Significance
The replication fork helicase unwinds double-stranded DNA at a replication fork, and assembly and activation of this helicase are tightly controlled. This paper describes a biological mechanism to link two essential functions related to helicase assembly and activation. Sld3, a protein required for initiation of DNA replication, coordinates the assembly of the replication fork helicase with the chemical modification of a helicase protein subunit. This chemical modification is important for subsequent activation of the helicase.
Keywords: DNA replication, kinase, phosphorylation, helicase, initiation
Abstract
Dbf4-dependent kinase (DDK) phosphorylates minichromosome maintenance 2 (Mcm2) during S phase in yeast, and Sld3 recruits cell division cycle 45 (Cdc45) to minichromosome maintenance 2-7 (Mcm2-7). We show here DDK-phosphoryled Mcm2 preferentially interacts with Cdc45 in vivo, and that Sld3 stimulates DDK phosphorylation of Mcm2 by 11-fold. We identified a mutation of the replication initiation factor Sld3, Sld3-m16, that is specifically defective in stimulating DDK phosphorylation of Mcm2. Wild-type expression levels of sld3-m16 result in severe growth and DNA replication defects. Cells expressing sld3-m16 exhibit no detectable Mcm2 phosphorylation in vivo, reduced replication protein A-ChIP signal at an origin, and diminished Go, Ichi, Ni, and San association with Mcm2-7. Treslin, the human homolog of Sld3, stimulates human DDK phosphorylation of human Mcm2 by 15-fold. DDK phosphorylation of human Mcm2 decreases the affinity of Mcm5 for Mcm2, suggesting a potential mechanism for helicase ring opening. These data suggest a conserved mechanism for replication initiation: Sld3/Treslin coordinates Cdc45 recruitment to Mcm2-7 with DDK phosphorylation of Mcm2 during S phase.
The replication fork helicase in eukaryotes is composed of Cdc45, the Mcm2-7 heterohexameric ATPase, and the tetrameric GINS (Go, Ichi, Ni, and San) complex (CMG assembly) (1). The replication fork helicase (CMG) assembles in S phase in a manner that is dependent upon the replication initiation factors Sld2, Sld3, and Dpb11 (2). Sld3 (Treslin/TICRR in humans), Sld2 (RecQL4/RecQ4 in humans), and Dpb11 (TopBP1 in humans) are required for the initiation of DNA replication, but these proteins do not travel with the replication fork (3). The S-phase-specific kinases, cyclin-dependent kinase (CDK) and the Dbf4-dependent kinase (DDK), are also required for CMG assembly and origin activation (4, 5). In late M and G1 phases, the Mcm2-7 complex loads to encircle dsDNA as a double hexamer (6, 7). During S phase, a single strand of DNA is extruded from the central channel of Mcm2-7, and this event is required because the CMG complex unwinds DNA by a steric exclusion mechanism (8).
Central to the initiation of DNA replication is the coordination of entry into S phase with origin firing (4, 5). Levels of the S phase-specific kinases, S-CDK and DDK, rise during the onset of S phase, and these two kinases are central to coordinating S-phase entry with origin firing (4, 5). S-CDK phosphorylates Sld2 and Sld3, and these phosphorylation events are the essential functions of S-CDK (9, 10). S-CDK phosphorylation of Sld3 is conserved in human Treslin (11). S-CDK phosphorylation of Sld2 promotes the association of Sld2 with yeast Dpb11 (12), and also the association of Sld2 with T-rich ssDNA (13). S-CDK phosphorylation of Sld3 stimulates the association of Sld3 with Dpb11 (9, 10). The associations of Sld2 with Dpb11 and Sld3 with Dpb11 have been proposed to be important for the recruitment of GINS to origins, through the generation of a preloading complex (Pre-LC), composed of Sld2, GINS, Polε, and Dpb11 (14). S-CDK–catalyzed formation of an Sld3-Dpb11-Sld2 complex has also been proposed to be important to generate a ternary ssDNA-binding complex of high affinity, because Sld2, Sld3, and Dpb11 bind to T-rich ssDNA (13, 15, 16).
The essential role of DDK in yeast cells is the phosphorylation of subunits of the Mcm2-7 complex (17). DDK phosphorylation of Mcm4 is important for cell growth, and this phosphorylation event alleviates an inhibitory function of the N terminus of Mcm4 (18). DDK phosphorylation of Mcm4 may also promote the interaction between Cdc45 and Mcm2-7 (18). DDK phosphorylation of Mcm6 may also be important for cell growth (19). Mcm2 is also a target for DDK (20), and DDK phosphorylation of Mcm2 is also required for DNA replication under normal growth conditions (21). Furthermore, expression of a mutant of mcm2 (mcm2-S164A,S170A) that is not phosphorylated by DDK exerts a dominant-negative severe growth defect in budding yeast that is bypassed by the mcm5-bob1 (mcm5-P83L) mutation (21). The biochemical mechanism of this genetic suppression has also been examined. DDK phosphorylation of Mcm2 reduces the affinity of budding yeast Mcm2 for Mcm5, and the mcm5-bob1 mutation also reduces this affinity (21). This reduced affinity may help open the “Mcm2-Mcm5 gate,” which may be important for the extrusion of ssDNA from the central channel of Mcm2-7 during S phase, a requirement for origin activation (22).
Cdc45 binds weakly to Mcm2-7 in the absence of accessory factors (23). Sld3 binds tightly to Mcm2-7 and Cdc45, and thus Sld3 recruits Cdc45 to Mcm2-7 complexes (2, 23). This step may further require DDK and involve the nonessential initiation factor Sld7 (24). During origin activation, Sld3 is removed from Mcm2-7, presumably through the exposure of sequestering T-rich ssDNA (16). GINS can substitute for Sld3 as a factor that promotes the association of Cdc45 with Mcm2-7, thereby forming the stable Cdc45-Mcm2-7-GINS (CMG) replicative helicase complex (23, 25).
The mechanism of GINS recruitment may involve the formation of the S-CDK–dependent preloading complex, wherein the pre-LC recruits GINS to Mcm2-7, analogous to how Sld3 recruits Cdc45 to Mcm2-7 (14). A second proposal posits that Sld3, Sld2, and Dpb11 compete with GINS for binding to Mcm2-7 before origin activation, blocking the premature interaction between GINS and Mcm2-7 before origin activation (15, 16, 26). However, when T-rich ssDNA is extruded from the central channel of Mcm2-7, an ssDNA binding surface for Sld3-Sld2-Dpb11 is generated (15, 16, 26). Sld3-Sld2-Dpb11 dissociates from Mcm2-7 once the origin is melted, because Sld3-Sld2-Dpb11 has a higher affinity for ssDNA then Mcm2-7 (15, 16, 26). The dissociation of Sld3-Sld2-Dpb11 from Mcm2-7 allows GINS to bind Mcm2-7 by a passive, sequestration mechanism (15, 16, 26). The two models are not incompatible with one another, and they may both be correct.
There is an excess of Mcm2-7 double hexamer complexes loaded onto dsDNA in M phase and G1 relative to the number of Mcm2-7 double hexamer complexes that actually fire. Remarkably, the activated Mcm2-7 complexes share several features in common: DDK phosphorylation of Mcm2-7, initiation factor (Sld3, Sld2, Dpb11, Mcm10, and Sld7) binding to Mcm2-7, and Cdc45/GINS attachment to Mcm2-7. What coordinates these different activities at a particular Mcm2-7 double hexamer? In other words, what prevents Cdc45 from binding to one Mcm2-7 double hexamer while DDK phosphorylates a different Mcm2-7 double hexamer? We sought to address this fundamental question in this paper.
