Significance
Understanding olfactory communication in natural vertebrate populations requires knowledge of how genes and the environment influence highly complex individual chemical fingerprints. To understand how relevant information is chemically encoded and may feed into mother–offspring recognition, we therefore generated chemical and genetic data for Antarctic fur seal mother–pup pairs. We show that pups are chemically highly similar to their mothers, reflecting a combination of genetic and environmental influences. We also reveal associations between chemical fingerprints and both genetic quality and relatedness, the former correlating positively with substance diversity and the latter encoded mainly by a small subset of substances. Dissecting apart chemical fingerprints to reveal subsets of potential biological relevance has broad implications for understanding vertebrate chemical communication.
Keywords: chemical communication, mother–offspring recognition, GC-MS, genotype, pinniped
Abstract
Chemical communication underpins virtually all aspects of vertebrate social life, yet remains poorly understood because of its highly complex mechanistic basis. We therefore used chemical fingerprinting of skin swabs and genetic analysis to explore the chemical cues that may underlie mother–offspring recognition in colonially breeding Antarctic fur seals. By sampling mother–offspring pairs from two different colonies, using a variety of statistical approaches and genotyping a large panel of microsatellite loci, we show that colony membership, mother–offspring similarity, heterozygosity, and genetic relatedness are all chemically encoded. Moreover, chemical similarity between mothers and offspring reflects a combination of genetic and environmental influences, the former partly encoded by substances resembling known pheromones. Our findings reveal the diversity of information contained within chemical fingerprints and have implications for understanding mother–offspring communication, kin recognition, and mate choice.
The chemical senses are the evolutionarily oldest and arguably most widespread means of interacting with the outside world. Olfaction in particular is fundamental to animal communication, mediating social interactions as varied as territorial behavior, kin recognition, and mate choice (1). Metabolomic tools, such as gas chromatography–mass spectrometry (GC-MS) have made it possible to generate individual-specific chemical “fingerprints.” By separating compounds and quantifying their relative abundances, these fingerprints provide a wealth of information, even though not all compounds can necessarily be identified. Both volatile and contact cues are potentially hidden within the extreme complexity of chemical profiles, which is why a mechanistic understanding of chemical communication is still lacking in natural vertebrate populations (2).
In particular, “surprisingly little progress” has been made in understanding the link between vertebrate chemical fingerprints and genotype (2). Experimental studies have shown that females of several species are capable of discriminating potential partners based on olfactory cues (3–5). However, very few studies have demonstrated a convincing link between the molecular composition of chemical fingerprints and genetic traits, such as heterozygosity (a measure of genetic quality) and relatedness (6–9). These studies were almost exclusively conducted on a captive population of lemurs, a species known for its conspicuous use of scent marking.
A functional understanding of how genotype is chemically encoded also requires knowledge of how many and which types of substances are involved. This is challenging because, especially in natural populations, an individual’s mixture of surface chemicals is not only the product of its genotype but may also be mediated by hormones, the microbial flora, body condition, and environmental factors (2). Thus, analyses based on overall chemical fingerprints may overlook subtle genetic signatures and make little if any headway toward identifying the specific substances involved. A second less-appreciated problem is that the modest panels of around 10–15 microsatellite loci typical of most studies may be underpowered to detect genetic associations because they provide relatively imprecise estimates of both heterozygosity and relatedness (10, 11).
In arguably the only study to report a convincing link between chemical fingerprints and genotype in a natural vertebrate population, Leclaire et al. (9) used principle component analysis (PCA) to reduce chemical complexity. The authors identified a principle component in kittiwakes that correlated significantly with heterozygosity in both sexes and another that correlated with relatedness, but only in adult males. However, PCA iteratively maximizes the explained variance per component instead of seeking to capture the underlying structure and dimensionality of the data, which makes the resulting components hard to interpret (12). A better approach could be factor analysis (FA), a method from the field of psychology that estimates the latent variable structure of a dataset by dividing the total variability into that common to variables and a residual value unique to each variable (13). Statistical developments that allow FA to be applied to data with more variables than observations (14) have only recently made this approach amenable to studying chemical fingerprints.
Pinnipeds are an important group of marine mammals that provide an unusual opportunity to reveal insights into the basis of chemical communication. Studies of Steller’s sea lions and harbor seals have revealed a large repertoire of functional olfactory receptor genes (15) and remarkably high olfactory sensitivity (16), respectively. Individuals of many pinniped species also have a strong musky smell that has been attributed to secretions of facial sebaceous and apocrine glands (17). These glands are known to hypertrophy during the mating season in at least two species (18), suggesting that olfactory cues may be particularly important during the reproductive phase of the life cycle.
Females of many otariid species breed in dense colonies and alternate lactation ashore with foraging trips at sea, necessitating accurate mechanisms for offspring localization and recognition (19). Although otariids use a combination of geographical, visual, auditory, and olfactory cues to find and recognize their pups (19), olfactory recognition is particularly important because females of many species accept or reject pups based on naso-nasal inspection (20, 21). Furthermore, a recent experiment on Australian sea lions (22) suggests that female pinnipeds are capable of discriminating filial from nonfilial pups using olfaction in the absence of other cues.
Antarctic fur seals (Arctocephalus gazella) provide a highly tractable model system for studying the importance of chemical cues in a free-ranging marine mammal. On Bird Island, South Georgia (Southwest Atlantic), a colony of fur seals has been studied intensively for over two decades (23). In this species, olfaction is known to be important for the close-range recognition of pups (20). However, females also show active mate choice for males who are both heterozygous and unrelated to themselves (24), raising the possibility that chemical cues might be involved not only in mother–offspring recognition, but also in mate choice.
Here, we combined GC-MS fingerprinting of skin swabs and genetic analysis to explore the chemical basis by which Antarctic fur seal mothers may recognize their pups. Because females of this species appear capable of choosing males based on heterozygosity and relatedness, we hypothesized that genotype should be chemically encoded and that this could provide a mechanism by which females could identify their pups. We therefore sampled mother–offspring pairs from two discrete but genetically indistinguishable colonies (see “colony differences” in Results), which in principle allows genetically encoded substances to be disentangled from those influenced by environmental differences between colonies. We also deployed over 40 microsatellite loci to enhance the power to detect associations between chemical fingerprints and genotype. Finally, we used FA together with a variety of nonparametric approaches to explore the structure of the chemical data and to uncover specific subsets of compounds associated with chemical differences between the colonies, mother–offspring similarity, and genetic relatedness.
Results
Chemical and Genetic Data.
Chemical fingerprints and multilocus microsatellite genotypes were obtained for 41 mother–offspring pairs from two breeding colonies at Bird Island, South Georgia (Fig. 1). After removing compounds present in the control sample or only in a single individual, the total number of substances in each individual’s chemical fingerprint averaged 35.9 and did not differ significantly between mothers and offspring (paired t test, t = −0.05, P = 0.96). All of the animals were genotyped at 43 highly polymorphic microsatellite loci, 41 of which did not deviate significantly from Hardy–Weinberg equilibrium (HWE) in either mothers or offspring after table-wide false-discovery rate (FDR) correction, and were therefore retained for subsequent analyses (Table S1). The mother-offspring pairs all had match probabilities of 100% (Table S2).
Fig. 1.
Map of the study area showing the two breeding colonies from which Antarctic fur seal mother–offspring pairs were sampled. The red and blue areas demarcate freshwater beach and the special study beach, respectively.
Table S1.
