Abstract
Principal component analysis was applied to a biomaterial library of poly(beta-amino ester)s, useful for non-viral gene delivery, to elucidate chemical parameters that drive biological function. Correlative relationships and principal components were analyzed between 24 physico-chemical polymer properties and 3 cell-based functional variables in human glioblastoma cells (transfection, uptake, and viability).
Viral methods for gene therapy have been actively investigated for many years in more than 2,000 world-wide clinical trials, but due in part to reported toxicological and immunological concerns, have not been approved for use in the United States.1 Polymeric vectors are an alternative for gene delivery worth investigating as they can be physico-chemically modified to enhance function and minimize toxicity. They also benefit by being easier and less expensive to manufacture than viruses and, unlike viruses, do not have a restriction to their nucleic acid cargo capacity. While high-throughput screening methods have recently been adapted to allow for evaluation of biomaterial libraries, it is difficult to use this data to isolate key structural drivers of biological activity or to predict characteristics of untested structures.2, 3 Understanding fundamental structure-function relationships for gene delivery polymers would allow for improved rational engineering and enhanced chemical delivery systems.
Principal Component Analysis (PCA) is a powerful tool for reducing complex data sets that contain many variables with unknown correlations. The data set is reduced into orthogonal, linearly uncorrelated variables, termed principal components (PC). PCs are useful in helping to determine underlying relationships between variables.4, 5 While to our knowledge these methods have not been previously used to elucidate how polymer structure can affect biological function including gene delivery efficacy, we hypothesized that we would find trends based on our recent work on evaluating how polymer structure can tune DNA binding and gene delivery.6 We chose hydrolytically degradable poly(beta-amino ester)s (PBAE) to study as a PBAE polymer library can be readily synthesized by semi-high throughput methods and we have previously shown utility of these polymers for both in vitro and in vivo gene therapy applications.7, 8 We report the use of PCA to aid our understanding of the physico-chemical properties of polymers that drive transfection, uptake, and viability in human cells.
A PBAE library consisting of polymers with varying backbone (B), sidechain (S), or endcap (E) was recently synthesized by our lab (Scheme S1).9 In brief, the base polymer was synthesized by mixing B and S monomers neat in 1.05:1, 1.1:1, or 1.2:1 B:S monomeric ratios and the reaction was allowed to stir for 24 hours at 90°C in the dark; after which the B-S base polymer was solvated in anhydrous dimethyl sulfoxide (DMSO) to 167 mg/mL. 480 μL of the 167 mg/mL base polymer was then endcapped in DMSO for 1 hr using an approximate 10:1 E (0.5 M solution in DMSO; 320 μL) to B-S ratio (Scheme S1).9
The B to S monomeric ratio (B:S) dictates the molecular weight of the polymer, with molecular weight increasing as the ratio approaches unity. The numbers associated with the B and the S monomer names are the number of carbons between the backbone's acrylate groups and the sidechain's amine and hydroxyl groups, respectively (Scheme S1). “B+S” refers to the sum of these numbers for an individual polymer, or the number of carbons in its repeating unit. As the carbons in the backbone and sidechain increase, the overall hydrophobicity of the polymer increases. The numbers associated with the “E” term are randomly assigned and are not indicative of endcap structure.
