Significance
Oosporein, a red 1,4-bibenzoquinone derivative, was first identified from fungi in the 1960s and exhibits antibiotic, antiviral, antifungal, and insecticidal activities. We report, to our knowledge, for the first time the novel pathway for oosporein biosynthesis in Beauveria bassiana that includes the polyketide synthase oosporein synthase 1 (OpS1) to produce the precursor orsellinic acid for OpS4 hydroxylation and then OpS7 oxidation to benzenetetrol, and the dimerization of the intermediate to oosporein is catalyzed by the catalase OpS5. The gene cluster is regulated by the transcription factor OpS3. We also found that oosporein is required for fungal virulence by inhibiting insect immunity. These results advance the knowledge of quinone biosynthetic machineries and demonstrate that a small molecule contributes to fungus–host interactions.
Keywords: Beauveria bassiana, bibenzoquinone, oosporein, biosynthesis, virulence
Abstract
Quinones are widely distributed in nature and exhibit diverse biological or pharmacological activities; however, their biosynthetic machineries are largely unknown. The bibenzoquinone oosporein was first identified from the ascomycete insect pathogen Beauveria bassiana >50 y ago. The toxin can also be produced by different plant pathogenic and endophytic fungi with an array of biological activities. Here, we report the oosporein biosynthetic machinery in fungi, a polyketide synthase (PKS) pathway including seven genes for quinone biosynthesis. The PKS oosporein synthase 1 (OpS1) produces orsellinic acid that is hydroxylated to benzenetriol by the hydroxylase OpS4. The intermediate is oxidized either nonenzymatically to 5,5′-dideoxy-oosporein or enzymatically to benzenetetrol by the putative dioxygenase OpS7. The latter is further dimerized to oosporein by the catalase OpS5. The transcription factor OpS3 regulates intrapathway gene expression. Insect bioassays revealed that oosporein is required for fungal virulence and acts by evading host immunity to facilitate fungal multiplication in insects. These results contribute to the known mechanisms of quinone biosynthesis and the understanding of small molecules deployed by fungi that interact with their hosts.
Quinones are ubiquitous in living organisms such as bacteria, fungi, plants, and humans, and a large number of derivatives containing either a 1,2-benzoquinone or 1,4-benzoquinone backbone have been shown to express an array of bioactive or pharmaceutical-relevant activities (1, 2). However, only a handful quinone biosynthetic pathways have been characterized. For example, compounds derived from chorismate formed via the shikimate pathway have been well established as precursors for the biosynthesis of isoprenoid quinones (including ubiquinones and menaquinone) (3). Indole pyruvic acid deaminated from l-tryptophan can be dimerized into terrequinone by a single-module nonribosomal peptide synthetase (4). The catalysis of the substrates tyrosine or Dopa by tyrosinase to form dopaquinone is a well-known pathway for Dopa–melanin biosynthesis (5). Similar to tyrosinase, catechol oxidase is a copper-containing enzyme that can oxidize catechol into 1,2-benzoquinone (2). In Streptomyces antibioticus, monooxygenation of tetrahydroxynaphthalene to naphthoquinone can be catalyzed by a cupin-family enzyme (6). In this study, we report the biosynthesis of bibenzoquinone through a polyketide synthase (PKS) pathway in fungi.
Oosporein (1), a red, symmetrical 1,4-bibenzoquinone derivative, was first identified in the 1960s from the ascomycete insect-pathogenic fungus Beauveria bassiana (7–9), as well as from Phlebia basidiomycetes (10). Oosporein is also produced by the insect pathogen Beauveria brongniartii (11) and various plant pathogenic and endophytic fungi (12–15). Oosporein is highly reactive in biological systems, including insecticidal activity (16), antibiotic activity against Gram-positive bacteria (17), and antiviral (18) activity, as well as an antagonistic effect against plant pathogenic oomycetes (13). The toxin can cause avian gout and mortality in chickens and turkeys (19), which raises the safety concerns of the application of Beauveria spp. to control insect pests (10). An understanding of the toxin biosynthetic machinery would be critical to manage the toxin production in biocontrol situations.
Chemical synthesis of oosporein from 2,5-dimethoxytoluene has been successful (20). Biologically, however, an early experiment demonstrated that 14C-labeled orsellinic acid (OA) (2) could be converted to bibenzoquinone oosporein in B. bassiana cultures (8). It is now clear that OA can be biosynthesized by PKSs in bacteria (21) and fungi (22–24). These findings suggest that oosporein may be biosynthesized through OA by a PKS gene cluster in Beauveria. Our previous analysis indicated that there are 12 type I PKS genes encoded in the B. bassiana genome (25). After the bioinformatics analysis, the putative PKS genes involved in OA biosynthesis were predicted and deleted. It was found that the BbPKS9, termed OpS1 (for oosporein synthase 1), is responsible for OA production leading to oosporein biosynthesis. In addition to the elucidation of the longstanding question about the nature of the oosporein biosynthetic machinery, this study identifies a PKS gene cluster including seven genes for quinone biosynthesis in fungi. Insect bioassays indicated that oosporein is required for full fungal virulence through evasion of host immune responses, thus facilitating fungal development within insects.
Results
Prediction of the PKS Gene Cluster.
Based on previous reports that OA can be converted into oosporein by B. bassiana (8) and that OA is biosynthesized by PKSs in fungi (22, 23), we performed a genome-wide modulation analysis of 12 PKSs encoded in the Beauveria genome (SI Appendix, Fig. S1A). Of these, two PKSs—BbPKS7 (BBA_06613) and BbPKS9 (BBA_08179)—have similar modules with the OrSA of Aspergillus nidulans (22), TerA of Aspergillus terreus (23), and FgPKS14 of Fusarium graminearum (24), which have been verified to produce OA (SI Appendix, Fig. S1B). Phylogenetic analysis using the ketosynthase (KS) domain sequences retrieved from the PKSs involved in the biosynthesis of quinone pigments indicated that BbPKS9, but not BbPKS7, is closely related to the counterparts responsible for OA biosynthesis (SI Appendix, Fig. S2).
