Abstract
Microvesicles have been shown to mediate varieties of intercellular communication. Work in murine species has shown that lung-derived microvesicles can deliver mRNA, transcription factors, and microRNA to marrow cells and alter their phenotype. The present studies evaluated the capacity of excised human lung cancer cells to change the genetic phenotype of human marrow cells. We present the first studies on microvesicle production by excised cancers from human lung and the capacity of these microvesicles to alter the genetic phenotype of normal human marrow cells. We studied 12 cancers involving the lung and assessed nine lung-specific mRNA species (aquaporin, surfactant families, and clara cell-specific protein) in marrow cells exposed to tissue in co-culture, cultured in conditioned media, or exposed to isolated lung cancer-derived microvesicles. We assessed two or seven days of co-culture and marrow which was unseparated, separated by ficoll density gradient centrifugation or ammonium chloride lysis. Under these varying conditions, each cancer derived from lung-mediated marrow expression of between one and seven lung-specific genes. Microvesicles were identified in the pellet of ultracentrifuged conditioned media and shown to enter marrow cells and induce lung-specific mRNA expression in marrow. A lung melanoma and a sarcoma also induced lung-specific mRNA in marrow cells. These data indicate that lung cancer cells may alter the genetic phenotype of normal cells and suggest that such perturbations might play a role in tumor progression, tumor recurrence, or metastases. They also suggest that the tissue environment may alter cancer cell gene expression.
Recent studies have indicated that membrane enclosed vesicles derived from a variety of cell types can alter the phenotype of adjacent cells. Microvesicles secreted by activated normal cells play a role in cellular communication [1]. They have been found to transfer CD41, integrin, or CXCR4 [2–5], as well as human immunodeficiency virus and Prions [6–9] between cells. Embryonic stem cell microvesicles have been reported to reprogram hematopoietic stem/progenitor cells via the horizontal transfer of mRNA and protein [10]. Similarly, tumor-derived microvesicles have been shown to carry several surface determinants and mRNA and to transfer some of these determinants to monocytes [11]. Apoptotic bodies from irradiated Epstein–Barr virus (EBV)-carrying cell lines have been seen to transfer DNA to a variety of co-cultured cells and integrated, but not episomal, copies of EBV resulted in expression of the EBV-encoded genes EBER and EBNAI in recipient cells at high copy number [12]. Extracts from T lymphocytes containing transcription factor complexes could induce fibroblasts to express lymphoid genes [13]. Investigators have evaluated the protein and mRNA content of microvesicles. In a recent study, RNA [14] was extracted from endothelial progenitor cell-derived microvesicles and microarray. They found a total of 298 transcripts. Our own work has shown the capacity of murine lung-derived microvesicles to alter the phenotype of murine marrow cells [15,16] (vide infra).
We have investigated the capacity of murine lung to alter the genetic phenotype of normal murine marrow cells. Using a co-culture system in which marrow cells were cultured across from normal or irradiated lungs, but separated from the lung by a cell impermeable (0•4 micron) membrane, we found that marrow cells expressed lung-specific mRNA, as detected by real-time polymerase chain reaction (PCR). Here, co-cultured marrow cells expressed surfactants A, B, C, and D, aquaporin-5, and clara cell-specific protein after 2 or 7 days of co-culture [15,16]. Conditioned media from lungs mediated the same genetic phenotype in incubated marrow cells, and we demonstrated that pelleted microvesicles had high levels of lung-specific mRNA. Incubation of marrow cells with fluorescence activated cell sorting-isolated lung-derived microvesicles also induced marked elevations of lung-specific mRNA and entry of the microvesicles into a minority of the marrow cells. Marrow cells co-cultured across from lungs also showed an increased capacity to convert to lung cells after transplantation into irradiated hosts, indicating that the induced mRNA caused functional changes in the marrow cells. Recent studies [16] showing that actinomycin and alpha-amanitin affects these phenotype shifts suggested that transcriptional mechanisms were involved in the finally observed genetic phenotype. This was essentially established, using cross species cultures of rat lung and mouse marrow with species-specific primers for rat and mouse surfactants B and C. When rat lung was cultured opposite mouse marrow, the induced surfactant mRNA was both rat and mouse. Further study of murine lung-derived microvesicles has shown the presence of both protein and micro-RNA. A working hypothesis here is that both lung-specific mRNA and a protein transcription factor are transferred to cells via microvesicles. In addition, transferred microRNA may, in turn, modulate mRNA levels.
These observations formed the basis for studies on the capacity of cancers derived from human lung to mediate phenotypic changes in target human marrow cells. We have used the same co-culture system as used for our murine studies, culturing specimens of human lung cancer, from operative samples, across from freshly harvested normal human marrow cells, but separated from them by a 0.4 micron cell impermeable membrane. Alternatively, we exposed human marrow cells to conditioned media from the cancers or to microvesicles derived from the lung cancers. We then evaluated the induction of lung-specific mRNA in the marrow cells by real-time reverse transcriptase polymerase chain reaction (RT-PCR) analysis.
Material and methods
Tumor collection
Consent was obtained according to Rhode Island Hospital’s Committee on Protection of Human Subjects (Institutional Review Board) for each of the patients involved. Tumors were surgically removed and taken to the Pathology Department for processing. A sample of each tumor was then brought to our laboratory for use in the present study. The sample was weighed and finely minced with a sterile scalpel into approximately one cm2 pieces.