Gel permeation chromatography (GPC; Waters, Milford, MA) was performed on the polymers using 94% tetrahydrofuran (THF), 5% DMSO, 1% piperidine with a few 100 mg of butylated hydroxytoluene. The solvated polymer was then filtered using a 0.2 μm polytetrafluoroethylene filter and compared against polystyrene standards to obtain the number- and weight-average molecular weights (Mn and Mw), the polydispersity indices (PDI) and the degree of polymerization (DP).9
PCA was performed utilizing recently reported biological in vitro data on glioblastoma cells (GBM319).9 Briefly, PBAE/eGFP DNA nanoparticles were ionically complexed for 10 minutes in 25 mM sodium acetate (NaAc) at room temperature to self-assemble into nanoparticles at a polymer to DNA mass ratio (w/w) of 60.9 The total incubation time for the polyplexes with the cells (final dose of 5 μg/mL in 100 μL for 15,000 cells/well in 96-well plates) was 2 hrs and to assess uptake, Cy™3 (Mirus Bio LLC; MIR 7020)-conjugated plasmid DNA was directly assessed via flow cytometry after a 2 hr incubation.9 A viability assay (Cell Titer 96®AQueous One) was used at 24 hrs to assess cell viability and flow cytometry to assess transfection efficacy at 48 hrs.9
Besides the GPC-obtained variables, other physical and chemical parameters were calculated with the aid of ChemDraw, the Joback fragmentation method, and the Crippen's fragmentation method. These included boiling point (BP); melting point (MP); critical volume (CV), which is the volume of 1 mole at the critical temperature and pressure; Gibb's free energy (GFE), which is the thermodynamic potential to perform work; LogP, the partition-coefficient between two immiscible phases at equilibrium which is proportional to hydrophobicity; molar refractivity (MR), which is a measurement of the total polarizability of 1 mole; the heat of formation (HtF), which is the change in enthalpy of 1 mole from the formation the elemental constituents; and the topological polar surface area (tPSA), which is the total area of all polar atoms (predominantly oxygen and nitrogen) including their affixed hydrogen atoms. Properties associated with the polymer repeating unit are differentiated from those of the full polymer, by the presence of an asterisk in their name (i.e., LogP* vs. LogP).
PCA was carried out using the standard “princomp” function in MATLAB to calculate the coefficients, scores, and variances. All included variables were first scaled from 0 to 1 for normalization and included the following 27 parameters: B, B:S, B+S, BP, BP*, CV, CV*, DP, GFE, GFE*, HtF, HtF*, LogP, LogP*, Mn, MP, MP*, MR, MR*, Mw, MW*, PDI, S, tPSA, transfection efficacy, cell uptake, and cell viability. 24 of the 27 are physico-chemical variables determined by the structure of the polymers; the remaining 3 are cell-based functional variables determined experimentally. As a control, a random variable, the “E” number assigned to each endcap but not meaningful when normalized from 0 to 1, was incorporated into the set of parameters.
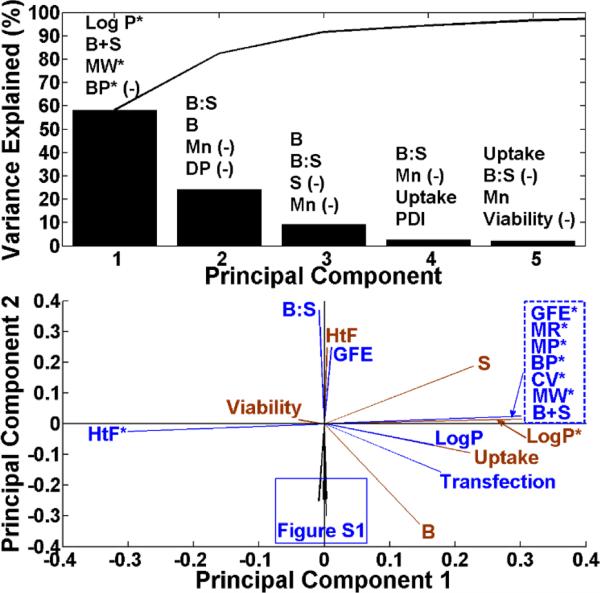
The variance of a particular PC divided by the sum of all of the PC variances multiplied by 100 is the percentage that a particular PC recapitulates the data set. We rank the variables by the degree to which they contribute positively or negatively to each PC using their associated coefficients. Although there are 27 variables, it is striking that 5 PCs can cover almost all the variance in the data and just the first two PCs cover 83% of this variance, significantly decreasing the complexity of this multivariate data. The first and second PCs were responsible for 58.2% and 24.3% of the variance in the data set, respectively (Figure 1, top). Cumulatively, the first 5 PCs capture 96.6% of the variance in the data set. The top four variables contributing to each of the first five PCs are listed above the variances of each PC (Figure 1; top) and are indicated in parentheses as “(-)” if it is a negative contribution. Table S1 contains a full list of the 27 variables ranked for the first five PCs.
