Significance
To our knowledge, this is the first detailed study of Golgi ultrastructure within unperturbed cells. Three intracisternal structures were identified, with implications for Golgi architecture and trafficking: (i) Bundles of filaments show how cargoes may oligomerize to increase their local concentration at trans-Golgi buds. (ii) Granular aggregates provide evidence for cisternal maturation, as they are likely too large to transit the Golgi via vesicles. (iii) Protein arrays link the membranes of the central trans-Golgi cisternae, simultaneously maintaining the narrow luminal spacing while promoting cargo exit from the Golgi periphery by excluding material from the center. The asymmetry of the array structure indicates that the apposing membranes of a single cisterna have distinct compositions. The assembly of arrays may also enhance glycosyltransferase kinetics.
Keywords: focused ion beam, cryo-electron tomography, Chlamydomonas, Golgi, glycosyltransferase
Abstract
We acquired molecular-resolution structures of the Golgi within its native cellular environment. Vitreous Chlamydomonas cells were thinned by cryo-focused ion beam milling and then visualized by cryo-electron tomography. These tomograms revealed structures within the Golgi cisternae that have not been seen before. Narrow trans-Golgi lumina were spanned by asymmetric membrane-associated protein arrays that had ∼6-nm lateral periodicity. Subtomogram averaging showed that the arrays may determine the narrow central spacing of the trans-Golgi cisternae through zipper-like interactions, thereby forcing cargo to the trans-Golgi periphery. Additionally, we observed dense granular aggregates within cisternae and intracisternal filament bundles associated with trans-Golgi buds. These native in situ structures provide new molecular insights into Golgi architecture and function.
Cryo-electron tomography (cryo-ET) provides the unique ability to visualize macromolecules and supramolecular structures within frozen hydrated cells (1–4). Biological material is immobilized in vitreous ice, preserving cellular structures in a near-native state. Compression-free thinning of these frozen samples by cryo-focused ion beam (cryo-FIB) milling offers unparalleled access to the cellular interior (5–7). The recent combination of cryo-FIB with the improved image quality of direct detection cameras has opened new frontiers for in situ structural biology, enabling the study of how molecular complexes establish cellular architecture.
The relationship between Golgi structure and function has been intensely debated since the first electron microscopy observations of this alluring organelle (8, 9). Over the last two decades, electron tomography of plastic sections has been applied extensively to characterize Golgi morphology within animals, plants, and single-celled organisms, including yeast and algae (10–17). Three-dimensional views of fenestrated, interconnected cisterna stacks interacting with a constellation of coated vesicles led to revised models of how Golgi structure directs cargo sorting through the organelle (18–20). However, these tomographic studies were restricted to descriptions of membrane architecture and, in the best cases, the classification of membrane coats, due to the resolution limitations imposed by conventional sample preparation, involving dehydration, plastic embedding, and staining with heavy-metal contrasting agents. To date, cryo-ET studies of the Golgi have been extremely limited (2, 21–23).
In this study, we used cryo-FIB of vitreous Chlamydomonas cells followed by cryo-ET to image the native molecular landscape of the Golgi with unprecedented resolution and sample integrity. Our tomograms revealed new structures within the Golgi cisternae, including ordered membrane-associated protein arrays, dark granular aggregates, and bundles of filaments near the trans-Golgi coated buds.
Results and Discussion
Trans-Golgi Intracisternal Protein Arrays with an Alternating Repeat.
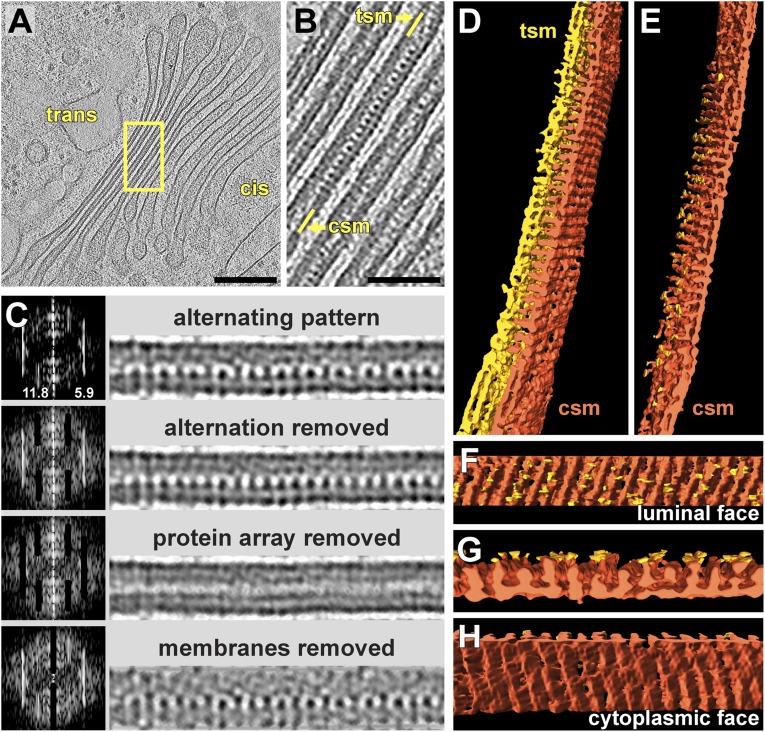
The Chlamydomonas Golgi has a characteristic morphology, with approximately nine parallel, disk-shaped cisternae (Fig. 1A) (19, 24). The centers of the four to five trans-most cisternae drastically narrow to a uniform thickness of 18–19 nm (Fig. 1 A and B). Within these narrow trans-cisternae, we often saw regularly spaced rows of luminal proteins that had lateral repeats of 5.5–6.8 nm (Fig. S1A). The longest and best resolved of these protein arrays (Fig. 1 B–H) was composed of alternating long and short luminal projections that appeared to be embedded within the membrane on the cis-side of the cisterna. The long projections spanned the entire width of the cisterna lumen, whereas the short projections only extended halfway across the lumen. A Fourier transform of the array revealed clear power spectrum signals for a 5.9-nm lateral repeat for all of the projections and an 11.8-nm lateral repeat for the alternation between short and long projections (Fig. 1C). Masking the 5.9-nm and 11.8-nm peaks in Fourier space removed the corresponding features from the real-space images.
Fig. 1.
Intracisternal membrane-associated protein arrays in the trans-Golgi. (A) Overview and (B) close-up tomogram slices showing a Golgi with an intracisternal protein array within the third cisterna from the left. B is enlarged from the box in A. tsm, trans-side membrane; csm, cis-side membrane. (C) Fourier analysis of the protein array. For each row, the frequency image (power spectrum) is on the Left, and the corresponding real-space image is on the Right. (Top row) Original image, rotated from B. (Second row) Masking the 11.8-nm peak in the power spectrum removes the alternation between long and short proteins projecting into the lumen. (Third row) Masking both the 11.8-nm and 5.9-nm peaks removes the protein array altogether. (Bottom row) Masking the central peak removes the cisterna’s lipid bilayers, but densities remain within the cis-side bilayer region that may be the array’s transmembrane domains. (D–H) Threshold-based segmentation of the cisterna containing the protein array. (D) Segmentation in the same orientation as B, separated into the cisterna’s trans-side (yellow) and cis-side (dark orange). (E) Segmentation with the trans-side removed, revealing ordered rows of luminal projections. (F) Orthogonal view of the cis-side’s luminal face. (G) Side view showing alternating rows of small projections that do not contact the trans-side and long projections with multiple contact sites (yellow). (H) View of the array’s cytoplasmic face, with ridges that correspond to the positions of the luminal projections. (Scale bars, 200 nm in A; 50 nm in B.)
