Significance
We demonstrate that the fission yeast telomerase RNA has a stem terminus element (STE) that it is essential for telomerase action in vivo and in vitro. Using a partial loss-of-function STE allele, we show that the STE is required for wild-type telomeric sequence. This is the first example, to our knowledge, of a sequence that is not part of the telomerase RNA core region that affects the sequence of telomeric DNA. Because mutating the STE has the same phenotypes as mutating the template boundary element (TBE), the STE promotes TBE function. The association of two sequence-specific telomere binding proteins is impaired in the STE mutant. Thus, the STE is critical to assemble the normal sequence and chromatin structure of fission yeast telomeres.
Keywords: telomerase RNA, telomere repeat, stem terminus element, Pot1, Taz1
Abstract
The stem terminus element (STE), which was discovered 13 y ago in human telomerase RNA, is required for telomerase activity, yet its mode of action is unknown. We report that the Schizosaccharomyces pombe telomerase RNA, TER1 (telomerase RNA 1), also contains a STE, which is essential for telomere maintenance. Cells expressing a partial loss-of-function TER1 STE allele maintained short stable telomeres by a recombination-independent mechanism. Remarkably, the mutant telomere sequence was different from that of wild-type cells. Generation of the altered sequence is explained by reverse transcription into the template boundary element, demonstrating that the STE helps maintain template boundary element function. The altered telomeres bound less Pot1 (protection of telomeres 1) and Taz1 (telomere-associated in Schizosaccharomyces pombe 1) in vivo. Thus, the S. pombe STE, although distant from the template, ensures proper telomere sequence, which in turn promotes proper assembly of the shelterin complex.
The specialized reverse-transcriptase telomerase compensates for the inability of the conventional semiconservative replication machinery to copy the very end of the chromosome. In addition to solving this “end replication problem,” telomeres perform other roles, such as protecting chromosomes from degradation and end-to-end fusions, a function that requires both telomeric DNA and telomere-associated proteins. In the fission yeast Schizosaccharomyces pombe, telomeric DNA is bound and protected by a six-protein complex that has many similarities to mammalian shelterin. S. pombe shelterin protects the telomere, inhibits nonhomologous end joining, facilitates telomere replication, and recruits telomerase (1–3).
The shelterin complex binds both double-stranded (ds) telomeric DNA and the terminal single-stranded (ss) guanine-rich overhang, known as the G-tail (3). The ds region of S. pombe telomeres is bound by the Myb domain of the S. pombe shelterin component Taz1 (telomere-associated in Schizosaccharomyces pombe 1), whereas the G-tail is bound by the OB domains of Pot1 (protection of telomeres 1). Both the Myb and OB (oligonucleotide/oligosaccharide-binding) domains are present in the mammalian homologs of these proteins, TRF1/TRF2 and POT1, respectively (1–3).
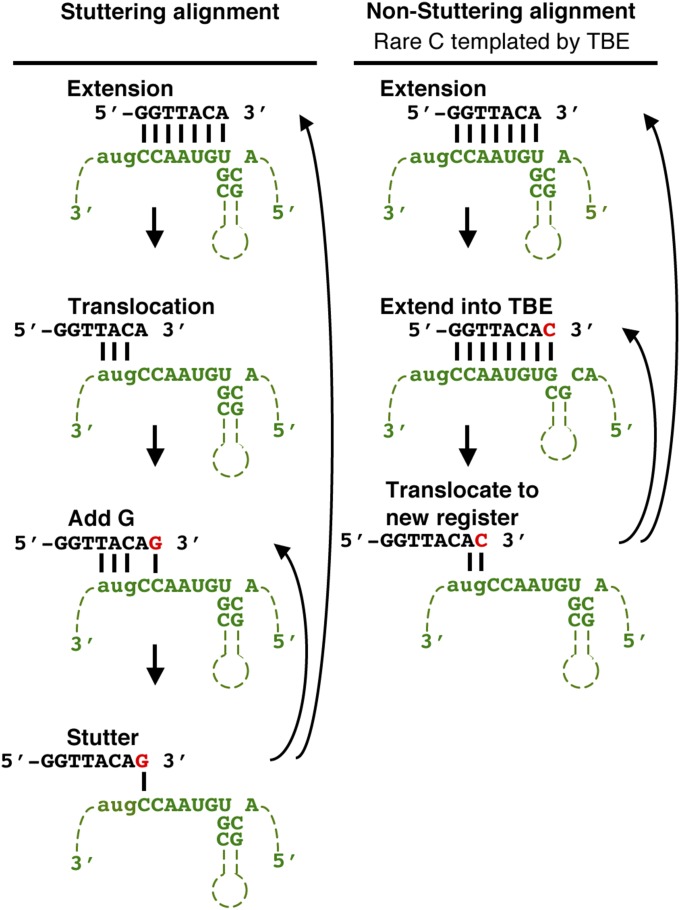
Although telomeric DNA almost always consists of short repeats, the fidelity of these repeats is not perfect in most organisms, including humans (4). In S. pombe, repeat heterogeneity is particularly high. Although the core S. pombe telomere repeat 5′-GGTTACA-3′ is incorporated at high levels, heterogeneity is created by the inclusion of 1–6 guanines before the core repeat and more rarely by the addition of a cytosine at the end of the repeat. These additions yield a telomere consensus sequence of 5′-(G)0–6GGTTACAC-3′ (rare cytosine is underlined throughout this paper). Almost half (42%) of the telomeric repeats are preceded by a guanine tract of 1–6 Gs, whereas the rare cytosine is present in ∼12% of the repeats (5). The (G)1–6 tracts result from template stuttering between the 3′ end of the telomerase RNA template and the end of the telomere (Fig. S1, Left). The rare cytosine is introduced by the inefficient action of the template boundary element (TBE), a short helix immediately adjacent to the core template region that prevents the catalytic subunit from copying a noncore templating sequence. When the TBE does not function properly, it allows reverse transcription past the core templating sequence into the TBE helix, which codes for the rare cytosine (6–8) (Fig. S1, Right).
Fig. S1.
Mechanism for incorporation of sequence heterogeneity into S. pombe telomeres (6–8). (Left) Telomere repeat addition is templated by 3 bp formed between the telomere end and the TER1 template. This alignment leaves the terminal adenine unpaired and contributes to the stuttering reaction that produces variable guanine tracts in S. pombe. (Right) If reverse transcription proceeds into the 5′ TBE, then a cytosine is added to the end of the core 5′-GGTTACA-3′ repeat to generate 5′-GGTTACAC-3′, which causes a nonstuttering register after translocation (underlined C is the rare cytosine incorporated by TBE read-through).
All telomerase RNAs contain a short sequence, the template, which is copied by the reverse-transcriptase subunit, called Trt1 (telomerase reverse transcriptase) in S. pombe, to lengthen the 3′ end of the chromosome. The TBE is also conserved. Likewise, most telomerase RNAs contain a pseudoknot that affects template use. Finally telomerase RNAs from yeast to vertebrates contain a structural element called the three way junction (TWJ), whose primary sequence and secondary structure are both conserved (9). Until this report, only the core region, consisting of the template, TBE, and pseudoknot, was implicated in establishing the sequence of telomeric DNA.
Here we identify the S. pombe TER1 (telomerase RNA 1) stem terminus element (STE) tetraloop, a region that is distant from the template, as an “enforcer” that limits the incorporation of atypical repeats into telomeric DNA. STEs are identifiable in ciliate, yeasts, and mammalian telomerase RNAs. Although STEs are critical for telomerase activity in ciliates, mammals, some yeasts, and (as shown here) S. pombe, the Saccharomyces cerevisiae STE is not (10, 11). However, a different region of TLC1 was recently identified that may perform similar functions to mammalian and S. pombe STEs (12).
The ciliate STE is a terminal stem loop structure, and the budding yeast STE is a TWJ with bulged nucleotides (11). Most mammalian STEs, however, contain a terminal stem loop called the P6.1 helix (13), which partially overlaps the TWJ (11, 13, 14). As in humans, the predicted secondary structure for the S. pombe TER1 STE contains both a TWJ and P6.1 helix and loop, making S. pombe an excellent model for the human STE (6, 15) (Fig. 1 A and B). In this study, we define the STE as the TWJ and P6.1 helix (as in ref. 9; our definition does not include the surrounding RNA).
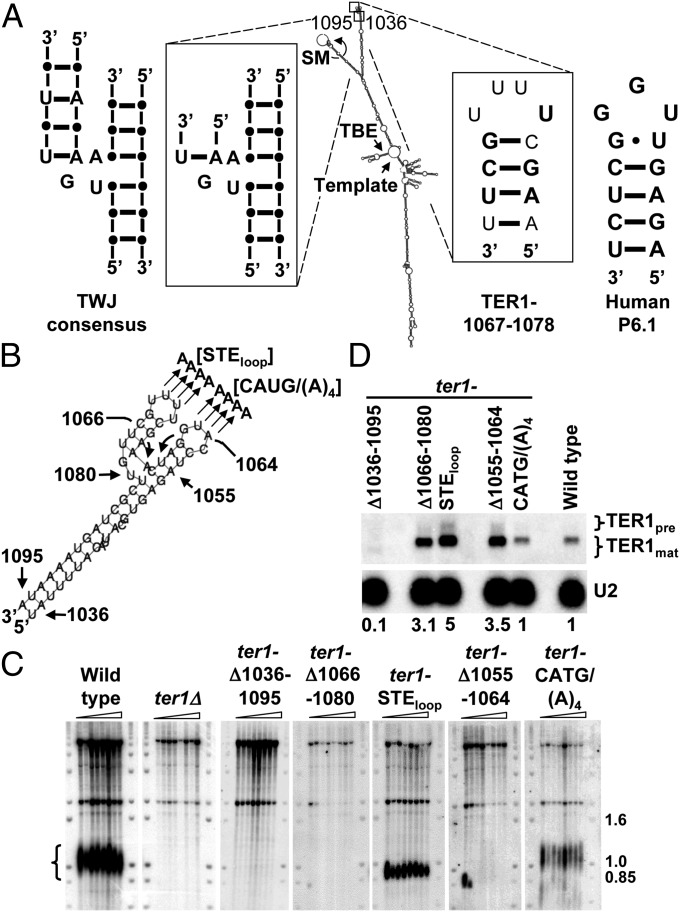
Fig. 1.