Bone marrow cells
Bone marrow was obtained from healthy volunteers with informed consent as per the Institutional Review Board. Bone marrow cells used in this study were either: (1) whole unseparated bone marrow cells; (2) Ficoll-separated bone marrow cells isolated by a Ficoll-Paque Premium (GE Healthcare, Uppsala, Sweden) density gradient according to the manufacturer’s instructions; and (3) bone marrow cells with red blood cells removed using an ammonium chloride-based (ACL2) lysis buffer (BD Pharm Lyse lysing buffer, Becton Dickinson, Franklin Lakes, NJ, USA) according to the manufacturer’s instructions.
Co-culture
Experiments were performed using six well plates (BD Falcon, San Jose, CA, USA) plated with 3 × 106 bone marrow cells per well. Fifty to 100 mg of minced tumor was placed into a Millipore 0.4 um Millicell Culture plate insert (Millipore, Billerica, MA, USA) with 3 mL of growth media consisting of Delbecco’s Modified Eagles Medium (Invitrogen, Carlsbad, CA, USA) supplemented with 10% fetal bovine serum (Hyclone, South Logan, UT, USA), 1% penicillin/streptomycin (Invitrogen), and 20 ng/mL human stem cell factor (R&D Systems, Minneapolis, MN, USA). Control wells were cultured with equal numbers of bone marrow cells plated without tumor cell inserts. Co-cultures were maintained for 2 or 7 days at 5% CO2 at 37°C. In the case of conditioned media, tumor pieces were cultured without bone marrow cells and, after 7 days of culture, the cell-free conditioned media was removed and cultured with 3 × 106 bone marrow cells for an additional 7 days.
Isolation of lung tumor microvesicles
Microvesicles were isolated from cell-free conditioned media after 2 or 7 days of culture. The conditioned media was first centrifuged at 300 × g for 10 min at 4°C. The supernatant was then ultracentrifuged (UCF) at 28,000 × g for 1 hour at 4°C in a Thermo Scientific Sorval WX Ultra series ultracentrifuge (Thermo Scientific, Waltham, MA, USA). This spin was repeated, and the resulting pellet re-suspended in one × Dulbecco’s phosphate-buffered saline (PBS; Invitrogen). An equal volume of the red fluorescent cell membrane dye PKH26 (Sigma, St. Louis, MO, USA), diluted 1:250 in diluent C (Sigma), and the cell cytoplasm dye CFSE [5-(and 60-carboxyfluorescein diacetate, succinimidyl ester)] (Invitrogen), at a final concentration of 0 02 uM, were incubated with the pellet for 15 min at 37°C. An equal volume of 10% fetal bovine serum solution in PBS was added and the samples were UCF as before. The UCF pellet was either resuspended in growth media and co-cultured with bone marrow cells, or further processed for electron microscopy.
Transmission electron microscopy
The UCF pellets were fixed with 3% glutaraldehyde in 0.15 M sodium cacodylate buffer (Electron Microscopy Sciences, Hatfield, PA, USA) for several days at 4°C. Following several rinses, the samples were post-fixed in 1% osmium tetroxide (Electron Microscopy Sciences) for 1 hour at 4°C. After post-fixation, samples were diced in 1.5 mm cubes and covered with a 3% agar solution (Electron Microscopy Sciences). Once hardened, excess agar was removed and the sample was dehydrated through a graded series of acetone washes and embedded in Spurr’s epoxy resin (Ladd Research Industries, Williston, VT, USA). Semi-thin sections (1 μm) were prepared using a Reichert Ultracut-S microtome, stained with methylene blue-azure II, and evaluated for areas of interest. Ultra-thin sections were prepared, retrieved onto 300 mesh-thin bar copper grids, and contrasted with uranyl acetate and lead citrate (Electron Microscopy Sciences). Sections were examined using a Morgagni 268-transmission electron microscope, and images were collected with an AMT-Advantage 542 CCD camera system.
Fluorescence microscopy
Bone marrow cells co-cultured for 2 days with isolated microvesicles were harvested by gently rinsing the cells off the bottom of a T-75 tissue culture flask (BD Falcon) with PBS and spun at 300 × g for 10 min at 4°C. Bone marrow cells containing fluorescent-labeled microvesicles were sorted on an Influx cell sorter (BD Bioscience, San Jose, CA, USA) equipped with the following lasers: 100 mW 488 nm sapphire, 100 mW UV, 405 nm violet, 635 nm red, and a 561 nm green/yellow. The PKH26 was excited by the 561 nm laser and emission was detected through a 624/40 filter. The CFSE was excited using the 488 nm laser and emission was detected through a 528/38 filter. Cytospin slides were made using 1 × 105 cells per slide with a Shandon Cytospin 4 (Thermo Scientific), spinning the cells 350 revolutions per minute for 10 min. Vectashield with 1.5 μg/mL DAPI (Vector Laboratories, Burlingame, CA, USA) was added to each slide and images were taken using a Zeiss Axio Imager Z1 fluorescent microscope and Axiovision 4.6.3 software. Digital images were acquired using an AxioCam HRm. Three dimensional images were taken using a Zeiss ApoTome.