Details of the 43 microsatellite loci used in this study together with their polymorphism characteristics in 41 mother–offspring pairs
| Locus | Sources | Mix | Ta | Number of alleles | Observed heterozygosity | HWE P value | |
| Mothers | Offspring | ||||||
| Pv9 | (61) | 1 | 53 | 10 | 0.691 | 0.037 | 0.129 |
| Hg6.1 | (61) | 7 | 60 | 13 | 0.888 | 0.691 | 0.991 |
| Hg6.3 | (61) | 1 | 53 | 12 | 0.901 | 0.942 | 0.074 |
| Hg8.10 | (61) | 1 | 53 | 2 | 0.407 | 0.399 | 1.000 |
| PvcA | (62) | 1 | 53 | 7 | 0.802 | 0.998 | 0.412 |
| PvcE | (62) | 2 | 60 | 13 | 0.926 | 0.836 | 0.755 |
| Aa4 | (63) | 4 | 60 | 6 | 0.720 | 0.685 | 0.419 |
| Hg1.3 | (63) | 1 | 53 | 11 | 0.815 | 0.136 | 0.443 |
| Pv11 | (64) | 8 | 60 | 11 | 0.329 | 0.399 | 0.000 |
| OrrFCB2 | (65) | 2 | 60 | 11 | 0.888 | 0.394 | 0.234 |
| OrrFCB7 | (65) | 2 | 60 | 10 | 0.813 | 0.391 | 0.649 |
| M11a | (66) | 4 | 60 | 17 | 0.867 | 0.331 | 0.654 |
| Lc28 | (67) | 4 | 60 | 9 | 0.875 | 0.988 | 0.542 |
| Lw10 | (67) | 2 | 60 | 15 | 0.938 | 0.605 | 0.525 |
| Zcc7t | (68) | 7 | 60 | 13 | 0.896 | 0.568 | 0.492 |
| ZcwCgDh1.8 | (68) | 3 | 60 | 9 | 0.744 | 0.608 | 0.020 |
| ZcwDh3.6 | (68) | 4 | 60 | 4 | 0.234 | 0.590 | 0.392 |
| ZcwCgDh4.7 | (68) | 3 | 60 | 13 | 0.924 | 0.842 | 0.336 |
| ZcwCgDh5.16 | (68) | 2 | 60 | 7 | 0.500 | 0.000 | 0.000 |
| ZcCgDh5.8 | (68) | 6 | 60 | 11 | 0.850 | 0.489 | 0.748 |
| ZcwCgDh7tg | (68) | 3 | 60 | 12 | 0.742 | 0.347 | 0.026 |
| ZcwCgDhB.14 | (68) | 2 | 60 | 6 | 0.747 | 0.287 | 0.587 |
| Zcwb09 | (69) | 6 | 60 | 12 | 0.864 | 0.652 | 0.933 |
| Zcwc03 | (69) | 6 | 60 | 11 | 0.813 | 0.124 | 0.544 |
| Zcwc11 | (69) | 6 | 60 | 14 | 0.875 | 0.100 | 0.678 |
| Zcwd02 | (69) | 3 | 60 | 13 | 0.878 | 0.346 | 0.596 |
| Zcwe03 | (69) | 7 | 60 | 9 | 0.838 | 0.602 | 0.919 |
| Ssl301 | (70) | 3 | 60 | 14 | 0.901 | 0.473 | 0.910 |
| Zcwa05 | (71) | 5 | 60 | 14 | 0.896 | 0.645 | 0.999 |
| Zcwb07 | (71) | 1 | 53 | 11 | 0.914 | 0.465 | 0.093 |
| Zcwc01 | (71) | 2 | 60 | 11 | 0.823 | 0.345 | 0.511 |
| Zcwe04 | (71) | 8 | 60 | 12 | 0.864 | 0.132 | 0.362 |
| Zcwe12 | (71) | 8 | 60 | 8 | 0.768 | 0.595 | 0.881 |
| Zcwf07 | (71) | 4 | 60 | 9 | 0.802 | 0.194 | 0.834 |
| Ag1 | (72) | 3 | 60 | 10 | 0.813 | 0.706 | 0.733 |
| Ag2 | (72) | 2 | 60 | 7 | 0.854 | 0.847 | 0.428 |
| Ag3 | (72) | 2 | 60 | 2 | 0.420 | 0.454 | 0.311 |
| Agaz2 | (73) | 1 | 53 | 8 | 0.802 | 0.215 | 0.394 |
| Agaz3 | (73) | 2 | 60 | 5 | 0.629 | 0.896 | 0.288 |
| Agaz5 | (73) | 2 | 60 | 3 | 0.469 | 0.641 | 0.174 |
| Agaz6 | (73) | 2 | 60 | 4 | 0.765 | 0.750 | 0.156 |
| Agaz10 | (73) | 2 | 60 | 11 | 0.767 | 0.572 | 0.493 |
| Zcwe05 | 3 | 60 | 9 | 0.866 | 0.999 | 0.564 | |
HWE P values are shown separately for mothers and offspring, with significant values highlighted in bold. Values that remained significant following table-wide correction for the FDR are underlined. Loci Pv11 and ZcwCgDh5.16 were excluded from further analyses because they deviated significantly from HWE after FDR correction in either or both mothers or offspring. “Mix” denotes the PCR mastermix into which each locus was multiplexed and “Ta” denotes the annealing temperature used. Locus Zcwe05 is unpublished.
Table S2.
Details of the mother-offspring pairs and their match probabilities calculated based on 41 microsatellite loci within Colony (46)
| Colony | Mother ID | Offspring ID | Probability (%) |
| Special study beach | AGF11002 | AGP11014 | 100 |
| Special study beach | AGF11003 | AGP11022 | 100 |
| Special study beach | AGF11004 | AGP11026 | 100 |
| Special study beach | AGF11005 | AGP11018 | 100 |
| Special study beach | AGF11006 | AGP11032 | 100 |
| Special study beach | AGF11007 | AGP11051 | 100 |
| Special study beach | AGF11008 | AGP11041 | 100 |
| Special study beach | AGF11009 | AGP11078 | 100 |
| Special study beach | AGF11010 | AGP11065 | 100 |
| Special study beach | AGF11011 | AGP11063 | 100 |
| Special study beach | AGF11012 | AGP11079 | 100 |
| Special study beach | AGF11014 | AGP11125 | 100 |
| Special study beach | AGF11015 | AGP11144 | 100 |
| Special study beach | AGF11016 | AGP11145 | 100 |
| Special study beach | AGF11018 | AGP11174 | 100 |
| Special study beach | AGF11019 | AGP11151 | 100 |
| Special study beach | AGF11020 | AGP11192 | 100 |
| Special study beach | AGF11021 | AGP11185 | 100 |
| Special study beach | AGF11022 | AGP11211 | 100 |
| Special study beach | AGF11023 | AGP11200 | 100 |
| Freshwater beach | W8913mum | W8913pup | 100 |
| Freshwater beach | W8914mum | W8914pup | 100 |
| Freshwater beach | W8915mum | W8915pup | 100 |
| Freshwater beach | W8916mum | W8916pup | 100 |
| Freshwater beach | W8918mum | W8918pup | 100 |
| Freshwater beach | W8920mum | W8920pup | 100 |
| Freshwater beach | W8921mum | W8921pup | 100 |
| Freshwater beach | W8922mum | W8922pup | 100 |
| Freshwater beach | W8923mum | W8923pup | 100 |
| Freshwater beach | W8924mum | W8924pup | 100 |
| Freshwater beach | W8925mum | W8925pup | 100 |
| Freshwater beach | W8927mum | W8927pup | 100 |
| Freshwater beach | W8928mum | W8928pup | 100 |
| Freshwater beach | W8552/8258mum | W8552/8258pup | 100 |
| Freshwater beach | W8930mum | W8930pup | 100 |
| Freshwater beach | W8931mum | W8931pup | 100 |
| Freshwater beach | W8933mum | W8933pup | 100 |
| Freshwater beach | W8935mum | W8935pup | 100 |
| Freshwater beach | W8936mum | W8936pup | 100 |
| Freshwater beach | W8937mum | W8937pup | 100 |
| Freshwater beach | W8939mum | W8939pup | 100 |
Colony Differences.
Multivariate statistical analysis of the relative proportions of each substance revealed highly significant differences between animals sampled from the two colonies, both overall (Fig. 2A) [analyses of similarities (ANOSIM), global R = 0.57, P < 0.0001] and separately for mothers (ANOSIM, global R = 0.58, P < 0.0001) and offspring (ANOSIM, global R = 0.56, P < 0.0001). Bayesian structure analyses of the genetic data yielded the highest average log-likelihood value for K = 1 in both mothers and pups (Fig. S1), indicating a lack of population structure. By implication, chemical differences between the colonies appear to reflect environmental influences (see Discussion).
Fig. 2.
Two-dimensional nonmetric multidimensional scaling plots of chemical fingerprints of 41 Antarctic fur seal mother–offspring pairs. Bray–Curtis similarity values were calculated from standardized and log(x+1)-transformed abundance data; (A) color-coded by colony (red points: freshwater beach; blue points: special study beach); (B) plotted by mother–offspring pair, with each pair being denoted by a different symbol/color combination. The scales of the two axes are arbitrary. The closer the symbols appear on the plot, the more similar the two chemical fingerprints are.
Fig. S1.
Results of Bayesian analyses of population structure. Mean ± SE Ln P(D) values are shown based on five replicates for each value of K, the hypothesized number of genetic clusters represented in the data, for (A) mothers and (B) pups.
Mother–Offspring Similarity.
Pups were significantly more similar to their mothers in their chemical fingerprints than expected by chance (Fig. 2B), both overall (ANOSIM, global R = 0.67, P < 0.0001) and within each of the colonies (special study beach: ANOSIM, global R = 0.53, P < 0.0001; freshwater beach: ANOSIM, global R = 0.45, P < 0.0001). Chemical similarities between mothers and offspring could be encoded by shared genes or might simply reflect their spatial proximity. However, we found no relationship between chemical similarity and geographic distance within the special study beach, where pupping locations are recorded to the nearest square meter, either for mothers (Mantel’s r = 0.008, n = 20, P = 0.44) or offspring (Mantel’s r = 0.06, n = 20, P = 0.31). This finding suggests that chemical similarity is not associated with geographic proximity per se.
Genotype and Overall Chemical Fingerprints.
To determine whether genetic relatedness is reflected in chemical similarity, we tested for an association between pairwise r (see Table S3 for summary statistics) and Bray–Curtis similarity. A highly significant relationship was obtained when all of the animals were analyzed together (Mantel’s r = 0.07 n = 82, P = 0.005) but nonindependence of both chemical and genetic data for mothers and offspring may introduce pseudoreplication. We therefore repeated the analysis separately for mothers and offspring, finding no significant relationships (mothers, Mantel’s r = 0.06 n = 41, P = 0.10; offspring, Mantel’s r = 0.030 n = 41, P = 0.25).