We show that DDK-phosphorylated Mcm2 preferentially interacts with Cdc45 in vivo compared with Mcm2 (all Mcm2 in cell, phosphorylated and unphosphorylated), suggesting that Cdc45 recruitment to Mcm2-7 is correlated with DDK phosphorylation of Mcm2. We also show that Sld3 substantially stimulates DDK phosphorylation of Mcm2 in vitro. We identified a mutant of Sld3, sld3-m16, that is defective in the stimulation of DDK phosphorylation of Mcm2. When sld3-m16 is expressed in budding yeast cells, a dominant negative severe growth defect is observed that is bypassed by mcm5-bob1. Wild-type expression levels of sld3-m16 confer a growth and DNA replication defect, with decreased DDK phosphorylation of Mcm2. Furthermore, expression of sld3-m16 results in diminished replication protein A (RPA)-ChIP signal at an origin, and decreased GINS interaction with Mcm2-7. Furthermore, human Treslin substantially stimulates DDK phosphorylation of Mcm2 at serines 53 and 108. Finally, a mutant of human Mcm2 that mimics DDK-phosphorylated Mcm2 exhibits diminished interaction with human Mcm5, suggesting a mechanism for “gate” opening. We conclude that Sld3 coordinates helicase phosphorylation with helicase assembly.
Results
Strain Harboring Constitutively Expressed DDK-Dead mcm2 Mutant (mcm2-S164A,S170A Expressed from Native Promoter) Exhibits Suppression.
We previously found that induced expression of mcm2-S164A,S170A, which is a DDK-dead mutant of mcm2, confers a dominant-negative severe growth defect in budding yeast. Researchers from a different laboratory found that expression of mcm2-S164A,S170A, when incorporated into the genome under constitutive expression by its native promoter, exhibited a defect only when exposed to DNA-damaging agents such as methyl methanesulfonate (27, 28). Given the dominant-negative phenotype that we observed, we predicted that this native promoter strain harbored a suppressor mutation. Thus, we obtained the native promoter strain from a coauthor on the paper and expressed the mcm2-S164A,S170A gene from a galactose-inducible promoter. Whereas wild-type yeast cells exhibit a severe growth defect upon expression of mcm2-S164A,S170A (Fig. S1, Top and ref. 21), the native promoter strain exhibited no growth defect (Fig. S1, Bottom). These results suggest that the native promoter strain harbors a suppressor mutation, explaining the differences in results between our two laboratories. These data also lend further support that expression of mcm2-S164A,S170A confers a dominant negative growth defect under normal growth conditions, and DDK phosphorylation of Mcm2 is required for growth under normal conditions.
Fig. S1.
Consequence of overexpression of mcm2-2A (mcm2-S164A,S170A) in normal, wild-type budding yeast (Top and ref. 21) and in the strain harboring mcm2-S160AS170A (28) (Bottom).
Mcm2 Association with Cdc45 Is Correlated with DDK Phosphorylation of Mcm2 During S Phase.
Mcm2-7 complexes are loaded in excess relative to those Mcm2-7 complexes in late M phase and G1 relative to those Mcm2-7 complexes that unwind DNA in S phase (29). Although there are many steps involved in activation of the helicase, one key to Mcm2-7 activation is the association of Mcm2 with Cdc45 (25), and a second key is the DDK phosphorylation of Mcm2 (20). We wondered whether these two events are correlated. Thus, we synchronized cells in G1 with α-factor, and then released into medium lacking α-factor for 0, 15, 30, and 45 min. We next immunoprecipitated wild-type yeast cell extracts with antibody directed against Mcm2 [anti–Mcm2-1–160, insensitive to DDK phosphorylation of Mcm2 (21)] or antibody directed against phospho-Mcm2 [anti–mcm2-phospho-S164-phospho-S170, sensitive to DDK phosphorylation of Mcm2 (21)]. We then examined the immunoprecipitate by Western analysis using antibodies directed against Cdc45-6HA (Fig. 1A). Anti–phospho-Mcm2 antibody yielded a substantially stronger signal at 15-, 30-, and 45-min time points compared with anti-Mcm2. These data suggest that DDK phosphorylation of Mcm2 is correlated with Cdc45 attachment to Mcm2 during S phase. Although a simple explanation is that DDK phosphorylation of Mcm2 increases the affinity of Mcm2 for Cdc45, our previously published in vitro experimental data argue against this idea (21). Thus, DDK phosphorylation of Mcm2 does not directly cause Cdc45 to associate with Mcm2-7. We therefore wondered whether Sld3, which recruits Cdc45 to Mcm2-7 (2), plays a role in coordinating Cdc45 recruitment to Mcm2-7 with DDK phosphorylation of Mcm2.
Fig. 1.
Sld3 substantially stimulates DDK phosphorylation of Mcm2. (A) Coimmunoprecipitation analysis of wild-type budding yeast cells using antibody against the N terminus of Mcm2 (residues 1–160) or an antibody directed against the DDK-phosphorylated Mcm2 (anti-phosphoserines 164 and 170). Cells were arrested in G1 with α-factor and release into medium lacking α-factor for the times incubated. (B) Three picomoles budding yeast DDK (Dbf4-Cdc7) was incubated with 3 pmol Mcm2 and [γ-32P]ATP as described in SI Materials and Methods. The amount of Sld3 added is indicated on top of the gel. The products were analyzed by SDS/PAGE followed by phosphorimaging. Molecular weight markers were used to identify the position of Mcm2 in the gel. A known amount of [γ-32P]ATP was also spotted on the gel to quantify the amount of phosphate incorporation. (C) Experiments similar to those in B were quantified and plotted. (D) Similar experiment to B, except the products were analyzed by Western analysis using antibody specific for DDK phosphorylation of Mcm2 at S164 and S170, as described previously (21). (E) Similar to D, except the Mcm2-7 complex was used as a substrate.
Sld3 Substantially Stimulates DDK Phosphorylation of Mcm2 in Vitro.
DDK phosphorylates Mcm2 weakly in vitro (30), and we next investigated whether Sld3 stimulates this phosphorylation event. DDK was incubated with Mcm2, [γ-32P]ATP, and increasing amounts of Sld3. The results were analyzed by SDS/PAGE followed by phosphorimaging and quantitation (Fig. 1 B and C). We found a substantial, 11-fold increase in DDK phosphorylation of Mcm2 with the addition of Sld3. These data suggest that Sld3 substantially stimulates DDK phosphorylation of Mcm2 in vitro. We next examined whether the anti–phospho-Mcm2 antibody, which recognizes Mcm2-phosphoserine-164-phosphoserine-170, detects higher levels of DDK phosphorylation of Mcm2 by Western analysis, using Mcm2 as a substrate (Fig. 1D). Indeed, there is a substantial increase in phospho-antibody signal, suggesting that Sld3 stimulates DDK phosphorylation of Mcm2 by stimulating phosphorylation at S164/S170. We then repeated the experiment reported in Fig. 1D, but used Mcm2-7 complex as a substrate (Fig. 1E). Western analysis demonstrates substantial stimulation of the DDK phosphorylation of Mcm2 at S164/170. These data suggest that Sld3 substantially stimulates DDK phosphorylation of Mcm2 when Mcm2 is incorporated into the Mcm2-7 complex.
Sld3-m16 Is a Separation-of-Function Mutant That Is Specifically Defective in Stimulating DDK Phosphorylation of Mcm2.