The RNA extraction and RT-PCR analysis
The RNA from cultured or co-cultured bone marrow cells was isolated using the RNeasy Mini or QIAmp RNA Blood Mini Kit (Qiagen, Valencia, CA, USA). The RNA was measured for quantity and quality (260/280) using a Nanodrop ND/1000 spectrophotometer (Thermo Scientific). Isolated RNA was used to amplify cDNA using High Capacity cDNA transcription kit (Applied Biosystems, Foster City, CA, USA) in a final volume of 20 μL. The amount of RNA used ranged from 10 ng to 500 ng; however, equal amounts of RNA were used for all samples in any given experiment. Amplification reactions were run on a 9800 Fast Thermal Cycler (Applied Biosystems) and consisted of one cycle for 10 min at 25°C, two 60 min cycles at 37°C, and one cycle for 5 sec at 85°C. Gene expression was analyzed by RT-PCR in 96-well plates on a 7900 HT-Fast Real Time PCR System (Applied Biosystems).The reactions were in a final volume of 25 μL consisting of: 20× assay mix (for either glyceraldehydes-3-phosphate dehydrogenase or one of the target genes), 2× TaqMan PCR Master Mix (Applied Biosystems), and predetermined amounts of cDNA. All 20× assay mixes were purchased from Applied Biosystems. Human lung assays were as follows: clara cell specific protein (CCSP, Hs00171092_m1), Surfactant A (SURF A, Hs 00359837_m1), Surfactant B (SURF B, Hs 01090667_m1), Surfactant C (SURF C, Hs 00161628_m1), Surfactant D (SURF D, Hs 00427523_m1), Aquaporin 1 (AQP1, Hs 00166067_m1), Aquaporin 3 (AQP3, Hs 00185020_m1), Aquaporin 4 (AQP4, Hs 00242342_m1), Aquaporin 5 (AQP5, Hs 00387048_m1), hTERT (Hs 00972646_m1), and glyceraldehydes-3-phosphate dehydrogenase (Hs 99999905_m1). The PCR reaction consisted of an initial enzyme activation step at 95°C for 10 min, followed by 40 cycles of 95°C for 15 sec, and 60°C for 1 min. A cycle threshold (CT) value was obtained for each sample, and triplicate sample values were averaged. The 2−ΔΔT method was used to calculate relative expression of each target gene. Briefly, mean CT value of target genes in each sample were normalized to its averaged housekeeping gene CT to give a ΔCT value. This was then normalized to the control samples (ΔΔCT), and the 2−ΔΔT value was obtained. To calculate 2−ΔΔT for target genes with no expression in the control group, a CT value of 40 was assigned to the control group so that a relative quantity of the target gene could be reported. The control group used for all comparisons was bone marrow cells co-cultured without lung tumor, conditioned media, or microvesicles for the same duration as the experimental groups.
Statistics
Data were analyzed using the Wilcoxon rank sum test. We considered results to be statistically significant only when p < 0.05 (two-sided).
Results
The co-culture system is illustrated in Figure 1. Fifty to 100 mg of lung tissue was co-cultured opposite 3 × 106 human bone marrow cells for 2 or 7 days, and real-time RT-PCR analysis was then performed on co-cultured marrow cells. Clinical samples were obtained from patients undergoing a variety of procedures to obtain tissue for diagnostic purposes. In most cases, the patients were undiagnosed. Informed consent was obtained and, after the Pathology Department obtained appropriate diagnostic material, aliquots of tumor tissue were provided for our experimental studies. The patients in this study had a variety of non-small-cell lung cancers; 1 patient had a lung melanoma, and 1 patient had a lung sarcoma. These patients also had a variety of comorbid medical conditions which could influence microvesicle generation. In addition, 2 patients had a history of bladder carcinoma, 1 patient had a history of Hodgkin’s lymphoma, and 1 patient was undergoing treatment for chronic lymphocytic leukemia. Patient demographics are outlined in Table 1.
Figure 1.
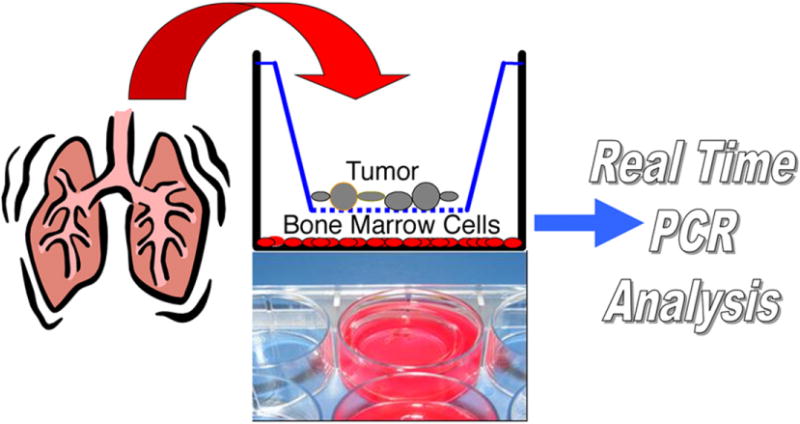
Experimental design. After surgery, the lung tumor sample was placed into a 0.4 um impermeable filter insert and co-cultured with bone marrow cells. After 2 or 7 days, the bone marrow cells were harvested and their RNA was isolated and analyzed using reverse transcriptase polymerase chain reaction (RT-PCR).
Table 1.