In vivo analysis of the STE region of TER1. (A) Model for the TER1 secondary structure (6). The TWJ and TER1-1067–1078 regions are enlarged to show detail. Nucleotides and base pairs that are identical to the TWJ consensus (11) or the human P6.1 sequence (13, 18) are in bold. SM, Sm binding site (6, 19); TBE, template boundary element (6, 7); Template, templating sequence; TWJ, three way junction. (B) TER1-1036–1095 enlarged. Mutants made for this study are indicated. (C) Telomere blots. Genomic DNA extracted from successively streaked spore products was digested with EcoRI and hybridized to a telomeric probe. Telomere signal is indicated by brackets. Molecular weight markers are in kb. (D) Northern blot. Total RNA extracted from ter1 mutants described in C were hybridized to TER1 and U2 probes. TER1 precursor and mature forms are indicated. Relative levels of TER1 normalized to the U2 signal are indicated below the blot.
We report that deletion of the S. pombe STE was incompatible with telomere maintenance and normal levels of TER1 RNA. However, a partial loss-of-function allele in the STE loop, hereafter called ter1-STEloop, was generated that maintained stable but short telomeres. The ter1-STEloop allele greatly reduced telomerase activity in vitro. Neither the association of this mutant TER1 with the telomerase complex nor the binding of the complex to telomeres was compromised. However, loss of TBE function in the TER1-STEloop RNA produced telomeres of atypical sequence that had more rare cytosines and fewer guanine tracts than wild-type telomeric DNA. These aberrant sequences reduced Pot1 and Taz1 telomere binding.
We conclude that in addition to its essential role for telomerase activity, the well-conserved STE is required for wild-type TBE function. When the STE and hence the TBE are not fully functional, telomeric DNA contains a high number of aberrant repeats, and these reduce shelterin binding. Because a human partial loss-of-function STE allele that is associated with aplastic anemia results in short telomeres and very low telomerase activity (13), it would not be surprising if the human STE also affects TBE function and the sequence of telomeric DNA.
Results
The Uncharacterized TER1 Arm Terminates in the STE, Which Promotes TER1 Stability and Telomere Maintenance.
We previously showed that the terminus of the long TER1 arm binds Est1 (ever shorter telomeres protein 1) (16). The other region splits into two arms: One arm binds Sm and Lsm proteins (17), and the other is uncharacterized. We undertook a mutational structure and function analysis of the end of this uncharacterized arm. We generated and analyzed a series of mutants and determined their in vivo effects on telomeres.
The first mutant removed the most distal 59 nt of the arm (ter1-Δ1036–1095) (Fig. 1 A and B). Cells expressing this mutation were streaked seven successive times, and DNA was prepared for Southern blotting. This analysis demonstrated that ter1-Δ1036–1095 cells lost telomeric DNA and senesced in a manner similar to that of ter1Δ cells (Fig. 1C). Because Northern analysis showed extremely low levels of TER1-Δ1036–1095 RNA (Fig. 1D), telomere loss in this mutant can be explained by insufficient amounts of telomerase RNA.
Next, we generated less severe mutations. The terminus of the arm consists of two stem loops, each comprised of a 4-bp helix with a 4-nt loop at the end (Fig. 1 A and B). Deletion of stem loop ter1-Δ1055–1064 or ter1-Δ1066–1080 resulted in the complete loss of telomeric DNA (Fig. 1C). Unlike mutant ter1-Δ1036–1095, which deleted the entire end of the arm, the telomere loss phenotypes of these alleles could not be attributed to RNA instability, as the steady-state level of each TER1 stem loop mutant was higher than that of wild-type TER1 (Fig. 1D). Examination of the TER1-1066–1080 sequence (Fig. 1 A and B) revealed that it is similar to the telomerase RNA TWJ consensus sequence and structure, which is found in >90% of vertebrate and yeast telomerase RNAs (11) as well as the human P6.1 helix (13, 18). Based on its sequence, secondary structure, and critical role in telomere maintenance, we conclude that the TER1-1066–1080 stem loop and TWJ is the highly conserved STE in S. pombe (9, 13).
To refine the contribution of each terminal stem loop to telomere maintenance, substitution alleles were constructed in which the sequences of the 4-nt loops were changed. Mutation of the loop in stem loop nucleotides 1055–1064 to adenines [ter1-CATG/(A)4] maintained near wild-type telomere length and RNA levels (Fig. 1 C and D). In contrast, adenine substitutions of the STE loop (ter1-STEloop) resulted in short but stable telomeres, ∼150 bp shorter than wild type and five times higher levels of telomerase RNA (Fig. 1 C and D). High expression of telomerase RNA does not explain the short telomere phenotype of ter1-STEloop cells, as, as previously reported (6), TER1 overexpression did not affect telomere length (Fig. S2A) even though its expression level was sevenfold higher than the endogenous level (Fig. S2B). Some STE mutants were semidominant, with heterozygous diploids having shorter telomeres than ter1+/− cells (Fig. S3). Because ter1-STEloop is a partial loss-of-function allele, it was used in subsequent experiments to assess the function of the S. pombe STE.
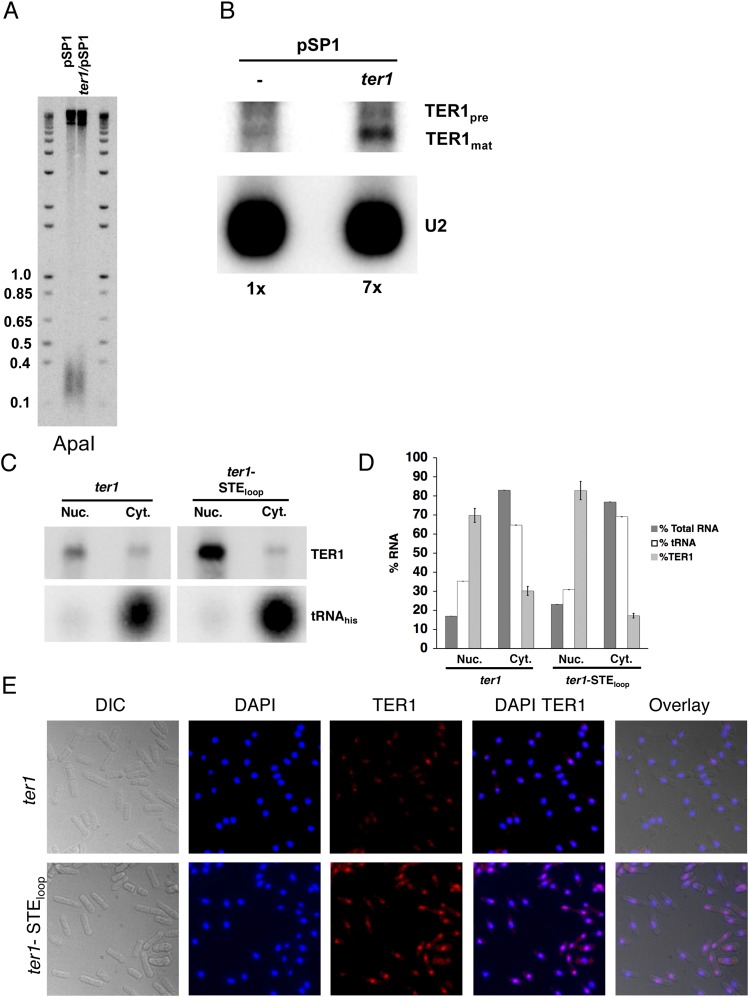
Fig. S2.
TER1 overexpression does not cause short telomeres. (A) Southern blots. Genomic DNA was extracted from cells harboring pSP1 and ter1/pSP1 after seven successive streaks. DNA was digested with ApaI and hybridized to a telomeric probe. Molecular weight markers are in kb. (B) Northern blot. Total RNA was extracted from wild-type cells containing either pSP1 vector or pSP1 carrying ter1 expressed from its own promoter [ter1/pSP1 (6)]. Northern blotting was performed as described in Fig. 1D. Relative levels of TER1 normalized to the U2 signal are indicated below the blot. (C) TER1 and TER1-STEloop RNAs localize to the nucleus. Northern blot of RNAs isolated from cytoplasmic and nuclear fractions. TER1 was identified by hybridization as in Fig. 1D. Effective fractionation was demonstrated by localization of the tRNAhis to the cytoplasmic fraction. (D) Nuclear and cytoplasmic RNAs analyzed in C were used as substrates for quantitative RT-PCR analysis. RNAs detected in each fraction are expressed as percent of the nuclear plus cytoplasmic RNAs. Error bars are SDs. The amounts of total fractionated RNA were determined by Nanodrop spectrophotometer. (E) FISH. Cells expressing TER1 or TER1-STEloop were fixed and hybridized with TER1 (Quasar 670) fluorescence probes (Biosearch Technologies). Nuclear DNA is indicated by DAPI staining.