We next worked to identify a separation-of-function mutant of Sld3 that is specifically defective in stimulating DDK phosphorylation of Mcm2, to pursue in vivo studies. We therefore divided Sld3 into two fragments, an N-terminal fragment (Sld3-1-510), and a C-terminal fragment (Sld3-511-C). We next repeated the phosphorylation assay described in Fig. 1 C and D. We found that the C-terminal region of Sld3 (Sld3-511-C) functions like wild-type Sld3 in stimulating DDK phosphorylation of Mcm2, whereas the N-terminal region of Sld3 (Sld3-1-510) had no effect (Fig. S2A). These results suggest that the C-terminal region of Sld3 is responsible for DDK phosphorylation of Mcm2. We next performed site-directed mutagenesis in the 511-C region of Sld3. We screened a large number of mutants, and the 16th mutant we tried, Sld3-m16 (Sld3-S556A,H557A,S558A,T559A), exhibited a substantial defect in stimulating DDK phosphorylation of Mcm2 (Fig. S2A).
Fig. S2.
sld3-m16 is specifically defective in stimulating DDK phosphorylation of Mcm2. (A) Similar to experiments in Fig. 1 B and C, except Sld3-full-length, Sld3-511-C, Sld3-1–510, or Sld3-m16 were used. (B) Three picomoles GST-Dpb11 was used to pull down CDK-phosphorylated and radiolabeled Sld3-wild-type or Sld3-m16. The products of the pulldown were analyzed by SDS/PAGE followed by phosphorimaging. Repeated experiments were quantified, averaged, and plotted. (C) Similar to B, except 3 pmol biotin-ssARS305 (an 80mer origin ssDNA described in ref. 16) was used with streptavidin beads to pull down Sld3. (D) Similar to B, except 3 pmol GST-Mcm2-7 was used to pull down Sld3. (E) Similar to B, except 3 pmol GST-Cdc45 was used to pull down Sld3.
We next determined whether Sld3-m16 was specifically defective for stimulating DDK phosphorylation of Mcm2, or whether it is defective in some other known function of Sld3. CDK-phosphorylated Sld3 binds Dpb11 (9, 10). We thus determined the interaction between CDK-phosphorylated Sld3-m16 and Dpb11 and found that it bound like wild-type Sld3 (Fig. S2B). Sld3 is also known to bind T-rich origin ssDNA, Mcm2-7, and Cdc45 (16, 23). Therefore, we determined the interaction between Sld3-m16 and origin ssDNA (Fig. S2C), Mcm2-7 (Fig. S2D), or Cdc45 (Fig. S2E). In each instance, Sld3-m16 bound to ssDNA, Mcm2-7, or Cdc45 like wild-type Sld3. These data suggest that Sld3-m16 is a separation-of-function mutant that is specifically defective in stimulating DDK phosphorylation of Mcm2.
Expression of sld3-m16 Results in a Dominant-Negative Severe Growth Defect with Substantially Decreased Phosphorylation of Mcm2.
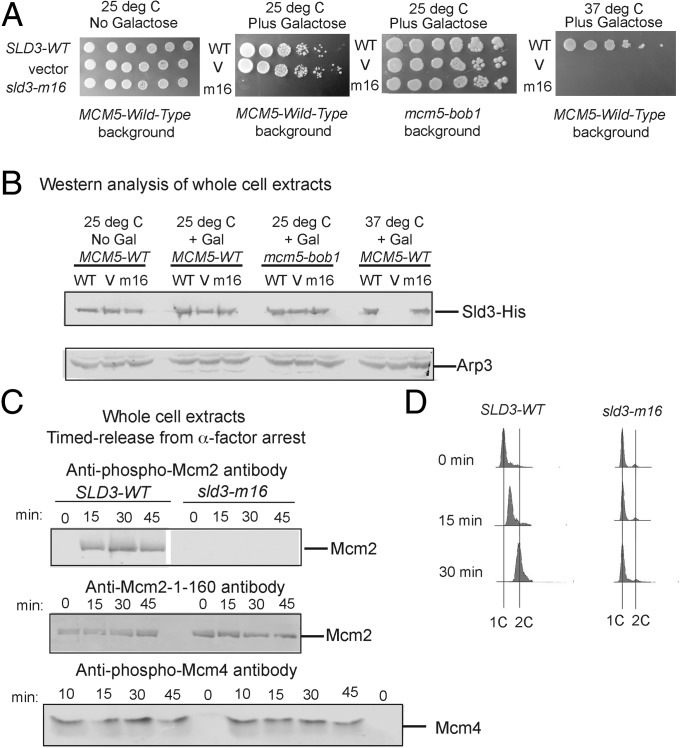
An sld3-7 td strain was generously provided by Karim Labib, University of Dundee, Dundee, Scotland, for our in vivo studies (3). In this strain, the genomic copy of SLD3 is degraded at the restrictive temperature (37 °C). Given that expression of mcm2-S164A,S170A confers a dominant-negative severe growth defect in budding yeast cells (21), we anticipated that expression of sld3-m16 may also confer a dominant negative phenotype. We thus expressed sld3-m16 from the GAL S inducible expression system, a low-copy inducible expression system, and varied the concentration of galactose until we achieved wild-type levels of Sld3-m16 at the restrictive temperature (Fig. 2B). In the absence of galactose, no sld3-m16 is induced, and the cells grow like wild-type SLD3 (Fig. 2A, first panel). However, once galactose is added at the permissive temperature (25 °C), the cells exhibit a severe growth defect (Fig. 2A, second panel). These results suggest that expression of sld3-m16 results in a dominant-negative severe growth defect.
Fig. 2.
Sld3 stimulation of DDK phosphorylation of Mcm2 is required for cell growth and DNA replication. (A) Tenfold serial dilution analysis of budding yeast sld3-7 td (sld3-temperature-sensitive degron) cells expressing SLD3-WT, vector, or sld3-m16 from the GAL-S plasmid inducible promoter system (pRS415). The growth conditions are described at the top of the image, and the strain background is described at the bottom of the image. (B) Western analysis of whole cell extracts from cells used in A, probing with antibody directed against Sld3. (C) Similar to B, except probing with antibody directed against DDK-phosphorylated Mcm2. A white line in the image indicates where the image was spliced from different gels. (D) FACS analysis of cells described in A, using propidium iodide as a stain for DNA content.
We next performed the same experiment, but in a mcm5-bob1 genetic background, because the mcm5-bob1 mutation suppresses the growth defect conferred by expression of mcm2-S164A,S170A (21). Indeed, the mcm5-bob1 mutation suppressed the growth defect conferred by expression of sld3-m16 (Fig. 2A, third panel), suggesting that the mechanism for growth inhibition conferred by expression of mcm2-S164A,S170A or sld3-m16 is similar. We next expressed sld3-m16 at the restrictive temperature at wild-type expression levels (Fig. 2B) and found a severe growth defect (Fig. 2A, fourth panel). These data suggest that under normal growth conditions and wild-type expression of sld3-m16 cell growth is severely impaired.
We next examined whether for the cells examined at the restrictive temperature there is a defect in Mcm2 phosphorylation by DDK (Fig. 2C). We thus performed whole-cell-extract analysis followed by blotting with antibody directed against DDK-phosphorylated Mcm2 (Mcm2-phospho-serine-164-phosphoserine-170). We synchronized cells in G1 with α-factor and then released in the absence of α-factor with no hydroxyurea for 0, 15, 30, and 45 min. Whereas wild-type cells exhibited an increase in phospho-Mcm2 as cells entered S phase, cells expressing sld3-m16 exhibited no detectable levels of phospho-Mcm2 during S phase. These results suggest that expression of sld3-m16 confers a substantial decrease in phospho-Mcm2 expression (Fig. 2C). This result is specific for phosho-Mcm2 expression, and not phospho-Mcm4 expression (Fig. 2C). Next, we examined whether there was a defect in DNA replication for cells examined at the restrictive temperature, and indeed we found slow progression through S phase for cells expressing sld3-m16 (Fig. 2D).