Gene Deletions and Verification of OpS1 Function.
To verify the functions of OpS1 and its tailoring enzymes, serial gene deletions were performed. We found that loss of BbPKS9 (i.e., OpS1), but not BbPKS7, led to the loss of oosporein production in B. bassiana (SI Appendix, Fig. S3A). Deletion of the OpS1 ortholog in B. brongniartii (BBO_00072; 92% identity at the amino acid sequence level) also abolished the production of oosporein (SI Appendix, Fig. S3B). Thus, the OpS1 gene cluster (Fig. 1A and SI Appendix, Fig. S4A) is responsible for oosporein biosynthesis. Not surprisingly, the cluster is highly conserved between the two Beauveria species, but is nonsyntenic between the OpS1 and BbPKS7 clusters (SI Appendix, Fig. S4B).
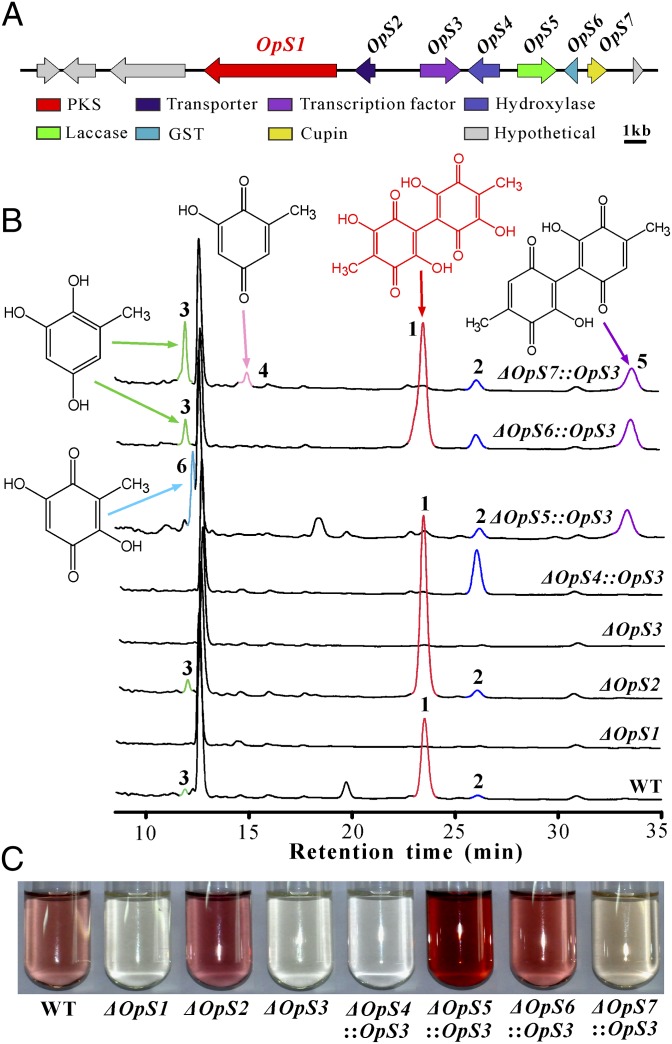
Fig. 1.
Prediction and functional verification of the gene cluster for oosporein biosynthesis in B. bassiana. (A) A schematic map of the oosporein biosynthetic gene cluster. The genes involved in oosporein production are termed OpS1–OpS7. (B) HPLC traces of the WT and different mutants showing the accumulation of different compounds. Peaks 1 and 2 are for oosporein (1) and OA (2). Peaks 3–6 are labeled with the corresponding metabolite structures. (C) Culture filtrate color variations among the WT and different mutants.
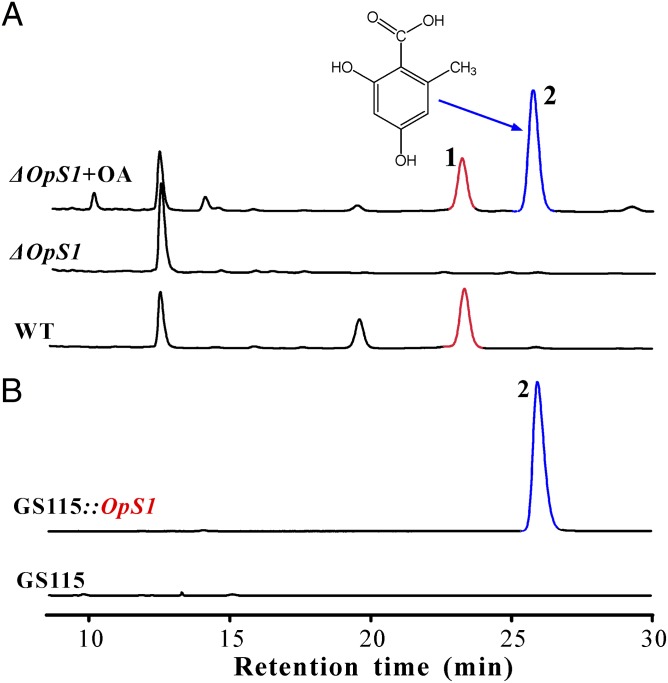
We found that six additional genes, termed OpS2–OpS7 (Fig. 1A), were associated with oosporein biosynthesis in B. bassiana (SI Appendix, Fig. S5 A and B). Unexpectedly, deletion of OpS2, which encodes a putative major facilitator superfamily transporter, significantly increased oosporein production in the mutant (48.71 ± 8.16 μg/mL) compared with the wild-type (WT; 31.34 ± 4.41 μg/mL) strain (t test, P = 0.039). Otherwise, oosporein was similarly nondetectable in the culture extracts of the null mutants of OpS3–OpS7 (SI Appendix, Fig. S5 A and B). To explore the function of PKS OpS1, substrate feeding and yeast heterologous expression experiments were conducted. Consistent with the above analysis, we found that the supplement of OA (2) in the ΔOpS1 culture restored the ability of Beauveria to produce oosporein (Fig. 2A). Expression of the OpS1 cDNA in Pichia pastoris enabled the yeast to produce OA (Fig. 2B). The results thereby confirmed that OpS1 is responsible for OA biosynthesis.
