Abstract
The child-killer and famously recalcitrant parasite Cryptosporidium is the latest organism to yield to the magic of CRISPR/Cas9. The ability to knockout genes and introduce markers promises a new heyday for drug discovery and vaccine development as well as the basic biology of these fascinating parasites.
Keywords: CRISPR/Cas9, gene knockout, reporter parasites, target validation
Cryptosporidium parasites have traveled a long road to recognition as an important human pathogen. Tyzzer discovered these protozoa at the beginning of the 20th century, believing them to be benign inhabitants of the gastrointestinal tract of most animals (see [1] for a history). It took seventy years to dispel this notion and recognize that Cryptosporidium infections cause severe diarrhea, and another thirty years before the recent Global Enteric Multicenter Study (GEMS) revealed that Cryptosporidium is the second leading cause of diarrheal mortality in small children after rotavirus [2]. Every year, diarrhea causes the deaths of 800 000 children under five, which is more than that well-recognized child-killer malaria. Alone among the top four diarrheal pathogens, which also include enterotoxigenic Escherichia coli and Shigella bacteria, no effective treatments or vaccines are on the horizon for Cryptosporidium [3,4].
Infection occurs when the spore-like Cryptosporidium oocysts are ingested with contaminated water, excyst in the gastrointestinal tract and invade epithelial cells, forming a unique extracytosolic compartment that protrudes into the intestinal lumen (Figure 1). The life cycle involves successive rounds of asexual and sexual multiplication eventually producing oocysts that are excreted. Cryptosporidium infections are self-limiting in immunocompetent patients, but can be chronic and fatal in immunosuppressed patients, such as those infected with HIV and in small children, in particular those who are malnourished. While the disease burden is mainly on the developing world, Cryptosporidium oocysts are resistant to commonly used water treatments, so the developed world is also beset by frequent outbreaks. Indeed, Cryptosporidium infections account for half of the illness associated with recreational water in the USA [5]. In addition, oocysts can be obtained relatively easily, posing a significant biowarfare threat. The tools to deal with Cryptosporidum outbreaks are woefully inadequate. Diagnosis requires specialized tests and treatment is limited to one approved drug, nitazoxanide, that at best it shortens the course of disease by a few days in immunocompetent patients. Perplexingly, Cryptosporidium is resistant to antifolates and the many commonly used antiprotozoal drugs. No vaccines are available and no effective treatment exists for immunocompromised patients.
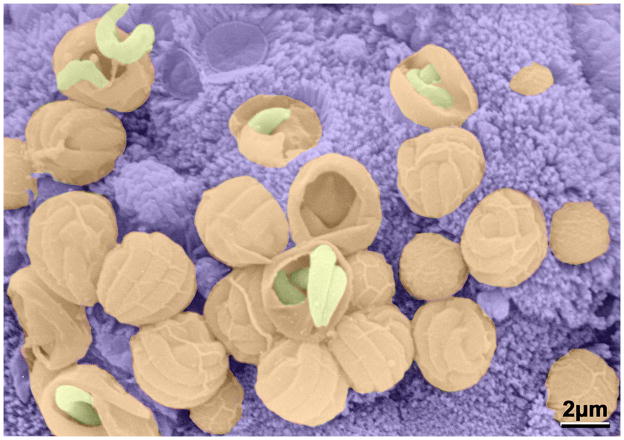
Figure 1. A Cryptoporidium infected intestine.
False color scanning electron micrograph of Cryptosporium parvum showing a group of merozoites emerging from schizonts attached to the epithelial cell surface of a mouse villus (courtesy of David Ferguson, Oxford University).
The Cryptosporidium field has long been defined by what cannot be done: no continuous culture, inadequate animal models and no genetic tools. Cryptosporidium oocysts can be excysted to release sporozoites that infect a mammalian cell monolayer, but undergo just a few rounds of replication before dying. This severely constrains investigation, limiting experiments to small numbers of parasites and a fraction of the life cycle. Parasites can be propagated in immunodeficient animals, though these experiments are largely limited to C. parvum rather than the primary human pathogen C. hominis. As a result, Cryptosporidium drug discovery efforts have been largely futile, with few compounds showing efficacy in animals, much less in humans. Since no truly effective drug exists, we do not even know what pharmacokinetic properties are required to eradicate infections [6].