Expression of sld3-m16 Results in Decreased RPA-ChIP Signal at an Origin of Replication.
We next determined whether cells expressing sld3-m16 at the restrictive temperature exhibit a defect in RPA-ChIP signal at an origin of replication. For this experiment, we synchronized cells in G1 with α-factor and then released into medium lacking α-factor for 20 min (S phase cells). RPA-ChIP signal at an origin increases in wild-type cells in S phase at the early origins ARS305 and ARS306, coincident with the formation of ssDNA at an origin of replication during S phase (origin melting and replication initiation, Fig. S3). In contrast, cells expressing sld3-m16 do not exhibit an increase in RPA-ChIP signal at an origin of replication as cells are released from α-factor arrest, suggesting that these cells have no origin melting or replication initiation. These results are similar to those observed for expression of mcm2-S164A,S170A (21), suggesting that Sld3-stimulated DDK phosphorylation of Mcm2 is required for origin melting and replication initiation. One possible mechanism for the lack of origin melting for cells expressing sld3-m16 may be that Sld3 stimulation of DDK phosphorylation of Mcm2 is required to disengage Mcm2 from Mcm5 during S phase, allowing for the extrusion of ssDNA from the central channel of Mcm2-7 (21).
Fig. S3.
Sld3 stimulation of DDK phosphorylation of Mcm2 is required for RPA-CHiP signal at an origin. Chromatin immunoprecipitation was performed using cells described in Fig. 2A, Right at the restrictive temperature, in the presence of galactose, and in a wild-type background. Cells were arrested with α-factor (G1 cells) and then released for 20 min (S phase cells). Cells extracts were fixed and immunoprecipitated with antibodies directed against RPA. The immunoprecipitate was probed for DNA sequence using quantitative PCR at two early origins (ARS305 or ARS306) and at a region midway between these origins. Results from repeated experiments were quantified and plotted.
Expression of sld3-m16 Results in Decreased GINS-Mcm2-7 Signal at an Origin of Replication.
We next examined the result of expressing sld3-m16 at the restrictive temperature on in vivo protein–protein interactions by coimmunoprecipitation analysis (Fig. 3). We first examined the effect of Sld3–Cdc45 interaction and Sld3–Dpb11 interaction using cross-linking, because cross-linking is required to detect these interactions (Fig. 3A). Cells were arrested in G1 with α-factor and then released into medium lacking α-factor for 0, 15, 30, and 45 min. We found no difference in Sld3–Cdc45 interaction (Fig. 3A, Top) or Sld3-Dpb11 interaction (Fig. 3A, Bottom) with this analysis. These data suggest that expression of sld3-m16 does not affect Sld3–Cdc45 or Sld3–Dpb11 interaction in vivo.
Fig. 3.
Sld3 stimulation of DDK phosphorylation of Mcm2 is required for GINS association with Mcm2-7 during S phase. (A) Cells were fixed and analyzed for interaction between Sld3 and Dpb11 or Sld3 and Cdc45. Cells were synchronized in G1 with α-factor and released into medium lacking hydroxyurea for the indicated times. (B) Cells were not fixed and analyzed for interaction between Mcm2-7 and Cdc45, GINS, or Sld3.
Next we performed experiments in the absence of cross-linking (Fig. 3B) to observe the interaction between Mcm2-7 and GINS (Psf2, Fig. 3B, Top), Mcm2-7 and Cdc45 (Fig. 3B, Middle), or Mcm2-7 and Sld3 (Fig. 3B, Bottom). No cross-linking is required to observe these interactions. Although there is no effect of sld3-m16 on the loading of Cdc45 with Mcm2-7 during S phase (Fig. 3B, Middle), there is a substantial decrease in GINS interaction with Mcm2-7 in cells expressing sld3-m16 (Fig. 3B, Top). These data suggest that Sld3 stimulation of DDK phosphorylation of Mcm2 is required for GINS assembly with Mcm2-7. Because there is no origin melting in cells expressing sld3-m16 (Fig. S3), the lack of GINS assembly with Mcm2-7 in mutant cells may reflect that origin melting is required for GINS assembly with Mcm2-7 (15, 16, 26). We also observed that Sld3–Mcm2-7 interaction is increased at 30- and 45-min time points in mutant cells (Fig. 3B, Bottom), suggesting that expression of sld3-m16 results in prolonged interaction between Sld3 and Mcm2-7. This observation is consistent with the idea that origin melting is required for Sld3 sequestration from Mcm2-7 (16).
Expression of sld3-m16 Yields a Phenotype Similar to Expression of mcm2-S164A,S170A.
Our data from this paper, compared with that of previous work from our laboratory (21), indicate that expression of sld3-m16 yields a phenotype similar to expression of mcm2-S164A,S170A. Expression of either mutation yields no growth on agar plates and is suppressed by mcm5-bob1. Furthermore, FACS analysis of S phase progression, RPA-ChIP analysis, and Co-IP analysis yield very similar results. To further compare the phenotypes of these two mutants, we analyzed the growth rate in solution of sld3-m16;mcm5-bob1 cells compared with mcm2-S164A,S170A;mcm5-bob1 cells (Fig. S4). The growth rates of these two strains are very similar, suggesting that the phenotypes of these two mutations are indeed very similar. These data lend further support to the idea that the primary defect in sld3-m16 cells is a lack of DDK phosphorylation of Mcm2. We thus sought to test whether expression of mcm2-S164D,S170D can suppress the growth defect conferred by expression of sld3-m16, but unfortunately cells expressing mcm2-S164D,S170D cells are dead on agar plates (Fig. S5). These data suggest that the unphosphorylated state of Mcm2 may be required for cell growth.
Fig. S4.
The growth rates of sld3-m16;mcm5-bob1 and mcm2-S164A,S170A;mcm5-bob1 are very similar. The growth of the indicated strains in liquid media was determined and plotted as a function of time.
Fig. S5.
Expression of mcm2-S164D,S170D is lethal. Tenfold dilution of budding yeast cells harboring a plasmid with galactose-induced expression of mcm2-S164D,S170D. In the presence of galactose, with induced expression of mcm2-S164D,S170D, cells are dead on agar plates.
Human Treslin Stimulates Human DDK Phosphorylation of Human Mcm2.
DDK phosphorylation of Mcm2 occurs in humans as well as yeast (31), and the human homolog of Sld3 is Treslin (TICRR) (11). We therefore wondered whether Sld3 stimulation of DDK phosphorylation of Mcm2 was conserved from yeast to humans. We purified human Mcm2, human DDK, and human Treslin. We then incubated human Mcm2 with human DDK, [γ-32P]ATP, and increasing concentrations of Treslin and analyzed the results by phosphorimaging and quantitation. We found that human Treslin stimulates human DDK phosphorylation of human Mcm2 by 15-fold (Fig. S6 A and B). Human DDK phosphorylates human Mcm2 at serines 40, 53, and 108 (31). We therefore wondered whether human Treslin stimulates DDK phosphorylation of these residues. We thus obtained antibodies specific for phophos-40/41, phospho-53, or phospho-108 from Bethyl laboratories. We found that human Treslin substantially stimulates human DDK phosphorylation of human Mcm2 at serines 53 (Fig. S6C) and 108 (Fig. S6D), but not at 40/41. These data suggest that Sld3/Treslin stimulation of DDK phosphorylation of Mcm2 is conserved from yeast to human.
Fig. S6.