Patient demographics
| Patient# | Gender | Age | Diagnosis | Current medications |
|---|---|---|---|---|
| 1 | F | 40 | Adenocarcinoma | Prozac, synthroid, glipizide, actos, Lipitor, wellbutrin, hydrocodone, lorazepam, Maxalt |
| 2 | F | 66 | Endobronchial adenocarcinoma | Advair, albuterol, Triametenane, aspirin |
| 3 | M | 60 | Bronchioloalveolar carcinoma | Albuterol, Lipitor, Flomax, Celexa, Coumadin |
| 4 | M | 70 | Non-small-cell lung cancer | Lisinopril, Prilosec, alprazolam |
| 5 | F | 60 | Squamous cell carcinoma of the lung | Lipitor, effexor, Ambien |
| 6 | M | 79 | Adenosquamous carcinoma | Plavix, Coreg, Renagel, Nephrocaps, insulin, Epogen, Proscar |
| 8 | M | 67 | Adenocarcinoma | Benicar, Rituxan |
| 9 | M | 72 | Adenocarcinoma | Cisplatin |
| 10 | M | 79 | Squamous cell carcinoma | Simvastatin, terazosin, Fluconazole, aspirin, niacin, isosorbide mononitrate, Amlodipine |
| 11 | M | 55 | Melanoma | Allopurinol |
| 12 | F | 60 | Sarcoma | Lipitor, Lisinopril, Trazodone |
| 13 | M | 59 | Squamous cell carcinoma | Aspirin, Colchicine, Lipitor, Lisinopril, Metoprolol |
F = Female; M = male.
We selected genes with relative specificity (except for hTERT) for lung cells. They are listed with a brief description of function in Table 2.
Table 2.
Genes with lung specificity and their function
| Gene | Name | Description | Cell specificity |
|---|---|---|---|
| QP 1 | Aquaporin 1 | Aquaporins are a family of intrinsic membrane proteins that function as water-selective channels in the plasma membrane. AQP 1 functions in alveolar hydration. | Type I pneumocyte |
| AQP 3 | Aquaporin 3 | AQP 3 is the channel for water out of the cell. | Type I pneumocyte |
| AQP 4 | Aquaporin 4 | AQP 4 functions on bronchial fluid secretion. | Type I pneumocyte |
| AQP 5 | Aquaporin 5 | AQP 5 plays a role in mediating the sheer effects on paracellular permeability. | Type I pneumocyte |
| Surf A | Surfactant A | The lipid rich material that prevent lung collapse by lowering surface tension at the air-liquid interface in the lung alveoli. | Type II pneumocyte |
| Surf B | Surfactant B | Surf B enhances the rate of spreading and increases the stability of surfactant monolayers. | Type II pneumocyte |
| Surf C | Surfactant C | Surf C functions in alveolar stability by lowering the surface tension at the air-liquid interface. | Type II pneumocyte |
| Surf D | Surfactant D | Surf D binds to a variety of lung pathogens and enhances the opsonization and killing by phagocytic cells. | Type II pneumocyte |
| hTERT | Human telomerase reverse transcriptase | hTERT is an enzyme that adds specific DNA sequence repeats (“TTAGGG”) to the three prime end of DNA strands in the telomere regions found at the ends of eukaryotic chromosomes. | |
| CCSP | Clara specific cell protein | CCSP functions to inhibit pulmonary inflammation of surfactant proteins, which prevent alveolar collapse. | Clara cells |
The results shown in Figure 2 summarize gene expression of marrow cells co-cultured with minced lung tumor tissue. Gene expression was considered positive if it was over twofold greater than that of control cells. In these experiments, co-cultured marrow cells demonstrated varied expression of lung-specific genes, but in every case there was expression of at least one of these genes. In co-cultures with tumor from patient #4 (non-small-cell carcinoma), after 2 days of co-culture, whole bone marrow cells showed increased expression of AQP1, 3, and 5, and SURF C, while Ficoll marrow cells showed increased expression of AQP1 and SURF D. Conversely, after 2 days of co-culture, the ACL2 sample showed expression of SURF B and SURF D. These were not seen in the 7-day co-culture samples. The SURF D expression persisted in the 7-day co-cultured Ficoll marrow cells. Marrow cells co-cultured with adenocarcinomas showed similar variation in gene expression as did co-cultures with other tumors. Marrow cells co-cultured with conditioned media and microvesicles for 7 days (Fig. 3A, B) expressed elevated levels of lung-specific genes; however, levels varied from sample to sample. Under different co-culture conditions and using different marrow cell preparations, marrow cells co-cultured with all of the lung-derived cancer samples showed increased expression of between one and seven lung-specific genes, with a range of expression of up to > 1,000-fold. Patients #11 and #12 had lung melanoma and sarcoma, respectively. Marrow cells co-cultured with these tumors also expressed lung-specific genes. The most consistently expressed lung-specific genes were AQP1 (10 patients) and SURF B (9 patients), while AQP 4 was only expressed in co-cultures derived from 2 patient tumor samples. Co-cultures derived from different patient tumor samples expressed a different number of lung-specific genes (patient #8, 7 genes; patients #4 and #5, 5 genes; patient #13, 1 gene). Altogether, expression of lung-specific genes in co-cultured marrow cells was statistically significant (p < 0.02).
Figure 2.