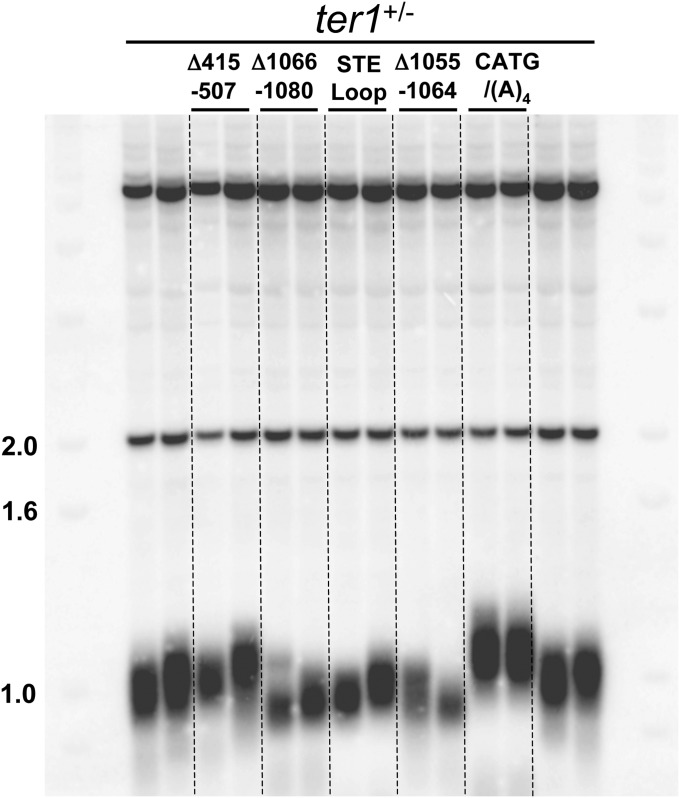
Fig. S3.
Telomere lengths in heterozygous ter1 diploid cells. Shown is the telomere blot. Genomic DNA from heterozygous diploid ter1+/MUTANT cells was isolated after cells were streaked >5 times. DNA was digested with ApaI and hybridized to a telomeric probe. The mutant allele in the heterozygous diploid is indicated above each lane. The first two and last two lanes contain DNA from ter1+/− diploid cells. Molecular weight markers are in kb.
TER1 and TER1-STEloop Localize to the Nucleus.
A defect in nuclear-cytoplasmic shuttling and/or in nuclear retention might explain the high cellular TER-STEloop steady-state levels (Fig. 1D). To address these possibilities, nuclear and cytoplasmic RNAs from WT and ter1-STEloop cells were fractioned. The fractions were analyzed by both Northern blotting (Fig. S2C) and quantitative RT-PCR (Fig. S2D). Like wild-type TER1 RNA, over 70% of TER1-STEloop RNA was in the nuclear fraction. Fluorescence in situ hybridization (FISH) confirmed that TER1 and TER1-STEloop RNA were mostly in the nucleus (Fig. S2E). Although we detected no defects in intracellular trafficking of the TER1-STEloop RNA, we speculate that the increase in its abundance reflects a processing bottleneck. For example, the human STE loop is pseudouridinylated in the nucleus (13). If this modification also occurs in S. pombe, the inability to modify the STE loop in TER1-STEloop RNA could slow RNA processing.
TER1-STEloop Does Not Affect Holoenzyme Formation or Telomerase–Telomere Interaction.
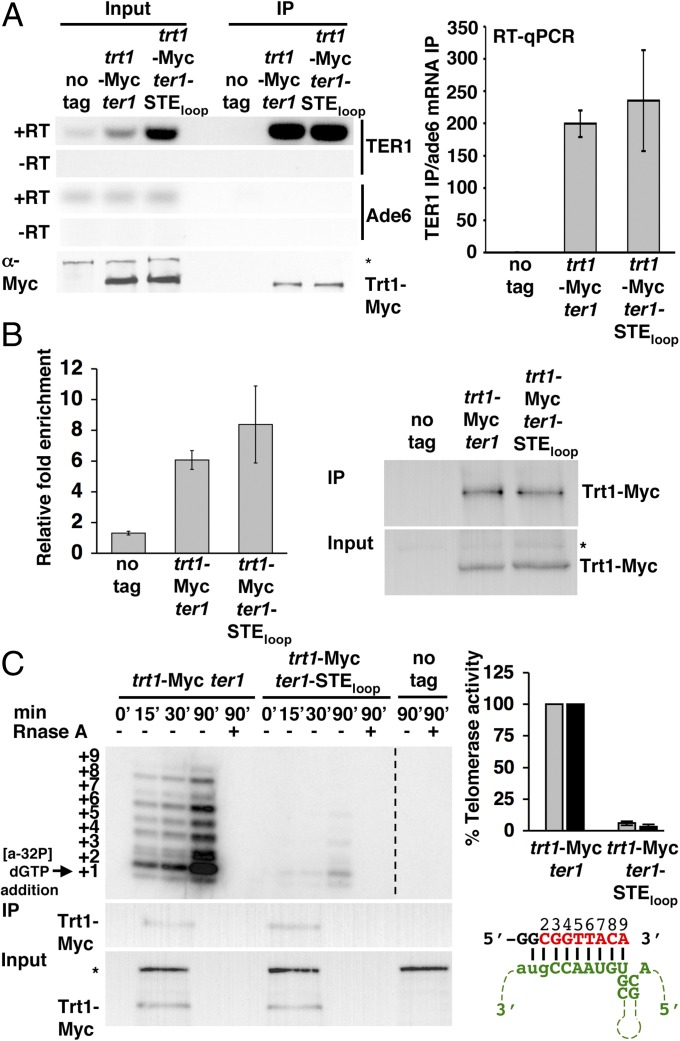
Another possibility is that the short telomeres in ter1-STEloop cells are due to defective holoenzyme formation. The TER1–Est1 interaction is STE-independent (16). However, it was not known if Trt1 requires the STE to interact with TER1. We used RNA immunoprecipitation and RT-PCR to determine if Trt1 interacts normally with TER1-STEloop RNA. As previously demonstrated (6, 19), Trt1-Myc efficiently immunoprecipitated wild-type TER1 (Fig. 2A). The same amount of TER1-STEloop was immunoprecipitated by Trt1-Myc (Fig. 2A, Right), even though TER1-STEloop was more abundant (Fig. 2 A, Left). Western blotting demonstrated that Trt1-Myc levels were the same in WT and ter1-STEloop cells (Fig. 2 A, Left). Thus, the short telomeres in ter1-STEloop cells are not due to a defective Trt1–TER1 interaction. This result also demonstrates that processing of the 3′ end of TER1, called slicing, is not impaired by the ter1-STEloop allele, as Trt1 does not interact with improperly sliced TER1 RNA (17).
Fig. 2.
The ter1-STEloop allele does not affect the Trt1–TER1 interaction or telomerase recruitment to telomeres but is necessary for normal catalytic activity. (A, Left) Co-immunoprecipitation of Ade6 mRNA and TER1 or TER1-STEloop with Trt1-G8-13Myc. Ade6 mRNA, TER1, and TER1-STEloop were detected in the immunoprecipitate (IP) and Input lysate by 22 cycles of RT-PCR. Absence of contaminating DNA was demonstrated by reactions without reverse transcriptase (–RT). α-Myc Western blotting was used to determine levels of Trt1-G8-13Myc in Input and IP. The asterisk indicates a nonspecific band. (A, Right) Relative fold enrichment of TER1 and TER1-STEloop by Trt1-G8-13Myc quantified by quantitative RT-PCR. The enrichment of TER1 or TER1-STEloop in the Trt1-G8-13Myc IP amplified by a common region of TER1 normalized to Ade6 mRNA in the Trt1-G8-13Myc IP is depicted on the y axis. Error bars represent SD (trt1-G8-13Myc TER1 compared with trt1-G8-13Myc TER1-STEloop; P = 0.596). (B, Left) Effects of TER1 and TER1-STEloop on Trt1-G8-13Myc–telomere interaction determined by ChIP. Shown is the relative fold enrichment of Trt1-G8-13Myc at the telomere in ter1 and ter1-STEloop cells. Relative fold enrichment compares telomere-adjacent STE1 IP/input signal with act1 IP/input signal. (B, Right) Western blot using α-Myc to detect levels of Trt1-G8-13Myc in Input and ChIP. The asterisk indicates a nonspecific band. Error bars represent SD (trt1-G8-13Myc compared with trt1-G8-13Myc ter1-STEloop; P = 0.195). (C, Left) In vitro telomerase assay. α-Myc antibodies conjugated to agarose beads were used to immunoprecipitate equivalent amounts of Trt1-G8-13Myc from ter1 and ter1-STEloop cells to perform an in vitro telomerase primer extension reaction. Extension products are visualized by sequencing gel electrophoresis. RNase A treatment demonstrates RNA dependence for primer extension activity. A dashed line indicates the removal of lanes from the otherwise unmodified gel image. α-Myc Western blotting monitored the levels of telomerase in the reaction and Input lysate. (C, Upper Right) Quantification of telomerase reactions. Incorporations at the +2 (gray) and +3 (black) positions are shown as a percentage of wild-type activity. (C, Lower Right) Schematic of pBoli14 primer (black) aligned with the TER1 templating region (green). The identity of de novo telomeric sequence (red) was reported in ref. 20. The position of each added nucleotide in relation to the sequencing gel extension products are numbered above the de novo sequence. The +1 position is due to primer degradation and dG misincorporation (20).
We used chromatin immunoprecipitation (ChIP) to determine if the short telomere phenotype of ter1-STEloop cells was due to reduced telomere binding of the telomerase complex. Because Trt1 interacted similarly with wild-type and ter1-STEloop telomeres (P = 0.596) (Fig. 2B), we conclude that the ter1-STEloop short telomeres are not due to defective recruitment of telomerase to telomeres. Furthermore, because Trt1:TER1-STEloop binding and holoenzyme–telomere interaction occurred normally, we conclude that the 4-nt TER1-STEloop substitution did not affect TER1 folding.
The STE Loop Is Required for Normal Levels of in Vitro Telomerase Activity.