Fig. S1.
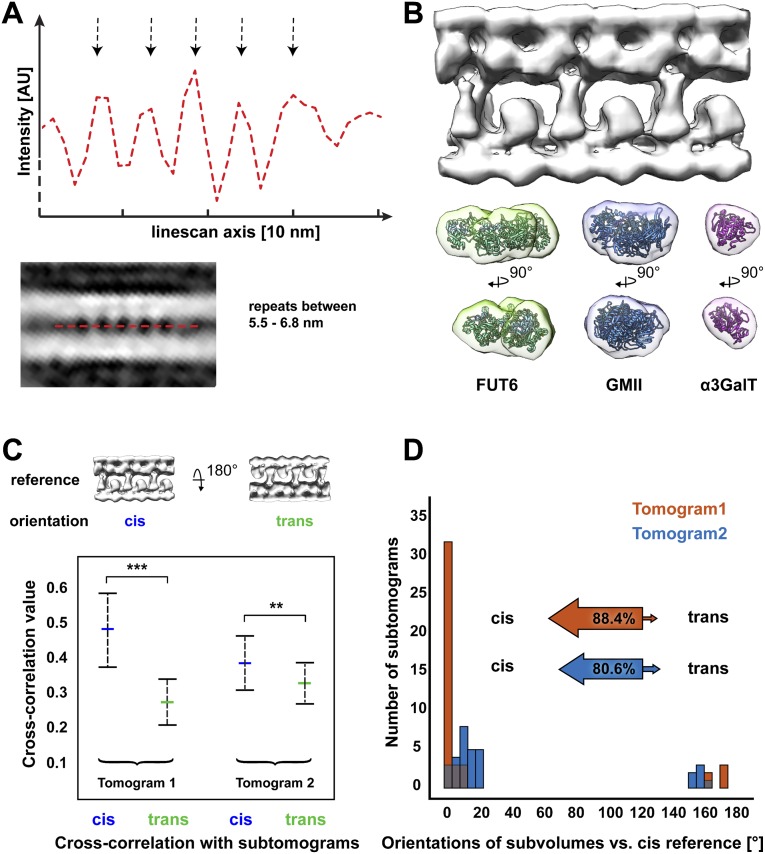
(A) Subtomogram average of extracted array structures from tomograms 2–8 (194 subvolumes), which were not used to generate the structure in Fig. 3. Mass is shown in white. A linescan through the central xy slice reveals an average lateral repeat in the range of 5.5–6.8 nm. This is consistent with the 5.9-nm repeat of the highly ordered array in tomogram 1 (Figs. 1 and 3). Arrows: intensity peak positions from the linescan corresponding to luminal densities in the average. (B) The subtomogram average of the array in tomogram 1 (Fig. 3) is displayed to scale with the crystal structures of three glycosyltransferase luminal catalytic domains, fucosyltransferase FUT6 (PDB: 2NZW) (78), mannosidase GMII (PDB: 1HTY) (79), and galactosyltransferase α3GaIT (PDB: 1G8O) (80). These structures are comparable in size with the luminal densities identified in the array average. (C) Cross-correlation values were calculated by comparing the reference structure (from Fig. 3) in the cis and trans orientations to the individual subtomograms from tomogram 1 (containing the highly ordered array, Figs. 1 and 3) and tomogram 2 (another high quality tomogram, Fig. 2). The arrays from both tomograms show a high degree of asymmetry, favoring the cis orientation- tomogram 1 at a significance level of 0.1% (***) and tomogram 2 at 1% (**). Error bars, SD. (D) Histograms showing the final orientations of each subvolume relative to the cis reference used for alignment; 88.4% of the arrays from tomogram 1 (red) and 80.6% of the arrays from tomogram 2 (blue) were in the cis orientation (larger projections from the cis-sides of the cisternae), the same orientation as the highly ordered array shown in Figs. 1 and 3.
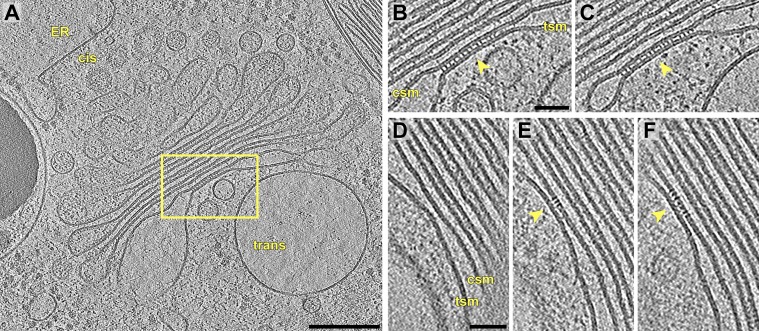
Threshold-based segmentation of the cisterna volume showed that the array’s projections formed extended rows in the z dimension along the luminal face of the cis-side membrane (Fig. 1 D–F). The long projections made frequent contacts with the cisterna’s trans-side, whereas the short projections terminated before reaching the trans-side (Fig. 1 F and G). In addition, the segmentation revealed rows of density on the cytoplasmic face of the cis-side membrane that were aligned with the luminal projections (Fig. 1 D and H). These cytoplasmic densities were likely connected to the luminal projections by transmembrane domains (Fig. 1C, Bottom). The protein arrays were only found within cisternae with uniform narrow lumina. In fact, we observed the formation of arrays exactly at the location where the cisterna membranes started to come into close apposition (Fig. 2). Thus, it is likely that the arrays perform bridging interactions between the cisterna membranes to determine the narrow luminal spacing of the trans-Golgi.
Fig. 2.
Protein arrays likely maintain the trans-Golgi’s narrow luminal spacing. (A) Overview and (B and C) two sequential slices through a tomogram showing a protein array (arrows) that is exclusively located where the trans-most cisterna’s membranes are closely apposed. B and C correspond to the box in A. (D–F) Three sequential slices through another tomogram showing a protein array that is only found where the trans-most cisterna’s membranes are closely apposed. tsm, trans-side membrane, csm, cis-side membrane. (Scale bars, 200 nm in A; 50 nm in B–F.)