Figure 1.
Top: the variances are explained for the first 5 PCs. The top 4 variables of the 27 are ranked by the degree to which they contribute to the PCs according to their associated coefficients and are listed above the variances. The “(-)” indicates a negative contribution; Bottom: The loading plot of all of the variables showing relative correlations.
The loading plot (Figure 1; bottom) was generated using the first and second coefficients associated with each variable. The loading plot reveals the correlative relationships between the variables. Variables within the same and opposite quadrants of the loading plot indicate that they are positively and inversely correlated, respectively. Variables in adjacent quadrants are positively correlated with respect to one PC but not the other.
Based on the loading plot, variables can be ranked according to the degree to which they correlate to a reference variable using Acos(Θ); where A is the magnitude of the vectors of the variables being compared to the reference and Θ is the angle between the variables and the reference. Variables corresponding to a positive Acos(Θ) value are positively correlated with the reference variable of interest; whereas negative Acos(Θ) values are negatively correlated. Thus, the most positive and the most negative variables drive the reference variable. The Acos(Θ) values near neutral contribute relatively little to the reference variable.
The scores plot was generated using the first and second scores of the polymers in the PBAE library associated with the 1st and 2nd PCs. The patterns within score plots can be assessed for self-assembling trends; the scores plot was plotted against transfection levels in the 3rd dimension. A supplemental auto-rotating video was created using MATLAB of this 3-D plot.
The loading plot is shown in Figure 1 (bottom) and clearly demonstrates graphically that LogP* is a leading driver of cellular uptake and transfection. Figure S1 shows other variables (PDI, Mn, Mw, MR, CV) also positively correlated to transfection and uptake. Mn, Mw and PDI are shown in Figure S2. Table 1 describes the biological variables of interest, transfection, uptake and viability, and the remaining 26 variables ranked accordingly. When the normalized “E” number, a random variable, was included for analysis in the data set, no correlation was found, as expected. The Acos(Θ) values associated with “E” for transfection, uptake, and viability were 0.02, 0.01, and −0.01, respectively. This validates that PCA successfully identified the normalized “E” number as a random variable and not a chemical parameter for analysis.
Table 1.
Ranking of variables for transfection, uptake, and viability. The value of Acos(Θ) indicates the strength of the correlation.
| Transfection | Uptake | Viability | ||||
|---|---|---|---|---|---|---|
| Variable | A(cosθ) | Variable | A(cosθ) | Variable | A(cosθ) | |
| 1 | B | 0.33 | LogP* | 0.27 | HtF* | 0.28 |
| 2 | Uptake | 0.23 | B+S | 0 27 | B:S | 0.13 |
| 3 | LogP* | 0.22 | GFE* | 0.27 | HtF | 0.08 |
| 4 | B+S | 0.21 | MW* | 0.27 | GFE | 0.07 |
| 5 | GFE* | 0.21 | BP* | 0.27 | PDI | −0.06 |
| 6 | MW* | 0.21 | MP* | 0.27 | DP | −0.07 |
| 7 | BP* | 0.21 | CV* | 0.27 | tPSA | −0.07 |
| 8 | MP* | 0.21 | MR* | 0.27 | MP | −0.08 |
| 9 | CV* | 0.21 | B | 0 26 | BP | −0.08 |
