Abstract
5-Fluorouracil (5-FU) is a key drug for the treatment of esophageal squamous cell carcinoma (ESCC); however, resistance to it remains a critical limitation to its clinical use. To clarify the mechanisms of 5-FU resistance of ESCC, we originally established 5-FU-resistant ESCC cells, TE-5R, by step-wise treatment with continuously increasing concentrations of 5-FU. The half maximal inhibitory concentration of 5-FU showed that TE-5R cells were 15.6-fold more resistant to 5-FU in comparison with parental TE-5 cells. TE-5R cells showed regional copy number amplification of chromosome 1p including the DPYD gene, as well as high mRNA and protein expressions of dihydropyrimidine dehydrogenase (DPD), an enzyme involved in 5-FU degradation. 5-FU treatment resulted in a significant decrease of the intracellular 5-FU concentration and increase of the concentration of α-fluoro-ureidopropionic acid (FUPA), a metabolite of 5-FU, in TE-5R compared with TE-5 cells in vitro. Conversely, gimeracil, a DPD inhibitor, markedly increased the intracellular 5-FU concentration, decreased the intracellular FUPA concentration, and attenuated 5-FU resistance of TE-5R cells. These results indicate that 5-FU resistance of TE-5R cells is due to the rapid degradation of 5-FU by DPD overexpression. The investigation of 5-FU-resistant ESCC with DPYD gene copy number amplification and consequent DPD overexpression may generate novel biological evidence to explore strategies against ESCC with 5-FU resistance.
Keywords: Esophageal squamous cell carcinoma, chemotherapy, 5-fluorouracil, drug resistance, dihydropyrimidine dehydrogenase
Introduction
Esophageal squamous cell carcinoma (ESCC) is one of the deadliest cancers worldwide [1-3]; the overall 5-year survival rate of patients with ESCC still ranges from 15 to 25% [3,4]. Chemotherapy is one of the main therapeutic strategies for advanced or metastatic ESCC [5-7]; however, the efficacy of chemotherapy is limited, with only a 6.7-8.9-month median OS [8-10]. To overcome these issues, the mechanism of chemo-resistance of ESCC should be clarified.
Genomic DNA copy number alterations are frequently found in solid tumors, and they are utilized as diagnostic markers as well as indicators of the prognosis and drug efficacy [11]. Recently, genome-wide array comparative genomic hybridization (aCGH) has facilitated the analysis of DNA copy number alterations at high resolution [12], and thereby several DNA copy number alterations, such as EIF2B5 (chromosome 3q), MYC (8q), CCND1 (11q), and PARP2 (14q), have been detected in patients with ESCC [13,14]; however, it has not been elucidated whether such alterations are involved in the drug resistance of ESCC.
5-Fluorouracil (5-FU) is a key drug in first-line therapy against ESCC [15]. 5-FU metabolism comprises anabolic and catabolic processes [16]. To exert its cytotoxicity, 5-FU enters an anabolic process in which it disrupts nucleic acids through various enzymes, such as thymidylate synthase (TS) [16]. On the other hand, 5-FU is degraded by dihydropyrimidine dehydrogenase (DPD), the initial and rate-limiting enzyme encoded by the DPYD gene located on the short arm of chromosome 1 (1p21.3) [16,17], to its metabolites including α-fluoro-ureidopropionic acid (FUPA). As DPD expression in tumors increases 5-FU resistance [18-22] and exacerbates the prognosis of patients treated with 5-FU [23,24], DPD is a critical mediator influencing the 5-FU resistance of cancers [25-27].
In this study, we established novel 5-FU-resistant ESCC cells, TE-5R, derived from parental TE-5 cells, and investigated the mechanisms of 5-FU resistance in these cells. We revealed that high DPD expression due to DPYD gene copy number amplification is involved in TE-5R cells acquiring 5-FU resistance.
Materials and methods
Establishment of 5-FU-resistant ESCC cells
Human ESCC cells, TE-5, were obtained from Riken BioResource Center (Ibaragi, Japan) [28]. The cells were cultured in RPMI1640 medium (Life Technologies Corp., Grand Island, NY, USA), supplemented with 10% fetal bovine serum (Life Technologies Corp.), 100 μg/mL of streptomycin, and 100 units/mL of penicillin (Life Technologies Corp.) at 37ºC in a 5% CO2 incubator. TE-5 cells were treated with continuous and step-wise concentrations of 5-FU (1, 2, 5, and 10 μM), based on previous reports [29,30]. Consequently 5-FU-resistant ESCC cells were established, and named TE-5R cells. Both TE-5 and TE-5R cells have been verified by short tandem repeat analysis and confirmed to be consistent with each other and with the original cell source from Riken BioResource Center.
WST-1 cell proliferation assays
5-FU resistance of TE-5R cells was assessed by the WST-1 assay. TE-5 and TE-5R cells (5 × 103 cells) were seeded in 96-well plates, and treated with the indicated concentrations of 5-FU for 72 h. Cell viability was measured with Cell Proliferation Reagent WST-1 (Roche Applied Science, Upper Bavaria, Germany) following the manufacturer’s instructions. All data were obtained in sextuplicate. The half maximal inhibitory concentration (IC50) of 5-FU in each cell was calculated by probit analysis [31], and the resistance ratio was determined by comparing to the IC50 values of parental cells.
Genomic DNA preparation, array-comparative genomic hybridization (aCGH) experiments, and copy number assays
To compare the genomic alterations between TE-5 and TE-5R cells, we performed aCGH analysis. Genomic DNA was extracted from TE-5 and TE-5R cells using AllPrep DNA/RNA/Protein Mini Kit (QIAGEN GmbH, Hilden, Germany) according to the manufacturer’s instructions. aCGH experiments were performed with SurePrint G3 Human CGH Microarray Kit 2 × 400K (G4448A, Agilent Technologies, Santa Clara, CA, USA) according to the manufacturer’s protocol. Raw aCGH data were analyzed and processed using CytoGenomics 3.0 software (Agilent Technologies). The aCGH data set is available at Gene Expression Omnibus (GEO; http://www.ncbi.nlm.nih.gov/geo/) under accession number GSE69494.