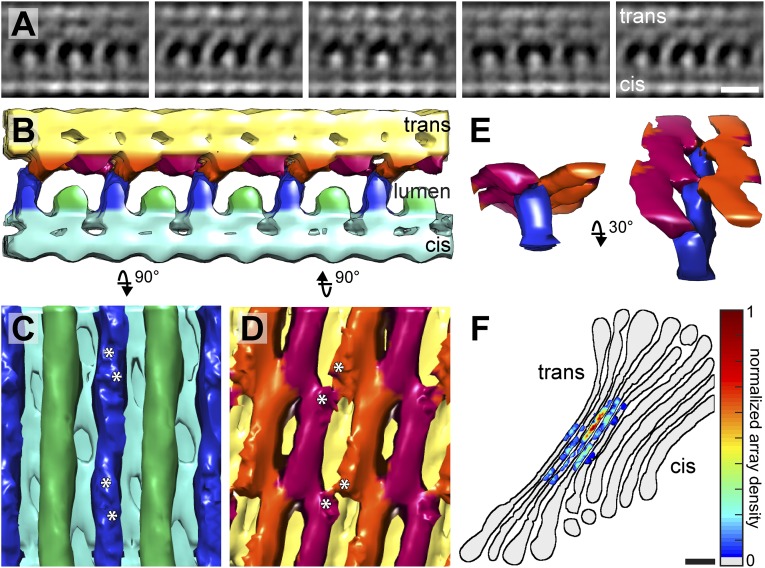
Subtomogram averaging displayed the structure of this large protein array in greater detail, revealing additional luminal projections from the trans-side membrane (Fig. 3 A–E and Movie S1, EMDB entry number: 3100). From cis to trans, the side view of the symmetrized average clearly showed 1-nm densities on the cytoplasmic face of the cis-side membrane, a 5-nm cis-side bilayer with embedded proteins, alternating long 5.5-nm and short 3.5-nm luminal projections from the cis-side bilayer, 2.5-nm luminal projections from the trans-side bilayer that interact with the long luminal cis-side projections, and a 5.5-nm trans-side bilayer with embedded proteins. The total cisterna width of the average, including the 1-nm cytoplasmic densities, was 19.5 nm. Template matching confirmed that the protein arrays were restricted to the narrow cisternae of the trans-Golgi, occupying 37% of the narrow cisterna surface area in the examined tomogram (Fig. 3F).
Fig. 3.
Subtomogram average and localization of the intracisternal protein arrays. (A–E) Symmetrized subtomogram average calculated from the array in Fig. 1. (A) Sequential side view slices every 3.4 nm through the average, showing the array’s periodicity and asymmetric structure. Mass is shown in white. (B) Segmentation of the average into the following regions: cis-side bilayer and embedded proteins (cyan), trans-side bilayer and embedded proteins (yellow), long 5.5-nm and short 3.5-nm luminal projections from the cis-side bilayer (blue and green, respectively), and 2.5-nm luminal projections from the trans-side bilayer (magenta and orange). (C) View of the cis-side’s luminal surface, rotated 90° from B, showing alternating rows of short and long projections. (D) View of the trans-side’s luminal surface, rotated 90° from B, showing an interlocking network of projections. Asterisks in C and D: contact sites between the cis-side and trans-side projections. The periodicity of contacts along the rows of projections is 10.3 nm. (E) Side views showing how the rows of long cis-side projections form y-shaped zipper-like interactions with the rows of trans-side projections. (F) Heat map displaying the density of protein arrays (gradient of gray to red, determined by template matching) within the Golgi from Fig. 1 (black outlines). (Scale bars, 10 nm in A; 50 nm in F.)
The most striking feature of the subtomogram average is the asymmetry of the structures linking the cisterna membranes (Fig. S1 C and D). While straight rows of alternating long and short proteins project from the cis-side bilayer (Fig. 3C), the projections from the trans-side bilayer are all of the same height and form an interlocking meshwork of short rows that are tilted by 10°–15° relative to the cis-side projections (Fig. 3D). The lateral periodicity of the trans-side matches that of the cis-side, as the alternating long cis-side projections contact every other junction in the network of trans-side projections (Fig. 3 B–D). Together, these cis- and trans-side projections form y-shaped structures that bridge the cisterna membranes (Fig. 3E). The y-shaped structures repeat laterally every 11.8 nm and also repeat every 10.3 nm along the rows of projections. Thus, the protein arrays appear to form asymmetric zipper-like interactions that propagate in two dimensions to link the cisterna membranes together. Although several symmetrical zippers are known to hold membranes together, including connexin-mediated gap junctions (25), myelin (26, 27), and aquaporin-0 (28), this is one of the first reported examples of an asymmetric membrane zipper. Other known asymmetric membrane adhesion interactions, such as neuronal and immunological synapses (29, 30), are likely less ordered than the arrays described in our study. While the specialization of different cisternae has been well characterized, the assembly of asymmetric intracisternal arrays indicates that the two membranes of each cisterna also have distinct compositions. It will be important to understand how this asymmetry is established and maintained.
Intercisternal linkers have been seen in a variety of cell types, including the alga Scherffelia dubia (19), the roots of maize (31–33), and rat liver (34). In contrast, to the best of our knowledge, intracisternal linkers have only been previously described in chemically-fixed plastic sections of Clivia and Lilium pollen tubes (35). However, these structures are difficult to discern and have received scrutiny due to the lack of supporting studies reporting similar structures and artifacts that can arise from traditional sample preparation methods. It has been proposed that intracisternal linkers may not be necessary for maintaining closely apposed cisterna membranes, as acidification of the trans-Golgi could provide an osmotic mechanism for compressing the cisternae (36).
The array structure identified in our study provides compelling evidence that intracisternal linkers are indeed present within the narrow trans-Golgi cisternae of Chlamydomonas. The array’s longer cis-side projections span the entire luminal space (Fig. 1) and make zipper-like contacts with the trans-side projections, linking the membranes together (Fig. 3). We propose that whereas osmotic forces may indeed compress the central regions of the trans-cisternae, once the membranes come into close apposition, the arrays link the membranes to maintain the narrow luminal spacing (Fig. 2). There may be positive feedback where the arrays help bring the membranes together, while the closely apposed membranes enable the arrays to form.
One interpretation of the trans-Golgi arrays is that they might be composed of glycosyltransferases, which add sugars to the hydroxyproline-rich glycoproteins that are trafficked to the cell wall (37). The structures of these enzymes are reasonably compatible with the array structure (Fig. S1B). Glycosyltransferases are type II transmembrane proteins consisting of a small cytoplasmic domain, a single transmembrane domain, a stem region, and a large catalytic domain in the Golgi lumen. These enzymes may become arrayed either by lateral interactions or by binding the same long glycoprotein substrates. Given the highly ordered array structure, the former hypothesis seems more plausible. Indeed, many Golgi-resident glycosyltransferases form homo-oligomeric and hetero-oligomeric complexes via interactions between their luminal domains (38–42). The hetero-oligomerization of two mammalian glycosyltransferases, EXT1 and EXT2, was shown to be required for both the Golgi localization and increased activity of the enzymes (38). Interestingly, our array structure also appears to be a hetero-oligomer.
If the arrays are indeed composed of glycosyltransferases, the interaction with the opposite cisterna membrane revealed in our tomograms must be explained. By oligomerizing into arrays, the glycosyltransferases may accomplish two synergistic functions. Linking the cisterna membranes to maintain a narrow lumen could increase the local substrate concentration and thus accelerate the kinetics of the glycosyltransferase reactions. Simultaneously, narrow lumina occupied by arrays would exclude larger complexes, forcing mature cargo, including the glycoproteins assembled by the glycosyltransferase arrays, to the cisterna periphery for Golgi exit. Ultimately, definitive answers will require molecular identification of the protein array components and subsequent deletion and complementation experiments to monitor changes in Golgi architecture and function.