| 10 | MR* | 0.21 | Transfection | 0.23 | Mw | −0.08 |
| 11 | Mn | 0.20 | LogP | 0.18 | MR | −0.08 |
| 12 | LogP | 0.17 | S | 0.14 | CV | −0.08 |
| 13 | CV | 0.16 | Mn | 0.12 | Mn | −0.10 |
| 14 | MR | 0.16 | CV | 0.10 | S | −0.15 |
| 15 | Mw | 0.16 | MR | 0.10 | LogP | −0.18 |
| 16 | BP | 0.16 | Mw | 0.10 | Transfection | −0.22 |
| 17 | MP | 0.16 | BP | 0.10 | Uptake | −0.24 |
| 18 | DP | 0.16 | MP | 0.10 | B | −0.25 |
| 19 | tPSA | 0.16 | tPSA | 0.09 | B+S | −0.28 |
| 20 | PDI | 0.13 | DP | 0.09 | GFE* | −0.28 |
| 21 | S | 0.05 | PDI | 0.08 | MW* | −0.28 |
| 22 | Viability | −0.04 | Viability | −0.04 | BP* | −0.28 |
| 23 | GFE | −0.16 | GFE | −0.09 | MP* | −0.28 |
| 24 | HtF | −0.16 | HtF | −0.09 | CV* | −0.28 |
| 25 | HtF* | −0.21 | B:S | −0.15 | MR* | −0.28 |
| 26 | B:S | −0.25 | HtF* | −0.27 | LogP* | −0.28 |
Parameters B, transfection, uptake, Mn, Mw, PDI, MR, CV and LogP* are all positively correlated in quadrant IV of Figure 1. Hydrophobicity, as measured quantitatively and in silico by LogP*, was found to positively correlate with transfection efficacy, and this finding supports a qualitative relationship between hydrophobicity and transfection as has been previously hypothesized.3 This result highlights the potential of calculating putative chemical parameters of biomaterial libraries in silico and using these chemical properties as design criteria prior to synthesizing a full biomaterial library. In this manner, a subset of the library can be focused on with the desired chemical properties hypothesized to make the greatest impact on biological function. Between the biological parameters, as expected, increased cellular uptake correlated strongly to higher transfection efficacy. Viability is negatively correlated with the variables in quadrant IV. For example, polymers that transfect strongly generally correlate to slightly lower cell viability.
Because uptake and transfection are highly positively correlated, it was expected that chemical variables would affect these two biological variables similarly, which is what was observed. In contrast, because viability is negatively correlated with transfection and uptake, it was expected that the same chemical variables would affect it negatively. This was observed as, for example, LogP* and B+S were highly positively correlated with transfection and uptake but were negatively correlated with viability. This data quantitatively demonstrates how the same chemical parameter can both positively and negatively drive biological functional outcomes.
PCA reduces the complexity of multiple variables and assembles correlated, non-orthogonal variables together. As an example, the B:S monomer ratio used during polymer synthesis as well as the molecular weights, Mn and Mw, and the degree of polymerization, DP, all relate to molecular weight and all lie along the PC2 axis. B:S is in an opposing direction as it is inversely correlated, as monomer ratios used during synthesis that are closer to unity lead to polymers with the highest molecular weight. Similarly, each of the PCs assemble correlated variables that vary together. In many ways, the two main drivers of transfection efficacy with this polymer library are PC1, which encompasses hydrophobicity, and PC2, which encompasses molecular weight.