The genomic sequences of both C. hominis and C. parvum were reported in 2004, ushering in a molecular era filled with promise [7,8]. The genome revealed that Cryptosporidium has numerous transporters but few enzymes. Not surprisingly given its nutrient rich environmental niche, Cryptosporidium has very streamlined biosynthetic pathways, instead relying heavily on salvage. Many of the genes appear to have been obtained by horizontal gene transfer from other gut organisms, and thus are very different from other apicomplexan parasites as well as the host [9]. Several promising drug targets emerged, including inosine monophosphate dehydrogenase, calcium dependent protein kinases, hexokinase, lactate dehydrogenase, and fatty acid synthetase. However, target validation required the arduous development of selective inhibitors, always with the fear that the process was not as essential as it appeared on paper. If only there was a way to knockout genes… and now there is!
Using the magic of CRISPR/Cas9, the Striepen laboratory from the University of Georgia now reports a series of elegant experiments resulting in the first gene knockout in C. parvum [10]. The team methodically overcame every hurdle, first identifying a construct that transiently expressed nanoluciferase in parasites, then developing a surgical procedure to directly introduce sporozoites into the mouse intestine and finally using CRISPR/Cas9 to introduce aminoglycoside resistance, enabling the selection of transgenic parasites in vivo. They exploited an apparent redundancy in thymidine biosynthesis, the presence of both thymidylate synthetase and thymidine kinase (TK), suggesting that perhaps neither gene was essential. Indeed, the TK gene could be disrupted with no apparent consequence-except that the parasite is now sensitive to antifolates!
The availability of nanoluciferase reporter parasites alone is a major step forward for drug discovery, greatly simplifying efficacy and screening assays. The prospects of using genetically encoded reporters to image infections in animals is even more exciting. A screen for essential genes would validate drug targets. The development of attenuated vaccine strains is also on the horizon.
And there is more good news: like its apicomplexan cousin, Plasmodium (the malaria parasite), Cryptosporidium has sexual stages. But whereas sex requires passage through mosquitos for Plasmodium, the entire Cryptosporidium life cycle occurs in the mammalian gut, so performing a genetic cross could be as simple as co-infecting strains. With the newfound ability to create parents carrying different genetic markers, isolation of recombinant progeny would be easy. It is no longer completely unimaginable that Cryptosporidium could someday become the model apicomplexan of choice.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Tzipori S, Widmer G. A hundred-year retrospective on cryptosporidiosis. Trends Parasitol. 2008;24:184–189. doi: 10.1016/j.pt.2008.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kotloff KL, et al. Burden and aetiology of diarrhoeal disease in infants and young children in developing countries (the Global Enteric Multicenter Study, GEMS): a prospective, case-control study. Lancet. 2013;382:209–222. doi: 10.1016/S0140-6736(13)60844-2. [DOI] [PubMed] [Google Scholar]
- 3.Striepen B. Time to tackle cryptosporidiosis. Nature. 2013;503:189–191. doi: 10.1038/503189a. [DOI] [PubMed] [Google Scholar]
- 4.Checkley W, et al. A review of the global burden, novel diagnostics, therapeutics, and vaccine targets for cryptosporidium. The Lancet Infectious diseases. 2014;15:85–94. doi: 10.1016/S1473-3099(14)70772-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hlavsa MC, et al. Outbreaks of illness associated with recreational water - United States, 2011–2012. MMWR. Morbidity and mortality weekly report. 2015;64:668–672. [PMC free article] [PubMed] [Google Scholar]
- 6.Gorla SK, et al. Validation of IMP dehydrogenase inhibitors in a mouse model of cryptosporidiosis. Antimicrob Agents Chemother. 2014;58:1603–1614. doi: 10.1128/AAC.02075-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Abrahamsen MS, et al. Complete genome sequence of the apicomplexan, Cryptosporidium parvum. Science. 2004;304:441–445. doi: 10.1126/science.1094786. [DOI] [PubMed] [Google Scholar]
- 8.Xu P, et al. The genome of Cryptosporidium hominis. Nature. 2004;431:1107–1112. doi: 10.1038/nature02977. [DOI] [PubMed] [Google Scholar]
- 9.Striepen B, et al. Gene transfer in the evolution of parasite nucleotide biosynthesis. Proc Natl Acad Sci U S A. 2004;101:3154–3159. doi: 10.1073/pnas.0304686101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Vinayak S, et al. Genetic modification of the diarrhoeal pathogen Cryptosporidium parvum. Nature. 2015;523:477–480. doi: 10.1038/nature14651. [DOI] [PMC free article] [PubMed] [Google Scholar]