An in vitro telomerase activity assay was performed to determine if TER1-STEloop compromised telomerase activity. Robust telomerase activity over a 90-min time course was observed in Trt1-Myc immunoprecipitated from wild-type cells (Fig. 2C). Even though we used the substrate that produces the best primer extension (20) (Fig. 2 C, Right Lower), after 90 min Trt1:TER1-STEloop incorporated only 6% and 3% of wild-type nucleotide incorporation at positions +2 and +3, respectively. Incorporation continued to decrease with each subsequent position and was undetectable after position +6 (Fig. 2 C, Right Upper). Thus, the ter1-STEloop short telomere phenotype is most likely due to telomerase catalytic defects. The conclusion that the STE loop affects catalysis is consistent with results showing that a 61-nt fragment that includes the STE can function in trans to restore telomerase activity in a rabbit reticulocyte lysate assay reconstituted with a mutant TER1 RNA that included the core region (15).
The STE Loop Is Required for Normal Telomere Sequence.
Unlike mammalian systems (9, 21), S. pombe in vitro telomerase assays cannot add multiple rounds of telomeric repeats to a substrate (15, 20, 22). The lack of repeat processivity suggests that the in vitro systems lack one or more components that are needed for full telomerase activity. In addition, the telomerase activity produced by ter1-STEloop telomerase complexes in vitro was so poor that it was not possible to assess nucleotide incorporation after the +3 position or the sequence of the added DNA (Fig. 2C).
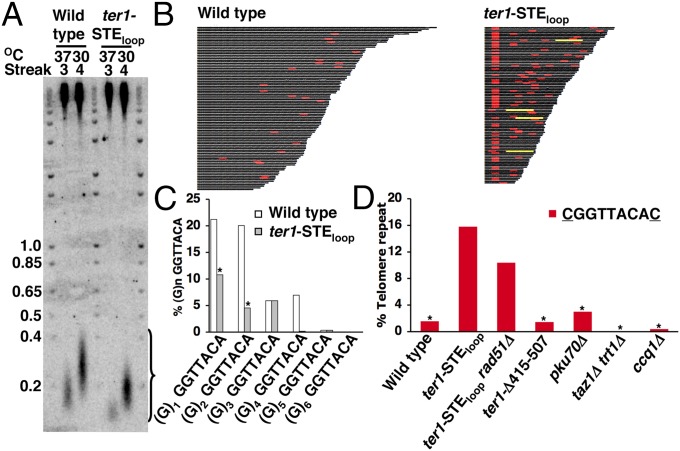
To determine the effects of the ter1-STEloop mutant on telomere sequence, we generated a new system to manipulate S. pombe telomere length that allowed us to determine the impact of the ter1-STEloop mutation on telomeric sequence directly. As in S. cerevisiae (23), telomeres in wild-type S. pombe shortened during growth at 37 °C. After cells were returned to 30 °C, telomeres returned to wild-type lengths within ∼25 generations (Fig. 3A). We exploited this temperature-dependent telomere length regulation to shorten and then reextend the telomeres in both wild-type and ter1-STEloop cells (Fig. 3A). After relengthening, telomeres from wild-type and mutant cells were cloned and sequenced. The most internal portions of the telomeric tracts, which were present at all telomeres, were not used in this analysis, as these must have been present before telomere shortening at 37 °C (Fig. 3B). All of the telomere sequence data in this study were obtained by this method.
Fig. 3.
Telomeric DNA from ter1-STEloop cells has increased rare cytosines and reduced guanine tracts. (A) Telomere blot of genomic DNA was extracted from wild-type and ter1-STEloop cells after three successive streaks at 37 °C and one subsequent streak at 30 °C. (B) Telomere sequences. Telomeric sequences recovered from wild-type and ter1-STEloop cells after three successive streaks at 37 °C and one subsequent streak at 30 °C. The 5′-CGGTTACAC-3′ repeats are in red, and the (5′-GGTTACAC-3′)4 tracts are in yellow. We analyzed 100 wild-type and 104 ter1-STEloop telomeres. (C) Bar chart. Core repeats (5′-GGTTACA-3′) preceded by variable length guanine tracts are normalized to core repeats not preceded by a guanine. The percentage of guanine tracts in ter1-STEloop telomeres compared with wild type was 40%. An asterisk above the bars indicates χ2 analysis (P < 0.0005) of the mutant compared with wild type. (D) Bar chart. The 5′-CGGTTACAC-3′ repeats are normalized to the core 5′-GGTTACA-3′ repeat for wild type and mutants after telomere shortening and extension. An asterisk indicates that the frequency of 5′-CGGTTACAC-3′ repeats was significantly different (P < 0.0005) from that in ter1-STEloop cells by χ2 analysis.
Although the sequence of wild-type telomeric DNA was similar to a previous report (5), the DNA from ter1-STEloop telomeres had a dramatic 60% reduction in guanine tracts of all lengths compared with wild type (Fig. 3C). Most S. pombe guanine tracts have either one or two extra guanines (5). However, in ter1-STEloop telomeric DNA, there was a highly significant 52% (P < 0.0005 by χ2 analysis) and 75% (P < 0.0005) reduction in guanine residues, respectively, at these two positions.
The levels of rare cytosines were also affected in the mutant. Although 9% of wild-type repeats contained the rare cytosine (5′-GGTTACAC-3′) [similar to the 12% previously reported (5)], 24% of the repeats in ter1-STEloop telomeric DNA were 5′-GGTTACAC-3′ repeats. Consistent with guanine tract loss being due to altered template–substrate alignment (7, 8) (Fig. S1, Right and Discussion), we observed a significant increase (P < 0.0005; Fig. 3D) in the density of 5′-CGGTTACAC-3′ normalized to the core 5′-GGTTACA-3′ sequence in ter1-STEloop telomeres relative to wild type. Thus, the sequence of ter1-STEloop telomeric DNA was significantly different from wild type in two ways, having both reduced guanine tracts and increased rare cytosines.
The sequence 5′-GGTTACAC-3′ is relatively rare in WT telomeric DNA, accounting for only 9% of the repeats. However, because ter1-STEloop telomeric DNA contained more rare cytosines and fewer guanine tracts, even tandem copies of 5′-GGTTACAC-3′ were seen. For example, in the 104 sequenced ter1-STEloop telomeres, there were four occurrences of (5′-GGTTACAC-3′)4 (Fig. 3B). We determined the expected probability of (5′-GGTTACAC-3′)4 in wild-type telomeres based on our analysis of the sequences of 1,453 wild-type telomere repeats. As only 9% of wild-type repeats were 5′-GGTTACAC-3′, the calculated frequency of four tandem repeats is 0.007%. In addition, 59.4% of wild-type repeats lacked a (G)1–6 tract. Thus, the chance of four tandem repeats without a (G)1–6 tract is 12.4%, which leads to a probability of only 0.0009% for (5′-GGTTACAC-3′)4 in wild-type telomeric DNA. We conclude that the frequency of (5′-GGTTACAC-3′)4 in 759 ter1-STEloop telomeric repeats (Fig. 3B) was ∼580 times higher than expected for the same number of wild-type repeats.
The Unusual ter1-STEloop Telomeric Sequence Is Not Due to Having Short Telomeres, Loss of Telomere Capping, or Replication Errors.
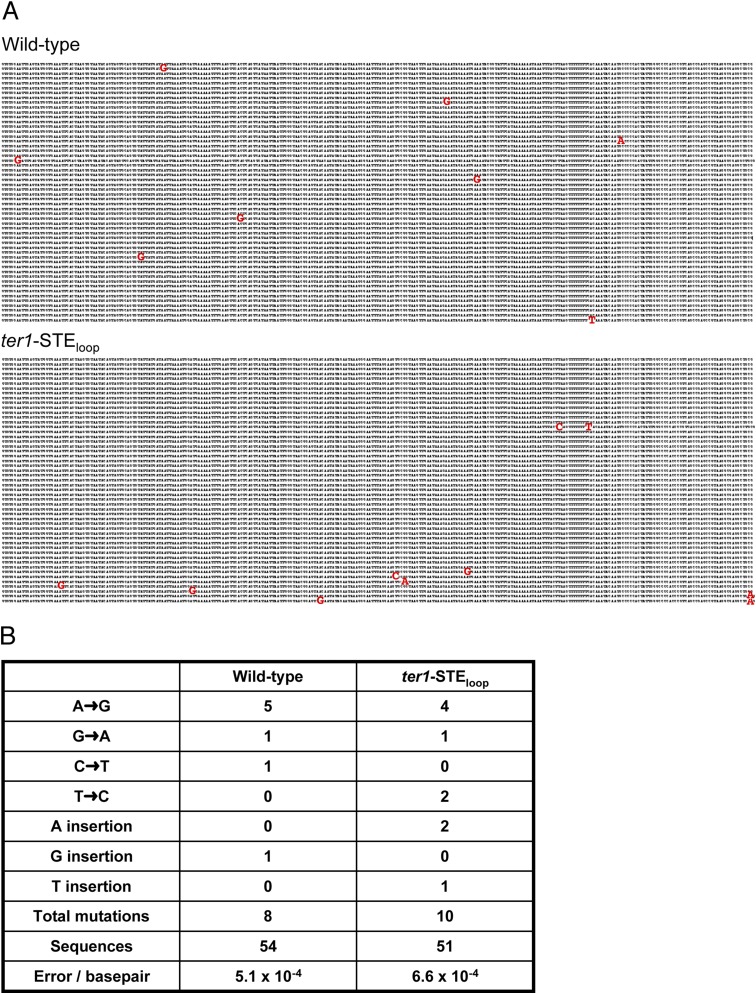
We tested the possibility that the unusual sequence of ter1-STEloop telomeres was a general feature of short telomeres. To this end, we analyzed telomeric DNA from two mutants, which have telomeres of a length similar to those in ter1-STEloop cells. The occurrences of 5′-CGGTTACAC-3′ in ter1-Δ415–507 (16) telomeres were significantly different from ter1-STEloop (Fig. 3D). Similarly, short telomeres produced by removal of the Ku heterodimer [pku70Δ (24)] had significantly fewer 5′-CGGTTACAC-3′ repeats than found in the ter1-STEloop. As Pku is required for telomere capping (24), we can conclude that the unusual sequence of ter1-STEloop telomeric DNA does not result from their being short or from their having impaired capping. Comparison of the DNA sequence in the region immediately adjacent to the telomere in ter1-STEloop and wild-type cells showed equivalent nucleotide misincorporation rates (Fig. S4), which demonstrated that the unusual ter1-STEloop repeats are unlikely to be due to decreased replication fidelity.
