Abstract
Background
In the isolated population of Sardinia, a Mediterranean island, ~25% of ALS cases carry either a p.A382T mutation of the TARDBP gene or a GGGGCC hexanucleotide repeat expansion in the first intron of the C9ORF72 gene.
Objective
To describe the co-presence of two genetic mutations in two Sardinian ALS patients.
Methods
We identified two index ALS cases carrying both the p.A382T missense mutation of TARDBP gene and the hexanucleotide repeat expansion of C9ORF72 gene.
Results
The index case of Family A had bulbar ALS and frontemporal dementia (FTD) at 43. His father, who carried the hexanucleotide repeat expansion of C9ORF72 gene, had spinal ALS and FTD at 64 and his mother, who carried the TARDBP gene p.A382T missense mutation, had spinal ALS and FTD at 69. The index case of Family B developed spinal ALS without FTD at 35 and had a rapid course to respiratory failure. His parents are healthy at 62 and 63. The two patients share the known founder risk haplotypes across both the C9ORF72 9p21 locus and the TARDBP 1p36.22 locus.
Conclusions
Our data show that in rare neurodegenerative causing genes can co-exist within the same individuals and are associated with a more severe disease course.
Introduction
Amyotrophic lateral sclerosis (ALS) is a fatal neurodegenerative disease characterised by upper and lower motor neuron dysfunction resulting in rapidly progressive paralysis and death from respiratory failure.1 Population-based epidemiological studies estimate 5% of cases to be familial in nature.2 Of these, approximately 30%–40% are caused by a large GGGGCC hexanucleotide repeat expansions of the C9ORF72 gene,3,4 15% by mutations in the SOD1 gene,2 and a further ~10% of cases are due to pathogenic variants in the TARDBP, FUS and VCP genes.5–8
We recently reported that two mutations, namely the pathogenic repeat expansion of C9ORF72 and the p.A382T missense mutation of TARDBP, together account for ~60% of familial ALS cases and ~25% of apparently sporadic ALS cases on the Mediterranean island of Sardinia.9,10 During those mutational screening projects, we identified two apparently unrelated ALS patients who carried both of these mutations. The purpose of the current manuscript is to provide a clinical description of the extended kindred of these unusual patients, highlighting the phenotypes associated with the different mutations observed within the families.
Case reports
Family A
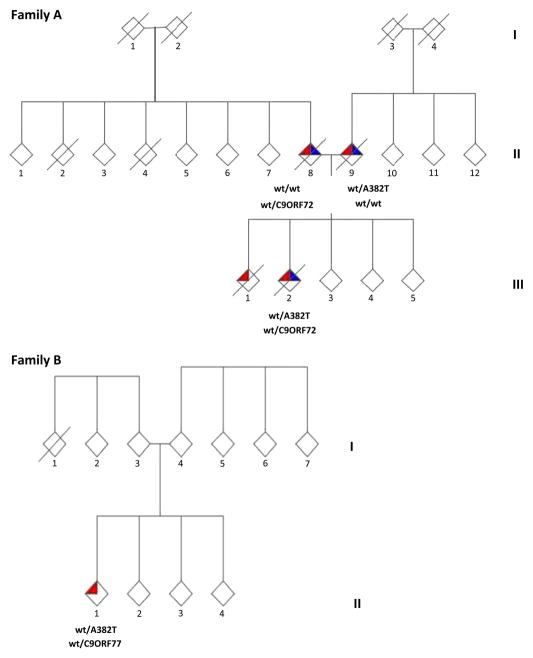
This family has been partially reported in a previous paper.11 The index case (II-2) developed bulbar-onset ALS at 43 years of age with frontotemporal dementia (FTD) occurring 1 year later. Detailed cognitive testing (see Supplementary data) performed 1 year after the onset of ALS demonstrated cognitive and behavioural dysfunction consistent with the Neary’s diagnostic criteria for FTD. The Frontal Systems Behaviour Scale (family rating) showed a prominent impairment of the apathy and executive functions domains. He died 34 months after symptom onset. He carried both the c.1144G→A (p.A382T) missense mutation of the TARDBP gene and a hexanucleotide repeat expansion of C9ORF72 gene. His father (II-8), who also carried a hexanucleotide repeat expansion of C9ORF72 gene, developed spinal onset ALS at 64, and behavioural FTD 2 years later. He died 78 months after the onset of ALS. His mother (II-9) developed spinal onset ALS at 69 and a mild FTD 6 months later. She died from respiratory failure 43 months after the onset of ALS. She carried the p.A382T missense mutation of the TARDBP gene. The proband’s brother (III-1) developed ALS at 33 years of age and died 75 months after the onset of ALS, without cognitive impairment. DNA was not available for this patient. The maternal grandfather (I-3) died at the age of 43 in a work-related accident, and the maternal grandmother (I-4) died at the age of 91 from a cerebrovascular accident. The paternal grandfather died at the age of 47 (I-1) also in a work-related accident, and the paternal grandmother (I-2) at the age of 74 from breast cancer (figure 1).
