Abstract
MiR-146a has been shown to play a critical role in cell immunity and phagocytosis, processes that require rearrangement of the cytoskeleton. However, the detailed mechanism by which miR-146a regulates these events remains elusive. Here, we used luciferase reporter and protein assays to show that the cytoskeleton-regulatingprotein verprolin-homologous protein 2 (WAVE2), is a direct target of miR-146a. MiR-146a overexpression resulted in a decrease in WAVE2 protein expression under endotoxin-free culture conditions. Unexpectedly, however, miR-146a activated rather than repressed the expression of WAVE2 in macrophage RAW264.7 cells when cultured continuously in the presence of endotoxin. Furthermore, we demonstrated that miR-146a induced WAVE2 expression and enhanced phagocytosis in lipopolysaccharide-stimulated RAW264.7 macrophages. Our study suggests that lipopolysaccharide- induced miR146a indirectly activates WAVE2 expression; thus, facilitating cytoskeletal reorganization and phagocytosis in lipopolysaccharide-stimulated macrophages.
Keywords: MiR-146a, endotoxin, WAVE2, phagocytosis, macrophage
Introduction
The immune system senses and responds to lipopolysaccharides (LPS) via Toll-like receptor 4(TLR4)-mediated activation of downstream signaling pathways to produce a proinflammatory cytokine storm. The release of these cytokines,including TNF-α and IL-6, primes the first line of host defense against microbial challenge. In healthy individuals, an inhibitory feedback mechanism is established resulting in tolerance to subsequent endotoxin stimuli and preventing excessive inflammation by suppressing TLR-mediated production of proinflammatory cytokines and increasing phagocytic ability. This feedbackmechanism prevents extensive damage to tissues and relateddiseases, such as sepsis, autoimmune diseases, and cancer [1].
Recently, miRNAs have emerged as key regulators of endotoxin immunitythrough inhibition of several important mediators of the TLR4-mediated signaling cascade [2]. MiR-132, miR-155, and miR-146a are rapidly induced in endotoxin-stimulated human monocytes to regulate proinflammatory cytokine production [3,4]. MiR-146a in particular, has been shown to repress IL-1 receptor-associated kinase (IRAK1) and tumor-necrosis factor receptor-associated factor 6 (TRAF6) in the TLR4-mediated pathway, leading to suppression of proinflammatory cytokines in endotoxin-stimulated macrophages [3,4]. Importantly, it has been reported that overexpression of miR-146a alone inducesendotoxin tolerance similar to that induced by LPS challenge [4]. Similarly, miR-146a knockout results in diminished LPS tolerance in mouse peritoneal monocytes [5]. These reports indicate that miR-146a plays a critical role in endotoxin tolerance in innate immune cells.
Increased phagocytic ability is a hallmark of endotoxin-tolerant macrophages [6,7] and requires reorganization of the cytoskeletal network. Verprolin-homologous protein 2 (WAVE2) has been validated as a downstream effector of small GTPases for reorganization of the cytoskeletal network during phagocytosis, and may be involved in macrophageimmunity [8,9]. However, the mechanism by which WAVE2 is regulated in endotoxin-stimulated macrophages remains to be elucidated.
In this study, we show that miR-146a targets WAVE2 directly and activates WAVE2 expression in endotoxin-stimulated macrophages. Both miR-146a and WAVE2 are correlated with differential phagocytic ability in endotoxin-stimulated macrophages. Furthermore, LPS-induced miR-146 activates WAVE2 expression leading to cytoskeletal reorganization and phagocytosis in endotoxin-stimulated macrophages.
Materials and methods
Cell culture
The murine macrophage cell line RAW264.7 and the human embryonic kidney cell line HEK293T were purchased from the Cell Resource Center, Shanghai Institute of Biochemistry and Cell Biology at the Chinese Academy of Sciences (China). RAW 264.7 and 293T cells were cultured in complete Dulbecco’s modified Eagle’s medium (DMEM) containing 4.5 g/L glucose and sodium pyruvate, supplemented with 10% (v/v) fetal bovine serum (Life Technologies) at 37°C under 5% carbon dioxide (CO2).