Fig. S4.
Comparison of wild-type and ter1-STEloop subtelomeric DNA sequence. (A) DNA sequences 293 bp immediately upstream of the first telomeric repeat were cloned and sequenced from wild-type and ter1-STEloop cells. We analyzed 54 wild-type and 51 ter1-STEloop subtelomeric sequences. Nucleotide misincorporations are shown in red. (B) Analysis of nucleotide misincorporations shown in A.
ter1-STEloop Telomeres Are Not Maintained by Recombination.
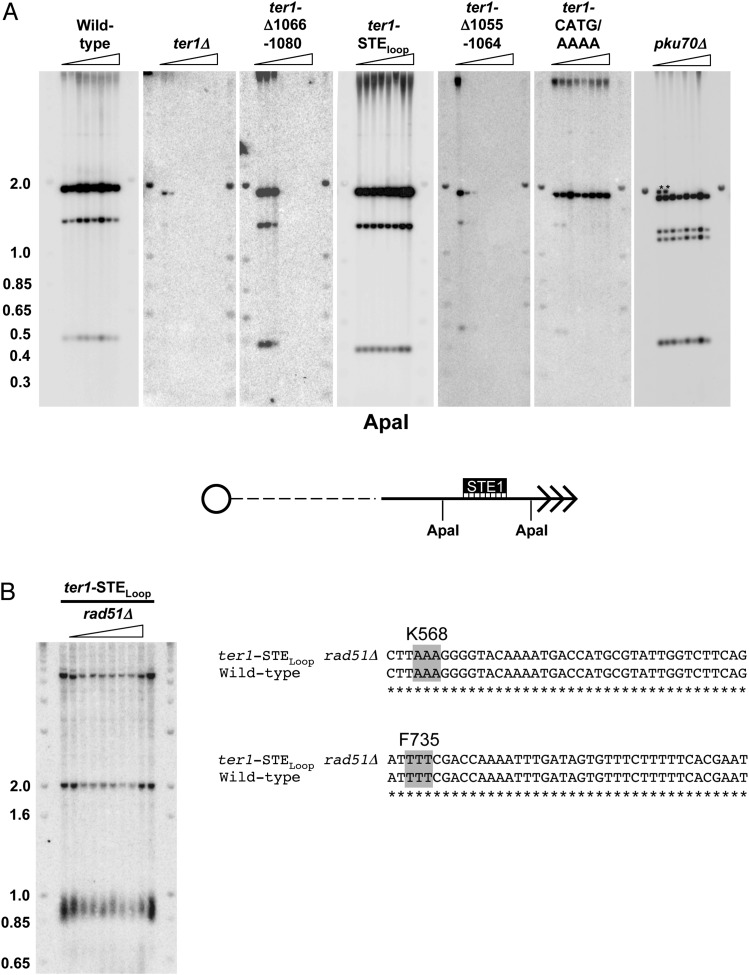
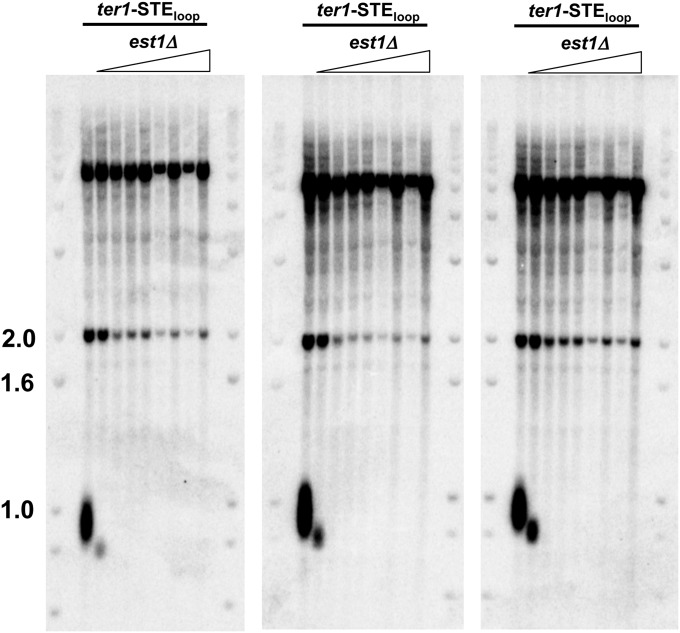
Although ter1-STEloop and ter1-CATG/(A)4 cells did not senesce, we considered the possibility that these mutants maintained telomeres by recombination. We did several experiments to test this possibility. First, we used Southern analysis to detect the rearrangements of subtelomeric DNA produced in cells that maintain telomeres by recombination (24, 25). Genomic DNA extracted from seven successively streaked ter1-STEloop and ter1-CATG/(A)4 colonies demonstrated that the subtelomeres were stable (Fig. S5A). Second, we determined the sequence of telomeric DNA in taz1Δ trt1Δ and ccq1Δ (coiled coil protein quantitatively enriched) cells, strains where telomeres are maintained solely by recombination (25, 26). However, neither taz1Δ trt1Δ nor ccq1Δ telomeric DNA had an increased content of aberrant repeats (Fig. 3D). Third, we generated a ter1-STEloop rad51Δ strain and monitored telomere length over multiple restreaks. By Southern analysis, ter1-STEloop rad51Δ (radiation sensitive 51) cells maintained stably short telomeres (Fig. S5B). As S. pombe Rad51 is required for most homologous recombination events including recombinational maintenance of telomeres (2, 3), we conclude that telomeres in ter1-STEloop cells can be maintained for hundreds of generations without recombination (Fig. S5B). Moreover, the telomeric sequence in ter1-STEloop rad51Δ cells was significantly different from wild-type telomeres (Fig. 3D). Finally, ter1-STEloop est1Δ cells rapidly lose telomeres and senesce, indicating that ter1-STEloop telomeres are maintained by telomerase, not recombination (Fig. S6). Thus, the TER1 templating function must be impaired in ter1-STEloop cells.
Fig. S5.
Recombination is not elevated in ter1-STEloop subtelomeric DNA. (A) Southern blots. Genomic DNA from wild-type and TER1 mutants was isolated from seven successive streaks as described in Fig. 1C (24). DNA was digested with ApaI and hybridized to the STE1 subtelomeric probe (25). Recombination is demonstrated by the transient appearance of novel subtelomeric fragments as in pku70Δ cells (marked by asterisks). Molecular weight markers are in kb. (Lower) Schematic of the subtelomeric region and telomere with ApaI sites and STE1 probe indicated. (B, Left) ter1-STEloop telomeres are maintained for many generations in the absence of homologous recombination. rad51 was disrupted in ter1Δ haploid cells that expressed ter1-STEloop from the leu1 locus (ter1-STEloop). Genomic DNA from ter1-STEloop rad51Δ and ter1-STEloop cells was isolated from seven successive streaks. DNA was digested with EcoRI and hybridized to a telomeric probe. Molecular weight markers are in kb. (B, Right) Sequence analysis. To demonstrate that the rad51Δ phenotype was not suppressed by fbh1 mutation, the fbh1 loci in ter1-STEloop rad51Δ cells were PCR amplified, sequenced, and compared with reported wild-type fbh1 sequence. The relevant sequence is depicted. Asterisks denote identical sequence.
Fig. S6.
Telomeric DNA is not maintained in ter1-STEloop cells that lack telomerase. The telomerase holoenzyme component est1 was deleted in ter1-STEloop cells. Genomic DNA from ter1-STEloop and three independently generated ter1-STEloop est1Δ mutants was isolated after seven successive streaks. DNA was digested with ApaI and hybridized to a telomeric probe. Molecular weight markers are in kb.
Pot1 Binding to ter1-STEloop Telomeres Is Reduced.