Figure 1.
(A) Pedigree of Family A. (B) Pedigree of Family B. C9ORF72 GGGGCC hexanucleotide repeat expansions are shown by C9, whereas wild-type alleles are indicated by wt. p.A382T missense mutations of TARDBP gene are indicated as A382T, whereas wild-type alleles are indicated by wt. Red triangles represent a diagnosis of ALS, blue triangles represent FTD and a diagonal slash indicates the subject is deceased. Gender of the pedigree members is obscured to protect privacy. Chromatograms of part of exon 6 of TARDBP showing c.1144G→A (p.A382T) mutations and of the C9ORF72 GGGGCC hexanucleotide repeat expansions are shown in Supplementary figures 1 and 2.
Family B
The proband (II-1) developed proximal muscle weakness and wasting of the upper limbs at 35 years of age. Neurological examination revealed diffuse hyperreflexia, and neurophysiological testing showed chronic and active denervation. Motor evoked potentials demonstrated increased central conduction time of both corticospinal tracts. His presentation was consistent with the flail arm variant of ALS and he was diagnosed with definite ALS according to the El Escorial diagnostic criteria. He had no evidence of cognitive impairment after formal testing. He developed respiratory failure and was electively tracheotomised 25 months after the onset of ALS. He remains alive, 42 months after the onset of ALS. Genetic analysis revealed that he carried both a hexanucleotide repeat expansion of C9ORF72 gene and the c.1144G→A (p.A382T) missense mutation of the TARDBP gene. His father (I-3) is 62 years of age and his mother (I-4) is 63; neither had signs of ALS, FTD or other neurodegenerative diseases. He has three healthy sibs (II-2/4) aged between 34 and 37. No other relative within the pedigree was known to have ALS or FTD. Both the parents and the siblings of the patient refused to undergo genetic analysis (figure 1).
Gene haplotype
The haplotype across the TARDBP gene on chromosome 1p36.22 and across the C9ORF72 gene on chromosome 9p21 (tables 1 and 2) of the two index cases (Family A, case II-1, and Family B, case II-1) demonstrated that they carried the known founder risk haplotypes for both genes.9,12
Table 1.