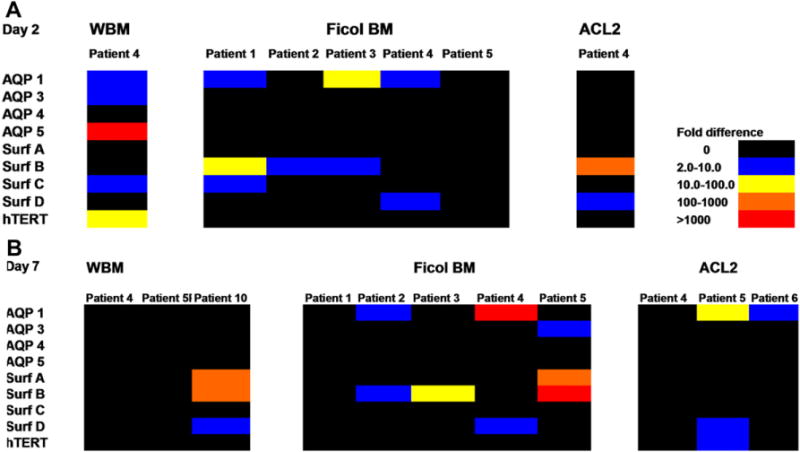
Lung tumor samples co-cultured with human bone marrow cells for 2 or 7 days. Colors indicate fold increases of lung-specific mRNA levels as determined by reverse transcriptase polymerase chain reaction (RT-PCR) when compared to marrow cells cultured without lung tumor. Graph illustrates bone marrow cells that were co-cultured with lung tumor for (A) 2 days or (B) 7 days. The following bone marrow cell populations were used in co-culture: WBM = Whole bone marrow; Ficoll BM = Ficoll-separated bone marrow cells; ACL2 = bone marrow cells with red blood cells removed using an ammonium chloride-based lysis buffer.
Figure 3.
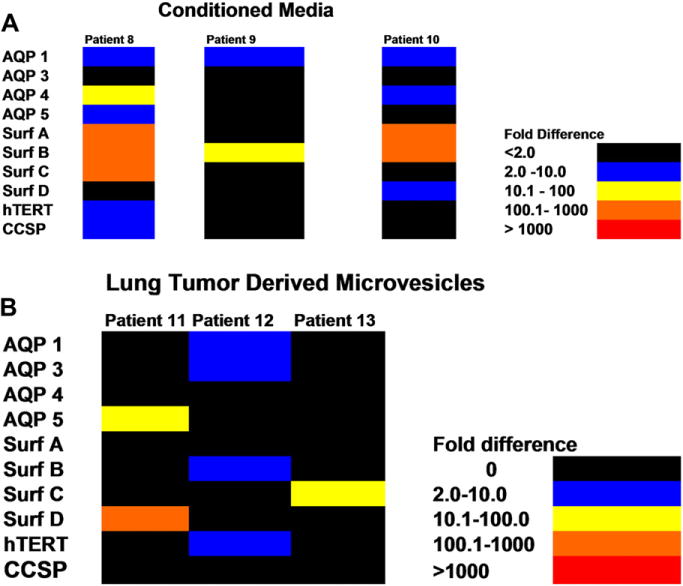
Conditioned media or lung tumor-derived microvesicles co-cultured with human bone marrow cells for 7 days. (A) Bone marrow cells co-cultured with conditioned media, or (B) lung tumor-derived microvesicles. Colors indicate fold increases of lung-specific RNA levels as determined by reverse transcriptase polymerase chain reaction (RT-PCR) when compared to marrow cells cultured without conditioned media or microvesicles.
Figure 4 shows marrow cells which have taken up PKH26 (red fluorescence) and CFSE (green fluorescence)-labeled microvesicles isolated from lung tumor tissue (melanoma) from patient #11 (Fig. 4A–C). Figure 4D represents a marrow cell which did not take up microvesicles. Figure 5 shows transmission electron microscopy of vesicles isolated from lung tumor tissue (adenocarcinoma) from patient #8 (Fig. 5A–C) reveal that they have a morphologic appearance consistent with microvesicles. The hematoxylin and eosin stained pathology is shown in Figure 5D.
Figure 4.
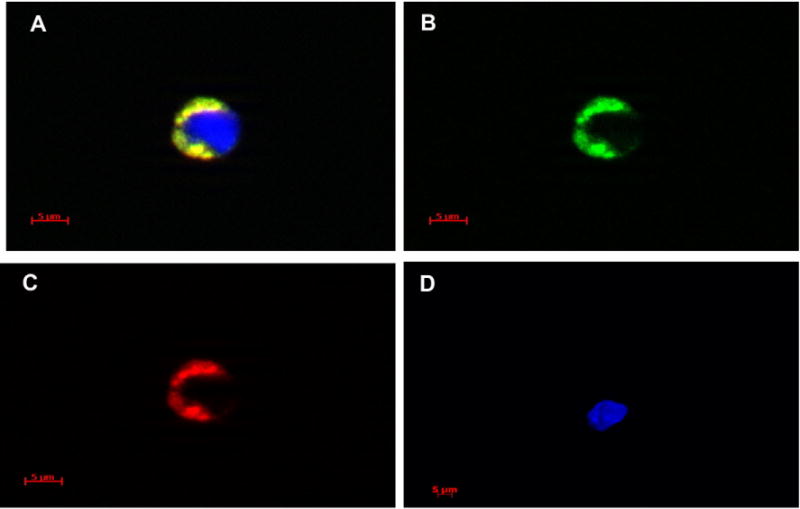
Digital images of bone marrow cells containing microvesicles. Examples of bone marrow cells containing PKH26 and CFSE labeled microvesicles are shown (A–C). (A) Composite image of one bone marrow cell taken through DAPI, FITC, and Texas Red filters. Blue color is DAPI (nuclear counter stain). (B, C) Images of the same cell taken through FITC and Texas Red filters, respectively. (D) Example of DAPI counterstained cell that has not taken up microvesicles image taken using DAPI, FITC, and Texas Red filters. Fluorescent images were taken with a Zeiss Axio Imager Z1 fluorescent microscope at 63 × magnification and Axiovision 4.6.3 software. Digital images were acquired using an AxioCam HRm. Three-dimensional images were taken utilizing a Zeiss ApoTome for structural imaging and a four-dimensional acquisition module.