Human Treslin stimulates human DDK phosphorylation of human Mcm2, promoting Mcm2 dissociation from Mcm5. (A) Three picomoles human DDK (Dbf4-Cdc7) was incubated with 3 pmol Mcm2 and [γ-32P]ATP as described in SI Materials and Methods. The amount of Treslin added is indicated on top of the gel. The products were analyzed by SDS/PAGE followed by phosphorimaging. Molecular weight markers were used to identify the position of Mcm2 in the gel. A known amount of [γ-32P]ATP was also spotted on the gel to quantify the amount of phosphate incorporation. (B) Experiments similar to A were quantified and plotted as a function of Treslin input. (C) Similar experiment to A, except the products were analyzed by Western analysis using an antibody specific for phosphorylation of Mcm2 at S53 (Left) or S108 (Middle) or Mcm2 (Right). (D) GST pulldown using 3 picomoles human GST-Mcm2, radiolabeled Mcm5, and 1 mM ATP. Mcm2-2D is Mcm2-S53D,S108D. (E) Similar to D, except radiolabeled Mcm6 was used as the input protein. (F) Similar to E, except radiolabeled Cdc45 was used as the input protein.
Treslin Stimulation of DDK Phosphorylation of Human Mcm2 May Weaken Mcm2/Mcm5 Interaction.
We also found previously that a mutant of yeast Mcm2 that mimics DDK phosphorylation (Mcm2-S164D,S170D) exhibits substantially diminished interaction with Mcm5 compared with wild-type Mcm2 (21). The Mcm2–Mcm5 interaction functions as a “gate” to allow the Mcm2-7 complex to encircle single- or double-stranded DNA (22). To determine whether DDK phosphorylation of Mcm2 inhibits the interaction with Mcm5 in humans, we incubated human GST-Mcm2 with radiolabeled Mcm5 in the presence of ATP (Fig. S6E). Whereas wild-type Mcm2 bound tightly to Mcm5, the DDK-phospho-mimic form of human Mcm2 (Mcm2-2D or Mcm2-S53D,S108D) exhibited substantially diminished interaction with Mcm5. Furthermore, Mcm2-2D bound to human Mcm6 (Fig. S6F) and human Cdc45 (Fig. S6G) like wild-type Mcm2, suggesting that the loss of interaction of Mcm2 with Mcm5 is specific. These data also suggest that Sld3/Treslin stimulation of DDK phosphorylation of Mcm2 reduces the interaction between Mcm2 and Mcm5 in a manner that is conserved from yeast to humans, and thus a potential regulatory mechanism may be conserved to open the Mcm2-7 “gate” during S phase. This mechanism may function along with other factors to allow for the extrusion of ssDNA from the central channel of Mcm2-7 during S phase (origin melting).
Discussion
DDK Phosphorylation of Mcm2 Is Coordinated with Cdc45 Recruitment to Mcm2-7.
We found that the yeast strain harboring genomic mcm2-S164A,S170A under control by native promoter (27, 28) exhibits normal growth upon galactose-induced overexpression of mcm2-S164A,S170A (Fig. S1). These data suggest that this native promoter strain harbors a suppressor mutation, and the data further support that DDK phosphorylation of Mcm2 is required for growth under normal growth conditions. We also found with coimmunoprecipitation analysis that DDK-phosphorylated Mcm2 is enriched for Cdc45 interaction in vivo compared with Mcm2 (Fig. 1A), suggesting that Mcm2-7 complexes that bind to Cdc45 are also phosphorylated by DDK. These data suggest that some mechanism exists to coordinate DDK phosphorylation of Mcm2 with Cdc45 recruitment to Mcm2-7.
Sld3 Stimulates DDK Phosphorylation of Mcm2.
We found that yeast Sld3 stimulates yeast DDK phosphorylation of yeast Mcm2 by 11-fold using purified proteins (Fig. 1 B and C). We also found that phosphorylation of serine 164 and serine 170 of yeast Mcm2 is substantially enhanced by the addition of Sld3 to DDK (Fig. 1D). This stimulation also occurs when Mcm2-7 is present as a hexameric complex (Fig. 1E). We found that the C-terminal region of Sld3 is responsible for the stimulation of DDK phosphorylation of Mcm2, and after extensive screening of the C-terminal region we identified a mutant of Sld3, Sld3-m16 (Sld3-S556A,H557A,S558A,T559A) that is specifically defective in stimulating DDK phosphorylation of Mcm2 (Fig. S2).
Sld3 Stimulation of DDK Phosphorylation of Mcm2 May Be Important for Origin Melting and GINS Assembly with Mcm2-7.
Expression of sld3-m16 confers a dominant negative severe growth defect in budding yeast cells that is suppressed by mcm5-bob1 (Fig. 2A). Expression of wild-type levels of sld3-m16 also results in a severe growth and DNA replication defect (Fig. 2 A, B, and D). DDK phosphorylation of Mcm2 is undetectable in cells expressing sld3-m16 compared with wild-type cells (Fig. 2C), suggesting that Sld3 substantially stimulates DDK phosphorylation of Mcm2 in vivo. Cells expressing sld3-m16 exhibit a substantially reduced RPA-ChIP signal in S phase (Fig. S3), suggesting that Sld3 stimulation of DDK phosphorylation of Mcm2 is important for origin melting and subsequent replication initiation. Coimmunoprecipitation analysis supports that Sld3 stimulation of DDK phosphorylation of Mcm2 is important for GINS association with Mcm2-7 and also timely disengagement of Sld3 from Mcm2-7 (Fig. 3). These results are consistent with previous data suggesting that DDK phosphorylation of Mcm2 is required for origin melting and subsequent sequestration of Sld3 from Mcm2-7, allowing the assembly of GINS with Mcm2-7 (21). Furthermore, the phenotypes of cells expressing sld3-m16 and mcm2-S164A,S170A are very similar, suggesting that the primary defect in cells expressing sld3-m16 is the lack of DDK phosphorylation of Mcm2.
Human Treslin Stimulates DDK Phosphorylation of Mcm2.
We also found that human Treslin stimulates human DDK phosphorylation of human Mcm2 by 15-fold (Fig. S6 A and B), and we found that serines 53 and 108 are phosphorylated in response to Treslin stimulation of DDK phosphorylation of Mcm2 (Fig. S6 C and D). Furthermore, the phosphomimic form of human Mcm2 (Mcm2-2D or Mcm2-S53D,S108D) binds substantially more weakly to Mcm5 compared with unmodified Mcm2, suggesting a potential mechanism for Mcm2-7 “gate” opening with subsequent origin melting.
A Conserved Mechanism to Couple Helicase Assembly with Helicase Phosphorylation.
A large excess of Mcm2-7 complexes are loaded onto dsDNA relative to the number of Mcm2-7 complexes that actually fire during replication initiation (29). Furthermore, Sld3 and DDK are two limiting factors for DNA replication, along with Sld2 and Dpb11 (32). Thus, a question arises: What orchestrates the set of events that are required for origin firing at an activated Mcm2-7 double hexamer? We show in the paper that Sld3 may be critical for this orchestration, because Sld3 may couple recruiting Cdc45 to Mcm2-7 with stimulation of DDK phosphorylation of Mcm2. It is important to emphasize that DDK phosphorylation of Mcm2 does not cause the Cdc45 recruitment to Mcm2-7. Instead, DDK phosphorylation of Mcm2 is correlated with Cdc45 recruitment to Mcm2-7. This correlation is the result of Sld3 binding to Mcm2-7. When Sld3 binds to Mcm2-7, Sld3 simultaneously recruits Cdc45 to Mcm2-7, and Sld3 stimulates DDK phosphorylation of Mcm2.