SNP genotype across the TARDBP gene on chromosome 1p36.22
| SNP | C hr | Position | Risk allele | Genotype | |
|---|---|---|---|---|---|
| Family A/II-2 | Family B/II-1 | ||||
| rs1925666 | 1 | 10999600 | A | AG | AA |
| rs6656310 | 1 | 11002488 | T | TT | TT |
| rs1281008 | 1 | 11011002 | G | AG | GG |
| rs11121663 | 1 | 11024395 | T | CT | TT |
| rs2003046 | 1 | 11032827 | C | AC | AC |
| rs12059717 | 1 | 11043485 | C | CC | CC |
| rs9430161 | 1 | 11046855 | G | GT | GT |
| rs6704113 | 1 | 11053101 | A | AA | AA |
| rs11121675 | 1 | 11060075 | T | GT | TT |
| rs11121676 | 1 | 11063043 | C | CC | CC |
| rs3765896 | 1 | 11074420 | A | AG | AA |
| rs2273348 | 1 | 11079077 | C | CT | CC |
| rs1033638 | 1 | 11086717 | C | CT | CC |
| rs12711521 | 1 | 11090916 | A | AC | AA |
| rs7548659 | 1 | 11107439 | T | GT | TT |
| rs12121344 | 1 | 11111629 | C | CC | CC |
| rs2802211 | 1 | 11132217 | A | AG | AA |
| rs2536 | 1 | 11166713 | T | TT | TT |
| rs2275525 | 1 | 11169676 | G | GG | GG |
| rs1057079 | 1 | 11205058 | A | AG | AA |
| rs7540001 | 1 | 11243833 | A | AA | AA |
| rs1074078 | 1 | 11326788 | C | CT | CC |
| rs3010223 | 1 | 11372638 | A | AG | AA |
| rs2982373 | 1 | 11377114 | G | AG | GG |
| rs909836 | 1 | 11398365 | C | CT | CT |
| rs10864498 | 1 | 11409469 | G | AG | AG |
| rs2982364 | 1 | 11432744 | C | CC | CT |
| rs2788557 | 1 | 11439588 | G | GG | AG |
| rs3010208 | 1 | 11444975 | T | TT | CT |
| rs6660541 | 1 | 11447805 | G | GG | AG |
| rs6685668 | 1 | 11447850 | C | CC | CC |
| rs12058244 | 1 | 11456027 | G | GG | GG |
| rs6671983 | 1 | 11461601 | G | GG | AG |
| rs10864504 | 1 | 11476203 | G | GG | AG |
| rs877309 | 1 | 11489678 | G | GG | AG |
| rs10864508 | 1 | 11490744 | T | TT | GT |
| rs2922240 | 1 | 11493832 | C | CC | CT |
| rs2379151 | 1 | 11494573 | G | GG | AG |
| rs7414229 | 1 | 11498299 | T | TT | TT |
| rs11806225 | 1 | 11499286 | T | TT | TT |
| rs3124627 | 1 | 11508016 | A | AA | AG |
| rs12026602 | 1 | 11518397 | T | TT | CT |
| rs11121742 | 1 | 11520653 | T | TT | GT |
| rs12046038 | 1 | 11525533 | A | AA | AA |
| rs17036930 | 1 | 11531918 | C | CC | CC |
| rs2007215 | 1 | 11537977 | G | GG | AG |
| rs7555562 | 1 | 11542957 | A | AA | AC |
| rs17036947 | 1 | 11575260 | G | GG | GG |
| rs3887379 | 1 | 11577831 | T | TT | TT |
| rs2072996 | 1 | 11578973 | A | AA | AA |
| rs2076469 | 1 | 11584164 | T | TT | TT |
| rs2076468 | 1 | 11589911 | C | CC | CC |
| rs2235660 | 1 | 11595331 | C | CC | CT |
| rs2817621 | 1 | 11601119 | T | TT | TT |
| rs11808316 | 1 | 11619154 | C | CC | CC |
| rs2817581 | 1 | 11622979 | C | CC | CC |
| rs2745285 | 1 | 11626858 | A | AA | AA |
| rs2745275 | 1 | 11645598 | C | CC | CC |
| rs7530509 | 1 | 11654491 | G | GG | GG |
The Risk allele column is the previously identified Sardinian founder risk haplotype across this locus as published in Chiò et al.9
The Genotype columns show the single-nucleotide polymorphism data for the probands of Family A and Family B.
The shared allele is highlighted in bold.
Table 2.