To determine the copy number, quantitative real-time PCR was performed with a PRISM 7900 sequence detection system (Applied Biosystems, Foster City, CA, USA) using a TaqMan Gene Expression Master Mix (Applied Biosystems) and TaqMan Copy Number Assays: LPHN2 (Hs00381445_cn), DPYD (Hs02103805_cn), and PALMD (Hs01617339_cn). Ribonuclease P RNA component H1 (RPPH1), and Human Genomic DNA (Cat No. G3041, Promega Inc.) were used as the endogenous control. The TaqMan copy number assay contained 1 µL of LPHN2, DPYD, and PALMD probes (20 ×, FAM-labeled), 1 µL of RPPH1 probe mix (20 ×, VIC-labeled), 10 µL of TaqMan Gene Expression Master Mix (2 ×), 4 µL of genomic DNA, and 4 µL of water. PCR cycling conditions were 95°C for 10 minutes, followed by 2-step cycling: 40 cycles of 95°C for 15 seconds and 60°C for 1 minute. A manual cycle threshold of 0.2 and an automatic baseline were used to detect the template quantity of target genes and RPPH1 gene by sequence detection system software (ABI, version 2.4). The target probes and endogenous control were loaded in the same well, and each reaction was performed in triplicate. CopyCaller software (ABI, version 2.0) was used to calculate the copy number of each probe based on real-time PCR data. Copy numbers of each target gene were calculated as the average of those based on two reference genes.
RNA isolation, cDNA synthesis, and real-time reverse transcriptase-PCR (RT-PCR)
RNA extraction and cDNA synthesis were conducted as previously described [32,33]. Real-time reverse transcriptase polymerase chain reaction (RT-PCR) was performed with LightCycler 480 Instrument II Real-Time PCR System (Roche Diagnostics Ltd., Rotkreuz, Switzerland). The relative level of each mRNA was normalized to ACTB as an internal control. The primers used in this study were as follows: DPYD (Hs_DPYD_1_SG, QuantiTect Primer Assay, QIAGEN GmbH) and ACTB (Hs_ACTB_1_SG, QuantiTect Primer Assay).
Protein extraction and Western blotting
Whole-cell lysates were collected according to prior reports [34-36], and Western blotting was performed as described previously [37]. Primary antibodies and their titers were as follows: rabbit monoclonal anti-DPD antibody (D35A8, #4654, Cell Signaling Technology, Inc., Danvers, MA, USA; 1:1,000) and rabbit monoclonal anti-β-actin antibody (13E5, #4970, Cell Signaling Technology, Inc.; 1:2,000). β-Actin served as a loading control for whole-cell lysates. The band intensity was quantified using Image Lab 4.1 software (Bio-Rad Laboratories, Hercules, CA, USA).
Measurement of intracellular concentrations of 5-FU and its metabolite
The intracellular concentrations of 5-FU and FUPA, a metabolite of 5-FU, were quantified by liquid chromatography-tandem mass spectrometry (LC-MS/MS) [38]. TE-5 and TE-5R cells (5 × 105 cells) were seeded in 6-well plates with 2 mL of RPMI1640 medium supplemented with 10% fetal bovine serum (Life Technologies Corp.), and treated with 5-FU (10 μM) for 24 h. Cells were harvested and homogenized in methanol (300 µL). The samples were centrifuged and supernatants were filtrated through a filter (SGJVL; 0.45 µm; Millipore, Billerica, MA, USA) and injected into the LC-MS/MS system. The LC-MS/MS assay was performed using a liquid chromatography system consisting of a Prominence series chromatograph (Shimadzu, Kyoto, Japan) coupled to an API 4000 triple-quadrapole tandem mass spectrometer (AB SCIEX, Foster City, CA, USA). Chromatographic separation was carried out on COSMOSIL® 5C18-MS-II (Nacalai Tesque, Kyoto, Japan). The MS/MS analysis was performed using an elector-spray ionization source in the negative mode. Detection was carried out by monitoring the ion transition of 5-FU from m/z 129.0 to 42.0 and FUPA from m/z 149.0 to 106.0, respectively [38].
DPD inhibitor treatment
A DPD inhibitor, 5-chloro-2,4-pyridinediol (gimeracil), was synthesized by Taiho Pharmaceutical Co., Ltd. (Tokyo, Japan). TE-5 and TE-5R cells (5 × 103 cells) were plated in a 96-well plate and cultured for 24 h, and then they were treated with the indicated concentrations of 5-FU in the presence of gimeracil for 72 h at a molar ratio of 1:0.2 (5-FU:gimeracil), referring to a previous report [39]. The effect of gimeracil was assessed by the WST-1 assay and measuring intracellular concentrations of 5-FU and its metabolite, as mentioned above.
Statistical analyses
All data were analyzed by the 2-tailed Student’s t-test, and are presented as the mean ± standard deviation (SD). P < 0.05 was considered significant. All statistical analyses were performed with SPSS 19 for Windows software (SPSS Inc., Chicago, IL, USA).
Results
Establishment of 5-FU-resistant ESCC TE-5R cells
TE-5R cells could grow exponentially in the presence of 5-FU (10 µM), although TE-5 cells could not grow under the same condition (data not shown). Dose-response curves indicated that the IC50 values of TE-5 and TE-5R cells were 3.6 ± 1.1 and 55.5 ± 10.1 μM, respectively (Figure 1), and TE-5R cells had a significantly high IC50 value (P < 0.001, vs TE-5 cells). Accordingly, TE-5R cells were 15.6-fold more resistant to 5-FU in comparison with parental TE-5 cells. Cross-resistance (e.g., cisplatin and docetaxel) was not noted in TE-5R cells (data not shown). The 5-FU resistance of TE-5R cells was not diminished even though they were cultured in a 5-FU-free medium for eight weeks (data not shown), and so TE-5R cells were incubated with 5-FU-free medium for at least one week prior to use in the subsequent experiments.