Fig. 2.
Verification of OA biosynthesis by OpS1. (A) HPLC profiles showing that the addition of OA (2) (used at a final concentration of 300 μg/mL) restored the ability of ΔOpS1 to produce oosporein (1) in B. bassiana. (B) Yeast production of OA when being engineered to express OpS1 cDNA.
Transcription Control of the Biosynthetic Gene Cluster.
Within the gene cluster, OpS3 encodes a Gal4-like Zn2Cys6 domain-containing transcription factor (SI Appendix, Fig. S4A). To verify whether the gene cluster is regulated by OpS3, the gene was made under the control of a constitutive gpdA gene (BBA_05480) promoter (26), and the cassette was used to engineer the WT strain. We performed semiquantitative reverse-transcription PCR (RT-PCR) analysis and found that the clustered genes were marginally transcribed by the fungus grown in a saprophytic medium. With the exception of OpS5, deletion of OpS3 disabled the expression of other genes associated with oosporein biosynthesis in the mutant strains. Relative to the parental WT strain, overexpression of OpS3 significantly increased the clustered gene expressions, including of OpS3 itself (SI Appendix, Fig. S6A). Thus, the production of oosporein was boosted up to fourfold higher in the transformant WT::OpS3 than in the WT (SI Appendix, Fig. S6B). Analysis of the OpS3 putative binding motif identified the presence of a highly conserved CGGA motif (different from the Gal-4 binding site CGGN11CCG) in the promoters of the clustered genes, except for in OpS5 (SI Appendix, Fig. S6C), which is consistent with the OpS5 unimpaired expression profile in ΔOpS3 (SI Appendix, Fig. S6A). This outcome indicates that the CGGA motif is the putative OpS3 binding site. The presence of this motif in the OpS3 promoter would suggest the formation of a positive feedback loop for self-activation. Additionally, we identified the presence of a yeast Msn2-like stress-response element AGGGG (27) in the promoter regions of the clustered genes, except for that of the transporter gene OpS2 (SI Appendix, Fig. S6D). Consistent with this finding, BbMsn2 (BBA_00971), a homolog of Msn2, has been identified as a negative regulator of oosporein production in B. bassiana (28). The regulation of oosporein biosynthesis thus represents a further example of fungal secondary metabolisms being cocontrolled by a pathway-specific transcription factor and a global regulator (29).
Elucidation of the Oosporein Biosynthetic Pathway.
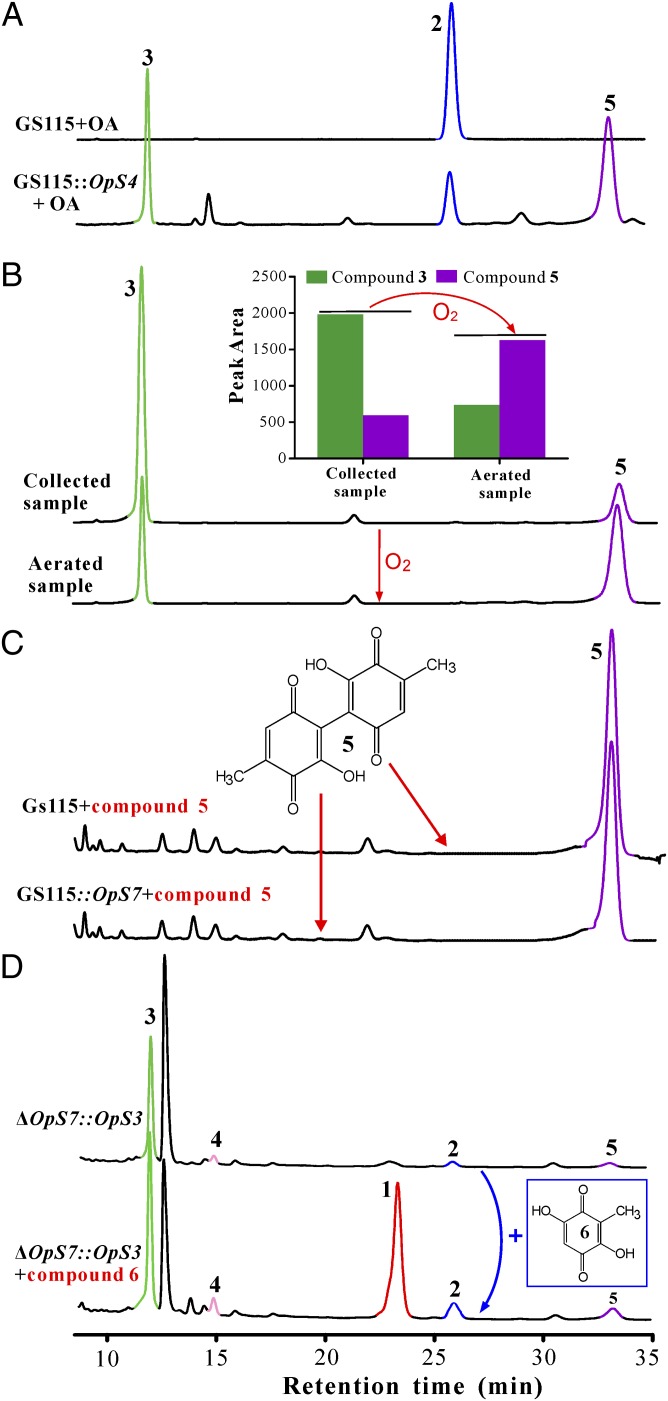
Based on the above information, we suspected that the similar chromatographic patterns of OpS3–OpS7 gene deletion mutants (SI Appendix, Fig. S5 A and B) might actually be due to the low level of gene expression after deletion of the alternate gene. To test this hypothesis, we engineered the mutants ΔOpS4–ΔOpS7 to overexpress OpS3 and directly analyzed the obtained culture filtrates of ΔOpS4::OpS3–ΔOpS7::OpS3 by HPLC in case of a loss in stability of intermediate metabolites during ethyl acetate extraction. Fortunately, altered chromatographic profiles (Fig. 1B) and colored culture filtrates (Fig. 1C) between the WT and mutant strains differing from the initial analyses were observed (SI Appendix, Fig. S5 A and B). The OpS4 gene encodes a putative salicylate hydroxylase (Fig. 1A). We detected the accumulation of OA (2) in the ΔOpS4::OpS3 mutant, confirming again that the function of OpS1 PKS is to form OA. Heterologous expression of OpS4 in yeast converted OA into the detectable 6-methyl-1,2,4-benzenetriol (3) and the product 5,5′-dideoxy-oosporein (5) (Fig. 3A), indicating the hydroxylation of OA to the metabolite 3 by OpS4 (Fig. 4). We hypothesized that benzenetriol 3 could be nonenzymatically oxidized into 5,5′-dideoxy-oosporein. To test this hypothesis, metabolite 3 was collected by using preparative HPLC. Examinations of both the newly collected sample and the sample aliquot aerated for 1 h led to the formation of 5 (Fig. 3B), a metabolite that has not been previously reported because it could not be produced by the WT strain (Fig. 1B).