Haplotype across the C9ORF72 gene on chromosome 9p21 in the two ALS patients
| SNP | C hr | Position | Risk allele | Genotype | |
|---|---|---|---|---|---|
| Family A/II-2 | Family B/II-1 | ||||
| rs765709 | 9 | 27451960 | A | AA | AA |
| rs702231 | 9 | 27588731 | A | AC | AA |
| rs2589054 | 9 | 27443802 | G | AG | AA |
| rs3849942 | 9 | 27543281 | A | AG | AA |
| rs1982915 | 9 | 27579560 | G | GG | AG |
| rs2453556 | 9 | 27586162 | G | GG | AG |
| rs1110155 | 9 | 27386505 | A | AA | CC |
| rs725804 | 9 | 27458939 | A | AC | CC |
| rs1977661 | 9 | 27502986 | C | CC | CC |
| rs1058326 | 9 | 27447089 | C | CT | CC |
| rs1822723 | 9 | 27478052 | C | CT | CC |
| rs774359 | 9 | 27561049 | C | CT | CC |
| rs1948522 | 9 | 27575785 | C | CT | CC |
| rs2150336 | 9 | 27402961 | C | TT | CC |
| rs944404 | 9 | 27450211 | T | TT | CC |
| rs868856 | 9 | 27489251 | T | CT | CT |
| rs7046653 | 9 | 27490967 | A | AG | AG |
| rs2814707 | 9 | 27536397 | A | AG | AG |
| rs1161680 | 9 | 27430232 | G | AA | GG |
| rs10122902 | 9 | 27556780 | G | AG | GG |
| rs1565948 | 9 | 27559733 | G | AG | GG |
| rs2282241 | 9 | 27572255 | G | GT | GG |
| rs903603 | 9 | 27529316 | C | CC | CT |
| rs2477518 | 9 | 27599746 | T | CT | TT |
| rs1330921 | 9 | 27367278 | T | TT | TT |
| rs10511816 | 9 | 27468461 | T | TT | TT |
| rs895023 | 9 | 27483959 | T | TT | TT |
| rs12349820 | 9 | 27553876 | T | TT | TT |
The Risk allele column is the previously identified Sardinian founder risk haplotype across this locus as published in Mok et al.12
The Genotype columns show the single-nucleotide polymorphism data for the probands of Family A and Family B.
The shared allele is highlighted in bold.
ALS, amyotrophic lateral sclerosis.
The ethical committees of all the institutions involved approved the study, and all patients and healthy controls gave written informed consent.
Discussion
Sardinia, with a population of 1.7 million, is the second largest Mediterranean island located 120 miles west of the main Italian coastline. Despite numerous invasions over the millennia, genetic, linguistic and archaeological studies indicate that Sardinians have a phylogeny distinct from other Europeans, including mainland Italians.13,14 We and others have reported that a large percentage of ALS patients of Sardinian ancestry carry the p c.1144G→A (p.A382T) missense mutation of the TARDBP gene, namely ~35% of FALS and ~19% of apparently sporadic cases.9,15 More recently, we determined that an additional ~35% of FALS and ~7% of apparently SALS of the same island population carry a hexanucleotide repeat expansion of C9ORF72 gene.10,16 These high mutational rates for both ALS-causing mutations most likely reflect genetic drift within a relatively small island population. Thus, it is perhaps not surprising that we found two patients carrying both ALS-related mutations. Nevertheless, it provides an opportunity to delineate inheritance patterns within these families and to examine the resulting phenotypes.
The two pedigrees described in this paper illustrate several important points about the genetics of ALS and its influence on disease phenotype. First, despite the relative rarity of ALS in the general population, our data prove that two late onset neurodegenerative genes can operate within the same pedigree. In our opinion, such an occurrence was an extremely rare event that only occurred in this instance because of the unique genetic architecture of the Sardinian population. It is much less likely to occur within outbred populations, a notion supported by the fact that there is only another report of a compound heterozygosity for two recessive SOD1 mutations (p.D90A and p.D96N) in a family with recessively transmitted ALS.17
Second, the presence of two different pathogenetic mutations in our cases indicates that a second unidentified cause of ALS may coexist in the same individuals even when a known pathogenetic mutation has been already identified. This could be the case of the four pedigrees in which SOD1 mutations did not segregate with ALS,18 and also of the finding of a recent paper which has described two apparently unrelated patients carrying both a SOD1 G93D missense mutation and the angiogenin (ANG) synonymous variant c.329 T→G, p.G110G, which was not found in healthy controls and was considered to be pathogenic in silico.19
Third, the penetrance of both the C9ORF72 repeat expansion and the p.A383T mutation of the TARDBP gene is not 100%, as illustrated by Family B, where both parents of the index case (II-1) were healthy and no other cases of ALS or FTD were detected within the pedigree. Low penetrance is, by definition, to be expected in a late-onset disease. The alternative possibility that both mutations arose de novo is improbable, especially as the known founder risk haplotypes across both the C9ORF72 locus on chromosome 9p and the TARDBP gene on chromosome 1p were present in both of the probands.