Figure 1.
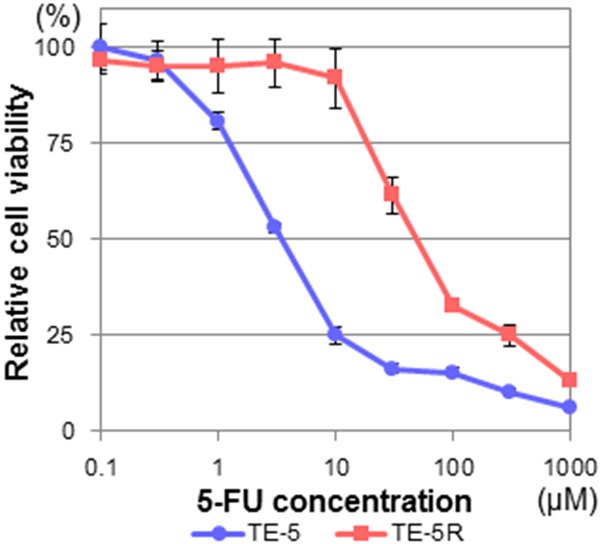
5-FU resistance of TE-5R cells. TE-5 and TE-5R cells were treated with the indicated concentrations of 5-FU for 72 h, and cell viability was assessed using the WST-1 assay. A viability of 100% was defined as the amount of absorption at 450 nm in untreated cells. The mean value ± S.D. of six replicate wells from a representative experiment is shown. Each experiment was repeated at least three times, and consistent results were obtained. IC50 values of TE-5 and TE-5R cells were 3.6 ± 1.1 and 55.5 ± 10.1 μM, respectively. Note that TE-5R cells were 15.6-fold more resistant to 5-FU in comparison with parental TE-5 cells.
Regional chromosome 1p amplification and DPD mRNA/protein overexpression in TE-5R cells
To compare the fundamental gene alterations between TE-5 and TE-5R cells, we performed aCGH analysis, and found that TE-5R cells acquired regional gene amplification in the short arm of chromosome 1 (1p) including the DPYD gene (Figure 2A). Quantitative PCR analysis revealed that DNA copies of the DPYD gene, which were located within chromosome 1p (Figure 2A), were highly amplified in TE-5R cells, but not the LPHN2 or PALMD gene (Figure 2B), located outside the specific amplified region (Figure 2A). Moreover, DPD mRNA as well as protein expression levels in TE-5R cells were also much higher than those in TE-5 cells (Figure 2C, 2D).
Figure 2.
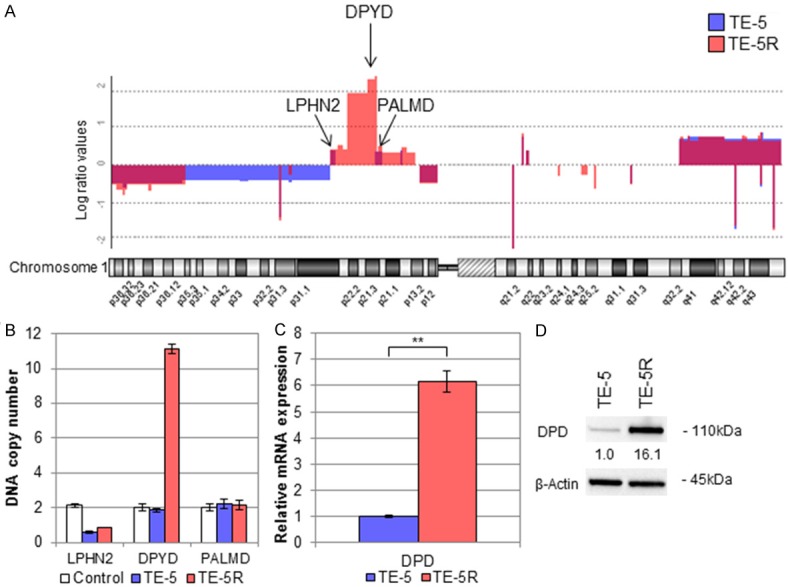
Amplification of DPYD gene and subsequent DPD expression in TE-5R cells. A. Chromosome 1 copy number alteration in aCGH analysis. The vertical axis indicates the log (base 2) ratio of DNA expression. TE-5R harbored specific regional amplification of the short arm of chromosome 1 around 1p2 including the DPYD gene (1p21.3). Note that LPHN2 and PALMD genes are not located in the specific amplified regions. B. qPCR analysis using LPHN2, DPYD and PALMD gene probes. The DPYD gene is amplified to produce 10 copies in DNA samples derived from TE-5R cells, but not non-cancerous human genomic DNA or parental TE-5 cells. On the other hand, LPHN2 and PALMD genes are not amplified in any samples. C. DPD mRNA expression levels in TE-5 and TE-5R cells. qPCR revealed significantly increased DPD mRNA expression in TE-5R compared with TE-5 cells. **P < 0.01. D. DPD protein expression levels in TE-5 and TE-5R cells. The relative density was calculated by densitometry. β-Actin served as a loading control. The DPD protein expression level in TE-5R cells was higher than in TE-5 cells.
Intracellular concentrations of 5-FU and its metabolite in TE-5 and TE-5R cells
To address the functional role of DPD overexpression in TE-5R cells, we measured intracellular concentrations of 5-FU and FUPA in TE-5 and TE-5R cells treated with 5-FU (10 µM) for 24 h. The intracellular 5-FU concentration in TE-5R cells (5.4 ± 0.3 pmol/106 cells) was markedly lower than in TE-5 cells (96.2 ± 3.3 pmol/106 cells; P < 0.001, Figure 3A). Conversely, FUPA concentrations in TE-5R cells (5.9 ± 1.6 pmol/106 cells) were significantly higher than in TE-5 cells (2.1 ± 0.2 pmol/106 cells, P = 0.014, Figure 3B).