DNA constructs
The lentivirus vector pEZX-MR03 expressing the murine miR-146a precursor (MmiR3434-MR03), precursor miRNA scramble (CmiR0001-MR03), and pEZX-MR04 expressing a miR-146a inhibitor (MmiR-AN0196-AM04) were obtained from GeneCopoeia (Guangzhou, China). The murine WAVE2 open reading frame (ORF) clone was purchased from Open Biosystems (without a native stop codon). Primers were designed for PCR amplification of the WAVE2 full-length ORF sequence, including a stop codon in the downstream primer. The primer sequences are as follows: upstream sequence 5’-CATAGAAGATTCTAGAATGCCGTTAGTAACGAGGAACATCGAG-3’, downstream sequence 5’-ATTTAAATTCGAATTTTAATCGGACCAGTCGTCCTCATCAAATT-3’. The product was subcloned into the pCDH-CMV-MCS-EF1-Purovector (System Biosciences, USA) according to the In-Fusion HD cloning kit protocol (TaKaRa Biomedical, China). MISSION lentiviral shRNAconstructstargeting mouse WAVE2 were obtained from Sigma (MISSION shRNA collection); clone used in this study was TRCN0000099310. A non-targeting scramble sequence lentiviral construct (5’-CCTAAGGTTAAGTCGCCCTCGCTCGAGCGAGGGCGACTTAACCTTAGG-3’) was used as the control.
Plasmid vectors for dual-luciferase reporter assays and mutagenesis
The full-length 3’UTR sequence of murine WAVE2 (NM_001201404) was cloned into the XhoI and NotI sites of the psiCHECK-2 vector after PCR amplification from mouse genomic DNA using the following primers: Cycle 1 primers: 5’-TCGCTCGAGATTTTTTTTTATCAAGAGGG-3’ and 5’-TCGGCGGCCGCGACAAGAATGAGACTTT AATC-3’; Cycle 2 primers: 5’-TCGCTCGAGCTCCGCCTGCTGCCCACATTCC-3’ and 5’-TCGGCGGCCGCGGGTACACATATGCCTAAG-3’. Specific nucleotides mutations within the putative miR-146a seed sequence binding region were created with a QuickChange site-directed mutagenesis kit (Stratagene). The mutagenic primers were as follows: 5’-GGGTTTCGATGTGCTGTGATATAATCCTACAGCTG-3’, 5’-CATTACCAAACCAGCTGTAGGATTATATCACAGC-3’. Sequencing was performed to confirm that all constructs contained the expected DNA sequence.
Lentivirus production and generation of stable cell lines
Briefly, 10 µg of the miR-146a precursor, scramble miRNA vector, or miR-146a Tough Decoy RNA (TuD)constructs and 10 μg of a plasmid mixture containing the psPAX2 packaging plasmid (7.5 μg) and the pMD2.G envelope plasmid (2.5 μg) were incubated with Fugene HD transfection reagent (Roche). Subsequently, the mixture was added to 293T cells cultured in a 10-cm dish. Lentiviral particles of the WAVE2 expression construct, shRNA construct, and non-targeting shRNA control, wereproduced by the same method. The pseudo-lentiviral supernatant was harvested after 48 h, and the cells were treated with pseudo-lentiviral supernatant (1 mL supernatant per 1×106 cells plus 8 μg/mL polybrene) in 6-well plates. Virus-containing supernatant was harvested after 48 h and used to infect cells. Stable cell lineswere obtained by puromycin (8.0 μg/mL) or hygromycin (8.0 mg/mL) selection for 24 h (Life Technologies).