Fourth, it is interesting to note that the affected child in Family A developed disease at a much younger age compared with either of his parents. Such observations suggest that the age of symptom onset of ALS is influenced by the genetic burden of the individual. By extension, it is interesting to speculate whether the age at onset within the more general ALS population is also driven by protective and damaging genetic variants lying outside of the primary genetic cause of disease.
Finally, the pattern of cognitive defects with Family A may be explainable in terms of the precise genetic mutation involved in each individual case. It is well established that the GGGGCC hexanucleotide repeat expansion of the C9ORF72 gene is frequently associated with FTD/cognitive impairment in addition to the motor dysfunction of ALS. This is clearly seen in the affected proband and in his father who carried the pathogenic expansion. In contrast, mutations of the TARDBP gene are only occasionally associated with cognitive defects; his mother was reported to have only mild cognitive compartment consistent with her carrier status of this mutation.
In summary, our data show that it is possible for rare neurodegenerative causing genes to coexist within the same family and illustrates the power of studying isolated populations with high prevalence of rare diseases. Such populations facilitate gene discovery by decreasing genetic heterogeneity and provide unique opportunities to study phenotypes associated with these genotypes. Genetic studies are on-going to identify additional causative genes within the Sardinian ALS population.
Supplementary Material
Acknowledgments
Adriano Chiò had full access to all of the data in the study and takes responsibility for the integrity of the data and the accuracy of the data analysis. We thank the patients and their families for having collaborated with this study.
Funding
This work was supported by the Intramural Research Programs of the NIH, National Institute on Aging (Z01-AG000949-02), the Packard Center for ALS Research at Hopkins, the ALS Association, Microsoft Research, Federazione Italiana Giuoco Calcio (grant 2010, #2) and European Community’s Health Seventh Framework Programme (FP7/2007-2013) under grant agreement 259867. Dr. Traynor reports that a patent is pending based on the discovery of the hexanucleotide repeat expansion of C9ORF72.
Footnotes
Competing interests
BJT has a patent pending based on the hexanucleotide repeat expansion of the C9ORF72 gene.
Ethics approval
Approval provided by the ethical committees of San Giovanni Battista Hospital, Torino; Azienda Universitaria-Ospedaliera di Cagliari; and Azienda Ospedaliera di Sassari.
References
- 1.Kiernan MC, Vucic S, Cheah BC, et al. Amyotrophic lateral sclerosis. Lancet. 2011;377:942–55. doi: 10.1016/S0140-6736(10)61156-7. [DOI] [PubMed] [Google Scholar]
- 2.Chiò A, Traynor BJ, Lombardo F, et al. Prevalence of SOD1 mutations in the Italian ALS population. Neurology. 2008;70:533–7. doi: 10.1212/01.wnl.0000299187.90432.3f. [DOI] [PubMed] [Google Scholar]
- 3.Renton AE, Majounie E, Waite A, et al. A hexanucleotide repeat expansion in C9ORF72 is the cause of chromosome 9p21-Linked ALS-FTD. Neuron. 2011;72:257–68. doi: 10.1016/j.neuron.2011.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dejesus-Hernandez M, Mackenzie IR, Boeve BF, et al. Expanded GGGGCC hexanucleotide repeat in noncoding region of C9ORF72 causes chromosome 9p-linked FTD and ALS. Neuron. 2011;72:245–56. doi: 10.1016/j.neuron.2011.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sreedharan J, Blair IP, Tripathi VB, et al. TDP-43 mutations in familial and sporadic myotrophic lateral sclerosis. Science. 2008;319:1668–72. doi: 10.1126/science.1154584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kwiatkowski TJ, Jr, Bosco DA, Leclerc AL, et al. Mutations in the FUS/TLS gene on chromosome 16 cause familial amyotrophic lateral sclerosis. Science. 2009;323:1205–8. doi: 10.1126/science.1166066. [DOI] [PubMed] [Google Scholar]