Fig. 3.
Verification of different gene functions by yeast heterologous expression and substrate feeding of gene deletion mutants of B. bassiana. (A) Conversion of OA by the yeast cells expressing the OpS4 gene led to the formations of 3 and 5. (B) Nonenzymatic conversion of 3 to 5. Inset shows the ratio change of the two compounds after an aeration treatment for 1 h. (C) Failure of conversion of 5,5′-dideoxy-oosporein (5) by OpS7. The yeast cells expressing or not expressing the OpS7 gene were supplemented with 5 (at a final concentration of ∼10 μg/mL) for 24 h. (D) Restoration of oosporein production by ΔOpS7::OpS3 after addition of compound 6.
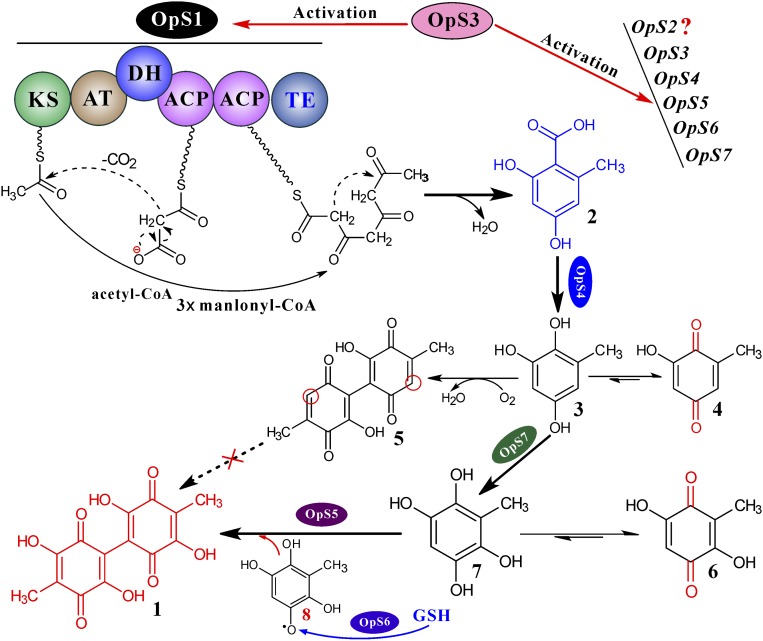
Fig. 4.
Oosporein biosynthetic machinery. The domains of OpS1 include: KS, β-ketoacyl synthase; AT, acyltransferase; DH, dehydrogenase; ACP, acyl carrier protein; TE, thioesterase. Compound 1, oosporein; 2, OA; 3, 6-methyl-1,2,4-benzenetriol; 4, 2-hydroxy-6-methyl-2,5-cyclohexadiene-1,4-dione; 5, 5,5′-dideoxy-oosporein; 6, 2,5-dihydroxy-3-methyl-2,5-cyclohexadiene-1,4-dione; 7, 6-methyl-1,2,4,5-benzenetetrol; 8, free radical form of 7. The question mark after OpS2 means the function of this putative MFS transporter remains unclear. GSH, glutathione.
The genes OpS5, OpS6, and OpS7 encode a putative laccase, GST, and cupin-family dioxygenase, respectively (Fig. 1A and SI Appendix, Fig. S4A). Oosporein was detected in ΔOpS6::OpS3, but not in the ΔOpS5::OpS3 and ΔOpS7::OpS3, cultures. Compounds 2, 3, 4, and 5 were detected in the ΔOpS7::OpS3 culture, and 2, 5, and 6 were detected in the ΔOpS5::OpS3 culture (Fig. 1B). We found that the yeast cells expressing OpS7 could not convert 5,5′-dideoxy-oosporein (5) into oosporein (Fig. 3C), suggesting a nonfunctional effect of OpS7 dioxygenation. In bacteria, a study demonstrated that, instead of dioxygenation, monooxygenation of metabolites could be catalyzed by the cupin-family enzyme (6). This finding gave rise to the idea that OpS7 may be responsible for de facto monooxygenation of metabolite 3 to 6-methyl-1,2,4,5-benzenetetrol (7) (Fig. 4). Thus, we performed a mixed fermentation of yeast mutants GS115::OpS4 and GS115::OpS7 with the addition of OA. As indicated above, OpS4 successfully converted OA to benzenetriol 3. However, benzenetetrol 7 was not detected, whereas 6 was observed together with other undetermined compounds in the mixed culture (SI Appendix, Fig. S7A). Because of its structural instability, compound 7 was either dimerized into oosporein by the catalase OpS5 through a free radical reaction or largely interconverted into compound 6 through keto-enol tautomerization because only the latter was detected in the ΔOpS5::OpS3 culture (Fig. 1B) or yeast mixed-fermentation culture (SI Appendix, Fig. S7).
