Significance
Within eastern North America, distributions of vector-transmitted haemosporidian blood parasites of birds, commonly known as “avian malaria parasites,” are associated with the distributions of their host species independently of direct effects of climate on potential vectors. Spatial analyses additionally indicated an absence of dispersal limitation for these parasites. Finally, host-breadth, ranging continuously from specialist to generalist, varies among parasite lineages and is dynamic within parasite assemblages over space and time. The distributions of avian haemosporidian parasites emphasize the ability of parasites to disperse across broad regions and to switch readily between hosts to become emerging infectious diseases.
Keywords: avian malaria, community assembly, emerging infectious disease, Haemosporida, parasite communities
Abstract
The drivers of regional parasite distributions are poorly understood, especially in comparison with those of free-living species. For vector-transmitted parasites, in particular, distributions might be influenced by host-switching and by parasite dispersal with primary hosts and vectors. We surveyed haemosporidian blood parasites (Plasmodium and Haemoproteus) of small land birds in eastern North America to characterize a regional parasite community. Distributions of parasite populations generally reflected distributions of their hosts across the region. However, when the interdependence between hosts and parasites was controlled statistically, local host assemblages were related to regional climatic gradients, but parasite assemblages were not. Moreover, because parasite assemblage similarity does not decrease with distance when controlling for host assemblages and climate, parasites evidently disperse readily within the distributions of their hosts. The degree of specialization on hosts varied in some parasite lineages over short periods and small geographic distances independently of the diversity of available hosts and potentially competing parasite lineages. Nonrandom spatial turnover was apparent in parasite lineages infecting one host species that was well-sampled within a single year across its range, plausibly reflecting localized adaptations of hosts and parasites. Overall, populations of avian hosts generally determine the geographic distributions of haemosporidian parasites. However, parasites are not dispersal-limited within their host distributions, and they may switch hosts readily.
A regional community can be thought of as a set of species whose distributions partially overlap within a large geographic area (1, 2). The structure of the regional community (i.e., the relative abundances of species across space and the degree to which populations cooccur) is governed by local (e.g., interspecific competition) and regional (e.g., species diversification and dispersal) processes (3). Although regional communities include all species, parasites and pathogens are rarely considered integral community members (4). Indeed, impacts of parasites on community structure are frequently associated with epidemics—often following introductions to nonnative regions—that have driven naïve hosts to extinction or near extinction (5–7). However, parasites likely play a critical role in shaping regional community structure. Parasites can comprise a large proportion of the community biomass (8), form the majority of links in a community food web (9), and influence regional diversity by variously accelerating (10) or slowing (11) host diversification.
Nevertheless, few studies have investigated the processes influencing the regional community structure of both parasites and their hosts. Parasite populations are integrated into community studies with difficulty, partly because these populations are distributed across multiple dimensions—space, host species, and host individuals (12)—and also because parasites are difficult to sample. Moreover, although parasites tend to specialize on one or a few host species, host-breadth may vary across a parasite’s range (13).
Regional studies of birds and their dipteran-vectored haemosporidian (“malaria”) blood parasites (14–19) have shown that many parasites are heterogeneously distributed across space despite the availability of suitable hosts. Specialized associations between specific parasites and vectors (20–22) may drive such heterogeneity, although a recent analysis suggests that parasite–host compatibility is also important (23), and local coevolutionary relationships between parasites and their hosts likely influence geographic distributions of both host and parasite populations (11, 14, 15). However, most regional studies of these parasites have focused on individual host species (24–30).
Here, we investigate the regional community structure of avian hosts and their haemosporidian parasites with respect to abiotic and biotic drivers of both host and parasite distributions. We surveyed local assemblages of avian haemosporidian parasites across eastern North America and related the distributions of individual parasite lineages to regional climate variation and to the distributions and abundances of their avian hosts. Community dissimilarities between sampling locations based on host assemblage structure (i.e., the relative abundances of potential host species) were positively correlated with those based on parasite assemblage structure, suggesting interdependence of host and parasite population distributions. However, when controlling statistically for that interdependence, local host assemblages responded strongly to environmental gradients and differed more with increasing geographic separation, whereas parasite assemblages did not. This finding suggests that haemosporidian parasites disperse readily across the distributions of their host populations in eastern North America, independently of difference in climate and geographic distance. The degree to which some parasite lineages specialized on particular hosts varied across years and locations, and the nonrandom parasite lineage turnover across the distribution of one well-sampled host species suggested that adaptations of hosts and parasites may also shape regional community structure. Despite evidence of pathogenicity of haemosporidian parasites in birds (31), correlations between host abundances and parasite relative abundances across the region were statistically indistinguishable from random. Taken together, these results suggest that the distributions of parasite populations largely follow the distributions of their hosts but that parasites readily switch hosts and may replace each other across the ranges of individual hosts, resulting in a complex and dynamic regional community.
Results
Parasite Populations Track Populations of Their Hosts.
We screened 5,867 individuals of 99 bird species, mostly from the order Passeriformes, from 13 locations in eastern North America (Fig. 1), and found 1,720 (29.3%) infected with haemosporidian parasites of the genera Plasmodium or Haemoproteus. Overall, we recovered 87 parasite lineages (see SI Appendix, Table S1 for lineage details; see Materials and Methods for lineage determination). We calculated pairwise dissimilarities between “community” sampling locations (i.e., sites where sampling was not restricted to focal species; Fig. 1 and SI Appendix, Table S2) separately by bird species abundances and by parasite lineage relative abundances (i.e., the number of infections of each lineage divided by the total number of infections). We used Bray–Curtis dissimilarity (32–34), which quantifies the difference between two locations based on the relative abundances of their species (Materials and Methods), to assess community difference. We restricted this analysis to 33 parasite lineages, sampled 10 or more times across the community samples, and to 64 host species infected at least once by any of the 87 parasite lineages within the region (Dataset S1; results are similar using all sampled host species). We compared dissimilarities between locations based on hosts and parasites with a Mantel test, which is equivalent to a correlation test between two distance matrices (34). The Mantel test revealed a significant, positive correlation between host and parasite dissimilarities (r = 0.45, P = 0.003), showing some interdependence between host and parasite populations across the region. When the same 33 parasite lineages were analyzed separately by genus, these correlations remained significant (Plasmodium: n = 15, r = 0.37, P = 0.026; Haemoproteus: n = 18, r = 0.29, P = 0.034).
