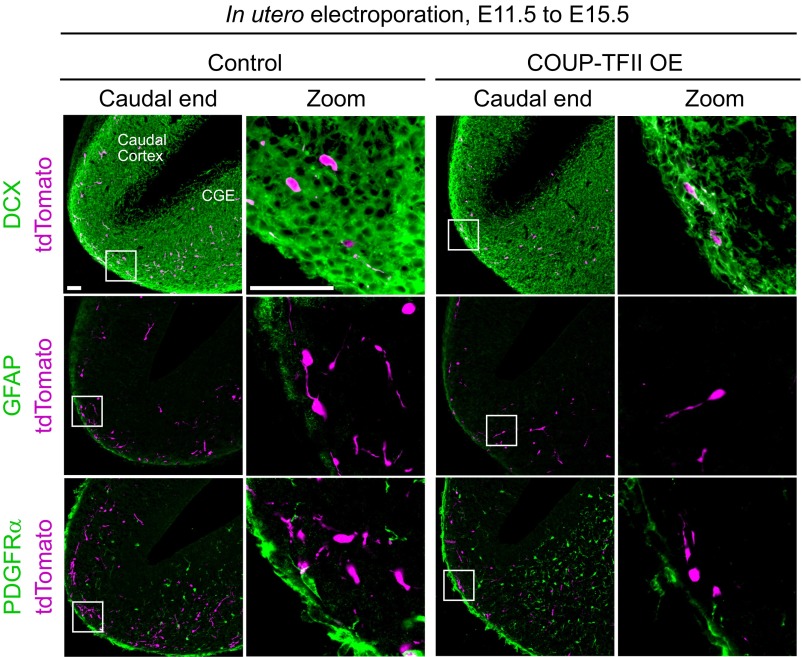
In utero electroporation overexpressing COUP-TFII at E11.5 was then used to investigate the migratory profile of POa-derived cells at E15.5 (Fig. 4E) and E13.5 (Fig. 5). In control embryos at E15.5, a large number of POa-derived cells was observed in the dorsal cortex and was distributed diffusely throughout the entire cortex (Fig. 4E). By contrast, E15.5 embryos (n = 4) with overexpressed (OE) COUP-TFII showed an accumulation of POa-derived cells in the medial amygdala and the caudal end of the cortex (Fig. 4E, arrow), whereas no cells had entered the dorsal cortex (Fig. 4E, Cortex zoom). When brains were analyzed at E13.5, a majority of the POa-derived cells overexpressing COUP-TFII had accumulated in the medial amygdala and the caudal end of the cortex (Fig. 5 A and B, COUP-TFII OE). COUP-TFII overexpression was confirmed as not affecting neuronal cell fate by immunostaining for a neuronal marker, doublecortin (DCX), and glial cell markers, glial fibrillary acidic protein (GFAP) and alpha-type platelet-derived growth factor receptor (PDGFRα) (Fig. S4). These results demonstrate that a lower expression level of COUP-TFII is required for the POa-derived cells to enter the dorsal cortex.
Fig. 5.
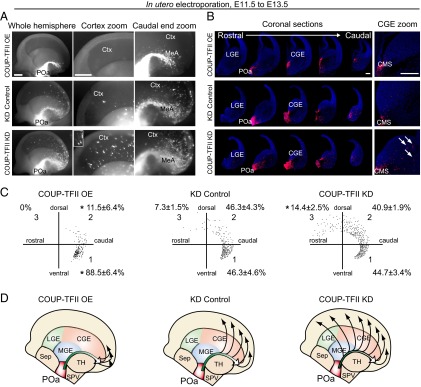
COUP-TFII inhibits detachment of POa-derived cells from the CMS. (A) Representative micrographs of an E13.5 brain electroporated into the POa at E11.5 with CAG-COUP-TFII (Upper), shRNA-control (Middle), or shRNA-COUP-TFII (Bottom). Median views and higher magnification of the hemisphere in each electroporated condition are shown. The thalamus and medial cortex were removed. The Inset shows a magnified image of representative cells. (B) A series of coronal sections of each electroporated hemisphere. The image of each section was prepared by tiling and combining multiple images to show the whole brain slice. Higher magnification of the CGE area (Right) shows that POa-derived cells overexpressing COUP-TFII localize in the CMS. By contrast more POa-derived cells migrate away from the CMS following knockdown of COUP-TFII (arrows) than following control knockdown. (C) Representative distributions of POa-derived cells for each of the electroporated conditions. The distribution of ∼200 cells in the cortex was dotted in the three sectors. Percentages correspond to the following: CAG-COUP-TFII, six brains, 173 cells; shRNA-control, 10 brains, 2,161 cells; shRNA COUP-TFII, six brains, 1,707 cells. The asterisk indicates a significant difference from the control [*P < 0.05; CAG-COUP-TFII, P = 0.022 (sector 1) and P = 0.029 (sector 2); shRNA COUP-TFII, P = 0.025 (sector 3); Tukey–Kramer test]. (D) Schematic diagram of the migratory profile of POa-derived cells in three different COUP-TFII expression conditions. Ctx, cortex; MeA, medial amygdala. [Scale bars, (A) 500 µm and (B) 250 µm.]
Fig. S4.
COUP-TFII overexpression does not affect the neuronal fate of the POa-derived cells. Shown is the immunohistochemical staining for DCX, GFAP, and PDGFRα in the E15.5 caudal cortex to label immature neurons, astroglia, and oligodendrocyte precursors, respectively. The POa-derived cells were labeled with tdTomato at E11.5. The expression levels of these markers are similar in control samples and in samples overexpressing COUP-TFII. (Scale bars, 50 µm.)
POa-derived cells overexpressing COUP-TFII in coronal sections taken at E13.5 showed a dramatically decreased scattered migration to the dorsal cortex and accumulation of cells in the CMS to a narrow bundle (Fig. 5B and Fig. 2G as the control). Quantitative analysis revealed that the percentage of cells in sector 1, which is the caudal region of the hemisphere, was significantly higher when overexpressing COUP-TFII (88.5 ± 6.4%, n = 173 and n = 6) compared with the control (62.5 ± 7.1%, n = 696 and n = 7) (Figs. 2 F and H and 5 C and D).
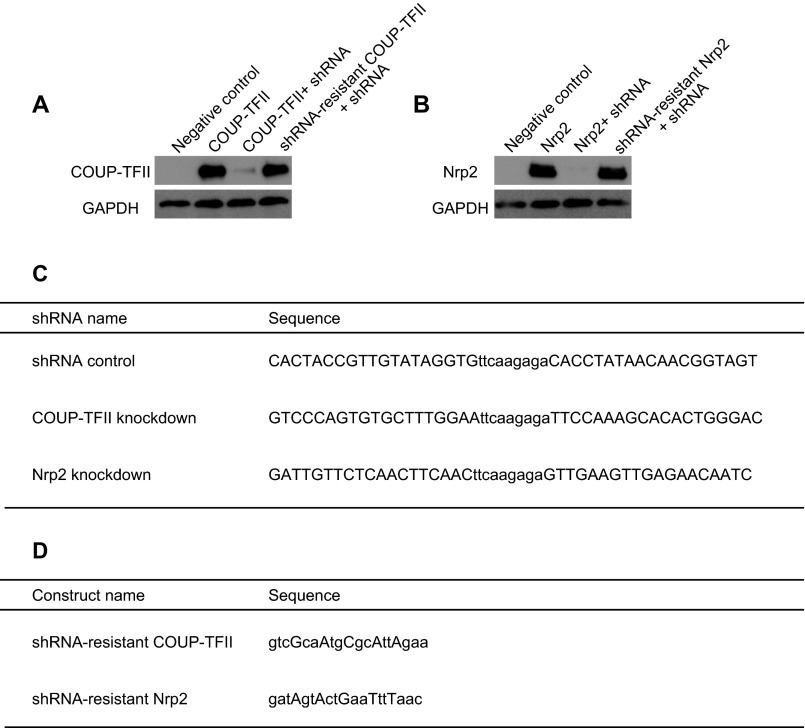
To knock down COUP-TFII in the POa-derived cells, a previously published target sequence for COUP-TFII was used (18). The knockdown efficiency was verified in Neuro2a cells (Fig. S5) and in flow cytometry-sorted CMS cells (Fig. 6 E and F). Cells in which COUP-TFII was depleted were distributed more rostrally in the cortex than those transfected with a scrambled control shRNA vector (Fig. 5, COUP-TFII KD). Coronal sections revealed more detached cells from the CMS, following COUP-TFII knockdown, compared with the control (Fig. 5B, arrows). Quantification showed a significant increase in the percentage of cells in sector 3, which is the rostral region of the hemisphere, when COUP-TFII was knocked down (14.4 ± 2.5%, n = 2,161 and n = 10), compared with the control (7.3 ± 1.5%, n = 1,707 and n = 6) (Fig. 5 C and D). Collectively, these results suggest that COUP-TFII regulates the caudal migration along the CMS and inhibits the detachment of cells from the CMS and their subsequent migration into the dorsal cortex.