Table S3.
Mean and SD of pairwise Queller and Goodnight (48) relatedness values
| Sample | All individuals | Mothers | Offspring |
| Entire sample | 0.009 ± 0.1 | 0.0008 ± 0.09 | 0.004 ± 0.09 |
| Special study beach | 0.016 ± 0.1 | −0.005 ± 0.09 | 0.012 ± 0.09 |
| Freshwater beach | 0.011 ± 0.1 | −0.004 ± 0.09 | 0.008 ± 0.10 |
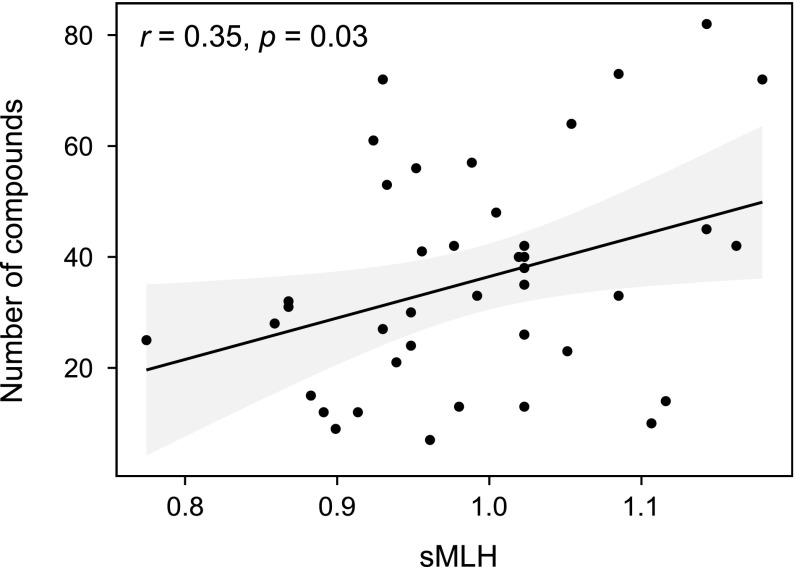
To test for a chemical signal of genetic quality, we regressed the number of compounds in an individual’s chemical fingerprint, a measure of chemical complexity, on standardized multilocus heterozygosity (sMLH). A significant positive correlation was found in mothers (Fig. 3) (F1,40 = 5.26, P = 0.026) but not in offspring (F1,40 = 0.50, P = 0.483). The strength of correlation also increased steadily with the number of microsatellites deployed in mothers and to a lesser extent in offspring (Fig. 4A). Conversely, the estimation error of the parameter g2, which quantifies the extent to which heterozygosities are correlated across loci, decreased with increasing marker number (Fig. S2). Overall, g2 was significantly positive (0.0022, P = 0.032 based on 1,000 iterations of the dataset), indicating that heterozygosity is correlated across the genome.
Fig. 3.
Relationship in mothers between sMLH and the number of compounds in an individual's chemical fingerprint.
Fig. 4.
Dependency of the strength of genetic associations on the number of randomly sampled microsatellite loci. Strength of association was quantified as the correlation coefficient (r) between (A) sMLH and the number of compounds in an individual's chemical fingerprint (gray symbols) and the sum of an individual's factor 1 and factor 2 values (black symbols), plotted separately for mothers (circles) and offspring (squares); (B) relatedness and Bray–Curtis similarity at the 10 best substances in mothers (see Methods for details). Mean ± SE of five resamplings of the data are shown for each point. The dashed lines represent significance thresholds.
Fig. S2.
Sensitivity of g2 to the number of microsatellite loci deployed. Different-sized subsets of loci were each resampled 1,000 times and the mean ± SD calculated.
Factor Analysis.
Chemical fingerprints are highly complex and may contain numerous compounds influenced by nongenetic factors. We therefore used principal axis FA to decompose the multidimensional chemical data into four factors (see Methods for details). Fitting the scores of all four factors together in a generalized linear model (GLM) of maternal heterozygosity, factors 1 and 2 were retained as significant predictor variables (Table 1, mother’s sMLH) and together explained almost twice as much deviance as the number of compounds in an individual’s chemical fingerprint (23.4% vs. 11.9%, respectively). A simple GLM of sMLH fitting the sum of the two factors as a single explanatory variable explained roughly the same amount of deviance (23.4%, F1,39 = 11.91, P = 0.001). In contrast, none of the factors were significantly associated with offspring heterozygosity.
Table 1.
Generalized linear models of sMLH in mothers and the colony from which an animal was sampled
| Term | Slope | F | df | P |
| Mother's sMLH (n = 41, total explained deviance = 23.40%) | ||||
| Factor 1 | 0.028 | 5.69 | 1 | 0.022 |
| Factor 2 | 0.028 | 5.83 | 1 | 0.021 |
| Colony (n = 82, total explained deviance = 56, 26%) | ||||
| Factor 4 | 0.38 | 102.88 | 1 | <0.0001 |
All four factors were initially fitted together as explanatory variables and the models were then reduced by sequentially removing the least-significant term until only significant terms were retained (see Methods for details). The F values for each term represent the change in deviance after removing that term from the model.
To test whether any of the factors are also associated with genetic relatedness, we used partial Mantel tests to derive the statistical significance of each factor while controlling for the others (see Methods for details). Factor 1 was significantly correlated with relatedness in mothers (Mantel’s r = −0.123, n = 41, P = 0.028) (Fig. S3) but not in offspring (Mantel’s r = 0.024, n = 41, P = 0.65). None of the other factors correlated significantly with relatedness in either mothers or offspring. As with the signal of heterozygosity, the strength of association between factor 1 and relatedness increased steadily with marker number (Fig. 4B).
Fig. S3.
Relationship between pairwise relatedness among mothers and the difference between individuals in their factor 1 scores. Mean and SEs are shown for the data partitioned into roughly equal-sized groups.
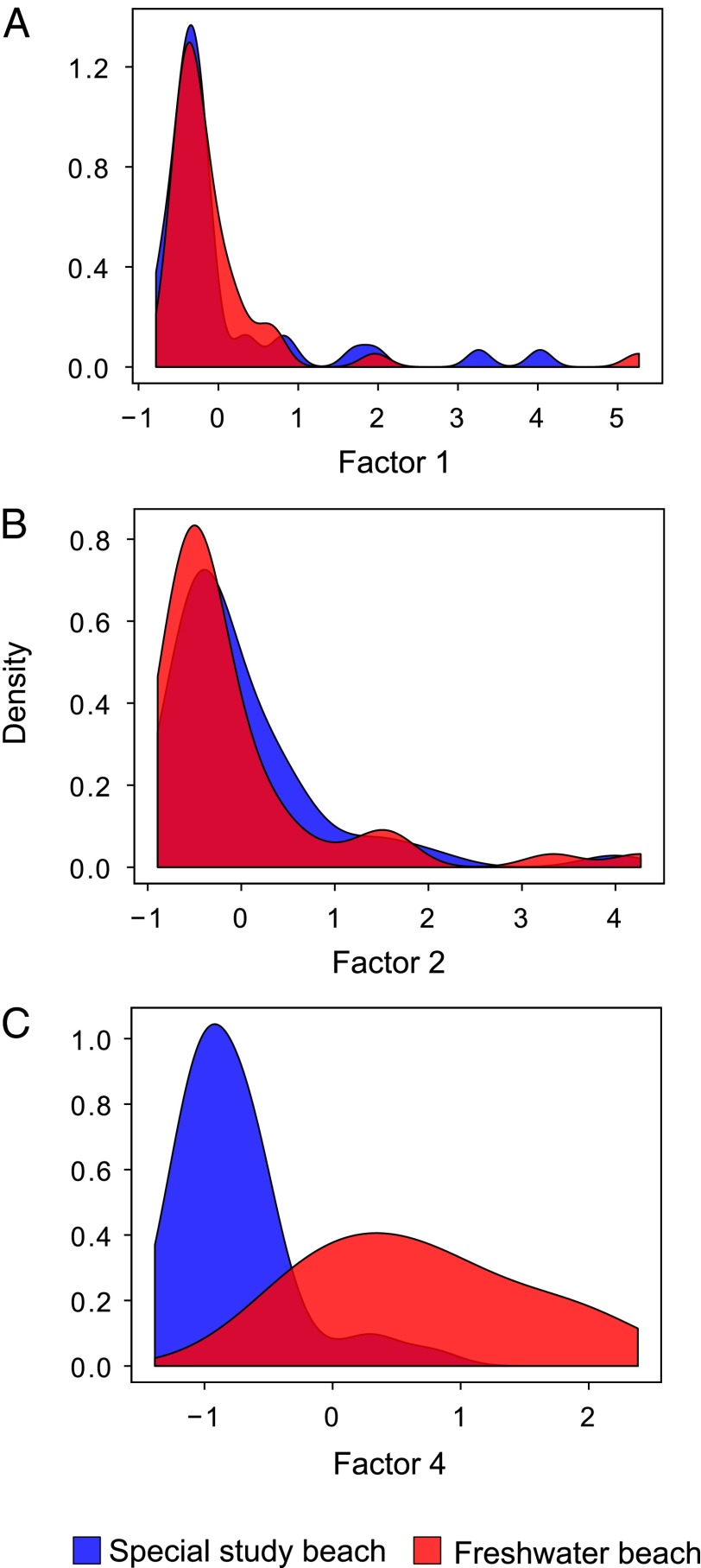
We next constructed a GLM to test for differences in the values of each of the four factors between the two colonies (Table 1, colony). Factors 1, 2, and 3 did not differ significantly, whereas factor 4 exhibited a highly significant difference between the colonies (Fig. 5). Thus, factors 1 and 2 both show correlations with genetic traits as well as overlapping distributions between colonies, factor 3 is not significantly associated with any of the variables we measured, and factor 4 represents substances that discriminate the two colonies and must therefore be environmentally influenced.