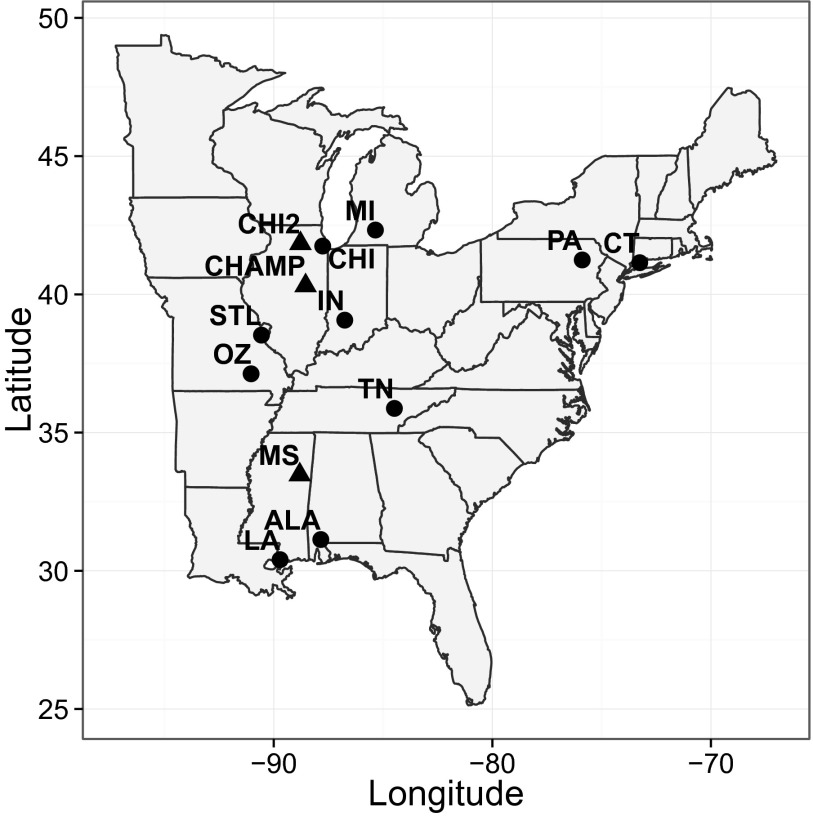
Fig. 1.
Sampling locations. Circles are community samples (i.e., sampling was not restricted to focal bird species), triangles are samples of one or a few bird species only (SI Appendix, Table S2). Location codes are as follows: ALA, Alabama; CHAMP, Champaign (Illinois); CHI, Chicago (Illinois); CHI2, western Chicago (Illinois); CT, Connecticut; IN, Indiana; LA, Louisiana; MI, Michigan; MS, Mississippi; OZ, Ozarks (Missouri); PA, Pennsylvania; STL, St. Louis (Missouri); and TN, Tennessee.
Differences between regional distributions of populations of hosts and parasites might reflect dispersal limitation (i.e., geographic distance), differences between local environments (e.g., climate or habitat variables), and interactions among hosts and parasites (Fig. 2). To evaluate these relationships, we calculated partial Mantel coefficients for the connections in Fig. 2 (Table 1; simple Mantel correlations are provided for comparison in SI Appendix, Table S3). Partial Mantel coefficients represent the strength of correlation between two distance matrices while controlling for the effect of a third (34). For example, the effect of the environment on hosts may be related to the geographic distance between localities (space). However, the correlation between hosts and environment can be controlled for the effect of geographic distance by computing a partial Mantel coefficient. Because geographic distances and climate differences are independent of the hosts and parasites at each locality, we tested their relationship with a standard Mantel test, which involves no control for a third variable. The distance matrix for the Mantel test comprised geographic distances between all pairs of sampling locations (space), and the elements of the climate matrix were the Euclidean distances between sampling locations based on the first five principal components scores for 19 climatic variables (worldclim.org) downloaded for each location (SI Appendix, Fig. S1 and Table S4). Additional Mantel tests were based on Bray–Curtis dissimilarities between sites in their parasite lineages or host assemblages as before. Partial Mantel tests between these distance matrices revealed that although host populations are related to variation in climate across eastern North America, parasite populations, when controlling for the effect of hosts, are not (Table 1). Furthermore, parasite community similarity does not decline with distance [i.e., parasite distributions were not spatially restricted (35) when controlling for hosts], suggesting that parasites disperse readily across the region within their host populations. These results generally held when the parasite genera were analyzed separately (SI Appendix, Table S5) and when using an alternative statistical approach (SI Appendix, Table S6).
Fig. 2.
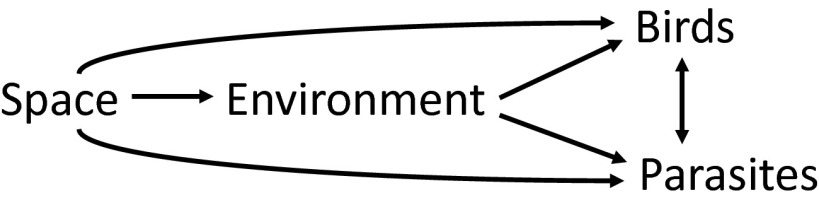
Path diagram of hypothesized interactions between space (i.e., geographic distance between sampling locations), environment (i.e., climatic differences between sampling locations), and bird and parasite communities (i.e., differences in species richness and abundances of birds and of parasite lineages, respectively, between sampling locations), all of which are represented as distance matrices. We tested these hypotheses with partial Mantel tests, which allow for the calculation of correlations between two distance matrices while controlling for the effect of a third. For example, bird and parasite assemblage distances were positively correlated (r = 0.335, P = 0.027) even when controlling for the effect of geographic distance (Table 1).
Table 1.