Stable expression of WAVE2 and miR-146a in RAW264.7 cells induced by LPS
Naïve RAW264.7 cells and RAW264.7 cells infected with lentivirus miR-146a TuD or scramble miRNA vectors were incubated with Escherichia coliLPS (100 ng/mL final concentration; Sigma-Aldrich) for 0 h, 4 h, 8 h, 12 h, and 24 h. Cell extracts were analyzed for WAVE2 and IRAK1 protein expression and miR-146a expression by Western blot and qRT-PCR, respectively.
Stable expression of miR-146a in RAW264.7 cells induced by LPS
MiRNA-enriched total RNA was isolated with a mirVana miRNA Isolation Kit (Ambion). MiR146a was determined using the comparative Ct method. A TaqMan microRNA assay was performed using the ABI 7900 PCR system. Quantification of mature miR-146a was performed using TaqMan MicroRNA assays, and reverse transcription using the MicroRNA Reverse Transcription Kit (Applied Biosystems). Mature miR-146a was reverse-transcribed using specific stem-loop primers which allow for generation of cDNA amenable to analysis by qPCR. Input was 1 ng of total miRNA per RT reaction. After real time qPCR, expression values were normalized to that of endogenous let-7a RNA [10].
Dual-luciferase assays
Dual-luciferase assayswere performed as described previously [11]. 293T cells were seeded in 96-well plates and transfected with 100 ng of the plasmid mixture (80 ng of the pre-miR-146a expression vector or scramble miRNA expression vectors and 20 ng of the WAVE2 wild-type or mutant 3’UTR psiCHECK-2 construct) using Fugene HD transfection reagent. A constitutively expressed firefly luciferase gene in psiCHECK-2 was used as a normalization control for transfection efficiency. After 48 h, both firefly and Renilla luciferases were detected with a Dual-Glo Luciferase assay kit (Promega). Luminescence was quantitated and Renilla luciferase readings were normalized against the firefly luciferase activity to determine the relative luciferase activity using an Orion II microplate luminometer (Berthold Detection Systems).
In vitro phagocytosis assays
Phagocytic function was analyzed using a Vybrant phagocytosis assay kit (V-6694; Molecular Probes), which allows quantification of phagocytosis by following the internalization of fluorescently labeled bacterial particles. Briefly, 1.0×105 cells/mL RAW264.7 cells in 96-well plate were incubated with fluorescein-labeled E. coli in the dark at 37°C for 2 h. After removal of the supernatant, the cells were treated with trypan blue to quench any remaining extracellular probe and then washed with PBS. The intracellular fluorescence emitted by the engulfed bacteria in 96-well plates was measured using a Fluoroskan Ascent fluorometer (Thermo Electron). Phagocytosis was expressed as the percentage increase in fluorescence over the baseline phagocytosis mediated by the untreated RAW 264.7 cells.
Western blot
The expression of WAVE2, IRAK1 or GAPDH proteins in RAW264.7 cell lysates was investigated by Western blot analysis. Cell lysates were loaded on an 8% SDS-PAGE gel and subsequently transferred to a PVDF membrane. The membranes were blocked with 5% driedskimmed milk and incubated with primary detection monoclonal antibodies overnight at 4°C (anti-WAVE2 and anti-GAPDH, Cell Signaling Technology; anti-IRAK1, AbgentCorporation). Membranes were then incubated with the HRP-conjugated secondary detection antibodies. Immunoreactive band detection was achieved via enzyme-linked chemiluminescence using theImageQuant LAS 4000 mini. The protein bands were quantified using ImageQuant TL software (GE Healthcare), and the data were normalized to GAPDH.
Statistical analysis
Statistical analyses were performed using Prism 6 software (GraphPad). All data are expressed as the mean ± SEM. The results were analyzed by ANOVA, followed by a Bonferroni correction post-hoc t-test. P < 0.05 were considered to indicate statistical significance.