Fig. 5.
Distribution of factor scores of individuals sampled from the two seal colonies. Factors 1, 2, and 4 are shown in A, B, and C, respectively. Freshwater beach is shown in red and the special study beach is shown in blue.
Identification of Important Substances.
To identify substances that contribute most strongly toward chemical similarity within mother–offspring pairs, we used the “similarity percentages” routine (SIMPER, see Methods). Selecting the two most important compounds for each of the 41 mother–offspring pairs, we identified a total of 12 substances (Table S4, mother–offspring similarity). These substances yield a much stronger pattern of within-pair mother–offspring similarity (ANOSIM, global R = 0.68, P < 0.0001) than was obtained for the full dataset. Similarly strong patterns were obtained separately for each of the colonies (special study beach: ANOSIM, global R = 0.53, P < 0.0001; freshwater beach: ANOSIM, global R = 0.31, P = 0.001).
Table S4.
List of putative substances identified as being important for chemical similarity within mother–offspring pairs, chemical dissimilarity between the colonies, and genetic relatedness
| Retention time (min) | Mean similarity explained (mother/offspring); Similarity contribution (colony); Occurrences in best subsets (relatedness) | Chemical name | Probability | Empirical Kovats Index | Kovats Index |
| Mother-offspring similarity | |||||
| 19.723 | 15.54 | Ethyl hexadecanoate (hexadecanoic acid ethyl ester) | 58.3 | 1,992 | 1,993 |
| 15.458 | 12.25 | 1-Hexadecene | 20 | 1,591 | 1,593 |
| 26.789 | 11.97 | Squalene | 46 | 2,815 | 2,790 |
| 16.397 | 11.30 | 8-Pentadecanone | 94 | 1,673 | 1,648 |
| 19.525 | 10.87 | Ethyl 9-hexadecenoate | 87 | 1,972 | 1,977 |
| 21.405 | 8.49 | Ethyl oleate | 66 | 2,175 | 2,171 |
| 37.564 | 6.48 | Not identified | — | — | — |
| 15.623 | 6.48 | Not identified | — | 1,606 | — |
| 33.637 | 6.28 | Campesterol | 71 | — | — |
| 30.804 | 6.03 | Cholestanol | 67 | — | — |
| 20.362 | 5.34 | Heptadecanoic acid | 69 | 2,086 | 2,067 |
| 17.409 | 4.79 | Not identified | — | 1,766 | — |
| Colony dissimilarity | |||||
| 15.458 | 3.01 | 1-Hexadecene | 20 | 1,591 | 1,593 |
| 16.397 | 2.42 | 8-Pentadecanone | 94 | 1,673 | 1,648 |
| 26.789 | 2.07 | Squalene | 46 | 2,815 | 2,790 |
| 19.525 | 1.97 | Ethyl 9-hexadecenoate | 87 | 1,972 | 1,977 |
| 21.405 | 1.89 | Ethyl oleate | 66 | 2,175 | 2,171 |
| 21.348 | 1.67 | Not identified | — | — | — |
| 19.723 | 1.67 | Ethyl hexadecanoate (hexadecanoic acid ethyl ester) | 58.3 | 1,992 | 1,993 |
| 30.804 | 1.48 | Cholestanol | 67 | — | — |
| 38.518 | 1.44 | Not identified | — | — | — |
| 17.409 | 1.33 | Not identified | — | 1,766 | — |
| 20.511 | 1.29 | Not identified | — | — | — |
| 33.637 | 1.27 | Campesterol | 71 | — | — |
| 21.575 | 1.21 | Octadecanoic acid ethyl ester | 85 | 2,194 | 2,194 |
| 15.742 | 1.18 | Not identified | — | — | — |
| 19.665 | 1.13 | Not identified | — | — | — |
| Relatedness | |||||
| 36.941 | 315,926 | Not identified | — | — | — |
| 19.525 | 250,140 | Ethyl 9-hexadecenoate | 87 | 1,972 | 1,977 |
| 13.124 | 245,569 | Not identified | — | — | — |
| 20.362 | 214,830 | Heptadecanoic acid | 69 | 2,086 | 2,067 |
| 14.699 | 207,155 | Not identified | — | — | — |
| 21.090 | 203,683 | Not identified | — | — | — |
| 21.575 | 198,366 | Octadecanoic acid ethyl ester | 85 | 2,194 | 2,194 |
| 37.049 | 192,000 | Not identified | — | — | — |
| 19.620 | 189,049 | Not identified | — | — | — |
| 37.074 | 185,017 | Not identified | — | — | — |
Substances are listed in decreasing order of importance, as measured by the mean proportion of mother-pup similarity explained in the SIMPER analysis, the percentage contribution toward dissimilarity between beaches, and the number of occurrences within the best subsets identified by the BIO-ENV bootstrap procedure (see Methods for details). The chemical name and assignment probability are derived by a comparison of the empirical mass spectra with the most similar substance in the NIST library. The Kovats Index (Methods) was calculated for all substances with a retention time less than 28 min. For comparison, we provide the Kovats indices (60) of the substances to which our compounds show the closest resemblance.
We also used SIMPER to search for substances accounting for most of the chemical dissimilarity between the two colonies. This approach identified a total of 15 substances (Table S4, colony dissimilarity) that collectively yield a much higher global R value (ANOSIM, global R = 0.77, P < 0.0001) than was obtained for all of the chemicals. To identify substances associated with genetic relatedness, we used the BIO-ENV procedure embedded in a bootstrap framework (see Methods for details). We obtained a subset of 10 substances (Table S4, relatedness) that consistently occurred within the “best” subsets (i.e., maximizing the relationship between chemical similarity and relatedness) over all 10 × 106 bootstrap samples and collectively maximized the relationship between chemical distance and relatedness (Fig. S4). Chemical similarity based on these 10 substances was significantly associated with genetic relatedness (Mantel’s r = 0.164, n = 41, P = 0.001).
Fig. S4.
Results of the BIO-ENV bootstrapping procedure. See Methods for details. The y axis shows the strength of correlation between genetic relatedness and pairwise Bray–Curtis similarity. On the x axis, the chemicals are shown in decreasing order of importance, given by the number of subsamples in which the chemical was retained in the “best” subset. Each chemical was progressively added to the calculation of Bray–Curtis similarity. The relationship between chemical similarity and relatedness is maximized for a subset of the 10 most important chemicals.
Finally, we cross-referenced the three lists of substances to evaluate any potential overlap. Of the 12 compounds carrying the strongest signal of mother–offspring similarity, 9 also occurred in the subset of chemicals that differ between the two colonies, implying that they may be influenced by environmental conditions ashore. Remarkably, an additional two compounds overlapped with the best subset of chemicals associated with genetic relatedness. The mass spectra and Kovats indices of these substances indicate close resemblance to the known pheromones ethyl-9-hexadecenoate and heptadecanoic acid (see Discussion and Table S4, relatedness).
Discussion
Although mother–offspring recognition is under strong selection in many species, little is known about its chemical basis, particularly in natural populations of nonmodel organisms. We show that fur seal pups are highly similar to their mothers in their chemical fingerprints and that this similarity is largely encoded by a handful of substances that also carry information about either colony or genotype. Our findings provide intriguing insights into how females could use chemical information to recognize their offspring and may also help to explain how fur seals appear capable of exercising mate choice for heterozygous and unrelated partners (24).
Our study was partly motivated by the discovery that female Australian sea lions can identify their pups using only olfactory cues (22). In most vertebrate species, chemical fingerprints show marked differences by sex, age, and reproductive status (25), a pattern that is partly reflected in our data because it is only the mother’s chemical fingerprints that encode genotype. However, the overall chemical fingerprints of mothers and offspring are still very similar, raising the possibility that self-referent phenotype matching (26) could be used in mother–pup recognition. This is a conceptually simple mechanism by which the own phenotype is a representation or template used for the recognition of relatives. Self-referent phenotype matching has been demonstrated in a variety of mammalian, bird, and fish species (27). However, further experimental evidence would be needed to show that mother–offspring recognition in fur seals relies on self-matching rather than social learning. Interestingly, allosuckling rates vary considerably among pinniped species, from 6% in New Zealand sea lions to up to 90% in Hawaiian monk seals, suggesting that mother–pup recognition abilities may vary among species (28). The Antarctic fur seal has one of the lowest observed rates of allosuckling (29), which is consistent with the strong pattern of chemical similarity we find between mothers and their pups.