Figure 3.
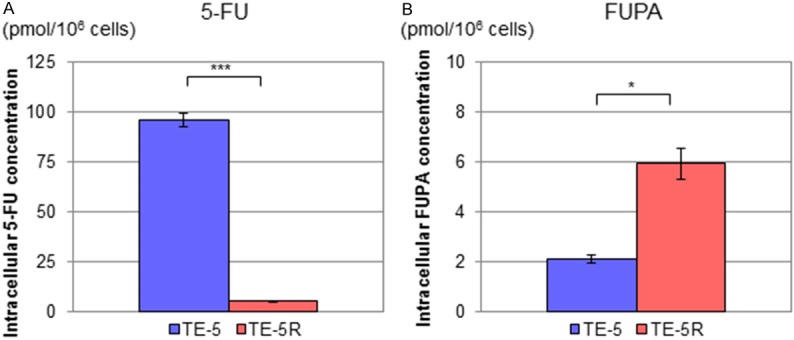
Intracellular 5-FU and FUPA concentrations in TE-5 and TE-5R cells treated with 5-FU. TE-5 and TE-5R cells (5 × 105 cells) were seeded in 6-well plates, and then treated with 5-FU (10 μM) for 24 h. Cells were harvested, and levels of 5-FU and the FUPA concentration were measured with liquid chromatography-tandem mass spectrometry. A. Intracellular 5-FU concentrations in TE-5 and TE-5R cells treated with 5-FU. Intracellular 5-FU concentrations in TE-5R cells were significantly lower than those in TE-5 cells (n = 3). ***P < 0.001. B. Intracellular FUPA concentrations in TE-5 and TE-5R cells treated with 5-FU. Intracellular FUPA concentrations in TE-5R cells were significantly higher than in TE-5 cells (n = 3). *P < 0.05.
Effects of a DPD inhibitor on 5-FU resistance of TE-5R cells
Furthermore, we assessed the effects of a DPD inhibitor, gimeracil, on 5-FU degradation in TE-5R cells. As shown in Figure 4, gimeracil markedly increased the intracellular 5-FU concentration in TE-5R cells from 5.4 ± 0.3 to 62.0 ± 0.3 pmol/106 cells (Figure 4A, P < 0.001) and decreased the FUPA concentration from 5.9 ± 1.6 to 0.6 ± 0.6 pmol/106 cells (Figure 4B, P = 0.006). Thus, gimeracil significantly reduced 5-FU degradation in TE-5R cells.
Figure 4.
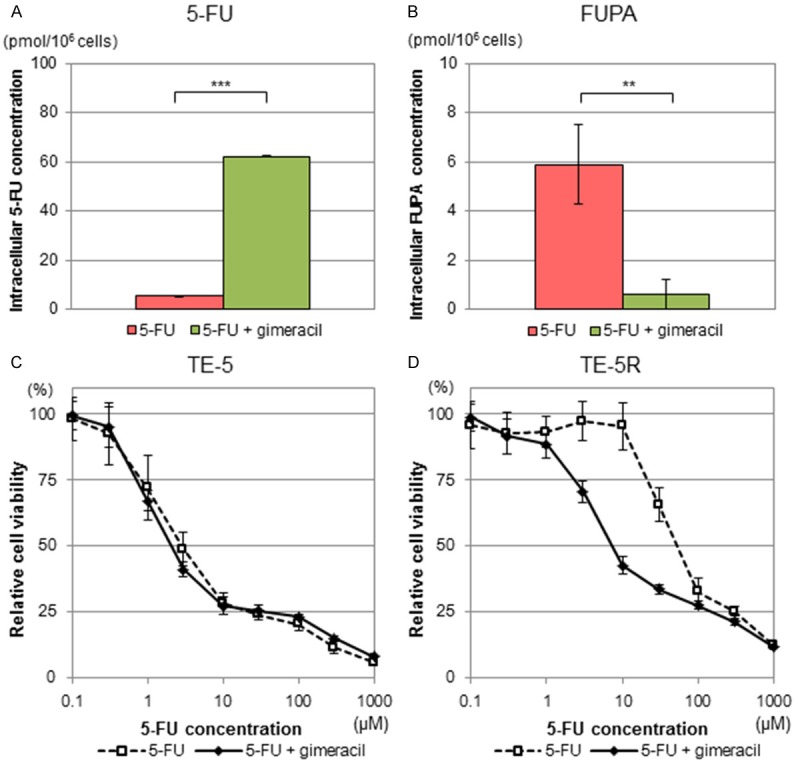
Effects of DPD inhibitor on TE-5R cells. A DPD inhibitor, gimeracil, was added to the culture medium with the indicated concentrations of 5-FU at a molar ratio of 1:0.2 (5-FU:gimeracil) for 24 h. (i.e., 2 μM gimeracil to 10 μM 5-FU). (A) Intracellular 5-FU concentrations in TE-5R cells in the presence or absence of gimeracil. Intracellular 5-FU concentrations in TE-5R cells treated with 5-FU and gimeracil were markedly higher than those treated with 5-FU alone. (n = 3). ***P < 0.001. (B) Intracellular FUPA concentrations in TE-5R cells in the presence or absence of gimeracil. Intracellular FUPA concentrations in TE-5R cells treated with 5-FU and gimeracil were significantly lower than those treated with 5-FU alone. (n = 3). **P < 0.01. (C, D) Effects of gimeracil on 5-FU sensitivity of TE-5 (C) and TE-5R (D) cells. Gimeracil was added to the culture medium with the indicated concentrations of 5-FU at a molar ratio of 1:0.2 (5-FU:gimeracil) for 72 h. Cell viability was assessed by the WST-1 assay. A viability of 100% was defined as the amount of absorption at 450 nm in untreated cells. Each point represents the mean ± S.D. of sextuplicate wells. Note that gimeracil significantly attenuated 5-FU resistance in TE-5R but not TE-5 cells. Representative data from three independent experiments are shown.