- 7.Vance C, Rogelj B, Hortobágyi T, et al. Mutations in FUS, an RNA processing protein, cause familial amyotrophic lateral sclerosis type 6. Science. 2009;323:1208–11. doi: 10.1126/science.1165942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Johnson JO, Mandrioli J, Benatar M, et al. Exome sequencing reveals VCP mutations as a cause of familial ALS. Neuron. 2010;68:857–64. doi: 10.1016/j.neuron.2010.11.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chiò A, Borghero G, Pugliatti M, et al. Large proportion of amyotrophic lateral sclerosis cases in Sardinia due to a single founder mutation of the TARDBP gene. Arch Neurol. 2011;68:594–8. doi: 10.1001/archneurol.2010.352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sabatelli M, Conforti LF, Zollino M, et al. C9ORF72 hexanucleotide repeat expansions in the Italian sporadic ALS population. Neurobiol Aging. 2012 doi: 10.1016/j.neurobiolaging.2012.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chiò A, Calvo A, Moglia C, et al. Amyotrophic lateral sclerosis-frontotemporal lobar dementia in 3 families with p.Ala382Thr TARDBP mutations. Arch Neurol. 2010;67:1002–9. doi: 10.1001/archneurol.2010.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mok K, Traynor BJ, Schymick J, et al. The chromosome 9 ALS and FTD locus is probably derived from a single founder. Neurobiol Aging. 2011;33:209.e3–8. doi: 10.1016/j.neurobiolaging.2011.08.005. Google Scholar. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Underhill PA, Shen P, Lin AA, et al. Y chromosome sequence variation and the history of human populations. Nat Genet. 2000;26:358–61. doi: 10.1038/81685. [DOI] [PubMed] [Google Scholar]
- 14.Piazza A, Mayr WR, Contu L, et al. Genetic and population structure of four Sardinian villages. Ann Hum Genet. 1985;49:47–63. doi: 10.1111/j.1469-1809.1985.tb01675.x. [DOI] [PubMed] [Google Scholar]
- 15.Orrù S, Manolakos E, Orrù N, et al. High frequency of the TARDBP p.Ala382Thr mutation in Sardinian patients with amyotrophic lateral sclerosis. Clin Genet. 2012;81:172–8. doi: 10.1111/j.1399-0004.2011.01668.x. [DOI] [PubMed] [Google Scholar]
- 16.Chiò A, Borghero G, Restagno G, et al. Clinical characteristics of familial ALS patients carrying the pathogenic GGGGCC hexanucleotide repeat expansion of the C9ORF72 gene. Brain. 2012;135:784–93. doi: 10.1093/brain/awr366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hand CK, Mayeux-Portas V, Khoris J, et al. Compound heterozygous D90A and D96N SOD1 mutations in a recessive amyotrophic lateral sclerosis family. Ann Neurol. 2001;49:267–71. doi: 10.1002/1531-8249(20010201)49:2<267::aid-ana51>3.0.co;2-d. [DOI] [PubMed] [Google Scholar]
- 18.Felbecker A, Camu W, Valdmanis PN, et al. Four familial ALS pedigrees discordant for two SOD1 mutations: are all SOD1 mutations pathogenic? J Neurol Neurosurg Psychiatry. 2010;81:572–7. doi: 10.1136/jnnp.2009.192310. [DOI] [PubMed] [Google Scholar]
- 19.Luigetti M, Lattante S, Zollino M, et al. SOD1 G93D sporadic amyotrophic lateral sclerosis (SALS) patient with rapid progression and concomitant novel ANG variant. Neurobiol Aging. 2011;32:1924.e15–18. doi: 10.1016/j.neurobiolaging.2011.04.004. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.