Intracisternal Filaments Associated with trans-Golgi Buds.
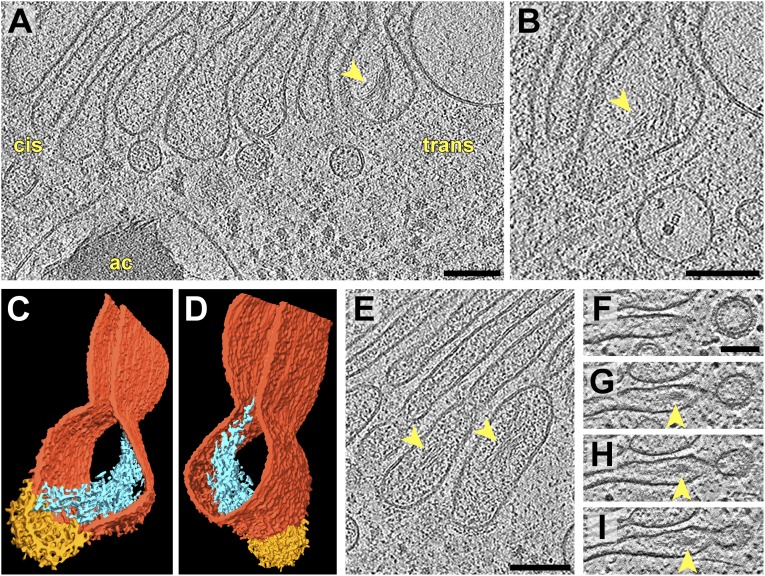
We frequently observed fine filaments (2–3 nm diameter) bundled within the cisternae of the trans-Golgi (Fig. 4 and Movie S2), most commonly within the final trans-cisterna (Fig. 4 A–I) but also in the penultimate cisterna (Fig. 4E). The bundles were always located within the swollen trans-Golgi periphery, with their filaments aligned roughly perpendicular to the membranes of nearby COPI-coated buds. Although the coalescence of cargo proteins at trans-Golgi buds has been previously observed (24, 33, 43), to the best of our knowledge, the only filamentous structure that has been described in the peripheral trans-Golgi is animal procollagen (44, 45).
Fig. 4.
Intracisternal filament bundles near COPI buds in the trans-Golgi. (A and B) Two sequential slices through a tomogram showing a filament bundle (arrows) within the trans-most cisterna, close to a coated bud. ac, an acidocalcisome containing a dense aggregate of polyphosphate filaments. In B, an actin filament can be seen below the cisterna, along with a vesicle surrounding a complex that resembles a 20S proteasome. (C and D) Segmentation of the cisterna from A and B showing the Golgi membrane (dark orange), the coated bud (yellow), and the filament bundle (blue). The view in D is flipped 180° from C. (E) A slice from a second tomogram showing filament bundles within the last two trans-Golgi cisterna, close to coated buds. Cis-Golgi is toward the left, trans-Golgi is toward the right. (F–I) Four sequential slices from a third tomogram showing a filament bundle within the terminal cisterna of the trans-Golgi. The bundle is adjacent to a coated bud that is undergoing scission to become a vesicle. Cis-Golgi is up, trans-Golgi is down. This tomogram was acquired with a Volta phase plate (68) to enhance contrast. (Scale bars, 100 nm.)
While the Chlamydomonas trans-Golgi filaments may be glycoproteins that are secreted to the cell wall, another possibility is that they are targeted to the dense granules of similar filaments in nearby vacuoles (“ac” in Fig. 4A). These vacuoles, called acidocalcisomes, are acidified compartments that store polyphosphate complexed with high concentrations of cations, including calcium, iron, zinc, and copper (46–48). Thus, an alternative candidate for the trans-Golgi filaments is polyphosphate, the major nonorganic component of acidocalcisomes (49–51). These filament bundles could also contain the enzyme polyphosphate kinase 2, which forms actin-like fibers concurrent with polyphosphate synthesis (52). Similar acidocalcisome compartments can be found in other single-celled eukaryotes, including trypanosomes, apicomplexans, and Dictyostelium, as well as evolutionarily distant organisms such as bacteria and humans (53). The mechanisms of polyphosphate traffic and storage may have clinical implications, as platelets release polyphosphate to stimulate blood coagulation (54, 55).
Dark Luminal Aggregates Within Golgi Cisternae.
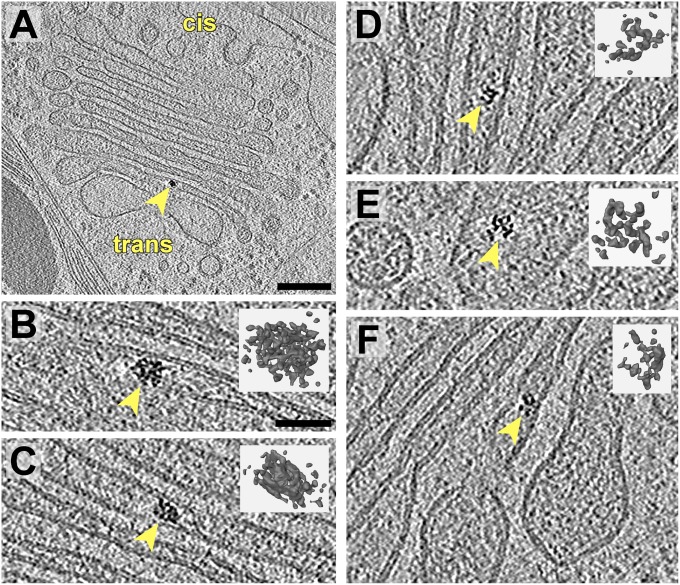
In the majority of our tomograms, we observed one or two dark granular aggregates within the Golgi cisternae (Fig. 5). These structures ranged from 15 to 30 nm in diameter and were composed of smaller 3–5-nm particles. The aggregates were found throughout the cis- and trans-Golgi, positioned toward the centers (Fig. 5 A–C) and periphery (Fig. 5 D–F) of cisternae. However, they were never found within the narrow central lumina of the trans-Golgi. The exceptionally high contrast of the aggregates indicates that they may contain compounds such as phosphate or metals.
Fig. 5.
Dark aggregates within Golgi cisterna. (A) Overview slice from a tomogram showing a Golgi with a dark intracisternal aggregate (arrow). A COPII bud can be seen emanating from the ER. This is a more peripheral Golgi section compared with Figs. 1 and 2. (B–F) Slices from several tomograms showing close-ups of intracisternal aggregates and corresponding segmentations in the same orientations (Insets). B is a magnified view of the aggregate in A. (Scale bars, 200 nm in A; 50 nm in B–F.)