The attempted expression of the laccase gene OpS5 in yeast was not successful; i.e., no detectable activity was observed. However, the functions of OpS5 and OpS7, as well as the keto-enol tautomerism, were confirmed by substrate feeding of 6 into the ΔOpS7::OpS3 (with functional catalase OpS5) culture and successfully produced oosporein (Fig. 3D). The experiment also suggested that up-regulation of OpS5 in WT::OpS3 (SI Appendix, Fig. S6A) would be due to the accumulation of a higher level of compound 6 or 7, a mechanism of substrate feedback activation. The primary role of GSTs is to detoxify reactive electrophilic compounds (30). In this respect, OpS6 would function en route for protecting cells against oxidative stress by scavenging any leaked free radical 8 that was present by activating the thiol group of glutathione. We thereby fully elucidated the pathway of oosporein biosynthesis in B. bassiana (Fig. 4). The structures of the metabolites, except 8, were alternatively verified by liquid chromatography (LC)-MS (1–7), NMR (3, 5, and 6), and/or LC-tandem MS fragmentation (3, 4, 6, and 7) analyses (SI Appendix, Figs. S8 and S9).
Effect of Oosporein on Fungal Virulence.
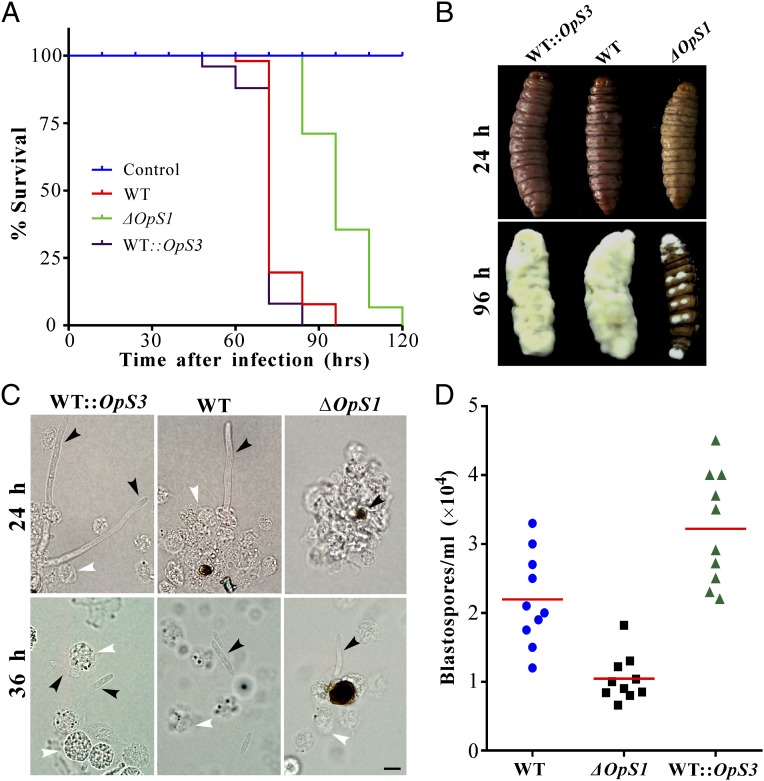
In contrast to the phenotypic alteration in ΔBbMsn2 of B. bassiana (28), deletion of OpS1 or overexpression of OpS3 had no apparent effect on fungal growth on artificial medium and did not cause any defect on antioxidative response compared with the WT (SI Appendix, Fig. S10). To examine whether the production of oosporein would contribute to fungal virulence, we performed insect bioassays using the spores of the WT, ΔOpS1, and WT::OpS3 strains against the wax moth larvae. A comparison of the insect survival curves (Fig. 5A) indicated significant differences between WT and ΔOpS1 (χ2 = 63.5; P < 0.0001), WT::OpS3 and ΔOpS1 (χ2 = 83.9; P < 0.0001), and WT and WT::OpS3 (χ2 = 7.42; P = 0.0065) strains. The data thereby confirmed that oosporein contributes to fungal virulence. We found that the insect cadavers killed by the WT and WT::OpS3 strains could be pigmented 24 h after insect death, but pigmentation was not observed for ΔOpS1-infected insects. In addition, we found that fungal mycosis of insect cadavers was delayed considerably after loss of the toxin-production ability (Fig. 5B). Microscopic observations indicated that oosporein contributed to the evasion of insect host immunity (Fig. 5C). After injection into the insect body cavity (hemocoel), the spores of the WT and WT::OpS3 germinated and quickly escaped from insect hemocyte encapsulation within 24 h; however, ΔOpS1 took ∼36 h. Thus, in contrast to the null mutant, many more (t test, P < 0.001) free-living blastospores were formed by the WT and WT::OpS3 48 h after injection. Relative to the WT, overexpression of OpS3 also significantly (P = 0.0108) promoted fungal propagation within insect hemocoels (Fig. 5D). Immune interference verified that, relative to the activations by fungal spores, injection of oosporein could inhibit prophenoloxidase (PPO) activity (SI Appendix, Fig. S11 A and B) and down-regulation of the antifungal peptide gallerimycin gene in insects (SI Appendix, Fig. S11C).
Fig. 5.
Contribution of oosporein to fungal virulence. (A) Survival of insects after injection with the spores of the WT, ΔOpS1, and WT::OpS3 strains. (B) Mycosis of insects by the WT and mutant strains. (C) Microscopic observation of fungal development and insect immune responses to the WT and mutant strains at different times after injection. Black arrows point to fungal cells and white arrows point to insect hemocytes. (Scale bar: 5 μm.) (D) Quantification of the free-living fungal blastospores in insect hemocoels 48 h after infection. Statistical analyses (t test) indicate a significant difference between the WT and ΔOpS1 strains (P = 8.58e−4), between the WT::OpS3 and ΔOpS1 strains (P = 3.55e−5), and between the WT and WT::OpS3 strains (P = 0.01084).