Results of partial Mantel tests comparing hypothesized relationships between space (i.e., geographic distance between sites), the environment (climate differences between sites), birds (host community dissimilarity between sites), and parasites (parasite community dissimilarity between sites) identified in Fig. 2
| Relationship between | And | Controlling for | rp | P |
| Space | Environment | None | 0.595 | 0.005 |
| Birds | Environment | Parasites | 0.772 | <0.001 |
| Birds | Space | Parasites | 0.504 | 0.012 |
| Birds | Environment | Space | 0.720 | <0.001 |
| Birds | Space | Environment | 0.185 | 0.137 |
| Parasites | Environment | Birds | 0.117 | 0.277 |
| Parasites | Space | Birds | 0.097 | 0.302 |
| Parasites | Environment | Space | 0.303 | 0.076 |
| Parasites | Space | Environment | 0.101 | 0.300 |
| Birds | Parasites | Environment | 0.191 | 0.144 |
| Birds | Parasites | Space | 0.335 | 0.027 |
We report the partial Mantel correlation coefficient (rp) and associated P value. The relationship between space and environment was tested with a standard Mantel test. Bolded values of rp represent P < 0.05.
Host Specialization.
The host-breadth of a parasite may vary geographically or temporally, and may also be limited by the phylogenetic relatedness of potential host species (13). For example, in the Chicago location, each Plasmodium parasite lineage was associated with a single host taxon at the superfamily level (23). To determine the importance of host phylogeny on parasite distributions across the region, we created a phylogenetic distance matrix for all hosts infected at least once by any of the 33 parasite lineages sampled 10 or more times (60 host species). We then calculated a second matrix by computing Bray–Curtis dissimilarities between those hosts based on the number of times each host species was infected with each of the 33 parasite lineages. A Mantel test comparing these two matrices showed a weak, but significant, correlation (r = 0.28, P = 0.002), indicating that parasite host distribution is constrained to more closely related hosts than expected by chance. Interestingly, this effect varied across locations in the region (SI Appendix, Table S7).
To quantify the host-breadth of each parasite, we used the Gini–Simpson index (36), which accounts for the number of infections recorded for each host species (13). We weighted the index by the phylogenetic distance between hosts using the formula for Rao’s quadratic entropy [Rao’s QE (37, 38); see Materials and Methods for formula; results did not change qualitatively if phylogenetic distances were not included in these analyses]. Although ecologists often distinguish generalist and specialist parasites, host-breadth in the 33 parasite lineages sampled 10 or more times was continuously distributed (SI Appendix, Fig. S2) and did not differ statistically from a unimodal distribution [Hartigan’s dip test: D33 = 0.047, P = 0.87 (39)]. Furthermore, we found no difference in the host-breadth of individual parasite lineages between the parasite genera (t31 = −1.1, P = 0.28).
When all years were pooled, parasite lineages recovered at least four times from each of at least four community sampling locations exhibited variation in local host-breadth across the region (Fig. 3). A linear mixed-effects model with parasite lineage as a random effect showed no influence of local phylogenetically weighted bird diversity (Rao’s QE, using host species infected at least once in the region) on parasite host-breadth (F1,21.4 = 1.26, P = 0.27), suggesting that variation in host-breadth is not simply attributable to the diversity of available hosts. Furthermore, local parasite diversity did not influence parasite host-breadth (F1,21.2 = 2.41, P = 0.14). For example, parasite lineage LA01 (Haemoproteus sp.) was recovered exclusively from Dumetella carolinensis in Chicago, IL (23/157 D. carolinensis hosts infected; years sampled 2006 and 2007); Connecticut (4/45; 2002 and 2003); and Michigan (11/94; 2012). However, in the 2013 Tennessee sample, LA01 was recovered from the hosts Mimus polyglottos (like D. carolinensis, in the family Mimidae; 2/9 infected), Cardinalis cardinalis (1/36), and Spinus tristis (1/19), whereas the two D. carolinensis hosts sampled in Tennessee were both uninfected. We also recovered LA01 from D. carolinensis in the western Chicago location (6/7) in 2014 and from D. carolinensis (2/6) and Toxostoma rufum (also in the family Mimidae; 1/7) in Champaign, IL, in the same year (although those were not community samples).
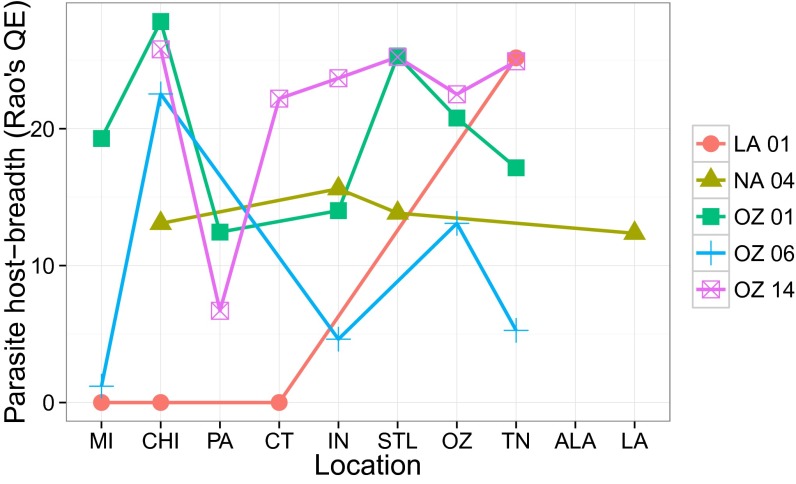
Fig. 3.
Parasite host-breadth (calculated as Rao’s QE) for parasite lineages sampled at least four times at each of at least four sampling locations, pooling data from all years, showing clear variation in host-breadth across the region. Locations are ordered from north to south.