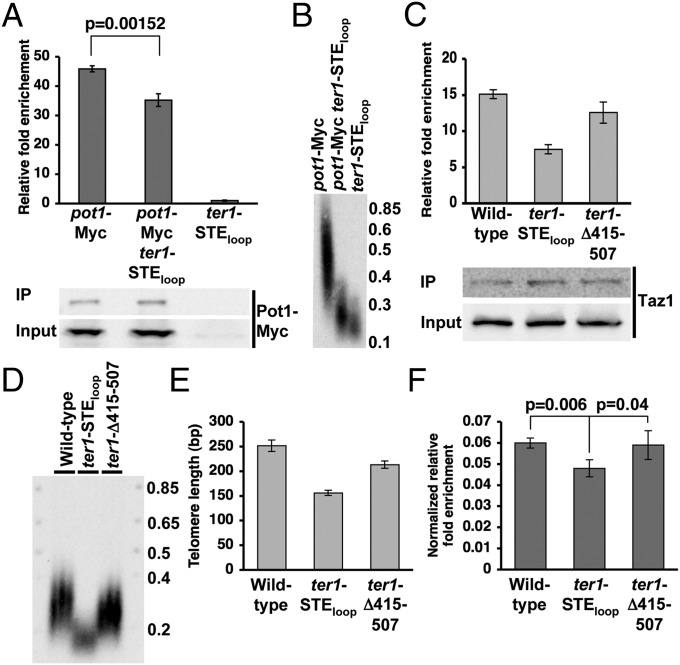
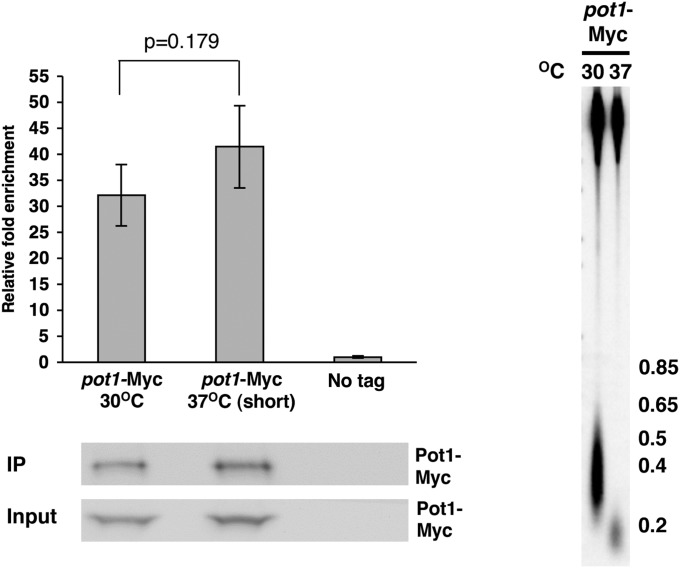
A ChIP assay was performed to determine if Pot1 binding to S. pombe telomeres in vivo was affected by the ter1-STEloop allele (Fig. 4A). Indeed, Pot1-Myc bound less well to ter1-STEloop than to wild-type telomeres (P = 0.00152). This difference was not due to reduced levels of Pot1-Myc in the ter1 mutant (Fig. 4A), nor can it be explained by the shorter length of ter1-STEloop telomeres (Fig. 4B), as Pot1 bound equally well to short (37 °C) and wild-type–length (30 °C) telomeres when both had wild-type sequence (Fig. S7).
Fig. 4.
Pot1 and Taz1 bind less well to ter1-STEloop telomeres compared with wild type. (A, Upper) ChIP assay demonstrating Pot1 relative fold enrichment at wild-type and ter1-STEloop telomeres. ChIP was performed as described in Fig. 2B. Error bars represent SD, and P values were determined by two-tailed Student’s t test. (A, Lower) Western blot showing levels of Pot1-Myc in input lysates and immunoprecipitates. (B) Telomere blot comparing telomere lengths of pot1-Myc, pot1-Myc ter1-STEloop, and ter1-STEloop cells. (C, Upper) ChIP assay demonstrating Taz1 relative fold enrichment at wild-type, ter1-STEloop, and ter1-Δ415–507 telomeres. (C, Lower) Western blot showing levels of Taz1 in input lysates and immunoprecipitates. (D) Telomere blot comparing telomere lengths of wild-type, ter1-STEloop, and ter1-Δ415–507 cells. (E) Quantification of telomere lengths shown in D. Lengths were determined by measured telomeric fragment length minus 45 bp, which is the distance from telomeric sequence to the ApaI site. (F) Normalized ChIP assay depicting Taz1 relative fold enrichment normalized to individual telomere lengths for wild-type, ter1-STEloop, and ter1-Δ415–507 cells. Error bars represent SD, and P values were determined by two-tailed Student’s t test.
Fig. S7.
Short telomeres are not sufficient to cause reduced Pot1 binding. (Upper Left) ChIP assay demonstrating Pot1 relative fold enrichment at wild-type (30 °C) and short (37 °C) telomeres. ChIP was performed as described in Fig. 4. (Bottom Left) Western blot showing levels of Pot1-Myc in input lysates and immunoprecipitates. (Right) Telomere blot comparing telomere lengths of pot1-Myc wild-type cells before and after growth at 37 °C to shorten telomeres.
ter1-STEloop Telomeric DNA Reduces Taz1 Binding.
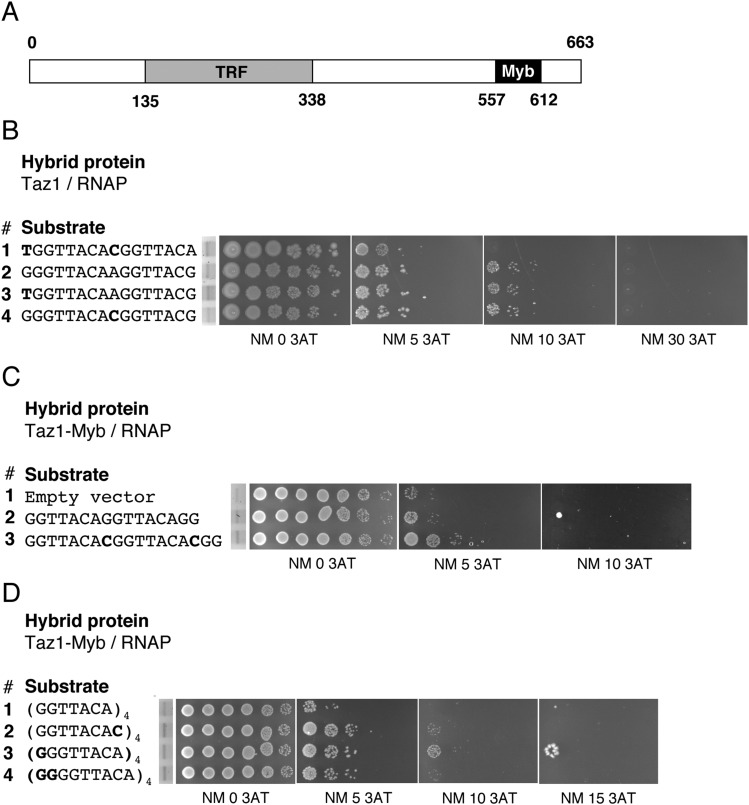
A bacterial one-hybrid screen was used to examine Taz1 binding to mutant substrates (27). The binding preferences as determined by the bacterial one-hybrid system were identical to those obtained with the S. cerevisiae one-hybrid screen that identified Taz1 (Fig. S8B, lines 1 and 2) (28). Next we showed that Taz1 telomeric repeat binding was not affected by the presence of the rare cytosine (Fig. S8B, lines 2 and 4). When we extended this analysis to multiple direct repeats, we again found that the rare cytosine did not affect Taz1-Myb binding, as Taz1-Myb bound at least as well to two or four direct repeats each containing the rare C as it did to core telomeric repeats (Fig. S8C, lines 2 and 3 and Fig. S8D, lines 1 and 2). However, the presence of one or two guanines before each of the four copies of the core repeat increased Taz1-Myb binding (Fig. S8D, lines 1, 3, and 4). Each additional guanine increased Taz1 binding compared with the core sequence, but the first guanine had the largest effect (Fig. S8D, lines 1 and 3). Thus, repeats preceded by one or two guanines were the preferred Taz1-Myb binding substrate.
Fig. S8.
Taz1–telomere repeat interaction. (A) Schematic of Taz1 structure. Myb, Myb/SANT domain; TRF, Telomere repeat binding factor domain, dimerization domain. Amino acid numbers are indicated. (B) Bacterial one-hybrid activity assay. Shown are interactions between Taz1/RNAP and telomeric repeats as assayed by growth of 10-fold dilution series on 0, 5, 10, and 30 mM 3-aminotriazole. DNA sequences are depicted with relevant mutations in bold. Dilution series preceded by EtBr-stained gels demonstrate equivalent substrate levels. (C) Bacterial one-hybrid activity assay. Shown are interactions between Taz1-Myb/RNAP (amino acids 557–612) and telomeric repeats as assayed by growth of 10-fold dilution series in 0, 5, 10, and 30 mM 3-aminotriazole. Taz1-Myb DNA binding domain was used to avoid protein dimerization, which interferes with the one-hybrid assay. DNA sequences are depicted with relevant mutations in bold. Dilution series preceded by EtBr-stained gels demonstrate equivalent substrate levels. (D) Taz1-Myb domain binding to four direct repeats of the indicted sequences. (D, Left) The same as described in C, except that four telomeric repeat tracts were assayed at 0, 5, 10, and 15 mM 3AT.
Based on the one-hybrid data (Fig. S8), we predicted that in vivo Taz1 would bind less well to ter1-STEloop telomeric DNA (Fig. 3C). To control for differences in telomere length, we normalized Taz1 ChIP signals (Fig. 4C) to telomere length (Fig. 4 D and E). Indeed, Taz1 binding to ter1-STEloop telomeres was significantly reduced compared with binding to wild-type telomeres (Fig. 4F). In contrast, Taz1 binding to the similarly short but wild-type sequence ter1-Δ415–507 telomeres was indistinguishable from wild type (Fig. 4F). We conclude that the aberrant sequence produced by the ter1-STE loop mutant compromises Taz1 telomere binding in vivo.
Discussion
Even though mammalian STEs have been known for over a decade to be essential for telomerase activity, their mechanism of action is unclear. Here we show that, as in mammals, deleting the S. pombe STE (ter1-Δ1066–1080) caused telomere loss and senescence (Fig. 1C). However, because we were able to isolate a partial loss-of-function STE allele, ter1-STEloop, we were able to determine the phenotypes of S. pombe cells with a defective STE. Isolation of this allele was undoubtedly facilitated by the atypically high heterogeneity of fission yeast telomeric DNA and somewhat flexible sequence requirements for S. pombe telomere binding proteins (29). Thus, although Pot1 (Fig. 4) and Taz1 (Fig. 4 and Fig. S8) had reduced binding to ter1-STEloop telomeres, the level of binding was sufficient to provide telomere functions, which may not be the case for a similar STE allele in an organism with more precise telomeric repeats.
ter1-STEloop cells had short telomeres (Fig. 1C), which can be explained by a combination of reduced telomerase activity (Fig. 2C) and reduced shelterin binding (Fig. 4 and Fig. S8). However, the most remarkable phenotype of ter1-STEloop cells is their altered telomere sequence. Two aspects of S. pombe telomeric DNA sequence were significantly perturbed: the number of repeats containing the rare cytosine, 5′-GGTTACAC-3′, was increased from 9% to 24%, and the occurrence of guanine tracts preceding the core repeat was reduced by 60%. These results are unprecedented, as they are the first example, to our knowledge, of a distant region (i.e., a region that is not part of the telomerase RNA core) that affects the sequence of telomeric DNA. The altered sequence is telomerase-generated and therefore a result of the STE mutation, as it arises in the absence of recombination and is not due to short telomeres (Fig. 3 and Figs. S5 and S6).