Discussion
Herein, we report, to our knowledge, the first full pathway by which PKS-produced OA is hydroxylated to form the intermediate hydroxytoluene, which undergoes further oxidation and dimerization to yield bibenzoquinone oosporein. This pathway is associated with the formation of different 1,4-benzoquinone derivatives, including the new bibenzoquinone 5,5′-dideoxy-oosporein in B. bassiana. Biosynthesis of OA by PKS has been reported in various bacteria and fungi (21). However, the divergent tailoring-enzyme genes within the PKS gene clusters lead to the formation of structurally diverse metabolites. For example, the OA precursor biosynthesized by OrsA (AN7909) in A. nidulans is further catalyzed into lecanoric acid or the analogs F-9775A and F-9775B, presumably through the functions of amdohydrolase (OrsB; AN7911), tyrosinase (OrsC; AN7912), and dehydrogenase (OrsE; AN7914) (22, 31). In A. terreus, the closely related PKS TerA (SI Appendix, Fig. S2) produces a mixture of OA, 4-hydroxy-6-methylpyranone, and 6,7-dihydroxymellein. The latter is further oxidized to terrein by monooxygenase and multicopper oxidase (23). The OA produced by PKS ArmB is esterified to a diverse class of sesquiterpene alcohols in the plant pathogen Armillaria mellea (32). It is noteworthy that the closely related PKSs containing an additional methyltransferase domain form 3-methyl or 3,5-dimethyl OA (SI Appendix, Fig. S2). For example, the precursor 3,5-dimethyl OA is produced by the PKS AusA (AN8383), and the metabolite is then catalyzed to meroterpenoids via sequential modification and oxidation by different tailoring enzymes in A. nidulans (33). Except for the cupin-like dioxygenase OpS7, the homologs of OpS2–OpS6 are present in the A. nidulans genome, however, not within the OrSA gene cluster (SI Appendix, Fig. S4A). Thus, the tailoring enzymes of OA-biosynthetic PKS determine the structure(s) of the end product(s). Future studies are still required regarding the function of putative transporter OpS2 and the mechanism of OpS5 dimerization reaction.
The OpS1 gene cluster is present in two Beauveria species; however, interestingly, it is not in the genomes of the closely related insect pathogens Cordyceps militaris (34) (SI Appendix, Fig. S4B) and Metarhizium species (35). This phenomenon raises a question of how the gene cluster has evolved. After its first identification in the 1960s from B. bassiana (7, 8), oosporein has been isolated from a range of fungal species, including from basidiomycetes (10, 36) to ascomycetes such as the mycoparasite Lecanicillium psalliotae (current name Verticillium psalliotae) (13) and the plant endophyte Cochliobolus kusanoi (15). It has been demonstrated that the insect pathogens Beauveria spp. and Metarhizium spp. evolutionarily diverged from the plant endophytes or pathogens (25, 35). Thus, similar to the acquisition of the cyclodepsipeptide destruxin biosynthetic gene cluster in Metarhizium species (35, 37), the oosporein biosynthesis genes could have been horizontally transferred to Beauveria from the plant endophytes or pathogens. In addition to killing insects, Beauveria species have been identified as plant endophytes, and they share similar habitats with other plant endophytes or pathogens (38), which is a possible mode of gene transfer. It could not be precluded that the ancestor of Beauveria/Cordyceps obtained the gene cluster, while the subsequent gene loss led to the vertical absence of OpS1-clustered genes in C. militaris. The genomes of other oosporein-producing fungi are still unavailable. Future investigations of the OpS1–OpS7 ortholog (if exactly present) functions in these fungi would facilitate the understanding of the evolution of the oosporein gene cluster.
Although 1,4-benzoquinones cause 100% mortality in subterranean termites (39), topical application of oosporein on the sap-sucking whitefly indicated that the use of toxin alone resulted in ∼20% insect mortality, and the use of the fungal spores alone led to ∼60% mortality. However, the percentage of insect death increased to 92% when the fungal spores were used in combination with the red pigment (16). The results suggest that oosporein promotes fungal infection, rather than directly killing the insects. Consistent with this result, we found that oosporein contributes to fungal virulence by inhibiting PPO activity and down-regulating antifungal gene expression, thereby facilitating fungal multiplication within the insects. However, in contrast to our observations, deletion of the negative regulator BbMsn2 increased oosporein production in B. bassiana, but the null mutant was impaired in virulence against the wax moth larvae (28). This outcome is due to the additional impairments on fungal growth, protease expression, and stress responses caused by deletion of BbMsn2. However, except for the effects on oosporein production, deletion of OpS1 or overexpression of OpS3 did not lead to any defects on fungal growth or antioxidative stress compared with the WT (SI Appendix, Fig. S10). Nevertheless, the target protein of oosporein remains unknown in insect hosts. Future study is also required to determine the function(s) of oosporein in plant pathogens and endophytes.
In conclusion, we report that the biosynthetic mechanism of bibenzoquinone oosporein in Beauveria species is a PKS pathway for quinone biosynthesis. It was found that the metabolite contributes to fungal virulence by inhibiting the host immune response, thus promoting fungal infection. The results obtained in this study expand our knowledge of quinone biosynthetic machinery, as well as illuminate the function of a small molecule in fungus–host interactions.
Materials and Methods
Fungal Strains and Maintenance.
Insect pathogenic fungi B. bassiana ARSEF 2860 and B. brongniartii RCEF 3172 were routinely maintained on potato dextrose agar (Difco). For liquid incubation, fungal spores were inoculated in Sabouraud dextrose broth (SDB; Difco) and incubated at 25 °C on a rotatory shaker. The Escherichia coli Top 10 bacterial strain (Invitrogen) was used for vector construction, and the Agrobacterium tumefaciens AGL-1 strain was used for transformations (40). The yeast strain P. pastoris GS115 was used for the heterologous expression of target genes from B. bassiana.
Bioinformatics and Phylogenetic Analyses.
Homologous PKS sequences of different fungal species that have been functionally verified to be involved in production of OA, pigments, and/or melanin were retrieved from the National Center for Biotechnology Information database. Modular analysis of different PKS enzymes, including the mapping of KS domain, was performed by using the program antiSMASH (Version 2.0) (41). For phylogenetic analysis, the KS domain sequences from functionally or putative PKSs involved in biosynthesis of pigments (including melanins) were aligned by using the program Clustal X (Version 2.0) (42), and a maximum-likelihood tree was generated by using MEGA (Version 6.0) software (43). Analysis of putative promoter binding sites was conducted by using the weight matrix-based program Match (Version 1.0) (44).