Next, we investigated the effects of gimeracil on 5-FU resistance in TE-5R cells. IC50 values with and without gimeracil in TE-5 cells were 4.9 ± 0.9 and 4.8 ± 0.9 μM, respectively (Figure 4C, P = 0.881), and those in TE-5R cells were 20.8 ± 9.2 and 59.0 ± 14.3 μM, respectively (Figure 4D, P = 0.023). Thus, gimeracil significantly reduced the 5-FU resistance of TE-5R cells, but did not affect the 5-FU sensitivity of TE-5 cells.
Discussion
In this study, we established novel 5-FU-resistant ESCC cells, TE-5R, by exposing parental TE-5 cells step-wisely to continuously increasing concentrations of 5-FU. TE-5R showed higher DPD expressions and a more rapid degradation of intracellular 5-FU than TE-5 cells. Furthermore, the DPD inhibitor, gimeracil, rapidly elevated the intracellular 5-FU concentration in TE-5R cells treated with 5-FU, and consequently reduced 5-FU resistance. These results indicate that the 5-FU resistance of TE-5R cells is based on DPD-dependent metabolism. To the best of our knowledge, this is the first report on elucidating the mechanism of 5-FU resistance in ESCC cells.
Our study showed that DPD played a key role in the acquisition of 5-FU resistance by TE-5R cells. DPD is produced in various tumors, including liver [40], breast [24], colorectal [41], and oral [42] cancers; however, the mechanisms of DPD production in cancer cells have not been fully clarified. In the present study, aCGH analysis revealed the regional gene amplification of chromosome 1p including the DPYD gene in TE-5R cells. Consistently, expression levels of DPD mRNA as well as protein were increased comparably to DPD copy number amplification. These findings support the importance of DPD in 5-FU resistance. As DPD expression was not enhanced by short-term (one week) treatment with 5-FU (data not shown), we suggest that long-term 5-FU exposure may lead to the selection of a unique subset of TE-5 cells expressing high levels of DPD.
This was an in vitro study of 5-FU resistance with DPD copy number amplification in ESCC cells. DNA is more stable than mRNA or protein expression [43], and so analysis of copy number alterations, such as of the DPYD gene, might be useful for predicting 5-FU resistance in ESCC patients. We should further examine whether DPD expression is increased by 5-FU treatment in clinical samples.
Consistent with the result that DPD is highly expressed in TE-5R cells, we demonstrated a lower intracellular 5-FU concentration, higher FUPA concentration, and weaker 5-FU cytotoxicity in TE-5R than TE-5 cells. These data suggest that high DPD expression contributes to the degradation of 5-FU in TE-5R cells, which consequently decreases the intracellular 5-FU concentration as well as 5-FU cytotoxicity. Furthermore, we showed that the DPD inhibitor, gimeracil, elevated intracellular 5-FU levels in TE-5R cells treated with 5-FU and attenuated 5-FU resistance. As gimeracil was effective for high DPD-overexpressing ESCC cells such as TE-5R cells, but not low or non-DPD-expressing ESCC cells (data not shown), it is considered to be specifically efficient for the high DPD-expressing ESCC cells. Gimeracil has already been integrated into S-1, a combination drug containing tegafur (5-FU prodrug), potassium oxonate, and gimeracil, and clinically utilized for several cancers, such as gastric and colon cancers [44]. Therefore, we suggest that the therapeutic utility of S-1 should be verified for ESCC patients with high DPD expression.
In the present study, the growth of TE-5R cells was not accelerated in comparison with that of TE-5 cells, and, moreover, TE-5R cells did not form xenografted tumors in nude mice (data not shown). Thus, high DPD expression did not influence cell growth or tumorigenicity, suggesting that DPD overexpression may not affect the biological characteristics of cancer cells directly. Furthermore, we analyzed other possible underlying mechanisms of 5-FU resistance, such as the modulation of 5-FU metabolism-related proteins [45], MDR1 overexpression [46], epithelial-mesenchymal transition [30], or deregulation of apoptosis [47]; however, none of them were associated with the 5-FU resistance of TE-5R cells (data not shown). As other anti-cancer drugs such as cisplatin and docetaxel were equally cytotoxic to TE-5R cells, the drug resistance of these cells might be limited to 5-FU.
A limitation of this study was that we established only a single 5-FU-resistant cell line; however, further examination of this cell line will contribute to the development of a novel therapeutic strategy against 5-FU-resistant ESCC.
Overall, we established unique 5-FU-resistant ESCC cells with DPD overexpression. Our findings provide not only useful knowledge to explore the mechanism of 5-FU resistance, but also suggest the possibility of combination therapy with gimeracil targeting 5-FU-resistant refractory ESCC.
Acknowledgements
The authors are grateful for the technical assistance of the Medical Research Support Center, Kyoto University Graduate School of Medicine, regarding the use of laboratory equipment. This work was supported by JSPS KAKENHI Grant Numbers 25460926 and 262741, in part by NIH Grants P30-DK050306, P01CA098101 and K26 RR032714, and in part by TAIHO Pharmaceutical Co., Ltd.
Disclosure of conflict of interest
None.