A pertinent question is whether these aggregates perform a biological task or whether they are accumulations of nonfunctional material. Protein aggregates have been shown to traffic through the Golgi en route to functional activity elsewhere inside or outside the cell. A well-characterized example is the transport of procollagen aggregates in mammalian cells, which helped prove the validity of the cisternal maturation model (56–58). The aggregates in our tomograms were more similar in appearance to the developing scales of the green alga Scherffelia dubia (17, 59, 60), which also traverse the Golgi by cisternal maturation before they are secreted to the cell wall and flagella (61). However, the Chlamydomonas cell wall is not composed of scales but rather a fibrillar meshwork of hydroxyproline-rich glycoproteins (62, 63). Nonetheless, granular structures are visible in some layers of the Chlamydomonas cell wall (64, 65). Insights into the identity of the Golgi aggregates could be gained by correlating their abundance with conditions that increase cell wall secretion, such as synchronized growth-phase cultures and recovery from treatment with the autolysin enzyme, which removes the cell wall (66, 67).
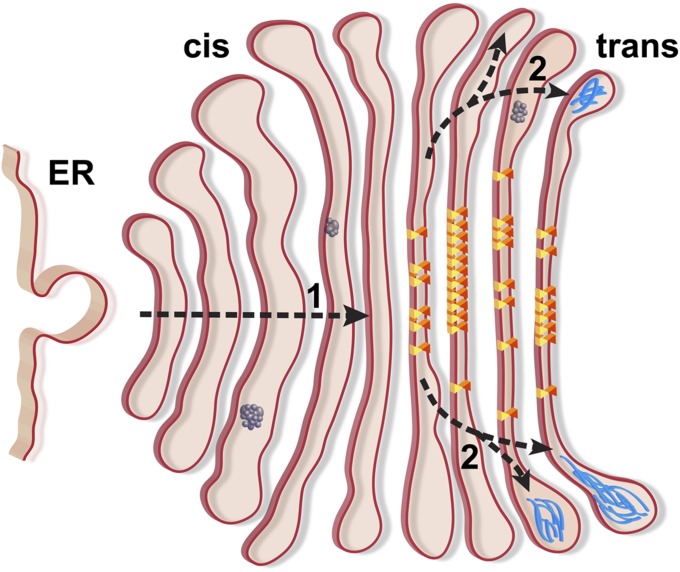
The three intracisternal structures revealed in this study by in situ cryo-ET provide insights into the mechanisms of cargo transport through the Golgi (Fig. 6). The large granular aggregates likely transit the Golgi via cisternal maturation, providing evidence for this mechanism in Chlamydomonas. The bundles of filaments near the trans-Golgi buds illustrate how specific cargo may accumulate via oligomerization to facilitate Golgi exit. Finally, the membrane-linking protein arrays likely contribute to the narrow central spacing of the trans-Golgi cisternae, thereby forcing cargo toward the bud sites at the trans-Golgi periphery. Future studies will focus on the molecular identification of these intracisternal structures to characterize how their modification affects Golgi architecture and function.
Fig. 6.
Model depicting how the intracisternal structures identified in this study may relate to traffic through the Golgi. Protein arrays (yellow and orange triangles), filament bundles (blue), and granular aggregates (dark gray) are illustrated alongside the proposed route of cargo transit (dashed arrows). (1) The aggregates likely proceed through the Golgi by cisternal maturation, although multiple transport mechanisms are possible for smaller cargo. (2) In the trans-Golgi, the protein arrays exclude cargo from the centers of cisternae, promoting cargo exit from the periphery.
Materials and Methods
A detailed description of cryo-FIB sample preparation, cryo-ET, and image analysis is found in SI Materials and Methods.
SI Materials and Methods
Cell Culture.
The Chlamydomonas reinhardtii mat3-4 strain CC-3994 (69) was acquired from the Chlamydomonas Resource Center (University of Minnesota) and grown with constant light and normal atmosphere aeration in Tris-acetate-phosphate (TAP) medium. The small size of mat3-4 cells facilitated complete vitrification by plunge-freezing.
Vitrification and cryo-FIB Milling.
Plunge-freezing and cryo-focused ion beam milling were performed as previously described (3, 70). Using a Vitrobot Mark 4 (FEI), cells were blotted onto carbon-coated 200-mesh copper grids (Quantifoil Micro Tools), which were immediately plunged into a liquid ethane/propane mixture. Vitrified grids were mounted into modified Autogrids (FEI), which were loaded either into a Scios FIB/SEM dual-beam microscope (FEI) or onto a custom-made 360° rotatable cryo-stage (6, 71) in a Quanta 3D FEG FIB/SEM dual-beam microscope (FEI). After coating samples with organometallic platinum (72) using the in situ gas injection system (GIS, FEI), thin lamellas were milled using the Ga+ ion beam at 30 kV and a shallow 8°–12° angle.
Cryo-ET.
Cryo-electron tomography was performed on a 300-kV Titan Krios microscope (FEI) with a postcolumn energy filter (968 Quantum, Gatan) and a direct detection camera (Gatan, K2 Summit). Low-dose tilt-series acquisition (<100 e/Å2 cumulative dose) was controlled by SerialEM software (73) with 2° tilt increments, −5 μm defocus, and 14,600× magnification (pixel size of 3.4 Å). The tomogram in Fig. 4 F–I was acquired with a Volta phase plate (68), providing enhanced contrast.
Tomogram Reconstruction and Segmentation.
IMOD software (74) was used for patch tracking alignment and weighted back-projection reconstruction of the tomograms. Fourier analysis of the intracisternal protein array was performed with Fiji (ImageJ, National Institutes of Health). Tomogram segmentation was performed with Amira software (FEI Visualization Sciences Group).
Template Matching and Subtomogram Averaging.
Template matching was performed with PyTom software (75). Tomographic volumes were searched with a low-pass–filtered average of the protein array structure, cross-correlation peaks were extracted, and corresponding subtomograms were generated. The initial template was a single subvolume containing two cisterna membranes connected by a protein array, whereas later iterations used a subtomogram average of the protein array as a template. The extracted subtomograms were visually checked in Matlab (MathWorks) with TOM software (76) and then aligned and averaged in real-space with PyTom.
The subtomogram average shown in Fig. 3 was calculated using 244 subvolumes from a single tomogram. Peaks in the autocorrelation of the subtomogram average revealed the periodicity of the subvolume. This information was then used to generate a subtomogram average with translational symmetry in both the x and y dimensions, which is displayed in Fig. 3 A–E. The average was rendered in 3D with UCSF Chimera (77) and segmented with Amira. Subvolumes from seven tomograms were used to produce the subtomogram average in Fig. S1A, which shows the average lateral repeat of all other arrays in our dataset.
The heat map in Fig. 3F was determined by template matching. At every position with a positive hit, a sphere with a diameter smaller than the subtomogram size was inserted into a volume and masked with a 3D segmentation of the Golgi cisternae. The result was then projected along the z axis and subsequently normalized. To ease visualization, this projection was overlaid on an outline of the central slice of the Golgi.
Supplementary Material
Acknowledgments
We thank L. Andrew Staehelin, Vivek Malhotra, Martin Jonikas, William Snell, Jan Arnold, and Karin Engel for their insightful advice and critical reading of the manuscript. This work was supported by an Alexander von Humboldt Foundation postdoctoral fellowship (to B.D.E.), the European Commission grant agreement ERC-2012-SyG_318987-ToPAG, the Deutsche Forschungsgemeinschaft Excellence Clusters CIPSM and SFB 1035, and the Max Planck Society.
Footnotes
The authors declare no conflict of interest.