Results and discussion
Cytoskeleton protein WAVE2 is a direct target regulated by miR-146a in macrophages
MiR-146a has previously been shown to be associated with enhanced phagocytic activity in patient monocytes [12]. Insilico target prediction using TargetScan software indicated that WAVE2 is a potential target of miR-146a, with a perfect match between the miR-146a ‘seed sequence’ and the 3’UTR of WAVE2 mRNA (Figure 1A). This prediction was validated experimentally using a dual-luciferase reporter system. As shown in Figure 1B, Renilla luciferase expression was repressed in 293T cells transfected with wild-type reporter plasmids, but not in nucleotide mutant 3’UTR transfected cells. These observation sindicate that reporter inhibition is mediated by direct targeting of the 3’UTR of WAVE2 by miR-146a. Furthermore, compared to naïve and scramble miRNA overexpressing RAW264.7 cells, Western blot analysis (Figure 1C and 1D) showed that miR-146a overexpression was associated with decreased WAVE2 expression under non-endotoxin conditions. These data support the prediction that WAVE2 is a direct target of miR-146a in RAW264.7 cells.
Figure 1.
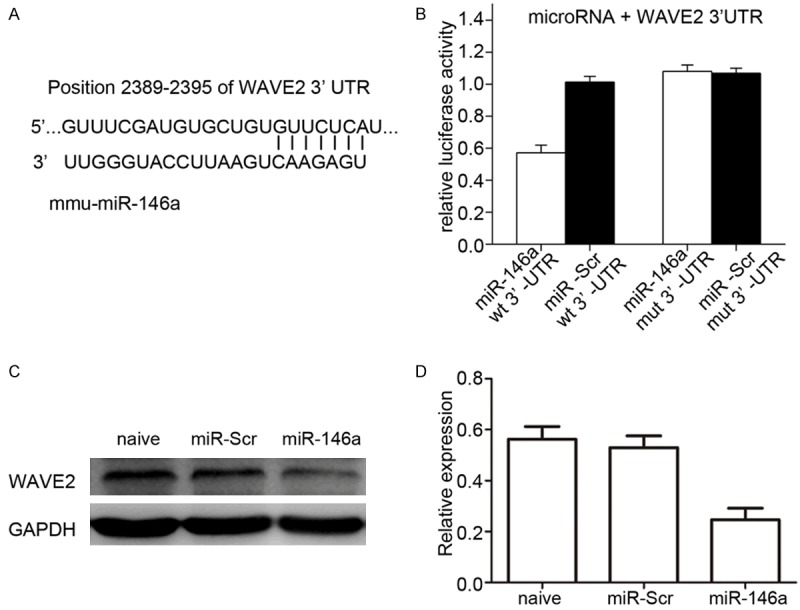
Both in silico analysis and a reporter assay show that WAVE2 is a direct target of miR-146a. A. The 3’UTR sequence of murine WAVE2 mRNA is completely complementary to the ‘seed sequence’ of miR-146a (indicated by the solid lines). B. Dual-luciferase assays were carried out using cell lysates of 293T cells 48 h after co-transfection with psiCHECK2-based plasmids, with either the wild-type or mutant 3’UTR of WAVE2 mRNA downstream of the Renilla luciferase (RLuc) reporter and miRNA expressing vectors. MiR-146a repressed the expression of the Rluc reporter under the control of the wild-type, but not the mutant 3’UTR of WAVE2 as indicated. Scramble microRNA was used as a control. Rluc activity was normalized to the activity of firefly luciferase (Fluc) encoded on the same psiCHECK2 vector. C, D. Western blotting results showing that the level of WAVE2 protein was significantly reduced in RAW264.7 cells infected by viruses derived from LV-miR-146a compared to those from scramble miRNA under normal culture conditions. GAPDH was used as the loading control.