Abbreviations
- ESCC
esophageal squamous cell carcinoma
- 5-FU
5-fluorouracil
- TS
thymidylate synthase
- DPD
dihydropyrimidine dehydrogenase
- FUPA
α-fluoro-ureidopropionic acid
- IC50
half maximal inhibitory concentration
- RT-PCR
reverse transcriptase-polymerase chain reaction
- aCGH
array comparative genomic hybridization
References
- 1.Napier KJ, Scheerer M, Misra S. Esophageal cancer: A Review of epidemiology, pathogenesis, staging workup and treatment modalities. World J Gastrointest Oncol. 2014;6:112–120. doi: 10.4251/wjgo.v6.i5.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pennathur A, Gibson MK, Jobe BA, Luketich JD. Oesophageal carcinoma. Lancet. 2013;381:400–412. doi: 10.1016/S0140-6736(12)60643-6. [DOI] [PubMed] [Google Scholar]
- 3.Enzinger PC, Mayer RJ. Esophageal cancer. N Engl J Med. 2003;349:2241–2252. doi: 10.1056/NEJMra035010. [DOI] [PubMed] [Google Scholar]
- 4.Pennathur A, Farkas A, Krasinskas AM, Ferson PF, Gooding WE, Gibson MK, Schuchert MJ, Landreneau RJ, Luketich JD. Esophagectomy for T1 esophageal cancer: outcomes in 100 patients and implications for endoscopic therapy. Ann Thorac Surg. 2009;87:1048–1054. doi: 10.1016/j.athoracsur.2008.12.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ancona E, Ruol A, Santi S, Merigliano S, Sileni VC, Koussis H, Zaninotto G, Bonavina L, Peracchia A. Only pathologic complete response to neoadjuvant chemotherapy improves significantly the long term survival of patients with resectable esophageal squamous cell carcinoma: final report of a randomized, controlled trial of preoperative chemotherapy versus surgery alone. Cancer. 2001;91:2165–2174. [PubMed] [Google Scholar]
- 6.Law S, Fok M, Chow S, Chu KM, Wong J. Preoperative chemotherapy versus surgical therapy alone for squamous cell carcinoma of the esophagus: a prospective randomized trial. J Thorac Cardiovasc Surg. 1997;114:210–217. doi: 10.1016/S0022-5223(97)70147-8. [DOI] [PubMed] [Google Scholar]
- 7.National Comprehensive Cancer Network (NCCN) Clinical Practice Guidelines in Oncology: Esophageal and Esophagogastric Junction Cancers Version.3.2015. J Natl Compr Canc Netw. 2015;13:194–227. doi: 10.6004/jnccn.2015.0028. [DOI] [PubMed] [Google Scholar]
- 8.Iizuka T, Kakegawa T, Ide H, Ando N, Watanabe H, Tanaka O, Takagi I, Isono K, Ishida K, Arimori M, Endo M, Fukushima M. Phase II evaluation of cisplatin and 5-fluorouracil in advanced squamous cell carcinoma of the esophagus: a Japanese Esophageal Oncology Group Trial. Jpn J Clin Oncol. 1992;22:172–176. [PubMed] [Google Scholar]
- 9.Kato K, Muro K, Ando N, Nishimaki T, Ohtsu A, Aogi K, Aoyama N, Nagai K, Kato H. A phase II study of nedaplatin and 5-fluorouracil in metastatic squamous cell carcinoma of the esophagus: The Japan Clinical Oncology Group (JCOG) Trial (JCOG 9905-DI) Esophagus. 2014;11:183–188. [Google Scholar]
- 10.Hayashi K, Ando N, Watanabe H, Ide H, Nagai K, Aoyama N, Takiyama W, Ishida K, Isono K, Makuuchi H, Imamura M, Shinoda M, Ikeuchi S, Kabuto T, Yamana H, Fukuda H. Phase II evaluation of protracted infusion of cisplatin and 5-fluorouracil in advanced squamous cell carcinoma of the esophagus: a Japan Esophageal Oncology Group (JEOG) Trial (JCOG9407) Jpn J Clin Oncol. 2001;31:419–423. doi: 10.1093/jjco/hye090. [DOI] [PubMed] [Google Scholar]
- 11.Albertson DG. Gene amplification in cancer. Trends Genet. 2006;22:447–455. doi: 10.1016/j.tig.2006.06.007. [DOI] [PubMed] [Google Scholar]
- 12.Michels E, De Preter K, Van Roy N, Speleman F. Detection of DNA copy number alterations in cancer by array comparative genomic hybridization. Genet Med. 2007;9:574–584. doi: 10.1097/gim.0b013e318145b25b. [DOI] [PubMed] [Google Scholar]
- 13.Shi ZZ, Shang L, Jiang YY, Hao JJ, Zhang Y, Zhang TT, Lin DC, Liu SG, Wang BS, Gong T, Zhan QM, Wang MR. Consistent and differential genetic aberrations between esophageal dysplasia and squamous cell carcinoma detected by array comparative genomic hybridization. Clin Cancer Res. 2013;19:5867–5878. doi: 10.1158/1078-0432.CCR-12-3753. [DOI] [PubMed] [Google Scholar]
- 14.Shi ZZ, Liang JW, Zhan T, Wang BS, Lin DC, Liu SG, Hao JJ, Yang H, Zhang Y, Zhan QM, Zhang KT, Wang MR. Genomic alterations with impact on survival in esophageal squamous cell carcinoma identified by array comparative genomic hybridization. Genes Chromosomes Cancer. 2011;50:518–526. doi: 10.1002/gcc.20875. [DOI] [PubMed] [Google Scholar]
- 15.Kuwano H, Nishimura Y, Ohtsu A, Kato H, Kitagawa Y, Tamai S, Toh Y, Matsubara H. Guidelines for diagnosis and treatment of carcinoma of the esophagus. Esophagus. 2008;5:117–132. [Google Scholar]
- 16.Rich TA, Shepard RC, Mosley ST. Four decades of continuing innovation with fluorouracil: current and future approaches to fluorouracil chemoradiation therapy. J. Clin. Oncol. 2004;22:2214–2232. doi: 10.1200/JCO.2004.08.009. [DOI] [PubMed] [Google Scholar]