To determine whether local host-breadth differed from a random expectation, we restricted our dataset to infected individuals of those five potential host species of LA01. We then shuffled all parasite lineages infecting those hosts within sampling locations and recalculated randomized host-breadths for LA01 (9,999 randomizations) and compared observed host-breadths to the distribution of randomized host breadths. In Chicago, the host-breadth of LA01 was lower than expected by chance (P < 0.001), whereas in Tennessee, this lineage's host-breadth was higher than expected by chance (P = 0.019). The host-breadth of LA01 did not differ from random in Connecticut and Michigan because there were no potential alternative hosts in either location. Lineage Ozarks 06 (OZ06) (Plasmodium sp.) also varied with respect to host-breadth (Fig. 3). The host-breadth of OZ06 was lower than expected based on a random distribution (again shuffling infections among potential hosts) in Michigan (P = 0.003), Indiana (P < 0.001), and Tennessee (P = 0.030) but did not differ from random in Chicago (P = 0.76) and the Ozarks (P = 0.94).
Because locations were sampled in different years, some variation in host-breadth between localities might reflect temporal change within localities. Within particular years, parasite lineages sampled more than three times at multiple locations mostly showed little variation in host-breadth. However, in 2013, OZ14 (Plasmodium sp.) infected three hosts in Pennsylvania (6/12 Melospiza melodia infected, also 1/3 Pipilo erythrophthalmus, and 1/1 Pheucticus ludovicianus) but infected a larger variety of species in Tennessee (6/50 Passerina cyanea individuals infected and 12 more infections in nine other species) and Indiana (six infections recovered across five species). Host-breadth of OZ14 was greater than expected based on a random distribution in Indiana (P = 0.050), no different from expected in Tennessee (P = 0.127), and, although low, still within the random expectation in Pennsylvania (P = 0.082). Overall, these results demonstrate that parasite host-breadth can vary geographically, independent of temporal variation, and that this variation does not merely reflect the array of potential host species available or the local diversity of parasites.
Nonrandom Parasite Turnover Across Space.
We found evidence of parasite lineage turnover across locations within our best-sampled host in 2013, C. cardinalis (Fig. 4). We restricted this analysis to the four locations in which C. cardinalis was well-sampled in 2013 and to parasite lineages recovered at least nine times from C. cardinalis across those locations. Prevalences of all parasite lineages of C. cardinalis, except for OZ14, were significantly heterogeneous across sampling locations (Table 2). Furthermore, at each location, C. cardinalis harbored a single dominant parasite lineage, making each location’s parasite assemblage distinct. To test whether these parasite assemblages differed more from each other than one would expect by chance, we calculated the mean Bray–Curtis dissimilarity between the four locations based on parasite lineage prevalence and compared it to a distribution of randomized average dissimilarities. We created a randomized parasite-by-location matrix by shuffling parasite lineages among infected birds, recalculating prevalence for each lineage at each site, and then calculating the randomized mean Bray–Curtis dissimilarity 9,999 times. Mean observed dissimilarity between sites based on parasite prevalence greatly exceeded the randomized average dissimilarities (P < 0.001; SI Appendix, Fig. S3), confirming that location–parasite combinations were more distinct than expected by chance, plausibly a result of localized host and parasite adaptations.
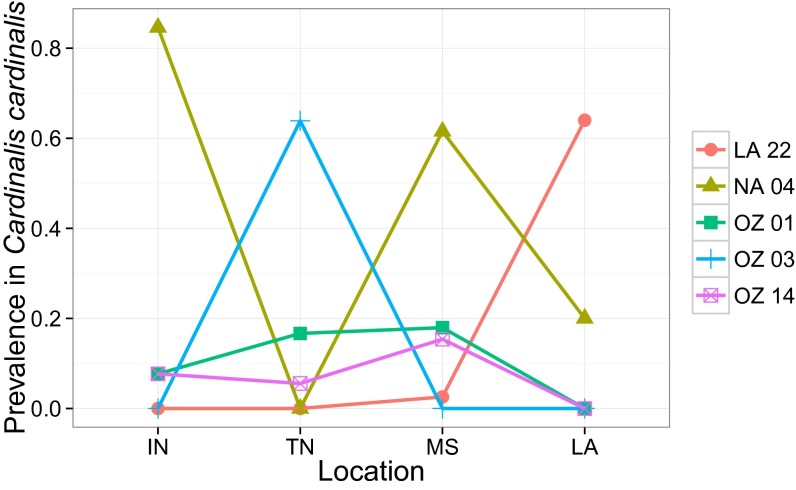
Fig. 4.
Prevalences of well-sampled parasite lineages on the avian host C. cardinalis at four locations in 2013. Prevalences of four of five parasites were significantly heterogeneous across space (Table 2) and parasite assemblages within this host exhibited significant spatial turnover (mean Bray–Curtis dissimilarity between sites was significantly greater than random, P < 0.001; SI Appendix, Fig. S3).
Table 2.
Results of G tests comparing the prevalence of each well-sampled parasite of the host C. cardinalis in 2013 across sampling locations
| Statistic | LA22 | NA04 | OZ01 | OZ03 | OZ14 |
| G(df = 3) | 53.73 | 58.72 | 8.46 | 67.1 | 6.82 |
| P | <0.001 | <0.001 | 0.037 | <0.001 | 0.078 |
Prevalence data are shown graphically in Fig. 4.
Parasites and Host Abundance.
We calculated pairwise Spearman rank correlation coefficients (ρ) between all host abundances and parasite relative abundances across the region, for which an excess of negative correlations would be consistent with pathogenic effects of these parasites (31). We restricted our analysis to community sampling locations and to parasite lineages sampled at least 10 times and hosts infected at least once (results did not differ qualitatively using the full dataset). Our analysis included the abundances of 64 host species and the relative abundances of 33 parasite lineages at each sampling location, resulting in 2,112 pairwise correlations. Mean observed ρ was −0.012 ±0.008 SE, which did not differ significantly from the distribution of mean ρ values obtained by randomizing the parasite frequency matrix (by row shuffling 9,999 times) and recalculating the mean ρ each time (P = 0.628). Furthermore, the observed SE did not differ from the distribution of randomized SEs (P = 0.147), and the proportion of correlations with P < 0.05 (141/2112) also did not differ from random (P = 0.236). Results were largely similar when analyzing the parasite genera separately (SI Appendix, Table S8).