Details of gene deletions, compound induction and structure analysis, target gene expression in yeasts, insect bioassays, and immune interference tests are provided in SI Appendix, SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank Mr. Fan Zhang and Drs. Yong-Jiang Xu and Yining Liu for their help for chemical analyses. This work was supported by Strategic Priority Research Program of Chinese Academy of Sciences Grant XDB11030100, National Nature Science Foundation of China Grant 31225023, and Chinese Academy of Sciences Grant KSZD-EW-Z-021-2-4.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1503200112/-/DCSupplemental.
References
- 1.Abraham I, Joshi R, Pardasani P, Pardasani RT. Recent advances in 1,4-benzoquinone chemistry. J Braz Chem Soc. 2011;22:385–421. [Google Scholar]
- 2.Solomon EI, Chen P, Metz M, Lee SK, Palmer AE. Oxygen binding, activation, and reduction to water by copper proteins. Angew Chem Int Ed Engl. 2001;40(24):4570–4590. doi: 10.1002/1521-3773(20011217)40:24<4570::aid-anie4570>3.0.co;2-4. [DOI] [PubMed] [Google Scholar]
- 3.Nowicka B, Kruk J. Occurrence, biosynthesis and function of isoprenoid quinones. Biochim Biophys Acta. 2010;1797(9):1587–1605. doi: 10.1016/j.bbabio.2010.06.007. [DOI] [PubMed] [Google Scholar]
- 4.Balibar CJ, Howard-Jones AR, Walsh CT. Terrequinone A biosynthesis through L-tryptophan oxidation, dimerization and bisprenylation. Nat Chem Biol. 2007;3(9):584–592. doi: 10.1038/nchembio.2007.20. [DOI] [PubMed] [Google Scholar]
- 5.Langfelder K, Streibel M, Jahn B, Haase G, Brakhage AA. Biosynthesis of fungal melanins and their importance for human pathogenic fungi. Fungal Genet Biol. 2003;38(2):143–158. doi: 10.1016/s1087-1845(02)00526-1. [DOI] [PubMed] [Google Scholar]
- 6.Funa N, Funabashi M, Yoshimura E, Horinouchi S. A novel quinone-forming monooxygenase family involved in modification of aromatic polyketides. J Biol Chem. 2005;280(15):14514–14523. doi: 10.1074/jbc.M500190200. [DOI] [PubMed] [Google Scholar]
- 7.Vining LC, Kelleher WJ, Schwarting AE. Oosporein production by a strain of Beauveria bassiana originally identified as Amanita muscaria. Can J Microbiol. 1962;8:931–933. [Google Scholar]
- 8.el-Basyouni SH, Vining LC. Biosynthesis of oosporein in Beauveria bassiana (Bals.) Vuill. Can J Biochem. 1966;44(5):557–565. doi: 10.1139/o66-067. [DOI] [PubMed] [Google Scholar]
- 9.Sohair HE, Basyouni SH, Brewer D, Vining LC. Pigments of the genus Beauveria. Can J Bot. 1968;46:441–448. [Google Scholar]
- 10.Takeshita H, Anchel M. Production of oosporein and its leuco form by Basidiomycete species. Science. 1965;147(3654):152–153. doi: 10.1126/science.147.3654.152. [DOI] [PubMed] [Google Scholar]
- 11.Seger C, Längle T, Pernfuss B, Stuppner H, Strasser H. High-performance liquid chromatography-diode array detection assay for the detection and quantification of the Beauveria metabolite oosporein from potato tubers. J Chromatogr A. 2005;1092(2):254–257. doi: 10.1016/j.chroma.2005.08.069. [DOI] [PubMed] [Google Scholar]
- 12.Cole RJ, Kirksey JW, Cutler HG, Davis EE. Toxic effects of oosporein from Chaetomium trilaterale. J Agric Food Chem. 1974;22(3):517–520. doi: 10.1021/jf60193a049. [DOI] [PubMed] [Google Scholar]
- 13.Wainwright M, Betts RP. Antibiotic activity of oosporein from Verticillium psalliotae. Trans Br Mycol Soc. 1986;86:168–170. [Google Scholar]
- 14.Nagaoka T, Nakata K, Kouno K, Ando T. Antifungal activity of oosporein from an antagonistic fungus against Phytophthora infestans. Z Naturforsch C. 2004;59(3-4):302–304. doi: 10.1515/znc-2004-3-432. [DOI] [PubMed] [Google Scholar]
- 15.Alurappa R, Bojegowda MRM, Kumar V, Mallesh NK, Chowdappa S. Characterisation and bioactivity of oosporein produced by endophytic fungus Cochliobolus kusanoi isolated from Nerium oleander L. Nat Prod Res. 2014;28(23):2217–2220. doi: 10.1080/14786419.2014.924933. [DOI] [PubMed] [Google Scholar]
- 16.Amin GA, Youssef NA, Bazaid S, Saleh WD. Assessment of insecticidal activity of red pigment produced by the fungus Beauveria bassiana. World J Microb Biot. 2010;26:2263–2268. [Google Scholar]
- 17.Brewer D, Jen WC, Jones GA, Taylor A. The antibacterial activity of some naturally occurring 2,5-dihydroxy-1,4-benzoquinones. Can J Microbiol. 1984;30(8):1068–1072. doi: 10.1139/m84-166. [DOI] [PubMed] [Google Scholar]
- 18.Terry BJ, et al. Inhibition of herpes simplex virus type 1 DNA polymerase by the natural product oosporein. J Antibiot (Tokyo) 1992;45(2):286–288. doi: 10.7164/antibiotics.45.286. [DOI] [PubMed] [Google Scholar]
- 19.Pegram RA, Wyatt RD. Avian gout caused by oosporein, a mycotoxin produced by Caetomium trilaterale. Poult Sci. 1981;60(11):2429–2440. doi: 10.3382/ps.0602429. [DOI] [PubMed] [Google Scholar]
- 20.Love BE, Bonner-Stewart J, Forrest LA. An efficient synthesis of oosporein. Tetrahedron Lett. 2009;50(35):5050–5052. [Google Scholar]
- 21.Shen B. Polyketide biosynthesis beyond the type I, II and III polyketide synthase paradigms. Curr Opin Chem Biol. 2003;7(2):285–295. doi: 10.1016/s1367-5931(03)00020-6. [DOI] [PubMed] [Google Scholar]