Fig. S5.
Characterization of shRNA knockdown efficiency in Neuro2a cells. (A and B) Western blot analyses of COUP-TFII (A) and Nrp2 (B). COUP-TFII shRNA and Nrp2 shRNA efficiently deplete COUP-TFII and Nrp2 protein expression, respectively. The shRNA vectors did not knock down shRNA-resistant COUP-TFII and Nrp2. (C) Target sequences of shRNA for the control, COUP-TFII, and Nrp2 vectors. (D) Sequences of shRNA-resistant COUP-TFII and Nrp2 vectors. Capital letters indicate mutations generated in the original COUP-TFII and Nrp2 sequences.
Fig. 6.
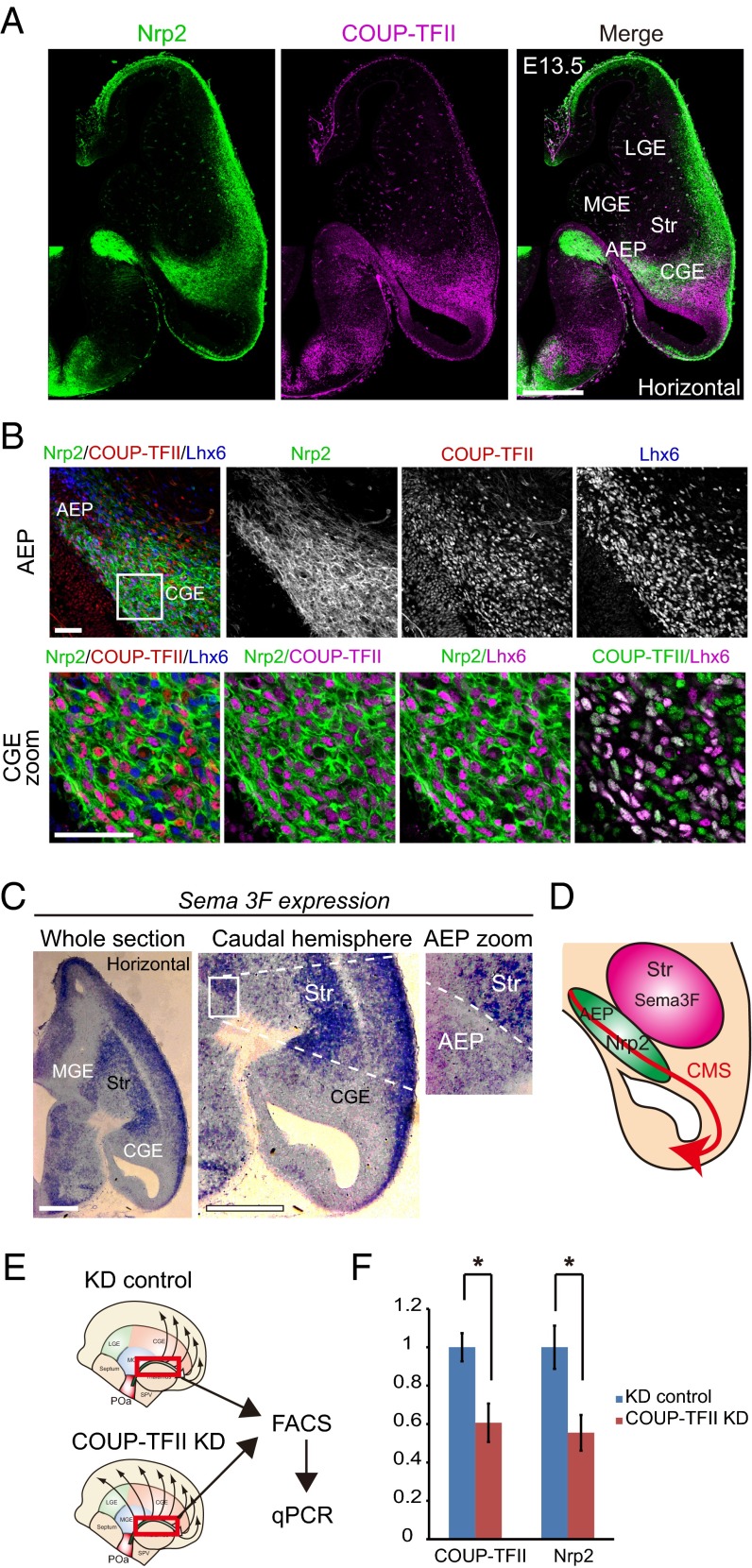
Nrp2 is preferentially expressed in the AEP and the CGE and is regulated by COUP-TFII. (A) Immunohistochemical staining for Nrp2-tauGFP (green), COUP-TFII (magenta) and Lhx6 (blue) in an E13.5 horizontal section of a heterozygous Nrp2-Δ mouse brain (+/−). Multiple images were tiled and combined to show the whole brain slice. Whole section micrographs of Nrp2 and COUP-TFII show that Nrp2 is preferentially expressed in the AEP and CGE. (B) Magnified views of the region of Nrp2/COUP-TFII/Lhx6 expression in the AEP and the CGE. Virtually all COUP-TFII–expressing cells in the CGE also express Nrp2. (C) In situ hybridization of Sema3F in an E13.5 horizontal section. Sema3F is highly expressed in the striatum region (Str) and weakly expressed in the CMS. A magnified image shows the Sema3F expression contrast between the AEP and striatum. (D) Schematic diagram of Nrp2 and Sema3F expression in the CMS. (E) Schematic representation of the experimental design for real-time PCR. POa-derived cells migrating into the CMS (red squared) were dissected from the electroporated brain, sorted by FACS, and analyzed by quantitative PCR (qPCR). (F) qPCR results following COUP-TFII knockdown. Transfection of shRNA-COUP-TFII significantly reduced the endogenous expression level of COUP-TFII and Nrp2 mRNA in migrating POa-derived cells (COUP-TFII, *P = 0.021; Nrp2, *P = 0.016; two-tailed Student’s t test). Str, striatum. [Scale bars, (A) 500 µm, (B) 50 µm, and (C) 500 µm.]
Nrp2 Is Preferentially Expressed in the CMS and Functions as a Downstream Target of COUP-TFII.
Our previous study shows that COUP-TFII overexpression induces the CMS in MGE cells and that COUP-TFII knockdown inhibits the CMS in CGE cells (3). This COUP-TFII function in the GE is analogous to that of the POa-derived cells, suggesting the possibility of a common molecular target(s) of COUP-TFII, which may be involved in CMS induction in the GE cells and the POa. Thus, to search for downstream target(s) of COUP-TFII that may be regulating the direction of the caudal migration from the POa to the dorsal cortex, we compared the transcriptome of MGE cells overexpressing COUP-TFII with that of CGE cells depleted of COUP-TFII. The screening of molecules that were up-regulated in the CMS-induced cells, and down-regulated in the CMS-inhibited cells, revealed Nrp2 as a candidate. Interestingly, analysis of the heterozygous Nrp2-Δ mouse brain (+/−), in which tauGFP was knocked in into the Nrp2 locus (19), revealed that the pattern of Nrp2 expression was similar to that of COUP-TFII (Fig. 6). Nrp2 was strongly expressed in the AEP and the CGE, with its caudally migrating COUP-TFII–expressing cells, and more weakly expressed in the caudal cortex (Fig. 6 A and B). Sema3F is a ligand for Nrp2 and works as a repulsive cue to cortical interneurons (20). In situ hybridization on E13.5 horizontal sections demonstrated a strong expression of Sema3F in the striatum, as previously reported (20, 21), and a faint expression along the route of the CMS (Fig. 6C). Intriguingly, Sema3F was markedly higher in the striatum than in the AEP (Fig. 6C, AEP zoom) and suggests that Sema3F acts as a beacon for POa-derived cells (Fig. 6D).