Discussion
We have found that the distributions of haemosporidian blood parasites of birds in eastern North America strongly mirror those of their hosts, with broad-scale climatic gradients and barriers to dispersal having little influence, even though the distributions of avian host populations were related to environmental gradients when controlling for the distributions of parasites (Table 1). Because parasite transmission takes place primarily during the warm summer months [as evidenced by infections in hatch-year birds in late summer (38, 40, 41)], haemosporidian parasites probably are buffered against variation in climate (average summer temperatures varied between 19.9 and 26.9 °C across our sampling locations). Although we do not know the extent to which the populations of the parasites’ dipteran vectors track hosts, in at least one location in the region (Chicago, IL), parasite–host associations were unrelated to vector–host encounter rates (23). Nevertheless, more studies are needed to identify vectors and reveal their contribution to host–parasite associations. Indeed, vector movement has been linked to pathogen spread in other systems (42), and such movement, along with changes in vector–host associations (43), might facilitate parasite host-switching and dispersal. Interestingly, regional studies of ectoparasites of small mammals have shown that flea assemblages can differ more with increasing distance between sampling locations and with increasing differences between local habitat and climate characteristics, even on the same host species (44, 45), perhaps because fleas are more exposed to the environment than haemosporidian parasites. Similar distance–decay relationships have been observed in trophically transmitted helminth parasites of both mammals and fishes as well (46). Thus, mode of transmission might play an important role in structuring regional parasite assemblages.
Distributions of parasite populations across the region also were characterized by localized host-switching and by geographic parasite turnover within host populations. Our best sampled host, C. cardinalis, supported statistically differentiated parasite assemblages at each of four sampling locations within a single year (Fig. 4). For example, parasite lineages LA22 and NA04 replaced each other as the most common parasites of C. cardinalis in Louisiana and Mississippi, respectively, although both lineages infect C. cardinalis in both locations. This finding raises the possibility that parasite distributions may be influenced by localized adaptations of hosts and parasites to each other across the region (47). Furthermore, parasite host-breadth can vary across time and space (Fig. 3), even when controlling for the local diversity of potential hosts and parasites, indicating the importance of host-switching in determining parasite distributions across the region.
Finally, although theoretical (48) and empirical (49) studies suggest that parasites may often limit host population size, the distributions of correlations between host and parasite populations across the region did not differ from random, suggesting that haemosporidian parasites do not impact the population densities of their hosts in eastern North America. Our analyses suggest that populations of haemosporidian parasites are largely structured by populations of their hosts, although parasite lineages change between nearby localities within host species distributions and over short intervals within localities.
Materials and Methods
Field Methods.
We captured birds with mist-nets at 13 locations across eastern North America (Fig. 1) during summer months (primarily late May to August, with minimal sampling in April and September; removal of April and September samples did not qualitatively change results) from 1999 to 2014 (SI Appendix, Table S2). We took a small (approximately 10-μL) blood sample from the brachial vein of each bird and stored the blood in Puregene or Longmire’s (50) lysis buffer. We collected all samples under appropriate state and federal permits and Institutional Animal Care and Use Committee (IACUC) protocols.
Laboratory Methods.
We extracted DNA from blood samples using an ammonium acetate-isopropanol precipitation protocol (51). We screened DNA samples for haemosporidian parasites using a PCR protocol designed to amplify a small section of parasite mitochondrial DNA (52). We then amplified a portion of the cytochrome b gene in positive samples using several primer pairs and protocols (15, 40, 53, 54). We identified unique parasite lineages based on their cytochrome b sequences and on their host and geographic distributions (55, 56). Multiple infections were separated by phasing (57) where possible. GenBank Accession numbers for all lineages can be found in SI Appendix, Table S1.
Statistical Analysis.
All analyses were performed in R v3.1.2 (58), and we report two-tailed P values for all tests. We calculated Bray–Curtis dissimilarities between locations with the “vegdist” function in the vegan package (59). Bray–Curtis dissimilarity between two sampling locations (1, 2) is calculated by
where y represents the number (or frequency) of individuals sampled of species j, and p represents the total number of species sampled over both locations (34).
We created a geographic distance matrix between locations with the “rdist.earth” function in the fields package (60) in R. We compared distance matrices with Mantel and partial Mantel tests using functions “mantel” and “mantel.partial” (method = “spearman”) in the vegan package. Mantel statistics were tested for significance by permutation (9,999 trials) according to ref. 34. Mantel tests have been criticized for lack of power, but they are appropriate when hypotheses can be formulated in terms of distances, as is the case here (61). We tested for a departure from unimodality in the frequency of host breadth values using Hartigan’s dip test (39) with the function “dip.test” in the diptest package (62) in R. Linear mixed effects models were run with the lme4 R package (63), and denominator degrees of freedom for F tests were calculated using the “Kenward–Roger” approach (64) implemented in the lmerTest (65) and pbkrtest (66) packages in R.
Host Abundance.
Our mist-net effort varied across locations and years and therefore provided unreliable estimates of avian abundance. To estimate avian abundance, we downloaded route data from the North American Breeding Bird Survey (https://www.pwrc.usgs.gov/bbs). We selected routes deemed acceptable by the survey organizers (i.e., routes that met all survey requirements in a particular year) located within 80 km of our sampling locations, and we used route data corresponding to the year each location was sampled, plus 1 y before and 1 y after our sample was taken. For example, Chicago, IL, was sampled in 2006 and 2007, so we used route data from 2005 to 2008 within the 80-km buffer (for the locations sampled in 2014, we used route data from years 2013 and 2014). We then averaged bird species abundances across routes and across years for each sampling location. We used these spatial and temporal buffers to account for potential variability in abundance estimates attributable to environmental heterogeneity within routes (67) and observer error (68), but our results did not change qualitatively with changes in the sizes of these buffers.
Parasite Host-Breadth.