The altered sequence of ter1-STEloop telomeric DNA caused defects in shelterin binding, which mediates all key telomere functions. Pot1 binding was significantly reduced at ter1-STEloop telomeres (Fig. 4), consistent with in vitro studies showing reduced Pot1 binding to repeats containing the rare cytosine (29). Taz1 content at ter1-STEloop telomeres was also reduced (Fig. 4 and Fig. S8), owing to their reduced content of guanine tracts (Fig. 3C). Reduced telomere binding of the sequence-specific telomere binding protein Taz1 (and probably Pot1) reduces the association of other shelterin components that bind telomeres via protein–protein interactions (1–3). Moreover, we suspect that the level of atypical repeats (Fig. 3 C and D) detected in ter1-STEloop telomeric DNA is an underestimation, as reduced Pot1 binding, which protects telomeres from degradation, will lead to loss of the most atypical repeats. This hypothesis is supported by the finding that at a low rate ter1-STEloop telomeres were vulnerable to abrupt loss in recombination-deficient cells (<0.57% telomere loss events per generation). This interpretation suggests that our sequencing data underestimate the extent of TBE failure and resulting sequence variation in ter1-STEloop telomeric DNA.
The two changes in ter1-STEloop telomeric DNA, more rare cytosines and fewer guanine tracts, are mechanistically linked (Fig. S1) (7, 8). In wild-type cells, after addition of the core repeat and subsequent TER1 template translocation along the DNA, the last adenine cannot pair with the TER1 template, a situation that frequently results in “stuttering” and guanine tract incorporation in wild-type cells (Fig. S1, Left). Addition of the rare cytosine in wild-type telomeric DNA arises from loss of TBE function (6, 7). When the TBE is not maintained, reverse transcription extends through the first nucleotide of the TBE to add a cytosine to the telomeric core repeat (Fig. S1, Right). Upon translocation, the rare cytosine causes a different telomere–template alignment. As a result, stuttering is less thermodynamically favorable and the next repeat begins with a cytosine and lacks a guanine tract (5′-CGGTTACA-3′).
Although it is possible that the STE affects both TBE function and another aspect of telomerase action, its effects on the TBE is sufficient to explain all of the ter1-STEloop phenotypes. Indeed, this possibility is consistent with existing data, as mutations that eliminate the function of the S. pombe TBE have identical phenotypes to our ter1-STEloop mutant. Specifically, both STE and TBE loss-of-function mutations cause telomere shortening (Fig. 1) (7), and both generate the same pattern of aberrant repeats by increasing the rare cytosine (6, 7), which reduces the frequency of guanine tracts (7). Moreover, both the TBE helix mutant (7) and the STEloop mutant (Fig. 2C) dramatically reduce in vitro telomerase activity. The confluence of in vivo and in vitro STE and TBE phenotypes argues that they perform similar functions during telomere addition. Therefore, the best model to explain our data are that the TER1 STE loop enforces addition of canonical repeats and telomerase activity by promoting TBE structure. Finally, the phenotypes of our STE mutant have striking similarities to the phenotypes of human cells with similar mutations. Both have short telomeres and reduced in vitro telomerase activity (13) (Figs. 1C and 2C). In addition, UV treatment cross-links the uridines in the human STE loop to the 5′ end of the template, which suggests that the STE and 5′ end of the template could be in close proximity (30). Therefore, our model may be relevant to mammalian STE function.
In summary, we show that in S. pombe cells, the STE is required for normal telomerase activity and telomeric sequence; that is, the core region of telomerase RNA plus the catalytic subunit of telomerase are not sufficient to generate wild-type telomeric DNA. The role performed by the S. pombe STE is reminiscent of that of exonic enhancers during pre-mRNA splicing. These RNA sequences enforce correct catalytic events at the ends of introns even though their positions in pre-mRNA are remote from the splice sites (31). Impaired shelterin assembly and hence loss of telomere function are downstream consequences of STE failure (Fig. S9). Thus, it is not surprising that the STE region arose early in telomerase RNA evolution and is essential for telomerase action and telomere integrity in diverse organisms.
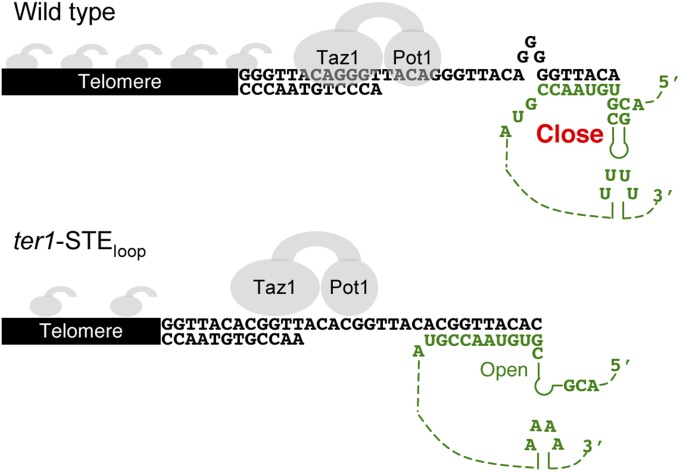
Fig. S9.
Model for STE action during telomere extension. (Upper) TER1 STE maintains the TBE to prevent telomerase reverse transcription into the TBE. Taz1 and Pot1 bind normally. TER1 RNA is depicted as a colored green line with dashes to denote additional linear distance. (Lower) TBE function is compromised in the TER1-STEloop mutant. Reverse transcription extends into the TBE to template addition of the rare cytosine. Inclusion of the rare cytosine prevents guanine tract addition in the next round of telomere extension. Increased incorporation of rare cytosines and reduced guanine tracts reduce binding of the shelterin components Pot1 and Taz1.
Materials and Methods
For more details, see SI Materials and Methods.
ter1 mutants were constructed by using standard molecular genetic techniques. Southern and Northern blotting were performed as previously described (16). RNA immunoprecipitation and ChIP were performed as previously described (16). In vitro telomerase assays were performed as in ref. 16. For telomere cloning, genomic DNA was extracted and cytosine tailed with terminal deoxynucleotidyl transferase (Roche). PCR was performed using Ex Taq (Takara) with a poly(G)18 oligo and an oligo designed to anneal to the subtelomeric element (5′-gtgtggaattgagtatggtgaa-3′). Both oligos included restriction sites, which were used to clone the amplified fragments. We used ∼100 sequences from each condition in the telomere analysis. Bacterial one-hybrid assays were performed as described (27).
SI Materials and Methods
DNA Manipulation and Strain Construction.
ter1 mutants were constructed by overlapping PCR mutagenesis and subcloned into the S. pombe integration plasmid pJK148. ter1/pJK148 mutants were integrated into the leu1-32 locus of the diploid h+/− ade6-M210/ade6-M216 his3-D1/his3-D1 leu1-32/leu1-32 ura4-D18/ura4-D18 ter1/ter1::his3 strain. Correct integration at leu1-32 was confirmed by Southern blotting. After sporulation, his+ leu1+ cells were identified and used in analyses. Because various N-terminal and C-terminal Pot1 epitopes caused extensive telomere lengthening, Pot1 was internally tagged after Q355 [pot1-(G8-9Myc-G8)] using the POM20 epitope tagging vector. Epitope tag sequences were confirmed by sequencing (Genewiz). The trt1-G8-13Myc strain has already been described (6).
Telomere Blots.
Wild-type and TER1 mutants were expressed from the leu1 locus in a ter1Δ background. Genomic DNA was extracted from the colony formed by the spore product from ter1/ter1::his3 ter1-mutant-leu1/leu1-32 diploids as well as from seven successive streaks of the spore clones. DNA was digested with EcoRI or ApaI and hybridized to a telomeric probe.
FISH.
Midlog cells were fixed with 3.7% (vol/vol) formaldehyde for 45 min at room temperature. After spheroplasting, cells were incubated with Custom Stellaris FISH probes (Biosearch Technologies Inc.) designed to hybridize across the ter1 ORF. Images were acquired with a Hamamatsu digital camera (C4742-98) and Slidebook 5.0 digital image acquisition software (Intelligent Imaging Innovations).
Nuclear and Cytoplasmic RNA Fractionation.
Nuclear and cytoplasmic RNA fractionation was performed as described, with the exception that spheroplasting was achieved with 20 mg Zymolyase and 20 mg Lysing Enzymes (Novozym 234) per gram of cells. Cells were lysed after spheroplasting by 7–10 strokes with a tight-fitting dounce and spun through a 0.5 M sucrose cushion at 8,000 × g for 10 min. The cytoplasmic RNA contained in the supernatant and nuclear pellet were extracted with 65 °C acidic phenol, precipitated, and resuspended in equal volumes. RNA in each fraction was quantified by loading equal volumes for Northern analysis and quantitative RT-PCR. Northern blotting and quantitative RT-PCR were performed as previously described (16).
In Vitro Telomerase Assay.
Assays were performed as described (20), with minor modifications. Cells were ground in a 6750 Freezer Mill (SPEX CertiPrep) six times for 1 min each with 2-min rest intervals. We used 9.2 mg of extract for each reaction. Washing was performed with a Nutator (BD Clay Adams) for 5 min. Reaction signals were acquired with a Typhoon 9410 and quantified by ImageQuant TL v.8.1.