It is well established that in lymphatic vessels, COUP-TFII binds the promoter region of Nrp2 via SP1 and induces Nrp2 expression (22). The same regulation process of the Nrp2 expression by COUP-TFII has recently been reported in the developing murine forebrain at E12.5 (23). To investigate whether Nrp2 expression is also regulated by COUP-TFII in the CMS, real-time PCR on fluorescence-activated cell sorted (FACS) POa-derived cells was performed. Knocking down of COUP-TFII significantly reduced Nrp2 mRNA expression to 55.5 ± 9.3% (shRNA-control, n = 4; COUP-TFII shRNA, n = 6) (Fig. 6 E and F). These results indicate that COUP-TFII regulates Nrp2 expression in the CMS.
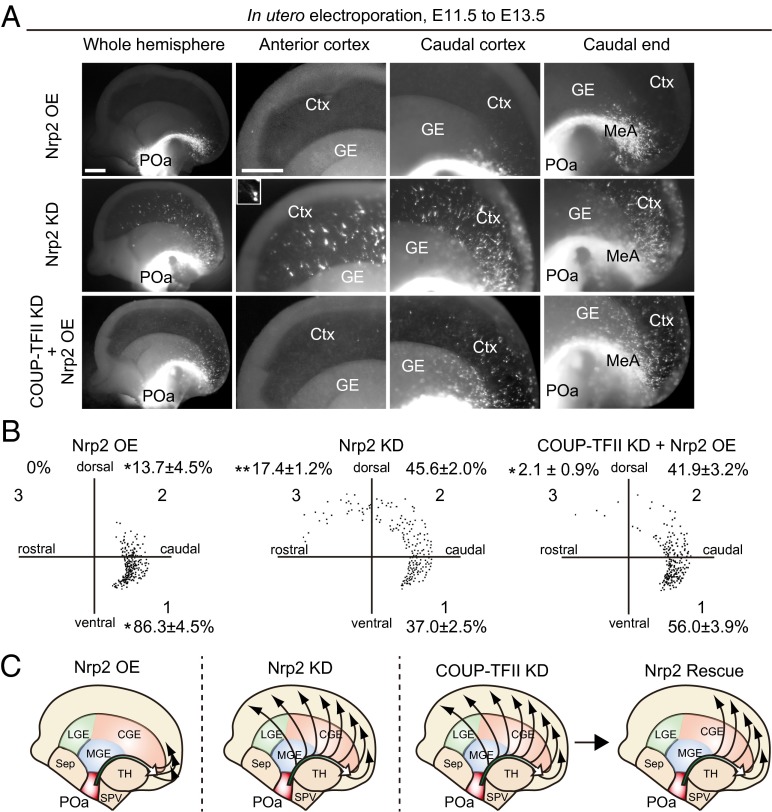
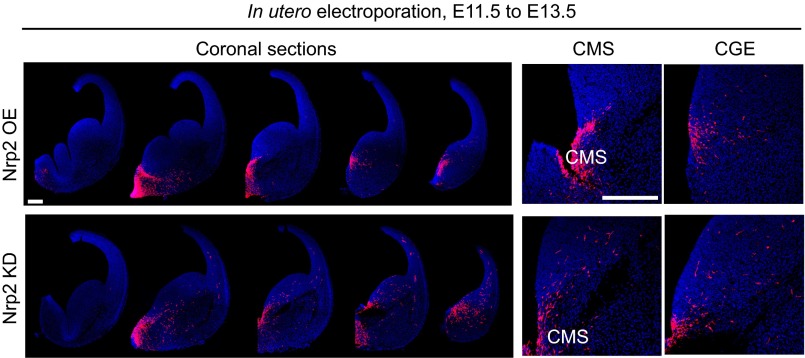
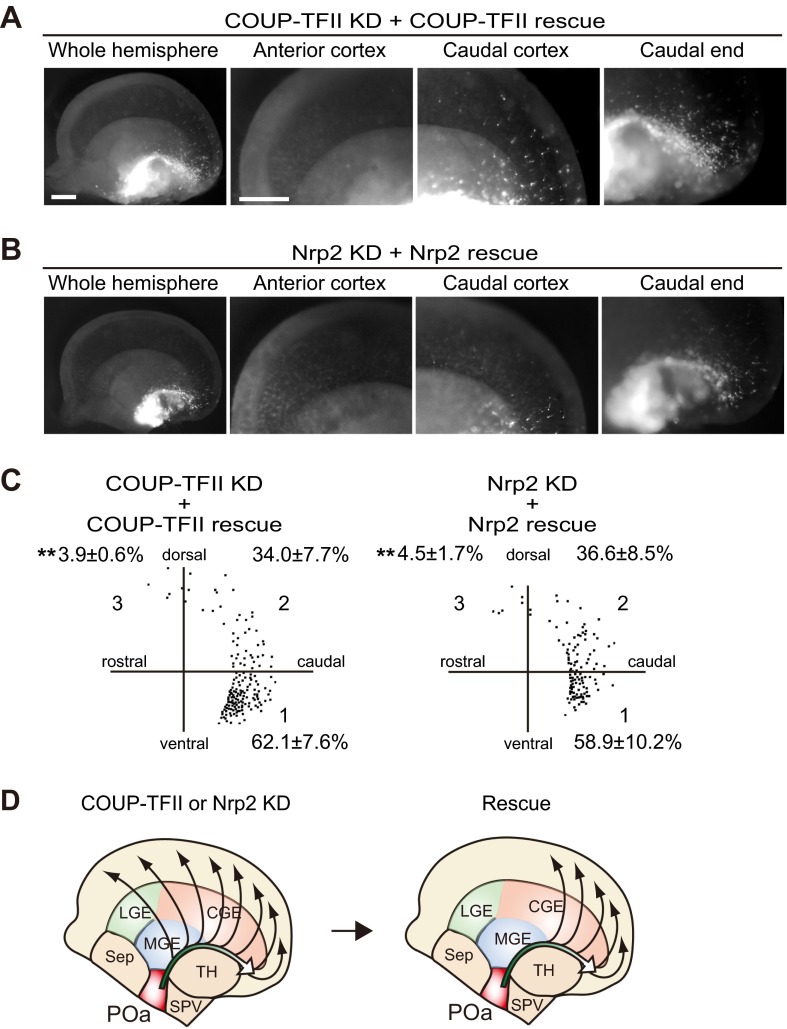
To examine the specific role of Nrp2 in regulating the migration of POa-derived cells, Nrp2 was OE or knocked down at E11.5. The efficiency of the Nrp2 knockdown was confirmed in Neuro2a cells (Fig. S5). Overexpression or knocking down of Nrp2 had similar effects on the migration profile as overexpressing or knocking down COUP-TFII, respectively (Fig. 5A, COUP-TFII OE and COUP-TFII KD and Fig. 7A, Nrp2 OE and Nrp2 KD). When Nrp2 was OE, cells were distributed in the narrow stream of the CMS and the medial amygdala (Fig. 7 A–C, Nrp2 OE), whereas depletion caused cells to migrate more rostrally and diffusely in the cortex (Fig. 7 A–C, Nrp2 KD). Thus, detachment of cells from the CMS and migration into the dorsal cortex was promoted and inhibited by Nrp2 knockdown and overexpression, respectively, similar to the observed effects of COUP-TFII knockdown and overexpression (Fig. S6). Interestingly, overexpressing Nrp2 significantly rescued the abnormal migration into the rostral cortex of cells in which COUP-TFII was knocked down (n = 938 and n = 6) (Fig. 7). The knockdown effects of COUP-TFII or Nrp2 were significantly rescued by coexpression of the shRNA-resistant mutant of COUP-TFII or Nrp2 (Fig. S7).
Fig. 7.