We calculated host-breadth for each parasite lineage using Rao’s QE (37) defined by the formula
where tij is a matrix of phylogenetic distances between host species i and j observed to be infected by a given parasite lineage (divided by two to obtain average phylogenetic distance), pi is the proportion of infections by the parasite in host species i (i.e., the number of individuals of host species i infected by the parasite divided by the total number of individuals infected by that parasite), pj is the proportion of infections by the parasite in host species j, and S is the total number of host species. Our parasite host-breadth score varies from zero (complete host specialization) to
which represents a maximally generalized parasite (i.e., a parasite that infects all hosts in the community equally; however, an alternative might be that a perfect generalist would infect all hosts in direct proportion to host abundance) and is equivalent to a phylogenetically weighted Gini–Simpson diversity index. We calculated Rao’s QE using the “raoD” function in the picante package (69) in R and report the “Dkk” value the function produces. We used the phylogeny of Jetz et al. (70) to estimate phylogenetic relationships between bird species. Based on parasites sampled at least 10 times over the community sampling locations, we showed a strong relationship between Rao’s QE and the Gini–Simpson index applied to parasite host-breadth (SI Appendix, Fig. S4). Because of the apparent effect of host phylogeny, we used Rao’s QE as a metric of parasite host-breadth for all analyses.
Supplementary Material
Acknowledgments
We thank Brandt Ryder, Pete Marra, Keegan Tranquillo, Peter Pyle, and the Indiana Monitoring Avian Productivity and Survivorship (MAPS) teams for assistance collecting Indiana samples; Alicia Burke for assistance collecting Ozarks samples; field biologist Brenda Keith and the Kalamazoo Valley Bird Observatory/Kalamazoo Nature Center for collecting Michigan samples; the Connecticut Audubon Society for samples from their banding station; Steve Latta for Alabama parasite sequences; Woody Walstrom and Diana Outlaw for Mississippi samples; and Emma Levenson and Kathleen Riley for assistance collecting Louisiana samples. Our work would not have been possible without the help of local government and conservation agencies. We also thank the thousands of volunteers and organizers of the Breeding Bird Survey. V.A.E. thanks Fabrício Santos for providing support and a place to write. Field and laboratory work was supported by the Harris World Ecology Center, the St. Louis and the Missouri Audubon Societies, the Curators of the University of Missouri, the National Science Foundation Malaria Research Coordination Network, and a University of Missouri–St. Louis Dissertation fellowship (to V.A.E.).
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1515309112/-/DCSupplemental.
References
- 1.Gleason HA. The individualistic concept of the plant association. Bull Torrey Bot Club. 1926;53(1):7–26. [Google Scholar]
- 2.Ricklefs RE. Disintegration of the ecological community. Am Nat. 2008;172(6):741–750. doi: 10.1086/593002. [DOI] [PubMed] [Google Scholar]
- 3.Ricklefs RE. Community diversity: Relative roles of local and regional processes. Science. 1987;235(4785):167–171. doi: 10.1126/science.235.4785.167. [DOI] [PubMed] [Google Scholar]
- 4.Poulin R. The functional importance of parasites in animal communities: Many roles at many levels? Int J Parasitol. 1999;29(6):903–914. doi: 10.1016/s0020-7519(99)00045-4. [DOI] [PubMed] [Google Scholar]
- 5.Day FP, Monk CD. Vegetation patterns on a southern Appalachian watershed. Ecology. 1974;55(5):1064–1074. [Google Scholar]
- 6.van Riper C, III, van Riper SG, Goff ML, Laird M. The epizootiology and ecological significance of malaria in Hawaiian land birds. Ecol Monogr. 1986;56(4):327–344. [Google Scholar]
- 7.Sinclair ARE, et al. Long-term ecosystem dynamics in the Serengeti: Lessons for conservation. Conserv Biol. 2007;21(3):580–590. doi: 10.1111/j.1523-1739.2007.00699.x. [DOI] [PubMed] [Google Scholar]
- 8.Kuris AM, et al. Ecosystem energetic implications of parasite and free-living biomass in three estuaries. Nature. 2008;454(7203):515–518. doi: 10.1038/nature06970. [DOI] [PubMed] [Google Scholar]
- 9.Lafferty KD, Dobson AP, Kuris AM. Parasites dominate food web links. Proc Natl Acad Sci USA. 2006;103(30):11211–11216. doi: 10.1073/pnas.0604755103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Page RDM. Tangled Trees, Phylogeny, Cospeciation, and Coevolution. Univ of Chicago Press; Chicago: 2003. [Google Scholar]
- 11.Ricklefs RE. Host-pathogen coevolution, secondary sympatry and species diversification. Philos Trans R Soc Lond B Biol Sci. 2010;365(1543):1139–1147. doi: 10.1098/rstb.2009.0279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Combes C. Parasitism: The Ecology and Evolution of Intimate Interactions. Univ of Chicago Press; Chicago: 2001. [Google Scholar]
- 13.Poulin R, Krasnov BR, Mouillot D. Host specificity in phylogenetic and geographic space. Trends Parasitol. 2011;27(8):355–361. doi: 10.1016/j.pt.2011.05.003. [DOI] [PubMed] [Google Scholar]
- 14.Apanius V, Yorinks N, Bermingham E, Ricklefs RE. Island and taxon effects in parasitism and resistance of Lesser Antillean birds. Ecology. 2000;81(7):1959–1969. [Google Scholar]
- 15.Fallon SM, Bermingham E, Ricklefs RE. Host specialization and geographic localization of avian malaria parasites: A regional analysis in the Lesser Antilles. Am Nat. 2005;165(4):466–480. doi: 10.1086/428430. [DOI] [PubMed] [Google Scholar]