Data deposition: The data reported in this paper has been deposited in the Electron Microscopy Data Bank (EMDB), www.emdatabank.org (accession no. EMD-3100).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1515337112/-/DCSupplemental.
References
- 1.Medalia O, et al. Macromolecular architecture in eukaryotic cells visualized by cryoelectron tomography. Science. 2002;298(5596):1209–1213. doi: 10.1126/science.1076184. [DOI] [PubMed] [Google Scholar]
- 2.Leforestier A, Lemercier N, Livolant F. Contribution of cryoelectron microscopy of vitreous sections to the understanding of biological membrane structure. Proc Natl Acad Sci USA. 2012;109(23):8959–8964. doi: 10.1073/pnas.1200881109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Engel BD, et al. Native architecture of the Chlamydomonas chloroplast revealed by in situ cryo-electron tomography. eLife. 2015;4:e04889. doi: 10.7554/eLife.04889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Asano S, et al. Proteasomes. A molecular census of 26S proteasomes in intact neurons. Science. 2015;347(6220):439–442. doi: 10.1126/science.1261197. [DOI] [PubMed] [Google Scholar]
- 5.Marko M, Hsieh C, Schalek R, Frank J, Mannella C. Focused-ion-beam thinning of frozen-hydrated biological specimens for cryo-electron microscopy. Nat Methods. 2007;4(3):215–217. doi: 10.1038/nmeth1014. [DOI] [PubMed] [Google Scholar]
- 6.Rigort A, et al. Focused ion beam micromachining of eukaryotic cells for cryoelectron tomography. Proc Natl Acad Sci USA. 2012;109(12):4449–4454. doi: 10.1073/pnas.1201333109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Villa E, Schaffer M, Plitzko JM, Baumeister W. Opening windows into the cell: Focused-ion-beam milling for cryo-electron tomography. Curr Opin Struct Biol. 2013;23(5):771–777. doi: 10.1016/j.sbi.2013.08.006. [DOI] [PubMed] [Google Scholar]
- 8.Dalton AJ, Felix MD. A comparative study of the Golgi complex. J Biophys Biochem Cytol. 1956;2(4) Suppl:79–84. doi: 10.1083/jcb.2.4.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Farquhar MG, Palade GE. The Golgi apparatus: 100 years of progress and controversy. Trends Cell Biol. 1998;8(1):2–10. doi: 10.1016/S0962-8924(97)01187-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ladinsky MS, Mastronarde DN, McIntosh JR, Howell KE, Staehelin LA. Golgi structure in three dimensions: Functional insights from the normal rat kidney cell. J Cell Biol. 1999;144(6):1135–1149. doi: 10.1083/jcb.144.6.1135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Marsh BJ, Mastronarde DN, Buttle KF, Howell KE, McIntosh JR. Organellar relationships in the Golgi region of the pancreatic beta cell line, HIT-T15, visualized by high resolution electron tomography. Proc Natl Acad Sci USA. 2001;98(5):2399–2406. doi: 10.1073/pnas.051631998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ladinsky MS, Wu CC, McIntosh S, McIntosh JR, Howell KE. Structure of the Golgi and distribution of reporter molecules at 20 degrees C reveals the complexity of the exit compartments. Mol Biol Cell. 2002;13(8):2810–2825. doi: 10.1091/mbc.01-12-0593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Marsh BJ, Volkmann N, McIntosh JR, Howell KE. Direct continuities between cisternae at different levels of the Golgi complex in glucose-stimulated mouse islet beta cells. Proc Natl Acad Sci USA. 2004;101(15):5565–5570. doi: 10.1073/pnas.0401242101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mogelsvang S, Gomez-Ospina N, Soderholm J, Glick BS, Staehelin LA. Tomographic evidence for continuous turnover of Golgi cisternae in Pichia pastoris. Mol Biol Cell. 2003;14(6):2277–2291. doi: 10.1091/mbc.E02-10-0697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zeuschner D, et al. Immuno-electron tomography of ER exit sites reveals the existence of free COPII-coated transport carriers. Nat Cell Biol. 2006;8(4):377–383. doi: 10.1038/ncb1371. [DOI] [PubMed] [Google Scholar]
- 16.Donohoe BS, Kang BH, Staehelin LA. Identification and characterization of COPIa- and COPIb-type vesicle classes associated with plant and algal Golgi. Proc Natl Acad Sci USA. 2007;104(1):163–168. doi: 10.1073/pnas.0609818104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Donohoe BS, et al. Cis-Golgi cisternal assembly and biosynthetic activation occur sequentially in plants and algae. Traffic. 2013;14(5):551–567. doi: 10.1111/tra.12052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mogelsvang S, Marsh BJ, Ladinsky MS, Howell KE. Predicting function from structure: 3D structure studies of the mammalian Golgi complex. Traffic. 2004;5(5):338–345. doi: 10.1111/j.1398-9219.2004.00186.x. [DOI] [PubMed] [Google Scholar]
- 19.Donohoe BS, Mogelsvang S, Staehelin LA. Electron tomography of ER, Golgi and related membrane systems. Methods. 2006;39(2):154–162. doi: 10.1016/j.ymeth.2006.05.013. [DOI] [PubMed] [Google Scholar]
- 20.Day KJ, Staehelin LA, Glick BS. A three-stage model of Golgi structure and function. Histochem Cell Biol. 2013;140(3):239–249. doi: 10.1007/s00418-013-1128-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Henderson GP, Gan L, Jensen GJ. 3-D ultrastructure of O. tauri: Electron cryotomography of an entire eukaryotic cell. PLoS One. 2007;2(8):e749. doi: 10.1371/journal.pone.0000749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bouchet-Marquis C, Starkuviene V, Grabenbauer M. Golgi apparatus studied in vitreous sections. J Microsc. 2008;230(Pt 2):308–316. doi: 10.1111/j.1365-2818.2008.01988.x. [DOI] [PubMed] [Google Scholar]
- 23.Han HM, Bouchet-Marquis C, Huebinger J, Grabenbauer M. Golgi apparatus analyzed by cryo-electron microscopy. Histochem Cell Biol. 2013;140(4):369–381. doi: 10.1007/s00418-013-1136-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Farquhar MG, Palade GE. The Golgi apparatus (complex)-(1954-1981)-from artifact to center stage. J Cell Biol. 1981;91(3 Pt 2):77s–103s. doi: 10.1083/jcb.91.3.77s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Maeda S, et al. Structure of the connexin 26 gap junction channel at 3.5 A resolution. Nature. 2009;458(7238):597–602. doi: 10.1038/nature07869. [DOI] [PubMed] [Google Scholar]
- 26.Shapiro L, Doyle JP, Hensley P, Colman DR, Hendrickson WA. Crystal structure of the extracellular domain from P0, the major structural protein of peripheral nerve myelin. Neuron. 1996;17(3):435–449. doi: 10.1016/s0896-6273(00)80176-2. [DOI] [PubMed] [Google Scholar]