Nrp2 regulates detachment of migrating POa-derived cells from the CMS, downstream of COUP-TFII. (A) Nrp2 overexpression, Nrp2 knockdown, or rescue of COUP-TFII knockdown by Nrp2 overexpression into the POa, using in utero electroporation at E11.5, followed by observation at E13.5. Shown are representative micrographs of E13.5 brains electroporated with CAG-tdTomato together with CAG-Nrp2 (Nrp2 OE) or shRNA Nrp2 (Nrp2 KD) or shRNA COUP-TFII and CAG-Nrp2 (COUP-TFII KD + Nrp2 OE). The Inset shows a magnified image of representative cells. (B) Representative distributions of POa-derived cells following Nrp2 OE, KD, and COUP-TFII KD rescued by Nrp2 OE. The distribution of ∼200 cells in the cortex was dotted in the three sectors. Percentages correspond to the following: Nrp2 OE, seven brains, 402 cells; Nrp2 KD, six brains, 1,445 cells; COUP-TFII + Nrp2 OE, six brains, 938 cells. Asterisks indicate a significant difference from the control [*P < 0.05, **P < 0.01; CAG-MCS2 control (Fig. 2 D and E) vs. CAG-Nrp2, P = 0.03 (sector 1) and P = 0.04 (sector 2); KD control vs. Nrp2 KD, P = 0.00059 (sector 3); COUP-TFII KD vs. COUP-TFII KD + Nrp2 OE, P = 0.00016 (sector 3), Tukey–Kramer test]. (C) Schematic diagram of POa-derived cell migration following Nrp2 overexpression, knockdown, and rescue of COUP-TFII knockdown by Nrp2 overexpression. The migratory profile of POa-derived cells in COUP-TFII–depleted brains was significantly rescued following overexpression of Nrp2, at levels comparable to those of the control. Ctx, cortex; GE, ganglionic eminence; MeA, medial amygdala. [Scale bar, (A) 500 µm.]
Fig. S6.
Detachment of POa-derived cells from the CMS and their lateral migration following overexpression or knockdown of Nrp2 is similar to that observed following overexpression or knockdown of COUP-TFII. A coronal section series of E13.5 brains following Nrp2 overexpression (Nrp2 OE) or knockdown (Nrp2 KD) via electroporation at E11.5. Higher magnification of the CMS and CGE area shows that POa-derived cells overexpressing Nrp2 localize in the CMS (Nrp2 OE), and POa-derived cells in which Nrp2 is depleted migrate away from the CMS (Nrp2 KD). (Scale bars, 250 µm.)
Fig. S7.
Migration defects caused by knockdown of COUP-TFII or Nrp2 in POa-derived cells are rescued by shRNA-resistant COUP-TFII or Nrp2 expression, respectively. (A and B) Representative micrographs of E13.5 brains electroporated with CAG-tdTomato together with shRNA COUP-TFII and CAG-shRNA–resistant COUP-TFII (COUP-TFII KD + COUP-TFII rescue) or shRNA Nrp2 and shRNA-resistant Nrp2 (Nrp2 KD + Nrp2 rescue). (C) Representative distributions of POa-derived cells under COUP-TFII or Nrp2 KD conditions rescued by the shRNA-resistant constructs. Percentages are based on the following number of samples: COUP-TFII KD + COUP-TFII, four brains, 371 cells; Nrp2 KD + Nrp2 rescue, three brains, 313 cells. Asterisks indicate a significant difference from the control [*P < 0.05, **P < 0.01; COUP-TFII KD (Fig. 5) vs. COUP-TFII + COUP-TFII rescue, P = 0.0050 (sector 3); Nrp2 KD (Fig. 7) vs. Nrp2 KD + Nrp2 rescue, P = 0.00042 (sector 3); Tukey–Kramer test]. (D) Schematic diagrams of COUP-TFII or Nrp2 knockdowns and their rescue by shRNA-resistant COUP-TFII or Nrp2 overexpression. [Scale bars, (A and B) 500 µm.]
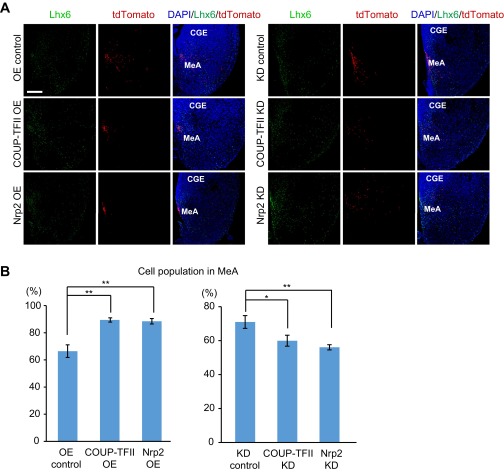
Finally, we analyzed whether the expression level of COUP-TFII or Nrp2 affected the distribution of POa-derived cells in the medial amygdala on coronal sections. Because no specific marker for the medial amygdala at E13.5 exists, we used Lhx6 as the reference, because Lhx6 was reported as a regional marker for the dorsal part of the adult medial amygdala (24, 25). As expected from the medial view of the transfected hemispheres, we found that more POa-derived cells populated the medial amygdala when COUP-TFII (n = 128 and n = 6) or Nrp2 (n = 184 and n = 6) was OE, compared with the control (n = 272 and n = 6) (Fig. S8). In contrast, when COUP-TFII (n = 275 and n = 6) or Nrp2 (n = 416 and n = 6) was knocked down, fewer cells were detected in the medial amygdala, compared with the control (n = 383 and n = 6) (Fig. S8).
Fig. S8.
Expression levels of COUP-TFII and Nrp2 affect the distribution of POa-derived cells into the medial amygdala. (A) Cell distribution of the POa-derived cells electroporated at E11.5 was examined in coronal sections of the medial amygdala at E13.5. Immunohistochemistry for Lhx6 was performed to visualize the dorsal part of the medial amygdala. When COUP-TFII or Nrp2 was OE, the POa-derived cells labeled with tdTomato at E11.5 accumulated in the medial part of the region with Lhx6-expressing cells. In contrast, the POa-derived cells sparsely distributed when COUP-TFII or Nrp2 was knocked down (KD). (B) Proportions of tdTomato-positive POa-derived cells in medial amygdala among all tdTomato-positive cells on each coronal section were calculated. Asterisks indicate a significant difference between the control and the OE or KD samples (*P < 0.05, **P < 0.01; COUP-TFII OE, P = 0.0002; Nrp2 OE, P = 0.0003; COUP-TFII KD, P = 0.036; Nrp2 KD, P = 0.006, six brains each, Dunnett’s test). CGE, caudal ganglionic eminence; MeA, medial amygdala. (Scale bars, 100 µm.)
Taken together, these results demonstrate that a precise regulation of the levels of COUP-TFII and Nrp2 controls the migratory profile of POa-derived GABAergic neurons.
Discussion
GABAergic neurons born in the forebrain migrate long distances to populate different areas of the brain, including the neocortex, the striatum, and the amygdala (26). To reach all these distant destinations, migrating neurons need to navigate through the different structures of the brain (27, 28). This study demonstrates a unique migratory profile of POa-derived GABAergic neurons and that dynamic COUP-TFII and Nrp2 expression functions as a molecular switch directing the cells toward either the amygdala or the neocortex in the developing mouse brain. Previously, our group reported a caudal migration profile of the CGE cells regulated by COUP-TFII and named it the CMS (7). The current study shows that the CMS contains POa-derived cells that migrate through the ventral CGE. Although it remains unclear to what extent each population contributes to the CMS, our results indicate that the CMS is a large migratory stream covering the diencephalon and the telencephalon and includes both POa-derived and CGE-derived cells.
The Migratory Profile of POa-Derived Neurons.
The migration of the POa-derived cells is poorly understood, although recently this population has been revealed to be the source of GABAergic neurons in the neocortex and the amygdala (8–10). The present findings show that the POa-derived cells that were migrating to the neocortex and the amygdala initially shared the migratory stream and then took separate paths controlled by the dynamic expression of COUP-TFII and Nrp2. A previous study by Soma et al. shows a sharp, bundle-like migratory stream from the third ventricle; however, it was reported without characterizing the origin of this stream (29). Our present findings, which are based on focal electroporation into specific regions of the hypothalamus, demonstrate that the bundle-like migratory stream originates mainly from the POa2 domain. This profile of migration appears different from the well-characterized tangential migration of MGE-derived cortical interneurons. Rather, this migration profile is yet another example of atypical tangential migration such as the rostral migratory stream (30) and the interneuron migration from ventral telencephalon to the thalamus in human (31). These results were confirmed by in utero electroporation, which revealed a clearly delineated migratory stream from the POa. The findings of the present study suggest that the CMS in the AEP originates predominantly from the POa domain and not from other hypothalamic regions of the diencephalon.