A Model for Replication Initiation.
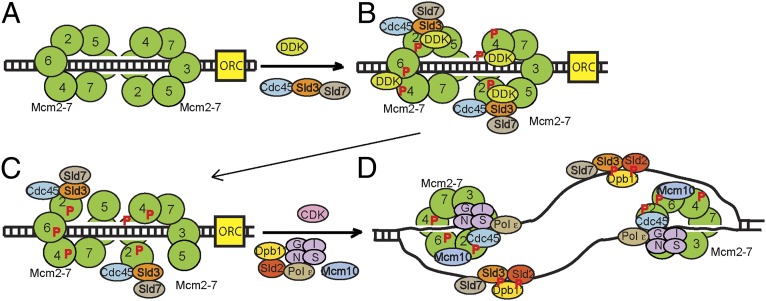
A model for the initiation of DNA replication is shown in Fig. 4. In late M and G1, the origin recognition complex (Orc), along with Cdc6 and Cdt1, promotes the loading of Mcm2-7 as a double hexamer to surround dsDNA (Fig. S3A) (6, 7, 33, 34). In S phase, Sld3-Sld7 recruits Cdc45 to Mcm2-7 (2, 17, 24, 35) (Fig. 4B). Sld3, once bound to Mcm2-7, substantially stimulates DDK phosphorylation of Mcm2 (this paper). Sld3 also blocks the premature interaction between GINS and Mcm2-7 (16, 23). DDK also phosphorylates Mcm4 and Mcm6 (18, 19, 36–38). The Mcm2-7 double hexamer then dissociates to single hexamers by an unknown mechanism (39). Once DDK phosphorylates Mcm2, and possibly with the aid of accessory proteins, Mcm2 loses its affinity for Mcm5 (Fig. 4C) (21). This allows for single-strand extrusion from the central channel of Mcm2-7 (i.e., origin melting). Origin melting generates ssDNA that sequesters the Sld3-Sld2-Dpb11 complex onto ssDNA, removing Sld3-Sld2-Dpb11 from Mcm2-7 (15, 16, 26). This sequestration allows GINS to bind Cdc45-Mcm2-7 (Fig. 4D). The CMG replication fork helicase is now fully assembled and activated.
Fig. 4.
Model for the initiation of DNA replication. (A) Mcm2-7 loads as double hexamer to encircle dsDNA during late M and G1 phases. (B) In S phase, Sld3, along with Sld7, recruits Cdc45 to Mcm2-7. Sld3, while bound to Mcm2-7, substantially stimulates DDK phosphorylation of Mcm2. DDK also phosphorylates Mcm4 and Mcm6. Sld3 also blocks the premature interaction between GINS and Mcm2-7. (C) Once Mcm2 is phosphorylated by DDK, the interaction between Mcm2 and Mcm5 weakens, allowing single-strand extrusion from the central channel of Mcm2-7 (origin melting). (D) Sld3-Sld2-Dpb11 form a CDK-dependent ternary complex in S phase. Once the origin is melted, Sld3-Sld2-Dpb11 is released from Mcm2-7, because Sld3-Sld2-Dpb11 binds preferentially to ssDNA. The sequestration of Sld3-Sld2-Dpb11 onto ssDNA allows GINS to engage with Cdc45-Mcm2-7, and the CMG helicase is assembled and activated for unwinding.
Materials and Methods
Antibodies were obtained from a commercial supplier (Pierce or Bethyl laboratories, see SI Materials and Methods for details). Plasmids and proteins were generated using established methods (see SI Materials and Methods for details). Yeast strains were generated from material supplied by Karim Labib; Robert Sclafani, University of Colorado, Denver, CO; and the Yeast Genetic Resource (see SI Materials and Methods for details). Kinase labeling of proteins were performed as described. DDK reactions were performed at 30 °C for 60 min (see SI Materials and Methods for details). Yeast dilutions were performed in 1:10 dilutions on agar plates (see SI Materials and Methods for details). FACS analysis was performed with propidium iodide staining (see SI Materials and Methods for details). Chromatin Immunoprecipitation was performed with cross-linking reagent as previously described (see SI Materials and Methods for details). Coimmunoprecipitation analyses were performed with no cross-linking agent and no hydroxyurea as described (see SI Materials and Methods for details).
SI Materials and Methods
Antibodies.
Antibodies directed against RPA were purchased from Pierce. Antibodies directed against Mcm2-1–160 and Mcm2-161–173-phosphoserine-164-phosphoserine-170 were validated as described (21). Antibodies directed against the Flag, HA, or His epitopes were commercially purchased. Antibodies directed against human Mcm2-phosphoserine-40/41, Mcm2-phosphoserine-53, or Mcm2-phosphoserine-108 were commercially purchased (Bethyl Laboratories). Antibodies against Mcm4-S171-phosphoserine-S174-phosphoserine were produced by Pierce.
Yeast Strains.
The sld3-7 td degron strain was obtained from Karim Labib. The epitope tags were generated using reagents from Yeast Genetic Resource Center and Karim Labib. The mcm5-bob1 mutation was introduced into the yeast strain by allelic replacement of the MCM5 endogenous locus (21).
MDY104 BY4743(MATa/α his3Δ1/his3Δ1 leu2Δ0/leu2Δ0 ura3Δ0/ura3Δ0 MET15/met15Δ0 LYS2/lys2Δ0 MCM2/mcm2AA (27)
MDY139 MATa his3Δ1/his3Δ1 leu2Δ0/leu2Δ0 MET15/met15Δ0 LYS2/lys2Δ0 mcm2AA (Ura3)
YMK517(2889) MATa ade2-1 ura3-1 his3-11,15 trp1-1 leu2-3,112 can1-100 GAL-UBR19HIS3 sld3-7td(kanMX) mcm4::Mcm4-5FLAG(k.l. TRP1), cdc45::Cdc45-6HA(hphNT), psf2::PSF2-5FLAG (hphNT), dpb11::DPB11-V5(Ura3)
YKL69 MATa ade2-1 ura3-1 his3-11,15 trp1-1 leu2-3,112 can1-100 GAL-UBR19HIS3 MCM2::mcm2-td(Ura3) mcm5::mcm5 bob1(TRP1)
Plasmids.
cDNA for human Mcm2, Mcm5, Mcm6, and Cdc45 and was a gift from Jerard Hurwitz, Memorial Sloan Kettering, NY. The genes were recloned from baculovirus vector to pET33b (PKA-Mcm5, PKA-Mcm6, or PKA-Cdc45) or pET41 (GST-Mcm2, wild-type and mutants). Human Treslin cDNA was a gift from William Dunphy, Caltech, Pasadena, CA. Full-length human Treslin was recloned into pET33b (PKA-Treslin). cDNA for human Dbf4 and Cdc7 was purchased from Thermo Scientific. Dbf4 was cloned into pET41a, and Cdc7 was cloned in to pETDuet. The following plasmids were used for experiments in this study: pIB401 (pRS415 CEN6/ARSH4 GALS::SLD3-6His LEU2),pIB402 (pRS415 CEN6/ARSH4 GALS::sld3S556A,H557A,S558A,T559A-6His LEU2).
Protein Purification.
Yeast Mcm2-7 subunits and complex, DDK, Sld3, Cdc45, Dpb11, and CDK were purified as described (15, 16, 21, 26). Human Mcm2, Mcm5, Mcm5, Cdc45, DDK (coexpression of GST-Dbf4 and Cdc7), and Treslin were subjected to nickel chromatography, anion exchange (Q Sepharose), and gel filtration. GST-Mcm2, wild-type and mutants, and DDK were subjected to additional glutathione Sepharose (GE Healthcare) chromatography. PKA was a gift from Susan Taylor, University of California, San Diego, La Jolla, CA.