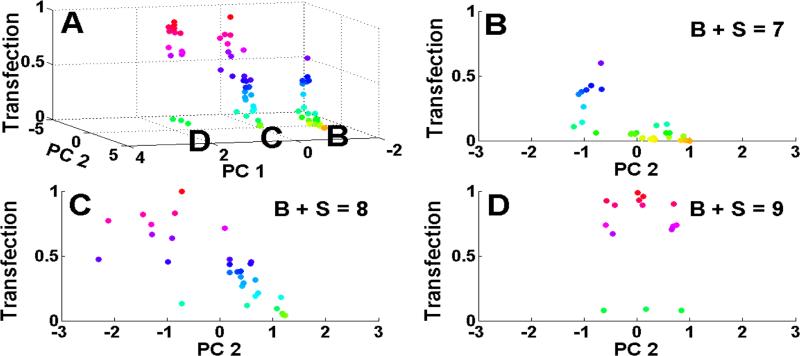
The scores plot associated with PC1 and PC2 is shown in Figure S3. The Mn and Mw are listed in kDa to the right of the polymer names (i.e., 447, 8.8, 28.3), respectively. Since there are 27 variables being analysed, there are 27 scores associated with each of the polymer samples within the PBAE library. The PC1 and PC2 scores for the polymers in Figure S2 were plotted against transfection efficacy in the 3rd dimension in Figure 2A. A supplemental auto-rotating video of Figure 2A created using MATLAB can also be found online. The colour in Figure 2 corresponds to the level of transfection with red being the highest and yellow the lowest. Figure 2A demonstrates that the polymers self-clustered into three groups along 3 specific regions of PC1. This self-clustering was not expected and validates the PCA approach at quantitatively elucidating the key chemical parameters of the polymer library at driving biological function. The 3 regions of PC1 were named as B, C, and D in Figure 2A and these three groups were plotted in 2-dimensions vs. PC2 in Figures 2B, 2C, and 2D, respectively. Each of these regions of PC1 matched exactly with the “B+S” number, or the number of carbons in a polymer's repeat unit.
Figure 2.
A: The scores plot versus transfection efficacy with red being the highest level of transfection; B: region B of 2A with a B+S (sum of carbons in backbone and sidechain) value of 7; C: region C of 2A with a B+S value of 8; D: region D of 2A with a B+S value of 9.
The top 4 positively correlated physico-chemical variables driving the biological parameters, transfection, uptake and viability were: B, uptake, LogP*, and B+S; LogP*, B+S, GFE*, and MW*; HtF*, B:S, HtF, and GFE, respectively. Whereas the top 4 negatively correlated variables driving transfection, uptake and viability were: B:S, HtF*, HtF, and GFE; HtF*, B:S, HtF, and GFE; LogP*, MR*, CV*, and MP*, respectively.
Table S1 ranks all of the variables according to the degree to which they contribute to each of the first 5 PCs. B+S contributes positively to PC1 and Mn and Mw contribute negatively to PC2, which is observed in the scores plot (Figure S3); as PC1 increases, B+S increases and as PC2 increases, the molecular weight generally decreases. Surprisingly, the self-clustered regions B, C, and D (Figure 2) correspond to B+S values equal to 7, 8, and 9, respectively. Intriguingly, the number of carbons within a polymer's repeat unit was found to group the polymer's behaviour more than any other parameter. This B+S grouping dictated the role of PC2 on transfection efficacy among the polymers within the group. For example, polymers in group B (B+S=7) had generally very low transfection efficacy, with transfection efficacy increasing to ~half the maximum for more negative PC2 values (indicating a higher molecular weight, higher degree of polymerization, and a smaller B:S ratio closer to unity). For polymers in group C (B+S=8), transfection is higher, reaching the maximum. Like with the B+S=7 group, lower values of PC2 (and higher MW) increased transfection efficacy. In both of these groups, a lower value of PC2 could increase transfection efficacy by larger than an order of magnitude. In contrast, group D (B+S=9) transfection was uniformly high near the maximum and PC2 did not influence transfection efficacy. This trend is also shown qualitatively in the fluorescence microscope images of representative polymers in Figure S4. Thus these two principal components, PC1 (hydrophobicity, B+S) and PC2 (molecular weight), were found to cluster and elucidate the polymer structures and their biological efficacy in new ways. As B+S increased from 7 to 9, a greater portion of polymers had higher transfection levels generally. Optimal transfections were associated with PC2 values of −1, −1, and 0 for groups B, C, and D, respectively.
These results demonstrate that in silico PCA analysis can reveal what chemical parameters are key drivers of biological activity. In this study, our analysis shows the LogP*, MW, and B+S are the polymer physico-chemical parameters that quantitatively drive polymer-mediated transfection efficacy of PBAEs. The effect of new polymer structures on tuning LogP* and B+S for PBAEs can be obtained computationally and, for a given B+S value, MW can be tuned during synthesis by varying B:S monomer ratio and through polymer purification.6 In this manner, guidelines can be developed for the design of next generation biomaterials.