Figure 5.
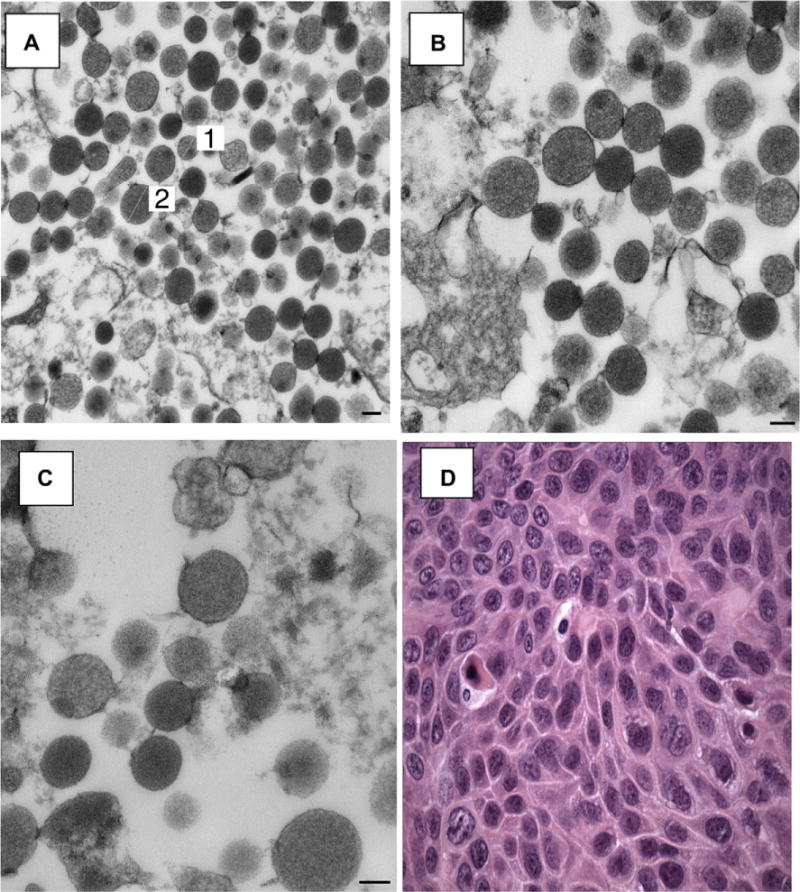
Transmission electron microscope images of lung tumor-derived microvesicles. Lung tumor sample from patient #8 was cultured without bone marrow cells for 7 days, the conditioned media removed and microvesicles were isolated by high speed centrifugation. (A) Microvesicles, 44,000 × magnification, (B) 56,000 × magnification, (C) 71,000× magnification. Magnification bars are all equal to 100 nm. (D) Hematoxylin and eosin (H&E) stained pathology slide showing lung tumor cells from the same patient, 600 × magnification. Electron microscope sections were examined using a Morgagni 268-transmission electron microscope, and images were collected with an AMT Advantage 542 CCD camera system.
Discussion
The present work is the first to show microvesicle evolution from explant fresh human lung cancer cells and microvesicle transfer of genetic phenotype to human marrow cells. In these studies, we have shown that cells from 12 lung-derived cancers induced lung-specific mRNA in target marrow cells, either by tissue co-culture with cells separated by a 0.4 μm-cell impermeable membrane, by incubation in conditioned media, or by exposure to pelleted microvesicles. These lung-specific mRNA increases, compared to control marrow cells, varied from each tumor co-culture, but all 12 cancers induced between one and seven of the designate lung-specific genes (p < 0.02). Increased expression of AQP1 was seen in 10 tumor co-culture samples and AQP4 in two samples. There was similar variability with the surfactant genes: SURF A was expressed in three of the tumor co-culture samples, SURF B in nine, SURF C in four, and SURF D in four. Increased expression of lung-specific mRNA in co-cultured marrow cells was seen with 2 or 7 day co-cultures, and co-culture with conditioned media for 7 days seemed to give the most robust expression. Expression of lung-specific mRNA in co-cultured marrow cells was also seen whether whole marrow, ammonium chloride lysed marrow, or Ficoll-separated marrow was evaluated. In general, tumors evaluated by pathology consisted predominantly of cancer cells, although contributions of microvesicles from associated normal cells cannot be excluded in these experiments. Microvesicles were identified in the pellet of an UCF conditioned media sample, and sizes ranged from those typical of exosomes to those typical of microvesicles. Finally, we also demonstrated that microvesicles purified by fluorescent staining and fluorescence activated cell sorting entered a minority of marrow cells.