WAVE2 is activated by miR-146a in endotoxin-stimulated macrophages
To determine a mechanistic link between WAVE2 and miR146a in endotoxin-stimulated macrophages, we examined the expression of both miR-146a and WAVE2 in RAW264.7 cells under LPS-stimulated conditions. In accordance with previous observations [10], miR-146a expression was continuously upregulated over time in LPS-stimulated scramble miRNA knockdown (KD) cells similar to the effects observed in naïve RAW264.7 cells. (Figure 2A). Upregulation of miR-146a was validated by miR-146a Tough Decoy (TuD) RNAs that specifically inhibited miR-146a in endotoxin-treated cells [13] (Figure 2A). Furthermore, WAVE2 protein was unexpectedly found to be upregulated in LPS-stimulated naïve and scramble KD RAW264.7 cells following induction of miR-146a (Figure 2B). In contrast, WAVE2 protein levels were not significantly affected in LPS-stimulated miR-146a KD cells (Figure 2B), suggesting that miR-146a activated, rather than inhibited WAVE2 expression in these cells. These findings were further confirmed by the observation that the protein levels of IRAK1, a well-known target of miR-146a [3,4], appeared to be decreased under conditions when both miR-146a and WAVE2 were increased (Figure 2A and 2B). Interestingly, this significant change in the protein levelsof both WAVE2 and IRAK1 was abolished in LPS-stimulated miR-146a TuD cells, suggesting opposing functions for miR-146a functionsin different targets under LPS-stimulation in macrophages. These data are consistent with a report that miRNAs upregulated translation of their target mRNAs on cell cycle arrest [14]. Similarly, miR-146a TuD treatment abolished WAVE2 upregulation (Figure 2B). These results suggest that WAVE2 is activated by miR-146a in endotoxin-stimulated macrophages.
Figure 2.
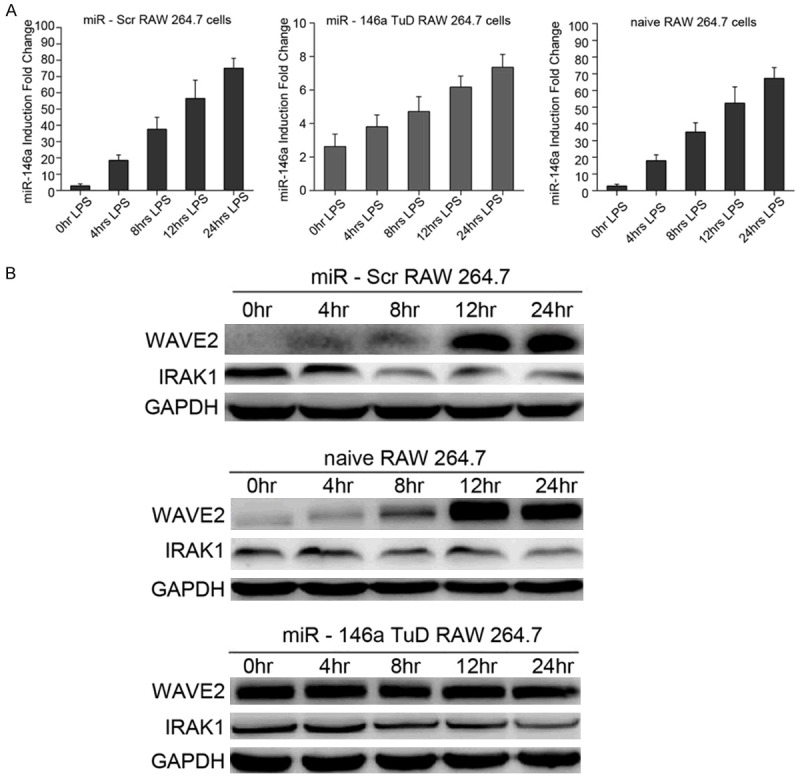
WAVE2 is activated by induced miR-146a in RAW246.7 cells under LPS-stimulated conditions. A. RAW246.7 cells treated with either scramble or miR-146a Tough Decoy (TuD) RNA were stimulated with LPS (100 ng/mL) for 0, 4, 8, 12, or 24 h. The relative levels of miR146a, determined using qRT-PCR followed by normalization to let-7a expression, was increased over time in scramble TuD treated cells with LPS-stimulation similar to that of naïve cells, while the levels of miR-146a in TuD treated cells barely changed under the same conditions. B. Western blotting results showing increased WAVE2 protein expression in scramble miRNA (miR-Scr), but not in miR-146 a treated RAW264.7 cells with LPS-stimulation. In contrast, the protein level of IRAK1, a well-known miR-146a target, was decreased in the same cells. GAPDH was used as the loading control.