Using Dbx1-Cre mice, Hirata et al. demonstrate that amygdala GABAergic neurons derived from the POa form a migratory stream called the POa amygdala migratory stream (10). The results presented here demonstrate the presence of a POa-derived CMS that supplies GABAergic neurons to the caudal telencephalon, including at least the medial amygdala and neocortex. Thus, it is likely that the POa-derived CMS includes the POa amygdala migratory stream. In another study using Dbx1-Cre mice, Dbx1-expressing cells were found in the intermediate zone of the caudal cortex (32). These observations are consistent with the results presented here. Gelman et al. reported that one-third of the cells from the Dbx1-expressing domain (POa2) express Lhx6 (9). Our results demonstrate that 48.5% of the POa-derived cells that reached the dorsal cortex expressed Lhx6. This finding verifies previous studies and supports the possibility that POa2 is the principal source of the CMS in the AEP.
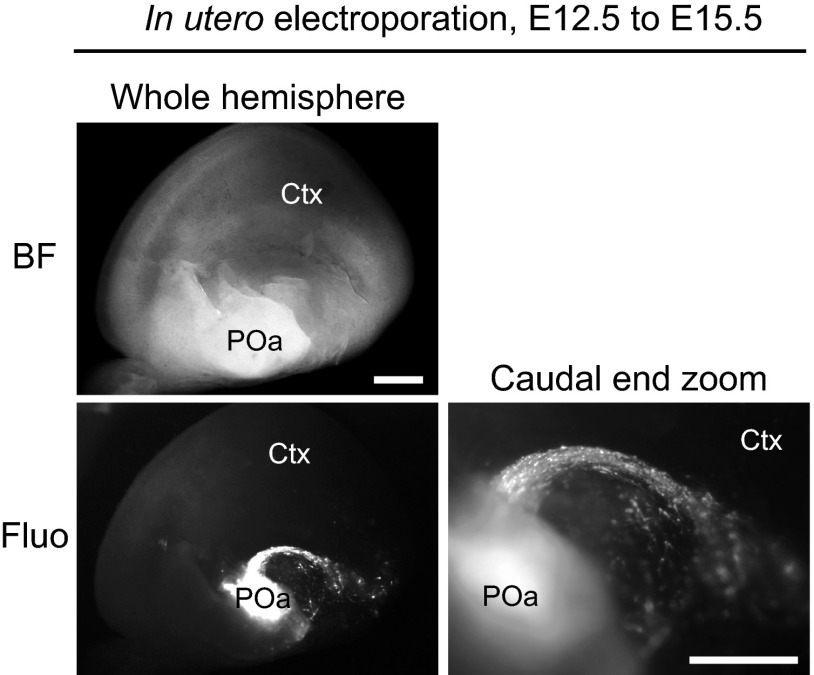
In this study, we extensively analyzed brains at E13.5 because detailed expression analysis data exist at this stage (16), and we found the CMS at E13.5. Hirata et al. analyzed birthdates of migrating neurons from the POa to the amygdala and found that POa-derived cells labeled by tamoxifen injection from E9.5 to E11.5 populated the medial amygdala in Dbx-1 CreERT2 mice with a peak at E10.5 and E11.5 (10). Our data showed that electroporation at E12.5 to the POa also gave rise to the CMS from the POa (Fig. S9). Thus, the POa-derived migrating cells in the CMS are likely to be generated over several days during development, including E13.5.
Fig. S9.
The CMS from the POa observed in the E15.5 brain electroporated at E12.5. In utero electroporation with CAG-tdTomato was performed into the POa at E12.5, and the brain was observed at E15.5. BF, bright field; Ctx, cortex; Fluo, fluorescence. (Scale bars, 500 µm.)
COUP-TFII Expression in the POa and in POa-Derived GABAergic Neurons.
The present study demonstrates that the dynamic expression of COUP-TFII functioned as a molecular switch for the POa-derived GABAergic neurons to change their migratory direction from caudal to dorsal. Although most POa-derived cells expressed COUP-TFII at E13.5, the results presented here suggest that almost none of the POa-derived neurons that had migrated to the dorsal cortex expressed COUP-TFII at E15.5. POa-derived cells in the caudal cortex expressed COUP-TFII at E13.5; therefore, we postulate that COUP-TFII is down-regulated by E15.5. However, it is also possible that a few POa-derived cells that do not express COUP-TFII are somehow able to migrate to the dorsal cortex. The present study indicates that COUP-TFII caused and/or maintained the caudal migration of POa-derived cells. In support of this hypothesis, a recent study demonstrated hypoplasia or complete loss of the medial amygdala in COUP-TFII conditional knockout mice (23), suggesting that COUP-TFII expression is necessary for caudal migration of POa-derived GABAergic neurons in the formation of the medial amygdala. By contrast, cortical GABAergic neurons derived from the COUP-TFII–expressing domain were reported to remain unaltered in COUP-TFII conditional knockout (RxCre;COUP-TFF/F) mice (23). However, given the results that (i) COUP-TFII expression in this conditional knockout mouse was weakly observed at E12.5 in the ventral telencephalon (23) and (ii) COUP-TFII expression was down-regulated in the POa-derived cortical GABAergic neurons, weak expression of COUP-TFII in POa-derived cells at early stages might be sufficient for their ultimate distribution in the cortex. Further studies are required to draw a final conclusion about the requirement of COUP-TFII for the migration of cortical interneurons.
In human GE, COUP-TFII is expressed in a gradient manner with a small increase in the CGE at gestational week 15 (33, 34). In the human cortex, it is shown that the COUP-TFII expression is abundant in the caudal cortex and moderate in the rostral cortex at gestational weeks 15–22 (33), which are reminiscent of the expression patterns in the mouse cortex at E13.5 shown in this study. Unfortunately, the COUP-TFII expression in the POa has not yet been examined in the developing human brain; however, a large population in the caudal MGE has been reported to coexpress COUP-TFII and Nkx2.1 (34), similar to the POa2 in the mouse brain shown in the present study. Indeed, it would be worth investigating whether the CMS is present in the developing human brain using double immunohistochemistry for COUP-TFII and Lhx6.
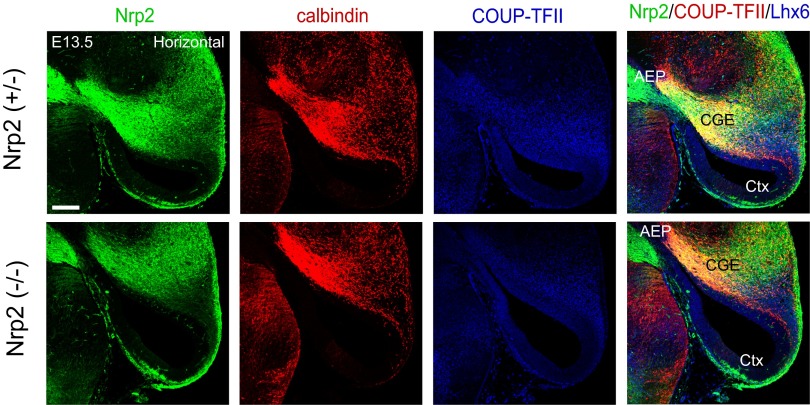
In our analysis, Nrp2-Δ mouse brain (−/−) failed to show significant aberrant migration, as assessed by GFP and calbindin immunohistochemistry at E13.5 (Fig. S10). However, it has been reported recently that Nrp1 expression is also directly regulated by COUP-TFII (23). We confirmed that Nrp1 has a similar function with that of Nrp2 in inhibiting the detachment of the POa-derived cells from the CMS and their accumulation in the amygdala (Fig. S11). This functional and expression compensation of Nrp1 could explain why the Nrp2-Δ mouse brain (−/−) did not show obvious disruption of the CMS.
Fig. S10.
Migration analysis of Nrp2 knockout mice. Immunohistochemical staining for Nrp2-tauGFP (green), calbindin (red), and COUP-TFII (blue) in an E13.5 horizontal section of a heterozygous (+/−) or homozygous (−/−) Nrp2-Δ mouse brain. No significant difference in migration was observed. Ctx, cortex; GE, ganglionic eminence. (Scale bars, 200 µm.)
Fig. S11.