Kinase Labeling of Proteins.
PKA, CDK, and DDK labeling of proteins was performed as described (16, 26, 30). Proteins containing a PKA tag at the N terminus were radiolabeled in a reaction volume of 100 μL that contained 20 mM PKA-tagged protein in kinase reaction buffer (5 mM Tris⋅HCl, pH 8.5, 10 mM MgCl2, 1 mM DTT, 500 μM ATP, and 500 μCi [γ-32P]ATP) containing 5 mg PKA, DDK, or CDK. Reactions were incubated for 1 h at 30 °C. The kinase was then removed from the mixture by affinity chromatography. DDK phosphorylation assays were performed as described (30). Briefly, DDK was added to Mcm2 in the presence of ATP and different amounts of Sld3 for 1 h at 30 °C.
Yeast Dilutions.
Tenfold serial dilutions were performed as described (26).
FACS Analysis.
FACS analysis was performed as described (26), and 6 × 106 cells per mL were treated with α-factor (Zymo Research) for 3 h. After extensive washes and the addition of 50 μg/mL Pronase, the cells were incubated for the indicated time. Cell cycle progression was then analyzed by flow cytometry (FACS) stained with propidium iodide with FACSAria.
Chromatin Immunoprecipitation.
Chromatin immunoprecipitation was performed as described (21), and 6 × 106 cells per mL were treated with α-factor (Zymo Research) for 3 h. Following extensive washes and the addition of 50 μg/mL Pronase, cells were further incubated for 0 or 20 min at the indicated temperature of the experiment. We performed PCR with [α-32P]dCTP as a component of the PCR to quantify the amplified DNA product. Formaldehyde cross-linked cells were lysed with glass beads in a BeadBeater. DNA was fragmented by sonication (Branson 450, six cycles of 15 s each). Antibody and magnetic protein A beads were added to the cleared lysate to immunoprecipitate the DNA. Immunoprecipitates were then washed extensively to remove nonspecific DNA. Eluted DNA was subjected to PCR analysis using primers directed against ARS305, ARS306, or a region midway between ARS305 and ARS306 as described (21). The radioactive band in the agarose gel, representing specific PCR-amplified DNA product, was quantified by phosphorimaging and normalized by a reference standard run in the same gel. The reference standard was a PCR accomplished with known quantity of template DNA replacing immunoprecipitate.
Coimmunoprecipitation.
Coimmunoprecipitation was performed as described (21), and 6 × 106 cells were treated with α-factor (Zymo Research) for 3 h. Cells were then subjected to extensive washes, followed by the addition of 50 μg/mL Pronase. Cells (4 × 108) were collected and lysed at 4 °C with glass beads (BeadBeater) in IP buffer (100 mM Hepes–KOH, pH 7.9, 100 mM potassium acetate, 10 mM magnesium acetate, 2 mM NaF, 1 mM PMSF, 0.1 mM Na3VO4, 10 mM β-glycerophosphate, 1% Triton X-100, leupeptin, pepstatin, 1% protease inhibitor mixture, and 1× complete protease inhibitor mixture without EDTA). Lysed material was treated with 200 U of Benzonase nuclease on ice for 1 h. Clarified extract was then mixed with 2 μL of specified antibody and rotated for 2 h in the cold room, and then 5 μL of Dynabeads Protein A beads equilibrated with IP buffer were added and further incubated for 2 h. Beads were then washed two times with 1 mL of IP buffer and resuspended in SDS-sample buffer. Western analysis was performed using the Odyssey system.
GST Pulldown.
The GST pulldown assays were performed as described (21). GST-pulldown reactions were in a volume of 100 μL and contained GST-tagged protein in GST-binding buffer (40 mM Tris⋅HCl, pH 7.5, 100 mM NaCl, 0.1 mM EDTA, 10% glycerol, 0.1% Triton X-100, 1 mM DTT, 0.7 mg/mL pepstatin, 0.1 mM PMSF, and 0.1 mg/mL BSA) and varying amounts of radiolabeled protein as described in each figure. Reactions were incubated at 25 °C for 1 h. Following incubation, reactions were added to 40 μL glutathione Sepharose and gently mixed. Binding of GST-tagged protein to the protein was performed for 20 min with gentle mixing every 2 min. When the binding was complete, the beads were allowed to settle, the supernatant was removed, and the glutathione beads were washed two times with 0.5 mL GST-binding buffer. After the last wash, 30 μL of 5× SDS sample buffer were added to each reaction, and the samples were heated to 95 °C for 10 min. Samples (20 μL) were then analyzed by SDS/PAGE followed by phosphorimaging and quantitation.
Acknowledgments
We thank Christopher Brandl for the yeast strain harboring mcm2-S164A,S170A regulated by native promoter; Jerard Hurwitz for cDNA for human Mcm2, Mcm5, Mcm6 and Cdc45; William Dunphy for cDNA for human Treslin; Robert Sclafani for the mcm5-bob1 strain; Karim Labib for the sld3-7 td strain; and Dr. Susan Taylor for supplying purified PKA. We thank the FACS facility at Florida State University College of Medicine for the use of the FACS equipment. Funding was provided by Florida State University and National Science Foundation Grant 1265431 (to D.L.K.).
Footnotes
Conflict of interest statement: Patent pending for assay of human Treslin-induced stimulation of human DDK phosphorylation of human Mcm2.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1509608112/-/DCSupplemental.