WAVE2 and miR-146a increase phagocytic activity of endotoxin-stimulated macrophages
To determine role of WAVE2 and miR-146a in the phagocytic function of endotoxin-tolerant macrophages, phagocytic activity of macrophages was quantified under various conditions using the Vybrant phagocytosis assay. Under non-endotoxin conditions, miR-146a overexpression resulted in decreased phagocytic activity compared to naïve and scramble miRNA overexpressing RAW 264.7 cells (Figure 3A). In contrast, phagocytic activity was increased in a time-dependent manner in endotoxin-stimulated macrophages (Figure 3B), suggesting that miR-146a upregulation is positively correlated with phagocytic activity in endotoxin-tolerant macrophages.
Figure 3.
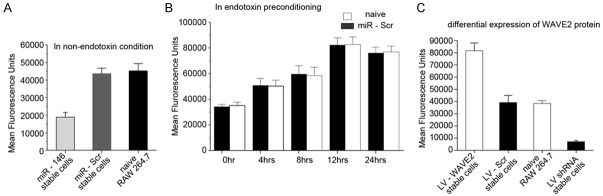
WAVE2 and miR-146a promote phagocytosis of bacteria in endotoxin-stimulated macrophages. A. The expression of miR-146a (LV-146a) resulted in decreased phagocytic activity as indicated by the signals of fluorescence-labeled bacteria compared to those of naïve and scramble miRNA treated RAW264.7 cells under non-endotoxin conditions. B. The phagocytic activity, indicated by the average fluorescence signal, increased similarly over time in both naïve and scramble miRNA treated RAW264.7 cells stimulated by LPS. C. The phagocytic activity, indicated by the average fluorescence signal, was increased in WAVE2 overexpressing cells and decreased in WAVE2 knockdown cells compared to those in naïve and scramble knockdown cells under LPS-stimulation conditions.
Next, we investigated the function of WAVE2 in phagocytosis by overexpression and knockdown of WAVE2 in LPS-treated RAW264.7 cells. Overexpression of WAVE2 enhanced phagocytosis in stimulated RAW264.7 cells (Figure 3C), while WAVE2 knockdown in stimulated RAW264.7 cells was associated with lower phagocytic activity than that of naïve and control knockdown RAW264.7 cells. Our results suggest that LPS-stimulated RAW264.7 cell induction of miR-146a resulted in activation of WAVE2 expression, therefore enhancing the phagocytic function of these cells.
While miRNAs have been generally thought to repress expression of their direct mRNA targets, miRNAs have also been reported to activate their targets directly under stress conditions, such as cell cycle arrest [14]. In this study, we showed that miR-146a activated WAVE2 expression in LPS-stimulated macrophages. Given that our data also show that WAVE2 is a direct target of miR-146a in vitro, it can be speculated that miR-146a activates WAVE2 expression directly in endotoxin-stimulated macrophages; however, we cannot rule out an indirect mechanism for this effect. Nevertheless, our results clearly demonstrate that miR-146a positively regulates WAVE2 expression and contributes to increased phagocytic ability in endotoxin-tolerant macrophages. Given the limitations of in silico prediction and luciferase reporter assays for ascertaining miRNA targets, alternative methods, such as tagged miRNA pull-down or miR-TRAP (miRNA target affinity purification), should be performed to demonstrate direct interaction between miR-146a and WAVE2. In combination with proteomics approaches, these methods may also elucidate the differential composition of miRNA ribonucleoprotein (miRNP) complexes (i.e., AGO2 and RNA binding proteins [FXR1]) under various conditions.