These data are strikingly similar to those we obtained studying mouse lung microvesicle alteration of mouse marrow genetic and functional phenotype [15,16] and, as with those studies, suggest that cancer cell phenotype might be exportable. It is of interest, but perhaps not too surprising, that marrow cells co-cultured with lung tumors, which turned out to be melanoma and sarcoma, expressed lung-specific genes. Marrow cells co-cultured with melanoma expressed AQP5 and SURF D, while marrow cells co-cultured with sarcoma expressed AQP 1 and 2 and SURF B. We assume that this reflects lung microenvironmental influences on the tumor situated in the lung. This raises interesting possibilities of bidirectional cross talk between cancers and the normal host tissue. Tumor-derived microvesicles were first noted in the 1970s [17] and, since this time, effects of normal or tumor cell-derived microvesicles on cancer cells or their environment have been noted to impact cell survival, escape of immune surveillance, degradation of extracellular matrix, angiogenesis, and metastases. More recent work with a murine melanoma model has shown that tumor-derived microvesicles can enhance metastatic potential of melanoma cells in vivo [18]. Work with human cancer cell lines has demonstrated the delivery of oncogenic estimated glomerular filtration rate via microvesicles to cultured endothelial cells [19], and microvesicles seem to be generated by human prostate [20] and colorectal cell lines [21] as well. A report by Skog et al. [22], who studied human glioblastoma tissues obtained from surgical resections, showed release of microvesicles containing mRNA, microRNA, and angiogenic proteins. These microvesicles were taken up by normal host cells, including brain microvascular endothelial cells. Furthermore, mRNA mutant variants and microRNAs characteristic of gliomas could be detected in microvesicles isolated from the serum of glioblastoma patients. Studies by Wysoczynski and Ratajczak [23] showed that both human and murine lung cancer cell lines secrete microvesicles, and that secretion was increased by irradiation and hypoxia. These tumor-derived microvesicles enhanced metastatic potential of both murine and human lung cancer cells in vivo. Our studies extend these observations to human explant lung cancer and show entrance of lung cancer-derived microvesicles into human marrow cells. They also indicate that this seems to be a universal phenomenon with lung cancers. The variability in lung cancer gene expression could be related to different conditions in specific experiments and the different tumors under study.
The relative stability of genetic cellular phenotype has been a staple of both cell and cancer biology. Our previous work with normal murine lung and marrow cell [17,18], and the present work with excised human lung cancer and human marrow, suggest that the systems are less stable than previously considered. The implications of this work are that lung cancer cells might alter blood cells or other normal cells toward a cancer cell phenotype. The possibility of stable alteration of genetic phenotype of normal cells by cancer cells introduces a number of potentially important concepts. The possible alteration of a normal cell to a neoplastic phenotype could be a means of local recurrence, progression, or distant dissemination. The observation that cancer tissue can alter the phenotype of adjacent normal marrow cells as shown here, and that these alterations may be due to transfer of cancer-derived microvesicles is, of course, a first step. The patient population in this study had many comorbid conditions including 4 patients with history of other cancers. The variables here are quite large and probably do influence the generation of microvesicles in this experimental setting. In addition, the excised cancer tissue studied will have associated normal lung cells. The possibility of a contribution by normal cells to the target cell phenotype cannot be ruled out, although given the pathology of the specimens which are largely tumor cells, this seems less likely.
Acknowledgments
Financial support: This publication was made possible by National Institute of Health Grant #1P20 RR025179-01, 1P20 RR017695-01 (Douglas C. Hixson), and 5KO8 HL086868-03 (Jason M. Aliotta) and its contents are solely the responsibility of the authors and do not necessarily represent the official views of National Institute of Health/National Center for Research Resources.