References
- 1.Ilves I, Petojevic T, Pesavento JJ, Botchan MR. Activation of the MCM2-7 helicase by association with Cdc45 and GINS proteins. Mol Cell. 2010;37(2):247–258. doi: 10.1016/j.molcel.2009.12.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kamimura Y, Tak YS, Sugino A, Araki H. Sld3, which interacts with Cdc45 (Sld4), functions for chromosomal DNA replication in Saccharomyces cerevisiae. EMBO J. 2001;20(8):2097–2107. doi: 10.1093/emboj/20.8.2097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kanemaki M, Labib K. Distinct roles for Sld3 and GINS during establishment and progression of eukaryotic DNA replication forks. EMBO J. 2006;25(8):1753–1763. doi: 10.1038/sj.emboj.7601063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Araki H. Cyclin-dependent kinase-dependent initiation of chromosomal DNA replication. Curr Opin Cell Biol. 2010;22(6):766–771. doi: 10.1016/j.ceb.2010.07.015. [DOI] [PubMed] [Google Scholar]
- 5.Labib K. How do Cdc7 and cyclin-dependent kinases trigger the initiation of chromosome replication in eukaryotic cells? Genes Dev. 2010;24(12):1208–1219. doi: 10.1101/gad.1933010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Remus D, et al. Concerted loading of Mcm2-7 double hexamers around DNA during DNA replication origin licensing. Cell. 2009;139(4):719–730. doi: 10.1016/j.cell.2009.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Evrin C, et al. A double-hexameric MCM2-7 complex is loaded onto origin DNA during licensing of eukaryotic DNA replication. Proc Natl Acad Sci USA. 2009;106(48):20240–20245. doi: 10.1073/pnas.0911500106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fu YV, et al. Selective bypass of a lagging strand roadblock by the eukaryotic replicative DNA helicase. Cell. 2011;146(6):931–941. doi: 10.1016/j.cell.2011.07.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zegerman P, Diffley JF. Phosphorylation of Sld2 and Sld3 by cyclin-dependent kinases promotes DNA replication in budding yeast. Nature. 2007;445(7125):281–285. doi: 10.1038/nature05432. [DOI] [PubMed] [Google Scholar]
- 10.Tanaka S, et al. CDK-dependent phosphorylation of Sld2 and Sld3 initiates DNA replication in budding yeast. Nature. 2007;445(7125):328–332. doi: 10.1038/nature05465. [DOI] [PubMed] [Google Scholar]
- 11.Kumagai A, Shevchenko A, Shevchenko A, Dunphy WG. Direct regulation of Treslin by cyclin-dependent kinase is essential for the onset of DNA replication. J Cell Biol. 2011;193(6):995–1007. doi: 10.1083/jcb.201102003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tak YS, Tanaka Y, Endo S, Kamimura Y, Araki H. A CDK-catalysed regulatory phosphorylation for formation of the DNA replication complex Sld2-Dpb11. EMBO J. 2006;25(9):1987–1996. doi: 10.1038/sj.emboj.7601075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kanter DM, Kaplan DL. Sld2 binds to origin single-stranded DNA and stimulates DNA annealing. Nucleic Acids Res. 2011;39(7):2580–2592. doi: 10.1093/nar/gkq1222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Muramatsu S, Hirai K, Tak YS, Kamimura Y, Araki H. CDK-dependent complex formation between replication proteins Dpb11, Sld2, Pol (epsilon, and GINS in budding yeast. Genes Dev. 2010;24(6):602–612. doi: 10.1101/gad.1883410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dhingra N, Bruck I, Smith S, Ning B, Kaplan DL. Dpb11 protein helps control assembly of the Cdc45·Mcm2-7·GINS replication fork helicase. J Biol Chem. 2015;290(12):7586–7601. doi: 10.1074/jbc.M115.640383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bruck I, Kaplan DL. Origin single-stranded DNA releases Sld3 protein from the Mcm2-7 complex, allowing the GINS tetramer to bind the Mcm2-7 complex. J Biol Chem. 2011;286(21):18602–18613. doi: 10.1074/jbc.M111.226332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yeeles JT, Deegan TD, Janska A, Early A, Diffley JF. Regulated eukaryotic DNA replication origin firing with purified proteins. Nature. 2015;519(7544):431–435. doi: 10.1038/nature14285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sheu YJ, Stillman B. The Dbf4-Cdc7 kinase promotes S phase by alleviating an inhibitory activity in Mcm4. Nature. 2010;463(7277):113–117. doi: 10.1038/nature08647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Masai H, et al. Phosphorylation of MCM4 by Cdc7 kinase facilitates its interaction with Cdc45 on the chromatin. J Biol Chem. 2006;281(51):39249–39261. doi: 10.1074/jbc.M608935200. [DOI] [PubMed] [Google Scholar]
- 20.Lei M, et al. Mcm2 is a target of regulation by Cdc7-Dbf4 during the initiation of DNA synthesis. Genes Dev. 1997;11(24):3365–3374. doi: 10.1101/gad.11.24.3365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bruck I, Kaplan DL. The Dbf4-Cdc7 kinase promotes Mcm2-7 ring opening to allow for single-stranded DNA extrusion and helicase assembly. J Biol Chem. 2015;290(2):1210–1221. doi: 10.1074/jbc.M114.608232. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 22.Bochman ML, Schwacha A. The Mcm2-7 complex has in vitro helicase activity. Mol Cell. 2008;31(2):287–293. doi: 10.1016/j.molcel.2008.05.020. [DOI] [PubMed] [Google Scholar]
- 23.Bruck I, Kaplan DL. GINS and Sld3 compete with one another for Mcm2-7 and Cdc45 binding. J Biol Chem. 2011;286(16):14157–14167. doi: 10.1074/jbc.M111.218305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tanaka S, Nakato R, Katou Y, Shirahige K, Araki H. Origin association of Sld3, Sld7, and Cdc45 proteins is a key step for determination of origin-firing timing. Curr Biol. 2011;21(24):2055–2063. doi: 10.1016/j.cub.2011.11.038. [DOI] [PubMed] [Google Scholar]
- 25.Costa A, et al. DNA binding polarity, dimerization, and ATPase ring remodeling in the CMG helicase of the eukaryotic replisome. Elife. 2014;Aug 12:e03273. doi: 10.7554/eLife.03273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bruck I, Kaplan DL. The replication initiation protein Sld2 regulates helicase assembly. J Biol Chem. 2014;289(4):1948–1959. doi: 10.1074/jbc.M113.532085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stead BE, Brandl CJ, Davey MJ. Phosphorylation of Mcm2 modulates Mcm2-7 activity and affects the cell’s response to DNA damage. Nucleic Acids Res. 2011;39(16):6998–7008. doi: 10.1093/nar/gkr371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stead BE, Brandl CJ, Sandre MK, Davey MJ. Mcm2 phosphorylation and the response to replicative stress. BMC Genet. 2012;13:36. doi: 10.1186/1471-2156-13-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ge XQ, Jackson DA, Blow JJ. Dormant origins licensed by excess Mcm2-7 are required for human cells to survive replicative stress. Genes Dev. 2007;21(24):3331–3341. doi: 10.1101/gad.457807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bruck I, Kaplan D. Dbf4-Cdc7 phosphorylation of Mcm2 is required for cell growth. J Biol Chem. 2009;284(42):28823–28831. doi: 10.1074/jbc.M109.039123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Montagnoli A, et al. Identification of Mcm2 phosphorylation sites by S-phase-regulating kinases. J Biol Chem. 2006;281(15):10281–10290. doi: 10.1074/jbc.M512921200. [DOI] [PubMed] [Google Scholar]
- 32.Mantiero D, Mackenzie A, Donaldson A, Zegerman P. Limiting replication initiation factors execute the temporal programme of origin firing in budding yeast. EMBO J. 2011;30(23):4805–4814. doi: 10.1038/emboj.2011.404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Frigola J, Remus D, Mehanna A, Diffley JF. ATPase-dependent quality control of DNA replication origin licensing. Nature. 2013;495(7441):339–343. doi: 10.1038/nature11920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Samel SA, et al. A unique DNA entry gate serves for regulated loading of the eukaryotic replicative helicase MCM2-7 onto DNA. Genes Dev. 2014;28(15):1653–1666. doi: 10.1101/gad.242404.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Heller RC, et al. Eukaryotic origin-dependent DNA replication in vitro reveals sequential action of DDK and S-CDK kinases. Cell. 2011;146(1):80–91. doi: 10.1016/j.cell.2011.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sheu YJ, Kinney JB, Lengronne A, Pasero P, Stillman B. Domain within the helicase subunit Mcm4 integrates multiple kinase signals to control DNA replication initiation and fork progression. Proc Natl Acad Sci USA. 2014;111(18):E1899–E1908. doi: 10.1073/pnas.1404063111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sheu Y-J, Stillman B. Cdc7-Dbf4 phosphorylates MCM proteins via a docking site-mediated mechanism to promote S phase progression. Mol Cell. 2006;24(1):101–113. doi: 10.1016/j.molcel.2006.07.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Francis LI, Randell JC, Takara TJ, Uchima L, Bell SP. Incorporation into the prereplicative complex activates the Mcm2-7 helicase for Cdc7-Dbf4 phosphorylation. Genes Dev. 2009;23(5):643–654. doi: 10.1101/gad.1759609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yardimci H, Loveland AB, Habuchi S, van Oijen AM, Walter JC. Uncoupling of sister replisomes during eukaryotic DNA replication. Mol Cell. 2010;40(5):834–840. doi: 10.1016/j.molcel.2010.11.027. [DOI] [PMC free article] [PubMed] [Google Scholar]