Although chemical fingerprints are widely assumed to encode genetic traits, such as relatedness and individual heterozygosity, only a handful of studies have reported the expected associations. Moreover, chemical profiles typically change with age and reproductive status (25) and genetic correlations have, to our knowledge, only been detected in breeding adults (6, 7). Analyzing the relationship between heterozygosity and chemical complexity separately for mothers and pups shows a clear correlation that increases with the number of loci for mothers, a pattern that is weak or lacking in pups (Fig. 4A). Because of the consistency of our results with the literature, we believe this reflects a genuine functional difference between the chemical fingerprints of mothers and pups.
We also find a marked difference in the way that heterozygosity and relatedness are encoded in chemical fingerprints. Heterozygosity is detectable in the overall fingerprint, as it is correlated with the number of chemicals, whereas relatedness is encoded by a small subset of chemicals, whose signal is diluted by analyzing the overall chemical fingerprint. The diversity of chemicals reflected in heterozygosity could be the result of genetic polymorphisms in the enzymes involved in the synthesis of semiochemicals (6) but may also be influenced by condition dependent factors (see below). In contrast, it makes sense that genetic relatedness could be encoded by a small subset of chemicals that potentially reflect certain genes, such as the MHC, a highly polymorphic cluster of immune genes detectable through scent (30, 31).
In natural populations, environmental effects on chemical fingerprints are likely to be particularly strong. The only study of a free-ranging, natural population to have detected an association with genotype used PCA to reduce the dimensionality of the chemical data (9). However, this approach is not ideally suited to detecting such signals because a principal component that explains maximal variance may not necessarily provide an optimal representation of the underlying genotype. We applied PCA to our dataset but obtained no significant correlations between any of the resulting principal components and relatedness, and a weaker signal of heterozygosity than was obtained using FA. This result could be because of the so called “simple structure” that is obtained by rotation of the factors within FA (32). This results in each substance loading primarily on a single factor and not on the others, meaning that the factors represent subsets of variables that covary and are therefore likely to have a shared basis, such as genes or the environment.
FA was considerably more successful than PCA at detecting patterns relating to genotype within our chemical dataset. Factors 1 and 2 together explained almost twice as much of the deviance in heterozygosity as a simple regression on the number of substances, and relatedness was significantly associated with factor 1 but not with Bray–Curtis similarity based on the overall fingerprints. Because each factor mostly represents a subset of the total pool of chemicals, this finding is consistent with Hurst and Beynon’s suggestion that the selective assessment of specific semiochemicals may allow individuals to assess genotype more accurately than from entire chemical fingerprints (2).
It is unclear why factor 1 carries information about both heterozygosity and relatedness, whereas factor 2 correlates only with heterozygosity. One possibility is that heterozygosity and relatedness are to some extent signaled by the same substances, potentially deriving from the MHC. As the substances loading on factor 2 are essentially uncorrelated with those loading on factor 1, we speculate that heterozygosity may influence the chemical fingerprint through two or more different pathways. Factor 1 could thus represent a direct pathway from genes to the chemical fingerprint, whereas factor 2 may represent an indirect pathway where body condition or the microbiome could be possible mediators. Future work will aim to explore these possibilities.
An important strength of our study was a sampling design that facilitated disentangling genetically encoded substances from those influenced by the environment. We found that factors 1 and 2, which both encode some aspect of genotype, did not differ significantly in the distribution of factor scores between the colonies, whereas factor 4, which carried no discernible genetic information, showed a highly significant difference. These differences could either be a result of environmental chemicals that directly contribute toward the profile, or could reflect alterations to the chemical fingerprint caused by different conditions on the beaches (e.g., temperature, wind, solar radiation). We would need to sample more colonies to determine the concrete causes.
Another important aspect of our study design was the unusually high genetic resolution provided by 41 microsatellites. Most studies use around 10–15 loci, which for our dataset was insufficient to detect a significant correlation between maternal heterozygosity and compound richness (Fig. 4A). However, the strength of correlation increased steadily as more microsatellites were deployed until a highly significant relationship was obtained with the full marker panel. Similarly, the error with which the parameter g2 was estimated from the genetic data decreased steadily with increasing marker number. This finding is consistent with the suggestion that, as long as heterozygosity is correlated across the genome (as is the case where appreciably inbred individuals are present), increasing the number of markers should improve the estimation accuracy of genome-wide heterozygosity, leading to a strengthening of effect size (10, 33). A similar pattern was also obtained for genetic relatedness, suggesting that, if many thousands of genetic markers could be deployed, an even greater proportion of the chemical variance should be explicable by genotype (11).
In many species, heterozygosity is associated with fitness (34). In Antarctic fur seals, multilocus heterozygosity at nine microsatellites correlates with early survivorship and breeding success in females (35), as well as reproductive success in males (36). Females of this species also appear to exert mate choice based on their partner’s genotype (24) but it is unclear how this could be achieved. The finding that heterozygosity and relatedness are both encoded in mother’s chemical fingerprints lends support to the hypothesis that chemical cues could be involved, although unfortunately we were not able to include adult males in this study because they are challenging to capture and sedate. Nevertheless, as male fur seals emit a strong musky odor (17), which has been proposed to attract females during the mating season (37), it seems plausible that genotype could also be encoded in adult male chemical fingerprints.
To explore the extent to which genes and the environment influence mother–offspring similarity, we first attempted to identify the most important substances associated with mother–offspring similarity, colony dissimilarity, and genetic relatedness. We obtained relatively small subsets of 12, 15, and 10 chemicals, respectively. In the case of mother–offspring similarity and relatedness, these subsets yielded much stronger associations than were obtained for the overall fingerprints. This result suggests that SIMPER and BIO-ENV were successful in identifying important chemicals within the total set of 213 substances, although this does not preclude additional chemicals playing a lesser role. It is also noteworthy that as many as 10 or more chemicals appear to encode relatedness, given that a single locus is expected to provide little power to distinguish anything other than close relatives (2).
Evaluating the overlap between the subsets of chemicals associated with mother–offspring similarity, colony dissimilarity, and genetic relatedness revealed an interesting pattern. Of the top 12 substances accounting for the similarity between mothers and their pups, 9 also occurred in the subset of chemicals that showed the greatest differences between the two colonies. Although our analysis is not exhaustive, as we focused only on the most important substances, this nevertheless suggests that chemical similarity within mother–offspring pairs is strongly influenced by the local environment. A further two substances also overlapped with the subset of chemicals associated with genetic relatedness, implying that mother–offspring similarity also has a genetic basis. Both of these substances reveal similarity to known pheromones, consistent with the previous suggestion that pheromone-like chemical signals may play an important role in mother–offspring recognition across a variety of taxa (38).
Little is currently known about the specific chemicals that signal genetic relatedness in vertebrates (2). Although we were only able to putatively identify three of the top 10 substances encoding relatedness using the National Institute of Standards and Technology (NIST) database, the mass spectra and Kovats indices of these compounds reveal close resemblance to the known pheromones ethyl 9-hexadecenoate, heptadecanoic acid and ethyl stearate (Table S4, relatedness). According to the pherobase database, all three of these substances are part of the chemical communication system of a variety of different taxa, ranging from bumblebees to badgers. Heptadecanoic acid, for example, is a known pheromone of 33 different species, including 26 vertebrate taxa. However, to act as a pheromone in a given species, a chemical must meet a number of strict criteria (39), which would require experimental evidence (see below).
Although we captured a large number of substances of varying volatility, we only recovered compounds soluble in ethanol and which could be detected by GC-MS. Extraction with other solvents was not possible because of logistic reasons. Nevertheless, even though our sampling of chemicals is likely to be incomplete, our analyses revealed a number of statistically significant and potentially biologically relevant patterns. In addition, we detected a number of chemicals that may carry important information. However, because some of these substances may have been further metabolized after extracting them from the skin (40), we cannot exclude the possibility that some of the putatively identified compounds could be breakdown products.
Finally, biologically relevant chemical cues can be transferred in a variety of ways, from volatile substances recognized by olfaction to chemicals that act when two individuals are in physical contact (41). Because adult female fur seals and their pups conduct naso-nasal inspections during the recognition procedure (20), it is possible that some of the chemicals may act through contact. To unequivocally determine the biological relevance of the chemicals we have identified, as well as their precise mode of action, would require behavioral assays in the field. This will be challenging, but our results provide the basis for testable hypotheses on potential chemical signals and the substances involved.
Methods
Study Site and Field Methods.
Forty-four mother–offspring pairs were sampled from two breeding colonies: freshwater beach and special study beach, separated by ∼200 m (Fig. 1) on Bird Island, South Georgia (54° 00′ S, 38° 02′ W). Breeding females and their pups were captured and restrained on land using standard methodology (42). Seal capture and restraint were part of annual routine procedures of the Long Term Monitoring and Survey program of the British Antarctic Survey. We obtained chemical samples by rubbing the cheek, underneath the eye, and behind the snout with a sterile cotton wool swab. Each swab was individually preserved in a glass vial in 60% (vol/vol) ethanol stored at −20 °C. All of the samples were obtained immediately after capture by the same team of two seal scientists at both colonies. Tissue samples for genetic analysis were collected as described by Hoffman et al. (43) and stored individually at –20 °C in the preservative buffer 20% (vol/vol) DMSO saturated with salt. Fieldwork was approved by the British Antarctic Survey Ethics Review Committee. Samples were collected and retained under permits issued by the Department for Environment, Food and Rural Affairs (DEFRA), and in accordance with the Convention on International Trade in Endangered Species of Wild Fauna and Flora (CITES).