- 27.Min Y, et al. Interaction forces and adhesion of supported myelin lipid bilayers modulated by myelin basic protein. Proc Natl Acad Sci USA. 2009;106(9):3154–3159. doi: 10.1073/pnas.0813110106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gonen T, Sliz P, Kistler J, Cheng Y, Walz T. Aquaporin-0 membrane junctions reveal the structure of a closed water pore. Nature. 2004;429(6988):193–197. doi: 10.1038/nature02503. [DOI] [PubMed] [Google Scholar]
- 29.Aricescu AR, Jones EY. Immunoglobulin superfamily cell adhesion molecules: Zippers and signals. Curr Opin Cell Biol. 2007;19(5):543–550. doi: 10.1016/j.ceb.2007.09.010. [DOI] [PubMed] [Google Scholar]
- 30.Frei JA, Stoeckli ET. SynCAMs extend their functions beyond the synapse. Eur J Neurosci. 2014;39(11):1752–1760. doi: 10.1111/ejn.12544. [DOI] [PubMed] [Google Scholar]
- 31.Mollenhauer HH. An intercisternal structure in the Golgi apparatus. J Cell Biol. 1965;24(3):504–511. doi: 10.1083/jcb.24.3.504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Craig S, Staehelin LA. High pressure freezing of intact plant tissues. Evaluation and characterization of novel features of the endoplasmic reticulum and associated membrane systems. Eur J Cell Biol. 1988;46(1):81–93. [PubMed] [Google Scholar]
- 33.Mollenhauer HH, Morré DJ. Perspectives on Golgi apparatus form and function. J Electron Microsc Tech. 1991;17(1):2–14. doi: 10.1002/jemt.1060170103. [DOI] [PubMed] [Google Scholar]
- 34.Cluett EB, Brown WJ. Adhesion of Golgi cisternae by proteinaceous interactions: Intercisternal bridges as putative adhesive structures. J Cell Sci. 1992;103(Pt 3):773–784. doi: 10.1242/jcs.103.3.773. [DOI] [PubMed] [Google Scholar]
- 35.Franke WW, et al. Inter- and intracisternal elements of the Golgi apparatus. A system of membrane-to-membrane cross-links. Z Zellforsch Mikrosk Anat. 1972;132(3):365–380. doi: 10.1007/BF02450714. [DOI] [PubMed] [Google Scholar]
- 36.Staehelin LA, Giddings TH, Jr, Kiss JZ, Sack FD. Macromolecular differentiation of Golgi stacks in root tips of Arabidopsis and Nicotiana seedlings as visualized in high pressure frozen and freeze-substituted samples. Protoplasma. 1990;157(1-3):75–91. doi: 10.1007/BF01322640. [DOI] [PubMed] [Google Scholar]
- 37.Oikawa A, Lund CH, Sakuragi Y, Scheller HV. Golgi-localized enzyme complexes for plant cell wall biosynthesis. Trends Plant Sci. 2013;18(1):49–58. doi: 10.1016/j.tplants.2012.07.002. [DOI] [PubMed] [Google Scholar]
- 38.McCormick C, Duncan G, Goutsos KT, Tufaro F. The putative tumor suppressors EXT1 and EXT2 form a stable complex that accumulates in the Golgi apparatus and catalyzes the synthesis of heparan sulfate. Proc Natl Acad Sci USA. 2000;97(2):668–673. doi: 10.1073/pnas.97.2.668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sasai K, et al. The critical role of the stem region as a functional domain responsible for the oligomerization and Golgi localization of N-acetylglucosaminyltransferase V. The involvement of a domain homophilic interaction. J Biol Chem. 2001;276(1):759–765. doi: 10.1074/jbc.M004972200. [DOI] [PubMed] [Google Scholar]
- 40.Harholt J, et al. ARAD proteins associated with pectic Arabinan biosynthesis form complexes when transiently overexpressed in planta. Planta. 2012;236(1):115–128. doi: 10.1007/s00425-012-1592-3. [DOI] [PubMed] [Google Scholar]
- 41.Chou Y-H, Pogorelko G, Zabotina OA. Xyloglucan xylosyltransferases XXT1, XXT2, and XXT5 and the glucan synthase CSLC4 form Golgi-localized multiprotein complexes. Plant Physiol. 2012;159(4):1355–1366. doi: 10.1104/pp.112.199356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hassinen A, Kellokumpu S. Organizational interplay of Golgi N-glycosyltransferases involves organelle microenvironment-dependent transitions between enzyme homo- and heteromers. J Biol Chem. 2014;289(39):26937–26948. doi: 10.1074/jbc.M114.595058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bainton DF, Farquhar MG. Origin of granules in polymorphonuclear leukocytes. Two types derived from opposite faces of the Golgi complex in developing granulocytes. J Cell Biol. 1966;28(2):277–301. doi: 10.1083/jcb.28.2.277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cho M, Garant PR. Sequential events in the formation of collagen secretion granules with special reference to the development of segment-long-spacing-like aggregates. Anat Rec. 1981;199(3):309–320. doi: 10.1002/ar.1091990302. [DOI] [PubMed] [Google Scholar]
- 45.Marchi F, Leblond CP. Radioautographic characterization of successive compartments along the rough endoplasmic reticulum-Golgi pathway of collagen precursors in foot pad fibroblasts of [3H]proline-injected rats. J Cell Biol. 1984;98(5):1705–1709. doi: 10.1083/jcb.98.5.1705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Scott DA, Docampo R, Dvorak JA, Shi S, Leapman RD. In situ compositional analysis of acidocalcisomes in Trypanosoma cruzi. J Biol Chem. 1997;272(44):28020–28029. doi: 10.1074/jbc.272.44.28020. [DOI] [PubMed] [Google Scholar]
- 47.Docampo R, Moreno SN. Acidocalcisomes. Cell Calcium. 2011;50(2):113–119. doi: 10.1016/j.ceca.2011.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hong-Hermesdorf A, et al. Subcellular metal imaging identifies dynamic sites of Cu accumulation in Chlamydomonas. Nat Chem Biol. 2014;10(12):1034–1042. doi: 10.1038/nchembio.1662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Komine Y, Eggink LL, Park H, Hoober JK. Vacuolar granules in Chlamydomonas reinhardtii: Polyphosphate and a 70-kDa polypeptide as major components. Planta. 2000;210(6):897–905. doi: 10.1007/s004250050695. [DOI] [PubMed] [Google Scholar]
- 50. Ruiz FA, Marchesini N, Seufferheld M, Govindjee, Docampo R (2001) The polyphosphate bodies of Chlamydomonas reinhardtii possess a proton-pumping pyrophosphatase and are similar to acidocalcisomes. J Biol Chem 276(49):46196–46203. [DOI] [PubMed]
- 51.Aksoy M, Pootakham W, Grossman AR. Critical function of a Chlamydomonas reinhardtii putative polyphosphate polymerase subunit during nutrient deprivation. Plant Cell. 2014;26(10):4214–4229. doi: 10.1105/tpc.114.129270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gómez-García MR, Kornberg A. Formation of an actin-like filament concurrent with the enzymatic synthesis of inorganic polyphosphate. Proc Natl Acad Sci USA. 2004;101(45):15876–15880. doi: 10.1073/pnas.0406923101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Docampo R, de Souza W, Miranda K, Rohloff P, Moreno SNJ. Acidocalcisomes - conserved from bacteria to man. Nat Rev Microbiol. 2005;3(3):251–261. doi: 10.1038/nrmicro1097. [DOI] [PubMed] [Google Scholar]