- 16.Hellgren O, Pérez-Tris J, Bensch S. A jack-of-all-trades and still a master of some: Prevalence and host range in avian malaria and related blood parasites. Ecology. 2009;90(10):2840–2849. doi: 10.1890/08-1059.1. [DOI] [PubMed] [Google Scholar]
- 17.Levin II, et al. Multiple lineages of avian malaria parasites (Plasmodium) in the Galapagos Islands and evidence for arrival via migratory birds. Conserv Biol. 2013;27(6):1366–1377. doi: 10.1111/cobi.12127. [DOI] [PubMed] [Google Scholar]
- 18.Ricklefs RE, Dodge Gray J, Latta SC, Svensson-Coelho M. Distribution anomalies in avian haemosporidian parasites in the southern Lesser Antilles. J Avian Biol. 2011;42(6):570–584. [Google Scholar]
- 19.Loiseau C, et al. Host and habitat specialization of avian malaria in Africa. Mol Ecol. 2012;21(2):431–441. doi: 10.1111/j.1365-294X.2011.05341.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gager AB, Del Rosario Loaiza J, Dearborn DC, Bermingham E. Do mosquitoes filter the access of Plasmodium cytochrome b lineages to an avian host? Mol Ecol. 2008;17(10):2552–2561. doi: 10.1111/j.1365-294X.2008.03764.x. [DOI] [PubMed] [Google Scholar]
- 21.Kimura M, Darbro JM, Harrington LC. Avian malaria parasites share congeneric mosquito vectors. J Parasitol. 2010;96(1):144–151. doi: 10.1645/GE-2060.1. [DOI] [PubMed] [Google Scholar]
- 22.Martínez-de la Puente J, Martínez J, Rivero-de Aguilar J, Herrero J, Merino S. On the specificity of avian blood parasites: Revealing specific and generalist relationships between haemosporidians and biting midges. Mol Ecol. 2011;20(15):3275–3287. doi: 10.1111/j.1365-294X.2011.05136.x. [DOI] [PubMed] [Google Scholar]
- 23.Medeiros MCI, Hamer GL, Ricklefs RE. Host compatibility rather than vector-host-encounter rate determines the host range of avian Plasmodium parasites. Proc Biol Sci. 2013;280(1760):20122947. doi: 10.1098/rspb.2012.2947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fallon SM, Fleischer RC, Graves GR. Malarial parasites as geographical markers in migratory birds? Biol Lett. 2006;2(2):213–216. doi: 10.1098/rsbl.2005.0429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ishtiaq F, et al. Prevalence and evolutionary relationships of haematozoan parasites in native versus introduced populations of common myna Acridotheres tristis. Proc Biol Sci. 2006;273(1586):587–594. doi: 10.1098/rspb.2005.3313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Durrant KL, et al. Parasite assemblages distinguish populations of a migratory passerine on its breeding grounds. J Zool (Lond) 2008;274(4):318–326. [Google Scholar]
- 27.Marzal A, et al. Diversity, loss, and gain of malaria parasites in a globally invasive bird. PLoS One. 2011;6(7):e21905. doi: 10.1371/journal.pone.0021905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sehgal RNM, et al. Spatially explicit predictions of blood parasites in a widely distributed African rainforest bird. Proc Biol Sci. 2011;278(1708):1025–1033. doi: 10.1098/rspb.2010.1720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Scordato ESC, Kardish MR. Prevalence and beta diversity in avian malaria communities: Host species is a better predictor than geography. J Anim Ecol. 2014;83(6):1387–1397. doi: 10.1111/1365-2656.12246. [DOI] [PubMed] [Google Scholar]
- 30.Swanson BL, Lyons AC, Bouzat JL. Distribution, prevalence and host specificity of avian malaria parasites across the breeding range of the migratory lark sparrow (Chondestes grammacus) Genetica. 2014;142(3):235–249. doi: 10.1007/s10709-014-9770-9. [DOI] [PubMed] [Google Scholar]
- 31.Asghar M, et al. Chronic infection. Hidden costs of infection: Chronic malaria accelerates telomere degradation and senescence in wild birds. Science. 2015;347(6220):436–438. doi: 10.1126/science.1261121. [DOI] [PubMed] [Google Scholar]
- 32.Odum EP. Bird populations of the highlands (North Carolina) plateau in relation to plant succession and avian invasion. Ecology. 1950;31(4):587–605. [Google Scholar]
- 33.Bray JR, Curtis JT. An ordination of the upland forest communities of southern Wisconsin. Ecol Monogr. 1957;27(4):325–349. [Google Scholar]
- 34.Legendre P, Legendre L. Numerical Ecology. 2nd English Ed Elsevier Science; Amsterdam: 1998. [Google Scholar]
- 35.Nekola JC, White PS. The distance decay of similarity in biogeography and ecology. J Biogeogr. 1999;26(4):867–878. [Google Scholar]
- 36.Jost L. Entropy and diversity. Oikos. 2006;113(2):363–375. [Google Scholar]
- 37.Rao CR. Diversity and dissimilarity coefficients: A unified approach. Theor Popul Biol. 1982;21(1):24–43. [Google Scholar]
- 38.Medeiros MCI, Ellis VA, Ricklefs RE. Specialized avian Haemosporida trade reduced host breadth for increased prevalence. J Evol Biol. 2014;27(11):2520–2528. doi: 10.1111/jeb.12514. [DOI] [PubMed] [Google Scholar]
- 39.Hartigan JA, Hartigan PM. The dip test of unimodality. Inst Math Stat. 1985;13(1):70–84. [Google Scholar]
- 40.Ricklefs RE, et al. Community relationships of avian malaria parasites in southern Missouri. Ecol Monogr. 2005;75(4):543–559. [Google Scholar]
- 41.Ellis VA, Kunkel MR, Ricklefs RE. The ecology of host immune responses to chronic avian haemosporidian infection. Oecologia. 2014;176(3):729–737. doi: 10.1007/s00442-014-3048-x. [DOI] [PubMed] [Google Scholar]