Chemical Analyses.
We first took 1 mL of each sample and allowed the ethanol to evaporate at room temperature under a fume hood for a maximum of 12 h before resuspending in 50 µL dichlormethane for subsequent processing. The samples were then analyzed on a GC equipped with a VF-5 ms capillary column (30 m size × 0.25 mm inner diameter, DF 0.25, 10-m guard column; Varian) and coupled to a quadrupole mass spectrometer (Focus GC-DSQ MS system, Thermo Electron). A blank sample (control with cotton wool and ethanol) and an alkane mix (C8–C28) were analyzed as well. One microliter of each sample was injected into a deactivated glass wool-packed liner at an inlet temperature of 225 °C and processed in a splitless mode. Carrier gas (He) flow rate was held at 1.2 mL/min. The GC run was initiated at 60 °C for 3 min then ramped at 10 °C/min to 280 °C, where it remained for 20 min. The transfer line temperature was set to 280 °C and mass spectra were taken in electron ionization mode at 70 eV with five scans per second in full-scan mode (50–500 m/z). GC-MS data were processed using the program Xcalibur (Thermo Scientific). To ensure that the scoring of compounds was as objective as possible, we wrote a custom R script (available on request) that compensated for minor shifts in retention times among chromatograms by maximizing the number of shared components between samples through very small (≤0.03 ms) shifts in the retention time. To double-check the reliability of the scoring, ∼10% of compounds were selected at random and scored by eye.
Genetic Analysis.
Total genomic DNA was extracted from each sample using a standard phenol-chloroform protocol and genotyped at 43 highly polymorphic microsatellite loci (see Table S1 for details). These were PCR-amplified in eight separate multiplexed reactions using a Type It Kit (Qiagen) as described in Table S1. The following PCR profile was used: one cycle of 5 min at 94 °C; 24 cycles of 30 s at 94 °C, 90 s at Ta °C and 30 s at 72 °C; and one final cycle of 15 min at 72 °C (see Table S1 for Ta). Fluorescently labeled PCR products were then resolved by electrophoresis on an ABI 3730xl capillary sequencer and allele sizes were scored automatically using GeneMarker v1.95. To ensure high genotype quality, all traces were manually inspected and any incorrect calls were adjusted accordingly.
Genepop (44) was used to calculate observed and expected heterozygosities and to test for deviations from HWE, separately for mothers and pups, specifying 10,000 dememorizations, 1,000 batches, and 10,000 iterations per batch. Two loci that deviated from HWE in either mothers or pups after table-wide correction for the FDR using Q-value (45) were excluded from subsequent analyses, leaving a total of 41 loci (Table S1). Because milk stealing is common in fur seals and can lead to errors in the assignment of mother–offspring pairs in the field (29), we used the program Colony v2.0.5.0 (46) to verify that all of our mother–offspring pairs were genuine. Coancestry v1.0.1.2 (47) was then used to generate a pairwise relatedness matrix based on Queller and Goodnight’s statistic, r (48). Each individual’s heterozygosity was expressed as sMLH, which is defined as the total number of heterozygous loci in an individual divided by the sum of average observed heterozygosities in the population over the subset of loci successfully typed in the focal individual (49). The two-locus heterozygosity disequilibrium g2, which measures the extent to which heterozygosities are correlated across loci, was then computed using the method of David et al. (50). Sensitivity of this estimate to the number of loci was explored by randomly selecting different sized subsets of loci and recalculating g2 1,000 times.
To test for population structure, Bayesian cluster analysis of the microsatellite dataset was implemented using Structure v2.3.3 (51). Structure uses a maximum-likelihood approach to determine the most likely number of genetically distinct clusters in a sample (K) by subdividing the dataset in a way that maximizes HWE and minimizes LD within the resulting clusters. Separately for mothers and pups, we ran five independent runs for each value of K ranging from 1 to 10 using 1 × 106 Markov chain Monte Carlo iterations after a burn-in of 1 × 105, specifying the correlated allele frequencies model and assuming admixture. The most likely K was then evaluated using the maximal average value of Ln P(D), a model-choice criterion that estimates the posterior probability of the data.
Statistical Analysis Framework.
Any chemicals appearing in the control sample or present in only one sample were excluded from further analyses, leaving a total of 213 substances. To explore the completeness of our sampling, we estimated the maximum number of substances present in the population using the Michaelis–Menten Function, based on a permutation procedure (9,999 iterations). Up to 229 substances might be expected in a larger sample of individuals, suggesting that we have sampled around 95% of all potential substances. Analyses were conducted on the relative proportion of each substance (%) to the total amount of substances (52). We then used a three-step analytical framework to: (i) visualize and statistically analyze overall patterns of chemical fingerprint similarity in relation to breeding colony, mother-offspring pair, relatedness, and heterozygosity; (ii) tease out subsets of chemicals containing genotypic and environmental information; and (iii) identify specific compounds involved. Computer code and documentation are provided as a PDF file written in Rmarkdown (Dataset S1) together with the data (Dataset S2).
Overall Patterns of Chemical Similarity.
The chemical fingerprint data were visualized using nonmetric multidimensional scaling (53) based on a matrix of pairwise Bray–Curtis similarity values calculated from the log(x+1)-transformed data. This approach allows visualization of a high-dimensional chemical similarity space by placing each individual in a 2D scatterplot such that ranked between-individual distances are preserved, points close together representing individuals with relatively high chemical similarity. Differences between a priori defined groups (i.e., the breeding colonies and mother–offspring pairs) were then analyzed through nonparametric ANOSIM (53) using 99,999 iterations of the dataset. ANOSIM is a permutation test that provides a way to evaluate whether there is a significant difference between two or more groups of sampling units without the need for assumptions concerning data distribution or homoscedasticity. These analyses were implemented in R using the vegan package (54).
Factor Analysis.
To dissect apart genetic from environmental components, we performed a principal axis FA on the chemical data. We used an oblique rotation technique (promax), which allows the factors to be correlated. This type of rotation was used because it is possible that certain compounds within the chemical fingerprint may encode more than one genetic characteristic (e.g., heterozygosity and relatedness) and could thus be correlated with more than one factor. FA cannot be applied when a dataset has more variables than observations (D>>N) because the covariance matrix is singular and an inverse cannot be computed. We therefore used the function factor.pa.ginv() from the R package HDMD, which uses a generalized inverse matrix (14). An important step in factor analysis is choosing a reasonable number of factors to represent the data (32). As our dataset is complex and contains many zero entries, some common methods like parallel analysis may lead to an impracticably large number of factors. Consequently, we applied two methods for determining the optimal number of factors. First, we used the Bayesian Information Criterion, which optimizes the trade-off between model complexity and model fit, and second we used a scree plot, which visually depicts the drop in the factor eigenvalue course (32, 55). Both methods suggested four factors.
Generalized Linear Models.
To explore the contributions of each of the four factors toward the signal of heterozygosity, we constructed separate GLMs of mother and offspring sMLH, in which we fitted all four factors together and specified a Gaussian error structure. We then tested for factors that differ significantly between the two colonies by constructing a GLM with colony as the response variable (modeled using a binomial error structure) and the values of the four factors fitted as predictors. For each GLM, we initially implemented a full model containing all of the predictor variables and then used standard deletion testing procedures based on F tests (56) to sequentially remove each term unless doing so significantly reduced the amount of deviance explained.
Partial Mantel Tests.
To test for associations between each of the factors and genetic relatedness, we used the relatedness matrix based on all 41 loci as the response variable and fitted as predictor variables matrices of pairwise similarity at each of the four factors using a Partial Mantel test implemented in the ecodist package (57). This randomizes the rows and columns of one dissimilarity matrix but leaves the others unpermuted. Separate models were constructed for mothers and offspring, each using 10,000 permutations of the dataset. Finally, we computed the Spearman rank correlation (Mantel’s r) and two-tailed P value for the association between relatedness and a factor matrix given the other factors as covariates.
Identification of Chemicals.
We next attempted to identify specific chemicals associated with breeding colony, mother–offspring similarity, and genetic relatedness. First, we assessed the contributions of specific substances to the similarity within groups, using the “similarity percentages” routine (SIMPER) (58). This process decomposes all Bray–Curtis similarities within a group into percentage contributions from each compound, listing the compounds in decreasing order of importance. As groups, we specified (i) the two breeding colonies and (ii) the 41 different mother–offspring pairs.