- 54.Smith SA, et al. Polyphosphate modulates blood coagulation and fibrinolysis. Proc Natl Acad Sci USA. 2006;103(4):903–908. doi: 10.1073/pnas.0507195103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Müller F, et al. Platelet polyphosphates are proinflammatory and procoagulant mediators in vivo. Cell. 2009;139(6):1143–1156. doi: 10.1016/j.cell.2009.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bonfanti L, et al. Procollagen traverses the Golgi stack without leaving the lumen of cisternae: Evidence for cisternal maturation. Cell. 1998;95(7):993–1003. doi: 10.1016/s0092-8674(00)81723-7. [DOI] [PubMed] [Google Scholar]
- 57.Mironov AA, et al. Small cargo proteins and large aggregates can traverse the Golgi by a common mechanism without leaving the lumen of cisternae. J Cell Biol. 2001;155(7):1225–1238. doi: 10.1083/jcb.200108073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Glick BS, Malhotra V. The curious status of the Golgi apparatus. Cell. 1998;95(7):883–889. doi: 10.1016/s0092-8674(00)81713-4. [DOI] [PubMed] [Google Scholar]
- 59.McFadden GI, Melkonian M. Golgi apparatus activity and membrane flow during scale biogenesis in the green flagellate Scherffelia dubia (Prasinophyceae). I: Flagellar regeneration. Protoplasma. 1986;130(2-3):186–198. [Google Scholar]
- 60.McFadden GI, Preisig HR, Melkonian M. Golgi apparatus activity and membrane flow during scale biogenesis in the green flagellate Scherffelia dubia (Prasinophyceae). II: Cell wall secretion and assembly. Protoplasma. 1986;131(2):174–184. [Google Scholar]
- 61.Becker B, Bölinger B, Melkonian M. Anterograde transport of algal scales through the Golgi complex is not mediated by vesicles. Trends Cell Biol. 1995;5(8):305–307. doi: 10.1016/s0962-8924(00)89047-9. [DOI] [PubMed] [Google Scholar]
- 62.Catt JW, Hills GJ, Roberts K. Cell wall glycoproteins from Chlamydomonas reinhardii, and their self-assembly. Planta. 1978;138(1):91–98. doi: 10.1007/BF00392922. [DOI] [PubMed] [Google Scholar]
- 63.Goodenough UW, Gebhart B, Mecham RP, Heuser JE. Crystals of the Chlamydomonas reinhardtii cell wall: Polymerization, depolymerization, and purification of glycoprotein monomers. J Cell Biol. 1986;103(2):405–417. doi: 10.1083/jcb.103.2.405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Roberts K, Gurney-Smith M, Hills GJ. Structure, composition and morphogenesis of the cell wall of Chlamydomonas reinhardi. I. Ultrastructure and preliminary chemical analysis. J Ultrastruct Res. 1972;40(5):599–613. doi: 10.1016/s0022-5320(72)80046-7. [DOI] [PubMed] [Google Scholar]
- 65.Goodenough UW, Heuser JE. The Chlamydomonas cell wall and its constituent glycoproteins analyzed by the quick-freeze, deep-etch technique. J Cell Biol. 1985;101(4):1550–1568. doi: 10.1083/jcb.101.4.1550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Matsuda Y, Yamasaki A, Tatsuaki S, Yamaguchi T. Purification and characterization of cell wall lytic enzyme released by mating gametes of Chlamydomonas reinhardtii. FEBS Lett. 1984;166(2):293–297. [Google Scholar]
- 67.Imam SH, Snell WJ. The Chlamydomonas cell wall degrading enzyme, lysin, acts on two substrates within the framework of the wall. J Cell Biol. 1988;106(6):2211–2221. doi: 10.1083/jcb.106.6.2211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Danev R, Buijsse B, Khoshouei M, Plitzko JM, Baumeister W. Volta potential phase plate for in-focus phase contrast transmission electron microscopy. Proc Natl Acad Sci USA. 2014;111(44):15635–15640. doi: 10.1073/pnas.1418377111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Umen JG, Goodenough UW. Control of cell division by a retinoblastoma protein homolog in Chlamydomonas. Genes Dev. 2001;15(13):1652–1661. doi: 10.1101/gad.892101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Schaffer M, et al. Cryo-focused ion beam sample preparation for imaging vitreous cells by cryo-electron tomography. Bio-protocol. 2015;5(17):e1575. doi: 10.21769/bioprotoc.1575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Rigort A, Villa E, Bäuerlein FJ, Engel BD, Plitzko JM. Integrative approaches for cellular cryo-electron tomography: Correlative imaging and focused ion beam micromachining. Methods Cell Biol. 2012;111:259–281. doi: 10.1016/B978-0-12-416026-2.00014-5. [DOI] [PubMed] [Google Scholar]
- 72.Hayles MF, Stokes DJ, Phifer D, Findlay KC. A technique for improved focused ion beam milling of cryo-prepared life science specimens. J Microsc. 2007;226(Pt 3):263–269. doi: 10.1111/j.1365-2818.2007.01775.x. [DOI] [PubMed] [Google Scholar]
- 73.Mastronarde DN. Automated electron microscope tomography using robust prediction of specimen movements. J Struct Biol. 2005;152(1):36–51. doi: 10.1016/j.jsb.2005.07.007. [DOI] [PubMed] [Google Scholar]
- 74.Mastronarde DN. Dual-axis tomography: An approach with alignment methods that preserve resolution. J Struct Biol. 1997;120(3):343–352. doi: 10.1006/jsbi.1997.3919. [DOI] [PubMed] [Google Scholar]
- 75.Hrabe T, et al. PyTom: A python-based toolbox for localization of macromolecules in cryo-electron tomograms and subtomogram analysis. J Struct Biol. 2012;178(2):177–188. doi: 10.1016/j.jsb.2011.12.003. [DOI] [PubMed] [Google Scholar]
- 76.Nickell S, et al. TOM software toolbox: Acquisition and analysis for electron tomography. J Struct Biol. 2005;149(3):227–234. doi: 10.1016/j.jsb.2004.10.006. [DOI] [PubMed] [Google Scholar]
- 77.Pettersen EF, et al. UCSF Chimera--a visualization system for exploratory research and analysis. J Comput Chem. 2004;25(13):1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 78.Sun HY, et al. Structure and mechanism of Helicobacter pylori fucosyltransferase. A basis for lipopolysaccharide variation and inhibitor design. J Biol Chem. 2007;282(13):9973–9982. doi: 10.1074/jbc.M610285200. [DOI] [PubMed] [Google Scholar]
- 79.van den Elsen JM, Kuntz DA, Rose DR. Structure of Golgi alpha-mannosidase II: A target for inhibition of growth and metastasis of cancer cells. EMBO J. 2001;20(12):3008–3017. doi: 10.1093/emboj/20.12.3008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Gastinel LN, et al. Bovine alpha1,3-galactosyltransferase catalytic domain structure and its relationship with ABO histo-blood group and glycosphingolipid glycosyltransferases. EMBO J. 2001;20(4):638–649. doi: 10.1093/emboj/20.4.638. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.