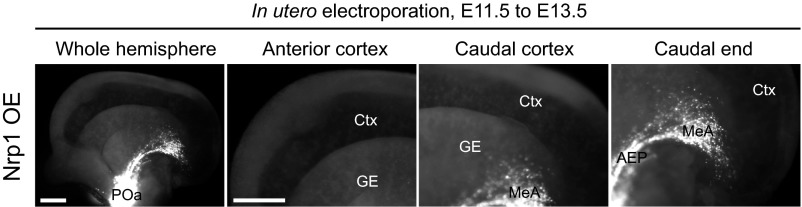
Nrp1 overexpression shows almost the identical effect with the Nrp2 overexpression in the POa-derived cells. In utero electroporation of Nrp1 overexpression in the POa at E11.5 and observed at E13.5 is shown. Representative micrographs of E13.5 brains electroporated with CAG-tdTomato together with CAG-Nrp1 are shown. Ctx, cortex; GE, ganglionic eminence; MeA, medial amygdala. (Scale bars, 500 µm.)
Our finding that COUP-TFII expression in the POa-derived cells is sustained in the CGE and the medial amygdala, but not in the dorsal cortex, raises the possibility that the expression dynamics of COUP-TFII and Nrp2 are related to the final destination of the POa-derived cells. Because the amygdala is highly related to autism spectrum disorders in humans (35) and the human NRP2 gene is associated with the clinical expression of autism in Chinese populations (36), the present study may provide a plausible link between Nrp2 and the development of the amygdala. Because COUP-TFII expression was also observed in other areas of the amygdala nucleus—for example, the basolateral complex, which was severely affected in COUP-TFII KO mice—the COUP-TFII/Nrp2 pathway may also regulate the caudal migration of neurons destined for other amygdala regions. Further studies of the formation of the CMS will contribute to our understanding of the development of the amygdala and the cerebral cortex, and their possible relation to autism.
Materials and Methods
Animals.
Pregnant ICR (Institute of Cancer Research) mice were purchased from Japan SLC, Inc. All animal experiments in Japan were performed under the control of the Keio University Institutional Animal Care and Use Committee in accordance with Institutional Guidelines on Animal Experimentation at Keio University. In Sweden, ethical approval was granted by the Stockholm’s Ethical Committee North (D.no N486/12). The day on which a vaginal plug was detected was recorded as E0.5. For the analysis of Nrp2 expression and migration analysis, we used heterozygous (+/−) and homozygous (−/−) Nrp2-Δ mice generated by crossing ubiquitous Cre-expressing mice with mice with an NP2-flox-neo mutant allele, including tauGFP, from the Jackson Laboratory (19) (STOCK Nrp2tm1.1Mom/MomJ, 006697).
Plasmid Vectors.
CAG–COUP-TFII derived from a FANTOM (functional annotation of mammalian genome) cDNA clone (clone ID B230309B05) was used as described previously (3, 15, 37). Nrp1 and Nrp2 cDNA was cloned into a CAG-GS vector (37) to generate the CAG-Nrp2 expression vector. tdTomato cDNA was isolated from the ptdTomato cDNA plasmid (Clontech, cat. no. 632531) and inserted into the pCAG-MCS2 plasmid to generate the CAG-tdTomato (38) expression vector. The shRNA-resistant expression vectors for COUP-TFII and Nrp2 (CAG–COUP-TFII resistant, CAG-Nrp2 resistant) were generated by a standard mutagenesis PCR method. The target sequences of shRNA-COUP-TFII and shRNA-Nrp2 and the sequence of shRNA-resistant COUP-TFII and Nrp2 are described in Fig. S5. The target sequence of COUP-TFII is identical to the sequence used in previous studies (18). A FITC-labeled oligonucleotide (oligo; 5′-FITC-TACGTACGTACGTA-3′) was used as a marker for the site of electroporation as described previously (7).
Focal Electroporation into the Telencephalic Hemisphere.
Focal electroporation and hemisphere culture were performed as reported previously (3, 7). Briefly, telencephalic hemispheres from E13.5 ICR mouse embryos were dissected and placed in PBS. The tdTomato expression vector containing a CAG promoter (37) was injected into each region of the POa1 and POa2. To monitor the injection site, a Fast Green solution (0.1%; Sigma Aldrich) was added to the plasmid solution at a ratio of 1:10, as reported previously (39). Approximately 0.1 µL of the plasmid solution (5 µg/µL) was injected into the POa with a glass micropipette. The telencephalic hemispheres were placed between a platinum plate electrode and a tungsten needle electrode. Electronic pulses (100 V, 5 ms) were discharged four times at 5 ms apart with an electroporator (CUY21E; NepaGene), and this series of four electroporations was repeated three times for each hemisphere.
Whole Telencephalic Hemisphere Culture.
Electroporated E13.5 telencephalic hemispheres were cultured for 48 h as previously described (3). Brains were rotated in 2 mL DMEM nutrient mixture/Ham’s F-12 (Sigma) containing the N-2 supplement (Invitrogen) under a continuous gas flow (95% O2 and 5% CO2 at 37 °C). Images of the whole mount telencephalic hemispheres were acquired with a cooled color CCD camera (VB-7010; KEYENSE).
In Situ Hybridization.
Whole mount and section in situ hybridization were performed as reported previously (3, 40). Digoxigenin (DIG)-labeled antisense RNA probes were synthesized with DIG-UTPs (Roche Diagnostics), using PCR products that had been amplified from the FANTOM clones (40, 41). The FANTOM cDNA clones were established by the Genome Exploration Research Group, RIKEN Genomics Science Center, using full-length technologies described elsewhere (15), and the clones were replicated and provided by K.K. Dnaform (Kanagawa, Japan). The clones used for in situ hybridization were as follows: COUP-TFII, B230309B05; Nkx2.1, 8430401I11; Er81, 9630017N10; Dbx1, 6230409C10; and Shh, 5730522J07. The Sema 3F probe was transcribed from a Sema3F cDNA fragment used in the previous work by Ito et al. (21).
Immunohistochemistry.
Immunohistochemical analyses were performed as described previously (7, 39). The primary antibodies were anti–COUP-TFII (mouse monoclonal, 1:100; PP-H7147-00, Perseus Proteomics Inc.), anti-Lhx6 [rabbit polyclonal, a gift from V. Pachnis, Medical Research Council National Institute for Medical Research, London, or rabbit polyclonal (H-75), 1:50, sc-98607, Santa Cruz Biotechnology], anti-DCX (rabbit polyclonal, 1:500, ab18723, Abcam), anti-GFAP (rabbit polyclonal, 1:1,000, Z0334, Dako), anti-PDGFRα [rabbit polyclonal (C-20), 1:100, sc-338, Santa Cruz Biotechnology], anti-calbindin D28k (rabbit polyclonal, 1:1,000, CB-38a, Swant), and anti-GFP (chicken polyclonal, 1:1,000, ab13970, Abcam). Embryonic mice brains were dissected out in ice-cold Dulbecco’s PBS and fixed in 0.1 M sodium phosphate buffer, pH 7.4, containing 4% (wt/vol) paraformaldehyde overnight. For immunostaining, horizontal sections (14 µm thick) were prepared with a cryostat (CM1900, CM3050S; Leica Microsytems) and incubated overnight with the primary antibodies. After washing, the sections were incubated with the appropriate species-specific secondary antibodies, as follows: donkey DyLight 488-, 549-, or 649-conjugated anti-mouse IgG, anti-rabbit IgG, or anti-chicken IgY (Jackson ImmunoResearch Laboratories). Images were acquired with a confocal microscope (FV300, FV1000; Olympus) and analyzed using Photoshop CS5 software (Adobe).
Targeted in Utero Electroporation into the POa.
The method of targeted in utero electroporation has been described previously (39, 42). Briefly, CAG-tdTomato alone (2.5 µg/µL) or CAG-tdTomato (5.0 µg/µL) mixed with CAG–COUP-TFII (5.0 µg/µL), shRNA–COUP-TFII (5.0 µg/µL), or shRNA-Nrp2 (5.0 µg/µL) at a ratio of 1:1 was used. Plasmids in a solution of 0.01% Fast Green were injected into the third ventricle around the POa, and electronic pulses (35 V, 50 ms, five times at E11.5; 40 V, 50 ms, five times at E12.5) were applied using an electroporator (CUY21; Nepa Gene), with a forceps-type electrode (CUY650P3). For rescue experiments, the Nrp2 expression vector (0.5 µg/µL) was added (the ratio of CAG-tdTomato:shRNA vector:CAG-expression vector was 4:5:1). Embryos were allowed to live within the uterine horn until the desired time of observation. The success rate of specifically labeling to the POa was approximately one embryo per two pregnant mice. Great care was paid for selecting the brains only labeled in the POa, not the GE area including the AEP. Images of the whole mount telencephalic hemispheres were acquired with a cooled color CCD camera (VB-7010; KEYENSE).