Second, to explore the contributions of individual chemicals to the signal of genetic relatedness, a continuously distributed variable, we used the BIO-ENV procedure (58) to identify the “best” subset of compounds within the chemical abundance matrix that maximizes the rank correlation between pairwise Bray–Curtis similarities and relatedness. However, with over 200 different chemicals being present in the chemical data matrix, it seems likely that this approach could yield spurious associations, especially given that some of the chemicals were present only in a few individuals. For this reason, we embedded the BIO-ENV procedure in a bootstrap analysis as follows: (i) we randomly subsampled 20 of the 41 mothers 20,000 times; (ii) for each subsample, we randomly selected 10 chemicals, each 500 times; (iii) for each of the resulting 10 × 106 subsamples, comprising 20 individuals and 10 compounds, we applied the BIO-ENV procedure and saved the compounds present in the best subset. We then summed up the occurrences of every chemical throughout all of the subsets and sorted them in decreasing order to represent their relative importance. The basic assumption of our approach is that random correlations will not be consistent over the different subsamples of individuals and compounds, whereas compounds that genuinely encode relatedness should be recovered consistently across many subsets. This procedure was conducted in R using the bio.env() function in the sinkr package (59).
Identification of putative substances encoding mother-pup similarity, colony differences, and relatedness were based on two steps: (i) comparing the mass spectrum of a specific substance with the best match of the NIST library (NIST 2005 and 2008) and (ii) calculating the Kovats Retention Index and comparing this to the literature value (obtained from www.Pherobase.com and www.chemspider.com). Kovats Indices (60) were calculated by running a sample of linear alkanes (C8–C28) under the identical GC-MS conditions as described above.
Supplementary Material
Acknowledgments
We thank Mareike Esser for guidance on the implementation of g2 in R. This research was supported by Marie Curie FP7-Reintegration-Grant within the 7th European Community Framework Programme PCIG-GA-2011-303618 (to J.I.H.) and Deutsche Forschungsgemeinschaft standard Grant HO 5122/3-1 (to J.I.H.). This article contributes to the Polar Science for Planet Earth Ecosystems and Long Term Monitoring and Survey Projects of the British Antarctic Survey, Natural Environment Research Council core funded science programme.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
See Commentary on page 11146.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1506076112/-/DCSupplemental.
References
- 1.Wyatt TD. Pheromones and Animal Behavior. 2nd Ed Cambridge Univ Press; Cambridge, UK: 2014. [Google Scholar]
- 2.Hurst JL, Beynon RJ. Making progress in genetic kin recognition among vertebrates. J Biol. 2010;9(2):13. doi: 10.1186/jbiol221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wedekind C, Füri S. Body odour preferences in men and women: Do they aim for specific MHC combinations or simply heterozygosity? Proc Biol Sci. 1997;264(1387):1471–1479. doi: 10.1098/rspb.1997.0204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Radwan J, et al. MHC and preferences for male odour in the bank vole. Ethology. 2008;114(9):827–833. [Google Scholar]
- 5.Olsson M, et al. Major histocompatibility complex and mate choice in sand lizards. Proc Biol Sci. 2003;270(Suppl 2):S254–S256. doi: 10.1098/rsbl.2003.0079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Boulet M, Charpentier MJ, Drea CM. Decoding an olfactory mechanism of kin recognition and inbreeding avoidance in a primate. BMC Evol Biol. 2009;9:281. doi: 10.1186/1471-2148-9-281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Charpentier MJE, Boulet M, Drea CM. Smelling right: The scent of male lemurs advertises genetic quality and relatedness. Mol Ecol. 2008;17(14):3225–3233. doi: 10.1111/j.1365-294X.2008.03831.x. [DOI] [PubMed] [Google Scholar]
- 8.Crawford JC, Boulet M, Drea CM. Smelling wrong: Hormonal contraception in lemurs alters critical female odour cues. Proc Biol Sci. 2011;278(1702):122–130. doi: 10.1098/rspb.2010.1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Leclaire S, et al. Semiochemical compounds of preen secretion reflect genetic make-up in a seabird species. Proc Biol Sci. 2012;279(1731):1185–1193. doi: 10.1098/rspb.2011.1611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Balloux F, Amos W, Coulson T. Does heterozygosity estimate inbreeding in real populations? Mol Ecol. 2004;13(10):3021–3031. doi: 10.1111/j.1365-294X.2004.02318.x. [DOI] [PubMed] [Google Scholar]
- 11.Hoffman JI, et al. High-throughput sequencing reveals inbreeding depression in a natural population. Proc Natl Acad Sci USA. 2014;111(10):3775–3780. doi: 10.1073/pnas.1318945111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fabrigar LR, et al. Evaluating the use of exploratory factor analysis in psychological research. Psychol Methods. 1999;4(3):272–299. [Google Scholar]
- 13.Eid M, et al. Statistik und Forschungsmethoden. Beltz; Weinheim, Germany: 2010. [Google Scholar]
- 14.McFerrin L. 2013 HDMD: Statistical Analysis Tools for High Dimension Molecular Data (HDMD). R package version 1.2. Avaliable at http://cran.r-project.org/web/packages/HDMD/index.html. Accessed April 17, 2015.
- 15.Kishida T, Kubota S, Shirayama Y, Fukami H. The olfactory receptor gene repertoires in secondary-adapted marine vertebrates: Evidence for reduction of the functional proportions in cetaceans. Biol Lett. 2007;3(4):428–430. doi: 10.1098/rsbl.2007.0191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kowalewsky S, Dambach M, Mauck B, Dehnhardt G. High olfactory sensitivity for dimethyl sulphide in harbour seals. Biol Lett. 2006;2(1):106–109. doi: 10.1098/rsbl.2005.0380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ling JK. The integument of marine mammals. In: Harrison RJ, editor. Functional Anatomy of Marine Mammals. Academic; London: 1972. pp. 1–44. [Google Scholar]
- 18.Hardy MH, et al. Facial skin glands of ringed and grey seals, and their possible function as odoriferous glands. Can J Zool. 1991;69(1):189–200. [Google Scholar]
- 19.Insley SJ, et al. A review of social recognition in pinnipeds. Aquat Mamm. 2003;29(2):181–201. [Google Scholar]
- 20.Dobson FS, Jouventin P. How mothers find their pups in a colony of Antarctic fur seals. Behav Processes. 2003;61(1-2):77–85. doi: 10.1016/s0376-6357(02)00164-x. [DOI] [PubMed] [Google Scholar]
- 21.Phillips AV. Behavioural cues used in reunions between mother and pup South American fur seals (Arctocephalus australis) J Mammal. 2003;84(2):524–535. [Google Scholar]
- 22.Pitcher BJ, Harcourt RG, Schaal B, Charrier I. Social olfaction in marine mammals: Wild female Australian sea lions can identify their pup’s scent. Biol Lett. 2011;7(1):60–62. doi: 10.1098/rsbl.2010.0569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Doidge DW, et al. Growth rates of Antarctic fur seal Arctocephalus gazella pups at South Georgia. J Zool. 1984;203(1):87–93. [Google Scholar]
- 24.Hoffman JI, Forcada J, Trathan PN, Amos W. Female fur seals show active choice for males that are heterozygous and unrelated. Nature. 2007;445(7130):912–914. doi: 10.1038/nature05558. [DOI] [PubMed] [Google Scholar]
- 25.Caspers BA, et al. The scent of adolescence: The maturation of the olfactory phenotype in a free ranging mammal. PLoS One. 2011;6:6. doi: 10.1371/journal.pone.0021162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Blaustein AR. Kin recognition mechanisms: Phenotypic matching or recognition alleles? Am Nat. 1983;121(5):749–754. [Google Scholar]
- 27.Hauber ME, Sherman PW. Self-referent phenotype matching: Theoretical considerations and empirical evidence. Trends Neurosci. 2001;24(10):609–616. doi: 10.1016/s0166-2236(00)01916-0. [DOI] [PubMed] [Google Scholar]
- 28.Pitcher BJ, et al. Allosuckling behavior in the Australian sea lion (Neophoca cinerea): An updated understanding. Mar Mamm Sci. 2011;27(4):881–888. [Google Scholar]
- 29.Hoffman JI, Amos W. Does kin selection influence fostering behaviour in Antarctic fur seals (Arctocephalus gazella)? Proc Biol Sci. 2005;272(1576):2017–2022. doi: 10.1098/rspb.2005.3176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yamazaki K, et al. Recognition among mice. Evidence from the use of a Y-maze differentially scented by congenic mice of different major histocompatibility types. J Exp Med. 1979;150(4):755–760. doi: 10.1084/jem.150.4.755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wedekind C, Seebeck T, Bettens F, Paepke AJ. MHC-dependent mate preferences in humans. Proc Biol Sci. 1995;260(1359):245–249. doi: 10.1098/rspb.1995.0087. [DOI] [PubMed] [Google Scholar]