- 22.Schroeckh V, et al. Intimate bacterial-fungal interaction triggers biosynthesis of archetypal polyketides in Aspergillus nidulans. Proc Natl Acad Sci USA. 2009;106(34):14558–14563. doi: 10.1073/pnas.0901870106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zaehle C, et al. Terrein biosynthesis in Aspergillus terreus and its impact on phytotoxicity. Chem Biol. 2014;21(6):719–731. doi: 10.1016/j.chembiol.2014.03.010. [DOI] [PubMed] [Google Scholar]
- 24.Jørgensen SH, et al. Fusarium graminearum PKS14 is involved in orsellinic acid and orcinol synthesis. Fungal Genet Biol. 2014;70:24–31. doi: 10.1016/j.fgb.2014.06.008. [DOI] [PubMed] [Google Scholar]
- 25.Xiao G, et al. Genomic perspectives on the evolution of fungal entomopathogenicity in Beauveria bassiana. Sci Rep. 2012;2:483. doi: 10.1038/srep00483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liao XG, et al. Characterization of a highly active promoter, PBbgpd, in Beauveria bassiana. Curr Microbiol. 2008;57(2):121–126. doi: 10.1007/s00284-008-9163-3. [DOI] [PubMed] [Google Scholar]
- 27.Görner W, et al. Nuclear localization of the C2H2 zinc finger protein Msn2p is regulated by stress and protein kinase A activity. Genes Dev. 1998;12(4):586–597. doi: 10.1101/gad.12.4.586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Luo Z, et al. Bbmsn2 acts as a pH-dependent negative regulator of secondary metabolite production in the entomopathogenic fungus Beauveria bassiana. Environ Microbiol. 2015;17(4):1189–1202. doi: 10.1111/1462-2920.12542. [DOI] [PubMed] [Google Scholar]
- 29.Yu JH, Keller N. Regulation of secondary metabolism in filamentous fungi. Annu Rev Phytopathol. 2005;43:437–458. doi: 10.1146/annurev.phyto.43.040204.140214. [DOI] [PubMed] [Google Scholar]
- 30.Hayes JD, Flanagan JU, Jowsey IR. Glutathione transferases. Annu Rev Pharmacol Toxicol. 2005;45:51–88. doi: 10.1146/annurev.pharmtox.45.120403.095857. [DOI] [PubMed] [Google Scholar]
- 31.Sanchez JF, et al. Molecular genetic analysis of the orsellinic acid/F9775 gene cluster of Aspergillus nidulans. Mol Biosyst. 2010;6(3):587–593. doi: 10.1039/b904541d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lackner G, Bohnert M, Wick J, Hoffmeister D. Assembly of melleolide antibiotics involves a polyketide synthase with cross-coupling activity. Chem Biol. 2013;20(9):1101–1106. doi: 10.1016/j.chembiol.2013.07.009. [DOI] [PubMed] [Google Scholar]
- 33.Lo HC, et al. Two separate gene clusters encode the biosynthetic pathway for the meroterpenoids austinol and dehydroaustinol in Aspergillus nidulans. J Am Chem Soc. 2012;134(10):4709–4720. doi: 10.1021/ja209809t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zheng P, et al. Genome sequence of the insect pathogenic fungus Cordyceps militaris, a valued traditional Chinese medicine. Genome Biol. 2011;12(11):R116. doi: 10.1186/gb-2011-12-11-r116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hu X, et al. Trajectory and genomic determinants of fungal-pathogen speciation and host adaptation. Proc Natl Acad Sci USA. 2014;111(47):16796–16801. doi: 10.1073/pnas.1412662111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.He G, Yan J, Wu XY, Gou XJ, Li WC. Oosporein from Tremella fuciformis. Acta Crystallogr Sect E Struct Rep Online. 2012;68(Pt 4):o1231. doi: 10.1107/S1600536812012950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang B, Kang Q, Lu Y, Bai L, Wang C. Unveiling the biosynthetic puzzle of destruxins in Metarhizium species. Proc Natl Acad Sci USA. 2012;109(4):1287–1292. doi: 10.1073/pnas.1115983109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Behie SW, Bidochka MJ. Nutrient transfer in plant-fungal symbioses. Trends Plant Sci. 2014;19(11):734–740. doi: 10.1016/j.tplants.2014.06.007. [DOI] [PubMed] [Google Scholar]
- 39.Mozaina K, et al. Activity of 1,4-benzoquinones against formosan subterranean termites (Coptotermes formosanus) J Agric Food Chem. 2008;56(11):4021–4026. doi: 10.1021/jf800331r. [DOI] [PubMed] [Google Scholar]
- 40.Duan Z, et al. Linkage of autophagy to fungal development, lipid storage and virulence in Metarhizium robertsii. Autophagy. 2013;9(4):538–549. doi: 10.4161/auto.23575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Blin K, et al. antiSMASH 2.0—a versatile platform for genome mining of secondary metabolite producers. Nucleic Acids Res. 2013;41(Web Server issue):W204-W212. doi: 10.1093/nar/gkt449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Larkin MA, et al. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23(21):2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 43.Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: Molecular evolutionary genetics analysis version 6.0. Mol Biol Evol. 2013;30(12):2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Matys V, et al. TRANSFAC and its module TRANSCompel: Transcriptional gene regulation in eukaryotes. Nucleic Acids Res. 2006;34(Database issue):D108–D110. doi: 10.1093/nar/gkj143. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.