Previous research in our lab6 has demonstrated that transfection levels can be biphasic with respect to binding constants and also that binding constants increase with increasing molecular weight. Our results in this current PCA study are consistent with these past results as the highest transfection efficacy among all polymers occurs at intermediate values of PC2 (between −1 and 0).
In this work we demonstrate the utility of PCA to investigate biomaterial structure and its functional effect on intracellular delivery to cells. This type of analysis could potentially be used across a broader spectrum of polymeric vectors10 (i.e., poly(l-lysine), polyethyleneimine, chitosan, dendrimers, and β-cyclodextrin-containing vectors) and various types of cargo (i.e., siRNA/miRNA, shRNA, mRNA). Such a large-scale analysis would undoubtedly further elucidate additional structure-function relationships allowing improved polymer and delivery system design.
In conclusion, we have been able to demonstrate that PCA is a useful tool for helping elucidate how physico-chemical properties of polymers drive transfection, uptake, and viability in human primary glioblastoma cells. We determined that for poly(beta-amino ester)-mediated transfection of glioblastoma cells, the leading PC was driven by hydrophobicity and the second PC by molecular weight. By determining the principal components, one can design next generation materials by tuning the chemical parameters that matter most in the particular ranges determined to lead to the desired biological functional outcomes (ie., high transfection).
Supplementary Material
Acknowledgments
We would like to acknowledge our funding sources: the NIH (1R01EB016721) and the NSF Research Fellowship awarded to CJB (DGE-0707427).
Footnotes
Electronic Supplementary Information (ESI) available: [details of any supplementary information available should be included here]. See DOI: 10.1039/x0xx00000x
References
- 1.Gene therapy clinical trials worldwide. http://www.abedia.com/wiley/index.html.
- 2.Anderson DG, Lynn DM, Langer R. Angewandte Chemie-International Edition. 2003;42:3153–3158. doi: 10.1002/anie.200351244. [DOI] [PubMed] [Google Scholar]
- 3.Sunshine JC, Akanda MI, Li D, Kozielski KL, Green JJ. Biomacromolecules. 2011;12:3592–3600. doi: 10.1021/bm200807s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bishop CJ, Mason NO, Kfoury AG, Lux R, Stoker S, Horton K, Clayson SE, Rasmusson B, Reid BB. J. Heart Lung Transplant. 2010;29:27–31. doi: 10.1016/j.healun.2009.08.027. [DOI] [PubMed] [Google Scholar]
- 5.Martinerie J, Adam C, Le Van Quyen M, Baulac M, Clemenceau S, Renault B, Varela FJ. Nat. Med. 1998;4:1173–1176. doi: 10.1038/2667. [DOI] [PubMed] [Google Scholar]
- 6.Bishop CJ, Ketola TM, Tzeng SY, Sunshine JC, Urtti A, Lemmetyinen H, Vuorimaa-Laukkanen E, Yliperttula M, Green JJ. J. Am. Chem. Soc. 2013;135:6951–6957. doi: 10.1021/ja4002376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bishop CJ, Tzeng SY, Green JJ. Acta Biomater. 2014 doi: 10.1016/j.actbio.2014.09.020. DOI: 10.1016/j.actbio.2014.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kamat CD, Shmueli RB, Connis N, Rudin CM, Green JJ, Hann CL. Mol. Cancer Ther. 2013;12:405–415. doi: 10.1158/1535-7163.MCT-12-0956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tzeng SY, Green JJ. Adv. Healthcare Mater. 2013;2:468–480. doi: 10.1002/adhm.201200257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yin H, Kanasty RL, Eltoukhy AA, Vegas AJ, Dorkin JR, Anderson DG. Nature Reviews Genetics. 2014;15:541–555. doi: 10.1038/nrg3763. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.