Verification of shRNA Efficiencies.
To confirm the knockdown efficiency, shRNA–COUP-TFII or shRNA-Nrp2 vectors were cotransfected with the COUP-TFII or HA-tagged Nrp2 expression vectors, which contain the CAG promoter, into the Neuro2a cell line using the GeneJuice Transfection reagent (EMD Millipore). To validate the expression of the shRNA-resistant COUP-TFII and Nrp2 vectors, the CAG-COUP-TFII–resistant or CAG-Nrp2–resistant HA vector was cotransfected with the shRNA–COUP-TFII or shRNA-Nrp2 vector. After 2 d, cells were lysed in RIPA buffer (Sigma Aldrich), and the protein lysate was used for SDS/PAGE and Western blot analysis. The anti–COUP-TFII (mouse monoclonal, 1:500; PP-H7147-00, Perseus Proteomics Inc.), anti-HA [mouse monoclonal (clone 16B12), 1:500, MMS-101P, Covance], anti-GAPDH [mouse monoclonal (6C5), 1:1,000, sc-32233, Santa Cruz Biotechnology], and anti-mouse HRP (goat polyclonal, 1:500, P0447, Dako) antibodies were used for Western blotting.
Cell Sorting and Real-Time PCR.
The CAG-EGFP and CAG-tdTomato vectors were coelectroporated into E11.5 embryos, and the major CMS was dissected at E13.5, as shown in Fig. 6. Dissected tissues were processed in 0.05% trypsin-EDTA for 5 min, after which trypsin was inhibited by the addition of FBS. Tissues were dissociated by gentle pipetting. Cells expressing EGFP were FACS-sorted by EPICS ALTRA (Beckman Coulter). Sorted cells were directly lysed with the RNAqueous-Micro Kit (Life Technologies). cDNA was synthesized using the SuperScript VILO cDNA Synthesis Kit (Life Technologies). Quantitative RT-PCR was performed using the ABI PRISM 7500 Fast Real-Time PCR system (Life Technologies). Expression data were normalized to the mean expression levels of 18S rRNA, β-actin, and GAPDH. The following TaqMan gene expression assays were used: Nr2f2 (Mm00772789_m1), Nrp2 (Mm00803099_m1), 18S rRNA (Mm03928990_g1), Actb (Mm00607939_s1), and GAPDH (Mm99999915_g1).
Downstream Analysis Regulated by COUP-TFII.
The central regions of the MGE and CGE were dissected out at E13.5 in Dulbecco’s PBS and dissociated as described in the cell sorting and real-time PCR. Primary MGEs were transfected with control vectors (CAG-EGFP and/or CAG-GS at a ratio of 2:1) or COUP-TFII overexpression vectors (CAG-EGFP and COUP-TFII at a ratio of 2:1) using Amaxa mouse neuron nucleofector kit (Amaxa Biosystems) according to the manufacturer’s instructions. Primary CGE cells were transfected with fluorescent protein and control or COUP-TFII siRNA (Dharmacon, SMARTpool reagent ON-TARGET plus) using the same protocol with the MGE cells. Transfected cells were cultured for 40 h on poly–l-lysine (Sigma)-coated culture dish in DMEM nutrient mixture/Ham’s F-12 (Sigma) containing the N-2 supplement (Invitrogen). Cultured cells were FACS-sorted by EPICS ALTRA (Beckman Coulter), and then their transcriptome was analyzed using Mouse Expression set 430 2.0 chips (Affymetrix) as described previously (3, 40).
Statistical Analysis.
All data are shown as means ± SEM. Normal distribution and equal variance were checked using Shapiro–Wilk’s test and F test of equality of variances. For single comparison, Student’s t test or Mann–Whitney rank sum test were performed. For multiple comparisons, data were evaluated with a one-way ANOVA, followed by a multiple comparison test (Tukey–Kramer or Dunnett’s test). For single comparisons, data were evaluated with a two-tailed Student’s t test.
Acknowledgments
We are grateful to Dr. Jun-ichi Miyazaki for the CAG-GS plasmid and Dr. Vassilis Pachnis for the Lhx6 antibody. This project was supported by the Ministry of Education, Culture, Sports, Science and Technology (MEXT) Strategic Research Program for Brain Sciences (“Understanding of Molecular and Environmental Bases for Brain Health”) and MEXT/Japan Society for the Promotion of Science (JSPS) KAKENHI Grants 22111004, 15H01586, 15H02355, 15K06745, 15K06746, 26430075, 15H01293, and 25640039. It was also partly supported by the JSPS Strategic Young Researcher Overseas Visits Program for Accelerating Brain Circulation, Keio Gijuku Academic Development Funds and Fukuzawa Memorial Fund for the Advancement of Education and Research, SENSHIN Medical Research Foundation, Terumo Life Science Foundation, Takeda Science Foundation, Life Science Foundation of Japan, the Swedish Research Council, The Swedish Brain Foundation, and Wenner-Gren Foundations.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1420701112/-/DCSupplemental.
References
- 1.Marín O. Interneuron dysfunction in psychiatric disorders. Nat Rev Neurosci. 2012;13(2):107–120. doi: 10.1038/nrn3155. [DOI] [PubMed] [Google Scholar]
- 2.Sussel L, Marin O, Kimura S, Rubenstein JL. Loss of Nkx2.1 homeobox gene function results in a ventral to dorsal molecular respecification within the basal telencephalon: Evidence for a transformation of the pallidum into the striatum. Development. 1999;126(15):3359–3370. doi: 10.1242/dev.126.15.3359. [DOI] [PubMed] [Google Scholar]
- 3.Kanatani S, Yozu M, Tabata H, Nakajima K. COUP-TFII is preferentially expressed in the caudal ganglionic eminence and is involved in the caudal migratory stream. J Neurosci. 2008;28(50):13582–13591. doi: 10.1523/JNEUROSCI.2132-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chédotal A, Rijli FM. Transcriptional regulation of tangential neuronal migration in the developing forebrain. Curr Opin Neurobiol. 2009;19(2):139–145. doi: 10.1016/j.conb.2009.04.005. [DOI] [PubMed] [Google Scholar]
- 5.Alifragis P, Liapi A, Parnavelas JG. Lhx6 regulates the migration of cortical interneurons from the ventral telencephalon but does not specify their GABA phenotype. J Neurosci. 2004;24(24):5643–5648. doi: 10.1523/JNEUROSCI.1245-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Grigoriou M, Tucker AS, Sharpe PT, Pachnis V. Expression and regulation of Lhx6 and Lhx7, a novel subfamily of LIM homeodomain encoding genes, suggests a role in mammalian head development. Development. 1998;125(11):2063–2074. doi: 10.1242/dev.125.11.2063. [DOI] [PubMed] [Google Scholar]
- 7.Yozu M, Tabata H, Nakajima K. The caudal migratory stream: A novel migratory stream of interneurons derived from the caudal ganglionic eminence in the developing mouse forebrain. J Neurosci. 2005;25(31):7268–7277. doi: 10.1523/JNEUROSCI.2072-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gelman DM, et al. The embryonic preoptic area is a novel source of cortical GABAergic interneurons. J Neurosci. 2009;29(29):9380–9389. doi: 10.1523/JNEUROSCI.0604-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gelman D, et al. A wide diversity of cortical GABAergic interneurons derives from the embryonic preoptic area. J Neurosci. 2011;31(46):16570–16580. doi: 10.1523/JNEUROSCI.4068-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hirata T, et al. Identification of distinct telencephalic progenitor pools for neuronal diversity in the amygdala. Nat Neurosci. 2009;12(2):141–149. doi: 10.1038/nn.2241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.García-López M, et al. Histogenetic compartments of the mouse centromedial and extended amygdala based on gene expression patterns during development. J Comp Neurol. 2008;506(1):46–74. doi: 10.1002/cne.21524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Legaz I, et al. Development of neurons and fibers containing calcium binding proteins in the pallial amygdala of mouse, with special emphasis on those of the basolateral amygdalar complex. J Comp Neurol. 2005;488(4):492–513. doi: 10.1002/cne.20608. [DOI] [PubMed] [Google Scholar]