- 32.Preacher KJ, et al. Choosing the optimal number of factors in exploratory factor analysis: A model selection perspective. Multivariate Behav Res. 2013;48(1):28–56. doi: 10.1080/00273171.2012.710386. [DOI] [PubMed] [Google Scholar]
- 33.Hoffman JI, Forcada J, Amos W. Exploring the mechanisms underlying a heterozygosity-fitness correlation for canine size in the Antarctic fur seal Arctocephalus gazella. J Hered. 2010;101(5):539–552. doi: 10.1093/jhered/esq046. [DOI] [PubMed] [Google Scholar]
- 34.Hansson B, Westerberg L. On the correlation between heterozygosity and fitness in natural populations. Mol Ecol. 2002;11(12):2467–2474. doi: 10.1046/j.1365-294x.2002.01644.x. [DOI] [PubMed] [Google Scholar]
- 35.Forcada J, Hoffman JI. Climate change selects for heterozygosity in a declining fur seal population. Nature. 2014;511(7510):462–465. doi: 10.1038/nature13542. [DOI] [PubMed] [Google Scholar]
- 36.Hoffman JI, Boyd IL, Amos W. Exploring the relationship between parental relatedness and male reproductive success in the Antarctic fur seal Arctocephalus gazella. Evolution. 2004;58(9):2087–2099. doi: 10.1111/j.0014-3820.2004.tb00492.x. [DOI] [PubMed] [Google Scholar]
- 37.Hamilton JE. Scent of otariids. Nature. 1956;177:900. [Google Scholar]
- 38.Vaglio S. Chemical communication and mother-infant recognition. Commun Integr Biol. 2009;2(3):279–281. doi: 10.4161/cib.2.3.8227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wyatt TD. The search for human pheromones: The lost decades and the necessity of returning to first principles. Proc Biol Sci. 2015;282(1804):20142994. doi: 10.1098/rspb.2014.2994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Theis KR, et al. Symbiotic bacteria appear to mediate hyena social odors. Proc Natl Acad Sci USA. 2013;110(49):19832–19837. doi: 10.1073/pnas.1306477110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wyatt TD. Proteins and peptides as pheromone signals and chemical signatures. Anim Behav. 2014;97:273–280. [Google Scholar]
- 42.Gentry RL, Holt JR. National Oceanic and Atmospheric Administration Technical Report, ed Sindermann CJ. National Oceanic and Atmospheric Administration; Seattle, WA: 1982. Equipment and techniques for handling Northern fur seals. [Google Scholar]
- 43.Hoffman JI, Boyd IL, Amos W. Male reproductive strategy and the importance of maternal status in the Antarctic fur seal Arctocephalus gazella. Evolution. 2003;57(8):1917–1930. doi: 10.1111/j.0014-3820.2003.tb00598.x. [DOI] [PubMed] [Google Scholar]
- 44.Raymond M, Rousset F. Genepop (Version 1.2)—Population genetics software for exact tests of ecumenicism. J Hered. 1995;86(3):248–249. [Google Scholar]
- 45.Storey JD. A direct approach to false discovery rates. J R Stat Soc, B. 2002;64(3):479–498. [Google Scholar]
- 46.Jones OR, Wang J. COLONY: A program for parentage and sibship inference from multilocus genotype data. Mol Ecol Resour. 2010;10(3):551–555. doi: 10.1111/j.1755-0998.2009.02787.x. [DOI] [PubMed] [Google Scholar]
- 47.Wang J. COANCESTRY: A program for simulating, estimating and analysing relatedness and inbreeding coefficients. Mol Ecol Resour. 2011;11(1):141–145. doi: 10.1111/j.1755-0998.2010.02885.x. [DOI] [PubMed] [Google Scholar]
- 48.Queller DC, Goodnight KF. Estimating relatedness using molecular markers. Evolution. 1989;43(2):258–275. doi: 10.1111/j.1558-5646.1989.tb04226.x. [DOI] [PubMed] [Google Scholar]
- 49.Coltman DW, et al. Parasite-mediated selection against inbred Soay sheep in a free-living, island population. Evolution. 1999;53(4):1259–1267. doi: 10.1111/j.1558-5646.1999.tb04538.x. [DOI] [PubMed] [Google Scholar]
- 50.David P, Pujol B, Viard F, Castella V, Goudet J. Reliable selfing rate estimates from imperfect population genetic data. Mol Ecol. 2007;16(12):2474–2487. doi: 10.1111/j.1365-294X.2007.03330.x. [DOI] [PubMed] [Google Scholar]
- 51.Pritchard JK, Stephens M, Donnelly P. Inference of population structure using multilocus genotype data. Genetics. 2000;155(2):945–959. doi: 10.1093/genetics/155.2.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sun L, Muller-Schwarze D. Anal gland secretion codes for family membership in beaver. Behav Ecol Sociobiol. 1998;44(3):199–208. [Google Scholar]
- 53.Clarke KR. Nonmetric multivariate analysis in community-level ecotoxicology. Environ Toxicol Chem. 2009;18(2):118–127. [Google Scholar]
- 54.Dixon P. VEGAN, a package of R functions for community ecology. J Veg Sci. 2003;14(6):927–930. [Google Scholar]
- 55.Cattel RB. The scree tests for the number of factors. Multivariate Behav Res. 1966;1(2):245–276. doi: 10.1207/s15327906mbr0102_10. [DOI] [PubMed] [Google Scholar]
- 56.Crawley MJ. Statistical Computing, an Introduction to Data Analysis Using S-plus. John Wiley and Sons; Chichester, UK: 2002. [Google Scholar]
- 57.Goslee SC, Urban DL. The ecodist package for dissimilarity-based analysis of ecological data. J Stat Softw. 2007;22(7):1–19. [Google Scholar]
- 58.Clarke KR, Warwick RM. Change in Marine Communities: An Approach to Statistical Analysis and Interpretation. 2nd Ed Plymouth Marine Laboratory; Plymouth, UK: 2001. [Google Scholar]
- 59.Taylor M. 2014 sinkr: A collection of functions featured on the blog 'Me nugget'. R package version 1.0. Available at https://github.com/marchtaylor/sinkr. Accessed July 2, 2015.
- 60.Kováts E. Gas-chromatogrpahische characterisierung organischer verbindungen Teil 1: Retentionsindices aliphatischer halogenoide, alkohole aldehyde und ketone. Helv Chim Acta. 1958;41(7):1915–1932. [Google Scholar]
- 61.Allen PJ, Amos W, Pomeroy PP, Twiss SD. Microsatellite variation in grey seals (Halichoerus grypus) shows evidence of genetic differentiation between two British breeding colonies. Mol Ecol. 1995;4(6):653–662. doi: 10.1111/j.1365-294x.1995.tb00266.x. [DOI] [PubMed] [Google Scholar]
- 62.Coltman DW, Bowen WD, Wright JM. PCR primers for harbour seal (Phoca vitulina concolour) microsatellites amplify polymorphic loci in other pinniped species. Mol Ecol. 1996;5(1):161–163. doi: 10.1111/j.1365-294x.1996.tb00303.x. [DOI] [PubMed] [Google Scholar]
- 63.Gemmell NJ, Allen PJ, Goodman SJ, Reed JZ. Interspecific microsatellite markers for the study of pinniped populations. Mol Ecol. 1997;6(7):661–666. doi: 10.1046/j.1365-294x.1997.00235.x. [DOI] [PubMed] [Google Scholar]
- 64.Goodman SJ. Dinucleotide repeat polymorphisms at seven anonymous microsatellite loci cloned from the European harbour seal (Phoca vitulina vitulina) Anim Genet. 1997;28(4):310–311. [PubMed] [Google Scholar]
- 65.Buchanan FC, Maiers LD, Thue TD, De March BG, Stewart RE. Microsatellites from the Atlantic walrus Odobenus rosmarus rosmarus. Mol Ecol. 1998;7(8):1083–1085. doi: 10.1046/j.1365-294x.1998.00401.x. [DOI] [PubMed] [Google Scholar]
- 66.Hoelzel AR, et al. Alpha-male paternity in elephant seals. Behav Ecol Sociobiol. 1999;46:298–306. [Google Scholar]
- 67.Davis CS, et al. Dinucleotide microsatellite markers from the Antarctic seals and their use in other pinnipeds. Mol Ecol Notes. 2002;2:203–208. [Google Scholar]
- 68.Hernandez-Velazquez F, et al. New polymorphic microsatellite markers for California sea lions (Zalophus californianus) Mol Ecol Notes. 2005;5:140–142. [Google Scholar]
- 69.Wolf JBW, et al. Development of new microsatellite loci and evaluation of loci from other pinniped species for the Galapagos sea lion (Zalophus californianus wollebaeki) Conserv Genet. 2006;7:461–465. [Google Scholar]
- 70.Huebinger RM, et al. Characterization of eight microsatellite loci in Steller sea lions (Eumetopias jubatus) Mol Ecol Notes. 2007;7:1097–1099. [Google Scholar]
- 71.Hoffman JI, et al. Ten novel dinucleotide microsatellite loci cloned from the Galápagos sea lion (Zalophus californianus wollebaeki) are polymorphic in other pinniped species. Mol Ecol Notes. 2007;7:103–105. [Google Scholar]
- 72.Hoffman JI, Dasmahapatra KK, Nichols HJ. PERMANENT GENETIC RESOURCES: Ten novel polymorphic dinucleotide microsatellite loci cloned from the Antarctic fur seal Arctocephalus gazella. Mol Ecol Resour. 2008;8(2):459–461. doi: 10.1111/j.1471-8286.2007.01993.x. [DOI] [PubMed] [Google Scholar]
- 73.Hoffman JI. A panel of new microsatellite loci for genetic studies of Antarctic fur seals and other otariids. Conserv Genet. 2009;10:989–992. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.