Telomere Cloning.
Cells were streaked three times onto YES (yeast extract with supplements) plates and grown at 37 °C for 3 d to shorten telomeres. The fourth streak was grown for 3 d at 30 °C to allow telomere lengthening. Genomic DNA was extracted and cytosine tailed with terminal deoxynucleotidyl transferase (Roche). PCR was performed using Ex Taq (Takara) with a poly(G)18 oligo and an oligo designed to anneal to the subtelomeric element (5′-gtgtggaattgagtatggtgaa-3′). Both oligos included restriction sites, which were used to clone the amplified fragments. We used ∼100 sequences from each condition in telomere analysis, except for in ter1-STEloop rad51Δ cells, where 49 telomeres were analyzed.
Bacterial One Hybrid.
USOW (omega knockout bacteria hybrid selection strain) cells were cotransformed with Taz1/LPPC or Taz1-Myb/LPPC and different DNA substrates cloned into the pH3U3 vector and selected on 2xYT kanamycin and ampicillin plates. Cells were grown in SOC (super optimal broth with catabolite repression) media with selection for ∼2 d. Cells were washed in H2O, and dilutions were grown in selective media containing uracil, 0.1% histidine, IPTG, kanamycin, and ampicillin for 1 h at 37 °C. A 10-fold dilution series was then spotted onto selective solid media containing uracil, IPTG, kanamycin, and ampicillin. The dilution series was made on identical media supplemented with increasing concentrations of the competitive inhibitor 3-aminotriazole.
Acknowledgments
We thank Julie Cooper for the Taz1 antibody (NIH); Marcus Noyes (Princeton University) for the bacterial one-hybrid system and advice; Sandy Silverman (Princeton University) for FISH expertise; David Robinson (Princeton University) for statistical assistance; Kathleen Collins (University of California, Berkeley), Shelly Lim, Kah-Wai Lin, Lindsey Williams, and Yun Wu (Protein Potential, LLC) for critical comments on the manuscript; and Toru Nakamura (University of Illinois at Chicago) and Peter Baumann (Stowers Institute) for strains. This research was supported by a US National Research Service Award and American Cancer Society Post-Doctoral Fellowships (to C.J.W.), NIH Grant GM043265 (to V.A.Z.), and an ARRA (American Recovery and Reinvestment Act) supplement to Grant GM043265.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1503157112/-/DCSupplemental.
References
- 1.Moser BA, Nakamura TM. Protection and replication of telomeres in fission yeast. Biochem Cell Biol. 2009;87(5):747–758. doi: 10.1139/o09-037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dehé PM, Cooper JP. Fission yeast telomeres forecast the end of the crisis. FEBS Lett. 2010;584(17):3725–3733. doi: 10.1016/j.febslet.2010.07.045. [DOI] [PubMed] [Google Scholar]
- 3.Webb CJ, Wu Y, Zakian VA. DNA repair at telomeres: Keeping the ends intact. Cold Spring Harb Perspect Biol. 2013;5(6):a012666. doi: 10.1101/cshperspect.a012666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Moyzis RK, et al. A highly conserved repetitive DNA sequence, (TTAGGG)n, present at the telomeres of human chromosomes. Proc Natl Acad Sci USA. 1988;85(18):6622–6626. doi: 10.1073/pnas.85.18.6622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hiraoka Y, Henderson E, Blackburn EH. Not so peculiar: Fission yeast telomere repeats. Trends Biochem Sci. 1998;23(4):126. doi: 10.1016/s0968-0004(98)01176-1. [DOI] [PubMed] [Google Scholar]
- 6.Webb CJ, Zakian VA. Identification and characterization of the Schizosaccharomyces pombe TER1 telomerase RNA. Nat Struct Mol Biol. 2008;15(1):34–42. doi: 10.1038/nsmb1354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Box JA, Bunch JT, Zappulla DC, Glynn EF, Baumann P. A flexible template boundary element in the RNA subunit of fission yeast telomerase. J Biol Chem. 2008;283(35):24224–24233. doi: 10.1074/jbc.M802043200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ares M, Jr, Chakrabarti K. Stuttering against marginotomy. Nat Struct Mol Biol. 2008;15(1):18–19. doi: 10.1038/nsmb0108-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Blackburn EH, Collins K. Telomerase: An RNP enzyme synthesizes DNA. Cold Spring Harb Perspect Biol. 2011;3(5):a003558. doi: 10.1101/cshperspect.a003558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zappulla DC, Goodrich K, Cech TR. A miniature yeast telomerase RNA functions in vivo and reconstitutes activity in vitro. Nat Struct Mol Biol. 2005;12(12):1072–1077. doi: 10.1038/nsmb1019. [DOI] [PubMed] [Google Scholar]
- 11.Brown Y, et al. A critical three-way junction is conserved in budding yeast and vertebrate telomerase RNAs. Nucleic Acids Res. 2007;35(18):6280–6289. doi: 10.1093/nar/gkm713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Laterreur N, Eschbach SH, Lafontaine DA, Wellinger RJ. A new telomerase RNA element that is critical for telomere elongation. Nucleic Acids Res. 2013;41(16):7713–7724. doi: 10.1093/nar/gkt514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang Q, Kim NK, Feigon J. Architecture of human telomerase RNA. Proc Natl Acad Sci USA. 2011;108(51):20325–20332. doi: 10.1073/pnas.1100279108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen JL, Opperman KK, Greider CW. A critical stem-loop structure in the CR4-CR5 domain of mammalian telomerase RNA. Nucleic Acids Res. 2002;30(2):592–597. doi: 10.1093/nar/30.2.592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Qi X, et al. The common ancestral core of vertebrate and fungal telomerase RNAs. Nucleic Acids Res. 2013;41(1):450–462. doi: 10.1093/nar/gks980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Webb CJ, Zakian VA. Schizosaccharomyces pombe Ccq1 and TER1 bind the 14-3-3-like domain of Est1, which promotes and stabilizes telomerase-telomere association. Genes Dev. 2012;26(1):82–91. doi: 10.1101/gad.181826.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tang W, Kannan R, Blanchette M, Baumann P. Telomerase RNA biogenesis involves sequential binding by Sm and Lsm complexes. Nature. 2012;484(7393):260–264. doi: 10.1038/nature10924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mitchell JR, Collins K. Human telomerase activation requires two independent interactions between telomerase RNA and telomerase reverse transcriptase. Mol Cell. 2000;6(2):361–371. doi: 10.1016/s1097-2765(00)00036-8. [DOI] [PubMed] [Google Scholar]
- 19.Leonardi J, Box JA, Bunch JT, Baumann P. TER1, the RNA subunit of fission yeast telomerase. Nat Struct Mol Biol. 2008;15(1):26–33. doi: 10.1038/nsmb1343. [DOI] [PubMed] [Google Scholar]
- 20.Haering CH, Nakamura TM, Baumann P, Cech TR. Analysis of telomerase catalytic subunit mutants in vivo and in vitro in Schizosaccharomyces pombe. Proc Natl Acad Sci USA. 2000;97(12):6367–6372. doi: 10.1073/pnas.130187397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Legassie JD, Jarstfer MB. The unmasking of telomerase. Structure. 2006;14(11):1603–1609. doi: 10.1016/j.str.2006.09.004. [DOI] [PubMed] [Google Scholar]
- 22.Lue NF, Peng Y. Identification and characterization of a telomerase activity from Schizosaccharomyces pombe. Nucleic Acids Res. 1997;25(21):4331–4337. doi: 10.1093/nar/25.21.4331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Paschini M, et al. A naturally thermolabile activity compromises genetic analysis of telomere function in Saccharomyces cerevisiae. Genetics. 2012;191(1):79–93. doi: 10.1534/genetics.111.137869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Baumann P, Cech TR. Protection of telomeres by the Ku protein in fission yeast. Mol Biol Cell. 2000;11(10):3265–3275. doi: 10.1091/mbc.11.10.3265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rog O, Miller KM, Ferreira MG, Cooper JP. Sumoylation of RecQ helicase controls the fate of dysfunctional telomeres. Mol Cell. 2009;33(5):559–569. doi: 10.1016/j.molcel.2009.01.027. [DOI] [PubMed] [Google Scholar]
- 26.Tomita K, Cooper JP. Fission yeast Ccq1 is telomerase recruiter and local checkpoint controller. Genes Dev. 2008;22(24):3461–3474. doi: 10.1101/gad.498608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Noyes MB. Analysis of specific protein-DNA interactions by bacterial one-hybrid assay. Methods Mol Biol. 2012;786:79–95. doi: 10.1007/978-1-61779-292-2_5. [DOI] [PubMed] [Google Scholar]
- 28.Cooper JP, Nimmo ER, Allshire RC, Cech TR. Regulation of telomere length and function by a Myb-domain protein in fission yeast. Nature. 1997;385(6618):744–747. doi: 10.1038/385744a0. [DOI] [PubMed] [Google Scholar]
- 29.Altschuler SE, Dickey TH, Wuttke DS. Schizosaccharomyces pombe protection of telomeres 1 utilizes alternate binding modes to accommodate different telomeric sequences. Biochemistry. 2011;50(35):7503–7513. doi: 10.1021/bi200826a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ueda CT, Roberts RW. Analysis of a long-range interaction between conserved domains of human telomerase RNA. RNA. 2004;10(1):139–147. doi: 10.1261/rna.5118104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Braunschweig U, Gueroussov S, Plocik AM, Graveley BR, Blencowe BJ. Dynamic integration of splicing within gene regulatory pathways. Cell. 2013;152(6):1252–1269. doi: 10.1016/j.cell.2013.02.034. [DOI] [PMC free article] [PubMed] [Google Scholar]