Interestingly, the activation of WAVE2 by miR-146a in endotoxin-stimulated macrophages is consistent with a previous report that a number of miRNAs activate their direct targets only on cell cycle arrest [14]. We found that miR-146a activated only WAVE2 in endotoxin-stimulated cells. In accordance with this, miR-146a enhanced phagocytosis capacity only in endotoxin-tolerant cells. These results suggest an interesting scenario in which at least some, if not all, miRNAs modulate their effects on target mRNAs based on endotoxin conditions, cell cycle status, or the developmental stage of a cell. The fine-tuning of the downstream effects of miRNAs on their target mRNAs is of great interest to researchers in related fields. Both miR-146a and WAVE2 have previously been shown to be involved in endotoxin tolerance [15,16]. Our results reveal an intriguing mechanistic link between these two key regulators in endotoxin-stimulated macrophages. Based on our findings, we propose a model in which miR-146a is induced by LPS-stimulation, leading to upregulation of the cytoskeleton regulator WAVE2, which in turn, promotes cytoskeletal reorganization and the formation of a phagocytic cup for pathogen engulfment in endotoxin-stimulated macrophages (Figure 4) [17]. Further studies are required to define the mechanism by which miR-146a activates WAVE2 and promotes phagocytosis in endotoxin-stimulated macrophages or other innate immune cells.
Figure 4.
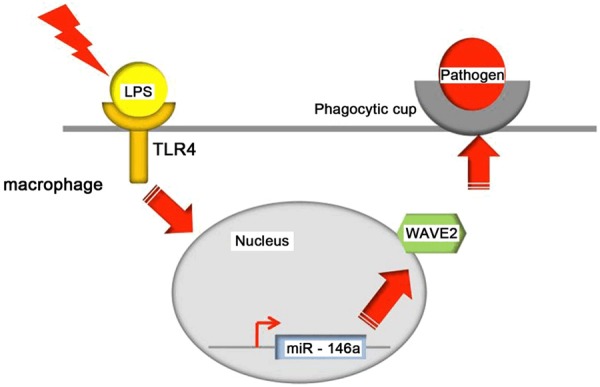
Proposed model for miR-146a-mediated activation of WAVE2 expression and enhancement of phagocytosis in endotoxin-stimulated macrophages. Under conditions of endotoxin stimulation, miR146a is rapidly induced by the TLR4-mediated signaling pathway in macrophages, leading to upregulation of WAVE2, which in turn promotes cytoskeletal reorganization/phagocytic cup formation and subsequent phagocytosis of pathogens.
Disclosure of conflict of interest
None.