- 13.Batista-Brito R, et al. The cell-intrinsic requirement of Sox6 for cortical interneuron development. Neuron. 2009;63(4):466–481. doi: 10.1016/j.neuron.2009.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lavdas AA, Grigoriou M, Pachnis V, Parnavelas JG. The medial ganglionic eminence gives rise to a population of early neurons in the developing cerebral cortex. J Neurosci. 1999;19(18):7881–7888. doi: 10.1523/JNEUROSCI.19-18-07881.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Carninci P, et al. FANTOM Consortium RIKEN Genome Exploration Research Group and Genome Science Group (Genome Network Project Core Group) The transcriptional landscape of the mammalian genome. Science. 2005;309(5740):1559–1563. doi: 10.1126/science.1112014. [DOI] [PubMed] [Google Scholar]
- 16.Flames N, et al. Delineation of multiple subpallial progenitor domains by the combinatorial expression of transcriptional codes. J Neurosci. 2007;27(36):9682–9695. doi: 10.1523/JNEUROSCI.2750-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tamamaki N, et al. Green fluorescent protein expression and colocalization with calretinin, parvalbumin, and somatostatin in the GAD67-GFP knock-in mouse. J Comp Neurol. 2003;467(1):60–79. doi: 10.1002/cne.10905. [DOI] [PubMed] [Google Scholar]
- 18.Naka H, Nakamura S, Shimazaki T, Okano H. Requirement for COUP-TFI and II in the temporal specification of neural stem cells in CNS development. Nat Neurosci. 2008;11(9):1014–1023. doi: 10.1038/nn.2168. [DOI] [PubMed] [Google Scholar]
- 19.Walz A, Rodriguez I, Mombaerts P. Aberrant sensory innervation of the olfactory bulb in neuropilin-2 mutant mice. J Neurosci. 2002;22(10):4025–4035. doi: 10.1523/JNEUROSCI.22-10-04025.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Marín O, Yaron A, Bagri A, Tessier-Lavigne M, Rubenstein JLR. Sorting of striatal and cortical interneurons regulated by semaphorin-neuropilin interactions. Science. 2001;293(5531):872–875. doi: 10.1126/science.1061891. [DOI] [PubMed] [Google Scholar]
- 21.Ito K, et al. Semaphorin 3F confines ventral tangential migration of lateral olfactory tract neurons onto the telencephalon surface. J Neurosci. 2008;28(17):4414–4422. doi: 10.1523/JNEUROSCI.0372-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lin F-J, et al. Direct transcriptional regulation of neuropilin-2 by COUP-TFII modulates multiple steps in murine lymphatic vessel development. J Clin Invest. 2010;120(5):1694–1707. doi: 10.1172/JCI40101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tang K, Rubenstein JLR, Tsai SY, Tsai M-J. COUP-TFII controls amygdala patterning by regulating neuropilin expression. Development. 2012;139(9):1630–1639. doi: 10.1242/dev.075564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Choi GB, et al. Lhx6 delineates a pathway mediating innate reproductive behaviors from the amygdala to the hypothalamus. Neuron. 2005;46(4):647–660. doi: 10.1016/j.neuron.2005.04.011. [DOI] [PubMed] [Google Scholar]
- 25.García-Moreno F, et al. A neuronal migratory pathway crossing from diencephalon to telencephalon populates amygdala nuclei. Nat Neurosci. 2010;13(6):680–689. doi: 10.1038/nn.2556. [DOI] [PubMed] [Google Scholar]
- 26.Marín O, Rubenstein JLR. Cell migration in the forebrain. Annu Rev Neurosci. 2003;26(1):441–483. doi: 10.1146/annurev.neuro.26.041002.131058. [DOI] [PubMed] [Google Scholar]
- 27.Ayala R, Shu T, Tsai L-H. Trekking across the brain: The journey of neuronal migration. Cell. 2007;128(1):29–43. doi: 10.1016/j.cell.2006.12.021. [DOI] [PubMed] [Google Scholar]
- 28.Tanaka DH, Nakajima K. Migratory pathways of GABAergic interneurons when they enter the neocortex. Eur J Neurosci. 2012;35(11):1655–1660. doi: 10.1111/j.1460-9568.2012.08111.x. [DOI] [PubMed] [Google Scholar]
- 29.Soma M, et al. Development of the mouse amygdala as revealed by enhanced green fluorescent protein gene transfer by means of in utero electroporation. J Comp Neurol. 2009;513(1):113–128. doi: 10.1002/cne.21945. [DOI] [PubMed] [Google Scholar]
- 30.Lois C, García-Verdugo J-M, Alvarez-Buylla A. Chain migration of neuronal precursors. Science. 1996;271(5251):978–981. doi: 10.1126/science.271.5251.978. [DOI] [PubMed] [Google Scholar]
- 31.Letinic K, Rakic P. Telencephalic origin of human thalamic GABAergic neurons. Nat Neurosci. 2001;4(9):931–936. doi: 10.1038/nn0901-931. [DOI] [PubMed] [Google Scholar]
- 32.Bielle F, et al. Multiple origins of Cajal-Retzius cells at the borders of the developing pallium. Nat Neurosci. 2005;8(8):1002–1012. doi: 10.1038/nn1511. [DOI] [PubMed] [Google Scholar]
- 33.Reinchisi G, Ijichi K, Glidden N, Jakovcevski I, Zecevic N. COUP-TFII expressing interneurons in human fetal forebrain. Cereb Cortex. 2012;22(12):2820–2830. doi: 10.1093/cercor/bhr359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ma T, et al. Subcortical origins of human and monkey neocortical interneurons. Nat Neurosci. 2013;16(11):1588–1597. doi: 10.1038/nn.3536. [DOI] [PubMed] [Google Scholar]
- 35.Amaral DG, Schumann CM, Nordahl CW. Neuroanatomy of autism. Trends Neurosci. 2008;31(3):137–145. doi: 10.1016/j.tins.2007.12.005. [DOI] [PubMed] [Google Scholar]
- 36.Wu S, et al. Association of the neuropilin-2 (NRP2) gene polymorphisms with autism in Chinese Han population. Am J Med Genet B Neuropsychiatr Genet. 2007;144B(4):492–495. doi: 10.1002/ajmg.b.30495. [DOI] [PubMed] [Google Scholar]
- 37.Niwa H, Yamamura K, Miyazaki J. Efficient selection for high-expression transfectants with a novel eukaryotic vector. Gene. 1991;108(2):193–199. doi: 10.1016/0378-1119(91)90434-d. [DOI] [PubMed] [Google Scholar]
- 38.Kawauchi T, Chihama K, Nishimura YV, Nabeshima Y, Hoshino M. MAP1B phosphorylation is differentially regulated by Cdk5/p35, Cdk5/p25, and JNK. Biochem Biophys Res Commun. 2005;331(1):50–55. doi: 10.1016/j.bbrc.2005.03.132. [DOI] [PubMed] [Google Scholar]
- 39.Tabata H, Nakajima K. Efficient in utero gene transfer system to the developing mouse brain using electroporation: Visualization of neuronal migration in the developing cortex. Neuroscience. 2001;103(4):865–872. doi: 10.1016/s0306-4522(01)00016-1. [DOI] [PubMed] [Google Scholar]
- 40.Tachikawa K, Sasaki S, Maeda T, Nakajima K. Identification of molecules preferentially expressed beneath the marginal zone in the developing cerebral cortex. Neurosci Res. 2008;60(2):135–146. doi: 10.1016/j.neures.2007.10.006. [DOI] [PubMed] [Google Scholar]
- 41.Ajioka I, Maeda T, Nakajima K. Identification of ventricular-side-enriched molecules regulated in a stage-dependent manner during cerebral cortical development. Eur J Neurosci. 2006;23(2):296–308. doi: 10.1111/j.1460-9568.2005.04544.x. [DOI] [PubMed] [Google Scholar]
- 42.Friocourt G, et al. Cell-autonomous roles of ARX in cell proliferation and neuronal migration during corticogenesis. J Neurosci. 2008;28(22):5794–5805. doi: 10.1523/JNEUROSCI.1067-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]