- 42.Venkatesan M, Rasgon JL. Population genetic data suggest a role for mosquito-mediated dispersal of West Nile virus across the western United States. Mol Ecol. 2010;19(8):1573–1584. doi: 10.1111/j.1365-294X.2010.04577.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hamer GL, et al. Fine-scale variation in vector host use and force of infection drive localized patterns of West Nile virus transmission. PLoS One. 2011;6(8):e23767. doi: 10.1371/journal.pone.0023767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Krasnov BR, Shenbrot GI, Medvedev SG, Vatschenok VS, Khokhlova IS. Host-habitat relations as an important determinant of spatial distribution of flea assemblages (Siphonaptera) on rodents in the Negev Desert. Parasitology. 1997;114(Pt 2):159–173. doi: 10.1017/s0031182096008347. [DOI] [PubMed] [Google Scholar]
- 45.Krasnov BR, Shenbrot GI, Mouillot D, Khokhlova IS, Poulin R. Spatial variation in species diversity and composition of flea assemblages in small mammalian hosts: Geographical distance or faunal similarity? J Biogeogr. 2005;32(4):633–644. [Google Scholar]
- 46.Poulin R. The decay of similarity with geographical distance in parasite communities of vertebrate hosts. J Biogeogr. 2003;30(10):1609–1615. [Google Scholar]
- 47.Bonneaud C, Pérez-Tris J, Federici P, Chastel O, Sorci G. Major histocompatibility alleles associated with local resistance to malaria in a passerine. Evolution. 2006;60(2):383–389. [PubMed] [Google Scholar]
- 48.Anderson RM, May RM. Regulation and stability of host-parasite population interactions: I. Regulatory processes. J Anim Ecol. 1978;47(1):219–247. [Google Scholar]
- 49.Hudson PJ, Dobson AP, Newborn D. Prevention of population cycles by parasite removal. Science. 1998;282(5397):2256–2258. doi: 10.1126/science.282.5397.2256. [DOI] [PubMed] [Google Scholar]
- 50.Longmire JL, Maltbie M, Baker RJ. Use of “‘lysis buffer’” in DNA isolation and its implication for museum collections. Occas Pap Mus Tex Tech Univ. 1997;163:1–4. [Google Scholar]
- 51. Svensson, MLE, Ricklefs RE (2009) Low diversity and high intra-island variation in prevalence of avian Haemoproteus parasites on Barbados, Lesser Antilles. Parasitology 136(10):1121–1131. [DOI] [PubMed]
- 52.Fallon SM, Ricklefs RE, Swanson BL, Bermingham E. Detecting avian malaria: An improved polymerase chain reaction diagnostic. J Parasitol. 2003;89(5):1044–1047. doi: 10.1645/GE-3157. [DOI] [PubMed] [Google Scholar]
- 53.Waldenström J, Bensch S, Hasselquist D, Östman O. A new nested polymerase chain reaction method very efficient in detecting Plasmodium and Haemoproteus infections from avian blood. J Parasitol. 2004;90(1):191–194. doi: 10.1645/GE-3221RN. [DOI] [PubMed] [Google Scholar]
- 54.Fecchio A, Lima MR, Svensson-Coelho M, Marini MÂ, Ricklefs RE. Structure and organization of an avian haemosporidian assemblage in a Neotropical savanna in Brazil. Parasitology. 2013;140(2):181–192. doi: 10.1017/S0031182012001412. [DOI] [PubMed] [Google Scholar]
- 55.Svensson-Coelho M, et al. Diversity, prevalence, and host specificity of avian Plasmodium and Haemoproteus in a Western Amazon assemblage. Ornithol Monogr. 2013;76(1):1–47. [Google Scholar]
- 56.Ricklefs RE, et al. Species formation by host shifting in avian malaria parasites. Proc Natl Acad Sci USA. 2014;111(41):14816–14821. doi: 10.1073/pnas.1416356111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Browning SR, Browning BL. Haplotype phasing: Existing methods and new developments. Nat Rev Genet. 2011;12(10):703–714. doi: 10.1038/nrg3054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.R Core Team 2014 R: A language and environment for statistical computing. Available at R-project.org.
- 59.Oksanen J, et al. 2015 vegan: Community Ecology Package. R package version 2.2-1. Available at: https://cran.r-project.org/web/packages/vegan/index.html.
- 60.Nychka D, Furrer R, Sain S. 2014 fields: Tools for spatial data. R package version 7.1. Available at: https://cran.r-project.org/web/packages/fields/index.html.
- 61.Legendre P, Fortin M-J. Comparison of the Mantel test and alternative approaches for detecting complex multivariate relationships in the spatial analysis of genetic data. Mol Ecol Resour. 2010;10(5):831–844. doi: 10.1111/j.1755-0998.2010.02866.x. [DOI] [PubMed] [Google Scholar]
- 62.Maechler M. 2014 diptest: Hartigan’s dip test statistic for unimodality - corrected code. R package version 0.75-6. Available at: https://cran.r-project.org/web/packages/diptest/index.html.
- 63.Bates D, Maechler M, Bolker B, Walker S. 2014 lme4: Linear mixed-effects models using Eigen and S4. R package version 1.1-7. Available at: https://cran.r-project.org/web/packages/lme4/index.html.
- 64.Kenward MG, Roger JH. Small sample inference for fixed effects from restricted maximum likelihood. Biometrics. 1997;53(3):983–997. [PubMed] [Google Scholar]
- 65.Kuznetsova A, Brockhoff PB, Christensen RHB. 2014 lmerTest: Tests in linear mixed effects models. R package version 2.0-20. Available at: https://cran.r-project.org/web/packages/lmerTest/index.html.
- 66.Halekoh U, Højsgaard S. A Kenward-Roger approximation and parametric bootstrap methods for tests in linear mixed models - the R package pbkrtest. J Stat Softw. 2014;59(9):1–30. [Google Scholar]
- 67.Bart J, Hopschen M, Peterjohn BG. Reliability of the Breeding Bird Survey: Effects of restricting surveys to roads. Auk. 1995;112(3):758–761. [Google Scholar]
- 68.Sauer JR, Peterjohn BG, Link WA. Observer differences in the North American breeding bird survey. Auk. 1994;111(1):50–62. [Google Scholar]
- 69.Kembel SW, et al. Picante: R tools for integrating phylogenies and ecology. Bioinformatics. 2010;26(11):1463–1464. doi: 10.1093/bioinformatics/btq166. [DOI] [PubMed] [Google Scholar]
- 70.Jetz W, Thomas GH, Joy JB, Hartmann K, Mooers AO. The global diversity of birds in space and time. Nature. 2012;491(7424):444–448. doi: 10.1038/nature11631. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.