- 17.Milano G, McLeod HL. Can dihydropyrimidine dehydrogenase impact 5-fluorouracilbased treatment? Eur J Cancer. 2000;36:37–42. doi: 10.1016/s0959-8049(99)00211-7. [DOI] [PubMed] [Google Scholar]
- 18.Diasio RB, Johnson MR. Dihydropyrimidine dehydrogenase: its role in 5-fluorouracil clinical toxicity and tumor resistance. Clin Cancer Res. 1999;5:2672–2673. [PubMed] [Google Scholar]
- 19.Jiang W, Lu Z, He Y, Diasio RB. Dihydropyrimidine dehydrogenase activity in hepatocellular carcinoma: implication in 5-fluorouracil-based chemotherapy. Clin Cancer Res. 1997;3:395–399. [PubMed] [Google Scholar]
- 20.Etienne MC, Cheradame S, Fischel JL, Formento P, Dassonville O, Renee N, Schneider M, Thyss A, Demard F, Milano G. Response to fluorouracil therapy in cancer patients: the role of tumoral dihydropyrimidine dehydrogenase activity. J. Clin. Oncol. 1995;13:1663–1670. doi: 10.1200/JCO.1995.13.7.1663. [DOI] [PubMed] [Google Scholar]
- 21.Beck A, Etienne MC, Cheradame S, Fischel JL, Formento P, Renee N, Milano G. A role for dihydropyrimidine dehydrogenase and thymidylate synthase in tumour sensitivity to fluorouracil. Eur J Cancer. 1994;30A:1517–1522. doi: 10.1016/0959-8049(94)00216-r. [DOI] [PubMed] [Google Scholar]
- 22.Ishikawa Y, Kubota T, Otani Y, Watanabe M, Teramoto T, Kumai K, Takechi T, Okabe H, Fukushima M, Kitajima M. Dihydropyrimidine dehydrogenase and messenger RNA levels in gastric cancer: possible predictor for sensitivity to 5-fluorouracil. Jpn J Cancer Res. 2000;91:105–112. doi: 10.1111/j.1349-7006.2000.tb00866.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tokunaga Y, Sasaki H, Saito T. Clinical role of orotate phosphoribosyl transferase and dihydropyrimidine dehydrogenase in colorectal cancer treated with postoperative fluoropyrimidine. Surgery. 2007;141:346–353. doi: 10.1016/j.surg.2006.06.025. [DOI] [PubMed] [Google Scholar]
- 24.Horiguchi J, Takei H, Koibuchi Y, Iijima K, Ninomiya J, Uchida K, Ochiai R, Yoshida M, Yokoe T, Iino Y, Morishita Y. Prognostic significance of dihydropyrimidine dehydrogenase expression in breast cancer. Br J Cancer. 2002;86:222–225. doi: 10.1038/sj.bjc.6600040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kimura M, Kuwabara Y, Mitsui A, Ishiguro H, Sugito N, Tanaka T, Shiozaki M, Naganawa Y, Takeyama H. Thymidylate synthetase and dihydropyrimidine dehydrogenase mRNA levels in esophageal cancer. Oncol Lett. 2011;2:297–301. doi: 10.3892/ol.2010.227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ando T, Ishiguro H, Kuwabara Y, Kimura M, Mitsui A, Sugito N, Mori R, Ogawa R, Katada T, Fujii Y. Relationship between expression of 5-fluorouracil metabolic enzymes and 5-fluorouracil sensitivity in esophageal carcinoma cell lines. Dis Esophagus. 2008;21:15–20. doi: 10.1111/j.1442-2050.2007.00700.x. [DOI] [PubMed] [Google Scholar]
- 27.Kobunai T, Ooyama A, Sasaki S, Wierzba K, Takechi T, Fukushima M, Watanabe T, Nagawa H. Changes to the dihydropyrimidine dehydrogenase gene copy number influence the susceptibility of cancers to 5-FU-based drugs: Data mining of the NCI-DTP data sets and validation with human tumour xenografts. Eur J Cancer. 2007;43:791–798. doi: 10.1016/j.ejca.2006.09.026. [DOI] [PubMed] [Google Scholar]
- 28.Nishihira T, Hashimoto Y, Katayama M, Mori S, Kuroki T. Molecular and cellular features of esophageal cancer cells. J Cancer Res Clin Oncol. 1993;119:441–449. doi: 10.1007/BF01215923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chung YM, Park S, Park JK, Kim Y, Kang Y, Yoo YD. Establishment and characterization of 5-fluorouracil-resistant gastric cancer cells. Cancer Lett. 2000;159:95–101. doi: 10.1016/s0304-3835(00)00535-8. [DOI] [PubMed] [Google Scholar]
- 30.Uchibori K, Kasamatsu A, Sunaga M, Yokota S, Sakurada T, Kobayashi E, Yoshikawa M, Uzawa K, Ueda S, Tanzawa H, Sato N. Establishment and characterization of two 5-fluorouracil-resistant hepatocellular carcinoma cell lines. Int J Oncol. 2012;40:1005–1010. doi: 10.3892/ijo.2011.1300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Finney DJ. The adjustment for a natural response rate in probit analysis. Ann Appl Biol. 1949;36:187–195. doi: 10.1111/j.1744-7348.1949.tb06408.x. [DOI] [PubMed] [Google Scholar]
- 32.Ohashi S, Natsuizaka M, Yashiro-Ohtani Y, Kalman RA, Nakagawa M, Wu L, Klein-Szanto AJ, Herlyn M, Diehl JA, Katz JP, Pear WS, Seykora JT, Nakagawa H. NOTCH1 and NOTCH3 coordinate esophageal squamous differentiation through a CSL-dependent transcriptional network. Gastroenterology. 2010;139:2113–2123. doi: 10.1053/j.gastro.2010.08.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ohashi S, Natsuizaka M, Naganuma S, Kagawa S, Kimura S, Itoh H, Kalman RA, Nakagawa M, Darling DS, Basu D, Gimotty PA, Klein-Szanto AJ, Diehl JA, Herlyn M, Nakagawa H. A NOTCH3-mediated squamous cell differentiation program limits expansion of EMTcompetent cells that express the ZEB transcription factors. Cancer Res. 2011;71:6836–6847. doi: 10.1158/0008-5472.CAN-11-0846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Takaoka M, Harada H, Deramaudt TB, Oyama K, Andl CD, Johnstone CN, Rhoades B, Enders GH, Opitz OG, Nakagawa H. Ha-Ras(G12V) induces senescence in primary and immortalized human esophageal keratinocytes with p53 dysfunction. Oncogene. 2004;23:6760–6768. doi: 10.1038/sj.onc.1207923. [DOI] [PubMed] [Google Scholar]