References
- 1.Morel O, Toti F, Hugel B, Freyssinet JM. Cellular microparticles: a disseminated storage pool of bioactive vascular effectors. Curr Opin Hematol. 2004;11:156–164. doi: 10.1097/01.moh.0000131441.10020.87. [DOI] [PubMed] [Google Scholar]
- 2.Janowska-Wieczorek A, Majka M, Kijowski J, et al. Platelet-derived microparticles bind to hematopoietic stem/progenitor cells and enhance their engraftment. Blood. 2001;98:3143–3149. doi: 10.1182/blood.v98.10.3143. [DOI] [PubMed] [Google Scholar]
- 3.Baj-Krzyworzeka M, Majka M, Pratico D, et al. Platelet-derived microparticles stimulate proliferation, survival, adhesion, and chemotaxis of hematopoietic cells. Exp Hematol. 2002;30:450–459. doi: 10.1016/s0301-472x(02)00791-9. [DOI] [PubMed] [Google Scholar]
- 4.Rozmyslowicz T. Platelet- and megakaryocyte-derived microparticles transfer CXCR4 receptor to CXCR4-null cells and make them susceptible to infection by X4-HIV. AIDS. 2003;17:33–42. doi: 10.1097/00002030-200301030-00006. [DOI] [PubMed] [Google Scholar]
- 5.Graves LE, Ariztia EV, Navari JR, Matzel HJ, Stack MS, Fishman DA. Proinvasive properties of ovarian cancer ascites-derived membrane vesicles. Cancer Res. 2004;64:7045–7049. doi: 10.1158/0008-5472.CAN-04-1800. [DOI] [PubMed] [Google Scholar]
- 6.Fackler OT, Peterlin BM. Endocytic entry of HIV-1. Curr Biol. 2000;10:1005–1008. doi: 10.1016/s0960-9822(00)00654-0. [DOI] [PubMed] [Google Scholar]
- 7.Fevrier B, Vilette D, Archer F, et al. Cells release prions in association with exosomes. Proc Natl Acad Sci U S A. 2004;101:9683–9688. doi: 10.1073/pnas.0308413101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Greco V, Hannus M, Eaton S. Argosomes: a potential vehicle for the spread of morphogens through epithelia. Cell. 2001;106:633–645. doi: 10.1016/s0092-8674(01)00484-6. [DOI] [PubMed] [Google Scholar]
- 9.Speck RF, Esser U, Penn ML, et al. A trans-receptor mechanism for infection of CD4-negative cells by human immunodeficiency virus type 1. Curr Biol. 1999;9:547–550. doi: 10.1016/s0960-9822(99)80241-3. [DOI] [PubMed] [Google Scholar]
- 10.Ratajczak J, Miekus K, Kucia M, et al. Embryonic stem cell-derived microvesicles reprogram hematopoietic progenitors: evidence for horizontal transfer of mRNA and protein delivery. Leukemia. 2006;20:847–856. doi: 10.1038/sj.leu.2404132. [DOI] [PubMed] [Google Scholar]
- 11.Baj-Krzyworzeka M, Szatanek R, Weglarczyk K, et al. Tumour-derived microvesicles carry several surface determinants and mRNA of tumour cells and transfer some of these determinants to monocytes. Cancer Immunol Immunother. 2006;55:808–818. doi: 10.1007/s00262-005-0075-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Holmgren L, Bergsmedh A, Spetz AL. Horizontal transfer of DNA by the uptake of apoptotic bodies. Vox Sang. 2002;83:305–306. doi: 10.1111/j.1423-0410.2002.tb05323.x. [DOI] [PubMed] [Google Scholar]
- 13.Hakelien AM, Landsverk HB, Robl JM, Skalhegg BS, Collas P. Reprogramming fibroblasts to express T-cell functions using cell extracts. Nature Biotechnol. 2002;20:460–466. doi: 10.1038/nbt0502-460. [DOI] [PubMed] [Google Scholar]
- 14.Deregibus MC, Cantaluppi V, Calogero R, et al. Endothelial progenitor cell-derived microvesicles activate an angiogenic program in endothelial cells by a horizontal transfer of mRNA. Blood. 2007;110:2440–2448. doi: 10.1182/blood-2007-03-078709. [DOI] [PubMed] [Google Scholar]
- 15.Aliotta JM, Sanchez-Guijo FM, Dooner GJ, et al. Alteration of marrow gene expression, protein production, and engraftment into lung by lung-derived microvesicles: a novel mechanism for phenotype modulation. Stem Cells. 2007;9:2245–2256. doi: 10.1634/stemcells.2007-0128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Aliotta JM, Pereira M, Johnson KW, et al. Microvesicle entry into marrow cells mediates tissue-specific changes in mRNA by direct delivery of mRNA and induction of transcription. Exp Hematol. 2010;38:233–245. doi: 10.1016/j.exphem.2010.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Friend C, Marovitz W, Henie G, et al. Observations on cell lines derived from a patient with Hodgkin’s disease. Cancer Res. 1978;38:2581–2591. [PubMed] [Google Scholar]
- 18.Lima LG, Chammas R, Monteiro RQ, Moreira ME, Barcinski MA. Tumor-derived microvesicles modulate the establishment of metastatic melanoma in a phosphatidyserine-dependent manner. Cancer Letters. 2009;283:168–175. doi: 10.1016/j.canlet.2009.03.041. [DOI] [PubMed] [Google Scholar]
- 19.Al-Nedawi K, Meehan B, Micallef J, et al. Intercellular transfer of the oncogenic receptor EGFRvIII by microvesicles derived from tumor cells. Nat Cell Biol. 2008;10:619–624. doi: 10.1038/ncb1725. [DOI] [PubMed] [Google Scholar]
- 20.Di Vizio D, Kim J, Hager MH, et al. Oncosome formation in prostate cancer: association with a region of frequent chromosomal deletion in metastatic disease. Cancer Res. 2009;69:5601–5609. doi: 10.1158/0008-5472.CAN-08-3860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Choi DS, Lee JM, Park GW, et al. Proteomic analysis of microvesicles derived from human colorectal cancer cells. J Proteome Res. 2007;6:4646–4655. doi: 10.1021/pr070192y. [DOI] [PubMed] [Google Scholar]
- 22.Skog J, Wurdinger T, van Rijn S, et al. Glioblastoma microvesicles transport RNA and proteins that promote tumour growth and provide diagnostic biomarkers. Nat Cell Biol. 2008;10:1470–1476. doi: 10.1038/ncb1800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wysoczynski M, Ratajczak MZ. Lung cancer secreted microvesicles: underappreciated modulators of microenvironment in expanding tumors. Int J Cancer. 2009;125:1595–1603. doi: 10.1002/ijc.24479. [DOI] [PMC free article] [PubMed] [Google Scholar]