References
- 1.Beutler B, Rietschel ET. Innate immune sensing and its roots: the story of endotoxin. Nat Rev Immunol. 2003;3:169–176. doi: 10.1038/nri1004. [DOI] [PubMed] [Google Scholar]
- 2.Nahid MA, Satoh M, Chan EK. MicroRNA in TLR signaling and endotoxin tolerance. Cell Mol Immunol. 2011;8:388–403. doi: 10.1038/cmi.2011.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Taganov KD, Boldin MP, Chang KJ, Baltimore D. NF-kappaB-dependent induction of microRNA miR-146, an inhibitor targeted to signaling proteins of innate immune responses. Proc Natl Acad Sci U S A. 2006;103:12481–12486. doi: 10.1073/pnas.0605298103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.El Gazzar M, Church A, Liu T, McCall CE. MicroRNA-146a regulates both transcription silencing and translation disruption of TNF-alpha during TLR4-induced gene reprogramming. J Leukoc Biol. 2011;90:509–519. doi: 10.1189/jlb.0211074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nahid MA, Pauley KM, Satoh M, Chan EK. miR-146a is critical for endotoxin-induced tolerance: IMPLICATION IN INNATE IMMUNITY. J Biol Chem. 2009;284:34590–34599. doi: 10.1074/jbc.M109.056317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Boldin MP, Taganov KD, Rao DS, Yang L, Zhao JL, Kalwani M, Garcia-Flores Y, Luong M, Devrekanli A, Xu J, Sun G, Tay J, Linsley PS, Baltimore D. miR-146a is a significant brake on autoimmunity, myeloproliferation, and cancer in mice. J Exp Med. 2011;208:1189–1201. doi: 10.1084/jem.20101823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Biswas SK, Lopez-Collazo E. Endotoxin tolerance: new mechanisms, molecules and clinical significance. Trends Immunol. 2009;30:475–487. doi: 10.1016/j.it.2009.07.009. [DOI] [PubMed] [Google Scholar]
- 8.Wheeler DS, Lahni PM, Denenberg AG, Poynter SE, Wong HR, Cook JA, Zingarelli B. Induction of endotoxin tolerance enhances bacterial clearance and survival in murine polymicrobial sepsis. Shock. 2008;30:267–273. doi: 10.1097/shk.0b013e318162c190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shi J, Scita G, Casanova JE. WAVE2 signaling mediates invasion of polarized epithelial cells by Salmonella typhimurium. J Biol Chem. 2005;280:29849–29855. doi: 10.1074/jbc.M500617200. [DOI] [PubMed] [Google Scholar]
- 10.Pauley KM, Satoh M, Pauley BA, Dominguez-Gutierrez PR, Wallet SM, Holliday LS, Cha S, Reeves WH, Chan EK. Formation of GW/P bodies as marker for microRNA-mediated regulation of innate immune signaling in THP-1 cells. Immunol Cell Biol. 2010;88:205–212. doi: 10.1038/icb.2009.84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Graff JW, Dickson AM, Clay G, McCaffrey AP, Wilson ME. Identifying functional microRNAs in macrophages with polarized phenotypes. J Biol Chem. 2012;287:21816–21825. doi: 10.1074/jbc.M111.327031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pauley KM, Stewart CM, Gauna AE, Dupre LC, Kuklani R, Chan AL, Pauley BA, Reeves WH, Chan EK, Cha S. Altered miR-146a expression in Sjogren’s syndrome and its functional role in innate immunity. Eur J Immunol. 2011;41:2029–2039. doi: 10.1002/eji.201040757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Haraguchi T, Ozaki Y, Iba H. Vectors expressing efficient RNA decoys achieve the long-term suppression of specific microRNA activity in mammalian cells. Nucleic Acids Res. 2009;37:e43. doi: 10.1093/nar/gkp040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vasudevan S, Tong Y, Steitz JA. Switching from repression to activation: microRNAs can up-regulate translation. Science. 2007;318:1931–1934. doi: 10.1126/science.1149460. [DOI] [PubMed] [Google Scholar]
- 15.Pils S, Kopp K, Peterson L, Delgado Tascon J, Nyffenegger-Jann NJ, Hauck CR. The adaptor molecule Nck localizes the WAVE complex to promote actin polymerization during CEACAM3-mediated phagocytosis of bacteria. PLoS One. 2012;7:e32808. doi: 10.1371/journal.pone.0032808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu M, John CM, Jarvis GA. Induction of endotoxin tolerance by pathogenic Neisseria is correlated with the inflammatory potential of lipooligosaccharides and regulated by microRNA-146a. J Immunol. 2014;192:1768–1777. doi: 10.4049/jimmunol.1301648. [DOI] [PubMed] [Google Scholar]
- 17.Swanson JA. Shaping cups into phagosomes and macropinosomes. Nat Rev Mol Cell Biol. 2008;9:639–649. doi: 10.1038/nrm2447. [DOI] [PMC free article] [PubMed] [Google Scholar]