- 35.Okawa T, Michaylira CZ, Kalabis J, Stairs DB, Nakagawa H, Andl CD, Johnstone CN, Klein-Szanto AJ, El-Deiry WS, Cukierman E, Herlyn M, Rustgi AK. The functional interplay between EGFR overexpression, hTERT activation, and p53 mutation in esophageal epithelial cells with activation of stromal fibroblasts induces tumor development, invasion, and differentiation. Genes Dev. 2007;21:2788–2803. doi: 10.1101/gad.1544507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Akakura N, Kobayashi M, Horiuchi I, Suzuki A, Wang J, Chen J, Niizeki H, Kawamura K, Hosokawa M, Asaka M. Constitutive expression of hypoxia-inducible factor-1alpha renders pancreatic cancer cells resistant to apoptosis induced by hypoxia and nutrient deprivation. Cancer Res. 2001;61:6548–6554. [PubMed] [Google Scholar]
- 37.Ohashi S, Kikuchi O, Tsurumaki M, Nakai Y, Kasai H, Horimatsu T, Miyamoto S, Shimizu A, Chiba T, Muto M. Preclinical validation of talaporfin sodium-mediated photodynamic therapy for esophageal squamous cell carcinoma. PLoS One. 2014;9:e103126. doi: 10.1371/journal.pone.0103126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Deenen MJ, Rosing H, Hillebrand MJ, Schellens JH, Beijnen JH. Quantitative determination of capecitabine and its six metabolites in human plasma using liquid chromatography coupled to electrospray tandem mass spectrometry. J Chromatogr B Analyt Technol Biomed Life Sci. 2013;913-914:30–40. doi: 10.1016/j.jchromb.2012.11.033. [DOI] [PubMed] [Google Scholar]
- 39.Takechi T, Fujioka A, Matsushima E, Fukushima M. Enhancement of the antitumour activity of 5-fluorouracil (5-FU) by inhibiting dihydropyrimidine dehydrogenase activity (DPD) using 5-chloro-2,4-dihydroxypyridine (CDHP) in human tumour cells. Eur J Cancer. 2002;38:1271–1277. doi: 10.1016/s0959-8049(02)00048-5. [DOI] [PubMed] [Google Scholar]
- 40.Li LH, Dong H, Zhao F, Tang J, Chen X, Ding J, Men HT, Luo WX, Du Y, Ge J, Tan BX, Cao D, Liu JY. The upregulation of dihydropyrimidine dehydrogenase in liver is involved in acquired resistance to 5-fluorouracil. Eur J Cancer. 2013;49:1752–1760. doi: 10.1016/j.ejca.2012.12.013. [DOI] [PubMed] [Google Scholar]
- 41.Salonga D, Danenberg KD, Johnson M, Metzger R, Groshen S, Tsao-Wei DD, Lenz HJ, Leichman CG, Leichman L, Diasio RB, Danenberg PV. Colorectal tumors responding to 5-fluorouracil have low gene expression levels of dihydropyrimidine dehydrogenase, thymidylate synthase, and thymidine phosphorylase. Clin Cancer Res. 2000;6:1322–1327. [PubMed] [Google Scholar]
- 42.Kawasaki G, Yoshitomi I, Yanamoto S, Mizuno A. Thymidylate synthase and dihydropyrimidine dehydrogenase expression in oral squamous cell carcinoma: an immunohistochemical and clinicopathologic study. Oral Surg Oral Med Oral Pathol Oral Radiol Endod. 2002;94:717–723. doi: 10.1067/moe.2002.126912. [DOI] [PubMed] [Google Scholar]
- 43.Frankel A, Nancarrow D, Wayte N, Barbour A. Clinical issues in oesophageal adenocarcinoma: could DNA copy number hold the key? ANZ J Surg. 2012;82:599–606. doi: 10.1111/j.1445-2197.2012.06144.x. [DOI] [PubMed] [Google Scholar]
- 44.Shirasaka T. Development history and concept of an oral anticancer agent S-1 (TS-1): its clinical usefulness and future vistas. Jpn J Clin Oncol. 2009;39:2–15. doi: 10.1093/jjco/hyn127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.van Triest B, Pinedo HM, van Hensbergen Y, Smid K, Telleman F, Schoenmakers PS, van der Wilt CL, van Laar JA, Noordhuis P, Jansen G, Peters GJ. Thymidylate synthase level as the main predictive parameter for sensitivity to 5-fluorouracil, but not for folate-based thymidylate synthase inhibitors, in 13 nonselected colon cancer cell lines. Clin Cancer Res. 1999;5:643–654. [PubMed] [Google Scholar]
- 46.Hu XF, Slater A, Rischin D, Kantharidis P, Parkin JD, Zalcberg J. Induction of MDR1 gene expression by anthracycline analogues in a human drug resistant leukaemia cell line. Br J Cancer. 1999;79:831–837. doi: 10.1038/sj.bjc.6990133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.De Angelis PM, Svendsrud DH, Kravik KL, Stokke T. Cellular response to 5-fluorouracil (5-FU) in 5-FU-resistant colon cancer cell lines during treatment and recovery. Mol Cancer. 2006;5:20. doi: 10.1186/1476-4598-5-20. [DOI] [PMC free article] [PubMed] [Google Scholar]


